- Department of Chemistry, Bose Institute, Kolkata, India
Mycobacterium tuberculosis can survive within its host for extended periods of time without any clinical symptoms of disease and reactivate when the immune system is weakened. A detailed understanding of how M. tuberculosis enters into and exits out of dormancy, is necessary in order to develop new strategies for tackling tuberculosis. Omics methodologies are unsupervised and unbiased to any hypothesis, making them useful tools for the discovery of new drug targets. This review summarizes the findings of transcriptomic and proteomic approaches toward understanding dormancy and reactivation of M. tuberculosis. Within the granuloma of latently infected individuals, the bacteria are dormant, with a marked slowdown of growth, division and metabolism. In vitro models have attempted to simulate these features by subjecting the bacterium to hypoxia, nutrient starvation, potassium depletion, growth in the presence of vitamin C, or growth in the presence of long-chain fatty acids. The striking feature of all the models is the upregulation of the DosR regulon, which includes the transcriptional regulator Rv0081, one of the central hubs of dormancy. Also upregulated are chaperone proteins, fatty acid and cholesterol degrading enzymes, the sigma factors SigE and SigB, enzymes of the glyoxylate and the methylcitrate cycle, the Clp proteases and the transcriptional regulator ClgR. Further, there is increased expression of genes involved in mycobactin synthesis, fatty acid degradation, the glyoxylate shunt and gluconeogenesis, in granulomas formed in vitro from peripheral blood mononuclear cells from latently infected individuals compared to naïve individuals. Genes linked to aerobic respiration, replication, transcription, translation and cell division, are downregulated during dormancy in vitro, but upregulated during reactivation. Resuscitation in vitro is associated with upregulation of genes linked to the synthesis of mycolic acids, phthiocerol mycocerosate (PDIM) and sulfolipids; ribosome biosynthesis, replication, transcription and translation, cell division, and genes encoding the five resuscitation promoting factors (Rpfs). The expression of proteases, transposases and insertion sequences, suggests genome reorganization during reactivation.
Introduction
Tuberculosis (TB) remains a global health problem with 1,000,000 new cases and 1,400,000 deaths in 2019 (World Health Organization, 2020). Drug resistance continues to be on the rise, with 465,000 reported cases in 2019. In the last 40 years, only three new drugs have been marketed for the treatment of TB, namely pretomanid, delamanid, and bedaquiline. This confirms the need for renewed efforts aimed at better understanding strategies for developing new chemotherapeutic agents as well as efficacious vaccines.
Latent TB is characterized by a positive tuberculin skin test without any symptoms of disease (Lordi and Reichman, 1988; Huebner et al., 1993). Houben and Dodd (2016) estimated in 2016 that out of the 1.7 billion people latently infected with Mycobacterium tuberculosis, 56 million could potentially reactivate into active disease upon weakening of the immune system. Dormancy is characterized by a reversible metabolic slowdown of the bacterium (Lewis, 2010). Kussell and Leibler (2005) and Kussell et al. (2005) show that even before sensing stress, a small fraction of the bacteria are in a non-growing state. The existence of a small subpopulation of cells with reduced metabolic activity, results in a given treatment (directed against growing cells) failing to kill this subpopulation giving rise to persisters, which can then resuscitate many years later. The molecular basis of latent TB infection remains incompletely understood. The shift to a dormant state is associated with transcriptomic and proteomic changes that reduce metabolic activity, increase the resistance of M. tuberculosis to environmental stresses and increase antibiotic tolerance (Caño-Muñiz et al., 2018). Once the stress is removed, dormant cells reactivate and replication is initiated. Treating the dormant state of M. tuberculosis requires a detailed understanding of the key processes and pathways that are adjusted when the bacteria transition into dormancy. It is equally important to understand the triggers of resuscitation so that these may be also be targeted.
Omics technologies free scientists from the biases of reductionist approaches. The unsupervised nature of omics technologies can open up new approaches for drug and vaccine development. Here we will discuss how omics technologies have been employed to understand dormancy and resuscitation of M. tuberculosis, focusing on transcriptomics and proteomics. We will first outline the models used for studying dormancy and resuscitation of M. tuberculosis, and then discuss the pathways, processes and targets which are differentially regulated in response to a shift toward dormancy and an exit out of dormancy. We will conclude by attempting to bring into perspective the implications of these dynamic shifts in gene and protein expression during the course of infection, our gaps in understanding of these shifts and how this knowledge may help in the combat of disease.
The Granuloma in Tuberculosis
During the course of human pulmonary tuberculosis, inhaled M. tuberculosis is transported across the alveolar epithelium, where the formation of granulomas is initiated (Ramakrishnan, 2012). It was widely held that the host attempts to control infection by segregating the bacteria in the granuloma. However, it is now established that the bacterium itself plays an active role in granuloma formation (Co et al., 2004; Saunders and Britton, 2007; Paige and Bishai, 2010). Elegant studies using zebrafish infection with M. marinum, have established that the 6 kDa early secretory antigenic target (ESAT-6) induces the production of matrix metalloproteinase-9 (MMP-9), activation of the epithelium, and recruitment of macrophages to the site of infection (Volkman et al., 2010). The granuloma also enables the dissemination of infected macrophages to new sites (Davis and Ramakrishnan, 2009). This is accomplished when M. tuberculosis spreads from dying macrophages into newly recruited ones. Initially, the granuloma is a loose aggregate composed of recruited innate immune cells such as macrophages and neutrophils. This is followed by the recruitment of antigen-specific T lymphocytes, and the activation of infected macrophages. The hallmark of the granuloma is a central core containing infected macrophages surrounded by foamy macrophages, epitheloid cells and T lymphocytes. It is the niche in which dormant bacilli reside for extended periods of time and are associated with latent infection. As the infection progresses to active disease, a caseous core forms, consisting of dead or dying macrophages with extracellular M. tuberculosis. The bacilli within the core of the granuloma in latent TB reside within a hypoxic environment in which lipids are the major source of nutrients. The caseating granuloma presents a less hypoxic core than that associated with latent disease.
Evidence of hypoxia in the core of the granuloma has been demonstrated in pulmonary granulomas in mouse, guinea pig, rabbits, and non-human primates using pimonidazole hydrochloride, an imaging agent that is bioreductively activated only under hypoxic conditions. Hypoxia has been confirmed by directly measuring oxygen partial pressure with a fiber optic oxygen probe inserted into the granuloma (Via et al., 2008). The lipid-rich environment contains cholesteryl ester, triacylglycerides, and lactosylceramide (Kim et al., 2010). The transcriptional signature of M. tuberculosis RNA isolated from the lungs of chronically infected mouse, is reminiscent of the signatures associated with environmental conditions such as low pH, oxygen depletion, iron limitation, nitrosative stress and nutrient starvation, suggesting that these conditions are probably associated with bacteria residing in granulomas (Timm et al., 2003). In summary, long-term residence of M. tuberculosis within the granuloma, requires that the bacterium be able to adapt to oxygen-limited conditions and a lipid-rich environment that is likely limited in terms of availability of other nutrients and metal ions, and a high concentration of nitric oxide (Nathan and Ehrt, 2004). In vitro or ex vivo models for studying dormancy, have therefore attempted to recreate some of these in vivo conditions that the bacterium must encounter over the duration of its residence within the host.
Models for Studying Dormancy and Reactivation
In vitro Models
Hypoxia, a defining characteristic of the granuloma, is one of the most widely documented conditions that is associated with the transition of M. tuberculosis to a dormant state. As M. tuberculosis senses a gradient of oxygen depletion, it prepares itself for transition into a metabolically dormant, non-replicating state, known as non-replicating persistence (NRP). The most widely used model for studying NRP is the in vitro Wayne model (Wayne and Hayes, 1996). Wayne (1976) observed that as the concentration of dissolved oxygen (DO) decreases, M. tuberculosis arrests growth over extended periods of time but retains the ability to continue exponential growth once the DO concentration rises. Wayne’s model attempts to simulate the gradual depletion of oxygen in the granuloma. Bacteria are grown in sealed containers with a controlled ratio of air to culture medium equal to 0.5 (called the head space ratio or HSR). The model defines two states. The first termed NRP1 occurs as the oxygen saturation reaches 1%. The bacteria can no longer replicate but still have high levels of ATP (Wayne and Sohaskey, 2001). At NRP2, the oxygen saturation falls below 0.06%. As oxygen depletes, the bacilli adjust metabolism and enter growth arrest (Boshoff and Barry, 2005). The Wayne model is restricted by the fact that it fails to account for conditions other than hypoxia, that prevail in the granuloma and likely have an effect on bacterial metabolism.
Bacon et al. (2004) have employed a chemostat model in which aerobic cultures [dissolved oxygen tension (DOT) of 50%] were transferred to new vessels and allowed to stabilize to a DOT of 20% for 2–3 days. Low-oxygen cultures were established by lowering the DOT from 20 to 1% (hypoxic) in a stepwise manner over a period of 5–7 days, and allowed to reach steady state growth (i.e., a constant turbidity) before collection for gene expression.
Peterson et al. (2020) have employed a more accurate technique for generating an oxygen gradient. A programmable multiplexed reactor system has been designed to precisely monitor oxygen levels within the growth media, with minimal disturbance to the bacteria. Air and nitrogen gas lines were connected to separate mass flow controllers allowing programmable gradients of gas mixtures to be streamlined into the headspace of spinner flasks containing M. tuberculosis cultures. The DO contents of the cultures were measured using non-invasive fiber-optic technology. The reproducibility of this method is superior to that of the Wayne model and dissolved oxygen concentrations can be measured in real time.
Other in vitro models have been developed in attempts to simulate the conditions prevailing within the granuloma. Betts et al. (2002) established a model in which M. tuberculosis was subjected to nutrient starvation, by suspending cells in phosphate-buffered saline (PBS) for extended periods of time. Under these conditions, the bacterium underwent growth arrest and a decrease in respiration rate. Salina et al. (2014, 2019) have studied growth in potassium-deficient media which generates non-culturable (NC) bacilli, tolerant to cell wall targeting antimicrobials. Potassium supplementation enables resuscitation of growth. Rodríguez et al. (2014) have analyzed adaptation of M. tuberculosis to long-chain fatty acids (the main energy source of M. tuberculosis within the host milieu). Taneja et al. (2010) have reported that vitamin C triggers growth arrest and a dormancy phenotype in M. tuberculosis.
The in vitro Granuloma Model
Granulomas contain macrophages, epitheloid cells and multinucleated giant cells surrounded by lymphocytes. Granulomas contain M. tuberculosis in a niche that is purported to be hypoxic. Conditions within the granuloma are intimately linked to the dormant state of M. tuberculosis. Guirado et al. (2015) have developed an in vitro granuloma model derived from human peripheral blood mononuclear cells from individuals with or without latent TB infection (LTBI).
In vivo Models
Several in vivo models have been used to understand M. tuberculosis dormancy and reactivation using animals such as mice (Dutta and Karakousis, 2014), guinea pigs (Ordway et al., 2010), rabbits (Manabe et al., 2008; Subbian et al., 2012) and non-human primates (Kaushal et al., 2012; Peña and Ho, 2015). However, each model has its own drawbacks, most significantly in terms of the ability to reproduce the human disease pathology, the composition of the granuloma and the necrosis that is the hallmark of pathological tuberculosis. In the light of these limitations, in vitro models attempting to simulate the conditions of latency and reactivation, remain in vogue.
