- 1Key Laboratory of Gastrointestinal Cancer (Fujian Medical University), Ministry of Education, Fuzhou, China
- 2Fujian Key Laboratory of Tumor Microbiology, Department of Medical Microbiology, Fujian Medical University, Fuzhou, China
- 3Fujian Medical University Union Hospital, Fuzhou, China
Helicobacter pylori harbors a dipeptide (Dpp) transporter consisting of a substrate-binding protein (DppA), two permeases (DppB and C), and two ATPases (DppD and F). The Dpp transporter is responsible for the transportation of dipeptides and short peptides. We found that its expression is important for the growth of H. pylori. To understand the role of the Dpp transporter in the pathogenesis of H. pylori, the expression of virulence factors and H. pylori-induced IL-8 production were investigated in H. pylori wild-type and isogenic H. pylori Dpp transporter mutants. We found that expression of CagA was downregulated, while expression of type 4 secretion system (T4SS) components was upregulated in Dpp transporter mutants. The DppA mutant strain expressed higher levels of outer membrane proteins (OMPs), including BabA, HopZ, OipA, and SabA, and showed a higher adhesion level to gastric epithelial AGS cells compared with the H. pylori 26695 wild-type strain. After infection of AGS cells, H. pylori ΔdppA induced a higher level of NF-κB activation and IL-8 production compared with wild-type. These results suggested that in addition to supporting the growth of H. pylori, the Dpp transporter causes bacteria to alter the expression of virulence factors and reduces H. pylori-induced NF-κB activation and IL-8 production in gastric epithelial cells.
Introduction
Helicobacter pylori is a microaerophilic, Gram-negative bacterium that is closely related to chronic gastritis, peptic ulcers, and gastric cancer (Kusters et al., 2006; Chmiela and Kupcinskas, 2019). To colonize the human stomach, H. pylori has to pass through the mucous layer to the surface of gastric mucosal epithelial cells via the movement of its flagella, and then colonizes the epithelial cells with the aid of adhesins (Sgouras et al., 2015). The pathogenesis of H. pylori is driven by several virulence factors that facilitate bacterial colonization, induce inflammation, and damage host cells. Among the virulence factors confirmed to function in H. pylori infection, Type 4 secretion system (T4SS) and its effector protein CagA encoded by cag pathogenicity island (cagPAI) are one of the most extensively studied H. pylori virulence factors (Guillemin et al., 2002; Sanchezzauco et al., 2013). cagPAI is about 40 kb in size and comprises 26 genes that encode the components of the T4SS. Relying on the T4SS, which binds to the α5β1 integrin expressed on the surface of gastric epithelial cells (Kwok et al., 2007), H. pylori delivers CagA, ADP-heptose (Pfannkuch et al., 2019), and peptidoglycan into host cells (Viala et al., 2004). H. pylori infection activates nuclear factor-kappa B (NF-κB) in gastric epithelial cells, inducing the release of proinflammatory factors such as interleukin 8 (IL-8) (Maeda et al., 2000; Backert and Naumann, 2010).
Successful colonization requires adaptation of the bacterium to the gastric environment. Environmental factors such as pH, reactive oxygen species, temperature, or nutrients can affect the expression of H. pylori virulence factors (Merrell et al., 2003a; Pflock et al., 2006; Augusto et al., 2007; Noto et al., 2015). Acidic pH highly stimulates the expression of antioxidant proteins, flagellar structural proteins, and T4SS component proteins in H. pylori (Marcus et al., 2018). Upregulation of vacA and downregulation of genes related to motility were observed under iron-restricted conditions (Merrell et al., 2003b). Iron deficiency enhances H. pylori virulence; thus, H. pylori isolated from iron-depleted gerbils expressed significantly higher levels of CagA, which induced more robust proinflammatory responses (Noto et al., 2012).
Considering that nutrients are important for the growth of bacteria, genes involved in the metabolism serve as targets for antimicrobial therapies. The peptide transporter systems have been extensively investigated in bacteria such as Escherichia coli and Lactococcus lactis (Sanz et al., 2001; Harder et al., 2008). Peptide transporters play an important role in nutritional supply by providing carbon sources or nitrogen sources for bacterial growth (Gilvarg, 1972). Three types of peptide transporters in bacteria have been found to date: oligopeptide (Opp) transporters, dipeptide (Dpp) transporters, and dip/tripeptide (Dtp) transporters (Garai et al., 2017). The Opp and Dpp transporters belong to the ATP-binding cassette (ABC) superfamily, while the Dtp transporter belongs to the proton-dependent oligopeptide transporter (POT) (Paulsen and Skurray, 1994). The Dpp transporters are responsible for transporting mainly dipeptides but also tripeptides into cells (Payne and Smith, 1994), while the Opp transporters are responsible for the import of oligopeptides. The Dpp transporter in H. pylori is composed of five proteins encoded by dppA, B, C, D, and F (Davis and Mobley, 2005; Weinberg and Maier, 2007). DppA is a periplasmic peptide-binding protein, DppB and DppC are integral membrane proteins that form permeases for substrates, while DppD and DppF are cytoplasmic proteins responsible for ATP hydrolysis.
Apart from being involved in the transport of nutrients, peptide transporters play a role in the virulence of various bacterial pathogens (Samen et al., 2004; Moraes et al., 2014). In E. coli, the Dpp transporter acts as a primary chemoreceptor, and its interaction with the membrane components for dipeptide chemotaxis initiates flagellar motion (Manson et al., 1986). In Borrelia burgdorferi, an opp mutant strain promotes the expression of the virulence factor OspC by regulating the Rrp2-RpoN-RpoS pathway (Zhou et al., 2018). In group A Streptococci, Dpp mutation results in a decreased expression of SpeB, a major cysteine protease (Podbielski and Leonard, 1998). In Pseudoalteromonas, DppA plays an important role in cold adaptation (Zhang et al., 2010). However, hitherto the role of Dpp transporters in the growth and pathogenesis of H. pylori remains unknown. A study has shown that expression of DppA in H. pylori was stimulated by gastric epithelial cells, suggesting that DppA might play an important role in the pathogenesis of H. pylori (Sharma et al., 2010).
In this work, we constructed Dpp transporter mutants in H. pylori and evaluated the effects of the Dpp system on growth, expression of virulence factors, and inflammatory responses of AGS cells stimulated by H. pylori.
Materials and Methods
Bacterial Strains and Cultivation Conditions
Helicobacter pylori 26695, NCTC11637 and Dpp transporter mutant strains were cultured in a microaerobic environment (5% O2, 10% CO2, and 85% N2) at 37°C on Columbia agar plates (Oxoid, Cambridge, United Kingdom) containing 7% sheep blood. For liquid cultivation of H. pylori, Brucella broth supplied with 10% fetal bovine serum (FBS) was used, and the strains were incubated in a shaker at 120 rpm and 37°C. A total of 5 μg/ml kanamycin (MP Biomedicals, CA, United States) was supplied when necessary.
Construction of Isogenic ΔdppA, ΔdppB, ΔdppC, ΔdppD, ΔdppF Mutants of H. pylori 26695 and Isogenic ΔdppA Mutant of NCTC11637
To construct a dppA knockout mutant of H. pylori 26695 (ΔdppA), a DNA fragment containing an upstream sequence of dppA was amplified with the primers DppA-up-F and DppA-up-R, a DNA fragment containing a downstream sequence of dppA was amplified with the primers DppA-down-F and DppA-down-R, and a DNA fragment containing AphA, which confers kanamycin resistance, was amplified with primers DppA-Kana-F and DppA-Kana-R. The dppA upstream sequence, dppA downstream sequence, and kanamycin resistance DNA fragments were ligated into a pBluescript II SK (–) vector (Novagen, Madison, WI, United States) using the ClonExpress MultiS One Step Cloning Kit (Vazyme, Nanjing, China), resulting in pBluscript-DppAKO, which was further transformed into E. coli DH5α. The plasmid sequence was then confirmed using colony PCR and Sanger sequencing. The pBluescript-DppAKO was then purified and subsequently transformed to H. pylori 26,695 by electroporation, and bacteria were then cultivated on agar plates containing kanamycin. dppA knockout mutants were further confirmed by colony PCR and Sanger sequencing. The construction of isogenic H. pylori 26695 mutants of ΔdppB, ΔdppC, ΔdppD, ΔdppF, and NCTC11637ΔdppA was conducted in a similar manner, and the primers used are listed in Table 1.
