
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 16 March 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.637656
This article is part of the Research TopicBacterial Pathogen Genomics: Recent Achievements, Current Applications and Future ChallengesView all 16 articles
Multidrug-resistant Staphylococcus and vancomycin-resistant Enterococcus (VRE) are important human pathogens that are resistant to most clinical antibiotics. Treatment options are limited and often require the use of ‘last-line’ antimicrobials such as linezolid, daptomycin, and in the case of Staphylococcus, also vancomycin. The emergence of resistance to these last-line antimicrobial agents is therefore of considerable clinical concern. This mini-review provides an overview of resistance to last-line antimicrobial agents in Staphylococcus and VRE, with a particular focus on how genomics has provided critical insights into the emergence of resistant clones, the molecular mechanisms of resistance, and the importance of mobile genetic elements in the global spread of resistance to linezolid.
Staphylococcus and Enterococcus are Gram-positive cocci that are recognized as globally important opportunistic pathogens that can cause serious infections in humans, especially in hospitalized patients (Otto, 2009; Gilmore et al., 2013; Lee A.S. et al., 2018). Over recent decades, there has been a significant increase in the rates of acquired antimicrobial resistance (AMR) in these species, either through the acquisition of resistance determinants by horizontal gene transfer of mobile genetic elements (MGEs), or through mutations that alter gene expression or binding sites in native genes. This has resulted in the emergence of polyclonal lineages that are resistant to front-line therapeutic agents. In staphylococci, this includes the development and global spread of methicillin-resistant Staphylococcus aureus (MRSA) (Enright et al., 2002) and the recent emergence of multidrug-resistant Staphylococcus epidermidis (MDRSE) (Lee J.Y.H. et al., 2018). In enterococci, the emergence and dissemination of vancomycin-resistant enterococci (VRE) (Arias and Murray, 2012), particularly in two healthcare-associated species, vancomycin-resistant Enterococcus faecalis (VREfs) and vancomycin-resistant Enterococcus faecium (VREfm), is of particular clinical concern.
As resistance to different antimicrobials increases in MRSA, MDRSE, and VRE, effective and appropriate treatment becomes increasingly difficult. Limited “last-line” therapeutic options, such as linezolid, daptomycin, and, in the case of staphylococci, vancomycin, remain to treat these infections. However, the high prevalence of these species in healthcare settings (Weiner et al., 2016; Weiner-Lastinger et al., 2020), the increasing clinical use of these last-resort antimicrobials, and the ability of these species to readily develop resistance, provides strong selective pressure for the emergence of clones that are resistant to these agents. Accordingly, resistance to these last-line agents with resultant treatment failure has been increasingly reported. Individual and multiple resistance to linezolid, daptomycin, and vancomycin have been reported in clinical MRSA and MDRSE. While combined resistance to linezolid and daptomycin has recently been reported in VREfm (Wardenburg et al., 2019).
Genomic data can be used to understand the evolution of resistance and investigate the underlying resistance mechanisms. Phylogenetic analyses based on whole-genome sequencing (WGS) can also identify the presence of clones associated with specific resistance determinants, and track their emergence and global spread. The utilization of WGS in hospital infection control to identify outbreaks and transmission events associated with resistant strains is increasingly common. In this mini-review we will provide an overview of the application of genomic analyses to provide critical insights into last-line AMR in MRSA, MDRSE, and VRE, with a particular focus on the emergence of resistant clones, the molecular mechanisms of resistance, and the importance of MGEs in the global spread of linezolid resistance.
Linezolid was the first oxazolidinone approved for clinical use due to its bacteriostatic activity against Gram-positive species. Its mechanism of action inhibits protein synthesis by preventing the formation of the ternary complex between tRNAfMet, mRNA, and the ribosome (Hashemian et al., 2018). Although preliminary research indicated that resistance to linezolid should be rare, resistance in clinical enterococci and staphylococci have been increasingly reported since 2001 (Zurenko et al., 1996; Xiong et al., 2000; Gu et al., 2013; Bender et al., 2018a; Kosecka-Strojek et al., 2020). Early studies using PCR amplicon sequencing in clinical VRE and S. aureus isolates identified mutations in the 23S rRNA genes, that form the binding pocket of the ribosomal peptidyl transferase centre (PTC) to which linezolid binds (Howe et al., 2003; Meka et al., 2004; Livermore et al., 2007). A number of resistance-associated mutations, some specific to staphylococci, others conserved across staphylococci and enterococci, have been identified within and outside the PTC; the G2576T (E. coli nucleotide numbering) mutation is most common (Figure 1). Conferring high-level resistance in MRSA, MDRSE, and VRE, the G2576T 23S rRNA mutation is associated with linezolid treatment failure. Additional single nucleotide polymorphisms (SNPs) in the ribosomal proteins L3 and L4 have been identified through WGS of linezolid-resistant clinical staphylococci (Locke et al., 2009a,b, 2010; Endimiani et al., 2011). Mutations in the L3 and L4 proteins appear to be less common in enterococci, but several reports have identified SNPs in L3 (S133L) and L4 (T35A, N79D, I98V, and N130K) (Mendes et al., 2014; Hua et al., 2019). Further, mutations (A138G, C141T, and G166A) in ribosomal protein L22 have also been identified in linezolid-resistant staphylococci (Bender et al., 2015), as well as a S77T mutation identified in one linezolid-intermediate E. faecium (Lee et al., 2017). The L3, L4, and L22 proteins are in close proximity to the linezolid binding site in the ribosomal PTC, with identified mutations typically adjacent to the PTC, which may affect linezolid binding (Long and Vester, 2012). Using WGS, several studies have documented the local emergence of linezolid resistance in response to linezolid treatment during individual infections (Seedat et al., 2006; Wong et al., 2010; Chen et al., 2018). These studies identified common mutations in 23S rRNA and/or L3, L4, or L22 proteins that independently and repeatedly arise in different species of staphylococci and enterococci from different patients, demonstrating the conserved nature of mutational linezolid resistance and suggesting that convergent evolution is occurring.
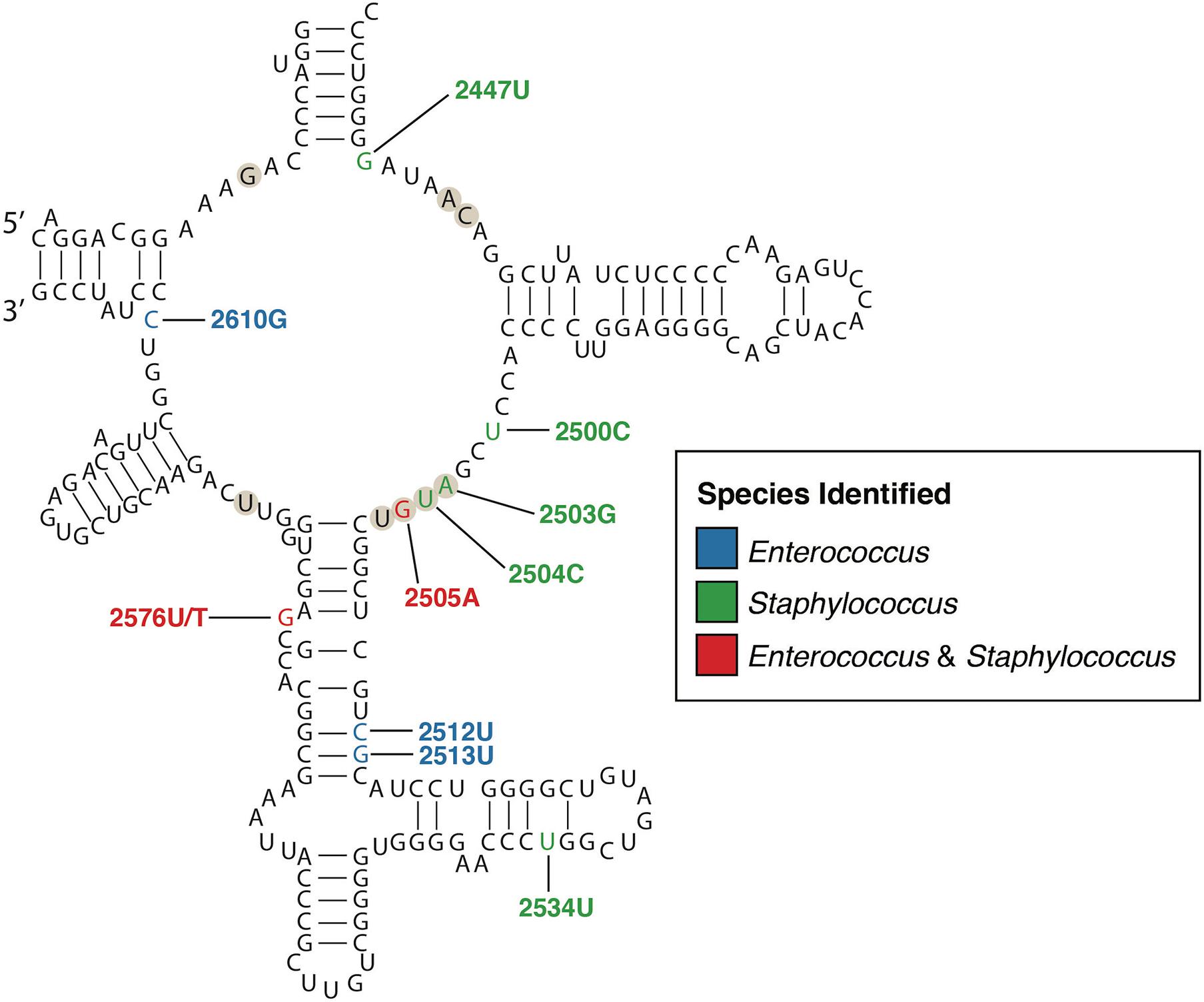
Figure 1. Secondary structure of the peptidyl transferase loop of domain V of 23S rRNA (E. coli numbering). The nucleotides that form the linezolid binding pocket are marked with circles. Nucleotide positions where mutations confer linezolid resistance are colored according to the species identified, blue for Enterococcus, green for Staphylococcus, and red for both. Only mutations with a published relationship in clinical isolates have been included (Wong et al., 2010; Long and Vester, 2012; Mendes et al., 2012; Chen et al., 2018; Wardenburg et al., 2019).
