
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 12 March 2021
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.632908
A gene encoding a protein similar to ethylene receptor was isolated from maize (Zea mays L.), which was named as ZmERS4.The gene was 1,905 bp in length with an open reading frame that encoded a protein consisting of 634 amino acids. The homologous analysis showed that ZmERS4 shared high similarity with the ethylene receptor protein, OsERS1, from rice (Oryza sativa L.). ZmERS4 grouped into the ETR1 subfamily of ethylene receptors based on its conserved domain and phylogenetic status. Tissue-specific and induced expression analyses indicated that ZmERS4 was differentially expressed in maize tissues, predominantly in the stems and leaves, and was induced by salicylic acid (SA). Overexpression of ZmERS4 in Arabidopsis improved resistance against the bacterial pathogen, PstDC3000, by inducing the expression of SA signaling-related genes. Moreover, treatment with flg22 induced the expression of the defense-related gene, PR1, in maize protoplasts that transiently expressed ZmERS4. Furthermore, the ultra-high-performance liquid chromatography (UPLC) analysis showed that the SA contents in ZmERS4-overexpressing Arabidopsis lines were significantly higher than the control lines. Additionally, the improved resistance of ZmERS4-overexpressing Arabidopsis against PstDC3000 was blocked after pretreatment with the SA biosynthetic inhibitor, ABT. Based on the collective findings, we hypothesize that ZmERS4 plays an important role in disease resistance through SA-mediated signaling pathways.
Plants were attacked of various pathogenic microorganisms, including fungi, bacteria, viruses, and nematodes, in their natural environment. Maize (Zea mays L.) is one of the most important food crops produced in the world and is a staple food for ~50% of the global population (Ranum et al., 2014). To date, major maize diseases, including southern corn leaf blight, brown leaf spot, northern corn leaf blight, Curvularia leaf spot, and rough dwarf, have caused severe yield losses in maize production worldwide (Subash, 2015). To address this problem, improving host resistance against pathogens is the most economical and environmentally friendly approach. To breed resistant cultivars, efforts have aimed to introduce resistance (R) genes into susceptible cultivars. R genes are the key components of disease resistance against particular pathogens, which are often associated with hypersensitive responses (HR). According to “gene-to-gene” hypotheses, the R-genes of host plants interact with pathogen virulence effect factors to produce disease resistant phenotypes in the hosts (Sekhwal et al., 2015). However, owing to variations in pathogen virulence, resistant cultivars with R genes are effective for only a few years in agricultural production (Wang et al., 2013). Moreover, although several quantitative trait loci (QTLs; or quantitative genes) that confer disease resistance have been identified in maize, these sources have not been used effectively for maize improvement due to the complexity of controlling quantitative resistance. Therefore, breeding resistance cultivars with broad-spectrum and durable disease resistance is a top priority in maize improvement efforts around the world.
To date, >300 R genes have been identified and studied in Arabidopsis, tomato, rice, and other species (Kourelis and van der Hoorn, 2018). However, only a few genes have been identified as genetic resources for broad-spectrum disease resistance in maize. For example, Hml was first found in corn and enhanced plant resistance against northern leaf spot (Johal and Briggs, 1992). The maize resistance gene, ZmRxo1, which has a nucleotide-binding site-leucine-rich repeat structure, confers resistance to rice bacterial streak disease (Zhao et al., 2005). Li et al. identified an F-box protein (ZmFBL41) that confers maize resistance to banded leaf and sheath blight through a genome-wide association study, and ZmFBL41-overexpressing rice exhibited increased susceptibility to Rhizoctonia solani. Two amino acid substitutions in this allele prevent its interaction with ZmCAD, which encodes the final enzyme in the monolignol biosynthetic pathway and results in the inhibition of ZmCAD degradation and, consequently, the accumulation of lignin and restriction of lesion expansion (Li et al., 2019). Thus, genetic improvement and disease resistance cultivar breeding are current viable alternative strategies for crops suffering from biotic stress.
Ethylene is a gaseous hormone that regulates various processes during plant development including seed germination, root hair formation, leaf and petal abscission, fruit ripening, and senescence (Abeles et al., 1992). Little is known about the process of ethylene perception prior to the isolation of ethylene receptor genes. Ethylene receptors are homologous to bacterial two-component histidine kinase receptors (Bleecker, 1999) associated with the endoplasmic reticulum (ER) membrane (Chen et al., 2002, 2007; Ma et al., 2006) and have been confirmed to function as negative regulators of ethylene responses, which are inactivated by ethylene binding (Hua and Meyerowitz, 1998). Ethylene receptor genes have been identified in many plant species and classified into two subfamilies (Zhou et al., 1996; Hua and Meyerowitz, 1998). Subfamily I contain three transmembrane domains in the amino-terminal region and a well-conserved histidine kinase domain in the carboxy-terminal region. Subfamily II has four membrane-spanning domains and a degenerate histidine kinase domain that lacks one or more of the conserved amino acids, which are believed to be necessary for catalytic activities. The expression patterns of ethylene receptor genes have been examined and were found to be generally regulated according to tissue, developmental stage, and environmental stimuli (Hua and Meyerowitz, 1998; Chen et al., 2005).
In our previous work, the endophytic strain, Bacillus subtilis DZSY21, was isolated from Eucommia ulmoides and effectively colonized maize leaves and enhanced disease resistance (Ding et al., 2017). Key disease-resistance genes in the interaction system between maize and endophytic DZSY21 were identified by whole genome bisulfite and transcriptome sequencing (Zhou et al., 2019) and ZmERS4 gene was one of them. In this study, the maize gene, ZmERS4, is isolated and characterized, and its expression patterns during development and under hormone treatments are investigated. The role of ZmERS4 in conferring plant disease resistance is clarified, thus laying a foundation for cultivating resistant maize varieties.
The coding sequences (CDS) and protein sequences of ZmERS4 were downloaded from Phytozome.1 The molecular weight (Ww) and isoelectric point (pI) were calculated by ExPASy.2 The conserved motifs in the ZmERS4 protein were detected by SMART.3 Then, protein sequences between ZmERS4 and homologous genes were aligned using MEGA v7.0 (a software). A phylogenetic tree was constructed using the neighbor-joining (NJ) method (bootstrap = 1,000; Saitou and Nei, 1987). Gene transcription data of ZmERS4 at various growth times and in different tissues were downloaded from the Corn database,4 which were used to draw a heat map using R software (Paradis et al., 2004).
ZmERS4 was cloned by RT-PCR from leaf mRNA from maize leaves by PCR using specific primers (forward: ATGGACGGATGCGATTGC and reverse: TCATACGCTTCTTTGGTACCG) and Primer STAR Mix (Vazyme Biotech Co., Ltd., Nanjing, China). The PCR products were sequenced successfully and constructed into the pCAMBIA 1301 vector to obtain the p35S: ZmERS4 fusion vector. It was also inserted into the pCAMBIA1305 vector, which included a Cauliflower Mosaic Virus 35S (CaMV35S) promoter to obtain the p35S: ZmERS4-GFP fusion vector for subcellular localization. Then, the pCUB-ZmERS4-GFP fusion vector and pCUB-GFP vector were constructed, which were transiently expressed in maize protoplasts by homologous recombination for analyzing disease resistance.
The pCAMBIA1301 vector carrying 35S: ZmERS4 was transformed into Agrobacterium tumefaciens GV3101, then transgenic Arabidopsis thaliana lines were generated by Agrobacterium-mediated genetic transformation (Clough and Bent, 1998). T1 seeds were screened on Murashige and Skoog (MS) plates containing 20 μg/ml hygromycin. The DNA of transgenic A. thaliana (T1) were extracted by the CTAB method (Clarke, 2009) and used in PCR assays to determine positive transgenic plants. Finally, the homozygous T3 generation derived from transgenic A. thaliana was used for subsequent experiments.
Pseudomonas syringae pv. tomato DC3000 (PstDC3000) plants were grown at 28°C for 24 h in beef-protein liquid medium. The bacterial suspension (OD600 = 0.2) was adjusted using 10 mM MgCl2 6H2O, which included 0.02% Silwet L-77.
Maize lines (B73) were germinated in sand for 2 days at 28°C, and then grown in pots at 25–30°C under a normal photoperiod. Plants at the three-leaf stage were sprayed with 1 mM salicylic acid (SA), 50 μM jasmonic acid (JA), 1 mM ethephon (ET), and the conidial suspension of Curvularia lunata (1×105 CFU/ml), and plant leaves at the three-leaf stage were injected with Pantoea stewartii suspensions (1×106 CFU/ml). After 0, 3, 6, 12, 24, and 48 h, leaves were harvested and stored at −80°C for RNA extraction. Fifty-four plants were present in each treatment, nine maize plants were harvested and divided into three replicates at different period, and each sample was mixtures of the leaves of the three maize plants. For the tissue-specific expression analysis, maize roots, stems, leaves, leaves, corn silk, and endosperm were obtained and stored at −80°C for RNA extraction. Nine maize plants were harvested and divided into three replicates at different period, and each sample was mixtures of the leaves of the three maize plants.
