- 1Department of Chemical Engineering, Tokyo University of Agriculture and Technology, Koganei, Japan
- 2Global Innovation Research Institute, Tokyo University of Agriculture and Technology, Fuchu, Japan
- 3Department of Civil and Environmental Engineering Science, Institute IWAR, Chair of Wastewater Engineering, Technical University of Darmstadt, Darmstadt, Germany
Upcycling wastes into valuable products by mixed microbial communities has recently received considerable attention. Sustainable production of high-value substances from one-carbon (C1) compounds, e.g., methanol supplemented as an external electron donor in bioreactors for wastewater treatment, is a promising application of upcycling. This study undertook a gene-centric approach to screen valuable production potentials from mixed culture biomass, removing organic carbon and nitrogen from landfill leachate. To this end, the microbial community of the activated sludge from a landfill leachate treatment plant and its metabolic potential for the production of seven valuable products were investigated. The DNA extracted from the activated sludge was subjected to shotgun metagenome sequencing to analyze the microbial taxonomy and functions associated with producing the seven products. The functional analysis confirmed that the activated sludge could produce six of the valuable products, ectoine, polyhydroxybutyrate (PHB), zeaxanthin, astaxanthin, acetoin, and 2,3-butanediol. Quantification of the detected functional gene hit numbers for these valuable products as a primary trial identified a potential rate-limiting metabolic pathway, e.g., conversion of L-2,4-diaminobutyrate into N-γ-acetyl-L2,4,-diaminobutyrate during the ectoine biosynthesis. Overall, this study demonstrated that primary screening by the proposed gene-centric approach can be used to evaluate the potential for the production of valuable products using mixed culture or single microbe in engineered systems. The proposed approach can be expanded to sites where water purification is highly required, but resource recovery, or upcycling has not been implemented.
Introduction
Microbial resource management (MRM), which enables a better understanding and harnessing of microorganisms in ecosystems for sustainable development, has been advocated and implemented in engineered systems (Verstraete, 2007). One application of MRM is recovery or upcycling of valuable resources from wastes and wastewater (McCarty et al., 2011). MRM does not hinge upon the use of one species, but rather a myriad of uncultured microorganisms with various functions that are not fully understood. Accordingly, exploring and comprehending microbial community compositions and functions could pave the way for the innovation of novel bioprocesses, contributing to resource recovery, production of valuable materials, and energy-saving by wastewater treatment systems.
Among targeted sites amenable to MRM applications, bioreactors treating industrial water and wastewater, e.g., waste produced during food processing and by livestock industries (Matassa et al., 2015; Spiller et al., 2020) and landfill leachate, are promising. From a societal perspective, the demand for appropriate leachate treatment is expected to surge as the amount of waste increases from 2.0 billion tons in 2016 to an expected 3.4 billion tons by 2050 (Kaza et al., 2018). Landfill leachate consists of small molecular organic substances and biodegradable volatile fatty acids in the early stages of the landfill process, while ammonia and persistent organic substances are the main components in the later stages following microbial decomposition of major organic waste constituents (Costa et al., 2019). Methanol is often used as a C1 electron donor to replenish insufficient supplies of electron donors for nitrogen oxides produced during ammonia oxidation in landfill leachate treatment after long-term implementation to facilitate denitrification (Ilies and Mavinic, 2001). Recent studies have shown that methanotrophs and methylotrophs, which are both C1-assimilating bacteria, synthesize valuable products including biodegradable polymers and active ingredients in cosmetics (Zamakhaeva et al., 2017). Given that leachate treatment plants likely harbor microbial consortia that assimilate C1 compounds and ammonia, leachate can be expected to function as a site at which these simple wastewater constituents can be converted into valuable products.
Methanol supplied to activated sludge of a wastewater treatment plant and indigenously high salinity (1–3%) triggers changes in microbial community compositions (Osaka et al., 2006). Moreover, microbial communities enriched by high methanol concentrations are reportedly capable of producing high-value products such as ectoine (Doronina et al., 2010), polyhydroxybutyrate (PHB; Zamakhaeva et al., 2017), acetoin (3-hydroxy-2-butanone; Schendel et al., 1990; Gogleva et al., 2010), and carotenoids (Berry et al., 2003; Lee et al., 2004; Guo et al., 2017). Despite their benefits (Supplementary Table 1), chemical industries face challenges with cost-effective and high-yield production of these products (Pastor et al., 2010; Xiao and Lu, 2014; Levett et al., 2016; Zhang, 2018). However, the development of a biorefinery platform based on the MRM concept has the potential to overcome the challenges. Therefore, bioprocesses based on mixed microbial communities are potential surrogates to commercial chemical processes. Accordingly, it is important to explore and harness effective microbes harboring the genetic potential to synthesize valuable products. However, the phylogeny and physiology of such microbes and varieties of targeted valuable products remain mostly unexploited.
The design and operation of bioreactors for treating wastewater has been reliant on an empirical top-down approach, in which macro-scale variables, e.g., pH and dissolved oxygen, are precisely controlled because of limited understanding of the intricate and diverse compositions of microbial communities in wastewater (Lawson et al., 2019). However, recent advances in high-throughput sequencing technologies, genome-centric with binning, and multi-omics approaches have enabled analysis of micro-scale microbiological processes, including metabolic networks, and interactions among microbes (Nobu et al., 2015; Lawson et al., 2017). Despite massive and invaluable information on functions and taxonomies of microbial communities, genome-centric and multi-omics approaches, however, are labor-intensive and time-consuming, and require strong expertise on microbial ecology and bioinformatics, which does not allow engineers to fully harness the approaches. Therefore, a simple platform to rapidly explore the possibility for a microbial consortium to produce valuable products is demanded.
This study advocates a simple gene-centric method without binning to screen the capability of synthesizing valuable products. Taking a landfill as an MRM site, the analysis was conducted to elucidate the microbial community composition and functions of activated sludge in a landfill leachate treatment system in which methanol was supplemented as an electron donor for nitrogen removal. Using shotgun metagenomics analysis, the metabolic potential for the production of valuable products was targeted. The specific goals of this study were: (i) to determine if an individual species of bacteria or microbial community in the activated sludge possesses the metabolic potential for the synthesis of targeted products at the gene level; and (ii) to infer potential pathways that could act as rate-limiting steps during production.
Materials and Methods
Sampling
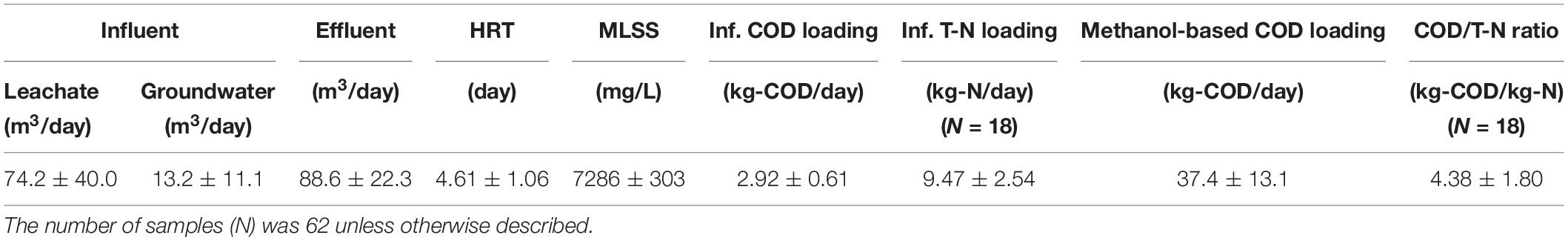
Activated sludge of a landfill leachate treatment plant at a completed landfill (Tokyo, Japan) was collected on 20 July 2018. The landfill was in operation for several decades, during which time the leachate was treated by an activated sludge system in a sequencing batch mode. The sequencing batch reactor (SBR) had an effective volume of 740 m3 and received 74.2 m3/day of leachate on average. One cycle of the SBR operation lasted 2 h and consisted of 10 min for inflow, 45 min for mixing to ensure anoxic conditions, 15 min for aeration, and 50 min for settling and outflow of the treated water. Prior to entry of the influent to the SBR, the leachate was mixed with groundwater at a volumetric ratio of 3 (leachate): 1 (groundwater). Tables 1, 2 summarize the operating conditions, and the leachate characteristics and SBR performance, respectively. The organic carbon concentration was low (BOD < 15 mg/L and COD < 50 mg/L), whereas the ammonium concentration in the leachate was as high as 150 mg-N/L. Because of the imbalance of the carbon/nitrogen ratio, about 5.3 L of 50% v/v methanol was added during each inflow period as an external electron donor for denitrification. The methanol amount allowed a relative loading of COD/T-N (kg-COD/kg-N) of 4.38 ± 1.80, which is higher than the denitrification stoichiometry with methanol.
DNA Extraction
Twenty liters of activated sludge were taken from the SBR and immediately transferred to the laboratory. After thoroughly mixing the activated sludge, 15 mL of wastewater containing 22.2 mg-wet weight-activated sludge were used for DNA extraction by a DNA extraction kit (Fast DNATM Spin Kit for Soil, MP-Biomedicals, Santa Ana, CA, United States) according to the manufacturer’s protocol. The DNA concentration was measured using a spectrophotometer (NanoDrop 2000c, Thermo Scientific, Wilmington, DE, United States).