Cell Lines, Cultivation, and Co-culture of AGS Cells and H. pylori Strains
The human gastric epithelial AGS cell line (derived from a human gastric adenocarcinoma) was cultured in a DMEM/F12 medium (HyClone Laboratories Inc., Logan, UT, United States), with supplementation of 10% FBS (PANS, Aidenbach, Bayern, Germany) at 37°C in a 5% CO2 humidified atmosphere. For H. pylori infection assays, AGS cells were grown in 6-well plates (NUNC, Thermo, DE, United States) until the confluence reached 75% in DMEM/F12 medium containing 10% FBS. Before infection, the supernatant was removed, and cells were washed twice with phosphate-buffered saline (PBS), followed by culture in FBS-free DMEM/F12 for 4 h. H. pylori strains were first cultivated on agar plates; then, the bacteria were collected and resuspended in Brucella broth at an initial OD600 of 0.1, followed by culture for 24 h. Bacterial cells were then pelleted and washed twice with DMEM/F12 medium, resuspended in DMEM/F12 medium, and added to the AGS cell culture at a multiplicity of infection (MOI) of 100.
Determination of Bacterial Growth Rates
To monitor the growth of H. pylori strains, bacteria were first cultivated on Columbia agar plates for 3 days, followed by collection of bacterial cells and resuspension in Brucella broth at an initial OD600 = 0.1. Next, the bacteria were cultured at 37°C with agitation. The OD600 values of the bacterial culture were recorded every 8 h. Each experiment was repeated at least three times.
RNA Sequencing and Data Analysis
To prepare total RNA for transcriptomic study, H. pylori 26,695 and ΔdppA cells were cultivated in Brucella broth containing 10% FBS for 20 h until reaching the exponential phase in a shaker at 120 rpm in a microaerobic environment (5% O2, 10% CO2, and 85% N2) at 37°C. Total RNA was isolated using the RNeasy Mini Kit (QIAGEN, Valencia, CA, United States). RNA degradation and contamination were monitored on 1% agarose gels. RNA sequencing was carried out at Novogene (Beijing, China), and RNA purity was confirmed using a NanoPhotometer spectrophotometer (IMPLEN, CA, United States). RNA concentration was measured using the Qubit RNA Assay Kit with a Qubit 2.0 Flurometer (Life Technologies, CA, United States). RNA integrity was assessed using the RNA Nano 6000 Assay Kit for the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, United States). Ribosomal RNA (rRNA) was then depleted using the Ribo-zero kit (Ambion, Thermo, DE, United States) in accordance with the manufacturer’s instructions. Sequencing libraries were generated using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, United States) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina, San Diego, CA, United States). After cluster generation, the library preparations were sequenced on an Illumina Hiseq platform, and paired-end reads were generated. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 found by DESeq were designated as differentially expressed. The data were deposited in the NCBI Gene Expression Omnibus database (GEO1) under accession number GSE164216.
RNA Isolation and Quantitative RT-PCR
To prepare bacterial RNA samples, bacteria were grown in Brucella broth containing 10% FBS for 20 h, which was followed by extraction of total RNA using a RNeasy Mini Kit (QIAGEN, Valencia, CA, United States) in line with the manufacturer’s instructions. In order to extract RNA from AGS cells or AGS cells infected with H. pylori, the cells were collected after co-culture with bacteria at 37°C in a 5% CO2 humidified atmosphere after infection with H. pylori using TRIzol reagent (Life Technologies, Carlsbad, CA, United States) according to the manufacturer’s instructions. RNA concentration and purity were then determined by spectrophotometry (NanoDrop One, Thermo, DE, United States). For quantitative RT-PCR (qPCR) analysis, cDNA was prepared through reverse transcription using 1 μg of total RNA and the HiScript II Q RT SuperMix for qPCR (+gDNA wiper) kit (Vazyme, Nanjing, China). qPCR assays were carried out using the SYBR qPCR Master Mix kit (Vazyme, Nanjing, China). Specific primers for each gene indicated were designed with Primer 5.0 and are listed in Table 1. Genes encoding 16S rRNA were used as endogenous controls, and relative RNA levels were calculated using the 2–ΔΔCt method. Experiments were performed in triplicate for each condition.
Bacterial Pulldown Assays
Helicobacter pylori was grown in Brucella broth containing 10% FBS for 20 h, after which the bacterial cells were washed twice and resuspended in PBS. Approximately 3 × 107 cells were incubated with α5β1 integrin (250 μg/mL) (R&D Systems, Minneapolis, MN, United States) for 30 min at 37°C with rotation. In order to measure the amount of α5β1 integrin bound by H. pylori, the samples were centrifuged at 6,000 rpm for 10 min. Bacterial cells were collected, washed twice with PBS, and then resuspended in 1 × SDS loading buffer (60 mM Tris-HCl [pH 6.8], 2% SDS 10 ml, 10% glycerol, 100 mM DTT, 0.01% bromophenol blue). After denaturation by boiling for 10 min, the samples were resolved on a 10% SDS-polyacrylamide gel (SDS-PAGE). Western blot with an anti-β1 Rabbit antibody (1:1000; Cell Signaling Technology, Danvers, MA, United States) was performed as described previously.
Bacteria Protein Extraction and Western Blotting
To determine the expression of CagA, H. pylori cells were harvested after 20 h culture in Brucella broth containing 10% FBS. The cells were then washed twice with PBS, and total lysates were obtained using RIPA lysis buffer (Beyotime, Shanghai, China). The concentration of proteins was determined using a BCA Protein Assay Kit (Beyotime, Shanghai, China). Protein samples were then subjected to electrophoresis using a 10% SDS-PAGE gel and subsequently transferred to PVDF membranes (Millipore, Darmstadt, Germany) for antibody blotting. Membranes were blocked with Tris buffered saline containing 0.1% Tween-20 (TBST) containing 5% BSA. The membranes were then probed with an anti-CagA (b-10) antibody (1:800; Santa Cruz Biotechnology, Dallas, TX, United States), followed by an m-IgGκBP-HRP secondary antibody (1:1500; Santa Cruz Biotechnology, Dallas, TX, United States).
Adhesion Tests
AGS cells were seeded in 6-well plates at a density of 3.5 × 105 cells/well with 2 ml of DMEM/F12 to form a confluent monolayer, and then infected with H. pylori at an MOI of 100 as described above. After 4 h of infection, the AGS cells were washed three times with PBS to remove any unattached bacteria. To determine the number of adherent H. pylori, the AGS cells were lysed using 0.1% saponin for 20 min at room temperature. After a serial dilution, 50 μl of each diluted cell lysate containing bacteria was placed on a Columbia sheep blood plate. Subsequently, the bacteria were incubated under microaerobic condition (5% O2, 15% CO2, and 75% N2) for 4 days, and colonies were counted.
Dual-Luciferase Reporter Assay
NF-κB activation was determined using luciferase reporter assays. AGS cells were seeded in 12-well plates at a density of 5 × 105 cells/well in 1 ml DMEM/F12 with FBS and cultured overnight. Subsequently, 1 μg of pNL3.2.NF-κB-RE (Promega, Madison, WI, United States) and 0.1 μg of pRL-TK (Promega, Madison, WI, United States) were co-transfected into these AGS cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instruction. After 48 h of culture, the cells were infected with H. pylori strains at an MOI = 100. After infection for 4 h, the AGS cells were harvested, and luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, United States) in accordance with the manufacturer’s instruction. Each result represents the mean of three independent experiments.
IL-8 Secretion Assays
The AGS cells were seeded in 6-well culture plates at a density of 3.5 × 105 per well in 2 ml DMEM/F12 medium with FBS to form a confluent monolayer. After 20 h of culture, the supernatant was replaced with fresh DMEM/F12 without serum after washing with 1 × PBS to starve cells for 4 h. Next, the cells were infected with H. pylori 26,695 at an MOI of 100 or infected with NCTC11637 at an MOI of 30. After 4 h of infection, the supernatant was harvested, and IL-8 concentration was measured using enzyme linked immunosorbent assay (ELISA) with a Human IL-8 ELISA kit (BD Biosciences, San Jose, CA, United States) in line with the manufacturer’s instructions.
Statistical Analysis
All data were presented as the mean ± the standard error of mean. An unpaired t-test was used for comparisons between the two groups. Graph-Pad Prism 7.0 (La Jolla, CA, United States) was used to plot the data, and P < 0.05 was considered statistically significant.
