- 1Muping Coastal Environment Research Station, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3School of Life Sciences, Nantong University, Nantong, China
- 4Key Laboratory of Coastal Environmental Processes and Ecological Remediation, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
- 5Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
Cnidarians have large surface areas available for colonization by microbial organisms, which serve a multitude of functions in the environment. However, relatively few studies have been conducted on scyphozoan-associated microbial communities. Blooms of scyphozoan species are common worldwide and can have numerous deleterious consequences on the marine ecosystem. Four scyphozoan species, Aurelia coerulea, Cyanea nozakii, Nemopilema nomurai, and Rhopilema esculentum, form large blooms in Chinese seas. In this study, we analyzed the bacterial communities associated with these four jellyfish based on 16S rRNA gene sequencing. We found that the bacterial communities associated with each scyphozoan species were significantly different from each other and from those of the surrounding seawater. There were no significant differences between the bacterial communities associated with different body parts of the four scyphozoan jellyfish. Core bacteria in various compartments of the four scyphozoan taxa comprised 57 OTUs (Operational Taxonomic Units), dominated by genera Mycoplasma, Vibrio, Ralstonia, Tenacibaculum, Shingomonas and Phyllobacterium. FAPROTAX function prediction revealed that jellyfish could influence microbially mediated biogeochemical cycles, compound degradation and transmit pathogens in regions where they proliferate. Finally, Six genera of potentially pathogenic bacteria associated with the scyphozoans were detected: Vibrio, Mycoplasma, Ralstonia, Tenacibaculum, Nautella, and Acinetobacter. Our study suggests that blooms of these four common scyphozoans may cause jellyfish species-specific impacts on element cycling in marine ecosystems, and serve as vectors of pathogenic bacteria to threaten other marine organisms and human health.
Introduction
The external and internal surfaces of marine animals are considered to be ubiquitously colonized by microorganisms (Grossart and Tang, 2010). Microbial associations are often dominated by bacteria but co-inhabited by fungi, protozoa, archaea, and viruses (Cárdenas et al., 2020; Deines et al., 2020). Microbial colonization of a certain surface is determined by the availability of nutrients, host immune responses, and competition between microbes from the surrounding environment for attachment space (Wilson et al., 2011). Microbial communities have therefore developed special mechanisms to interfere with the ability of adverse bacteria to colonize surfaces and acquire nutrients (Vezzulli et al., 2012). Microorganisms are vital to marine ecology, being involved in a multitude of exchange processes with the environment: respiration, exudation of wastes and secondary metabolites, absorption of energetic irradiation or informational signals, uptake of nutrients, and gases, and so on (Wahl et al., 2010). Microbes may influence developmental programs of hosts with complex life history strategies, such as inducing normal developmental changes in morphology and maturation of the immune system (Schuh et al., 2020). Changes in composition and function of microbiomes have been linked to disorders in many organisms (McKenney and Pamer, 2015; Contijoch et al., 2019). A recent study suggested microbes could directly affect the functional activity of the nervous system by controlling spontaneous body contractions (Klimovich et al., 2020). Development of multicellularity in eukaryotes may never have been autonomous, rather requiring transient or persistent interactions with the microbial world (Bosch and McFall-Ngai, 2021).
Bacterial communities associated with jellyfish mainly consisted of Alpha- and Gammaproteobacteria, Bacteroidetes, Tenericutes, and Cyanobacteria (Weiland-Bräuer et al., 2015; Daley et al., 2016; Basso et al., 2019; Kramar et al., 2019; Jaspers et al., 2020; Stabili et al., 2020). Certain jellyfish increase in numbers and form blooms or aggregations during certain periods, during which the population density of the pelagic medusae peaks distinctly over broad temporal and regional scales (Purcell et al., 2007; Condon et al., 2012; Purcell, 2012). It is well documented that jellyfish as voracious predators may graze upon a variety of plankton taxa (Colin et al., 2010; Granhag et al., 2011) and cause cascading effects on the food web (Zoccarato et al., 2016; Xiao et al., 2019). Jellyfish can stimulate bacterioplankton growth by direct release of nutrients from tissue, mucus secretion, excretion, and release of food particles when feeding (Titelman et al., 2006; Pitt et al., 2009; Condon et al., 2012). Increased bacterial abundance has been observed in the vicinity of decaying jellyfish (Titelman et al., 2006; Tinta et al., 2010; Blanchet et al., 2015; Guy-Haim et al., 2020; Tinta et al., 2020), such as the genera Vibrio and Pseudoalteromonas (Tinta et al., 2012). However, even more importantly, growth of bacterioplankton surrounding live jellyfish may be stimulated by the release of nutrients and bioavailable carbon (Titelman et al., 2006; Tinta et al., 2010; Condon et al., 2011). Hence, bacterial communities associated with jellyfish need to be resolved in order to understand the interaction between jellyfish and marine microbial communities, and its impact on biogeochemical cycles and marine ecosystems.
Jellyfish-associated bacterial communities have been found to play an important role in the life cycle of jellyfish. Planulae and propagules of some scyphozoans have been successfully induced to enter metamorphosis using bacteria found in the environment of settled polyps; in particular, larvae of Cassiopea andromeda responded to a species of Vibrio alginolyticus (Neumann, 1979; Hofmann et al., 1996), and the propagules (pedal stolons) of Aurelia aurita to a species of Micrococcaceae (Schmahl, 1985). The asexual reproduction and strobilation of A. aurita was severely inhibited in the absence of the native microbiota (Weiland-Bräuer et al., 2020). Moreover, Lee et al. (2018) identified OTUs within Chrysaora plocamia and A. aurita most closely related to a suite of metabolically and physiologically diverse microorganisms capable of mediating the interrelated pathways of carbon, nitrogen, sulfur, and phosphorus. Several jellyfish-associated bacteria were previously also associated with the processing of peculiar substances, such as polycyclic aromatic hydrocarbons (PAHs), plastics, and xenobiotics found in the ocean, with possible benefits for the host. For example, Almeda et al. (2013) suggested the presence of PAH-degrading bacteria in A. aurita; Kramar et al. (2019) detected PAH and plastic-degrading bacteria (Burkholderia, Achromobacter and Kocuria) within the gastric cavity of A. aurita.
Recent studies have indicated that scyphozoan jellyfish may be key vectors of bacterial pathogens, and may thus harm cultured organisms and human well-being (Ferguson et al., 2010; Schuett and Doepke, 2010; Delannoy et al., 2011; Småge et al., 2017; Basso et al., 2019; Clinton et al., 2020). An outbreak of disease in farmed Atlantic salmon has been linked to Tenacibaculum maritimum isolated within the adherent microbial communities of the jellyfish Phialella quadrata and Pelagia noctiluca (Ferguson et al., 2010; Delannoy et al., 2011). Therefore, it is essential to identify the bacteria associated with the scyphozoans to understand the role scyphozoan jellyfish may play in the spread of bacterial disease. Recently, a number of potential pathogens of commercial aquaculture have been identified, including Moritella viscosa in the jellyfish Cyanea lamarckii (Schuett and Doepke, 2010); Chryseobacterium, Flavobacterium, Tenacibaculum, Coxiella and Vibrio in the blooming scyphozoan Rhizostoma pulmo (Basso et al., 2019); and Aeromonas salmonicida, A. molluscorum, Pseudomonas fluorescens, P. fulva and Vibrio splendidus in the Lion’s Mane jellyfish Cyanea capillata (Clinton et al., 2020).
The association of bacteria with jellyfish is highly dynamic and complex, specifically expressed as distinction between jellyfish microbiomes and the ambient water, as well as the population specificity, taxa specificity, life stages specificity and body part specificity of the microbial groups associated with jellyfish (Schuett and Doepke, 2010; Tinta et al., 2012; Cortés-Lara et al., 2015; Weiland-Bräuer et al., 2015; Viver et al., 2017; Lee et al., 2018; Basso et al., 2019; Hao et al., 2019; Kramar et al., 2019; Tinta et al., 2019). To the authors’ knowledge, no studies to date have focused on the microbiomes associated with jellyfish species in Chinese Seas. The present study focuses on the bacterial communities associated with Aurelia coerulea, Cyanea nozakii and Nemopilema nomurai, as the scyphozoans with the highest frequencies of outbreaks in Chinese seas (Dong et al., 2010; Wang P.P. et al., 2020), as well as Rhopilema esculentum, which is an economically valuable edible jellyfish and is artificially proliferated and released to increase its population. For example, the biomass of A. coerulea can reach 45.45 tons km–2 in Chinese local outbreak areas (Wang P.P. et al., 2020), C. nozakii has been proven to be able to reach a biomass of 4000-6000 individuals km–2 during bloom periods (Zhang et al., 2012), and N. nomurai during the outbreak periods may reach a maximum abundance of 15-75 tons km–2 in some China waters (Wang P.P. et al., 2020). The total harvest of R. esculentum peaked at 0.43 million tons in 1998 (Dong et al., 2014). The aims of our research were to characterize the differences and similarities of the bacterial communities associated with the four blooming scyphozoans in Chinese Seas based on 16S rRNA gene sequencing. Ecological functions and potentially pathogenic bacteria within scyphomedusa-associated bacterial communities are also discussed to provide insights into the potential consequences for marine organisms and humans health.
Materials and Methods
Sample Collection and Processing
Scyphomedusa specimens were carefully collected from Jiaozhou Bay, Qingdao (120.298 E, 36.066 N; QD) and Shidao Bay, Rongcheng (122.414 E, 36.918 N; RC), in Shandong province, in the Northern Yellow Sea of China (Figure 1). All samples were collected at the peak abundance at which A. coerulea (Au) and C. nozakii (Cy) occurred in Qingdao, and N. nomurai (Ne) and R. esculentum (Rh) occurred in Rongcheng in August 2018 (Figure 1). Scyphomedusae were temporarily placed in 20 L aerated tanks with ambient seawater which were transported to the laboratory in 4 hours. Five samples of each species which had empty guts were individually washed three times with sterile seawater (0.22 μm pre-filtered and autoclaved) to remove loosely associated microorganisms and bacteria from the surrounding seawater. After rinsing, each individual was dissected into the following parts with sterilized scalpels: umbrella (U), oral arms (O), stomach (S) and gonad (G). Umbrella, oral arms and stomach were collected from the individuals of A. coerulea; umbrella, oral arms and gonad were collected from the other three scyphomedusa species (C. nozakii, N. nomurai and R. esculentum). Specimens were then given a final rinse with sterile seawater, flash-frozen with liquid nitrogen, and stored at −80°C. Bulk seawater samples (500 mL) collected at the same time as scyphomedusa collection from each location were subjected to sequential filtration. Particle-associated bacteria were removed with 3 μm-pore size filters (TCTP, 47 mm, Millipore, Germany). To collect free-living bacteria, the resulting filtrate was subsequently filtered on a 0.22 μm-pore size filter (GTTP, 47 mm, Millipore, Germany). This was repeated three times for each location to obtain bacteria from the water column for comparison with scyphomedusa-associated bacteria. In total, 60 scyphomedusa samples (4 species × 3 body parts × 5 individuals) and 6 seawater samples (2 locations × 3 replicates) were prepared for DNA extraction.