Sequence Processing and Metagenomic Assembly
The sequencing and metagenomic assembly have been described in detail in a previous study (Yasuda et al., 2020). Briefly, sequencing was performed with an Ion Torrent (ION Torrent Ion S5) using an Ion 530TM Chip. Torrent Suite v 4.4.2 (Thermo Fisher Scientific, Germany) was used for base calling and to run multiplexing according to the manufacturer’s protocol. After quality filtering of metagenomic reads and trimming both sides of the reads to ensure a Q20 quality score, 20.24 million reads were processed, giving 13,005,150 reads. Megahit v 1.1.3 (Li et al., 2015) was used at the minimum contig length of 1,000 bp for assembly, resulting in acquisition of 104.86 Mbp contigs. Mapping the reads to the assembly was accomplished with Bowtie2 v 2.3.4.2 (Langmead and Salzberg, 2013), and the subsequent identification of single-copy bacterial and archaeal genes was achieved using HMMER v 3.2 (Finn et al., 2011). Taxonomy was assigned to contigs using Centrifuge v 1.0.3 (Kim et al., 2016). The latest taxonomic information was used for classification (Waite et al., 2017, 2020; Parks et al., 2018). Default parameters were used for all software unless otherwise specified.
Functional Genes and Metabolic Pathways of Valuable Products
Amino-acid sequence data obtained by the HMMER procedure were used to query the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa and Goto, 2000) with GhostKOALA v 2.1 (Kanehisa et al., 2016). The existence of functional genes and metabolic pathways associated with the production of valuable products by either microbial communities or a single species of indigenous bacteria were confirmed. Based on a literature survey for valuable products microbiologically synthesized from C1 compounds, the metabolic pathways and related functional genes for the production of ectoine (Pastor et al., 2010), PHB (Orita et al., 2014), carotenoids (astaxanthin and zeaxanthin; Ye and Bhatia, 2012; Zhang, 2018), and acetoin (Nielsen et al., 2010) were selected. Carotenoids are synthesized from acetyl-CoA to terpenoid backbone synthesis via either a mevalonate pathway or a non-mevalonate pathway (Supplementary Figure 1 and Supplementary Table 1). The majority of bacteria possess the latter, whereas some use the former pathway (Hoshino and Gaucher, 2018; Rico et al., 2019). Therefore, this study investigated both pathways.
This research assumed that all eubacteria possess central metabolisms, e.g., glycolysis and the TCA cycle. Essential functional genes and enzymes regulating biosynthesis of each valuable product are listed in Table 3. In addition to these compounds that C1-assimilating bacteria are potentially able to synthesize, 2,3-butanediol, which is a promising fuel that is chemically produced from acetoin (Nielsen et al., 2010; Ji et al., 2011), and lutein, which is a carotenoid mainly produced by plants and is known to play a role in vision enhancement (Ye and Bhatia, 2012; Nwachukwu et al., 2016; Zhang, 2018), were also selected.
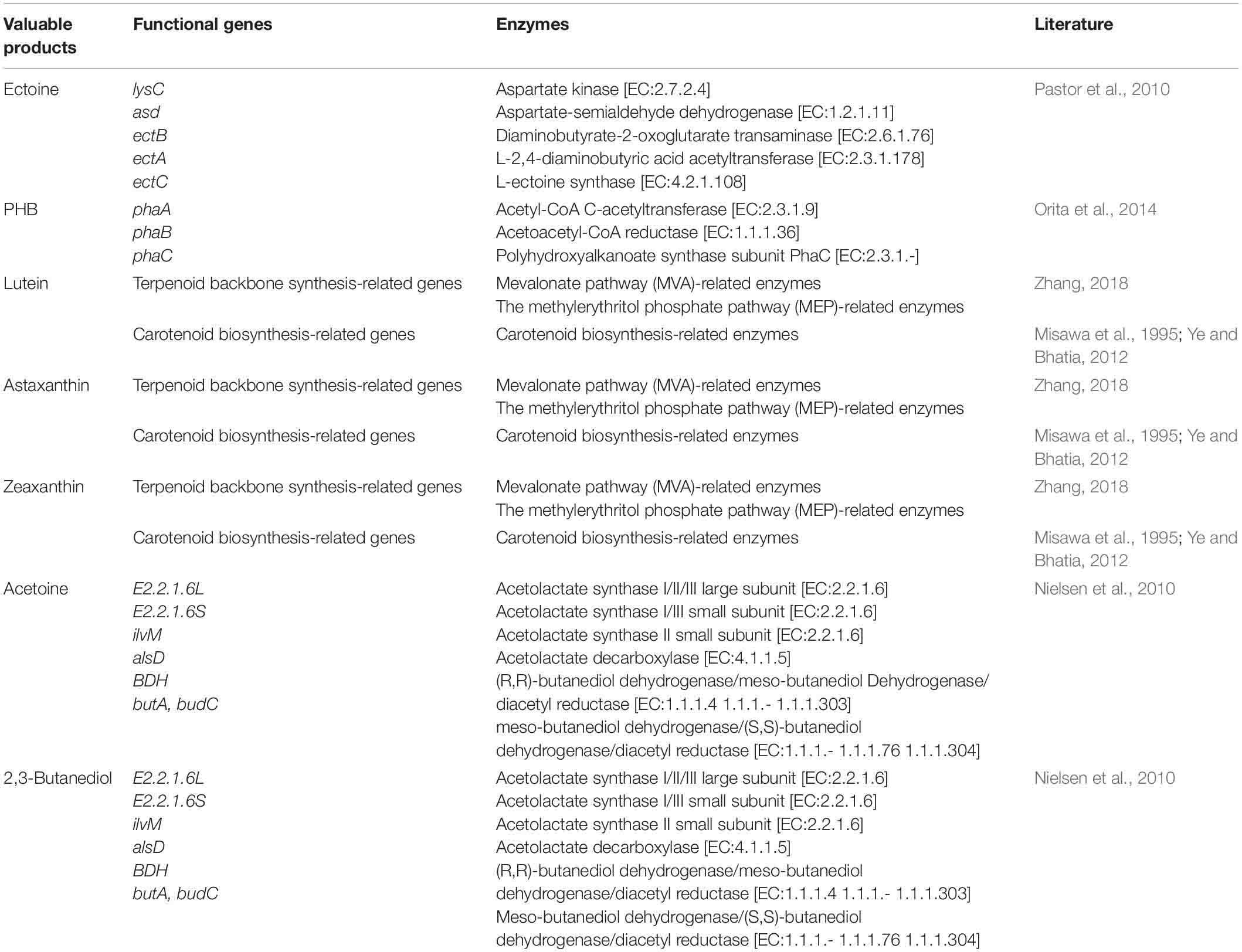
Table 3. Functional genes and enzymes related to production of valuable products. The functional genes of the metabolic pathways, consist of terpenoid backbone synthesis and carotenoid biosynthesis, are shown in Supplementary Table 1.
Visualization of Figures
Taxonomic classification results (Figures 1, 2 and Supplementary Figure 1) and repertories of metabolic pathways (Figure 3) were visualized with R (R Core Team, 2020), Rstudio (RStudio_Team, 2020), and Tidyverse package (Wickham et al., 2019).
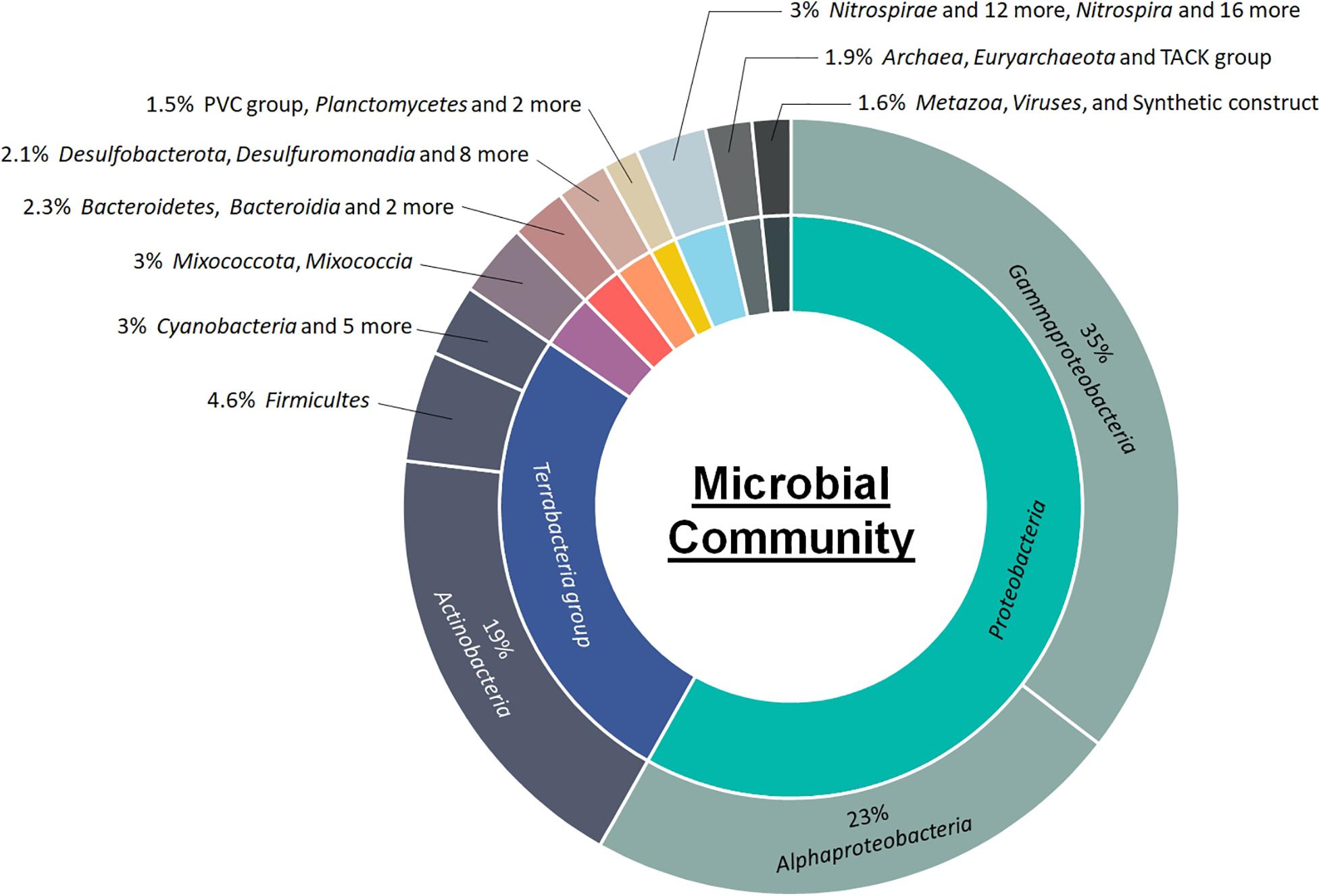
Figure 1. Class-level microbial community composition and relative abundance in activated sludge of landfill leachate.
