- 1Department of Medical Microbiology and Infectious Diseases, University of Manitoba, Winnipeg, MB, Canada
- 2National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, MB, Canada
Outer membrane vesicles (OMVs) produced by Gram-negative bacteria are mediators of cell survival and pathogenesis by facilitating virulence factor dissemination and resistance to antimicrobials. Studies of OMV properties often focus on hypervesiculating Escherichia coli mutants that have increased OMV production when compared to their corresponding wild-type (WT) strains. Currently, two conventional techniques, ultracentrifugation (UC) and ultradiafiltration (UF), are used interchangeably to isolate OMVs, however, there is concern that each technique may inadvertently alter the properties of isolated OMVs during study. To address this concern, we compared two OMV isolation methods, UC and UF, with respect to final OMV quantities, size distributions, and morphologies using a hypervesiculating Escherichia coli K-12 ΔtolA mutant. Nanoparticle tracking analysis (NTA) indicated that UC techniques result in lower vesicle yields compared to UF. However, UF permitted isolation of OMVs with smaller average sizes than UC, highlighting a potential OMV isolation size bias by each technique. Cryo-transmission electron microscopy (cryo-TEM) visualization of isolated OMVs revealed distinct morphological differences between WT and ΔtolA OMVs, where ΔtolA OMVs isolated by either UC or UF method possessed a greater proportion of OMVs with two or more membranes. Proteomic OMV analysis of WT and ΔtolA OMVs confirmed that ΔtolA enhances inner plasma membrane carryover in multi-lamellar OMVs. This study demonstrates that UC and UF are useful techniques for OMV isolation, where UF may be preferable due to faster isolation, higher OMV yields and enrichment of smaller sized vesicles.
Introduction
Bacterial outer membrane vesicles (OMVs) are spherical membrane structures typically ranging from 20-200 nm in diameter that are released from the outer membrane (OM) of Gram-negative bacteria into the extracellular milieu (Mashburn-Warren et al., 2008; Schwechheimer and Kuehn, 2015). OMVs are constitutively released from bacteria, in culture and during host infection, where they transport cargo such as toxins, virulence factors, autolysins, DNA and RNA (Jan, 2017; Cecil et al., 2019). OMVs play a critical role in promoting bacterial survival in stressful conditions, intercellular communication between bacteria, and by modulating host-pathogen interactions (Mashburn-Warren et al., 2008; Kulkarni and Jagannadham, 2014; Schwechheimer and Kuehn, 2015; Jan, 2017; Cecil et al., 2019). For example, OMVs from a variety of bacterial species can modulate the host immune response by activating immune cells and promoting cytokine secretion (Cecil et al., 2017), by delivering cytotoxic factors that induce apoptosis after internalization into host cells (Chmiela et al., 2018), and by secreting substances that damage surrounding tissues (O’Donoghue and Krachler, 2016; Cecil et al., 2019). OMVs have been proposed as specialized delivery vehicles, with their lipid bilayer topology ideal for transporting therapeutics to specific host cells (O’Donoghue and Krachler, 2016; Cecil et al., 2019). They have been incorporated into vaccine preparations due to their immunogenicity and ability to display antigens without the accompanying risk posed by metabolically active bacterial cells (Cecil et al., 2019). However, an important drawback for these applications is the low yield of vesicles that can be recovered from in vitro culture supernatants.
Previous studies seeking to identify genes associated with higher OMV production by Escherichia coli have involved gene knockout and gene disruption screens (McBroom et al., 2006; Kulp et al., 2015). Based on these studies, it was shown that mutations in certain membrane protein genes altered the OM architecture of E. coli, leading to hypervesiculation phenotypes with increased OMV production (Bernadac et al., 1998; Moon et al., 2012; Kulp et al., 2015; Turner et al., 2015; Pérez-Cruz et al., 2016). An important example is the E. coli Tol-Pal proteins, which are encoded within a seven gene cluster (ybgC, tolQ, tolR, tolA, tolB, pal, and cpoB) expressed from promoters upstream of ybgC and tolB; these proteins are vital for membrane maintenance and integrity of Gram-negative bacteria (Webster, 1991; Vianney et al., 1996; Bernadac et al., 1998; Lazzaroni et al., 1999; Lloubès et al., 2001; Cascales et al., 2002). The Tol-Pal system is composed of five interacting proteins that form a trans-membrane protein complex in the periplasmic space and associate with OmpA and Lpp in the OM (Lloubès et al., 2001). Mutations in any of the Tol-Pal genes can confer defects in the OM that lead to the activation of regulatory cascades responsible for extra-cytoplasmic stress responses, hypersensitivity to drugs and detergents, release of periplasmic proteins into the medium, and increased formation of OMVs (Bernadac et al., 1998; Lloubès et al., 2001; Vinés Marolda et al., 2005; Turner et al., 2015; Pérez-Cruz et al., 2016). Most recently, ΔtolB mutants of aquatic bacteria Buttiauxella agrestis and other Gram-negative species demonstrated that the loss of TolB enhanced the formation of multi-lamellar/multi-vesicular OMVs, referred to as M-OMVs (Takaki et al., 2020). As a result, the Tol-Pal system is of particular interest and importance to researchers seeking to better understand E. coli OMV morphology, formation and production.
One of the main limitations involved in studying OMVs is the challenging isolation and purification methods required to obtain sufficient quantities of these small vesicular structures. Techniques cited by most authors include ultracentrifugation and ultrafiltration (Horstman and Kuehn, 2000; Wai et al., 2003; Lee et al., 2007; Chutkan et al., 2013). It is important to note that the isolation method may affect an OMV’s morphology and total yield, promote aggregation of OMVs, and/or collect lipoproteins and other unwanted cell debris (Witwer et al., 2013; Yuana et al., 2014). Thus, an ideal OMV isolation method should provide high OMV yields without damaging vesicles for downstream experimental analyses or biotechnological applications. At the present time, comparative studies of OMV isolation methods and OMV quantifications are lacking, but both are important for improved in-depth analyses of OMVs.
The goal of our study was to evaluate two of the most commonly used OMV isolation techniques, ultracentrifugation (UC) and ultradiafiltration (UF) with an E. coli K-12 BW25113 strain (WT) and JW0729 (ΔtolA), a mutant containing a single-gene deletion of TolA component in the Tol-Pal system. ΔtolA was selected for comparison as previous studies (Bernadac et al., 1998; McBroom et al., 2006) identified that this deletion mutant confers a hypervesiculating phenotype when compared to the WT E. coli strain. In our study, we compared OMVs isolated from both strains grown under identical growth conditions to assess the yield and size of vesicles with nanoparticle tracking analysis (NTA). NTA rapidly detects nanoparticles in solution by combining laser light scattering microscopy with a charge-coupled device camera to visualize particles. These detected nanoparticles are assessed with software to relate the rate of particle movement by Brownian motion to its particle size in nm according to the Stokes–Einstein equation (Filipe et al., 2010; Gardiner et al., 2013; Gerritzen et al., 2017). Using NTA is advantageous to other OMV quantification methods as it allows for direct measurement of polydisperse samples, while the flow mode allows a large number of particles to be measured in a small timeframe, resulting in more accurate measurements with less variance (Filipe et al., 2010; Gerritzen et al., 2017). Cryo-transmission electron microscopy (cryo-TEM) analysis was used to visually determine OMV morphology and verify OMV size and total quantity produced by each isolation technique. Comparing OMV isolations from a WT strain as well as a hypervesiculating ΔtolA strain allowed us to explore the limits of each technique. We also employed Nano-LC/MS/MS proteomic analysis to compare the protein compositions of WT and ΔtolA vesicles. The outcome of this analysis revealed that UC and UF methods are similar with the exception of OMV minimal size limits. It allowed us to provide the first in-depth characterization of ΔtolA mutant OMVs, which revealed not only an increase in ΔtolA OMV quantity but also ΔtolA vesicles with two (outer-inner membrane vesicles; O-IMVs) or more membranes (multi-lamellar outer membrane vesicles; M-OMVs, grouped outer membrane vesicles; G-OMVs) by cryo-TEM visualization. Proteomic analysis of WT and ΔtolA OMVs demonstrated that ΔtolA OMVs possess more inner membrane (IM), periplasmic, and cytoplasmic proteins than WT, indicating that the loss of TolA may decrease linkages between the outer and inner membranes and result in the formation of these unique vesicle morphologies, similar to phenotypes recently described in ΔtolB and ΔtolR mutants (Pérez-Cruz et al., 2016; Takaki et al., 2020).
Results
ΔtolA Produces Significantly More Vesicles Than WT
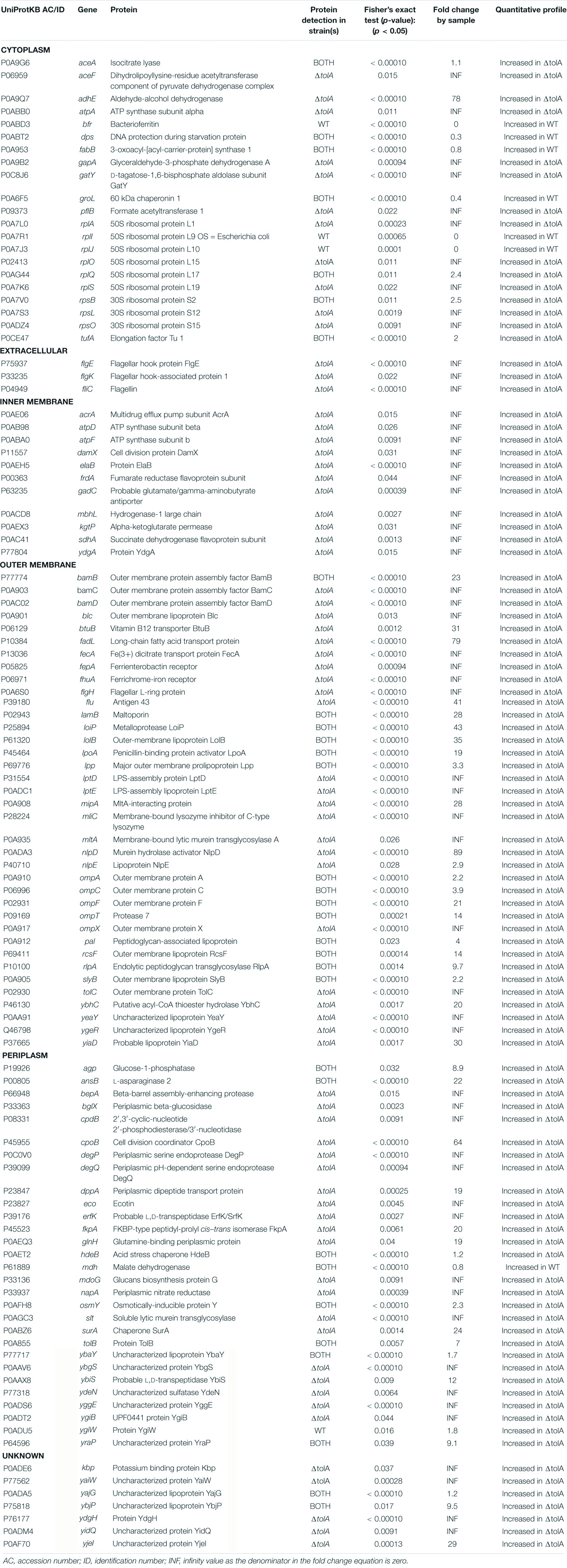
The primary aim of this study was to compare two of the most commonly used UC and UF OMV isolation methods and in doing so, provide an opportunity to examine OMV production differences between an E. coli K-12 BW25113 (WT) strain and its hypervesiculating gene deletion mutant ΔtolA (JW0729). Prior to UC and UF OMV isolations, we wanted to ensure that OMV formation from WT and ΔtolA was proportional to the total quantity of cells grown in culture; this measurement was important to account for potential cell titer differences caused by growth rate differences between the mutant and WT. To accomplish this, we measured growth curves of each strain prior to OMV isolation (Figure 1A). WT and ΔtolA growth rates were significantly different (p < 0.05) in optical density at 600 nm (OD600 nm) unit values for all time points, and the maximum OD600 nm units for WT was 1.11 ± 0.03 and ΔtolA was 0.98 ± 0.02 after 24 h (Figure 1A). Due to lower OD600 nm values of ΔtolA, we calculated OMV production yields based on total cells in colony forming units (CFU)/mL from OD600 nm measurements of each culture. This allowed a more accurate compare comparison of WT and ΔtolA OMV formation and quantity differences by UC and UF methods, and these values are listed in Figure 1C. For all comparisons made between UC and UF, a single large-scale bacterial culture was grown, and equally divided for UC and UF OMV isolations in order to minimize differences in OMV populations caused by batch growth effects.