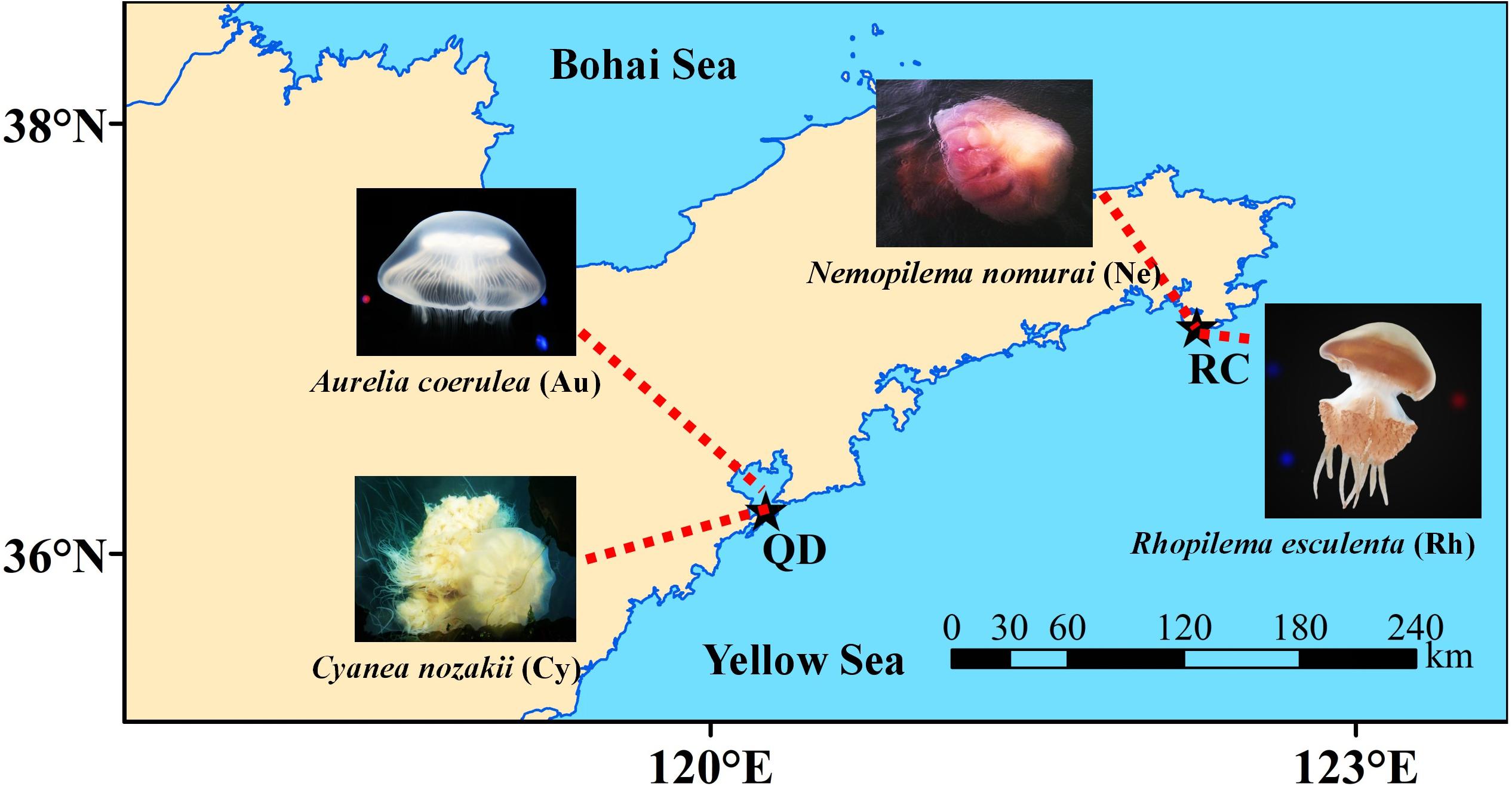
Figure 1. Sampling sites for four blooming scyphozoan jellyfish. Note: Abbreviations: QD, Jiaozhou Bay in Qingdao; RC, Shidao Bay in Rongcheng.
DNA Extraction and 16S rRNA Gene Amplicon Sequencing
one hundred milli gram of each scyphomedusa sample were homogenized for bacterial DNA extraction using CTAB (cetyl-trimethyl-ammonium bromide), as described by Hao (2014) with slight modifications. The whole filter membranes of each seawater sample were used to extract the total DNA of seawater samples using a PowerWater DNA Isolation Kit (MOBIO, United States) according to the manufacturer’s instructions. DNA was diluted to 1 ng/μL using sterile water. The variable 4 and 5 regions of 16S rRNA genes were amplified used specific primers 515F (5′-GTG CCA GCM GCC GCG GTA A-3′) and 907R (5′-CCG TCA ATT CCT TTG AGT TT-3′) (Walters et al., 2015) with the forward primer modified to contain a unique 6 nt barcode at the 5′ end. All PCR reactions were carried out using the Phusion High-Fidelity PCR Master Mix (New England Biolabs) with 0.2 μM of each primer and 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s, and finally 72°C for 5 min. All PCRs were performed in triplicate, and no-template controls were included in all steps of the process. PCR products were detected by electrophoresis in a 2% (w/v) agarose gel. PCR amplicons of each sample with bright bands were mixed in equal density ratios and purified with GeneJETTM Gel Extraction Kit (Thermo Scientific). The amplicon libraries were generated using Ion Plus Fragment Library Kit 48 rxns (Thermo Scientific) and sequenced using the Ion S5TM XL platform at Novogene Bioinformatics Technology Co., Ltd. (Beijing, China).
Sequence Analysis of the 16S rRNA Amplicons
Single-end reads were assigned to samples based on unique barcode and truncated by cutting off the barcode and primer sequence. Quality filtering on the raw reads was performed under specific filtering conditions to obtain the high-quality clean reads according to the Cutadapt (v1.9.1) quality control process (Martin, 2011). All raw reads were compared with the SILVA 1321 (Quast et al., 2013) using the UCHIME algorithm (Robert et al., 2011) to detect chimera sequences, and then the chimera sequences were removed to obtain the Clean Reads (Brian et al., 2011).
Sequences with ≥ 97% similarity were assigned to the same operational taxonomic units (OTUs) and the representative sequence for each OTU was screened for further annotation with Uparse software (v7.0.1001) (Robert, 2013). For each representative sequence, assignments of taxonomic annotations from the phylum to species levels to the clean OTU sequences were performed using the Silva Database based on the Mothur algorithm (Quast et al., 2013). The abundance data for the OTUs were normalized using a standard of sequence number corresponding to the sample with the least sequences. Then the original OTU data of the repeated samples of each body part of each scyphozoan as well as seawater samples were combined by averaging. Subsequently, the combined OTU data of the three body parts of each scyphozoan were averaged and merged to obtain the OTU data of a single total scyphozoan.
Data Analysis
Multiple Alpha diversity indices (Observed OTUs, Chao1, ACE, Shannon, Simpson and Good’s coverage) were calculated based on 37847 reads per sample (minimum number of sequences required to normalize the differences in sequencing depth) using QIIME and a box plot of the Shannon index was visualized using R software (V3.6.2). Statistically significant differences of the Shannon indices among the microbiota of the four scyphozoan taxa, the microbiota of the scyphozoans and ambient seawater, and the microbiota of the three compartments of each scyphozoan taxa were set at p < 0.05 using a Wilcoxon test based on IBM SPSS Statistics software (V22). The Principal Co-ordinates Analysis (PCoA) was performed to determine the Beta diversity of samples based on unweighted Unifrac distance at OTU level and visualized using ggplot2 and vegan package of R software (V3.6.2). Permutational analysis of molecular variance (PERMANOVA) was conducted to test statistical differences in the Beta diversity of the microbiota of the four scyphozoan taxa, the microbiota of the scyphozoans and ambient seawater, and the microbiota of the three compartments of each scyphozoan taxa using Primer software (Version 6, PRIMER-E Ltd., Lutton, United Kingdom), with 999 random permutations of the appropriate units. The relative abundances of dominant bacteria at distinct classification levels among scyphozoan groups and seawater groups were demonstrated using stacked bar plots, and visualized with Origin software (Version Pro 2018). The core bacterial communities associated with all the studied body parts of the four scyphozoan taxa were selected based on the relative abundance information of the OTUs, shown in a bubble plot using R software (V3.6.2). Functional Annotation of Prokaryotic Taxa (FAPROTAX) (Louca et al., 2016) was applied to predict the main ecological functions of the microbial communities associated with four scyphozoan species and ambient seawater. The cumulative absolute abundances of the OTUs contributing to functional groups were square root transformed after normalization and visualized in a heat-map with R software (Version 3.6.2). Then a Mann-Whitney U-test was carried out to determine significantly different functional groups of bacteria between scyphozoan and seawater, while a Kruskal-Wallis test was conducted to verify significant differences between functional groups of the four scyphozoan species, both with IBM SPSS Statistics software (V22). A phylogenetic tree was constructed by the Neighbor-joining method using Mega software (Version X) based on the V4-V5 region of the 16S rRNA gene (Kumar et al., 2018). Sequence data from identified pathogenic strains deposited in the NCBI database for 16S rRNA gene were used in this study.
Results
The bacterial communities associated with four scyphomedusae and surrounding seawater were analyzed by Ion S5TM XL sequencing which generated 4,351,887 filtered high-quality sequences with an average of 69,078 sequences for each sample. In total, 1888 OTUs were defined by 97% sequence similarity, clustered into 2 Kingdoms (Bacteria and Archaea), 41 phyla, 53 classes, 115 orders, 228 families, and 547 genera. The coverage of all samples in this study was ≥ 99.7% and Shannon curves showed saturation for all samples, indicating good coverage of the bacterial communities in the present study (Supplementary Table 1 and Supplementary Figure 1). In the following analysis, only bacteria with a relative abundance higher than 1% in at least one group were considered.
Bacterial Community Composition
The relative abundance of taxa identified to the most resolvable taxa (phylum, class, order, and genus) were shown as stacked bar plots for the analyzed groups (Figure 2). At the level of phylum and class, the bacterial communities associated with A. coerulea was dominated by Proteobacteria (82.27%), which mainly consisted of the class Gammaproteobacteria (80.57%), followed by Firmicutes (of which Bacilli was the most common class, 6.63%). Proteobacteria (45.75%) and Tenericutes (28.50%) were predominant in the bacterial communities of C. nozakii, with Gammaproteobacteria (19.04%) and Alphaproteobacteria (26.67%) the most common classes. Oxyphotobacteria (34.67%), Actinobacteria (33.79%), Bacteroidetes (21.94%) and Proteobacteria (7.78%) were most common in the surrounding seawater of Qingdao. Tenericutes were the most abundant bacteria in N. nomurai and R. esculentum with 47.10% and 25.14%, respectively, followed by Proteobacteria, accounting for 22.35% and 30.37%, respectively. Proteobacteria (57.46%) and Bacteroidetes (39.78%) were dominant in the surrounding seawater of Rongcheng.
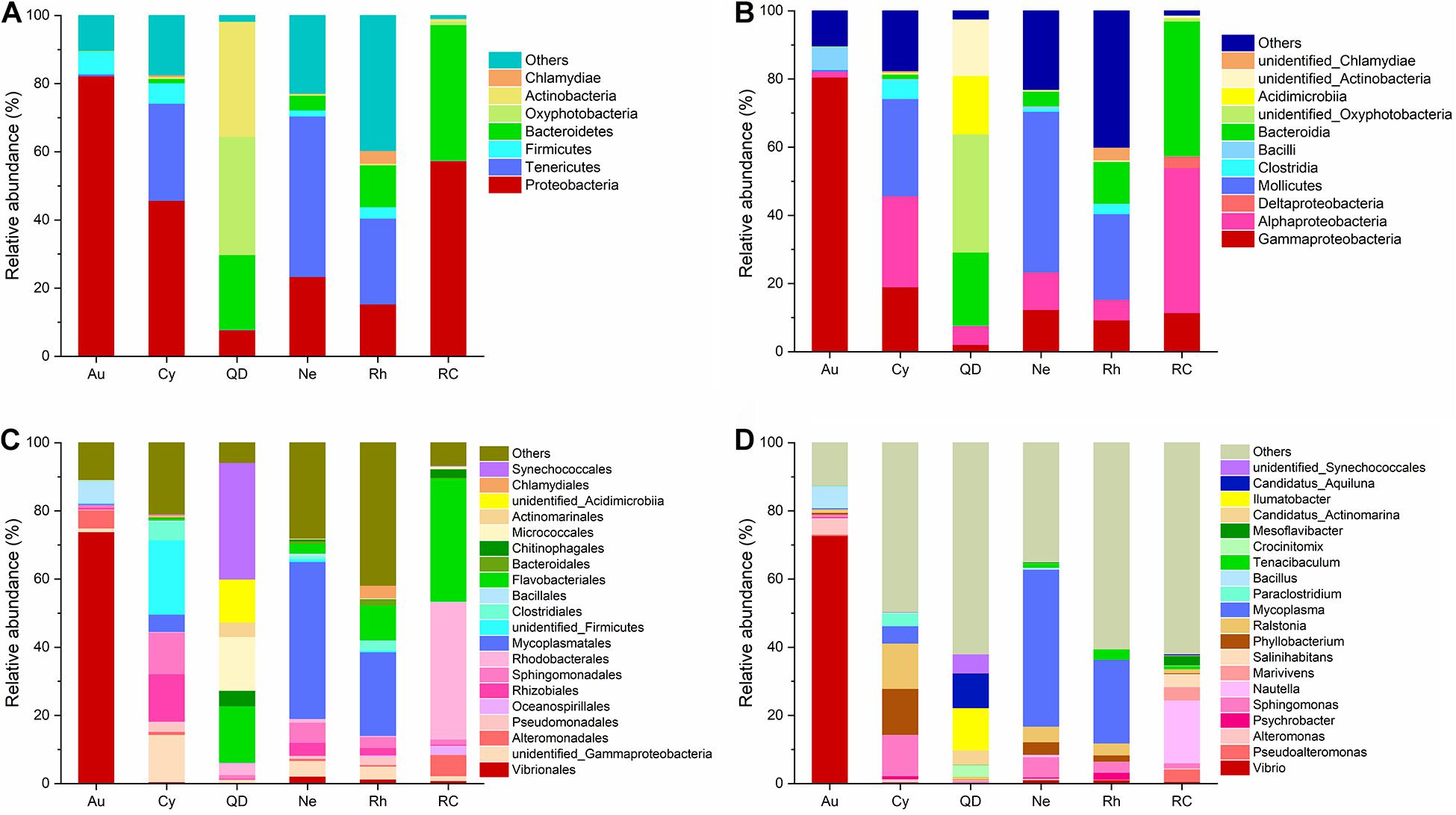
Figure 2. Stacked Bar plots of bacterial taxa at (A) phylum level, (B) class level, (C) order level, and (D) genus level, in scyphozoan-associated bacterial communities and free-living bacteria in surrounding seawater. Note: Abbreviations: Au, A. coerulea from Jiaozhou Bay in Qingdao; Cy, C. nozakii from Jiaozhou Bay in Qingdao; Ne, N. nomurai from Shidao Bay in Rongcheng; Rh, R. esculentum from Shidao Bay in Rongcheng; QD, ambient seawater from Jiaozhou Bay in Qingdao; RC, ambient seawater from Shidao Bay in Rongcheng. (A) Phylum and (B) class levels list taxa with mean relative abundance > 1% in at least one group, and those with mean relative abundance of < 1% across all groups were subsumed under “others.” (C) Order and (D) genus levels list the most abundant 20 bacterial taxa with mean relative abundance > 1% in at least one group, with less abundant taxa included under “others.”