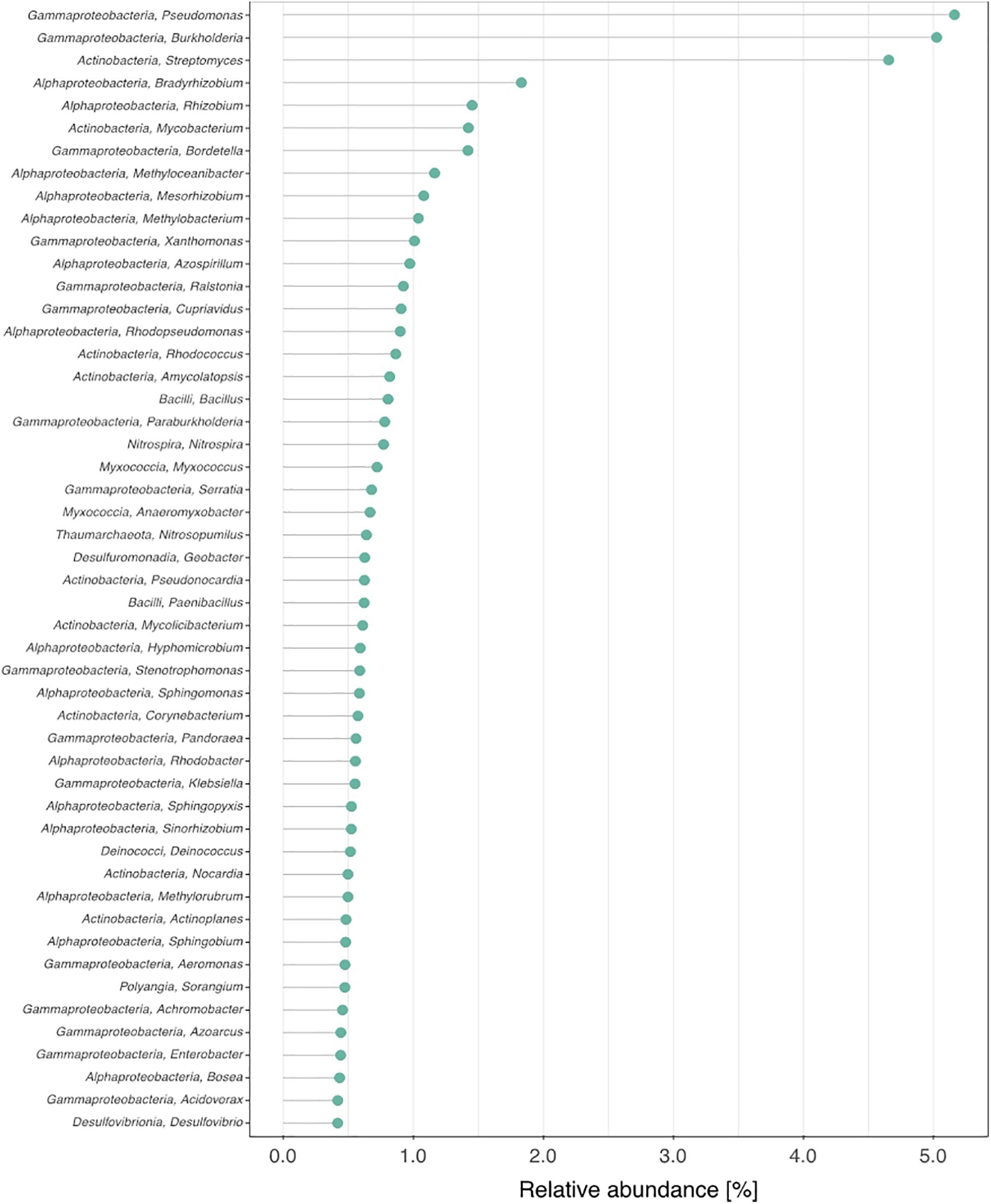
Figure 2. Fifty most abundant microbial genera and their relative abundances in the activated sludge sample.
Nucleotide Accession Numbers
The sequencing data generated by metagenomic analysis have been deposited in DDBJ with the following accession numbers: DRA009549, BioProject: PRJDB9252, and BioSample: SAMD00202538.
Results
Landfill Leachate Compositions and Reactor Performances
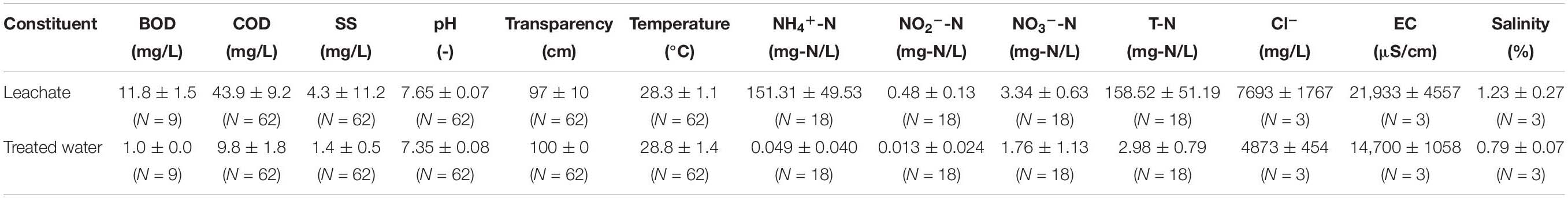
The SBR-based activated sludge system in this study intermittently received methanol as an external electron donor to replenish the limited amount of organic carbon constituents (average COD concentration of 43.9 ± 9.2 mg/L) from the closed landfill. The leachate contained a chloride concentration of 7,693 ± 1,767 mg/L, and the salinity was calculated at 1.23% by the correlation with electron conductivity and temperature (APHA et al., 1992), which is classified as brackish (Table 2). The average removal efficiencies of COD (including the added methanol) and nitrogen were 95.5 ± 1.6 and 97.2 ± 2.1%, and the volumetric removal rates of COD and T-N were 26.1 ± 8.8 g-COD/m3/day and 16.6 ± 3.1 g-N/m3/day, respectively. Nitrification was nearly complete in the SBR, resulting in an effluent ammonium concentration of below 0.05 mg N/L on average.
Microbial Community of Mixed Culture Biomass in Activated Sludge
The class-level taxonomic composition of the activated sludge biomass is summarized in Figure 1. The class Gamma-proteobacteria was most abundant, accounting for up to 35% of the total population. This was followed by Alpha-proteobacteria (23%), and Actinobacteria (19%). The top 50 most abundant microbes at the genus and species levels and the corresponding relative abundances are shown in Figure 2 and Supplementary Figure 1, respectively. Methyloceanibacter caenitepidi, which is a marine methanol-assimilating bacterium, was the most abundant in the biomass (Supplementary Figure 1). Based on read numbers, its relative abundance was 1.3 times (1.2%) higher than that of the second most predominant species, Rhodopseudomonas palustris (0.9%). As for major guilds for ammonia oxidation, the genera Candidatus Nitrosopumilus and Nitrospira, known as ammonia-oxidizing archaea and complete ammonia-oxidizing bacteria, respectively, were detected (Figure 2 and Supplementary Figure 1).
