
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 April 2021
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.646273
This article is part of the Research Topic Next-Generation Sequencing and CRISPR-Cas Editing in Plant Virology View all 17 articles
Citrus dwarfing viroid (CDVd) induces stunting on sweet orange trees [Citrus sinensis (L.) Osbeck], propagated on trifoliate orange rootstock [Citrus trifoliata (L.), syn. Poncirus trifoliata (L.) Raf.]. MicroRNAs (miRNAs) are a class of non-coding small RNAs (sRNAs) that play important roles in the regulation of tree gene expression. To identify miRNAs in dwarfed citrus trees, grown in high-density plantings, and their response to CDVd infection, sRNA next-generation sequencing was performed on CDVd-infected and non-infected controls. A total of 1,290 and 628 miRNAs were identified in stem and root tissues, respectively, and among those, 60 were conserved in each of these two tissue types. Three conserved miRNAs (csi-miR479, csi-miR171b, and csi-miR156) were significantly downregulated (adjusted p-value < 0.05) in the stems of CDVd-infected trees compared to the non-infected controls. The three stem downregulated miRNAs are known to be involved in various physiological and developmental processes some of which may be related to the characteristic dwarfed phenotype displayed by CDVd-infected C. sinensis on C. trifoliata rootstock field trees. Only one miRNA (csi-miR535) was significantly downregulated in CDVd-infected roots and it was predicted to target genes controlling a wide range of cellular functions. Reverse transcription quantitative polymerase chain reaction analysis performed on selected miRNA targets validated the negative correlation between the expression levels of these targets and their corresponding miRNAs in CDVd-infected trees. Our results indicate that CDVd-responsive plant miRNAs play a role in regulating important citrus growth and developmental processes that may participate in the cellular changes leading to the observed citrus dwarf phenotype.
Small RNAs (sRNAs) can be divided into several categories, which include small-interfering (si)RNAs, trans-acting (ta)-siRNAs, microRNAs (miRNAs), natural-antisense siRNAs (nat-siRNAs), and Piwi-interacting RNAs (piwi-RNAs) (Borges and Martienssen, 2015; Czech et al., 2018; Zhu et al., 2018; Treiber et al., 2019). One of the major components of endogenous plant sRNAs are miRNAs. miRNAs are encoded by plant MIR genes and have independent transcriptional units with their own regulatory promoters. They form double stranded stem loop structures that are processed to produce single stranded transcripts, typically 21–24 nucleotides (nt) in length (Wang et al., 2019).
MicroRNAs have essential functions in plant development and are involved in regulating a myriad of plant processes such as leaf, root, stem, and floral organ morphogenesis and development, biosynthesis, metabolism, homeostasis, vegetative to reproductive growth transition, senescence, signal transduction, and response to biotic and abiotic stress. Upon expression and processing, plant miRNAs are incorporated into the activated RNA-induced silencing complex (RISC) to target RNAs which are complementary to the miRNA guide strand. Once the activated miRNA–RISC complex finds the complementary plant mRNA, it silences the target via RNA degradation or translational repression (Wang et al., 2019) (Figure 1A).
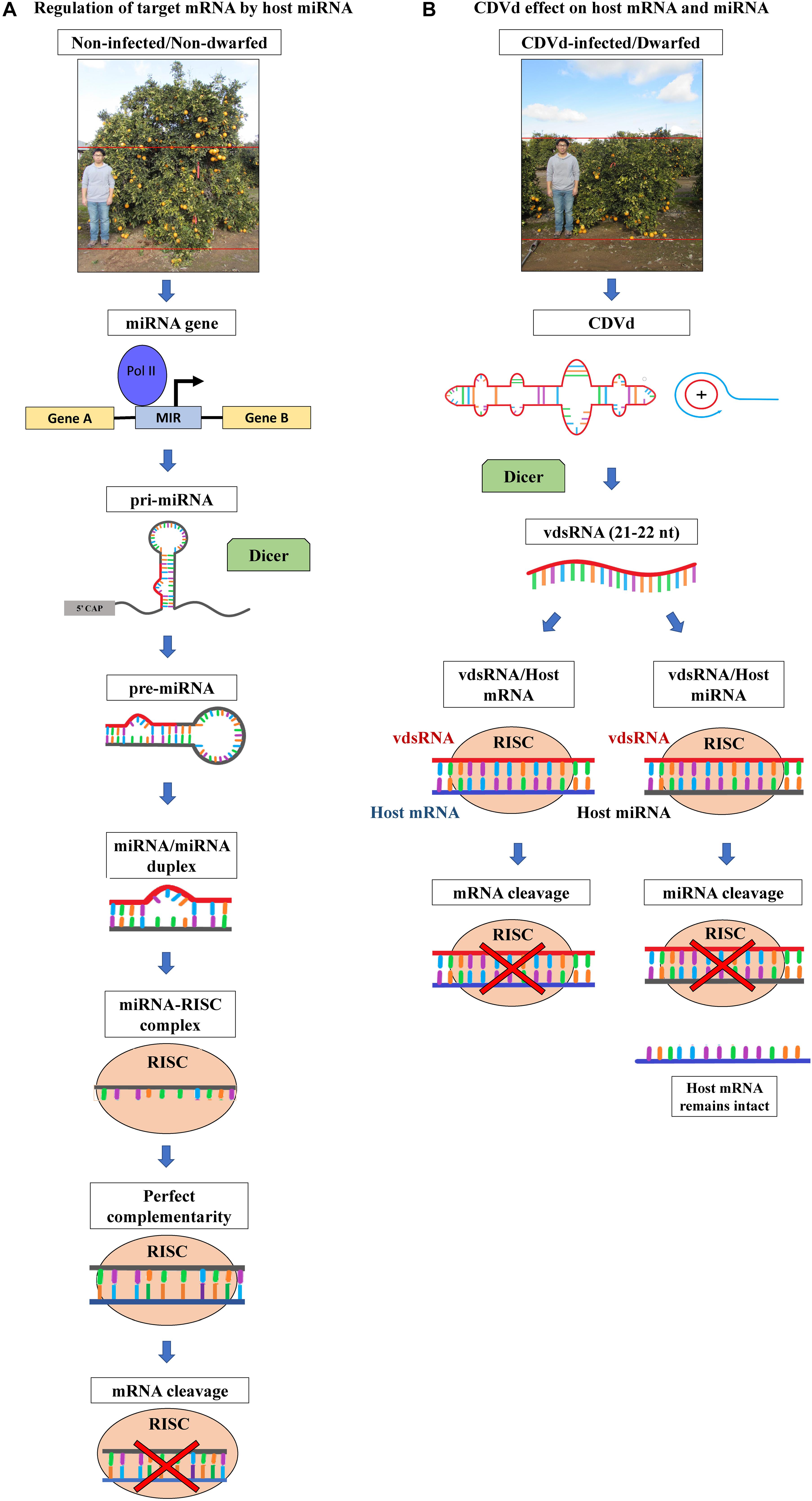
Figure 1. Citrus phenotypes and schematic representation of host microRNA (miRNA)-based gene expression regulatory pathway and the hypothesized effects of citrus dwarfing viroid (CDVd) on the expression of host target genes. (A) Non-viroid-infected, non-dwarf phenotype (∼3 m height) of navel orange [Citrus sinensis (L.) Osbeck] on trifoliate orange [Citrus trifoliata (L.) Raf.] rootstock, citrus tree and regulation of target messenger RNA (mRNA) by host miRNA. miRNA genes are transcribed by RNA polymerase II into primary miRNA (pri-miRNA) that folds into self-complementary stem-loop structures. The pri-RNA is then processed by the dicer-like (DCL) RNAse III endonucleases to form precursor miRNA (pre-miRNA), and the pre-miRNA is cleaved again to generate a miRNA/miRNA duplex. One strand of the duplex is degraded and the guide strand is incorporated into the RNA-inducing silencing effector complex (RISC). RISC, with RNAse H activity, targets RNA complementary to the miRNA guide strand. Once the complex finds the complementary plant mRNA, cleaves it, thus regulating its gene expression levels (for a review see Wang et al., 2019). (B) Viroid-infected, dwarf phenotype (∼2 m height) of navel orange trees on trifoliate orange rootstock and the production of viroid derived small RNAs (vdsRNAs). The highly structured CDVd RNA molecule and double stranded intermediate RNAs produced during the viroid rolling-circle replication, are processed by DCL into 21–22 nt vdsRNAs. vdsRNAs are incorporated into RISC and can be guided to form either vdsRNA/host mRNA or vdsRNA/host miRNA complexes. As a result, CDVd vdsRNAs could alter the expression levels of plant mRNA (directly or indirectly via cleavage of plant miRNAs), therefore resulting in the observed citrus tree dwarf phenotype in the field (for a review see Dadami et al., 2017).
Viroid derived sRNAs (vdsRNA) are products of the RNA interference (RNAi) basal plant antiviral defense response (Navarro et al., 2012; Dadami et al., 2013, 2017; Eamens et al., 2014; Adkar-Purushothama et al., 2015, 2017; Reis et al., 2015; Taliansky et al., 2021). Viroids (246–401 nt), highly structured, autonomously replicating RNA plant pathogenic agents, and trigger RNAi during their replication due to the formation of double stranded intermediate RNAs (Flores et al., 2009; Dadami et al., 2017). Similarly to plant endogenous sRNAs, vdsRNAs are 21–22 and 24 nt in length and have been detected in plants infected by several different viroids (Navarro et al., 2009; Bolduc et al., 2010; Diermann et al., 2010; Tsushima et al., 2011). vdsRNAs play an important role in viroid-mediated biological and pathogenic activities by guiding the RISC-mediated cleavage of host RNAs (Figure 1B) (Wassenegger et al., 1994; Itaya et al., 2007; Navarro et al., 2012; Dadami et al., 2013, 2017; Eamens et al., 2014; Adkar-Purushothama et al., 2015, 2017; Reis et al., 2015; Flores et al., 2017; Ramesh et al., 2020). Viroid infection might cause symptoms through the action of vdsRNAs which alter the expression levels of plant miRNAs, which in turn affects the expression levels of the plant mRNA targets of those plant miRNAs. It was reported that potato spindle tuber viroid (PSTVd) infection of tomato affects host miRNA production (Diermann et al., 2010) and host mRNA production (Wang et al., 2011; Owens et al., 2012). It was also reported, citrus bark cracking viroid infection was shown to affect plant miRNA regulation of plant transcription factors regulating leaf, cone and root growth and development of hop plants (Mishra et al., 2016).
The Citrus genus (family Rutaceae), includes several cultivars of high economic value including oranges, mandarins, grapefruits and lemons (2018–2019 US citrus crop packinghouse-door equivalent $3.35 billion) (USDA-NASS, 2019). Citrus flavors and aromas are among the most recognizable and preferred worldwide. In addition, citrus fruits are a rich source of vitamins, antioxidants, minerals, and dietary fiber essential for overall nutritional wellbeing (Van Duyn and Pivonka, 2000; Yao et al., 2004). Citrus trees are produced by grafting a desired scion variety onto a suitable rootstock species that then are planted in commercial citrus orchards. Tree spacing in citrus orchards has varied depending upon the cultivated species and a variety of factors such as soil type, climatic conditions and available farming equipment (Platt, 1973; Tucker and Wheaton, 1978). The historical global trend of citrus orchard spacing has been toward higher tree densities to maintain yield on the reduced available agricultural land and to increase economic returns.
Citrus dwarfing viroid (CDVd) infection of navel orange trees [Citrus sinensis (L.) Osb.] propagated on trifoliate orange [Citrus trifoliata (L.), syn. Poncirus trifoliata (L.) Raf.] rootstock has been previously reported to reduce canopy volume by approximately 50% (Figure 1B) (Vidalakis et al., 2011) and we recently demonstrated that the observed reduction in tree size results from a > 20% reduction in the apical growth of individual shoots within the tree canopy (Lavagi-Craddock et al., 2020). Understanding the molecular mechanism of the CDVd-induced citrus tree size reduction, will be most valuable as it could provide information on how to systematically produce dwarf trees for high density plantings.