Transferable or acquired resistance to linezolid has been identified in both staphylococcal and enterococcal clinical isolates. The first transferable resistance gene identified was the multi-resistance gene cfr, that encodes a rRNA methyltransferase (Arias et al., 2008). Cfr catalyzes the post-transcriptional methylation of nucleotide A2503 in the 23S rRNA, which confers combined resistance to five different classes of antimicrobials: phenicols, lincosamides, oxazolidinones, pleuromutilins, and Streptogramin A; that bind at overlapping non-identical sites at the PTC, known as the PhLOPSA phenotype (Long et al., 2006). The first linezolid-resistant clinical isolate bearing cfr was reported in 2005, in which cfr had chromosomally integrated into a MRSA strain, within an IS21-558 MGE together with ermB, resulting in resistance to all clinically relevant antibiotics that inhibit protein synthesis (Toh et al., 2007). The presence of a cfr gene has subsequently been identified in staphylococci, including MRSA (Locke et al., 2010; Morales et al., 2010; Antonelli et al., 2016) and MDRSE (Bonilla et al., 2010; Lazaris et al., 2017), and in Enterococcus (Liu et al., 2014; Bender et al., 2016; Fioriti et al., 2020; Ruiz-Ripa et al., 2020) isolates worldwide, although the overall prevalence and distribution in different sequence types (STs) is poorly characterized. In MDRSE, phylogenetic analysis suggests that cfr carriage is most commonly associated with healthcare-associated, clonal complex 2 strains (ST22, ST2, ST5, and ST168), where carriage is associated with a multidrug-resistant phenotype (Mendes et al., 2012).
Several recent studies have identified novel variants of the cfr gene [cfr(B) (Bender et al., 2016), cfr(C) (Candela et al., 2017), and cfr(D) (Guerin et al., 2020)], which are present within a variety of MGEs including multiple plasmids, insertion sequences, and transposons (summarized in Sadowy, 2018 for enterococci). Furthermore, a diverse range of resistance determinants have been co-located with cfr, suggesting that broader antimicrobial use may co-select for the spread of these MGEs. Variation in the genes surrounding cfr in clinical strains suggests that cfr-mediated linezolid resistance has been independently acquired on multiple occasions in clinical staphylococci and enterococci isolates worldwide. Although the reservoir for cfr remains unknown, one study using WGS data from cfr-positive E. faecium suggests that E. faecium may be an important reservoir of cfr(D), which has clear implications for infection control (Guerin et al., 2020). Alternatively, cfr variants may originate from Bacillales, since three cfr-like genes have been identified (Hansen et al., 2012).
Resistance to linezolid also arises through the acquisition of the transferable ribosomal protection genes, optrA, and poxtA. OptrA and PoxtA are part of the F-lineage in the ATP-binding cassette (ABC) superfamily of proteins that are associated with AMR (Jones et al., 2009). These ABC-F proteins mediate resistance to several different classes of anti-ribosomal antimicrobials. For instance, optrA confers resistance to oxazolidinones and phenicols. While, poxtA confers resistance to phenicols, oxazolidinones, and tetracyclines. The family of ABC-F proteins have been shown to mediate resistance by displacing the antibiotic from the ribosome to provide ribosomal protection within the Staphylococcus genus (Sharkey et al., 2016). OptrA was first detected in 2009, on pE349 conjugative plasmids found within three linezolid-resistant E. faecalis clinical isolates (Wang et al., 2015). A phylogenetic analysis of 43 optrA-carrying enterococci revealed nine different variants of the optrA gene (optrAWT, optrA1-5, optrA6, optrA7, and optrA8). These variants were distributed throughout diverse genetic backgrounds of E. faecium and E. faecalis, indicating that multiple independent acquisitions of optrA have occurred in these species (Bender et al., 2018b). The presence of optrA has been subsequently observed in both vancomycin-resistant and vancomycin-susceptible E. faecium and E. faecalis from clinical and food-producing animal isolates globally; but only reported in food-producing animal isolates of S. aureus (Zhu et al., 2020). This suggests that food-producing animals may serve as a reservoir for optrA transfer. Phylogenetic analysis of optrA-carrying enterococci, including some strains from a hospital outbreak, in Europe and South America led to the frequent identification of ST480 E. faecalis, suggesting a potential association between ST480 E. faecalis strains and optrA carriage (Sassi et al., 2019; Egan et al., 2020b; Freitas et al., 2020a; Moure et al., 2020). OptrA has also been reported in several different clonal backgrounds of E. faecalis (Bender et al., 2019) and has been identified in pets carrying E. faecalis (Wu et al., 2019). In E. faecium, optrA has been identified in several different lineages, including ST17, ST80, ST117, ST412, and ST650, indicating that optrA carriage is present in phylogenetically diverse strains in E. faecium (Morroni et al., 2018; Sassi et al., 2019).
The poxtA linezolid resistance gene was first detected in a MRSA clinical strain in 2016 (Antonelli et al., 2018). The poxtA gene exhibits genetic similarity (32% identity and 95% coverage) to optrA and possesses conserved features with other members of the ABC-F family (Antonelli et al., 2018). Carriage of the poxtA gene appears to occur sporadically in MRSA and VREfm clinical isolates (Papagiannitsis et al., 2019), with the gene most commonly identified in food-producing animals (Huang et al., 2017; Elghaieb et al., 2019; Hao et al., 2019; Lei et al., 2019; Fioriti et al., 2020; Freitas et al., 2020b; Na et al., 2020).
Since their initial discovery, the prevalence of cfr, optrA, and poxtA genes in clinical multidrug-resistant enterococcal and staphylococcal isolates has increased (Figure 2; Bi et al., 2018). To date, the highest prevalence of optrA and/or poxtA (35/154; 23%) were reported in E. faecium and E. faecalis in a retrospective Irish study from 2016 to 2019, suggesting significant selection pressure for gene maintenance (Egan et al., 2020b). Genomics has provided critical insights into the genetic relatedness of resistant strains, the structure of MGEs and their role in the mobilization of linezolid resistance determinants. Examples include: an analysis of optrA-carrying plasmids from E. faecalis, which found optrA flanked by two active copies of IS1216 that can facilitate its spread (Shang et al., 2019); the widespread identification of optrA located in a Tn6674-like element in E. faecalis isolated from different countries and sources (Freitas et al., 2020b); IS1216E elements that have been shown to mediate the insertion of poxtA into a pT-E1077-31 plasmid in E. faecium (Shan et al., 2020); while in MRSA, a Tn6349 composite transposon carrying both poxtA and cfr was found to possess a modular structure consisting of fragments from other previously described MGEs (D’Andrea et al., 2019). The presence of these MGEs in cfr, poxtA, and optrA carrying multi-resistance plasmids likely aids in the persistence and dissemination of linezolid resistance genes among these Gram-positive pathogens.
