
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 March 2021
Sec. Evolutionary and Genomic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.647119
This article is part of the Research Topic Evolution of Animal Microbial Communities in Response to Environmental Stress View all 11 articles
 Yao-Dong Wu1
Yao-Dong Wu1 Li Li1
Li Li1 Yan-Lei Fan1,2
Yan-Lei Fan1,2 Xing-Wei Ni1,3
Xing-Wei Ni1,3 John Asekhaen Ohiolei1
John Asekhaen Ohiolei1 Wen-Hui Li1
Wen-Hui Li1 Jian-Qiu Li1
Jian-Qiu Li1 Nian-Zhang Zhang1
Nian-Zhang Zhang1 Bao-Quan Fu1
Bao-Quan Fu1 Hong-Bin Yan1*
Hong-Bin Yan1* Wan-Zhong Jia1,4*
Wan-Zhong Jia1,4*The larva of Taeniidae species can infect a wide range of mammals, causing major public health and food safety hazards worldwide. The Qinghai-Tibet Plateau (QTP), a biodiversity hotspot, is home to many species of rodents, which act as the critical intermediate hosts of many Taeniidae species. In this study, we identified two new larvae of Taenia spp., named T. caixuepengi and T. tianguangfui, collected from the plateau pika (Ochotona curzoniae) and the Qinghai vole (Neodon fuscus), respectively, in QTP, and their mitochondrial genomes were sequenced and annotated. Phylogenetic trees based on the mitochondrial genome showed that T. caixuepengi has the closest genetic relationship with T. pisiformis, while T. tianguangfui was contained in a monophyletic group with T. crassiceps, T. twitchelli, and T. martis. Biogeographic scenarios analysis based on split time speculated that the speciation of T. caixuepengi (∼5.49 Mya) is due to host switching caused by the evolution of its intermediate host. Although the reason for T. tianguangfui (∼13.11 Mya) speciation is not clear, the analysis suggests that it should be infective to a variety of other rodents following the evolutionary divergence time of its intermediate host and the range of intermediate hosts of its genetically close species. This study confirms the species diversity of Taeniidae in the QTP, and speculates that the uplift of the QTP has not only a profound impact on the biodiversity of plants and animals, but also that of parasites.
The most recent molecular phylogenetic analysis has suggested that the family Taeniidae (Eucestoda: Cyclophyllidea) should be composed of four genera: Taenia, Echinococcus, Hydatigera, and Versteria (Nakao et al., 2013). Among them, Taenia and Echinococcus species pose a serious public health threat to humans and animals globally. Terrestrial mammals are crucial to the life cycle of taeniids. Most adult tapeworms parasitize the intestines of carnivores while the intermediate hosts harbor the larva stage that develops from ingested eggs, causing severe health effects (Jia et al., 2012; Nakao et al., 2013; Lymbery, 2017; Deplazes et al., 2019).
Before the new classification recommendation of Nakao et al. (2013), two genera (Taenia and Echinococcus) were generally accepted. Taenia was constituted by about 42 valid species and three subspecies based on morphology (Hoberg et al., 2000; Hoberg, 2006; Nakao et al., 2013). As for Echinococcus, a total of 16 species and 13 subspecies were described based on morphology, but most of these taxa were subsequently invalidated following widespread application of molecular genetic methods (Lymbery, 2017). It is difficult to distinguish taeniid species according to their morphological characteristics at different stages of their life cycle, even by specialists (Flisser et al., 2005; Mathis and Deplazes, 2006; Jia et al., 2012). Sometimes, morphological characteristics are substantially influenced by the different intermediate host origins (Lymbery, 1998).
Mitochondrial (mt) DNA sequence has been recognized among the most suitable molecular markers of molecular ecology, population genetics, evolutionary biology and biological differentiation due to its high mutation rate and maternal inheritance (Hebert and Gregory, 2005; Will et al., 2005; Hajibabaei et al., 2007; Jia et al., 2012). In the last two decades, comparative analyses of taeniid mtDNAs have been increasingly applied in phylogenetic studies, for inferring evolutionary relationship, new species identification, species reclassification, phylogeography, genetic diversity, and tracing of evolutionary origins of related and identical species (Xiao et al., 2005; Nakao et al., 2007; Nakao et al., 2013; Terefe et al., 2014; Kinkar et al., 2018). Among the taeniid family, mt genomes of 36 species and genotypes have been sequenced and are available on GenBank1, providing valuable data support for phylogenetic studies of Taeniidae.
The shrinkage and fragmentation of wildlife habitats due to human activities can lead to increased contact between humans or livestock and wildlife, which potentially increases the risk of transmission of natural focal disease (Suzán et al., 2008). Rodents, the largest (∼43% of all mammal species) and most widely distributed group of mammals, act as major vectors of human and domestic animal diseases (Singla et al., 2008; Wu et al., 2018). The Qinghai-Tibet Plateau (QTP), one of the biodiversity hotspots on earth, is habitat to a rich diversity of wild rodent species (Zhou and Ma, 2002), as well as many rodent-eating carnivores (Smith et al., 2019), creating the conditions for various taeniid species to complete their life cycles. The high altitude geographic isolation combined with the geological complexity of the QTP increases the opportunities for genetic variation and speciation, leading to the continuous discovery of new species of rodents and Taeniidae (Xiao et al., 2005; Dahal et al., 2017). However, few studies have involved the population structure and biodiversity of taeniid species in QTP, except for Echinococcus.
As endangered or protected carnivores are difficult to sample, we collected metacestode samples of rodents to investigate the biodiversity and distribution of taeniid species in QTP. In this study, two new mt genomes of the metacestode samples were firstly sequenced and annotated. Through the phylogenetic analysis of mt genomes with species in the four different genera of taeniids, the validity of these two new Taenia spp., named T. caixuepengi and T. tianguangfui larvae, were confirmed and their phylogenetic relationship and evolutionary origin were analyzed.
All animals were handled in strict accordance with good animal practice according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China, and the study was approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (No. LVRIAEC2012-007).
Plateau pikas (Ochotona curzoniae) and Qinghai voles (Neodon fuscus) were trapped in Darlag county (33°43′N; 99°38′E; altitude at 4,068 m) and Jiuzhi county (33°19′N; 100°32′E; altitude at 3,832 m) of Qinghai province, the People’s Republic of China in July 2013. Following ethical approval, all trapped pikas and voles were dissected regarding the enterocoelia, chest and cranial cavities. Many banded cysticerci were collected in the enterocoelia of Plateau pika (Supplementary Figures S1A,B) and numerous lenticular cysticerci were collected in the enterocoelia and chest of Qinghai vole (Supplementary Figures S1C,D). Detailed sample collection data can be found in Supplementary Table S1. After detaching the lesions, the cysticerci were put into 75% (v/v) ethanol for molecular and morphological identification. Cysticerci from pikas and voles were photographed by Thermo ScientificTM Apreo S SEM, and their hooks were hand-drawn using Point 3D of Microsoft.
Cysticercus DNA was extracted using a commercial kit as instructed by the manufacturer (Blood and Tissue Kit, Qiagen, Germany). The mt genomes of Taenia spp., whose intermediate hosts include rodents, downloaded from GenBank (Supplementary Table S3) were aligned by using MEGA 7.0. Nine overlapping primers targeting the complete mt genome were designed using Oligo 6.0 at relatively conserve regions observed on alignment of the mt genome sequences. The primer sequences (Supplementary Table S2) were synthesized by Genewiz Biotech (Beijing, China). A standard 50 μl PCR protocol was used to amplify the mtDNA fragments. PCR products were purified directly from an agarose gel (1%) using an Axy PrepTM DNA Gel Extraction Kit (AXVGEN, United States) and then sent to the company Genewiz Biotech for sequencing.