To date, very few studies exist of miRNAs in citrus and even fewer in navel orange trees (Lu et al., 2015; Ma et al., 2016; Liang et al., 2017; Xie et al., 2017; Huang et al., 2019) and to our knowledge, there are no published studies on citrus miRNAs in response to viroid infection of citrus field trees. To explore the effect of CDVd-infection on citrus miRNAs and gain insight into the symptom development mechanism leading to the dwarfed phenotype observed in field plantings, we analyzed the effect of CDVd infection using next-generation sequencing (NGS) approach. The increasing number of miRNAs deposited in the miRBase database (Kozomara and Griffiths-Jones, 2014; Kozomara et al., 2019) from a wide range of species (<200), including C. sinensis, enables the discovery of novel miRNAs and their responses to pathogen infection, which may account for the observed species specific reactions and symptom development. Many plant miRNAs are conserved (Axtell and Bartel, 2005) but some are species specific (Moxon et al., 2008) and expressed at lower levels, thus making NGS the ideal approach to discover them and study their expression profiles (Jagadeeswaran et al., 2010; Motameny et al., 2010). Indeed, miRNAs from different plant species such as maize (Zhang et al., 2009), potato (Zhang et al., 2013), peanut (Zhang et al., 2017), barley (Ferdous et al., 2017), soybean (Zhang et al., 2008), and hop (Mishra et al., 2016), have been identified using NGS approaches.
In this study, we analyzed sRNA libraries prepared from field grown CDVd-infected navel orange and non-infected control trees to characterize miRNAs in the C. sinensis (stems) and C. trifoliata (roots) genomes and their expression profile in response to CDVd infection. This work provides valuable information at the molecular level and establishes the foundational framework that is necessary to dissect the subcellular mechanisms responsible for the observed citrus dwarf phenotype in the field.
Plant material (stems and roots) was collected in January 2016 from six 18-years-old “Parent Washington” navel [C. sinensis (L.) Osbeck] on “Rich 16-6” trifoliate orange [C. trifoliata (L.), syn. Poncirus trifoliata (L.) Raf.] rootstock infected (n = 3) and non-infected (n = 3) with CDVd, respectively. Trees were planted in an East–West running orchard located at the University of California (UC), Agriculture and Natural Resources, Lindcove Research and Extension Center (Exeter, CA, United States). CDVd-infected trees were planted at high density (3 × 6.7 m), whereas non-infected control trees were spaced at standard density (6.1 × 6.7 m).
Stem and root samples were processed in the field and immediately frozen in liquid nitrogen. For each tree, eight stem samples from around the canopy were collected. Leaves and petioles were removed, the stems were roughly chopped into approximately 0.5–1 cm pieces, placed into 50 ml conical tubes, and flash frozen. Root samples were collected from around the tree, at approximately 1 m away from the trunk and 20 cm deep, near the irrigation emitters, using a corer. The roots from eight core soil samples were washed thoroughly with water, gently blotted dry with paper towels, chopped into 0.5–1 cm pieces, placed into 50 ml conical tubes, and flash frozen. In between each sample collection and processing, cutting tools, and working surfaces were sanitized with 10% bleach solution (0.5–1% sodium hypochlorite) and rinsed with water and new sterile disposable plasticware and razor blades were used. Samples were transported into the Citrus Clonal Protection Program (CCPP), Citrus Diagnostic Therapy and Research Laboratory at the UC Riverside (Riverside, CA, United States) on dry ice and stored at −80°C until analysis.
Total RNA was isolated using the InvitrogenTM TRIzolTM (Thermo Fisher Scientific, Waltham, MA, United States) reagent. For each sample, 300 mg of frozen tissue were ground in liquid nitrogen with mortar and pestle. The ground material was transferred to a 5 ml Eppendorf tube and 3 ml of TRIzolTM reagent was added immediately. RNA extraction was performed according to the manufacturer’s instructions. The eluted RNA was aliquoted into four 1.5 ml microcentrifuge tubes to prevent freezing-thawing cycles during downstream analysis. The RNA concentration and quality was assessed with a spectrophotometer and the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, United States) using the Plant RNA Nano assay (RIN values were between 7.9 and 8.6).
The presence or absence of CDVd in each sample was confirmed by reverse transcription quantitative polymerase chain reaction (RT-qPCR) using a CCPP developed and validated assay [F: 5′-AACTTACCTGTCGTCGTC-3′; R: 5′-CGTGTTTTACCCTGGAGG-3′; Probe (FAM): 5′-CTCCGCTAGTCGGAAAGACTCCGC-3′]. The assay was performed using the iTaq Universal Probes One-Step Kit (Bio-Rad, Hercules, CA, United States) in 20 μL reactions with 10 μL of iTaq universal probe reaction mix, 0.5 μL of reverse transcriptase, 0.6 μL of forward primer (300 nM final concentration), 1.2 μL reverse primer (600 nM final concentration), 0.4 μL of probe (200 nM final concentration), 1 μL of RNA template, and 6.3 μL of water. The RT-qPCR was performed in the Bio-Rad CFX-96 and the reaction conditions were as follows: 30 min at 50°C, 5 min at 95°C, followed by 45 cycles of 10 s 95°C, 30 s at 59°C.
The sRNA libraries were prepared using the Illumina TruSeq Small RNA Kit (San Diego, CA, United States) following the manufacturer’s recommended protocol. The libraries were sequenced using an Illumina HiSeqTM 2500 instrument with single-end 50 bp reads (SeqMatic, Fremont, CA, United States). Raw reads were trimmed to remove low quality bases and adapters using cutadapt v. 1.15 (Martin, 2011) to generate clean sRNAs reads ranging from 18 to 28 nt in length.
The clean reads were then filtered for rRNA, tRNA, snRNA, snoRNA, repeat sequences, and other ncRNAs, using Rfam v.13.0 (Kalvari et al., 2018) with default parameters. The remaining reads were mapped to known miRNAs from the miRBase database (release 21, June 2014) to identify conserved miRNAs (Kozomara and Griffiths-Jones, 2011, 2014). The reads were further analyzed to predict potential novel miRNAs using miR-PREFer v. 0.24 using default parameters (Lei and Sun, 2014).
The conserved and novel miRNA sequences were analyzed against C. sinensis mRNA transcripts and C. trifoliata coding sequences (CDS) using psRNATarget v. 2.0 (Dai et al., 2018) to predict potential miRNA-mRNA interactions. DESeq2 v. 1.18 (Love et al., 2014) was used for the differential expression analyses of the miRNAs. The annotated mRNA targets were identified using the Blast2Go (Götz et al., 2008) tool within the OmicsBox software suite v. 1.4.11 (Cambridge, MA, United States). Figures were created using GraphPad Prism v. 9.0 (San Diego, CA, United States).
To validate the expression levels of conserved and novel miRNAs, custom stem-loop RT-qPCR assays (catalog number: 4398987) were designed by Thermo Fisher Scientific based on the sequences provided in Supplementary Table 1. For the relative expression quantification, U6 spliceosomal RNA was used as an internal control gene to normalize the efficiency between the target and internal control using the comparative Cq method (Schmittgen and Livak, 2008; Kou et al., 2012). The assay was carried out based on the manufacturer’s recommended protocol and all samples were standardized to the same concentration to ensure equal representation. The reverse transcription reactions were performed in a total volume of 15 μL with the TaqManTM MicroRNA Reverse Transcription Kit (Thermo Fisher) which contained 0.15 μL of 100 mM dNTP, 1 μL of MultiScribe Reverse transcriptase, 1.5 μL of 10x RT Buffer, 0.19 μL of RNase Inhibitor, 4.16 μL of nuclease-free water, 5 μL of total RNA, and 3 μL of 5x RT primer. The reverse transcription reactions were performed with the ProFlex PCR System (Thermo Fisher) as follows: 16°C for 30 min, 42°C for 30 min, 85°C for 5 min, and 4°C hold. The endpoint qPCR was performed in triplicates, according to the MIQE guidelines (Bustin et al., 2009), on a QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher) with the TaqManTM Fast Advanced Master Mix (Thermo Fisher) in a total of 20 μL reactions which included: 10 μL of master mix, 7.67 μL of nuclease-free water, 1 μL of TaqMan Small RNA Assay, 1.33 μL of the cDNA template. The endpoint PCR conditions were as follows: 50°C for 2 min, 95°C for 20 s, followed by 40 cycles of 95°C for 1 s, and 60°C for 20 s.
To verify the relative expression levels of the miRNA target genes, primers for the predicted target genes of miRNAs, were designed for RT-qPCR (Supplementary Table 2). Actin2 was used as an internal control gene to determine the relative abundance of the target mRNA expression levels by the comparative Cq method (Schmittgen and Livak, 2008; Mafra et al., 2012). Reverse transcription was performed using the InvitrogenTM SuperScriptTM II Reverse Transcriptase (RT) (Carlsbad, CA, United States). The reaction was performed using the manufacturer’s recommended protocol as follows: 1μL of olig (dT) (500 μg/mL), 1 μL of dNTP (10 mM), 2 μL of total RNA and 8 μL of nuclease-free water. The mixture was incubated for 5 min at 65°C and subsequently chilled on ice. The reaction was prepared with 4 μL of 5x First-Strand Buffer, 2 μL of 0.1M DTT, and 1 μL of RNaseOUT (40 units/μL) and then incubated for 2 min at 42°C. Finally, 1 μL of SuperScriptTM II RT (200 units) was added and the reaction was incubated at 42°C for 50 min followed by 70°C for 15 min. Downstream qPCR was also performed in triplicates, according to the MIQE guidelines, using the iTaq Universal SYBR Supermix (Bio-Rad): 10 μL of iTaq Universal SYBR Supermix, 1 μL of cDNA, 0.6 μL of each forward and reverse primers and 7.8 μL of nuclease-free water. The qPCR was performed on the Bio-Rad CFX-96 with the following conditions: 95°C for 1 min, followed by 40 cycles of 95°C for 10 s, and 60°C for 15 s.
To characterize citrus miRNAs and their expression profile in response to CDVd infection, we prepared and analyzed two sRNA libraries from stems and root samples of CDVd-infected and non-infected controls of navel orange citrus trees on trifoliate orange rootstock (Table 1).