Metabolic Pathways and Genetic Production Potential of the Mixed Culture
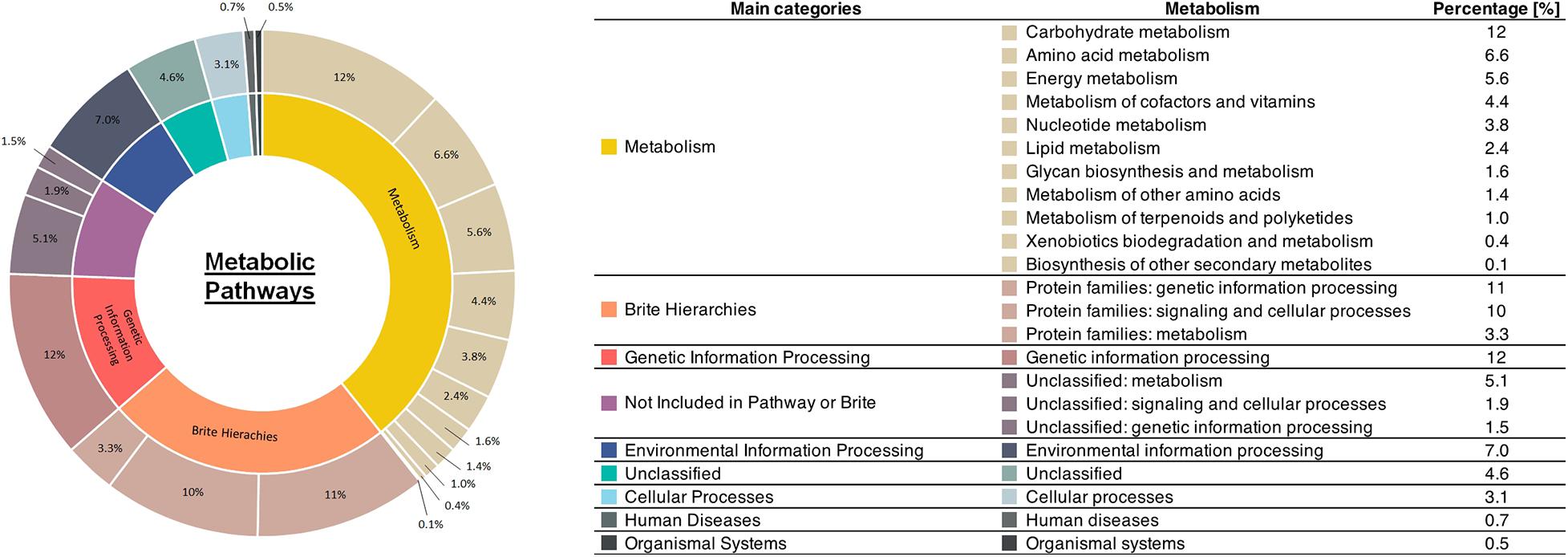
The results of mapping our sample to reference metabolic pathways in the KEGG are summarized in Figure 3. The highest relative gene abundance was associated with carbohydrate metabolism (12%) and genetic information processing (12%), followed by protein families: genetic information processing (11%), and protein families: signaling and cellular processes (10%). The biosynthesis pathways for each valuable product after mapping with mixed culture data are illustrated in Figure 4. The mixed culture biomass in the activated sludge possessed a full set of functional genes to synthesize six of the valuable products, ectoine, PHB, zeaxanthin, astaxanthin, acetoin, and 2,3-butanediol (Figure 4 and Table 4). Comparing the hit numbers of the functional genes for ectoine, acetoin, and 2,3-butanediol for each enzymatic reaction step (Figures 4A,D,E) revealed a substantial difference in hit numbers. Specifically, the lowest and highest gene hit numbers for ectoine biosynthesis were ectA (177) and lysC (12,563) for conversion of L-2,4-diaminobutyrate into N-γ-acetyl-L2,4,-diaminobutyrate and of aspartate into aspartyl phosphate, respectively. These hit numbers were used to designate the degree of the rate-limiting reaction for biosynthesis of a valuable product (e.g., 0.014 = 177/12,563). Acetoin and 2,3-butanediol had the common functional gene alsD, which had a considerably lower hit number (115) than the others, leading to a relative abundance of the lowest/highest gene hit number being 0.0045. In contrast, PHB, zeaxanthin, and astaxanthin had relatively evenly-distributed gene hit numbers for their enzymatic reactions. For PHB biosynthesis, the hit numbers of phaA (2,532) and phaB (2,476), which encode acetyl-CoA acetyltransferase and acetoacetyl-CoA reductase, respectively, were lower than the one of phaC (14,491), which encodes poly (3-hydroxyalkanoate) polymerase. These hit numbers resulted in relative abundances of the lowest/highest gene hit number of 0.175 (phaA/phaC) and 0.171 (phaB/phaC), respectively. The same trend appeared with zeaxanthin and astaxanthin syntheses, resulting in a relative abundance of the lowest/highest hit number of 0.11. Terpenoid backbone biosynthesis pathways after gene-mapping are shown in Supplementary Figure 2. In addition, the results of gene-mapping of each valuable product to the KEGG reference pathways are shown in Supplementary Figures 3–5 and those of the terpenoid backbone biosynthesis are shown in Supplementary Figure 6. The mapping results shown in the figures indicate that the pathways for the production of the compounds are connected.
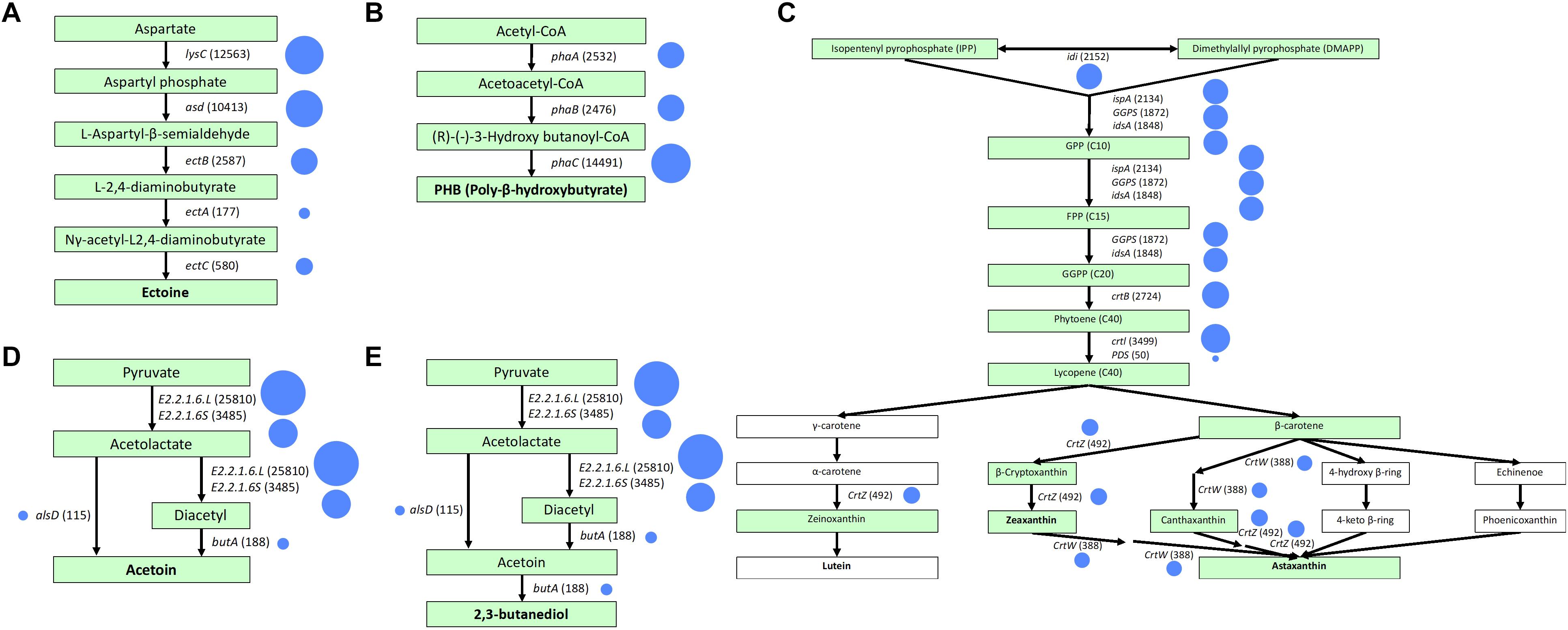
Figure 4. Gene-mapping to KEGG reference pathways for valuable products including (A) ectoine, (B) PHB, (C) lutein, zeaxanthin, and astaxanthin, (D) acetoin, and (E) 2,3-butanediol. A continuous arrow shows biosynthesis pathways. Chemical substances in boxes are generated by microbial reactions. Boxes in green indicate the presence of the metabolic reaction. Functional genes and the hit numbers are indicated next to the arrows. Circle size corresponds to the number of hits.