These two mtDNAs were assembled manually, and annotated preliminarily by Geseq2 with the reference of related species, T. pisiformis and T. crassiceps, identified by the cox1 gene alignment of Neighbor-Joining method in MEGA 7.0 (data not shown). Putative tRNA genes were identified using ARWEN3 (Laslett and Canbäck, 2008). The positions of their open reading frames and rRNA genes were also further checked and modified based on alignment with the mt genomes annotation of T. pisiformis and T. crassiceps, respectively. SnapGene (v3.2.1) was used to translate the amino acid sequence of the protein-coding genes with Echinoderm, Flatworm Mitochondrial genetic code and map the annular diagram of the mt genomes.
To determine the phylogenetic status of these two Taenia spp., the phylogenetic trees were constructed using Bayesian methods in MrBayes v3 with the tandem DNA sequences and amino acid sequences of 12 encoding genes in their mt genomes and other 32 taeniid mt genome sequences downloaded from GenBank, while the sequences of Schistosoma japonicum was used as outgroup (Supplementary Table S2). For the amino acid data set, the mixed model was applied (prset aamodelpr = mixed); two chains (temp = 0.2) were run for 3,000,000 generations and sampled every 1,000 generations (Rota-Stabelli et al., 2009). For the nucleotide data set, Modeltest 3.7 maxX (Posada and Crandall, 1998) was used to estimate a suitable model for nucleotide substitution; this was equivalent to GTR + I + G and settings were nst = 6, rates = invgamma, ngammacat = 4. Four chains (temp = 0.2) were run for 1,000,000 generations and sampled every 1,000 generations. The first 25% of trees were omitted as burn-in and the remaining trees were used to calculate Bayesian posterior probabilities. The best Bayesian tree was then compiled and processed by FigTree.v1.4.4.
The phylogenetic trees were used as a reference for species selection in divergence times analysis. Echinococcus multilocularis and E. shiquicus were also selected because the parasitism of their larvae is also found in Plateau pika (O. curzoniae) (Wang et al., 2018), besides E. multilocularis is the sister species of E. shiquicus (Lymbery, 2017). Divergence times were calculated from the concatenated CDS alignment of the 12 mitochondrial protein-coding genes by BEAST2 (v2.6.2). The Strict Clock model was chosen to ignore the rate differences between the branches in the mode. The gamma category count was set to 4, and HKY substitution model was selected with the empirical setting from the frequencies in site model. Other settings, such as substitution rate and shape, in the site model were evaluated in the analysis. The calibrated Yule model was used as the tree prior. Time calibration was calibrated with the previously estimated date between T. saginata and T. asiatica (∼1.14 Mya) (Michelet and Dauga, 2012; Wang et al., 2016). Samples from the posterior were drawn every 1,000 steps over a total of 10,000,000 steps per MCMC run. Other options were run on their default values. The convergence of likelihood values was determined by Tracer (v1.7.1). Trees were annotated by TreeAnnotator (v2.1.2) using maximum clade credibility tree and median heights settings with 50% burn-in. The evolutionary divergence time of the intermediate host, Qinghai vole (N. fuscus), was also calculated with the concatenated CDS alignment of 13 mt protein-coding genes of the rodents, and the species involved were selected from our previous report (Li et al., 2019; Supplementary Table S3). The time calibration was based on the divergence time of Mus and Rattus (11–13 Mya) (Wang et al., 2020), and other parameters were the same as above.
A total of 300 pika (125) and voles (175) were examined. Overall, 7.3% were infected with cysticerci (see Supplementary Table S1 for more details). Amplification and sequencing of a fragment of the cox1 gene using the conserved JB3 and JB4.5 primers (Bowles et al., 1992) confirmed their identity. From both study sites, cysticerci from pikas were the same new species, named T. caixuepengi larva, meanwhile, the cysticerci from voles were morphologically different from T. caixuepengi larva and were named T. tianguangfui larva. Their complete mt genomes were sequenced and spliced, and were 13,747 bp (GenBank ID: MT882036) and 13,522 bp (GenBank ID: MT882037) in length, respectively. Both contain 2 rRNA genes [the small (rrnS) and large (rrnL) subunits of rRNA], 12 protein-encoding genes (atp6, cytb, nad4L, cox1-3, and nad1-6) and 22 tRNA genes, but lack atp8 gene, which are typical of cestode mt genomes (Figure 1). The inferred gene boundaries and their lengths are shown in Table 1.
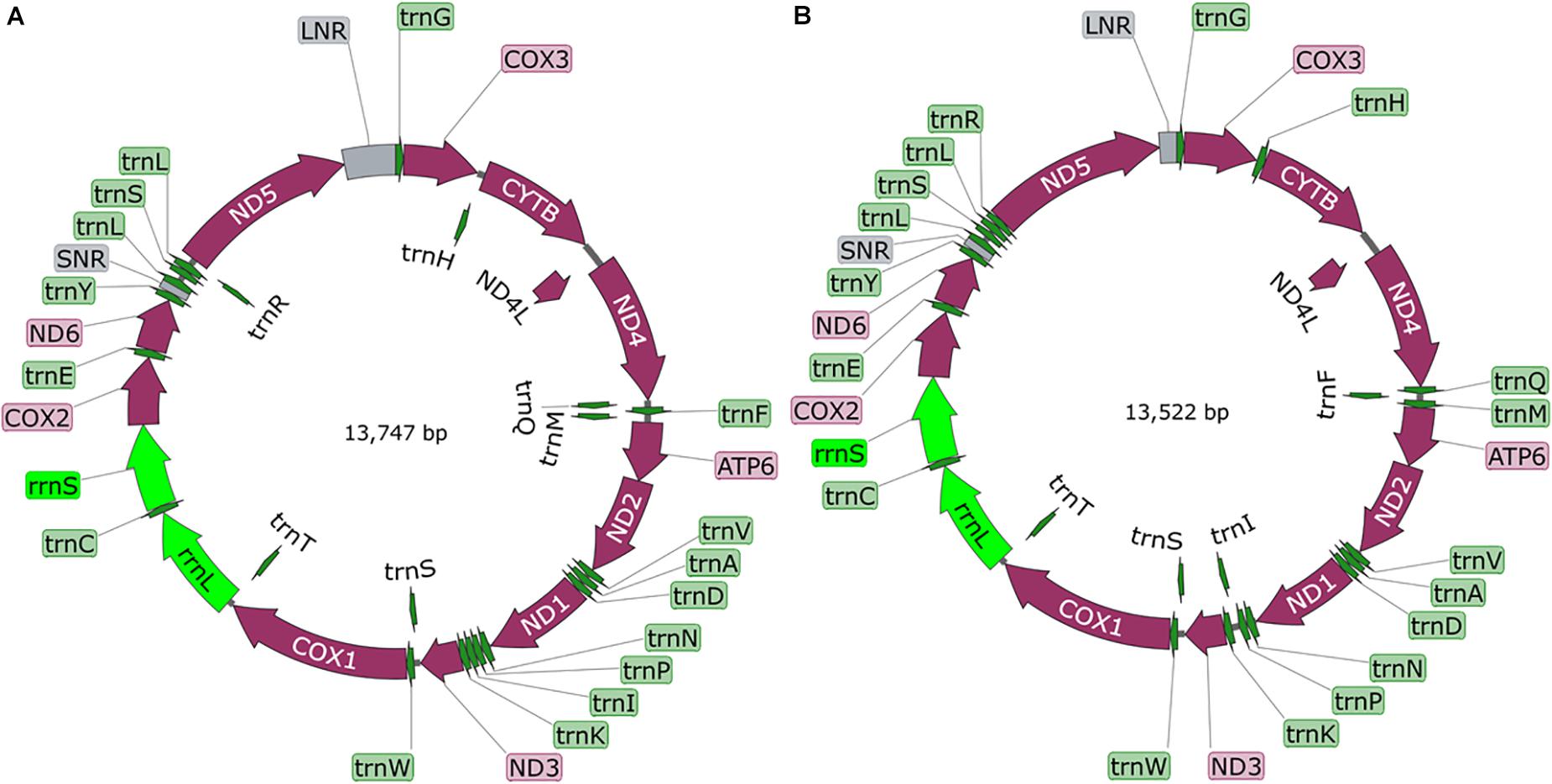
Figure 1. The diagram of complete mitochondrial genome of Taenia tianguangfui (A) and Taenia caixuepengi (B). The protein-encoding genes are depicted in plum, the tRNAs are depicted in green, the rRNAs are depicted in light green and the non-coding mitochondrial regions (NCRs including LNR and SNR) are depicted in gray. The inferred gene boundaries of them are shown in Table 1.