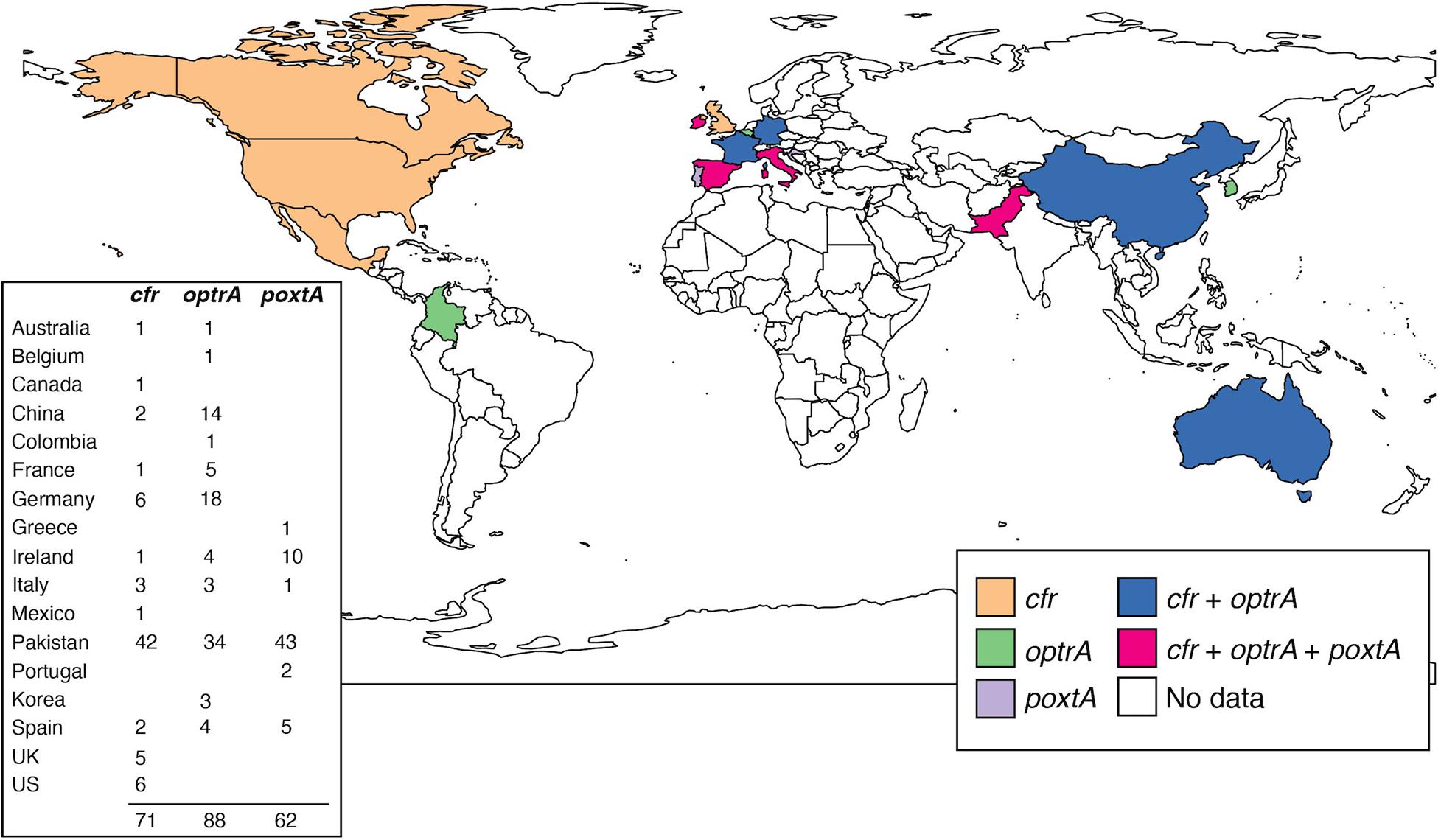
Figure 2. Snapshot of reported cfr, optrA, and poxtA genes in human, clinical, linezolid-resistant E. faecium. Each country is colored based on which gene(s) have been identified. The number of reported strains for each country is shown. It should be noted that the identification of each gene within regions may not accurately represent the genomic epidemiology in E. faecium, due to both collection and availability biases. Only published reports have been included (Bender et al., 2016, 2018b; Lazaris et al., 2017; Wardenburg et al., 2019; Egan et al., 2020a,b; Moure et al., 2020). Map was generated in R (v. 4.0.3) using the ggplot2 package (v. 1.3.0).
Recent studies have also used genomics to identify linezolid-resistant strains for surveillance and infection control purposes. These studies utilized different bioinformatic tools, including LRE-finder (Hasman et al., 2019) and the Resistance Gene Identifier using the Comprehensive Antibiotic Resistance Database (Alcock et al., 2020), to detect cfr, optrA, and poxtA resistance genes in WGS data, enabling rapid genotypic surveillance (Wardenburg et al., 2019; Eisenberger et al., 2020; Moure et al., 2020) and phylogenetic inference (Kerschner et al., 2019). For instance, WGS of E. faecium during an Irish hospital outbreak identified 19 nearly identical ST80 clinical isolates (allelic difference range < 10 SNPs) all carrying optrA-encoded within conjugative plasmid pEfmO_03. Subsequent implementation of enhanced infection control interventions successfully eliminated the outbreak (Egan et al., 2020a).
Shortly after FDA approval of daptomycin for use in MRSA and in the context of off-label use in VRE, resistance, and corresponding clinical treatment failures were reported in both species (Skiest, 2006; Arias et al., 2007; Boyle-Vavra et al., 2011). The development of daptomycin resistance commonly coincides with prior patient exposure, indicating that therapeutic use of daptomycin is likely the main driver of resistance. In S. aureus, the emergence of daptomycin resistance has also been associated with the use of un-related antimicrobials, including vancomycin (Tran et al., 2015) and rifampicin (Guérillot et al., 2018b). Importantly, horizontally acquired and transferable resistance to daptomycin has not been reported to date. The worldwide prevalence of daptomycin resistance is poorly defined since few large-scale surveillance studies exist. A recent meta-analysis in S. aureus, suggested that only 0.1% of S. aureus and 0.1% of MRSA were daptomycin-resistant (Shariati et al., 2020). In enterococci, the prevalence of daptomycin resistance was estimated to be 2.6% in VREfm and 0.1% in VREfs, based on a systemic review of previous studies (Kelesidis et al., 2011). Little is known about which lineages of staphylococci and enterococci are associated with the development daptomycin resistance. In S. aureus, genomic analyses indicate that daptomycin resistance has arisen in ST5, ST398, and ST22 isolates, based on data from the United States and South America (Dabul and Camargo, 2014; Damasco et al., 2019). In S. epidermidis, no comprehensive phylogenetic analyses of daptomycin-resistant strains have been completed, but resistance has been reported in ST2, internationally (Lee J.Y.H. et al., 2018). Similarly, no phylogenetic studies have been completed for E. faecalis, but a novel ST736 clone associated with daptomycin resistance has been identified for E. faecium (Wang et al., 2014) in some United States hospitals (Wang et al., 2018), as has daptomycin-resistant ST80 E. faecium (Udaondo et al., 2020).
Daptomycin’s mode of action involves binding to the anionic phosphoglycerol and various undecaprenyl-coupled cell envelope precursors within the bacterial cell membrane in the presence of Ca2+, blocking cell wall synthesis and triggering delocalization of biosynthetic proteins from the membrane, resulting in bacterial cell death (Grein et al., 2020). Although the specific mechanisms leading to resistance are not fully understood, comparative genomic studies have been pivotal in identifying genetic determinants of daptomycin resistance. Genetically, a critical step toward developing a resistant phenotype appears to involve mutations in two groups of genes: (1) bacterial regulatory systems that function in cell wall homeostasis and (2) enzymes involved in membrane phospholipid metabolism.
In S. aureus and S. epidermidis, mutations in mprF (S295L, S337L, T345L, I420N, and L826F) and walKR (yycFG) (WalKV500F and WalRR216S) are associated with the development of daptomycin resistance in clinical isolates and laboratory-derived strains (Howden et al., 2011; Yang et al., 2009, 2013; Jiang et al., 2019). The MprF enzyme produces cationic phospholipid lysylphophatidyl-glyercol (LysPG) and flips it from the inner to outer leaflet of the cytoplasmic membrane, which decreases the negative cell surface charge (Oku et al., 2004). Although not characterized in S. epidermidis, the mutations in S. aureus likely increase the enzymatic activity of MprF since daptomycin-resistant isolates display increased levels of LysPG in their membrane, which variably leads to a more cationic cell surface that electrostatically repels the daptomycin-Ca2+ complex (Mishra et al., 2011). For mutations in walKR, the mechanism leading to resistance is distinct in S. aureus and S. epidermidis, since S. aureus strains carrying mutations in walKR display increased surface charge while S. epidermidis strains display decreased biofilm formation, compared to the wild-type strains (Jiang et al., 2019). Comparative genomic studies in S. aureus have also identified mutations in the cardiolipin synthase gene cls2 and RNA polymerase β and β’ subunits (rpoB and rpoC) that are associated with resistance to daptomycin, although these are less frequently observed than mutations in mprF and walKR (Miller et al., 2016). Transposon-sequencing in S. aureus has also identified multi-component sensing genes, specifically graRS and vraFG (which regulate cell envelope processes), as being associated with daptomycin resistance, with these genes being important for survival following daptomycin stress (Rajagopal et al., 2016; Coe et al., 2019). However, graRS and vraFG mutations have so far not been described in clinical daptomycin-resistant S. aureus isolates.