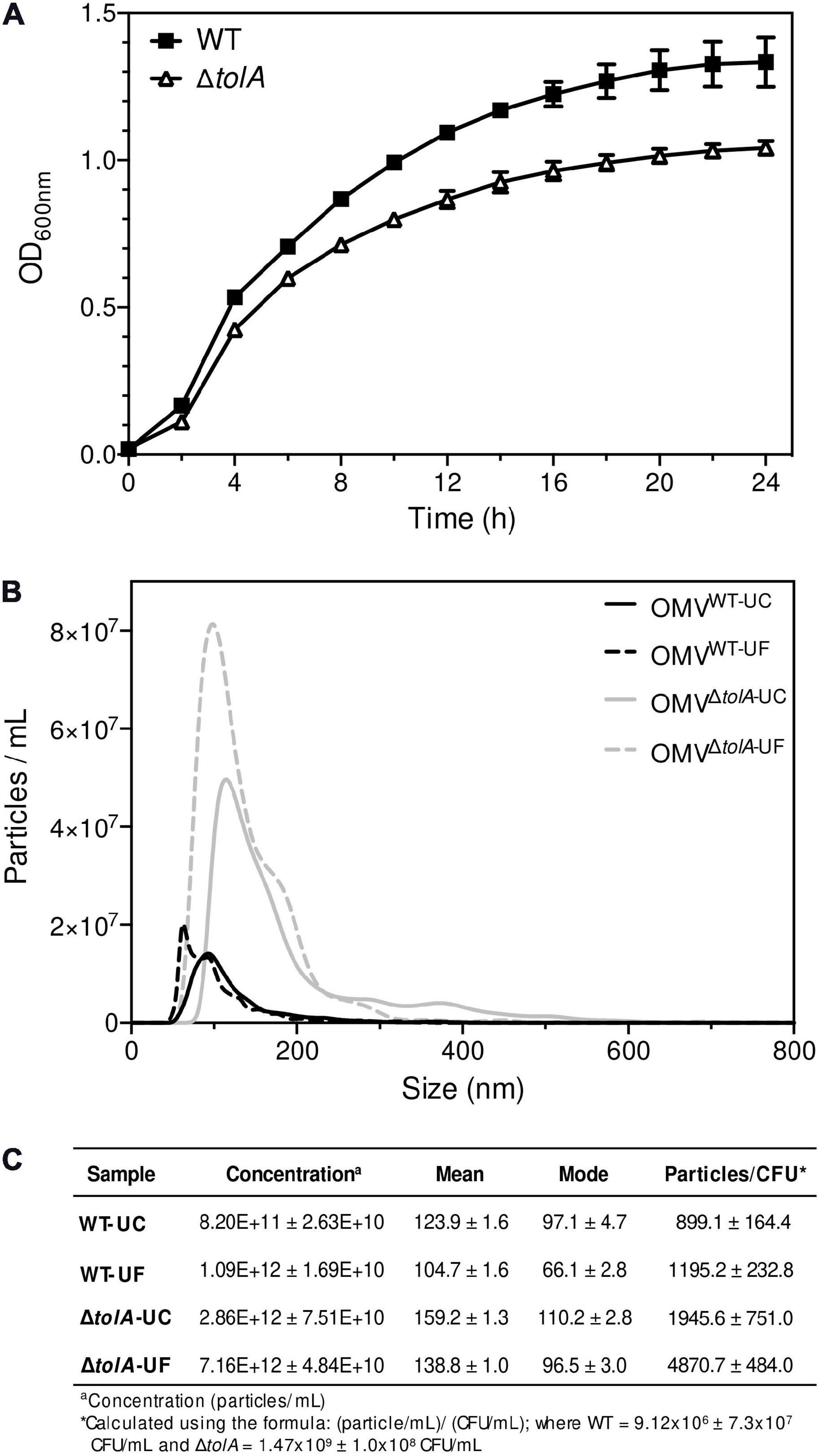
Figure 1. Growth curve analysis of Escherichia coli wild-type and ΔtolA and NTA measurements of their isolated OMVs. (A) Growth curves of BW25113 (WT) and JW0729 (ΔtolA) E. coli strains. OD600 nm measurements (y-axis) are shown over time (h; x-axis) for WT (squares) and ΔtolA (triangles) strains grown in LB broth at 37°C plates every 2 h for 24 h at 37°C. Growth curves represent the mean of two biological samples measured in triplicate (n = 6), where error bars represent the standard deviation of mean value. WT and ΔtolA growth were significantly different for each time point except 0 h according to a Mann–Whitney U test (p < 0.05). (B) OMVs isolated from WT and ΔtolA strains using either UC or UF methods (refer to panel legend) were diluted 1,000-fold and measured using a Nanosight NS500. All data represent the mean of triplicate experiments ± standard error. (C) A summary of NTA data collected, listing WT and ΔtolA mean vesicle size (nm), mode size (nm), concentration (particles/mL), and particles/CFU for each isolation method.
After factoring in the cell growth differences of each strain, we compared differences in OMV production by each strain when isolated by UC and UF methods using NTA (Figures 1B,C). NTA demonstrated statistically significant differences between WT and ΔtolA strains with respect to vesicle production. Both UC and UF methods confirmed greater ΔtolA OMVs quantities when compared to the WT based on particle/CFU calculations, corroborating previous studies on OMV production in Tol-Pal mutants (Bernadac et al., 1998; McBroom et al., 2006; Kulp et al., 2015; Pérez-Cruz et al., 2016; Takaki et al., 2020). Specifically, UC had a 2.2 (±0.45)-fold increase in ΔtolA OMVs when compared to WT and UF had a 4.1 (±0.41)-fold increase in ΔtolA OMVs compared to WT. NTA results also showed fewer WT and ΔtolA OMVs were recovered by UC methods when compared to UF (WT; 24.8% reduction, ΔtolA 60.0% reduction; Figures 1B,C). Hence, isolating OMVs using UC and UF methods confirmed that the ΔtolA strain hypervesiculates when compared to WT grown under the same conditions, but UF methods recover more OMVs as compared to UC.
OMVs Isolated by UC and UF Show no Differences in OmpA Abundance
To compare the differences in OMV content that may occur due to the isolation methods themselves, we performed Tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Tricine SDS-PAGE) analysis to determine if any OMV protein content was noticeably altered (Figure 2A). There were no significant differences in densitometry of stained protein bands between UC and UF OMV preparations for either strain. To determine if there were differences in key OM porins, Western blot analysis was performed to compare OmpA content ratios in OMVs. OmpA porin proteins are abundant and located in the OM, making them a reliable OMV detection marker (Bielaszewska et al., 2017). Based on this analysis, both UC and UF methods showed enrichment of OmpA in ΔtolA OMVs compared to WT based on net ΔtolA OmpA pixel density/WT OmpA pixel density (UC; 1.27, UF; 1.85, Figure 2B), and no significant differences in OmpA protein abundance between UC and UF-isolated OMV samples for either ΔtolA or WT (Figure 2B). This suggests that OmpA proteins present in OMVs can be accurately detected in both UC and UF isolation methods, indicating that the isolation method does not influence OmpA protein detection accuracy. This result indicates that OmpA could be a reliable detection marker for OMV production, as the ratio of OmpA present in our WT and ΔtolA OMV samples were comparable to OMV concentration ratios of WT and ΔtolA from NTA analysis.