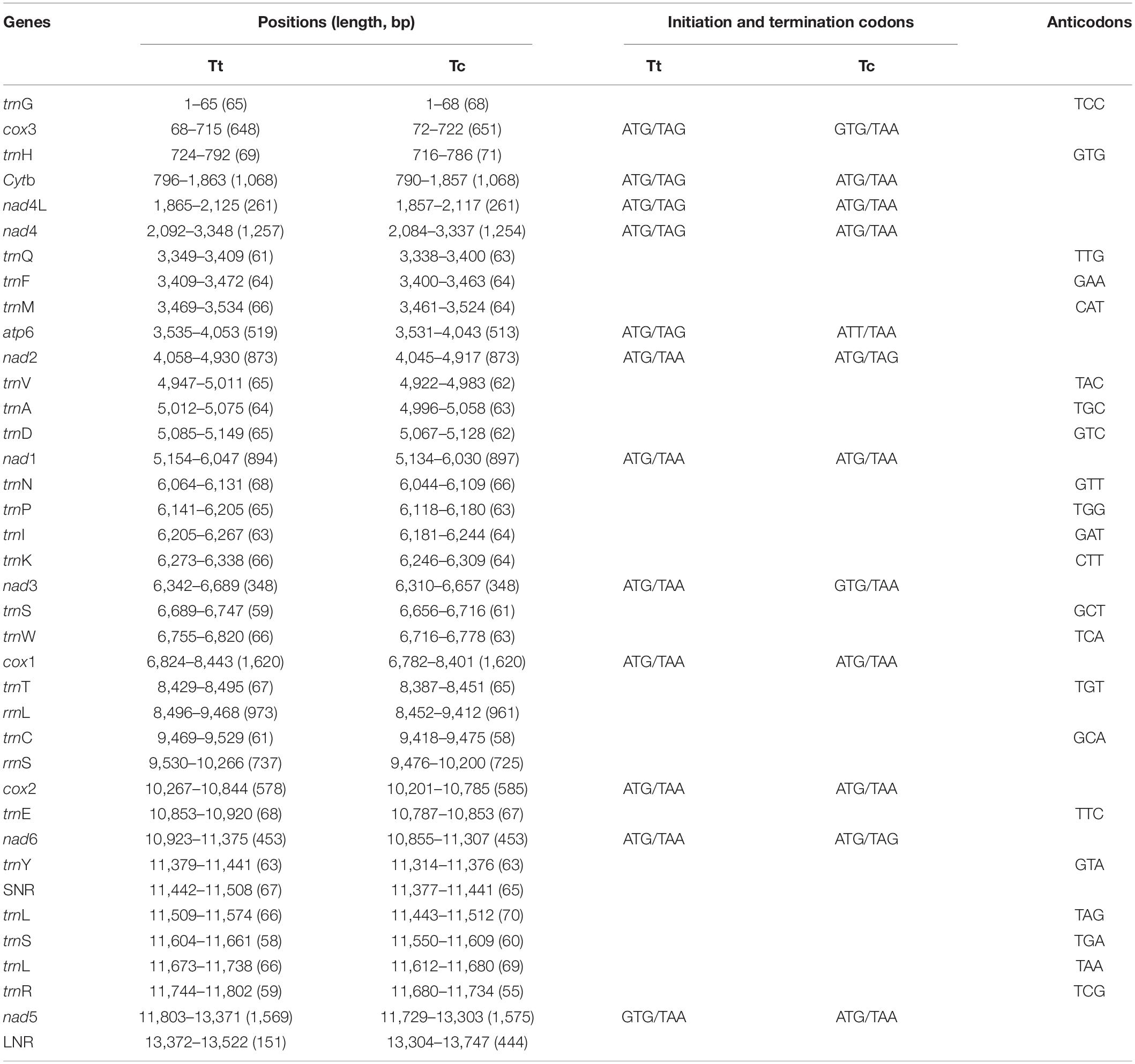
Table 1. Positions and gene lengths in the mitochondrial genomes of Taenia tianguangfui (Tt), T. caixuepengi (Tc).
In accordance with other mtDNAs of flatworms sequenced to date (Jia et al., 2010; Liu et al., 2011), the nucleotide compositions are mostly biased toward T, while least favored toward C. AT-richness of mtDNAs in T. caixuepengi and T. tianguangfui are 71.96% (45.00% T, 26.97% A, 19.17% G, 8.87% C) and 73.48% (46.35% T, 27.13% A, 18.61% G, 7.91% C), respectively.
Flatworms use an unusual mt code to exert protein translation (Nakao et al., 2000; Telford et al., 2000). GTG was used as an alternative initiation codon in cox3 and nad3 genes of T. caixuepengi and nad5 gene of T. tianguangfui. Furthermore, the codon ATT was inferred as a more unusual start codon of atp6 gene in T. caixuepengi. The termination codon was mostly TAA, and the ending codon TGA was deprecated (Table 1).
By gross observation and measurement, T. caixuepengi larva was translucent and stripped, 33-36 mm long, and contains cystic fluid (Supplementary Figures S1A,B); T. tianguangfui larva on the other hand was opaque, bean-shaped and about 4–10 mm in length (Supplementary Figures S1C,D). Scanning electron microscope observation of their scoleces showed that T. caixuepengi larva had four suckers, 16 large hooks and 18 small hooks, the average length of which was about 250 and 100 μm, respectively (Figures 2A–C); T. tianguangfui larva also had four suction cups, and 31–33 large and small hooks of about 100 and 80 μm in length, respectively (Figures 2D–F).