In enterococci, comparative genomic studies have shown that mutations in the three-component regulatory system liaFSR and cardiolipin synthase cls are associated with the development of clinical daptomycin resistance. Mutations within the LiaFSR three-component system, which regulates the cell envelope response to cationic host defense peptides and antimicrobial stress, have been frequently identified in daptomycin-resistant clinical isolates, with the LiaST120A and LiaRW73C mutations being of particular importance (Arias et al., 2011). A genomic analysis of E. faecium clinical isolates found that isolates with higher daptomycin MICs (≥ 4 mg/L) usually contained mutations in the liaFSR system (Diaz et al., 2014). Mutations in Cls (H215R and R218Q), encoding a transmembrane protein that catalyzes the synthesis of cardiolipin from phosphoglycerol, are also frequently associated with daptomycin resistance (Arias et al., 2011; Palmer et al., 2011). However, molecular studies have thus far not demonstrated a causal relationship between cls and daptomycin resistance in E. faecium (Tran et al., 2013). Since cls mutations are often found in conjunction with substitutions in the liaFSR regulon, they likely contribute to the progression of an isolate developing a daptomycin-resistant phenotype rather than independently causing resistance (Davlieva et al., 2013). Recent genomic studies in enterococci have also identified daptomycin-resistant isolates that contain wild-type liaFSR and cls alleles, indicating that other unknown genetic determinants of daptomycin resistance exist that require further investigation (Wang et al., 2018; Prater et al., 2019). Indeed, putative associations with daptomycin resistance and mutations within mprF, cfa, pgsA, and dlt have been suggested, but have not yet been verified.
Vancomycin is a critical last-line antibiotic for treating infections caused by multidrug-resistant Staphylococcus. Thirty-nine years after FDA approval of vancomycin, the first instances of S. aureus with intermediate vancomycin resistance (MICs 4 to 8 mg/L) were reported (Sieradzki et al., 1999; Smith et al., 1999). Soon after, the first clinical case of a vancomycin-resistant S. aureus isolate (MIC 1024 mg/L), carrying the vanA vancomycin resistance operon, was reported (Chang et al., 2003). Subsequent molecular studies demonstrated that the vanA operon originated from a co-infecting VREfs strain, raising significant concerns about the potential dissemination of vancomycin resistance in S. aureus from VRE reservoirs (de Niederhäusern et al., 2011). Fortunately, this has not eventuated. Detailed genomic studies have demonstrated that the transfer of the van operon to S. aureus rarely occurs, with very few reports worldwide (McGuinness et al., 2017). The rare occurrence of VRSA may be due to the fitness cost associated with carriage of the van genes, as a longer lag phase was observed during in vitro growth with vancomycin, and the plasmid carrying the van genes was found to be genetically unstable. Further, a MRSA PBP2a strain was unable to utilize lipid II with the D-Alanyl-D-lactate alteration conferred by the van operon (Foster, 2017). A single study identified S. aureus clonal complex 5 (CC5) as the genetic background in which the van operon is most commonly acquired. This study found that CC5 vancomycin-resistant S. aureus (VRSA) strains were genetically distinct, indicating independent acquisition of the vanA operon had occurred (Kos et al., 2012). This suggests that the spread of van operons within S. aureus may occur should appropriate selective conditions develop. Continued vigilance is therefore paramount.
Vancomycin-intermediate S. aureus (VISA) resulting from the progressive accumulation of mutations in key essential and regulatory genes are clinically more common than true vancomycin-resistant S. aureus. VISA isolates are commonly characterized by a thickened cell wall, slower growth, and increased autolysis (reviewed in Howden et al., 2010). Genetically, VISA strains typically contain mutations in two-component regulators vraRS (Cui et al., 2009), graRS (Cui et al., 2009), and walKR (Howden et al., 2011), which function in cell-wall synthesis, but also may contain mutations in the rifampicin resistance determining region of rpoB (Cui et al., 2010). A convergent evolution analysis of 7,099 S. aureus genomes suggested that clinical rifampicin use may promote the emergence of multidrug-resistant lineages of S. aureus that contain rifampicin resistance mutations in rpoB that also confer intermediate resistance to vancomycin (Guérillot et al., 2018a). Although VISA can be found in any genetic background, ST5 is most commonly associated with VISA (Howden et al., 2014), although ST239 isolates tend to display the highest vancomycin MICs (Holmes et al., 2014). Genome-wide association studies (GWAS) performed in ST239 S. aureus have identified mutations in rpoB (H481Y/L/N) (Alam et al., 2014) and the walKR genes (Baines et al., 2015) as being most strongly associated with the development of the VISA phenotype.
In S. epidermidis, the global dissemination of three nearly pan-drug-resistant lineages (two ST2 and one ST23) with heteroresistance to vancomycin was recently demonstrated (Lee J.Y.H. et al., 2018). Analyses of WGS data from 419 clinical isolates from 96 institutions in 24 countries, sampling at least 77 ST types, identified dual D471E and I527M RpoB mutations to be the most common cause of rifampicin resistance in S. epidermidis, accounting for 86.6% of mutations. Of note, the D471E and I527M combination occurred almost exclusively in isolates from the ST2 and ST23 lineages. Through site-specific mutagenesis, the dual RpoB mutation was shown to confer high-level rifampicin resistance as well as reduced susceptibility to vancomycin and teicoplanin. The presence of these same mutations in multiple genetically diverse backgrounds was consistent with their independent emergence with subsequent fixation, presumably due to their advantageous antimicrobial resistant phenotype. Furthermore, acquisition of additional resistance to linezolid (mediated by cfr carrying plasmids and/or mutations in 23S rRNA, L3 and L4) and daptomycin (substitutions in mprF) rendered some European S. epidermidis essentially untreatable. Further examples of convergent evolution include the presence of H481Y/L/N substitutions in RpoB that confer vancomycin resistance in both S. aureus (Guérillot et al., 2018a) and S. epidermidis (Gao et al., 2013; Lee J.Y.H. et al., 2018; Dao et al., 2020).
Collectively, MRSA, MDRSE, and VRE pose a significant clinical and economic healthcare burden. This has been compounded by increasing resistance to the key last-line agents daptomycin, linezolid and in the case of Staphylococcus, vancomycin. Genomic analyses have been pivotal in providing evolutionary insights into the underlying genetic mechanisms of resistance that have arisen in all three species. In particular, the identification of transferable and acquired linezolid resistance determinants through genomic studies, has been critical for understanding the global dissemination and genetic mobility of these genes. Genomic analyses using WGS data is therefore a powerful method for understanding last-line AMR in these and other clinically relevant multidrug-resistant species.
GC and AT conceptualized the mini-review topic. All authors contributed to writing the mini-review and editing the final version.