Genes/Proteins Differentially Regulated During Dormancy
Two Component Systems: The DosR Regulon
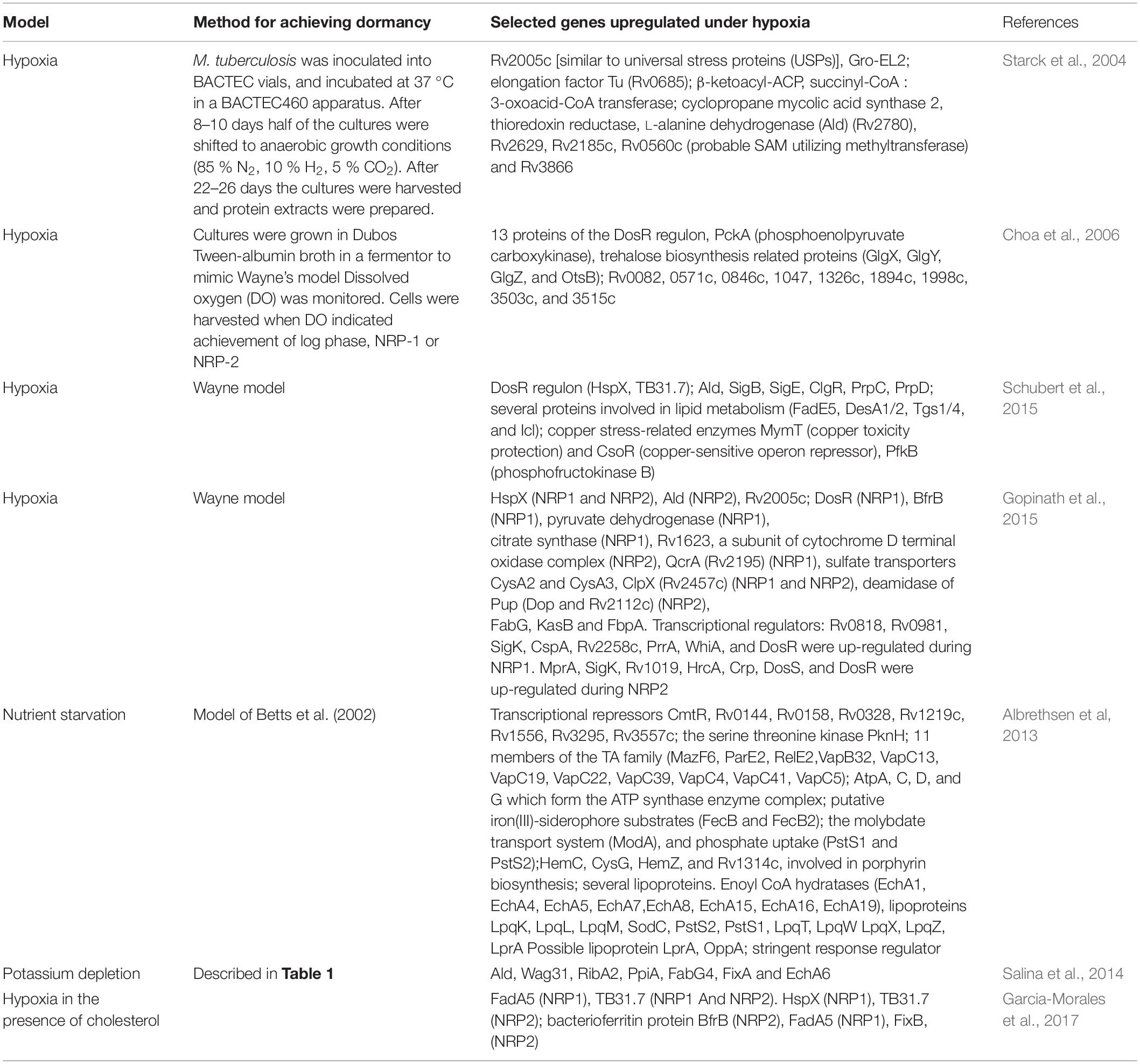
Out of the 190 regulatory proteins encoded by M. tuberculosis, 11 form the paired two-component systems (TCSs). In a typical TCS, a membrane-bound sensor kinase senses the environment signal which is subsequently transmitted through a phosphorelay to a cytoplasmic response regulator which regulates a subset of genes. These TCSs respond to environmental cues such as inorganic phosphate, SDS, oxygen, pH and nutrient limitation (Kundu, 2018). Of the paired TCSs, the DosR/DosS TCS is best characterized as the responder to hypoxia in the mycobacterial environment. The genes Rv3132c and Rv3133c encode a 578 amino acid histidine kinase protein (termed DosS) and a 217 amino acid response regulator protein (termed DosR). The signature feature of hypoxia-associated dormancy, is the upregulation of DosR and its regulon. Transcriptome analysis showed that dosR itself, and a dosR-regulated cluster of genes is induced early during hypoxia (Bacon et al., 2004; Muttucumaru et al., 2004; Voskuil et al., 2004; Iona et al., 2016). A partial list of dosR-dependent genes induced under hypoxia is given in Table 1. Intriguingly, Del Portillo et al. (2019) have reported that there are no genes in common between the dosR regulon and NRP1 during growth in fatty acid-containing medium. Under hypoxia, M. tuberculosis induces reduction of nitrate (NO3–) to nitrite (NO2–) to control redox homeostasis and energy production (Sohaskey and Wayne, 2003). The mycobacterial nitrate reductase (encoded by narGHIJ) as well as narK2, a nitrate transporter and narX (nitrate-reductase-like protein), members of the DosR regulon, are induced under hypoxia. These facilitate accumulation of nitrite under hypoxic conditions. In harmony with in vitro transcriptome and proteome analysis, transcripts from narG and narX, have been identified within granulomas of human TB samples (Rachman et al., 2006). Proteome analysis has also confirmed the induction of DosR during NRP1 (Gopinath et al., 2015) (Table 2). The induction of DosR was also evident in other models of dormancy, such as vitamin C exposure (Nandi et al., 2019).
Intragranulomatous lesions are believed to harbor dormant M. tuberculosis populations. The granulomata from different TB pathologies provide distinct microenvironments for M. tuberculosis. The lesions associated with active TB (ATB) are less hypoxic than those associated with LTBI. Hypoxia within the LTBI lesions is therefore a trigger for the bacterium to enter dormancy. Hudock et al. (2017) have analyzed gene expression in defined microanatomic compartments of the lungs of non-human primates with active or latent TB. A core group of 633 genes were identified associated with both ATB and LTBI, suggesting that these are required for M. tuberculosis survival. The dosR regulon was expressed at the lowest level in the least hypoxic lesions, and at the highest level in the most hypoxic lesions (Hudock et al., 2017).
Other Two-Component Systems
mprA and regX3 were identified as components of a core temporal regulatory response during 0.25–8 h of vitamin C treatment (Nandi et al., 2019). phoP was induced at an intermediate temporal window during vitamin C treatment. Potassium depletion was also associated with the induction of the mprAB TCS. In addition, in this model, the sensor kinase kdpD of the TCS kdpDE (linked to potassium transport) and the response regulator prrA of the TCS prrAB (required for macrophage infection), were induced. kdpDE was also induced by nutrient starvation (Betts et al., 2002). Among the less well understood TCSs, narL and tcrA were upregulated following vitamin C treatment of M. tuberculosis. Proteomics confirmed the induction of DosR/DosS, MprA, and PrrA under hypoxia (Gopinath et al., 2015).
Sigma Factors
In vitro models of dormancy (most notably hypoxia, vitamin C exposure, potassium depletion and nutrient starvation) have reported the induction of several of the transcripts of extracytoplasmic function (ECF) sigma factors. Almost all the in vitro models have confirmed that sigE, sigH, and sigB are induced during dormancy (Betts et al., 2002; Voskuil et al., 2004; Salina et al., 2014, 2019; Iona et al., 2016; Nandi et al., 2019). In addition, the upregulation of sigF and sigD and several other sigma factors, has also been reported in multiple studies. These are summarized in Table 1. sigB was induced at 10 days of hypoxia and remained induced upto 30 days. sigE and sigC were induced early during hypoxia and fell off at day 10 and day 12, respectively. sigH was induced throughout hypoxia (Voskuil et al., 2004). In vitro studies have also shown that sigE, sigB, and sigH respond to surface stress and oxidative stress, respectively, and that the transcription of sigB under surface stress is dependent on sigE (Manganelli et al., 2001, 2002). sigB, sigH, sigF, sigM, and sigE were enriched during vitamin C treatment of M. tuberculosis (Nandi et al., 2019). In the potassium depletion model of dormancy, genes sigA, sigB, sigE, sigF, sigG, sigH, sigI, sigL, and sigM were induced; sigD, sigJ, and sigK were repressed (Salina et al., 2014). Proteomic studies also confirmed the induction of SigK during NRP1 (Gopinath et al., 2015) (Table 2).
Intragranulomatous lesions from different TB pathologies in non-human primates [i.e., active TB (ATB) or latent TB (LTBI)] (Hudock et al., 2017) showed the expression of sigB, sigD, sigI, sigJ, and sigF in all the lesions. LTBI was associated with sigL and sigM (caseum), as well as sigK and sigG (granuloma) expression. These results corroborated at least in part, some of the observations made from the in vitro models of dormancy.
Transcriptional Regulators
A lipid-rich microenvironment is the hallmark of the granuloma. In vitro models have therefore tested the response of M. tuberculosis to growth in lipid-rich medium. During growth in the presence of lipids, ClgR appeared to be the predominantly upregulated transcriptional regulator under NRP1 (Del Portillo et al., 2019). On the other hand, Rv0081 was the more predominantly expressed transcriptional regulator during hypoxia in dextrose medium, although it is also induced in fatty acid-grown cells at stationary phase (Rodríguez et al., 2014). A core of 185 genes is upregulated during growth in the presence of long chain fatty acids (Aguilar-Ayala et al., 2017). Among these are the transcriptional regulators Rv3160c and Rv0678. Rv3160c is a TetR-like transcriptional repressor that regulates expression of the putative oxygenase Rv3161c (Tükenmez et al., 2021). Rv0678 encodes the MmpR repressor protein, responsible for regulating the transcription of MmpL5 and MmpS5 protein which together make up the MmpL5-MmpS5 efflux pump associated with cross-resistance between clofazimine and bedaquiline (Hartkoorn et al., 2014).
The WhiB family of proteins of M. tuberculosis represent a group of iron-sulfur containing redox sensing transcriptional regulators which respond to stress and maintain redox homeostasis (Singh et al., 2009; Larsson et al., 2012). Among these, whiB1 was upregulated during vitamin C treatment (Nandi et al., 2019) and potassium depletion (Salina et al., 2014); whiB2 was upregulated during nutrient starvation (Betts et al., 2002); whiB3 was upregulated in fatty acid grown cells at stationary phase (Rodríguez et al., 2014), and during treatment of M. tuberculosis with vitamin C (Nandi et al., 2019) and whiB6 was upregulated during potassium depletion (Salina et al., 2014, 2019).
Analysis of the transcriptional regulatory network that underlies the response to vitamin C, showed an early response (upto 1 h), an intermediate response (between 2 and 8 h) and a late response (24 h). The early response regulators induced included Rv0348, hrcA, and Rv0845. The late response included the regulators lsr2, Rv0081, Rv0678, trcR, and Rv0047. Nutrient starvation resulted in the upregulation of Rv2034, Rv1152, Rv3291c (Betts et al., 2002). kstR, a transcriptional repressor controlling cholesterol catabolism was induced in non-culturable bacilli resulting from potassium depletion (Salina et al., 2014).