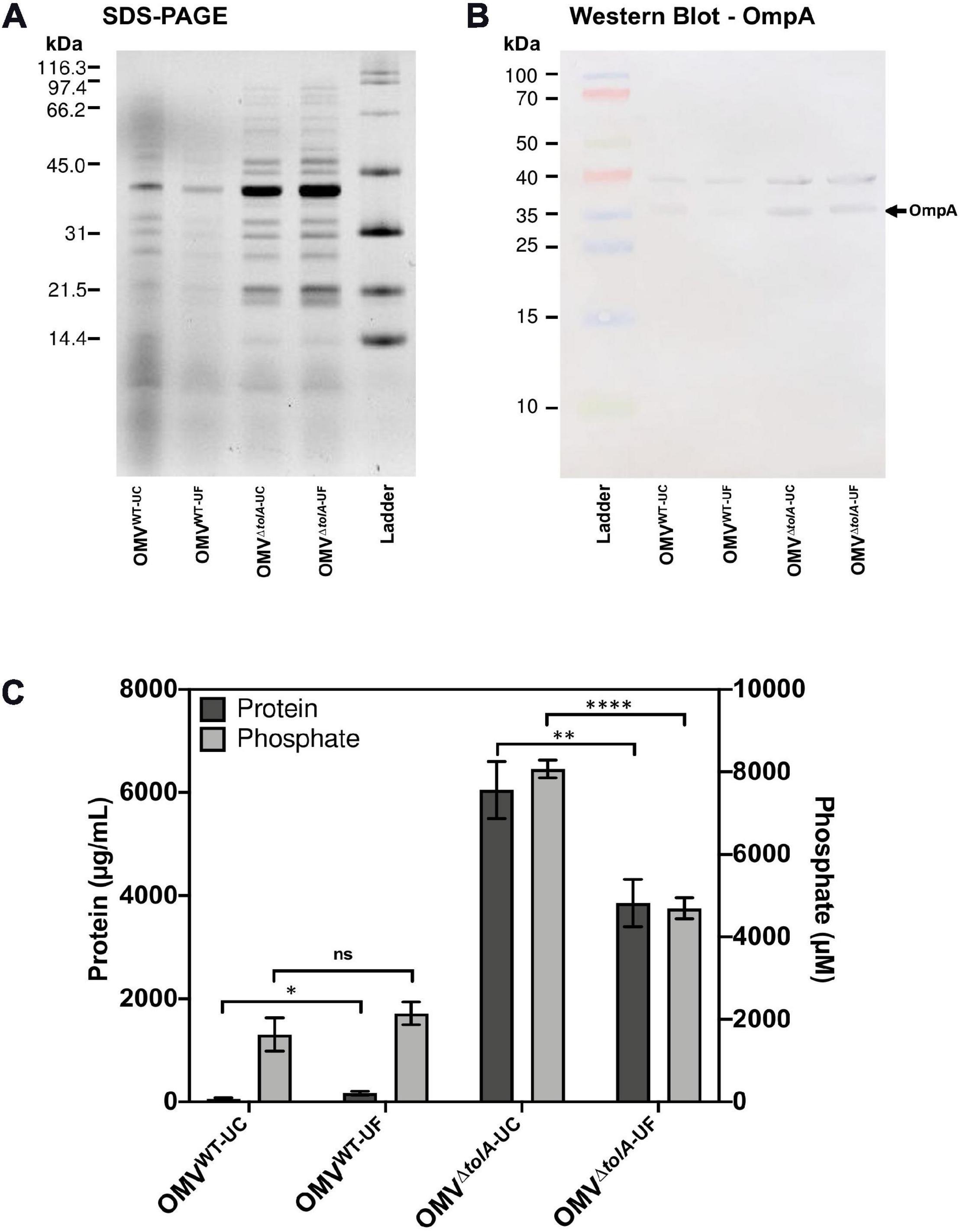
Figure 2. Protein and phosphorous profiles of isolated OMVs. (A) 12% acrylamide Tricine SDS-PAGE gel image of UC and UF OMV isolations for WT and ΔtolA E. coli strains detected with TCE. (B) Western blot of isolated OMV samples from WT and ΔtolA E. coli strains detected with anti-OmpA antibodies (1:25,000). (C) OMV sample total protein and total phosphorous content for each OMV isolation method. Values were obtained by BCA assay (protein) or malachite green assay (phosphate) and adjusted based on dilution factor and OD600 nm of the original culture. All data represent the mean of triplicate measurements, and error bars represent standard deviation. Significant differences between WT and ΔtolA samples or between UC and UF samples were determined using the Mann–Whitney U test at p-values of <0.05 (*), <0.005 (**), <0.00005 (****).
UC and UF Isolation Methods Differ in Concentration and Size of Recovered OMVs
In addition to NTA analysis, UC and UF OMVs isolated from WT and ΔtolA cultures were quantified by total protein bicinchoninic acid (BCA), and total lipid phosphorous (Malachite Green) assays to determine if UC and UF methods influence total protein or lipid phosphate contents in OMVs. As noted in our OMV OmpA protein detection experiments above, we wanted to determine if total protein and lipid OMV contents were altered specifically by each isolation method, as each method may differentially shift protein and lipid content carry-over. Total protein and lipid assays are routinely used to enumerate OMVs and to quantify protein–lipid content ratios of OMVs (McBroom et al., 2006; Orench-Rivera and Kuehn, 2016; Roier et al., 2016). Discordant results for WT and ΔtolA total protein and total phosphate were noted for UC and UF methods (Figure 2C). Significantly higher protein (p < 0.005) and phosphate (p < 0.00005) concentrations were detected in the UC-isolated ΔtolA sample as compared to ΔtolA OMVs isolated by UF, indicating greater protein and phosphate content in OMVs from these preparations (Figure 2C). The UF-isolated WT samples had significantly higher protein concentration (p < 0.05) than the UC-isolated WT, but no significant concentration differences between UC and UF WT isolations for phosphate concentration (Figure 2C). When considered with NTA data (Figures 1B,C), these results suggest that UF OMV isolation may enhance vesicle isolation yields as compared to UC but may also affect the WT and ΔtolA total protein and phosphate content.
Next, we determined the average vesicle sizes of WT and ΔtolA by NTA to determine if either method significantly altered the size of OMVs recovered (Figure 1C). The average size of UC-isolated OMVs was 123.9 ± 1.6 nm [mean ± standard error of the mean (SEM)] in dia for WT and 159.2 ± 1.3 nm dia for ΔtolA. UF-isolated OMVs had smaller average sizes of WT and ΔtolA vesicles at 104.7 ± 1.6 nm dia and 138.8 ± 1.0 nm dia, respectively. Thus, OMVs formed by the ΔtolA strain were larger in size as compared to the WT control by both methods (Figure 1B, p < 0.0001). When analyzing OMV particle size distributions, we also noticed that all UF-isolated OMV samples had a larger proportion of smaller sized vesicles when compared to vesicles isolated by UC which was enriched with larger sized vesicles. OMVs with diameter sizes between 0 and 100 nm were greatly enriched in UF isolations (WT UF; 74.3%, ΔtolA UF; 50.4%) as compared to UC (WT UC; 58.1%, ΔtolA UC; 38.3%; Supplementary Figure 1A). The opposite was true for UC-isolated vesicles, which had vesicles predominating at larger sizes ranging between 200 and 550 nm (Supplementary Figure 1B). OMVs with diameters over 100 nm corresponded to 41.9% of the total OMVs in UC-isolated WT samples and 61.7% in UC-isolated ΔtolA samples, whereas this range was 25.7% in UF-isolated WT samples and 49.6% in UF-isolated ΔtolA samples. These findings indicate that a size isolation bias exists for each method, where UF enriches for smaller sized particles when compared to the UC method.
Cryo-TEM of ΔtolA OMVs Reveals Distinct Morphological Differences From WT OMVs
OMV morphology analysis of each vesicle isolation method and strain type was performed using cryo-TEM analysis to establish any vesicle size and heterogeneity alterations. Statistical analysis of OMV measurements from cryo-TEM photomicrographs of WT and ΔtolA strains was performed, where representative examples are shown in Figures 3A–D, and revealed significant differences in vesicle size as summarized in Figure 3E. Measurements from cryo-TEM vesicle images identified a range of OMV sizes (40–400 nm dia) for each strain and methodology used (Figure 3E), supporting our NTA findings (Figure 1B). However, based this image analysis, all OMVs isolated by either UC or UF had a smaller size distribution range when compared to the same preparations analyzed by NTA (IQR; WT UC; 72.3–107.8 nm, WT UF; 75.7–107.7 nm, ΔtolA UC; 93.1–141.4 nm, ΔtolA UF; 86.2–133.7 nm). Additionally, cryo-TEM imaged vesicle diameters of ΔtolA isolated by UC and UF methods demonstrated significant differences in size, where average vesicle size of UC-isolated ΔtolA OMVs was 125.2 nm and UF-isolated ΔtolA OMVs was 116.7 nm (p = 0.0097; Figure 3C). Cryo-TEM average measurements of WT OMV diameters from either isolation method were not significantly different (UC; 93.55 nm, UF; 95.58 nm, p = 0.3123; Figure 3C). Hence, NTA and cryo-TEM measurements are generally in agreement with respect to UC and UF OMV size ranges and size averages, but when comparing vesicle size distributions by NTA and cryo-TEM techniques, cryo-TEM measurements suggest smaller diameter vesicle sizes and averages for both WT and ΔtolA by both isolation techniques. This disparity is likely due to differences in the number of vesicles counted by each method, indicating that NTA may be more precise due to the quantity and range of particle sizes that are accurately measurable.
Figure 3. Cryo-TEM images of vesicle types in WT and ΔtolA strains. Representative cryo-TEM images at 14,500× magnification of WT OMVs isolated by UC (A), WT OMVs isolated by UF (B), ΔtolA OMVs isolated by UC (C), ΔtolA OMVs isolated by UF (D). In each panel, enlarged images of representative OMVs frequently observed (based on table values shown in panel F) in WT and ΔtolA are highlighted as inset panel images in dashed boxes in the upper left-hand corner. Conventional OMVs as well as vesicles with two membranes (O-IMVs) and multi-layered vesicles (M-OMVs, G-OMVs) are shown in these inset images in panels (A–D). (E) Scatter plot summaries of vesicle diameters measured from cryo-TEM images of OMVs isolated from the WT and ΔtolA strains using either ultracentrifugation (UC) or ultradiafiltration (UF) at N = 250 vesicles/strain. Lines on each plot represent the mean value, and error bars represent standard deviations. Diameters of vesicles from cryo-TEM images were measured using ImageJ particle analysis software. Shape and color of data points represent the types of vesicle visualized (refer to in panel legend). Significance was determined using Mann–Whitney U test ∗p < 0.01, ****p < 0.0001, N = 250 vesicles). (F) Summary table of membrane vesicle architectures manually identified from cryo-TEM images using ImageJ. Values listed in the table represent the percent total number of vesicles manually assessed representing each shape/architecture type (N = 250 vesicles/strain).
Lastly, cryo-TEM highlighted stark differences between WT and ΔtolA OMVs with respect to their overall morphology. WT OMV morphologies were characteristic of previously described OMVs by either UC or UF methods (Koning et al., 2013; Pérez-Cruz et al., 2016; Thoma et al., 2018). Based on our cryo-TEM images, nearly all WT OMVs had a single membrane, presumably composed of the OM bilayer (Figures 3A,C). In contrast, the ΔtolA mutant had OMVs with variable single and multi-membranous structures when isolated by either UC or UF technique (Figures 3B,D). The ΔtolA strain had a high proportion of OMVs with two or more membranes by both methods (UC; 39.6, UF; 42.4%; Figure 3F), which included double-bilayer outer-inner OMVs (O-IMVs), multi-layered vesicles (M-OMVs) (≥3 layers), as well as grouped encapsulated OMVs (G-OMVs) surrounded by a larger extramembrane layer (Figure 3B). These altered vesicle morphologies produced by the ΔtolA mutant likely account for the larger average sized vesicles detected by NTA. ΔtolA vesicles with multiple membranes had a significantly larger average size (UC; 146.7 nm, UF; 143.2 nm) than WT OMVs when measured by cryo-TEM (UC; 110.5 nm, UF; 98.5 nm; Figure 3D). Taken altogether with the results from total protein/total phosphorous, NTA size distributions, and our cryo-TEM vesicle morphology analyses, we can state that tolA mutations considerably alter OMV formation and morphology. These analyses also reveal that neither UC or UF significantly altered the recoverable amount of WT and ΔtolA OMV content or morphology, highlighting both as useful OMV isolation techniques.
Proteomic Analysis Confirms IM Proteins in ΔtolA OMVs Which Were Absent From the WT
In an effort to further investigate the membrane contents of both WT and ΔtolA OMVs we used a proteomic approach to identify altered or unique OMV proteins (Figure 4). We performed in-depth nano-LC MS/MS analysis on WT and ΔtolA UF OMV preparations only, since these preparations produced greater yields of OMVs, at size ranges also present in UC methods. Our preliminary analyses of WT and ΔtolA OMVs proteomes including SDS-PAGE (Figure 2) did not reveal any significant differences in UC or UF proteins, which is not surprising given these OMVs were isolated from the same starting cultures. A total of 109 proteins were identified in this UF OMV proteomic analysis, where only 31 proteins were detected in both the WT and the tolA mutant (Figure 4A and Table 1). Only 5 proteins were exclusively over-accumulated in WT OMVs, whereas 73 proteins were exclusively enriched in ΔtolA OMVs (Figure 4A and Table 1). This initial analysis indicates that the mutant has a larger number of proteins sequestered in its vesicles as compared to WT, as we expected from its M-OMV morphology visualized in cryo-TEM images.
