- 1Department of Clinical Laboratory, The Second Affiliated Hospital of Soochow University, Suzhou, China
- 2Hackensack Meridian Health Center for Discovery and Innovation, Nutley, NJ, United States
- 3Department of Medical Sciences, Hackensack Meridian School of Medicine, Nutley, NJ, United States
Hypervirulent Klebsiella pneumoniae (hvKP) has spread globally since first described in the Asian Pacific Rim. It is an invasive variant that differs from the classical K. pneumoniae (cKP), with hypermucoviscosity and hypervirulence, causing community-acquired infections, including pyogenic liver abscess, pneumonia, meningitis, and endophthalmitis. It utilizes a battery of virulence factors for survival and pathogenesis, such as capsule, siderophores, lipopolysaccharide, fimbriae, outer membrane proteins, and type 6 secretion system, of which the former two are dominant. This review summarizes these hvKP-associated virulence factors in order to understand its molecular pathogenesis and shed light on new strategies to improve the prevention, diagnosis, and treatment of hvKP-causing infection.
Introduction
Klebsiella pneumoniae is a common opportunistic pathogen that frequently causes nosocomial infections, including pneumonia, meningitis, bloodstream and urinary tract infection (Togawa et al., 2015; Ku et al., 2017). In addition, K. pneumoniae has the potential to cause community associated infection, such as liver abscess, endophthalmitis, and meningitis, in healthy individuals (Russo and Marr, 2019). An unique case of K. pneumoniae caused liver abscess along with endophthalmitis was reported for the first time in Taiwan in the 1980s, and the causative organism was designated as hypervirulent K. pneumoniae (hvKP) (Liu et al., 1986). Since then, hvKP has been recognized as another circulating pathotype in addition to classical K. pneumoniae (cKP), associated with high pathogenicity and mortality due to hypervirulence (Lan et al., 2020). Factors contributing to the hypervirulence mainly include capsule, siderophores, lipopolysaccharide (LPS) and fimbriae (Parrott et al., 2020).
The primary differences between cKP and hvKP are summarized in Table 1. The string test based on hypermucoviscous phenotype (string ≥5 mm) was widely used as a marker for hvKp, with ∼90% predicted accuracy for clinical hvKp strains (Russo et al., 2018). This semi-qualitative assay is easily influenced by colony conditions and the user’s technique (Tan et al., 2014). Also, cKP strains with mucoviscosity and hvKP strains without hypermucoviscosity have been identified (Catalán-Nájera et al., 2017). Another mucoviscosity assay was quantified by comparing the absorbance of supernatants collected by low-speed centrifugation because hypermucoviscous strains cannot sediment sufficiently, leaving the supernatants turbid (Walker and Miller, 2020).
Except for intrinsic resistance to ampicillin, hvKp strains are commonly susceptible to a variety of antibiotics, including cephalosporins and carbapenems; however, multidrug-resistant and hypervirulent (MDR-hv) strains have recently emerged, primarily due to horizontal transfer of plasmid-mediated resistance or virulence (Liu C. et al., 2020; Tang et al., 2020). The spread of hvKP, including MDR-hvKP strains, poses a serious threat to the public health, and studies on hvKP virulence are therefore imperative to comprehend the underlying pathogenesis and provide a theoretical basis for the treatment and prevention of infection. In this review, we discuss hvKP relevant virulence elements, with reference to the discovery and epidemiology.
Discovery and Epidemiology
In 1986, a striking case of K. pneumoniae liver abscess (KLA) with extrahepatic complications such as purulent meningitis, prostate abscess, and pyogenic ophthalmia, was recorded in Taiwan, wherein patients eventually went blind despite receiving active treatment for the bacteria (Liu et al., 1986). Historically, pyogenic liver abscess was regarded as a polymicrobial infection and its predominant bacterial cause was Escherichia coli (Wang et al., 1998; Lederman and Crum, 2005). A single microorganism K. pneumoniae caused liver abscess occurred in this case, which was alarming as new invasive syndrome. Coincidentally, several studies by Nassif and Sansonetti (1986) and Nassif et al. (1989a;b) reported the existence of a ∼180-kbp plasmid in K. pneumoniae strains, contained the genes encoding aerobactin and its receptor, as well as rmpA conferring a mucoid phenotype. A comprehensive hvKp definition refers to hypermucoviscous phenotype, the corresponding virulence gene genotype and clinical manifestation of metastatic infection (Catalán-Nájera et al., 2017). For some variants with ambiguous pathogenic potential, murine but not Galleria mellonella models are more appropriate to evaluate the hypervirulence phenotype accurately (Russo and MacDonald, 2020), although the G. mellonella lethality assay combined with the string test has been demonstrated to improve clinical hvKP identification significantly (Li et al., 2020).
Due to increasing population mobility, hvKP infections have spread worldwide, throughout Asia (Chung et al., 2007; Shankar et al., 2018), Europe (Moore et al., 2013; Cubero et al., 2016; Rossi et al., 2018; Sánchez-López et al., 2019), Oceania (Foo et al., 2018; Sturm et al., 2018), North-America (Rahimian et al., 2004; Pastagia and Arumugam, 2008; Fazili et al., 2016), South-America (Coutinho et al., 2014), and Africa (Yu et al., 2007), with the highest prevalence in the Asian Pacific Rim. Genetic factors and geographic conditions are likely to be relative risk factors for this situation (Sellick and Russo, 2018). The geographical distribution of MDR-hvKP also showed similar pattern that they were mostly found in Asian countries, although global spread has been documented (Li et al., 2014; Tang et al., 2020). The genetic backgrounds of hvKP varied, among which clonal group (CG) 23 strains are the most predominant (Liao et al., 2014; Shi et al., 2018). However, novel hvKp sequence types continue to emerge, indicated the ongoing evolution of hvKP (Shankar et al., 2018). MDR-hvKP strains mainly belonged to ST11, and carbapenem-resistant K. pneumoniae (CR-KP) MDR-hvKp ST11 strains have been described in China (Gu et al., 2018; Tang et al., 2020). Both ST11 and ST258 belong the CG258 clone and ST11 is a single locus variant to ST258. Despite the high prevalence of MDR-hvKp in ST11 strains, ST258 MDR-hvKP strains have not yet been detected (Ernst et al., 2020).
An epidemic of infectious diseases requires three indispensable segments: source, route, and susceptible population. In terms of habitat associations, K. pneumoniae have been frequently found in human, animal, sewage, polluted water, and soil samples (Bagley, 1985; Podschun et al., 2001); however, the origin of hvKP is yet to be ascertained. Liu et al. (2018) recently found ST23 hvKP in a cucumber sample along with E. coli carrying blaNDM when analyzing ready-to-eat vegetables. In addition, three MDR-hvKP strains have been obtained from public aquatic environments in Brazil, suggesting the contaminated water source may potentially increase the risk of hvKP spread (Furlan et al., 2020). These results suggested that environment may serve as a reservoir for the dissemination of hvKp and MDR-hvKP strains.
Klebsiella pneumoniae isolates require suitable areas for colonization before setting off infections, and the gastrointestinal tracts are regarded as major reservoirs for K. pneumoniae colonization (Gorrie et al., 2017; Sun et al., 2019). A previous study investigated K. pneumoniae serotypes of stool specimens collected from healthy Chinese in Asian countries. The data presented that intestinal colonization rates of cKP and hvKP (K1/K2 serotypes) were 18.8–87.7 and 0–16.7%, respectively (Lin et al., 2012), while in Europe, the carriage rate of cKP carriers is 5.9–19.3% (Smith et al., 1997; Gorrie et al., 2017). Nevertheless, whether the difference in bacteria load among different worldwide regions could be ascribed to the incidence of highly invasive diseases caused by hvKP is yet to be confirmed. Additionally, Fung et al. (2012) discovered that serotypes and genotypes of bacterial isolates from healthy carriers’ fecal samples are identical to those of K. pneumoniae liver abscess (KLA) patients, which was in line with a family-clustered case report in Japan that the stool cultures from two healthy relatives of KLA patients were hvKP-positive, rendering them as carriers (Harada et al., 2011). Intestinal tract colonization by carbapenemase-producing K. pneumoniae has also been documented (Gorrie et al., 2017). For example, ST11 CR-hvKP strains have been isolated from stool samples of acute diarrhea inpatients in Zhejiang Province, China (Zheng et al., 2020). These findings indicated that guts might be a significant reservoir for hvKP strains as they can break the intestinal barrier and move to the liver to cause KLA and metastatic spread. Additionally, person-to-person transmission via a possible fecal-oral cycle is also possible (Siu et al., 2012), but the trigger for the occurrence of infection after colonization is not yet clarified.
Unlike cKP caused nosocomial infections, hvKP results in community-acquired infections (Ikeda et al., 2018; Harada et al., 2019). Once nosocomial cKP strains acquire hypervirulence plasmid and colonize in the intestine, healthcare-associated hvKP infections rise indiscriminately (Russo and Marr, 2019). A retrospective study conducted in Beijing by Liu C. et al. (2020) found that >90% hvKP-causing infections are nosocomial, suggesting the likelihood of hvKP evolved as a healthcare-associated pathogen. Retrospective studies showed hvKP infections mainly occur in susceptible individuals, male and aged over 50-years-old, and with an underlying condition of diabetes among (Fazili et al., 2016). Jin et al. (2020) reported that the bactericidal capacity of diabetic neutrophil extracellular traps (NETs) to hvKP was impaired, which partly explained why diabetics were vulnerable to hvKP infections. Another potential risk factor, co-existing hepatobiliary disease, was identified in a recent study (Parrott et al., 2020). In-depth studies analyzing the host factors to hvKP infections remain limited, while this knowledge could significantly contribute in unraveling the underlying epidemiology and implement appropriate prevention and control measures.