Proteomic analyses confirmed the upregulation of the stringent response regulator RelA (Rv2583c), and Rv1019 (a tetR family transcriptional regulator), during hypoxia (Gopinath et al., 2015).
Peterson et al. (2020) reported a transcriptional program that coordinates sequential state transitions to drive M. tuberculosis in and out of hypoxia-induced dormancy. This model employed an accurate technique for generating a defined oxygen gradient, where dissolved oxygen levels can be monitored. Non-overlapping sets of differentially expressed genes (DEGs) were associated with each of the following states: normoxia (81 genes), depletion (446 genes), early hypoxia (328 genes), mid hypoxia (320 genes), late hypoxia (978 genes), and resuscitation (429 genes). The hypoxia dataset was linked to a transcription factor (TF) gene network derived from chromatin immunoprecipitation sequencing (ChIP-seq) (Minch et al., 2015) in order to understand the transcriptional regulation of the transition between the aforesaid states. Network motifs such as feed forward loops (FFLs) and TFs which figure as central players in these FFLs such as Rv0081, were identified. The Rv0081-Rv0324 FFL was predicted to be responsible for controlling the late hypoxia genes. The regulatory activity of Rv0081 appeared to be oxygen-dependent, and the oscillations of state observed as oxygen dropped below 3%, could be due to Rv0081-directed incoherent FFLs (I-FFLs). The I-FFL controlled by Rv0081 probably regulates the transition to late hypoxia and imparts robustness into the hypoxic response.
Genes/Proteins Linked to Replication, Transcription, and Translation
Ribosomal protein encoding genes rpsR2, rpsN2 and rpmG1 were induced early during hypoxia, but repressed at later stages (after day 20) (Voskuil et al., 2004). In line with reduced requirements for mRNA and protein synthesis during dormancy, the 30s and 50s ribosomal protein genes (rpsJ-rpsQ, rplN-rpsN, and rpsH-rpmD) were downregulated in the vitamin C-induced model of dormancy (Nandi et al., 2019). rrnAP1 and rrnAPCL1 involved in ribosomal RNA synthesis were downregulated during dormancy (Iona et al., 2016). Genes linked to chromosome partitioning (parA and parB), 15 genes in the cluster Rv0700 to Rv0723, involved in ribosomal protein synthesis, rpoA and rpoC (subunits of RNA polymerase), aminoacyl tRNA synthases (gltS and trpS) were all downregulated during nutrient starvation (Betts et al., 2002). The vitamin C induced dormancy model showed the downregulation of GreA transcription elongation factor, 30s and 50s ribosomal proteins (rpsL, rpsL, rpsJ, rplC, rpsQ, rplN, rplX, rpmE, and rpmB2), and dnaA and dnaB (which interact with the origin of replication) (Nandi et al., 2019). Rodríguez et al. (2014) reported that bacteria grown in the presence of even chain fatty acids (F) till stationary (S) phase (FS phase) show a remarkable overexpression of tRNAs, a probable reflection of low translation activity. tRNA-Lys, tRNA-Ala, and tRNA-Arg neutralize the negative charge on the polar head groups of phosphatidylglycerol. These tRNAs were overexpressed in FS compared to DS. This could possibly be linked to changes in membrane permeability and decreased susceptibility to antibacterial cationic drugs.
Electron Transfer Processes and Aerobic Respiration
Cytochrome bd oxidase (encoded by cydABCD), and cytochrome bc1-aa3 regulate respiratory functions in M. tuberculosis. The cytochrome bd oxidase encoded by the cydABDC cluster consists of cydAB (encoding cytochrome bd oxidase) and cydCD (encoding the ABC transporter). Voskuil et al. (2004) have shown that the cydABCD cluster is induced early during hypoxia, consistent with a role of cytochrome bd oxidase as an alternative terminal oxidase for the aerobic respiratory chain that functions under low oxygen levels. On the other hand cydAB as well as cytochrome c reductase (qcrA and qcrC), and cytochrome c oxidase (ctaC and ctaE) were repressed under potassium depletion-induced dormancy. NADH dehydrogenase genes (nuoA-N) were repressed in non-culturable bacteria under potassium depletion (Salina et al., 2014) and under nutrient starvation (Betts et al., 2002), whereas the uncoupled non-proton pumping NADH dehydrogenase (ndh) was induced in non-culturable bacteria (Salina et al., 2014). This suggested that during potassium depletion induced dormancy, the bacilli switch from using proton motive force generated by respiration to using NADH and alternative electron acceptors. Genes of the NADH dehydrogenase complex and the ATP synthase were also downregulated when bacteria were treated with vitamin C (nuoH-nuoN, nuoC-G, and atpC-H) (Nandi et al., 2019) or subjected to nutrient starvation (nuoA-M and atpA-H) (Betts et al., 2002).
Proteomic studies further confirmed the decreased abundance of proteins involved in aerobic respiration (NuoE, NuoF, and NuoG), and quinolate synthase (NadA) (involved in the biosynthesis of NAD) during nutrient starvation (Albrethsen et al, 2013). Intriguingly, Gopinath et al. (2015) could not demonstrate diminished levels of the subunits of ATP synthase during hypoxia. FixB, an electron acceptor flavoprotein of dehydrogenases at complex II of the cell respiratory chain was overexpressed in NRP2 when cells were grown in the presence of cholesterol as carbon source (Garcia-Morales et al., 2017), most likely to maintain redox balance inside the cell.
Genes and Proteins Linked to Metal Ion Storage and Acquisition, and Transport of Inorganic Ions
The bacterium needs to increase iron stores during dormancy. Expectedly, the iron storage protein bacterioferritin (bfrB) (Voskuil et al., 2004) and mycobactin synthesizing gene cluster (Rv2377c to Rv2386c, mbtA-I) were induced during hypoxia (Bacon et al., 2004; Voskuil et al., 2004). Muttucumaru et al. (2004) confirmed the upregulation of the mycobactin biosynthesizing operon, and ferredoxin A (fdxA) during hypoxia. Copper is required by M. tuberculosis for survival, but copper overload can be toxic (Rowland and Niederweis, 2012). ctpV (an efflux pump) and ctpG (a P-type ATPase), which prevent copper toxicity, were upregulated during vitamin C treatment (Taneja et al., 2010; Sikri et al., 2015). ctpF, a cation transport ATPase was upregulated during hypoxia (Voskuil et al., 2004). The molybdate transport system (modA) and phosphate uptake system (pstS1 and psS2) were increased during nutrient starvation suggesting increased transport of iron, molybdate and phosphate under these conditions. However, following vitamin C treatment, the phosphate-specific transporter operon pstB-pstC1-pstA2, was downregulated. The sulfate transporters csyT–cysW–cysA are induced under nutrient starvation (Betts et al., 2002).
Schubert et al. (2015) have shown through proteomic analyses, that the copper stress-related enzymes MymT (copper toxicity protection) and CsoR (copper-sensitive operon repressor) were induced during hypoxia. Gopinath et al. (2015) have shown that the sulfate transporters CysA2 and CysA3 were upregulated in both dormancy and reactivation. Sulfate is required for the production of methionine and cysteine. Therefore it is possible that sulfate transport regulates reductive stress. Putative iron (III)-siderophore substrates (FecB and FecB2) the molybdate transport system (ModA), and phosphate uptake (PstS1 and PstS2), suggesting increased transport of iron, molybdate, and phosphate during starvation (Albrethsen et al, 2013). Gopinath et al. (2015) have reported the upregulation of the iron storage protein BfrB during hypoxia.
Central Carbon Metabolism
Several independent studies have confirmed that central carbon metabolism slows down during dormancy or a shift of the bacterium to a non-culturable state. When fatty acids or cholesterol is the sole carbon source for M. tuberculosis, acetyl-CoA and propionyl-CoA must be metabolized via the glyoxylate and methylcitrate cycle, respectively. The glyoxylate cycle requires the enzyme isocitrate lyase 1 (icl1). Growth in the presence of long chain fatty acids has shown a shift in metabolism to increased expression of icl1 (Rodríguez et al., 2014). pckA (required for growth on fatty acids) (Marrero et al., 2010) was also upregulated. Vitamin C treatment was associated with overexpression of icl1 (Nandi et al., 2019). Genes linked to glycolysis and the tricarboxylic acid (TCA) cycle (fum, acn, and icd1) were downregulated under nutrient starvation (Betts et al., 2002). The genes pdhABC encoding subunits of the pyruvate dehydrogenase enzyme complex, the fumarate reductase complex (frdABCD),were upregulated under nutrient starvation.
Salina et al. (2014, 2019) have analyzed transcriptional signatures generated for bacilli cultured in potassium-depleted medium to show that genes linked to glycolysis and gluconeogenesis are repressed in the non-culturable bacilli, with pgi, fba, tpi, gap, pgk, pgmA, eno, pykA, aceE, and lpdC downregulated compared to bacteria grown in potassium-sufficient media (Salina et al., 2014, 2019). Four genes implicated in the pentose phosphate shunt were also repressed (fgd1, zwf2, tkt, and tal). The tricarboxylic acid cycle (TCA) genes citA, acn, icd1, icd2, korA, korB, sucC, sucD, shdA, shdB, shdD, Rv0248c (probable succinate dehydrogenase), fumC and gltA2 were downregulated. icl1 was induced as was mutA of the methylmalonyl pathway. Bacon et al. (2004) have reported the induction of prpD, which is involved in the methylcitrate cycle, under hypoxic conditions. Eoh and Rhee (2013) have suggested that Icl-mediated synthesis of succinate may afford M. tuberculosis an efficient means of entry into and exit from hypoxia-induced dormancy.
Proteome analysis of cells under hypoxia showed the upregulation of pyruvate dehydrogenase and citrate synthase at NRP1 (Gopinath et al., 2015). However, increased expression of Icl was not observed. Choa et al. (2006) have reported the upregulation of PckA (phosphoenolpyruvate carboxykinase) in NRP-1 when mycobacteria are grown on fatty acid substrates, suggesting a shift to gluconeogenesis from lipid precursors.
Lipid Metabolizing Genes/Proteins
During entry into dormancy, mycobacteria maintain an equilibrium between fatty acid biosynthesis and degradation, while fatty acid biosynthesis surges during reactivation. Mycolic acid-biosynthesizing genes were generally observed to be downregulated in different models of dormancy. accA2 and accD2 (Barry et al., 2007), were downregulated during vitamin C treatment (Taneja et al., 2010) and hypoxia (Bacon et al., 2004). mmA3 (a methyltransferase that generates mycolates), (Behr et al., 2000), was downregulated during vitamin C treatment (Taneja et al., 2010). tgs1 (involved in triglyceride synthesis), fadE5, fadE13, and fadD19 (involved in fatty acid degradation) (Muñoz-Elías and McKinney, 2005); fadE6, fadE28, and fadE32 (putative acyl-CoA dehydrogenases), scoA and scoB (involved in the utilization of ketones) were upregulated during vitamin C treatment. desA3 a desaturase involved in oleic acid synthesis (Walker et al., 1970), was also downregulated. While several genes of fatty acid degradation were upregulated during dormancy, selected genes (fadD5, fadD10, fadD22, fadD29, and fadD30) were dowregulated in the vitamin C model. During nutrient starvation, desA3 and desA2, the polyketide synthase type I system, ppsABCDE, the type II polyketide synthase, mas, which are involved in the synthesis and transport of phthiocerol dimycocerosates (PDIMs), and the mycolyl transferases fbpA and fbpB, were downregulated (Betts et al., 2002). Genes implicated in the catabolism of branched-chain keto acids (bkdA, bkdB, bkdC, fadE2, fadE13, and accD2) were upregulated. Fatty acid and mycolic acid biosynthetic pathway genes (fabG1 and inhA from FASII, fas, accD4, mmaA2, mmaa4, cmaA2, umaA, and Rv2509) were repressed during potassium depletion induced dormancy (Salina et al., 2014, 2019). fadD26 a fatty acyl AMP ligase involved in the biosynthesis of PDIMs (Siméone et al., 2010) was upregulated in NRP1 and NRP2 (Muttucumaru et al., 2004).
