
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 28 January 2021
Sec. Virology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.638094
This article is part of the Research Topic Phage Biology and Phage Therapy View all 13 articles
 Nannan Wu1*
Nannan Wu1* Tongyu Zhu1,2*
Tongyu Zhu1,2*Nosocomial infections (NIs) are hospital-acquired infections which pose a high healthcare burden worldwide. The impact of NIs is further aggravated by the global spread of antimicrobial resistance (AMR). Conventional treatment and disinfection agents are often insufficient to catch up with the increasing AMR and tolerance of the pathogenic bacteria. This has resulted in a need for alternative approaches and raised new interest in therapeutic bacteriophages (phages). In contrast to the limited clinical options available against AMR bacteria, the extreme abundance and biodiversity of phages in nature provides an opportunity to establish an ever-expanding phage library that collectively provides sustained broad-spectrum and poly microbial coverage. Given the specificity of phage-host interactions, phage susceptibility testing can serve as a rapid and cost-effective method for bacterial subtyping. The library can also provide a database for routine monitoring of nosocomial infections as a prelude to preparing ready-to-use phages for patient treatment and environmental sterilization. Despite the remaining obstacles for clinical application of phages, the establishment of phage libraries, pre-stocked phage vials prepared to good manufacturing practice (GMP) standards, and pre-optimized phage screening technology will facilitate efforts to make phages available as modern medicine. This may provide the breakthrough needed to demonstrate the great potential in nosocomial infection management.
Nosocomial infections (NIs), also known as hospital-acquired infections (HAI), are infections that are newly acquired in a hospital or other healthcare facility. They arise from various sources including healthcare staff, contaminated equipment, bed linen, and air droplets. NIs represent the most frequent adverse events during care delivery and typically result in prolonged hospital stays. There can be massive additional costs for health systems and patients, and unnecessary deaths (Allegranzi et al., 2011).
The misuse and overuse of antibiotics are accelerating the creation of antimicrobial resistance (AMR), which has become a major public health problem. In 2017, the World Health Organization (WHO) highlighted, as a global priority, a list of multidrug-resistant (MDR) bacterial pathogens (informally termed “superbugs”) to help in prioritizing the research and development of new and effective antibiotic treatments (World Health Organization, 2017). These healthcare-associated superbugs usually cause nosocomial outbreaks and opportunistic infections in hospitalized patients. The most prominent causative agents are carbapenem-resistant Enterobacteriaceae (CRE), carbapenem-resistant Acinetobacter baumannii (CRAB), MDR Pseudomonas aeruginosa, methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant Enterococcus (VRE). In the absence of effective treatment for bacterial infections that have AMR, modern medical care becomes inadequate and the infections persist in the body, increasing the risk of spread to others. Even worse, increasing global connectivity facilitates the rapid transport of infectious agents and their resistance genes through the world (Mellon et al., 2020).
NIs pose a high healthcare burden in both developed and developing countries (Allegranzi et al., 2011), this problem is further aggravated by the global spread of multidrug-resistant organisms (MDROs), complicating infection management, limiting therapy options and resulting in poorer outcomes (Laxminarayan et al., 2013; Chng et al., 2020). The increasing AMR in bacteria has entailed a need for alternative approaches to therapy and prophylaxis of these infections and raised new interest in therapeutic bacteriophages (phages) for these purposes. Phages are viruses that exclusively infect bacteria and were used therapeutically against bacterial diseases almost immediately after their discovery. The extreme abundance and biodiversity of phages in our surroundings and inside our bodies constitute a source of natural precision agents for bactericidal use, which might play a major role in tackling the global AMR crisis. Despite the remaining obstacles for phage therapy such as regulatory issues and bacterial anti-phage resistance, efforts to make it available as modern medicine are ongoing. In fact, the potential of phages in clinical practice goes beyond therapeutic applications, it could be involved in the whole process of nosocomial infection management. In this review, we discuss the potential of therapeutic phages in nosocomial infection monitoring, phage therapy, and environmental disinfection. We also propose a practical model of nosocomial infection management that uses phages.
A key precondition in phage application is an eligible phage library (phage bank) with broad-spectrum, poly antimicrobial properties, where many well-characterized phages are constantly collected and arranged according to the host species. Although an increasing number of phage libraries has been established recently, the overall scale is small and the information in most of the existing phage collections is incomplete (Table 1) (Yerushalmy et al., 2020). This strongly limits wide access to phage therapy since not all patients can afford the time-consuming process of mailing pathogenic bacteria from a healthcare facility to a phage repository, followed by screening lytic phages and mailing therapeutic phages back; neither can they afford time for de novo phage screening from nature. For this reason, both the quantity and quality of phage libraries need enhancement.
A first step to build a phage library is to prepare a library of pathogenic bacteria that provides the baits for phage hunting. Phages can be isolated from diverse environmental samples, however, not all phages are useful for therapeutic use. Criteria of eligible phage candidates have been extensively discussed and, although estimates of index values vary among groups, there’s a general consensus that the key features that must be characterized include: host species, complete genome sequence (to exclude or to allow modification of phages with known integration/toxic/resistance genes), host range testing, and high lytic activity (Gill and Hyman, 2010; Letarov and Kulikov, 2018; Merabishvili et al., 2018). Some other factors such as high yield, identified receptor, morphology, anti-biofilm activity, low resistance induction, stability during storage, and pre-clinical evaluation (e.g., safety, efficacy, pharmacodynamics and pharmacokinetics, human immune responses) are generally considered to be important during optimization of phages for therapeutics (Malik et al., 2017; D´abrowska and Abedon, 2019; Hesse and Adhya, 2019; Kortright et al., 2019; Yerushalmy et al., 2020).