Results
Dpp Transporters Are Important for the Growth of H. pylori
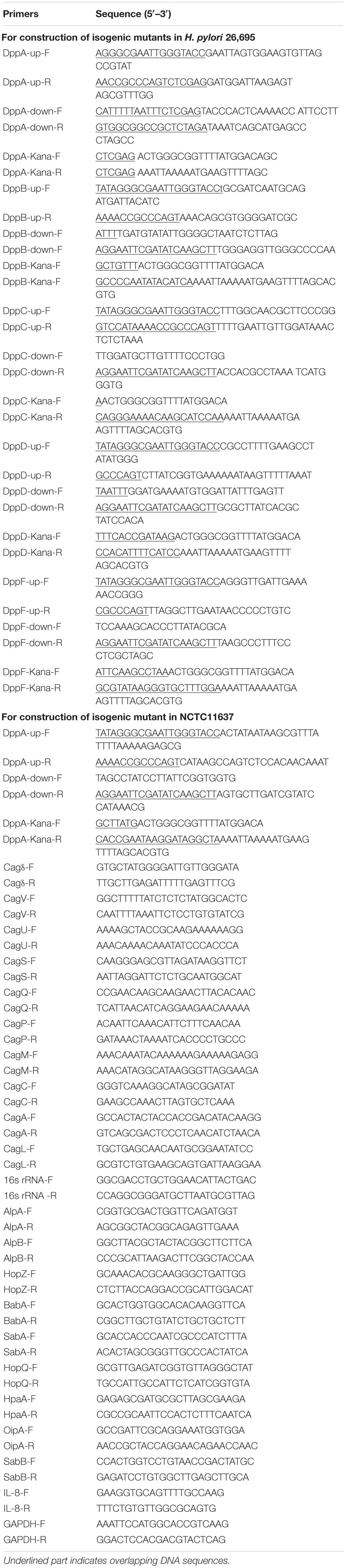
Five genes were studied, dppA, B, C, D, and F, which encode a dipeptide (Dpp) transporter comprising two permeases, two ATPases, and a substrate-binding protein (Davis and Mobley, 2005). To study the role of Dpp transporters in the virulence of H. pylori, we first constructed isogenic mutants of dppA, dppB, dppC, dppD, and dppF in H. pylori 26,695, and then assessed the impact of the Dpp transporter on bacterial growth. Growth curves showed that H. pylori 26,695 ΔdppA, ΔdppB, ΔdppC, ΔdppD, and ΔdppF strains proliferated slower than a wild-type strain (Figures 1A–E). Specifically, DppF had the strongest effect on the growth of H. pylori, suggesting its important role in the bacterial growth. We also constructed a ΔdppA mutant in the H. pylori strain NCTC11637 and conducted the same experiment. The ΔdppA mutant grew slower compared with the NCTC11637 wild type (Figure 1F). The results indicated that the importance of the Dpp system for the growth of H. pylori.
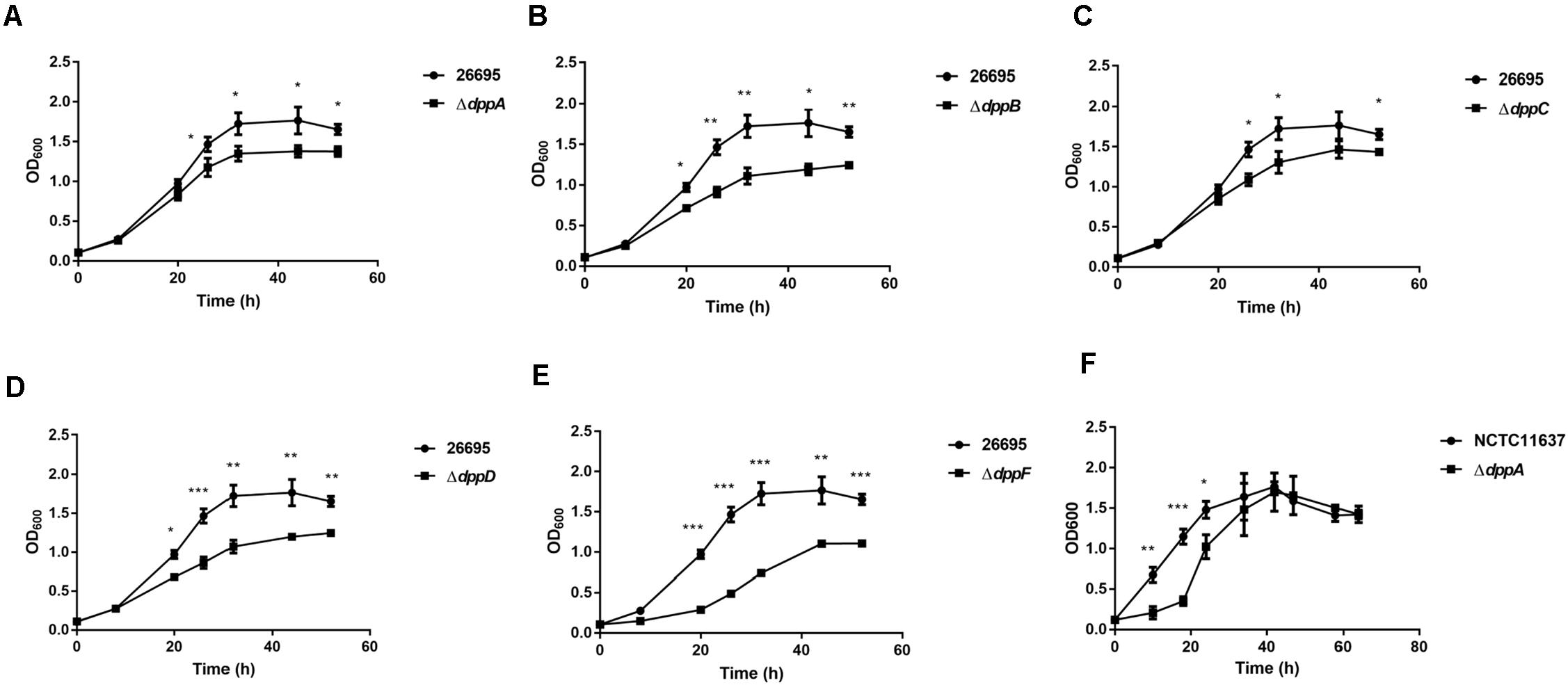
Figure 1. Growth of H. pylori wild-type strains and Dpp transporter mutant strains. Growth curves of H. pylori 26695 compared to its isogenic mutants ΔdppA (A), ΔdppB (B), ΔdppC (C), ΔdppD (D), and ΔdppF (E). (F) Growth curve of H. pylori NCTC11637 compared with its isogenic mutant ΔdppA. Data shown represent average means from three independent experiments, and standard deviations are also indicated. ***P < 0.001, **P < 0.01, *P < 0.05.
Transcriptomic Profiling of Gene Expression in H. pylori Wild-Type and ΔdppA Strains
To further examine the roles of DppA, we performed RNA-seq analysis and investigated the genes expressed differentially between H. pylori 26695 and a ΔdppA strain. We found that 253 genes were differentially expressed with a | Log2 (fold change)| > 1, including 116 genes that were upregulated and 137 genes that were downregulated in ΔdppA (P < 0.05) (Figure 2A). These genes are listed in Table 2. We also performed functional classification of the genes upregulated and downregulated in ΔdppA (Figure 2B). We found that genes involved in energy metabolism, cellular processes, transportation, and translation were significantly downregulated in ΔdppA, which might contribute to the decreased growth rate of H. pylori in this genetic background. Genes involved in DNA metabolism, bacterial pathogenesis, and motility were upregulated in this strain, and they might contribute to the virulence of H. pylori.
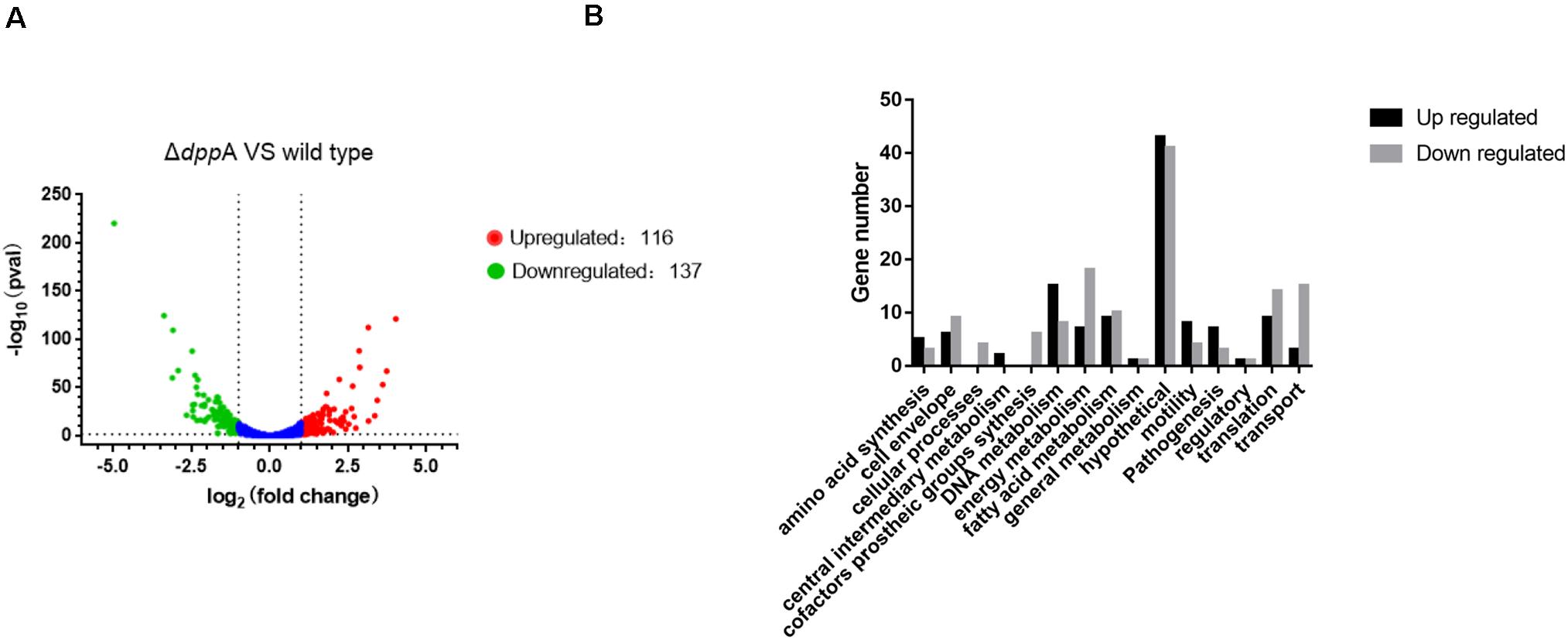
Figure 2. Differentially expressed genes between H. pylori 26695 and ΔdppA by RNA sequencing. (A) Volcano plot of gene expression in H. pylori 26695 and ΔdppA. The Y-axis represents -log10 (P-value), and X-axis represents log2 (fold change). Positive values represent genes upregulated in ΔdppA, while negative values represent genes downregulated in ΔdppA. The horizontal dashed line represents P = 0.05. Red dots represent those genes with expression in ΔdppA higher than wild type, with Log2 (fold change) > 1 and P < 0.05. Green dots represent genes with lower expression in ΔdppA compared with wild-type, with Log2 (fold change) < –1 and P < 0.05. (B) Functional annotation of genes differentially expressed in H. pylori 26,695 and ΔdppA. Black bars represent genes expressed higher in ΔdppA compared with wild type, while gray bars represent genes expressed lower in ΔdppA compared to wild type.
