Corrigendum: First Report of a Foodborne Salmonella enterica Serovar Gloucester (4:i:l,w) ST34 Strain Harboring blaCTX–M−55 and qnrS Genes Located in IS26-Mediated Composite Transposon
- 1Institute of Food Safety and Nutrition, Jinan University, Guangzhou, China
- 2Department of Veterinary and Animal Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
- 3Guangzhou Food Inspection Institute, Guangzhou, China
- 4Shandong New Hope Liuhe Group Ltd., Qingdao, China
- 5School of Food Science and Engineering, South China University of Technology, Guangzhou, China
Extended-spectrum β-lactamases (ESBLs) production and (fluoro)quinolone (FQ) resistance among Salmonella pose a public health threat. The objective of this study was the phenotypic and genotypic characterization of an ESBL-producing and nalidixic acid-resistant Salmonella enterica serovar Gloucester isolate (serotype 4:i:l,w) of sequence type 34 (ST34) from ready-to-eat (RTE) meat products in China. Whole-genome short and long read sequencing (HiSeq and MinION) results showed that it contained blaCTX–M–55, qnrS1, and tetB genes, with blaCTX–M–55 and qnrS1 located in chromosomal IS26-mediated composite transposon (IS26–qnrS1–IS3–Tn3–orf–blaCTX–M–55–ISEcp1–IS26). The same genetic structure was found in the chromosome of S. enterica subsp. enterica serovar Typhimurium strain and in several plasmids of Escherichia coli, indicating that the IS26-mediated composite transposon in the chromosome of S. Gloucester may originate from plasmids of E. coli and possess the ability to disseminate to Salmonella and other bacterial species. Besides, the structural unit qnrS1–IS3–Tn3–orf–blaCTX–M–55 was also observed to be linked with ISKpn19 in both the chromosomes and plasmids of various bacteria species, highlighting the contribution of the insertion sequences (IS26 and ISKpn19) to the co-dissemination of blaCTX–M–55 and qnrS1. To our knowledge, this is the first description of chromosomal blaCTX–M–55 and qnrS in S. Gloucester from RTE meat products. Our work expands the host range and provides additional evidence of the co-transfer of blaCTX–M–55 and qnrS1 among different species of Salmonella through the food chain.
Introduction
Salmonella enterica is a leading cause of global bacterial foodborne gastroenteritis (Kirk et al., 2015; Lokken et al., 2016). Severely invasive salmonellosis is recommended to be treated with (fluoro)quinolones (FQs) and extended-spectrum cephalosporin (ESC) antimicrobials. Over the past two decades, S. enterica strains that are resistant to ESCs and FQs have emerged among humans, animals, and animal products, which is a worldwide public health concern (Wong et al., 2013; Brown et al., 2018; Lu et al., 2019; Zhang et al., 2019).
Cephalosporin resistance is mediated predominantly by extended-spectrum β-lactamases (ESBLs), AmpC β-lactamases, and carbapenemase (Arlet et al., 2006). These β-lactamases confer resistance to a wide range of β-lactam antibiotics, including penicillins, cephalosporins, and carbapenems. Besides, they are typically associated with multiple antibiotic resistance, such as quinolones, aminoglycosides, chloramphenicol, tetracycline, and trimethoprim–sulfamethoxazole and can transfer together among different bacteria species via mobile genetic elements (MGEs), leaving few therapeutic choices (Thomson, 2010; Iwamoto et al., 2017; Qiao et al., 2017; Nadimpalli et al., 2019).
Reduced susceptibility to FQs is associated with chromosomal mutations and the acquisition of antibiotic resistance genes, such as efflux pumps and plasmid-mediated quinolone resistance (PMQR) genes [qnr, aac(6′)-Ib-cr, oqxAB, and qepA genes] (Cuypers et al., 2018). PMQR genes confer only a low-level resistance to quinolones. However, they would provide a favorable background for higher resistance to occur (Hooper and Jacoby, 2015). Moreover, the ability to be spread by horizontal gene transfer constitutes a serious concern that should be addressed (Poirel et al., 2012).
Food-producing animals are considered to be the primary reservoir of ESBL-producing and quinolone-resistant Salmonella (Nadimpalli et al., 2019; Zhang et al., 2019). Contaminated raw or undercooked meat products are primary vehicles for Salmonella transmission to humans (Fearnley et al., 2011; Qiao et al., 2017). ESBL-producing and quinolone-resistant Salmonella strains have been detected in various Salmonella serotypes from clinical settings and raw meat samples (Wu et al., 2015; Qiao et al., 2017; Zhang et al., 2019). However, to our knowledge, this has not been reported in S. enterica serovar Gloucester from ready-to-eat (RTE) food products. Here, we characterize the first ESBL-producing and quinolone-resistant S. Gloucester strain from an RTE duck product carrying blaCTX–M–55 and qnrS genes, isolated during routine surveillance in Guangdong Province, China.
