- Institute of Science, Nirma University, Ahmedabad, India
The plant growth-promoting Acinetobacter sp. SK2 isolated from Vigna radiata rhizosphere was characterized for mineral phosphate solubilization (MPS). To understand the contribution of the membrane glucose dehydrogenase (mGDH) and soluble glucose dehydrogenase (sGDH) in glucose oxidation and MPS, insertional inactivation of the corresponding genes was carried out. The disruption of mGDH encoding gene gdhA resulted in complete loss of mGDH activity, which confirmed its role in periplasmic glucose oxidation and gluconate-mediated MPS phenotype. The inactivation of sGDH encoding gene gdhB resulted in loss of sGDH activity, which did not alter the MPS or mGDH activity. Thus, it was also concluded that the sGDH was dispensable in gluconate-mediated MPS. Supplementation of succinate in glucose-containing medium suppressed the activity of mGDH (and sGDH) and therefore repressed the MPS phenotype. The catabolite repression control protein (Crc) of Pseudomonas was implicated in Acinetobacter sp. for a similar function in the presence of preferred and non-preferred carbon sources. To understand the regulatory linkage between Crc and genes for glucose oxidation, crc mutants were generated. The inactivation of crc resulted in increased activity of the mGDH in glucose + succinate-grown cells, indicating derepression. An increase in phosphate solubilization up to 44% in glucose + succinate-grown crc– compared with glucose-grown cells was recorded, which was significantly repressed in the wild-type strain under similar conditions. It is therefore proposed that in Acinetobacter sp. SK2, Crc is involved in the succinate-provoked repression of the MPS phenotype. The gene expression data indicated that Hfq may also have a regulating role in preferential utilization of carbon source by perhaps modulating Crc–Hfq functionality. V. radiata plants inoculated with the wild type improved both root and shoot length by 1.3 to 1.4-fold. However, crc– increased the root and shoot length by 1.6-fold, compared with the uninoculated controls. In mimicking the soil condition (in the presence of multiple carbon sources, e.g., succinate along with glucose), the crc– strain of Acinetobacter sp. SK2 performed better in supporting the growth of V. radiata in pot experiments.
Introduction
The metabolically versatile genus Acinetobacter can thrive in diverse environments including rhizosphere and can be exploited to support plant growth (Young et al., 2005). Acinetobacter sp. SK2 isolated from mung bean rhizosphere released soluble P from complex sources. Besides being a phosphate solubilizer, it was also reported for other plant growth promotion (PGP) activities like indole-3-acetic acid (IAA), siderophore, and hydrogen cyanide (HCN) production (Bharwad and Rajkumar, 2020). In a previous study, we have reported that the mineral phosphate solubilization (MPS) ability of the isolate is attributed to gluconate secretion when grown on glucose. However, the acid production and hence the MPS phenotype were repressed by succinate when present alone or in combination with glucose. The gluconate production was facilitated by membrane glucose dehydrogenase (mGDH) and possibly had a contribution from soluble glucose dehydrogenase (sGDH). The activity of enzymes (mGDH and sGDH) and the expression of the corresponding genes were significantly lower in the presence of succinate or succinate along with glucose. The preference of succinate over glucose was effected by repression of glucose oxidation genes in the isolate Acinetobacter sp. SK2 (Bharwad and Rajkumar, 2020).
Rhizosphere, the intended niche of the inoculated phosphate solubilizing bacteria (PSB), have plentiful mixtures of organic compounds like sugars, organic acids, and amino acids exuded by the roots (Kamilova et al., 2006). In the presence of multiple carbon sources, carbon catabolite repression (CCR) is plausible to run the preferential utilization of most efficient carbon source in rhizobacteria. Such preferences are important for the fitness of organisms in their natural environments for optimizing growth rate via controlling carbon flow. Though the mechanisms of CCR regulatory networks differ among bacterial species, many of these functions at the transcriptional or post-transcriptional level, controlling substrate uptake or expression of various catabolic enzymes (Rojo, 2010). The metabolically versatile pseudomonads use organic acids as the preferred carbon source over glucose. Succinate is known to inhibit the expression of many transporters and catabolic genes involved in the metabolism of several other carbon sources, for example, those required for the utilization of glucose and mannitol (MacGregor et al., 1991; Hester et al., 2000). The CCR regulatory network in pseudomonads is governed by several potential regulator proteins such as CyoB, PtsN, and PtsO and the Crc (catabolite repression control) protein (Görke and Stülke, 2008; Rojo, 2010).
Catabolite repression control protein was originally reported in Pseudomonas aeruginosa, and a mutation in the gene locus resulted in a strain that could utilize glucose and mannitol even in the presence of tricarboxylic acid (TCA) intermediates (MacGregor et al., 1991; Wolff et al., 1991). The protein sequence of Crc resembles nuclease family, though it did not show any nuclease or DNA-binding activity (MacGregor et al., 1996). The mechanism by which Crc regulates expression of carbohydrate metabolism remains unclear. The existing models in P. aeruginosa, Pseudomonas putida, and Pseudomonas syringae indicate that CCR takes place at the post-transcriptional level and arbitrate it to complex interactions among numerous regulatory components (Sonnleitner et al., 2009; Filiatrault et al., 2013; Hernández-Arranz et al., 2013). Besides Crc, another key player known for its regulatory role in overall bacterial physiology is the RNA chaperone Hfq (Vogel and Luisi, 2011). The Hfq protein was first found in Escherichia coli as a host factor required for replication of Qβ bacteriophage (thus regarded as Hfq) (Franze de Fernandez et al., 1968). Hfq is considered a global transcriptional controller participating in various cellular processes like outer membrane biogenesis, stress tolerance, virulence, and iron homeostasis (Chao and Vogel, 2010). The influence of Hfq on physiology and virulence in a number of Gram-negative and in Gram-positive bacteria has been reported (Simonsen et al., 2011; Wang et al., 2014; Kuo et al., 2017).
Regardless of the study of CCR in many gammaproteobacteria, attempts to unravel the key components and the molecular mechanism of CCR in bacteria other than members of the Pseudomonas group are limited. Zimmermann et al. (2009) identified that Crc protein is involved in the CCR in Acinetobacter baylyi. The key enzyme of protocatechuate degradation, protocatechuate 3,4-dioxygenase, was strongly reduced in the crc– strain, while in the wild-type strain, it endured strong catabolite repression by acetate and succinate. Here, we describe CCR of the MPS phenotype in the plant growth-promoting bacteria Acinetobacter sp. SK2. This work specifically focuses on succinate-induced repression of gdhA and gdhB genes, causing diminished gluconate production. The role of Crc as putative regulator of gdhA and gdhB was investigated by generating insertionally inactivated mutants of Crc. Although Acinetobacter sp. SK2 is phylogenetically related to Pseudomonas species, it possesses a distinctive feature, i.e., the presence of sGDH, which has a role in glucose oxidation and was repressed in the presence of succinate (or along with glucose) similar to mGDH (Bharwad and Rajkumar, 2020). The glucose utilization and gluconate-mediated MPS of Acinetobacter sp. SK2 were investigated by functional characterization of the two key enzymes involved in glucose oxidation. To this purpose, the mGDH encoding gene gdhA and the sGDH encoding gene gdhB of Acinetobacter sp. SK2 were inactivated systematically. The physiological and metabolic attributes of the respective gene mutation on the MPS phenotype were investigated thereafter. The expression profile of Hfq in glucose and glucose + succinate opened another avenue of investigating Hfq as the next putative regulator of carbon source utilization along with the Crc in Acinetobacter sp. SK2.
Materials and Methods
Bacterial Strains, Plasmids, and Chemicals Used
The list of bacterial strains and plasmids used in the study is shown in Table 1. Acinetobacter sp. SK2 is a previously characterized mineral phosphate solubilizing isolate (Bharwad and Rajkumar, 2020). The suicide vector, pKnockoutΩ, was obtained from Dr. Filiatrault, Cornell University (Windgassen et al., 2000). E. coli DH5α was used to harbor the native suicide vector and the newly constructed knockout plasmids. Antibiotics added to the medium, for selection and maintenance of the plasmid, were in the following concentration unless otherwise mentioned: 100 μg ml–1 of ampicillin, 50 μg ml–1 of streptomycin, and 50 μg ml–1 of spectinomycin. For the post-pulse recovery of electoporated Acinetobacter cells, super optimal broth (SOC) medium was used. All the plasmids were isolated using HiPurA Plasmid DNA miniprep purification kit (HiMedia, Mumbai, India) and adjusted to the final concentration of 50 ng μl–1.
Construction and Confirmation of Knockout Plasmids
Acinetobacter sp. SK2 mutants were constructed using suicide vector pKnockoutΩ. Linearized vector was generated using the three combinations of restriction enzyme; (1) KpnI + XbaI, (2) SalI + PstI, and (3) KpnI + SalI for insertion of crc, gdhA, and gdhB, respectively. The 650-bp crc, 500-bp gdhA, and 550-bp gdhB insert fragments were amplified from chromosomal DNA of Acinetobacter sp. SK2 using oligonucleotide primers described in Table 2. In-Fusion HD Plus Cloning kit (Takara Bio Inc., Kusatsu, Shiga, Japan) was used for the ligation of the linearized vector with the insert fragment, and the ligation mixture was used for transformation of the competent E. coli DH5α cells. The recombinant clones were selected through blue-white screening on Luria–Bertani (LB) agar containing ampicillin, streptomycin, and spectinomycin. Digestion of plasmids isolated from the recombinant white colonies was performed using restriction enzyme combinations of the respective gene inserts. Insert release of 650, 500, and 550 bp from the crc, gdhA, and gdhB knockout constructs confirmed a successful insertion of desired gene into the knockout vector. These constructs were named pKOcrc, pKOgdhA, and pKOgdhB, respectively, and were used for electrotransformation of Acinetobacter sp. SK2.
