- 1Department of Microbiology and Immunology, Montana State University, Bozeman, MT, United States
- 2Department of Land Resources and Environmental Sciences, Montana State University, Bozeman, MT, United States
- 3Department of Chemistry and Biochemistry, Montana State University, Bozeman, MT, United States
The microbial ars operon encodes the primary bacterial defense response to the environmental toxicant, arsenic. An important component of this operon is the arsR gene, which encodes ArsR, a member of the family of proteins categorized as DNA-binding transcriptional repressors. As currently documented, ArsR regulates its own expression as well as other genes in the same ars operon. This study examined the roles of four ArsR proteins in the well-developed model Gram-negative bacterium Agrobacterium tumefaciens 5A. RNASeq was used to compare and characterize gene expression profiles in ± arsenite-treated cells of the wild-type strain and in four different arsR mutants. We report that ArsR-controlled transcription regulation is truly global, extending well beyond the current ars operon model, and includes both repression as well as apparent activation effects. Many cellular functions are significantly influenced, including arsenic resistance, phosphate acquisition/metabolism, sugar transport, chemotaxis, copper tolerance, iron homeostasis, and many others. While there is evidence of some regulatory overlap, each ArsR exhibits its own regulatory profile. Furthermore, evidence of a regulatory hierarchy was observed; i.e. ArsR1 represses arsR4, ArsR4 activates arsR2, and ArsR2 represses arsR3. Additionally and unexpectedly, aioB (arsenite oxidase small subunit) expression was shown to be under partial positive control by ArsR2 and ArsR4. Summarizing, this study demonstrates the regulatory portfolio of arsenite-activated ArsR proteins and includes essentially all major cellular functions. The broad bandwidth of arsenic effects on microbial metabolism assists in explaining and understanding the full impact of arsenic in natural ecosystems, including the mammalian gut.
Introduction
Arsenic (As) is rated by the United States Environmental Protection Agency as a priority environmental toxin because of its occurrence, toxicity, and potential for human exposure (ASTDR, 2016), and it is ranked among the top 10 environmental threats to human health (WHO, 2016). Arsenic contamination derives from natural geologic sources and anthropogenic inputs, but regardless of source, it is recognized that microorganisms are major drivers of As chemical speciation, which influences As toxicity and mobility (Inskeep et al., 2002; Rosen, 2002; Stolz et al., 2006; Kruger et al., 2013). Consequently, understanding how and why microbes react to As in their environment is important with respect to issues involving land resource management (Inskeep et al., 2002), as well as human medicine (i.e. gastrointestinal tract) (Coryell et al., 2018; McDermott et al., 2019).
A well-characterized microbial response to As involves resistance encoded by the ars operon, which at the minimum is composed of arsR, arsB (alternatively acr3), and arsC (Mukhopadhyay et al., 2002; Rosen, 2002; Slyemi and Bonnefoy, 2012). ArsR, encoded by arsR, is a DNA binding transcriptional repressor that regulates its own expression and that of the other genes in the same ars operon; arsB or acr3 encodes active efflux that is the primary mechanism for removing arsenite [As(III)] from the cell; and arsC encodes an arsenate [As(V)] reductase. When As(III) enters the cell through aquaglyceroporins, it interacts with basal (non-induced) levels of ArsR in the cell, causing ArsR to undergo a conformational change that results in release from its DNA binding site and thus opening the ars operon for transcription. This leads to significantly increased levels of ArsB/Acr3 and ArsC that constitutes the basic As resistance response (Mukhopadhyay et al., 2002; Rosen, 2002). Many other ars genes with variable roles in arsenic resistance have been recently reviewed (Andres and Bertin, 2016; Fekih et al., 2018).
In addition to the ars operon response, the microbial response to As is considered global (Andres and Bertin, 2016), yet the regulatory basis remains to be fully elucidated. In addition to ArsR control of the ars operon, the PhoRB two-component phosphate stress response (PSR) system has been shown to be the master regulator of the AioSR two-component system (Wang et al., 2018) that, along with AioX (Liu et al., 2012), controls expression of As(III) oxidase. We have also just recently reported how AioS regulatory controls extend beyond As(III) oxidation per se (Rawle et al., 2019). It is important to distinguish the ArsR-based response from the PhoRB–AioSR regulatory network that is induced when the cell senses low phosphate in its environment in addition to sensing As(III). This contrasts with ArsR-based control, which only requires As(III) and is insensitive to environmental phosphate.
Agrobacterium tumefaciens strain 5A carries two recognizable ars loci [Supplementary Figure 1, previously published by Rawle et al. (2019), but provided here for reader convenience], with two arsR genes located at each (Kang et al., 2016). These loci/genes in strain 5A have been a focal point in our past reports (Kang et al., 2012, 2015, 2016; Rawle et al., 2019) and continue to be of particular interest in this study because of significant regulatory and functional activity encoded by key genes that are of importance to arsenic-linked gene transcriptional regulation, arsenite oxidation, and phosphorus acquisition. ArsR1 and ArsR4 share 93% identity and 96% similarity, but are different from ArsR2 and ArsR3, which are more closely related to each other (78% identity and 86% similarity) (Kang et al., 2016). ArsR1 represses the nearby phoB-1 and pstS-1 genes (Kang et al., 2016) that are involved in the cell response to As(III), implying that the ArsR regulatory roles extend beyond that suggested by the decades of literature regarding ArsR function (Rosen, 2002). To further examine this, the current study utilized RNASeq to assess the regulatory bandwidth of each of the four characterized ArsR proteins in this bacterium. Wild-type cells were compared to isogenic arsR mutants grown under high phosphate culture conditions so as to distinguish the regulatory roles of these ArsRs from the PhoRB-AioXSR controls. In contrast to the current paradigm that describes ArsR proteins as only being repressors of ars operons, this study shows that these proteins are involved in regulating an extensive range of cell functions, and as such assists in explaining the regulatory underpinnings of bacterial global As responses.
Materials and Methods
Strains and Plasmids
Previously constructed in-frame deletion arsR mutants and lacZ reporter constructs in pLSP-KT2lacZ (Kang et al., 2012) were used for this study (Supplementary Table 1). β-Galactosidase reporter assays were conducted as previously described (Kang et al., 2012, 2016); however, prior efforts showed that an arsR3:lacZ construct does not up-regulate in response to As(III) (Kang et al., 2016) and so consequently no reporter assays were conducted for this gene. A lacZ:aioB construct was also used for aioB induction assays, and its construction was also as previously described (Kang et al., 2012).
Bacterial Growth Conditions
Agrobacterium tumefaciens strain 5A wild-type and ΔarsR mutants were maintained on minimal mannitol (MMNH4) agar or cultured in liquid MMNH4 (Somerville and Kahn, 1983). Reporter constructs were maintained by adding 500 μg/ml kanamycin. For RNASeq work, cultures were grown and incubated in liquid MMNH4 at 30°C in water bath incubators with aeration by shaking. As(III) was added at the concentrations indicated to initiate inductions. The growth of bacterial cultures was monitored as culture optical density by a SpectraMax microtiter plate reader (Molecular Devices, California). For these experiments, the MMNH4 was modified to contain 10-fold reduced iron to accommodate RNA yields. Normal MMNH4 iron levels (62 μM) were found to interfere with the RNA extraction and purification kit protocol, reducing RNA yields to unworkable levels. Reducing initial Fe3+ content to 6.2 μM for the 40-min inductions at the cell densities used does not constitute an iron starvation scenario (Hamza et al., 1998; Small et al., 2009; Strange et al., 2011), which would otherwise require Fe-specific chelators (e.g., 2,2′-dipyridyl) (Bollinger et al., 2001). Further, in the current study, transcription of genes encoding iron acquisition/metabolism functions in the wild-type strain displayed patterns that were roughly balanced between up-regulation and down-regulation at the highest As(III) or were specific to a particular ArsR protein in the mutant analyses. We interpreted this as meaning that under the experimental short induction conditions employed, the cells were responding to As(III) and not activating an iron starvation response.
RNA Extraction and Purification
Overnight cultures were diluted to OD595 = 0.1 and allowed to grow to an OD595 of 0.2 to ensure cells were in the growth phase when As(III) was added. Each treatment was incubated at 30°C in a water bath incubator aerated by shaking with the appropriate As(III) concentration for 40 min. Cells were collected by centrifugation (8000 × g for 5 min at 4°C), resuspended in 1 ml of Qiagen RNAprotect to protect cellular RNA and then centrifuged at 5000 × g at 4°C for 10 min. Cell pellets were suspended by repeated gentle pipetting in 200 μl of cold TE buffer (10 mM Tris–HCl and 1 mM EDTA, pH 8.0) containing 1 mg/ml lysozyme. RNA was then extracted using the Qiagen RNeasy Minikit following the manufacturer’s protocol, including on-column DNase digestion. RNA concentrations were measured with the SpectraMax microtiter plate reader, and RNA quality and purity were determined with an Agilent 2100 Bioanalyzer (Agilent Technologies). RNA was stored at −80°C until sequencing.
RNA Sequencing
Sequencing was completed at Brigham Young University (Provo, UT, United States). Briefly, ribosomal RNA was removed using the Illumina Ribo-Zero rRNA removal kit for Gram-negative bacteria. The resulting RNA was prepared for sequencing using the Illumina TruSeq Stranded Total RNA Sample Prep and fragmented into 50-bp segments and reverse transcribed into cDNA. Sequence adapters used to initiate DNA polymerase binding were ligated to the cDNA, and the sequences were amplified with PCR. The cDNA library was sequenced in high output mode using Illumina sequencing technology (Illumina HiSeq 2500 sequencing platform), generating ∼12 million reads per sample. All sequence reads have been submitted to Genbank and can be found as BioSample accession: SAMN10686493, BioProject ID: PRJNA512538.