Virulence-Associated Factors
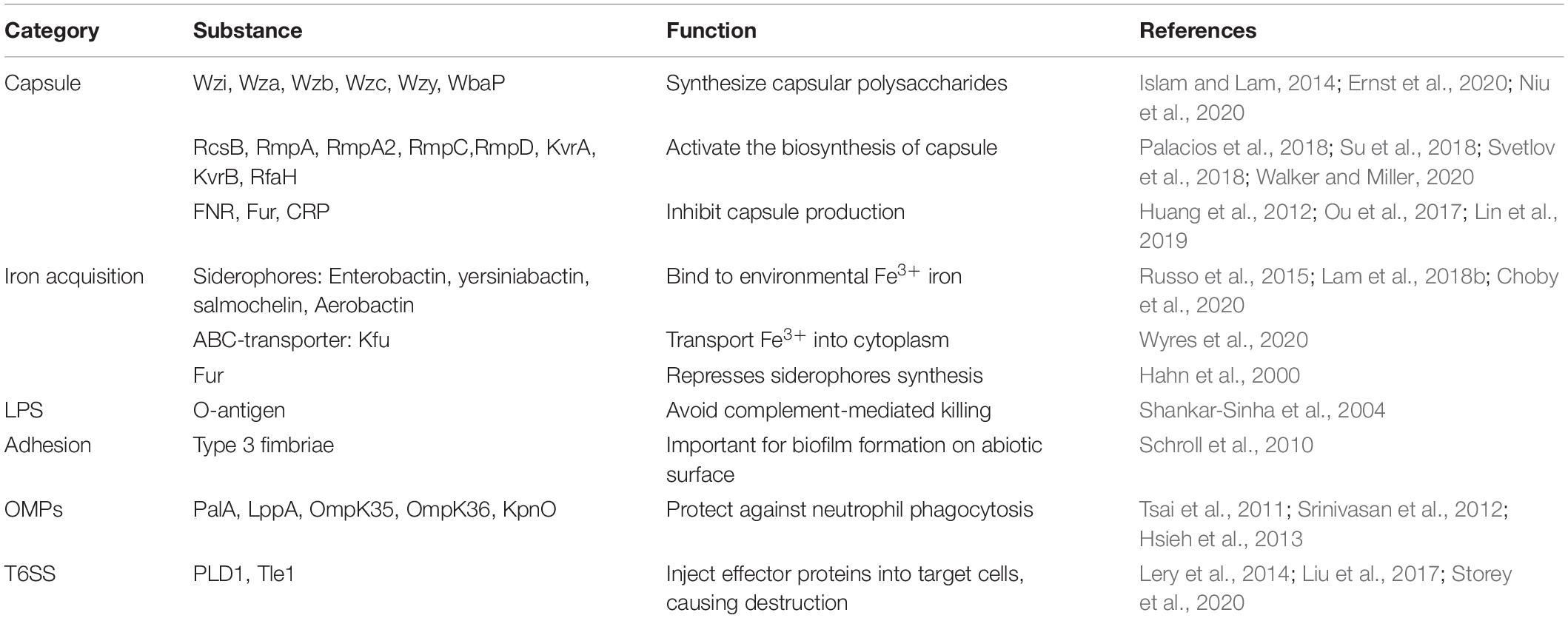
Capsule
The correlation between hypermucoviscosity and hypervirulence is widely accepted, albeit with a few exceptions. The capsule surrounding the surface of K. pneumoniae serves as main virulence factors associated with the viscous phenotype. The characterization of an intranasal infection model revealed that mice infected with acapsular strains of K. pneumoniae had longer survival significantly and lower bacterial density in lung and blood than those infected with capsular strains, as the capsule might provide protection from the immunological responses (Yoshida et al., 2000; Lawlor et al., 2005). Capsule-mediated resistance to bactericidal actions is inclined to be defensive rather than offensive. For example, K. pneumoniae evades phagocytosis, complement, antimicrobial peptides, and specific antibodies using capsules to make it difficult for bacteria to be bound, while active suppression and attack on immune cells through capsule are rarely observed (Domenico et al., 1994; Alvarez et al., 2000; Llobet et al., 2011; Paczosa and Mecsas, 2016). Furthermore, the capsule size of hvKP strains is above the basal level of cKP, and hvKP isolated from the cerebrospinal fluid is highly prevalent in a mean capsule size of >2 mm (Ku et al., 2017). Thus, these isolates have robust coats expressing hypermucoviscous phenotype that might enhance their viability. On the other side, the thick hypercapsule could serve as a physical barrier and impair the DNA uptake, consequently, limiting the horizontal gene transfer, which may partially explain why hvKp strains are less likely to harbor antimicrobial resistant genes than those of cKp strains (Wyres et al., 2019).
Based on the diversity of the polysaccharide components of the capsule and different structures and antigens, K. pneumoniae can be divided into at least 79 serotypes (Pan et al., 2015). Eight types, K1, K2, K5, K16, K20, K54, K57, and KN1 have been described in hvKP (Pan et al., 2008; Lee et al., 2016); with K1 and K2 the most common. Multilocus sequence typing (MLST) showed that the diversity of sequence-type (ST) was greater in capsule type K2 than K1 serotype that K1 was strongly related to ST23 while K2 was associated with ST25, ST86, ST375, and ST380, respectively (Struve et al., 2015; Lee et al., 2016). The prevalent serotypes differ with the geographic regions: K1 is the most predominant type in Asia, while K2 is more frequent found in Europe and North America (Catalán-Nájera et al., 2017; Remya et al., 2018). For MDR-hvKp, serotypes also depend on ST and region, as an example, serotypes K64 and K47, associated with the ST11 strains, are most common in China (Yang et al., 2021).
Several studies have suggested that K1 and K2 strains have higher virulence than the strains of other serotypes (Yeh et al., 2007; Wang et al., 2017), and hence, serotype is considered as one of the potential virulence factors, which could be attributed to the distinct compositions that confer survival advantages. A previous study showed that K1 and K2 capsule types lacked mannose and rhamnose, which could be recognized by macrophage lectin receptors to induce phagocytosis (Athamna et al., 1991; Fung et al., 2002). In addition, a surface-exposed protein, fructose-1, 6-bisphosphate aldolase (FBA), was identified in K1 hvKP, and its enhanced expression could protect bacteria against neutrophil killing under high glucose condition (Lee et al., 2020). Moreover, sialic acid, as a constituent of K1 and K2 capsular polysaccharide, contributes to hypermucoviscous phenotype, and is thus responsible for the anti-phagocytic activity directly or indirectly (Lee et al., 2014; Opal, 2014). These reports do not indicate equal hypervirulence among these serotypes, and K1/K2 cKP strains without increased viscosity and additional virulence factors usually fail to express hypervirulence phenotype. However, serotyping reports under the diagnosis of pyogenic liver abscess are valuable and could guide to take precautions to prevent further episodes (Fung et al., 2002).
Genes involving capsule production are located on the capsular polysaccharide synthesis (cps) region of chromosome. The region of cps cluster (from galF to ugd) harbors over 20 genes, largely driving by three promotors located upstream of genes, galF, wzi, and manC, respectively (Figure 1A). Among these genes, wbaP encoded protein mediated the first step of capsule biosynthesis, and the deletion of this gene confers impaired capsule production. Interestingly, a recent study found that certain wbaP-mutations enhanced the pathogenicity by improving biofilm formation and bladder epithelial cells invasion in urinary tract infections (Ernst et al., 2020). The function of wzx in capsule-biosynthesis is flipping the polymer from the cytoplasm to the periplasm of the inner membrane. The Wzy protein is involved in the polymerization via catch-and-release mechanism (Islam and Lam, 2014). Whilst proteins encoded by wza and wzc genes form a translocation complex responsible for assembling capsular polysaccharides and transporting them from the periplasm to the surface of the bacteria (Pan et al., 2013). The Wzb protein, as the cognate phosphatase of Wzc, combines with the catalytic domain on Wzc, and in turn dephosphorylates Wzc (Reid and Whitfield, 2005; Niu et al., 2020). A study concluded that wza knockout strains obstruct the polysaccharide transport, decrease the expression of wzb, wzc, and wzi genes to avoid overaccumulation of polysaccharides in the cells, and ultimately lead to capsule deficiency (Niu et al., 2020). The serotype-specific gene wzi encodes an outer membrane protein that assists in the formation of the bacterial capsule by anchoring the capsular polysaccharides to the outer membrane, and wzi-deficiency confers acapsular phenotype (Rahn et al., 2003; Brisse et al., 2013; Bushell et al., 2013). The wzy gene is regarded as a serotype-specific target which has been frequently used as a target to characterize different cps types. Intriguingly, the chromosomal mucoviscosity-associated gene A (magA) discovered in 2004 was later found to be the wzy gene in K1 locus (i.e., K1_wzy) (Fang et al., 2004, 2010; Chuang et al., 2006). In addition, magA (K1_wzy) is essential for the synthesis of capsular polysaccharide and plays an important role in hypermucoviscous and resistance to complement-mediated lysis, while ΔmagA mutants lose these features (Wu et al., 2009). Therefore, it is postulated that magA acts as a virulence-associated factor but is restricted to K1 isolates.
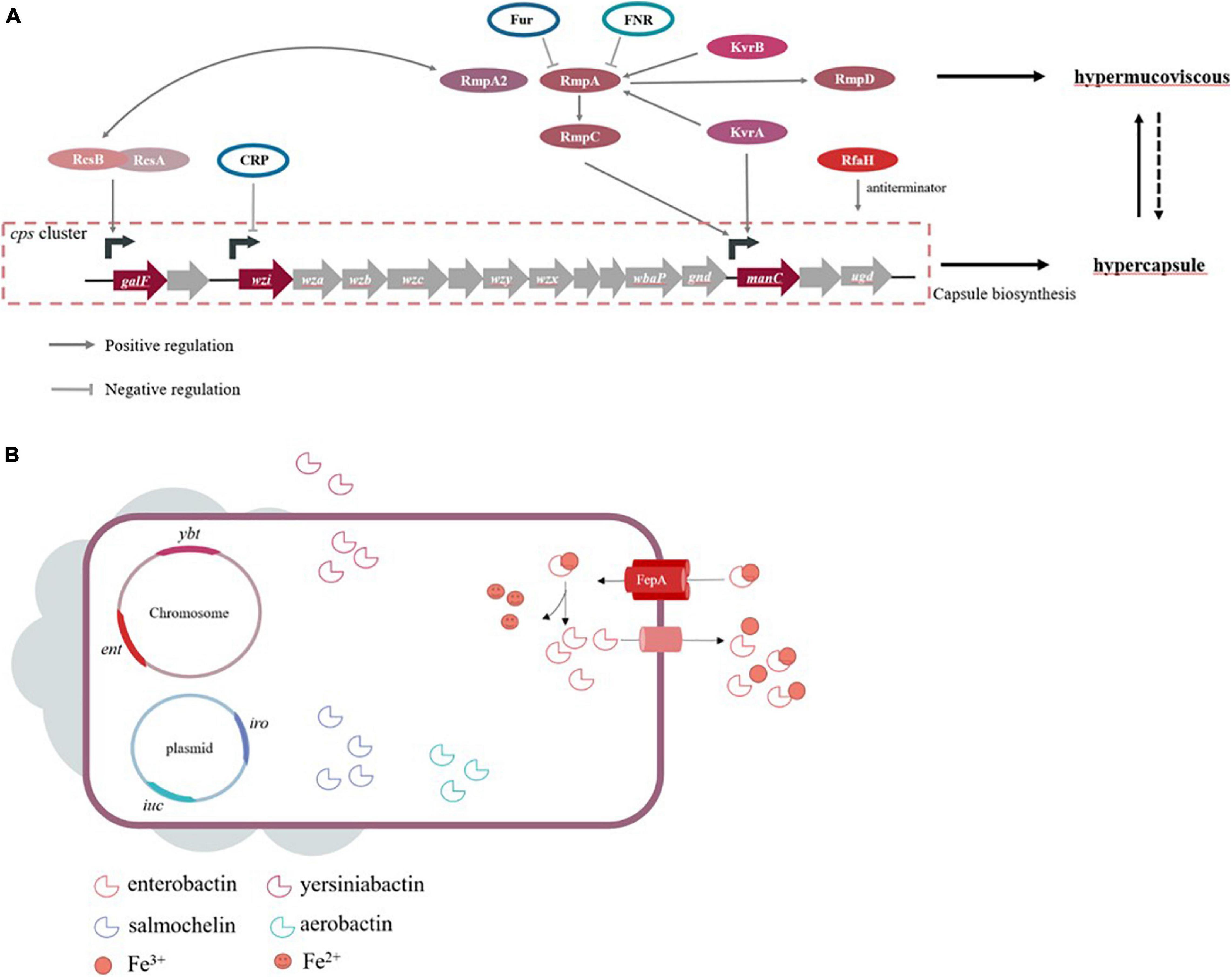
Figure 1. (A) Genes are known to influence capsule biosynthesis, including genes at cps locus and regulators affecting the cps expression directly or indirectly. The corresponding promoters that some regulators act on refer to studies of Walker and Miller (2020). Hypercapsule is sufficient but not necessary for hypermucoviscous phenotype, demonstrating that hypermucoviscous phenotype is not solely due to excessive capsule expression. (B) The siderophore systems enterobactin (ent), yersiniabactin (ybt), salmochelin (iro) and aerobactin (iuc) are present. Take enterobactin as an example to show the process of iron acquisition, which FepA is the transporter specific to enterobactin.