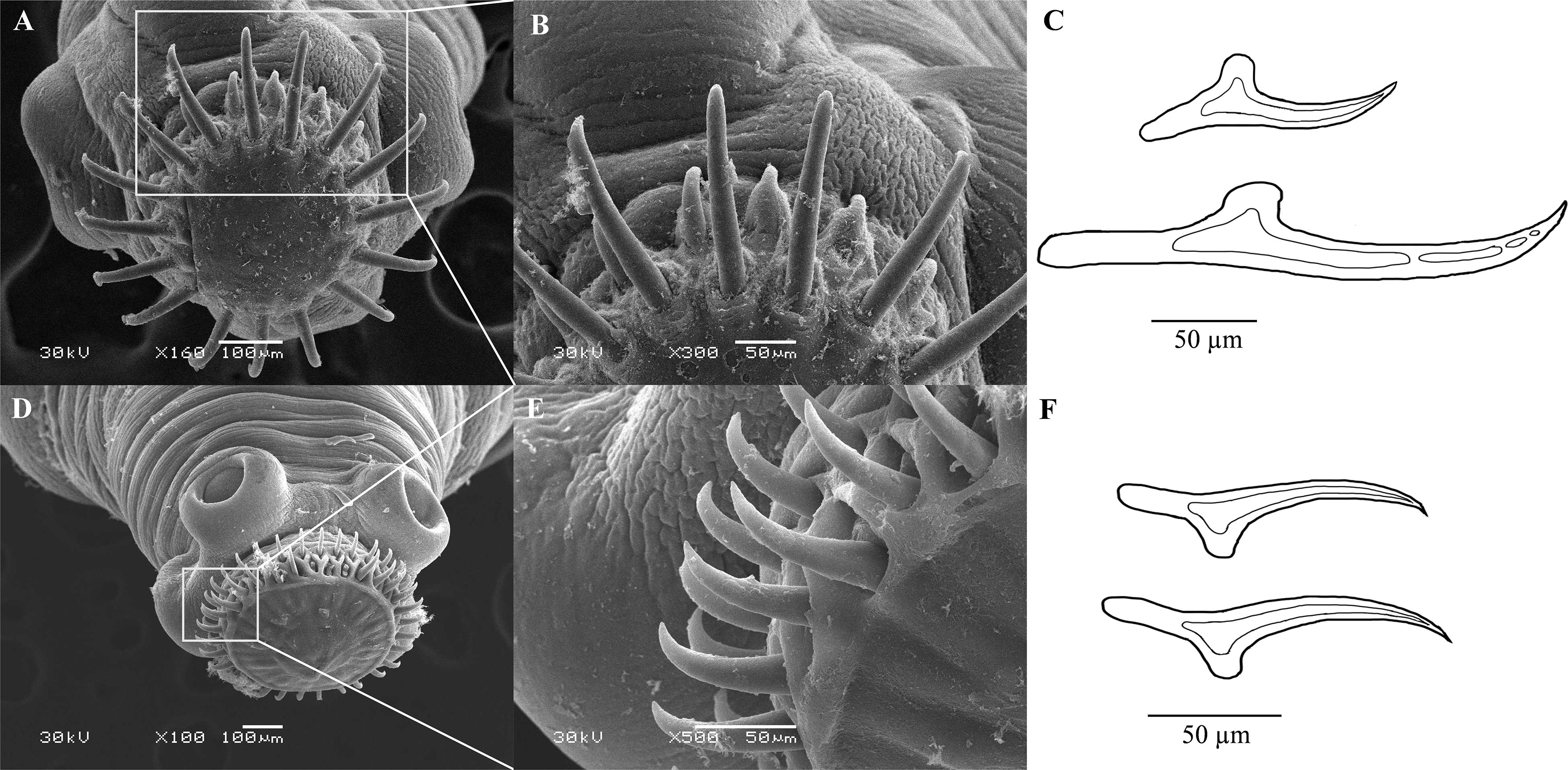
Figure 2. Scanning electron micrographs of scoleces and line drawing of hooks of Taenia tianguangfui larva (A–C) and Taenia caixuepengi larva (D–F).
Phylogenies inferred from both tandem amino acid sequences and DNA sequences of the 12 mt protein-encoding genes demonstrated T. caixuepengi in a monophyletic group with T. pisiformis and T. laticollis, with the closest genetic relative being T. pisiformis; T. tianguangfui was also found in a monophyletic group with T. crassiceps, T. twitchelli, and T. martis, and has a distant genetic relationship with T. caixuepengi (Figure 3).
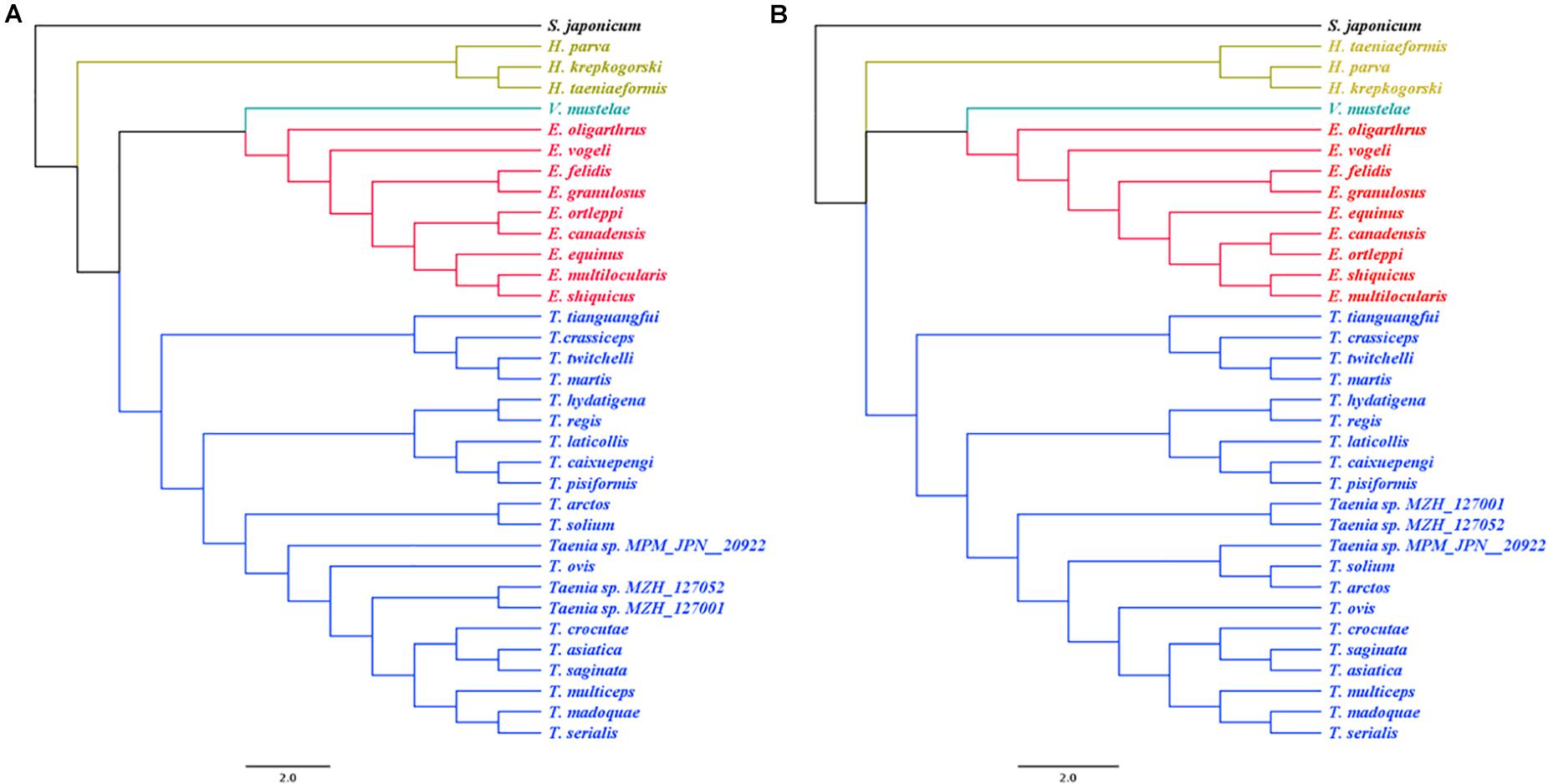
Figure 3. The phylogenetic relationship of Taenia tianguangfui and Taenia caixuepengi, with other 32 tapeworm species inferred from a Bayesian method based on the concatenated amino acid (A) and CDS alignments (B) of mitochondrial 12 protein-encoding genes. The species’ name corresponding to the GenBank ID is given in the Supplementary Table S2. The Echinococcus spp. are depicted in red, the Taenia spp. are depicted in blue, the Hydatigera spp. are depicted in yellow and the only one Versteria species, Versteria mustelae, is depicted in green. The Schistosoma japonicum depicted in black was chosen as outgroup.