Proteomics revealed the upregulation of FadA5 during hypoxia in the presence of cholesterol (Garcia-Morales et al., 2017). In the potassium-depleted non-culturable state (Salina et al., 2014), KasB (involved in fatty acid biosynthesis) was decreased compared to exponentially growing cells. FadE5, DesA1/2 and Tgs1/4 were induced during hypoxia-induced dormancy (Schubert et al., 2015). Starck et al. (2004) grew bacteria aerobically for 8–10 days and then shifted to anaerobic growth conditions and analyzed the cytosolic proteome of M. tuberculosis. The L-alanine dehydrogenase (Ald, Rv2780) (which converts pyruvate to alanine, and glyoxylate to glycine concurrent with the oxidation of NADH to NAD) (Giffin et al., 2016) was upregulated under hypoxic conditions. Succinyl-CoA : 3-oxoacid-CoA transferase (ScoB, Rv2503c), which catalyses the reversible conversion of succinyl-CoA to succinate, was upregulated, as was β-ketoacyl-ACP synthase (KasB and Rv2246) which is involved in the synthesis of mycolic acids (Schaeffer et al., 2001). A total of about 50 proteins were unique under anaerobic conditions and low ATP levels. A selected list of proteins upregulated during dormancy is given in Table 2. Gopinath et al. (2015) observed that six proteins linked to fatty acid degradation were upregulated at NRP1 and NRP2 and confirmed that FabG, KasB and FbpA were upregulated during hypoxia. EchA6 and FabG4, involved in fatty acid oxidation and fatty acid biosynthesis, respectively were induced during potassium depletion.
Trehalose
Trehalose can serve as a carbon source, as a storage carbohydrate, and as an osmoprotectant in the non-replicating bacteria. It also regulates the host immune response. The OtsAB pathway is the dominant pathway for trehalose synthesis and is required for M. tuberculosis grown in culture and for virulence in a mouse model. otsB, a trehalose 6 phosphate phosphatase was induced during dormancy (Voskuil et al., 2004). Choa et al. (2006) have shown that four of the trehalose biosynthesis related proteins (GlgX, GlgY, GlgZ, and OtsB) are upregulated in NRP-2.
ESX Secretion Systems
Albrethsen et al (2013) have analyzed the secreted proteome of M. tuberculosis under nutrient starvation. The Esx secretion system members, EsxA, EsxB, EsxJ/EsxK, EsxL, and EsxO, all showed decreased abundance. Iona et al. (2016) have reported downregulation of EsxA and EsxB during hypoxia-induced dormancy.
Proteases and Peptidases
The Clp protease is a key regulator of the response to stress. Proteomic analyses have shown that ClpX (Rv2457c) is upregulated during NRP1 and NRP2, whereas ClpP1 and ClpP2 are present at normal levels (Albrethsen et al, 2013). Mycobacteria possess a prokaryotic ubiquitin-like protein (Pup) proteasome pathway. The enzyme deamidase of Pup (Dop, Rv2112c) deamidates the C-terminal glutamine of Pup to form glutamate, thereby activating Pup. Dop was dramatically upregulated during NRP2, and its level returned to normal as the number of proteins began to increase during re-aeration. During potassium depletion, pepD (encoding an HtrA-like serine protease), htrA and clpC2 were upregulated (Salina et al., 2019).
Chaperones
Chaperone-encoding genes such as hspX, dnaK, dnaJ1, grpE, clpB, hsp (or acr2), and htpX were upregulated after Vitamin C treatment (Taneja et al., 2010; Sikri et al., 2015; Nandi et al., 2019). Also induced was trxB1 (encoding thioredoxin).
Proteome analyses showed the upregulation of the universal stress protein Rv2005c and HspX (Rv2031c) during dormancy (Gopinath et al., 2015; Garcia-Morales et al., 2017). HspX also exhibited high expression in LTBI and ATB caseum-derived samples obtained from the lungs of non-human primates (Hudock et al., 2017).
Toxin-Antitoxin Systems
Type II toxin–antitoxin (TA) systems are widely spread among bacteria and archaea. Type II TA systems are involved in persistence regulation, antibiotic tolerance, stress adaptation and virulence (Maisonneuve et al., 2011; Leung and Lévesque, 2012; De la Cruz et al., 2013). Aguilar-Ayala et al. (2017) reported the overexpression of toxin/antitoxins vapB9/vapC9 and vapB22/vapC22 during hypoxia in the presence of long chain fatty acids. Stationary phase in the presence of long chain fatty acids and cholesterol was associated with overexpression of vapC1, vapC22, vapB1, vapB10, vapB46, and vapB48. Bacon et al. (2004) reported the hypoxia-induced induction of higBA3 (Rv3182–Rv3183) (Zaychikova et al., 2015). vapB21, an antitoxin of the TA system, was induced approximately 100-fold in the granulomatous lesions derived from non-human primates with active TB (Hudock et al., 2017). Proteomic analysis by Del Portillo et al. (2019) showed the induction of the TA module proteins VapB10, VapC37, and VapC20 in dextrose-grown cells under hypoxia.
Non-coding RNAs
Rodríguez et al. (2014) suggest that adaptation of M. tuberculosis during growth in the presence long chain fatty acids as sole carbon sources, leads to a slow growth and drug-tolerant phenotype, characteristic of the dormant state. Gene expression in M. tuberculosis grown in a fatty acid (F) environment was compared with growth on dextrose containing (D) medium, using strand-specific RNA sequencing (Wang et al., 2009). Genes with higher expression during growth on LC-FAs were compared with the highest scoring dormancy-associated genes obtained in the meta-analysis of published microarray data by Murphy and Brown (2007), and a high degree of overlap was observed. Mycobacterial small RNAs modulate the response of mycobacteria to the environment (Arnvig and Young, 2009). Most of the more than 200 sRNAs of mycobacteria are ncRNAs (Haning et al., 2014). The most highly expressed ncRNA, MTS2823 (ncRv13661A) is upregulated in fatty acid grown cells. Del Portillo et al. (2019) established hypoxic cultures of M. tuberculosis after exponential phase growth in the presence of long chain fatty acids or dextrose. Two non-coding RNAs (ncRNAs), MTS1338 and MTS0194 were upregulated in the fatty acid grown cells only. MTS2823 and the stable 10S RNA were expressed in high levels in NRP2 in both carbon sources. Aguilar-Ayala et al. (2017) also observed that non-coding RNA MTS2823 is upregulated during growth in the presence of fatty acids.
Genes and Proteins Differentially Expressed During Reactivation From Dormancy
Transcriptional Regulators
The DosRST regulon and the MprAB regulon genes which play important roles in the response to stress (He et al., 2006; Pang et al., 2007) were downregulated during reactivation from hypoxia, including Rv0081 and its subnetwork. Among the sigma factors, the sigH and sigE network genes were downregulated, including ClgR and its targets Rv2743c and Rv2744c. This was in harmony with the role of the ClgR regulon in maintaining cell envelope functions under stress (Estorninho et al., 2010; Datta et al., 2015). These findings were in contrast to those of Sherrid et al. (2010), who subjected bacteria to hypoxic conditions for a shorter period of 7 days. In their study, a number of genes of the MprA subnetwork were upregulated.
The TetR-like transcriptional repressor KstR that represses the expression of a cluster of mycobacterial genes involved in cholesterol catabolism (Kendall et al., 2007; Van der Geize et al., 2007), was downregulated, consistent with heightened cholesterol catabolism under stress such as hypoxia.
The SWATH analysis (Schubert et al., 2015) identified a cluster of proteins that were transiently upregulated within 6 h of re-aeration, including the sigma factors SigE and SigB and the transcriptional regulator ClgR, suggesting their involvement early in resuscitation, but not at the later stages.
During reactivation from potassium depletion, the transcriptional regulators WhiB6 [which regulates cell division (Chen et al., 2016)] and TetR family regulators Rv3830c and Rv3160c which regulate multidrug efflux pumps, as well as the response to osmotic stress and toxic chemicals, were upregulated.
Energy Metabolism and Electron Transport Chain
Du et al. (2016) analyzed reactivation of M. tuberculosis after 25 days in the Wayne model of hypoxia. Early during reactivation, there is a dramatic increase in cellular ATP levels. In preparation for replication, M. tuberculosis upregulates pathways involved in ribosome biosynthesis and amino acid biosynthesis. Genes for TCA cycle and oxidative phosphorylation enzymes such as NADH:quinine oxidoreductase and F-type ATPase were upregulated. Sherrid et al. (2010) have shown that among genes of the MprA subnetwork, those encoding components of the NADH dehydrogenase complex (nuo) and ATP synthase (atp) were upregulated, consistent with increase in aerobic activity and ATP synthesis during reactivation.
The SWATH proteome analysis (Schubert et al., 2015), showed that the FoF1 ATP synthase levels did not change significantly over the time course of the experiment, underlining the requirement of this enzyme in maintaining ATP homeostasis in dormant M. tuberculosis (Gengenbacher et al., 2010; Leistikow et al., 2010). In contrast, components of the electron transport chain and energy generating machinery, changed in response to hypoxia, suggesting a reorganization of energy metabolism.
Central Carbon Metabolism
Genes involved in respiration, TCA cycle activity and translation were activated after 4 days of resuscitation from potassium-depleted to potassium-sufficient conditions. The SWATH analysis (Schubert et al., 2015) identified a cluster of proteins that were transiently upregulated within 6 h of re-aeration, but returned to pre-aeration levels within 2 days. This included PrpC and PrpD, two enzymes of the methylcitrate cycle.
Lipid Biosynthesis
There was upregulation of key enzymes involved in the biosynthesis of mycolic acids and sulfolipids during reaeration after hypoxia. The gene fabJ which links the FAS-I and FAS-II systems (Choi et al., 2000), was upregulated in the initial phase of reaeration. The genes fabG1 and inhA were also induced, indicative of a shift toward mycolic acid biosynthesis. The genes mas, faD26, and ppsA-E, involved in the synthesis and translocation of PDIM, were upregulated, in line with the observations of Sherrid et al. (2010). Several pks genes (pks1, psk4, pks7, pks8, and pks15) encoding polyketide synthases that participate in the synthesis of complex lipids including sulfolipids were induced. Taken together, the observations pointed to enhanced synthesis of major cell wall lipids in M. tuberculosis during the emergence from dormancy and preparation for cell division. Lipid catabolism is associated with entry of M. tuberculosis to the persistent state. Cholesterol uptake and utilization are required for M. tuberculosis survival during persistence (Pandey and Sassetti, 2008). Expectedly, genes associated with fatty acid beta-oxidation and degradation pathways were downregulated in the reaeration phase. Also downregulated were the Mce transport systems which encode ABC transporters involved in transport of diverse lipids across the cell wall (Casali and Riley, 2007; Pandey and Sassetti, 2008). Genes involved in glyoxylate and dicarboxylate metabolism, a canonical pathway for lipid utilization, were downregulated.