Sequence Analysis
BowTie (Langmead and Salzberg, 2012) was used to remove any remaining ribosomal RNA sequences. Quality control was completed with FASTQC (Version 0.11.9; Andrews, 2014) and adaptors trimmed with Trimmomatic (Version 0.36; Bolger et al., 2014). mRNA sequences were aligned and quantified using Kallisto (Version 0.43; Bray et al., 2016), including 100 bootstraps for assessment of technical variance. Parameters for alignment were as follows: kmer size of 31; sequence length 180 and standard deviation of 20; and the Ensembl Bacteria genome for A. tumefaciens 5A (ASM23612v2) (Kersey et al., 2018). The R package Sleuth (Pimentel et al., 2017) was used to perform differential gene expression analysis using the bootstraps performed in Kallisto to adjust for technical variance. Groups were compared using the Wald Test, and p values were adjusted for multiple comparisons using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995). Reads were normalized to transcripts per kilobase million (TPM) by dividing transcript number by gene length (in kilobases), and then reads per kilobase were counted within a sample and that number was divided by 1 million for a per-million scaling factor. Only differentially expressed genes with TPM > 1, a fold change greater than ± 2, and a q value ≤ 0.05 were included in the analysis. There were numerous instances where expression changes were statistically significant but failed to reach the fold change criterion and sometimes observed for genes within (apparent) operons in which other genes changed expression by > 2.0. Gene IDs were converted using UniProt (Consortium, 2019) to access available E.C. numbers, gene ontology terms, and gene names. Transcript names from Sleuth were input to UniProt for gene annotation purposes, and any genes annotated as encoding “unidentified proteins” were individually BLAST searched (Coordinators, 2016). The assignment of the function category was also based on UniProt annotation.
Motif Analysis
The MEME suite of tools (Bailey et al., 2009) was used for identifying DNA sequences as putative ArsR1 binding sites to facilitate repression. Initial searches identified putative binding motifs in a DNA region spanning the pstS1 and phoB1 genes (Supplementary Figure 1) wherein we previously used electrophoretic mobility shift assays to show strong evidence of at least two ArsR1 binding sites (Kang et al., 2016). These motifs were then used as queries to scan the upstream 300 nucleotides of genes (i.e. an area similar in size to the pstS1–phoB1 region) that exhibited transcriptional patterns consistent with them being transcriptionally repressed by ArsR1. This was done using the Find Individual Motif Occurrence algorithm in MEME Suite (5.0.1). FIMO utilizes log-posterior odds scoring and position-specific priors to search for a given motif (Grant et al., 2011; Cuellar-Partida et al., 2012). Motif search and queries were conducted using default settings and a p value ≤ 0.001 indicating a significant match. Representative sequence logos were constructed using WebLogo (2.8.2, Berkeley, CA, United States).
Results
Induction Optimization
arsR β-galactosidase reporter constructs were used to optimize As(III) concentration and induction time for each arsR. These data were then used for characterizing their regulatory activities by RNASeq (below). Each reporter had a distinct profile and level of expression. The non-induced (constitutive) expression levels ranged from 26 Miller units (MU) for arsR1:lacZ to 190 MU for arsR2:lacZ (Figure 1A). Reporter response varied (Figure 1A), with arsR4:lacZ showing the largest response relative to the other reporters. Other assays examined optimal induction times (Figure 1B). After considering reporter responses in both assays, As(III) concentrations selected for the induction treatment were 100 μM for arsR1:lacZ, 50 μM for arsR2:lacZ, and 75 μM for arsR4:lacZ and induction periods of 40 min. By limiting the As(III) exposure period, the goal was to induce an As(III) transcriptional response but constrain/avoid potential secondary wave(s) of transcriptional responses from other potential transcriptional regulators that might be controlled by these ArsRs. Further, extending incubation times did not appreciably enhance arsR reporter expression levels. Since ArsR proteins are typically viewed to autoregulate their own gene, we inferred that optimized induction of these arsR genes would also reflect near-optimal As(III)-triggered release of these ArsR from other similar DNA binding sites in the genome.
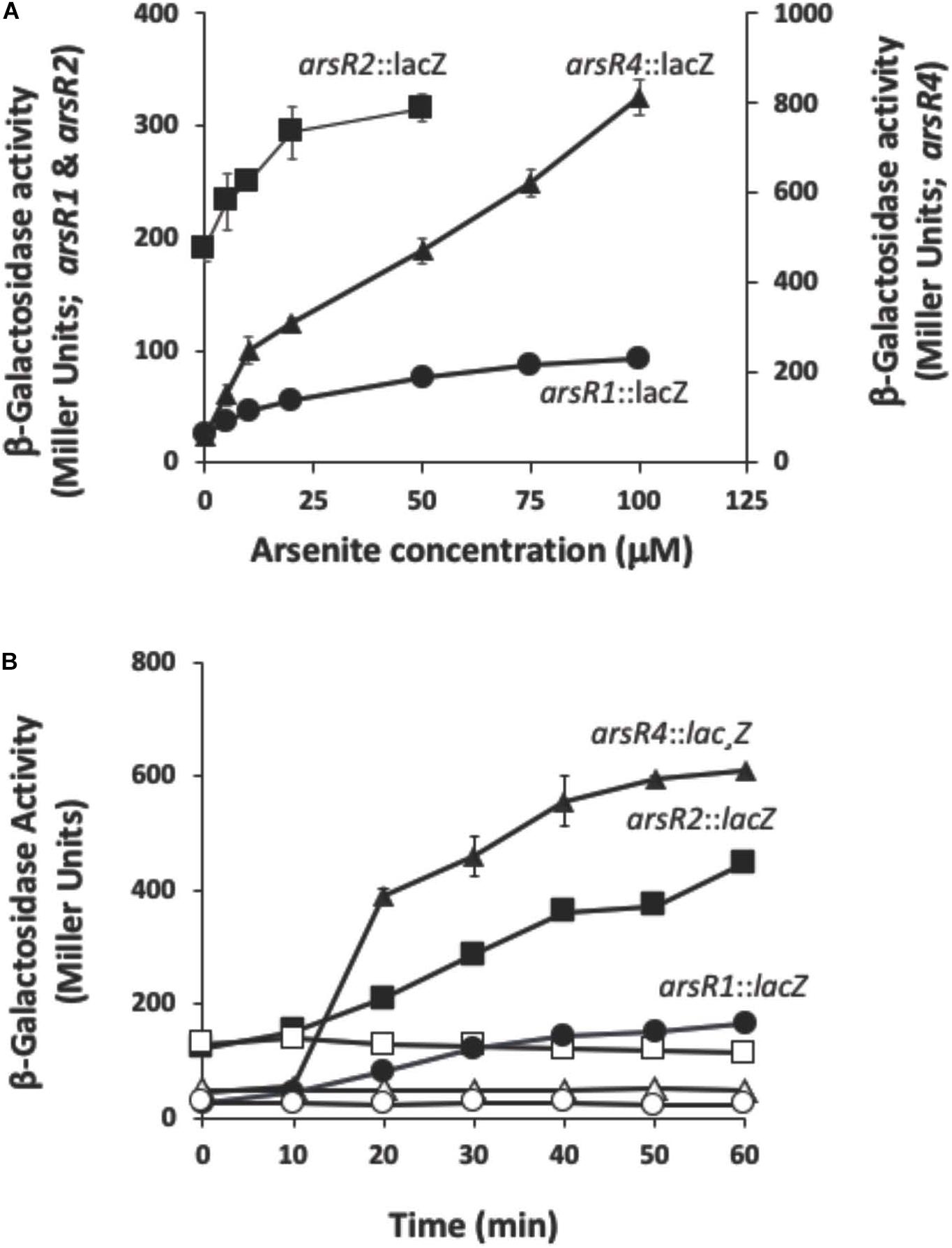
Figure 1. Reporter gene assays monitoring expression of arsRl, arsR2, and arsR4 as a function of As(lll) exposure and time. (A) Reporter gene assay monitoring expression as a function of As(lll) exposure. (B) Reporter gene assay monitoring expression as a function of time. Filled symbols, +As(lll); open symbols, –As(lll) controls. Where visible, error bars indicate ± 1 standard deviation.
Wild-Type Gene Transcription Changes
Based on the reporter gene assays, initial RNASeq experiments compared the response of −As(III) and +As(III) wild-type A. tumefaciens cells exposed to 50, 75, and 100 μM As(III) for a 40-min incubation period. A total of 138 genes were up-regulated and 111 genes were down-regulated in the wild-type strain exposed to As(III). Table 1 emphasizes selected functions that were represented by multiple genes (i.e. suggests non-random, organized, cell-wide response) as well as genes/operons depicted in Supplementary Figure 1 (see Supplementary Table 2 for a complete list of genes with UniProt gene identifiers). For reader convenience, expression data in Table 1 also include expression data for each of the four arsR mutants to facilitate a direct contrast to the wild-type strain and will be further discussed as total responses below. As expected based on prior research of numerous organisms (Andres and Bertin, 2016; Fekih et al., 2018) and the reporter assays (Figure 1), ars genes involved in As resistance showed increased expression in response to As(III) (Table 1). Up-regulation of the ars genes was consistent with what we have previously documented for strain 5A (Kang et al., 2012, 2016) using an orthogonal technique (lacZ reporters and reverse transcriptase PCR for a subset of the genes shown in Supplementary Figure 1) (Kang et al., 2012, 2016). Since these RNASeq-based ars gene transcriptional responses were observed in the same batches of mRNA used in the RNASeq libraries for all other transcriptional responses, they serve as internal controls to validate the entire data set. Except for aioB (see below), the aio genes [involved in As(III) oxidation] were not up-regulated because the cultures were incubated in high phosphate media, which inhibits their induction (Kang et al., 2012; Wang et al., 2018). This is consistent with prior work and thus also provides validation.