In contrast to the limited number of antibiotics available, the extreme abundance and biodiversity of phages in nature make it possible to build an ever-expanding phage library that collectively provides sustained broad-spectrum and poly microbial coverage against the most common nosocomial pathogens such as the ESKAPE group (E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter species) (Rice, 2008). However, given the diversity of phage-host interactions, isolation of suitable phages for some group of pathogenic bacteria has been found to be difficult (Mattila et al., 2015). For instance, almost all known phages against Helicobacter Pylori and Clostridia difficile are temperate (Hargreaves et al., 2015; Muñoz et al., 2020). Phage engineering and technical improvement on new phage isolation will be helpful to overcome these obstacles. The collaboration and sharing of phage libraries initiated by the Phage Directory (https://phage.directory) are also conducive to pool a global-scale phage library and to connect demands for therapeutic phages.
Rapid tracking of pathogen sources with appropriate subtyping tools and immediate initiation of appropriate antibacterial treatment are crucial in the management of nosocomial infections. However, conventional bacterial genotyping methods such as pulse-field gel electrophoresis (PFGE), multi-locus variable tandem repeat analysis (MLVA), multilocus-sequence typing (MLST), and whole-genome sequencing (WGS) are time-consuming, normally needing more than three workdays to yield results. In contrast, bacterial phenotypic technics such as antibiotic susceptibility test, immunological methods (serotyping), or phage-typing are limited in discriminatory power but advantageous to save time (Table 2; Ferrari et al., 2017; Quainoo et al., 2017; Sloan et al., 2017; Vijayakumar et al., 2019). For the most common nosocomial pathogens which grow fast, a phage-typing can be achieved within six to eight hours by phage susceptibility testing.
A phage susceptibility test is a method followed by bacterial diagnosis, to simultaneously test the contents of a sub-library (specific to the target bacterial species) of phage candidates against the bacteria isolated from a patient, thereby identifying phages for patient-specific precision therapy. Laboratory-developed phage susceptibility tests are widely used for therapeutic phage screening (Aslam et al., 2019; Dedrick et al., 2019; Kuipers et al., 2019; Nir-Paz et al., 2019; Bao et al., 2020; Petrovic Fabijan et al., 2020). Additionally, given the specificity of phage-host interactions, the distinguished phage susceptibility profiles can enable rapid fingerprinting of bacterial substrains. Several laboratories have applied this method to investigate outbreaks caused by nosocomial or foodborne bacterial pathogens (Demczuk et al., 2003; Boxrud et al., 2007; Cooke et al., 2010; Barco et al., 2013; Wuyts et al., 2015). However, due to the variable sizes and components among phage libraries, phage-typing is limited in discriminatory power than genotyping approaches. It needs standardization to guarantee comparability among laboratories, as well as methodology optimization to reduce labor costs (Ferrari et al., 2017).
Nevertheless, on the basis of known epidemiological situations and pre-established phage collections, phage susceptibility testing may serve as a rapid and cost-effective method for bacterial subtyping to routinely monitor nosocomial infections. It may also be a prelude to preparing ready-to-use phages for patient treatment and environmental sterilization. Data from several preliminary studies have shown good concordance of profiles between phage-typing and other subtyping methods in distinguishing bacterial isolates (Demczuk et al., 2003; Crabb et al., 2019; Sundell et al., 2019). However, the possibility of frequent shifts in phage susceptibility profiles of phage-resistant strains should be allowed for, especially by the medical establishments with an ongoing phage therapy practice. Nevertheless, a database of routine phage-typing profiles helps by providing an expandable reference map that can be adjusted based on conventional molecular genotyping methods. With further improvements in the phage library diversity and in understanding phage-bacteria interaction, hospital-wide surveys employing the phage-typing database will be increasingly feasible, providing valuable information for infection control and eventually becoming part of routine practice.
Fixed composition (prêt-à-porter) or customized screened (sur-mesure), that is the question for therapeutic phage selection (Pirnay et al., 2011). Fixed composition, which is a stable and widely distributed phage preparation, is more adaptive for the modern pharmaceutical regulations than a customized screened phage preparation (sur-mesure). However, due to the high species/strain specificity of phages and the frequently evolving phage resistance in bacteria, it is difficult for a fixed phage cocktail to provide broad coverage on clinical strains. There have been some well-conducted clinical trials on the effectiveness of fixed phage cocktails but they have failed to obtain positive results (Markoishvili et al., 2002; Wright et al., 2009; Gindin et al., 2019; Jault et al., 2019; Leitner et al., 2020).
In parallel with full-scale clinical trials, lessons learned from sur-mesure case studies have been reported by numerous groups all over the world. These efforts have advanced medicinal phage technologies and highlighted safety and efficacy of phage therapy (Table 3; Luong et al., 2020; Pires et al., 2020). However, the sur-mesure approach is not well compatible with modern licensing processes. The European Medicines Agency (EMA) and the US Food and Drug Administration (FDA) placed phages under the medicinal (biologicals) product regulation; this means that marketing a phage product requires proof of safety and efficacy, also of quality by manufacture under Good Manufacturing Practice (GMP) regulations (Pires et al., 2020).