Materials and Methods
Strain Isolation and Identification
During our routine surveillance of foodborne pathogens on various food products, a Salmonella isolate (named GSJ/2017-Sal.-014, hereafter 17Sal014) was recovered from a roasted duck product in Guangzhou, southern China, in 2017. The isolate was first identified by biochemical confirmation using API 20E test identification test strips (bioMérieux, France) and further by 16S ribosomal RNA (rRNA) gene sequencing using the universal primers 27F (5′-AGAGTTTGATCCTG GCTCAG-3′) and 1492R (5′-GGCTACCTTGTTACGACTT-3′). The serotype was determined by the slide agglutination test using Salmonella antisera (SSI Diagnostica, Denmark) according to the Kauffmann–White scheme.
The strain was routinely grown in Luria–Bertani (LB; Guangdong Huankai Microbial Sci. & Tech., Guangzhou, China) broth or agar plates at 37°C for 12–24 h. Escherichia coli J53 was cultured in LB broth or agar plates with 150 μg/ml sodium azide (Sigma–Aldrich, St. Louis, MO, United States) and incubated at 37°C for 12–24 h.
Antibiotic Susceptibility Testing
The susceptibility of 17Sal014 to a panel of antimicrobial drugs (Hangzhou Microbial Reagent Co., Ltd., China)—including ciprofloxacin, cephalosporin II (cefuroxime), cephalosporin III (cefotaxime and ceftazidime), cephalosporin IV (cefepime), tetracycline, doxycycline, cefazolin, gentamicin, trimethoprim, tigecycline, chloramphenicol, fosfomycin, tobramycin, amikacin, netilmicin, piperacillin, ertapenem, imipenem, meropenem, cefoxitin, ampicillin–sulbactam sodium, aztreonam and ampicillin, and amoxicillin–clavulanic acid—was determined by disk diffusion antibiotic susceptibility testing (CLSI, 2016). The minimal inhibitory concentrations (MICs) of 17Sal014 to polymyxin B, cefotaxime, ciprofloxacin, and nalidixic acid (Sigma–Aldrich, St. Louis, MO, United States) were determined by broth microdilution (CLSI, 2016). The production of ESBL was confirmed by the disk diffusion clavulanate inhibition test using ceftazidime and cefotaxime. Reference strain E. coli ATCC 25922 served as a quality control.
Whole-Genome Sequencing and Annotation
The genomic DNA of isolate 17Sal014 was extracted using a commercial DNA extraction kit (Magen, Guangzhou, China) following the manufacturer’s recommendations. The whole genome of the isolate was sequenced on Illumina HiSeq X Ten with 150-bp paired-end reads (MajorBio Co., Shanghai, China) and MinION (Oxford Nanopore, Oxford, United Kingdom). For the MinION platform, the library was prepared using the ONT 1D ligation sequencing kit (SQK-LSK109) with the native barcoding expansion kit (EXP-NBD104). The genome was assembled using a combination of short and long reads by SPAdes (Bankevich et al., 2012) and the Unicycler hybrid assembler (Wick et al., 2017) and annotated by Prokka (Seemann, 2014).
Clonal analysis was assessed by MLST 2.01. PlasmidFinder v2.1 (Carattoli and Hasman, 2020) was used to identify plasmid replicon types. The presence of acquired antibiotic resistance genes and mutations in the quinolone resistance-determining regions (QRDRs) (gyrA, gyrB, parC, and parE) was assessed by ResFinder (Zankari et al., 2012) and further determined by BLASTn2.
Conjugation
IS26 plays a critical role in the dissemination of antibiotic resistance genes in Gram-negative bacteria (Harmer et al., 2014; Harmer and Hall, 2015, 2016). To reveal the transferability of the chromosomally located blaCTX–M–55 and qnrS1, which is possibly mediated by an IS26-flanked composite transposon, a conjugation experiment was conducted by solid mating on a filter (Whatman, Maidstone, United Kingdom) using sodium azide-resistant E. coli J53 as a recipient and a selection of transconjugants on LB agar containing 150 μg/ml sodium azide and 4, 8, and 16 μg/ml cefotaxime, respectively. The transfer of plasmid to the transconjugants and possible monomeric circular intermediates of the IS26 in the transconjugants was confirmed by PCR with a set of specific primers (Supplementary Table 1).
Nucleotide Sequence Accession Number
The assembly genome sequence of S. Gloucester 17Sal014 was deposited in the NCBI database under the accession number SAMN14178317. The Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number VFRK00000000. The 8,993-bp composite transposon containing blaCTX–M–55 and qnrS1 was deposited in GenBank under the accession number MN619286.1.
Results
Identification of Salmonella and Antibiotic Susceptibility
The isolate was confirmed as S. enterica serovar Gloucester, serotype 4:i:l,w, by biochemical confirmation, 16S rRNA gene sequencing, and serotyping.