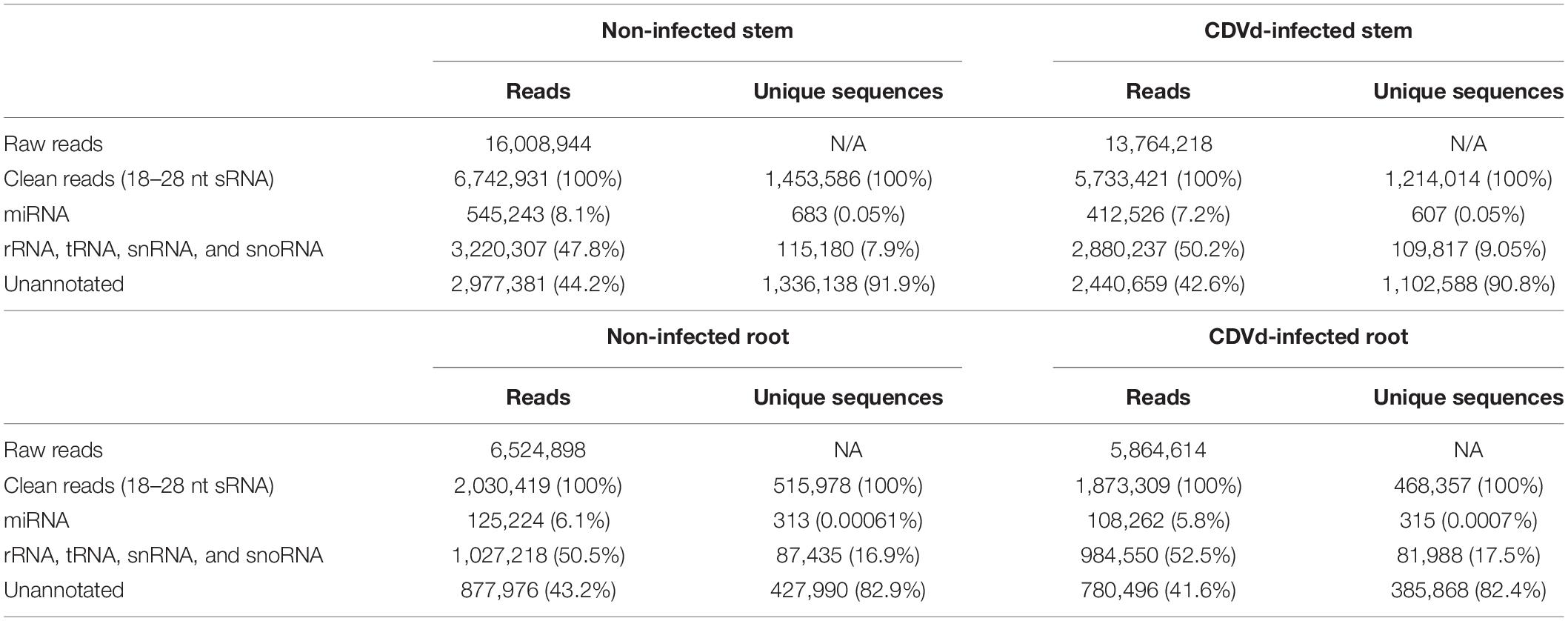
Table 1. Statistical summary of small RNA (sRNA) sequences from non-infected and citrus dwarfing viroid (CDVd)-infected libraries from stem (Citrus sinensis) and root (Citrus trifoliata) tissues.
From the non-infected trees, 8.1% of the stem and 6.1% of the root were classified as miRNAs. Similarly, for the CDVd-infected stems and roots, 7.2 and 5.8% of the reads, respectively, were classified as miRNAs. The unique unannotated sequences in both the non-infected and CDVd-infected stems represented at least 90% of the total reads while for the roots they represented over 82% (Table 1). The total unique miRNA reads for both non-infected and CDVd-infected stems represented 0.05% of the reads, while both non-infected and CDVd-infected roots represented 0.0006% of the reads (Table 1).
The most common size among the total mapped miRNAs sequences ranged between 20 and 24 nt in length, with 21-nt being the predominant miRNA class across different treatments and tissue types. This is consistent with plant antiviral RNAi responses and DCL-mediated processing of dsRNA producing 21 nt siRNAs.
The miRNA sequencing from non-infected control and CDVd-infected stems and roots identified 60 unique conserved miRNAs that ranged from 20 to 24 nt (Supplementary Tables 3, 4). Based on differential expression analysis, four conserved miRNAs (three in the stems and one in the roots) were found to be significantly altered in response to CDVd-infection (P-value and adjusted P-value < 0.05) (Table 2). Our results indicated that different members of the three miRNA families of interest had different expression levels between the non-infected and the CDVd-infected trees. The conserved miRNA families in the stems included csi-miR156, csi-miR171b, and csi-miR479, while csi-miR535 was the only conserved miRNA found in the roots. The conserved stem miRNAs were moderately more abundant compared to the conserved root miRNAs (Table 2). All four conserved miRNAs had higher expression levels in the non-infected control than the CDVd-infected trees (Table 2).
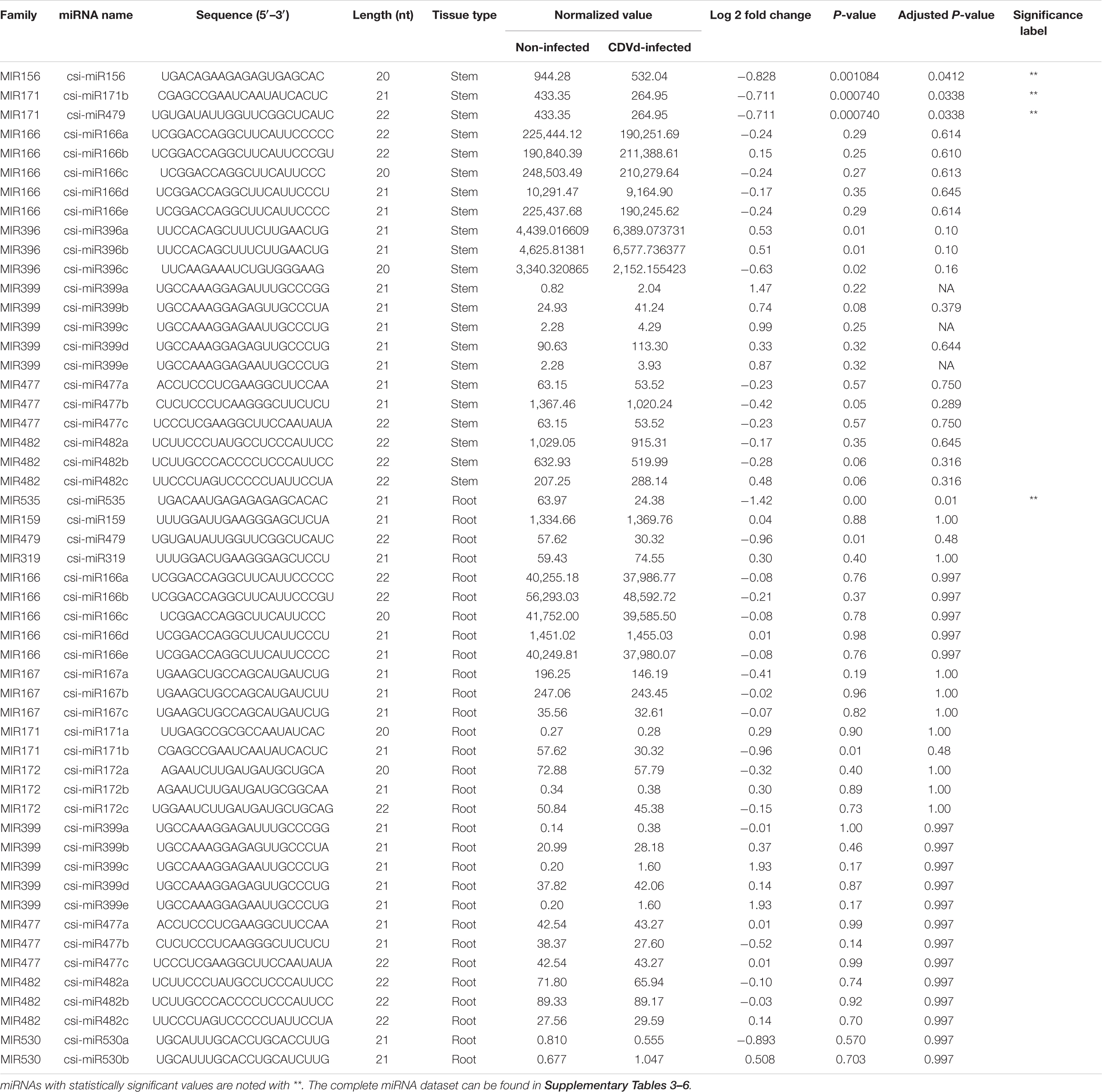
Table 2. Subsets of the conserved microRNAs (miRNAs) and their recovery profile in response to citrus dwarfing viroid (CDVd)-infection in stem (Citrus sinensis) and root (C. trifoliata) tissues.
Five miRNA families present in both stem and root tissues were identified: miR166, miR171b, miR399, miR477, and miR482. In stems, the highest represented miRNA families were miR166, and miR399, with five members each, followed by miR171b with four members, miR396, miR477, and miR482 with three members and the remaining 40 miRNAs were represented by a single member (Supplementary Table 3). In the roots, two miRNA families (miR166 and miR399) were represented by five members, five miRNA families (miR167, miR172, miR396, miR477, and miR482) were represented by three members, two miRNA families (miR530 and miR171b) contained two members, and the remaining 40 root miRNA families were represented by a single member (Supplementary Table 4).
Stem-loop RT-qPCR analysis was performed on the four conserved miRNAs in root and stem tissues (csi-miR479, csi-miR156, csi-miR171b, and csi-miR535) from non-infected and CDVd-infected trees to determine their relative abundance. The expression levels of the four conserved miRNAs were significantly altered as a result of CDVd infection. In the stems, csi-miR479’s expression decreased 3.55 fold and csi-miR171b had a fold decrease of 2.24, while csi-miR156 had the smallest negative fold change (0.11) (Figure 2). In the roots, csi-miR535′s expression decreased 1.12 fold (Figure 2). The results obtained from the RT-qPCR analysis (Figure 2) were consistent with the NGS read frequencies (Table 2) indicating a strong correlation between the RT-qPCR analysis and read frequencies obtained through small sRNA sequencing.
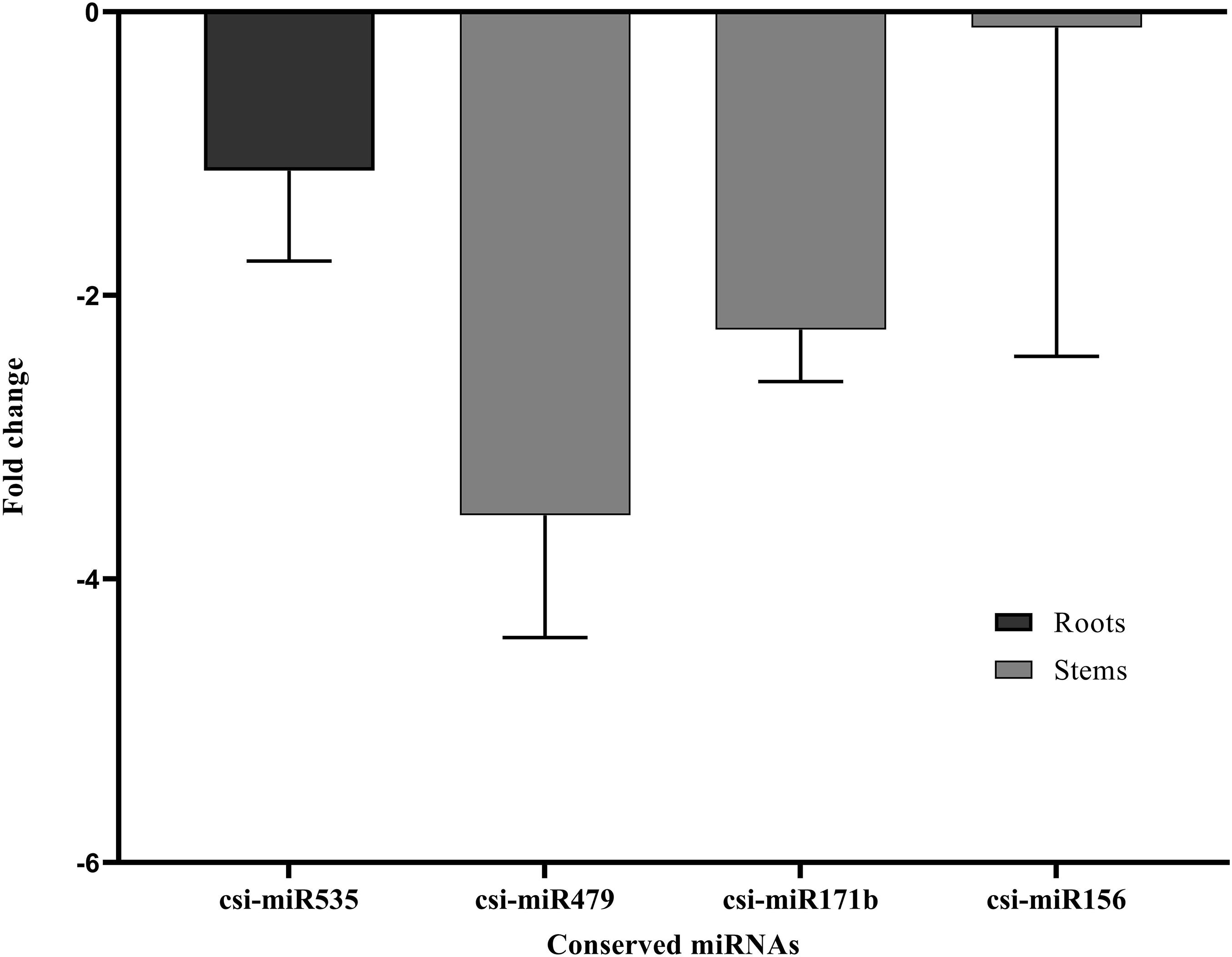
Figure 2. Differential expression analysis of four identified conserved miRNAs in response to CDVd infection. The relative abundance of each analyzed miRNA in CDVd-infected and non-infected trees was determined using the comparative Cq method by normalization to the U6 spliceosomal RNA. Conserved miRNAs with a significant change (P-value and adjusted P-value < 0.05) were considered to be differentially expressed. The bar graph shows the log2 fold change of expression levels of the miRNAs in CDVd-infected samples relative to non-infected samples in stem and root tissues.