High capsule productivity is also attributed to a number of virulence genes, in addition to those present in the cps cluster, including the regulation of the capsule synthesis B genes (rcsB), a regulator of mucoid phenotype A and A2 (rmpA and rmpA2) and Klebsiella virulence regulators (kvrA and kvrB) (Cheng et al., 2010; Palacios et al., 2018; Su et al., 2018). These genes are positive transcriptional regulation factors for the cps locus. The Rcs phosphorelay system is a two-component signal transduction system, which was originally identified as a regulator for the synthesis of colonic acid in E. coli and composed of three core proteins, RcsC, RcsD, and RcsB (Wall et al., 2018). RcsC protein is a transmembrane sensor kinase that can undertake autophosphorylation after sensing extra-cytoplasmic stimulus and then transfer the stimuli to RcsD. Finally, the signal is transmitted to RcsB, which is the response regulator forming homodimer or heterodimer with the auxiliary proteins. Subsequently, the phosphorylated RcsB interacts with RcsA, and this dimer binds to promoters in the cps cluster, which activates the biosynthesis of the capsule (Guo and Sun, 2017). This mechanism of regulation in E. coli is applicable to other Gram-negative bacteria, but capsular polysaccharide-related genes controlled by auxiliary regulator RcsA are limited in comparison to RcsB in K. pneumoniae (Su et al., 2018). On the other hand, the rmpA gene of K. pneumoniae expresses rcsA-like activity and complements a rcsA mutant of E. coli to positively control the mucoid phenotype (Nassif et al., 1989b; Arakawa et al., 1991; Gottesman and Stout, 1991). Similar to RcsA, RmpA depends on RcsB for activating the biosynthesis of capsular polysaccharides. Together, rmpA+rcsB– strains and rmpA–rcsB+ strains are unable to increase capsule production (Cheng et al., 2010). Consistent with this theory, RcsB seems vital for base-level capsule expression, while the regulator RmpA elevates the capsule production (Walker et al., 2019). Moreover, partial homology was also detected in rmpA2, rmpA, and rcsA (Wacharotayankun et al., 1993). While RmpA2 is directly bound to the cps gene promoter to regulate capsule productivity, rcsB– K. pneumoniae strain carrying rmpA2 recovers the mucoid phenotype (Lai et al., 2003). Furthermore, RmpA and RmpA2 are known regulators required for hypermucoviscous phenotype in hvKP strains. Different from rcsA, rcsB, and rcsC that were found in chromosome, both rmpA and rmpA2 could be carried by the plasmid or chromosome, wherein chromosomal rmpA (c-rmpA) was located in an integrative and conjugative element (ICEKp1), and plasmidic rmpA (p-rmpA) was most prevalent. Hsu et al. (2011) found that the K1 strain NTUH-K2044 contained three rmpA/rmpA2 (p-rmpA, p-rmpA2, and c-rmpA), only p-rmpA could enhance the expression of cps genes, while p-rmpA2 and c-rmpA had inhibitory and little effects, respectively. Notably, not all rmpA/rmpA2 genes can upregulate the capsule expression, and hence, rmpA/rmpA2 genes in K. pneumoniae strains lacking a hypermucoviscous phenotype are also isolated. Yu et al. (2015a;b) showed that p-rmpA/rmpA2 could occur in concurrence to frame-shift mutations in absence of c-rmpA. In addition, the process of capsule production needs an important transcription factor, RfaH, which actives the cps operon by inhibiting transcription termination (Bachman et al., 2015; Svetlov et al., 2018). The newly described regulators, kvrA and kvrB, affect virulence of K1/K2 hvKP strains due to the activation of capsule gene expression, which is not present on cKP strains. Also, the impact of KvrB on capsule expression may be dependent on its effect at the rmpA promoter in contrast to that of KvrA (Palacios et al., 2018; Walker and Miller, 2020).
The factors that influence capsule production involve various environmental stimuli in addition to the elements mentioned above. K. pneumoniae strains are facultative anaerobes growing under aerobic or anaerobic conditions, and hence, oxygen availability is crucial for their survival. Fumarate nitrate reduction regulator (FNR), an oxygen-responsive transcriptional regulator, contains a [4Fe-4S] cluster in the N-terminal sensory domain regulating the C-terminal DNA binding domain. FNR can be activated via the [4Fe-4S] cluster binding to DNA, specifically during anaerobic environment, while the [4Fe-4S] cluster is oxidized, causing the loss of FNR activity under aerobic conditions. Lin et al. (2019) found that activated FNR inhibits the transcription of rmpA/rmpA2 and decreases the amount of capsular polysaccharide during anaerobic growth. Capsular polysaccharide biosynthesis is also related to environmental iron availability. Some studies reported that the ferric uptake regulator Fur participates in the repression of RmpA, RmpA2, and RcsA that are responsible for capsule production (Lin et al., 2011). Moreover, Fur directly inhibits the transcription of non-coding RNA RyhB via binding to its promoter region, which reduces the amount of capsule independent of RmpA, RmpA2, and RcsA (Huang et al., 2012). The functional role of Fur entails regulated production of siderophores and capsules. cAMP receptor protein (CRP) decreases the expression of cps, which is a consequence of direct interaction between manC and wzi promoters (Ou et al., 2017). In response to exogenous glucose, K. pneumoniae produces capsular polysaccharide via cAMP-dependent carbon catabolite repression, which might be a probable cause of high KLA prevalence among diabetic patients (Lin et al., 2013).
The regulation of most genes mentioned is aimed at the cps cluster to affect capsule synthesis; however, the correlation between hypermucoviscosity of hvKP and capsule expression has not yet been clarified. Recently, Walker et al. (2019, 2020) identified two new genes (rmpC and rmpD) neighboring rmpA on the virulence plasmid. The ΔrmpC maintained the downregulated expression of capsule genes but preserved hypermucoviscosity, while rmpD mutants were hypermucoviscosity-negative and had no change in capsule production. As rmpA-deficient stains showed reduced cps expression and the loss of hypermucoviscosity phenotype, the author speculated that expressions of rmpD and rmpC are both driven by RmpA but regulate hypermucoviscosity phenotype and cps expression separately (Walker et al., 2019, 2020). Strikingly, hypercapsule is not a sole cause of hypermucoviscosity phenotype, and early reports have already proposed that the generation process of hypercapsule and hypermucoviscosity is different (Nassif et al., 1989a,b). The hypermucoviscosity phenotype might be attributed to the heterogeneity of capsule composition in addition to thick capsules, which described that KLA isolations contain fucose synthesized by genes wcaG and gmd, while the absence of these genes was verified in the classical strains of K. pneumoniae causing urinary tract infection (Wu et al., 2008). Pan et al. (2011) made a further exploration via disrupting the fucose synthesis gene in this KLA isolate; the results showed that the mutant displayed less mucoid than the wild-type, but the capsules were still visible. Briefly, hypercapsule is an indispensable virulence factor, but insufficient; either unknown functions of reported genes or additional regulatory factors could be the possible causes of hypermucoviscosity that require further exploration.
Siderophore
For hosts and bacteria, the metal iron is a critical element required for essential metabolic processes, and the limited availability of iron within extracellular fluid poses a difficulty for bacterial iron acquisition, rendering it a form of non-specific immune defense. If bacteria intend to survive and grow, they need to employ tactics to overcome this difficulty of iron acquisition. In human hosts, the iron ingestion is generally in need of a siderophore-dependent iron acquisition system as the result of the insolubility of free iron Fe3+ during physiological conditions. Similarly, the majority of the bacteria, including K. pneumoniae, possess this system with a higher affinity for iron than the host, which serves as a predominant strategy of acquiring iron in order to survive the competition with hosts (Miethke and Marahiel, 2007), while another solution employed by only a minority of pathogens, such as Borrelia burgdorferi and Treponema pallidum, is avoiding the need for iron by eliminating genes encoding iron-dependent proteins (Posey and Gherardini, 2000). Siderophores are small iron-binding molecules that are synthesized inside the bacteria and secreted outside the cells, following which, they bind to environmental iron and transport it back to the cells. Subsequently, iron-siderophore complexes are recognized by the specific outer-membrane receptors transporting the corresponding material to the periplasm, where siderophores combine with the periplasm proteins to transport them to the inner membrane. Finally, iron passes through the passage mediated by an ABC-transporter into the bacterial cytoplasm, where the ferric iron is reduced into ferrous iron accessible to bacteria (Brown and Holden, 2002; Khan et al., 2018). Previous studies by Rogers (1973) described that iron-binding chelators were considered as the true virulence factors in E. coli owing to their key roles in rapid bacterial growth. This hypothesis is commonly accepted and applicable to other pathogens. Previous studies have shown that hvKP presents a distinguishing 6- to 10-fold increase in siderophore production as compared to cKP (Russo et al., 2014). Four siderophores are expressed in hvKP strains, including enterobactin, yersiniabactin, salmochelin, and aerobactin; these improve the growth efficiency of the bacteria.
Among four siderophores, the conserved enterobactin contains three catechol rings that confer the highest binding affinity for Fe3+, which renders them essential and primary for both cKP and hvKP in iron uptake (Bachman et al., 2012; Palmer and Skaar, 2016). The genes required for enterobactin biosynthesis constitute the ent cluster localized on the chromosome, while its corresponding receptor is encoded by fepA (Baghal et al., 2010) (Figure 1B). In response, hosts produce the siderophore-binding protein lipocalin-2 that have antimicrobial capabilities by hampering iron acquisition rather than killing pathogens directly (Holmes et al., 2005). K. pneumoniae strains producing only enterobactin will cause them to fail owing to the presence of lipocalin-2 (Bachman et al., 2011). Smartly, a subset of K. pneumoniae strains evolves stealth siderophores to evade lipocalin-2 binding, such as the highly glycosylated enterobactin, salmochelin, and the alternative non-catecholate siderophore, yersiniabactin. In other words, the strains with salmochelin and yersiniabactin acquire iron to survive despite the presence of linpocalin-2 (Bachman et al., 2012; Holden et al., 2014). The synthesis, excretion, and uptake of salmochelin are carried out by genes of the iroA locus, iroBCDN, among which IroN and IroB are similar to the enterobactin receptor FepA and glycosyltransferases, respectively (Lin et al., 2005; Müller et al., 2009). In addition, a chromosomal copy of iro (ICEKp1) is detected in the DNA fragment in the strain NTUH-K2044 (Lin et al., 2008). Yersiniabactin, a part of Yersinia high-pathogenicity island (HPI) present in chromosomal structures ICEs, is synthesized by the ybt locus (Bach et al., 2000). ICEs are mobile genetic elements (MGEs) transmitting these virulence determinants within the bacterial populations via horizontal gene transfer. Lam et al. (2018a) analyzed 2,498 genomes and found that ybt loci were divided into 17 distinct lineages and associated with 14 distinct ICEKp structures (ICEKp1 ∼ ICEKp14). In addition to ICEKp1 of NTUH-K2044, the most common ICEs in CG23 hvKP appear to be ICEKp10, resembling the genomic island KPHPI208 in liver abscess strain 1084. A unique region located in this element is the synthesis locus of colibactin and microcin E492, which may play a role in the pathogenicity of hvKP (Struve et al., 2015). Colibactin is necessary for the full virulence of strain 1084 to develop meningitis, and microcin E492 presents its antibacterial capacity during gastrointestinal colonization by attaching to salmochelin (Lagos et al., 2001; Lu et al., 2017). Recently, a novel ICE variant named ICEKpSL1 with high homology to Yersinia HPI was detected in a hvKP strain SCsl1 isolated from the liver of infected pigs (Liu D. et al., 2019). Typically, all the four siderophore determinants can be plasmid-borne and potentially transferable. As an example, the FIBk plasmid-borne ybt is identified as a novel mechanism for ybt dispersal (Lam et al., 2018a).
