- 1Shanghai Municipal Center for Disease Control and Prevention, Shanghai, China
- 2The Jinshan District Center for Disease Control and Prevention, Shanghai, China
- 3Institute of Food Quality and Safety, University of Shanghai for Science and Technology, Shanghai, China
Listeria monocytogenes is the etiologic agent of listeriosis, which remains a significant public health concern in many countries due to its high case-fatality rate. The constant risk of L. monocytogenes transmission to consumers remains a central challenge in the food production industry. At present, there is very little known about L. monocytogenes contamination in ready-to-eat (RTE) processing plants in China. In this study, L. monocytogenes in an RTE meat processing plant in Shanghai municipality was characterized using pulsed-field gel electrophoresis (PFGE) and whole genome sequencing (WGS). Furthermore, the biofilm formation ability of the pathogen was also tested. Results revealed that L. monocytogenes isolates were present in 12 samples out of the 48 samples investigated. Most of them (66.7%, 8/12) were identified from the processing facilities irrespective of observed hygiene levels of aerobic plate count (APC) and coliforms. Coliforms were present in only one processing area. ST5 (1/2b) isolates were predominant (83.3%, 10/12) and were identified in two dominant pulsotypes (PTs) (three in PT3 and seven in PT4, respectively). Results of the core-genome multi-locus sequence typing (cgMLST) showed that ST5 in three PTs (PT1, PT3, and PT4) had 0–8 alleles, which confirmed that clonal transmission occurred in the RTE meat processing facilities. In addition, the biofilm formation test confirmed that the isolates from the processing facilities could form biofilms, which helped them colonize and facilitate persistence in the environment. These results indicated that common sanitation procedures regularly applied in the processing environment were efficient but not sufficient to remove L. monocytogenes isolates, especially biofilm of L. monocytogenes. Furthermore, the ST5 isolates in this study exhibited 12 alleles with one ST5 clinical isolate, which contributes to the understanding of the potential pathogenic risk that L. monocytogenes in RTE meat processing equipment posed to consumers. Therefore, strong hygienic measures, especially sanitation procedures for biofilms eradication, should be implemented to ensure the safety of raw materials. Meanwhile, continuous surveillance might be vital for the prevention and control of listeriosis caused by L. monocytogenes.
Introduction
Listeria monocytogenes is an important foodborne pathogen, which can cause severe human listeriosis, particularly in older adults, newborns, pregnant women, and immune-compromised individuals (Lomonaco et al., 2009). Listeriosis remains a significant public health concern due to the high case-fatality rate (Thomas et al., 2015). Various types of meat, especially ready-to-eat (RTE) meat products, are often vehicles of listeriosis outbreaks (Currie et al., 2015; Jensen et al., 2016; Thomas et al., 2020). A risk assessment report from the United States in 2003 attributed 90% of listeriosis cases to the consumption of contaminated RTE deli meats (Richard Whiting, 2003). In the United States, L. monocytogenes must not present in RTE foods at any point (USDA FSIS, 2003). A similar requirement in China is that L. monocytogenes is not detectable in 25 g or 25 ml RTE food according to GB29921 (GB29921-2013,2013).
L. monocytogenes survives well in the environment and can even colonize food production facilities for extended periods (Leong et al., 2017). L. monocytogenes is known to colonize niche areas such as drains and hard-to-clean surfaces, which allows the bacteria to survive or even proliferate and thus make it challenging to completely eradicate it (Ferreira et al., 2014). An epidemiological investigation of listeriosis outbreaks revealed significant lack of hygiene in processing facilities (Angelo et al., 2017). Several reported outbreaks caused by L. monocytogenes have been linked to contaminated food-contact surfaces, packing lines, and processing environments (McCollum et al., 2013; Angelo et al., 2017). A high prevalence of L. monocytogenes in food processing environments is often reported. However, a smaller number of studies have evaluated the incidence and identified the potential of L. monocytogenes contamination in RTE meat processing plants (Nastasijevic et al., 2017). The risk of L. monocytogenes transmission to consumers remains a central challenge for the food industry (Almeida et al., 2013; Malley et al., 2015).