Wild-type (Col-0) and transgenic A. thaliana were sterilized and placed in MS plates at 4°C for 3 days, then transferred to a culture room at 22°C for 10 days. Subsequently, 14-day-old Arabidopsis seedlings were transferred to nutritive soil under a 16/8 h light/dark cycle. Then, 4-week-old A. thaliana were sprayed with the PstDC3000 suspension; plants treated with 10 mM MgCl2 6H2O were used as negative controls. Thirty plants were present in one treatment. Disease severity was recorded 7-day post-infection based on a rating scale of 1–5 (Zha et al., 2019). Among them, 1/2/3/4/5 indicates that the area of sick leaves was 0–20, 21–40, 41–60, 61–80, and more than 80%. The disease index was calculated according to a previously described formula (Ding et al., 2017). Thirty plants in each treatment were divided into three replicates, 10 plants for each replicate. Then plant leaves were harvested and stored at −80°C for RNA extraction at 24 h post-infection (h).
Meanwhile, the bacterium number inside mesophyll tissues was harvested at 2, 4, and 6 days after inoculation and were estimated using the dilution and spread plate method. Nine maize plants were harvested and divided into three replicates at different period, and each sample was mixtures of the leaves of the three maize plants.
To further confirm that ZmERS4 enhanced plant resistance through the SA signaling pathway, 4-week-old wild and transgenic A. thaliana were sprayed with 100 μmol/L 1-aminobenzotriazole (ABT). After 24 h, plants were sprayed with the PstDC3000 suspension; plants pretreated with sterilized distilled water were used as negative controls.
RNA was extracted from maize and Arabidopsis tissues by the TRIzol method (Thermo Fisher Scientific, United States). RNA quality was evaluated based on the concentrations and electrophoresis strips. In the qRT-PCR assays, each reaction (20 μl total volume) consisted of 2 μl diluted cDNA, 10 μl AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Ltd., Nanjing, China), 1 μl forward and reverse primers each, and 6 μl RNA-free water. Three technical replicates were performed per sample. The qRT-PCR cycling program for validation was as follows: 95°C for 5 min, followed by 40 cycles at 95°C for 10 s, and 60°C for 30 s. Primers were designed using Primer Premier Software v5. 0 (Zha et al., 2019; Supplementary Tables S1 and S2). The relative expression levels of the genes were calculated by the 2-ΔΔCt method, the geometric mean of CT of Actin and GAPDH was used in calculation (Livak and Schmittgen, 2001) and were displayed using SigmaPlot v10.0 (Livak and Schmittgen, 2001).
Maize B73 plants were grown in pots at 28°C for 13–14 days in the dark. The third leaf was removed, cut up, and transferred to enzymolysis solution for 6 h. Maize protoplasts were obtained through centrifugation (followed by 100 g for 2 min) and suspended. The p35S: ZmERS4-GFP fusion vector and GFP control vector were transformed into protoplasts after 30 min. The reaction was terminated using W5 (Self configuration by the laboratory) solution (154 mmol/L NaCl, 125 mmol/L CaCl2, 5 mmol/L KCl, 5 mmol/L glucose, 0.03% MES, pH = 5.8). Then, protoplasts were transferred to round dishes and incubated for 36 h in the dark at room temperature. The fluorescence of protoplasts was observed under a ZEISS LSM 880 Airyscan microscope (Zeiss, Germany).
To detect the extent of damage in ZmERS4-overexpressing Arabidopsis leaves treated with the PstDC3000 suspension, trypan blue stain was applied to the leaves. Leaves treated with the PstDC3000 suspension were completely immersed in 2.5 mg/ml trypan blue solution, vacuum-infiltrated for 10 min, and transferred to a water bath at 100°C for 2 min. Then, leaves wrapped in tinfoil were gently placed on a horizontal shaker at 45 rpm for 8–12 h. At the end of the dying process, samples were distained with 2.5 g/ml chloral hydrate solution until colorless (Bhadauria et al., 2010). Finally, distained leaves were affixed for phenotypic observation and photography. Transgenic and wild-type leaves treated with 10 mM MgCl2 6H2O or sterilized distilled water were used as negative control.
After germination, maize plants were grown for 14 days in the dark. Then, the third leaf was removed and cut up for protoplast preparation (Sheen, 2001). Maize protoplasts in which the pCUB-ZmERS4-GFP and pCUB-GFP vectors were transiently expressed successfully were subsequently treated with bacterial PAMP flg22 (0.5 μM) for 2 h. The flg22 peptide, QRLSTGSRINSAKDDAAGLQIA (Guan et al., 2013), was synthetized from the Sangon Biotech (Shanghai) Co., Ltd. Gene primers were designed using Primer Premier Software v5.0 (Zha et al., 2019; Supplementary Table S2). RNA of maize protoplasts was extracted to analyze the expression of key genes.
The SA contents of wild and transgenic A. thaliana before and at 24 h were monitored by ultra-high-performance liquid chromatography (UPLC), which was performed using an UltiMate3000C18 column (100 mm × 2.6 mm, 2.1 μm; Thermo Scientific, United States). SA extraction was performed following previously described methods (Allasia et al., 2018). Samples were detected in the mobile phase [H2O (pH = 4): MeOH (20:80)] at a flow rate of 0.2 ml/min and were analyzed at 230 nm. The analysis was performed at the Biotechnology Center of Anhui Agricultural University (China). The total SA content was equal to the sum of free and bound SA. Nine maize plants were harvested and divided into three replicates at different period, and each sample was mixtures of the leaves of the three maize plants.
Statistical significance of results was calculated using one-way ANOVA followed by least significant difference Duncan’s test at a significant difference level of p < 0.05.
The full length of the ZmERS4 (NP_001295562.1) CDS obtained from the Phytozome database was 1905 bp in length, encoded 634 amino acids, and was located on chromosome 5. The pI and Ww of the ZmERS4 protein were predicted to be 7.07 and 70.5 3 kDa, respectively. It contained a typical conserved domain of an ethylene receptor protein, including three transmembrane domains, GAF, and HisKA (Figure 1A; Supplementary Figure S1), showing similarity to bacterial two-component His kinases (Chang et al., 1993; Sakai et al., 1998). The amino acid sequences of ZmERS4 were compared to the ethylene receptor protein families of A. thaliana, Oryza sativa, Z. mays, and Solanum lycopersicum. According to the phylogenetic relationships and bootstrap values, the ethylene receptor proteins in the four surveyed species divided into two subfamilies. Furthermore, ZmERS4 belonged to the first subfamily of ethylene receptor proteins and was highly homologous with ZmERS1 from Z. mays (Figure 1B).
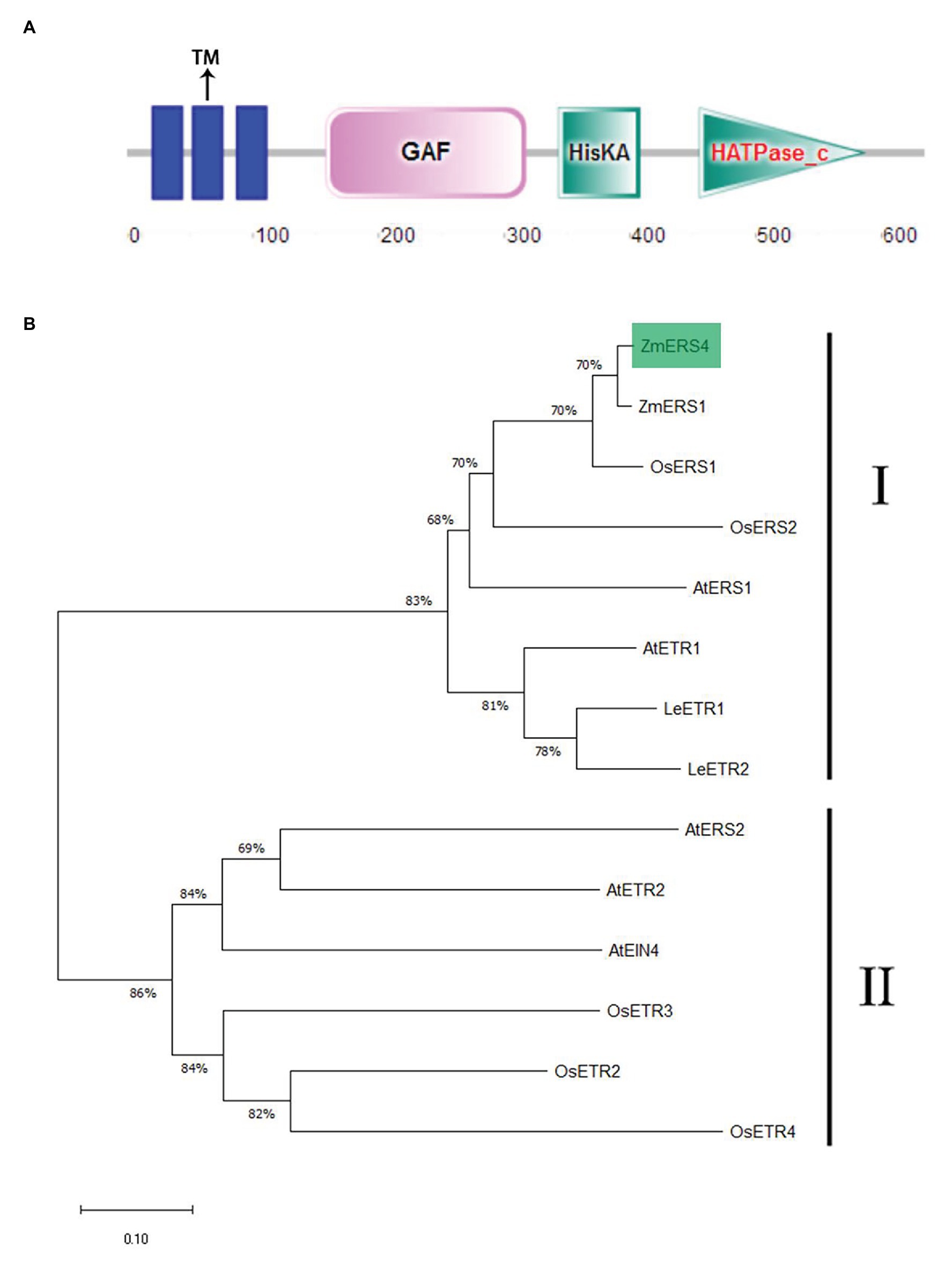
Figure 1. Characterization of ZmERS4. (A) The conserved motifs in the ZmERS4 protein were detected by SMART. (B) The NJ phylogenetic tree was constructed using ZmERS4 and other ethylene receptor proteins from model species. The phylogenetic tree divided into two clades. ZmERS4 is highlighted in green. Bootstrap values (1,000 replicates) are shown as percentages at the branch nodes. Bar = 0.05. The GenBank accession numbers are as follows: ZmERS4 (NP_001295562.1), ZmERS1 (AY359578) from Zea mays, AtERS1 (AT2G40940), AtERS2 (AT1G04310), AtETR1 (AT1G66340), AtETR2 (AT3G23150), and AtEIN4 (AT3G04580) from Arabidopsis thaliana, OsERS1 (AY043031.1), OsERS2 (AF460181.1), OsETR2 (AY136816.2), OsETR3 (AY434735.1), and OsETR4 (AY434734.1) from Oryza sativa, LeETR1 (U41103), LeETR2(U47279) from Solanum lycopersicum.