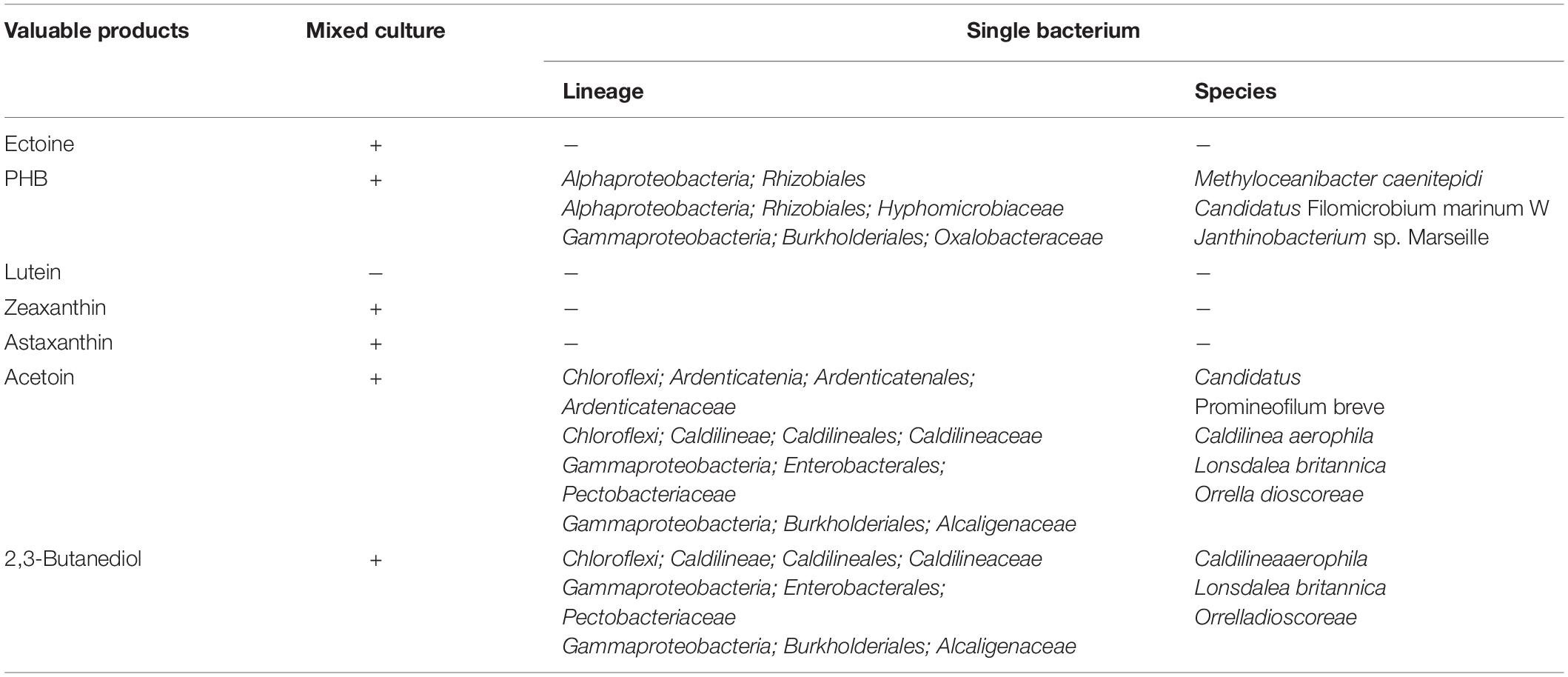
Table 4. Potential for production of valuable products by mixed microbial communities and a single species of bacteria.
Genetic Potential of a Single Bacterium to Produce Valuable Compounds
Linking the phylogeny with functions revealed that PHB, acetoin, and 2,3-butanediol can be genetically produced by a single bacterial species (Table 4). However, individual species of bacteria lack certain functional genes for the production of ectoine, lutein, zeaxanthin, and astaxanthin, implying that syntrophic interactions between different microbial species are needed for synthesis of these compounds. M. caenitepidi, Janthinobacterium sp. Marseille, and Ca. Filomicrobium marinum W harbored phaABC genes encoding enzymes for PHB biosynthesis, while Ca. promineofilum breve, Caldilinea aerophila, Lonsdalea britannica, and Orrella dioscoreae, possessed genes essential to acetoin production, i.e., ilvM, alsD, and butA (Figure 4). Among the bacteria capable of producing acetoin, Caldilinea aerophila, Lonsdalea britannica, and Orrella dioscoreae harbor butA and BDH, which genetically produce 2,3-butanediol from acetoin.
Discussion
Gene-Centric Approach That Facilitates the Development of Upcycling Bioprocesses From Landfill Leachates
The MRM concept (Verstraete, 2007) paves the way for a paradigm shift in which wastewater can be regarded as a resource for valuable products. Applying a bottom-up approach based on gene-centric analysis to the MRM concept is essential for the feasibility of valuable product biosynthesis based on metabolic network information. Therefore, feasibility assessments based on the metagenome will help to establish innovative bioprocesses for valuable product biosynthesis. However, this approach has not been applied to a bioreactor for nitrification and denitrification in landfill leachate containing high concentrations of ammonia (e.g., several hundred mg N/L) and salt (above > 1%; Díaz et al., 2019; Shu et al., 2019). This study, therefore, advocates a rapid screening procedure based on the gene-centric analysis from metagenomic data in a simple manner. To the best of our knowledge, this is the first metagenomics study that investigates the metabolic potential for valuable product biosynthesis by activated sludge used for leachate treatment at a closed landfill site. The application of gene-centric metagenomic analysis confirmed that the activated sludge possessed the metabolic potential for the biosynthesis of six targeted products. The holistic and exclusive screening of metabolic pathway networks for valuable products based on gene-centric metagenomic analysis revealed (i) the potential for valuable product biosynthesis (Table 4) and (ii) inferred potential rate-limiting steps within the metabolic pathways (Figure 4). Although the taxonomic and functional information attained by the gene-centric analysis is not as profound as that by the genome-centric counterpart, the information provides the implication if a biomass sample possesses metabolic potentials for valuable product production or not. The gene-centric approach implemented in this study will facilitate the development of upcycling bioprocesses from landfill leachates in which organic carbon and nitrogen are abundant.
The gene hit number analysis suggests a potential rate-limiting step during the bio-transformations for value-added compounds, e.g., the conversion from L-2,4-diaminobutylate to N-γ-acetyl-L2,4-diaminobutyrate in a series of ectoine production from aspartate (Figure 4). A challenge in this study stems from the authenticity of the gene hit number as an indicator for rate-limiting steps in the metabolic pathways because the differences in gene expression affecting RNA and protein abundances, and the existence of pseudogenes may also determine enzymatic reactions. Although this challenge is under debate, comparing the gene hit numbers may provide rough measures of bottleneck enzymes in metabolic reactions. The credibility of quantifying gene hit numbers warrants future investigation.
Metabolic Potentials of Activated Sludge Biomass at a Landfill Leachate Treatment Plant
This study demonstrated that the mixed microbial community in a landfill leachate treatment site harbored functional genes associated with the syntheses of ectoine, PHB, zeaxanthin, astaxanthin, acetoin, and 2,3-butanediol (Table 4). The presence of these genes suggests the potential for use of this activated sludge biomass in biosynthesis. As previously demonstrated, microbial consortia in activated sludge have been shown to produce short-chain fatty acids (Li et al., 2014), enzymes (Yu et al., 2009), polyhydroxyalkanoate (PHA; Lee and Yu, 1997), bio-flocculants (Wang et al., 2007), and fuels (Salama et al., 2017). As for ectoine synthesis, the analysis of the hit numbers of each functional gene suggested the conversion of L-2,4-diaminobutylate into N-γ-acetyl-L2,4-diaminobutyrate mediated by L-2,4-diaminobutyric acid acetyltransferase (EctA) as a rate-limiting step. Previous studies of EctA purified from halophilic bacteria revealed that suitable conditions for this process were pH 8.2–9.5, a temperature of 20°C, and a salt concentration of 2.3% (Ono et al., 1999; Richter et al., 2020), which do not coincide with the SBR conditions in this study (pH 7.35 ± 0.08, temperature of 28.8°C ± 1.4°C, and salinity of 1.23%). Moreover, caution should be taken when considering these findings because the conditions required to activate EctA in vitro and in vivo may not be the same. Accordingly, future studies to identify suitable environmental conditions for bacteria possessing EctA are needed.
Potential Valuable Products Produced by a Single Microorganism
The metagenomic analysis of the activated sludge used to treat landfill leachate allowed classification of the production potential for valuable products by (i) individual species of bacteria or (ii) mixed microbial communities (Table 4). The results demonstrated that individual species of bacteria present in the activated sludge of the landfill leachate treatment had the genetic potential to synthesize PHB, acetoin, and 2,3-butanediol among the seven products listed in Table 4.
Our metagenomic analysis unraveled that the species proximal to M. caenitepidi, Ca. Filomicrobium marinum W, and Janthinobacterium sp. Marseille were all found to possess the entire set of genes encoding for PHB synthesis. The metagenomic analysis of the taxonomy and functions revealed for the first time that M. caenitepidi, a marine methanol-assimilating bacterium, was dominant in the activated sludge of the landfill leachate treatment facility (Supplementary Figure 1). The presence of M. caenitepidi aside from the ocean has not been reported, except for one study (Yasuda et al., 2020). M. caenitepidi Gela4 was isolated from marine sediment near a hydrothermal vent containing methane in Kagoshima Bay, Japan (Takeuchi et al., 2014) and from a Belgian North Sea sediment (Vekeman et al., 2016). The genus Filomicrobium was detected in a methanol-fed denitrifying bioreactor from saline water (Rissanen et al., 2016). Ca. Filomicrobium marinum W (Henriques and De Marco, 2015), which was first isolated from North Atlantic surface seawater, is a methylotroph capable of potentially synthesizing PHB. Janthinobacterium isolates acquired from the Antarctic possess functional genes for PHB biosynthesis. Although they thrive in marine environments (Tan et al., 2020), the detection of Janthinobacterium sp. as a PHB producer in the landfill leachate treatment site makes sense as the presence of methanol and salinity provide a comparable environment. These species require greater ecophysiological descriptions to leverage their PHB synthesis potentials, which will be a follow-up aim.