The disk diffusion antibiotic susceptibility testing showed that the isolate was resistant to tetracycline, doxycycline, cefazolin, aztreonam, ampicillin, amoxicillin clavulanic acid, and cephalosporins II (cefuroxime) and III (cefotaxime and ceftazidime), intermediate resistant to cephalosporin IV (cefepime), and produces ESBL. The isolate exhibited MIC values of ciprofloxacin, nalidixic acid, and cefotaxime of 0.25, 64, and 128 mg/L, respectively.
General Features of the S. Gloucester 17Sal014 Genome
The complete genome sequence of S. Gloucester 17Sal014 contained a circular 4,986,395-bp chromosome with a G + C content of 52.2%. There were 5,089 predicted genes and 4,960 predicted coding sequences (CDs) in the whole genome sequence, including 85 RNA genes consisting of 80 transfer RNAs (tRNAs) and four rRNAs. Multilocus sequence typing analysis showed that 17Sal014 belongs to sequence type 34 (ST34).
Acquired resistance genes against tetracycline (tetB), quinolones (qnrS1), and cephalosporins (blaCTX–M–55) were identified. In addition, four more substitutions in parC (T255S, A395S, S469A, and A620T) were detected, which were not identified before.
The blaCTX–M–55 and qnrS1 genes were located on chromosome within 8,993 bp composite transposon, IS26–qnrS1–IS3–Tn3–orf–blaCTX–M–55–ISEcp1–IS26, and flanked by genes encoding DNA cytosine methyltransferase (orf1), type II site-specific deoxyribonuclease (orf2), hypothetical protein (orf3), 3′–5′ exoribonuclease (orf4), DUF4224 domain-containing protein (orf5), and tyrosine-type recombinase/integrase (orf6). blaCTX–M–55 was linked with incomplete ISEcp1. The 8,993-bp genetic structure was identical to the corresponding region on the chromosome of Salmonella Typhimurium strain S441 isolated from a human stool sample in Hangzhou, China (GenBank accession no. CP061122.1) and with 99% identity (with 100% query coverage) to pPN156 (GenBank accession number MT449721.1) of an E. coli strain from human intestine and pPN24 (GenBank accession number MT449722.1) of an E. coli strain from duck feces in Thailand, as well as pCHL5009T-102k-mcr3 (GenBank accession number CP032937.1) of an E. coli strain from urine samples in New Zealand. The plasmid p08-5333.1 of S. enterica subsp. enterica strain (GenBank accession number CP039562.1) shared a similar genetic environment, but differed in orf-blaCTX–M–55–ISEcp1–IS26 insertion site and orientation. The pPN42 of the E. coli strain (GenBank accession number MT449720.1) also contained a similar region, but lacked ISEcp1. Besides, the structural unit qnrS1–IS3–Tn3–orf–blaCTX–M–55 was observed in various bacterial species, such as the chromosome of E. coli strains and plasmids in E. coli, Klebsiella pneumoniae, Salmonella Typhi, and S. enterica (Figure 1 and Supplementary Table 2). Of note is that the unit was identified in either the chromosome or on plasmids in four Salmonella and two E. coli strains of animal, chicken meat, food, and human sources in China. In these isolates from China and various species from other countries, the unit qnrS1–IS3–Tn3–orf–blaCTX–M–55 was linked with ISKpn19 and/or IS26 (Table 1 and Figure 1). These findings indicate that the structural unit qnrS1–IS3–Tn3–orf–blaCTX–M–55 was transferred among different bacterial species with the help of the insertion sequences (IS26 and ISKpn19). The transfer of blaCTX–M–55 and qnrS1 to E. coli was not detected under laboratory experimental conditions in this study. However, it is possible that the IS26-mediated composite transposon maintains the transfer capacity under adverse environments.