This work was supported by the Australian Government Research Training Program (AMT). GC was supported by a National Health and Medical Research Council (NHMRC) of Australia Ideas grant (GNT1185213). BH was supported by a NHMRC Practitioner Fellowship (GNT1105905).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alam, M. T., Petit, R. A., Crispell, E. K., Thornton, T. A., Conneely, K. N., Jiang, Y., et al. (2014). Dissecting vancomycin-intermediate resistance in Staphylococcus aureus using genome-wide association. Genome Biol. Evol. 6, 1174–1185. doi: 10.1093/gbe/evu092
Alcock, B. P., Raphenya, A. R., Lau, T. T., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistance surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Antonelli, A., D’Andrea, M. M., Brenciani, A., Galeotti, C. L., Morroni, G., Pollini, S., et al. (2018). Characterization of poxtA, a novel phenicol–oxazolidinone–tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemother. 73, 1763–1769. doi: 10.1093/jac/dky088
Antonelli, A., D’Andrea, M. M., Galano, A., Borchi, B., Brenciani, A., Vaggelli, G., et al. (2016). Linezolid-resistant cfr-positive MRSA, Italy. J. Antimicrob. Chemother. 71, 2349–2351. doi: 10.1093/jac/dkw108
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
Arias, C. A., Panesso, D., McGrath, D. M., Qin, X., Mojica, M. F., Miller, C., et al. (2011). Genetic basis for in vivo daptomycin resistance in Enterococci. N. Engl. J. Med. 365, 892–900. doi: 10.1056/NEJMoa1011138
Arias, C. A., Torres, H. A., Singh, K. V., Panesso, D., Moore, J., Wanger, A., et al. (2007). Failure of daptomycin monotherapy for endocarditis caused by an Enterococcus faecium strain with vancomycin-resistant and vancomycin-susceptible subpopulations and evidence of in vivo loss of the vanA gene cluster. Clin. Infect. Dis. 45, 1343–1346. doi: 10.1086/522656
Arias, C. A., Vallejo, M., Reyes, J., Panesso, D., Moreno, J., Castañeda, E., et al. (2008). Clinical and microbiological aspects of linezolid resistance mediated by the cfr gene encoding a 23S rRNA methyltransferase. J. Clin. Microbiol. 46, 892–896. doi: 10.1128/JCM.01886-07
Baines, S. L., Holt, K. E., Schultz, M. B., Seemann, T., Howden, B. O., Jensen, S. O., et al. (2015). Convergent adaptation in the dominant global hospital clone ST239 of methicillin-resistant Staphylococcus aureus. mBio 6:e00080-15. doi: 10.1128/mBio.00080-15
Bender, J. K., Cattoir, V., Hegstad, K., Sadowy, E., Coque, T. M., Westh, H., et al. (2018a). Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: towards a common nomenclature. Drug Resist. Updat. 40, 25–39. doi: 10.1016/j.drup.2018.10.002
Bender, J. K., Fleige, C., Klare, I., Fiedler, S., Mischnik, A., Mutters, N. T., et al. (2016). Detection of a cfr(B) variant in German Enterococcus faecium clinical isolates and the impact on linezolid resistance in Enterococcus spp. PLoS One 11:e0167042. doi: 10.1371/journal.pone.0167042
Bender, J. K., Fleige, C., Klare, I., and Werner, G. (2019). Development of a multiplex-PCR to simultaneously detect acquired linezolid resistance genes cfr, optrA and poxtA in enterococci of clinical origin. J. Microbiol. Methods 160, 101–103. doi: 10.1016/j.mimet.2019.03.025
Bender, J. K., Fleige, C., Lange, D., Klare, I., and Werner, G. (2018b). Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int. J. Antimicrob. Agents 52, 819–827. doi: 10.1016/j.ijantimicag.2018.09.009
Bender, J. K., Strommenger, B., Steglich, M., Zimmermann, O., Fenner, I., Lensing, C., et al. (2015). Linezolid resistance in clinical isolates of Staphylococcus epidermidis from German hospitals and characterization of two cfr-carrying plasmids. J. Antimicrob. Chemother. 70, 1630–1638. doi: 10.1093/jac/dkv025
Bi, R., Qin, T., Fan, W., Ma, P., and Gu, B. (2018). The emerging problem of linezolid-resistant enterococci. J. Global Antimicrob. Resist. 13, 11–19. doi: 10.1016/j.jgar.2017.10.018
Bonilla, H., Huband, M. D., Seidel, J., Schmidt, H., Lescoe, M., McCurdy, S. P., et al. (2010). Multicity outbreak of linezolid-resistant Staphylococcus epidermidis associated with clonal spread of a cfr-containing strain. Clin. Infect. Dis. 51, 796–800. doi: 10.1086/656281
Boyle-Vavra, S., Jones, M., Gourley, B. L., Holmes, M., Ruf, R., Balsam, A. R., et al. (2011). Comparative genome sequencing of an isogenic pair of USA800 clinical methicillin-resistant Staphylococcus aureus isolates obtained before and after daptomycin treatment failure. Antimicrob. Agents Chemother. 55, 2018–2025. doi: 10.1128/AAC.01593-10
Candela, T., Marvaud, J.-C., Nguyen, T. K., and Lambert, T. (2017). A cfr-like gene cfr(C) conferring linezolid resistance is common in Clostridium difficile. Int. J. Antimicrob. Agents 50, 496–500. doi: 10.1016/j.ijantimicag.2017.03.013
Chang, S., Sievert, D. M., Hageman, J. C., Boulton, M. L., Tenover, F. C., Downes, F. P., et al. (2003). Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348, 1342–1347. doi: 10.1056/NEJMoa025025
Chen, M., Pan, H., Lou, Y., Wu, Z., Zhang, J., Huang, Y., et al. (2018). Epidemiological characteristics and genetic structure of linezolid-resistant Enterococcus faecalis. Infect. Drug Resist. 11, 2397–2409. doi: 10.2147/IDR.S181339
Coe, K. A., Lee, W., Stone, M. C., Komazin-Meredith, G., Meredith, T. C., Grad, Y. H., et al. (2019). Multi-strain Tn-Seq reveals common daptomycin resistance determinants in Staphylococcus aureus. PLoS Pathog. 15:e1007862. doi: 10.1371/journal.ppat.1007862
Cui, L., Isii, T., Fukuda, M., Ochiai, T., Neoh, H., Camargo, I. L. B., et al. (2010). An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 54, 5222–5233. doi: 10.1128/AAC.00437-10
Cui, L., Neoh, H., Shoji, M., and Hiramatsu, K. (2009). Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 53, 1231–1234. doi: 10.1128/AAC.01173-08
Dabul, A. N. G., and Camargo, I. L. B. C. (2014). Molecular characterization of methicillin-resistant Staphylococcus aureus resistant to tigecycline and daptomycin isolated in a hospital in Brazil. Epidemiol. Infect. 142, 479–483. doi: 10.1017/S0950268813001325
Damasco, A. P., Costa, T. M., da Morgado, P. G. M., Guimarães, L. C., Cavalcante, F. S., Nouér, S. A., et al. (2019). Daptomycin and vancomycin non-susceptible methicillin-resistant Staphylococcus aureus clonal lineages from bloodstream infection in a Brazilian teaching hospital. Braz. J. Infect. Dis. 23, 139–142. doi: 10.1016/j.bjid.2019.03.003
D’Andrea, M. M., Antonelli, A., Brenciani, A., Di Pilato, V., Morroni, G., Pollini, S., et al. (2019). Characterization of Tn6349, a novel mosaic transposon carrying poxtA, cfr and other resistance determinants, inserted in the chromosome of an ST5-MRSA-II strain of clinical origin. J. Antimicrob. Chemother. 74, 2870–2875. doi: 10.1093/jac/dkz278
Dao, T. H., Alsallaq, R., Parsons, J. B., Ferrolino, J., Hayden, R. T., Rubnitz, J. E., et al. (2020). Vancomycin heteroresistance and clinical outcomes in bloodstream infections caused by coagulase-negative staphylococci. Antimicrob. Agents Chemother. 64:e00944-20. doi: 10.1128/AAC.00944-20
Davlieva, M., Zhang, W., Arias, C. A., and Shamoo, Y. (2013). Biochemical characterization of cardiolipin synthase mutations associated with daptomycin resistance in enterococci. Antimicrob. Agents Chemother. 57, 289–296. doi: 10.1128/AAC.01743-12
de Niederhäusern, S., Bondi, M., Messi, P., Iseppi, R., Sabia, C., Manicardi, G., et al. (2011). Vancomycin-resistance transferability from VanA enterococci to Staphylococcus aureus. Curr. Microbiol. 62, 1363–1367. doi: 10.1007/s00284-011-9868-6