The Dpp Transporter Activates the Expression of CagA
The transcriptomic study revealed differential expression of H. pylori virulence genes, especially those involved with cagPAI, between the wild-type and ΔdppA strain. In this study, we focused on those virulence factors that are closely related to the cellular inflammatory response. We first investigated the expression of CagA at both the mRNA and protein levels. The qPCR results showed that CagA mRNA levels in ΔdppA, ΔdppB, ΔdppC, ΔdppD, and ΔdppF strains were lower than in the H. pylori 26,695 wild-type strain (Figure 3A). CagA protein expression was also investigated, and similar results were obtained, i.e., CagA expression was repressed in ΔdppA, ΔdppB, ΔdppC, ΔdppD, and ΔdppF strains (Figures 3B,C). This was also shown in the NCTC11637 background, as CagA expression was significantly lower in ΔdppA compared with the wild type (Figures 3D,E). This result demonstrated that the Dpp transporter is important for the expression of CagA, and that DppA, DppB, DppC, DppD, and DppF are all critical for the function of the Dpp transporter to regulate the expression of CagA.
Figure 3. Effects of DppA on the expression of CagA. (A) mRNA level of CagA expressed in H. pylori 26,695 and Dpp transporter mutant strains. Values represent the relative mRNA level of CagA normalized to H. pylori 26,695. (B–D) CagA expression level determined by Western blot. Total protein represents the cell lysate resolved by SDS-PAGE. Protein bands representing CagA are indicated, and the position of a 130 kDa size marker is indicated by an arrow. (C,E) Quantification analysis of CagA bands. Densitometry was normalized to total protein. Values are shown as averages ± SD (n = 3). ***P < 0.001, **P < 0.01, *P < 0.05.
The Dpp Transporter Causes the Inhibition of the Expression of Genes Encoding T4SS
Our RNA sequencing data suggested that components of T4SS, including Cag3, Cag5, Cagα, CagZ, Cag7, and Cag22, were upregulated in ΔdppA mutants (Table 2). Thus, we analyzed all 26 genes related to T4SS; surprisingly, most of these genes were expressed relatively highly in ΔdppA mutants (Figure 4A). Next, we performed qPCR to confirm this result. T4SS genes comprise nine operons, according to a previous study (Kabamba et al., 2018). To evaluate the effects of Dpp on the expression of T4SS genes, we compared the mRNA levels of Cagζ, CagV, CagU, CagS, CagQ, CagP, CagL, CagY, CagM, CagE, and CagC from each operon. Except Cagζ and CagC, whose expression showed no difference between wild type and ΔdppA, the expression of CagV, CagU, CagS, CagQ, CagP, CagM, and CagL was significantly higher in ΔdppA compared with H. pylori 26695 (Figure 4B). This was consistent with our transcriptomic data. We also tested the ΔdppC background, and found that all 10 genes from each operon were expressed more in ΔdppC than in wild-type strain. During infection, CagA or LPS metabolites were delivered through the T4SS to gastric epithelial cells, and this was dependent on the direct interaction between T4SS proteins and α5β1 integrins (Kwok et al., 2007). To investigate if the Dpp transporter also influenced the binding of T4SS to α5β1 integrin, a bacterial pulldown assay using purified α5β1 integrin was performed, and we measured the amount of α5β1 integrin bound by H. pylori. The results showed that the ΔdppA strain bound significantly more α5β1 integrin compared with H. pylori 26,695 wild-type strain (Figures 4D,E). These results suggested that deficiency in Dpp transporter resulted in a higher expression of T4SS genes and an increase in T4SS binding to α5β1 integrin.
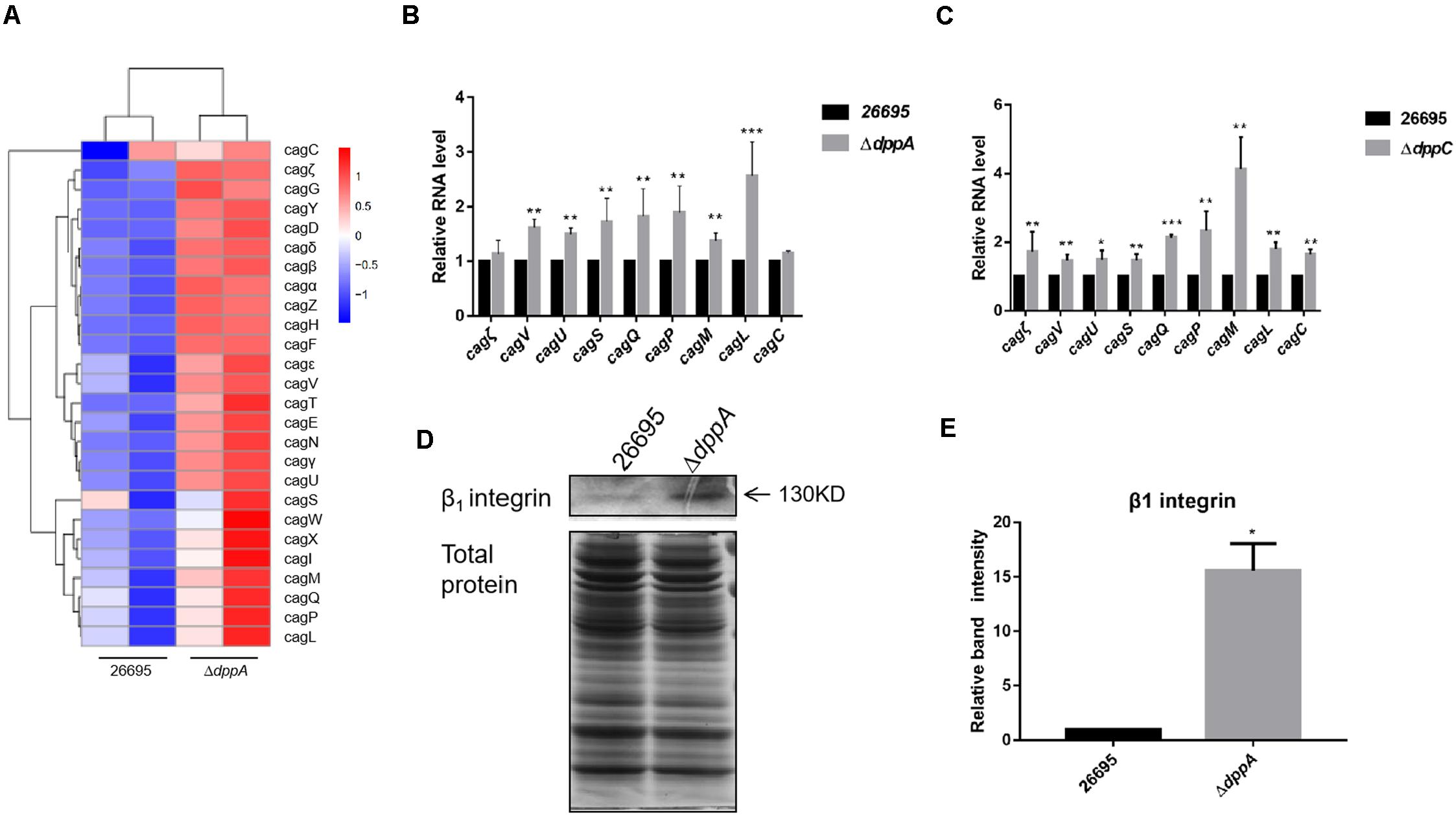
Figure 4. Effects of DppA on the expression of CagT4SS. (A) Hierarchical cluster analysis of T4SS gene expression in 26,695 and ΔdppA strains. (B) Determination of mRNA levels of T4SS components in H. pylori 26,695, ΔdppA, and ΔdppC (C). Values represent the relative mRNA level of each gene normalized to H. pylori 26,695. (D) α5β1 integrin bound by H. pylori 26,695 and ΔdppA. Bands representing β1 integrin are indicated, and total bacterial protein load is shown. (E) Quantification analysis of β1 integrin bands. Densitometry was normalized to total protein. ***P < 0.001, **P < 0.01, *P < 0.05.