Electrotransformation of Acinetobacter sp. SK2
Members of the genera Acinetobacter are naturally competent and can take up high-molecular-weight extracellular DNA from a source in the natural environments (Bruns et al., 1992). However, to increase the transformation efficiency, electrotransformation was employed (Lorenz et al., 1992; Yildirim et al., 2016), in which 200 μl of overnight grown cells was transferred into 5 ml of fresh mineral medium supplied with 10 mM of succinate and incubated for 2 h at 37°C. In sterile, ice-cold electroporation cuvettes (Eppendorf, Hamburg, Germany), 500 μl of these cells was added with ∼100–250 ng of recombinant plasmids (constructs: pKOcrc, pKOgdhA, and pKOgdhB) as individual treatments. All electroporations were conducted in Eporator (Eppendorf), set at 1.8 kV for 3 s to transform DNA into the bacterial cells. The electroporated cells were added to a fresh, sterile microfuge tube containing 500 μl of SOC medium followed by 2 h incubation at 37°C. For the selection of tranformants, 100 μl of cell culture was spread on LB medium supplemented with streptomycin and spectinomycin and was incubated at 37°C. Colonies were transferred on fresh selective media plates several times to ensure a pure culture.
Confirmation of Insertion of Suicide Vector in crc–, gdhA–, and gdhB– Strains
The gdhA mutants were spot inoculated on YGC medium (yeast–glucose–calcium carbonate medium containing g L–1: yeast extract 20; glucose 50; CaCO3 10; and agar power 15) (Zhu et al., 2011) to observe phenotypic change caused by the inactivation of the gene. However, since no known visual phenotypic character of crc and gdhB is known, only molecular methods were used for confirmation of the successful gene disruption. The presence of omega (Ω) cassette in the genome of clones was confirmed through a colony PCR using oligonucleotide primer (Table 2). Integration of omega cassette into the correct chromosomal location was verified by PCR using a set of primers hybridizing the gene of interest and the omega cassette (Table 2). An increase in the amplicon size was indicative of the successful insertion of the omega cassette into the gene of interest, which was further confirmed by DNA sequencing.
Growth Conditions
Strains of Acinetobacter sp. SK2 were grown on LB medium at 37°C on an orbital shaker incubator (Rojas-Tapias et al., 2014). For the cell growth experiments, enzyme assays, and expression analysis, all the strains were grown under identical conditions in M9 minimal medium at 37°C with a continuous agitation (Mara et al., 2012), where the carbon sources added individually were at the following final concentrations unless otherwise indicated: succinate, 50 mM; glucose, 50 mM; and glucose + succinate, 25 mM each. Specifications pertaining to preparation of cells for enzyme assay are described in the following sections. To ensure chromosomal integration of the antibiotic-encoding omega cassette, the cells were grown in the absence of antibiotics to ensure optimal growth, after which inoculation on the media containing the appropriate antibiotics was carried out to check the presence of the resistance omega cassette. Instability of the chromosomally integrated resistance cassettes was not observed. E. coli transformed with plasmid constructs were grown in LB medium at 37°C, supplemented with ampicillin (100 μg ml–1), streptomycin (50 μg ml–1), and spectinomycin (50 μg ml–1).
Enzyme Activities of Membrane Glucose Dehydrogenase and Soluble Glucose Dehydrogenase
Glucose dehydrogenase activity in the wild-type and mutants was done using a chromogenic assay involving 2,6-dichlorophenolindophenol (DCIP; HiMedia) and phenazine methosulfate (PMS; SRL Chemicals, Mumbai, India). Cells were grown in M9 minimal medium with 50 mM of glucose; 50 mM of succinate; and 25 mM glucose + 25 mM succinate. For mGDH, a whole cell pellet was obtained through centrifugation at 12,000 g at 4°C for 5 min. The pellet was resuspended in 50 mM of potassium phosphate buffer (pH = 7.5) with MgCl2. For assay of the sGDH, the cell pellet was suspended in a small volume of same buffer containing 20% glycerol and 1 mM of DTT and sonicated for 5 min at a pulse rate of 30 s at 50 Hz on ice. The suspension was centrifuged at 11,000 g at 4°C for 30 min in order to remove cell debris. Furthermore, to separate out the crude soluble fraction, the supernatant was subjected to ultracentrifugation at 120,000 g for 90 min and was immediately used for the sGDH enzyme assay (Matsushita et al., 1989).
Expression Analysis of crc, gdhA, and gdhB Genes in Wild Type and Mutants
The relative expression of genes was estimated using quantitative real-time PCR (qRT-PCR). cDNA was synthesized from RNA isolated from the wild type and mutants grown in M9 minimal medium containing glucose, succinate, and glucose + succinate. Cells were harvested at different growth stages and subjected to total RNA isolation using Hybrid-RTM Kit (GeneAll®), as per manufacturer’s instructions. Residual genomic DNA contamination was removed using DNase I (Thermo Fisher Scientific, Waltham, MA, United States) followed by DNase inactivation by adding 2 μl of stop solution containing EDTA and heating at 65°C for 10 min. Finally, the DNA contamination was checked by PCR using 16S rRNA universal primers F27 (5′-AGAGTTTGATCATGGCTCAG-3′) and R1492 (5′-TACGGTTACCTTGTTACGACTT-3′), and the RNA integrity was determined on agarose gel electrophoresis. cDNAs from each sample were obtained using PrimeScriptTM 1st strand cDNA synthesis kit (Takara Bio Inc.) following the manufacturer’s instructions. qPCR was performed on an Applied Biosystems QuantStudio4 system using a SYBR Premix Ex Taq II (Tli RNaseH Plus) kit (Takara Bio Inc.). Amplification and detection of the specific products were performed with three technical replicates per sample under the following conditions: one cycle at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 3 min, and annealing and extension at 60°C for 1 min. Cycle threshold (Ct or Cq value) values were recorded with QuantStudioTM Design and Analysis Software (Thermo Fisher Scientific). Ct values of 35 were considered beyond the limit of detection (Chen et al., 2009). Fold changes were determined using the relative quantification comparative threshold cycle (Ct) method (DDCt) (Livak and Schmittgen, 2001), and rpoD housekeeping gene was used as a reference for normalization. Oligonucleotide primers used for the qRT-PCR are shown in Table 2. For each gene tested, a standard curve was plotted using serial dilutions from 50 to 0.005 ng of Acinetobacter sp. SK2 genomic DNA in order to quantify the efficiency of each PCR. The PCR products ranged between 100 and 150 bp.
Effect of Insertional Inactivation on Mineral Phosphate Solubilization Phenotype
Qualitative estimation of MPS in the wild type and mutants was performed on Pikovskaya’s agar (Pikovskaya, 1948) and Tris-buffered rock phosphate (TRP) agar, as described by Rajput et al. (2013). Quantitative estimation of soluble P was carried out in Pikovskaya’s agar and TRP broth containing glucose (50 mM), succinate (50 mM), or equimolar mixture of glucose + succinate (25 mM each; repression medium). P released in the medium was quantified by a reaction of soluble P with ammonium molybdate to form phosphomolybdate, which when reduced by ascorbic acid forms a blue colored complex that is quantified at 820 nm (Ames, 1966).
Effect of Wild-Type and Mutant Acinetobacter sp. SK2 on Growth of Vigna radiata
Plant growth experiments were carried out in pots sown with mung bean (Vigna radiata) as reported by Kumari et al. (2018). Soil samples were collected from the Nirma herbal health garden, air dried, cleaned, and sterilized by three repeats of autoclave cycles. Soil was allowed to cool down, and 1 kg of soil was filled in each pot. V. radiata seeds were surface sterilized using 95% ethanol and then 0.2% HgCl2 surface sterilant for 3 min each. Sterilant was totally removed by washing seeds 10 times with the sterile distilled water. Eight to ten seeds were sown in each pot at a depth of 1–5 cm. Post germination, the plantlets were kept in sunlight and watered daily. Plantlets were inoculated with the wild-type, gdhA–, gdhB–, and crc– strains (107−108 cfu ml–1) on the seventh day of germination, and the uninoculated plantlets served as a control. Plants were gently uprooted on the 31st day post germination and scored for plant parameters like root length (cm), shoot length (cm), root:shoot ratio, and dry mass (g).
Results
Generation of gdhA, gdhB, and crc Mutants of Acinetobacter sp. SK2
To evaluate the role of mGDH, sGDH, and Crc in the MPS of Acinetobacter sp. SK2, the individual mutant strains defective in gdhA, gdhB, and crc were constructed. Fragments of 500-bp gdhA, 540-bp gdhB, and 650-bp crc were cloned in a suicide vector pKnockoutΩ near the omega fragment (streptomycin–spectinomycin cassette). The resulting plasmids were transferred into the wild type via electroporation. As pKnockoutΩ cannot replicate in Acinetobacter sp. SK2, single-crossover integrants were selected by resistance to streptomycin–spectinomycin. An integrant lacking the gdhA that produced no or little gluconate on glucose was selected on YGC medium. The selected strains with insertional inactivation were named gdhA–, gdhB–, and crc–. The correctness of the insertion of the respective genes in the integrants was verified by appropriate PCR amplifications (Supplementary Figure 1). The vector construction and general strategy employed for mutant generation are described in Supplementary Figures 2, 3.
P Solubilization by gdhA–, gdhB–, and crc– Strains
The MPS ability of the wild type was significantly reduced in medium containing succinate. The addition of succinate to glucose-containing medium repressed the glucose utilization and oxidation to gluconate, which is achieved by the mGDH activity. The sGDH, along with mGDH, was also found to be under the repressing influence of succinate (Bharwad and Rajkumar, 2020). To evaluate the function of mGDH and sGDH in glucose oxidation and relevance of the Crc protein in this succinate-mediated repression, we compared the MPS ability of the wild type and the gdhA–, gdhB–, and crc– strains on PVK and TRP media containing glucose, succinate, and glucose + succinate. The wild-type, gdhB–, and crc– strains showed tricalcium phosphate (TCP) solubilization (halo zone around the colony) and rock phosphate (RP) (pink coloration around the colony) in the presence of glucose; however, gdhA– failed to grow on the media (Figures 1A,C). The wild-type, gdhA–, and gdhB– failed in MPS on glucose + succinate, while the phenotype was observed in crc– (Figures 1B,D).