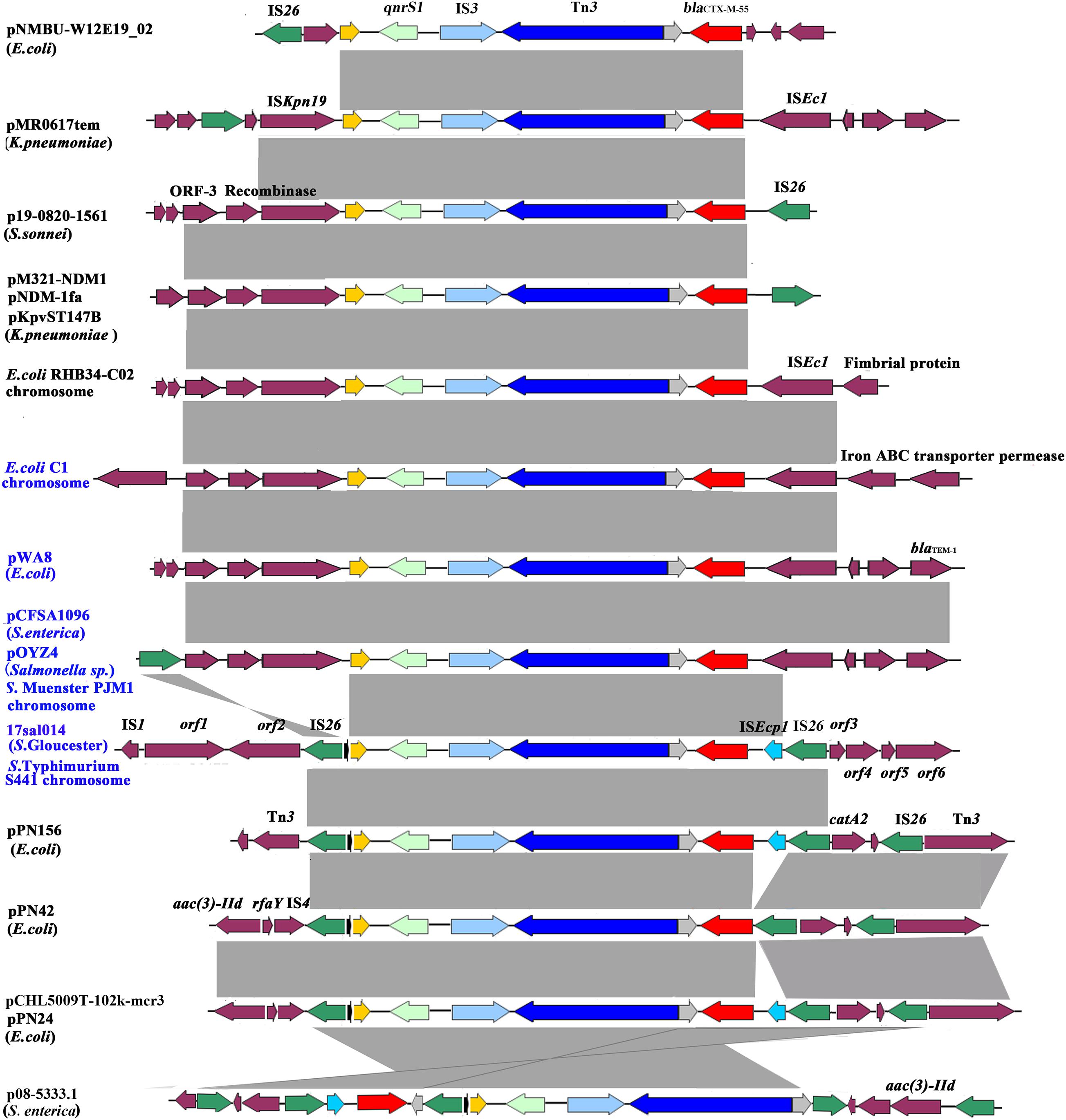
Figure 1. Genetic environment of the blaCTX–M–55 and qnrS1 genes from the Salmonella Gloucester isolate 17Sal014 and bacterial species from different regions. The arrows indicate open reading frames. Light gray shading denotes homology regions. The orf1–orf6 genes encode DNA cytosine methyltransferase, type II site-specific deoxyribonuclease, hypothetical protein, 3′–5′ exoribonuclease, DUF4224 domain-containing protein, and tyrosine-type recombinase/integrase, respectively. Strains from China are in blue font.
Discussion
ESBL-producing and quinolone-resistant strains of Salmonella, which constitute a great public health concern, have been increasingly reported throughout the world (Castellanos et al., 2018; Nadimpalli et al., 2019; Zhang et al., 2019). Previous studies have reported the presence of ESBL-producing and quinolone-resistant Salmonella isolates from humans (Wong et al., 2014), healthy pigs and chickens (de Jong et al., 2014), pig feces and tonsils at slaughter (Van Damme et al., 2017), and retail pork and chicken meat (Qiao et al., 2017; Brown et al., 2018; Nadimpalli et al., 2019). These data highlight that the food of animals is an important reservoir of ESBL-producing and quinolone-resistant Salmonella. However, ESBL-producing and quinolone-resistant Salmonella strains have only rarely been reported in RTE meat products. In this study, we identified an S. Gloucester strain isolated from an RTE duck product in China harboring the ESBL-encoding blaCTX–M–55 gene conferring ESC resistance and qnrS1 encoding quinolone resistance.