The Dpp Transporter Causes Lower Expression of Outer Membrane Proteins and Reduces the Adhesion of H. pylori to AGS Cells
Our transcriptomic study revealed that several outer membrane proteins related to adhesion were differentially expressed in ΔdppA strain. This suggested that the Dpp transporter might play an important role in bacterial adhesion. To test this hypothesis, we first confirmed the expression of the OMPs involved in bacterial adhesion. Our results showed that, compared with H. pylori 26,695, the expression of adhesion genes (babA, hopZ, oipA, and sabA) was higher in a ΔdppA strain, while alpAB, hpaA, hopQ, and sabB showed similar expression levels between the wild-type and ΔdppA strains (Figure 5A). This suggested that DppA caused a lower expression of OMPs. We also investigated the expression of OMPs in ΔdppB, ΔdppC, ΔdppD, and ΔdppF strains, and showed higher expression of BabA, HopZ, OipA, and SabA than in wild-type (data not shown), which confirmed that the Dpp transporter causes a lower expression of OMPs. Next, to verify whether the Dpp transporter altered the adhesion of H. pylori to AGS cells, AGS cells were infected with H. pylori 26,695 wild-type and ΔdppA cells; subsequently, we investigated the number of bacteria bound to AGS cells. Our results showed that ΔdppA cells had a higher binding capacity compared with wild-type 26,695 cells (Figure 5B). H. pylori NCTC11637 and its isogenic mutant ΔdppA were also analyzed, and we found that in the H. pylori NCTC11637 strain, deletion of dppA also resulted in a higher bacterial adhesion level (Figure 5C). This suggested that in H. pylori, the Dpp transporter caused a reduced expression level of OMPs, including babA, hopZ, oipA, and sabA, thereby reducing the adhesion of H. pylori to AGS cells.
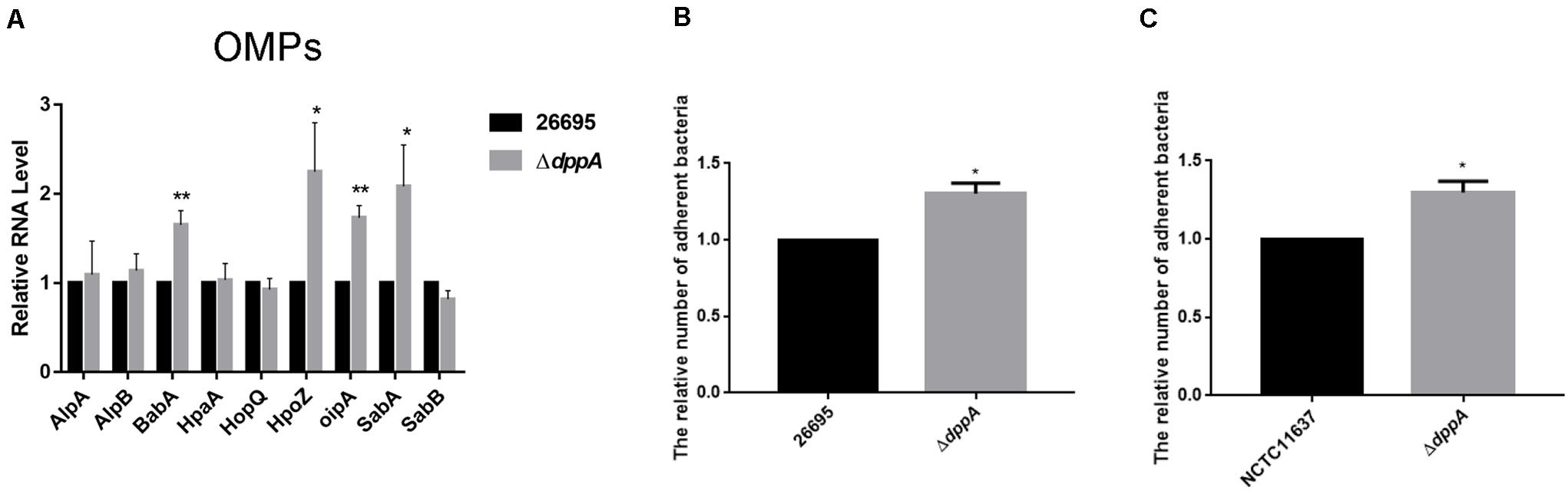
Figure 5. Effects of DppA on the adherence of H. pylori to AGS cells. (A) qPCR study of the expression of OMPs in H. pylori 26,695 and its isogenic mutant ΔdppA. Adherence of H. pylori to AGS cells. Relative adherence represents the number of ΔdppA cells adherent to AGS cells normalized to H. pylori 26,695 (B) and NCTC11637 (C). Data shown are the average values from three independent experiments, and bars represent standard deviations. **P < 0.01, *P < 0.05.
The Dpp Transporter Inhibits H. pylori Activation of Gastric Epithelial NF-κB
Upon adhesion to AGS cells, H. pylori directly activates NF-κB through the T4SS, which delivers the effector protein CagA, peptidoglycan, or ADP-heptose to cells. We investigated the effect of the Dpp transporter on H. pylori-induced NF-κB activation in AGS cells. We performed a dual-luciferase reporter assay using an NF-κB-luc reporter plasmid. After 4 h of infection with H. pylori, NF-κB was activated in wild-type infected cells. We also found that ΔdppA, ΔdppB, ΔdppC, and ΔdppD infection activated NF-κB to a level 50% higher than infection with a wild-type strain (Figure 6A). However, infection with the ΔdppF strain failed to activate NF-κB, likely due to low activity of ΔdppF for its low growth ability as shown in Figure 1E. We also checked H. pylori NCTC11637 and its isogenic mutant ΔdppA, and found that NF-κB-luc was expressed at a level 70% higher than in wild type (Figure 6B). This suggested that the Dpp transporter in H. pylori reduced the ability to activate NF-κB in AGS cells. Activation of NF-κB directly induces the expression of the inflammatory factors, including IL-8 (Brandt et al., 2005). Thus, we next utilized by ELISA and qPCR to examine IL-8 expression in AGS cells infected by H. pylori. Our results showed that IL-8 expression in AGS cells induced by H. pylori ΔdppA, ΔdppB, ΔdppC, and ΔdppD was significantly higher than that induced by wild-type H. pylori 26,695 (Figures 6C,D). As shown in Figure 6A, ΔdppF failed to activate the expression of IL-8 in AGS cells. In H. pylori 11,637, ΔdppA also induced a higher level of IL-8 expression compared with wild-type H. pylori NCTC11637 (Figures 6E,F). This indicated that the Dpp transporter repressed NF-κB and IL-8, thereby reducing the inflammatory response of AGS cells induced by H. pylori.