The divergence time analysis based on mitochondrial protein-coding genes suggested that T. saginata and T. asiatica should diverge at 1.10 Mya (0.80–1.41, 95% highest probability density) in the early Pleistocene period, which is consistent with the previous reports based on genomic genes (Michelet and Dauga, 2012; Wang et al., 2016); T. caixuepengi should diverge from T. pisiformis 5.49 Mya (3.87–7.19, 95% highest probability density) in the initial Pliocene period, which is close to the divergence time between E. shiquicus and E. multilocularis (4.12 Mya, 2.81–5.32, 95% highest probability density); T. tianguangfui on the other hand, originated 13.11 Mya (9.36–17.18, 95% highest probability density) in the middle Miocene period, which was earlier than the differentiation of its intermediate host, N. fuscus (4.98 Mya, 4.08–5.90, 95% highest probability density) (Figure 4 and Supplementary Figure S2).
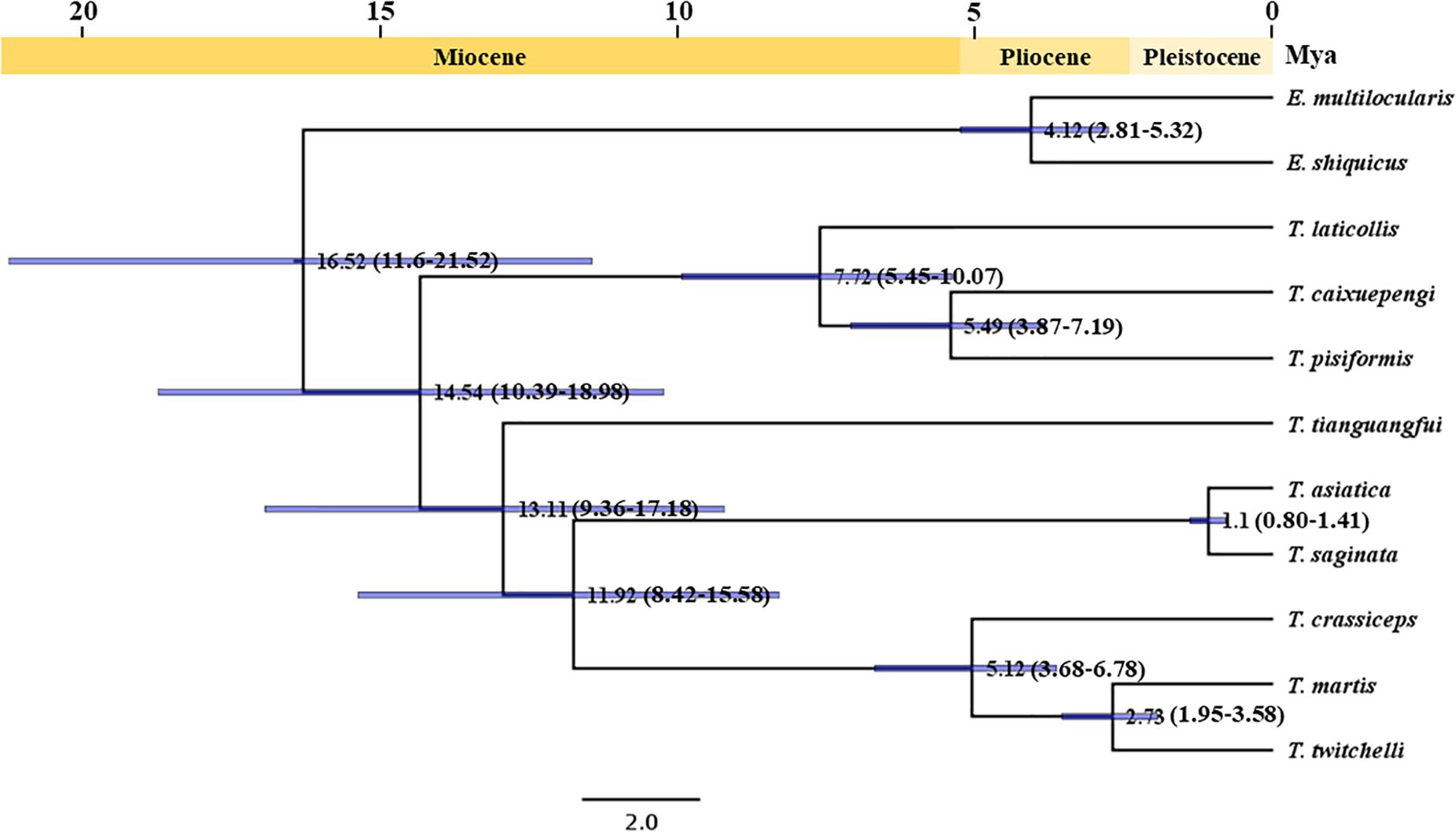
Figure 4. Divergence times construction for Taenia tianguangfui and Taenia caixuepengi based on the concatenated CDS alignments of mitochondrial 12 protein-encoding genes. The number at the node represents the divergence time between two lineages. The blue bar represents interval of 95% highest probability density, and the number in brackets represents the detailed time interval of 95% highest probability density of divergence time between two lineages. A time scale shows the extent of the Miocene, Pliocene and Pleistocene period.
The discovery of these two new parasites, T. caixuepengi and T. tianguangfui, highlights the species diversity of the family Taeniide, and further proved the true biodiversity characteristic of the QTP. Given the lack of human intervention and the rich diversity of wild host species, the present understanding of the species diversity within this family in QTP is apparently just a tip of the iceberg. This is not surprising, given the appreciable cryptic diversity so far uncovered within the taeniid family in Africa and northern latitudes (Lavikainen et al., 2011, 2013; Terefe et al., 2014).
Morphological features traditionally used to distinguish the cysticerci, including the number of hooks, the length of large hooks and small hooks (Loos-Frank, 2000), are insufficient in inferring evolutionary lineages. This is because homoplasy of morphological characters can represent a serious obstacle in taxonomic investigation (Scholz et al., 2020). Here the whole mt genomes of both species were sequenced, and clearly different from all available Taenia mt genome sequences, verifying the validity of their species status. Their mt genomes were similar as those of other sequenced tapeworms with respect to length, nucleotide bias, and their tRNA, rRNA and protein-encoding genes composition (Figure 1; Le et al., 2000; Nakao et al., 2003; Jeon et al., 2005; Jeon et al., 2007). Furthermore, the codon ATT was inferred as a more unusual start codon for the atp6 gene of T. caixuepengi (Table 1), which is a common start codon used by Caenorhabditis elegans and Ascaris suum (Okimoto et al., 1990).
T. caixuepengi larva is so far undetected in other animals, except plateau pika (O. curzoniae), meanwhile, no other cysticerci have been found in plateau pika hitherto. Lagomorph is the intermediate host of T. pisiformis and T. laticollis (Valdmann et al., 2004; Hallal-Calleros et al., 2016). Although similar in appearance and size to the vole, the plateau pika belongs to Lagomorpha (Smith et al., 2019). The close phylogenetic relatedness of these three Taenia species (Figure 3) is further highlighted by their high preference for lagomorphs as an intermediate host. Based on the divergence time and phylogeographic analyses, the extent pikas (genus Ochotona) originated on the QTP in the middle Miocene, ∼14 Mya (Wang et al., 2020). However, the rapid speciation of many Ochotona species, including O. curzoniae, occurred during the late Miocene and early Pliocene period (Wang et al., 2020), which almost coincided with the rapid uplift of the QTP (An et al., 2006; Li et al., 2007; Shi et al., 2015). Coincidentally, the evolutionary divergence time analysis in this study also suggests that both T. caixuepengi and E. shiquicus had evolved in the early Pliocene epoch, about 5.49 and 4.12 Mya, respectively (Figure 4). These almost synchronous events may not have happened by chance. Large-scale diversification of species is often provoked by abiotic factors, such as changes in the living environment and food supply (Benton, 2009). The uplift of the QTP from south to north provided climatic opportunities and food supply for the diversification of cold temperature-preferring pikas but led to the extinction of other warm temperature-preferring rodents (Wang et al., 2020).