Bacteria harboring phaABC store PHB intracellularly under feast-famine conditions (Inoue et al., 2018). Because the SBR in this study may induce feast–famine conditions over one cycle, these three bacteria accumulated PHB to attain energy for their preferential growth. Accordingly, the conditions likely exerted high selection pressure to preferentially enrich the sludge with these three potential PHB producers. Identification of their presence and functions paves the way for developing a PHB-producing process in a water/wastewater treatment system. However, this preliminary study was designed to identify the functions and compositions of an activated sludge microbiome treating landfill leachate, not to quantify water and intracellular constituents in the SBR. Therefore, future investigations to evaluate long-term SBR operation and the associated rates of production of valuable products in accordance with microbial population dynamics in the activated sludge are warranted.
This study also revealed that the bacteria listed in Table 4 harbor genes capable of producing acetoin and 2,3-butanediol. However, the ecological niches of taxonomic proxies for these bacteria are not consistent. Ca. Promineofilum and Caldilinea aerophile, which belong to the phylum Chloroflexi, are commonly found in municipal wastewater treatment plants (McIlroy et al., 2016) and hot spring sulfur turf mats (Sekiguchi et al., 2003), while Lonsdalea Britannica and Orrella dioscoreae are Gamma-proteobacteria isolated from the leaves of Quercus robur (Li et al., 2017) and Dioscorea sansibarensis (Carlier et al., 2017), respectively. Despite their genetic potentials to produce valuable products, in-depth physiological activity tests for actual production have not been conducted (Sekiguchi et al., 2003; Brady et al., 2012; McIlroy et al., 2016; Carlier et al., 2017; Li et al., 2017). Future research will be undertaken to confirm the product availability, yield, and production rate by bacterial species harboring genes related to PHB, acetoin, and 2,3-butanediol production.
Metagenomic Screening of Promising Bacteria Capable of Producing Valuable Products
In-depth analysis of mixed microbial community compositions and functions by gene-centric metagenomic analysis, proposed in this study, could facilitate a primary screening strategy for exploration and leveraging of promising microbial communities or species. Our gene-centric analysis confirmed that the activated sludge investigated did not harbor the functional genes encoding enzymes to synthesize lutein (Supplementary Figure 5). Moreover, inoculating the activated sludge would not succeed in enrichment with a microbial guild producing lutein. Pre-screening by gene-centric metagenomic analysis allows determination of whether a single microbe (I), a mixed microbial community (II), or no microbes (III) synthesize a valuable product (Figure 5). Such classifications may enable identification of the conditions required for a key mixed microbial community or an individual microbe to be enriched (Cases I and II Figure 5), ultimately allowing the development of an innovative microbial process. Accordingly, implementing bioaugmentation strategies (Laocharoen et al., 2015; Lin et al., 2018) or retrofitting treatment processes for valuable product biosynthesis, analogous to the concept of bioaugmentation technology where nitrifying bacteria are enriched in a tank installed in a sludge recycle line (Salem et al., 2003), are potential applications of this screening process (Figure 5). These findings will also enable research toward isolation or selective enrichment of promising microbes using suitable environmental conditions identified from metagenomic data. Moreover, if screening reveals no microbes have the desired properties, the method developed here will save a great deal of time and labor. The advocated scheme could be beneficial especially for engineers to bridge fundamental and applied research toward resource recovery and upcycling.
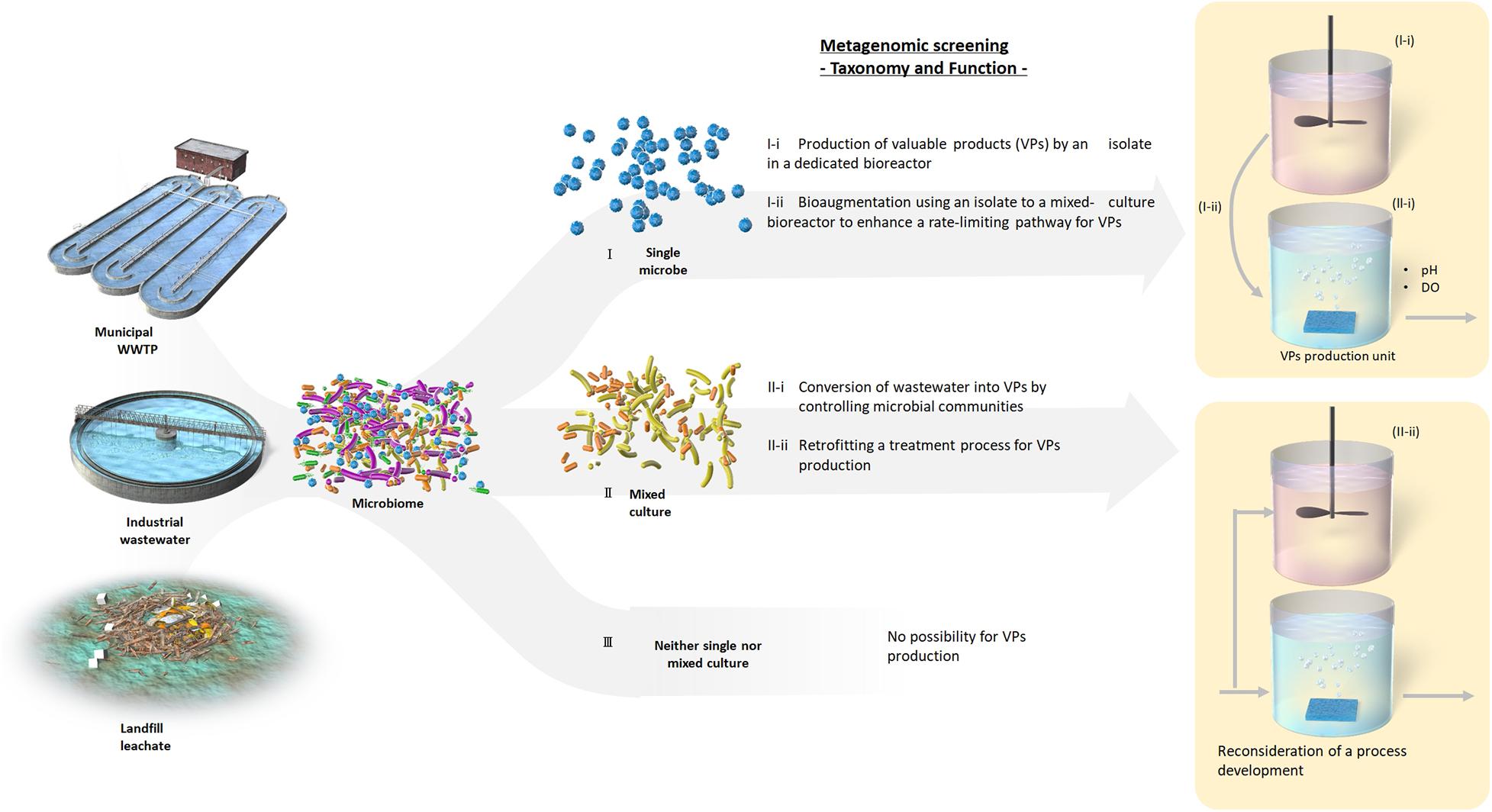
Figure 5. A workflow of a gene-based metagenomic screening method for production of valuable products.
Conclusion
In this study, gene-centric metagenomic analysis of an activated sludge used to treat landfill leachate with a high nitrogen concentration and methanol supplementation revealed the genetic potential for biosynthesis of valuable products by microorganisms in the activated sludge. Specifically, it was confirmed that PHB, ectoine, astaxanthin, zeaxanthin, acetoin, and 2,3-butanediol could be biosynthesized by the mixed microbial community in the activated sludge. Moreover, the pathway-based analysis implied a rate-limiting step for each product. PHB, acetoin and 2,3-butanediol also had the biosynthetic potential to be produced by individual species of microorganisms in the activated sludge. Furthermore, a marine methanol-assimilating bacterium with the genetic potential for PHB biosynthesis, M. caenitepidi, was shown to be dominant in the activated sludge. Our results demonstrate the potential for application of this method as tool for primary screening of promising microbial species in development of bioprocesses for valuable product biosynthesis.
Overall, primary screening by gene-centric metagenomic analysis can be used to evaluate the potential for valuable compound production by microbial communities or individual species in engineered systems was demonstrated. The analysis specifically provides engineers with an opportunity to preliminarily test a possibility of mixed culture biomass to produce valuable products. Future studies to link the screening by metagenomic analysis as described in this study with enrichment and isolation of promising microbial species are warranted.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
AT designed and led the study. SY and AT took samples and extracted DNA. SY, LO, and SA conducted the genomic and metabolic pathway analyses. SY, TS, LO, and SA performed laboratory works for metagenomic analysis. SY wrote the manuscript with major edits. SL and AT discussed the outline of the manuscript and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a Grant-in-Aid for Scientific Research (17H01893 and 20H04362), a Grant-in-Aid for Research Activity Start-up (19K21555), and the Open Partnership Joint Research Projects (Japan-Germany) from the Japan Society for the Promotion of Science (JSPS), as well as the visiting scholar fund from the Institute of Global Innovation Research of Tokyo University of Agriculture and Technology.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Edanz Group (https://en-author-services.edanz.com/ac) for editing a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.640848/full#supplementary-material
References
APHA, AWWA, and WEF (1992). Standard Methods for the Examination of Water and Wastewater, 18th Edn. Washington, DC: American Public Health Association.