Publicly available microarray data of maize were used to detect tissue-specific expression patterns of ZmERS4. Data from the transcriptome database indicated that ZmERS4 was expressed in different tissues at distinct stages and the expression levels were higher in the leaves, corn silks, and stems (Figure 2A). Furthermore, the tissue-specific expression profiles were verified by qRT-PCR using six tissues from the inbred maize B73 line. Results revealed that the expression levels of ZmERS4 in the six tissues were consistent with the chip data and ZmERS4 was expressed at higher levels in the stems, leaves, and corn silks (Figure 2B). These results suggested that ZmERS4 was expressed in all corn growth stages.
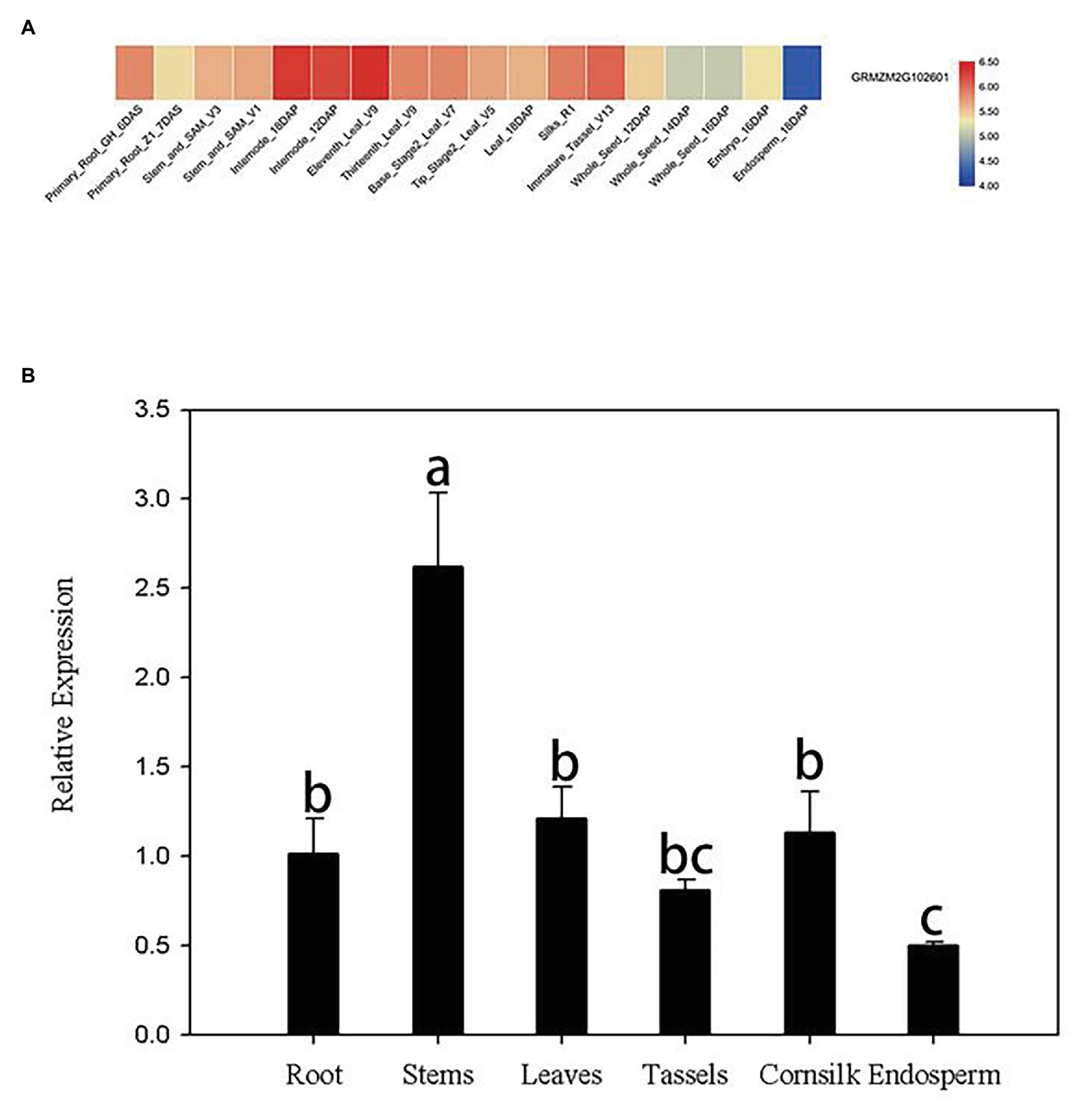
Figure 2. Tissue-specific expression patterns of ZmERS4. (A) Heat map of ZmERS4 gene expression in maize. High, medium, and low expression levels are indicated in red, gray, and blue, respectively. GH, greenhouse; V, vegetative growth stage; R, reproductive growth stage; DAS, days after sowing; DAP, days after pollination. (B) Spatial expression patterns of ZmERS4 in maize. Data are presented as the mean ± SD.
SA, JA, and ethylene, three typical stress hormones that mediate plant defenses in response to biotic stress, were applied to determine whether ZmERS4 is involved in pathogen resistance. At the three-leaf stage, the transcript levels of ZmERS4 gradually increased 3–12 h after SA application and its expression was increased by 1.70-fold in SA-treated plants after 12 h compared to the pre-treatment 0 h control group (Figure 3A). ZmERS4 gradually increased 0–3 h after JA inoculation and its transcripts were increased by 1.75-fold 3 h after JA application. Specifically, ZmERS4 transcripts were increased by 1.46-, 1.37-, 1.47-, and 1.43-fold after 6, 12, 24, and 48 h in JA-treated plants, respectively (Figure 3B). Moreover, the expression of ZmERS4 in ET-treated plants after 3 and 6 h were not significantly different when compared to 0 h, and the expression of ZmERS4 was 1.29-fold higher at 12 h after ET application, and then slowly declined (Figure 3C). It seemed that there was an early response of ZmERS4 to JA (3 h), later to SA (12 h) and after to ET (12–48 h), the results indicated that ZmERS4 might respond to all three hormones with different timing of pattern and expression.