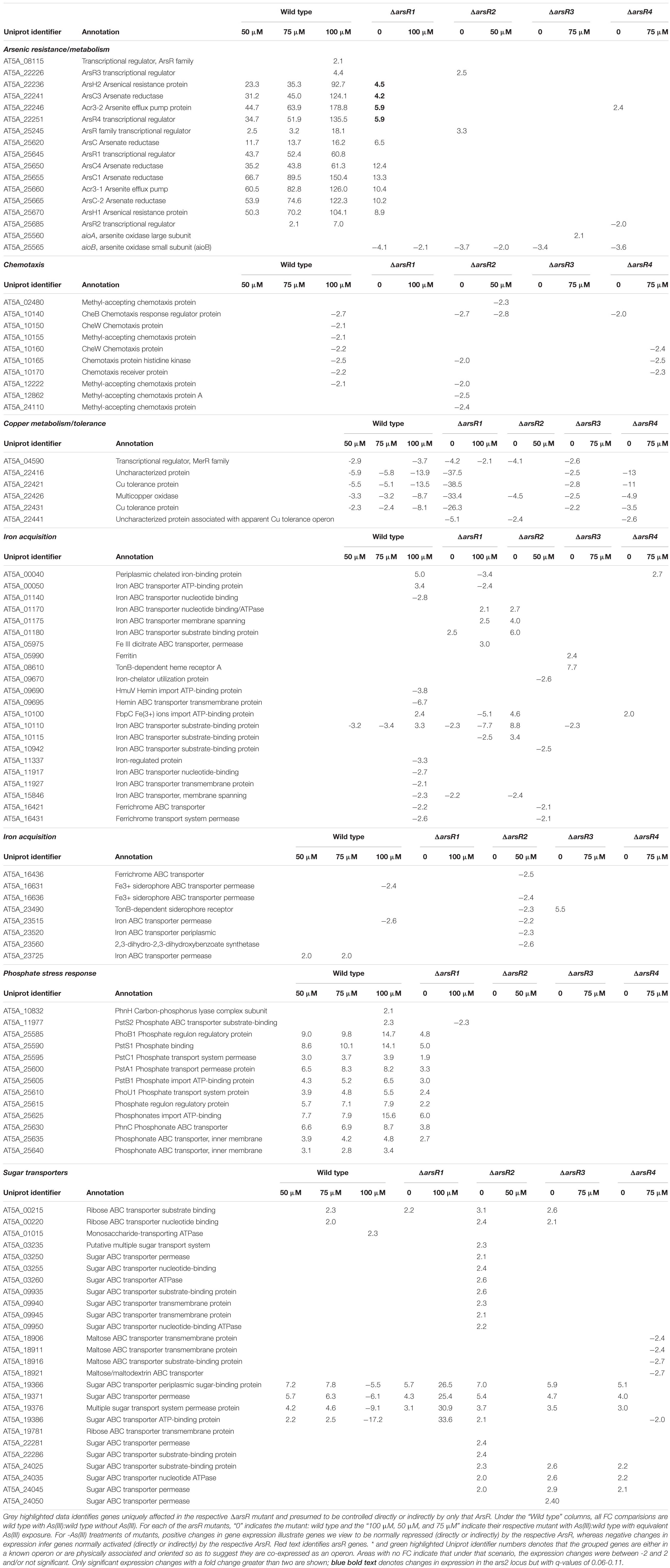
Table 1. Gene expression profiles of selected major functional groups in the wild type strain 5A and the four arsR mutant derivatives as a function of As(III) exposure.
The group of validation genes includes pst/pho/phn1 that encode aspects of phosphate and phosphonate uptake/transport and are proximal to the ars1 locus (Supplementary Figure 1). These genes were up-regulated as observed previously for pstS1 and phoB1 (Kang et al., 2012, 2016), although, by contrast, no changes were observed for the separate pho/pst2 locus that controls the formal PSR and is not responsive to As(III) (results not shown). Other affected functional categories include the up-regulation of genes involved in DNA replication (Supplementary Table 2), although these responses occurred primarily at the highest level of As(III) applied (100 μM). Several genes annotated as membrane proteins were also affected, including numerous ABC-type transporters (Supplementary Table 2). For genes up-regulated by the lowest As(III) level, transcription levels often increased with increasing As(III) exposure levels; e.g., all ars genes (As resistance) and more subtly for many pst/pho/phn1 locus genes (Table 1 and Supplementary Table 2).
Many genes/functions were down-regulated in As(III)-exposed cells. However, this primarily occurred at the highest As(III) level, 100 μM (Table 1 and Supplementary Table 3). Interesting examples include copper metabolism/tolerance and chemotaxis (Table 1). Many iron transport/regulation genes were primarily down-regulated at the highest As(III) level as well (Table 1). We interpreted this to mean that the cells were not iron-stressed in the 40-min inductions, but rather that As(III) exposure interacts with iron homeostasis functions (see more below, arsR3 mutant). Other down-regulated functions include membrane proteins, β-lactam resistance (five genes), transcription regulation (four genes), and molybdenum transport (four genes, Supplementary Table 2). There were also instances of As(III) concentration having reversible effects; e.g., one sugar transporter operon is up-regulated at the lower As(III) levels, but then repressed at the highest As(III) level (Supplementary Table 2, rows 326–329).
Altered Gene Expression in the arsR Deletion Mutants
The next phase of analysis assessed regulatory control by the different ArsRs. Each mutant was compared to wild type. The mutant transcriptomics experiments were optimized for As(III) concentrations and exposure times as determined above (Figure 1). Since the arsR3 reporter does not respond to As(III), 75 μM As(III) was selected as an average used for the other arsR genes when characterizing its regulatory profile.
Based on current models of how ArsR-type repressors control gene expression, removing the repressor proteins (i.e. deletion mutations as in the current study) should result in constitutive expression of genes that are normally repressed by that protein in the absence of the de-repressing ligand [As(III) in this instance]. In the context of this study where As(III) was not added, increased transcription in the mutant relative to the wild type would indicate that the gene is normally repressed by the specific ArsR, whereas decreased expression in the mutant suggests that the gene is activated (directly or indirectly) by the respective ArsR. Overall, there were 482 genes significantly influenced by one or more of the ArsRs in the absence of As(III). A total of 295 genes were classified as repressed and 187 genes were classified as activated, spanning a broad range of cell physiology. The volcano plots (Figure 2) offer a comparative visual assessment of total gene expression patterns for each mutant relative to the wild type in the absence of As(III). Both positive and negative changes in gene transcription were observed for all four mutants, with ArsR2 exhibiting patterns that suggest it has the greatest influence over cellular gene expression (220 genes total), whereas ArsR3 influences the fewest (61 genes) (Figure 2).
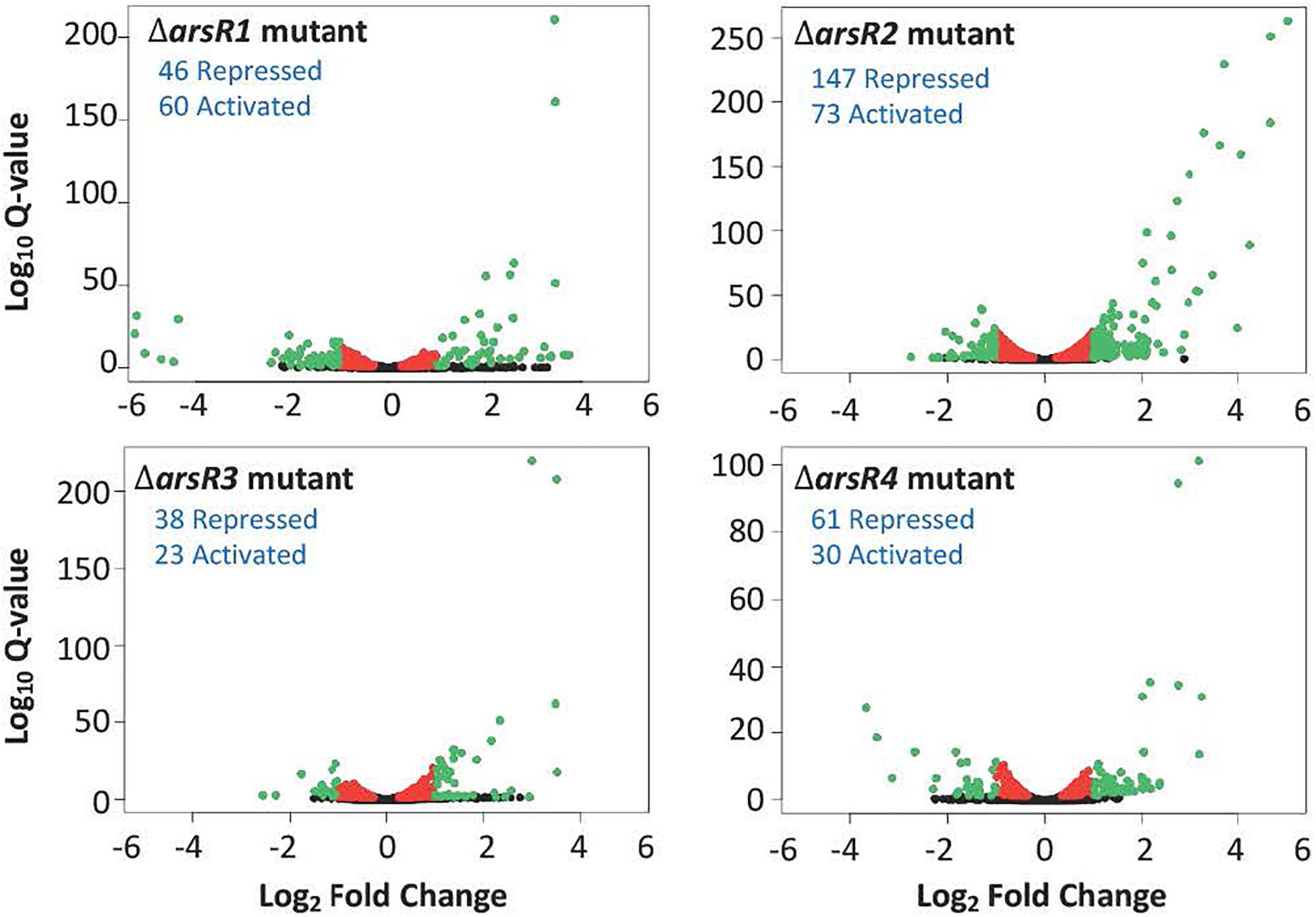
Figure 2. Volcano plots illustrating the influence of each ArsR on genome-wide transcription in the absence of As(lll). In each case, expression changes are calculated as the ratio of the mutant compared to the wild type. Red colored dots indicate genes that were significantly changed (q-value < 0.05), but less than two-fold. Green dots indicate genes that were significantly changed (q-value < 0.05) and had a fold change > + 2.0. Blue text indicates the number of genes with increased or decreased expression and are presumed to be activated either directly or indirectly by the respective ArsR in the wild type cell.