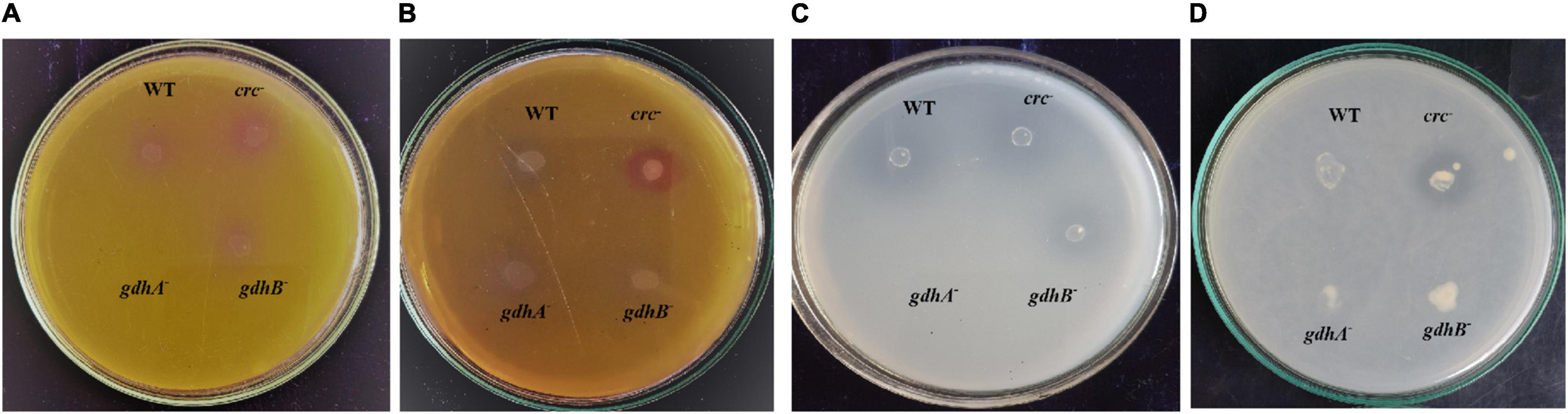
Figure 1. MPS phenotype of wild-type, gdhA–, gdhB–, and crc– mutants on TRP agar (A) with glucose and (B) with glucose + succinate; Pikovskaya agar (C) with glucose and (D) with glucose + succinate.
The soluble P released by the wild-type and mutant strains in PVK and TRP broth media supplemented with glucose, succinate, and glucose + succinate in the treatments is presented (Table 3). The free P measured in succinate and glucose + succinate in all the treatments was comparable with the uninoculated medium (pH 7 or 8) or the soluble P measured on day 0. The maximum P released by the wild type in the media containing glucose + succinate was only 7% of the P released in the glucose-containing media. The repression of P solubilization in PVK medium was 90–91% in glucose + succinate- and succinate-containing media. The amount of soluble P released during the course of growth in glucose + succinate and succinate was insignificant. A similar trend was observed for RP solubilization where soluble P decreased up to 97–99% in glucose + succinate and succinate (Table 3).
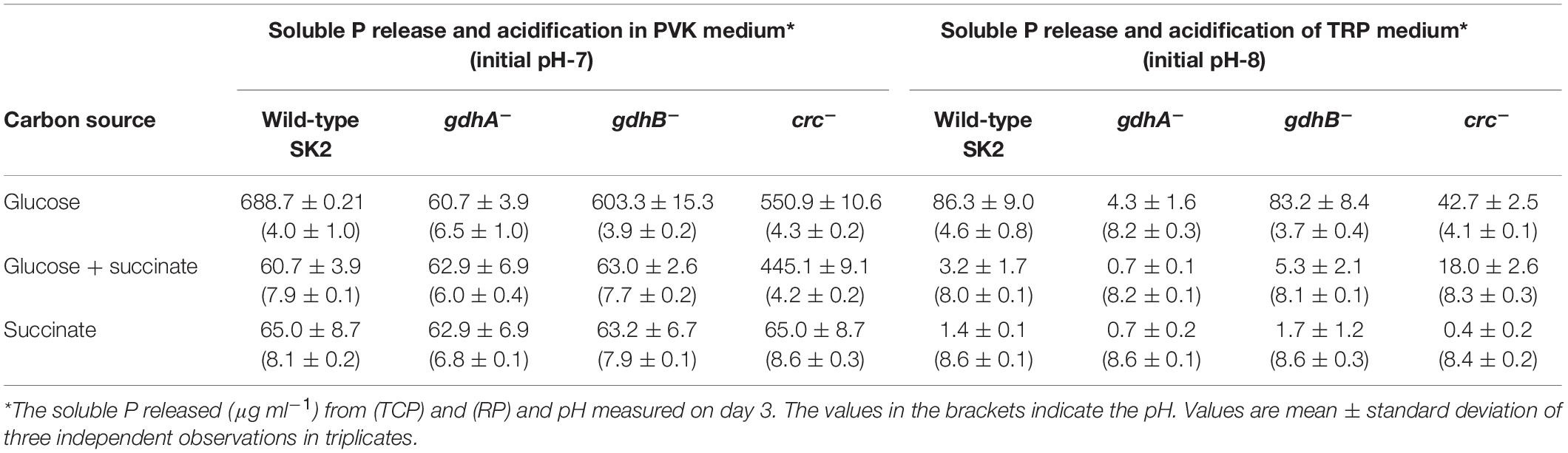
Table 3. Phosphate solubilization by Acinetobacter sp. SK2 wild-type, gdhA–, gdhB–, and crc– strains in PVK and TRP broth medium.
The P release was almost completely abolished in gdhA– as compared with the wild type. The amount of P released by the mutant was similar to that of the wild-type in glucose + succinate- and succinate-containing media. The absence of gdhA and abolition of MPS also correlated to the pH of the medium that remained unchanged.
The P release and its repression in gdhB– in glucose + succinate were similar to those of the wild type. Disruption of gdhB– influenced neither P solubilization nor mGDH-mediated gluconate production. Likewise, the P released by crc– strain in PVK was measured in medium supplemented with glucose, succinate, and glucose + succinate. The P release in the glucose media was 550 μg ml–1, which was similar to that of the wild type grown in glucose. An enhanced P release was observed in glucose + succinate medium, reaching up to 44% of the wild-type grown in glucose, indicating derepression. A concomitant decrease in pH up to 4.3 was the indication of acidification of the medium and P release in glucose + succinate media. It can therefore be concluded that gdhA was solely responsible for periplasmic oxidation of glucose to gluconate, which leads to the MPS (Table 3). When RP solubilization by gdhA–, gdhB–, and crc– was compared with the wild type in TRP medium, a trend similar to TCP solubilization was observed (Table 3).
Changes in Growth Profiles of gdhA–, gdhB–, and crc– Strains
Since the wild type preferentially utilized succinate over glucose in medium containing glucose + succinate affecting gluconate-mediated MPS phenotype (Bharwad and Rajkumar, 2020), the differences in the growth of the three mutants in medium containing glucose, succinate, and glucose + succinate were observed.
Though there were no differences in the growth of the wild type, gdhB–, and crc– on glucose, significant deviation in cell growth on glucose was observed in the case of gdhA– strain (Figure 2A). This observation suggested that gdhA contributed mostly to glucose utilization or oxidation in the wild type. The inactivation of sGDH encoding gene gdhB did not influence the growth of the mutant on glucose. The growth of all the three mutants in succinate was similar to that of the wild type (Figure 2B). The wild-type and crc– strains took 20 h for the complete glucose consumption, while gdhB– required 15 h to completely exhaust glucose from the medium. All the strains grew on glucose after a lag phase of 8–10 h, the growth being slower and significantly lower than on succinate. A prolonged lag phase in glucose compared with succinate indicated that succinate was more rapidly utilized in the wild type (Figure 2).
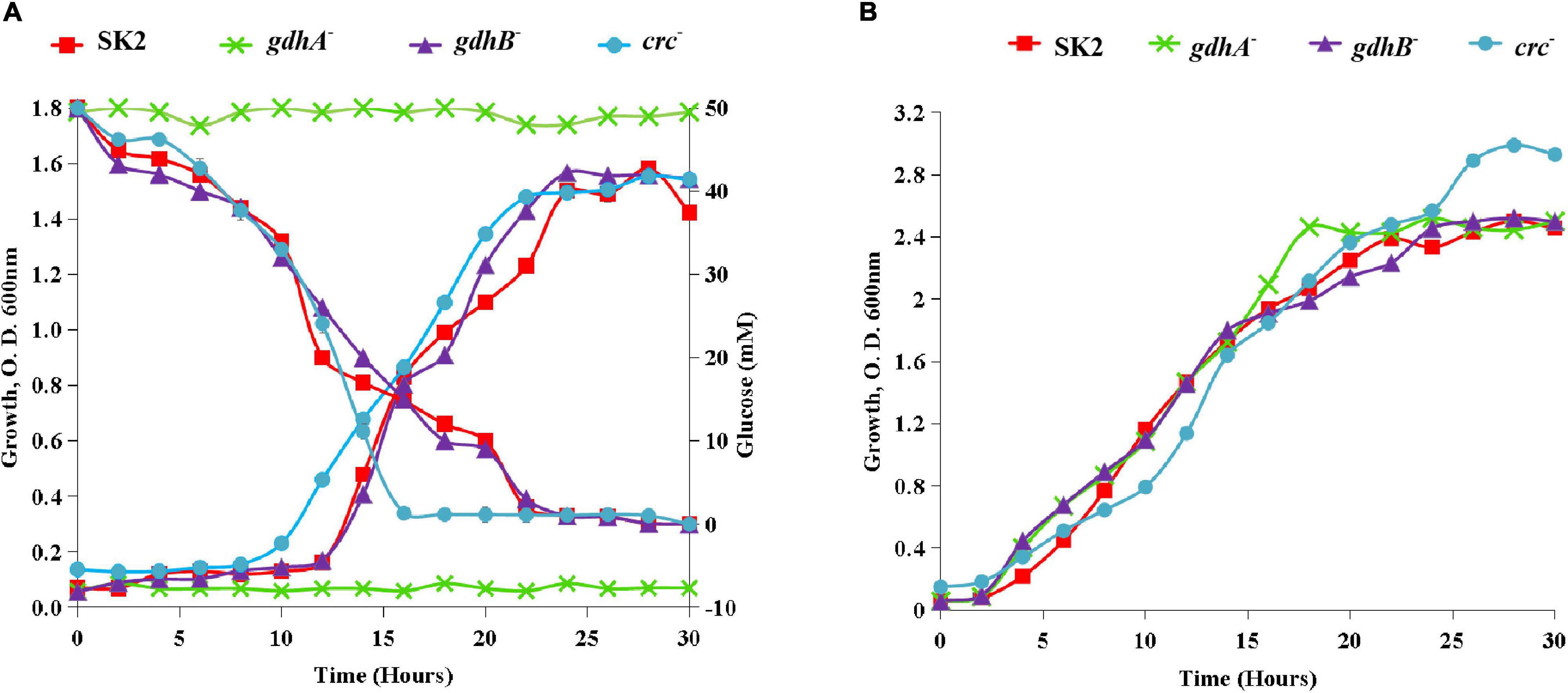
Figure 2. Monoauxic growth profile of wild-type SK2 and mutants on (A) glucose and (B) succinate. SK2 ( ), gdhA– (
), gdhA– ( ), gdhB– (
), gdhB– ( ), and crc– (
), and crc– ( ).