The good news is that some small companies focused on phage-specific therapies are growing fast and some authorities have been open to rigorously controlled clinical phage applications. Despite the long way to go before general approval is reached for the use of phage therapy, recent progress in phage manufacturing and quality control has revitalized phage therapy worldwide. Notably, the FDA has approved several investigational phage therapy pipelines targeting prosthetic joint infection, urinary/genital tract infection, chronic wound infection, and secondary bacterial pneumonia under the Investigational New Drug (IND) allowance (Adaptive Phage Therapeutics, 2020b; Armata Therapeuticals, 2020; Intralytix, 2020; Phagelux, 2020).
Good infection prevention practices such as hand hygiene, daily and terminal cleaning, and disinfection have been demonstrated to reduce the incidence of healthcare-associated infections (Ariza-Heredia and Chemaly, 2018; Mitchell et al., 2019). However, conventional disinfection methods such as ultraviolet and chemical agents (e.g., hydrogen peroxide, chlorine-derivatives) are often insufficient to cope with the increasing complexity and tolerance of the target bacteria. The application context and frequency are limited by the presence of the patient in the room. The concentration of chemical disinfectants needs to be constrained to a narrow range to balance the disinfection effectiveness and the level of harmful residuals and byproducts (Dong et al., 2019). In consequence, pathogenic bacteria can rapidly recolonize the environmental surface after disinfection (Laxminarayan et al., 2013; Weber et al., 2019).
Meanwhile, there has been great interest in developing novel sterilizing methods, such as “self-disinfecting” surfaces, continuous room disinfection with diluted chemical sterilant, and other reagents (Weber et al., 2019). Among these, the potential of phages in environmental disinfection has been long neglected but is now being applied in food processing workshops, livestock farms, and croplands (Holtappels et al., 2020; Pinto et al., 2020; Żbikowska et al., 2020). Phage disinfectants possess multiple advantages over conventional ones: first, phages are a very abundant microbial resource that holds great promise and options for targeting problematic bacteria such as those with multiple AMR or biofilm formation properties. Second, phages are species-specific viruses of prokaryotes and do not infect mammalian cells, which allows disinfection unlimited by frequency or by the presence of the patient in the room. Consequently, phage application has been proposed as a potential decontamination method for hospital surfaces and water systems (Jensen et al., 2015; Mathieu et al., 2019).
Recently, there has been a trickle of studies that demonstrated the efficacy of phages to rapidly decrease the load of pathogens commonly associated with nosocomial infections when they are present on different types of surfaces and aqueous systems in healthcare facilities (Ho et al., 2016; D’Accolti et al., 2019; Tseng et al., 2019). Ho et al reported an investigation of phage intervention in the intensive care unit (ICU) rooms. They applied 500 ml of customized phage-containing preparation with a concentration of 107 PFU (plaque forming unit)/ml for a ∼ 27m3 room. A significant reduction in CRAB-associated NIs was observed when adding a single aerosolized phage treatment to the conventional chemical-based disinfection performed in ICU rooms at the patient discharge (Ho et al., 2016). D’Accolti et al added phages to probiotic-based sanitation (PCHS) in the Staphylococcal-contaminated bathrooms (∼4.5 m2, applied 2 × 108 PFU phages for a multiplicity of infection ∼1000) of General Medicine wards, reporting a rapid and significant decrease in Staphylococcus spp. load on treated surfaces, up to 97% more than PCHS alone (D’Accolti et al., 2019). Tseng et al developed a chamber study model to analyze the potential of aerosolized phage BTCU-1 in reducing airborne Mycobacterium smegmatis in different multiplicity of infection (MOI) and aerosol generation time. The result showed that, MOI of 10 000 for 10 seconds significantly blocked recovery of M. smegmatis colony from the culture medium (Tseng et al., 2019).
Notably, several challenges to balance between benefits and costs of implementing phage disinfection cannot be neglected. In particular, storage condition and content of phage library depend on the stability and consumption of stocked phages; the absent of the quality control specification (QCS) for environmental applied phages should be addressed. It is certain that manufacture and QCS of phage disinfectants do not require as much as therapeutic phages (GMP-approved, endotoxin removal), however, this strategy needs mass consumption of phage and thus is production-capacity dependent. Although phages should not replace broad-spectrum agents in terminal disinfection, they offer great potential for continuous target-specific applications where the target is drug-resistant and antimicrobial chemicals are either relatively ineffective or their use would result in unintended detrimental consequences.
The use of phages is consistent with the need of nosocomial infection control to diminish both bacterial infection and transmission, as well as to relieve the overuse of antibiotics. However, little has been described regarding the systemic application of phages in infection management. Nevertheless, a similar concept has been successfully practiced in several other settings (e.g., food, animals, crops) (Holtappels et al., 2020; Pinto et al., 2020; Żbikowska et al., 2020), where a few phage-based products have been approved by the supervisory authorities in the United States, Canada, Israel, Europe, Australia and New Zealand (Pinto et al., 2020).