Surprisingly, aioB (AT5A_25565) expression was altered in all arsR mutants, and in all cases expression was reduced, indicating that these ArsrR proteins have an activating influence (Table 1). This suggests that in the wild-type strain, one or more of these ArsR proteins have some type of positive influence over aioB expression. This was unexpected because strong experimental evidence has shown that the aioBA genes are controlled by AioXSR (reviewed by Andres and Bertin, 2016). Therefore, additional experiments were conducted under full induction potential conditions [i.e. low phosphate plus 100 μM As(III)] to determine if any of these ArsRs exert control over aioB. As can be seen in Figure 3 and as we have reported previously (Kashyap et al., 2006; Kang et al., 2012), the aioB:lacZ reporter in the wild-type strain failed to induce in the absence of As(III); however, it was induced at the expected times and levels when the cells were incubated with As(III). The same induction profiles were observed in the arsR1 and arsR3 mutants; however, full induction was not achieved in the arsR2 and arsR4 mutants, demonstrating ArsR2 and ArsR4 are involved at some level in positively regulating aioB (Figure 3 and Table 1).
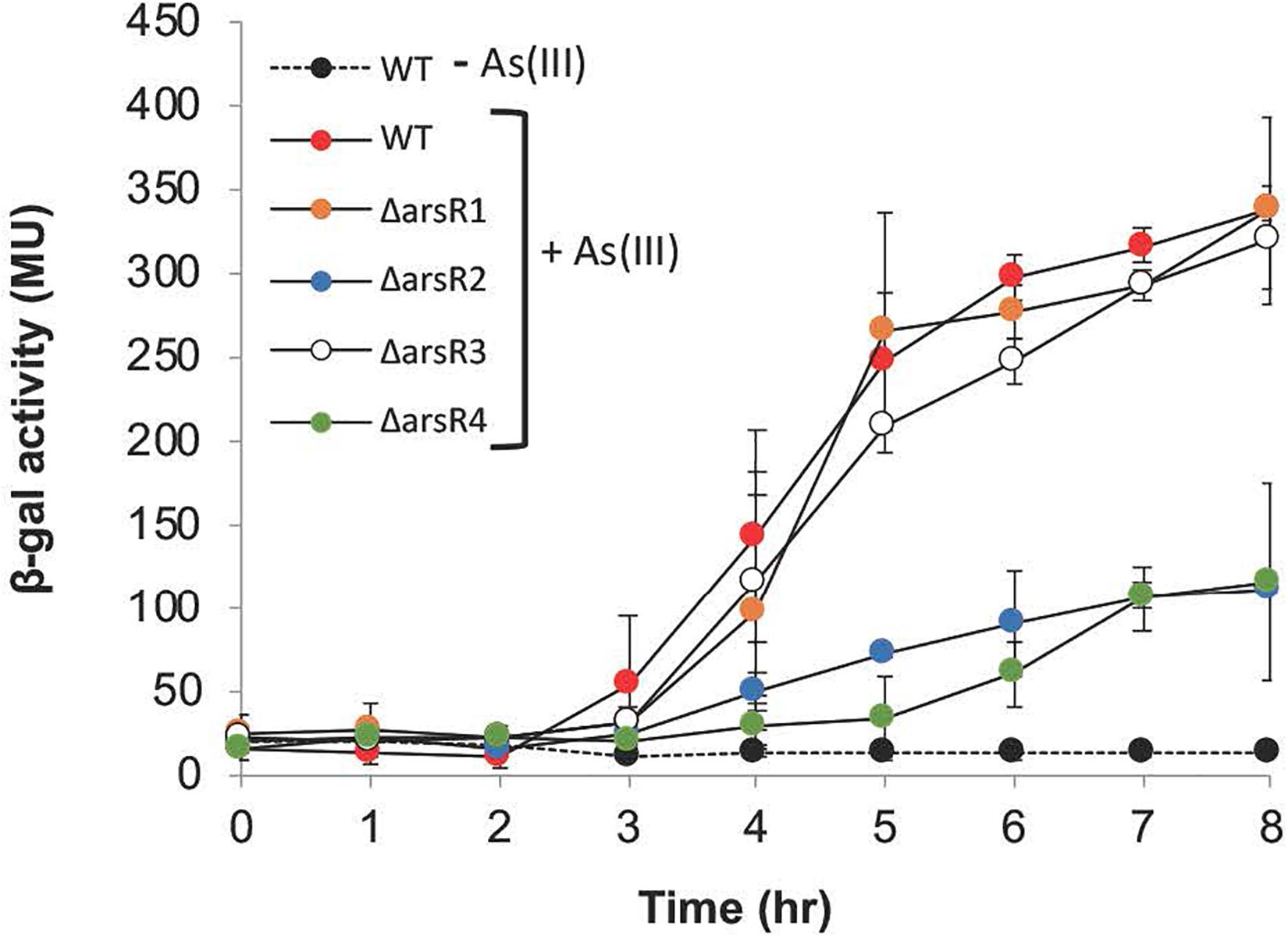
Figure 3. Reporter gene (aioB:lacZ) assays monitoring aioB induction as a function of arsR genotype, As(lll) exposure and time. Where visible, error bars indicate ± 1 standard deviation.
Specific Functions Associated With Individual ArsRs
Table 1 highlights mutant:wild-type comparisons with respect to what appears to be major, coordinated transcriptional changes for several cellular functions. Comprehensive lists for each arsR mutant are provided in Supplementary Tables 3–6, and demonstrate that virtually every aspect of cell physiology is influenced. ArsR1 appears to be the major repressor of ars1 arsenic resistance locus/operon (Supplementary Table 3) and the PSR related activities encoded by the pho/pst1 operon (Supplementary Figure 1), which is physically located near the ars1 locus (Supplementary Figure 1). ArsR1 control over the pst/pho/phn1 locus (Table 1) is consistent with our prior DNA binding experiments, which showed that purified ArsR1 would bind at two distinct locations between pstS1 and phoB1 (Kang et al., 2012) (please refer to Supplementary Figure 1). Expression changes of genes in the distal ars2 locus, including arsR4, also reflect repression by ArsR1 (Supplementary Table 3). Additionally, ArsR1 appears to be involved at some level in activating (directly or indirectly) functions such as amino acid metabolism, copper tolerance, iron acquisition, nickel transport, pilus assembly/Type VI secretion, and succinoglycan biosynthesis (Supplementary Table 3).
ArsR2 exhibits repressor activity controlling cobalamin synthesis, heat/cold shock responses, iron acquisition, and six different sugar transport operons. Additionally, it appears to be a repressor of arsR3. Indeed, some of the genes apparently activated by ArsR2 are also activated by ArsR3 (compare Supplementary Tables 4, 5). ArsR2 regulatory control of ars genes is greatly reduced relative to ArsR1, but ArsR2 does exclusively control the uncharacterized arsR gene (AT5A_25245). This demonstrates regulatory interaction between these ArsR proteins, as was observed for ArsR1 exerting control over arsR4 (Table 1). Other genes up-regulated in the ΔarsR2 mutant (presumed repressed in wild type) encode transporters for oligonucleotides, amino acids, iron, and six disparately located ABC-type sugar transporters (Supplementary Table 4). Functions apparently activated by ArsR2 in the absence of As(III) include various other transport activities (amino acid, nitrogen, iron, and sulfate), copper tolerance, chemotaxis, queuosine biosynthesis, purine metabolism, membrane proteins, amino acid biosynthesis, synthesis of ribosomal proteins, glycosyl transfer, and signal transduction (Supplementary Table 4).