Salmonella Gloucester is a rarely reported S. enterica serovar, only sporadically found in human and retail chicken meat samples (Hoque et al., 1994; Azemi et al., 2013; Tîrziu et al., 2020). Of the reported S. Gloucester isolates from clinical cases, resistance to ampicillin, chloramphenicol, trimethoprim–sulfamethoxazole, and ceftriaxone was reported (Rahman et al., 2001). However, no sequence information of the genetic determinants of these strains is publicly available. There are only two whole-genome sequences of S. Gloucester strains from clinical samples in the United Kingdom deposited in NCBI and EnteroBase (accession number SRR7842640). The two isolates contained no acquired antibiotic resistance gene by searching against the ResFinder database. In contrast, the S. Gloucester 17sal1004 strain isolated in this study showed a multiple antibiotic resistance phenotype and contained multiple resistance genes. Of note is that this strain exhibited high-level resistance to third-generation cephalosporins, produced ESBL, and are resistant to nalidixic acid, which represents a public health concern due to the possibility of dissemination through the food chain. Improved sanitary practices are important to help control the transmission of the ESBL-producing S. Gloucester to humans through the food chain.
Salmonella resistance to ESCs is reported to be associated with cross-resistance to FQs (Lu et al., 2019; Zhang et al., 2019). In the current study, S. Gloucester strain was susceptible to ciprofloxacin, but resistant to nalidixic acid, and it harbored PMQR genes qnrS1 as well as contained resistance-associated mutations in parC. The existence of qnrS and the mutations in parC provide strains with a selective advantage under quinolone exposure and can accelerate the development of chromosome-mediated quinolone resistance (Jacoby et al., 2014; Rodríguez-Martínez et al., 2016).
CTX-M-55 is an emerging ESBL type with enhanced cephalosporin-hydrolyzing activity (He et al., 2015). Since initially identified in E. coli of human origin in the United States and China in 2011, blaCTX–M–55 has been increasingly identified in E. coli, K. pneumonia, and in several Salmonella serovars from humans, animals, and food of animals, as well as in the environment (Shi et al., 2008; Lupo et al., 2018; Nadimpalli et al., 2019; Dong et al., 2020; Li et al., 2021). Recent studies in China (Zhang et al., 2019), Cambodia (Nadimpalli et al., 2019), and Thailand (Luk-In et al., 2018) showed the high frequency of the CTX-M-55 type in different Salmonella serovars among food animals and retail meats, which suggests that CTX-M-55 transferred rapidly cross Salmonella species and could be stably persistent in Salmonella in food animals.
The blaCTX–M–55 gene is reported to be mostly located on epidemic self-mobilizable plasmids, such as IncF, IncI1 and IncHI2, and IncA/C2 (Wang et al., 2018). However, a recent study from China demonstrated a high frequency of chromosomal copies of blaCTX–M–55 in CTX-M-55-producing Salmonella strains and the transferability of the chromosomal blaCTX–M–55 to E. coli by conjugation (Zhang et al., 2019). The blaCTX–M–55 gene was also found in the chromosome of S. Gloucester 17Sal014 in the present study. Moreover, the blaCTX–M–55 gene was identified to be located in an IS26-mediated composite transposon, IS26–qnrS1–IS3–Tn3–orf–blaCTX–M–55–ISEcp1–IS26, with the structural unit qnrS1–IS3–Tn3–orf–blaCTX–M–55–ISEcp1 similar to the corresponding region in the chromosome of S. Muenster strain PJM1 (GenBank accession number CP045038.1) and the plasmid of a Salmonella sp. isolate (GenBank accession number MN539018.1), as described in Zhang et al. (2019). Similar structure units linked with ISKpn19, instead of IS26, were seen in the chromosomes and plasmids of different bacterial species from China and other countries, suggesting that both IS26 and ISKpn19 contributed to the transfer of the qnrS1- and blaCTX–M–55-carrying unit to chromosomes and plasmids among different bacterial species.
The blaCTX–M–55 gene was reported to be often encoded in typical ISEcp1–blaCTX–M–55–ORF477 format on various plasmids, such as pSTH21, pHN1122-1, and p1081-CTXM (Lv et al., 2013; Qu et al., 2014; Wong et al., 2015; Li et al., 2021). However, we found that ISEcp1 was incomplete and inserted with IS26, which was also inserted on the other end of the structural unit qnrS1–IS3–Tn3–orf–blaCTX–M–55–ISEcp1. This configuration might have resulted from the insertion of the ISEcp1–blaCTX–M–55 transposition unit into a plasmid backbone and capture of the adjacent fragment, as well as IS26 in a subsequent transposition event (Lv et al., 2013; Qu et al., 2014; Wong et al., 2015). The unit, combined with the bounded two copies of IS26, constitutes a composite transposon which was able to transfer by forming a circular molecule (Harmer et al., 2014; Harmer and Hall, 2015, 2016). In particular, the identical IS26-mediated composite transposon was observed to exist mainly on plasmids of E. coli. It is likely that the IS26-mediated composite transposon in the chromosome of S. Gloucester in this study originated from the plasmid of E. coli and possesses the ability to disseminate to Salmonella and other bacterial species. The IS26-mediated composite transposon IS26–qnrS1–IS3–Tn3–orf–blaCTX–M–55–ISEcp1–IS26 was found in Salmonella and E. coli strains of animal, chicken meat, food, RTE food, and human sources in China, suggesting that it has spread along the food chain, which may contribute to the development of antibiotic resistance to cephalosporin and fluoroquinolones.