How might phages be integrated into the clinical workflow at a healthcare establishment? First, a system of cooperation between different departments (e.g., clinical, infectious diseases, laboratory, infection control, phage, pharmacy, and GMP factory) is essential. Second, an established phage library is needed for screening lytic phages against a given pathogenic bacterium isolated from a patient or from a hospital surface. Third, ready-to-use phage vials need to be properly manufactured prior to application to humans (GMP-approved) or environments; qualified phage vials should be pre-stocked to accelerate phage delivery. In parallel to use in phage therapy, the same phage formula can be aerosol-applied to the room or even the entire ward of the treated patient to prevent nosocomial transmission. Fourth, the accumulated phage-typing database of many bacteria can serve to monitor nosocomial infection and as a prelude to preparing phage stocks for patient treatment and environmental sterilization. Fifth, both retrospective and prospective phage susceptibility tests can be united (adjusted by conventional bacterial-typing methods and epidemiological links) to design fixed-composition phage cocktails with broad-spectrum, poly antimicrobial properties against the epidemic strains; the fixed-composition cocktails could then be applied in routine environmental sterilization or in treatment of serious infections such as septic shock or severe pneumonia that call for rapid initiation of treatment (Figure 1).
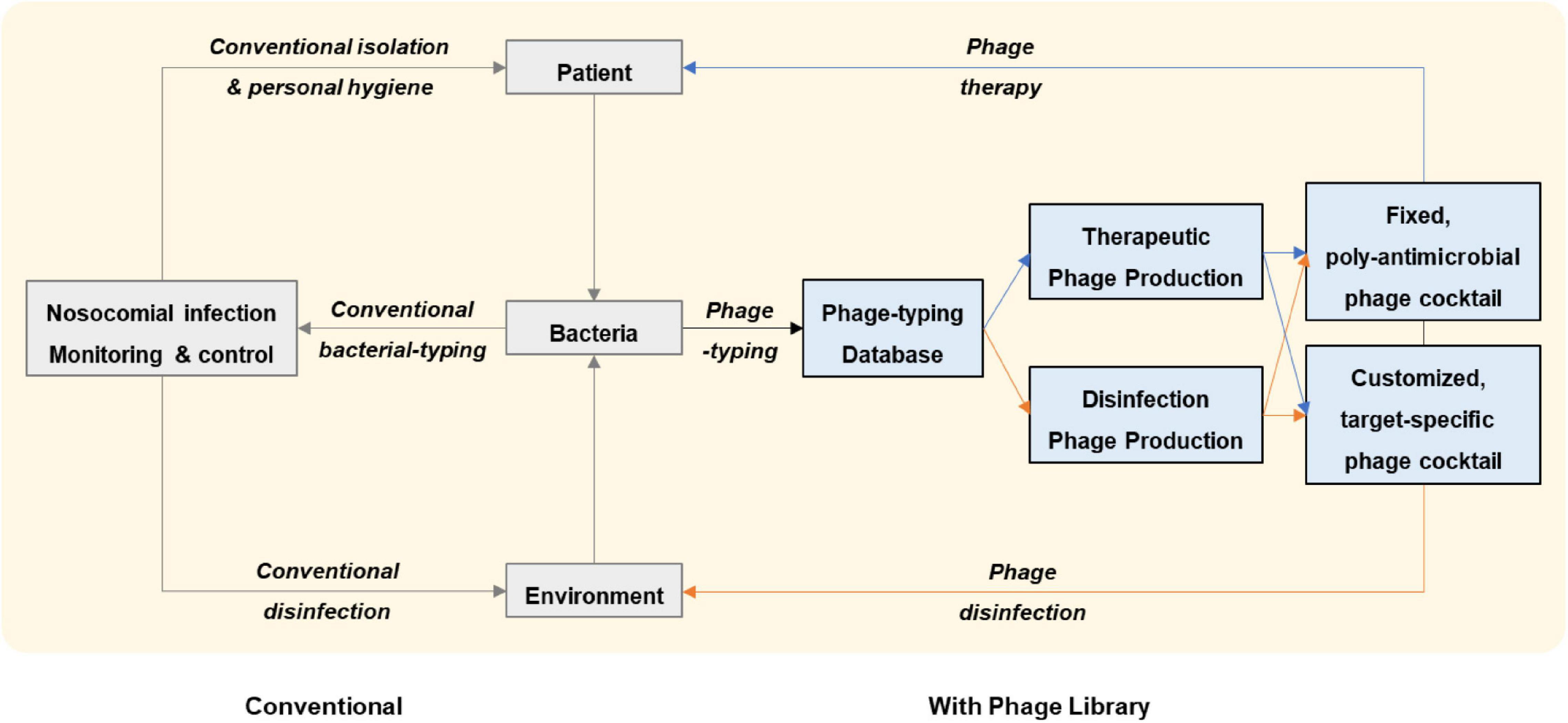
Figure 1. Workflow of nosocomial infection management proposed in this study. With an established phage library, lytic phages against a given pathogenic bacterium that has been isolated from a patient or from a hospital surface are screened and pre-stocked phage vials are delivered for application to the patient or to the environment. Retrospective and prospective phage susceptibility tests on hospital bacterial isolates can be united in a phage-typing database which can be further adjusted by conventional bacterial-typing methods and epidemiological links. Engaging the phage-typing database serves as a complementary strategy to conventional nosocomial infection management. By monitoring nosocomial infections in this way, fixed-composition phage cocktails with broad-spectrum, poly antimicrobial properties against the epidemic strains can be rapidly applied to the patient or the environment.