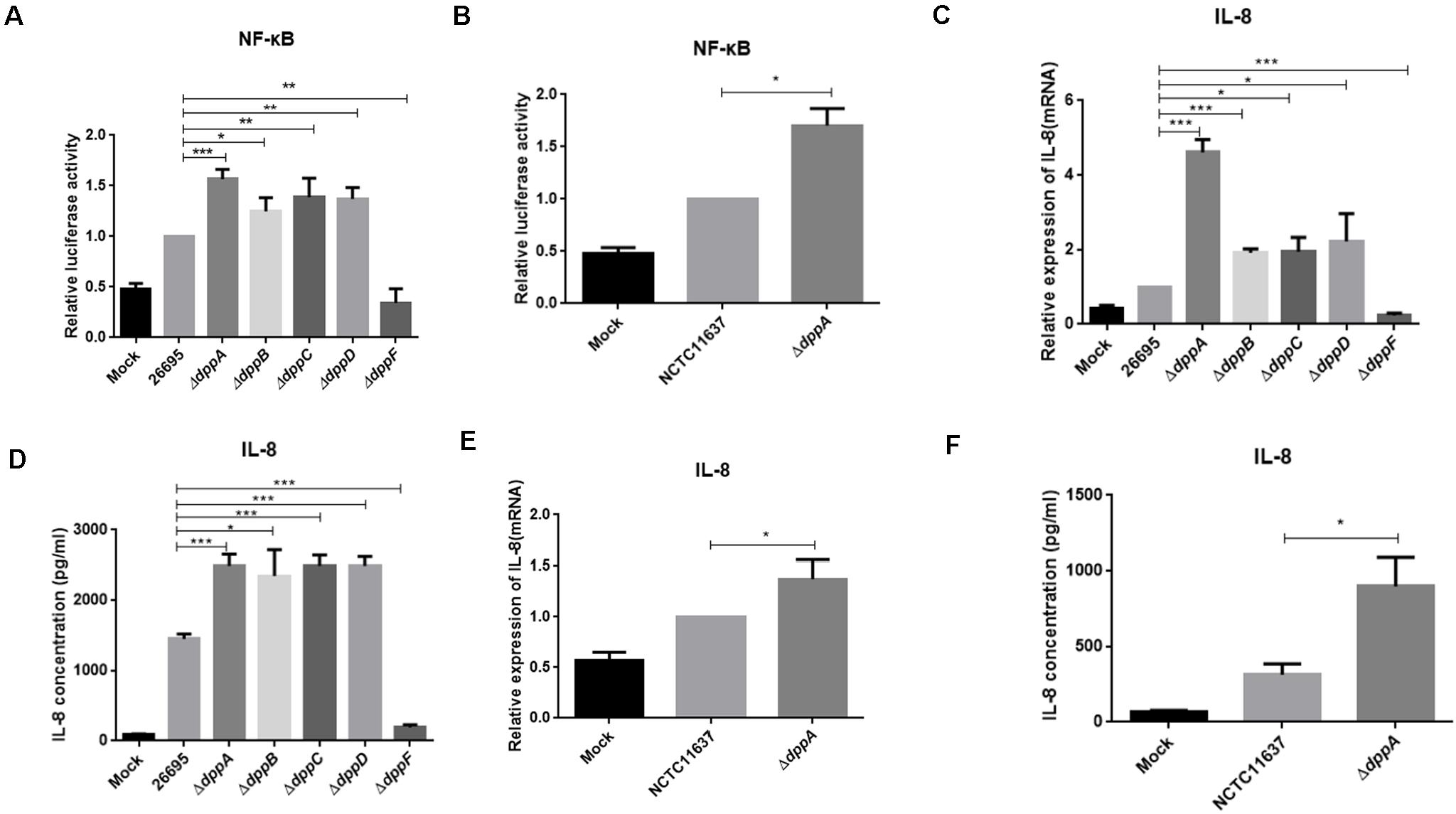
Figure 6. NF-κB activation and IL-8 production induced by H. pylori and Dpp transporter mutant strains. (A) NF-κB activity determined by luciferase activity of NF-κB-luc. AGS cells were transfected with a NF-κB luciferase reporter and were infected with H. pylori 26695, ΔdppA, ΔdppB, ΔdppC, ΔdppD, or ΔdppF, as well as being infected with H. pylori NCTC11637 and its isogenic mutant ΔdppA (B). Relative luciferase activity represents luciferase activity normalized to mock samples. (C,E) Expression of IL-8 in AGS cells as determined by qPCR or by ELISA (D,F). AGS cells were infected with H. pylori 26,695 and its isogenic Dpp transporter mutant strains, or H. pylori NCTC11637 and ΔdppA for 4 h with an MOI of 100. Data represent averages normalized to mock controls and are shown as the mean ± SD (n = 3). ***P < 0.001, **P < 0.01, *P < 0.05.
Taken together, this work was the first study to show the role of the Dpp transporter in the regulation of virulence of H. pylori. Our study demonstrated that the Dpp transporter is important for the growth of H. pylori, suggesting that dipeptides might serve as an important nutrient source for this bacterium. Although Dpp transporter-deficient strains proliferated slower, they were associated with higher bacterial adhesion and T4SS expression and induced a stronger inflammatory response in AGS cells. The Dpp transporter also activated the expression of CagA, illustrating the complex role of Dpp transporters in subtle control of bacterial virulence.
Discussion
The host tissue is a rich source of nutrients for bacteria, providing nutrients such as sugars and amino acids. To acquire the nutrients from host, pathogens produce specific virulence factors and causes host damage. It is important to understand the interaction between metabolism and bacterium pathogenesis since bacterial growth is the main goal for the pathogen to colonize in the host (Rohmer et al., 2011). Peptide transporters are important to acquire carbon from host sources for pathogen’s growth. Moreover, these transporters are also responsible for importing environmental cues to coordinate bacterial behavior (Garai et al., 2017). Human pathogens always face various environmental stresses, such as temperature variation, pH, nutrient changes, and oxidative stress (Shao et al., 2005). Understanding these transporters and their cognate substrates may help in unraveling the mechanisms of bacterial adaptation through changes in bacterial behavior, including virulence.
In this study, we found that the Dpp transporter was important for the bacterial growth (Figure 1). However, H. pylori ΔdppD and ΔdppF grew significantly slower compared to wild type, ΔdppA, ΔdppB, or ΔdppC cells. DppD and DppF are both dipeptide ABC transporter ATP binding subunits, which suggests that ΔdppD and ΔdppF might completely abolish the function of Dpp transporters, resulting in a shortage of nutrients. Specifically, we noticed that the ΔdppF strain grew much slower compared with other strains. DppF acts as an ABC transporter ATP binding subunit, which might be critical for the growth of H. pylori, causing ΔdppF with its decreased capacity to induce an inflammatory response in AGS cells (Figures 6A,C,D).
In this study, we found that the CagA expression was largely dependent on the expression of the Dpp transporter (Figure 3). During the infection of gastric epithelial cells, H. pylori translocates CagA using the T4SS. Through interactions with SH2 domains, CagA activates their function to promote the Ras-Erk signaling pathway to activate oncogenesis of gastric epithelial cells (Tohidpour, 2016; Hatakeyama, 2017; Naumann et al., 2017). The CagA protein is strongly associated with development of gastric cancer, and regulation of CagA expression is closely related to gastric cancer development (Hatakeyama, 2017). Studies have shown that CagA expression varies depending on the growth stage and conditions (Karita et al., 1996). It has also been shown that high-salt concentrations induce the expression of CagA, which is related to gastric cancer development. Iron and pH also regulate CagA expression (Odenbreit et al., 1999). Our study provided new evidence on the regulation of CagA expression in H. pylori, suggesting that the nutrient status of the environment affects CagA expression and H. pylori-related gastric cancer. Specifically, when grown in environments with abundant nutrients, H. pylori might express high levels of CagA.
In the early stages of infection, H. pylori activates NF-κB in a CagT4SS-dependent manner. The regulation of T4SS expression might contribute to H. pylori-induced inflammatory response in AGS cells. In this study, we found upregulated expression of T4SS components in the ΔdppA strain (Figures 4A,B), which was also replicated in a ΔdppC strain (Figure 4C). Among these genes, RNA sequencing data and qPCR results showed that genes were upregulated in ΔdppA strain to a different degree, which suggested that although these genes are all responsible for the T4SS apparatus, they might be regulated by different mechanisms. A previous study investigated the expression of T4SS and found that these genes responded differently to growth phase, temperature, pH, iron, and cell contact (Yamaoka et al., 2000). Some environmental signals even exert pleiotropic effects on these genes. This suggests that T4SS expression and assembly are controlled by sophisticated mechanisms, but more studies are necessary. The T4SS machinery translocates CagA, ADP-heptose, peptidoglycan, and other substrates to host cells (Mobley et al., 1991; Peck et al., 1999; Pfannkuch et al., 2019). Recent studies have shown that ADP-heptose is a novel pathogen-associated molecular marker in H. pylori, and it is the main factor activating NF-κB in a T4SS-dependent manner (Pfannkuch et al., 2019). We speculate that T4SS expression is repressed under rich nutrient conditions, and H. pylori reduces the translocation of ADP-heptose or other effector molecules. Under poor nutrient condition, T4SS expression is activated and results in a high level of activation of the NF-κB response.
After H. pylori passes through the mucous layer and reaches the gastric mucosa via flagellar movements, OMPs promote close contact between H. pylori and gastric epithelial cells. OMPs play important roles in the establishment of colonization (Yuichi et al., 2017; Terbenc et al., 2019). The OMPs in H. pylori have been gradually unveiled, and their cognate interaction partners have been identified. In this study, we found that expression of BabA, HopZ, OipA, and SabA, and bacterial adhesion were upregulated in the ΔdppA strain (Figure 6). BabA was the first OMP identified to be involved in the adhesion of H. pylori and important for inducing severe inflammation in the stomach. Moreover, studies have shown that T4SS function and CagA translocation are enhanced by BabA (Boren et al., 1993; Ilver et al., 1998; Aspholm-Hurtig et al., 2004). Some studies have shown that the adhesion ability is significantly decreased in oipA mutant strain and hopZ mutant strain in AGS cells (Peck et al., 1999; Dossumbekova et al., 2006). SabA is also important for colonization and induction of inflammation in the stomach (Aspholm et al., 2006). SabA expression is regulated by a pH-responsive ArsRS two-component signal transduction system (Goodwin et al., 2008). The HopZ gene is involved in the adhesion of H. pylori to gastric epithelial AGS cell line in vitro, but it did not show any influence on the ability of colonization in the stomachs of guinea pigs (Peck et al., 1999). However, the cognate receptor of HopZ remains unknown. Our study suggested that the Dpp transporter in H. pylori plays an important role in the colonization of the stomach.