For most free-living organisms, speciation is usually the result of genetic drift or adaptive differentiation between geographically separate populations (Turelli et al., 2001). For parasites, however, it has long been thought that sympatric speciation of parasites is common, mediated by ecological isolation caused by host switching within the same geographic region (de Meeûs et al., 1998; Paul, 2002; Huyse et al., 2005). Therefore, we speculate that T. pisiformis in the QTP (Li et al., 2013) may share a common ancestor with T. caixuepengi; the split of the pika population caused the ecological isolation between their ancestral populations, which further resulted in the lack of gene flow between them due to intermediate host switching, and the eventual formation of two different species. It can also be speculated that the differentiation pattern between E. shiquicus and E. multilocularis is similar as that of T. caixuepengi and T. pisiformis.
Our evolutionary divergence time analysis suggests that the speciation of T. tianguangfui occurred in the middle Miocene period (∼13.11 Mya) (Figure 4) when the QTP was undergoing a slow uplift period (An et al., 2006). The timing of the divergence of N. fuscus evolved from ∼4.98 Mya (Supplementary Figure S2), which also coincided with the rapid uplift of the QTP (An et al., 2006; Li et al., 2007; Shi et al., 2015). As the species spread in the QTP and Himalaya (Pradhan et al., 2019), the evolutionary origin of the Neodon spp., like the plateau pika, may well be due to changes in climate and food supply caused by the uplift of the QTP and Himalaya. The speciation of T. tianguangfui was earlier than that of its intermediate host, indicating that T. tianguangfui did not differentiate into a new Taenia species in order to adapt to the intermediate host, rather, it suggests that T. tianguangfui larva might not be limited to N. fuscus. Taenia crassiceps and T. martis have similar intermediate hosts range, infecting a variety of rodents, even humans and other primates (Deplazes et al., 2019). Given the close relationship between T. tianguangfui, T. crassiceps, and T. martis, it also cannot be excluded that T. tianguangfui may be infective to a variety of rodents other than N. fuscus, as well as humans and other primates. So far, a clear understanding of their evolutionary origin from these clues is elusive, thus, more data and investigation are needed to provide further insight.
Adult worms of the T. tianguangfui and T. caixuepengi have not yet been collected due to the difficulty in sampling endangered or protected carnivores. However, nucleic acid from feces of wolves, foxes and dogs found at the sampling sites were examined, so far, no positive feces samples for these two species have been found. Plateau pikas and voles are the primary food source for wild canids across the QTP. Tibetan foxes are the obligate predator of plateau pikas, as their remains (plateau pikas) are often encountered in 99% of their feces (Smith et al., 2019). Wild canids, especially the red fox and the Tibetan fox, may well be important definitive hosts for T. tianguangfui and T. caixuepengi.
Adult or larval samples of tapeworm are easily damaged in the process of collection, freeze-thaw and processing, and the morphological features are mostly unidentifiable (Lavikainen et al., 2013). While mt genome data alone may not fully answer the scientific questions surrounding their evolutionary origins, it is the most cost-effective and accurate method. Recently, although laborious and costly, there have been an increasing whole genome sequencing and analyses for many tapeworm species. This kind of investigation, not only is it important to provide insights into their host adaptation and switching, evolution mechanisms through gene groups amplification, hosts-parasites interaction, immune regulation and nutrition, it also provides urgently needed resources for the identification of drug target and diagnostic molecular markers (Wang et al., 2016; International Helminth and Genomes Consortium, 2019). In the future, a lot of genomic data will be needed to study this fascinating group.
In conclusion, the mitochondrial genome sequence data adequately confirm the validity of the two new Taenia species named T. caixuepengi and T. tianguangfui, we have previously reported. The phylogenetic trees and divergence times analyses suggest that T. caixuepengi evolve from its closest relative, T. pisiformis, in the initial Pliocene period (∼5.49 Mya), due to the intermediate host switching caused by the rapid uplift of the QTP; T. tianguangfui is probably parasitic in a wide variety of rodents, and share a common ancestor with T. crassiceps, T. twitchelli and T. martis, splitting in middle Miocene period (∼13.11 Mya).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nucleotide. GenBank ID: MT882036 and MT882037.
The animal study was reviewed and approved by the Animal Ethics Procedures and Guidelines of the People’s Republic of China and Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (No. LVRIAEC2012-007).
Y-DW, LL, H-BY, and W-ZJ conceived and designed the experiments. Y-DW, Y-LF, and X-WN performed the experiments. Y-DW and Y-LF performed the data analyses. Y-DW prepared the figures and wrote the manuscript. JO provided improving paragraphs and language editing. W-HL, N-ZZ, and B-QF provided very constructive suggestions for revisions. All authors read and approved the final manuscript.
This work was funded by the National Key Research and Development Plan (2018YFC1602504 and 2017YFD0501301), Central Public-Interest Scientific Institution Basal Research Fund (1610312020016), and Cultivation of Achievements of State Key Laboratory of Veterinary Etiological Biology (SKLVEB2020CGPY01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.647119/full#supplementary-material
Supplementary Figure 1 | The larvae of Taenia caixuepengi and Taenia tianguangfui were found in Plateau pikas (Ochotona curzoniae) and Qinghai voles (Neodon fuscus), respectively. The banded larvae of T. caixuepengi in the plate (A) were picked out from the enterocoelia of Plateau pikas (B), and the granular larvae of T. tianguangfui in the plate (C) were picked out from the enterocoelia and chest of Qinghai voles (D). The blue arrow indicates the parasitic larvae.
Supplementary Figure 2 | Divergence times construction for Neodon fuscus based on the concatenated CDS alignments of mitochondrial 13 protein-encoding genes. The number at the node represents the divergence time between two lineages. The blue bar represents interval of 95% highest probability density, and the number in brackets represents the detailed time interval of 95% highest probability density of divergence time between two lineages. A time scale shows the extent of the Miocene, Pliocene and Pleistocene period.
Supplementary Table 1 | Detailed sample collection data.
Supplementary Table 2 | The primers for amplifying the complete mitochondrial genomes of cysticercus.
Supplementary Table 3 | GenBank accession numbers of mitochondrial genome sequences used for phylogenetic analyses and divergence times analyses in this study.
An, Z. S., Zhang, P. Z., Wang, E. Q., Wang, S. M., Qiang, X. K., Li, L., et al. (2006). Changes of the monsoon−arid environment in China and growth of the Tibetan Plateau since the Miocene. Q. Sci. 26, 678–693.