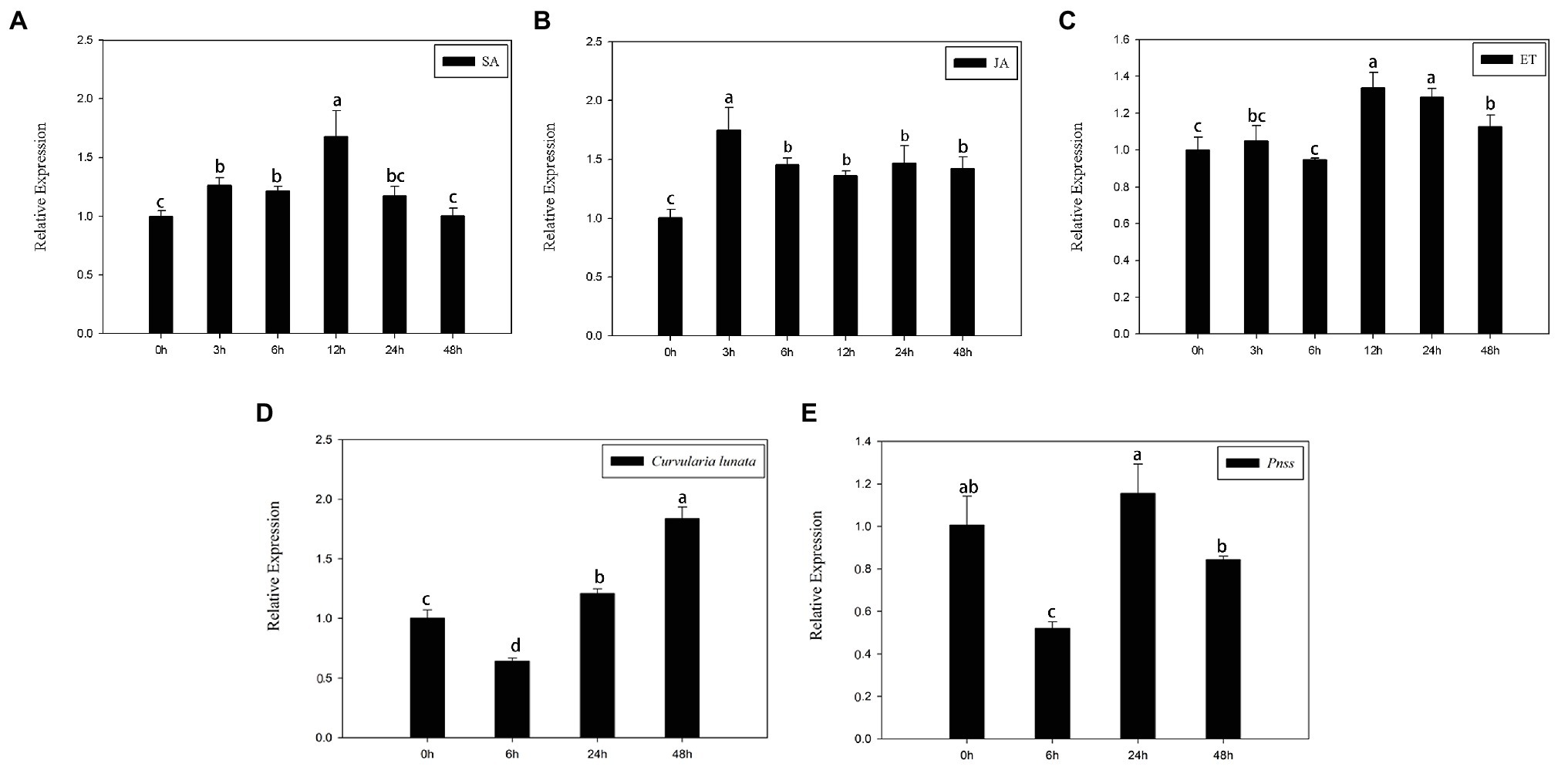
Figure 3. Relative expression levels of ZmERS4 under three different treatments at six time points. (A) Salicylic acid (SA) spraying, (B) jasmonic acid (JA) spraying, (C) ethephon (ET) spraying, (D) The conidial suspension of Curvularia lunata (1 × 105 CFU/ml) spraying, and (E) Pantoea stewartii suspensions (1 × 106 CFU/ml). ZmActin1 and GADPH were used as reference genes for normalization. The expression levels at 0 h were used as the control and assigned a value of 1. Data are presented as the mean ± SD. Three biological replicates were performed.
Meanwhile, the transcript levels of ZmERS4 gradually increased 6–48 h after C. lunata application (Figure 3D), and the expression of ZmERS4 in C. lunata-treated after 48 h were 1.84-fold higher compared to 0 h. However, the expression of ZmERS4 in P. stewartii-treated plants were 1.16-fold higher at 24 h (Figure 3E). These results showed that ZmERS4 may participate in plant disease resistance, but the underlying mechanisms require further investigation.
To verify the location of ZmERS4 in cells, the p1305-ZmERS4-GFP fusion vector was constructed, in which the ZmERS4 CDS without the termination codon was fused upstream of the GFP reporter under control of the CaMV35S promoter (Supplementary Figure S2). The recombinant plasmid was introduced into maize protoplasts with a nuclear localization plasmid, and then observed under a laser confocal microscope. The green fluorescent signal of ZmERS4-GFP was observed around the plasma membrane without overlapping the nuclear marker. Because the cytoplasm can be “pushed” against the plasma membrane in vacuolated cells, the ZmERS4 protein was localized in plasma membrane or cytoplasm (Figure 4).
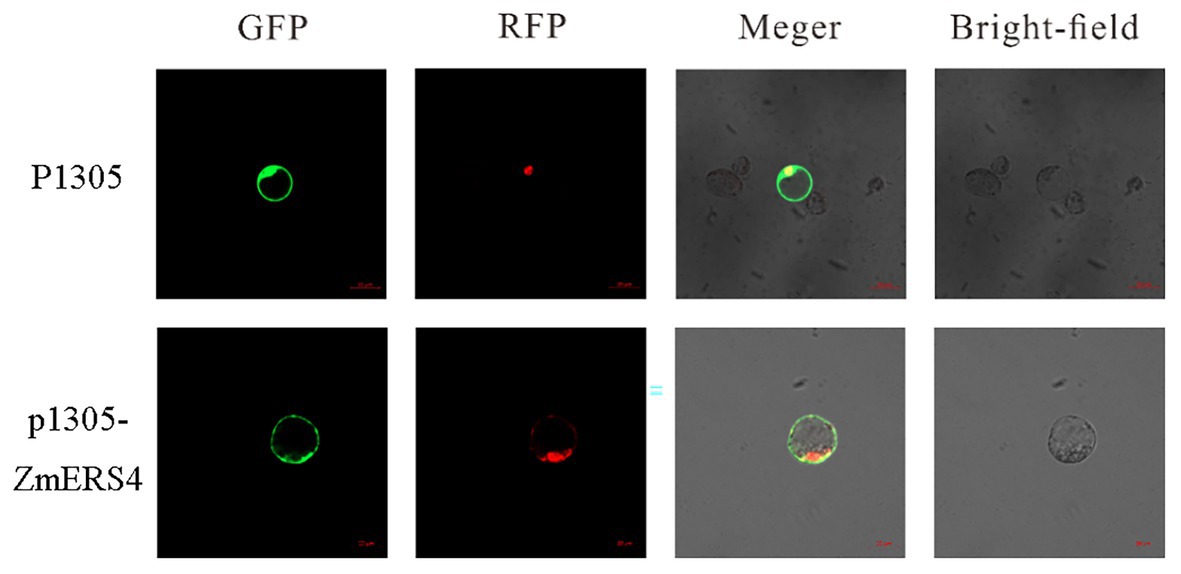
Figure 4. Subcellular localization of the ZmERS4-GFP fusion protein in maize protoplasts. GFP: green fluorescent protein; RFP: nuclear marker pSAT6-mCherry-VirD2NLS. Scale bar = 20 μm.
To further investigate the function of ZmERS4, the p1301-ZmERS4 fusion expression vector was constructed (Supplementary Figure S3). Agrobacterium tumefaciens cells containing 35S: ZmERS4 were transformed into wild-type Arabidopsis (Supplementary Figure S4). Two independent T3 homozygous lines (L1 and L7) were selected for ZmERS4. Under normal growth conditions, L1 and L7 exhibited similar growth behavior to the background wild-type control, Col-0 (Figure 5A). PstDC3000 suspension was sprayed on the leaves of L1 and L7 (2 ml per plant). Symptoms appeared in all groups at 3 days. At 7 days, small tan necrotic spots on the leaves were observed. Compared to the negative control (Figure 5A), the L1 and L7 had less spots (Figures 5A,B). The plant disease severity assessment was also conducted using the disease index 7 days with PstDC3000. The disease indexes of L1 and L7 were 35.94 and 39.09, respectively. L1 had a significantly lower disease index (47.17) compared to wild-type Arabidopsis (Figure 5C). These results indicated that the overexpressing lines conferred enhanced resistance compared to wild-type Arabidopsis.