In an hvKP background, the data shows that the proportion of aerobactin in the total siderophore production is about 90% and that it has the predominant contribution to hypervirulence under experimental conditions (Russo et al., 2014, 2015). A previous study demonstrated that hvKP mutants with aerobactin deficience, but not enterobactin, salmochelin, and yersiniabactin, decreased virulence in mice after pneumonic and subcutaneous infection (Russo et al., 2014, 2015). Aerobactin is expressed in >90% hvKP strains but only in 6% cKP strains, and aerobactin is more specific for hvKP as compared to enterobactin and yersiniabactin, suggesting it may be a reliable biomarker for hvKP (Paczosa and Mecsas, 2016; Russo et al., 2018). Similarly, aerobactin is more sensitive in defining hvKP than hypermucoviscosity (Zhang et al., 2016; Lin et al., 2020). The genes responsible for aerobactin biosynthesis and transport, iucABCD and iutA, are found on the virulence plasmid (Bailey et al., 2018; Russo and Gulick, 2019). The iuc locus structure in K. pneumoniae presented genetic diversity, resolving into six iuc lineages. Most of these lineages were mobilized by diverse plasmids, with the exception of iuc4 that was localized on the chromosome of K. pneumoniae subspecies rhinoscleromatis (Lam et al., 2018b). Interestingly, sequence analysis revealed that pLVPK, a plasmid initially described in the K2 strain K. pneumoniae CG43, simultaneously contains genetic loci iucABCDiutA and iroBCDN, along with rmpA encoding hypermucoviscous phenotype, which is uncommon in cKP strains (Chen et al., 2004; Tang et al., 2010; Liu and Guo, 2019). Yu et al. (2007) found that aerobactin producer was observed in 96% of rmpA gene-positive isolates after analyzing 455 K. pneumoniae bacteremia in seven countries. The phenomenon that aerobactin production is concomitant with the mucoid phenotype might imply the potential correlation between areobaction and RmpA, but needs to be substantiated with additional studies.
The analysis of the genomic sequence of hvKP strain NTUH-K2044 identified an additional iron transport system, kfu, localized on the chromosome, which is also prevalent in hvKP as compared to the cKP strains. It is also regarded as a clone-specific marker because it is not conserved in other clones except CG23 (Wyres et al., 2020). Kfu was shown to be a potential virulence factor, and murine experiments demonstrated that ΔkfuABC mutants presented lower virulence than wild-type strain (Ma et al., 2005). Interestingly, this effect only occurs in mice infected intragastrically with the kfu mutant, but not in intraperitoneal infection, which indicates that kfu might contribute to intestinal colonization. Furthermore, the discrepancy among iroA, Yersinia HPI, iucABCDiutA, and kfu is that the first three systems are TonB-dependent (Hsieh et al., 2008). Based on of these findings, the presence of these multiple and redundant iron transport systems is reasonable and complementary because they can function in different organs or microenvironmental conditions during infection.
Although iron is necessary for bacterial growth, based on the Fenton reaction, superfluous Fe2+ ions catalyze the generation of hydroxyl radicals, i.e., reactive oxygen species (ROS) oxidizing lipids and macromolecules, thereby damaging the cells (Lloyd et al., 1997). The above-mentioned Fur regulates the expression of rmpA and siderophore synthetic gene expression. Under iron-rich conditions, Fur binding to Fe2+ represses siderophore synthesis by affecting the Fur box in the promoters of genes related to iron uptake, thereby avoiding iron overload (Hahn et al., 2000). For example, in K. pneumoniae CG43, Fur could bind to the Fur box sequences in the coding region of iucA, iroB, and entC to play its regulatory role, as described by Lin et al. (2011). The promoter sequences of kfu operon also contain the conserved Fur box (Ma et al., 2005). Emerging evidence supports that Fur acts as a transcriptional activator, increasing the expression of active enzymes against toxic ROS. Thus, Fur contributes to virulence, and fur mutants exhibit a decrease in virulence and inability to cause disease within the animal host (Troxell and Hassan, 2013).
Other Virulence Factors
Lipopolysaccharide consists of lipid A, oligosaccharide core, and O antigen and is known as the endotoxin of all Gram-negative bacteria, both cKP and hvKP. Presently, it is unclear whether LPS produced by hvKP strains has a unique role in hypervirulence. As the outermost subunit of LPS, O-antigen is the first molecule to be encountered by the host’s innate immune system and protect the pathogens against complement-mediated killing. Specifically, the O antigen binds a complement component C3b involving pore-formation to prevent drilling of the bacterial membrane (Shankar-Sinha et al., 2004). The number of O serotypes is estimated to be eight, and O1 antigen is the most common among clinical K. pneumoniae strains (Hansen et al., 1999; Follador et al., 2016). Previous studies have shown that O-antigen in O1:K2 cKP might contribute to bacterial virulence and lethality by weakening macrophage activation and promoting bacteremia (Lugo et al., 2007). Hsieh et al. (2012) demonstrated that the prevalence of O1 serotype in KLA isolates is higher than that in non-tissue-invasive isolates, and KLA strains with defect O1 antigen were less virulent than parental strains. However, its function has not yet been embodied in K1 hvKP strains, in which O1 antigen is masked by K1 capsule (Merino et al., 1992; Hsieh et al., 2012).
The types of fimbriae characterized experimentally for K. pneumoniae are type 1 and type 3 fimbriae. In the genome of hvKP NTUH-2044, seven novel fimbrial gene clusters, kpa, kpb, kpc, kpd, kpe, kpf, and kfg, were identified, and the Kpc fimbriae are highly associated with K1 serotype hvKP (Wu et al., 2010). Type 3 fimbriae constitute well-known bacterial virulence factors, mediating enhanced biofilm formation on abiotic surfaces, however, only little is known about their role in hvKP strains (Schroll et al., 2010). Using K. pneumoniae CG43, Wu et al. (2012) proved that type 3 fimbriae activity and biofilm formation could be influenced by iron availability and positively controlled by Fur, suggesting that type 3 fimbriae might be less important for hvKP infection in vivo under the condition of limited iron (Shon et al., 2013). A recent study also reported that hypermucoviscous isolates with serotype K1, despite harboring mrkD and fimH genes encoding type 3 and type 1 fimbriae, respectively, presented low initial adhesion because the presence of hypercapsule could hide these fimbriae (Cubero et al., 2019). Another adhesive structure, a non-fimbrial protein CF29K, is prevalent in hvKP strains, indicating its involvement in KLA pathogenesis or association with other pathogenicity factors in the same plasmid (Darfeuille-Michaud et al., 1992; Brisse et al., 2009).
Several outer membrane proteins (OMPs), including OmpA, peptidoglycan-associated lipoprotein (Pal), and murein lipoprotein (LppA), contribute to K. pneumoniae virulence. In response to intraperitoneal infection with ompA-, pal-, and lppA-deficient NTUH-20444, only the pal and lppA mutants yielded high LD50 values (50% lethal dose) than wild type strains, meaning lower virulence. The mechanisms behind the decreased virulence were likely to be that these mutants were easily killed by serum and neutrophil. Remarkably, pal and lppA mutants with low virulence retained intact K1 and O1 antigen inducing antibodies production and mice immunization, which indicated that these mutant strains could be meaningful for vaccine development (Hsieh et al., 2013). Similarly, K. pneumoniae also produces two major porins, OmpK35 and OmpK36. The downregulation of these molecules could be ascribed to antibiotic resistance and virulence sacrifice of the bacteria. In a mouse peritonitis model, the loss of both ompK35 and ompK36 in KLA isolate NVT2001 significantly reduced the pathogenicity and growth rate, thereby leading its elimination from the immune system (Tsai et al., 2011). This phenomenon is consistent with a previous report that disruption of ompK36 failed to colonize the liver persistently due to the rapid clearance of bacterial burden (Chen et al., 2010). Srinivasan et al. (2012) found a novel porin in K. pneumoniae NTUH-2044, termed as KpnO. With respect to virulence, ΔkpnO produced lesser capsular polysaccharide and killed Caenorhabditis elegans slowly than wild-type strains. However, mechanisms underlying the attenuated virulence due to absence of these OMPs are yet to be elucidated.
In recent years, a new factor associated with virulence, type 6 secretion system (T6SS), has been identified in hvKP strains by in silico analysis. T6SS functions as a nanosyringe that injects effector proteins into target cells to destroy eukaryotic cells or contend against other prokaryotic cells (Sarris et al., 2011). Although >25% Gram-negative bacteria encode T6SSs, wild-type NTUH-2044 has been confirmed to outcompete Salmonella Typhimurium, E. coli, and T6SS-null K. pneumoniae in a T6SS-dependent manner, and the frequency of T6SSs in KLA strains is significantly higher compared to that in the intestinal-colonizing strains, suggesting that T6SSs may confer an advantage for hvKP survival (Clemens et al., 2018; Hsieh et al., 2019). Another study reported that in bacterial competition, K. pneumoniae T6SSs were activated by the PhoPQ system, followed by the use of effectors to kill E. coli prey that lack T6SS immunity genes (Storey et al., 2020). Moreover, the overexpression of a T6SS effector in the carbapenemase-producing K. pneumoniae strain, the phospholipase Tle1, was found to inhibit the growth of E. coli (Liu et al., 2017). Furthermore, in a hypervirulent strain Kp52.145, another T6SS effector gene pld1, encoding a phospholipase D family protein (PLD1), located within the T6SS locus, was reported as a virulence factor, exhibiting that Δpld1 was avirulent in comparison to wild-type in a mouse model of pneumonia (Lery et al., 2014). The previously sequenced genome showed that T6SS genes are clustered in two contiguous loci, while Barbosa and Lery (2019) later analyzed the neighboring genes and found that the genes coding for iron uptake systems is adjacent to T6SS-related genes: tssD in Kp52.145. Thus, it was hypothesized that T6SS is involved in iron import in hvKP (Barbosa and Lery, 2019).
Convergence of Hypervirulence and Multidrug Resistance in K. pneumoniae
Increasing reports have described the emergence of multidrug-resistant hypervirulent K. pneumoniae strains (Hao et al., 2020; Shankar et al., 2020). MDR-hvKP can arise by two different mechanisms. In one case, hvKP strains can acquire antimicrobial resistance genes or plasmids through horizontal transfer, therefore becoming MDR-hvKP, which was tentatively designated as type I MDR-hvKP. For example, two carbapenemase plasmids, blaNDM–1-bearing IncN plasmid and blaKPC-2-carrying IncFIIK plasmid, were recovered together from a K2 ST86 MDR-hvKP strain (Liu Y. et al., 2019). Additionally, MDR-hvKP strains can also arise from the transfer of a pLVPK-like virulence plasmid into a classic MDR K. pneumoniae strain, named as type II MDR-hvKP. As an example, a recent study in China described a fatal outbreak caused by a KPC-producing ST11 strain acquiring a pLVPK-like virulence plasmid (Gu et al., 2018). Type I MDR-hvKP strains share the same virulence determinants as the hvKP strains described above, while the hypervirulence in type II MDR-hvKP is mainly attributed to the over production of capsule and siderophore, as a result of the acquisition of plasmid-mediated virulence determinants, including rmpA/rmpA2, iut and iro. A recent study validated that five biomarkers (peg344, iroB, iucA, prmpA, and prmpA2), and ≥30 μg/mL siderophore production could accurately identify hvKP (Russo et al., 2018). The gene peg-344 present on the virulence plasmid in hvKP including both type I and type II MDR-hvKP strains, had potential application in rapid diagnosis of hvKP, and a peg-344 loop-mediated isothermal amplification technology had recently been established (Liao et al., 2020). PEG344 acted as a transporter in the inner membrane, but its specific role in virulence is unclear. Interestingly, Bulger et al. (2017) found that PEG344 was required for maximal virulence in pulmonary challenge model, but not in subcutaneous challenge.