The lengths of the predicted novel miRNAs from stems and roots ranged between 19 and 24 nt. A total of 646 stem and 108 root novel miRNAs were identified (Supplementary Tables 5, 6). No novel root miRNAs had significant differential expression levels. On the other hand, three novel stem miRNAs (csi-miRNA-75, csi-miRNA-114, and csi-miRNA-435) had significantly different expression levels in response to CDVd infection (P-value and adjusted P-value < 0.05). All three novel stem miRNAs had higher expression levels in the non-infected trees than in the CDVd-infected trees (Table 3).
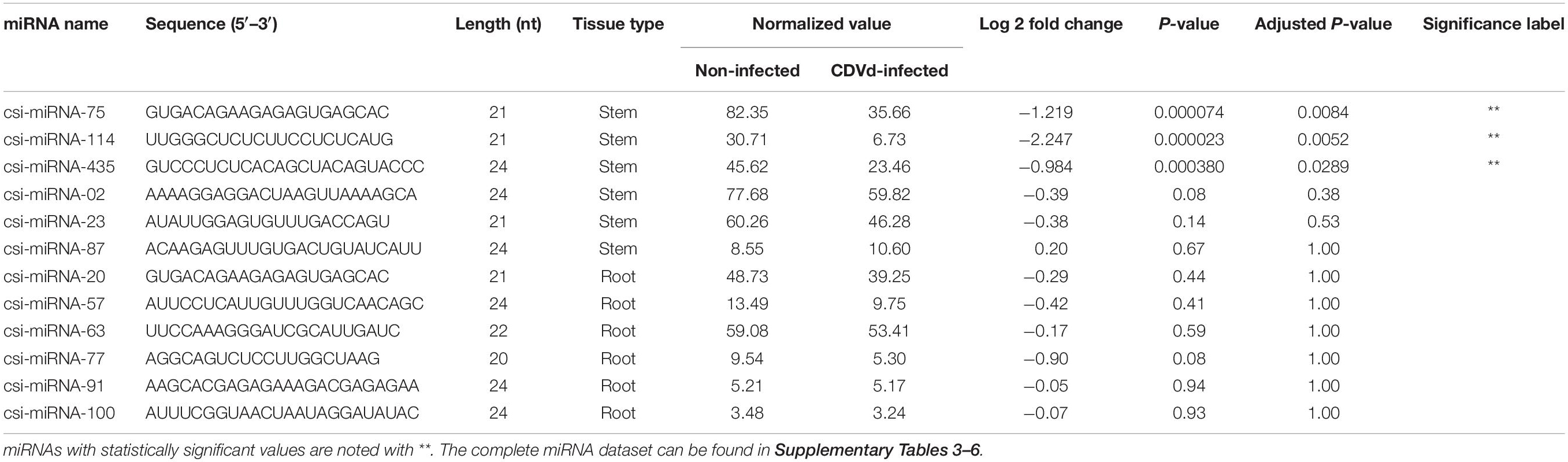
Table 3. Subsets of the novel microRNAs (miRNAs) and their recovery profile in response to citrus dwarfing viroid (CDVd)-infection in stem (Citrus sinensis) and root (C. trifoliata) tissues.
Stem-loop RT-qPCR was performed to confirm the NGS read frequency results of the identified novel miRNAs in response to CDVd-infection. csi-miRNA-114 showed the largest expression fold change (−9.23), while csi-miRNA-75 and csi-miRNA-435 showed similar fold changes (−1.44 and −1.22, respectively) (Figure 3). These results also support the reliability of RT-qPCR (Figure 3) and NGS read frequencies (Table 3) as measures of the expression levels of miRNAs.
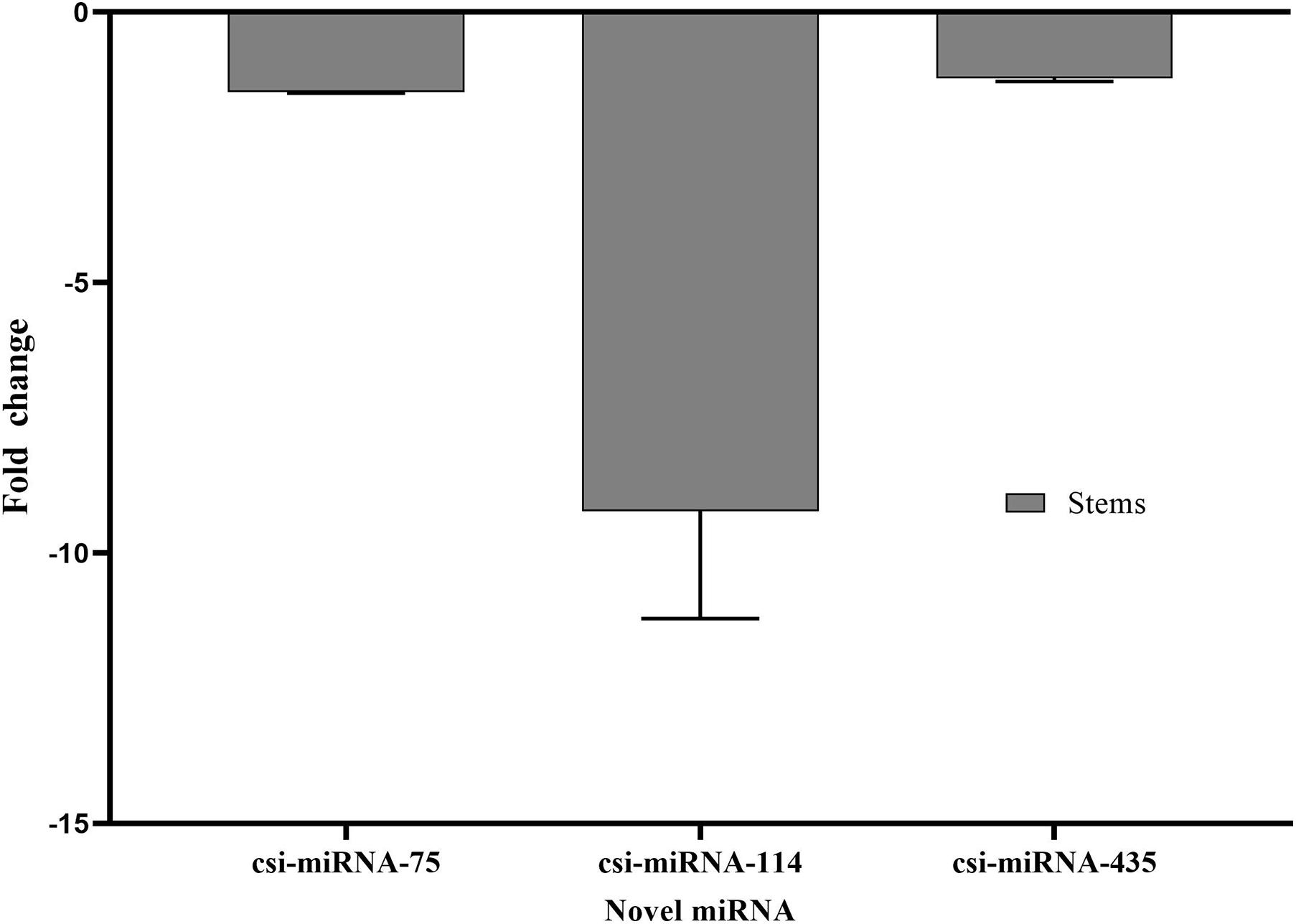
Figure 3. Differential expression analysis of three predicted novel miRNAs in response to CDVd infection. The relative abundance of each analyzed miRNA in CDVd-infected compared to non-infected trees was determined using the comparative Cq method by normalization to the U6 spliceosomal RNA (U6). Novel miRNAs with significant fold changes (P-value and adjusted P-value < 0.05) were considered to be differentially expressed. The bar graph shows log2 fold changes in expression levels of miRNAs in CDVd-infected samples relative to non-infected samples from stems.
To understand the function of the identified citrus miRNAs, host target genes were analyzed using the psRNATarget program by cross referencing the results against the C. sinensis genome for the stems (ref: GCF_000317415.1) and the C. trifoliata CDS for the roots (Kawahara et al., 2020). For both conserved and novel stem miRNAs, 83.1% of the miRNA targets were predicted to be regulated by cleavage and 16.9% by translational inhibition (Supplementary Tables 7, 8). Similarly, in the roots, 86.4% of the miRNA targets were predicted to be regulated by cleavage and 13.6% by translational inhibition (Supplementary Tables 9, 10).
Based on the extent of sequence complementarity between miRNAs and their targets, a total of 5,542 potential targets were predicted for the conserved and novel stem and root miRNAs (conserved: stem 63 and root 64; novel: stem 647 and root 109). Of the 5,542 potential miRNA target genes, 494 and 3,926 were targets of the conserved and novel stem miRNAs while 495 and 627 were targets of the conserved and novel root miRNAs, respectively.
The conserved stem and root miRNAs (miR479, mi171b, miR156, and miR535) analyzed in this study were associated with 10 different groups of target genes while the novel stem miRNAs (csi-miRNA-75, csi-miRNA-114, and csi-miR435) associated with three different target genes (see section “Discussion”) (Supplementary Table 11).
Clusters of orthologous groups (COG) functional classification of the targets of conserved and novel miRNAs revealed that the highest proportion of the genes were associated with (i) the nucleus (21% conserved and 11% novel); (ii) the integral component of membrane (14% conserved and 22% novel); and (iii) ATP binding (11% conserved and 10% novel) (Figures 4A,B).