Diaz, L., Tran, T. T., Munita, J. M., Miller, W. R., Rincon, S., Carvajal, L. P., et al. (2014). Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob. Agents Chemother. 58, 4527–4534. doi: 10.1128/AAC.02686-14
Egan, S. A., Corcoran, S., McDermott, H., Fitzpatrick, M., Hoyne, A., McCormack, O., et al. (2020a). A hospital outbreak of linezolid-resistant and vancomycin-resistant ST80 Enterococcus faecium harbouring an optrA-encoding conjugative plasmid investigated by whole-genome sequencing. J. Hosp. Infect. 105, 726–735. doi: 10.1016/j.jhin.2020.05.013
Egan, S. A., Shore, A. C., O’Connell, B., Brennan, G. I., and Coleman, D. C. (2020b). Linezolid resistance in Enterococcus faecium and Enterococcus faecalis from hospitalized patients in Ireland: high prevalence of the MDR genes optrA and poxtA in isolates with diverse genetic backgrounds. J. Antimicrob. Chemother. 75, 1704–1711. doi: 10.1093/jac/dkaa075
Eisenberger, D., Tuschak, C., Werner, M., Bogdan, C., Bollinger, T., Hossain, H., et al. (2020). Whole-genome analysis of vancomycin-resistant Enterococcus faecium causing nosocomial outbreaks suggests the occurrence of few endemic clonal lineages in Bavaria, Germany. J. Antimicrob. Chemother. 75, 1398–1404. doi: 10.1093/jac/dkaa041
Elghaieb, H., Freitas, A. R., Abbassi, M. S., Novais, C., Zouari, M., Hassen, A., et al. (2019). Dispersal of linezolid-resistant enterococci carrying poxtA or optrA in retail meat and food-producing animals from Tunisia. J. Antimicrob. Chemother. 74, 2865–2869. doi: 10.1093/jac/dkz263
Endimiani, A., Blackford, M., Dasenbrook, E. C., Reed, M. D., Bajaksouszian, S., Hujer, A. M., et al. (2011). Emergence of linezolid-resistant Staphylococcus aureus after prolonged treatment of cystic fibrosis patients in Cleveland, Ohio. Antimicrob. Agents Chemother. 55, 1684–1692. doi: 10.1128/AAC.01308-10
Enright, M. C., Robinson, D. A., Randle, G., Feil, E. J., Grundmann, H., and Spratt, B. G. (2002). The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. U.S.A. 99, 7687–7692. doi: 10.1073/pnas.122108599
Fioriti, S., Morroni, G., Coccitto, S. N., Brenciani, A., Antonelli, A., Di Pilato, V., et al. (2020). Detection of oxazolidinone resistance genes and characterization of genetic environments in Enterococci of swine origin, Italy. Microorganisms 8:2021. doi: 10.3390/microorganisms8122021
Foster, T. J. (2017). Antibiotic resistance in Staphylococcus aureus. current status and future prospects. FEMS Microbiol. Rev. 41, 430–499. doi: 10.1093/femsre/fux007
Freitas, A. R., Tedim, A. P., Novais, C., Lanza, V. F., and Peixe, L. (2020a). Comparative genomics of global optrA-carrying Enterococcus faecalis uncovers a common chromosomal hotspot for optrA acquisition within a diversity of core and accessory genomes. Microb. Genom. 6:e000350. doi: 10.1099/mgen.0.000350
Freitas, A. R., Tedium, A. P., Duarte, B., Elghaieb, H., Abbassi, M. S., Hassen, A., et al. (2020b). Linezolid-resistant (Tn6346::fexB-poxtA) Enterococcus faecium strains colonizing humans and bovines on different continents: similarity without epidemiological link. J. Antimicrob. Chemother. 75, 2416–2423. doi: 10.1093/jac/dkaa227
Gao, W., Cameron, D. R., Davies, J. K., Kostoulias, X., Stepnell, J., Tuck, K. L., et al. (2013). The RpoB H481Y rifampicin resistance mutation and an active stringent response reduce virulence and increase resistance to innate immune responses in Staphylococcus aureus. J. Infect. Dis. 207, 929–939. doi: 10.1093/infdis/jis772
Gilmore, M. S., Lebreton, F., and van Schaik, W. (2013). Genomic transition of Enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr. Opin. Microbiol. 16, 10–16. doi: 10.1016/j.mib.2013.01.006
Grein, F., Müller, A., Scherer, K. M., Liu, X., Ludwig, K. C., Klöckner, A., et al. (2020). Ca 2+-daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids. Nat. Commun. 11, 1455. doi: 10.1038/s41467-020-15257-1
Gu, B., Kelesidis, T., Tsiodras, S., Hindler, J., and Humphries, R. M. (2013). The emerging problem of linezolid-resistant Staphylococcus. J. Antimicrob. Chemother. 68, 4–11. doi: 10.1093/jac/dks354
Guérillot, R., Gonçalves da Silva, A., Monk, I., Giulieri, S., Tomita, T., Alison, E., et al. (2018a). Convergent evolution driven by rifampin exacerbates the global burden of drug-resistant Staphylococcus aureus. mSphere 3, e00550-17. doi: 10.1128/mSphere.00550-17
Guérillot, R., Li, L., Baines, S., Howden, B., Schultz, M., Seemann, T., et al. (2018b). Comprehensive antibiotic-linked mutation assessment by resistance mutation sequencing (RM-seq). Genome Med. 10:63. doi: 10.1186/s13073-018-0572-z
Guerin, F., Sassi, M., Dejoies, L., Zouari, A., Schutz, S., Potrel, S., et al. (2020). Molecular and functional analysis of the novel cfr(D) linezolid resistance gene identified in Enterococcus faecium. J. Antimicrob. Chemother. 75, 1699–1703. doi: 10.1093/jac/dkaa125
Hansen, L. H., Planellas, M. H., Long, K. S., and Vester, B. (2012). The order Bacillales hosts functional homologs of the worrisome cfr antibiotic resistance gene. Antimicrob. Agents Chemother. 56, 3563–3567. doi: 10.1128/AAC.00673-12
Hao, W., Shan, X., Li, D., Schwarz, S., Zhang, S.-M., Li, X.-S., et al. (2019). Analysis of a poxtA- and optrA-co-carrying conjugative multiresistance plasmid from Enterococcus faecalis. J. Antimicrob. Chemother. 74, 1771–1775. doi: 10.1093/jac/dkz109
Hashemian, S. M. R., Farhadi, T., and Ganjparvar, M. (2018). Linezolid: a review of its properties, function, and use in critical care. Drug Des. Dev. Ther. 12, 1759–1767. doi: 10.2147/DDDT.S164515
Hasman, H., Clausen, P. T. L. C., Kaya, H., Hansen, F., Knudsen, J. D., Wang, M., et al. (2019). LRE-Finder, a web tool for detection of the 23S rRNA mutations and the optrA, cfr, cfr(B) and poxtA genes encoding linezolid resistance in Enterococci from whole-genome sequences. J. Antimicrob. Chemother. 74, 1473–1476. doi: 10.1093/jac/dkz092
Holmes, N. E., Turnidge, J. D., Munckhof, W. J., Robinson, J. O., Korman, T. M., O’Sullivan, M. V. N., et al. (2014). Genetic and molecular predictors of high vancomycin MIC in Staphylococcus aureus bacteremia isolates. J. Clin. Microbiol. 52, 3384–3393. doi: 10.1128/JCM.01320-14
Howden, B. P., Davies, J. K., Johnson, P. D. R., Stinear, T. P., and Grayson, M. L. (2010). Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23, 99–139. doi: 10.1128/CMR.00042-09
Howden, B. P., McEvoy, C. R. E., Allen, D. L., Chua, K., Gao, W., Harrison, P. F., et al. (2011). Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Path. 7:e1002359. doi: 10.1371/journal.ppat.1002359
Howden, B. P., Peleg, A. Y., and Stinear, T. P. (2014). The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect. Genet. Evol. 21, 575–582. doi: 10.1016/j.meegid.2013.03.047
Howe, R. A., Wootton, M., Noel, A. R., Bowker, K. E., Walsh, T. R., and MacGowan, A. P. (2003). Activity of AZD2563, a novel oxazolidinone, against Staphylococcus aureus strains with reduced susceptibility to vancomycin or linezolid. Antimicrob. Agents. Chemother. 47, 3651–3652. doi: 10.1128/AAC.47.11.3651-3652.2003
Hua, R., Xia, Y., Wu, W., Yang, M., and Yan, J. (2019). Molecular epidemiology and mechanisms of 43 low-level linezolid-resistant Enterococcus faecalis strains in Chongqing. China. Ann. Lab. Med. 39, 36–42. doi: 10.3343/alm.2019.39.1.36
Huang, J., Chen, L., Wu, Z., and Wang, L. (2017). Retrospective analysis of genome sequences revealed the wide dissemination of optrA in Gram-positive bacteria. J. Antimicrob. Chemother. 72, 614–616. doi: 10.1093/jac/dkw488