Proteome analysis during a shift from hypoxia to reaeration showed that four proteins involved in mycolic acid biosynthesis were upregulated at day 6 of reaeration, whereas cyclopropane mycolic acid synthase 2 (CmaA2) was downregulated (Gopinath et al., 2015). At day 24 of reaeration, 20 proteins were at normal levels and three (KasA, BacA, and FbpA) were upregulated.
Cell Wall Synthesis, Division, DNA Replication, and Repair
Iona et al. (2016) have shown that when cells subjected to a 25 days period of dormancy were reactivated, ftsZ and dnaA associated with cell division and replication, were upregulated. The resuscitation promoting factors (Rpfs), rpfB, rpfC went up early, followed by the other rpfs. Shift from potassium depletion to potassium sufficient conditions showed that most of the rpf encoding genes were activated only after the onset of cell division. Only rpfE was upregulated at day 7, coinciding with the onset of multiplication. rpfB was upregulated after 8 days of resuscitation (Salina et al., 2014).
At least seven proteins involved in DNA replication and repair, (single-stranded DNA binding protein (SSB), Hns, FtsE, ParB, DNA polymerase I, PolA, DNA topoisomerase I TopA omega, and DNA polymerase III beta chain DnaN) were present at normal levels at day 6 of reaeration (Gopinath et al., 2015). At day 24, a further set of proteins appeared, including the NrdEF system which encodes an enzyme that catalyzes the formation of deoxyribonucleotides from ribonucleotides. Rv2817c (putative Cas1, a CRISPR-associated endonuclease) was detected in the early stages of reactivation, suggesting a possible role of genome editing during reactivation of M. tuberculosis from dormancy.
ESX Secretion Systems
Peterson et al. (2020) have shown that a shift from late hypoxia into resuscitation was characterized by expression of the ESX-5 export system (Gey van Pittius et al., 2001).
Toxin–Antitoxins
A few toxins and antitoxins also showed expression specific to the reactivation [Rv0299 and RelE, in the early period of reactivation and Rv0298 in the later phase (Gopinath et al., 2015)].
Chaperones
The universal stress protein Rv2005c was upregulated about 40-fold immediately upon reaeration, but decreased to 17-fold at R24. HspX (Rv2031c) was decreased 175-fold during R6 and 64-fold during R24 (Gopinath et al., 2015).
Proteases, Transposases, and Insertion Sequences
Proteases, transposases and insertion sequences were also expressed during resuscitation, prompting the speculation that there is genome reorganization to facilitate the chances of the bacterium to survive and to transmit to a new host. The endopeptidase clpB which is required for recovery from the stationary phase or antibiotic exposure (Vaubourgeix et al., 2015) was upregulated during resuscitation from potassium depletion (Salina et al., 2014).
Conclusion
M. tuberculosis remains in a dormant state, in latently infected individuals, with reactivation occurring when the immune system weakens. Targeting the dormant bacterium or triggering reactivation, represent important strategies for combating the disease. Omic approaches have provided valuable insights into how the bacterium remodels its transcriptome and its proteome during dormancy and reactivation. A feature of most of the models of dormancy was the upregulation of dosR and DosR regulon genes. A summary of selected genes or processes that are differentially regulated during dormancy is provided in Table 3. Besides dosR, the implications of upregulation of other components of TCSs during various time windows of dormancy, such as mprA, regX3, prrA, kdpDE, and phoP, remain poorly understood till date. Upregulation of the narX-narK2 operon, suggests that nitrate transport into the cell occurs during dormancy. The Rv0079 to Rv0087 cluster, featuring the transcriptional regulator Rv0081 which likely directs FFLs which are crucial to the adaptation to hypoxia, the chaperones acr or hspX, acr2, tgs1, the regulator clgR, cation transporting ATPases, thioredoxin (trxB1), genes involved in fatty acid degradation, sigma factors such as SigE, SigB, and SigH, and genes/proteins of the mycobacterial Clp protease. Among the sigma factors that are induced under hypoxia, SigE regulates genes involved in fatty acid degradation and the glyoxylate cycle, such as icl1 (Muñoz-Elías and McKinney, 2005). Genes regulated by sigH include sigE, sigB, DNA repair proteins, stress response proteins, and enzymes involved in thiol metabolism such as thioredoxin and thioredoxin reductase. A cyclopeptide lassomycin active against stationary phase bacteria, interacts with ClpC1 (Gavrish et al., 2014), making the Clp proteases, promising targets in dormant bacteria. Understanding the importance of the regulator ClgR during dormancy in a lipid-rich environment, is an important area for future investigation. ClgR is induced in response to both hypoxia and reaeration (McGillivray et al., 2015). It is a regulator of Clp proteases which are crucial for the degradation of misfolded proteins and therefore, for stress management. ClgR is likely a crucial regulator of the ability of M. tuberculosis to survive within the host under stress as well as to reactivate under suitable conditions.
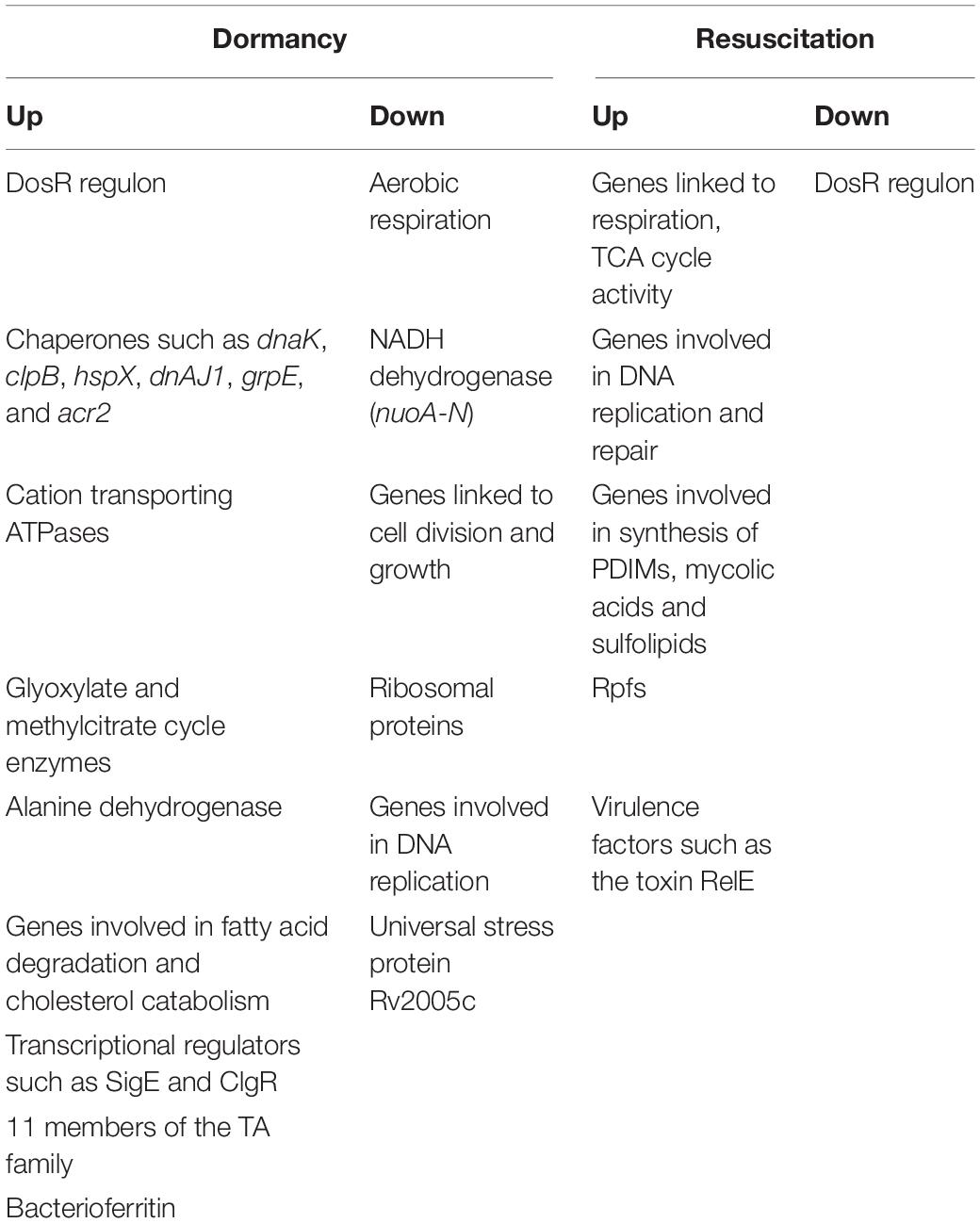
Table 3. Summary of selected genes that are differentially regulated in various models of dormancy and resuscitation.
Genes linked to aerobic respiration and glycolysis, replication, transcription, translation and cell division, and NADH dehydrogenase (nuo A-G; nuoH-N) were repressed during dormancy. Degradation of even-chain-length or odd-chain-length fatty acids leads to the formation of acetyl-CoA, or propionyl-CoA, respectively. Degradation of the cholesterol side chain or ring structure also yields acetyl-CoA and propionyl-CoA (Ouellet et al., 2011). Acetyl-CoA and propionyl-CoA are metabolized via the glyoxylate and methylcitrate cycle, respectively (McKinney et al., 2000; Muñoz-Elías and McKinney, 2005; Muñoz-Elías et al., 2006), making these pathways important for bacterial survival in vivo. Rv0467 encodes icl1 which acts as an isocitrate as well as methylisocitrate lyase, and is involved in both glyoxylate and methylcitrate cycles, (Gould et al., 2006). The prpDC (Rv1130–1131) operon encodes two enzymes, methylcitrate dehydratase and methylcitrate synthase, which are involved in the methylcitrate pathway. Mutations in prpR, a transcriptional activator of the prpDC operon, have been associated with drug tolerance in M. tuberculosis (Hicks et al., 2018). As expected icl1 and prpDC were induced in dormancy.
Proteomic studies have corroborated several of the findings from transcriptomics under dormancy, while raising some questions. The enzyme, Ald is induced under hypoxia (Starck et al., 2004; Gopinath et al., 2015; Schubert et al., 2015) as well as potassium depletion (Salina et al., 2014). This enzyme is suggested to generate alanine for protein and peptidoglycan synthesis. Alanine synthesis is coupled to NADH oxidation (Hutter and Dick, 1998). It is possible that the induction of Ald activity supports the maintenance of the NAD pool when oxygen becomes limiting. Tgs1 (Schubert et al., 2015), PckA (phosphoenolpyruvate carboxykinase), and trehalose biosynthesis related genes/proteins (Voskuil et al., 2004; Choa et al., 2006) were induced under hypoxia. The bacterioferritin protein BfrB (involved in iron storage), was induced in NRP1 (Voskuil et al., 2004; Gopinath et al., 2015), and then declined. Garcia-Morales et al. (2017) reported its induction during hypoxia in cholesterol-containing medium. Whereas several models have shown that prpDC, sigE, sigB, and clgR are induced during dormancy (Table 1), Schubert et al. (2015) have reported their induction at the protein level during reactivation as well, suggesting probable roles of these molecules during both dormancy and resuscitation. This could be an interesting avenue for future investigations.