The extreme abundance and biodiversity of phages in our surrounding environment and inside our body provides an opportunity to establish an ever-expanding phage library that collectively provides a sustainable broad-spectrum and poly microbial coverage. This library might play a major role in tackling the global AMR crisis. Whilst acknowledging the remaining obstacles for clinical phage application, we believe that by taking advantage of pre-established phage libraries, pre-stocked phage productions, and pre-optimized phage screening technology, phages can become available as modern medicine. This availability may constitute a breakthrough in both clinical treatment and environmental disinfection in the context of nosocomial infection management.
NW and TZ reviewed the literature, summarized the tables, conceptualized the figure, wrote the manuscript, and acquired funding. Both authors contributed to the article and approved the submitted version.
This work was financially supported by the National Major Science and Technology Projects of China (grant 2020ZX09201001-005-003 to NW), Natural Science Foundation of Shanghai (20Y11900300 to NW), National Natural Science Foundation of China (82070772 to TZ), and the Clinical Research Plan of SHDC (SHDC2020CR2028B to TZ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank all members from the Shanghai Institute of Phage for thoughtful discussions.
Adaptive Phage Therapeutics (2020a). Adaptive Phage Therapeutics Enters Agreement with Mayo Clinic to Commercialize a Phage Susceptibility Test. Available online at: https://www.aphage.com/adaptive-phage-therapeutics-enters-agreement-with-mayo-clinic-to-commercialize-a-phage-susceptibility-test/ (accessed June 9, 2020).
Adaptive Phage Therapeutics (2020b). Development Pipeline. Available online at: https://www.aphage.com/science/pipeline/ (accessed December 3, 2020).
Allegranzi, B., Bagheri Nejad, S., Combescure, C., Graafmans, W., Attar, H., Donaldson, L., et al. (2011). Burden of endemic health-care-associated infection in developing countries: systematic review and meta-analysis. Lancet 377, 228–241. doi: 10.1016/s0140-6736(10)61458-4
ANI (2020). In a first, Dr Dangs Lab begins bacteriophage susceptibility testing to combat antibiotic resistance. Available online at: https://www.aninews.in/news/health/in-a-first-dr-dangs-lab-begins-bacteriophage-susceptibility-testing-to-combat-antibiotic-resistance20201022154604/ (accessed October 22, 2020).
Ariza-Heredia, E. J., and Chemaly, R. F. (2018). Update on infection control practices in cancer hospitals. CA Cancer J. Clin. 68, 340–355. doi: 10.3322/caac.21462
Armata Therapeuticals (2020). Pipeline Overview. Available online at: https://www.armatapharma.com/pipeline/pipeline-overview/ (accessed December 3, 2020).
Aslam, S., Courtwright, A. M., Koval, C., Lehman, S. M., Morales, S., Furr, C. L., et al. (2019). Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. Am. J. Transplant. 19, 2631–2639. doi: 10.1111/ajt.15503
Bao, J., Wu, N., Zeng, Y., Chen, L., Li, L., Yang, L., et al. (2020). Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerg. Microbes Infect. 9, 771–774. doi: 10.1080/22221751.2020.1747950
Barco, L., Barrucci, F., Olsen, J. E., and Ricci, A. (2013). Salmonella source attribution based on microbial subtyping. Int. J. Food Microbiol. 163, 193–203. doi: 10.1016/j.ijfoodmicro.2013.03.005
Boxrud, D., Pederson-Gulrud, K., Wotton, J., Medus, C., Lyszkowicz, E., Besser, J., et al. (2007). Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype Enteritidis. J. Clin. Microbiol. 45, 536–543. doi: 10.1128/jcm.01595-06
Chan, B. K., Turner, P. E., Kim, S., Mojibian, H. R., Elefteriades, J. A., and Narayan, D. (2018). Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 60–66. doi: 10.1093/emph/eoy005
Chng, K. R., Li, C., Bertrand, D., Ng, A. H. Q., Kwah, J. S., Low, H. M., et al. (2020). Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment. Nat. Med. 26, 941–951. doi: 10.1038/s41591-020-0894-4
Cooke, F. J., Gkrania-Klotsas, E., Howard, J. C., Stone, M., Kearns, A. M., Ganner, M., et al. (2010). Clinical, molecular and epidemiological description of a cluster of community-associated methicillin-resistant Staphylococcus aureus isolates from injecting drug users with bacteraemia. Clin. Microbiol. Infect. 16, 921–926. doi: 10.1111/j.1469-0691.2009.02969.x
Crabb, H. K., Allen, J. L., Devlin, J. M., Firestone, S. M., Stevenson, M., Wilks, C. R., et al. (2019). Traditional Salmonella Typhimurium typing tools (phage typing and MLVA) are sufficient to resolve well-defined outbreak events only. Food Microbiol. 84:103237. doi: 10.1016/j.fm.2019.06.001
D´abrowska, K., and Abedon, S. T. (2019). Pharmacologically aware phage therapy: pharmacodynamic and pharmacokinetic obstacles to phage antibacterial action in animal and human bodies. Microbiol. Mol. Biol. Rev. 83:e00012-19.