Benton, M. J. (2009). The Red Queen and the Court Jester: species diversity and the role of biotic and abiotic factors through time. Science 323, 728–732. doi: 10.1126/science.1157719
Bowles, J., Blair, D., and McManus, D. P. (1992). Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol. Biochem. Parasitol. 54, 165–173. doi: 10.1016/0166-6851(92)90109-w
Dahal, N., Lissovsky, A. A., Lin, Z., Solari, K., Hadly, E. A., Zhan, X., et al. (2017). Genetics, morphology and ecology reveal a cryptic pika lineage in the Sikkim Himalaya. Mol. Phylogenet. Evol. 106, 55–60. doi: 10.1016/j.ympev.2016.09.015
de Meeûs, T., Michalakis, Y., and Renaud, F. (1998). Santa rosalia revisited: or why are there so many kinds of parasites in; the garden of earthly delights’? Parasitol. Today 14, 10–13. doi: 10.1016/s0169-4758(97)01163-0
Deplazes, P., Eichenberger, R. M., and Grimm, F. (2019). Wildlife-transmitted Taenia and Versteria cysticercosis and coenurosis in humans and other primates. Int. J. Parasitol. Parasites Wildl. 9, 342–358. doi: 10.1016/j.ijppaw.2019.03.013
Flisser, A., Correa, D., Avilla, G., and Marvilla, P. (2005). “Biology of Taenia solium, Taenia saginata and Taenia saginata asiatica,” in WHO/FAO/OIE Guidelines for the Surveillance, Prevention and Control of Taeniosis/Cysticercosis, eds K. D. Murrell Paris (Geneva: World Health Organization), 14–22.
Hajibabaei, M., Singer, G. A., Hebert, P. D., and Hickey, D. A. (2007). DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 23, 167–172. doi: 10.1016/j.tig.2007.02.001
Hallal-Calleros, C., Morales-Montor, J., Orihuela-Trujillo, A., Togno-Peirce, C., Murcia-Mejía, C., Bielli, A., et al. (2016). Taenia pisiformis cysticercosis induces decreased prolificacy and increased progesterone levels in rabbits. Vet. Parasitol. 229, 50–53. doi: 10.1016/j.vetpar.2016.09.015
Hebert, P. D., and Gregory, T. R. (2005). The promise of DNA barcoding for taxonomy. Syst. Biol. 54, 852–859. doi: 10.1080/10635150500354886
Hoberg, E. P., Jones, A., Rausch, R. L., Eom, K. S., and Gardner, S. L. (2000). A phylogenetic hypothesis for species of the genus Taenia (Eucestoda : Taeniidae). J. Parasitol. 86, 89–98. doi: 10.1645/0022-33952000086[0089:APHFSO]2.0.CO;2
Hoberg, E. P. (2006). Phylogeny of Taenia: Species definitions and origins of human parasites. Parasitol. Int. 55, S23–S30. doi: 10.1016/j.parint.2005.11.049
Huyse, T., Poulin, R., and Théron, A. (2005). Speciation in parasites: a population genetics approach. Trends Parasitol. 21, 469–475. doi: 10.1016/j.pt.2005.08.009
International Helminth and Genomes Consortium. (2019). Comparative genomics of the major parasitic worms. Nat. Genet. 51, 163–174. doi: 10.1038/s41588-018-0262-1
Jeon, H. K., Kim, K. H., and Eom, K. S. (2007). Complete sequence of the mitochondrial genome of Taenia saginata: comparison with T. solium and T. asiatica. Parasitol. Int. 56, 243–246. doi: 10.1016/j.parint.2007.04.001
Jeon, H. K., Lee, K. H., Kim, K. H., Hwang, U. W., and Eom, K. S. (2005). Complete sequence and structure of the mitochondrial genome of the human tapeworm. Taenia asiatica (Platyhelminthes; Cestoda). Parasitology 130, 717–726. doi: 10.1017/s0031182004007164
Jia, W. Z., Yan, H. B., Guo, A. J., Zhu, X. Q., Wang, Y. C., Shi, W. G., et al. (2010). Complete mitochondrial genomes of Taenia multiceps, T. hydatigena and T. pisiformis: additional molecular markers for a tapeworm genus of human and animal health significance. BMC Genomics 11:447. doi: 10.1186/1471-2164-11-447
Jia, W., Yan, H., Lou, Z., Ni, X., Dyachenko, V., Li, H., et al. (2012). Mitochondrial genes and genomes support a cryptic species of tapeworm within Taenia taeniaeformis. Acta Trop. 123, 154–163. doi: 10.1016/j.actatropica.2012.04.006
Kinkar, L., Laurimäe, T., Acosta-Jamett, G., Andresiuk, V., Balkaya, I., Casulli, A., et al. (2018). Global phylogeography and genetic diversity of the zoonotic tapeworm Echinococcus granulosus sensu stricto genotype G1. Int. J. Parasitol. 48, 729–742. doi: 10.1016/j.ijpara.2018.03.006
Laslett, D., and Canbäck, B. (2008). ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24, 172–175. doi: 10.1093/bioinformatics/btm573
Lavikainen, A., Haukisalmi, V., Deksne, G., Holmala, K., Lejeune, M., Isomursu, M., et al. (2013). Molecular identification of Taenia spp. in the Eurasian lynx (Lynx lynx) from Finland. Parasitology 140, 653–662. doi: 10.1017/S0031182012002120
Lavikainen, A., Laaksonen, S., Beckmen, K., Oksanen, A., Isomursu, M., and Meri, S. (2011). Molecular identification of Taenia spp. in wolves (Canis lupus), brown bears (Ursus arctos) and cervids from North Europe and Alaska. Parasitol. Int. 60, 289–295. doi: 10.1016/j.parint.2011.04.004
Le, T. H., Blair, D., Agatsuma, T., Humair, P. F., Campbell, N. J., Iwagami, M., et al. (2000). Phylogenies inferred from mitochondrial gene orders-a cautionary tale from the parasitic flatworms. Mol. Biol. Evol. 17, 1123–1125. doi: 10.1093/oxfordjournals.molbev.a026393
Li, D. P., Zhao, Y., Hu, J. M., Wan, J. L., Li, X. L., Zhou, X. K., et al. (2007). Fission track thermochronologic constraints on plateau surface and geomorphic relief formation in the northwestern margin of the Tibetan Plateau. Acta Petrologica Sinica 23, 900–910.