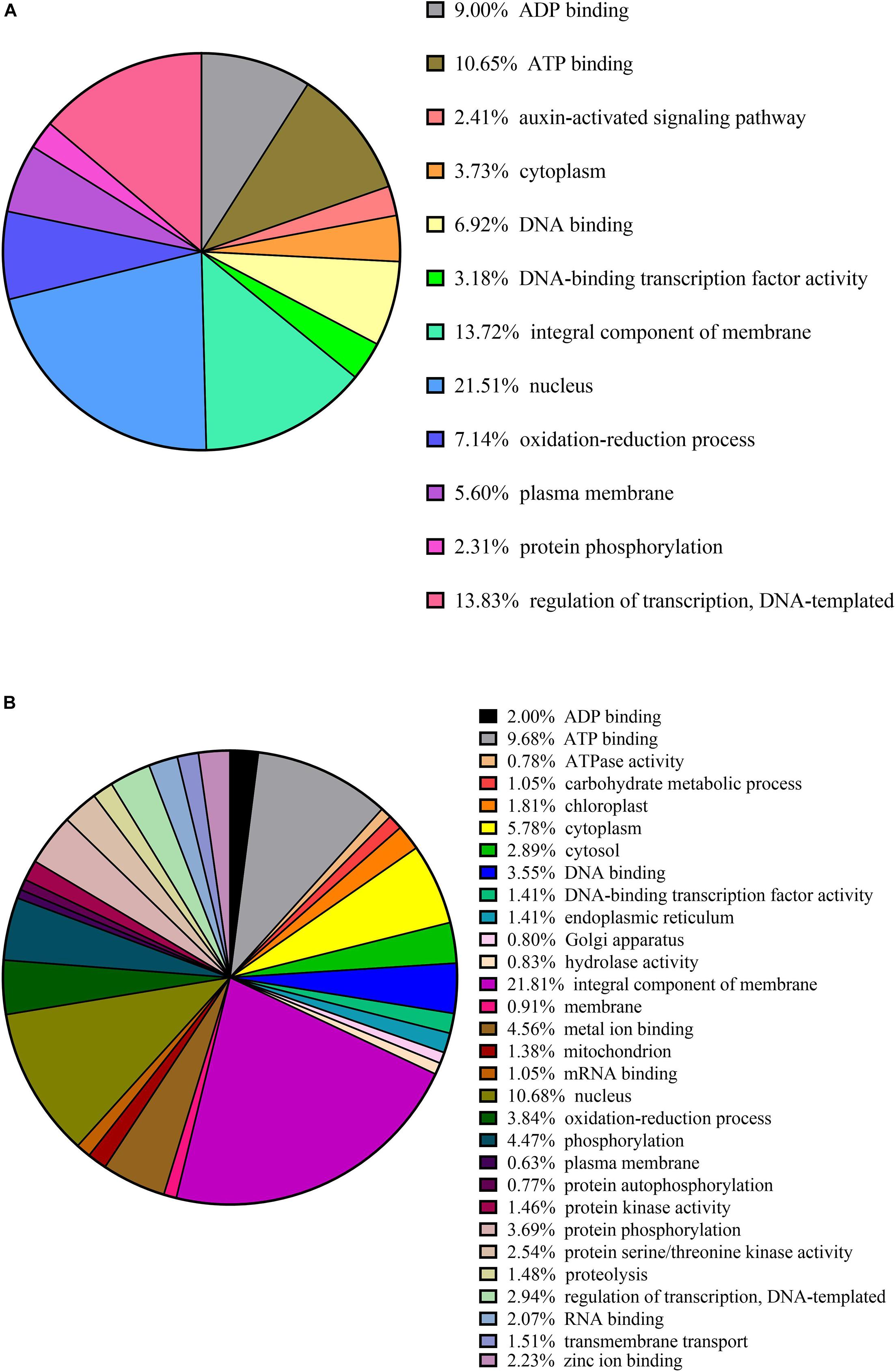
Figure 4. Cluster orthologous groups (COG) function calcification of predicted citrus target genes of conserved in panel (A) and novel in panel (B) miRNAs.
Other miRNA targets shared by the conserved and novel miRNAs include (i) ADP binding (9% conserved and 2% novel); (ii) cytoplasm (4% conserved and 6% novel); (iii) DNA-binding transcription factor activity (3% conserved and 1% novel); (vi) oxidation-reduction processes (7% conserved and 4% novel); (v) plasma membrane (6% conserved and 1% novel); (vi) protein phosphorylation (2% conserved and 4% novel); and (vii) regulation of transcription (14% conserved and 3% novel) (Figures 4A,B).
Our data were further annotated based on ontological definitions of the gene ontology (GO) terms, which categorized the predicted targets of the conserved miRNAs differentially expressed in response to CDVd infection into various biological, molecular and cellular processes (Figure 5A). Under the biological process, the predicted targets of conserved miRNA responsive to CDVd infection were subcategorized to (i) metabolic process; (ii) cellular process; (iii) biological regulation; and (iv) regulation of biological processes. The number of sequences associated with these four biological process subcategories had similar values in the stems and roots with the exception of the metabolic process that was higher in the roots (Figure 5A). For the molecular process, the majority of the predicted target genes of conserved miRNAs responsive to CDVd infection in the roots were subcategorized to catalytic activity while the targets in the stems were mostly subcategorized to binding. Targets belonging to the cellular components category were subcategorized to (i) cellular anatomical entity; and (ii) intracellular subcategories (Figure 5A) (Supplementary Table 11).

Figure 5. Clusters of orthologous groups functional classification of predicted target genes of conserved in panel (A) and novel in panel (B) CDVd-responsive miRNAs. The bar graph shows the number of sequences and distribution in different functional categories of the predicted miRNA targets at gene ontology (GO) level 2.
The targets of the predicted novel miRNAs displaying significant differential expression in response to CDVd infection (csi-miRNA-75, csi-miRNA-114, and csi-miRNA-435) were categorized to (i) biological process; (ii) molecular process; and (iii) cellular components. The cellular components subcategories (i) cellular anatomical entity; and (ii) intracellular contained most of the sequences (Figure 5B) (Supplementary Table 11).
The expression levels of eight predicted mRNA targets of the conserved CDVd-responsive miRNAs was determined via RT-qPCR. The results indicate that the expression of the miRNA target genes correlates negatively with the expression of their corresponding miRNA (Figure 6), thus confirming the relationship between CDVd-infection and altered expression levels of specific miRNA targets. Targets of miRNAs belonging to the same miRNA family showed variable results. For example, orange1.1g011651m (Figure 6, bar #5), orange1.1g032310m (Figure 6, bar #6), and orange1.1g008776m (Figure 6, bar #7), which are all targets of different members of the csi-miR156 family, were not uniform and showed fold-change differences.
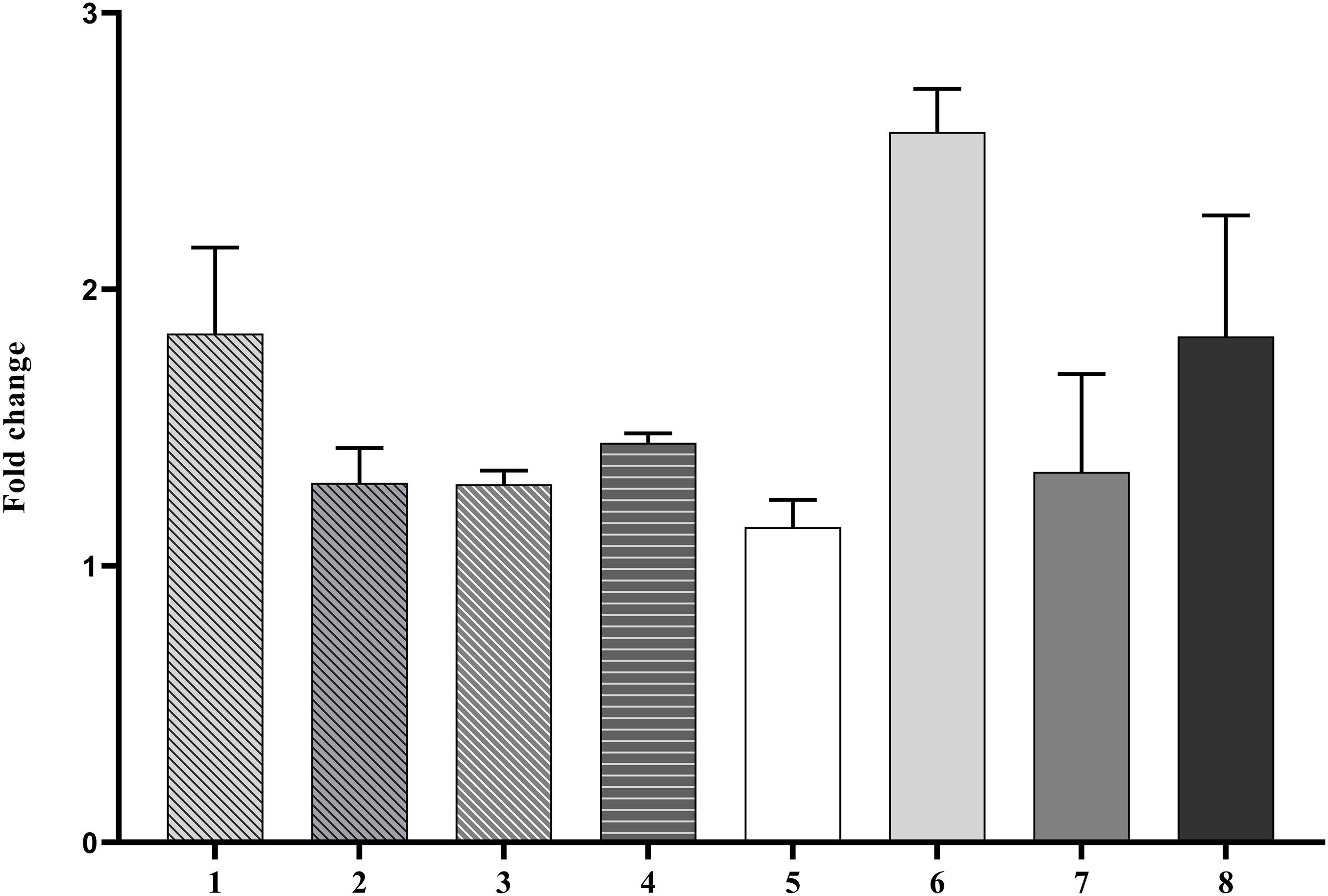
Figure 6. Differential expression profile of selected miRNA target genes. The relative gene expression was evaluated by the comparative Cq method using actin2 as a reference gene. The bar graph shows log2 fold changes of expression levels of target genes in CDVd-infected stems and roots relative to non-infected tissues. The predicted target genes used in the analysis were (1) UDP-glucose flavonoid glucosyl-transferase (orange1.1g033614m, target of csi-miR479-1-stem); (2) DEAD/DEAH box helicase (orange1.1g028826m, target of csi-miR479-2-stem); (3) DEAD/DEAH box helicase (orange1.1g026925m, target of csi-miR479-3-stem); (4) glutathione S-transferase (orange1.1g033674m, target of csi-miR171b-stem); (5) squamosa promoter binding protein-like 2 (orange1.1g011651m, target of csi-miR156-1-stem); (6) squamosa promoter binding protein-like 3 (orange1.1g032310m, target of csi-miR156-2-stem); (7) squamosa promoter-binding protein-like transcription factor family protein (orange1.1g008776m, target of csi-miR156-3-stem); and (8) RHOMBOID-like protein, P_trifoliata_00066_mRNA_51.1, target of csi-miR535-root).
Citrus production exceeded 157.9 million tons in over 9.8 million hectares worldwide for 20191, while in California alone the citrus industry is valued at $3.4 billion dollars with an estimated total economic impact of $7.1 billion (Babcock, 2018). Global decrease in farmland availability, increasing land, water and labor costs, and the continued spread of the deadly Huanglongbing (HLB) disease of citrus, make it imperative to develop tools that allow for high-density citrus plantings for maximization of yields and economic returns per land surface unit. In addition, these factors have forced the citrus industry toward the implementation of novel cultivation practices that would allow for mechanized citrus production under protective structures (Gottwald, 2010; Vidalakis et al., 2011; Lambin, 2012; Verburg et al., 2013; da Graça et al., 2016).