Molecular typing of L. monocytogenes isolates can help to establish links between isolates from different sources and assist in tracing the original source of contamination (Chen et al., 2013). It has been reported that certain serotypes and clonal complexes (CCs) are more commonly encountered in clinical cases (Chenal-Francisque et al., 2011; Maury et al., 2016). In China, ST87 and ST8 were the most prevalent types of isolates from patients (Li et al., 2019). However, ST9 was the most common type of isolates from foods (Wang et al., 2012).
Pulsed-field gel electrophoresis (PFGE), as the “gold standard” typing method, has been used to characterize clusters of L. monocytogenes isolates through the National Molecular Tracing Network for Foodborne Disease Surveillance (TraNet) in China (Li et al., 2019). Whole genome sequencing (WGS) is a powerful tool for obtaining genomic data, which can help to determine sequence types (STs), serogroups, virulence, and resistance gene profiles (Li et al., 2017). More importantly, several molecular typing methods have been developed using WGS data such as core-genome multiple locus sequence typing (cgMLST) and single nucleotide polymorphism (SNP). Recently, WGS has been used to determine the contamination and/or colonization routes of pathogens within food processing environments (Food and Agriculture Organization of the United Nations, 2016; Wang et al., 2016). The aim of this study was (1) to identify the transmission routes of L. monocytogenes using PFGE and WGS via tracking L. monocytogenes isolates in an RTE meat processing plant in Shanghai, as well as (2) to provide a basis for measures to prevent and control the transmission of L. monocytogenes in RTE meat processing plants.
Materials and Methods
Processing of RTE Meat Products
An RTE cooked meat product processing plant in Shanghai was used to investigate L. monocytogenes contamination in RTE-processing environments and products as well as to track its transmission route. The plant is the one of the most important companies producing RTE meat products in Shanghai, and the RTE meat products are common food in Shanghai. The RTE meat products were processed as follows. Firstly, frozen meats were bought from trade companies as raw materials. Secondly, water thawing took place, and meats were pickled in a pickling liquid with many accessory materials including oil, salt, sauce, vinegar, herbs, and spices. Thirdly, the pickled meat products were boiled to produce intermediate products. These intermediate products were eventually processed by activities such as weighing and cutting into shapes to create end products. Finally, end products were packaged and transported to retail stores to be eventually purchased and used by consumers.