).
Though the molecular mechanism of preferential utilization of succinate over glucose is not clear in Acinetobacter sp., similarities can be drawn from the earlier studies on gammaproteobacteria Pseudomonas sp. The intracellular phosphorylative pathway for glucose utilization was suppressed in P. putida CSV86 when grown in the presence of succinate (Basu et al., 2006). The glucose catabolic enzymes were found to be repressed by succinate in Rhizobium and Bradyrhizobium (Mandal and Chakrabartty, 1993). How the preferential utilization of succinate over glucose and absence of gdhA, gdhB, and crc influence growth of Acinetobacter sp. was investigated (Figure 3). The wild type showed diauxic growth curve on glucose + succinate (Figure 3A), which had a distinct first log, second lag, and second log phases. Succinate utilization during the first log phase was followed by glucose utilization, which was consumed after 7–8 h of incubation, when succinate was completely exhausted from the media. The diauxic growth of the wild type was absent in gdhA–. Though gdhA– could not utilize glucose, it continued to grow due to presence of succinate in the medium (Figure 3B). The absence of gdhA gene or loss of mGDH caused no impairment in succinate uptake. The gdhB– when grown on glucose + succinate had a profile similar to that of the wild type. This observation implied that the absence of gdhB might not have any major role in glucose uptake in the wild type (Figure 3C). In crc–, glucose utilization and growth started from the late lag phase and continued with gradual decrease of glucose from the medium throughout the growth phase. The loss of diauxie in crc– confirmed that Crc was involved in regulating the preferential utilization of succinate over glucose, as crc– continued utilization of glucose regardless of the presence of succinate (Figure 3D).
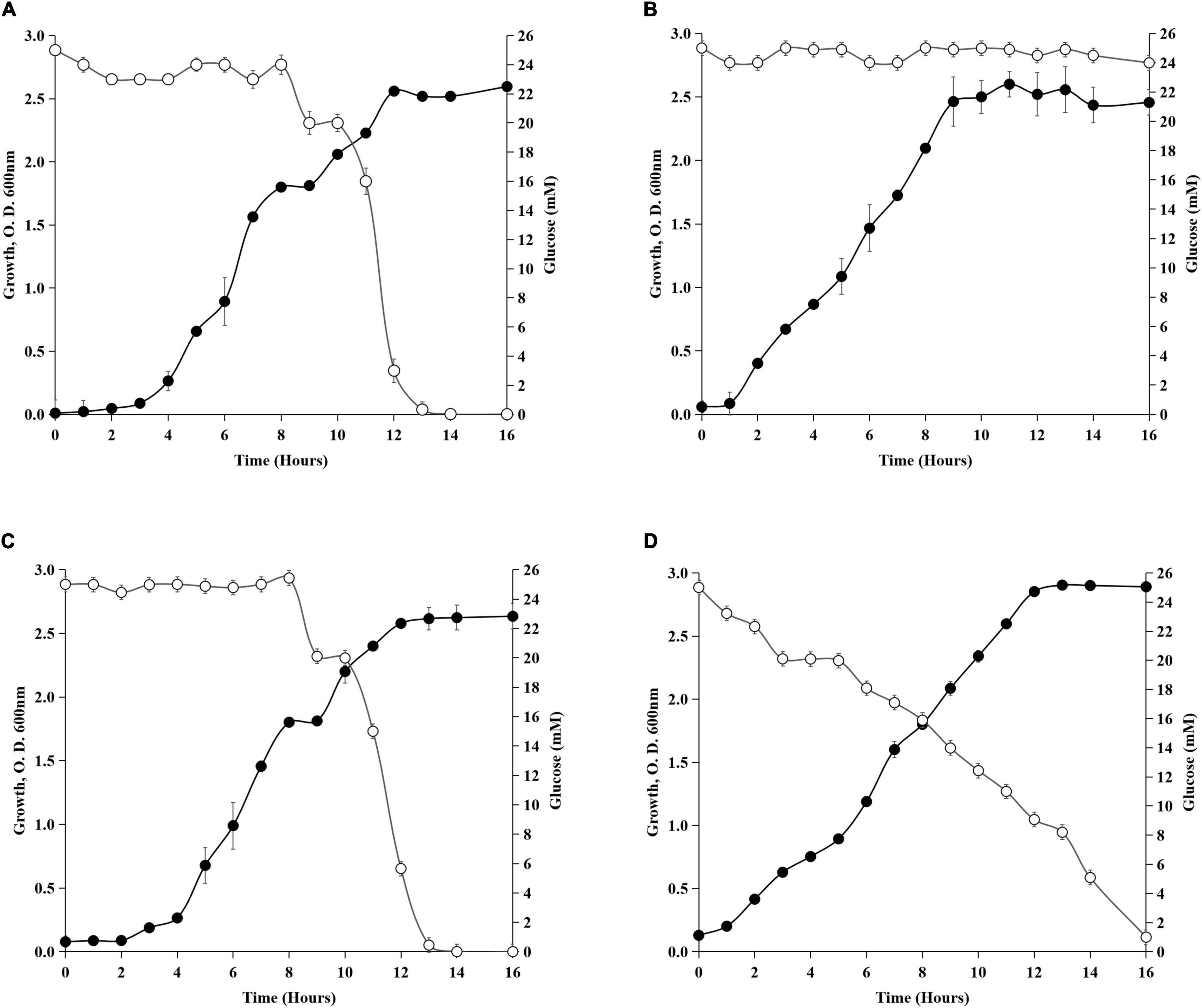
Figure 3. Diauxic growth profile on glucose + succinate. (A) Wild-type SK2, (B) gdhA–, (C) gdhB–, and (D) crc–. Filled circles represent the growth and open circles represent the glucose concentration (mM) in the medium.
Alteration of GDH Activity in gdhA–, gdhB–, and crc– Strains
To correlate the biochemical functions of the gdhA and gdhB genes, enzyme activities of mGDH and sGDH were assayed in the crude cell extracts of the wild-type and mutant strains grown in glucose, succinate, and glucose + succinate-containing media (Figure 4). Since succinate is not a substrate of GDH, no or insignificant mGDH or sGDH activity was observed in the wild-type or mutant strains as expected. In contrast to the high activity of the wild type in glucose, the mGDH activity of gdhA– was abolished (Figure 4A). The mGDH activity of gdhB– remained similar to that of the wild type. Under repression conditions, i.e., in glucose + succinate, the mGDH activity was repressed in the wild type as well as in gdhA– and gdhB–. To assess the involvement of the Crc protein in repression, the GDH activities in the wild-type and the crc– strains were compared. The crc inactivation resulted in an increase of mGDH activity by 30% in glucose + succinate-grown crc– cells as compared with the wild type, indicating derepression. The mGDH and sGDH activities of succinate-grown wild-type cells were only 4–6% of those of the glucose-grown cells. The sGDH activity was completely abolished in glucose-grown gdhB–, which remained unaltered in gdhA– and crc– (Figure 4B). The sGDH activity in glucose + succinate-grown cells of gdhA– and crc– was repressed to the extent of the wild type. The results indicated that the gdhA and gdhB genes in Acinetobacter sp. SK2 encoded enzymes functionally equal to the mGDH and sGDH, respectively.
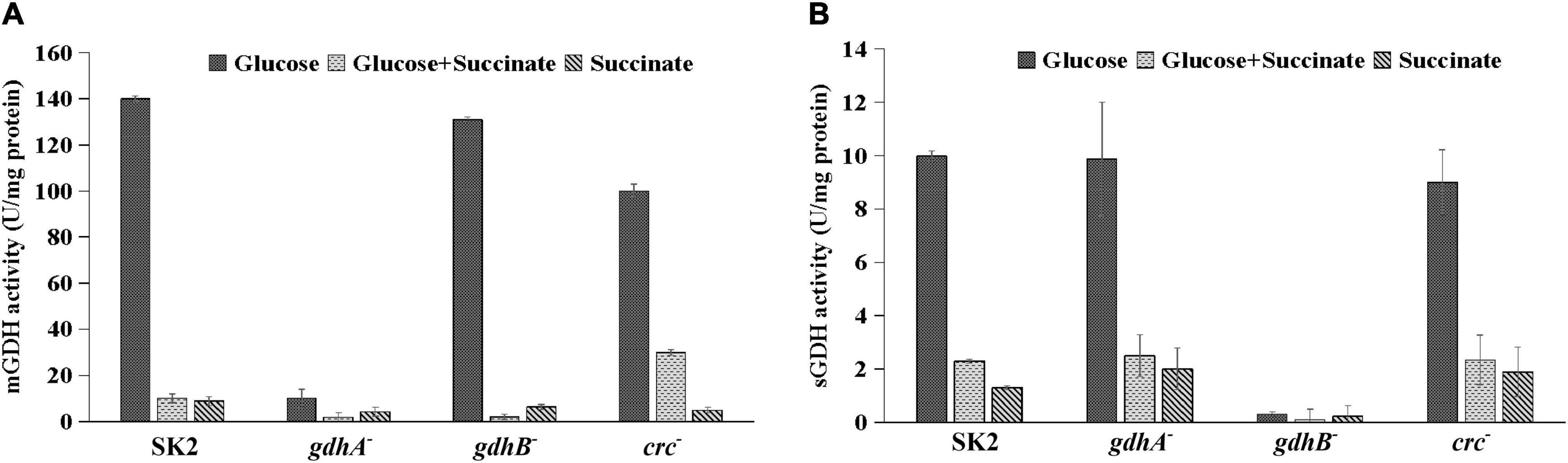
Figure 4. Enzyme activity of glucose, glucose + succinate, and succinate-grown Acinetobacter sp. SK2, gdhA–, gdhB–, and crc– (A) membrane glucose dehydrogenase(mGDH) and (B) soluble glucose dehydrogenase (sGDH).