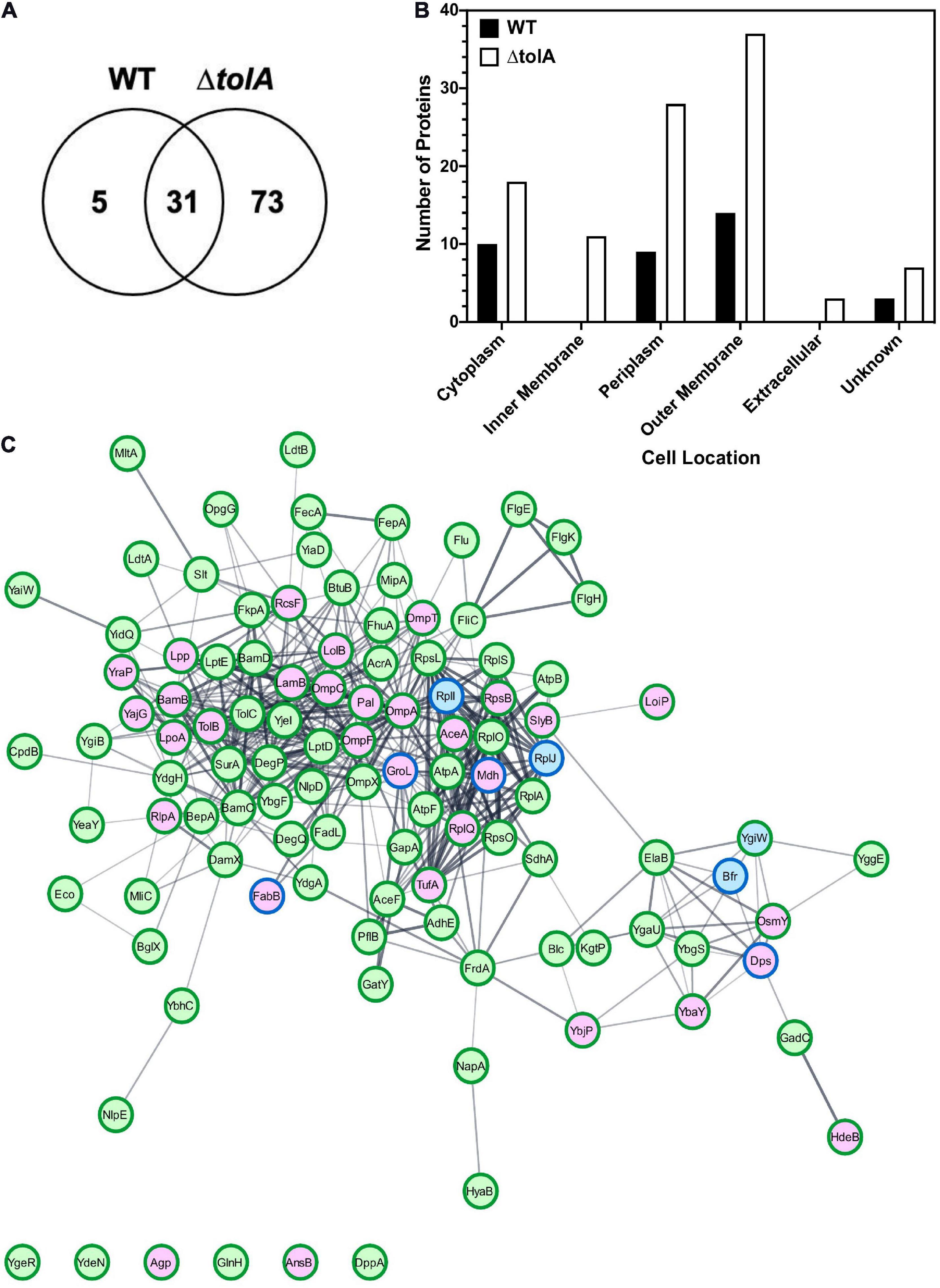
Figure 4. Proteomic characterization of WT and ΔtolA vesicles isolated by UF. (A) Venn-diagram of identified proteins showing overlap between WT and ΔtolA vesicle samples. (B) Cellular localization of identified proteins from WT (black) and ΔtolA (white) vesicle samples. (C) String network of interacting proteins from WT (blue), ΔtolA (green), or both (purple) vesicle samples. Border colors represent protein abundance, with dark green borders denoting upregulation of the protein in the ΔtolA sample, and dark blue borders denoting upregulation of the protein in the WT sample. The network diagram was generated using the StringApp v1.5.0 in Cytoscape v.3.8.0 (Doncheva et al., 2019). All data used in this analysis are summarized in Table 1.
To determine the membrane location of proteins we detected in each strain’s OMV preparation, we annotated the identified proteins and predicted their subcellular localization using the pSORTb algorithm (Yu et al., 2010), as shown in Figure 4B. 61% of the proteins we identified in both strains were classified as either OM proteins (37 proteins) or periplasmic proteins (29 proteins), whereas a smaller proportion of proteins from the IM (10 proteins) and cytoplasm (21 proteins) were detected primarily in ΔtolA mutant vesicles (Figure 4B). Of the remaining 10% of subcellularly localized proteins, 3 were predicted to be secreted proteins, and 8 had an unknown localization. From this analysis, we noted that all proteins associated with the IM and the extracellular space were exclusively identified in ΔtolA OMVs and at two to three-fold higher quantities than in WT OMVs (Figure 4B). This strongly supports the presence of greater IM, periplasmic, and extracellular protein carryover in ΔtolA OMVs, in agreement with our cryo-TEM multi-lamellar vesicle images. Additionally, many of the overlapping proteins identified in both WT and ΔtolA vesicle proteomes were located in the OM, including porins (OmpA, OmpC, and OmpF), lipoproteins (LolB, Lpp, RcsF, RlpA, and SlyB), and membrane assembly proteins (BamB and LpoA) (Figure 4C and Table 1). Membrane integrity proteins (Pal and TolB) as well as stress-related proteins (Dps, HdeB, and OsmY) were noted in both WT and ΔtolA OMV proteomes (Table 1), indicating that membrane components involved in stress and membrane maintenance were present even in the WT vesicles. Some of the proteins we detected were previously identified in other OMV proteomic studies (Lee et al., 2007; Berlanda Scorza et al., 2008; Aguilera et al., 2014; Kim et al., 2018; Hong et al., 2019), as well as others implicated in studies pertaining to OMV formation (McBroom et al., 2006; McBroom and Kuehn, 2007; Schwechheimer et al., 2015). Such proteins included periplasmic chaperone/protease DegP and the OM-anchored lipoprotein NlpE, which were found exclusively in the ΔtolA OMV sample (Table 1).
Functional analysis of OMV proteomes from WT and ΔtolA strains was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Supplementary Figure 2). This analysis revealed enrichment of a number of proteins associated with two-component regulatory systems, flagellar proteins, ribosomal proteins as well as several metabolic pathways, predominantly from the ΔtolA OMV sample (Supplementary Figure 2). Together, this proteomic analysis shows that both WT and ΔtolA strains produce vesicles enriched with proteins that are highly membrane-associated, that are responsible for membrane trafficking and assembly, and play a role in membrane integrity and bacterial stress response. It also verifies that deletion of tolA results in OMV formation with much greater periplasmic, IM and extracellular content carryover than WT OMVs as suggested by cryo-TEM imaging.
Discussion
This study determined that both UC and UF OMV isolation methods were effective for isolating intact OMVs, as both methods yielded comparable WT and ΔtolA vesicle populations (Figure 3). It did reveal that UC and UF vesicles differed with respect to their specific OMV size ranges and size distributions (Figures 1B, 3). It also revealed some differences in total protein and phosphorous content quantifications of WT and ΔtolA OMVs (Figure 2). These findings are important when considering how any isolated OMVs will be experimentally studied. OMVs isolation is a time-consuming process that requires large volumes of culture to overcome lower yields of vesicles naturally produced by cultured bacteria. While several authors have sought to optimize current methods to achieve the highest yields of vesicles (Cvjetkovic et al., 2014; Klimentová and Stulík, 2015; van der Pol et al., 2015), to our knowledge this is the first study to directly compare two of the most widely used isolation methods for their experimental OMV analysis applications. Despite both methods isolating high purity OMVs, our findings show that UF improves the recovery of OMVs as compared to UC isolations, without qualitatively altering vesicle contents or morphology. In addition, the UC approach is more time-consuming as compared to UF due to the duration of centrifugation runs and was shown in this study to reduce the recoverable quantity of smaller sized vesicles (Supplementary Figure 1). However, it is important to note that UC did enhance isolation of larger sized vesicles (>100 nm) (Supplementary Figure 1). As a result, our study offers more insights into the benefits and limitations of each technique, which should be considered in future OMV isolation experiments. It is important to note that many OMV isolation methods involving either UC or UF must be performed under sterile conditions to avoid foreign particle contamination which may obscure downstream analyses. Additionally, many OMV isolation studies include an extra OMV purification step such as gel filtration or gradient ultracentrifugation to further purify and enrich for vesicles of defined sizes or molecular weights as reviewed by Klimentová and Stulík (2015). We chose to omit gradient ultracentrifugation and gel filtration method assessments herein, as these methods require some prior knowledge of vesicle size distributions and would have prevented our unbiased assessment of vesicle size ranges obtainable by these initial UC or UF OMV isolation techniques.
Smaller Sized Vesicles Are Enriched by UF
The average vesicle size and size distribution as determined by NTA was generally in agreement with the cryo-TEM analysis data for both UC and UF OMVs, but we did notice differences in the size of particles isolated by each method. Based on NTA results, UF-isolated vesicles from WT and ΔtolA had a higher proportion of smaller (<100 nm) vesicles, while UC-isolated samples from WT and ΔtolA had a higher proportion of vesicles of larger (>100 nm) sizes (Supplementary Figure 1A). This trend in size distribution was also observed when we compared the ΔtolA vesicles in cryo-TEM measurements, where UF samples had a smaller average size, however, no significant differences were found between WT UC and UF sizes according to cryo-TEM images (Supplementary Figure 1B). This result suggests that NTA may be detecting small fragments of particles rather than fully formed OMVs in the UF-isolated samples. In a previous study it was found that NTA can measure vesicles as small as ∼50 nm (Dragovic et al., 2011), however, we noticed that smaller particles were being measured by the NTA NanoSight instrument in both UC and UF samples. Therefore, the smaller particles (0–50 nm in diameter) could be undissolved salts/media components and/or cell debris particles that were carried over from cell cultures in UF isolations, potentially skewing the size distribution of vesicles in samples with smaller contaminating particles. We suspect that the UF procedure may naturally bias isolation of smaller sized particles as the medium is membrane filtered, and may be prone to some filter blockage by medium components and culture carryover over time. Filter blockage may cause the MW cut-off to become lower over time and allow smaller sized particles, such as salt or cell debris, to build up and enrich in UF techniques. Sucrose-gradient cushions have been used in past experiments to help eliminate these carryover particles (Alexander et al., 2016) but it is unclear if cushions further bias the recovery of heterogenous OMV populations.