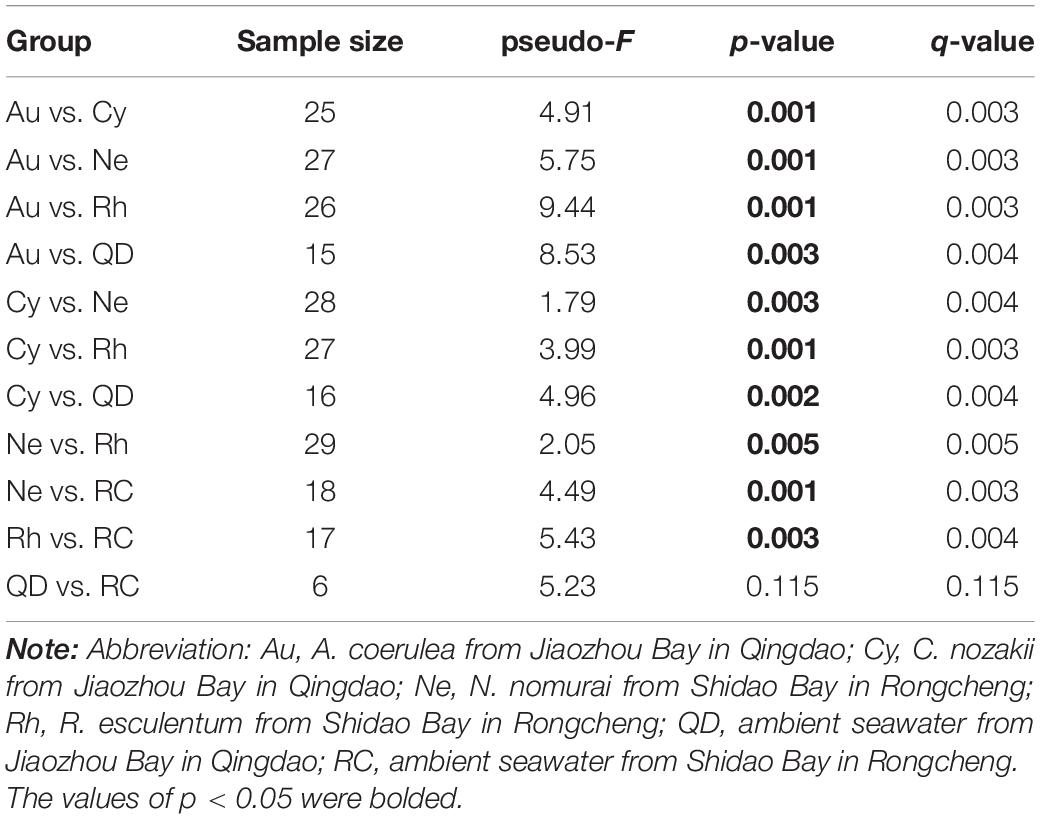
At the level of order and genus, the most abundant bacteria in A. coerulea were Vibrionales (73.90%, dominated by genus Vibrio), Bacillales (6.48%, dominated by genus Bacillus) and Alteromonadales (5.31%, dominated by genus Alteromonas). Prevailing bacteria in C. nozakii were Rhizobiales (13.92%, dominated by genus Phyllobacterium), Sphingomonadales (12.16%, dominated by genus Sphingomonas), Clostridiales (5.46%, dominated by genus Paraclostridium), Mycoplasmatales (5.08%, dominated by genus Mycoplasma) and genus Ralstonia (13.28%). The ambient water at the Qingdao location contained mainly Synechococcales (34.22%), Flavobacteriales (16.53%), Micrococcales (15.79%), Chitinophagales (4.51%), Actinomarinales (4.22%) and Rhodobacterales (3.56%). The bacterial communities associated with N. nomurai and R. esculentum were relatively similar, with high relative abundances of order Mycoplasmatales, 46.04% and 24.54% respectively, of which Mycoplasma was the most abundant genus in both scyphozoans. Rhizobiales, Sphingomonadales (mostly genus Sphingomonas), Flavobacteriales (mostly genus Tenacibaculum), Vibrionales (mostly genus Vibrio), and genera Phyllobacterium and Ralstonia were also detected in N. nomurai and R. esculentum. In addition, the bacterial communities of the Rongcheng seawater were characterized by high relative abundances of order Rhodobacterales (40.33%), Flavobacteriales (36.17%), Alteromonadales (6.25%), Oceanospirillales (2.66%) and Chitinophagales (2.56%). The composition and relative abundance of bacterial communities in scyphomedusae were significantly different from those in the ambient seawater (Kruskal-Wallis test, p < 0.05) (Figure 2). Furthermore, the composition and relative abundance of the four scyphomedusa-associated bacterial communities were significantly different from each other (Kruskal-Wallis test, p < 0.05) (Figure 2).
Bacterial Community Diversity
The α-diversities (Shannon indices) of the bacterial communities associated with the four scyphomedusae, A. coerulea (1.5), C. nozakii (2.4), N. nomurai (2.4) and R. esculentum (3.0), were lower than those of the water column sampled at both locations (5.3 in Qingdao and 5.8 in Rongcheng) according to the 16S rRNA gene sequencing. The statistical analysis showed significant differences between scyphomedusae and ambient seawater, and between scyphomedusae A. coerulea and C. nozakii (Wilcoxon test, p < 0.05) (Figure 3). However, there were no significant differences between the Shannon indices of scyphomedusae N. nomurai and R. esculentum in Rongcheng (Wilcoxon test, p > 0.05) (Figure 3).
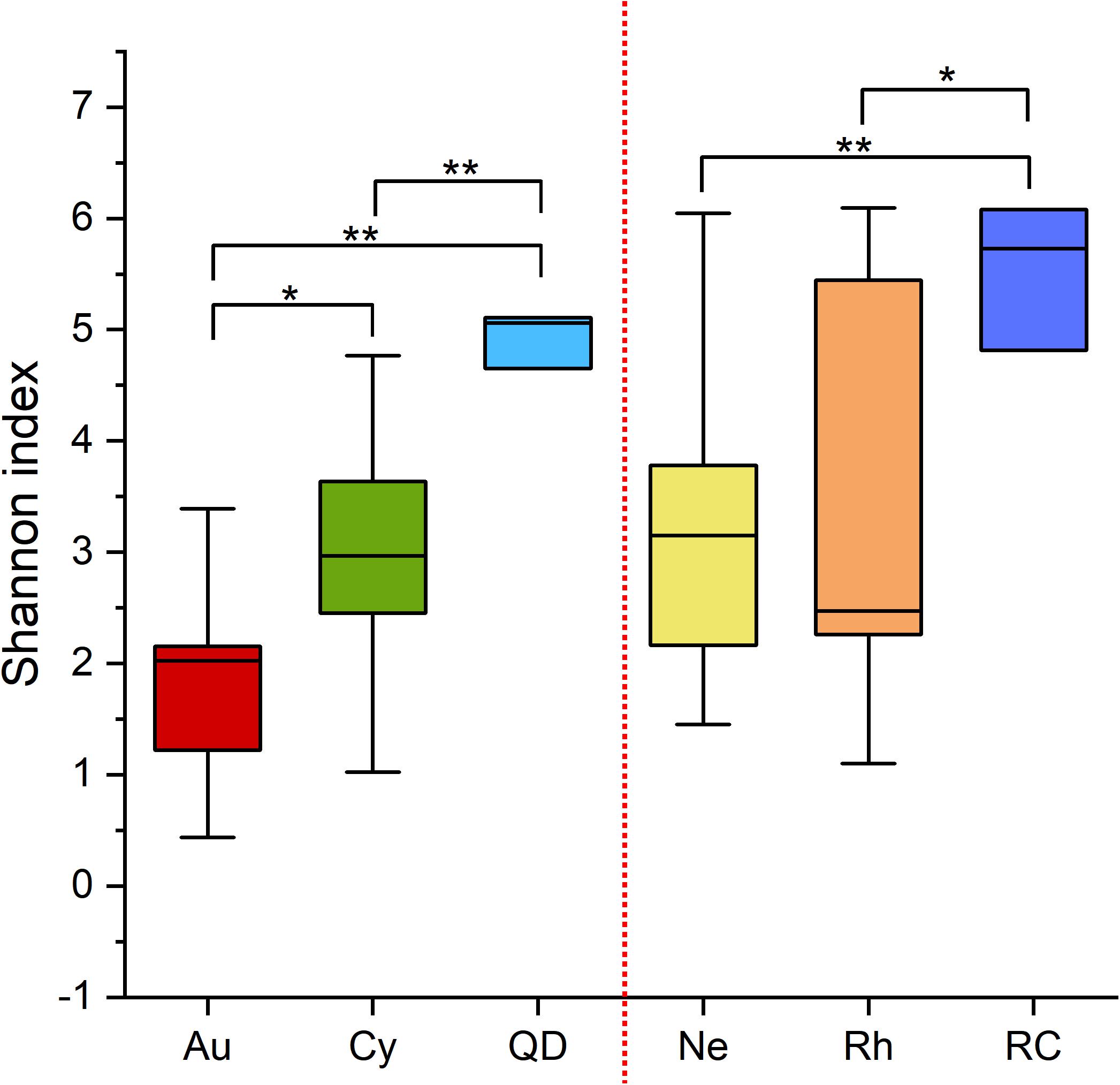
Figure 3. Boxplot of Shannon indices of scyphozoan-associated bacterial communities and free-living bacteria in surrounding seawater. Note: Abbreviations: Au, A. coerulea from Jiaozhou Bay in Qingdao; Cy, C. nozakii from Jiaozhou Bay in Qingdao; Ne, N. nomurai from Shidao Bay in Rongcheng; Rh, R. esculentum from Shidao Bay in Rongcheng; QD, ambient seawater from Jiaozhou Bay in Qingdao; RC, ambient seawater from Shidao Bay in Rongcheng. Significant differences were determined using the Wilcoxon test, *: p < 0.05, **: p < 0.01.
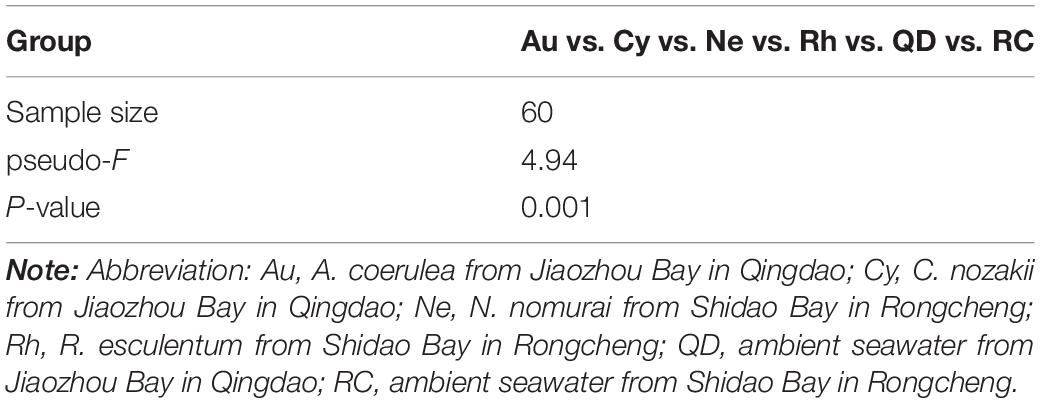
Principal Co-ordinates Analysis demonstrated that the bacterial communities associated with the four scyphomedusa taxa were significantly different from the free-living bacterial communities in the seawater at the two locations (Qingdao and Rongcheng) according to PERMANOVA main test (p < 0.01) (Figure 4 and Tables 1, 2). A. coerulea and C. nozakii-associated bacterial communities were relatively separate from those of N. nomurai and R. esculentum, while those of the two scyphomedusa species sampled from the same seawater were closely clustered. The significance analysis revealed that the bacterial communities associated with the four scyphomedusa taxa all had extremely significant differences from each other (PERMANOVA, p < 0.01) (Figure 4 and Tables 1, 2).