Conclusion
To the best of our knowledge, we describe for the first time an ESBL-producing and quinolone-resistant S. Gloucester (4:i:l,w) ST34 strain from an RTE duck product co-harboring the chromosomally located blaCTX–M–55 and qnrS1. These strains represent potential clinical and food safety issues since they may transmit to humans through the food chain and may lead to a reduced susceptibility of Salmonella to critical antibiotics, cephalosporins and fluoroquinolones, that are front-line drugs of choice for treating severe Salmonella infections. The structural unit qnrS1–IS3–Tn3–orf–blaCTX–M–55, combined with IS26 and ISKpn19, contributed to the co-transfer of the blaCTX–M–55 and qnrS genes into the chromosomes or plasmids of different bacterial species, which may accelerate the development and dissemination of isolates co-resistant to cephalosporins and fluoroquinolones. This warrants continuous investigation of the coexistence and the co-transfer mechanisms of blaCTX–M–55 and qnrS among different bacterial species from various sources in China.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
AS and JX conceptualized and designed the study. LL and HM performed the experiment and wrote the manuscript. CW was involved in the generation of short and long read sequencing of the genomes. LS and RO assisted in revising the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 32001796 and 31901789), Applied Basic Research Foundation of Guangdong Province (Grant No. 2020A1515010218), and the Guangzhou Science and Technology Planning Project (Grant No. 202002030145).
Conflict of Interest
CW was employed by the company Shandong New Hope Liuhe Group Ltd., Qingdao.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.646101/full#supplementary-material
Footnotes
References
Arlet, G., Barrett, T. J., Butaye, P., Cloeckaert, A., Mulvey, M. R., and White, D. G. (2006). Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes. Infect. 8, 1945–1954. doi: 10.1016/j.micinf.2005.12.029
Azemi, M., Ismaili-Jaha, V., Kolgreci, S., Berisha, M., Jakupi, X., Gashi, S., et al. (2013). Causes of infectious acute diarrhea in infants treated at pediatric clinic. Med. Arch. 67, 17–21. doi: 10.5455/medarh.2013.67.17-21
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Brown, A. C., Chen, J. C., Watkins, L., Campbell, D., Folster, J. P., Tate, H., et al. (2018). CTX-M-65 extended-spectrum β-lactamase-producing Salmonella enterica serotype Infantis, United States. Emerg. Infect. Dis. 24, 2284–2291. doi: 10.3201/eid2412.180500
Carattoli, A., and Hasman, H. (2020). PlasmidFinder and In Silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol. Biol. 2075, 285–294. doi: 10.1007/978-1-4939-9877-7_20
Castellanos, L. R., van der Graaf-van Bloois, L., Donado-Godoy, P., León, M., Clavijo, V., Arévalo, A., et al. (2018). Genomic characterization of extended-spectrum cephalosporin-resistant Salmonella enterica in the Colombian poultry chain. Front. Microbiol. 9:2431. doi: 10.3389/fmicb.2018.02431
CLSI (2016). M100-S26 Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Sixth Informational Supplement. Wayne, PA: Clinical an Laboratory Standards Institute.