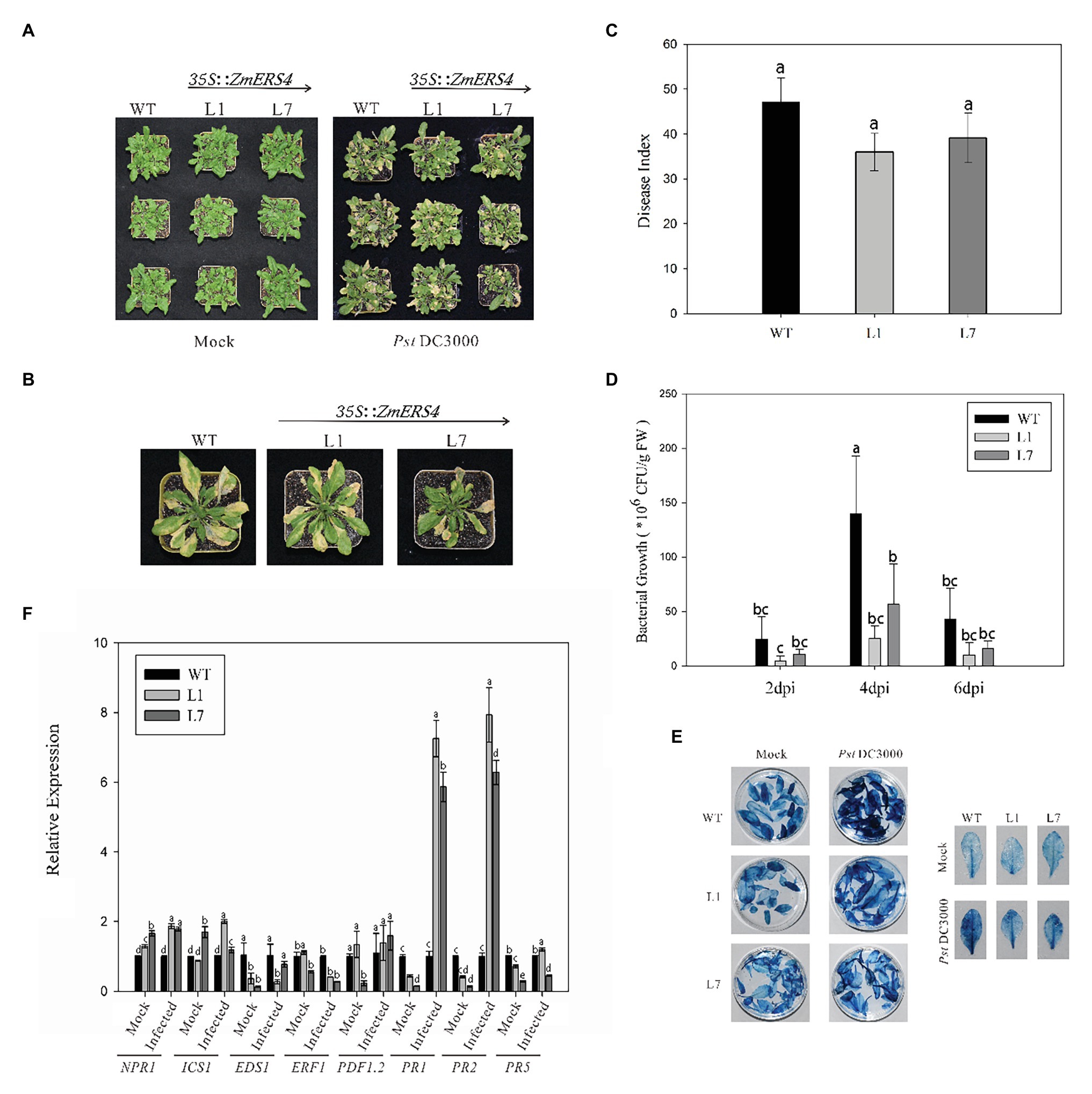
Figure 5. ZmERS4 transgenic Arabidopsis plants conferred enhanced resistance to PstDC3000. (A,B) Phenotypes of ZmERS4 transgenic and wild-type Arabidopsis at 7 days. (C) The disease indexes of different groups. (D) The population density of bacterium in Arabidopsis leaves of different groups at 2, 4, and 6 days. Values of p were determined by a two-tailed Student’s t test assuming equal variances (*p < 0.05); three biological replicates were performed. (E) Trypan blue staining in Arabidopsis leaves of different groups at 7 days. (F) Expression levels of marker genes in SA, JA, and ET-mediated signaling pathways at 24 h. AtActin2 and AtTUB4 were used as reference genes for normalization. The expression levels in wild-type plants were used as the control and assigned a value of 1. Data are presented as the mean ± SD based on three biological replicates.
To determine the relationship between the number of bacteria in Arabidopsis thaliana overexpressing ZmERS4 and plant disease resistance, the dilution plating method was used and the population density of bacterium in Arabidopsis leaves was detected. The population densities in L1 and L7 were consistently lower than wild-type Arabidopsis at 2, 4, and 6 days, respectively. The population density of L1 reached 2.52 × 107 CFU/g in leaf tissues at 4 days and remained at 1.37 × 107 CFU/g until 6 days compared to 1.39 × 108 CFU/g at 4 days and 3.71 × 107 CFU/g at 6 days in wild-type Arabidopsis, respectively (Figure 5D). These results showed that ZmERS4, when over-expressed in A. thaliana, partially increases resistance to PstDC3000.
Plant hypersensitive cell death is an important event in early plant pathogen invasion. Infection by site and adjacent site of programed cell death to restrict pathogen expansion causes plants to develop local acquired resistance (LAR) and SAR and plays an important role in plant resistance to pathogen invasion. After staining WT plant and transgenic A. thaliana lines L1 and L7 inoculated with PstDC3000 using trypan blue, findings showed that the transgenic A. thaliana lines and wild-type plants exhibited necrotic cell accumulation and appeared dark blue (Figure 5E). However, after PstDC3000 treatment, wild-type plants inoculated with PstDC3000 were dark blue compared to the light blue leaves of the transgenic lines (Figure 5E). According to the disease indexes of different treatment groups (Figure 5C), the leaf damage degree of transgenic plants was lower than wild-type plants and the trypan blue staining results confirmed these findings. These results suggest that HR was induced in heterologous expression of ZmERS4- in A. thaliana in response to PstDC3000.
To investigate the signaling pathways in which ZmERS4 was involved in and conferred enhanced resistance in transgenic Arabidopsis, the expression levels of target plant genes that are known to function in the SA, JA, or ET pathways were analyzed at 24 h after (Dinolfo et al., 2017; Zheng et al., 2019).when the control groups were treated with 10 mM MgCl2 6H2O, the EDS1, PR1, PR2, and PR5 were expressed at lower levels in L1 and L7 than wild-type plants, the NPR1 were expressed at higher levels in L1 and L7 than wild-type plants (Figure 5F). However, after the control groups were infected with PstDC3000, the PR1 was strongly induced in L1 and L7, and was 7.26- and 5.87-fold higher compared to wild-type plants, respectively (Figure 5F). PR2 expression in L1 and L7 were similar to PR1 and was induced 7.93-fold and 6.28-fold higher, respectively (Figure 5F). Moreover, NPR1, ICS1, and PDF1.2 were slightly induced in L1 and L7 at 24 h (Figure 5F). EDS1 and PR5 transcripts did not exhibit apparent changes before and after inoculation (Figure 5F). And the expression levels of Ethylene Response Factor1 (ERF1) in L1 and L7 were lower than the negative controls (Figure 5F). These results showed that PR1 and PR2 were concurrently highly expressed in ZmERS4-overexpressing lines after pathogen inoculation, suggesting that ZmERS4 may be involved in plant defense mechanisms by activating SA-dependent signaling pathways.
The pCUB-ZmERS4-GFP and pCUB-GFP (control) vectors were constructed and transiently expressed in maize protoplasts (Figure 6A). The expression levels of ZmERS4 increased by 50-fold in maize protoplasts that transiently expressed the pCUB-ZmERS4-GFP vector compared to the control (Figure 6B). The expression of PR1 (a SA-responsive marker gene), lipoxygenase 1(LOX1; a JA-responsive marker gene) and ERF1, which are expressed in plant defense mechanisms (Erb et al., 2009; Xie et al., 2019), were used in this study. As demonstrated by qRT-PCR, the expression of PR1 was not significantly different between protoplasts that transiently expressed pCUB-ZmERS4-GFP or empty vectors without pathogen inoculation, while the expression of ERF1 and LOX1 in protoplasts that expressed pCUB-ZmERS4-GFP was slightly higher than protoplasts that expressed empty vectors. However, the expression of PR1 in pCUB-ZmERS4-GFP protoplasts treated with 0.5 μM flg22 peptide, an elicitor of P. syringae (Guan et al., 2013), was strongly induced 8-fold higher compared to the pCUB-GFP vector (Figure 6C). Additionally, the expression of ERF1 and LOX1 in pCUB- ZmERS4-GFP protoplasts treated with 0.5 μM flg22 peptide was lower than the pCUB-GFP vector (Figure 6C). These results further corroborated that ZmERS4 participates in plant defense responses through the SA signaling pathway.