Summary
Following the early reports of hvKP in Asia, hvKP strains have spread globally, causing both community associated and hospital associated infections. The convergence of hypervirulence and antimicrobial resistance in hvKp further exacerbates this public crisis, and challenges the clinical treatment options for hvKp infections. Epidemiologically, the source and route of hvKp infection remain poorly understood, but intestinal colonization appears to be a critical step prior to infection, while susceptible factors include diabetes mellitus, male gender, and Asian ethnicity. Capsule and siderophores are predominant virulence factors that play a major role in the hypermucoviscosity phenotype of hvKP. However, molecular mechanisms under hypermucoviscosity remain to be explored as hypervirulent strains without hypermucoviscosity phenotype are frequently identified. The current definition of hvKP considers clinical characteristics, phenotypes, and genotypes, however, microbiologically determination of hvKp strains in clinical lavatories remains to be a challenge. Additional virulence factors include LPS, fimbriae, outer membrane proteins, and T6SS, whose functions in hvKP need to be further explored (Table 2). These factors are coordinated and regulated when bacteria orchestrate a series of events causing infection. Moreover, with the emergency of MDR-hvKP, the development of non-antimicrobial therapy targeting virulence factors is indispensable. Taken together, further studies on hvKp are needed to improve the prevention, diagnosis, and treatment of hvKP caused infections.
Author Contributions
JZ drafted the manuscript. TW, LC, and HD revised the manuscript critically for important intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Gusu Key Health Talent of Suzhou, the Jiangsu Youth Medical Talents Program (QN-867), Science Foundation of Jiangsu Province Health Department (ZDB2020014), the Science and Technology Program of Suzhou (SZS201715), and the Discipline Construction Program of the Second Affiliated Hospital of Soochow University (XKTJ-TD202001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alvarez, D., Merino, S., Tomás, J. M., Benedí, V. J., and Albertí, S. (2000). Capsular polysaccharide is a major complement resistance factor in lipopolysaccharide O side chain-deficient Klebsiella pneumoniae clinical isolates. Infect. Immun. 68, 953–955. doi: 10.1128/iai.68.2.953-955.2000
Arakawa, Y., Ohta, M., Wacharotayankun, R., Mori, M., Kido, N., Ito, H., et al. (1991). Biosynthesis of Klebsiella K2 capsular polysaccharide in Escherichia coli HB101 requires the functions of rmpA and the chromosomal cps gene cluster of the virulent strain Klebsiella pneumoniae Chedid (O1:K2). Infect. Immun. 59, 2043–2050. doi: 10.1128/iai.59.6.2043-2050.1991
Athamna, A., Ofek, I., Keisari, Y., Markowitz, S., Dutton, G. G., and Sharon, N. (1991). Lectinophagocytosis of encapsulated Klebsiella pneumoniae mediated by surface lectins of guinea pig alveolar macrophages and human monocyte-derived macrophages. Infect. Immun. 59, 1673–1682. doi: 10.1128/iai.59.5.1673-1682.1991
Bach, S., De Almeida, A., and Carniel, E. (2000). The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol. Lett. 183, 289–294. doi: 10.1111/j.1574-6968.2000.tb08973.x
Bachman, M. A., Breen, P., Deornellas, V., Mu, Q., Zhao, L., Wu, W., et al. (2015). Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. mBio 6:e00775.
Bachman, M. A., Lenio, S., Schmidt, L., Oyler, J. E., and Weiser, J. N. (2012). Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. mBio 3:e00224-11.
Bachman, M. A., Oyler, J. E., Burns, S. H., Caza, M., Lépine, F., Dozois, C. M., et al. (2011). Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 79, 3309–3316. doi: 10.1128/iai.05114-11
Baghal, S. M. L., Gargari, S. L. M., and Rasooli, I. (2010). Production and immunogenicity of recombinant ferric enterobactin protein (FepA). Int. J. Infect. Dis. 14, (Suppl. 3), e166–e170.
Bagley, S. T. (1985). Habitat association of Klebsiella species. Infect. Control IC 6, 52–58. doi: 10.1017/s0195941700062603
Bailey, D. C., Alexander, E., Rice, M. R., Drake, E. J., Mydy, L. S., Aldrich, C. C., et al. (2018). Structural and functional delineation of aerobactin biosynthesis in hypervirulent. J. Biol. Chem. 293, 7841–7852. doi: 10.1074/jbc.ra118.002798
Barbosa, V. A. A., and Lery, L. M. S. (2019). Insights into Klebsiella pneumoniae type VI secretion system transcriptional regulation. BMC Genomics 20:506.
Brisse, S., Fevre, C., Passet, V., Issenhuth-Jeanjean, S., Tournebize, R., Diancourt, L., et al. (2009). Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. doi: 10.1371/journal.pone.0004982
Brisse, S., Passet, V., Haugaard, A. B., Babosan, A., Kassis-Chikhani, N., Struve, C., et al. (2013). wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J. Clin. Microbiol. 51, 4073–4078. doi: 10.1128/jcm.01924-13
Brown, J. S., and Holden, D. W. (2002). Iron acquisition by Gram-positive bacterial pathogens. Microbes Infect. 4, 1149–1156. doi: 10.1016/s1286-4579(02)01640-4
Bulger, J., Macdonald, U., Olson, R., Beanan, J., and Russo, T. A. (2017). Metabolite transporter PEG344 is required for full virulence of hypervirulent Klebsiella pneumoniae Strain hvKP1 after pulmonary but not subcutaneous challenge. Infect. Immun. 85:e00093-17.
Bushell, S. R., Mainprize, I. L., Wear, M. A., Lou, H., Whitfield, C., and Naismith, J. H. (2013). Wzi is an outer membrane lectin that underpins group 1 capsule assembly in Escherichia coli. Structure 21, 844–853. doi: 10.1016/j.str.2013.03.010
Catalán-Nájera, J. C., Garza-Ramos, U., and Barrios-Camacho, H. (2017). Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence 8, 1111–1123. doi: 10.1080/21505594.2017.1317412
Chen, J.-H., Siu, L. K., Fung, C.-P., Lin, J.-C., Yeh, K.-M., Chen, T.-L., et al. (2010). Contribution of outer membrane protein K36 to antimicrobial resistance and virulence in Klebsiella pneumoniae. J. Antimicrob. Chemother. 65, 986–990. doi: 10.1093/jac/dkq056
Chen, Y.-T., Chang, H.-Y., Lai, Y.-C., Pan, C.-C., Tsai, S.-F., and Peng, H.-L. (2004). Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337, 189–198. doi: 10.1016/j.gene.2004.05.008
Cheng, H. Y., Chen, Y. S., Wu, C. Y., Chang, H. Y., Lai, Y. C., and Peng, H. L. (2010). RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 192, 3144–3158. doi: 10.1128/jb.00031-10
Choby, J. E., Howard-Anderson, J., and Weiss, D. S. (2020). Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J. Intern. Med. 287, 283–300.
Chuang, Y.-P., Fang, C.-T., Lai, S.-Y., Chang, S.-C., and Wang, J.-T. (2006). Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 193, 645–654. doi: 10.1086/499968
Chung, D. R., Lee, S. S., Lee, H. R., Kim, H. B., Choi, H. J., Eom, J. S., et al. (2007). Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J. Infect. 54, 578–583. doi: 10.1016/j.jinf.2006.11.008
Clemens, D. L., Lee, B.-Y., and Horwitz, M. A. (2018). The Francisella type VI secretion system. Front. Cell. Infect. Microbiol. 8:121. doi: 10.3389/fcimb.2018.00121
Coutinho, R. L., Visconde, M. F., Descio, F. J., Nicoletti, A. G., Pinto, F. C., Silva, A. C., et al. (2014). Community-acquired invasive liver abscess syndrome caused by a K1 serotype Klebsiella pneumoniae isolate in Brazil: a case report of hypervirulent ST23. Mem. Inst. Oswaldo Cruz 109, 970–971. doi: 10.1590/0074-0276140196
Cubero, M., Grau, I., Tubau, F., Pallarés, R., Dominguez, M. A., Liñares, J., et al. (2016). Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013). Clin. Microbiol. Infect. 22, 154–160. doi: 10.1016/j.cmi.2015.09.025
Cubero, M., Marti, S., Domínguez, M., González-Díaz, A., Berbel, D., and Ardanuy, C. (2019). Hypervirulent Klebsiella pneumoniae serotype K1 clinical isolates form robust biofilms at the air-liquid interface. PLoS One 14:e0222628. doi: 10.1371/journal.pone.0222628
Darfeuille-Michaud, A., Jallat, C., Aubel, D., Sirot, D., Rich, C., Sirot, J., et al. (1992). R-plasmid-encoded adhesive factor in Klebsiella pneumoniae strains responsible for human nosocomial infections. Infect. Immun. 60, 44–55. doi: 10.1128/iai.60.1.44-55.1992
Domenico, P., Salo, R. J., Cross, A. S., and Cunha, B. A. (1994). Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect. Immun. 62, 4495–4499. doi: 10.1128/iai.62.10.4495-4499.1994
Ernst, C. M., Braxton, J. R., Rodriguez-Osorio, C. A., Zagieboylo, A. P., Li, L., Pironti, A., et al. (2020). Adaptive evolution of virulence and persistence in carbapenem-resistant Klebsiella pneumoniae. Nat. Med. 26, 705–711. doi: 10.1038/s41591-020-0825-4
Fang, C.-T., Chuang, Y.-P., Shun, C.-T., Chang, S.-C., and Wang, J.-T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705. doi: 10.1084/jem.20030857
Fang, C.-T., Lai, S.-Y., Yi, W.-C., Hsueh, P.-R., and Liu, K.-L. (2010). The function of wzy_K1 (magA), the serotype K1 polymerase gene in Klebsiella pneumoniae cps gene cluster. J. Infect. Dis. 201, 1268–1269. doi: 10.1086/652183
Fazili, T., Sharngoe, C., Endy, T., Kiska, D., Javaid, W., and Polhemus, M. (2016). Klebsiella pneumoniae liver abscess: an emerging disease. Am. J. Med. Sci. 351, 297–304.
Follador, R., Heinz, E., Wyres, K. L., Ellington, M. J., Kowarik, M., Holt, K. E., et al. (2016). The diversity of Klebsiella pneumoniae surface polysaccharides. Microb. Genom. 2:e000073.
Foo, J. M., Chandra, J., and Fung, A. T. (2018). Rising trends of endogenous Klebsiella pneumoniae endophthalmitis in Australia: comment. Clin. Exp. Ophthalmol. 46, 575–576. doi: 10.1111/ceo.13118
Fung, C. P., Chang, F. Y., Lee, S. C., Hu, B. S., Kuo, B. I. T., Liu, C. Y., et al. (2002). A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50, 420–424. doi: 10.1136/gut.50.3.420
Fung, C.-P., Lin, Y.-T., Lin, J.-C., Chen, T.-L., Yeh, K.-M., Chang, F.-Y., et al. (2012). Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg. Infect. Dis. 18, 1322–1325.