Another explanation for why UF isolates smaller OMVs than UC methods is that UC methods may promote aggregation of vesicles due to repeated pelleting steps. Vesicle pelleting may result in larger average sized OMV distributions as we observed in our study. Aggregation of vesicles and contamination with extravesicular protein complexes or aggregates is a commonly reported UC occurrence (for examples see Théry et al., 2006; Issman et al., 2013; Witwer et al., 2013; Linares et al., 2015). Due to the use of fixed-angle ultracentrifuge rotors during UC, material pelleting and deposition against the wall of the centrifuge tube may be physically damaging, potentially promoting vesicle aggregation, which favors the fusion of vesicles with weakened or altered membranes (Witwer et al., 2013; Klimentová and Stulík, 2015). In our study, we detected a small fraction of UC vesicles larger than 450 nm by NTA, which suggests this form of aggregation was occurring, as the 0.45 μm filtration step prior to UC/UF should remove all particles above this size. Thus, while UC and UF methods are both comparable, it appears that each method alters the size distribution of OMVs, with UF selecting for smaller particles and UC promoting the aggregation of vesicles into larger-sized particles.
ΔtolA OMVs Are Multilamellar and Enriched With IM-Associated Proteins
Mutations of the tol-pal system genes have been well documented in OMV studies and mutants are often used study hypervesiculation phenotypes (Bernadac et al., 1998; Kulp et al., 2015), however, our study is the first to examine the morphology and content of a tolA mutant in detail. Our study determined that the ΔtolA strain not only produced more OMVs as compared to the WT strain, but that these vesicles were larger in size and displayed M-OMV, G-OMV, and O-IMV membrane morphology as observed for tolB mutant of B. agrestis in a recent study (Takaki et al., 2020). Additionally, we determined that ΔtolA vesicles with multiple membranes were larger in size than WT OMVs. The presence of these unique O-IMV and multi-lamellar vesicle types (M-OMV and G-OMV) has also been observed for many Gram-negative species such as E. coli Nissle 1917, Helicobacter pylori strain 60190, Pseudomonas aeruginosa PAO1, Acinetobacter baumannii AB41, and Neisseria gonorrhoeae DSM15130 strains, but their functional significance to these species has yet to be determined (Fiocca et al., 1999; Pérez-Cruz et al., 2015, 2016). This altered ΔtolA vesicle morphology suggests that the loss of TolA in E. coli promotes the carryover of IM, possibly to compensate for loss of membrane integrity caused by reduced Tol-Pal inter-membrane connections. Tol-Pal mutants have been previously shown to cause cell division impairments, resulting in increased distance between the IM and the peptidoglycan layer and enhancing defects in peptidoglycan-cleaving enzymes (Takaki et al., 2020; Yakhnina and Bernhardt, 2020). Our proteomic data reveals that the peptidoglycan degrading enzymes NlpD, MltA, Slt, and RlpA were significantly enriched in the ΔtolA strain (Table 1), supporting the idea that the Tol-Pal system plays a role in promoting glycan cleavage. Similar to the recent ΔtolB study (Takaki et al., 2020), we suspect that increased OMV formation by the ΔtolA mutant is the result of incomplete tethering of the IM and OM, which would result in IM and cytoplasmic proteins being carried over into vesicles more frequently. This is supported by our proteomic analysis, which determined that more cytoplasmic proteins were detected in ΔtolA than WT, and IM proteins were found exclusively in the ΔtolA vesicles (Figure 4B). It is important to note that the detection of cytoplasmic proteins in both the WT and ΔtolA OMVs is not unexpected, since cytoplasmic protein detection in OMV isolations has been previously shown to occur in many past OMV studies, as reviewed by Nagakubo et al. (2020). There are two explanations for this, the first is that there are typically a small fraction of vesicles produced by a bacterium (∼0.1%) that contain both inner and outer membranes as well as carry over cytosolic proteins that remain detectable by proteomic analyses (Pérez-Cruz et al., 2013). Since many cytoplasmic proteins are present in the cell at higher amounts than many membrane proteins, cytoplasmic protein detection in OMV preparations is not surprising. Secondly, Pseudomonas OMV studies also suggest that cytosolic protein carryover may be the result of transient autolysin-induced peptidoglycan breakage which not only promotes some inner membrane carry over into vesicles but also likely carries over cytoplasmic proteins (Kadurugamuwa and Beveridge, 1995; Clarke, 2018). Hence, the detection of some cytoplasmic proteins even in the WT OMV samples of our study was expected.
Our isolated ΔtolA vesicles also appeared to be enriched with structural OMPs, cell membrane assembly proteins, and cell division proteins. Our proteomic analysis determined that ΔtolA OMVs were enriched with proteins involved in membrane biogenesis and degradation, including the OM assembly proteins (BamC and BamD), LPS assembly proteins (LptD, LptE, and FadL), cell division proteins (CpoB, DamK, and NlpD), and murein degrading proteins (MltA, MipA, and Slt) (Figure 4C and Table 1). When compared to the WT, this suggests that the ΔtolA mutant may have to increase membrane biogenesis as well as membrane turnover in order to keep up with the high levels of vesiculation and loss of both IM and OM to vesicles caused by Tol-Pal complex disruption. Our measurements of total phosphate and protein in the ΔtolA strain by both isolation methods indicated that more protein was detected relative to total lipid phosphate, which further supports this explanation. Additionally, many envelope stabilizing proteins were also enriched in ΔtolA, specifically NlpE, OmpX, and TolC, which support increased membrane biogenesis in tolA mutant vesicles (Figure 4C and Table 1). The increased prevalence of these proteins in ΔtolA OMVs is not surprising, as previous studies have shown that mutations in tolA can be partially compensated by expressing other OM-associated proteins that act to stabilize the OM in the absence of TolA (Godlewska et al., 2009; Bager et al., 2013; Pérez-Cruz et al., 2016). An increase in the amount of stabilizing proteins in the OM could conceivably compensate for the increased vesiculation in mutant strains and is worth further study. We also observed that the ΔtolA strain grows slower than the WT (Figure 1A), which underscores the fitness costs associated with loss of TolA and inter-membrane integrity in E. coli.
The Role of the Tol-Pal System in Vesicle Production
The modulation of cell envelope intermembrane layer crosslinks is a strong correlate of increased OMV production in Gram-negative species. Proteins intricately involved in linking the OM to the IM include: (i) OmpA, an OM porin that spans the periplasmic space and can bind to peptidoglycan. (ii) The Tol-Pal complex, a cell-division component that aids in invagination of the OM and membrane stability. (iii) Lpp, an OM lipoprotein that covalently crosslinks with the peptidoglycan (Schwechheimer and Kuehn, 2015). Studies have shown that mutants of E. coli, Salmonella, and A. baumannii lacking OmpA display increased OMV production (Deatherage et al., 2009; Schwechheimer and Kuehn, 2015). Mutations in the Tol-Pal genes are associated with increased vesicle production in E. coli and Salmonella, specifically deletions in pal, tolA, and tolB (Cascales et al., 2002; Deatherage et al., 2009; Jagannadham and Chattopadhyay, 2015). Our study corroborates these findings, with OmpA, Lpp, TolB, and Pal all enriched in the ΔtolA mutant (Table 1), suggesting that the hypervesiculation phenotype exhibited by our ΔtolA strain is a result of generalized membrane instability and incomplete membrane linkage.
Proteomic analysis of ΔtolA OMVs also identified the involvement of σE and Cpx envelope stress response pathways (Figure 4C and Table 1). In the presence of misfolded proteins and extracellular stress, the σE response is activated, and contributes to DNA repair, metabolism, OM biogenesis, and periplasmic homeostasis (Hews et al., 2019). σE-regulated chaperones and proteases (SurA, FkpA, and DegP) and several members of the BAM complex (BamB, BamC, and BamD) were all significantly enriched in ΔtolA, indicating that this pathway is highly active in the ΔtolA mutant. The chaperone Skp was also found in both WT and ΔtolA, although we did not significantly detect over-accumulation in either sample. The Cpx envelope stress response is crucial for mitigating envelope stress caused by misfolded proteins in the periplasm, and Cpx regulated members are involved in protein folding and degradation primarily within the IM (Danese et al., 1995; Snyder et al., 1995; Danese and Silhavy, 1998). NlpE, an activator of this system, was found to be significantly enriched in ΔtolA only (Figure 4C and Table 1). DegP, a periplasmic serine protease was the only significantly enriched protein regulated by the Cpx regulon. It is important to note, that the Cpx regulon has been associated with IM-associated proteins and functions (Raivio et al., 2013), as was seen in our ΔtolA proteomic dataset. Together, this suggests that mutations in the Tol-Pal system are intricately involved with envelope stress responses as a compensatory mechanism against envelope instability.
Conclusion
The study of OMVs is a rapidly expanding research area, so understanding the isolation method limitations improves our ability to modulate OMV production using the fewest genetic alterations. Better understanding of OMV recovery by common vesicle isolation methods aids ongoing and future biotechnological OMV applications, helping to standardize and improve efforts to enhance the overall recovery of OMVs from bacterial cultures. Our analyses suggest that UF may be an improved method for isolating OMVs, due to its faster isolation time and higher yield of smaller and averaged-sized vesicles. Depending on the OMV sizes desired, UC applications may be a desired methodology and both methods should be carefully considered based on the type of downstream experimental analysis needed. Our study also provides the first in-depth characterization of ΔtolA OMVs, which revealed a multi-lamellar membrane morphology similar to recent studies of tolB (Takaki et al., 2020). Our proteomic analysis highlighted the impact that the Tol-Pal system has on cell membrane content released into secreted vesicles and identified protein components worth following up on in future studies.
Materials and Methods
Bacterial Strains
The parental Keio collection strain E. coli K-12 BW25113 (WT) and its single gene deletion mutant JW0729 (ΔtolA) were obtained from the Coli Genetic Stock Center (Yale University, New Haven, CT, United States). All strains were grown in Luria-Bertani (LB) broth (Cold Spring Harbor Protocols, 2006) in a shaking incubator 170 RPM at 37°C from overnight cultures of cryopreserved dimethylsulfoxide (DMSO) stocks. Growth was monitored by measuring optical density (OD) at 600 nm (OD600 nm). All procedures involved the use of a biosafety cabinet to maintain sterility of bacterial and OMV isolates and prevent their contamination.
Growth Rate Measurements
To ensure that OMV production was not associated with impaired growth phenotypes, growth curves of the WT and ΔtolA strain were performed. Bacterial cells were inoculated into LB broth from frozen cryostocks and grown overnight. The resulting culture was standardized to 1 OD, and diluted 1/100 in LB into flat-bottom 96-well NUNC microtiter plates (Thermo Fisher Scientific, United States). Strains were grown for 24 h (h) in LB media at 37°C with continuous shaking, where OD600 nm was measured every 2 h in a BioTek EL808 microplate reader (BioTek, Winooski, VT, United States). Growth of each strain was measured in triplicate from 6 biological replicates (n = 6) and Mann–Whitney U tests were performed to determine OD values that differed significantly (p < 0.05) between WT and ΔtolA at all time points.