Besides the virulence factors investigated in this study, our transcriptomic study by RNA sequencing also indicated that the expression of other virulence genes is also altered in the ΔdppA background. Flagellar coding genes, including flaA, flaB, fliD, flaG, and flgK, were significantly upregulated in ΔdppA cells (Table 2). Flagellar movement is critical for the initial colonization of H. pylori by penetrating gastric mucus layer (Gu, 2017). Indeed, flagellar movement of H. pylori is an important factor in mediating high density colonization and severe inflammation. Studies have shown that FlaA and FlaB are necessary for H. pylori colonization of animals (Josenhans et al., 1995). ADP-heptose is a lipopolysaccharide synthesis intermediate, which is responsible for H. pylori-induced NF-κB activation (Pfannkuch et al., 2019). RNAseq data showed that upregulated expression of majority of LPS-related metabolic genes and the ADP-heptose synthesis gene gmhB in the ΔdppA strain (Table 2). GmhB (Hp0860) is an important synthase gene for the synthesis of ADP-heptose by dephosphorylation of D-glycero-β-d-manno-heptose-1,7-bisphosphate (HBP) (Stein et al., 2017). This suggests that LPS synthesis and ADP-heptose production might be upregulated in ΔdppA, thereby enhancing H. pylori-induced IL-8 production and NF-κB activation.
In conclusion, we have demonstrated that the Dpp transporter affects the expression of virulence factors such as CagA, T4SS, and OMPs. The Dpp transporter might enable the bacteria to recognize environmental nutrient conditions and change virulence factors such as adhesion and stimulate the release of other virulence factors. Since H. pylori causes a chronic infection and is closely related to gastric cancer, our study suggests that when nutrients are limited, and the Dpp transporter fails to transport dipeptides, H. pylori enhances its ability to colonize and stimulates an inflammatory response to acquire nutrients from the host. Thus, H. pylori tends to repress its ability to stimulate an inflammatory response in gastric epithelial cells while delivering the oncoprotein CagA, which induces gastric cancer.
Data Availability Statement
The original contributions generated for this study are publicly available. This data can be found here: NCBI Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession number GSE16421 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164216).
Author Contributions
YW and FS designed the study. XX, JC, and SF performed the experiments. XH analyzed the data. YW, XX, and FS wrote the manuscript. All authors have read and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81701980 and 82072316), the Natural Science Foundation of Fujian Province, China (Grant No. 2019J01295), the Key Projects of Youth Natural Science Foundation of Fujian Colleges and Universities (Grant No. JZ160440), and the Fujian Medical University Talent Startup Fund (XRCZX2017008, XRCZX2017027, and 2017XQ1008).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Footnotes
References
Aspholm, M., Olfat, F., Nordén, J., Sondén, B., Lundberg, C., Sjöström, R., et al. (2006). SabA is the Helicobacter pylori hemagglutinin and is polymorphic in binding to sialylated glycans. PLoS Pathog. 2:e110. doi: 10.1371/journal.ppat.0020110
Aspholm-Hurtig, M., Dailide, G., Lahmann, M., Kalia, A., Ilver, D., Roche, N., et al. (2004). Functional adaptation of BabA, the Helicobacter pylori ABO blood group antigen binding adhesin. Science 305, 519–522. doi: 10.1126/science.1098801
Augusto, A. C., Miguel, F., Mendonça, S., Pedrazzoli, J. Jr., and Gurgueira, S. A. (2007). Oxidative stress expression status associated to Helicobacter pylori virulence in gastric diseases. Clin. Biochem. 40, 615–622. doi: 10.1016/j.clinbiochem.2007.03.014
Backert, S., and Naumann, M. (2010). What a disorder: proinflammatory signaling pathways induced by Helicobacter pylori. Trends Microbiol. 18, 479–486. doi: 10.1016/j.tim.2010.08.003
Boren, T., Falk, P., Roth, K., Larson, G., and Normark, S. (1993). Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262, 1892–1895. doi: 10.1126/science.8018146
Brandt, S., Kwok, T., and Hartig, R., et al. (2005). NF-kB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. P. Natl. Acad. Sci. USA. 102. 9300–9305. doi: 10.1073/pnas.0409873102
Chmiela, M., and Kupcinskas, J. (2019). Review: Pathogenesis of Helicobacter pylori infection. Helicobacter 24 Suppl 1(Suppl. Suppl. 1):e12638. doi: 10.1111/hel.12638
Davis, G. S., and Mobley, H. L. (2005). Contribution of dppA to urease activity in Helicobacter pylori 26695. Helicobacter 10, 416–423. doi: 10.1111/j.1523-5378.2005.00348.x
Dossumbekova, A., Prinz, C., Mages, J., Lang, R., Kusters, J. G., Van Vliet, A. H., et al. (2006). Helicobacter pylori HopH (OipA) and bacterial pathogenicity: genetic and functional genomic analysis of hopH gene polymorphisms. J. Infect. Dis. 194, 1346–1355. doi: 10.1086/508426
Garai, P., Chandra, K., and Chakravortty, D. (2017). Bacterial peptide transporters: Messengers of nutrition to virulence. Virulence 8, 297–309. doi: 10.1080/21505594.2016.1221025
Gilvarg, C. (1972). “Peptide transport in bacteria,” in Peptide transport in bacteria and mammalian gut, Ciba Foundation Symposium, eds K. Elliott and M. O’Connor (Amsterdam: Elsevier).
Goodwin, A. C., Weinberger, D. M., Ford, C. B., Nelson, J. C., Snider, J. D., Hall, J. D., et al. (2008). Expression of the Helicobacter pylori adhesin SabA is controlled via phase variation and the ArsRS signal transduction system. Microbiol 154, 2231–2240. doi: 10.1099/mic.0.2007/016055-0
Gu, H. (2017). Role of Flagella in the Pathogenesis of Helicobacter pylori. Curr. Microbiol. 74, 863–869. doi: 10.1007/s00284-017-1256-4
Guillemin, K., Salama, N. R., Tompkins, L. S., and Falkow, S. (2002). Cag pathogenicity island-specific responses of gastric epithelial cells to Helicobacter pylori infection. Proc. Natl. A Sci. Ind. B 99, 15136–15141. doi: 10.1073/pnas.182558799
Harder, D., Stolz, J., Casagrande, F., Obrdlik, P., Weitz, D., Fotiadis, D., et al. (2008). DtpB (YhiP) and DtpA (TppB, YdgR) are prototypical proton−dependent peptide transporters of Escherichia coli. FEBS J. 275, 3290–3298. doi: 10.1111/j.1742-4658.2008.06477.x
Hatakeyama, M. (2017). Structure and function of Helicobacter pylori CagA, the first-identified bacterial protein involved in human cancer. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 93, 196–219. doi: 10.2183/pjab.93.013
Ilver, D., Arnqvist, A., and Ogren, J. (1998). Helicobacter pylori Adhesin Binding Fucosylated Histo-Blood Group Antigens Revealed by Retagging. Science 279, 373–377. doi: 10.1126/science.279.5349.373
Josenhans, C., Labigne, A., Suerbaum, S. (1995). Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 177, 3010–3020. doi: 10.1007/BF0000007
Kabamba, E. T., Tuan, V. P., and Yamaoka, Y. (2018). Genetic populations and virulence factors of Helicobacter pylori. Infect. Genet. Evol. 60, 109–116. doi: 10.1016/j.meegid.2018.02.022
Karita, M., Tummuru, M. K., Wirth, H. P., and Blaser, M. J. (1996). Effect of growth phase and acid shock on Helicobacter pylori cagA expression. Infect. Immun. 64, 4501–4507. doi: 10.1016/S1380-2933(96)00052-8
Kusters, J. G., van Vliet, A. H., and Kuipers, E. J. (2006). Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 19, 449–490. doi: 10.11209/jim.20.309
Kwok, T., Zabler, D., Urman, S., Rohde, M., Hartig, R., Wessler, S., et al. (2007). Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449:862. doi: 10.1038/nature06187