Furlan, J. P. R., Savazzi, E. A., and Stehling, E. G. (2020). Genomic insights into multidrug-resistant and hypervirulent Klebsiella pneumoniae co-harboring metal resistance genes in aquatic environments. Ecotoxicol. Environ. Saf. 201:110782. doi: 10.1016/j.ecoenv.2020.110782
Gorrie, C. L., Mirceta, M., Wick, R. R., Edwards, D. J., Thomson, N. R., Strugnell, R. A., et al. (2017). Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 65, 208–215. doi: 10.1093/cid/cix270
Gottesman, S., and Stout, V. (1991). Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol. Microbiol. 5, 1599–1606. doi: 10.1111/j.1365-2958.1991.tb01906.x
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/s1473-3099(17)30489-9
Guo, X.-P., and Sun, Y.-C. (2017). New insights into the non-orthodox two component rcs phosphorelay system. Front. Microbiol. 8:2014. doi: 10.3389/fmicb.2017.02014
Hahn, J. S., Oh, S. Y., and Roe, J. H. (2000). Regulation of the furA and catC operon, encoding a ferric uptake regulator homologue and catalase-peroxidase, respectively, in Streptomyces coelicolor A3(2). J. Bacteriol. 182, 3767–3774. doi: 10.1128/jb.182.13.3767-3774.2000
Hansen, D. S., Mestre, F., Alberti, S., Hernández-Allés, S., Alvarez, D., Doménech-Sánchez, A., et al. (1999). Klebsiella pneumoniae lipopolysaccharide O typing: revision of prototype strains and O-group distribution among clinical isolates from different sources and countries. J. Clin. Microbiol. 37, 56–62. doi: 10.1128/jcm.37.1.56-62.1999
Hao, M., Shi, X., Lv, J., Niu, S., Cheng, S., Du, H., et al. (2020). In vitro activity of apramycin against carbapenem-resistant and hypervirulent Klebsiella pneumoniae isolates. Front. Microbiol. 11:425. doi: 10.3389/fmicb.2020.00425
Harada, S., Aoki, K., Yamamoto, S., Ishii, Y., Sekiya, N., Kurai, H., et al. (2019). Clinical and molecular characteristics of Klebsiella pneumoniae isolates causing bloodstream infections in Japan: occurrence of hypervirulent infections in health care. J. Clin. Microbiol. 57:e01206-19.
Harada, S., Tateda, K., Mitsui, H., Hattori, Y., Okubo, M., Kimura, S., et al. (2011). Familial spread of a virulent clone of Klebsiella pneumoniae causing primary liver abscess. J. Clin. Microbiol. 49, 2354–2356. doi: 10.1128/jcm.00034-11
Holden, V. I., Lenio, S., Kuick, R., Ramakrishnan, S. K., Shah, Y. M., and Bachman, M. A. (2014). Bacterial siderophores that evade or overwhelm lipocalin 2 induce hypoxia inducible factor 1α and proinflammatory cytokine secretion in cultured respiratory epithelial cells. Infect. Immun. 82, 3826–3836. doi: 10.1128/iai.01849-14
Holmes, M. A., Paulsene, W., Jide, X., Ratledge, C., and Strong, R. K. (2005). Siderocalin (Lcn 2) also binds carboxymycobactins, potentially defending against mycobacterial infections through iron sequestration. Structure 13, 29–41. doi: 10.1016/j.str.2004.10.009
Hsieh, P.-F., Lin, T.-L., Lee, C.-Z., Tsai, S.-F., and Wang, J.-T. (2008). Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 197, 1717–1727. doi: 10.1086/588383
Hsieh, P.-F., Lin, T.-L., Yang, F.-L., Wu, M.-C., Pan, Y.-J., Wu, S.-H., et al. (2012). Lipopolysaccharide O1 antigen contributes to the virulence in Klebsiella pneumoniae causing pyogenic liver abscess. PLoS One 7:e33155. doi: 10.1371/journal.pone.0033155
Hsieh, P.-F., Liu, J.-Y., Pan, Y.-J., Wu, M.-C., Lin, T.-L., Huang, Y.-T., et al. (2013). Klebsiella pneumoniae peptidoglycan-associated lipoprotein and murein lipoprotein contribute to serum resistance, antiphagocytosis, and proinflammatory cytokine stimulation. J. Infect. Dis. 208, 1580–1589. doi: 10.1093/infdis/jit384
Hsieh, P.-F., Lu, Y.-R., Lin, T.-L., Lai, L.-Y., and Wang, J.-T. (2019). Klebsiella pneumoniae Type VI secretion system contributes to bacterial competition, cell invasion, type-1 fimbriae expression, and in vivo colonization. J. infect. Dis. 219, 637–647. doi: 10.1093/infdis/jiy534
Hsu, C.-R., Lin, T.-L., Chen, Y.-C., Chou, H.-C., and Wang, J.-T. (2011). The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 157, 3446–3457. doi: 10.1099/mic.0.050336-0
Huang, S.-H., Wang, C.-K., Peng, H.-L., Wu, C.-C., Chen, Y.-T., Hong, Y.-M., et al. (2012). Role of the small RNA RyhB in the Fur regulon in mediating the capsular polysaccharide biosynthesis and iron acquisition systems in Klebsiella pneumoniae. BMC Microbiol. 12:148. doi: 10.1186/1471-2180-12-148
Ikeda, M., Mizoguchi, M., Oshida, Y., Tatsuno, K., Saito, R., Okazaki, M., et al. (2018). Clinical and microbiological characteristics and occurrence of infection in Japan. Int. J. Gen. Med. 11, 293–299. doi: 10.2147/ijgm.s166940
Islam, S. T., and Lam, J. S. (2014). Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can. J. Microbiol. 60, 697–716. doi: 10.1139/cjm-2014-0595
Jin, L., Liu, Y., Jing, C., Wang, R., Wang, Q., and Wang, H. (2020). Neutrophil extracellular traps (NETs)-mediated killing of carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP) are impaired in patients with diabetes mellitus. Virulence 11, 1122–1130. doi: 10.1080/21505594.2020.1809325
Khan, A., Singh, P., and Srivastava, A. (2018). Synthesis, nature and utility of universal iron chelator - siderophore: a review. Microbiol. Res. 212-213, 103–111. doi: 10.1016/j.micres.2017.10.012
Ku, Y. H., Chuang, Y. C., Chen, C. C., Lee, M. F., Yang, Y. C., Tang, H. J., et al. (2017). Klebsiella pneumoniae Isolates from meningitis: epidemiology, virulence and antibiotic resistance. Sci. Rep. 7:6634.
Lagos, R., Baeza, M., Corsini, G., Hetz, C., Strahsburger, E., Castillo, J. A., et al. (2001). Structure, organization and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol. Microbiol. 42, 229–243. doi: 10.1046/j.1365-2958.2001.02630.x
Lai, Y.-C., Peng, H.-L., and Chang, H.-Y. (2003). RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J. Bacteriol. 185, 788–800. doi: 10.1128/jb.185.3.788-800.2003
Lam, M. M. C., Wick, R. R., Wyres, K. L., Gorrie, C. L., Judd, L. M., Jenney, A. W. J., et al. (2018a). Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb. Genom. 4:e000196.
Lam, M. M. C., Wyres, K. L., Judd, L. M., Wick, R. R., Jenney, A., Brisse, S., et al. (2018b). Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 10:77.
Lan, P., Shi, Q., Zhang, P., Chen, Y., Yan, R., Hua, X., et al. (2020). Core genome allelic profiles of clinical Klebsiella pneumoniae strains using a random forest algorithm based on multilocus sequence typing scheme for hypervirulence analysis. J. Infect. Dis. 221, S263–S271.
Lawlor, M. S., Hsu, J., Rick, P. D., and Miller, V. L. (2005). Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol. Microbiol. 58, 1054–1073. doi: 10.1111/j.1365-2958.2005.04918.x
Lederman, E. R., and Crum, N. F. (2005). Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am. J. Gastroenterol. 100, 322–331. doi: 10.1111/j.1572-0241.2005.40310.x
Lee, C.-H., Chang, C.-C., Liu, J.-W., Chen, R.-F., and Yang, K. D. (2014). Sialic acid involved in hypermucoviscosity phenotype of Klebsiella pneumoniae and associated with resistance to neutrophil phagocytosis. Virulence 5, 673–679. doi: 10.4161/viru.32076
Lee, C. H., Chuah, S. K., Chang, C. C., and Chen, F. J. (2020). The surface protein fructose-1, 6 bisphosphate aldolase of Klebsiella pneumoniae serotype K1: role of interaction with Neutrophils. Pathogens 9:1009. doi: 10.3390/pathogens9121009
Lee, I. R., Molton, J. S., Wyres, K. L., Gorrie, C., Wong, J., Hoh, C. H., et al. (2016). Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci. Rep. 6:29316.
Lery, L. M. S., Frangeul, L., Tomas, A., Passet, V., Almeida, A. S., Bialek-Davenet, S., et al. (2014). Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 12:41. doi: 10.1186/1741-7007-12-41
Li, G., Shi, J., Zhao, Y., Xie, Y., Tang, Y., Jiang, X., et al. (2020). Identification of hypervirulent Klebsiella pneumoniae isolates using the string test in combination with Galleria mellonella infectivity. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1673–1679. doi: 10.1007/s10096-020-03890-z
Li, W., Sun, G., Yu, Y., Li, N., Chen, M., Jin, R., et al. (2014). Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin. Infect. Dis. 58, 225–232. doi: 10.1093/cid/cit675
Liao, C. H., Huang, Y. T., Chang, C. Y., Hsu, H. S., and Hsueh, P. R. (2014). Capsular serotypes and multilocus sequence types of bacteremic Klebsiella pneumoniae isolates associated with different types of infections. Eur. J. Clin. Microbiol. Infect. Dis. 33, 365–369. doi: 10.1007/s10096-013-1964-z
Liao, W., Long, D., Huang, Q., Wei, D., Liu, X., Wan, L., et al. (2020). Rapid detection to differentiate hypervirulent Klebsiella pneumoniae (hvKp) from classical K. pneumoniae by Identifying peg-344 With loop-mediated isothermal amplication (LAMP). Front. Microbiol. 11:1189. doi: 10.3389/fmicb.2020.01189
Lin, C.-T., Chen, Y.-C., Jinn, T.-R., Wu, C.-C., Hong, Y.-M., and Wu, W.-H. (2013). Role of the cAMP-dependent carbon catabolite repression in capsular polysaccharide biosynthesis in Klebsiella pneumoniae. PLoS One 8:e54430. doi: 10.1371/journal.pone.0054430
Lin, C.-T., Wu, C.-C., Chen, Y.-S., Lai, Y.-C., Chi, C., Lin, J.-C., et al. (2011). Fur regulation of the capsular polysaccharide biosynthesis and iron-acquisition systems in Klebsiella pneumoniae CG43. Microbiology 157, 419–429. doi: 10.1099/mic.0.044065-0
Lin, H., Fischbach, M. A., Liu, D. R., and Walsh, C. T. (2005). In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 127, 11075–11084. doi: 10.1021/ja0522027
Lin, T.-H., Wu, C.-C., Kuo, J.-T., Chu, H.-F., Lee, D.-Y., and Lin, C.-T. (2019). FNR-dependent RmpA and RmpA2 regulation of capsule polysaccharide biosynthesis in Klebsiella pneumoniae. Front. Microbiol. 10:2436. doi: 10.3389/fmicb.2019.02436
Lin, T.-L., Lee, C.-Z., Hsieh, P.-F., Tsai, S.-F., and Wang, J.-T. (2008). Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J. Bacteriol. 190, 515–526. doi: 10.1128/jb.01219-07
Lin, Y.-T., Siu, L. K., Lin, J.-C., Chen, T.-L., Tseng, C.-P., Yeh, K.-M., et al. (2012). Seroepidemiology of Klebsiella pneumoniae colonizing the intestinal tract of healthy Chinese and overseas Chinese adults in Asian countries. BMC Microbiol. 12:13. doi: 10.1186/1471-2180-12-13
Lin, Z.-W., Zheng, J.-X., Bai, B., Xu, G.-J., Lin, F.-J., Chen, Z., et al. (2020). Characteristics of hypervirulent : does low expression of contribute to the absence of hypervirulence? Front. Microbiol. 11:436. doi: 10.3389/fmicb.2020.00436
Liu, B.-T., Zhang, X.-Y., Wan, S.-W., Hao, J.-J., Jiang, R.-D., and Song, F.-J. (2018). Characteristics of carbapenem-resistant in ready-to-eat vegetables in China. Front. Microbiol. 9:1147. doi: 10.3389/fmicb.2018.01147
Liu, C., Du, P., Xiao, N., Ji, F., Russo, T. A., and Guo, J. (2020). Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Beijing, China. Virulence 11, 1215–1224. doi: 10.1080/21505594.2020.1809322
Liu, C., and Guo, J. (2019). Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann. Clin. Microbiol. Antimicrob. 18:4.