D’Accolti, M., Soffritti, I., Lanzoni, L., Bisi, M., Volta, A., Mazzacane, S., et al. (2019). Effective elimination of Staphylococcal contamination from hospital surfaces by a bacteriophage-probiotic sanitation strategy: a monocentric study. Microb. Biotechnol. 12, 742–751. doi: 10.1111/1751-7915.13415
Dedrick, R. M., Guerrero-Bustamante, C. A., Garlena, R. A., Russell, D. A., Ford, K., Harris, K., et al. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733. doi: 10.1038/s41591-019-0437-z
Demczuk, W., Soule, G., Clark, C., Ackermann, H. W., Easy, R., Khakhria, R., et al. (2003). Phage-based typing scheme for Salmonella enterica serovar Heidelberg, a causative agent of food poisonings in Canada. J. Clin. Microbiol. 41, 4279–4284. doi: 10.1128/jcm.41.9.4279-4284.2003
Dong, F., Chen, J., Li, C., Ma, X., Jiang, J., Lin, Q., et al. (2019). Evidence-based analysis on the toxicity of disinfection byproducts in vivo and in vitro for disinfection selection. Water Res. 165:114976. doi: 10.1016/j.watres.2019.114976
Duplessis, C. A., Stockelman, M., Hamilton, T., Merril, G., Brownstein, M., Bishop-Lilly, K., et al. (2019). A case series of emergency investigational new drug applications for bacteriophages treating recalcitrant multi-drug resistant bacterial infections: confirmed safety and a signal of efficacy. J. Intensive Crit. Care 5:11.
Ferrari, R. G., Panzenhagen, P. H. N., and Conte-Junior, C. A. (2017). Phenotypic and genotypic eligible methods for Salmonella typhimurium source tracking. Front. Microbiol. 8:2587. doi: 10.3389/fmicb.2017.02587
Ferry, T., Batailler, C., Petitjean, C., Chateau, J., Fevre, C., Forestier, E., et al. (2020). The potential innovative use of bacteriophages within the DAC(®) hydrogel to treat patients with knee megaprosthesis infection requiring “debridement antibiotics and implant retention” and soft tissue coverage as salvage therapy. Front. Med. (Lausanne) 7:342. doi: 10.3389/fmed.2020.00342
Ferry, T., Boucher, F., Fevre, C., Perpoint, T., Chateau, J., Petitjean, C., et al. (2018). Innovations for the treatment of a complex bone and joint infection due to XDR Pseudomonas aeruginosa including local application of a selected cocktail of bacteriophages. J. Antimicrob. Chemother. 73, 2901–2903. doi: 10.1093/jac/dky263
Gill, J. J., and Hyman, P. (2010). Phage choice, isolation, and preparation for phage therapy. Curr. Pharm. Biotechnol. 11, 2–14. doi: 10.2174/138920110790725311
Gindin, M., Febvre, H. P., Rao, S., Wallace, T. C., and Weir, T. L. (2019). Bacteriophage for gastrointestinal health (PHAGE) study: evaluating the safety and tolerability of supplemental bacteriophage consumption. J. Am. Coll. Nutr. 38, 68–75. doi: 10.1080/07315724.2018.1483783
Hargreaves, K. R., Otieno, J. R., Thanki, A., Blades, M. J., Millard, A. D., Browne, H. P., et al. (2015). As clear as mud? determining the diversity and prevalence of prophages in the draft genomes of estuarine isolates of clostridium difficile. Genome Biol. Evol. 7, 1842–1855. doi: 10.1093/gbe/evv094
Hesse, S., and Adhya, S. (2019). Phage therapy in the twenty-first century: facing the decline of the antibiotic Era; is it finally time for the age of the phage? Annu. Rev. Microbiol. 73, 155–174. doi: 10.1146/annurev-micro-090817-062535
Ho, Y. H., Tseng, C. C., Wang, L. S., Chen, Y. T., Ho, G. J., Lin, T. Y., et al. (2016). Application of bacteriophage-containing aerosol against nosocomial transmission of carbapenem-resistant Acinetobacter baumannii in an intensive care unit. PLoS One 11:e0168380. doi: 10.1371/journal.pone.0168380
Holtappels, D., Fortuna, K., Lavigne, R., and Wagemans, J. (2020). The future of phage biocontrol in integrated plant protection for sustainable crop production. Curr. Opin. Biotechnol. 68, 60–71. doi: 10.1016/j.copbio.2020.08.016
Hoyle, N., Zhvaniya, P., Balarjishvili, N., Bolkvadze, D., Nadareishvili, L., Nizharadze, D., et al. (2018). Phage therapy against Achromobacter xylosoxidans lung infection in a patient with cystic fibrosis: a case report. Res. Microbiol. 169, 540–542. doi: 10.1016/j.resmic.2018.05.001
Intralytix (2020). Human Therapeutics-Intralytix, A Pioneer In Phage-Based Regulatory Approvals. Available online at: http://intralytix.com/index.php?page=hum (accessed December 3, 2020).
Jault, P., Leclerc, T., Jennes, S., Pirnay, J. P., Que, Y. A., Resch, G., et al. (2019). Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 19, 35–45. doi: 10.1016/s1473-3099(18)30482-1
Jensen, K. C., Hair, B. B., Wienclaw, T. M., Murdock, M. H., Hatch, J. B., Trent, A. T., et al. (2015). Isolation and host range of bacteriophage with lytic activity against methicillin-resistant Staphylococcus aureus and potential use as a fomite decontaminant. PLoS One 10:e0131714. doi: 10.1371/journal.pone.0131714
Kortright, K. E., Chan, B. K., Koff, J. L., and Turner, P. E. (2019). Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25, 219–232. doi: 10.1016/j.chom.2019.01.014
Kuipers, S., Ruth, M. M., Mientjes, M., De Sévaux, R. G. L., and Van Ingen, J. (2019). A dutch case report of successful treatment of chronic relapsing urinary tract infection with bacteriophages in a renal transplant patient. Antimicrob. Agents Chemother. 64:e01281-19.