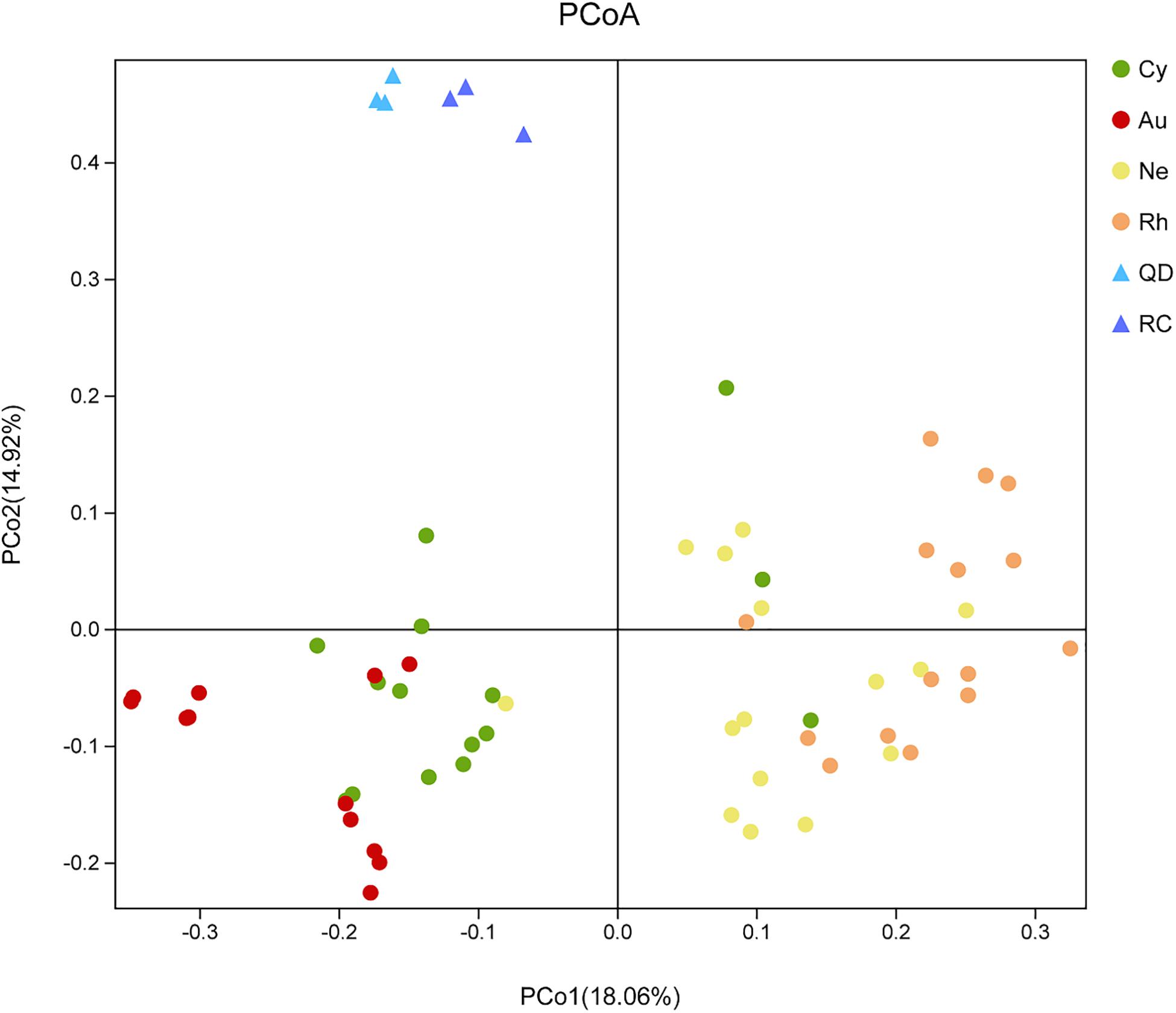
Figure 4. Unweighted unifrac PCoA plot of scyphozoan-associated bacterial communities and free-living bacteria in surrounding seawater. Note: Abbreviations: Au, A. coerulea from Jiaozhou Bay in Qingdao; Cy, C. nozakii from Jiaozhou Bay in Qingdao; Ne, N. nomurai from Shidao Bay in Rongcheng; Rh, R. esculentum from Shidao Bay in Rongcheng; QD, ambient seawater from Jiaozhou Bay in Qingdao; RC, ambient seawater from Shidao Bay in Rongcheng.
Core Bacterial Community Across Four Blooming Scyphozoans
To further analyze the bacterial variation, Alpha and Beta diversities of the bacterial communities of the three body parts of each scyphozoan were analyzed, represented by the Shannon index and PCoA analysis respectively. Results of Shannon indices demonstrated low divergence between the three body parts of each scyphozoan (Wilcoxon test, p > 0.05) (Supplementary Figure 2). PCoA analysis gave similar results (as shown in Supplementary Figure 3 and Supplementary Table 2). There was no significant difference between the three compartments of each scyphozoan taxa (PERMANOVA, p > 0.05).
In total, 57 OTUs were present in all scyphomedusa species and all compartments, which could represent core bacteria associated with the four scyphozoans (Figure 5). The core OTUs were composed mainly of Mycoplasma, Vibrio, Ralstonia, Tenacibaculum, Shingomonas and Phyllobacterium (Figure 5). These frequently occurring OTUs accounted for more than 80.99% of the core bacterial communities of each body part of each scyphomedusa species, except for the gonads (40.69%) and oral arms (65.67%) of C. nozakii, mainly because there were highly abundant unidentified Entomoplasmatales (OTU5) in these two compartments of C. nozakii (Figure 5).
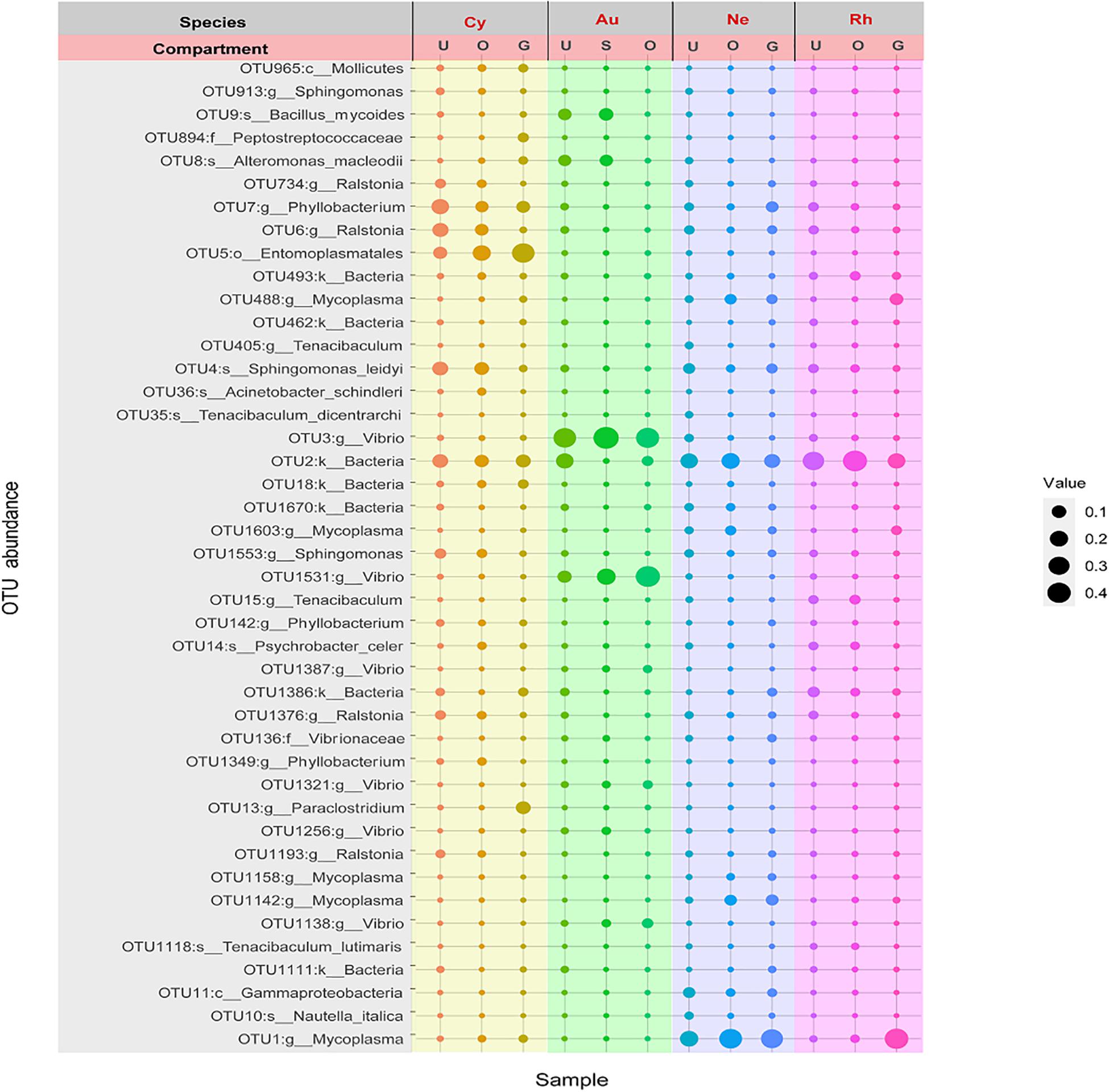
Figure 5. Bubble plot of core OTU members associated with four scyphozoan species with relative abundance greater than 1% in at least one body part of at least one scyphozoan species. Note: Abbreviations: Au, A. coerulea from Jiaozhou Bay in Qingdao; Cy, C. nozakii from Jiaozhou Bay in Qingdao; Ne, N. nomurai from Shidao Bay in Rongcheng; Rh, R. esculentum from Shidao Bay in Rongcheng; U, umbrella, O, oral arms, S, stomach and G, gonad. Bubble size indicates relative abundance.
Functional Prediction of the 16S Genes Using FAPROTAX
Functional annotation of the genomic information was performed based on FAPROTAX. A total of 91 functional groups were obtained, comprising 8279 members (5011 unique members), and some OTUs were predicted to be included in multiple functional groups. 11 functional groups showed significant differences between scyphomedusa-associated bacterial communities and free-living bacterial communities in seawater in the top 40 FAPROTAX functional groups, of which 8 were significantly more abundant in scyphomedusae, and 3 (xylanolysis, predatory or exoparasitic, and chloroplasts) were significantly more abundant in seawater (Mann-Whitney U-test, p < 0.05) (Figure 6). The functional groups which were significantly more abundant in scyphomedusa-associated bacterial communities consisted of nitrogen cycle-related functions (nitrate respiration, and nitrogen respiration), and heterotrophic-dominated functions (aerobic chemoheterotrophy, chemoheterotrophy, invertebrate parasites, all human pathogens, human associated and animal parasites or symbionts) (Figure 6). Nitrate respiration mainly included Vibrio, Bacillus, Arenibacter, Acinetobacter, and Stenotrophomonas; the latter two genera were also the dominant players in nitrogen respiration (Supplementary Material 1). Aerobic chemoheterotrophy and chemoheterotrophy had the highest abundances of accumulated OTUs both in scyphomedusae and seawater, including Vibrio, Bacillus, Stenotrophomonas, Arenibacter, Acinetobacter, Mycoplasma, Chryseobacterium, Tenacibaculum, Phyllobacterium, Sphingomonas, etc. (Supplementary Material 1).