Cuypers, W. L., Jacobs, J., Wong, V., Klemm, E. J., Deborggraeve, S., and Van Puyvelde, S. (2018). Fluoroquinolone resistance in Salmonella: insights by whole-genome sequencing. Microb. Genom. 4:e000195. doi: 10.1099/mgen.0.000195
de Jong, A., Smet, A., Ludwig, C., Stephan, B., De Graef, E., Vanrobaeys, M., et al. (2014). Antimicrobial susceptibility of Salmonella isolates from healthy pigs and chickens (2008-2011). Vet. Microbiol. 171, 298–306. doi: 10.1016/j.vetmic.2014.01.030
Dong, N., Li, Y., Zhao, J., Ma, H., Wang, J., Liang, B., et al. (2020). The phenotypic and molecular characteristics of antimicrobial resistance of Salmonella enterica subsp. enterica serovar Typhimurium in Henan Province. China. BMC Infect. Dis. 20:511. doi: 10.1186/s12879-020-05203-3
Fearnley, E., Raupach, J., Lagala, F., and Cameron, S. (2011). Salmonella in chicken meat, eggs and humans; Adelaide, South Australia, 2008. Int. J. Food Microbiol. 146, 219–227. doi: 10.1016/j.ijfoodmicro.2011.02.004
Harmer, C. J., and Hall, R. M. (2015). IS26-mediated precise excision of the IS26-aphA1a translocatable unit. MBio 6:e01866-15. doi: 10.1128/mBio.01866-15
Harmer, C. J., and Hall, R. M. (2016). IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1:e00038-16. doi: 10.1128/mSphere.00038-16
Harmer, C. J., Moran, R. A., and Hall, R. M. (2014). Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. MBio 5:e01801-14. doi: 10.1128/mBio.01801-14
He, D., Chiou, J., Zeng, Z., Liu, L., Chen, X., Zeng, L., et al. (2015). Residues distal to the active site contribute to enhanced catalytic activity of variant and hybrid β-Lactamases derived from CTX-M-14 and CTX-M-15. Antimicrob. Agents Chemother. 59, 5976–5983. doi: 10.1128/AAC.04920-14
Hooper, D. C., and Jacoby, G. A. (2015). Mechanisms of drug resistance: quinolone resistance. Ann. N.Y.Acad. Sci. 1354, 12–31. doi: 10.1111/nyas.12830
Hoque, S. S., Salam, M. A., Faruque, A. S., and Albert, M. J. (1994). Multiple-drug-resistant Salmonella gloucester infections in Bangladesh. Diagn. Microbiol. Infect. Dis. 20, 209–211. doi: 10.1016/0732-8893(94)90005-1
Iwamoto, M., Reynolds, J., Karp, B. E., Tate, H., Fedorka-Cray, P. J., Plumblee, J. R., et al. (2017). Ceftriaxone-resistant nontyphoidal Salmonella from humans, retail meats, and food animals in the United States, 1996–2013. Foodborne Pathog. Dis. 14, 74–83. doi: 10.1089/fpd.2016.2180
Jacoby, G. A., Strahilevitz, J., and Hooper, D. C. (2014). Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2:10.1128/microbiolspec.PLAS-0006-2013. doi: 10.1128/microbiolspec.PLAS-0006-2013
Kirk, M. D., Pires, S. M., Black, R. E., Caipo, M., Crump, J. A., Devleesschauwer, B., et al. (2015). World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 12:e1001921. doi: 10.1371/journal.pmed.1001921
Li, C., Zhang, Z., Xu, X., He, S., Zhao, X., Cui, Y., et al. (2021). Molecular characterization of cephalosporin-resistant Salmonella Enteritidis ST11 isolates carrying blaCTX-M from children with diarrhea. Foodborne Pathog. Dis. doi: 10.1089/fpd.2020.2878 ∗[Epub ahead of print].
Lokken, K. L., Walker, G. T., and Tsolis, R. M. (2016). Disseminated infections with antibiotic-resistant non-typhoidal Salmonella strains: contributions of host and pathogen factors. Pathog. Dis. 74:ftw103. doi: 10.1093/femspd/ftw103
Lu, X., Zeng, M., Xu, J., Zhou, H., Gu, B., Li, Z., et al. (2019). Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006-2016. EBio Med. 42, 133–144. doi: 10.1016/j.ebiom.2019.03.006
Luk-In, S., Chatsuwan, T., Pulsrikarn, C., Bangtrakulnonth, A., Rirerm, U., and Kulwichit, W. (2018). High prevalence of ceftriaxone resistance among invasive Salmonella enterica serotype choleraesuis isolates in thailand: the emergence and increase of CTX-M-55 in ciprofloxacin-resistant S. choleraesuis isolates. Int. J. Med. Microbiol. 308, 447–453. doi: 10.1016/j.ijmm.2018.03.008
Lupo, A., Saras, E., Madec, J. Y., and Haenni, M. (2018). Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J. Antimicrob. Chemother. 73, 867–872. doi: 10.1093/jac/dkx489
Lv, L., Partridge, S. R., He, L., Zeng, Z., He, D., Ye, J., et al. (2013). Genetic characterization of IncI2 plasmids carrying blaCTX-M-55 spreading in both pets and food animals in China. Antimicrob. Agents Chemother. 57, 2824–2827. doi: 10.1128/AAC.02155-12
Nadimpalli, M., Fabre, L., Yith, V., Sem, N., Gouali, M., Delarocque-Astagneau, E., et al. (2019). CTX-M-55-type ESBL-producing Salmonella enterica are emerging among retail meats in Phnom Penh. Cambodia. J. Antimicrob. Chemother. 74, 342–348. doi: 10.1093/jac/dky451
Poirel, L., Cattoir, V., and Nordmann, P. (2012). Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front. Microbiol. 3:24. doi: 10.3389/fmicb.2012.00024
Qiao, J., Zhang, Q., Alali, W. Q., Wang, J., Meng, L., Xiao, Y., et al. (2017). Characterization of extended-spectrum β-lactamases (ESBLs)-producing Salmonella in retail raw chicken carcasses. Int. J. Food Microbiol. 248, 72–81. doi: 10.1016/j.ijfoodmicro.2017.02.016
Qu, F., Ying, Z., Zhang, C., Chen, Z., Chen, S., Cui, E., et al. (2014). Plasmid-encoding extended-spectrum β-lactamase CTX-M-55 in a clinical Shigella sonnei strain, China. Future microbiol. 9, 1143–1150. doi: 10.2217/fmb.14.53
Rahman, M., Islam, H., Ahmed, D., and Sack, R. B. (2001). Emergence of multidrug-resistant Salmonella gloucester and Salmonella typhimurium in Bangladesh. J. Health Popul. Nutr. 19, 191–198.