Genes/proteins involved in cell division, replication, ribosomal protein synthesis, transcription and translation were expectedly downregulated during dormancy and upregulated during various phases of reactivation.
The role of ncRNAs in dormancy and resuscitation, is another area which is largely unexplored. Several studies have reported the upregulation of the ncRNA MTS2823 in different models of dormancy (Rodríguez et al., 2014; Aguilar-Ayala et al., 2017; Del Portillo et al., 2019). Arnvig et al. (2011) have suggested that the prpCD operon is one of the major targets of MTS2823. Deregulation of the RNA polymerase complex occurs when it is released from sequestration by MTS2823, leading to overexpression of the PrpBCD system (Płociński et al., 2019). MTS2823 is linked to the slow-down of M. tuberculosis growth (Arnvig and Young, 2012). Taken together, these observations suggest that MTS2823 could play a role in dormancy by regulating propionate metabolism.
There have been fewer omic studies capturing the transcriptional and proteomic network of M. tuberculosis during reactivation, compared to studies in various models of dormancy. Genes involved in respiration, TCA cycle activity, DNA replication and repair, synthesis of PDIMs, mycolic acids and sulfolipids, were activated during resuscitation (Table 3). The five genes encoding Rpfs were activated at different stages of reactivation. The DosRST regulon was downregulated. Proteases, transposases and insertion sequences were also expressed, suggesting genome reorganization during reactivation.
The genes and/or processes that are differentially regulated during dormancy and reactivation, are summarized in Figure 1. There are contradictions in the reports from different laboratories, and between transcriptomic and proteomic data. For example, Bacon et al. (2004) reported that bfrB is unchanged during hypoxia, whereas other studies have reported its induction under hypoxia (Voskuil et al., 2004; Gopinath et al., 2015). Contrary to other reports, Gopinath et al. (2015) failed to detect induction of Icl1 under hypoxia. While Voskuil et al. (2004) reported the induction of nrdZ (a ribonucleotide reductase class II enzyme that converts nucleoside triphosphates to deoxynucleoside triphosphates) during prolonged hypoxia, this could not be corroborated by Bacon et al. (2004). Voskuil et al. (2004) have reported that the four genes encoding the cytochrome bd oxidase were induced early during hypoxia whereas cytochrome bd oxidase was repressed in the potassium depletion model of dormancy (Salina et al., 2014, 2019). Most mycolic acid synthesizing/modifying genes were downregulated under dormancy (Sikri et al., 2015). However, Sikri et al. (2015) observed that accA2 and accD2 which are believed to be involved in the early steps of mycolic acid biosynthesis (Barry et al., 2007), were upregulated in the vitamin C model of dormancy. The KasB protein also involved in mycolic acid biosynthesis was upregulated during dormancy in at least two studies (Starck et al., 2004; Gopinath et al., 2015), but downregulated during potassium depletion (Salina et al., 2014). While some genes associated with fatty acid degradation were upregulated in the vitamin C model, others were downregulated. These contradictions may be attributed to the use of different in vitro models of dormancy, different time points of sampling of bacteria and methods with differing sensitivities for analysis of gene or protein expression. It must also be mentioned that a considerably large proportion of the observations of omics studies, await validation.
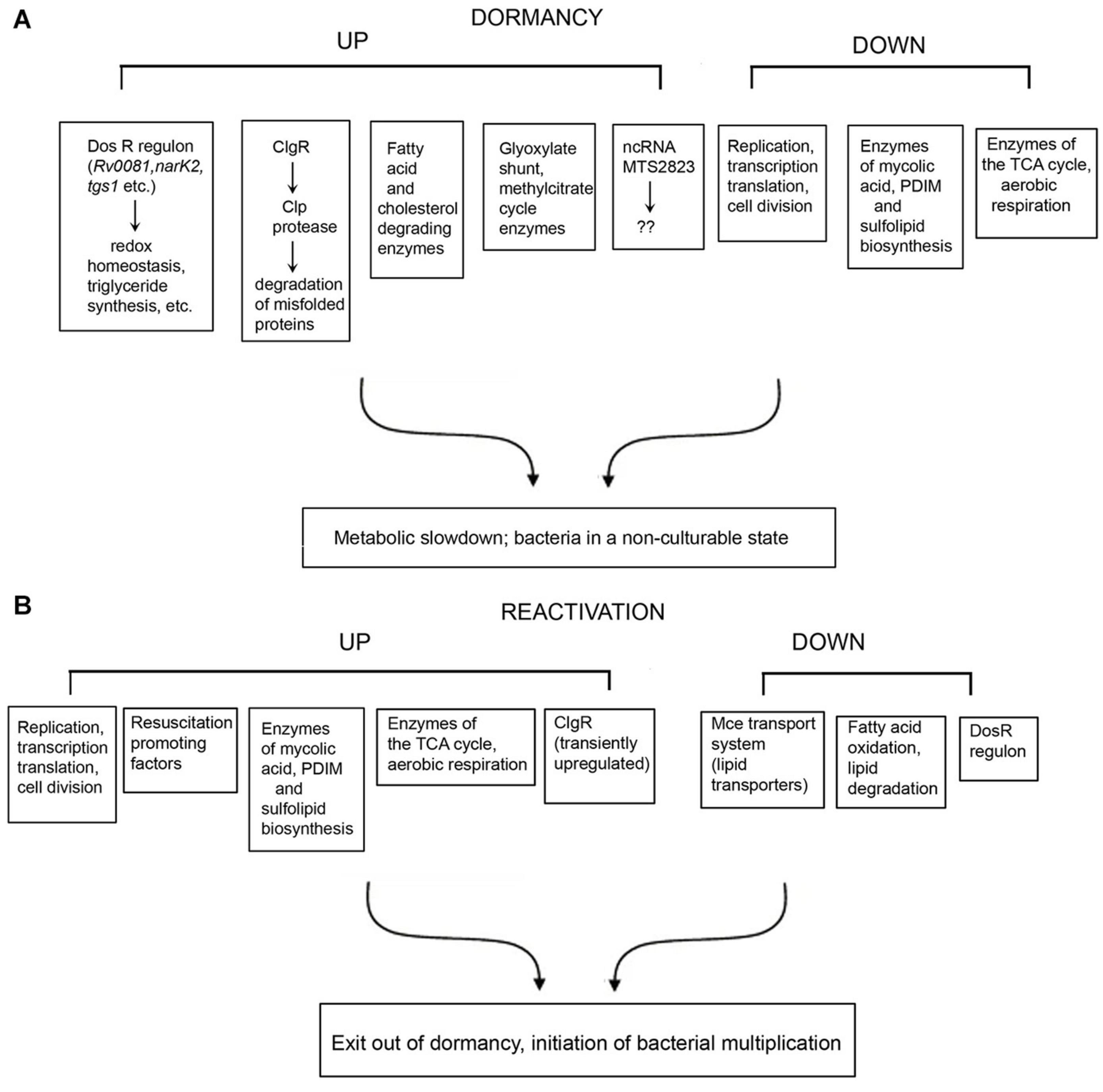
Figure 1. Summary of selected genes, proteins and pathways that are differentially regulated under dormancy (A) and reactivation (B) of Mycobacterium tuberculosis.
Areas for further investigation include understanding the differential expression of various Esx secretion systems members, chaperones, proteases and peptidases, during dormancy and reactivation, making these areas for future exploration. While the differential regulation of components of toxin/antitoxin modules have been reported, their significance remains unclear.
Overall, the better we understand dormancy and reactivation at a system level, the better will be the possibility of targeting these processes, and possibly shortening treatment regimens. As an example, omics approaches have unveiled a likely role of the transcriptional regulator ClgR in dormancy as well as in resuscitation. Further studies are required to understand the ClgR regulon and its role in dormancy and resuscitation. There is also an urgent need to develop better in vivo models for understanding dormancy and reactivation, so that the end goal of successfully containing TB is achieved in the shortest possible time.
Author Contributions
JB and MK reviewed the literature and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
JB was supported by funding from the J. C. Bose Fellowship (SB/S2/JCB-049/2016). MK was supported by the Council for Scientific and Industrial Research, Emeritus Scientist Scheme [21(1088)/19/EMR-II].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the contributions of all the scientists in this area and apologize for failing to cite any work due to constraints of space.