Quantitative Real-Time PCR Analysis of Glucose Oxidation and Carbon Catabolite Repression-Related Genes
Glucose and succinate are abundant in the rhizosphere; however, the preference among the two will differ among the different groups of bacteria. Therefore, it is probable that the expression of genes required for metabolism of non-preferred carbon source is subjected to repression by the preferred carbon substrates. The nature of CCR has not been examined in Acinetobacter while mimicking the rhizosphere conditions in context to sugars (glucose) and organic acids (succinate). To this end, insertional mutants of glucose oxidation and CCR-related genes were generated. The transcriptional profiles of gdhA, gdhB, crc, and hfq were measured in cells grown on glucose, succinate, and glucose + succinate. Gene transcriptions in the glucose-grown cells were used as a control. The data acquired indicated strong inhibition of gdhA and gdhB by succinate in the wild type (Figures 5A,B). High transcriptional levels of gdhA in glucose (Figure 5A) along with the high enzyme activity (Figure 4A) supported that glucose oxidation was catalyzed by the gdhA gene encoding mGDH enzyme in the wild type. A 1.9-fold downregulation of gdhA in the repression medium was an evidence of succinate-mediated catabolite repression (SMCR) of mGDH in the wild-type strain. A significant reduction of gdhA expression was observed in the presence of glucose + succinate in the wild-type but not in the crc– strain (Figure 5A). These data clearly indicate relaxation of succinate-provoked CCR in crc– strain, which was supported by soluble P estimation (Table 3) and enzyme assay data (Figure 4A) in repression condition. Surprisingly, the expression and activity of gdhA in gdhB– remained unaltered (Figure 5A). Notably, a more pronounced SMCR was detected in the wild type, and the inactivation of crc caused a significant increase in phosphate solubilization (Table 3) and mGDH activity (Figure 4A) under the CCR conditions. However, as expected, insertional inactivation of crc did not abolish the CCR completely, which entails the participation of other yet-unknown CCR regulators.
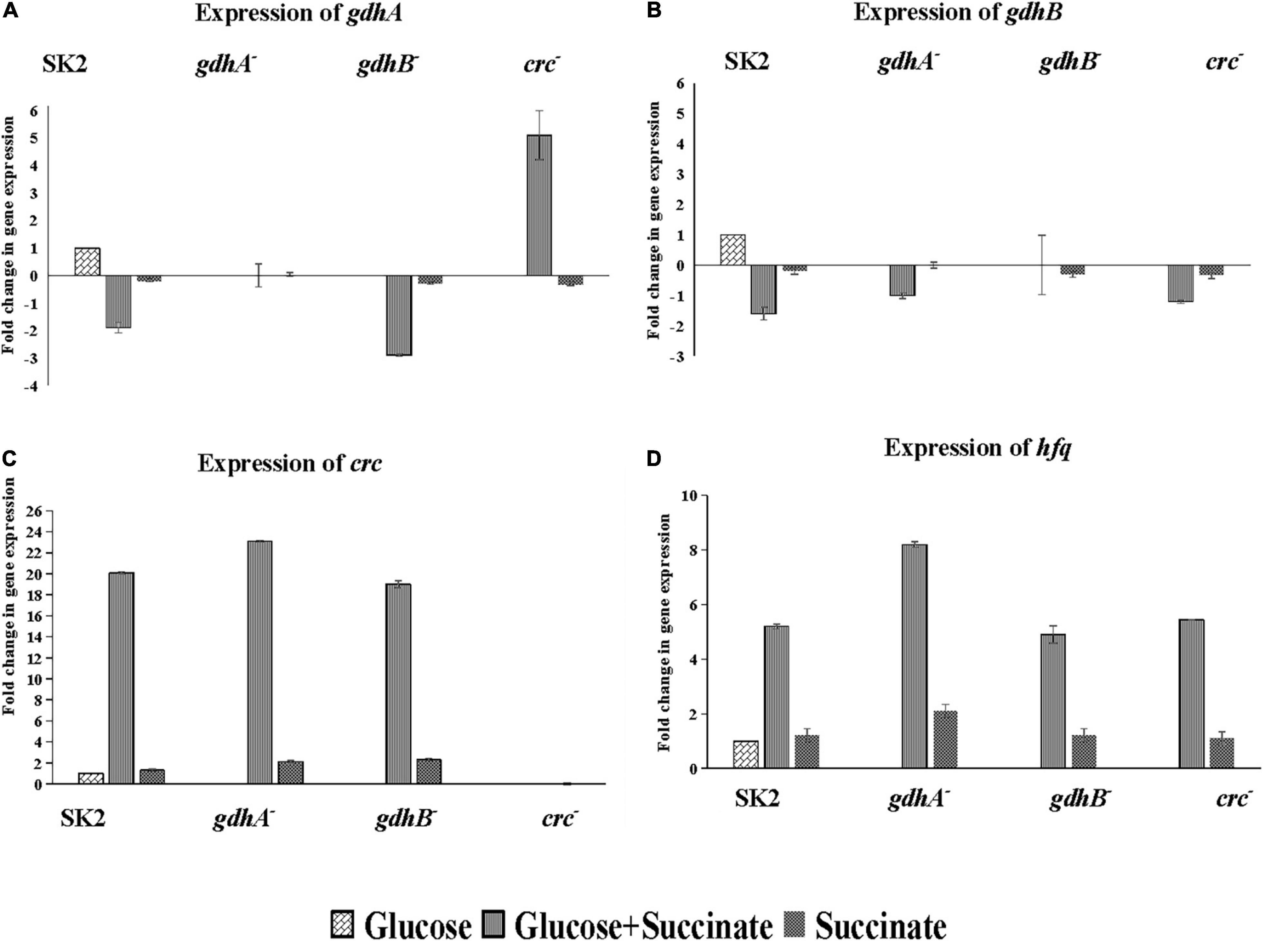
Figure 5. Relative expression of glucose oxidation and CCR genes in Acinetobacter sp. SK2 and mutants on glucose, succinate, and glucose + succinate. Expression of (A) gdhA, (B) gdhB, (C) crc, and (D) hfq. All cells grown to the exponential growth phase were harvested for total RNA isolation. Glucose grown wild-type cells were considered control for calculation of relative expression of all the genes in mutants.
The expression of gdhB in all the mutants and wild type was downregulated irrespective of the genes mutated with the fold change of 1.2–1.6 (Figure 5B). No expression of gdhA and gdhB in both gdhA– and gdhB– grown in succinate was in line with the enzyme activity (Figure 4).
The expression of crc gene was induced in the CCR condition, i.e., in glucose + succinate medium in the wild type as well as in gdhA– and gdhB–. Since crc was expressed only in the presence of secondary carbon source, it is implicated in CCR of the MPS (Figure 5C), and its mutation increased the mGDH activity (Figure 4A). A similar trend in expression of hfq gene in the cells grown in glucose + succinate was observed with lower fold changes as compared with crc (Figure 5D). Succinate being the preferred carbon source induced a basal expression of crc in the wild type, gdhA–, and gdhB– (Figure 5C), while the expression of hfq was recorded in the wild type and the three mutants (Figure 5D).
Effect of Wild-Type and Mutant Strains on the Growth of Vigna radiata Plant
Since phosphate solubilization is one of the important plant growth-promoting traits in the bacteria used as PGPR, the ability of the wild type and mutants in promoting growth of V. radiata was tested. V. radiata inoculated with bacteria were monitored for plant growth parameters in a pot experiment (Table 4). Plants inoculated with the wild-type, gdhA–, gdhB–, and crc– increased the root length and shoot length when compared with the control. The growth parameters in crc– treatments were significantly higher compared with those of the wild type, gdhA–, and gdhB–. crc– treatment significantly improved the root length and shoot length. An increased root length of ∼1.3-fold and shoot length of ∼1.4-fold was recorded in the plants inoculated with the wild type, gdhA–, and gdhB–. The crc– treatment increased the root length by ∼1.5-fold and the shoot length by ∼1.4-fold compared with the uninoculated control. The treatment of plants with the wild-type, gdhA–, gdhB–, and crc– significantly increased the fresh weight (∼1.2- to 1.6-fold) and dry mass (∼1- to 1.8-fold).
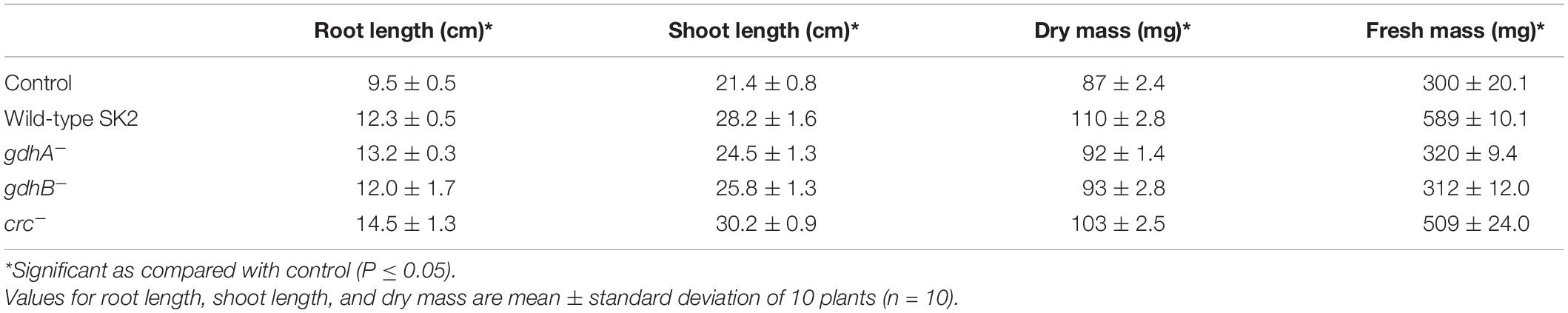
Table 4. Growth parameters of Vigna radiata inoculated with Acinetobacter sp. SK2 and mutant strains.