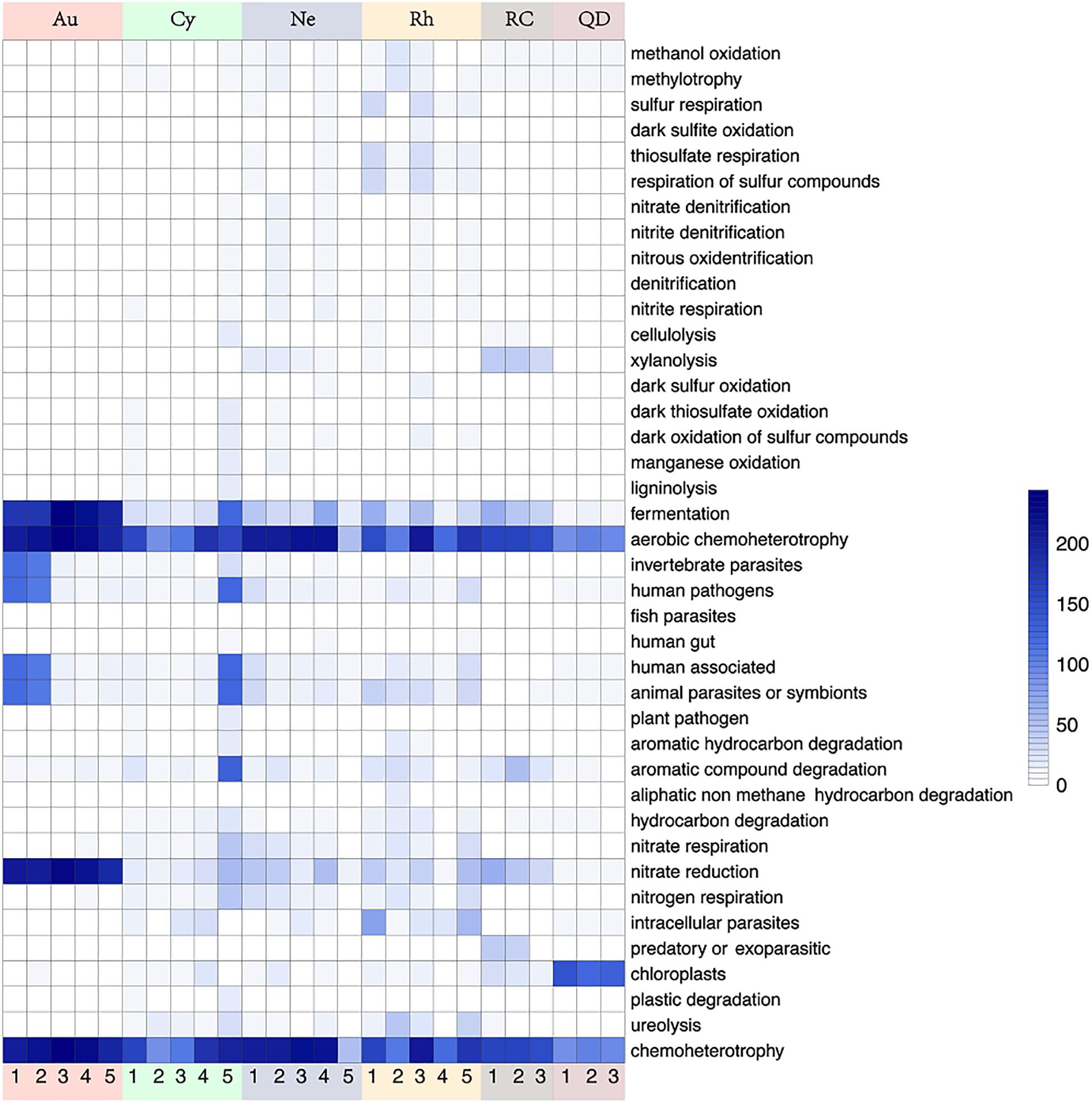
Figure 6. Heatmap of 40 most abundant functions based on FAPROTAX analysis in four scyphozoan-associated bacterial communities and free-living bacteria in surrounding seawater. Note: Abbreviations: Au, A. coerulea from Jiaozhou Bay in Qingdao; Cy, C. nozakii from Jiaozhou Bay in Qingdao; Ne, N. nomurai from Shidao Bay in Rongcheng; Rh, R. esculentum from Shidao Bay in Rongcheng; QD, ambient seawater from Jiaozhou Bay in Qingdao; RC, ambient seawater from Shidao Bay in Rongcheng. The values in the heatmap came from the square root transformation of the absolute abundance of OTUs contributing to functional groups after normalization.
Twenty-one significantly distinct functional groups were tested from top 40 functional groups of the four scyphomedusa-associated bacterial communities. The significant differences between 15 of those 21 functional groups existed primarily between A. coerulea and R. esculentum (Kruskal-Wallis test, p < 0.05) (Figure 6 and Supplementary Table 3). The cumulative abundances of functional groups were usually lower in A. coerulea than the other three scyphomedusa species, with the exception of the following functional groups: fermentation, invertebrate parasites, aerobic chemoheterotrophy, chemoheterotrophy, and nitrate reduction, as well as plastic degradation. At least one of Bacillus, Alteromonas, family Vibrionaceae, and Vibrio held a larger proportion in the assigned OTUs of the former five functional groups, and these OTUs happened to be more abundant in A. coerulea than in the other three scyphomedusa species (Figures 2, 6, and Supplementary Table 3, Supplementary Material 1). Plastic degradation was richer in C. nozakii, inferred to be related to the high relative abundance of Pseudomonas in the C. nozakii-associated bacterial communities (Figures 2, 6, and Supplementary Table 3, Supplementary Material 1). In addition, cellulolysis, aromatic hydrocarbon degradation, hydrocarbon degradation and ureolysis functional groups were well-represented among C. nozakii-associated bacteria. Interestingly, these functional groups were all related to material decomposition (Figure 6 and Supplementary Table 3). The two functional groups xylanolysis and nitrate respiration were most abundant in N. nomurai among the four scyphomedusa taxa (Figure 6 and Supplementary Table 3). R. esculentum had the highest cumulative abundances of 13 functional groups, covering the sulfur cycle, compound degradation and parasites, indicating that the FAPROTAX functions of R. esculentum-associated bacteria communities were more diverse and enriched than those of the other scyphomedusae.
Potentially Pathogenic Bacteria Associated With Four Blooming Scyphozoans
Six genera of potentially pathogenic bacteria were detected in the scyphozoan-associated bacterial communities based on phylogenetic analysis: Vibrio, Mycoplasma, Ralstonia, Tenacibaculum, Nautella, and Acinetobacter (Figure 7). Potentially pathogenic Vibrio, clustered with Vibrio alginolyticus and Vibrio parahaemolyticus, was the dominant genus in A. coerulea (> 48.43%), and its relative abundance was highest in the oral arms of A. coerulea (91.62%), followed by the umbrellas of N. nomurai (2.71%) and R. esculentum (2.06%). Mycoplasma was the second most abundant potentially pathogenic genus in the scyphozoan-associated bacterial communities, maintaining high relative abundances in all three body components (umbrella, oral arms, and gonad) of the other three scyphomedusae (C. nozakii, N. nomurai, and R. esculentum), and was the most abundant bacteria in the gonads of R. esculentum (65.93%), as well as the oral arms (57.14%), gonads (49.92%), and umbrellas (26.11%) of N. nomurai. Ralstonia, closely related to Ralstonia syzygii, was the most widely distributed potentially pathogenic genus, with relatively high abundances in all four scyphomedusa taxa, among which the relative abundances were particularly high in the umbrellas (23.78%) and oral arms (13.77%) of C. nozakii.

Figure 7. Neighbor-joining phylogenetic tree generated from the V4-V5 region of the 16S rRNA gene for scyphozoan-associated bacterial communities and reference sequences of identified pathogenic bacteria. The bootstrap was performed with 1000 replicates. Operational taxonomic units are from this study. Reference sequences include NCBI accession numbers.
N. nomurai harbored the most diverse potential pathogens, with 5 genera (Vibrio, Mycoplasma, Ralstonia, Nautella and Tenacibaculum). R. esculentum was associated with 4 potentially pathogenic genera (Vibrio, Mycoplasma, Ralstonia, and Tenacibaculum). C. nozakii contained 3 genera of potentially pathogenic bacteria (Mycoplasma, Ralstonia and Acinetobacter), of which Acinetobacter was only present in the oral arms in high relative abundance (1.79%), while A. coerulea was associated with Vibrio and Ralstonia. Genera Tenacibaculum, Nautella, and Acinetobacter were clearly identified at the species level: fish pathogens Tenacibaculum dicentrarchi and Tenacibaculum lutimaris, red alga pathogen Nautella italica, and human pathogen Acinetobacter schindleri (Figure 7).
Discussion
Jellyfish are conspicuous marine zooplankton that play critical roles in the exchange of material and energy in marine ecosystems (Riemann et al., 2006; Pitt et al., 2009; West et al., 2009; Condon et al., 2011; Blanchet et al., 2015; Chelsky et al., 2015; Sweetman et al., 2016; Guy-Haim et al., 2020). The four scyphomedusae in this study (A. coerulea, C. nozakii, N. nomurai, and R. esculentum) can all reach considerable biomass in coastal waters of China during June to September (Dong et al., 2010, 2014; Zhang et al., 2012; Wang P.P. et al., 2020). Blooming jellyfish may convert large amounts of carbon into gelatinous biomass, which may limit the transfer of carbon in the food web (Condon and Steinberg, 2008; Sweetman et al., 2016). Organic and inorganic matters released by jellyfish could influence microbial nutrient cycling and alter microbial structure and function (Riemann et al., 2006; Condon et al., 2011; Guy-Haim et al., 2020). Therefore, bacterial communities associated with the blooming cnidarian jellyfish were speculated to be major participants in the biogeochemical cycling of carbon, nitrogen, sulfur, and phosphorus (Lee et al., 2018) and represent key vectors of potentially bacterial pathogens (Ferguson et al., 2010; Schuett and Doepke, 2010; Delannoy et al., 2011; Basso et al., 2019; Clinton et al., 2020).
The Composition and Potential Functions of the Bacterial Communities Associated With the Four Scyphozoans
Scyphomedusa-associated bacterial communities presented different composition and lower diversities compared with the free-living bacterial communities in the ambient seawater, consistent with previous studies (Weiland-Bräuer et al., 2015; Daley et al., 2016; Kramar et al., 2019; Daniel and Ana, 2020). Most previous studies on scyphozoan-associated microbiomes focused on one single scyphozoan species (e.g., Cortés-Lara et al., 2015; Weiland-Bräuer et al., 2015; Viver et al., 2017; Lee et al., 2018; Basso et al., 2019; Kramar et al., 2019; Stabili et al., 2020), with few studies investigating the differences between the bacteria associated with various scyphozoan species. In the present study, the composition and structure of the microbiomes associated with four scyphomedusae were compared and found to be significantly different from each other, indicating that the scyphomedusa-associated bacteria were scyphozoan taxa-specific (as Cleary et al., 2016; Hao et al., 2019; and Daniel and Ana, 2020 reported). The A. coerulea-associated bacterial communities were dominated by Vibrio, as has previously been shown for the communities associated with A. aurita (Weiland-Bräuer et al., 2015; Kramar et al., 2019; Jaspers et al., 2020). The bacterial communities associated with N. nomurai and R. esculentum (family Rhizostomatidae) were similarly dominated by a single genus, Mycoplasma, which was consistent with previous results for R. pulmo (family Rhizostomatidae) (Basso et al., 2019; Stabili et al., 2020). Interestingly, three bacterial genera presented relatively similar abundances associated with C. nozakii including Shingomonas, Phyllobacterium, and Ralstonia. Schuett and Doepke (2010) first reported both Cyanea species (C. capillata and C. lamarckii) which were dominated by Gammaproteobacteria, exhibiting > 97% similarity to the Vibrio group based on culturing and culture-independent approaches. This might be due to the different methodologies applied, e.g., preculturing step, increasing the bottle effects and affecting the relevance of the resulting data.
A total of 57 core OTUs were identified in our study of the bacterial communities of the scyphozoan body parts, predominantly genera Mycoplasma, Vibrio, Ralstonia, Tenacibaculum, and Phyllobacterium. Comparison of the core bacteria in this study with previous reports of scyphozoan-associated bacteria found several commonalities. Several studies found Mycoplasma to be the prevailing taxa in the microbiomes of the scyphozoans A. aurita (Weiland-Bräuer et al., 2015; Daley et al., 2016; Jaspers et al., 2020), and R. pulmo (Basso et al., 2019; Stabili et al., 2020), similar to the present study. Vibrio has previously been found to be the dominant genus in A. aurita (Weiland-Bräuer et al., 2015; Kramar et al., 2019; Jaspers et al., 2020), A. coerulea (Chen et al., 2020), R. pulmo (Basso et al., 2019; Stabili et al., 2020), Cotylorhiza tuberculata (Cortés-Lara et al., 2015), C. lamarckii (Schuett and Doepke, 2010), and C. capillata (Schuett and Doepke, 2010; Clinton et al., 2020), and was abundant in A. coerulea, N. nomurai and R. esculentum in this study. Genus Tenacibaculum was dominant in N. nomurai and R. esculentum in this study, and has been suggested as a key part of the bacterial communities of A. aurita (Jaspers et al., 2020), C. tuberculata (Cortés-Lara et al., 2015; Viver et al., 2017) and P. noctiluca (Delannoy et al., 2011). These common genera might occupy important micro-niches in the bacterial communities of scyphozoans.