Maeda, S., Yoshida, H., Ogura, K., Mitsuno, Y., Hirata, Y., Yamaji, Y., et al. (2000). Helicobacter pylori activates NF-κB through a signaling pathway involving IκB kinases, NF-κB-inducing kinase, TRAF2, and TRAF6 in gastric cancer cells. Gastroenterology 119, 97–108. doi: 10.1053/gast.2000.8540
Manson, M. D., Blank, V., Brade, G., and Higgins, C. F. (1986). Peptide chemotaxis in Escherichia coli involves the Tap signal transducer and the dipeptide permease. Nature 321, 253–256. doi: 10.1038/321253a0
Marcus, E. A., Sachs, G., and Scott, D. R. (2018). Acid−regulated gene expression of Helicobacter pylori: Insight into acid protection and gastric colonization. Helicobacter 23:e12490. doi: 10.1111/hel.12490
Merrell, D. S., Goodrich, M. L., Otto, G., Tompkins, L. S., and Falkow, S. (2003a). pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71, 3529–3539. doi: 10.1128/iai.71.6.3529-3539.2003
Merrell, D. S., Thompson, L. J., Kim, C. C., Mitchell, H., Tompkins, L. S., Lee, A., et al. (2003b). Growth phase-dependent response of Helicobacter pylori to iron starvation. Infect. Immun. 71, 6510–6525. doi: 10.1128/iai.71.11.6510-6525.2003
Mobley, H., Hu, L.-T., and Foxall, P. (1991). Helicobacter pylori urease: properties and role in pathogenesis. Scand. J. Gastroenterol. 26, 39–46. doi: 10.3109/00365529109098223
Moraes, P. M., Seyffert, N., Silva, W. M., Castro, T. L., Silva, R. F., Lima, D. D., et al. (2014). Characterization of the Opp peptide transporter of Corynebacterium pseudotuberculosis and its role in virulence and pathogenicity. BioMed Res. Int. 2014:489782. doi: 10.1155/2014/489782
Naumann, M., Sokolova, O., Tegtmeyer, N., and Backert, S. (2017). Helicobacter pylori: A paradigm pathogen for subverting host cell signal transmission. Trends Microbiol. 25, 316–328. doi: 10.1016/j.tim.2016.12.004
Noto, J. M., Gaddy, J. A., Lee, J. Y., Piazuelo, M. B., Friedman, D. B., Colvin, D. C., et al. (2012). Iron deficiency accelerates Helicobacter pylori–induced carcinogenesis in rodents and humans. J. Clin. Invest. 123, 479–492. doi: 10.1172/JCI64373
Noto, J. M., Lee, J. Y., Gaddy, J. A., Cover, T. L., Amieva, M. R., and Peek, R. M. Jr. (2015). Regulation of Helicobacter pylori virulence within the context of iron deficiency. J. Infect. Dis. 211, 1790–1794. doi: 10.1093/infdis/jiu805
Odenbreit, S., Till, M., Hofreuter, D., Faller, G., and Haas, R. (1999). Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 31, 1537–1548. doi: 10.1046/j.1365-2958.1999.01300.x
Paulsen, I. T., and Skurray, R. A. (1994). The POT family of transport proteins. Trends Biochem. Sci. 19:404. doi: 10.1016/0968-0004(94)90087-6
Payne, J. W., and Smith, M. W. (1994). Peptide Transport by Micro-organisms. Adv. Microb. Physiol. 36, 1–80. doi: 10.1016/S0065-2911(08)60176-9
Peck, B., Ortkamp, M., Diehl, K. D., Hundt, E., and Knapp, B. (1999). Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 27, 3325–3333. doi: 10.1093/nar/27.16.3325
Pfannkuch, L., Hurwitz, R., Traulsen, J., Sigulla, J., Poeschke, M., Matzner, L., et al. (2019). ADP heptose, a novel pathogen-associated molecular pattern identified in Helicobacter pylori. FASEB J. 33, 9087–9099. doi: 10.1096/fj.201802555R
Pflock, M., Kennard, S., Finsterer, N., and Beier, D. (2006). Acid-responsive gene regulation in the human pathogen Helicobacter pylori. J. Biotechnol. 126, 52–60. doi: 10.1016/j.jbiotec.2006.03.045
Podbielski, A., and Leonard, B. A. (1998). The group A streptococcal dipeptide permease (Dpp) is involved in the uptake of essential amino acids and affects the expression of cysteine protease. Mol. Microbiol. 28, 1323–1334. doi: 10.1046/j.1365-2958.1998.00898.x
Rohmer, L., Hocquet, D., and Miller, S. I. (2011). Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19, 341–348. doi: 10.1016/j.tim.2011.04.003
Samen, U., Gottschalk, B., Eikmanns, B. J., and Reinscheid, D. J. (2004). Relevance of peptide uptake systems to the physiology and virulence of Streptococcus agalactiae. J. Bacteriol. 186, 1398–1408. doi: 10.1128/JB.186.5.1398-1408.2004
Sanchezzauco, N., Torres, J., Perezfigueroa, G. E., Alvarezarellano, L., Camorlingaponce, M., Gomez, A., et al. (2013). Impact of cagPAI and T4SS on the Inflammatory Response of Human Neutrophils to Helicobacter pylori Infection. PLoS One 8:e64623. doi: 10.1371/journal.pone.0064623
Sanz, Y., Lanfermeijer, F. C., Renault, P., Bolotin, A., Konings, W. N., and Poolman, B. (2001). Genetic and functional characterization of dpp genes encoding a dipeptide transport system in Lactococcus lactis. Arch. Microbiol. 175, 334–343. doi: 10.1007/s002030100270
Sgouras, D. N., Trang, T. T. H., and Yamaoka, Y. J. H. (2015). Pathogenesis of Helicobacter pylori infection. Helicobacter 20, 8–16. doi: 10.1111/hel.12251
Shao, S.-H., Wang, H., Chai, S.-G., and Liu, L.-M. (2005). Research progress on Helicobacter pylori outer membrane protein. World J. Gastroenterol. WJG 11:3011. doi: 10.3748/wjg.v11.i20.3011
Sharma, C., Hoffmann, S., Darfeuille, F., et al. (2010). The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255. doi: 10.1038/nature08756
Stein, S. C., Faber, E., Bats, S. H., Murillo, T., Speidel, Y., Coombs, N., et al. (2017). Helicobacter pylori modulates host cell responses by CagT4SS-dependent translocation of an intermediate metabolite of LPS inner core heptose biosynthesis. PLoS Pathog. 13:e1006514. doi: 10.1371/journal.ppat.1006514
Terbenc, A., Jarc, E., Poljak, M., and Homan, M. (2019). Helicobacter pylori virulence genes. World J. Gastroenterol. 25, 4870–4884. doi: 10.3748/wjg.v25.i33.4870
Tohidpour, A. (2016). CagA-mediated pathogenesis of Helicobacter pylori. Microb. Pathog. 93, 44–55. doi: 10.1016/j.micpath.2016.01.005
Viala, J., Chaput, C., Boneca, I. G., Cardona, A., Girardin, S. E., Moran, A. P., et al. (2004). Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 5, 1166–1174. doi: 10.1038/ni1131
Weinberg, M. V., and Maier, R. J. (2007). Peptide transport in Helicobacter pylori: roles of dpp and opp systems and evidence for additional peptide transporters. J. Bacteriol. 189, 3392–3402. doi: 10.1128/JB.01636-06
Yamaoka, Y., Kwon, D. H., and Graham, D. Y. (2000). A Mr 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. A Sci. Ind. B 97, 7533–7538. doi: 10.1073/pnas.130079797
Yuichi, M., Yasutoshi, K., and Yoshio, Y. (2017). Helicobacter pylori Outer Membrane Protein-Related Pathogenesis. Toxins 9:101. doi: 10.3390/toxins9030101
Zhang, W.-X., Xie, B.-B., Chen, X.-L., Dong, S., Zhang, X.-Y., Zhou, B.-C., et al. (2010). Cold-adaptation of the Periplasmic Dipeptide-Binding Protein (DppA) from the Deep-sea Psychrophilic Bacterium Pseudoalteromonas sp. SM9913: Domains III and I-2α at the Entrance of Binding Cleft Play an Important Role. Appl. Environ. Microb. 76, 4354–4361. doi: 10.1128/AEM.02884-09
Keywords: Helicobacter pylori, dipeptide transporter, NF-κB, T4SS, outer membrane proteins
Citation: Xu X, Chen J, Huang X, Feng S, Zhang X, She F and Wen Y (2021) The Role of a Dipeptide Transporter in the Virulence of Human Pathogen, Helicobacter pylori. Front. Microbiol. 12:633166. doi: 10.3389/fmicb.2021.633166
Received: 24 November 2020; Accepted: 05 February 2021;
Published: 25 February 2021.
Edited by:
Maurizio Sanguinetti, Catholic University of the Sacred Heart, ItalyReviewed by:
Nagendran Tharmalingam, Rhode Island Hospital, United StatesYundong Sun, Shandong University, China
Copyright © 2021 Xu, Chen, Huang, Feng, Zhang, She and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yancheng Wen, aGl0d3ljQHFxLmNvbQ==; Feifei She, c2hlZmVpZmVpQHllYWgubmV0
†These authors have contributed equally to this work
 Xiaohong Xu1,2,3†
Xiaohong Xu1,2,3† Yancheng Wen
Yancheng Wen