Li, J. Q., Li, L., Fu, B. Q., Yan, H. B., and Jia, W. Z. (2019). Complete mitochondrial genomes confirm the generic placement of the plateau vole. Neodon fuscus. Biosci. Rep. 39:BSR20182349. doi: 10.1042/BSR20182349
Li, W., Guo, Z., Duo, H., Fu, Y., Peng, M., Shen, X., et al. (2013). Survey on helminths in the small intestine of wild foxes in Qinghai. China. J. Vet. Med. Sci. 75, 1329–1333. doi: 10.1292/jvms.13-0187
Liu, G. H., Lin, R. Q., Li, M. W., Liu, W., Liu, Y., Yuan, Z. G., et al. (2011). The complete mitochondrial genomes of three cestode species of Taenia infecting animals and humans. Mol. Biol. Rep. 38, 2249–2256. doi: 10.1007/s11033-010-0355-0
Loos-Frank, B. (2000). An up-date of Verster’s (1969) ‘Taxonomic revision of the genus Taenia Linnaeus’ (Cestoda) in table format. Syst. Parasitol. 45, 155–183. doi: 10.1023/a:1006219625792
Lymbery, A. J. (1998). Combining data from morphological traits and genetic markers to determine transmission cycles in the tape worm, Echinococcus granulosus. Parasitology 117, 185–192. doi: 10.1017/s0031182098002911
Lymbery, A. J. (2017). Phylogenetic Pattern, Evolutionary Processes and Species Delimitation in the Genus Echinococcus. Adv. Parasitol. 95, 111–145. doi: 10.1016/bs.apar.2016.07.002
Mathis, A., and Deplazes, P. (2006). Copro-DNA tests for diagnosis of animal taeniid cestodes. Parasitol. Int. 55, S87–S90. doi: 10.1016/j.parint.2005.11.012
Michelet, L., and Dauga, C. (2012). Molecular evidence of host influences on the evolution and spread of human tapeworms. Biol. Rev. Camb. Philos. Soc. 87, 731–741. doi: 10.1111/j.1469-185X.2012.00217.x
Nakao, M., Lavikainen, A., Iwaki, T., Haukisalmi, V., Konyaev, S., Oku, Y., et al. (2013). Molecular phylogeny of the genus Taenia (Cestoda: Taeniidae): proposals for the resurrection of Hydatigera Lamarck, 1816 and the creation of a new genus Versteria. Int. J. Parasitol. 43, 427–437. doi: 10.1016/j.ijpara.2012.11.014
Nakao, M., McManus, D. P., Schantz, P. M., Craig, P. S., et al. (2007). A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes. Parasitology 134, 713–722. doi: 10.1017/S0031182006001934
Nakao, M., Sako, Y., and Ito, A. (2003). The mitochondrial genome of the tapeworm Taenia solium: a finding of the abbreviated stop codon U. J. Parasitol. 89, 633–635. doi: 10.1645/0022-3395(2003)089[0633:TMGOTT]2.0.CO;2
Nakao, M., Sako, Y., Yokoyama, N., Fukunaga, M., and Ito, A. (2000). Mitochondrial genetic code in cestodes. Mol. Biochem. Parasitol. 111, 415–424. doi: 10.1016/s0166-6851(00)00334-0
Okimoto, R., Macfarlane, J. L., and Wolstenholme, D. R. (1990). Evidence for the frequent use of TTG as the translation initiation codon of mitochondrial protein genes in the nematodes, Ascaris suum and Caenorhabditis elegans. Nucleic Acids Res. 18, 6113–6118. doi: 10.1093/nar/18.20.6113
Paul, R. (2002). Species concepts versus species criteria. Trends Parasitol. 18, 439–440. doi: 10.1016/s1471-4922(02)02319-x
Posada, D., and Crandall, K. A. (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Pradhan, N., Sharma, A. N., Sherchan, A. M., Chhetri, S., Shrestha, P., and Kilpatrick, C. W. (2019). Further assessment of the Genus Neodon and the description of a new species from Nepal. PLoS One 14:e0219157. doi: 10.1371/journal.pone.0219157
Rota-Stabelli, O., Yang, Z., and Telford, M. J. (2009). MtZoa: A general mitochondrial amino acid substitutions model for animal evolutionary studies. Mol. Phylogenet. Evol. 52, 268–272. doi: 10.1016/j.ympev.2009.01.011
Scholz, T., Waeschenbach, A., Oros, M., Brabec, J., and Littlewood, D. T. J. (2020). Phylogenetic reconstruction of early diverging tapeworms (Cestoda: Caryophyllidea) reveals ancient radiations in vertebrate hosts and biogeographic regions [published online ahead of print, 2020 Dec 2]. Int. J. Parasitol. 2020:009. doi: 10.1016/j.ijpara.2020.09.009
Shi, Z. G., Liu, X. D., and Sha, Y. Y. (2015). Did northern Tibetan Plateau uplift during Pliocene? A modeling test. J. Earth Env. 6, 67–80.
Singla, L. D., Singla, N., Parshad, V. R., Juyal, P. D., and Sood, N. K. (2008). Rodents as reservoirs of parasites in India. Integr. Zool. 3, 21–26. doi: 10.1111/j.1749-4877.2008.00071.x
Smith, A. T., Badingqiuying, Wilson, M. C., and Hogan, B. W. (2019). Functional-trait ecology of the plateau pika Ochotona curzoniae in the Qinghai-Tibetan Plateau ecosystem. Integr. Zool. 14, 87–103. doi: 10.1111/1749-4877.12300
Suzán, G., Marcé, E., Giermakowski, J. T., Armién, B., Pascale, J., Mills, J., et al. (2008). The effect of habitat fragmentation and species diversity loss on hantavirus prevalence in Panama. Ann. N. Y. Acad. Sci. 1149, 80–83. doi: 10.1196/annals.1428.063
Telford, M. J., Herniou, E. A., Russell, R. B., and Littlewood, D. T. (2000). Changes in mitochondrial genetic codes as phylogenetic characters: two examples from the flatworms. Proc. Natl. Acad. Sci. U. S. A. 97, 11359–11364. doi: 10.1073/pnas.97.21.11359
Terefe, Y., Hailemariam, Z., Menkir, S., Nakao, M., Lavikainen, A., Haukisalmi, V., et al. (2014). Phylogenetic characterisation of Taenia tapeworms in spotted hyenas and reconsideration of the “Out of Africa” hypothesis of Taenia in humans. Int. J. Parasitol. 44, 533–541. doi: 10.1016/j.ijpara.2014.03.013
Turelli, M., Barton, N. H., and Coyne, J. A. (2001). Theory and speciation. Trends Ecol. Evol. 16, 330–343. doi: 10.1016/s0169-5347(01)02177-2
Valdmann, H., Moks, E., and Talvik, H. (2004). Helminth fauna of Eurasian lynx (Lynx lynx) in Estonia. J. Wildl. Dis. 40, 356–360. doi: 10.7589/0090-3558-40.2.356
Wang, S., Wang, S., Luo, Y., Xiao, L., Luo, X., Gao, S., et al. (2016). Comparative genomics reveals adaptive evolution of Asian tapeworm in switching to a new intermediate host. Nat. Commun. 7:12845. doi: 10.1038/ncomms12845
Wang, X., Liang, D., Jin, W., Tang, M., Liu, S., and Zhang, P. (2020). Out of Tibet: Genomic Perspectives on the Evolutionary History of Extant Pikas. Mol. Biol. Evol. 37, 1577–1592. doi: 10.1093/molbev/msaa026
Wang, X., Liu, J., Zuo, Q., Mu, Z., Weng, X., Sun, X., et al. (2018). Echinococcus multilocularis and Echinococcus shiquicus in a small mammal community on the eastern Tibetan Plateau: host species composition, molecular prevalence, and epidemiological implications. Parasit. Vectors 11:302. doi: 10.1186/s13071-018-2873-x
Will, K. W., Mishler, B. D., and Wheeler, Q. D. (2005). The perils of DNA barcoding and the need for integrative taxonomy. Syst. Biol. 54, 844–851. doi: 10.1080/10635150500354878
Wu, Z., Lu, L., Du, J., Yang, L., Ren, X., Liu, B., et al. (2018). Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 6, 178. doi: 10.1186/s40168-018-0554-9
Xiao, N., Qiu, J., Nakao, M., Li, T., Yang, W., Chen, X., et al. (2005). Echinococcus shiquicus n. sp., a taeniid cestode from Tibetan fox and plateau pika in China. Int. J. Parasitol. 35, 693–701. doi: 10.1016/j.ijpara.2005.01.003
Keywords: mtDNA, Qinghai-Tibet Plateau, phylogeny, divergence time, Taenia spp.
Citation: Wu Y-D, Li L, Fan Y-L, Ni X-W, Ohiolei JA, Li W-H, Li J-Q, Zhang N-Z, Fu B-Q, Yan H-B and Jia W-Z (2021) Genetic Evolution and Implications of the Mitochondrial Genomes of Two Newly Identified Taenia spp. in Rodents From Qinghai-Tibet Plateau. Front. Microbiol. 12:647119. doi: 10.3389/fmicb.2021.647119
Received: 29 December 2020; Accepted: 23 February 2021;
Published: 23 March 2021.
Edited by:
Teresa Nogueira, Instituto Nacional Investigaciao Agraria e Veterinaria (INIAV), PortugalReviewed by:
Hamed Mirjalali, Shahid Beheshti University of Medical Sciences, IranCopyright © 2021 Wu, Li, Fan, Ni, Ohiolei, Li, Li, Zhang, Fu, Yan and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Bin Yan, eWFuaG9uZ2JpbkBjYWFzLmNu; Wan-Zhong Jia, amlhd2FuemhvbmdAY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.