References
Aguilar-Ayala, D. A., Tilleman, L., Van Nieuwerburgh, F., Deforce, D., Palomino, J. C., Vandamme, P., et al. (2017). The transcriptome of Mycobacterium tuberculosis in a lipid-rich dormancy model through RNAseq analysis. Sci. Rep. 7:17665. doi: 10.1038/s41598-017-17751-x
Albrethsen, J., Agner, J., Piersma, S. R., Højrup, P., Pham, T. V., Weldingh, K., et al. (2013). Proteomic profiling of Mycobacterium tuberculosis identifies nutrient-starvation-responsive Toxin-antitoxin systems. Mol. Cell. Proteom. 12, 1180–1191. doi: 10.1074/mcp.M112.018846
Arnvig, K. B., Comas, I., Thomson, N. R., Houghton, J., Boshoff, H. I., Croucher, N. J., et al. (2011). Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 7:e1002342. doi: 10.1371/journal.ppat.1002342
Arnvig, K. B., and Young, D. B. (2009). Identification of small RNAs in Mycobacterium tuberculosis. Mol. Microbiol. 73, 397–408. doi: 10.1111/j.1365-2958.2009.06777.x
Arnvig, K. B., and Young, D. B. (2012). Non-coding RNA and its potential role in Mycobacterium tuberculosis pathogenesis. RNA Biol. 9, 427–436. doi: 10.4161/rna.20105
Bacon, J., James, B. W., Wernisch, L., Williams, A., Morley, K. A., Hatch, J., et al. (2004). The influence of reduced oxygen availability on pathogenicity and gene expression in Mycobacterium tuberculosis. Tuberculosis 84, 205–217. doi: 10.1016/j.tube.2003.12.011
Barry, C. E., Crick, D. C., and McNeil, M. R. (2007). Targeting the formation of the cell wall core of M. tuberculosis. Infect. Disord. Drug. Targets 7, 182–202. doi: 10.2174/187152607781001808
Behr, M. A., Schroeder, B. G., Brinkman, J. N., Slayden, R. A., and Barry, C. E. III (2000). A point mutation in the mma3 gene is responsible for impaired methoxymycolic acid production in Mycobacterium bovis BCG strains obtained after 1927. J. Bacteriol. 182, 3394–3399. doi: 10.1128/JB.182.12.3394-3399.2000
Betts, J., Lukey, P., Robb, L., McAdam, R., and Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731. doi: 10.1046/j.1365-2958.2002.02779.x
Boshoff, H. I. M., and Barry, C. E. III (2005). Tuberculosis - metabolism and respiration in the absence of growth. Nat. Microbiol. 3, 70–80. doi: 10.1038/nrmicro1065
Caño-Muñiz, S., Anthony, R., Niemann, S., and Alffenaar, J.-W. C. (2018). New approaches and therapeutic options for Mycobacterium tuberculosis in a dormant state. Clin. Microbiol. Rev. 31:e0060-17. doi: 10.1128/CMR.00060-17
Casali, N., and Riley, L. W. (2007). A phylogenomic analysis of the Actinomycetales mce operons. BMC Genom. 8:60. doi: 10.1186/1471-2164-8-60
Chen, Z., Hu, Y., Cumming, B. M., Lu, P., Feng, L., Deng, J., et al. (2016). Mycobacterial WhiB6 differentially regulates ESX-1 and the dos regulon to modulate granuloma formation and virulence in zebrafish. Cell Rep. 16, 2512–2524. doi: 10.1016/j.celrep.2016.07.080
Choa, S. H., Goodlett, D., and Franzblau, S. (2006). ICAT-based comparative proteomic analysis of non-replicating persistent Mycobacterium tuberculosis. Tuberculosis 86, 445–460. doi: 10.1016/j.tube.2005.10.002
Choi, K. H., Kremer, L., Besra, G. S., and Rock, C. O. (2000). Identification and substrate specificity of beta -ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis. J. Biol. Chem. 275, 28201–28207. doi: 10.1074/jbc.M003241200
Co, D. O., Hogan, L. H., Kim, S.-I., and Sandor, M. (2004). Mycobacterial granulomas: keys to a long-lasting host-pathogen relationship. Clin. Immunol. 113, 130–136. doi: 10.1016/j.clim.2004.08.012
Datta, P., Ravi, J., Guerrini, V., Chauhan, R., Neiditch, M. B., Shell, S. S., et al. (2015). The Psp system of Mycobacterium tuberculosis integrates envelope stress-sensing and envelope-preserving functions. Mol. Microbiol. 97, 408–422. doi: 10.1111/mmi.13037
Davis, J. M., and Ramakrishnan, L. (2009). The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell 136, 37–49. doi: 10.1016/j.cell.2008.11.014
De la Cruz, M. A., Zhao, W., Farenc, C., Gimenez, G., Raoult, D., Cambillau, C., et al. (2013). A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog. 9:e1003827. doi: 10.1371/journal.ppat.1003827
Del Portillo, P., García-Morales, L., Menéndez, M. C., Anzola, J. M., Rodríguez, J. G., Helguera-Repetto, A. C., et al. (2019). Hypoxia is not a main stress when Mycobacterium tuberculosis is in a dormancy-like long-chain fatty acid environment. Front. Cell. Infect. Microbiol. 8:449. doi: 10.3389/fcimb.2018.00449
Du, P., Sohaskey, C. D., and Shi, L. (2016). Transcriptional and physiological changes during Mycobacterium tuberculosis reactivation from non-replicating persistence. Front. Microbiol. 7:1346. doi: 10.3389/fmicb.2016.01346
Dutta, N. K., and Karakousis, P. C. (2014). Latent tuberculosis infection: myths, models, and molecular mechanisms. Microbiol. Mol. Biol. Rev. 78, 343–371. doi: 10.1128/MMBR.00010-14
Eoh, H., and Rhee, K. Y. (2013). Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, 6554–6559. doi: 10.1073/pnas.1219375110
Estorninho, M., Smith, H., Thole, J., Harders-Westerveen, J., Kierzek, A., Butler, R. E., et al. (2010). ClgR regulation of chaperone and protease systems is essential for Mycobacterium tuberculosis parasitism of the macrophage. Microbiology 156, 3445–3455. doi: 10.1099/mic.0.042275-0
Garcia-Morales, L., Leon-Soli, L., Monroy-Muñoz, I. E., Talavera-Paulin, M., Serafin-López, J., Estrada-Garcia, I., et al. (2017). Comparative proteomic profiles reveal characteristic Mycobacterium tuberculosis proteins induced by cholesterol during dormancy conditions. Microbiology 163, 1231–1237. doi: 10.1099/mic.0.000512
Gavrish, E., Sit, C. S., Cao, S., Kandror, O., Spoering, A., Peoples, A., et al. (2014). Lassomycin, a sibosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 21, 509–518. doi: 10.1016/j.chembiol.2014.01.014
Gengenbacher, M., Rao, S. P. S., Pethe, K., and Dick, T. (2010). Nutrient starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology 156, 81–87. doi: 10.1099/mic.0.033084-0
Gey van Pittius, N. C., Gamieldien, J., Hide, W., Brown, G. D., Siezen, R. J., and Beyers, A. D. (2001). The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2:RESEARCH0044. doi: 10.1186/gb-2001-2-10-research0044
Giffin, M. M., Shi, L., Gennaro, M. L., and Sohaskey, C. D. (2016). Role of alanine dehydrogenase of Mycobacterium tuberculosis during recovery from hypoxic nonreplicating persistence. PLoS One 11:e0155522. doi: 10.1371/journal.pone.0155522
Gopinath, V., Raghunandan, S., Gomez, R. L., Jose, L., Surendran, A., Ramachandran, R., et al. (2015). Profiling the proteome of Mycobacterium tuberculosis during dormancy and reactivation. Mol. Cell Proteom. 14, 2160–2175. doi: 10.1074/mcp.M115.051151
Gould, T. A., van de Langemheen, H., Munoz-Elias, E. J., McKinney, J. D., and Sacchettini, J. C. (2006). Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in Mycobacterium tuberculosis. Mol. Microbiol. 61, 940–947. doi: 10.1111/j.1365-2958.2006.05297.x
Guirado, E., Mbawuike, U., Keiser, T. L., Arcos, J., Azad, A. K., Wang, S.-H., et al. (2015). Characterization of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. mBio 6:e002537-15. doi: 10.1128/mBio.02537-14
Haning, K., Cho, S. H., and Contreras, L. M. (2014). Small RNAs in mycobacteria: an unfolding story. Front. Cell. Infect. Microbiol. 4:96. doi: 10.3389/fcimb.2014.00096
Hartkoorn, R. C., Uplekar, S., and Cole, S. T. (2014). Cross-Resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 2979–2981. doi: 10.1128/AAC.00037-14
He, H., Hovey, R., Kane, J., Singh, V., and Zahrt, T. C. (2006). MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 188, 2134–2143. doi: 10.1128/JB.188.6.2134-2143.2006
Hicks, N. D., Yang, J., Zhang, X., Zhao, B., Grad, Y., Liu, L., et al. (2018). Clinically prevalent mutations in Mycobacterium tuberculosis alter propionate metabolism and mediate multidrug tolerance. Nat. Microbiol. 3, 1032–1042. doi: 10.1038/s41564-018-0218-3
Houben, R., and Dodd, P. J. (2016). The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med. 13:e1002152. doi: 10.1371/journal.pmed.1002152
Hudock, T. A., Foreman, T. W., Bandyopadhyay, N., Gautam, U. S., Veatch, A. V., LoBato, D. N., et al. (2017). Hypoxia sensing and persistence genes are expressed during the intragranulomatous survival of Mycobacterium tuberculosis. Am. J. Respir. Cell Mol. Biol. 56, 637–647. doi: 10.1165/rcmb.2016-0239OC
Huebner, R. E., Schein, M. F., and Bass, J. B. Jr. (1993). The tuberculin skin test. Clin. Infect. Dis. 17, 968–975. doi: 10.1093/clinids/17.6.968
Hutter, B., and Dick, T. (1998). Increased alanine dehydrogenase activity during dormancy in Mycobacterium smegmatis. FEMS Microbiol. Lett. 167, 7–11. doi: 10.1111/j.1574-6968.1998.tb13200.x
Iona, E., Pardini, M., Mustazzolu, A., Piccaro, G., Nisini, R., Fattorini, L., et al. (2016). Mycobacterium tuberculosis gene expression at different stages of hypoxia-induced dormancy and upon resuscitation. J. Microbiol. 54, 565–572. doi: 10.1007/s12275-016-6150-4
Kaushal, D., Mehra, S., Didier, P. J., and Lackner, A. A. (2012). The nonhuman primate model of tuberculosis. J. Med. Primatol. 41, 191–201. doi: 10.1111/j.1600-0684.2012.00536.x
Kendall, S. L., Withers, M., Soffair, C. N., Moreland, N. J., Gurcha, S., Sidders, B., et al. (2007). A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol. Microbiol. 65, 684–699. doi: 10.1111/j.1365-2958.2007.05827.x
Kim, M. J., Wainwright, H. C., Locketz, M., Bekker, L. G., Walther, G. B., Dittrich, C., et al. (2010). Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol. Med. 2, 258–274. doi: 10.1002/emmm.201000079
Kundu, M. (2018). The role of two-component systems in the physiology of Mycobacterium tuberculosis. IUBMB Life 70, 710–717. doi: 10.1002/iub.1872
Kussell, E., Kishony, R., Balaban, N. Q., and Leibler, S. (2005). Bacterial persistence a model of survival in changing environments. Genetics 169, 1807–1814. doi: 10.1534/genetics.104.035352
Kussell, E., and Leibler, S. (2005). Phenotypic diversity, population growth, and information in fluctuating environments. Science 309, 2075–2078.