Berry, A., Janssens, D., Hu, M., Jore, J. P. M., Hoste, B., Cleenwerck, I., et al. (2003). Paracoccus zeaxanthinifaciens sp. nov., a zeaxanthin-producing bacterium. Int. J. Syst. Evol. Microbiol. 53, 231–238. doi: 10.1099/ijs.0.02368-0
Brady, C. L., Cleenwerck, I., Denman, S., Venter, S. N., Rodríguez-Palenzuela, P., Coutinho, T. A., et al. (2012). Proposal to reclassify Brenneria quercina (Hildebrand and Schroth 1967) Hauben et al. 1999 into a new genus, Lonsdalea gen. nov., as Lonsdalea quercina comb. nov., descriptions of Lonsdalea quercina subsp. quercina comb. nov., Lonsdalea quercina subsp. ib. Int. J. Syst. Evol. Microbiol. 62, 1592–1602. doi: 10.1099/ijs.0.035055-0
Carlier, A., Cnockaert, M., Fehr, L., Vandamme, P., and Eberl, L. (2017). Draft genome and description of Orrella dioscoreae gen. nov. sp. nov., a new species of Alcaligenaceae isolated from leaf acumens of Dioscorea sansibarensis. Syst. Appl. Microbiol. 40, 11–21. doi: 10.1016/j.syapm.2016.10.002
Costa, A. M., Alfaia, R. G., de, S. M., and Campos, J. C. (2019). Landfill leachate treatment in Brazil – an overview. J. Environ. Manage. 232, 110–116. doi: 10.1016/j.jenvman.2018.11.006
Díaz, A. I., Oulego, P., Laca, A., González, J. M., and Díaz, M. (2019). Metagenomic analysis of bacterial communities from a nitrification–denitrification treatment of landfill leachates. Clean Soil Air Water 47, 1–8. doi: 10.1002/clen.201900156
Doronina, N. V., Ezhov, V. A., Beschastnyi, A. P., and Trotsenko, Y. A. (2010). Biosynthesis of the bioprotectant ectoin by aerobic methylotrophic bacteria from methanol. Appl. Biochem. Microbiol. 46, 173–176. doi: 10.1134/S0003683810020080
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, 29–37. doi: 10.1093/nar/gkr367
Gogleva, A. A., Kaparullina, E. N., Doronina, N. V., and Trotsenko, Y. A. (2010). Methylophilus flavus sp. nov. and Methylophilus luteus sp. nov., aerobic, methylotrophic bacteria associated with plants. Int. J. Syst. Evol. Microbiol. 60, 2623–2628. doi: 10.1099/ijs.0.019455-0
Guo, W., Li, D., He, R., Wu, M., Chen, W., Gao, F., et al. (2017). Synthesizing value-added products from methane by a new Methylomonas. J. Appl. Microbiol. 123, 1214–1227. doi: 10.1111/jam.13581
Henriques, A. C., and De Marco, P. (2015). Complete genome sequences of two strains of “Candidatus Filomicrobium marinum,” a methanesulfonate-degrading species. Genome Announc. 3, 1–2. doi: 10.1128/genomeA.00160-15
Hoshino, Y., and Gaucher, E. A. (2018). On the origin of isoprenoid biosynthesis. Mol. Biol. Evol. 35, 2185–2197. doi: 10.1093/molbev/msy120
Ilies, P., and Mavinic, D. S. (2001). The effect of decreased ambient temperature on the biological nitrification and denitrification of a high ammonia landfill leachate. Water Res. 35, 2065–2072. doi: 10.1016/S0043-1354(00)00477-2
Inoue, D., Suzuki, Y., Sawada, K., and Sei, K. (2018). Polyhydroxyalkanoate accumulation ability and associated microbial community in activated sludge-derived acetate-fed microbial cultures enriched under different temperature and pH conditions. J. Biosci. Bioeng. 125, 339–345. doi: 10.1016/j.jbiosc.2017.09.008
Ji, X., Huang, H., and Ouyang, P. (2011). Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol. Adv. 29, 351–364. doi: 10.1016/j.biotechadv.2011.01.007
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30.
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kaza, S., Yao, L., Bhada-Tata, P., and Van Woerden, F. (2018). What a Waste 2.0: A Global Snapshot of Solid Waste Management to 2050. Washington, DC: The World Bank.
Kim, D., Song, L., Breitwieser, F. P., and Salzberg, S. L. (2016). Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 26, 1721–1729. doi: 10.1101/gr.210641.116
Langmead, B., and Salzberg, S. (2013). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923.Fast
Laocharoen, S., Reungsang, A., and Plangklang, P. (2015). Bioaugmentation of Lactobacillus delbrueckii ssp. bulgaricus TISTR 895 to enhance bio-hydrogen production of Rhodobacter sphaeroides KKU-PS5. Biotechnol. Biofuels 8, 1–16. doi: 10.1186/s13068-015-0375-z
Lawson, C. E., Harcombe, W. R., Hatzenpichler, R., Lindemann, S. R., Löffler, F. E., O’Malley, M. A., et al. (2019). Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741. doi: 10.1038/s41579-019-0255-9
Lawson, C. E., Wu, S., Bhattacharjee, A. S., Hamilton, J. J., McMahon, K. D., Goel, R., et al. (2017). Metabolic network analysis reveals microbial community interactions in anammox granules. Nat. Commun. 8, 1–12. doi: 10.1038/ncomms15416
Lee, J. H., Kim, Y. S., Choi, T. J., Lee, W. J., and Kim, Y. T. (2004). Paracoccus haeundaensis sp. nov., a Gram-negative, halophilic, astaxanthin-producing bacterium. Int. J. Syst. Evol. Microbiol. 54, 1699–1702. doi: 10.1099/ijs.0.63146-0
Lee, S., and Yu, J. (1997). Production of biodegradable thermoplastics from municipal sludge by a two-stage bioprocess. Resour. Conserv. Recycl. 19, 151–164. doi: 10.1016/S0921-3449(96)01157-3
Levett, I., Birkett, G., Davies, N., Bell, A., Langford, A., Laycock, B., et al. (2016). Techno-economic assessment of poly-3-hydroxybutyrate (PHB) production from methane - the case for thermophilic bioprocessing. J. Environ. Chem. Eng. 4, 3724–3733. doi: 10.1016/j.jece.2016.07.033
Li, D., Liu, C., Luo, R., Sadakane, K., and Lam, T. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Li, X. L., Peng, Y. Z., Ren, N. Q., Li, B. K., Chai, T. Z., and Zhang, L. (2014). Effect of temperature on short chain fatty acids (SCFAs) accumulation and microbiological transformation in sludge alkaline fermentation with Ca(OH)2 adjustment. Water Res. 61, 34–45. doi: 10.1016/j.watres.2014.03.030
Li, Y., Xue, H., Guo, L. M., Koltay, A., Palacio-Bielsa, A., Chang, J., et al. (2017). Elevation of three subspecies of Lonsdalea quercina to species level: Lonsdalea britannica sp. nov., Lonsdalea iberica sp. nov. and Lonsdalea populi sp. nov. Int. J. Syst. Evol. Microbiol. 67, 4680–4684. doi: 10.1099/ijsem.0.002353
Lin, Z., Sun, D., Dang, Y., and Holmes, D. E. (2018). Significant enhancement of nitrous oxide energy yields from wastewater achieved by bioaugmentation with a recombinant strain of Pseudomonas aeruginosa. Sci. Rep. 8:11916. doi: 10.1038/s41598-018-30326-8
Matassa, S., Batstone, D. J., Hülsen, T., Schnoor, J., and Verstraete, W. (2015). Can direct conversion of used nitrogen to new feed and protein help feed the world? Environ. Sci. Technol. 49, 5247–5254. doi: 10.1021/es505432w
McCarty, P. L., Bae, J., and Kim, J. (2011). Domestic wastewater treatment as a net energy producer-can this be achieved? Environ. Sci. Technol. 45, 7100–7106. doi: 10.1021/es2014264
McIlroy, S. J., Karst, S. M., Nierychlo, M., Dueholm, M. S., Albertsen, M., Kirkegaard, R. H., et al. (2016). Genomic and in situ investigations of the novel uncultured Chloroflexi associated with 0092 morphotype filamentous bulking in activated sludge. ISME J. 10, 2223–2234. doi: 10.1038/ismej.2016.14
Misawa, N., Satomi, Y., Kondo, K., Yokoyama, A., Kajiwara, S., Saito, T., et al. (1995). Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J. Bacteriol. 177, 6575–6584. doi: 10.1128/jb.177.22.6575-6584.1995
Nielsen, D. R., Yoon, S., Yuan, C. J., and Prather, K. L. J. (2010). Metabolic engineering of acetoin and meso-2,3-butanediol biosynthesis in E. coli. Biotechnol. J. 5, 274–284. doi: 10.1002/biot.200900279
Nobu, M. K., Narihiro, T., Rinke, C., Kamagata, Y., Tringe, S. G., Woyke, T., et al. (2015). Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J. 9, 1710–1722. doi: 10.1038/ismej.2014.256
Nwachukwu, I. D., Udenigwe, C. C., and Aluko, R. E. (2016). Lutein and zeaxanthin: production technology, bioavailability, mechanisms of action, visual function, and health claim status. Trends Food Sci. Technol. 49, 74–84. doi: 10.1016/j.tifs.2015.12.005
Ono, H., Sawada, K., Khunajakr, N., Tao, T., Yamamoto, M., Hiramoto, M., et al. (1999). Characterization of biosynthetic enzymes for ectoine as a compatible solute in a moderately halophilic eubacterium, Halomonas elongata. J. Bacteriol. 181, 91–99. doi: 10.1128/jb.181.1.91-99.1999
Orita, I., Nishikawa, K., Nakamura, S., and Fukui, T. (2014). Biosynthesis of polyhydroxyalkanoate copolymers from methanol by Methylobacterium extorquens AM1 and the engineered strains under cobalt-deficient conditions. Appl. Microbiol. Biotechnol. 98, 3715–3725. doi: 10.1007/s00253-013-5490-9
Osaka, T., Yoshie, S., Tsuneda, S., Hirata, A., Iwami, N., and Inamori, Y. (2006). Identification of acetate- or methanol-assimilating bacteria under nitrate-reducing conditions by stable-isotope probing. Microb. Ecol. 52, 253–266. doi: 10.1007/s00248-006-9071-7
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36:996. doi: 10.1038/nbt.4229
Pastor, J. M., Salvador, M., Argandoña, M., Bernal, V., Reina-Bueno, M., Csonka, L. N., et al. (2010). Ectoines in cell stress protection: uses and biotechnological production. Biotechnol. Adv. 28, 782–801. doi: 10.1016/j.biotechadv.2010.06.005
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna: R Foundation Statistical Computing.