Discussion
The pyrroloquinoline quinone (PQQ)-dependent GDH has been reported in a wide variety of bacterial species, for example, Acinetobacter calcoaceticus, Gluconobacter suboxydans, and P. aeruginosa, which can carry out periplasmic glucose oxidation. GDH belongs to the largest group of quinoproteins, which require redox cofactor PQQ for its activity (Duine et al., 1982). In vitro studies have shown that the GDH has binding sites for Mg+2; and in vivo, it can bind to Ca+2, PQQ, and especially the substrate glucose (Matsushita et al., 1989). Two types of GDH have been reported based on their localization within the cell. The membrane-bound GDH (mGDH or GDH A) has been reported in many Gram-negative bacteria like Gluconobacter, Pseudomonas (An and Moe, 2016; Suleman et al., 2018), and Acinetobacter species (Rasul et al., 2019); however, sGDH is less common. The atypical existence of sGDH or GDHB is unique to Acinetobacter sp. (Duine et al., 1982; Cleton-Jansen et al., 1988; Matsushita et al., 1995). This atypical character is used as a taxonomical marker for the characterization of A. calcoaceticus. The sGDH of A. calcoaceticus is a homodimer consisting of identical subunits of 55 kDa, each binding to one molecule of PQQ. The amino acid sequences among the mGDH and sGDH are homologous only at the PQQ binding C-terminal region of the protein (Duine et al., 1982; Matsushita et al., 1989). Furthermore, the mechanistic properties of sGDH and mGDH from A. calcoaceticus are shown to be distinct. Since both sGDH and mGDH catalyze similar reactions involving same prosthetic group, their role in glucose oxidation, gluconate-mediated MPS, and its succinate-mediated repression was reported (Bharwad and Rajkumar, 2020). It was hypothesized that the Crc protein, which is known to govern carbon source utilization in Pseudomonas and related species, has a similar role in Acinetobacter sp. SK2. The putative role of Crc in regulating in the SMCR of the MPS phenotype was investigated by generating insertionally inactivated mutants of crc (Figure 1). The genome sequence of Acinetobacter pittii PHAE2 was used to generate mutants of gdhA, gdhB, and crc because its 16S rDNA sequence was 98.42%, similar to that of Acinetobacter sp. SK2.
In this study, the mutant strains of mGDH and sGDH were constructed to determine the role of GDHs in the MPS of Acinetobacter sp. SK2 (Supplementary Figures 1–3). It was evident from the growth profile that gdhA inactivation (gdhA–) abolished glucose oxidation as glucose remained in the spent medium (Figure 2A). The growth profile of gdhA– mutants in succinate was similar to that of the wild-type strain (Figure 2B). The diauxic growth and glucose utilization pattern in glucose + succinate that indicated preferential utilization of succinate over glucose was also identical to that of the wild type (Figure 3). These results are in line with those of Pseudomonas sp. where organic acids have been reported to suppress the uptake and activity of glucose-catabolizing enzymes (Rojo, 2010). Repression of phenanthrene degradation in P. putida by the plant root extract and exudates containing glucose, acetate, and amino acids has also been reported (Rentz et al., 2004).
The complete absence of mGDH activity (Figure 4) and P release was also evident in the gdhA– mutant; and therefore, it was concluded that the mGDH was solely responsible for gluconate production and MPS (Table 3). Similar results were obtained in Gluconobacter oxydans, which is broadly used for various biotechnological applications. In a previous study, to achieve improved growth properties on glucose, a gdhA– strain was constructed, which led to loss of periplasmic glucose oxidation and gluconate production (Zhu et al., 2011). Deletion of GDH in Serratia marcescens resulted in loss of acidification of the medium, which confirmed its role in gluconate production (Fender et al., 2012). In this study, gdhA inactivation completely abolished the MPS and glucose utilization throughout the growth (Figures 1, 3). It is assumed that the wild-type cells might be converting total glucose into gluconate, which is further internalized by the cell for growth when glucose is the sole carbon source. It is possible that most of the glucose available to the cell (20 out of 50 mM of glucose) in the medium was used for MPS (Bharwad and Rajkumar, 2020). These results were in correlation with those of Gyaneshwar et al. (1999) where MPS mutants of Enterobacter asburiae deficient in the GDH activity failed to release the phosphate from alkaline soils, indicating that the GDH activity was required to release the soluble P.
The inactivation of gdhB did not alter the growth of cells in glucose or glucose + succinate when compared with the wild type (Figure 2). Absence of sGDH activity in glucose confirmed gdhB disruption (Figure 4B); however, the unaltered levels of P released or pH confirmed that the extracellular oxidation of glucose to gluconate did not employ intracellular glucose dehydrogenation by sGDH. The gluconate produced and detected in high-performance liquid chromatography (HPLC) analysis (Bharwad and Rajkumar, 2020) was solely due to the activity of mGDH. Cleton-Jansen et al. (1988) showed the presence of a second glucose dehydrogenase apart from mGDH and explained the difference between the in vivo and in vitro substrate specification. The soluble and membrane-bound enzymes have different properties including molecular size, substrate specificity, and kinetics in A. calcoaceticus LMD 79.41 (Matsushita et al., 1988). sGDH is currently used in the accurate monitoring of blood glucose using diabetic control test strips (Yamazaki et al., 2008). Being different from mGDH, sGDH was postulated to be involved in an electron transport to glucose oxidation, but a mutant strain containing only sGDH did not show any in vivo activity of glucose oxidation (Cleton-Jansen et al., 1988). In this study, gdhB– did not show sGDH activity in glucose-containing medium; therefore, it was concluded that gdhB was an indispensable gene for sGDH activity and the oxidation of glucose leading to P solubilization did not employ sGDH. sGDH has been reported to bind PQQ rapidly, with high affinity and with larger conformational changes than mGDH (Cleton-Jansen et al., 1988; Matsushita et al., 1995). Therefore, it was proposed by Matsushita et al. (1995) that the function of sGDH could be of a PQQ carrier protein. sGDH is situated in the cytoplasm, and it could be accumulating PQQ from the external or internal milieu of the cells and may transfer PQQ to the target mGDH situated on the outer membrane to make it functional. Certainly, this was one of the assumptions made for the physiological role of sGDH, but more supporting experimental evidences are required for verification of the function of sGDH, as gdhB inactivation had no influence on the mGDH activity (Figure 4A). Wei et al. (2014) reported similar results upon deletion of sGDH gene in G. oxydans 621H. However, these results were contradictory from those reported for G. oxydans N44-1 where an improved cell growth rate was observed in glucose with the gdhA and gdhB double-gene knockout strain in comparison with the mGDH single-gene-deficient strain. The growth results of G. oxydans N44-1 double-gene deletion strain also suggested that glucose metabolism in this strain was not limited by the deficiency of the sGDH gene (Krajewski et al., 2010). Furthermore, the MPS of the gdhB– in glucose was similar to the P release of the wild type (Table 3). This disproved that sGDH had a role in extracellular glucose oxidation, gluconate secretion, or the MPS phenotype.
Several lines of evidence suggest that Crc acts post-transcriptionally in many Gram-negative bacteria like P. aeruginosa and P. putida to regulate the repressed utilization of polycyclic aromatic hydrocarbons (PAHs) in the presence of succinate (Hester et al., 2000; Yuste and Rojo, 2001). The deletion of crc in A. baylyi did not alter the growth on a number of carbon sources. However, the repression caused on enzyme of protocatechuate breakdown in succinate and acetate was strongly reduced in the crc deficient strain (Zimmermann et al., 2009). Therefore, it was hypothesized that the succinate-mediated repression of mGDH and possibly sGDH leading to the MPS phenotype was under the control of Crc, which upon inactivation may lead to derepression of the MPS phenotype. To assess the hypothesis, the Crc encoding gene crc was disrupted, and its impact on the MPS phenotype was explored. The growth of crc– cells on glucose + succinate was similar to that of the wild type (Figure 1), with loss of diauxie in the repression medium (Figure 3). Glucose utilization from the lag phase of the growth indicated co-utilization of glucose and succinate. Being a global regulator, hindrance in cell growth could be expected upon crc disruption; however, no such negative influence on the growth of the cells was recorded. The mGDH activity of crc– in succinate-containing medium was derepressed up to 40% (Figure 4A), while the sGDH activity remained unaltered. The derepression of mGDH activity increased MPS in the crc– mutants under repression conditions (Table 3), which again indicated that mGDH was solely responsible for P solubilization, while sGDH had no significant contribution to the phenotype. This report confirms that Crc has a role in regulation of the succinate-mediated repression of mGDH and the MPS phenotype in Acinetobacter sp. SK2. The expression of crc varies according to growth conditions and carbon sources. In Pseudomonas fluorescens SBW25, “xylose + succinate” created CCR condition, while xylose and glycerol were co-utilized. The expression of crc was higher only in the CCR condition, whereas when the cells were shifted to non-CCR condition, the expression level of crc decreased, implicating that Crc was one of the top candidates of the CCR regulatory hierarchy for the perception of nutrient signals (Liu et al., 2017). In P. aeruginosa, the inactivation of crc reportedly modified the expression of at least 57 genes when grown in LB medium, whereas in basal salt medium with succinate, the expression of 95 transcripts was altered. Among these genes, many were involved in the transport and assimilation of sugars and amino acids (Sonnleitner et al., 2012). The Crc-mediated regulation is not limited to carbon metabolism and may influence several other properties. In P. aeruginosa, Crc regulates the Lon protease for rhamnolipid production and rhl quorum sensing (Yang et al., 2015), and its role has also been implicated in virulence, biofilm formation, and antibiotic resistance (O’Toole et al., 2000; Yeung et al., 2011).
Apart from Crc, Hfq (RNA chaperone) is involved in the riboregulation of diverse metabolic genes via Crc-small RNAs. An attempt to explore the differences in hfq expression during the CCR conditions was also made in this study. It was observed that the expression pattern of hfq was similar to that of the crc. Apparent expression of hfq in glucose + succinate (the CCR condition) suggested its involvement in carbon source uptake and metabolism (Figure 5). As Crc functions to bring down catabolism of non-preferred carbon sources in the presence of preferred substrates (Sonnleitner et al., 2009; Rojo, 2010; Moreno et al., 2012), the catabolite repression in P. putida requires the Hfq along with the Crc protein. Crc and Hfq operate CCR networks by mediating efficient binding on “catabolite activity motifs” (CA-motifs; consensus AAnAAnAA) of RNAs required for the expression of the target carbon source (Madhushani et al., 2015; Moreno et al., 2015). In P. putida CF600, the dimethylphenol dmp system is required for the catabolism of phenol and (methyl) phenols, which requires Hfq and Crc for controlling DmpR levels via binding to the CA-motif of dmpRm RNA under the CCR conditions (Madhushani et al., 2015). Though the preliminary results implicate Hfq in the sequential utilization of carbon sources, the characterization of the detailed regulatory mechanisms of Hfq and Hfq-regulated pathways of Acinetobacter sp. SK2 needs experimental evidence for the idea to be substantiated.