The idea of using “graft-transmissible dwarfing agents” for tree size reduction has been continuously investigated in citrus since originally proposed by Cohen (1968) and Mendel (1968) and countries such as Australia and Israel have explored the application of such technology in commercial settings (Broadbent et al., 1992; Bar-Joseph, 1993; Hutton et al., 2000; Semancik, 2003). The observation that CDVd significantly reduced C. sinensis canopy volume on C. trifoliata rootstock (Semancik et al., 1997; Vidalakis et al., 2011); by reducing vegetative growth (Lavagi-Craddock et al., 2020) indicated that CDVd may be used as a possible tool for high-density plantings of citrus, and provided key information on the possible biological mechanism through which CDVd affects specific rootstock-scion combinations to reduce tree canopy volume (Vidalakis et al., 2011). Furthermore, understanding the detailed molecular regulatory mechanisms that lead to a reduction in tree canopy volume in response to CDVd infection would provide the necessary knowledge to produce reduced-size citrus trees without the need of a -graft-transmissible viroid agent.
Small RNAs play an essential regulatory role in cellular and plant development functions including antiviral host responses and potentially viroid pathogenesis (Borges and Martienssen, 2015; Dadami et al., 2017; Flores et al., 2017; Czech et al., 2018; Zhu et al., 2018; Treiber et al., 2019; Wang et al., 2019; Ramesh et al., 2020). Two models have been proposed to explain the involvement of RNAi in the pathogenic process induced by viroid infections and both involve vdsRNAs. In the first model, vdsRNAs might act as miRNAs, downregulating the expression of physiologically important host genes, thus inducing disease associated symptoms. vdsRNAs are expected to contain significant identity to a region of the host genome for this model to work and resistance of viroids to RNAi is a feature of the viroid genome (Wang et al., 2004). According to the second model, disease symptoms caused by the nucleus replicating pospiviroids might result from the incorporation of viroid replication intermediates into the trans-acting small interfering RNA (ta-siRNA) biogenesis pathway. The nucleolus is a ta-siRNA free zone, and mature viroid forms produced in the nucleolus are resistant to degradation. In contrast, (vd)ta-siRNA produced in the nucleus from replication intermediates can then translocate to the cytoplasm where they guide the cleavage of target host mRNA leading to observed symptoms (Gómez et al., 2009). Both models involve viroid secondary structures as a key element that can therefore be interpreted as the evolutionary compromise between the need to interact with host factors and the necessity to survive RNAi. Regardless of whether vdsRNAs are produced according to the first or second model, it is also important to point out that rather than acting directly on host mRNA, vdsRNAs may affect host mRNA targets genes via host miRNAs as previously described (Mishra et al., 2016; Dadami et al., 2017).
Plant disease resistance gene families are typically very large with thousands of members and are commonly considered the putative targets of sRNAs (Chen et al., 2010), thus making the study of sRNAs in response to viroid infection a valid approach to investigate the biological mechanisms associated with symptoms. The systematic profiling of sRNAs in CDVd-infected trees, using NGS technologies, was the next logical step to gain insight into the function and regulatory mechanisms of miRNAs through which CDVd may reduce tree canopy size. In this study, we identified conserved and novel miRNAs in citrus and their response to CDVd infection. Consistently with the distribution patterns of sRNAs in other plant species, most sRNAs from both the CDVd-infected and non-infected libraries were found in the 21 and 24 nt classes (Jia et al., 2014; Gao et al., 2015; Mishra et al., 2016; Farooq et al., 2017; Zhang et al., 2018). CDVd-infected stems produced higher frequencies of the 21-nt class than their non-infected counterparts, indicating a CDVd induction of the 21-nt class since the other sRNA classes remained at comparable levels with the non-infected libraries. The increased abundance of the 21-nt class of sRNAs in response to viroid infection observed here is in agreement with previous reports for viral (viroid and virus) infections (Minoia et al., 2014; Zavallo et al., 2015).
All identified differentially expressed conserved and novel miRNAs, in this study, displayed overall reduced expression levels in response to CDVd-infection (Figures 2, 3). Several evolutionary deeply-conserved miRNAs have been shown to retain homologous targets across plant phyla (Axtell et al., 2007) and these include miR156 (stem), miR535 (roots), and miR171b (stem), which represent three out of the four conserved miRNA with differential expression levels in response to CDVd infection identified in this study. In agreement with previous studies, miR156 was shown here to direct the cleavage of squamosa-promoter binding-like protein (SBP) box genes (orange1.1g011651m; orange1.1g032310m; and orange1.1g008776m) (Cardon et al., 1999; Rhoades et al., 2002; Wu and Poethig, 2006; Xie et al., 2006; Gandikota et al., 2007; Riese et al., 2007) (Supplementary Table 7 and Figure 6). Members of this transcription factor family are known to play important roles in flower and fruit development, plant architecture, and in the transitions from juvenile to adult stages and to flowering (Chen et al., 2010; Yu et al., 2012). Even though miR535 is also known to target squamosa promoter-binding-like protein 3 (Shi et al., 2017; Zhou et al., 2020), our study identified rhomboid-like protein 1 as the target of miR535 in the roots (P_trifoliata_00066_mRNA_51.1). In Arabidopsis, a rhomboid-like protein was identified, providing evidence for the existence of regulated intramembrane proteolysis (RIP), a fundamental mechanism for controlling a wide range of cellular functions, in plants (Kanaoka et al., 2005). miR171b directs the cleavage of GRAS domain transcription factor genes (Ma et al., 2014). However, in our study, we found that miR171b’s target, a probable glutathione-S-transferase, was altered in response to CDVd infection (orange1.1g033674m). Glutathione-S-transferases are ubiquitous and multifunctional enzymes encoded by large gene families that can be highly induced by biotic stress including bacterial, fungal, and viral infection (Gullner et al., 2018). We found that the less conserved miR479 cleaves the UDP-glucose flavonoid glucosyl-transferase (orange1.1g033614m) (Vogt and Jones, 2000; Offen et al., 2006) which is involved in the process of conjugating hormones, stabilizing secondary metabolites, solubility, transport, and regulating bioavailability of compounds for other metabolic process in Arabidopsis thaliana and DEAD/DEAH box helicases which play an important in regulatory events such as organ maturation and cellular growth and differentiation (orange1.1g028826m and orange1.1g026925m) (Macovei et al., 2012). Both target genes may be related to the observed dwarf phenotype (Supplementary Table 11).
The predicted target genes of the novel miRNAs identified in this study (csi-miRNA-75, csi-miRNA-114, and csi-miRNA-435) include proteins were associated with target genes including SBP proteins (orange1.1g030599m, orange1.1g029650m, orange1.1g021420m, orange1.1g017256m, orange1.1g016971m, orange1.1g032310m orange1.1g046416m, orange1.1g011662m, orange1.1g010865m, and orange1.1g010591m; target of csi-miRNA-75); plastid-lipid associated protein (orange1.1g030218m, orange1.1g025746m, orange1.1g030180m, and orange1.1g020639m; target of csi-miRNA-114) which are structures that contain lipids and proteins that sequester the overaccumulation of carotenoids during flower development and fruit ripening (Moriguchi et al., 1998; Leitner-Dagan et al., 2006); and vacuolar protein sorting-associated proteins (orange1.1g021304m and orange1.1g017530m; target of csi-miR435), which direct protein cargo from the Golgi apparatus to the vacuoles (Xiang et al., 2013) and have been shown to be important in plant development (Cai et al., 2014) (Supplementary Table 11). The results suggest that CDVd-infection affects a wide range of biological functions via different miRNAs. In addition, the GO distribution analysis performed in this study, identified targets of conserved and novel CDVd-responsive miRNAs involved in various processes (Figure 5). Taken together, our findings might suggest that CDVd-infection could lead to developmental reprogramming and growth alterations of citrus trees, leading to the observed symptoms of reduced vegetative growth and overall smaller tree size. Future transcriptome studies could provide additional evidence to elucidate the molecular details in support of this hypothesis.
In this study, the overall miRNA profile of roots (trifoliate orange, C. trifoliata) was not altered in response to CDVd infection to the same extent demonstrated by the stem (navel orange, C. sinensis) miRNA profile. Although CDVd-derived sRNAs were detected in the roots, the trifoliate orange rootstock does not display major symptoms in response to CDVd infection (Vidalakis et al., 2004; Vernière et al., 2006; Murcia et al., 2009). This observation is consistent with the findings that the striking dwarfed citrus tree phenotype caused by CDVd infection results from the reduced vegetative growth of the stems, supporting the hypothesis that the molecular mechanisms responsible for this reprograming must be primarily active in the stems. Future greenhouse studies could provide additional evidence to support this hypothesis.
Finally, the term “Transmissible small nuclear ribonucleic acids” (TsnRNAs) was coined to identify those viroids that do not express a disease syndrome, but rather act as modifying agents of tree performance that result in desirable agronomic traits with potential economic advantages (Semancik et al., 1997; Semancik, 2003). In our study, we did not observe induction and regulation of defense genes via the sRNA pathway in response to CDVd infection, a finding that concurs with the hypothesis that some viroid species might be considered as RNAs modifying cellular functions and plant performance.
The expression profile of CDVd-responsive miRNAs indicates that these miRNAs play a role in regulating important citrus tree growth and development processes that may participate in the cellular changes leading to the observed C. sinensis on C. trifoliata rootstock dwarf phenotype.
The datasets presented in this study can be found in the NCBI Sequence Read Archive under accession numbers SRS8100788-SRS8100791 and PRJNA693870.
TD, IL-C, and GV conceived and designed the experiments, analyzed the data, and wrote the manuscript. TD and IL-C performed the experiments with the assistance of SB. All authors contributed to the article and approved the submitted version.
This research was funded by the Citrus Research Board project “Citrus Dwarfing of Commercial Varieties using TsnRNAs” (project 5100-154) awarded to GV. Additional support was provided in part by the Citrus Research Board (project 6100), the USDA National Institute of Food and Agriculture, Hatch project 1020106.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge the assistance of the dedicated personnel of the CCPP and the UC-ANR Lindcove Research and Extension Center. We would also like to acknowledge Prof. Danelle Seymour (University of California, Riverside) for providing advice regarding Citrus trifoliata genome. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the University of California. UC is an equal opportunity provider and employer.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.646273/full#supplementary-material
Supplementary Table 1 | Sequences submitted for stem-loop RT-qPCR of miRNAs.
Supplementary Table 2 | Specific primers used for RT-qPCR relative quantification of miRNA target genes.
Supplementary Table 3 | List of all conserved miRNAs found in Citrus sinensis stems.
Supplementary Table 4 | List of all conserved miRNAs found in Citrus trifoliata roots.
Supplementary Table 5 | List of all novel miRNAs found in stems.
Supplementary Table 6 | List of all novel miRNAs found in roots.
Supplementary Table 7 | List of predicted targets for conserved miRNAs in stems.
Supplementary Table 8 | List of predicted targets for novel miRNAs in stems.
Supplementary Table 9 | List of predicted targets for conserved miRNA in roots.
Supplementary Table 10 | List of predicted targets for novel miRNAs in roots.
Supplementary Table 11 | List of target genes of citrus dwarfing viroid-responsive conserved and novel miRNAs.
Adkar-Purushothama, C. R., Brosseau, C., Giguère, T., Sano, T., Moffett, P., and Perreault, J.-P. (2015). Small RNA derived from the virulence modulating region of the potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell 27, 2178–2194. doi: 10.1105/tpc.15.00523
Adkar-Purushothama, C. R., Iyer, P. S., and Perreault, J.-P. (2017). Potato spindle tuber viroid infection triggers degradation of chloride channel protein CLC-b-like and Ribosomal protein S3a-like mRNAs in tomato plants. Sci. Rep. 7:8341.
Axtell, M. J., and Bartel, D. P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17, 1658–1673. doi: 10.1105/tpc.105.032185
Axtell, M. J., Snyder, J. A., and Bartel, D. P. (2007). Common functions for diverse small RNAs of land plants. Plant Cell 19, 1750–1769. doi: 10.1105/tpc.107.051706
Bar-Joseph, M. (1993). Citrus viroids and citrus dwarfing in Israel. Acta Hortic. 349, 271–276. doi: 10.17660/actahortic.1993.349.45
Bolduc, F., Hoareau, C., St-Pierre, P., and Perreault, J.-P. (2010). In-depth sequencing of the siRNAs associated with peach latent mosaic viroid infection. BMC Mol. Biol. 11:16. doi: 10.1186/1471-2199-11-16
Borges, F., and Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Broadbent, P., Forsyth, J. B., Hutton, R. J., and Bevington, K. B. (1992). “Guidelines for the commercial use of graft-transmissible dwarfing in Australia–potential benefits and risks,” in Proceedings of the International Society of Citriculture, (Acireale: International Society of Citriculture), 697–701.