OMV Isolation
Culture Supernatant Separation Prior to UC and UF
Prior to UC or UF OMV isolation, both methods separated supernatants prepared from large scale LB cultures as described previously (Horstman and Kuehn, 2000; McBroom et al., 2006; Lee et al., 2007) with slight modifications. Briefly, bacterial cells were inoculated into LB broth from frozen cryostocks and grown overnight. The resulting culture was standardized to 1 OD600 nm unit, washed two times to prevent carryover of OMVs, and diluted 1/100 into 1 L of LB broth. This culture was incubated at 37°C for 18 h with constant shaking (160 RPM). OD600 nm measurements were taken to confirm early stationary growth phase OD600 nm values in reference to the growth curves performed in Section “Growth Rate Measurements.” Cells were separated from the culture by centrifugation at 6,000 RPM for 15 min at 4°C in an JLA9.1000 rotor using an Avanti J-E high speed centrifuge (VWR Part of Avantor, United States). The collected supernatant was filtered with a 0.45 μm polyethersulfone (PES) vacuum filter (MilliporeSigma, United States) to remove any residual bacteria. Filtered supernatant aliquots from each strain preparation were spread plated onto LB agar and incubated at 37°C for 24 h to confirm the absence of intact, viable cells. The resulting filtrate was divided into two equal parts to be assessed by both ultracentrifugation (UC) and ultradiafiltration (UF) methods.
Ultracentrifugation (UC) OMV Isolation
The designated UC filtered supernatant portion was centrifuged at 40,000 RPM in polycarbonate tubes for 2 h at 4°C in a Ti70 rotor using a Beckman Coulter® Optima XPN Ultracentrifuge (VWR Part of Avantor, United States). The supernatant was carefully decanted to prevent pellet disruption. After all the filtrate had been centrifuged, the pellets in each tube were resuspended in 50 mM HEPES buffer (Fisher Scientific, NH, United States) then stored at −20°C (Yaron et al., 2000).
Ultradiafiltration (UF) OMV Isolation
The designated UF filtered supernatant portion was concentrated 50-fold in 50 mM HEPES buffer in a 400 ml capacity Amicon® stirred cell (MilliporeSigma, United States) ultradiafiltration system using a 500 kiloDalton (kDa) molecular weight cut off (MWCO) polyethersulfone (PES) ultrafiltration disk (MilliporeSigma, United States). The concentrated retentate was collected and divided into polycarbonate centrifuge tubes (Beckman) before ultracentrifugation at 40,000 RPM for 2 h at 4°C. The pellet was resuspended in 50 mM HEPES buffer and stored at −20°C until further use (Yaron et al., 2000).
Protein and Lipid Quantification of OMVs
Outer membrane vesicles produced by WT and ΔtolA bacteria were assessed using both protein and phosphate assays to quantify the amount of OMVs produced by each strain and each method. Protein concentration was measured by a bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, United States). Samples were measured in triplicate and compared with a standard curve plotted using serial dilutions of bovine serum albumin (BSA). Lipid content was inferred by measuring the total phosphate content using a malachite green phosphate assay (MilliporeSigma, United States). This assay quantifies the amount of phosphate in phospholipids, protein, and DNA. Assays were performed in triplicate for each OMV isolation. Protein- or phosphate-based measurements were adjusted for the amount of bacteria in the culture and for the vesicle production of the wild-type culture according to the following two equations.
EQ1a: [Protein]Sample = (OD562 nm − B)/A
EQ1b: [PO43–]Sample = (OD620 nm − B)/A
Equation 1 (EQ1) calculates the OMV sample’s protein and phosphate concentrations based on standard curves of bovine serum albumin (BSA) protein (EQ1a) and potassium phosphate (EQ1b) titrations, where the sample OD unit value at the measured wavelength (562 nm or 620 nm) is subtracted from the respective absorbance y-intercept (B) value from the standard curve, which is divided by the slope of the respective standard curve (A).
EQ2a: Adjusted [Protein]Sample = [Protein]Sample/original culture CFU/mL
EQ2b: Adjusted [PO43–]Sample = [PO43–]Sample/original culture CFU/mL
Equation 2 (EQ2) adjusts for the number of cells in the original culture by dividing the OMV sample’s protein or phosphate concentrations determined from EQ1 by the OD 600 nm of the original culture converted to colony forming units (CFU) per mL that the OMV isolations were obtained from.
Tricine Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (Tricine SDS-PAGE) and Western Blot Analysis of OMV Proteins
To determine whether the protein content of OMVs isolated by each method may differ due to the isolation methods themselves, Tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Tricine SDS-PAGE) was used. OMV preparations were denatured for 10 min at 65°C in 2 × Laemmli buffer [100 mM dithiothreitol, 150 mM Tris (pH 7), 12% w/v SDS, 30% w/v glycerol, 0.05% w/v Coomassie Brilliant Blue G-250] and equal amounts with respect to total protein quantities from both preparations were separated by 12% Tricine SDS-PAGE gels. Proteins were visualized with 0.5% v/v 2,2,2-trichloroethanol (TCE) by ultraviolet detection (Ladner et al., 2004). For Western blot, proteins were transferred to nitrocellulose and then blocked with 5% milk powder in TBS for 1 h. The membrane was incubated with anti-Gram-negative bacterial OmpA primary antibody at 1:25000 dilution (1.2 μg/mL; Antibody Research Corporation, Cottleville, MO, United States) overnight at 4°C followed by incubation with goat anti-rabbit IgG (Heavy + Light) HRP conjugate antibody at a 1:500 dilution (0.4 μ/mL; Life Technologies, United States) for 1 h. Proteins were detected using an enhanced chemiluminescence (ECL) detection system kit (Thermo Fisher Scientific, United States). Blot band densities were densitometrically analyzed using ImageJ software version 1.511 and quantified as described in Davarinejad (2020).
Nanoparticle Tracking Analysis (NTA) of OMVs
Outer membrane vesicles quantities and sizes were determined using a NanoSight NS500 nanoparticle tracking (NTA) instrument (Malvern Instruments Ltd., United Kingdom) equipped with a 488 nm blue laser and a complementary metal-oxide semiconductor (CMOS) image sensor camera. OMV samples were thawed to room temperature prior to analysis and diluted 1/1000 in 50 mM HEPES buffer at pH 7.4. Polystyrene beads (100 nm diameter) and HEPES buffer alone were run as positive and negative control standards, respectively. Samples were infused into the NanoSight instrument using a syringe pump set at ‘20’ speed setting (in arbitrary units). Measurements were captured in five 60 s reads at ambient room temperature (23.9–25.2°C), with instrument-optimized settings, where ‘blur,’ ‘minimum track length,’ and ‘minimum expected size’ options were set to “automatic” and viscosity was set to “water” (0.883–0.911 cP). Automated image setup (camera level and focus) was chosen whenever available for video enhancement. A total of 1,498 frames per sample were analyzed with NTA software version 2.3 (Malvern Instruments Ltd., United Kingdom) with a detection threshold of 5 (in arbitrary units). Mean size (nm), mode size (nm), and concentration (particles/mL) were tabulated, and the average of five reads was calculated and plotted as particle size versus number of particles per mL.
Cryo-TEM Analysis of Isolated OMVs
Samples for cryo-TEM were prepared as described above for OMV isolation, with the exception that UC and UF ΔtolA OMV samples were diluted 10-fold in 50 mM HEPES buffer (pH 7.4) due to their higher concentration of OMVs when compared to WT samples. All sets of samples were combined with 10 nm BSA-labeled gold tracer in a 6:1 ratio to assist with automated focusing; 3 μL of this suspension was applied to freshly glow-discharged Quantifoil R 2/2 grids (Quantifoil Micro Tools GmbH, Germany). This suspension was allowed to adhere, and the excess liquid was blotted with standard Vitrobot filter paper (Ted Pella Inc., United States) using a Vitrobot Mark IV (Thermo Fisher Scientific, United States), operating at 5°C and 100% humidity. Grids were then frozen in liquid ethane cooled by liquid nitrogen. Samples were transferred to a Tecnai F20 transmission electron microscope (Thermo Fischer Scientific, United States) using a Gatan 626 DH low-temperature specimen holder (Gatan Inc., United States), and images were recorded using an Eagle 4k CCD camera (Thermo Fischer Scientific, United States). Images were taken in low-dose imaging conditions (10 e/Å2) at both 5,000 and 14,500× magnifications, and vesicle sizes and morphologies were analyzed using ImageJ software version 1.512.
Proteomic Analysis and Gene Ontology
Sample Preparation and Nano LC-MS/MS
Samples for proteomic analysis were prepared from 1 L cultures as described for OMV isolation using the UF method for concentration. Protein from the outer membrane vesicles (OMV) were quantified using a bicinchoninic acid (BCA) protein assay kit, with bovine serum albumin (BSA) as the standard (Thermo Fisher Scientific, United States). SDS was added to 100 μg of OMV protein at a final concentration of 2%, and then heated at 95°C for 5 min. Upon cooling to room temperature, dithiothreitol was added to a final concentration of 100 mM, and heated at 95°C for 5 min. Samples were frozen at −80°C until ready for use. A total of 100 μg of protein from each OMV sample was used for each digestion. Protein samples were digested with trypsin (Promega, United States) overnight using a filter-assisted sample preparation (FASP) method described previously (Wiśniewski et al., 2009) using Nanosep 30K Omega Centrifugal Devices (Pall Corporation, United States). Following digestion, all samples were dried down and reconstituted using mass spectrometry grade water to a final concentration of 0.5μg/μl prior to mass spectrometry analysis.
Samples were each separately analyzed using a nano-flow Easy nLC II connected in-line to an LTQ Orbitrap Velos mass spectrometer with a nanoelectrospray ion source at 2.2 (Thermo Fisher Scientific, United States). Peptide samples (2 μl) were loaded onto a C18-reversed phase trap column (3 cm long, 100 μm inner diameter, 5 μm particles) with 100% buffer A (2% acetonitrile, 0.1% formic acid) for a total volume of 30 μl, and then separated on a C18-reversed phase column (15 cm long, 75 μm inner diameter, 2.4 μm particles). Both columns were packed in-house with ReproSil-Pur C18-AQ resin (Dr. Maisch) and fritted with Kasil. Peptides were eluted using a linear gradient of 5–25% buffer B (98% acetonitrile, 0.1% formic acid) over 120 min, 25–40% buffer B for 5 min, 40–80% buffer B for 5 min and a wash at 80% B for 8 min at a constant flow rate of 250 nl/min. Total LC/MS/MS run-time was about 165 min, including the loading, linear gradient, column wash, and the equilibration.
Data was acquired using these settings: dynamically choosing the top 10 most abundant precursor ions from each survey scan, each isolated with a width 2.0 m/z and fragmentation by CID. The survey scans were acquired in the Orbitrap over m/z 300–1,700 with a target resolution of 60,000 at m/z 400, and the subsequent fragment ion scans were also acquired in the iontrap at a normal scan rate. The lower threshold for selecting a precursor ion for fragmentation was 2000. Dynamic exclusion was enabled using a m/z tolerance of 15 ppm, a repeat count of 1, and an exclusion duration of 30 s.