Richter, A. A., Kobus, S., Czech, L., Hoeppner, A., Zarzycki, J., Erb, T. J., et al. (2020). The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine. J. Biol. Chem. 295, 2822–2838. doi: 10.1074/jbc.RA119.011277
Rico, J., Duquesne, K., Petit, J. L., Mariage, A., Darii, E., Peruch, F., et al. (2019). Exploring natural biodiversity to expand access to microbial terpene synthesis. Microb. Cell Fact. 18, 1–10. doi: 10.1186/s12934-019-1074-4
Rissanen, A. J., Ojala, A., Dernjatin, M., Jaakkola, J., and Tiirola, M. (2016). Methylophaga and Hyphomicrobium can be used as target genera in monitoring saline water methanol-utilizing denitrification. J. Ind. Microbiol. Biotechnol. 43, 1647–1657. doi: 10.1007/s10295-016-1839-2
Salama, E. S., Kurade, M. B., Abou-Shanab, R. A. I., El-Dalatony, M. M., Yang, I. S., Min, B., et al. (2017). Recent progress in microalgal biomass production coupled with wastewater treatment for biofuel generation. Renew. Sustain. Energy Rev. 79, 1189–1211. doi: 10.1016/j.rser.2017.05.091
Salem, S., Berends, D. H. J. G., Heijnen, J. J., and Van Loosdrecht, M. C. M. (2003). Bio-augmentation by nitrification with return sludge. Water Res. 37, 1794–1804. doi: 10.1016/S0043-1354(02)00550-X
Schendel, F. J., Bremmon, C. E., Flickinger, M. C., Guettler, M., and Hanson, R. S. (1990). L-lysine production at 50°C by mutants of a newly isolated and characterized methylotrophic Bacillus sp. Appl. Environ. Microbiol. 56, 963–970. doi: 10.1128/aem.56.4.963-970.1990
Sekiguchi, Y., Yamada, T., Hanada, S., Ohashi, A., Harada, H., and Kamagata, Y. (2003). Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain bacteria at the subphylum level. Int. J. Syst. Evol. Microbiol. 53, 1843–1851. doi: 10.1099/ijs.0.02699-0
Shu, D., Guo, J., Zhang, B., He, Y., and Wei, G. (2019). rDNA- and rRNA-derived communities present divergent assemblage patterns and functional traits throughout full-scale landfill leachate treatment process trains. Sci. Total Environ. 646, 1069–1079. doi: 10.1016/j.scitotenv.2018.07.388
Spiller, M., Muys, M., Papini, G., Sakarika, M., Buyle, M., and Vlaeminck, S. E. (2020). Environmental impact of microbial protein from potato wastewater as feed ingredient: comparative consequential life cycle assessment of three production systems and soybean meal. Water Res. 171, 1–12. doi: 10.1016/j.watres.2019.115406
Takeuchi, M., Katayama, T., Yamagishi, T., Hanada, S., Tamaki, H., Kamagata, Y., et al. (2014). Methyloceanibacter caenitepidi gen. nov., sp. nov., a facultatively methylotrophic bacterium isolated from marine sediments near a hydrothermal vent. Int. J. Syst. Evol. Microbiol. 64, 462–468. doi: 10.1099/ijs.0.053397-0
Tan, I. K. P., Foong, C. P., Tan, H. T., Lim, H., Zain, N. A. A., Tan, Y. C., et al. (2020). Polyhydroxyalkanoate (PHA) synthase genes and PHA-associated gene clusters in Pseudomonas spp. and Janthinobacterium spp. isolated from Antarctica. J. Biotechnol. 313, 18–28. doi: 10.1016/j.jbiotec.2020.03.006
Vekeman, B., Kerckhof, F., Cremers, G., Vos, P., De, Vandamme, P., et al. (2016). New methyloceanibacter diversity from North Sea sediments includes methanotroph containing solely the soluble methane monooxygenase. Environ. Microbiol. 18, 4523–4536. doi: 10.1111/1462-2920.13485
Verstraete, W. (2007). Microbial ecology and environmental biotechnology. ISME J. 1, 4–8. doi: 10.1038/ismej.2007.7
Waite, D. W., Chuvochina, M., Pelikan, C., Parks, D. H., Yilmaz, P., Wagner, M., et al. (2020). Proposal to reclassify the proteobacterial classes deltaproteobacteria and oligoflexia, and the phylum thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 70, 5972–6016. doi: 10.1099/ijsem.0.004213
Waite, D. W., Vanwonterghem, I., Rinke, C., Parks, D. H., Zhang, Y., Takai, K., et al. (2017). Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to epsilonbacteraeota (phyl. nov.). Front. Microbiol. 8:682. doi: 10.3389/fmicb.2017.00682
Wang, S. G., Gong, W. X., Liu, X. W., Tian, L., Yue, Q. Y., and Gao, B. Y. (2007). Production of a novel bioflocculant by culture of Klebsiella mobilis using dairy wastewater. Biochem. Eng. J. 36, 81–86. doi: 10.1016/j.bej.2007.02.003
Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L., François, R., et al. (2019). Welcome to the tidyverse. J. Open Source Softw. 4:1686. doi: 10.21105/joss.01686
Xiao, Z., and Lu, J. R. (2014). Strategies for enhancing fermentative production of acetoin: a review. Biotechnol. Adv. 32, 492–503. doi: 10.1016/j.biotechadv.2014.01.002
Yasuda, S., Suenaga, T., Orschler, L., Agrawal, S., Lackner, S., and Terada, A. (2020). Identification of a metagenome-assembled genome of an uncultured Methyloceanibacter sp. strain acquired from an activated sludge system used for landfill leachate treatment. Microbiol. Resour. Announc. 9, 1–2. doi: 10.1128/MRA.00771-20
Ye, V. M., and Bhatia, S. K. (2012). Pathway engineering strategies for production of beneficial carotenoids in microbial hosts. Biotechnol. Lett. 34, 1405–1414. doi: 10.1007/s10529-012-0921-8
Yu, G., He, P., Shao, L., and Zhu, Y. (2009). Enzyme extraction by ultrasound from sludge flocs. J. Environ. Sci. 21, 204–210. doi: 10.1016/S1001-0742(08)62252-4
Zamakhaeva, S. A., Fedorov, D. N., and Trotsenko, Y. A. (2017). Methylotrophic producers of bioplastics (Review). Appl. Biochem. Microbiol. 53, 389–400. doi: 10.1134/S0003683817040147
Keywords: landfill, leachate, activated sludge, metagenome, Methyloceanibacter, metagenomic screening
Citation: Yasuda S, Suenaga T, Orschler L, Agrawal S, Lackner S and Terada A (2021) Metagenomic Insights Into Functional and Taxonomic Compositions of an Activated Sludge Microbial Community Treating Leachate of a Completed Landfill: A Pathway-Based Analysis. Front. Microbiol. 12:640848. doi: 10.3389/fmicb.2021.640848
Received: 12 December 2020; Accepted: 01 April 2021;
Published: 30 April 2021.
Edited by:
Simona Rossetti, Water Research Institute, National Research Council (CNR), ItalyReviewed by:
Leonardo Erijman, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaNikolaos Remmas, Democritus University of Thrace, Greece
Copyright © 2021 Yasuda, Suenaga, Orschler, Agrawal, Lackner and Terada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akihiko Terada, YWt0ZUBjYy50dWF0LmFjLmpw
 Shohei Yasuda1
Shohei Yasuda1 Toshikazu Suenaga
Toshikazu Suenaga Akihiko Terada
Akihiko Terada