Transcript profile comparisons showed that loss of ArsR3 was associated with the fewest changes; 38 repressed and 23 activated (Table 1 and Supplementary Table 5). Functions apparently normally repressed by ArsR3 involve nucleotide triphosphate metabolism and transport of different sugars (Supplementary Table 5). Cell activities (in)directly activated by ArsR3 include benzoate degradation, copper tolerance, and chemotaxis. Of particular note, we draw attention to the very large number of iron acquisition genes down-regulated in the arsR3 mutant exposed to As(III); 34 genes in total, including seven full operons (Supplementary Table 5).
A comparison of the transcriptional patterns of the arsR4 mutant indicated that expression of 91 genes was altered (Supplementary Table 6). Sixty-one showed increased expression, suggesting that they are normally repressed by arsR4. This includes acr3-2 directly downstream from arsR4. The profile of regulated genes was similar to the other ArsR proteins, with the exception of universal stress response and a Fix operon associated with N2 fixation (Supplementary Table 6). It is worth noting that expression of arsR2 decreased, indicating that ArsR4 activates ArsR2, either directly or via a secondary response. Further, chemotaxis and copper tolerance are a regulatory target for ArsR4, again as an activator as well as for a large number of genes/operons encoding aspects of iron acquisition or iron-related regulatory activity (e.g., FecR).
Genes Regulated by Multiple ArsRs
There are numerous instances of the same gene (or apparent operon) being influenced in two or more arsR mutants. Examples here include evidence of (i) Acr3-2 arsenite efflux pump being primarily repressed by ArsR1, but ArsR4 also influences expression; (ii) the arsenite oxidase small subunit (aioB) expression was decreased in all four ArsR mutants; (iii) expression of several chemotaxis genes activated by both ArsR2 and ArsR4; and (iv) several genes involved in copper resistance/metabolism being apparently activated by three different ArsRs in the absence of As(III), with the influence by ArsR1 being strongest (Table 1).
Mutant Responses to As(III)
The next step was to analyze the response of the arsR mutant strains to As(III). As one would expect given the regulatory overlap described to this point, the observed patterns of gene regulation were complex. The majority of the apparent activation effects for ArsR1 were exhibited only in As(III)-treated cultures (Supplementary Table 3), whereas in contrast very few of the ArsR4-controlled genes displayed any response to As(III) (Supplementary Table 6). Overall, four basic patterns were observed. First, expression of some genes/operons increased relative to wild type in the absence of As(III) but were the same as wild type in As(III)-exposed cultures. This expression pattern is consistent with constitutive transcription in the absence of the relevant ArsR repressor leading to fully open transcription and is not affected by the ArsR ligand, As(III), per se. Prominent examples of this pattern include the arsenic response and pho/pst/phn1 locus, copper tolerance genes in the arsR1 mutant (Table 1 and Supplementary Table 3), and nearly all of the gene/operons controlled by ArsR4 (Supplementary Table 6). A second general pattern involved genes/operons exhibiting altered expression (relative to wild type) in the absence of As(III), that was changed even more in the same direction upon As(III) exposure. An example of this pattern is the sugar transporter system (AT5A_19366, AT5A_19371, AT5A_19376, AT5A_19386) in the arsR1 mutant (Supplementary Table 3). The third pattern involved no deviation from wild type in the absence of As(III), followed by a significant change in expression with As(III). The best examples of this pattern occurred in the As(III)-treated arsR3 mutant wherein decreased expression was observed for iron acquisition/homeostasis (33 genes) and chemotaxis/motility (11 genes) (Supplementary Table 5). This suggests that ArsR3 normally is somehow involved in the activation of these genes, but only in As(III)-exposed cells; however, we note that expression of these same genes was not significantly altered in the wild-type cultures, regardless of As(III) treatment level (Table 1 and Supplementary Table 2). Finally, a fourth pattern observed in all mutants involved genes exhibiting increased expression in the absence of As(III) and an additional increase upon As(III) exposure. This fourth pattern implies the involvement of the relevant ArsR protein, potentially working with another, weaker regulator whose expression is As(III) sensitive.
Motif Analysis
Our prior work found that ArsR1 exerts repressive control over pstS1 and phoB1 (please refer to Supplementary Figure 1) and showed that purified ArsR1 binds two sites in the DNA region between these genes (Kang et al., 2012). These genes are divergently transcribed, and their ATG translational start sites are separated by 363 nucleotides. In the current study, this region was examined to identify potential ArsR1 binding site motifs and then used these motifs as queries to scan the DNA upstream of each gene exhibiting transcriptional behavior consistent with being repressed by ArsR1 (identified in Table 1 and Supplementary Table 3). MEME software (Bailey et al., 2009) identified three distinct motifs that vary between 9 and 29 bp in length and in their degeneracy (Supplementary Figure 2). The two more conserved motifs occurred twice in the 363-nucleotide region whereas the more degenerate motif was found in four locations (Supplementary Figure 2A). Using these motifs as queries resulted in a total of 40 significant matches being found associated with the upstream regions of scanned genes (Supplementary Table 3). Based on location, orientation, and prior research (Kang et al., 2012; Kang et al., 2016), it is reasonable to predict that there are 12 operons repressed by ArsR1 (accounting for 47 genes; Supplementary Table 3). We found at least one or more of these motifs located within the upstream DNA of the promoter proximal gene for nine of these operons (Supplementary Table 3). This includes arsR1 and the phoB1 and pstS1 genes that we have studied extensively and shown to be under the control of ArsR1 (Kang et al., 2012; Kang et al., 2016). Alignment of the predicted motifs revealed significant conservation for the shorter predicted motifs, whereas the longer motif was much more variable with the exception of what may be key nucleotides sharing the same spacing (Supplementary Figure 2B).
Discussion
Agrobacterium tumefaciens or very closely related taxa have been frequently isolated from arsenic-contaminated soils throughout the world, including Korea (Chang et al., 2010), Australia (Santini et al., 2000), China (Cai et al., 2009a), India (Sarkar et al., 2013), and the United States (Rhine et al., 2007). A. tumefaciens strain 5A, used in this study, came from arsenic-contaminated soils in Montana, United States (Macur et al., 2004). Strain 5A has been used as a model organism to identify the AioSR two-component signal transduction system (Kashyap et al., 2006), the periplasmic As(III) binding protein AioX (Liu et al., 2012), co-regulation of aioBA induction and elements of the PSR (pst1-phoB1 locus) (Kang et al., 2012), ars locus comparisons (Kang et al., 2016), and PhoR regulation of the aioSRBA (Wang et al., 2018), and to define the PhoR and AioS regulation profiles (Rawle et al., 2019). As such, this organism can reasonably be considered an ecologically relevant model to understand and predict how bacterial metabolism and function are influenced or disrupted by As(III).
ArsR redundancy raises obvious questions regarding the specific role of each in regulating gene expression. Questions regarding ars gene/operon redundancy have been posed previously. Páez-Espino et al. (2015) were able to show that temperature influenced the expression of two different ars operons in Pseudomonas putida KT2440; however, we were unable to link temperature to the expression of the ars genes in A. tumefaciens 5A (Kang et al., 2016). Evolution models would argue that genetic and functional redundancy requires some level of positive selective pressure (Francino, 2005). Sorting out explanations for such apparent paradoxes, in particular regulators such as ArsR, is not easy but we took the most direct approach by mutating each arsR associated with the ars1 and ars2 loci (Supplementary Figure 1). This allowed for a systematic parsing of the role(s) each ArsR might contribute to regulation at a minimum of three levels: (i) contributing to the global response to As(III); (ii) regulation within the ars1/pho/pst/phn1/aio island; and (iii) cross-regulation between the ars1 and ars2 loci. Each ArsR is involved in controlling its own suite of genes, though there appears to be some regulatory overlap (Table 1 and Supplementary Tables 2–5), which might be particularly predicted for ArsR1 and ArsR4, which are highly related phylogenetically, sharing 93% identity and 96% similarity (Kang et al., 2016).
Each of these ArsRs affects a large number of cellular functions (Table 1 and Supplementary Tables 2–5) and thus offers an initial, reasonable explanation for global As(III) responses not controlled by AioS or PhoR (see below). Noteworthy are functions represented by numerous genes or whole operons dispersed throughout the genome, implying an organized, non-random cell response. Prominent examples include regulation of copper tolerance, chemotaxis, and iron acquisition/homeostasis (Table 1 and Supplementary Tables 1, 5). Also noteworthy is that the mutant transcriptional patterns suggest that an ArsR regulatory heirarchy may be in place (Figure 4), wherein ArsR1 is a repressor of arsR4, ArsR4 positively influences the expression of arsR2, and ArsR2 represses arsR3 (Table 1 and Figure 4).