Jiang, J.-H., Dexter, C., Cameron, D. R., Monk, I. R., Baines, S. L., Abbott, I. J., et al. (2019). Evolution of daptomycin resistance in coagulase-negative staphylococci involves mutations of the essential two-component regulator WalKR. Antimicrob. Agents Chemother. 63:e01926-18. doi: 10.1128/AAC.01926-18
Jones, P. M., O’Mara, M. L., and George, A. M. (2009). ABC transporters: a riddle wrapped in a mystery inside an enigma. Trends Biochem. Sci. 34, 520–531. doi: 10.1016/j.tibs.2009.06.004
Kelesidis, T., Humphries, R., Uslan, D. Z., and Pegues, D. A. (2011). Daptomycin nonsusceptible Enterococci: an emerging challenge for clinicians. Clin. Infect. Dis. 52, 228–234. doi: 10.1093/cid/ciq113
Kerschner, H., Cabal, A., Hartl, R., Machherndl-Spandl, S., Allerberger, F., Ruppitsch, W., et al. (2019). Hospital outbreak caused by linezolid resistant Enterococcus faecium in Upper Austria. Antimicrob. Resist. Infect. Control 8:150. doi: 10.1186/s13756-019-0598-z
Kos, V. N., Desjardins, C. A., Griggs, A., Cerqueira, G., Tonder, A. V., Holden, M. T. G., et al. (2012). Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with methicillin-resistant S. aureus hospital-acquired infection in the United States. mBio 3:e00112-12. doi: 10.1128/mBio.00112-12
Kosecka-Strojek, M., Sadowy, E., Gawryszewska, I., Klepacka, J., Tomasik, T., Michalik, M., et al. (2020). Emergence of linezolid-resistant Staphylococcus epidermidis in the tertiary children’s hospital in Cracow, Poland. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1717–1725. doi: 10.1007/s10096-020-03893-w
Lazaris, A., Coleman, D. C., Kearns, A. M., Pichon, B., Kinnevey, P. M., Earls, M. R., et al. (2017). Novel multiresistance cfr plasmids in linezolid-resistant methicillin-resistant Staphylococcus epidermidis and vancomycin-resistant Enterococcus faecium (VRE) from a hospital outbreak: co-location of cfr and optrA in VRE. J. Antimicrob. Chemother. 72, 3252–3257. doi: 10.1093/jac/dkx292
Lee, A. S., de Lencastre, H., Garau, J., Kluytmans, J., Malhotra-Kumar, S., Peschel, A., et al. (2018). Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. 4, 1–23. doi: 10.1038/nrdp.2018.33
Lee, J. Y. H., Monk, I. R., da Silva, A. G., Seemann, T., Chua, K. Y. L., Kearns, A., et al. (2018). Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat. Microbiol. 3, 1175–1185. doi: 10.1038/s41564-018-0230-7
Lee, S. M., Huh, J. H., Song, D. J., Shim, H. J., Park, K. S., Kang, C. I., et al. (2017). Resistance mechanisms of linezolid-nonsusceptible Enterococci in Korea: low rate of 23S rRNA mutation in Enterococcus faecium. J. Med. Microbiol. 66, 1730–1735. doi: 10.1099/jmm.0.00063
Lei, C.-W., Kang, Z.-Z., Wu, S.-K., Chen, Y.-P., Kong, L.-H., and Wang, H.-N. (2019). Detection of the phenicol–oxazolidinone–tetracycline resistance gene poxtA in Enterococcus faecium and Enterococcus faecalis of food-producing animal origin in China. J. Antimicrob. Chemother. 74, 2459–2461. doi: 10.1093/jac/dkz198
Liu, Y., Wang, Y., Schwarz, S., Wang, S., Chen, L., Wu, C., et al. (2014). Investigation of a multiresistance gene cfr that fails to mediate resistance to phenicols and oxazolidinones in Enterococcus faecalis. J. Antimicrob. Chemother. 69, 892–898. doi: 10.1093/jac/dkt459
Livermore, D. M., Warner, M., Mushtaq, S., North, S., and Woodford, N. (2007). In vitro activity of the oxazolidinone RWJ-416457 against linezolid-resistant and -susceptible Staphylococci and Enterococci. Antimicrob. Agents Chemother. 51, 1112–1114. doi: 10.1128/AAC.01347-06
Locke, J. B., Hilgers, M., and Shaw, K. J. (2009a). Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob. Agents Chemother. 53, 5275–5278. doi: 10.1128/AAC.01032-09
Locke, J. B., Hilgers, M., and Shaw, K. J. (2009b). Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob. Agents Chemother. 53, 5265–5274. doi: 10.1128/AAC.00871-09
Locke, J. B., Morales, G., Hilgers, M., C, K. G., Rahawi, S., Picazo, J. J., et al. (2010). Elevated linezolid resistance in clinical cfr-positive Staphylococcus aureus isolates is associated with co-occurring mutations in ribosomal protein L3. Antimicrob. Agents Chemother. 54, 5352–5355. doi: 10.1128/AAC.00714-10
Long, K. S., Poehlsgaard, J., Kehrenberg, C., Schwarz, S., and Vester, B. (2006). The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50, 2500–2505. doi: 10.1128/AAC.00131-06
Long, K. S., and Vester, B. (2012). Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 56, 603–612. doi: 10.1128/AAC.05702-11
McGuinness, W. A., Malachowa, N., and DeLeo, F. R. (2017). Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 90, 269–281.
Meka, V. G., Pillai, S. K., Sakoulas, G., Wennersten, C., Venkataraman, L., DeGirolami, P. C., et al. (2004). Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA. J. Infect. Dis. 190, 311–317. doi: 10.1086/421471
Mendes, R. E., Deshpande, L. M., Costello, A. J., and Farrell, D. J. (2012). Molecular epidemiology of Staphylococcus epidermidis clinical isolates from U.S. Hospitals. Antimicrob. Agents Chemother. 56, 4656–4661. doi: 10.1128/AAC.00279-12
Mendes, R. E., Deshpande, L. M., and Jones, R. N. (2014). Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist. Updat. 17, 1–12. doi: 10.1016/j.drup.2014.04.002
Miller, W. R., Bayer, A. S., and Arias, C. A. (2016). Mechanism of action and resistance to daptomycin in Staphylococcus aureus and Enterococci. Cold Spring Harb. Perspect. Med. 6:a026997. doi: 10.1101/cshperspect.a026997
Mishra, N. N., McKinnell, J., Yeaman, M. R., Rubio, A., Nast, C. C., Chen, L., et al. (2011). In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 55, 4012–4018. doi: 10.1128/AAC.00223-11
Morales, G., Picazo, J. J., Baos, E., Candel, F. J., Arribi, A., Peláez, B., et al. (2010). Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 50, 821–825. doi: 10.1086/650574
Morroni, G., Brenciani, A., Antonelli, A., Maria, D., Andrea, M., Di Pilato, V., et al. (2018). Characterization of a multiresistance plasmid carrying the optrA and cfr resistance genes from an Enterococcus faecium clinical isolate. Front. Microbiol. 9:2189. doi: 10.3389/fmicb.2018.02189
Moure, Z., Lara, N., Marín, M., Sola-Campoy, P. J., Bautista, V., Gómez-Bertomeu, F., et al. (2020). Interregional spread in Spain of linezolid-resistant Enterococcus spp. isolates carrying the optrA and poxtA genes. Int. J. Antimicrob. Agents 55, 105977. doi: 10.1016/j.ijantimicag.2020.105977
Na, S. H., Moon, D. C., Kim, M. H., Kang, H. Y., Kim, S. J., Choi, J. H., et al. (2020). Detection of the phenicol-oxazolidinone resistance gene poxtA in Enterococcus faecium and Enterococcus faecalis from food-producing animals during 2008-2018 in Korea. Microorganisms 8:1839. doi: 10.3390/microorganisms8111839
Oku, Y., Kurokawa, K., Ichihashi, N., and Sekimizu, K. (2004). Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 150, 45–51. doi: 10.1099/mic.0.26706-0
Otto, M. (2009). Staphylococcus epidermidis — the “accidental” pathogen. Nat. Rev. Microbiol. 7, 555–567. doi: 10.1038/nrmicro2182
Palmer, K. L., Daniel, A., Hardy, C., Silverman, J., and Gilmore, M. S. (2011). Genetic basis for daptomycin resistance in Enterococci. Antimicrob. Agents Chemother. 55, 3345–3356. doi: 10.1128/AAC.00207-11
Papagiannitsis, C. C., Tsilipounidaki, K., Malli, E., and Petinaki, E. (2019). Detection in Greece of a clinical Enterococcus faecium isolate carrying the novel oxazolidinone resistance gene poxtA. J. Antimicrob. Chemother. 74, 2461–2462. doi: 10.1093/jac/dkz155
Prater, A. G., Mehta, H. H., Kosgei, A. J., Miller, W. R., Tran, T. T., Arias, C. A., et al. (2019). Environment shapes the accessible daptomycin resistance mechanisms in Enterococcus faecium. Antimicrob. Agents Chemother. 63:e00790-19. doi: 10.1128/AAC.00790-19