The wild-type and mutant strains improved the plant growth parameters of V. radiata (Table 4). Significant improvement was observed in plants inoculated with crc–. Derepression of TCP solubilization by crc– in glucose + succinate (Table 3) suggested secretion of gluconate by the mutant strain even under repression condition. Hence, the disruption of crc would likely help in better performance of PGP bacteria in the rhizosphere. The PSB like wild-type SK2 when applied to the field may face constrains due to the multi-tiered regulation by Hfq and Crc, which may influence their performance. Crc deficiency may relieve such bacteria from the metabolic struggle in the environment and help them execute the customized traits or functions.
Conclusion
The natural habitat of Acinetobacter sp. SK2, a promising phosphate solubilizer is the rhizosphere, a nutritionally complex environment. The presence, composition, and abundance of various nutritional substrates change according to the stages of the plant development. When inoculated, it would require continuous changes in gene expression, particularly for those involved in phosphate acquisition. When the organism utilizes root exudates, CCR is most likely to operate due to the presence of glucose and succinate. CCR governed by the Crc protein and Hfq controls the availability of several enzymes and transporters involved in the assimilation of secondary carbon sources. Yet the regulation exerted by Crc on utilization of secondary substrates (for example, glucose, leading to the MPS phenotype) in Acinetobacter sp. had until now been unexplored. In this study, we explored the role of the MPS-mediating enzymes (mGDH and sGDH) and Crc in Acinetobacter sp. SK2. Loss of the MPS ability in gdhA– confirmed the role of mGDH in periplasmic oxidation of glucose to gluconate. Disruption of gdhB– led us to some speculations about the physiological functions of sGDH and unraveled its non-involvement in gluconate production or MPS. By adopting a combination of quantitative physiology experiments and transcriptional patterns of key genes in the wild-type, crc–, gdhA–, and gdhB– strains, we demonstrate that Crc (directly or indirectly) orchestrates the MPS phenotype of Acinetobacter sp. SK2 when glucose + succinate are present. The expression of hfq in the CCR condition implicated the role of this protein in glucose and succinate utilization. Further investigations would be required to confirm participation of other regulatory genes in the regulation of MPS and other PGP traits in CCR (Hfq/sRNAs/CbrAB).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
KB and SR conceived the idea. KB and SR designed the experimental work and performed the experiments with assistance from NG. KB and SR wrote the manuscript with support from NG. KB and NG assisted SR in critically revising the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from Department of Science and Technology (Ministry of Science, India) (Project Sanction number EMR/2017/001464). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Nirma University for providing infrastructure facility for the research work. The authors are very grateful to Dr. Filiatrault, Cornell University for providing the suicide vector, pKnockoutΩ.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.641119/full#supplementary-material
Supplementary Figure 1 | Screening and confirmation of Acinetobacter sp. SK2 mutants (A) Screening of mutants on LB agar with streptomycin and spectinomycin. (B) Selection of gdhA– colonies on YGC medium. The gdhA– cells lost the function of gluconic acid production, leaving no clear zone around the colonies (dashed circle). The false positive colonies still produced clear zones (solid circle). PCR confirmation. (C) Absence or presence of omega in wild-type and mutant strains. A band size of 2 kb consistent with the insertion of omega fragment was obtained from the mutant strains but not from the wild-type. (D) Verification of insertion of omega using primers designed to bind gene of interest and omega (MINF-Omega, SINF-Omega and CINF-Omega combination of primers were used to amplify omega in gdhA, gdhB, and crc, respectively). An increased band of approximately 2.7 to 3 kb was obtained due to the insertion of omega.
Supplementary Figure 2 | Plasmid maps of recombinant vectors pKOgdhA, pKOgdhB, and pKOcrc constructed from pKnockoutΩ. pKOgdhA was created by insertion of ∼500 bp gdhA DNA fragment (amplified from wild-type Acinetobacter sp. SK2) at SalI:PstI site of pKnockoutΩ. pKOgdhB was created by insertion of ∼540 bp gdhB DNA fragment at KpnI:SalI site of pKnockoutΩ. pKOcrc contained ∼650 bp fragment of crc at KpnI:XbaI sites of pKnockoutΩ.
Supplementary Figure 3 | Gene replacement strategy via single strand allelic exchange at the location of gene of interest (indicated as GeneX that can be gdhA, gdhB or crc). (A) Selection of insertional inactivated clones of GeneX (gdhA or gdhB or crc). The insertional inactivated mutants were screened on media containing streptomycin + spectinomycin and further confirmed by PCR. (B) Amplification of omega cassette from the chromosome of wild-type and mutants using primers that anneal to the omega cassette (red arrows). (C) Confirmation of insertion of omega cassette replacing the gene of interest. PCR amplification using primers that overlap with gene of interest (black arrow) and omega (red arrow) yield band of 2.7 to 3 kb.
References
Ames, B. N. (1966). [10] Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8, 115–118.
An, R., and Moe, L. A. (2016). Regulation of pyrroloquinoline quinone-dependent glucose dehydrogenase activity in the model rhizosphere-dwelling bacterium Pseudomonas putida KT2440. Appl. Environ. Microbiol. 82, 4955–4964. doi: 10.1128/AEM.00813-16
Basu, A., Apte, S. K., and Phale, P. S. (2006). Preferential utilization of aromatic compounds over glucose by Pseudomonas putida CSV86. Appl. Environ. Microbiol. 72, 2226–2230. doi: 10.1128/AEM.72.3.2226-2230.2006
Bharwad, K., and Rajkumar, S. (2020). Modulation of PQQ-dependent glucose dehydrogenase (mGDH and sGDH) activity by succinate in phosphate solubilizing plant growth promoting Acinetobacter sp. SK2. 3 Biotech 10:5. doi: 10.1007/s13205-019-1991-2
Bruns, S., Reipschläger, K., Lorenz, M. G., and Wackernagel, W. (1992). “Characterization of natural transformation of the soil bacteria Pseudomonas stutzeri and Acinetobacter calcoaceticus by chromosomal and plasmid DNA,” in Gene Transfers and Environment, ed. M. J. Gauthier (Berlin: Springer), 115–126. doi: 10.1007/978-3-642-77450-8_13
Chao, Y., and Vogel, J. (2010). The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13, 24–33. doi: 10.1016/j.mib.2010.01.001
Chen, Y., Gelfond, J. A., McManus, L. M., and Shireman, P. K. (2009). Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics 10:407. doi: 10.1186/1471-2164-10-407
Cleton-Jansen, A. M., Goosen, N. O. R. A., Wenzel, T. J., and van de Putte, P. I. E. T. E. R. (1988). Cloning of the gene encoding quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus: evidence for the presence of a second enzyme. J. Bacteriol. 170, 2121–2125. doi: 10.1128/jb.170.5.2121-2125.1988
de Fernandez, M. F., Eoyang, L., and August, J. T. (1968). Factor fraction required for the synthesis of bacteriophage Qβ-RNA. Nature 219, 588–590. doi: 10.1038/219588a0
Duine, J. A., Jzn, J. F., and Van der Meer, R. (1982). Different forms of quinoprotein aldose-(glucose-) dehydrogenase in Acinetobacter calcoaceticus. Arch. Microbiol. 131, 27–31. doi: 10.1007/BF00451494
Fender, J. E., Bender, C. M., Stella, N. A., Lahr, R. M., Kalivoda, E. J., and Shanks, R. M. (2012). Serratia marcescens quinoprotein glucose dehydrogenase activity mediates medium acidification and inhibition of prodigiosin production by glucose. Appl. Environ. Microbiol. 78, 6225–6235. doi: 10.1128/AEM.01778-12
Filiatrault, M. J., Stodghill, P. V., Wilson, J., Butcher, B. G., Chen, H., Myers, C. R., et al. (2013). CrcZ and CrcX regulate carbon source utilization in Pseudomonas syringae pathovar tomato strain DC3000. RNA Biol. 10, 245–255. doi: 10.4161/rna.23019
Görke, B., and Stülke, J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624. doi: 10.1038/nrmicro1932
Grant, S. G., Jessee, J., Bloom, F. R., and Hanahan, D. (1990). Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U.S.A. 87, 4645–4649. doi: 10.1073/pnas.87.12.4645
Gyaneshwar, P., Parekh, L. J., Archana, G., Poole, P. S., Collins, M. D., Hutson, R. A., et al. (1999). Involvement of a phosphate starvation inducible glucose dehydrogenase in soil phosphate solubilization by Enterobacter asburiae. FEMS Microbiol. Lett. 171, 223–229. doi: 10.1111/j.1574-6968.1999.tb13436.x
Hernández-Arranz, S., Moreno, R., and Rojo, F. (2013). The translational repressor Crc controls the Pseudomonas putida benzoate and alkane catabolic pathways using a multi-tier regulation strategy. Environ. Microbiol. 15, 227–241. doi: 10.1111/j.1462-2920.2012.02863.x
Hester, K. L., Madhusudhan, K. T., and Sokatch, J. R. (2000). Catabolite repression control by crc in 2xYT medium is mediated by posttranscriptional regulation of bkdR expression in Pseudomonas putida. J. Bacteriol. 182, 1150–1153. doi: 10.1128/JB.182.4.1150-1153.2000
Kamilova, F., Kravchenko, L. V., Shaposhnikov, A. I., Azarova, T., Makarova, N., and Lugtenberg, B. (2006). Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol. Plant Microbe Interact. 19, 250–256. doi: 10.1094/MPMI-19-0250
Krajewski, V., Simić, P., Mouncey, N. J., Bringer, S., Sahm, H., and Bott, M. (2010). Metabolic engineering of Gluconobacter oxydans for improved growth rate and growth yield on glucose by elimination of gluconate formation. Appl. Environ. Microbiol. 76, 4369–4376. doi: 10.1128/AEM.03022-09