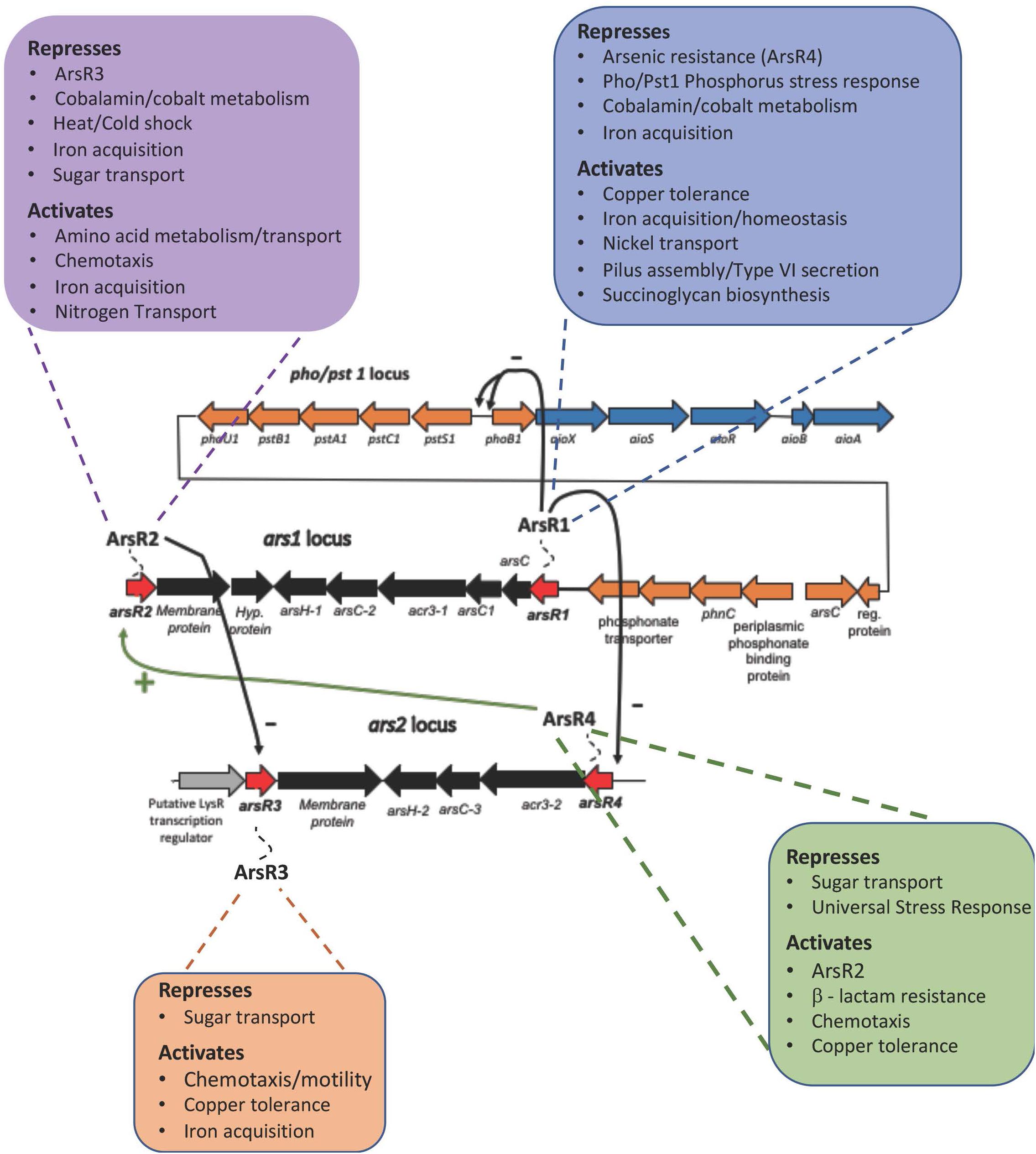
Figure 4. ArsR Regulatory Hierarchy. Transcription patterns are consistent with a regulatory hierarchy where ArsRl represses arsR4, ArsR4 activates arsR2, and ArsR2 represses arsR3. Only select, subset of functions are included in this figure to emphasize the broad bandwidth. The functions that are shown in this figure are exclusive to the ArsR protein; i.e. As response is exclusively regulated by ArsRl and displayed no changes due to ArsR2, ArsR3, and ArsR4. The “–” symbol indicates repression and the “+” symbol indicates activation.
Of the known As(III)-sensitive response systems, ArsR has been well studied by Rosen and colleagues for its role in regulating arsenic resistance encoded by the ars operon (Wu and Rosen, 1993; Rosen, 1995; Oremland and Stolz, 2003; Andres and Bertin, 2016). However, given the significant cellular response to As(III) (reviewed by Andres and Bertin, 2016), it was important to examine the ArsR repressor to determine the full extent of its regulatory activity. As we have clearly demonstrated here, ArsR-regulated gene expression in response to arsenic extends well beyond the current ars operon control paradigm. We present a new model that is considerably more complex, encompassing a wide range of cellular functions to varying degrees (Figure 4). ars operon redundancy has previously drawn interest (Ordóñez et al., 2005; Branco et al., 2008; Fernández et al., 2014; Páez-Espino et al., 2015; Kang et al., 2016), but functional explanations have been limited. This ars operon redundancy is additive for As(III) resistance (Kang et al., 2016) and temperature specificity has been shown to be relevant in Pseudomonas putida (Páez-Espino et al., 2015), although not in A. tumefaciens 5A (Kang et al., 2016).
Motif searches were conducted over a relatively broad span of 300 nucleotides upstream from the annotated translational start sites to determine if the ArsR1-regulated genes shared commonality in regard to potential targets for transcriptional regulatory elements. Query sequences for these searches were based on potential motifs identified within the pstS1–phoB1 spanning region wherein we previously identified at least two ArsR1 binding sites using electrophoretic mobility shift assays (Kang et al., 2012). We note that with the exception of the TCGTCG–TGTC motif, which was near the promoter regions of both pstS1 and phoB1, none of these motifs were located within a region closely associated with what would be reasonably predicted to be the -10/-35 region for these gene/operons; most were at least 100–150 nucleotides upstream. While there was significant sequence conservation for the two smaller motifs, none of these motifs correspond to the imperfect 12-2-12 inverted repeats model (Busenlehner et al., 2003; Supplementary Figure 2).
Co-localization of ars/pho/pst/aio genes (Supplementary Figure 1) is not unique to strain 5A, as it has been documented in many bacteria and referred to as an “arsenic island” (Li et al., 2013). This implies the likelihood of interrelatedness between arsenic and phosphorus metabolisms and that some level of regulatory synergy might be anticipated (Li et al., 2012, 2013). Our prior work concerning this island indicated that this occurs in strain 5A and the demonstrated connectivity in the current study now comprehensively confirms these prior observations and serves to validate the transcriptional changes observed throughout the current data set. In this context, we note that ArsR regulation only concerns the pho/pst1 locus, which is up-regulated primarily by As(III), not the pho/pst2 operon located elsewhere in the genome (Supplementary Figure 1) and should be regarded as the true phosphorus-stress response operon found and characterized in other Gram-negative bacteria (Hsieh and Wanner, 2010). We hasten to add that co-regulation between ars, aio, and pho/pst1 does not support the claim (Wolfe-Simon et al., 2010) that arsenate can functionally replace phosphate in structures such as nucleic acids or ATP metabolism (Rosen et al., 2011 Bioessays).
While induction of As(III) oxidation is controlled by the PhoRB and AioXSR systems (Kang et al., 2012; Wang et al., 2018; Rawle et al., 2019) and requires phosphate-limiting conditions, ArsR-based regulation is not influenced by phosphate levels (nor redox conditions) and consequently the role(s) of the ArsRs in this organism can be separated from these other As(III)-sensitive regulatory systems. Hence, the high phosphorus media used here (12 mM) avoids complications resulting from the global nature of the PSR (Hsieh and Wanner, 2010; Rawle et al., 2019) as well as transcriptional controls expected from AioXSR (Kang et al., 2012; Wang et al., 2018; Rawle et al., 2019). At a very general level, some As(III)-affected gene functions in A. tumefaciens 5A were also observed in microarray studies of “early phase” H. arsenicoxydans described by Cleiss-Arnold et al. (2010), which were not initially exposed to PSR conditions (as opposed to the “late phase” cells, which were). However, there are also many differences observed between H. arsenicoxydans and strain 5A, which may derive from the increased sensitivity of RNASeq in the current study as compared to microarrays. Some examples include perturbation of many sugar transporters, opposite responses for copper tolerance, ± regulation of iron acquisition, and down-regulating molybdenum transport (Supplementary Table 2).
The structural genes for As(III) oxidase (aioBA) have thus far been observed to be co-expressed in wild-type organisms as a two-gene operon – as would be expected (e.g., Kashyap et al., 2006; Cai et al., 2009b). However, altered aioB expression in all four arsR mutants in the absence of As(III) was unexpected and suggests low-level ArsR-linked constitutive expression of aioB in the absence of As(III). At this juncture, it is unclear why/how these genes are differentially expressed. Studies describing aioBA expression in strain 5A have consistently shown the requirement of As(III) and to be under the control of AioXSR (Kang et al., 2012; Wang et al., 2018). Here, aioB transcription was observed under high P conditions, without aioSRX up-regulation, in the absence of As(III) (Table 1), excluded aioA, and with ArsR2 and ArsR4 apparently normally playing some type of activation role(s) when the cell is exposed to As(III) (Table 1 and Figure 3). At present, we are unable to suggest a mechanism to account for this apparent ArsR positive regulatory effect. Unlinked expression of aioB from aioA could potentially be explained by a repressor that binds directly downstream of aioB and that blocks transcription readthrough to aioA – but apparently only under high phosphate conditions. Clearly, significant work remains.
The inference of ArsR positive regulator of gene function is novel. The regulatory profiles of all ArsRs in strain 5A include various transcriptional regulators (Supplementary Tables 3–6), including genes annotated as activators and thus are likely involved in this context; i.e. de-repression of a transcriptional activator would lead to secondary level activation. These putative regulators might also be active as transcriptional modulators. An example of this can be seen when comparing As(III)-exposed wild type against the ΔarsR4 mutant. Genes involved in chemotaxis and iron acquisition/homeostasis were down-regulated in the mutant (Table 1 and Supplementary Table 6), which would lead to the expectation that they are up-regulated in the wild-type cell containing ArsR4. However, the expected up-regulation appears modulated, by perhaps another regulator, such that there is no change in the wild-type cell at lower As(III) levels or indeed down-regulated at the highest As(III) level. The biological significance and cellular response logic is obscure even though the data demonstrating the possibility of it occurring is not.