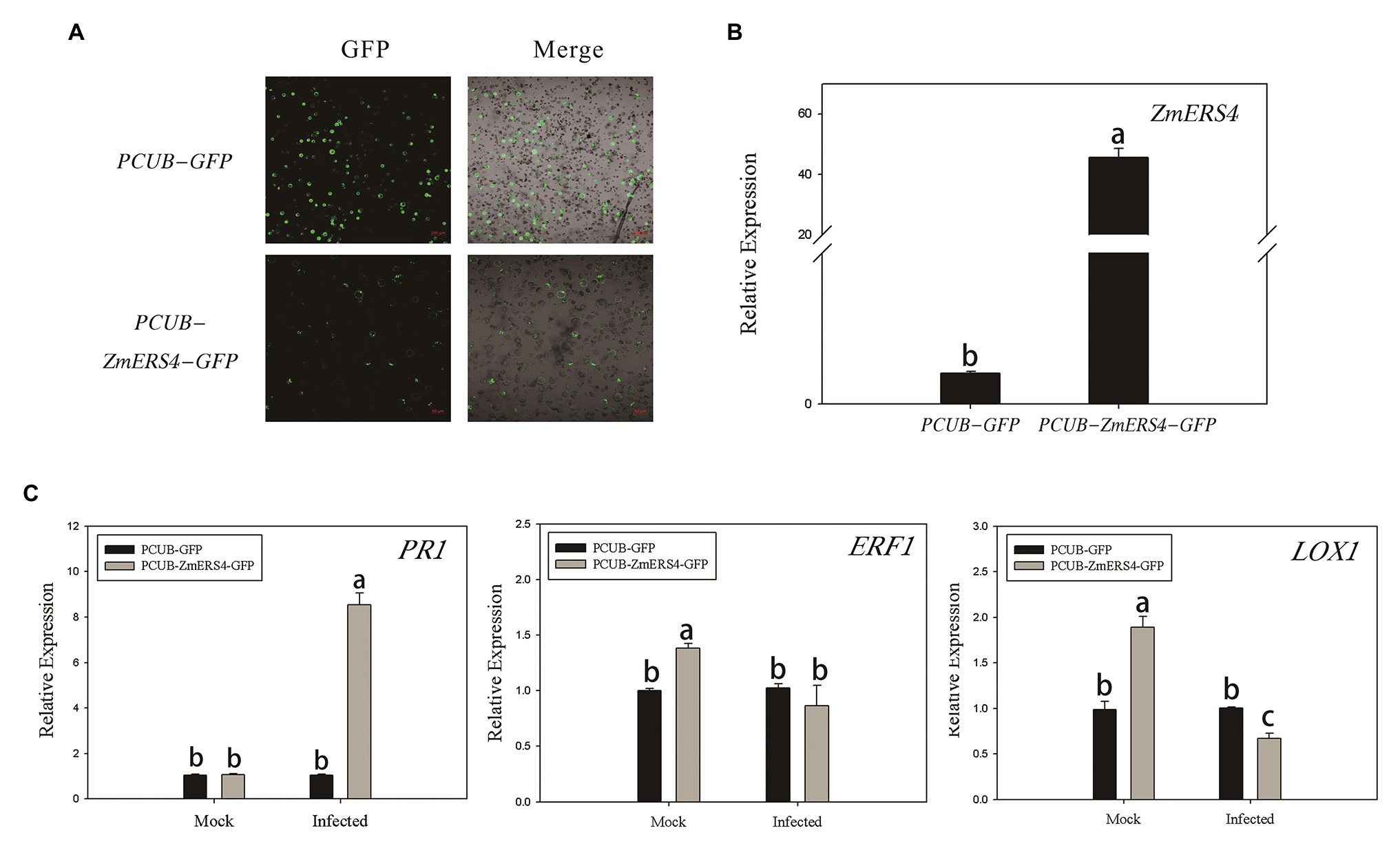
Figure 6. The transient overexpression of ZmERS4 in maize protoplasts was involved in plant defense responses through the SA signaling pathway. (A) The pCUB-ZmERS4-GFP and pCUB-GFP vectors were transiently expressed in maize protoplasts with a transformation efficiency >50%. GFP, green fluorescent protein. Scale bar = 100 μm. (B) Expression levels of ZmERS4 extracted from maize protoplasts that did and did not overexpress ZmERS4. (C) Expression levels of the marker genes in the three signaling pathways induced by 0.5 μM flg22 after 2 h. ZmActin and ZmGADPH were used as reference genes for normalization. The expression levels of the protoplasts that expressed pCUB-GFP were used as the control and were assigned a value of 1. The data are presented as the mean ± SD based on three biological replicates.
The preliminary results showed that ZmERS4 played an important role in plant defense responses through the SA signaling pathway; however, changes in the SA contents of transgenic plants after PstDC3000 infection were unclear. Thus, UPLC was used to detect the SA contents in ZmERS4-overexpressing Arabidopsis. The absorption peaks of the SA extracts from L1, L7, and wild-type Arabidopsis appeared at ~1.43 s (Supplementary Figure S5), which was consistent with the peak time of the standard SA. The total SA contents in L1 and L7 were generally higher than wild-type plants, which were treated with and without PstDC3000. Findings showed the SA contents in L1 and L7, which were 4.512 and 4.901 μg/g FW, were higher than that of the wild-type plants (3.848 μg/g FW) without pathogen infection. However, when the plants were infected by PstDC3000, the SA contents in L1 rapidly accumulated at 24 h (Figure 7A). The total SA contents in L1 and L7 reached 7.826 and 5.640 μg/g FW at 24 h compared to wild-type plants (4.54 μg/g FW) inoculated with the pathogen, respectively (Figure 7A). These results indicated that the resistance to PstDC3000 in L1 and L7 was enhanced due to SA accumulation.
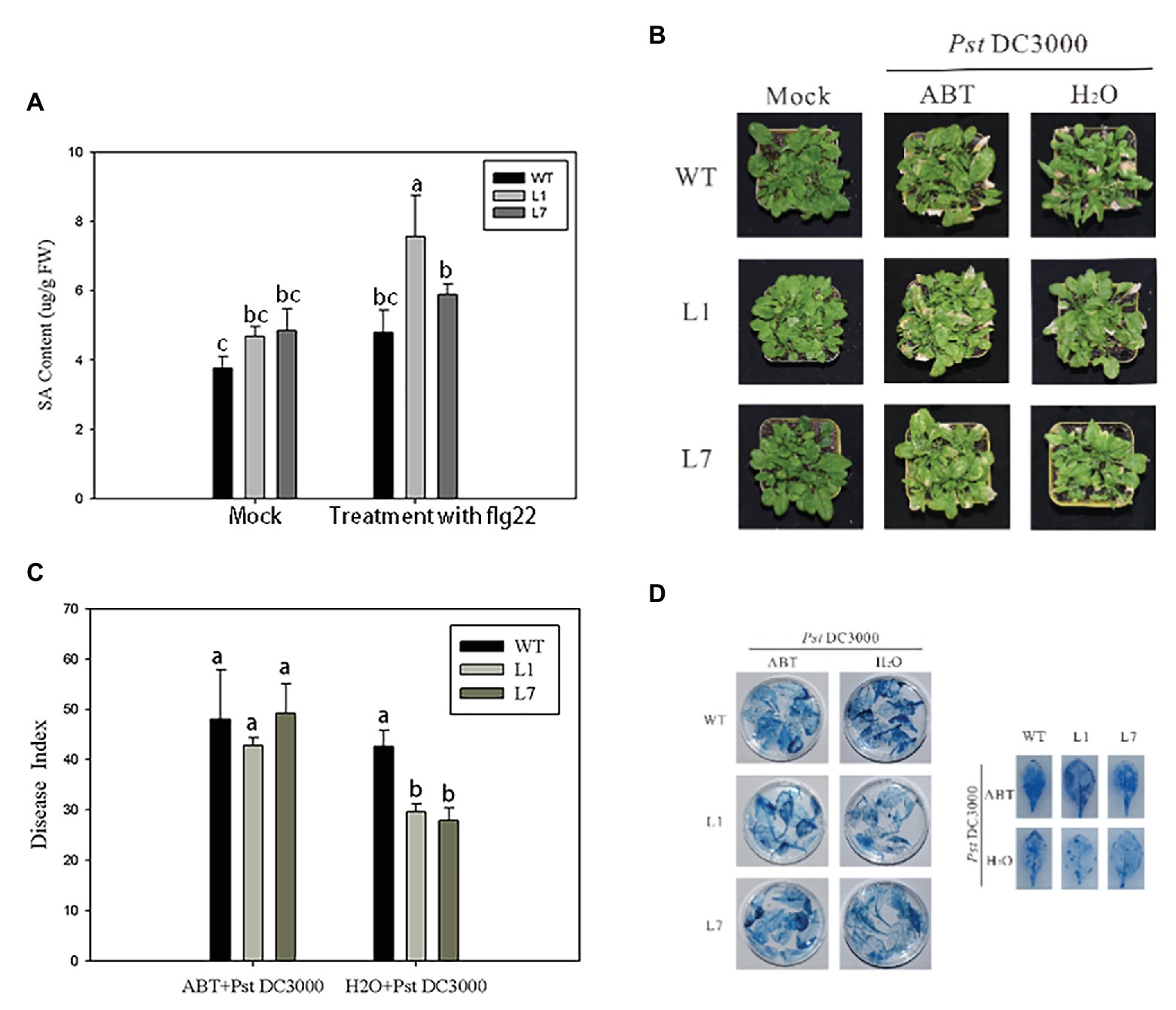
Figure 7. ZmERS4 transgenic Arabidopsis thaliana exhibited improved resistance to PstDC3000 by the SA-mediated signaling pathway. (A) SA contents were extracted from wild-type and ZmERS4 transgenic Arabidopsis at 24 h. (B) Phenotypes of wild-type and ZmERS4 transgenic Arabidopsis at 7 days; plants were pretreated with ABT for 24 h. (C) Disease indexes of the different groups shown in (B). (D) Trypan blue staining of the Arabidopsis leaves in the different groups shown in (B).