Data Processing
All spectra were processed using MaxQuant (v1.6.7, Max Plank Institute) using the imbedded Andromeda search engine. Searches were performed against a subset of the SwissProt database set to E. coli K12 (4519 sequences). The following search parameters were used: Carbamidomethyl (C) was selected as a fixed modification, Oxidation (M) and Acetyl (Protein N-term) as a variable modifications, fragment ion mass tolerance of 0.5 Da, parent ion tolerance of 20 ppm, and trypsin enzyme with up to 2 missed cleavage. False discovery rates were set up using 0.01 for peptides, 0.01 for proteins, and at least 1 razor peptide per protein. LFQ was enabled for Quantitation. Resulting LFQ intensities were imported into Perseus v1.6.5 (Max Plank Institute). In Perseus the data was Log2 transformed. Then all the proteins that did not have a least 3 valid log2 LFQ intensities from ID were filtered out.
All spectra were also processed using Proteome Discoverer (v2.2, Thermo Fisher Scientific) and database searching was done with Mascot v2.6 (Matrix Science). Searches were performed against the SwissProt database (2020_01) (5461,911 sequences) The decoy database option was selected, and the following search parameters were used: Carbamidomethyl (C) was selected as a fixed modification, Oxidation (M) as a variable modification, fragment ion mass tolerance of 0.5 Da, parent ion tolerance of 10 ppm, and trypsin enzyme with up to 1 missed cleavage. Mascot search results were imported into Scaffold Q + (v4.11.0). Proteins were filtered using a 1.0% false discovery rate and assessed for significance using Fisher’s exact test (p-value < 0.05). All significant proteins were annotated by their subcellular localization using the pSORTb algorithm (Yu et al., 2010). String protein networks were constructed using Cytoscape (version 3.8.03), and functional protein maps were constructed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database through the ClueGo plugin (v2.5.64) with default settings. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD022786 and doi: 10.6019/PXD022786.
Statistical Analysis
All data was analyzed using Graph Pad Prism 8 software (v8.4.2). Normality of data was assessed for cryo-TEM and NTA data using the Shapiro–Wilk and Kolmogorov–Smirnov test. Statistical significance for all data was determined by the Mann–Whitney U test. For all analyses, differences between either WT and ΔtolA preparations or between the same strain isolated by UC or UF were statistically compared, and results with a p-value of less than 0.05 were considered statistically significant due to sample numbers compared and their degrees of freedom.
Data Availability Statement
The mass spectrometry proteomic data presented in the study are deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD022786 and doi: 10.6019/PXD022786.
Author Contributions
DCB and SR designed the study. SR isolated the OMVs and performed growth curves, protein/phosphate assays, and western blots. SR completed the cryo-TEM imaging and NanoSight experiments with DRB, SH, and TB. SR prepared proteomic sample preparations for LC MS/MS collection by PC and GW. SR analyzed all data and prepared manuscript figures. SR wrote the manuscript drafts in consultation with DCB, where DCB and GGZ edited. All authors read and approved the final manuscript.
Funding
Funding for this study was provided by a Research Manitoba New Investigator Award [2017-3099] operating grant to DCB.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Stuart McCorrister and the Mass Spectrometry and Proteomics Core Facility group (National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, MB, Canada) for help with proteomic data analysis. Thanks also to the Diagnostic Microscopy and Imaging group (National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, MB, Canada) for assistance with cryo-transmission electron microscopy and NanoSight experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.628801/full#supplementary-material
Supplementary Figure 1 | Size of isolated OMVs by NanoSight NTA and cryo-TEM. OMVs isolated from WT and ΔtolA were binned in 50 nm size ranges and represented as a percentage of the total OMV population for (A) NTA, (B) Cryo-TEM.
Supplementary Figure 2 | KEGG network of WT and ΔtolA OMV-associated proteins. Proteins were functionally annotated using the ClueGO (v.2.5.6) add-in for Cytoscape (v.3.8.0) (Bindea et al., 2009). Bolded black terms with black nodes represent functional categories, and colored nodes represent proteins with the color of each node indicating the OMV sample it was found in. Proteins were exclusive to WT (blue), ΔtolA (green) or present in both samples (purple). Connecting edge lines represent multiple functional categorizations of proteins. Default parameters were used for network specificity.
Footnotes
- ^ https://imagej.nih.gov/ij/
- ^ https://imagej.nih.gov/ij
- ^ https://cytoscape.org
- ^ http://apps.cytoscape.org/apps/cluego
References
Aguilera, L., Toloza, L., Giménez, R., Odena, A., Oliveira, E., Aguilar, J., et al. (2014). Proteomic analysis of outer membrane vesicles from the probiotic strain Escherichia coli Nissle 1917. Proteomics 14, 222–229. doi: 10.1002/pmic.201300328
Alexander, R. P., Chiou, N.-T., and Ansel, K. M. (2016). Improved exosome isolation by sucrose gradient fractionation of ultracentrifuged crude exosome pellets. Protoc. Exch. Preprint.
Bager, R. J., Persson, G., Nesta, B., Soriani, M., Serino, L., Jeppsson, M., et al. (2013). Outer membrane vesicles reflect environmental cues in Gallibacterium anatis. Vet. Microbiol. 167, 565–572. doi: 10.1016/j.vetmic.2013.09.005
Berlanda Scorza, F., Doro, F., Rodríguez-Ortega, M. J., Stella, M., Liberatori, S., Taddei, A. R., et al. (2008). Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli ΔtolR IHE3034 mutant. Mol. Cell Proteomics 7, 473–485. doi: 10.1074/mcp.m700295-mcp200
Bernadac, A., Gavioli, M., Lazzaroni, J. C., Raina, S., and Lloubès, R. (1998). Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180, 4872–4878. doi: 10.1128/jb.180.18.4872-4878.1998
Bielaszewska, M., Rüter, C., Bauwens, A., Greune, L., Jarosch, K. A., Steil, D., et al. (2017). Host cell interactions of outer membrane vesicle-associated virulence factors of enterohemorrhagic Escherichia coli O157: Intracellular delivery, trafficking and mechanisms of cell injury. PLoS Pathog. 13:e1006159. doi: 10.1371/journal.ppat.1006159
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. doi: 10.1093/bioinformatics/btp101
Cascales, E., Bernadac, A., Gavioli, M., Lazzaroni, J.-C., and Lloubes, R. (2002). Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184, 754–759. doi: 10.1128/jb.184.3.754-759.2002
Cecil, J. D., O’Brien-Simpson, N. M., Lenzo, J. C., Holden, J. A., Singleton, W., Perez-Gonzalez, A., et al. (2017). Outer membrane vesicles prime and activate macrophage inflammasomes and cytokine secretion in vitro and in vivo. Front. Immunol. 8, 1–22. doi: 10.3389/fimmu.2017.01017
Cecil, J. D., Sirisaengtaksin, N., O’Brien-Simpson, N. M., and Krachler, A. M. (2019). Outer Membrane Vesicle-Host Cell Interactions. Microbiol. Spectr. 7:2018.
Chmiela, M., Walczak, N., and Rudnicka, K. (2018). Helicobacter pylori outer membrane vesicles involvement in the infection development and Helicobacter pylori-related diseases. J. Biomed. Sci. 25, 1–11.
Chutkan, H., MacDonald, I., Manning, A., and Kuehn, M. J. (2013). Quantitative and qualitative preparations of bacterial outer membrane vesicles. Methods Mol. Biol. 966, 259–272. doi: 10.1007/978-1-62703-245-2_16
Clarke, A. J. (2018). The “hole” story of predatory outer-membrane vesicles. Can. J. Microbiol. 64, 589–599. doi: 10.1139/cjm-2017-0466
Cold Spring Harbor Protocols (2006). LB (Luria-Bertani) liquid medium. Cold Spring Harb. Protoc. 2006:8141. doi: 10.1101/pdb.rec8141
Cvjetkovic, A., Lötvall, J., and Lässer, C. (2014). The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J. Extracell Vesicles 3:1.
Danese, P. N., and Silhavy, T. J. (1998). CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180, 831–839. doi: 10.1128/jb.180.4.831-839.1998
Danese, P. N., Snyder, W. B., Cosma, C. L., Davis, L. J. B., and Silhavy, T. J. (1995). The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9, 387–398. doi: 10.1101/gad.9.4.387
Davarinejad, H. (2020). Quantifications of Western Blots with ImageJ. Heslington: University of York.
Deatherage, B. L., Lara, J. C., Bergsbaken, T., Barrett, S. L. R., Lara, S., and Cookson, B. T. (2009). Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72, 1395–1407.
Doncheva, N. T., Morris, J. H., Gorodkin, J., and Jensen, L. J. (2019). Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 18, 623–632. doi: 10.1021/acs.jproteome.8b00702
Dragovic, R. A., Gardiner, C., Brooks, A. S., Tannetta, D. S., Ferguson, D. J. P., Hole, P., et al. (2011). Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed. Nanotechnol. Biol. Med. 7, 780–788. doi: 10.1016/j.nano.2011.04.003
Filipe, V., Hawe, A., and Jiskoot, W. (2010). Critical evaluation of nanoparticle tracking analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm. Res. 27, 796–810. doi: 10.1007/s11095-010-0073-2
Fiocca, R., Necchi, V., Sommi, P., Ricci, V., Telford, J., Cover, T. L., et al. (1999). Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J. Pathol. 188, 220–226. doi: 10.1002/(sici)1096-9896(199906)188:2<220::aid-path307=3.0.co;2-c
Gardiner, C., Ferreira, Y. J., Dragovic, R. A., Redman, C. W. G., and Sargent, I. L. (2013). Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell Vesicles 2:1.
Gerritzen, M. J. H., Martens, D. E., Wijffels, R. H., and Stork, M. (2017). High throughput nanoparticle tracking analysis for monitoring outer membrane vesicle production. J. Extracell Vesicles 6:1.
Godlewska, R., Wiśniewska, K., Pietras, Z., and Jagusztyn-Krynicka, E. K. (2009). Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: Function, structure, role in pathogenesis and potential application in immunoprophylaxis: Minireview. FEMS Microbiol. Lett. 298, 1–11. doi: 10.1111/j.1574-6968.2009.01659.x
Hews, C. L., Cho, T., Rowley, G., and Raivio, T. L. (2019). Maintaining integrity under stress: Envelope stress response regulation of pathogenesis in Gram-negative bacteria. Front. Cell Infect. Microbiol. 9:313. doi: 10.3389/fcimb.2019.00313
Hong, J., Dauros-Singorenko, P., Whitcombe, A., Payne, L., Blenkiron, C., Phillips, A., et al. (2019). Analysis of the Escherichia coli extracellular vesicle proteome identifies markers of purity and culture conditions. J. Extracell Vesicles 8:1.