Liu, D., Yang, Y., Gu, J., Tuo, H., Li, P., Xie, X., et al. (2019). The Yersinia high-pathogenicity island (HPI) carried by a new integrative and conjugative element (ICE) in a multidrug-resistant and hypervirulent Klebsiella pneumoniae strain SCsl1. Vet. Microbiol. 239:108481. doi: 10.1016/j.vetmic.2019.108481
Liu, L., Ye, M., Li, X., Li, J., Deng, Z., Yao, Y.-F., et al. (2017). Identification and characterization of an antibacterial type VI secretion system in the carbapenem-resistant strain HS11286. Front. Cell. Infect. Microbiol. 7:442. doi: 10.3389/fcimb.2017.00442
Liu, Y., Long, D., Xiang, T. X., Du, F. L., Wei, D. D., Wan, L. G., et al. (2019). Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J. Antimicrob. Chemother. 74, 1233–1240. doi: 10.1093/jac/dkz023
Liu, Y. C., Cheng, D. L., and Lin, C. L. (1986). Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146, 1913–1916. doi: 10.1001/archinte.146.10.1913
Llobet, E., Campos, M. A., Giménez, P., Moranta, D., and Bengoechea, J. A. (2011). Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect. Immun. 79, 3718–3732. doi: 10.1128/iai.05226-11
Lloyd, R. V., Hanna, P. M., and Mason, R. P. (1997). The origin of the hydroxyl radical oxygen in the Fenton reaction. Free Radic. Biol. Med. 22, 885–888. doi: 10.1016/s0891-5849(96)00432-7
Lu, M. C., Chen, Y. T., Chiang, M. K., Wang, Y. C., Hsiao, P. Y., Huang, Y. J., et al. (2017). Colibactin contributes to the hypervirulence of pks(+) K1 CC23 Klebsiella pneumoniae in mouse meningitis infections. Front. Cell Infect. Microbiol. 7:103. doi: 10.3389/fcimb.2017.00103
Lugo, J. Z., Price, S., Miller, J. E., Ben-David, I., Merrill, V. A., Merrill, V. J., et al. (2007). Lipopolysaccharide O-antigen promotes persistent murine bacteremia. Shock 27, 186–191. doi: 10.1097/01.shk.0000238058.23837.21
Ma, L.-C., Fang, C.-T., Lee, C.-Z., Shun, C.-T., and Wang, J.-T. (2005). Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J. Infect. Dis. 192, 117–128. doi: 10.1086/430619
Merino, S., Camprubí, S., Albertí, S., Benedí, V. J., and Tomás, J. M. (1992). Mechanisms of Klebsiella pneumoniae resistance to complement-mediated killing. Infect. Immun. 60, 2529–2535. doi: 10.1128/iai.60.6.2529-2535.1992
Miethke, M., and Marahiel, M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. MMBR 71, 413–451. doi: 10.1128/mmbr.00012-07
Moore, R., O’shea, D., Geoghegan, T., Mallon, P. W. G., and Sheehan, G. (2013). Community-acquired Klebsiella pneumoniae liver abscess: an emerging infection in Ireland and Europe. Infection 41, 681–686. doi: 10.1007/s15010-013-0408-0
Müller, S. I., Valdebenito, M., and Hantke, K. (2009). Salmochelin, the long-overlooked catecholate siderophore of Salmonella. Biometals 22, 691–695. doi: 10.1007/s10534-009-9217-4
Nassif, X., Fournier, J. M., Arondel, J., and Sansonetti, P. J. (1989a). Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect. Immun. 57, 546–552. doi: 10.1128/iai.57.2.546-552.1989
Nassif, X., Honoré, N., Vasselon, T., Cole, S. T., and Sansonetti, P. J. (1989b). Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol. Microbiol. 3, 1349–1359. doi: 10.1111/j.1365-2958.1989.tb00116.x
Nassif, X., and Sansonetti, P. J. (1986). Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 54, 603–608. doi: 10.1128/iai.54.3.603-608.1986
Niu, T., Guo, L., Luo, Q., Zhou, K., Yu, W., Chen, Y., et al. (2020). Wza gene knockout decreases virulence and affects Wzy-dependent capsular polysaccharide synthesis. Virulence 11, 1–13. doi: 10.1007/978-3-319-23534-9_1
Opal, S. M. (2014). Significance of sialic acid in Klebsiella pneumoniae K1 capsules. Virulence 5, 648–649. doi: 10.4161/viru.34349
Ou, Q., Fan, J., Duan, D., Xu, L., Wang, J., Zhou, D., et al. (2017). Involvement of cAMP receptor protein in biofilm formation, fimbria production, capsular polysaccharide biosynthesis and lethality in mouse of Klebsiella pneumoniae serotype K1 causing pyogenic liver abscess. J. Med. Microbiol. 66, 1–7. doi: 10.1099/jmm.0.000391
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. MMBR 80, 629–661. doi: 10.1128/mmbr.00078-15
Palacios, M., Miner, T. A., Frederick, D. R., Sepulveda, V. E., Quinn, J. D., Walker, K. A., et al. (2018). Identification of two regulators of virulence that are conserved in classical and hypervirulent strains. mBio 9:e01443-18.
Palmer, L. D., and Skaar, E. P. (2016). Transition metals and virulence in bacteria. Annu. Rev. Genet. 50, 67–91. doi: 10.1146/annurev-genet-120215-035146
Pan, P.-C., Chen, H.-W., Wu, P.-K., Wu, Y.-Y., Lin, C.-H., and Wu, J. H. (2011). Mutation in fucose synthesis gene of Klebsiella pneumoniae affects capsule composition and virulence in mice. Exp. Biol. Med. 236, 219–226. doi: 10.1258/ebm.2010.010193
Pan, Y.-J., Fang, H.-C., Yang, H.-C., Lin, T.-L., Hsieh, P.-F., Tsai, F.-C., et al. (2008). Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J. Clin. Microbiol. 46, 2231–2240. doi: 10.1128/jcm.01716-07
Pan, Y.-J., Lin, T.-L., Chen, C.-T., Chen, Y.-Y., Hsieh, P.-F., Hsu, C.-R., et al. (2015). Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 5:15573.
Pan, Y.-J., Lin, T.-L., Chen, Y.-H., Hsu, C.-R., Hsieh, P.-F., Wu, M.-C., et al. (2013). Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PLoS One 8:e80670. doi: 10.1371/journal.pone.0080670
Parrott, A. M., Shi, J., Aaron, J., Green, D. A., Whittier, S., and Wu, F. (2020). Detection of multiple hypervirulent Klebsiella pneumoniae strains in a New York City hospital through screening of virulence genes. Clin. Microbiol. Infect. [Epub ahead of print].
Pastagia, M., and Arumugam, V. (2008). Klebsiella pneumoniae liver abscesses in a public hospital in Queens, New York. Travel Med. Infect. Dis. 6, 228–233. doi: 10.1016/j.tmaid.2008.02.005
Podschun, R., Pietsch, S., Höller, C., and Ullmann, U. (2001). Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl. Environ. Microbiol. 67, 3325–3327. doi: 10.1128/aem.67.7.3325-3327.2001
Posey, J. E., and Gherardini, F. C. (2000). Lack of a role for iron in the Lyme disease pathogen. Science 288, 1651–1653. doi: 10.1126/science.288.5471.1651
Rahimian, J., Wilson, T., Oram, V., and Holzman, R. S. (2004). Pyogenic liver abscess: recent trends in etiology and mortality. Clin. Infect. Dis. 39, 1654–1659. doi: 10.1086/425616
Rahn, A., Beis, K., Naismith, J. H., and Whitfield, C. (2003). A novel outer membrane protein, Wzi, is involved in surface assembly of the Escherichia coli K30 group 1 capsule. J. Bacteriol. 185, 5882–5890. doi: 10.1128/jb.185.19.5882-5890.2003
Reid, A. N., and Whitfield, C. (2005). functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J. Bacteriol. 187, 5470–5481. doi: 10.1128/jb.187.15.5470-5481.2005
Remya, P., Shanthi, M., and Sekar, U. (2018). Occurrence and characterization of hyperviscous K1 and K2 serotype in. J. Lab. Phys. 10, 283–288. doi: 10.4103/jlp.jlp_48_18
Rogers, H. J. (1973). Iron-binding catechols and virulence in Escherichia coli. Infect. Immun. 7, 445–456. doi: 10.1128/iai.7.3.445-456.1973
Rossi, B., Gasperini, M. L., Leflon-Guibout, V., Gioanni, A., De Lastours, V., Rossi, G., et al. (2018). Hypervirulent Klebsiella pneumoniae in cryptogenic liver abscesses, Paris, France. Emerg. Infect. Dis. 24, 221–229.
Russo, T. A., and Gulick, A. M. (2019). Aerobactin synthesis proteins as antivirulence targets in hypervirulent. ACS Infect. Dis. 5, 1052–1054. doi: 10.1021/acsinfecdis.9b00117
Russo, T. A., and MacDonald, U. (2020). The galleria mellonella infection model does not accurately differentiate between hypervirulent and classical Klebsiella pneumoniae. mSphere 5:e00850-19.
Russo, T. A., and Marr, C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32:e00001-19.
Russo, T. A., Olson, R., Fang, C.-T., Stoesser, N., Miller, M., Macdonald, U., et al. (2018). Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J. Clin. Microbiol. 56:e00776-18.