The FAPROTAX method is most suitable for annotating and predicting ecological functions such as biogeochemical cycling processes (Louca et al., 2016). Our results suggested that some bacteria associated with the scyphozoans, such as Vibrio, Bacillus, Arenibacter, Acinetobacter and Stenotrophomonas, might play important roles in marine biogeochemical cycles. For example, Vibrio was involved in the nitrogen cycle, while Bacillus (mainly B. megaterium) was involved in the nitrogen and sulfur cycle. Such bacteria are likely to provide their hosts with unavailable nutrients (Lee et al., 2018). Ammonia-, nitrite- and sulfur-oxidizing microbes may be scavengers of toxic ammonia, nitrite and sulfide in the host (Tian et al., 2016). Moreover, during the decomposition of jellyfish after blooms, certain bacteria (e.g., Vibrionaceae) could grow rapidly, causing the accumulation of a large amount of inorganic nutrients and participating in the regeneration of elements (Tinta et al., 2012).
The genera Alteromonas, Tenacibaculum, Mycoplasma and Sphingomonas had only a single function, chemoheterotrophy, in the present study. Of the functional groups identified, chemoheterotrophy, including aerobic chemoheterotrophy and chemoheterotrophy, had the highest cumulative abundance of OTUs, both in scyphozoans and seawater, indicating that they played key roles both in scyphozoan-associated and free-living bacterial communities. Chemoheterotrophy has been reported as an essential ecological function of bacterioplankton and sediments (Zeng et al., 2014; He et al., 2020). Some scyphomedusa-associated bacteria displayed pollutant degradation including Alteromonas (Bakunina et al., 2013; Lopardo and Urakawa, 2019), Sphingomonas (Walayat et al., 2018; Carmen Garcia et al., 2019; Menon et al., 2019; Yun et al., 2019) and Ralstonia (Wang J. et al., 2020). It has been reported that scyphozoans are highly tolerant of certain pollutants (e.g., heavy metals and crude oil), and some of the most toxic PAHs of crude oil can be bioaccumulated in gelatinous zooplankton and potentially be transferred through the food web to contaminate apex predators (Almeda et al., 2013; Lucas and Horton, 2014). We speculated that these bacteria might be related to the pollution-tolerance of scyphomedusae. This might indicate that the jellyfish-associated bacterial communities plays an important ecological function in assisting longer tolerant of jellyfish in the environment pollution.
Potentially Pathogenic Bacteria Associated With the Four Scyphozoan Microbiomes
Since we applied the function prediction based on FAPROTAX which mainly focused on the biogeochemical cycle, the potential pathogens we mentioned here mainly focused on human pathogens. The database for pathogens of marine economic species is still missing. Here, we specifically discussed the potentially pathogenic bacteria associated with four scyphomedusa taxa in this study based on phylogenetic analysis. It has been shown that certain jellyfish-associated microbial communities contain pathogens, such as Tenacibaculum, Vibrio, Pseudomonas, Aeromonas, Photobacterium, Flavobacterium, Mycoplasma, and Chryseobacterium, among others (Ferguson et al., 2010; Schuett and Doepke, 2010; Delannoy et al., 2011; Cortés-Lara et al., 2015; Småge et al., 2017; Basso et al., 2019; Clinton et al., 2020; Jaspers et al., 2020; Stabili et al., 2020). Some researchers have expressed concern that jellyfish may threaten aquaculture and human health by spreading pathogens (Ferguson et al., 2010; Schuett and Doepke, 2010; Delannoy et al., 2011).
In this study, high relative abundances of Vibrio associated with A. coerulea, N. nomurai and R. esculentum were closely related to V. alginolyticus and V. parahaemolyticus. V. alginolyticus and V. parahaemolyticus have been determined to infect marine animals (e.g., fish, shrimp, shellfish) and even humans, and are common pathogens in aquaculture (Joshi et al., 2014; Lee et al., 2015; Escobedo-Hinojosa and Pardo-Lopez, 2017; Aly et al., 2021; Hsu et al., 2021). Genus Mycoplasma has been reported to be associated with algae (Altamiranda et al., 2011; Davis et al., 2013), as well as invertebrates such as bivalves (Fernandez-Piquer et al., 2012), crustaceans (Liang et al., 2011), ctenophores (Hao et al., 2015), and other cnidarians (Cortés-Lara et al., 2015; Weiland-Bräuer et al., 2015; Viver et al., 2017). To date, the function of Mycoplasma bacteria on gelatinous zooplankton remains unknown. Some Mycoplasma species are classified as common pathogens, such as M. mobile, M. penetrans, M. pneumoniae (Escobedo-Hinojosa and Pardo-Lopez, 2017). In this study, abundant Mycoplasma were detected in N. nomurai, R. esculentum and C. nozakii. Although we did not find homologous certified pathogens of Mycoplasma in this study, we cannot rule out the possibility that the unclassified Mycoplasma in this study included pathogenic species.
Genus Tenacibaculum currently includes a total of 21 species pathogenic to fish (Habib et al., 2014). T. dicentrarchi and T. lutimaris in this study was mainly associated with N. nomurai and R. esculentum, and is known to be pathogenic to fish (Avendano-Herrera et al., 2016; Slinger et al., 2020). Unclassified Tenacibaculum was clustered with two other fish pathogens, T. adriaticum and T. litoreum, which may pose a threat to fish populations and aquaculture industry (Habib et al., 2014; Fernandez-Alvarez et al., 2017). Moreover, Tenacibaculum spp. may be a key part of the digestive system of jellyfish, playing an important role in their immune defense and nutrition (Ferguson et al., 2010; Cortés-Lara et al., 2015).
Ralstonia was the most widely distributed potentially pathogenic genus among the four scyphomedusa species. Ralstonia spp. are non-fermenting gram-negative bacteria that have recently emerged as opportunistic pathogens (Alasehir et al., 2020), with Ralstonia picketti, R. mannitolilytica and R. insidiosa implicated particularly in nosocomial infections in immunocompromised patients (Fang et al., 2019). R. syzygii, one of the phylotypes of R. solanacearum, is believed to cause plant disease (Etminani et al., 2020). The widespread distribution of Ralstonia in various scyphomedusa taxa may present a risk for the spread of human and foodborne pathogens.
Genus Nautella, identified in the umbrella of N. nomurai, was dominated by N. italica, an etiological agent of bleaching in the red alga Delisea pulchra (Case et al., 2011; Gardiner et al., 2017; Hudson et al., 2018). The most abundant species of genus Acinetobacter, detected in the oral arms of C. nozakii, was A. schindleri, a human pathogen (McGann et al., 2013; Jani et al., 2019).
Due to the short amplified sequence and lack of verification by culture methods in our study, the pathogenic bacteria discussed in this study are only potential pathogens. To determine its pathogenicity and virulence, it is necessary to carry out long-range amplification sequencing, and isolation and culture of target bacteria. Some researchers put forward the hypothesis that scyphomedusae, carrying potential pathogens, might drift with marine currents over long distances and spread to large sea areas (Dong et al., 2010; Tinta et al., 2019). Outbreaks of the four scyphomedusae in this study commonly occur in areas of Chinese coastal waters with large aquaculture industries. China, as the largest exporter of aquatic products in the world, uses vast areas of sea for aquaculture, diverse aquaculture models (e.g., raft culture, cage culture, and pond culture) and numerous species of aquaculture organisms (e.g., fish, shellfish, shrimp, and sea cucumber) (Cao et al., 2015; Zhou et al., 2020). In such conditions, high occurrence of potentially pathogenic bacteria in common scyphomedusae is extremely concerning. Living scyphomedusae may pass pathogenic bacteria to other organisms including humans through contact or attack, causing bacterial diseases and even death (Basso et al., 2019; Clinton et al., 2020). During scyphomedusa degradation, certain pathogenic bacteria may be released into the surrounding seawater, increasing the risk of infection of other organisms in the surrounding area (Tinta et al., 2012). Furthermore, jellyfish carcasses might trigger or support growth of potential pathogens in ambient seawater (Dinasquet et al., 2012). Farmed organisms may suffer greater dangers compared to others due to their high population densities and habitats that overlap with blooming scyphomedusae. This has the potential to cause immense socio-economic losses to the aquaculture industry.
Data Availability Statement
The raw sequence data generated herein have been uploaded to NCBI’s Sequence Read Archive (SRA) under BioProject accession number PRJNA718816.
Author Contributions
SP did the formal analysis, data analysis, writing – original draft, and writing – review and editing. WH did the formal analysis, data analysis, and writing – review and editing. YL did the formal analysis and data analysis. LW and TS did the investigation and sample collection. JZ did the conceptualization and funding acquisition. ZD did the conceptualization, sample collection, data analysis, funding acquisition, and writing – review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA23050301), the National Key Research and Development Program of China (2018YFC1406501), the Instrument Developing Project of the Chinese Academy of Sciences (No. YJKYYQ20180047), the National Natural Science Foundation of China (Nos. 41876138; 42003064), and the Shandong Provincial Key Laboratory of Restoration for Marine Ecology (201921).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.647089/full#supplementary-material
Footnotes
References
Alasehir, E. A., Ipek, B. O., Thomas, D. T., Sitar, M. E., and Ercan, T. E. (2020). Ralstonia insidiosa neonatal sepsis: a case report and review of the literature. J. Pediatr. Inf. Dis. Ger. 15, 148–151.
Almeda, R., Wambaugh, Z., Chai, C., Wang, Z., Liu, Z., and Buskey, E. J. (2013). Effects of crude oil exposure on bioaccumulation of polycyclic aromatic hydrocarbons and survival of adult and larval stages of gelatinous zooplankton. PLoS One 8:e74476. doi: 10.1371/journal.pone.0074476
Altamiranda, M., Acuicultor, M. S. V., and Boris Briñez, R. (2011). Presence of Spiroplasma penaei in plankton, benthos and fauna in shrimps farms of Colombia. Rev. MVZ Cordoba 16, 2576–2583. doi: 10.21897/rmvz.1030