Laxminarayan, R., Duse, A., Wattal, C., Zaidi, A. K., Wertheim, H. F., Sumpradit, N., et al. (2013). Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098.
Leitner, L., Ujmajuridze, A., Chanishvili, N., Goderdzishvili, M., Chkonia, I., Rigvava, S., et al. (2020). Intravesical bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: a randomised, placebo-controlled, double-blind clinical trial. Lancet Infect. Dis. S1473-3099(20)30330-3. doi: 10.1016/S1473-3099(20)30330-3
Letarov, A. V., and Kulikov, E. E. (2018). Determination of the bacteriophage host range: culture-based approach. Methods Mol. Biol. 1693, 75–84. doi: 10.1007/978-1-4939-7395-8_7
Luong, T., Salabarria, A. C., and Roach, D. R. (2020). Phage therapy in the resistance Era: where do we stand and where are we going? Clin. Ther. 42, 1659–1680. doi: 10.1016/j.clinthera.2020.07.014
Maddocks, S., Fabijan, A. P., Ho, J., Lin, R. C. Y., Ben Zakour, N. L., Dugan, C., et al. (2019). Bacteriophage therapy of ventilator-associated pneumonia and empyema caused by Pseudomonas aeruginosa. Am. J. Respir. Crit. Care Med. 200, 1179–1181. doi: 10.1164/rccm.201904-0839le
Malik, D. J., Sokolov, I. J., Vinner, G. K., Mancuso, F., Cinquerrui, S., Vladisavljevic, G. T., et al. (2017). Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv. Colloid Interface Sci. 249, 100–133. doi: 10.1016/j.cis.2017.05.014
Markoishvili, K., Tsitlanadze, G., Katsarava, R., Morris, J. G. Jr., and Sulakvelidze, A. (2002). A novel sustained-release matrix based on biodegradable poly(ester amide)s and impregnated with bacteriophages and an antibiotic shows promise in management of infected venous stasis ulcers and other poorly healing wounds. Int. J. Dermatol. 41, 453–458. doi: 10.1046/j.1365-4362.2002.01451.x
Mathieu, J., Yu, P., Zuo, P., Da Silva, M. L. B., and Alvarez, P. J. J. (2019). Going viral: emerging opportunities for phage-based bacterial control in water treatment and reuse. Acc. Chem. Res. 52, 849–857. doi: 10.1021/acs.accounts.8b00576
Mattila, S., Ruotsalainen, P., and Jalasvuori, M. (2015). On-Demand isolation of bacteriophages against drug-resistant bacteria for personalized phage therapy. Front. Microbiol. 6:1271. doi: 10.3389/fmicb.2015.01271
McCallin, S., Sarker, S. A., Sultana, S., Oechslin, F., and Brüssow, H. (2018). Metagenome analysis of Russian and Georgian Pyophage cocktails and a placebo-controlled safety trial of single phage versus phage cocktail in healthy Staphylococcus aureus carriers. Environ. Microbiol. 20, 3278–3293. doi: 10.1111/1462-2920.14310
Mellon, G., Turbett, S. E., Worby, C., Oliver, E., Walker, A. T., Walters, M., et al. (2020). Acquisition of antibiotic-resistant bacteria by U.S. International Travelers. N. Engl. J. Med. 382, 1372–1374. doi: 10.1056/nejmc1912464
Merabishvili, M., Pirnay, J. P., and De Vos, D. (2018). Guidelines to compose an ideal bacteriophage cocktail. Methods Mol. Biol. 1693, 99–110. doi: 10.1007/978-1-4939-7395-8_9
Mitchell, B. G., Hall, L., White, N., Barnett, A. G., Halton, K., Paterson, D. L., et al. (2019). An environmental cleaning bundle and health-care-associated infections in hospitals (REACH): a multicentre, randomised trial. Lancet Infect. Dis. 19, 410–418. doi: 10.1016/s1473-3099(18)30714-x
Muñoz, A. B., Stepanian, J., Trespalacios, A. A., and Vale, F. F. (2020). Bacteriophages of Helicobacter pylori. Front Microbiol 11:549084. doi: 10.3389/fmicb.2020.549084
Nir-Paz, R., Gelman, D., Khouri, A., Sisson, B. M., Fackler, J., Alkalay-Oren, S., et al. (2019). Successful treatment of antibiotic-resistant, poly-microbial bone infection with bacteriophages and antibiotics combination. Clin. Infect. Dis. 69, 2015–2018. doi: 10.1093/cid/ciz222
Petrovic Fabijan, A., Lin, R. C. Y., Ho, J., Maddocks, S., Ben Zakour, N. L., and Iredell, J. R. (2020). Safety of bacteriophage therapy in severe Staphylococcus aureus infection. Nat. Microbiol. 5, 465–472.
Phagelux (2020). Summary of Clinical Trials. Available online at: http://en.phageluxbio.com/phageluxbioen/vip_doc/22746970_0_0_1.html (accessed December 3, 2020).