Russo, T. A., Olson, R., Macdonald, U., Beanan, J., and Davidson, B. A. (2015). Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect. Immun. 83, 3325–3333. doi: 10.1128/iai.00430-15
Russo, T. A., Olson, R., Macdonald, U., Metzger, D., Maltese, L. M., Drake, E. J., et al. (2014). Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 82, 2356–2367. doi: 10.1128/iai.01667-13
Sánchez-López, J., García-Caballero, A., Navarro-San Francisco, C., Quereda, C., Ruiz-Garbajosa, P., Navas, E., et al. (2019). Hypermucoviscous : a challenge in community acquired infection. IDCases 17:e00547. doi: 10.1016/j.idcr.2019.e00547
Sarris, P. F., Zoumadakis, C., Panopoulos, N. J., and Scoulica, E. V. (2011). Distribution of the putative type VI secretion system core genes in Klebsiella spp. Infect. Genet. Evol. 11, 157–166. doi: 10.1016/j.meegid.2010.09.006
Schroll, C., Barken, K. B., Krogfelt, K. A., and Struve, C. (2010). Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10:179. doi: 10.1186/1471-2180-10-179
Sellick, J. A., and Russo, T. A. (2018). Getting hypervirulent Klebsiella pneumoniae on the radar screen. Curr. Opin. Infect. Dis. 31, 341–346. doi: 10.1097/qco.0000000000000464
Shankar, C., Jacob, J. J., Vasudevan, K., Biswas, R., Manesh, A., Sethuvel, D. P. M., et al. (2020). Emergence of multidrug resistant hypervirulent ST23 Klebsiella pneumoniae: multidrug resistant plasmid acquisition drives evolution. Front. Cell Infect. Microbiol. 10:575289. doi: 10.3389/fcimb.2020.575289
Shankar, C., Veeraraghavan, B., Nabarro, L. E. B., Ravi, R., Ragupathi, N. K. D., and Rupali, P. (2018). Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 18:6. doi: 10.1186/s12866-017-1148-6
Shankar-Sinha, S., Valencia, G. A., Janes, B. K., Rosenberg, J. K., Whitfield, C., Bender, R. A., et al. (2004). The Klebsiella pneumoniae O antigen contributes to bacteremia and lethality during murine pneumonia. Infect. Immun. 72, 1423–1430. doi: 10.1128/iai.72.3.1423-1430.2004
Shi, Q., Lan, P., Huang, D., Hua, X., Jiang, Y., Zhou, J., et al. (2018). Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol. 18:94. doi: 10.1186/s12866-018-1236-2
Shon, A. S., Bajwa, R. P. S., and Russo, T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107. doi: 10.4161/viru.22718
Siu, L. K., Yeh, K.-M., Lin, J.-C., Fung, C.-P., and Chang, F.-Y. (2012). Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis. 12, 881–887. doi: 10.1016/s1473-3099(12)70205-0
Smith, G. W., Blackwell, C. C., and Nuki, G. (1997). Faecal flora in spondyloarthropathy. Br. J. Rheumatol. 36, 850–854. doi: 10.1093/rheumatology/36.8.850
Srinivasan, V. B., Venkataramaiah, M., Mondal, A., Vaidyanathan, V., Govil, T., and Rajamohan, G. (2012). Functional characterization of a novel outer membrane porin KpnO, regulated by PhoBR two-component system in Klebsiella pneumoniae NTUH-K2044. PLoS One 7:e41505. doi: 10.1371/journal.pone.0041505
Storey, D., Mcnally, A., Strand, M., Sa-Pessoa Graca Santos, J., Rodriguez-Escudero, I., Elmore, B., et al. (2020). Klebsiella pneumoniae type VI secretion system-mediated microbial competition is PhoPQ controlled and reactive oxygen species dependent. PLoS Pathog. 16:e1007969. doi: 10.1371/journal.ppat.1007969
Struve, C., Roe, C. C., Stegger, M., Stahlhut, S. G., Hansen, D. S., Engelthaler, D. M., et al. (2015). Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 6:e00630.
Sturm, E., Tai, A., Lin, B., Kwong, J., Athan, E., Howden, B. P., et al. (2018). Bilateral osteomyelitis and liver abscess caused by hypervirulent Klebsiella pneumoniae- a rare clinical manifestation (case report). BMC Infect. Dis. 18:380. doi: 10.1186/s12879-018-3277-4
Su, K., Zhou, X., Luo, M., Xu, X., Liu, P., Li, X., et al. (2018). Genome-wide identification of genes regulated by RcsA, RcsB, and RcsAB phosphorelay regulators in Klebsiella pneumoniae NTUH-K2044. Microb. Pathog. 123, 36–41. doi: 10.1016/j.micpath.2018.06.036
Sun, Q.-L., Gu, D., Wang, Q., Hu, Y., Shu, L., Hu, J., et al. (2019). Dynamic colonization of isolates in gastrointestinal tract of intensive care patients. Front. Microbiol. 10:230. doi: 10.3389/fmicb.2019.00230
Svetlov, D., Shi, D., Twentyman, J., Nedialkov, Y., Rosen, D. A., Abagyan, R., et al. (2018). In silico discovery of small molecules that inhibit RfaH recruitment to RNA polymerase. Mol. Microbiol. 110, 128–142. doi: 10.1111/mmi.14093
Tan, T. Y., Cheng, Y., Ong, M., and Ng, L. S. (2014). Performance characteristics and clinical predictive value of the string test for detection of hepato-virulent Klebsiella pneumoniae isolated from blood cultures. Diagn. Microbiol. Infect. Dis. 78, 127–128. doi: 10.1016/j.diagmicrobio.2013.10.014
Tang, H. L., Chiang, M. K., Liou, W. J., Chen, Y. T., Peng, H. L., Chiou, C. S., et al. (2010). Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur. J. Clin. Microbiol. Infect. Dis. 29, 689–698. doi: 10.1007/s10096-010-0915-1
Tang, M., Kong, X., Hao, J., and Liu, J. (2020). Epidemiological characteristics and formation mechanisms of multidrug-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 11:581543. doi: 10.3389/fmicb.2020.581543
Togawa, A., Toh, H., Onozawa, K., Yoshimura, M., Tokushige, C., Shimono, N., et al. (2015). Influence of the bacterial phenotypes on the clinical manifestations in Klebsiella pneumoniae bacteremia patients: a retrospective cohort study. J. Infect. Chemother. 21, 531–537. doi: 10.1016/j.jiac.2015.04.004
Troxell, B., and Hassan, H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. doi: 10.3389/fcimb.2013.00059
Tsai, Y.-K., Fung, C.-P., Lin, J.-C., Chen, J.-H., Chang, F.-Y., Chen, T.-L., et al. (2011). Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 55, 1485–1493. doi: 10.1128/aac.01275-10
Wacharotayankun, R., Arakawa, Y., Ohta, M., Tanaka, K., Akashi, T., Mori, M., et al. (1993). Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect. Immun. 61, 3164–3174. doi: 10.1128/iai.61.8.3164-3174.1993
Walker, K. A., and Miller, V. L. (2020). The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr. Opin. Microbiol. 54, 95–102. doi: 10.1016/j.mib.2020.01.006
Walker, K. A., Miner, T. A., Palacios, M., Trzilova, D., Frederick, D. R., Broberg, C. A., et al. (2019). A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. mBio 10: e00089-19.
Walker, K. A., Treat, L. P., Sepúlveda, V. E., and Miller, V. L. (2020). The small protein RmpD drives hypermucoviscosity in Klebsiella pneumoniae. mBio 11:e01750-20.
Wall, E., Majdalani, N., and Gottesman, S. (2018). The complex Rcs regulatory cascade. Annu. Rev. Microbiol. 72, 111–139. doi: 10.1146/annurev-micro-090817-062640
Wang, J. H., Liu, Y. C., Lee, S. S., Yen, M. Y., Chen, Y. S., Wang, J. H., et al. (1998). Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 26, 1434–1438.
Wang, L., Shen, D., Wu, H., and Ma, Y. (2017). Resistance of hypervirulent Klebsiella pneumoniae to both intracellular and extracellular killing of neutrophils. PLoS One 12:e0173638. doi: 10.1371/journal.pone.0173638
Wu, C.-C., Huang, Y.-J., Fung, C.-P., and Peng, H.-L. (2010). Regulation of the Klebsiella pneumoniae Kpc fimbriae by the site-specific recombinase KpcI. Microbiology 156, 1983–1992. doi: 10.1099/mic.0.038158-0
Wu, C.-C., Lin, C.-T., Cheng, W.-Y., Huang, C.-J., Wang, Z.-C., and Peng, H.-L. (2012). Fur-dependent MrkHI regulation of type 3 fimbriae in Klebsiella pneumoniae CG43. Microbiology 158, 1045–1056. doi: 10.1099/mic.0.053801-0
Wu, J. H., Wu, A. M., Tsai, C. G., Chang, X.-Y., Tsai, S.-F., and Wu, T.-S. (2008). Contribution of fucose-containing capsules in Klebsiella pneumoniae to bacterial virulence in mice. Exp. Biol. Med. 233, 64–70. doi: 10.3181/0706-rm-170
Wu, M.-F., Yang, C.-Y., Lin, T.-L., Wang, J.-T., Yang, F.-L., Wu, S.-H., et al. (2009). Humoral immunity against capsule polysaccharide protects the host from magA+ Klebsiella pneumoniae-induced lethal disease by evading Toll-like receptor 4 signaling. Infect. Immun. 77, 615–621. doi: 10.1128/iai.00931-08
Wyres, K. L., Lam, M. M. C., and Holt, K. E. (2020). Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359.
Wyres, K. L., Wick, R. R., Judd, L. M., Froumine, R., Tokolyi, A., Gorrie, C. L., et al. (2019). Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 15:e1008114. doi: 10.1371/journal.pgen.1008114
Yang, X., Dong, N., Chan, E. W., Zhang, R., and Chen, S. (2021). Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 29, 65–83. doi: 10.1016/j.tim.2020.04.012
Yeh, K.-M., Kurup, A., Siu, L. K., Koh, Y. L., Fung, C.-P., Lin, J.-C., et al. (2007). Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J. Clin. Microbiol. 45, 466–471. doi: 10.1128/jcm.01150-06
Yoshida, K., Matsumoto, T., Tateda, K., Uchida, K., Tsujimoto, S., and Yamaguchi, K. (2000). Role of bacterial capsule in local and systemic inflammatory responses of mice during pulmonary infection with Klebsiella pneumoniae. J. Med. Microbiol. 49, 1003–1010. doi: 10.1099/0022-1317-49-11-1003
Yu, V. L., Hansen, D. S., Ko, W. C., Sagnimeni, A., Klugman, K. P., Von Gottberg, A., et al. (2007). Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg. Infect. Dis. 13, 986–993.
Yu, W.-L., Lee, M.-F., Chang, M.-C., and Chuang, Y.-C. (2015a). Intrapersonal mutation of rmpA and rmpA2: a reason for negative hypermucoviscosity phenotype and low virulence of rmpA-positive Klebsiella pneumoniae isolates. J. Glob. Antimicrob. Resist. 3, 137–141. doi: 10.1016/j.jgar.2015.03.008
Yu, W.-L., Lee, M.-F., Tang, H.-J., Chang, M.-C., and Chuang, Y.-C. (2015b). Low prevalence of rmpA and high tendency of rmpA mutation correspond to low virulence of extended spectrum β-lactamase-producing Klebsiella pneumoniae isolates. Virulence 6, 162–172. doi: 10.1080/21505594.2015.1016703
Zhang, Y., Zhao, C., Wang, Q., Wang, X., Chen, H., Li, H., et al. (2016). High Prevalence of Hypervirulent Klebsiella pneumoniae infection in china: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob. Agents Chemother. 60, 6115–6120. doi: 10.1128/aac.01127-16
Keywords: Klebsiella pneumoniae, hypermucoviscous, capsule, siderophores, virulence factors, hypervirulence
Citation: Zhu J, Wang T, Chen L and Du H (2021) Virulence Factors in Hypervirulent Klebsiella pneumoniae. Front. Microbiol. 12:642484. doi: 10.3389/fmicb.2021.642484
Received: 17 December 2020; Accepted: 11 March 2021;
Published: 08 April 2021.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
Ning Dong, City University of Hong Kong, Hong KongDavid A. Rosen, Washington University in St. Louis, United States
Balaji Veeraraghavan, Christian Medical College & Hospital, India
Copyright © 2021 Zhu, Wang, Chen and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Du, aG9uZ19kdUAxMjYuY29t; aG9uZ2R1QHN1ZGEuZWR1LmNu
 Jie Zhu
Jie Zhu Tao Wang1
Tao Wang1 Liang Chen
Liang Chen Hong Du
Hong Du