Larsson, C., Luna, B., Ammerman, N. C., Maiga, M., Agarwal, N., and Bishai, W. R. (2012). Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1-7 in redox environments. PLoS One 7:e37516. doi: 10.1371/journal.pone.0037516
Leistikow, R. L., Morton, R. A., Bartek, I. L., Frimpong, I., Wagner, K., and Voskuil, M. I. (2010). The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J. Bacteriol. 192, 1662–1670. doi: 10.1128/JB.00926-09
Leung, V., and Lévesque, C. M. (2012). A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J. Bacteriol. 194, 2265–2274. doi: 10.1128/JB.06707-11
Lewis, K. (2010). Persister cells. Annu. Rev. Microbiol. 64, 357–372. doi: 10.1146/annurev.micro.112408.134306
Lordi, G. M., and Reichman, L. B. (1988). “Tuberculin skin testing,” in Tuberculosis, ed. D. Schlossberg (New York, NY: Springer), 33–38. doi: 10.1007/978-1-4684-0305-3_4
Maisonneuve, E., Shakespeare, L. J., Jørgensen, M. G., and Gerdes, K. (2011). Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 108, 13206–13211. doi: 10.1073/pnas.1100186108
Manabe, Y. C., Kesavan, A. K., Lopez-Molina, J., Hatem, C. L., Brooks, M., Fujiwara, R., et al. (2008). The aerosol rabbit model of TB latency, reactivation and immune reconstitution inflammatory syndrome. Tuberculosis 88, 187–196. doi: 10.1016/j.tube.2007.10.006
Manganelli, R., Voskuil, M. I., Schoolnik, G. K., Dubnau, E., Gomez, M., and Smith, I. (2002). Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45, 365–374. doi: 10.1046/j.1365-2958.2002.03005.x
Manganelli, R., Voskuil, M. I., Schoolnik, G. K., and Smith, I. (2001). The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41, 423–437. doi: 10.1046/j.1365-2958.2001.02525.x
Marrero, J., Rhee, K. Y., Schnappinger, D., Pethe, K., and Ehrt, S. (2010). Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U.S.A. 107, 9819–9824. doi: 10.1073/pnas.1000715107
McGillivray, A., Golden, N. A., and Kaushal, D. (2015). The Mycobacterium tuberculosis Clp gene regulator is required for in vitro reactivation from hypoxia-induced dormancy. J. Biol. Chem. 290:2531. doi: 10.1074/jbc.M114.615534
McKinney, J. D., Honer Zu Bentrup, K., Muñoz-Elías, E. J., Miczak, A., and Chen, B. (2000). Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738. doi: 10.1038/35021074
Minch, K. J., Rustad, T. R., Peterson, E. J., Winkler, J., Reiss, D. J., Ma, S., et al. (2015). The DNA-binding network of Mycobacterium tuberculosis. Nat. Commun. 6:5829. doi: 10.1038/ncomms6829
Muñoz-Elías, E. J., and McKinney, J. D. (2005). Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11, 638–644. doi: 10.1038/nm1252
Muñoz-Elías, E. J., Upton, A. M., Cherian, J., and McKinney, J. D. (2006). Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol. Microbiol. 60, 1109–1122. doi: 10.1111/j.1365-2958.2006.05155.x
Murphy, D. J., and Brown, J. R. (2007). Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect. Dis. 7:84. doi: 10.1186/1471-2334-7-84
Muttucumaru, D. G. N., Roberts, G., Hinds, J., Stabler, R. A., and Parish, T. (2004). Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis 84, 239–246. doi: 10.1016/j.tube.2003.12.006
Nandi, M., Sikri, K., Chaudhary, N., Mande, S. C., Sharma, R. D., and Tyagi, J. S. (2019). Multiple transcription factors co-regulate the Mycobacterium tuberculosis adaptation response to vitamin C. BMC Genom. 20:887. doi: 10.1186/s12864-019-6190-3
Ordway, D. J., Shanley, C. A., Caraway, M. L., Orme, E. A., Bucy, D. S., Hascall Dove, L., et al. (2010). Evaluation of standard chemotherapy in the guinea pig model of tuberculosis. Antimicrob. Agents Chemother. 54, 1820–1833. doi: 10.1128/AAC.01521-09
Ouellet, H., Johnston, J. B., and de Montellano, P. R. (2011). Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trend Microbiol. 19, 530–539. doi: 10.1016/j.tim.2011.07.009
Paige, C., and Bishai, W. R. (2010). Penitentiary or penthouse condo: the Tuberculous granuloma from the microbe’s point of view. Cell. Microbiol. 12, 301–309. doi: 10.1111/j.1462-5822.2009.01424.x
Pandey, A. K., and Sassetti, C. M. (2008). Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380. doi: 10.1073/pnas.0711159105
Pang, X., Vu, P., Byrd, T. F., Ghanny, S., Soteropoulos, P., Mukamolova, G. V., et al. (2007). Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 153, 1229–1242. doi: 10.1099/mic.0.29281-0
Peña, J. C., and Ho, W. Z. (2015). Monkey models of tuberculosis: lessons learned. Infect. Immun. 83, 852–862. doi: 10.1128/IAI.02850-14
Peterson, E. J., Abidi, A. A., Arrieta-Ortiz, M. L., Aguilar, B., Yurkovich, J. T., Kaur, A., et al. (2020). Intricate genetic programs controlling dormancy in Mycobacterium tuberculosis. Cell Rep. 31, 33–38. doi: 10.1016/j.celrep.2020.107577
Płociński, P., Macios, M., Houghton, J., Niemiec, E., Płocińska, R., Brzostek, A., et al. (2019). Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis. Nucleic Acids Res. 47, 5892–5905. doi: 10.1093/nar/gkz251
Rachman, H., Strong, M., Ulrichs, T., Grode, L., Schuchhardt, J., Mollenkopf, L., et al. (2006). Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect. Immun. 74, 1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006
Ramakrishnan, L. (2012). Revisiting the role of the granuloma in tuberculosis. Nat. Rev. Immunol. 12, 352–366. doi: 10.1038/nri3211
Rodríguez, J. G., Hernández, A. C., Helguera-Repetto, C., Ayala, D. A., Guadarrama-Medina, R., Anzóla, J. M., et al. (2014). Global adaptation to a lipid environment triggers the dormancy-related phenotype of Mycobacterium tuberculosis. mBio 53:e01125-14. doi: 10.1128/mBio.01125-14
Rowland, J. L., and Niederweis, M. (2012). Resistance mechanisms of Mycobacterium tuberculosis against phagosomal copper overload. Tuberculosis 92, 202–210. doi: 10.1016/j.tube.2011.12.006
Salina, E. G., Grigorov, A. S., Bychenko, O. S., Skvortsova, Y. V., Mamedov, I. Z., Azhikina, T. L., et al. (2019). Resuscitation of dormant “non-culturable” Mycobacterium tuberculosis is characterized by immediate transcriptional burst. Front. Cell. Infect. Microbiol. 9:272. doi: 10.3389/fcimb.2019.00272
Salina, E. G., Waddell, S. J., Hoffmann, N., Rosenkrands, I., Butcher, P. D., and Kaprelyants, A. S. (2014). Potassium availability triggers Mycobacterium tuberculosis transition to, and resuscitation from, non-culturable (dormant) states. Open Biol. 4:140106. doi: 10.1098/rsob.140106
Saunders, B. M., and Britton, W. J. (2007). Life and death in the granuloma: immunopathology of tuberculosis. Immunol. Cell. Biol. 85, 103–111. doi: 10.1038/sj.icb.7100027
Schaeffer, M. L., Agnihotri, G., Volker, C., Kallender, H., Brennan, P. J., and Lonsdale, J. T. (2001). Purification and biochemical characterization of the Mycobacterium tuberculosis β-ketoacyl-acyl carrier protein synthases KasA and KasB. J. Biol. Chem. 276, 47029–47037. doi: 10.1074/jbc.M108903200
Schubert, O. T., Ludwig, C., Kogadeeva, M., Zimmermann, M., Rosenberger, G., Gengenbacher, M., et al. (2015). Absolute proteome composition and dynamics during dormancy and resuscitation of Mycobacterium tuberculosis. Cell Host Microb. 18, 96–108. doi: 10.1016/j.chom.2015.06.001
Sherrid, A. M., Rustad, T. R., Cangelosi, G. A., and Sherman, D. R. (2010). Characterization of a Clp protease gene regulator and the reaeration response in Mycobacterium tuberculosis. PLoS One 5:e11622. doi: 10.1371/journal.pone.0011622
Sikri, K., Batra, S. D., Nandi, M., Kumari, P., Taneja, N. K., and Tyagi, J. S. (2015). The pleiotropic transcriptional response of Mycobacterium tuberculosis to vitamin C is robust and overlaps with the bacterial response to multiple intracellular stresses. Microbiology 161, 739–753. doi: 10.1099/mic.0.000049
Siméone, R., Léger, M., Constant, P., Malaga, W., Marrakchi, H., Daffé, M., et al. (2010). Delineation of the roles of FadD22, FadD26 and FadD29 in the biosynthesis of phthiocerol dimycocerosates and related compounds in Mycobacterium tuberculosis. FEBS J. 277, 2715–2725. doi: 10.1111/j.1742-4658.2010.07688.x
Singh, A., Crossman, D. K., Mai, D., Guidry, L., Voskuil, M. I., Renfrow, M. B., et al. (2009). Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 5:e1000545. doi: 10.1371/journal.ppat.1000545
Sohaskey, C. D., and Wayne, L. G. (2003). Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J. Bacteriol. 185, 7247–7256. doi: 10.1128/JB.185.24.7247-7256.2003
Starck, J., Källenius, G., Marklund, B.-I., Andersson, D. I., and Åkerlund, T. (2004). Comparative proteome analysis of Mycobacterium tuberculosis grown under aerobic and anaerobic conditions. Microbiology 150, 3121–3129. doi: 10.1099/mic.0.27284-0
Subbian, S., Tsenova, L., O’Brien, P., Yang, G., Kushner, N. L., Parsons, S., et al. (2012). Spontaneous latency in a rabbit model of pulmonary tuberculosis. Am. J. Pathol. 181, 1711–1724. doi: 10.1016/j.ajpath.2012.07.019
Taneja, N. K., Dhingra, S., Mittal, A., Naresh, M., and Tyagi, J. S. (2010). Mycobacterium tuberculosis transcriptional adaptation, growth arrest and dormancy phenotype development is triggered by vitamin C. PLoS One 5:e10860. doi: 10.1371/journal.pone.0010860
Timm, J., Post, F. A., Bekker, L. G., Walther, G. B., Wainwright, H. C., Manganelli, R., et al. (2003). Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. U.S.A. 100, 14321–14326. doi: 10.1073/pnas.2436197100
Tükenmez, H., Sarkar, S., Anoosheh, S., Kruchanova, A., Edström, I., Harrison, G. A., et al. (2021). Mycobacterium tuberculosis Rv3160c is a TetR-like transcriptional repressor that regulates expression of the putative oxygenase Rv3161c. Sci. Rep. 11:1523.
Van der Geize, R., Yam, K., Heuser, T., Wilbrink, M. H., Hara, H., Anderton, M. C., et al. (2007). A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952. doi: 10.1073/pnas.0605728104
Vaubourgeix, J., Lin, G., Dhar, N., Chenouard, N., Jiang, X., Botella, H., et al. (2015). Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells. Cell Host Microb. 17, 178–190. doi: 10.1016/j.chom.2014.12.008
Via, L. E., Lin, P. L., Ray, S. M., Carrillo, J., Allen, S. S., Eum, S. Y., et al. (2008). Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76, 2333–2340. doi: 10.1128/IAI.01515-07
Volkman, H. E., Pozos, T. C., Zheng, J., Davis, J. M., Rawls, J. F., and Ramakrishnan, L. (2010). Tuberculous granuloma induction via interaction of a bacterial secreted protein with host epithelium. Science 327, 466–469. doi: 10.1126/science.1179663
Voskuil, M. I., Visconti, K. C., and Schoolnik, G. K. (2004). Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis 84, 218–227. doi: 10.1016/j.tube.2004.02.003
Walker, R. W., Barakat, H., and Hung, J. G. (1970). The positional distribution of fatty acids in the phospholipids and triglycerides of Mycobacterium smegmatis and M. bovis BCG. Lipids 5, 684–691. doi: 10.1007/BF02531435
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wayne, L. G. (1976). Dynamics of submerged growth of Mycobacterium tuberculosis under aerobic and microaerophilic conditions. Am. Rev. Respir. Dis. 114, 807–811.
Wayne, L. G., and Hayes, L. G. (1996). An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64, 2062–2069. doi: 10.1128/IAI.64.6.2062-2069.1996
Wayne, L. G., and Sohaskey, C. D. (2001). Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55, 139–163. doi: 10.1146/annurev.micro.55.1.139
Zaychikova, M. V., Zakharevich, N. V., Sagaidak, M. O., Bogolubova, N. A., Smirnova, T. G., Andreevskaya, S. N., et al. (2015). Mycobacterium tuberculosis Type II toxin-antitoxin systems: genetic polymorphisms and functional properties and the possibility of their use for genotyping. PLoS One 10:e0143682. doi: 10.1371/journal.pone.0143682
Keywords: dormancy, resuscitation, Mycobacterium tuberculosis, transcriptomics, proteomics
Citation: Kundu M and Basu J (2021) Applications of Transcriptomics and Proteomics for Understanding Dormancy and Resuscitation in Mycobacterium tuberculosis. Front. Microbiol. 12:642487. doi: 10.3389/fmicb.2021.642487
Received: 16 December 2020; Accepted: 19 February 2021;
Published: 31 March 2021.
Edited by:
Andre G. Loxton, South African Medical Research Council, South AfricaReviewed by:
Wonsik Lee, Sungkyunkwan University, South KoreaEvgeniya V. Nazarova, Genentech, United States
Copyright © 2021 Kundu and Basu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manikuntala Kundu, bWFuaWt1bnRhbGEua3VuZHVAZ21haWwuY29t; Joyoti Basu, am95b3RpYmFzdUBnbWFpbC5jb20=
 Manikuntala Kundu
Manikuntala Kundu Joyoti Basu
Joyoti Basu