One final point bears consideration. The novel observations reported in this study have relevance beyond soil or water environments where microbe–arsenic interactions have traditionally been studied. Initial work examining the impact of arsenic on the gut microbiome has clearly established that there are numerous and significant effects, but thus far the basis for these effects is unknown. An initial fate of ingested arsenic is to interact with the gastrointestinal tract microbiome wherein numerous transformations are possible (Coryell et al., 2018; McDermott et al., 2019) and that will have consequences for the host as well as other microbiome members (McDermott et al., 2019). Initial work examining the impact of arsenic on the gut microbiome have clearly established that there are numerous and significant effects, including altered microbiome structure and composition as well as metabolomic profiles (Lu et al., 2014; Chi et al., 2017, 2019a,b; Liu et al., 2019); however, thus far the basis for these changes is unknown. The ars genes/operons of Gram-positive and Gram-negative organisms are quite common in the human gut microbiome (McDermott et al., 2019), and there is no reason to view their transcriptional control to be any different to that shown in numerous pure culture isolates from virtually every other environment. Now, in addition to the specific ars-encoded functions, the very extensive profile of functions influenced by the ArsR protein documented herein offers a qualitative indication of what might be expected in the human gut microbiome and assists in explaining the very significant microbiome metabolic changes observed in different mouse studies (Lu et al., 2013; Guo et al., 2014; Lu et al., 2014; Richardson et al., 2018; Chi et al., 2019c; Liu et al., 2019).
Summarizing, the RNASeq experiments described herein illustrate that arsenic-stimulated ArsR regulation extends well beyond current paradigms of bacterial arsenic resistance (i.e. the ars operon model). Rather, ArsR regulation involves genes/operons representing an extraordinary array of cell functions (Figure 4), leading us to suggest that it is reasonable to conclude that the overall impact(s) of ArsR regulation will include altering community structure and function, not just resistance.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
TS conducted culturing, nucleic acid extraction, and some transcriptomics analyses, and provided edits. RR conducted the transcriptomics analyses and provided edits. Y-SK generated the mutants. QW conducted data analysis and editing. SW and BB provided intellectual input and manuscript editing. TM conceived the project, designed the study, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Primary support for this research was provided by the National Science Foundation Systems and Synthetic Biology Program (MCB-1413321 and MCB-1714556) and the Montana Agricultural Experiment Station (923310), and additional support came from the National Institutes of Health (R01CA215784).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.630562/full#supplementary-material
References
Andres, J., and Bertin, P. N. (2016). The microbial genomics of arsenic. FEMS Microbiol. Rev. 40, 299–322. doi: 10.1093/femsre/fuv050
Andrews, S. (2014). FastQC A Quality Control Tool for High Throughput Sequence Data. Burlington, MA: ScienceOpen, Inc.
ASTDR (2016). Priority List of Hazardous Substances. Available online at: http://www.atsdr.cdc.gov/spl/ (accessed 2019).
Bailey, T. L., Bodén, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208.
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bollinger, N., Hassett, D. J., Iglewski, B. H., Costerton, J. W., and McDermott, T. R. (2001). Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 183, 1990–1996. doi: 10.1128/jb.183.6.1990-1996.2001
Branco, R., Chung, A.-P., and Morais, P. V. (2008). Sequencing and expression of two arsenic resistance operons with different functions in the highly arsenic-resistant strain Ochrobactrum tritici SCII24T. BMC Microbiol. 8:95. doi: 10.1186/1471-2180-8-95
Bray, N. L., Pimentel, H., Melsted, P., and Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi: 10.1038/nbt.3519
Busenlehner, L. S., Pennella, M. A., and Giedroc, D. P. (2003). The SmtB/ArsR family ofmetalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27, 131–143. doi: 10.1016/s0168-6445(03)00054-8
Cai, L., Liu, G., Rensing, C., and Wang, G. (2009a). Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol. 9:4. doi: 10.1186/1471-2180-9-4
Cai, L., Rensing, C., Li, X., and Wang, G. (2009b). Novel gene clusters involved in arsenite oxidation and resistance in two arsenite oxidizers: Achromobacter sp. SY8 and Pseudomonas sp. TS44. Appl. Microbiol. Biotechnol. 83, 715–725. doi: 10.1007/s00253-009-1929-4
Chang, J.-S., Yoon, I.-H., Lee, J.-H., Kim, K.-R., An, J., and Kim, K.-W. (2010). Arsenic detoxification potential of aox genes in arsenite-oxidizing bacteria isolated from natural and constructed wetlands in the Republic of Korea. Environ. Geochem. Health 32, 95–105. doi: 10.1007/s10653-009-9268-z
Chi, L., Bian, X., Gao, B., Tu, P., Ru, H., and Lu, K. (2017). The effects of an environmentally relevant level of arsenic on the gut microbiome and its functional metagenome. Toxicol. Sci. 160, 193–204. doi: 10.1093/toxsci/kfx174
Chi, L., Lai, Y., Tu, P., Liu, C. W., Xue, J., Ru, H., et al. (2019a). Lipid and cholesterol homeostasis after arsenic exposure and antibiotic treatment in mice: potential role of the microbiota. Environ. Health Perspect. 127:97002.
Chi, L., Tu, P., Liu, C. W., Lai, Y., Xue, J., Ru, H., et al. (2019b). Chronic arsenic exposure induces oxidative stress and perturbs serum lysolipids and fecal unsaturated fatty acid metabolism. Chem. Res. Toxicol. 32, 1204–1211. doi: 10.1021/acs.chemrestox.9b00039
Chi, L., Xue, J., Tu, P., Lai, Y., Ru, H., and Lu, K. (2019c). Gut microbiome disruption altered the biotransformation and liver toxicity of arsenic in mice. Arch. Toxicol. 93, 25–35. doi: 10.1007/s00204-018-2332-7
Cleiss-Arnold, J., Koechler, S., Proux, C., Fardeau, M.-L., Dillies, M.-A., Coppee, J.-Y., et al. (2010). Temporal transcriptomic response during arsenic stress in Herminiimonas arsenicoxydans. BMC Genomics 11:709. doi: 10.1186/1471-2164-11-709
Consortium, U. (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515. doi: 10.1093/nar/gky1049
Coordinators, N. R. (2016). Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 44, D7–D19.
Coryell, M., McAlpine, M., Pinkham, N. V., McDermott, T. R., and Walk, S. T. (2018). The gut microbiome is required for full protection against acute arsenic toxicity in mouse models. Nat. Commun. 9:5424.
Cuellar-Partida, G., Buske, F. A., McLeay, R. C., Whitington, T., Noble, W. S., and Bailey, T. L. (2012). Epigenetic priors for identifying active transcription factor binding sites. Bioinformatics 28, 56–62. doi: 10.1093/bioinformatics/btr614
Fekih, I. B., Zhang, C., Li, Y. P., Zhao, Y., Alwathnani, H. A., Saquib, Q., et al. (2018). Distribution of arsenic resistance genes in prokaryotes. Front. Microbiol. 9:2473. doi: 10.3389/fmicb.2018.02473
Fernández, M., Udaondo, Z., Niqui, J.-L., Duque, E., and Ramos, J.-L. (2014). Synergic role of the two ars operons in arsenic tolerance in Pseudomonas putida KT2440. Environ. Microbiol. Rep. 6, 483–489. doi: 10.1111/1758-2229.12167
Francino, M. P. (2005). An adaptive radiation model for the origin of new gene functions. Nat. Genet. 37, 573–577. doi: 10.1038/ng1579
Grant, C. E., Bailey, T. L., and Noble, W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. doi: 10.1093/bioinformatics/btr064
Guo, X., Liu, S., Wang, Z., Zhang, X. X., Li, M., and Wu, B. (2014). Metagenomic profiles and antibiotic resistance genes in gut microbiota of mice exposed to arsenic and iron. Chemosphere 112, 1–8. doi: 10.1016/j.chemosphere.2014.03.068
Hamza, I., Chauhan, S., Hassett, R., and O’Brian, M. R. (1998). The bacterial irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 273, 21669–21674. doi: 10.1074/jbc.273.34.21669
Hsieh, Y.-J., and Wanner, B. L. (2010). Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13, 198–203. doi: 10.1016/j.mib.2010.01.014
Inskeep, W., McDermott, T., and Fendorf, S. (2002). “Arsenic (V)(III) cycling in soils and natural waters: chemical and microbiological processes,” in Environmental Chemistry of Arsenic, eds W. F. Frankenberger Jr. and J. M. Macy (New York, NY: Marcel Dekker), 183–216.