Aly, S. M., Eissa, A. E., ElBanna, N. I., and Albutti, A. E. (2021). Efficiency of monovalent and polyvalent Vibrio alginolyticus and Vibrio Parahaemolyticus vaccines on the immune response and protection in gilthead sea bream, Sparus aurata (L.) against vibriosis. Fish Shellfish Immun. 111, 145–151. doi: 10.1016/j.fsi.2020.10.011
Avendano-Herrera, R., Irgang, R., Sandoval, C., Moreno-Lira, P., Houel, A., Duchaud, E., et al. (2016). Isolation, characterization and virulence potential of Tenacibaculum dicentrarchi in Salmonid cultures in Chile. Transbound. Emerg. Dis. 63, 121–126. doi: 10.1111/tbed.12464
Bakunina, I., Nedashkovskaya, O., Balabanova, L., Zvyagintseva, T., Rasskasov, V., and Mikhailov, V. (2013). Comparative analysis of glycoside hydrolases activities from phylogenetically diverse marine bacteria of the genus Arenibacter. Mar. Drugs 11, 1977–1998. doi: 10.3390/md11061977
Basso, L., Rizzo, L., Marzano, M., Intranuovo, M., Fosso, B., Pesole, G., et al. (2019). Jellyfish summer outbreaks as bacterial vectors and potential hazards for marine animals and humans health? The case of Rhizostoma pulmo (Scyphozoa, Cnidaria). Sci. Total Environ. 692, 305–318. doi: 10.1016/j.scitotenv.2019.07.155
Blanchet, M., Pringault, O., Bouvy, M., Catala, P., Oriol, L., Caparros, J., et al. (2015). Changes in bacterial community metabolism and composition during the degradation of dissolved organic matter from the jellyfish Aurelia aurita in a Mediterranean coastal lagoon. Environ. Sci. Pollut. Res. 22, 13638–13653. doi: 10.1007/s11356-014-3848-x
Bosch, T. C. G., and McFall-Ngai, M. (2021). Animal development in the microbial world: re-thinking the conceptual framework. Curr. Top. Dev. Biol. 141, 399–427. doi: 10.1016/bs.ctdb.2020.11.007
Brian, J. H., Dirk, G., Ashlee, M. E., Mike, F., Doyle, V. W., Georgia, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Cao, L., Naylor, R., Henriksson, P., Leadbitter, D., Metian, M., Troell, M., et al. (2015). China’s aquaculture and the world’s wild fisheries. Science 347, 133–135. doi: 10.1126/science.1260149
Cárdenas, A., Ye, J., Ziegler, M., Payet, J. P., McMinds, R., Vega Thurber, R., et al. (2020). Coral-associated viral assemblages from the central red sea align with host species and contribute to holobiont genetic diversity. Front. Microbiol. 11:572534. doi: 10.3389/fmicb.2020.572534
Carmen Garcia, M., Trujillo, L. A., Carmona, J. A., Munoz, J., and Carmen Alfaro, M. (2019). Flow, dynamic viscoelastic and creep properties of a biological polymer produced by Sphingomonas sp. as affected by concentration. Int. J. Biol. Macromol. 125, 1242–1247. doi: 10.1016/j.ijbiomac.2018.09.100
Case, R. J., Longford, S. R., Campbell, A. H., Low, A., Tujula, N., Steinberg, P. D., et al. (2011). Temperature induced bacterial virulence and bleaching disease in a chemically defended marine macroalga. Environ. Microbiol. 13, 529–537. doi: 10.1111/j.1462-2920.2010.02356.x
Chelsky, A., Pitt, K. A., and Welsh, D. T. (2015). Biogeochemical implications of decomposing jellyfish blooms in a changing climate. Estuar. Coast. Shelf Sci. 154, 77–83. doi: 10.1016/j.ecss.2014.12.022
Chen, X. T., Li, X. Y., Xu, Z., Liu, Q., Peng, Z. Y., Zhu, Y. N., et al. (2020). The distinct microbial community in Aurelia coerulea polyps versus medusae and its dynamics after exposure to Co-60-gamma radiation. Environ. Res. 188:109843. doi: 10.1016/j.envres.2020.109843
Cleary, D. F. R., Becking, L. E., Polónia, A. R. M., Freitas, R. M., and Gomes, N. C. M. (2016). Jellyfish-associated bacterial communities and bacterioplankton in Indonesian Marine lakes. FEMS Microbiol. Ecol. 92:fiw064. doi: 10.1093/femsec/fiw064
Clinton, M., Kintnera, A. H., Delannoyc, C. M. J., Brierleya, A. S., and Ferrier, D. E. K. (2020). Molecular identification of potential aquaculture pathogens adherent to cnidarian zooplankton. Aquaculture 518:734801. doi: 10.1016/j.aquaculture.2019.734801
Colin, S. P., Costello, J. H., Hansson, L. J., Titelman, J., and Dabiri, J. O. (2010). Stealth predation and the predatory success of the invasive ctenophore Mnemiopsis leidyi. Proc. Natl. Acad. Sci. U.S.A. 107, 17223–17227. doi: 10.1073/pnas.1003170107
Condon, R. H., and Steinberg, D. K. (2008). Development, biological regulation, and fate of ctenophore blooms in the York River estuary, Chesapeake Bay. Mar. Ecol. Prog. Ser. 369, 153–168. doi: 10.3354/meps07595
Condon, R. H., Graham, W. M., Duarte, C. M., Pitt, K. A., Lucas, C. H., Haddock, S. H. D., et al. (2012). Questioning the rise of gelatinous zooplankton in the world’s oceans. Bioscience 62, 160–169. doi: 10.1525/bio.2012.62.2.9
Condon, R. H., Steinberg, D. K., del Giorgio, P. A., Bouvier, T. C., Bronk, D. A., Graham, W. M., et al. (2011). Jellyfish blooms result in a major microbial respiratory sink of carbon in marine systems. Proc. Natl. Acad. Sci. U.S.A. 108, 10225–10230. doi: 10.1073/pnas.1015782108
Contijoch, E. J., Britton, G. J., Yang, C., Mogno, I., Li, Z., Ng, R., et al. (2019). Gut microbiota density influences host physiology and is shaped by host and microbial factors. Elife 8:337. doi: 10.7554/eLife.40553
Cortés-Lara, S., Urdiain, M., Mora-Ruiz, M., Prieto, L., and Rosselló-Móra, R. (2015). Prokaryotic microbiota in the digestive cavity of the jellyfish Cotylorhiza tuberculata. Syst. Appl. Microbiol. 38, 494–500. doi: 10.1016/j.syapm.2015.07.001
Daley, M. C., Urban-Rich, J., and Moisander, P. H. (2016). Bacterial associations with the hydromedusa Nemopsis bachei and scyphomedusa Aurelia aurita from the North Atlantic Ocean. Mar. Biol. Res. 12, 1088–1100. doi: 10.1080/17451000.2016.1228974
Daniel, F. R. C., and Ana, R. M. P. (2020). Marine lake populations of jellyfish, mussels and sponges host compositionally distinct prokaryotic communities. Hydrobiologia 847, 3409–3425. doi: 10.1007/s10750-020-04346-3
Davis, J., Fricke, W. F., Hamann, M. T., Esquenazi, E., Dorrestein, P. C., and Hill, R. T. (2013). Characterization of the bacterial community of the chemically defended Hawaiian sacoglossan Elysia rufescens. Appl. Environ. Microbiol. 79, 7073–7081. doi: 10.1128/AEM.01568-13
Deines, P., Hammerschmidt, K., and Bosch, T. C. G. (2020). Microbial species coexistence depends on the host environment. mBio 11, e807–e820. doi: 10.1128/mBio.00807-20
Delannoy, C. M. J., Houghton, J. D. R., Flemin, N. E. C., and Ferguson, H. W. (2011). Mauve stingers (Pelagia noctiluca) as carriers of the bacterial fish pathogen Tenacibaculum maritimum. Aquaculture 311, 255–257. doi: 10.1016/j.aquaculture.2010.11.033
Dinasquet, J., Granhag, L., and Riemann, L. (2012). Stimulated bacterioplankton growth and selection for certain bacterial taxa in the vicinity of the ctenophore Mnemiopsis leidyi. Front. Microbiol. 3:302. doi: 10.3389/fmicb.2012.00302
Dong, Z. J., Liu, D. Y., and Keesing, J. K. (2010). Jellyfish blooms in China: dominant species, causes and consequences. Mar. Pollut. Bull. 60, 954–963. doi: 10.1016/j.marpolbul.2010.04.022
Dong, Z. J., Liu, D. Y., and Keesing, J. K. (2014). “Contrasting trends in populations of Rhopilema esculentum and Aurelia aurita in Chinese waters,” in Jellyfish Blooms, eds K. Pitt and C. Lucas (Berlin: Springer-Verlag), 207–218.
Escobedo-Hinojosa, W., and Pardo-Lopez, L. (2017). Analysis of bacterial metagenomes from the Southwestern Gulf of Mexico for pathogens detection. Pathog. Dis. 75:ftx058. doi: 10.1093/femspd/ftx058
Etminani, F., Yousefvand, M., and Harighi, B. (2020). Phylogenetic analysis and molecular signatures specific to the Ralstonia solanacearum species complex. Eur. J. Plant Pathol. 158, 261–279. doi: 10.1007/s10658-020-02073-y
Fang, Q. Q., Feng, Y., Feng, P., Wang, X. H., and Zong, Z. Y. (2019). Nosocomial bloodstream infection and the emerging carbapenem-resistant pathogen Ralstonia insidiosa. BMC Infect. Dis. 19:334. doi: 10.1186/s12879-019-3985-4
Ferguson, H. W., Delannoy, C. M. J., Hay, S., Nicolson, J., Sutherland, D., and Crumlish, M. (2010). Jellyfish as vectors of bacterial disease for farmed salmon (Salmo salar). J. Vet. Diagn. Invest. 22, 376–382. doi: 10.1177/104063871002200305
Fernandez-Alvarez, C., Torres-Corral, Y., Saltos-Rosero, N., and Santos, Y. (2017). MALDI-TOF mass spectrometry for rapid differentiation of Tenacibaculum species pathogenic for fish. Appl. Microbiol. Biol. 101, 5377–5390. doi: 10.1007/s00253-017-8324-3
Fernandez-Piquer, J., Bowman, J. P., Ross, T., and Tamplin, M. L. (2012). Molecular analysis of the bacterial communities in the live Pacific oyster (Crassostrea gigas) and the influence of postharvest temperature on its structure. J. Appl. Microbiol. 112, 1134–1143. doi: 10.1111/j.1365-2672.2012.05287.x
Gardiner, M., Bournazos, A. M., Maturana-Martinez, C., Zhong, L., and Egan, S. (2017). Exoproteome analysis of the seaweed pathogen Nautella italica R11 reveals temperature-dependent regulation of RTX-Like proteins. Front. Microbiol. 8:1203. doi: 10.3389/fmicb.2017.01203
Granhag, L., Moller, L. F., and Hansson, L. J. (2011). Size-specific clearance rates of the ctenophore Mnemiopsis leidyi based on in situ gut content analyses. J. Plankton Res. 33, 1043–1052. doi: 10.1093/plankt/fbr010
Grossart, H. P., and Tang, K. W. (2010). www.aquaticmicrobial.net. Commun. Integr. Biol. 3, 491–494. doi: 10.4161/cib.3.6.12975
Guy-Haim, T., Rubin-Blum, M., Rahav, E., Belkin, N., Silverman, J., and Sisma-Ventura, G. (2020). The effects of decomposing invasive jellyfish on biogeochemical fluxes and microbial dynamics in an ultra-oligotrophic sea. Biogeosciences 17, 5489–5511. doi: 10.5194/bg-17-5489-2020
Habib, C., Houel, A., Lunazzi, A., Bernardet, J. F., Olsen, A. B., Nilsen, H., et al. (2014). Multilocus sequence analysis of the marine bacterial genus Tenacibaculum suggests parallel evolution of fish pathogenicity and endemic colonization of aquaculture systems. Appl. Environ. Microbiol. 80, 5503–5514. doi: 10.1128/AEM.01177-14
Hao, W. J., Gerdts, G., Holst, S., and Wichels, A. (2019). Bacterial communities associated with scyphomedusae at Helgoland Roads. Mar. Biodivers. 49, 1489–1503. doi: 10.1007/s12526-018-0923-4
Hao, W. J., Gerdts, G., Peplies, J., and Wichels, A. (2015). Bacterial communities associated with four ctenophore genera from the German Bight (North Sea). FEMS Microbiol. Ecol. 91, 1–11. doi: 10.1093/femsec/fiu006
He, H., Li, M. Y., Zhen, Y., Mi, T. Z., and Yu, Z. G. (2020). Bacterial and archaeal communities in sediments from the adjacent waters of Rushan Bay (China) revealed by Illumina sequencing. Geomicrobiol. J. 37, 86–100. doi: 10.1080/01490451.2019.1666193
Hofmann, D. K., Fitt, W. K., and Fleck, J. (1996). Checkpoints in the life-cycle of Cassiopea spp.: control of metagenesis and metamorphosis in a tropical jellyfish. Int. J. Dev. Biol. 40, 331–338.