ABT is an inhibitor of SA biosynthesis and can effectively control SA biosynthesis in plants (Wang et al., 2019). To confirm that ZmERS4 enhanced A. thaliana resistance to PstDC3000 through the SA signaling pathway, ZmERS4-overexpressing lines and wild-type plants were pretreated with 100 μmol/L ABT for 24 h. Then, plants were sprayed with the PstDC3000 suspension. The disease indexes were estimated at 7 days. As expected, both overexpressing and wild-type plants pretreated with ABT exhibited more severe symptoms (Figure 7B). The disease indexes of L1, L7, and wild-type Arabidopsis were 42.76, 49.26, and 48.06, respectively (Figure 7C), while the disease indexes of the lines pretreated with sterilized distilled water were 29.65, 27.87, and 42.67, respectively (Figure 7C). Dead cells in the leaves of plants pretreated with ABT and sterilized distilled water were further detected by trypan blue staining. Compared to the negative controls, the dead cells in the treatment groups pretreated with ABT were dark blue due to a higher number of larger areas of these spots (Figure 7D). Clearly, ABT application reduced plant resistance to pathogen infection, further indicating that the overexpression of ZmERS4 in Arabidopsis enhanced resistance to PstDC3000 by activating the SA signaling pathway.
A candidate disease-resistant gene, ZmERS4, was explored in our previous study. According to the protein domain prediction and sequence homology analysis, our results showed that ZmERS4 could encode a protein similar to ethylene receptor, showing the highest sequence similarity with OsERS1 in rice. Ethylene receptor genes have unique expression patterns in different plant tissues. Previous studies reported that ETR1 and ERS1, members of the ethylene receptor ETR1 subfamily, are expressed ubiquitously in Arabidopsis plants. The expression of AtETR1 is constitutive and ethylene independent, while AtERS1 is tissue specific, ethylene dependent, and developmentally regulated, which was previously observed in the leaves, stems, floral organs, and especially the procambium cells, and very strong expression was observed in the locules of the anthers (Hua et al., 1998). Our results demonstrated that ZmERS4 was expressed in maize tissues and predominantly expressed in the stems and leaves, suggesting that ZmERS4 was expressed in all stages of maize growth and development. The results were consistent with the findings of ETR1 and ERS1.
Ethylene receptor genes play important roles in plant responses to hormones. Different types of ethylene receptor genes exhibit different expression patterns in response to hormones. It was reported that ETR- and ERS-type genes, PpETR1 and PpERS1, of peach fruit showed different expression patterns in response to ethylene and propylene, such that propylene treatment did not affect the transcript levels of PpETR1, while PpERS1 was upregulated in the pit hardening and climacteric ripening stages (Rasori et al., 2002). Ethylene receptor proteins, which perceive ethylene signals, are located upstream of signal transduction and play negative regulatory roles in the transmission of ethylene signals (Hua et al., 1998). In this study, the expression of ZmERS4 was induced after SA or JA application. However, as an ethylene receptor protein, its transcript levels did not change after ET treatment, suggesting that ET did not affect the transcript levels of ZmERS4, which was consistent with the findings of previous studies.
To date, research on plant ethylene receptor proteins has mainly focused on the regulation of growth and development, changing the flowering period, and delaying senescence and death (Gallie, 2015; Chen et al., 2020). A few reports are known about the roles of ethylene receptor genes in plant disease resistance. Here, we identified some characteristic genes using ZmERS4-overexpressing Arabidopsis inoculated with PstDC3000, which are similar to known or deduced functions involved in SA, JA, and ET pathways (Niu et al., 2011). These genes are also suspected to play roles in generating signals for the activation of certain defense responses and protecting plants from damage associated with these defense responses. We found that the defense-related genes, PR1 and PR2, were highly expressed in the leaves of ZmERS4-overexpressing Arabidopsis, while the expression of other genes did not increase. PR1 and PR2 are marker genes of the SA-mediated signaling pathway, and SA can activate the expression of PR genes. Thus, we hypothesized that ZmERS4-overexpressing Arabidopsis confer enhanced pathogen resistance by activating an induced response through the SA signaling pathway. Interestingly, the SA contents in ZmERS4-overexpressing plants rapidly accumulated at 24 h and the improved resistance of ZmERS4-overexpressing Arabidopsis was repressed by the application of 100 μmol/L ABT, a SA biosynthetic inhibitor. Moreover, the expression of PR1 in pCUB-ZmERS4-GFP protoplasts treated with 0.5 μM flg22 peptide was strongly induced expression by 8-fold higher. These results showed that the overexpression of ZmERS4 elevated SA levels after pathogen infection and increased the expression of PR genes, suggesting that ZmERS4 was involved in plant defense processes through the SA signaling pathway and was subjected to SA feedback regulation.
According to a previous study, SA affects plant development and growth, and activates HR to promote cell death, resulting in pathogen resistance (Vlot et al., 2009; Kazan and Lyons, 2014; Han and Kahmann, 2019). SA also plays a role in abiotic stress responses (Alonso-Ramírez et al., 2009). Previous studies reported the SA was associated with ethylene signaling (Devadas et al., 2002; Yin et al., 2013). For example, SA and ethylene coordinate plant pathogen responses (Devadas et al., 2002), and many ethylene signaling components, such as tomato Pti4 (Gu et al., 2000) and apple MdETR1, MdERS1, and MdCTR1 (Li et al., 2006), are transcriptionally regulated after SA treatment. Yin et al. investigated the effects of acetylsalicylic acid (ASA) on kiwifruit ethylene biosynthesis and signaling, and found that the ERFs, AdERF1, AdERF3, and AdERF12, were characterized as ASA-responsive genes, suggesting that the effect of SA on fruit ripening may involve the regulation of genes associated with ethylene signaling (Yin et al., 2013). The results of this study indicated that ZmERS4 overexpressing plants conferred enhanced disease resistance to PstDC3000 by promoting SA accumulation, thus supporting previous findings (Devadas et al., 2002; Yin et al., 2013).
However, information regarding the molecular mechanisms of ZmERS4 overexpression in the modulation of SA signaling components in plants remains lacking. It was reported that the ethylene receptor protein, OsERS1, in O. sativa may interact with the rice protein, OsTPR1, and this interaction plays a key role in rice ethylene signaling, as well as influences rice growth and development (Yi, 2013). The Arabidopsis protein, AtTRP1, interacts preferentially with the Arabidopsis ethylene receptor, ERS1, and the overexpression of AtTRP1 enhances some ethylene responses (Lin et al., 2009). Therefore, we speculate that ZmERS4 may interact with certain factors or proteins to respond to ethylene signals. However, this notion requires further investigation in order to elucidate the mechanisms of ethylene signal responses in the ZmERS4 gene.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
TH has made major contributions to the conception and design of the study. TH and XL contributed to the acquisition, analysis, or interpretation of the data. TH, XL, SX, CH, HJ, and TD performed the experiments and participated to the interpretation of data. TH, XL, and TD wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by the General Program of Natural Science Foundation of Anhui Province, China (No. 2008085MC93).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.632908/full#supplementary-material
1. http://phytozome.jgi.doe.gov/pz/portal.html
2. https://web.expasy.org/compute_pi/
Abeles, F. B., Morgan, P. W., and Saltveit, M. E. Jr. (1992). Ethylene in plant biology. London: Academic Press Limited.