Kang, Y.-S., Brame, K., Jetter, J., Bothner, B. B., Wang, G., Thiyagarajan, S., et al. (2016). Regulatory activities of four ArsR proteins in Agrobacterium tumefaciens 5A. Appl. Environ. Microbiol. 82, 3471–3480. doi: 10.1128/aem.00262-16
Kang, Y.-S., Heinemann, J., Bothner, B., Rensing, C., and McDermott, T. R. (2012). Integrated co-regulation of bacterial arsenic and phosphorus metabolisms. Environ. Microbiol. 14, 3097–3109. doi: 10.1111/j.1462-2920.2012.02881.x
Kang, Y.-S., Shi, Z., Bothner, B., Wang, G., and Mcdermott, T. R. (2015). Involvement of the Acr3 and DctA anti-porters in arsenite oxidation in Agrobacterium tumefaciens 5A. Environ. Microbiol. 17, 1950–1962. doi: 10.1111/1462-2920.12468
Kashyap, D. R., Botero, L. M., Franck, W. L., Hassett, D. J., and McDermott, T. R. (2006). Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188, 1081–1088. doi: 10.1128/jb.188.3.1081-1088.2006
Kersey, P. J., Allen, J. E., Allot, A., Barba, M., Boddu, S., Bolt, B. J., et al. (2018). Ensembl genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res 46, D802–D808. doi: 10.1093/nar/gkx1011
Kruger, M. C., Bertin, P. N., Heipieper, H. J., and Arsène-Ploetze, F. (2013). Bacterial metabolism of environmental arsenic - mechanisms and biotechnological applications. Appl. Microbiol. Biotechnol. 97, 3827–3841. doi: 10.1007/s00253-013-4838-5
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, H., Li, M., Huang, Y., Rensing, C., and Wang, G. (2013). In silico analysis of bacterial arsenic islands reveals remarkable synteny and functional relatedness between arsenate and phosphate. Front. Microbiol. 4:347. doi: 10.3389/fmicb.2013.00347
Li, X., Hu, Y., Gong, J., Lin, Y., Johnstone, L., Rensing, C., et al. (2012). Genome sequence of the highly efficient arsenite-oxidizing bacterium Achromobacter arsenitoxydans SY8. J Bacteriol. 194, 1243–1244. doi: 10.1128/jb.06667-11
Liu, C. W., Chi, L., Tu, P., Xue, J., Ru, H., and Lu, K. (2019). Isobaric labeling quantitative metaproteomics for the study of gut microbiome response to arsenic. J. Proteome Res. 18, 970–981. doi: 10.1021/acs.jproteome.8b00666
Liu, G., Liu, M., Kim, E.-H., Maaty, W. S., Bothner, B., Lei, B., et al. (2012). A periplasmic arsenite-binding protein involved in regulating arsenite oxidation. Environ. Microbiol. 14, 1624–1634. doi: 10.1111/j.1462-2920.2011.02672.x
Lu, K., Abo, R. P., Schlieper, K. A., Graffam, M. E., Levine, S., Wishnok, J. S., et al. (2014). Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 122, 284–291. doi: 10.1289/ehp.1307429
Lu, K., Cable, P. H., Abo, R. P., Ru, H., Graffam, M. E., Schlieper, K. A., et al. (2013). Gut microbiome perturbations induced by bacterial infection affect arsenic biotransformation. Chem. Res. Toxicol. 26, 1893–1903.
Macur, R. E., Jackson, C. R., Botero, L. M., McDermott, T. R., and Inskeep, W. P. (2004). Bacterial populations associated with the oxidation and reduction of arsenic in an unsaturated soil. Environ. Sci. Technol. 38, 104–111. doi: 10.1021/es034455a
McDermott, T. R., Stolz, J. F., and Oremland, R. S. (2019). Arsenic and the gastrointestinal tract microbiome. Environ. Microbiol. Rep. 12, 136–159. doi: 10.1111/1758-2229.12814
Mukhopadhyay, R., Rosen, B. P., Phung, L. T., and Silver, S. (2002). Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol. Rev. 26, 311–325. doi: 10.1111/j.1574-6976.2002.tb00617.x
Ordóñez, E., Letek, M., Valbuena, N., Gil, J. A., and Mateos, L. M. (2005). Analysis of genes involved in arsenic resistance in Corynebacterium glutamicum ATCC 13032. Appl. Environ. Microbiol. 71, 6206–6215. doi: 10.1128/aem.71.10.6206-6215.2005
Oremland, R. S., and Stolz, J. F. (2003). The ecology of arsenic. Science 300, 939–944. doi: 10.1126/science.1081903
Páez-Espino, A. D., Durante-Rodríguez, G., and de Lorenzo, V. (2015). Functional coexistence of twin arsenic resistance systems in Pseudomonas putida KT2440. Environ. Microbiol. 17, 229–238. doi: 10.1111/1462-2920.12464
Pimentel, H., Bray, N. L., Puente, S., Melsted, P., and Pachter, L. (2017). Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690. doi: 10.1038/nmeth.4324
Rawle, R. A., Kang, Y.-S., Bothner, B., Wang, G., and McDermott, T. R. (2019). Transcriptomics analysis defines global cellular response of Agrobacterium tumefaciens 5A to arsenite exposure regulated through the histidine kinases PhoR and AioS. Environ. Microbiol. 12, 2659–2676. doi: 10.1111/1462-2920.14577
Rhine, E. D., Ní Chadhain, S. M., Zylstra, G. J., and Young, L. Y. (2007). The arsenite oxidase genes (aroAB) in novel chemoautotrophic arsenite oxidizers. Biochem. Biophys. Res. Commun. 354, 662–667. doi: 10.1016/j.bbrc.2007.01.004
Richardson, J. B., Dancy, B. C. R., Horton, C. L., Lee, Y. S., Madejczyk, M. S., Xu, Z. Z., et al. (2018). Exposure to toxic metals triggers unique responses from the rat gut microbiota. Sci. Rep. 8:6578.
Rosen, B. P. (1995). Resistance mechanisms to arsenicals and antimonials. J. Basic Clin. Physiol. Pharmacol. 6, 251–264.
Rosen, B. P. (2002). Biochemistry of arsenic detoxification. FEBS Lett. 529, 86–92. doi: 10.1016/s0014-5793(02)03186-1
Rosen, B. P., Ajees, A. A., and McDermott, T. R. (2011). Life and death with arsenic. Arsenic life: an analysis of the recent report “A bacterium that can grow by using arsenic instead of phosphorus”. Bioessays 33, 350–357. doi: 10.1002/bies.201100012
Santini, J. M., Sly, L. I., Schnagl, R. D., and Macy, J. M. (2000). A new chemolithoautotrophic arsenite-oxidizing bacterium isolated from a gold mine: phylogenetic, physiological, and preliminary biochemical studies. Appl. Environ. Microbiol. 66, 92–97. doi: 10.1128/aem.66.1.92-97.2000
Sarkar, A., Kazy, S. K., and Sar, P. (2013). Characterization of arsenic resistant bacteria from arsenic rich groundwater of West Bengal, India. Ecotoxicology 22, 363–376. doi: 10.1007/s10646-012-1031-z
Slyemi, D., and Bonnefoy, V. (2012). How prokaryotes deal with arsenic. Environ. Microbiol. Rep. 4, 571–586.
Small, S. K., Puri, S., Sangwan, I., and O’Brian, M. R. (2009). Positive control of ferric siderophore receptor gene expression by the irr protein in Bradyrhizobium japonicum. J. Bacteriol. 191, 1361–1368. doi: 10.1128/jb.01571-08
Somerville, J. E., and Kahn, M. L. (1983). Cloning of the glutamine synthetase I gene from Rhizobium meliloti. J. Bacteriol. 156, 168–176. doi: 10.1128/jb.156.1.168-176.1983
Stolz, J. F., Basu, P., Santini, J. M., and Oremland, R. S. (2006). Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60, 107–130. doi: 10.1146/annurev.micro.60.080805.142053
Strange, H. R., Zola, T. A., and Cornelissen, C. N. (2011). The fbpABC operon is required for ton-independent utilization of xenosiderophores by neisseria gonorrhoeae strain FA19. Infect. Immun. 79, 267–278. doi: 10.1128/iai.00807-10
Wang, Q., Kang, Y.-S., Alowaifeer, A., Shi, K., Fan, X., Wang, L., et al. (2018). Phosphate starvation response controls genes required to synthesize the phosphate analog arsenate. Environ. Microbiol. 20, 1782–1793. doi: 10.1111/1462-2920.14108
WHO (2016). Ten Chemicals of Major Public Health Concern. Available online at: http://www.who.int/ipcs/assessment/public_health/chemicals_phc/en/ (accessed 2018).
Wolfe-Simon, F., Blum, J. S., Kulp, T. R., Gordon, G. W., Hoeft, S. E., Pett-Ridge, J., et al. (2010). A bacterium that can grow by using arsenic instead of phosphorus. Science 332, 1163–1166.
Keywords: ArsR, arsenite, regulation, transcriptomics, global
Citation: Rawle R, Saley TC, Kang Y-S, Wang Q, Walk S, Bothner B and McDermott TR (2021) Introducing the ArsR-Regulated Arsenic Stimulon. Front. Microbiol. 12:630562. doi: 10.3389/fmicb.2021.630562
Received: 17 November 2020; Accepted: 18 January 2021;
Published: 03 March 2021.
Edited by:
Felipe Cava, Umeå University, SwedenReviewed by:
Christopher Rensing, Fujian Agriculture and Forestry University, ChinaGabriella Fiorentino, University of Naples Federico II, Italy
Copyright © 2021 Rawle, Saley, Kang, Wang, Walk, Bothner and McDermott. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Timothy R. McDermott, dGltbWNkZXJAbW9udGFuYS5lZHU=
†These authors have contributed equally to this work
‡Present address: Yoon-Suk Kang, Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, United States
 Rachel Rawle1†
Rachel Rawle1† Tara C. Saley
Tara C. Saley Yoon-Suk Kang
Yoon-Suk Kang Qian Wang
Qian Wang Seth Walk
Seth Walk Brian Bothner
Brian Bothner Timothy R. McDermott
Timothy R. McDermott