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cai, Y., Zhuang, X., Gao, C., Wang, X., and Jiang, L. (2014). The Arabidopsis endosomal sorting complex required for transport III regulates internal vesicle formation of the prevacuolar compartment and is required for plant development. Plant Physiol. 165, 1328–1343. doi: 10.1104/pp.114.238378
Cardon, G., Höhmann, S., Klein, J., Nettesheim, K., Saedler, H., and Huijser, P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237, 91–104. doi: 10.1016/s0378-1119(99)00308-x
Chen, H.-M., Chen, L.-T., Patel, K., Li, Y.-H., Baulcombe, D. C., and Wu, S.-H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. U.S.A. 107, 15269–15274. doi: 10.1073/pnas.1001738107
Chen, X., Zhang, Z., Liu, D., Zhang, K., Li, A., and Mao, L. (2010). SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J. Integr. Plant Biol. 52, 946–951. doi: 10.1111/j.1744-7909.2010.00987.x
Cohen, M. (1968). Exocortis virus as a possible factor in producing dwarf citrus trees. Proc. Fla. State Hort. Soc. 81, 115–119.
Czech, B., Munafò, M., Ciabrelli, F., Eastwood, E. L., Fabry, M. H., Kneuss, E., et al. (2018). piRNA-Guided genome defense: from biogenesis to silencing. Annu. Rev. Genet. 52, 131–157. doi: 10.1146/annurev-genet-120417-031441
da Graça, J. V., Douhan, G. W., Halbert, S. E., Keremane, M. L., Lee, R. F., Vidalakis, G., et al. (2016). Huanglongbing: an overview of a complex pathosystem ravaging the world’s citrus. J. Integr. Plant Biol. 58, 373–387. doi: 10.1111/jipb.12437
Dadami, E., Boutla, A., Vrettos, N., Tzortzakaki, S., Karakasilioti, I., and Kalantidis, K. (2013). DICER-LIKE 4 but not DICER-LIKE 2 may have a positive effect on potato spindle tuber viroid accumulation in Nicotiana benthamiana. Mol. Plant 6, 232–234. doi: 10.1093/mp/sss118
Dadami, E., Dalakouras, A., and Wassenegger, M. (2017). “Viroids and RNA Silencing,” in Viroids and Satellites, eds A. Hadidi, R. Flores, J. W. Randles, and P. Palukaitis (Boston: Academic Press), 115–124. doi: 10.1016/b978-0-12-801498-1.00011-5
Dai, X., Zhuang, Z., and Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54.
Diermann, N., Matoušek, J., Junge, M., Riesner, D., and Steger, G. (2010). Characterization of plant miRNAs and small RNAs derived from potato spindle tuber viroid (PSTVd) in infected tomato. Biol. Chem. 391, 1379–1390.
Eamens, A. L., Smith, N. A., Dennis, E. S., Wassenegger, M., and Wang, M.-B. (2014). In Nicotiana species, an artificial microRNA corresponding to the virulence modulating region of Potato spindle tuber viroid directs RNA silencing of a soluble inorganic pyrophosphatase gene and the development of abnormal phenotypes. Virology 450-451, 266–277. doi: 10.1016/j.virol.2013.12.019
Farooq, M., Mansoor, S., Guo, H., Amin, I., Chee, P. W., Azim, M. K., et al. (2017). Identification and characterization of miRNA transcriptome in asiatic cotton (Gossypium arboreum) using high throughput sequencing. Front. Plant Sci. 8:969. doi: 10.3389/fpls.2017.00969
Ferdous, J., Sanchez-Ferrero, J. C., Langridge, P., Milne, L., Chowdhury, J., Brien, C., et al. (2017). Differential expression of microRNAs and potential targets under drought stress in barley. Plant Cell Environ. 40, 11–24. doi: 10.1111/pce.12764
Flores, R., Di Serio, F., Navarro, B., and Owens, R. A. (2017). “Viroid Pathogenesis,” in Viroids and Satellites, eds A. Hadidi, R. Flores, J. W. Randles, and P. Palukaitis (Boston: Academic Press), 93–103. doi: 10.1016/b978-0-12-801498-1.00009-7
Flores, R., Gas, M.-E., Molina-Serrano, D., Nohales, M. -Á, Carbonell, A., Gago, S., et al. (2009). Viroid replication: rolling-circles, enzymes and ribozymes. Viruses 1, 317–334. doi: 10.3390/v1020317
Gandikota, M., Birkenbihl, R. P., Höhmann, S., Cardon, G. H., Saedler, H., and Huijser, P. (2007). The miRNA156/157 recognition element in the 3’ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49, 683–693. doi: 10.1111/j.1365-313x.2006.02983.x
Gao, J., Yin, F., Liu, M., Luo, M., Qin, C., Yang, A., et al. (2015). Identification and characterisation of tobacco microRNA transcriptome using high-throughput sequencing. Plant Biol. 17, 591–598. doi: 10.1111/plb.12275
Gómez, G., Martínez, G., and Pallás, V. (2009). Interplay between viroid-induced pathogenesis and RNA silencing pathways. Trends Plant Sci. 14, 264–269. doi: 10.1016/j.tplants.2009.03.002
Gottwald (2010). Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 48, 119–139. doi: 10.1146/annurev-phyto-073009-114418
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Gullner, G., Komives, T., Király, L., and Schröder, P. (2018). Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 9:1836. doi: 10.3389/fpls.2018.01836
Huang, J.-H., Lin, X.-J., Zhang, L.-Y., Wang, X.-D., Fan, G.-C., and Chen, L.-S. (2019). MicroRNA sequencing revealed citrus adaptation to long-term boron toxicity through modulation of root development by miR319 and miR171. Int. J. Mol. Sci. 20:1422. doi: 10.3390/ijms20061422
Hutton, R. J., Broadbent, P., and Bevington, K. (2000). Viroid dwarfing for high density citrus planting. Hortic Rev. 24, 277–317. doi: 10.1002/9780470650776.ch6
Itaya, A., Zhong, X., Bundschuh, R., Qi, Y., Wang, Y., Takeda, R., et al. (2007). A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 81, 2980–2994. doi: 10.1128/jvi.02339-06
Jagadeeswaran, G., Zheng, Y., Sumathipala, N., Jiang, H., Arrese, E. L., Soulages, J. L., et al. (2010). Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics 11:52. doi: 10.1186/1471-2164-11-52
Jia, L., Zhang, D., Qi, X., Ma, B., Xiang, Z., and He, N. (2014). Identification of the conserved and novel miRNAs in Mulberry by high-throughput sequencing. PLoS One 9:e104409. doi: 10.1371/journal.pone.0104409
Kalvari, I., Argasinska, J., Quinones-Olvera, N., Nawrocki, E. P., Rivas, E., Eddy, S. R., et al. (2018). Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46, D335–D342.
Kanaoka, M. M., Urban, S., Freeman, M., and Okada, K. (2005). An Arabidopsis rhomboid homolog is an intramembrane protease in plants. FEBS Lett. 579, 5723–5728. doi: 10.1016/j.febslet.2005.09.049
Kawahara, Y., Endo, T., Omura, M., Teramoto, Y., Itoh, T., Fujii, H., et al. (2020). Mikan Genome Database (MiGD): integrated database of genome annotation, genomic diversity, and CAPS marker information for mandarin molecular breeding. Breed. Sci. 70, 200–211. doi: 10.1270/jsbbs.19097
Kou, S.-J., Wu, X.-M., Liu, Z., Liu, Y.-L., Xu, Q., and Guo, W.-W. (2012). Selection and validation of suitable reference genes for miRNA expression normalization by quantitative RT-PCR in citrus somatic embryogenic and adult tissues. Plant Cell Rep. 31, 2151–2163. doi: 10.1007/s00299-012-1325-x
Kozomara, A., and Griffiths-Jones, S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157.
Kozomara, A., and Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73.
Kozomara, A., Birgaoanu, M., and Griffiths-Jones, S. (2019). miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162.
Lambin, E. F. (2012). Global land availability: malthus versus Ricardo. Glob. Food Sec. 1, 83–87. doi: 10.1016/j.gfs.2012.11.002
Lavagi-Craddock, I., Campos, R., Pagliaccia, D., Kapaun, T., Lovatt, C., and Vidalakis, G. (2020). Citrus dwarfing viroid reduces canopy volume by affecting shoot apical growth of navel orange trees grown on trifoliate orange rootstock. J. Citrus Pathol. 7, 1–6.
Lei, J., and Sun, Y. (2014). miR-PREFeR: an accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics 30, 2837–2839. doi: 10.1093/bioinformatics/btu380
Leitner-Dagan, Y., Ovadis, M., Shklarman, E., Elad, Y., Rav David, D., and Vainstein, A. (2006). Expression and functional analyses of the plastid lipid-associated protein CHRC suggest its role in chromoplastogenesis and stress. Plant Physiol. 142, 233–244. doi: 10.1104/pp.106.082404
Liang, W.-W., Huang, J.-H., Li, C.-P., Yang, L.-T., Ye, X., Lin, D., et al. (2017). MicroRNA-mediated responses to long-term magnesium-deficiency in Citrus sinensis roots revealed by Illumina sequencing. BMC Genomics 18:657. doi: 10.1186/s12864-017-3999-5
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.
Lu, Y.-B., Qi, Y.-P., Yang, L.-T., Guo, P., Li, Y., and Chen, L.-S. (2015). Boron-deficiency-responsive microRNAs and their targets in Citrus sinensis leaves. BMC Plant Biol. 15:271. doi: 10.1186/s12870-015-0642-y
Ma, C.-L., Qi, Y.-P., Liang, W.-W., Yang, L.-T., Lu, Y.-B., Guo, P., et al. (2016). MicroRNA regulatory mechanisms on citrus sinensis leaves to magnesium-deficiency. Front. Plant Sci. 7:201. doi: 10.3389/fpls.2016.00201
Ma, Z., Hu, X., Cai, W., Huang, W., Zhou, X., Luo, Q., et al. (2014). Arabidopsis miR171-targeted scarecrow-like proteins bind to GT cis-elements and mediate gibberellin-regulated Chlorophyll biosynthesis under light conditions. PLoS Genet. 10:e1004519. doi: 10.1371/journal.pgen.1004519
Macovei, A., Vaid, N., Tula, S., and Tuteja, N. (2012). A new DEAD-box helicase ATP-binding protein (OsABP) from rice is responsive to abiotic stress. Plant Signal. Behav. 7, 1138–1143. doi: 10.4161/psb.21343
Mafra, V., Kubo, K. S., Alves-Ferreira, M., Ribeiro-Alves, M., Stuart, R. M., Boava, L. P., et al. (2012). Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One 7:e31263. doi: 10.1371/journal.pone.0031263
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mendel, K. (1968). “Interrelations between tree performance and some virus diseases,” in Proceedings of the 4th Conference IOCV, (Riverside, CA: IOCV), 310–313.