Sampling
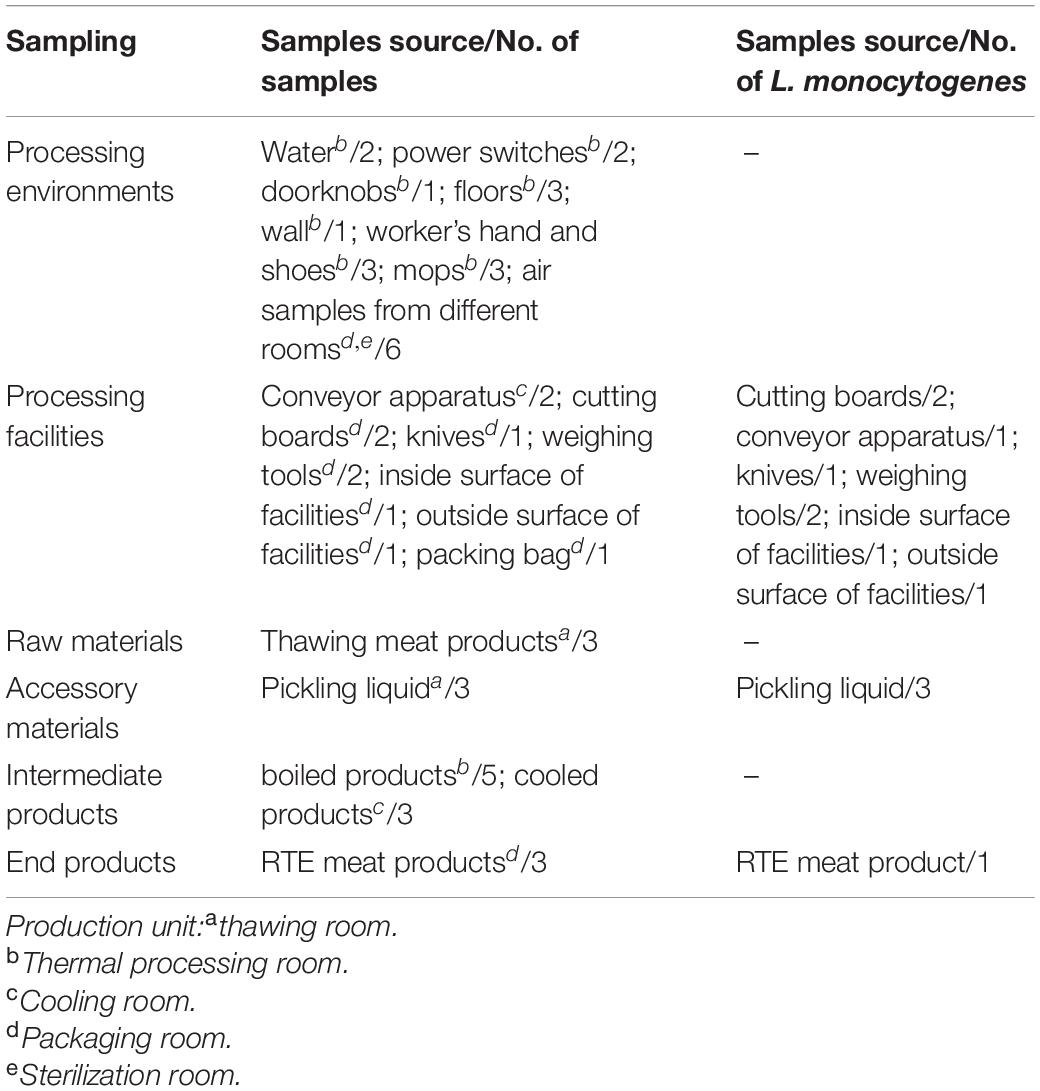
As shown in Table 1, a total of 48 samples were collected during one visit in July 2019, including 21 processing environmental samples, 10 processing facility samples, 3 raw materials, 3 accessory materials, 8 intermediate products, and 3 end products. After the cleaning and sanitation procedures were complete, the processed environmental samples and facility samples were collected from associated surfaces using pre-moistened swabs (Table 1). Air samples were collected using Anderson six-stage sampler. The raw materials were frozen meat products bought from trade companies. Intermediate products during the processing stage included pickled products and boiled products. The end products were RTE meat products, which were destined for retail and consumption. Samples of these products were delivered to the laboratory within 2 h, in a cold chain. Here, the levels of aerobic plate count (APC) and coliforms were determined, as well as the presence of L. monocytogenes.
Microbiological Analysis
Enumeration of APC and coliforms were determined according to the food safety national standards GB4789.2 (2016) and GB4789.3 (2016), respectively. L. monocytogenes was determined according to the food safety national standard GB4789.30 (2016). Regarding the standard, environmental swabs and raw and accessory materials were placed into L. monocytogenes Broth 1 (LB1) for pre-enrichment at 30°C for 24 h. Afterward, 100 μl LB1 was placed into L. monocytogenes Broth 2 (LB2) at 30°C for 24 h. One inoculation loop of LB2 was streaked on Polymyxin Acriflavine Licl Ceftazidime Esculin Mannitol Agar Plate (PALCAM). The isolates were identified as L. monocytogenes by standard biochemical tests (catalase; fermentation of dextrose, xylose, rhamnose, and mannitol; β-hemolysis; motility; and gram-staining). The positive control strain used in this study was L. monocytogenes ATCC 19114.
PFGE
PFGE for L. monocytogenes was performed with the PulseNet International protocol (Centers for Disease Control and Prevention, 2013). Based on the protocol, L. monocytogenes isolates were embedded into agarose plugs. Afterward, slices of the agarose plugs were digested using AscI (Takara, Dalian, China) for 3 h at 37°C. XbaI-digested Salmonella Braenderup H9812 DNA was used as a molecular size marker, and electrophoresis was conducted using the CHEF-DRII apparatus (Bio-Rad Laboratories, Hercules, CA, United States). Images were captured using the Gel Doc 2000 system (Bio-Rad) and were converted to TIFF files, which were analyzed by the BioNumerics software (version 7.7 Applied Maths, Kortrijk, Belgium). Finally, clustering was performed using the unweighted pair group method with arithmetic mean (UPGMA).