Allasia, V., Industri, B., Ponchet, M., Quentin, M., Favery, B., and Keller, H. (2018). Quantification of salicylic acid (SA) and SA-glucosides in Arabidopsis thaliana. Bio-protocol. 8:e2844. doi: 10.21769/BioProtoc.2844
Alonso-Ramírez, A., Rodríguez, D., Reyes, D., Jiménez, J. A., Nicolás, G., López-Climent, M., et al. (2009). Evidence for a role of gibberellins in salicylic acid-modulated early plant responses to abiotic stress in Arabidopsis seeds. Plant Physiol. 150, 1335–1344. doi: 10.1104/pp.109.139352
Bhadauria, V., Miraz, P., Kennedy, R., Banniza, S., and Wei, Y. (2010). Dual trypan-aniline blue fluorescence staining methods for studying fungus-plant interactions. Biotech. Histochem. 85, 99–105. doi: 10.3109/10520290903132196
Bleecker, A. B. (1999). Ethylene perception and signaling: an evolutionary perspective. Trends Plant Sci. 7, 269–274. doi: 10.1016/s1360-1385(99)01427-2
Chang, C., Kwok, S. F., Bleecker, A. B., and Meyerowitz, E. M. (1993). Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262, 539–544. doi: 10.1126/science.8211181
Chen, Y. F., Etheridge, N., and Schaller, G. E. (2005). Ethylene signal transduction. Ann. Bot. 6, 901–915. doi: 10.1093/aob/mci100
Chen, Y., Hu, G., Rodriguez, C., Liu, M., Binder, B. M., and Chervin, C. (2020). Roles of SlETR7, a newly discovered ethylene receptor, in tomato plant and fruit development. Hortic. Res. 7, 17–29. doi: 10.1038/s41438-020-0239-y
Chen, Y. F., Randlett, M. D., Findell, J. L., and Schaller, G. E. (2002). Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J. Biol. Chem. 22, 19861–19866. doi: 10.1074/jbc.M201286200
Chen, Y. F., Shakeel, S. N., Bowers, J., Zhao, X. C., Etheridge, N., and Schaller, G. E. (2007). Ligand-induced degradation of the ethylene receptor ETR2 through a proteasome-dependent pathway in Arabidopsis. J. Biol. Chem. 34, 24752–24758. doi: 10.1074/jbc.M704419200
Clarke, J. D. (2009). Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb. Protoc. 2009:5177. doi: 10.1101/pdb.prot5177
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Devadas, S. K., Enyedi, A., and Raina, R. (2002). The Arabidopsis hrl1 mutation reveals novel overlapping roles for salicylic acid jasmonic acid and ethylene signalling in cell death and defence against pathogens. Plant J. 30, 467–480. doi: 10.1046/j.1365-313X.2002.01300.x
Ding, T., Su, B., Chen, X., Xie, S., Gu, S., Wang, Q., et al. (2017). An endophytic bacterial strain isolated from Eucommia ulmoides inhibits southern corn leaf blight. Front. Microbiol. 8:903. doi: 10.3389/fmicb.2017.00903
Dinolfo, M. I., Castanares, E., and Stenglein, S. A. (2017). Resistance of Fusarium poae in Arabidopsis leaves requires mainly functional JA and ET signaling pathways. Fungal Biol. 121, 841–848. doi: 10.1016/j.funbio.2017.06.001
Erb, M., Flors, V., Karlen, D., De Lange, E., Planchamp, C., D’alessandro, M., et al. (2009). Signal signature of aboveground-induced resistance upon belowground herbivory in maize. Plant J. 59, 292–302. doi: 10.1111/j.1365-313X.2009.03868.x
Gallie, D. R. (2015). Ethylene receptors in plants—why so much complexity? F1000Prime Rep. 7, 39–51. doi: 10.12703/P7-39
Gu, Y. Q., Yang, C. M., Thara, V. K., Zhou, J. M., and Martin, G. B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12, 771–785. doi: 10.1105/tpc.12.5.771
Guan, X., Buchholz, G., and Nick, P. (2013). The cytoskeleton is disrupted by the bacterial effector HrpZ, but not by the bacterial PAMP flg22, in tobacco BY-2 cells. J. Exp. Bot. 64, 1805–1816. doi: 10.1093/jxb/ert042
Han, X., and Kahmann, R. (2019). Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front. Plant Sci. 10:822. doi: 10.3389/fpls.2019.00822
Hua, J., and Meyerowitz, E. M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94, 261–271. doi: 10.1016/S0092-8674(00)81425-7
Hua, J., Sakai, H., Nourizadeh, S., Chen, Q. G., Bleecker, A. B., Ecker, J. R., et al. (1998). EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10, 1321–1332.
Johal, G. S., and Briggs, S. P. (1992). Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258, 985–987. doi: 10.1126/science.1359642
Kazan, K., and Lyons, R. (2014). Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26, 2285–2309. doi: 10.1105/tpc.114.125419
Kourelis, J., and van der Hoorn, R. A. L. (2018). Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299. doi: 10.1105/tpc.17.00579
Li, N., Lin, B., Wang, H., Li, X. M., Yang, F. F., Ding, X. H., et al. (2019). Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat. Genet. 51, 1540–1548. doi: 10.1038/s41588-019-0503-y
Li, D. P., Xu, Y. F., Sun, L. P., Liu, L. X., Hu, X. L., Li, D. Q., et al. (2006). Salicylic acid, ethephon, and methyl jasmonate enhance ester regeneration in 1-MCP-treated apple fruit after long-term cold storage. J. Agric. Food Chem. 54, 3887–3895. doi: 10.1021/jf060240j
Lin, Z., Ho, C. W., and Grierson, D. (2009). AtTRP1 encodes a novel TPR protein that interacts with the ethylene receptor ERS1 and modulates development in Arabidopsis. J. Exp. Bot. 60, 3697–3714. doi: 10.1093/jxb/erp209
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, B., Cui, M. L., Sun, H. J., Takada, K., Mori, H., Kamada, H., et al. (2006). Subcellular lacalization and membrance topology of the melon ethylene receptor cmERS1. Plant Physiol. 141, 587–597. doi: 10.1104/pp.106.080523
Niu, D. D., Liu, H. X., Jiang, C. H., Wang, Y. P., Wang, Q. Y., Jin, H. L., et al. (2011). The plant growth-promoting rhizobacterium Bacillus cereus AR156 induce systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol. Plant Microbe Interact. 24, 533–542. doi: 10.1094/MPMI-09-10-0213
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Ranum, P., Pena-Rosas, J. P., and Garcia-Casal, M. N. (2014). Global maize production, utilization, and consumption. Ann. N. Y. Acad. Sci. 1312, 105–112. doi: 10.1111/nyas.12396
Rasori, A., Ruperti, B., Bonghi, C., Tonutti, P., and Ramina, A. (2002). Characterization of two putative ethylene receptor genes expressed during peach fruit development and abscission. J. Exp. Bot. 53, 2333–2339. doi: 10.1093/jxb/erf097
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sakai, H., Hua, J., Chen, Q. G., Chang, C., Medrano, L. J., Bleecker, A. B., et al. (1998). ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 95, 5812–5817. doi: 10.1073/pnas.95.10.5812
Sekhwal, M. K., Li, P., Lam, I., Wang, X., Cloutier, S., and You, F. M. (2015). Disease resistance gene analogs (RGAs) in plants. Int. J. Mol. Sci. 16, 19248–19290. doi: 10.3390/ijms160819248
Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475. doi: 10.1104/pp.010820
Subash, S. (2015). A review on important maize diseases and their management in Nepal. J. maize Res. Dev. 1, 28–52. doi: 10.3126/jmrd.v1i1.14242
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, J. C., Jia, Y., Wen, J. W., Liu, W. P., Li, X. M., Li, L., et al. (2013). Identification of rice blast resistance genes using international monogenic differentials. Crop Prot. 45, 109–116. doi: 10.1016/j.cropro.2012.11.020
Wang, L., Wang, H., He, S., Meng, F., Zhang, C., Fan, S., et al. (2019). GmSnRK1.1, a sucrose non-fermenting-1(SNF1)-related protein kinase, promotes soybean resistance to Phytophthora sojae. Front. Plant Sci. 10:996. doi: 10.3389/fpls.2019.00996
Xie, S., Yu, H., Li, E., Wang, Y., and Jiang, H. (2019). Identification of miRNAs involved in Bacillus velezensis FZB42-activated induced systemic resistance in maize. Int. J. Mol. Sci. 20, 5057–5071. doi: 10.3390/ijms20205057
Yi, W. K. (2013). Preliminary research of OsTPR1 modulating the ethylene signaling transduction pathway associated with OsERS1 in rice (Oryza sativa) [D] : HuNan Agricultural University.
Yin, X. R., Zhang, Y., Zhang, B., Yang, S. L., Shi, Y. N., Ferguson, L. B., et al. (2013). Effects of acetylsalicylic acid on kiwifruit ethylene biosynthesis and signaling components. Postharvest Biol. Technol. 83, 27–33. doi: 10.1016/j.postharvbio.2013.03.012
Zha, K., Xie, H., Ge, M., Wang, Z., Wang, Y., Si, W., et al. (2019). Expression of maize MADS transcription factor ZmES22 negatively modulates starch accumulation in rice endosperm. Int. J. Mol. Sci. 20, 483–499. doi: 10.3390/ijms20030483
Zhao, B., Lin, X., Poland, J., Trick, H., Leach, J., and Hulbert, S. (2005). A maize resistance gene functions against bacterial streak disease in rice. Proc. Natl. Acad. Sci. U. S. A. 102, 15383–15388. doi: 10.1073/pnas.0503023102
Zheng, X., Xing, J., Zhang, K., Pang, X., Zhao, Y., Wang, G., et al. (2019). Ethylene response factor ERF11 activates BT4 transcription to regulate immunity to Pseudomonas syringae. Plant Physiol. 180, 1132–1151. doi: 10.1104/pp.18.01209
Zhou, Q., Gu, S. Y., Xu, L., and Tan, G. J. (2019). Effects of endophyte DZSY21 colonization on DNA methylation levels in maize. J. Hefei Norm. Univ. 3, 32–37.
Keywords: maize, ZmERS4, ethylene receptor, disease-resistant, salicylic acid
Citation: Hang T, Ling X, He C, Xie S, Jiang H and Ding T (2021) Isolation of the ZmERS4 Gene From Maize and Its Functional Analysis in Transgenic Plants. Front. Microbiol. 12:632908. doi: 10.3389/fmicb.2021.632908
Received: 25 November 2020; Accepted: 15 February 2021;
Published: 12 March 2021.
Edited by:
Massimo Reverberi, Sapienza University of Rome, ItalyReviewed by:
Lei Huang, Zhejiang University, ChinaCopyright © 2021 Hang, Ling, He, Xie, Jiang and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting Ding, ZGluZ3Rpbmc5OEAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.