Minoia, S., Carbonell, A., Di Serio, F., Gisel, A., Carrington, J. C., Navarro, B., et al. (2014). Specific argonautes selectively bind small RNAs derived from potato spindle tuber viroid and attenuate viroid accumulation in vivo. J. Virol. 88, 11933–11945. doi: 10.1128/jvi.01404-14
Mishra, A. K., Duraisamy, G. S., Matoušek, J., Radisek, S., Javornik, B., and Jakse, J. (2016). Identification and characterization of microRNAs in Humulus lupulus using high-throughput sequencing and their response to Citrus bark cracking viroid (CBCVd) infection. BMC Genomics 17:919. doi: 10.1186/s12864-016-3271-4
Moriguchi, T., Kita, M., Endo-Inagaki, T., Ikoma, Y., and Omura, M. (1998). Characterization of a cDNA homologous to carotenoid-associated protein in citrus fruits1The nucleotide sequence reported in this paper has been submitted to DDBJ under accession No. AB011797 (CitPAP). Contribution No. 1112 of the NIFTS.1. Biochim. Biophys. Acta 1442, 334–338.
Motameny, S., Wolters, S., Nürnberg, P., and Schumacher, B. (2010). Next generation sequencing of miRNAs - strategies, resources and methods. Genes 1, 70–84. doi: 10.3390/genes1010070
Moxon, S., Jing, R., Szittya, G., Schwach, F., Rusholme Pilcher, R. L., Moulton, V., et al. (2008). Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 18, 1602–1609. doi: 10.1101/gr.080127.108
Murcia, N., Bernad, L., Serra, P., Hashemian, S. M. B., and Duran-Vila, N. (2009). Molecular and biological characterization of natural variants of Citrus dwarfing viroid. Arch. Virol. 154, 1329–1334. doi: 10.1007/s00705-009-0430-9
Navarro, B., Gisel, A., Rodio, M. E., Delgado, S., Flores, R., and Di Serio, F. (2012). Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 70, 991–1003. doi: 10.1111/j.1365-313x.2012.04940.x
Navarro, B., Pantaleo, V., Gisel, A., Moxon, S., Dalmay, T., Bisztray, G., et al. (2009). Deep sequencing of viroid-derived small RNAs from grapevine provides new insights on the role of RNA silencing in plant-viroid interaction. PLoS One 4:e7686. doi: 10.1371/journal.pone.0007686
Offen, W., Martinez-Fleites, C., Yang, M., Kiat-Lim, E., Davis, B. G., Tarling, C. A., et al. (2006). Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25, 1396–1405. doi: 10.1038/sj.emboj.7600970
Owens, R. A., Tech, K. B., Shao, J. Y., Sano, T., and Baker, C. J. (2012). Global analysis of tomato gene expression during Potato spindle tuber viroid infection reveals a complex array of changes affecting hormone signaling. Mol. Plant. Microbe. Interact. 25, 582–598. doi: 10.1094/mpmi-09-11-0258
Platt, R. G. (1973). Treatment of Frost-Inured Citrus, Avocados. California Citrograph. Available online at: http://agris.fao.org/agris-search/search.do?recordID=US201303229376 (accessed November 15, 2020).
Ramesh, S. V., Yogindran, S., Gnanasekaran, P., Chakraborty, S., Winter, S., and Pappu, H. R. (2020). Virus and viroid-derived small RNAs as modulators of host gene expression: molecular insights into pathogenesis. Front. Microbiol. 11:614231. doi: 10.3389/fmicb.2020.614231
Reis, R. S., Eamens, A. L., Roberts, T. H., and Waterhouse, P. M. (2015). Chimeric DCL1-partnering proteins provide insights into the MicroRNA pathway. Front. Plant Sci. 6:1201. doi: 10.3389/fpls.2015.01201
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520.
Riese, M., Höhmann, S., Saedler, H., Münster, T., and Huijser, P. (2007). Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene 401, 28–37. doi: 10.1016/j.gene.2007.06.018
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Semancik, J. S. (2003). “Considerations for the introduction of viroids for economic advantage,” in Viroids, eds A. Hadidi, R. Flores, J. W. Randles, and J. S. Semancik (Clayton VIC: CSIRO Publishing), 357–362.
Semancik, J. S., Rakowski, A. G., Bash, J. A., and Gumpf, D. J. (1997). Application of selected viroids for dwarfing and enhancement of production of “Valencia” orange. J. Hortic. Sci. 72, 563–570. doi: 10.1080/14620316.1997.11515544
Shi, M., Hu, X., Wei, Y., Hou, X., Yuan, X., Liu, J., et al. (2017). Genome-wide profiling of small RNAs and degradome revealed conserved regulations of miRNAs on auxin-responsive genes during fruit enlargement in peaches. Int. J. Mol. Sci. 18:2599. doi: 10.3390/ijms18122599
Taliansky, M., Samarskaya, V., Zavriev, S. K., Fesenko, I., Kalinina, N. O., and Love, A. J. (2021). RNA-Based technologies for engineering plant virus resistance. Plants 10, 82–101. doi: 10.3390/plants10010082
Treiber, T., Treiber, N., and Meister, G. (2019). Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 20, 5–20. doi: 10.1038/s41580-018-0059-1
Tsushima, T., Murakami, S., Ito, H., He, Y.-H., Charith Raj, A. P., and Sano, T. (2011). Molecular characterization of Potato spindle tuber viroid in dahlia. J. Gen. Plant Pathol. 77, 253–256.
Tucker, D. P. H., and Wheaton, T. A. (1978). Trends in higher citrus planting densities. Proc. Fla. State Hort. Soc. 91, 36–40.
USDA-NASS (2019). Citrus Fruits 2019 Summary August 2019. Available online at: https://www.nass.usda.gov/Publications/Todays_Reports/reports/cfrt0819.pdf. (accessed November 15, 2020).
Van Duyn, M. A., and Pivonka, E. (2000). Overview of the health benefits of fruit and vegetable consumption for the dietetics professional: selected literature. J. Am. Diet. Assoc. 100, 1511–1521. doi: 10.1016/s0002-8223(00)00420-x
Verburg, P. H., Mertz, O., Erb, K.-H., Haberl, H., and Wu, W. (2013). Land system change and food security: towards multi-scale land system solutions. Curr. Opin. Environ. Sustain. 5, 494–502. doi: 10.1016/j.cosust.2013.07.003
Vernière, C., Perrier, X., Dubois, C., Dubois, A., Botella, L., Chabrier, C., et al. (2006). Interactions between citrus viroids affect symptom expression and field performance of clementine trees grafted on trifoliate orange. Phytopathology 96, 356–368. doi: 10.1094/phyto-96-0356
Vidalakis, G., Gumpf, D. J., Bash, J. A., and Semancik, J. S. (2004). Finger imprint of Poncirus trifoliata: a specific interaction of a viroid, a host, and irrigation. Plant Dis. 88, 709–713. doi: 10.1094/pdis.2004.88.7.709
Vidalakis, G., Pagliaccia, D., Bash, J. A., Afunian, M., and Semancik, J. S. (2011). Citrus dwarfing viroid: effects on tree size and scion performance specific to Poncirus trifoliata rootstock for high-density planting. Ann. Appl. Biol. 158, 204–217. doi: 10.1111/j.1744-7348.2010.00454.x
Vogt, T., and Jones, P. (2000). Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 5, 380–386. doi: 10.1016/s1360-1385(00)01720-9
Wang, J., Mei, J., and Ren, G. (2019). Plant microRNAs: biogenesis. Front. Plant Sci. 10:360. doi: 10.3389/fpls.2019.00360
Wang, M.-B., Bian, X.-Y., Wu, L.-M., Liu, L.-X., Smith, N. A., Isenegger, D., et al. (2004). On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. U.S.A. 101, 3275–3280. doi: 10.1073/pnas.0400104101
Wang, Y., Shibuya, M., Taneda, A., Kurauchi, T., Senda, M., Owens, R. A., et al. (2011). Accumulation of Potato spindle tuber viroid-specific small RNAs is accompanied by specific changes in gene expression in two tomato cultivars. Virology 413, 72–83. doi: 10.1016/j.virol.2011.01.021
Wassenegger, M., Heimes, S., and Sänger, H. L. (1994). An infectious viroid RNA replicon evolved from an in vitro-generated non-infectious viroid deletion mutant via a complementary deletion in vivo. EMBO J. 13, 6172–6177. doi: 10.1002/j.1460-2075.1994.tb06964.x
Wu, G., and Poethig, R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547. doi: 10.1242/dev.02521
Xiang, L., Etxeberria, E., and Van den Ende, W. (2013). Vacuolar protein sorting mechanisms in plants. FEBS J. 280, 979–993. doi: 10.1111/febs.12092
Xie, K., Wu, C., and Xiong, L. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 142, 280–293. doi: 10.1104/pp.106.084475
Xie, R., Zhang, J., Ma, Y., Pan, X., Dong, C., Pang, S., et al. (2017). Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep. 7:42094.
Yao, L. H., Jiang, Y. M., Shi, J., Tomás-Barberán, F. A., Datta, N., Singanusong, R., et al. (2004). Flavonoids in food and their health benefits. Plant Foods Hum. Nutr. 59, 113–122.
Yu, S., Galvão, V. C., Zhang, Y.-C., Horrer, D., Zhang, T.-Q., Hao, Y.-H., et al. (2012). Gibberellin regulates the Arabidopsis floral transition through miR156-Targeted SQUAMOSA PROMOTER BINDING–LIKE transcription factors. Plant Cell 24, 3320–3332. doi: 10.1105/tpc.112.101014
Zavallo, D., Debat, H. J., Conti, G., Manacorda, C. A., Rodriguez, M. C., and Asurmendi, S. (2015). Differential mRNA accumulation upon early Arabidopsis thaliana infection with ORMV and TMV-Cg Is associated with distinct endogenous small RNAs level. PLoS One 10:e0134719. doi: 10.1371/journal.pone.0134719
Zhang, B., Pan, X., and Stellwag, E. J. (2008). Identification of soybean microRNAs and their targets. Planta 229, 161–182. doi: 10.1007/s00425-008-0818-x
Zhang, L., Chia, J.-M., Kumari, S., Stein, J. C., Liu, Z., Narechania, A., et al. (2009). A genome-wide characterization of microRNA genes in maize. PLoS Genet. 5:e1000716. doi: 10.1371/journal.pgen.1000716
Zhang, R., Marshall, D., Bryan, G. J., and Hornyik, C. (2013). Identification and characterization of miRNA transcriptome in potato by high-throughput sequencing. PLoS One 8:e57233. doi: 10.1371/journal.pone.0057233
Zhang, T., Hu, S., Yan, C., Li, C., Zhao, X., Wan, S., et al. (2017). Mining, identification and function analysis of microRNAs and target genes in peanut (Arachis hypogaea L.). Plant Physiol. Biochem. 111, 85–96.
Zhang, Y., Wang, Y., Gao, X., Liu, C., and Gai, S. (2018). Identification and characterization of microRNAs in tree peony during chilling induced dormancy release by high-throughput sequencing. Sci. Rep. 8:4537.
Zhou, Y., Luo, S., Hameed, S., Xiao, D., Zhan, J., Wang, A., et al. (2020). Integrated mRNA and miRNA transcriptome analysis reveals a regulatory network for tuber expansion in Chinese yam (Dioscorea opposita). BMC Genomics 21:117. doi: 10.1186/s12864-020-6492-5
Keywords: gene regulation, miRNA, sRNA, RNAi, vdsRNA, siRNA, gene silencing, plant antiviral response
Citation: Dang T, Lavagi-Craddock I, Bodaghi S and Vidalakis G (2021) Next-Generation Sequencing Identification and Characterization of MicroRNAs in Dwarfed Citrus Trees Infected With Citrus Dwarfing Viroid in High-Density Plantings. Front. Microbiol. 12:646273. doi: 10.3389/fmicb.2021.646273
Received: 05 January 2021; Accepted: 06 April 2021;
Published: 30 April 2021.
Edited by:
Ahmed Hadidi, Agricultural Research Service, United States Department of Agriculture, United StatesReviewed by:
Teruo Sano, Hirosaki University, JapanCopyright © 2021 Dang, Lavagi-Craddock, Bodaghi and Vidalakis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georgios Vidalakis, Z2Vvcmdpb3MudmlkYWxha2lzQHVjci5lZHU=; dmlkYWxnQHVjci5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.