Horstman, A. L., and Kuehn, M. J. (2000). Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275, 12489–12496. doi: 10.1074/jbc.275.17.12489
Issman, L., Brenner, B., Talmon, Y., and Aharon, A. (2013). Cryogenic transmission electron microscopy nanostructural study of shed microparticles. PLoS One 8:e83680. doi: 10.1371/journal.pone.0083680
Jagannadham, M. V., and Chattopadhyay, M. K. (2015). Role of outer membrane vesicles of bacteria. Resonance 8, 711–725. doi: 10.1007/s12045-015-0228-x
Jan, A. T. (2017). Outer membrane vesicles (OMVs) of Gram-negative bacteria: A perspective update. Front. Microbiol. 8, 1–11. doi: 10.3389/fmicb.2017.01053
Kadurugamuwa, J. L., and Beveridge, T. J. (1995). Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: A novel mechanism of enzyme secretion. J. Bacteriol. 177, 3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995
Kim, S. W., Park, S., Bin, Im, S. P., Lee, J. S., Jung, J. W., et al. (2018). Outer membrane vesicles from β-lactam-resistant Escherichia coli enable the survival of β-lactam-susceptible E. coli in the presence of β-lactam antibiotics. Sci. Rep. 8, 1–13.
Klimentová, J., and Stulík, J. (2015). Methods of isolation and purification of outer membrane vesicles from gram-negative bacteria. Microbiol. Res. 170, 1–9. doi: 10.1016/j.micres.2014.09.006
Koning, R. I., de Breij, A., Oostergetel, G. T., Nibbering, P. H., Koster, A. J., and Dijkshoorn, L. (2013). Cryo-electron tomography analysis of membrane vesicles from Acinetobacter baumannii ATCC19606T. Res. Microbiol. 164, 397–405. doi: 10.1016/j.resmic.2013.02.007
Kulkarni, H. M., and Jagannadham, M. V. (2014). Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology 160, 2109–2121. doi: 10.1099/mic.0.079400-0
Kulp, A. J., Sun, B., Ai, T., Manning, A. J., Orench-Rivera, N., Schmid, A. K., et al. (2015). Genome-wide assessment of outer membrane vesicle production in Escherichia coli. PLoS One 10:139200. doi: 10.1371/journal.pone.0139200
Ladner, C. L., Yang, J., Turner, R. J., and Edwards, R. A. (2004). Visible fluorescent detection of proteins in polyacrylamide gels without staining. Anal. Biochem. 326, 13–20. doi: 10.1016/j.ab.2003.10.047
Lazzaroni, J. C., Germon, P., Ray, M.-C., and Vianney, A. (1999). The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol. Lett. 177, 191–197. doi: 10.1111/j.1574-6968.1999.tb13731.x
Lee, E. Y., Joo, Y. B., Gun, W. P., Choi, D. S., Ji, S. K., Kim, H. J., et al. (2007). Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 7, 3143–3153. doi: 10.1002/pmic.200700196
Linares, R., Tan, S., Gounou, C., Arraud, N., and Brisson, A. R. (2015). High-speed centrifugation induces aggregation of extracellular vesicles. J. Extracell Vesicles 4:29509. doi: 10.3402/jev.v4.29509
Lloubès, R., Cascales, E., Walburger, A., Bouveret, E., Lazdunski, C., Bernadac, A., et al. (2001). The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152, 523–529. doi: 10.1016/s0923-2508(01)01226-8
Mashburn-Warren, L., Mclean, R. J. C., and Whiteley, M. (2008). Gram-negative outer membrane vesicles: Beyond the cell surface. Geobiology 6, 214–219. doi: 10.1111/j.1472-4669.2008.00157.x
McBroom, A. J., and Kuehn, M. J. (2007). Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63, 545–558. doi: 10.1111/j.1365-2958.2006.05522.x
McBroom, A. J., Johnson, A. P., Vemulapalli, S., and Kuehn, M. J. (2006). Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188, 5385–5392. doi: 10.1128/jb.00498-06
Moon, D. C., Choi, C. H., Lee, J. H., Choi, C.-W., Kim, H.-Y., Park, J. S., et al. (2012). Acinetobacter baumannii outer membrane protein a modulates the biogenesis of outer membrane vesicles. J. Microbiol. 50, 155–160. doi: 10.1007/s12275-012-1589-4
Nagakubo, T., Nomura, N., and Toyofuku, M. (2020). Cracking Open Bacterial Membrane Vesicles. Front. Microbiol. 10:3026. doi: 10.3389/fmicb.2019.03026
O’Donoghue, E. J., and Krachler, A. M. (2016). Mechanisms of outer membrane vesicle entry into host cells. Cell Microbiol. 18, 1508–1517. doi: 10.1111/cmi.12655
Orench-Rivera, N., and Kuehn, M. J. (2016). Environmentally-controlled bacterial vesicle-mediated export. Cell Microbiol. 18, 1525–1536. doi: 10.1111/cmi.12676
Pérez-Cruz, C., Cañas, M. A., Giménez, R., Badia, J., Mercade, E., Baldomà, L., et al. (2016). Membrane vesicles released by a hypervesiculating Escherichia coli Nissle 1917 tolR mutant are highly heterogeneous and show reduced capacity for epithelial cell interaction and entry. PLoS One 11:169186. doi: 10.1371/journal.pone.0169186
Pérez-Cruz, C., Carrión, O., Delgado, L., Martinez, G., López-Iglesias, C., and Mercade, E. (2013). New type of outer membrane vesicle produced by the gram-negative bacterium Shewanella vesiculosa M7T: Implications for DNA content. Appl. Environ. Microbiol. 79, 1874–1881. doi: 10.1128/aem.03657-12
Pérez-Cruz, C., Delgado, L., López-Iglesias, C., and Mercade, E. (2015). Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS One 10:e0116896. doi: 10.1371/journal.pone.0116896
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450. doi: 10.1093/nar/gky1106
Raivio, T. L., Leblanc, S. K. D., and Price, N. L. (2013). The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J. Bacteriol. 195, 2755–2767. doi: 10.1128/jb.00105-13
Roier, S., Zingl, F. G., Cakar, F., Durakovic, S., Kohl, P., Eichmann, T. O., et al. (2016). A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 7, 1–13. doi: 10.1016/j.micres.2014.09.006
Schwechheimer, C., and Kuehn, M. J. (2015). Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 13, 605–619. doi: 10.1038/nrmicro3525
Schwechheimer, C., Rodriguez, D. L., and Kuehn, M. J. (2015). NlpI-mediated modulation of outer membrane vesicle production through peptidoglycan dynamics in Escherichia coli. Microbiologyopen 4, 375–389. doi: 10.1002/mbo3.244
Snyder, W. B., Davis, L. J. B., Danese, P. N., Cosma, C. L., and Silhavy, T. J. (1995). Overproduction of nlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic lacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177, 4216–4223. doi: 10.1128/jb.177.15.4216-4223.1995
Takaki, K., Tahara, Y. O., Nakamichi, N., Hasegawa, Y., Shintani, M., Ohkuma, M., et al. (2020). Multilamellar and multivesicular outer membrane vesicles produced by a Buttiauxella agrestis tolB mutant. Appl. Environ. Microbiol. 86, 1131–1151.
Théry, C., Amigorena, S., Raposo, G., and Clayton, A. (2006). Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 30, 3.22.1–3.22.29.
Thoma, J., Manioglu, S., Kalbermatter, D., Bosshart, P. D., Fotiadis, D., and Müller, D. J. (2018). Protein-enriched outer membrane vesicles as a native platform for outer membrane protein studies. Commun. Biol. 1:23.
Turner, L., Praszkier, J., Hutton, M. L., Steer, D., Ramm, G., Kaparakis-Liaskos, M., et al. (2015). Increased Outer Membrane Vesicle Formation in a Helicobacter pylori tolB Mutant. Helicobacter 20, 269–283. doi: 10.1111/hel.12196
van der Pol, L., Stork, M., and van der Ley, P. (2015). Outer membrane vesicles as platform vaccine technology. Biotechnol. J. 10, 1689–1706. doi: 10.1002/biot.201400395
Vianney, A., Michelle Muller, M., Clavel, T., Lazzaroni, J. C., Portalier, R., and Webster, R. E. (1996). Characterization of the tol-pal region of Escherichia coli K-12: Translational control of tolR expression by tolQ and identification of a new open reading frame downstream of pal encoding a periplasmic protein. J. Bacteriol. 178, 4031–4038. doi: 10.1128/jb.178.14.4031-4038.1996
Vinés, Marolda, C. L., Balachandran, A., and Valvano, M. A. (2005). Defective O-antigen polymerization in tolA and pal mutants of Escherichia coli in response to extracytoplasmic stress. J. Bacteriol. 187, 3359–3368. doi: 10.1128/jb.187.10.3359-3368.2005
Wai, S. N., Lindmark, B., Söderblom, T., Takade, A., Westermark, M., Oscarsson, J., et al. (2003). Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115, 25–35. doi: 10.1016/s0092-8674(03)00754-2
Webster, R. E. (1991). The tol gene products and the import of macronmolecules into Escherichia coli. Mol. Microbiol. 5, 1005–1011. doi: 10.1111/j.1365-2958.1991.tb01873.x
Wiśniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Witwer, K. W., Buzás, E. I., Bemis, L. T., Bora, A., Lässer, C., Lötvall, J., et al. (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell Vesicles 2:1.
Yakhnina, A. A., and Bernhardt, T. G. (2020). The Tol-Pal system is required for peptidoglycan-cleaving enzymes to complete bacterial cell division. Proc. Natl. Acad. Sci. U S A. 117, 6777–6783. doi: 10.1073/pnas.1919267117
Yaron, S., Kolling, G. L., Simon, L., and Matthews, K. R. (2000). Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 66, 4414–4420. doi: 10.1128/aem.66.10.4414-4420.2000
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. doi: 10.1093/bioinformatics/btq249
Keywords: outer membrane vesicles, Escherichia coli, ultracentrifugation, ultradiafiltration, hypervesiculation, Tol-Pal system, nanoparticle tracking analysis, LC-MS/MS
Citation: Reimer SL, Beniac DR, Hiebert SL, Booth TF, Chong PM, Westmacott GR, Zhanel GG and Bay DC (2021) Comparative Analysis of Outer Membrane Vesicle Isolation Methods With an Escherichia coli tolA Mutant Reveals a Hypervesiculating Phenotype With Outer-Inner Membrane Vesicle Content. Front. Microbiol. 12:628801. doi: 10.3389/fmicb.2021.628801
Received: 12 November 2020; Accepted: 08 February 2021;
Published: 05 March 2021.
Edited by:
Daniela De Biase, Sapienza University of Rome, ItalyReviewed by:
Chang-Ro Lee, Myongji University, South KoreaDirk Linke, University of Oslo, Norway
Araceli Contreras-Rodriguez, Instituto Politécnico Nacional (IPN), Mexico
Copyright © 2021 Reimer, Beniac, Hiebert, Booth, Chong, Westmacott, Zhanel and Bay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Denice C. Bay, ZGVuaWNlLmJheUB1bWFuaXRvYmEuY2E=
 Shelby L. Reimer
Shelby L. Reimer Daniel R. Beniac
Daniel R. Beniac Shannon L. Hiebert
Shannon L. Hiebert Timothy F. Booth
Timothy F. Booth Patrick M. Chong
Patrick M. Chong Garrett R. Westmacott
Garrett R. Westmacott George G. Zhanel
George G. Zhanel Denice C. Bay
Denice C. Bay