Rajagopal, M., Martin, M. J., Santiago, M., Lee, W., Kos, V. N., Meredith, T., et al. (2016). Multidrug intrinsic resistance factors in Staphylococcus aureus identified by profiling fitness within high-diversity transposon libraries. mBio 7:e00950-16. doi: 10.1128/mBio.00950-16
Ruiz-Ripa, L., Feßler, A. T., Hanke, D., Eichhorn, I., Azcona-Gutiérrez, J. M., Pérez-Moreno, M. O., et al. (2020). Mechanisms of linezolid resistance among Enterococci of clinical origin in Spain – detection of optrA- and cfr(D)-carrying E. faecalis. Microorganisms 8:1155. doi: 10.3390/microorganisms8081155
Sadowy, E. (2018). Linezolid resistance genes and genetic elements enhancing their dissemination in Enterococci and streptococci. Plasmid 99, 89–98. doi: 10.1016/j.plasmid.2018.09.011
Sassi, M., Guérin, F., Zouari, A., Beyrouthy, R., Auzou, M., Fines-Guyon, M., et al. (2019). Emergence of optrA-mediated linezolid resistance in Enterococci from France, 2006–16. J. Antimicrob. Chemother. 74, 1469–1472. doi: 10.1093/jac/dkz097
Seedat, J., Zick, G., Klare, I., Konstabel, C., Weiler, N., and Sahly, H. (2006). rapid emergence of resistance to linezolid during linezolid therapy of an Enterococcus faecium infection. Antimicrob. Agents Chemother. 50, 4217–4219. doi: 10.1128/AAC.00518-06
Shan, X., Li, X.-S., Wang, N., Schwarz, S., Zhang, S.-M., Li, D., et al. (2020). Studies on the role of IS1216E in the formation and dissemination of poxtA-carrying plasmids in an Enterococcus faecium clade A1 isolate. J. Antimicrob. Chemother. 75, 3126–3130. doi: 10.1093/jac/dkaa325
Shang, Y., Li, D., Shan, X., Schwarz, S., Zhang, S.-M., Chen, Y.-X., et al. (2019). Analysis of two pheromone-responsive conjugative multiresistance plasmids carrying the novel mobile optrA locus from Enterococcus faecalis. Infect. Drug Resist. 12, 2355–2362. doi: 10.2147/IDR.S206295
Shariati, A., Dadashi, M., Chegini, Z., van Belkum, A., Mirzaii, M., Khoramrooz, S. S., et al. (2020). The global prevalence of daptomycin, tigecycline, quinupristin/dalfopristin, and linezolid-resistant Staphylococcus aureus and coagulase–negative staphylococci strains: a systematic review and meta-analysis. Antimicrob. Resist. Infect. Control. 9:56. doi: 10.1186/s13756-020-00714-9
Sharkey, L. K. R., Edwards, T. A., and O’Neill, A. J. (2016). ABC-F proteins mediate antibiotic resistance through ribosomal protection. mBio 7:e01975. doi: 10.1128/mBio.01975-15
Sieradzki, K., Roberts, R. B., Haber, S. W., and Tomasz, A. (1999). The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340, 517–523. doi: 10.1056/NEJM199902183400704
Skiest, D. J. (2006). treatment failure resulting from resistance of Staphylococcus aureus to daptomycin. J. Clin. Microbiol. 44, 655–656. doi: 10.1128/JCM.44.2.655-656.2006
Smith, T. L., Pearson, M. L., Wilcox, K. R., Cruz, C., Lancaster, M. V., Robinson-Dunn, B., et al. (1999). Emergence of vancomycin resistance in Staphylococcus aureus. N. Engl. J. Med. 340, 493–501. doi: 10.1056/NEJM199902183400701
Toh, S.-M., Xiong, L., Arias, C. A., Villegas, M. V., Lolans, K., Quinn, J., et al. (2007). Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64, 1506–1514. doi: 10.1111/j.1365-2958.2007.05744.x
Tran, T. T., Munita, J. M., and Arias, C. A. (2015). Mechanisms of drug resistance: daptomycin resistance. Ann. N. Y. Acad. Sci. 1354, 32–53. doi: 10.1111/nyas.12948
Tran, T. T., Panesso, D., Gao, H., Roh, J. H., Munita, J. M., Reyes, J., et al. (2013). Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob. Agents Chemother. 57, 261–268. doi: 10.1128/AAC.01454-12
Udaondo, Z., Jenjaroenpun, P., Wongsurawat, T., Meyers, E., Anderson, C., Lopez, J., et al. (2020). Two cases of vancomycin-resistant Enterococcus faecium bacteremia with development of daptomycin-resistant phenotype and its detection using oxford nanopore sequencing. Open Forum Infect. Dis. 7:ofaa180. doi: 10.1093/ofid/ofaa180
Wang, G., Kamalakaran, S., Dhand, A., Huang, W., Ojaimi, C., Zhuge, J., et al. (2014). Identification of a novel clone, ST736, among Enterococcus faecium clinical isolates and its association with daptomycin nonsusceptibility. Antimicrob. Agents Chemother. 58, 4848–4854. doi: 10.1128/AAC.02683-14
Wang, G., Yu, F., Lin, H., Murugesan, K., Huang, W., Hoss, A. G., et al. (2018). Evolution and mutations predisposing to daptomycin resistance in vancomycin-resistant Enterococcus faecium ST736 strains. PLoS One 13:e0209785. doi: 10.1371/journal.pone.0209785
Wang, Y., Lv, Y., Cai, J., Schwarz, S., Cui, L., Hu, Z., et al. (2015). A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 70, 2182–2190. doi: 10.1093/jac/dkv116
Wardenburg, K. E., Potter, R. F., D’Souza, A. W., Hussain, T., Wallace, M. A., Andleeb, S., et al. (2019). Phenotypic and genotypic characterization of linezolid-resistant Enterococcus faecium from the USA and Pakistan. J. Antimicrob. Chemother. 74, 3445–3452. doi: 10.1093/jac/dkz367
Weiner, L. M., Webb, A. K., Limbago, B., Dudeck, M. A., Patel, J., Kallen, A. J., et al. (2016). Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 37, 1288–1301. doi: 10.1017/ice.2016.174
Weiner-Lastinger, L. M., Abner, S., Edwards, J. R., Kallen, A. J., Karlsson, M., Magill, S. S., et al. (2020). Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: summary of data reported to the National Healthcare Safety Network, 2015–2017. Infect. Control Hosp. Epidemiol. 41, 1–18. doi: 10.1017/ice.2019.296
Wong, A., Reddy, S. P., Smyth, D. S., Aguero-Rosenfeld, M. E., Sakoulas, G., and Robinson, D. A. (2010). Polyphyletic emergence of linezolid-resistant staphylococci in the United States. Antimicrob. Agents. Chemother. 54, 742–748. doi: 10.1128/AAC.00621-09
Wu, Y., Fan, R., Wang, Y., Lei, L., Feßler, A. T., Wang, Z., et al. (2019). Analysis of combined resistance of oxazolidinones and phenicols among bacteria from dogs fed with raw meat/vegetables and the respective food items. Sci. Rep. 9:15500. doi: 10.1038/s41598-019-51918-y
Xiong, L., Kloss, P., Douthwaite, S., Andersen, N. M., Swaney, S., Shinabarger, D. L., et al. (2000). Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J. Bacteriol. 182, 5325–5331. doi: 10.1128/JB.182.19.5325-5331.2000
Yang, S.-J., Mishra, N. N., Rubio, A., and Bayer, A. S. (2013). Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob. Agents Chemother. 57, 5658–5664. doi: 10.1128/AAC.01184-13
Yang, S.-J., Xiong, Y. Q., Dunman, P. M., Schrenzel, J., François, P., Peschel, A., et al. (2009). Regulation of mprF in Daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob. Agents Chemother. 53, 2636–2637. doi: 10.1128/AAC.01415-08
Zhu, Y., Zhang, W., Wang, C., Liu, W., Yang, Q., Luan, T., et al. (2020). Identification of a novel optrA-harbouring transposon, Tn6823, in Staphylococcus aureus. J. Antimicrob. Chemother. 75, 3395–3397. doi: 10.1093/jac/dkaa323
Keywords: linezolid, daptomycin, vancomycin, genomics, Enterococcus, Staphylococcus
Citation: Turner AM, Lee JYH, Gorrie CL, Howden BP and Carter GP (2021) Genomic Insights Into Last-Line Antimicrobial Resistance in Multidrug-Resistant Staphylococcus and Vancomycin-Resistant Enterococcus. Front. Microbiol. 12:637656. doi: 10.3389/fmicb.2021.637656
Received: 11 December 2020; Accepted: 25 February 2021;
Published: 16 March 2021.
Edited by:
Guido Werner, Robert Koch Institute (RKI), GermanyReviewed by:
Ana P. Tedim, Institute of Health Sciences Studies of Castilla y León (IECSCYL), SpainCopyright © 2021 Turner, Lee, Gorrie, Howden and Carter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Glen P. Carter, Z2xlbi5jYXJ0ZXJAdW5pbWVsYi5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.