Kumari, P., Meena, M., Gupta, P., Dubey, M. K., Nath, G., and Upadhyay, R. S. (2018). Plant growth promoting rhizobacteria and their biopriming for growth promotion in mung bean (Vigna radiata (L.) R. Wilczek). Biocatal. Agric. Biotechnol. 16, 163–171. doi: 10.1016/j.bcab.2018.07.030
Kuo, H. Y., Chao, H. H., Liao, P. C., Hsu, L., Chang, K. C., Tung, C. H., et al. (2017). Functional characterization of Acinetobacter baumannii lacking the RNA chaperone Hfq. Front. Microbiol. 8:2068. doi: 10.3389/fmicb.2017.02068
Liu, Y., Gokhale, C. S., Rainey, P. B., and Zhang, X. X. (2017). Unravelling the complexity and redundancy of carbon catabolic repression in Pseudomonas fluorescens SBW25. Mol. Microbiol. 105, 589–605. doi: 10.1111/mmi.13720
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lorenz, M. G., Reipschläger, K., and Wackernagel, W. (1992). Plasmid transformation of naturally competent Acinetobacter calcoaceticus in non-sterile soil extract and groundwater. Arch. Microbiol. 157, 355–360. doi: 10.1007/BF00248681
MacGregor, C. H., Arora, S. K., Hager, P. W., Dail, M. B., and Phibbs, P. V. (1996). The nucleotide sequence of the Pseudomonas aeruginosa pyrE-crc-rph region and the purification of the crc gene product. J. Bacteriol. 178, 5627–5635. doi: 10.1128/jb.178.19.5627-5635.1996
MacGregor, C. H., Wolff, J. A., Arora, S. K., and Phibbs, P. V. (1991). Cloning of a catabolite repression control (crc) gene from Pseudomonas aeruginosa, expression of the gene in Escherichia coli, and identification of the gene product in Pseudomonas aeruginosa. J. Bacteriol. 173, 7204–7212. doi: 10.1128/jb.173.22.7204-7212.1991
Madhushani, A., del Peso-Santos, T., Moreno, R., Rojo, F., and Shingler, V. (2015). Transcriptional and translational control through the 5′-leader region of the dmpR master regulatory gene of phenol metabolism. Environ. Microbiol. 17, 119–133. doi: 10.1111/1462-2920.12511
Mandal, N. C., and Chakrabartty, P. K. (1993). Succinate-mediated catabolite repression of enzymes of glucose metabolism in root-nodule bacteria. Curr. Microbiol. 26, 247–251. doi: 10.1007/BF01575912
Mara, K., Decorosi, F., Viti, C., Giovannetti, L., Papaleo, M. C., Maida, I., et al. (2012). Molecular and phenotypic characterization of Acinetobacter strains able to degrade diesel fuel. Res. Microbiol. 163, 161–172. doi: 10.1016/j.resmic.2011.12.002
Matsushita, K., Shinagawa, E., Adachi, O., and Ameyama, M. (1988). Quinoprotein D-glucose dehydrogenases in Acinetobacter calcoaceticus LMD 79. 41: purification and characterization of the membrane-bound enzyme distinct from the soluble enzyme. Antonie Van Leeuwenhoek 56, 63–72. doi: 10.1007/978-94-009-0957-1_11
Matsushita, K., Shinagawa, E., Adachi, O., and Ameyama, M. (1989). Quinoprotein D-glucose dehydrogenase of the Acinetobacter calcoaceticus respiratory chain: membrane-bound and soluble forms are different molecular species. Biochemistry 28, 6276–6280. doi: 10.1021/bi00441a020
Matsushita, K., Toyama, H., Ameyama, M., Adachi, O., Dewanti, A., and Duine, J. A. (1995). Soluble and membrane-bound quinoprotein D-glucose dehydrogenases of the Acinetobacter calcoaceticus: the binding process of PQQ to the apoenzymes. Biosci. Biotechnol. Biochem. 59, 1548–1555. doi: 10.1271/bbb.59.1548
Moreno, R., Fonseca, P., and Rojo, F. (2012). Two small RNAs, CrcY and CrcZ, act in concert to sequester the Crc global regulator in Pseudomonas putida, modulating catabolite repression. Mol. Microbiol. 83, 24–40. doi: 10.1111/j.1365-2958.2011.07912.x
Moreno, R., Hernández-Arranz, S., La Rosa, R., Yuste, L., Madhushani, A., Shingler, V., et al. (2015). The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ. Microbiol. 17, 105–118. doi: 10.1111/1462-2920.12499
O’Toole, G. A., Gibbs, K. A., Hager, P. W., Phibbs, P. V., and Kolter, R. (2000). The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182, 425–431. doi: 10.1128/JB.182.2.425-431.2000
Pikovskaya, R. I. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17, 362–370.
Rajput, M. S., Kumar, G. N., and Rajkumar, S. (2013). Repression of oxalic acid-mediated mineral phosphate solubilization in rhizospheric isolates of Klebsiella pneumoniae by succinate. Arch. Microbiol. 195, 81–88. doi: 10.1007/s00203-012-0850-x
Rasul, M., Yasmin, S., Suleman, M., Zaheer, A., Reitz, T., Tarkka, M. T., et al. (2019). Glucose dehydrogenase gene containing phosphobacteria for biofortification of Phosphorus with growth promotion of rice. Microbiol. Res. 223, 1–12. doi: 10.1016/j.micres.2019.03.004
Rentz, J. A., Alvarez, P. J., and Schnoor, J. L. (2004). Repression of Pseudomonas putida phenanthrene-degrading activity by plant root extracts and exudates. Environ. Microbiol. 6, 574–583. doi: 10.1111/j.1462-2920.2004.00589.x
Rojas-Tapias, D. F., Bonilla, R., and Dussán, J. (2014). Effect of inoculation and co-inoculation of Acinetobacter sp. RG30 and Pseudomonas putida GN04 on growth, fitness, and copper accumulation of maize (Zea mays). Water Air Soil Pollut. 225:2232. doi: 10.1007/s11270-014-2232-2
Rojo, F. (2010). Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbial. Rev. 34, 658–684. doi: 10.1111/j.1574-6976.2010.00218.x
Simonsen, K. T., Nielsen, G., Bjerrum, J. V., Kruse, T., Kallipolitis, B. H., and Møller-Jensen, J. (2011). A role for the RNA chaperone Hfq in controlling adherent-invasive Escherichia coli colonization and virulence. PLoS One 6:e16387. doi: 10.1371/journal.pone.0016387
Sonnleitner, E., Abdou, L., and Haas, D. (2009). Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 106, 21866–21871. doi: 10.1073/pnas.0910308106
Sonnleitner, E., Valentini, M., Wenner, N., el Zahar Haichar, F., Haas, D., and Lapouge, K. (2012). Novel targets of the CbrAB/Crc carbon catabolite control system revealed by transcript abundance in Pseudomonas aeruginosa. PLoS One 7:e44637. doi: 10.1371/journal.pone.0044637
Suleman, M., Yasmin, S., Rasul, M., Yahya, M., Atta, B. M., and Mirza, M. S. (2018). Phosphate solubilizing bacteria with glucose dehydrogenase gene for phosphorus uptake and beneficial effects on wheat. PLoS One 13:e0204408. doi: 10.1371/journal.pone.0204408
Vogel, J., and Luisi, B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589. doi: 10.1038/nrmicro2615
Wang, M. C., Chien, H. F., Tsai, Y. L., Liu, M. C., and Liaw, S. J. (2014). The RNA chaperone Hfq is involved in stress tolerance and virulence in uropathogenic Proteus mirabilis. PLoS One 9:e85626. doi: 10.1371/journal.pone.0085626
Wei, L., Zhu, D., Zhou, J., Zhang, J., Zhu, K., Du, L., et al. (2014). Revealing in vivo glucose utilization of Gluconobacter oxydans 621H Δmgdh strain by mutagenesis. Microbiol. Res. 169, 469–475. doi: 10.1016/j.micres.2013.08.002
Windgassen, M., Urban, A., and Jaeger, K. E. (2000). Rapid gene inactivation in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 193, 201–205. doi: 10.1111/j.1574-6968.2000.tb09424.x
Wolff, J. A., MacGregor, C. H., Eisenberg, R. C., and Phibbs, P. V. (1991). Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J. Bacteriol. 173, 4700–4706. doi: 10.1128/jb.173.15.4700-4706.1991
Yamazaki, T., Okuda-Shimazaki, J., Sakata, C., Tsuya, T., and Sode, K. (2008). Construction and characterization of direct electron transfer-type continuous glucose monitoring system employing thermostable glucose dehydrogenase complex. Anal. Lett. 41, 2363–2373. doi: 10.1080/00032710802350567
Yang, N., Ding, S., Chen, F., Zhang, X., Xia, Y., Di, H., et al. (2015). The C rc protein participates in down-regulation of the L on gene to promote rhamnolipid production and rhl quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 96, 526–547. doi: 10.1111/mmi.12954
Yeung, A. T., Bains, M., and Hancock, R. E. (2011). The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J. Bacteriol. 193, 918–931. doi: 10.1128/JB.00911-10
Yildirim, S., Thompson, M. G., Jacobs, A. C., Zurawski, D. V., and Kirkup, B. C. (2016). Evaluation of parameters for high efficiency transformation of Acinetobacter baumannii. Sci. Rep. 6:22110. doi: 10.1038/srep22110
Young, D. M., Parke, D., and Ornston, L. N. (2005). Opportunities for genetic investigation afforded by Acinetobacter baylyi, a nutritionally versatile bacterial species that is highly competent for natural transformation. Annu. Rev. Microbiol. 59, 519–551. doi: 10.1146/annurev.micro.59.051905.105823
Yuste, L., and Rojo, F. (2001). Role of the crc gene in catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 183, 6197–6206. doi: 10.1128/JB.183.21.6197-6206.2001
Zhu, K., Lu, L., Wei, L., Wei, D., Imanaka, T., and Hua, Q. (2011). Modification and evolution of Gluconobacter oxydans for enhanced growth and biotransformation capabilities at low glucose concentration. Mol. Biotechnol. 49, 56–64. doi: 10.1007/s12033-011-9378-6
Keywords: Acinetobacter sp. SK2, succinate, MPS, sGDH, mGDH, Crc
Citation: Bharwad K, Ghoghari N and Rajkumar S (2021) Crc Regulates Succinate-Mediated Repression of Mineral Phosphate Solubilization in Acinetobacter sp. SK2 by Modulating Membrane Glucose Dehydrogenase. Front. Microbiol. 12:641119. doi: 10.3389/fmicb.2021.641119
Received: 13 December 2020; Accepted: 30 April 2021;
Published: 12 July 2021.
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Martin Jemo, Mohammed VI Polytechnic University, MoroccoMatthew McIntosh, Justus-Liebig-University Giessen, Germany
Praveen Rahi, National Centre for Cell Science, India
Copyright © 2021 Bharwad, Ghoghari and Rajkumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shalini Rajkumar, c2hhbGluaS5yamtAbmlybWF1bmkuYWMuaW4=
 Krishna Bharwad
Krishna Bharwad Niharika Ghoghari
Niharika Ghoghari Shalini Rajkumar
Shalini Rajkumar