Rodríguez-Martínez, J. M., Machuca, J., Cano, M. E., Calvo, J., Martínez-Martínez, L., and Pascual, A. (2016). Plasmid-mediated quinolone resistance: two decades on. Drug. Resist. Updat. 29, 13–29. doi: 10.1016/j.drup.2016.09.001
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shi, W., Qin, J., and Mi, Z. (2008). A Klebsiella pneumoniae sputum culture isolate from China carrying blaOXA-1, blaCTX-M-55 and aac(6’)-Ib-cr. J. Med. Microbiol. 57, 1588–1589. doi: 10.1099/jmm.0.2008/000950-0
Thomson, K. S. (2010). Extended-spectrum-beta-lactamase, AmpC, and carbapenemase issues. J. Clin. Microbiol. 48, 1019–1025. doi: 10.1128/JCM.00219-10
Tîrziu, E., Bărbălan, G., Morar, A., Herman, V., Cristina, R. T., and Imre, K. (2020). Occurrence and antimicrobial susceptibility profile of Salmonella spp. in raw and ready-to-eat foods and campylobacter spp. in retail raw chicken meat in Transylvania, Romania. Foodborne Pathog. Dis. 17, 479–484. doi: 10.1089/fpd.2019.2738
Van Damme, I., Garcia-Graells, C., Biasino, W., Gowda, T., Botteldoorn, N., and De Zutter, L. (2017). High abundance and diversity of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli in faeces and tonsils of pigs at slaughter. Vet. Microbiol. 208, 190–194. doi: 10.1016/j.vetmic.2017.08.009
Wang, J., Zeng, Z. L., Huang, X. Y., Ma, Z. B., Guo, Z. W., Lv, L. C., et al. (2018). Evolution and comparative genomics of F33:A-:B- plasmids carrying blaCTX-M-55 or blaCTX-M-65 in Escherichia coli and Klebsiella pneumoniae isolated from animals, food products, and humans in China. mSphere 3:e00137-18. doi: 10.1128/mSphere.00137-18
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wong, M. H., Liu, L., Yan, M., Chan, E. W., and Chen, S. (2015). Dissemination of IncI2 plasmids that harbor the blaCTX-M element among clinical Salmonella isolates. Antimicrob. Agents Chemother. 59, 5026–5028. doi: 10.1128/AAC.00775-15
Wong, M. H., Yan, M., Chan, E. W., Biao, K., and Chen, S. (2014). Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob. Agents Chemother. 58, 3752–3756. doi: 10.1128/AAC.02770-13
Wong, M. H., Zeng, L., Liu, J. H., and Chen, S. (2013). Characterization of Salmonella food isolates with concurrent resistance to ceftriaxone and ciprofloxacin. Foodborne Pathog. Dis. 10, 42–46. doi: 10.1089/fpd.2012.1266
Wu, H., Wang, Y., Wu, Y., Qiao, J., Li, H., Zheng, S., et al. (2015). Emergence of β-lactamases and extended-spectrum β-lactamases (ESBLs) producing Salmonella in retail raw chicken in China. Foodborne Pathog. Dis. 12, 228–234. doi: 10.1089/fpd.2014.1859
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: blaCTX–M–55, qnrS1, Salmonella Gloucester, ready-to-eat, IS26
Citation: Li L, Olsen RH, Song A, Xiao J, Wang C, Meng H and Shi L (2021) First Report of a Foodborne Salmonella enterica Serovar Gloucester (4:i:l,w) ST34 Strain Harboring blaCTX–M–55 and qnrS Genes Located in IS26-Mediated Composite Transposon. Front. Microbiol. 12:646101. doi: 10.3389/fmicb.2021.646101
Received: 25 December 2020; Accepted: 16 March 2021;
Published: 20 April 2021.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Jing Han, National Center for Toxicological Research (FDA), United StatesJason Patrick Folster, Centers for Disease Control and Prevention (CDC), United States
Copyright © 2021 Li, Olsen, Song, Xiao, Wang, Meng and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hecheng Meng, ZmVtZW5naGNAc2N1dC5lZHUuY24=
 Lili Li
Lili Li Rikke Heidemann Olsen2
Rikke Heidemann Olsen2 Anhua Song
Anhua Song Jian Xiao
Jian Xiao Chong Wang
Chong Wang Hecheng Meng
Hecheng Meng