Pinto, G., Almeida, C., and Azeredo, J. (2020). Bacteriophages to control Shiga toxin-producing E. coli - safety and regulatory challenges. Crit. Rev. Biotechnol. 40, 1081–1097. doi: 10.1080/07388551.2020.1805719
Pires, D. P., Costa, A. R., Pinto, G., Meneses, L., and Azeredo, J. (2020). Current challenges and future opportunities of phage therapy. FEMS Microbiol. Rev. 44, 684–700. doi: 10.1093/femsre/fuaa017
Pirnay, J. P., De Vos, D., Verbeken, G., Merabishvili, M., Chanishvili, N., Vaneechoutte, M., et al. (2011). The phage therapy paradigm: prêt-à-porter or sur-mesure? Pharm. Res. 28, 934–937. doi: 10.1007/s11095-010-0313-5
Quainoo, S., Coolen, J. P. M., Van Hijum, S., Huynen, M. A., Melchers, W. J. G., Van Schaik, W., et al. (2017). Whole-Genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin. Microbiol. Rev. 30, 1015–1063. doi: 10.1128/cmr.00016-17
Rice, L. B. (2008). Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197, 1079–1081. doi: 10.1086/533452
Rostkowska, O. M., Miêdzybrodzki, R., Miszewska-Szyszkowska, D., Górski, A., and Durlik, M. (2020). Treatment of recurrent urinary tract infections in a 60-year-old kidney transplant recipient. The use of phage therapy. Transpl. Infect. Dis. e13391. doi: 10.1111/tid.13391
Sarker, S. A., Berger, B., Deng, Y., Kieser, S., Foata, F., Moine, D., et al. (2017). Oral application of Escherichia coli bacteriophage: safety tests in healthy and diarrheal children from Bangladesh. Environ. Microbiol. 19, 237–250. doi: 10.1111/1462-2920.13574
Schooley, R. T., Biswas, B., Gill, J. J., Hernandez-Morales, A., Lancaster, J., Lessor, L., et al. (2017). Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61: e00954-17.
Sloan, A., Wang, G., and Cheng, K. (2017). Traditional approaches versus mass spectrometry in bacterial identification and typing. Clin. Chim. Acta 473, 180–185. doi: 10.1016/j.cca.2017.08.035
Sundell, K., Landor, L., Nicolas, P., Jørgensen, J., Castillo, D., Middelboe, M., et al. (2019). Phenotypic and genetic predictors of pathogenicity and virulence in Flavobacterium psychrophilum. Front. Microbiol. 10:1711. doi: 10.3389/fmicb.2019.01711
Tseng, C. C., Chang, D. C., and Chang, K. C. (2019). Development of a biocontrol method applying bacteriophage-containing aerosol against Mycobacterium tuberculosis using the bacteriophage BTCU-1 and M. smegmatis as models. Microorganisms 7:237. doi: 10.3390/microorganisms7080237
Vijayakumar, S., Veeraraghavan, B., Pragasam, A. K., and Bakthavachalam, Y. D. (2019). Genotyping of Acinetobacter baumannii in nosocomial outbreak and surveillance. Methods Mol. Biol. 1946, 17–22. doi: 10.1007/978-1-4939-9118-1_2
Weber, D. J., Rutala, W. A., Sickbert-Bennett, E. E., Kanamori, H., and Anderson, D. (2019). Continuous room decontamination technologies. Am. J. Infect. Control 47s, A72–A78.
World Health Organization (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Available online at: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed February 27, 2017).
Wright, A., Hawkins, C. H., Anggård, E. E., and Harper, D. R. (2009). A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 34, 349–357. doi: 10.1111/j.1749-4486.2009.01973.x
Wuyts, V., Denayer, S., Roosens, N. H., Mattheus, W., Bertrand, S., Marchal, K., et al. (2015). Whole genome sequence analysis of salmonella enteritidis PT4 outbreaks from a national reference laboratory’s viewpoint. PLoS Curr. 7, 1–14. doi: 10.1371/currents.outbreaks.aa5372d90826e6cb0136ff66bb7a62fc
Yerushalmy, O., Khalifa, L., Gold, N., Rakov, C., Alkalay-Oren, S., Adler, K., et al. (2020). The israeli phage bank (IPB). Antibiotics (Basel) 9:269. doi: 10.3390/antibiotics9050269
Żaczek, M., Weber-Da̧browska, B., Miêdzybrodzki, R., Łusiak-Szelachowska, M., and Górski, A. (2020). Phage therapy in poland – a centennial journey to the first ethically approved treatment facility in Europe. Front. Microbiol. 11:1056. doi: 10.3389/fmicb.2020.01056
Keywords: antimicrobial resistance, nosocomial infections, phage library, phage therapy, disinfection
Citation: Wu N and Zhu T (2021) Potential of Therapeutic Bacteriophages in Nosocomial Infection Management. Front. Microbiol. 12:638094. doi: 10.3389/fmicb.2021.638094
Received: 05 December 2020; Accepted: 11 January 2021;
Published: 28 January 2021.
Edited by:
Xuesong He, The Forsyth Institute, United StatesReviewed by:
Hang Yang, Chinese Academy of Sciences, ChinaCopyright © 2021 Wu and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nannan Wu, d3VuYW5uYW5Ac2hwaGMub3JnLmNu; Tongyu Zhu, dHl6aHVAc2hwaGMub3JnLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.