Hsu, C. H., Chen, J. C., Lin, Y. C., Chen, Y. Y., Liu, P. C., Lin, B. W., et al. (2021). White shrimp Litopenaeus vannamei that have received mixtures of heat-killed and formalin-inactivated Vibrio alginolyticus and V. harveyi exhibit recall memory and show increased phagocytosis and resistance to Vibrio infection. Fish Shellfish Immun. 112, 151–158. doi: 10.1016/j.fsi.2020.11.013
Hudson, J., Gardiner, M., Deshpande, N., and Egan, S. (2018). Transcriptional response of Nautella italica R11 towards its macroalgal host uncovers new mechanisms of host-pathogen interaction. Mol. Ecol. 27, 1820–1832. doi: 10.1111/mec.14448
Jani, K., Bandal, J., Rale, V., Shouche, Y., and Sharma, A. (2019). Antimicrobial resistance pattern of microorganisms isolated and identified from Godavari River across the mass gathering event. J. Biosci. 44:121. doi: 10.1007/s12038-019-9941-z
Jaspers, C., Weiland-Bräuer, N., Rühlemannc, M. C., Baines, J. F., Schmitz, R. A., and Reusch, T. B. H. (2020). Differences in the microbiota of native and non-indigenous gelatinous zooplankton organisms in a low saline environment. Sci. Total Environ. 734:139471. doi: 10.1016/j.scitotenv.2020.139471
Joshi, J., Srisala, J., Truong, V. H., Chen, I. T., Nuangsaeng, B., Suthienkul, O., et al. (2014). Variation in Vibrio parahaemolyticus isolates from a single Thai shrimp farm experiencing an outbreak of acute hepatopancreatic necrosis disease (AHPND). Aquaculture 428, 297–302. doi: 10.1016/j.aquaculture.2014.03.030
Klimovich, A., Giacomello, S., Bjorklund, A., Faure, L., Kaucka, M., Giez, C., et al. (2020). Prototypical pacemaker neurons interact with the resident microbiota. Proc. Natl. Acad. Sci. U.S.A. 117, 17854–17863. doi: 10.1073/pnas.1920469117
Kramar, M. K., Tinta, T., Lucic, D., Malej, A., and Turk, V. (2019). Bacteria associated with moon jellyfish during bloom and post-bloom periods in the Gulf of Trieste (northern Adriatic). PLoS One 14:e0198056. doi: 10.1371/journal.pone.0198056
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, C. T., Chen, I. T., Yang, Y. T., Ko, T. P., Huang, Y. T., Huang, J. Y., et al. (2015). The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. U.S.A. 112, 10798–10803. doi: 10.1073/pnas.1517100112
Lee, D. L., Kling, J. D., Araya, R., and Ceh, J. (2018). Jellyfish life stages shape associated microbial communities, while a core microbiome is maintained across all. Front. Microbiol. 9:1534. doi: 10.3389/fmicb.2018.01534
Liang, T. M., Li, X. L., Du, J., Yao, W., Sun, G. Y., Dong, X. H., et al. (2011). Identification and isolation of a spiroplasma pathogen from diseased freshwater prawns, Macrobrachium rosenbergii, in China: a new freshwater crustacean host. Aquaculture 318, 1–6. doi: 10.1016/j.aquaculture.2011.03.018
Lopardo, C. R., and Urakawa, H. (2019). Performance and microbial diversity of bioreactors using polycaprolactone and polyhydroxyalkanoate as carbon source and biofilm carrier in a closed recirculating aquaculture system. Aquac. Int. 27, 1251–1268. doi: 10.1007/s10499-019-00383-5
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Lucas, C. H., and Horton, A. A. (2014). Short-term effects of the heavy metals, silver and copper, on polyps of the common jellyfish, Aurelia aurita. J. Exp. Mar. Biol. Ecol. 461, 154–161. doi: 10.1016/j.jembe.2014.08.003
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
McGann, P., Milillo, M., Clifford, R. J., Snesrud, E., Stevenson, L., Backlund, M. G., et al. (2013). Detection of New Delhi metallo-beta-lactamase (encoded by blaNDM-1) in Acinetobacter schindleri during routine surveillance. J. Clin. Microbiol. 51, 1942–1944. doi: 10.1128/JCM.00281-13
McKenney, P. T., and Pamer, E. G. (2015). From hype to hope: the gut microbiota in enteric infectious disease. Cell 163, 1326–1332. doi: 10.1016/j.cell.2015.11.032
Menon, R. R., Kumari, S., Kumar, P., Verma, A., Krishnamurthi, S., and Rameshkumar, N. (2019). Sphingomonas pokkalii sp. nov., a novel plant associated rhizobacterium isolated from a saline tolerant pokkali rice and its draft genome analysis. Syst. Appl. Microbiol. 42, 334–342. doi: 10.1016/j.syapm.2019.02.003
Neumann, R. (1979). Bacterial induction of settlement and metamorphosis in the planula larvae of Cassiopea andromeda (Cnidaria, Scyphozoa, Rhizostomeae). Mar. Ecol. Prog. Ser. 1, 21–28. doi: 10.3354/meps001021
Pitt, K. A., Welsh, D. T., and Condon, R. H. (2009). Influence of jellyfish blooms on carbon, nitrogen and phosphorus cycling and plankton production. Hydrobiologia 616, 133–149. doi: 10.1007/s10750-008-9584-9
Purcell, J. E. (2012). Jellyfish and ctenophore blooms coincide with human proliferations and environmental perturbations. Annu. Rev. Mar. Sci. 4, 209–235. doi: 10.1146/annurev-marine-120709-142751
Purcell, J. E., Uye, S. I., and Lo, W. T. (2007). Anthropogenic causes of jellyfish blooms and their direct consequences for humans: a review. Mar. Ecol. Prog. Ser. 350, 153–174. doi: 10.3354/meps07093
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Riemann, L., Titelman, J., and Bamstedt, U. (2006). Links between jellyfish and microbes in a jellyfish dominated fjord. Mar. Ecol. Prog. Ser. 325, 29–42. doi: 10.3354/meps325029
Robert, C. E. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/NMETH.2604
Robert, C. E., Brian, J. H., Jose, C. C., Christopher, Q., and Rob, K. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Schmahl, G. (1985). Bacterially induced stolon settlement in the scyphopolyp of Aurelia aurita (Cnidaria, Scyphozoa). Helgol. Meeresuntersuchungen 39, 33–42. doi: 10.1007/BF01997517
Schuett, C., and Doepke, H. (2010). Endobiotic bacteria and their pathogenic potential in cnidarian tentacles. Helgol. Mar. Res. 64, 205–212. doi: 10.1007/s10152-009-0179-2
Schuh, N. W., Carrier, T. J., Schrankel, C. S., Reitzel, A. M., Heyland, A., and Rast, J. P. (2020). Bacterial exposure mediates developmental plasticity and resistance to lethal Vibrio lentus infection in purple sea urchin (Strongylocentrotus purpuratus) larvae. Front. Immunol. 10:3014. doi: 10.3389/fimmu.2019.03014
Slinger, J., Adams, M. B., and Wynne, J. W. (2020). Bacteriomic profiling of branchial lesions induced by Neoparamoeba perurans challenge reveals commensal dysbiosis and an association with Tenacibaculum dicentrarchi in AGD-Affected Atlantic Salmon (Salmo salar L.). Microorganisms 8:1189. doi: 10.3390/microorganisms8081189
Småge, S. B., Brevik, ØJ., Frisch, K., Watanabe, K., Duesund, H., and Nylund, A. (2017). Concurrent jellyfish blooms and tenacibaculosis outbreaks in Northern Norwegian Atlantic salmon (Salmo salar) farms. PLoS One 12:e0187476. doi: 10.1371/journal.pone.0187476
Stabili, L., Rizzo, L., Basso, L., Marzano, M., Fosso, B., Pesole, G., et al. (2020). The microbial community associated with Rhizostoma pulmo: ecological significance and potential consequences for marine organisms and human health. Mar. Drugs 18:437. doi: 10.3390/md18090437
Sweetman, A. K., Chelsky, A., Pitt, K. A., Andrade, H., van Oevelen, D., and Renaud, P. E. (2016). Jellyfish decomposition at the seafloor rapidly alters biogeochemical cycling and carbon flow through benthic food-webs. Limnol. Oceanogr. 61, 1449–1461. doi: 10.1002/lno.10310
Tian, R. M., Sun, J., Cai, L., Zhang, W. P., Zhou, G. W., Qiu, J. W., et al. (2016). The deep-sea glass sponge Lophophysema eversa harbours potential symbionts responsible for the nutrient conversions of carbon, nitrogen and sulfur. Environ. Microbiol. 18, 2481–2494. doi: 10.1111/1462-2920.13161
Tinta, T., Kogovšek, T., Klun, K., Malej, A., Herndl, G. J., and Turk, V. (2019). Jellyfish-associated microbiome in the marine environment: exploring its biotechnological potential. Mar. Drugs 17:94. doi: 10.3390/md17020094
Tinta, T., Kogovšek, T., Malej, A., and Turk, V. (2012). Jellyfish modulate bacterial dynamic and community structure. PLoS One 7:11. doi: 10.1371/journal.pone.0039274
Tinta, T., Malej, A., Kos, M., and Turk, V. (2010). Degradation of the adriatic medusa Aurelia sp. by ambient bacteria. Hydrobiologia 645, 179–191. doi: 10.1007/s10750-010-0223-x
Tinta, T., Zhao, Z. H., Escobar, A., Klun, K., Bayer, B., Amano, C., et al. (2020). Microbial processing of jellyfish detritus in the ocean. Front. Microbiol. 11:590995. doi: 10.3389/fmicb.2020.590995
Titelman, J., Riemann, L., Sornes, T. A., Nilsen, T., Griekspoor, P., and Bamstedt, U. (2006). Turnover of dead jellyfish: stimulation and retardation of microbial activity. Mar. Ecol. Prog. Ser. 325, 43–58. doi: 10.3354/meps325043
Vezzulli, L., Brettar, I., Pezzati, E., Reid, P. C., Colwell, R. R., Hoefle, M. G., et al. (2012). Long-term effects of ocean warming on the prokaryotic community: evidence from the vibrios. ISME J. 6, 21–30. doi: 10.1038/ismej.2011.89
Viver, T., Orellana, L. H., Hatt, J. K., Urdiain, M., Díaz, S., Richter, M., et al. (2017). The low diverse gastric microbiome of the jellyfish Cotylorhiza tuberculata is dominated by four novel taxa. Environ. Microbiol. 19, 3039–3058. doi: 10.1111/1462-2920.13763
Wahl, K. L., Colburn, H. A., Wunschel, D. S., Petersen, C. E., Jarman, K. H., and Valentine, N. B. (2010). Residual agar determination in bacterial spores by electrospray ionization mass spectrometry. Anal. Chem. 82, 1200–1206. doi: 10.1021/ac901491c
Walayat, S., Malik, A., Hussain, N., and Lynch, T. (2018). Sphingomonas paucimobilis presenting as acute phlebitis: a case report. IDCases 11, 6–8. doi: 10.1016/j.idcr.2017.11.006
Walters, W., Hyde, E. R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A., et al. (2015). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009. doi: 10.1128/mSystems.00009-15
Wang, J., She, J. Y., Zhou, Y. C., Tsang, D. C. W., Beiyuan, J. Z., Xiao, T. F., et al. (2020). Microbial insights into the biogeochemical features of thallium occurrence: a case study from polluted river sediments. Sci. Total Environ. 739:139957. doi: 10.1016/j.scitotenv.2020.139957
Wang, P. P., Zhang, F., Sun, S., and Yang, T. (2020). Distribution of giant jellyfish in the Bohai sea in june 2018. Oceanol. Limnol. Sin. 51, 85–94. (in Chinese with English abstract),Google Scholar
Weiland-Bräuer, N., Neulinger, S. C., Pinnow, N., Künzel, S., Baines, J. F., and Schmitz, R. A. (2015). Composition of bacterial communities associated with Aurelia aurita changes with compartment, life stage, and population. Appl. Environ. Microbiol. 81, 6038–6052. doi: 10.1128/AEM.01601-15
Weiland-Bräuer, N., Pinnow, N., Langfeldt, D., Roik, A., Güllert, S., Chibani, C. M., et al. (2020). The native microbiome is crucial for offspring generation and fitness of Aurelia aurita. mBio 11:e0233620. doi: 10.1128/mBio.02336-20
West, E. J., Pitt, K. A., Welsh, D. T., Koop, K., and Rissik, D. (2009). Top-down and bottom-up influences of jellyfish on primary productivity and planktonic assemblages. Limnol. Oceanogr. 54, 2058–2071. doi: 10.4319/lo.2009.54.6.2058
Wilson, G. S., Raftos, D. A., and Nair, S. V. (2011). Antimicrobial activity of surface attached marine bacteria in biofilms. Microbiol. Res. 166, 437–448. doi: 10.1016/j.micres.2010.08.003
Xiao, W. P., Zeng, Y., Liu, X., Huang, X. G., Chiang, K. P., Mi, T. Z., et al. (2019). The impact of giant jellyfish Nemopilema nomurai blooms on plankton communities in a temperate marginal sea. Mar. Pollut. Bull. 149:110507. doi: 10.1016/j.marpolbul.2019.110507
Yun, L., Yu, Z., Li, Y., Luo, P., Jiang, X., Tian, Y., et al. (2019). Ammonia nitrogen and nitrite removal by a heterotrophic Sphingomonas sp. strain LPN080 and its potential application in aquaculture. Aquaculture 500, 477–484. doi: 10.1016/j.aquaculture.2018.10.054
Zeng, Y. X., Yu, Y., Qiao, Z. Y., Jin, H. Y., and Li, H. R. (2014). Diversity of bacterioplankton in coastal seawaters of Fildes Peninsula, King George Island, Antarctica. Arch. Microbiol. 196, 137–147. doi: 10.1007/s00203-013-0950-2
Zhang, F., Sun, S., Jin, X. S., and Li, C. L. (2012). Associations of large jellyfish distributions with temperature and salinity in the Yellow Sea and East China Sea. Hydrobiologia 690, 81–96. doi: 10.1007/s10750-012-1057-5
Zhou, A. G., Zhang, Y., Xie, S. L., Chen, Y. L., Li, X., Wang, J., et al. (2020). Microplastics and their potential effects on the aquaculture systems: a critical review. Rev. Aquac. 13, 719–733. doi: 10.1111/raq.12496
Keywords: jellyfish blooms, scyphomedusae, jellyfish microbiome, pathogenic bacteria, high-throughput sequencing
Citation: Peng S, Hao W, Li Y, Wang L, Sun T, Zhao J and Dong Z (2021) Bacterial Communities Associated With Four Blooming Scyphozoan Jellyfish: Potential Species-Specific Consequences for Marine Organisms and Humans Health. Front. Microbiol. 12:647089. doi: 10.3389/fmicb.2021.647089
Received: 29 December 2020; Accepted: 14 April 2021;
Published: 05 May 2021.
Edited by:
Lasse Riemann, University of Copenhagen, DenmarkReviewed by:
Julie Dinasquet, University of California, San Diego, United StatesTinkara Tinta, Marine Biology Station Piran, National Institute of Biology (Slovenia), Slovenia
Copyright © 2021 Peng, Hao, Li, Wang, Sun, Zhao and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijun Dong, empkb25nQHlpYy5hYy5jbg==
†These authors have contributed equally to this work and share first authorship
 Saijun Peng
Saijun Peng Wenjin Hao
Wenjin Hao Yongxue Li
Yongxue Li Lei Wang
Lei Wang Tingting Sun
Tingting Sun Jianmin Zhao
Jianmin Zhao Zhijun Dong
Zhijun Dong