WGS
Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (QIAGEN, Germany) according to the manufacturer’s protocol except that the cells were pre-lysed with lysozyme for 30 min at 37°C and the proteinase K treatment was extended to 30 min. A Qubit Fluorometer (Invitrogen, United States) and a NanoDrop Spectrophotometer (Thermo Fisher Scientific, United States) were used to determine the concentration, quality, and integrity of the DNA. Sequencing libraries were generated using the TruSeq DNA Sample Preparation Kit (Illumina, United States). Afterward, genome sequencing was performed using the Illumina Hiseq platform (Illumina). Finally, the reads were trimmed and assembled using the CLC Genomics Workbench v7.0 (CLC Bio, Aarhus, Denmark), and the assembled contigs were exported as FASTA files for further analysis.
Ten ST5 L. monocytogenes isolates from foods and three isolates from patients were analyzed using WGS to be compared to 10 isolates in this study.
Serotypes, MLST, and Pathogenic Island Determination
Serotypes of L. monocytogenes isolates were identified using a commercially available L. monocytogenes antiserum test kit (Denka Seiken, Tokyo, Japan).
Multi-locus sequence typing (MLST) was defined by the Pasteur scheme (Burall et al., 2011) and based on the sequence analysis of the seven housekeeping genes, which were extracted from WGS data using the BioNumerics software.
The virulence associated genes extracted from WGS data using BioNumerics software were put into the Virulence Factor Database (VFDB) (MOH Key Laboratory of Systems Biology of Pathogen, Institute of Pathogen Biology, Beijing, China)1 in order to identify LIPI-1, LIPI-2, LIPI-3, and LIPI-4.
cgMLST Characterization
cgMLST typing was conducted based on the profile of 1,748 coding loci in the BigsDB Pasteur cgMLST2. Cluster analysis was conducted by applying a complete linkage using the BioNumerics software.
Biofilm Formation
The biofilm formation test was conducted according to Pang and Yuk (2018) with minor modifications. Stainless steel (304, Tull Metals Company, Atlanta, GA) coupons (2 cm × 1 cm × 0.2 cm) were soaked in an acetone solution for 3 h. After being wiped clean, these coupons were soaked in 70% (v/v) ethanol overnight and then rinsed with distilled water. After being air-dried and autoclaved at 121°C for 15 min, the coupons were ready for use.
Biofilm was developed on sterile coupons in tryptic soy broth with yeast extract (TSBYE). Firstly, the L. monocytogenes were inoculated into TSBYE and cultured overnight at 26°C. Secondly, the inoculum was washed two times with 0.9% sodium chloride and then added to 5 ml TSBYE at the final concentration of 105 CFU/ml for use on the coupons (one coupon in one tube). Afterward, the inoculated TSBYE samples were incubated at 26°C. After incubation for 24 and 48 h, the coupons were removed and placed into 5 ml 0.9% sodium chloride and vortexed for 10 s. Next, the liquid was discarded, and the coupons were placed into a centrifuge (12,000 rpm) with 10 ml 0.9% sodium chloride and glass beads. Finally, after vortexing for 2 min, the liquid was used to enumerate the biofilm cells.
Statistical Analysis
Each set of experiments was repeated three times, and its mean values (APC, coliforms) and standard deviation were calculated using the SPSS software (17.0, IBM, United States).
Results
APC and coliforms testing in various samples revealed the hygiene levels within the RTE meat processing plant. APC levels were the highest in raw and accessory materials, while coliforms levels were the highest in one processing facility (Table 2). The level of APC in air samples from processing rooms was 33 CFU/plate.
The L. monocytogenes isolate occurrence rate was 25% (12/48) (Table 1). Briefly, eight (8/12, 66.7%) L. monocytogenes isolates were detected in the processing equipment, which included nearly all production area that had direct contact with products, e.g., the cutting board, conveyor apparatus, knives, inside and outside surfaces of the equipment, and weighing tools (Table 1). The presence of L. monocytogenes was confirmed in all RTE processing facilities irrespective of the observed hygiene levels (Table 2).
The PFGE analysis of the comprised AscI divided the 12 isolates into 4 pulsotypes (PT1–4) (Figure 1). PT3 and PT4 values were similarly high (97.9%). They accounted for 83.3% (10/12) of the isolates. Two PTs (PT1, PT2) were presented by only one single isolate. Seven isolates in PT3 and PT4 were processing facility samples, two were from accessory material, and one was from an end product.
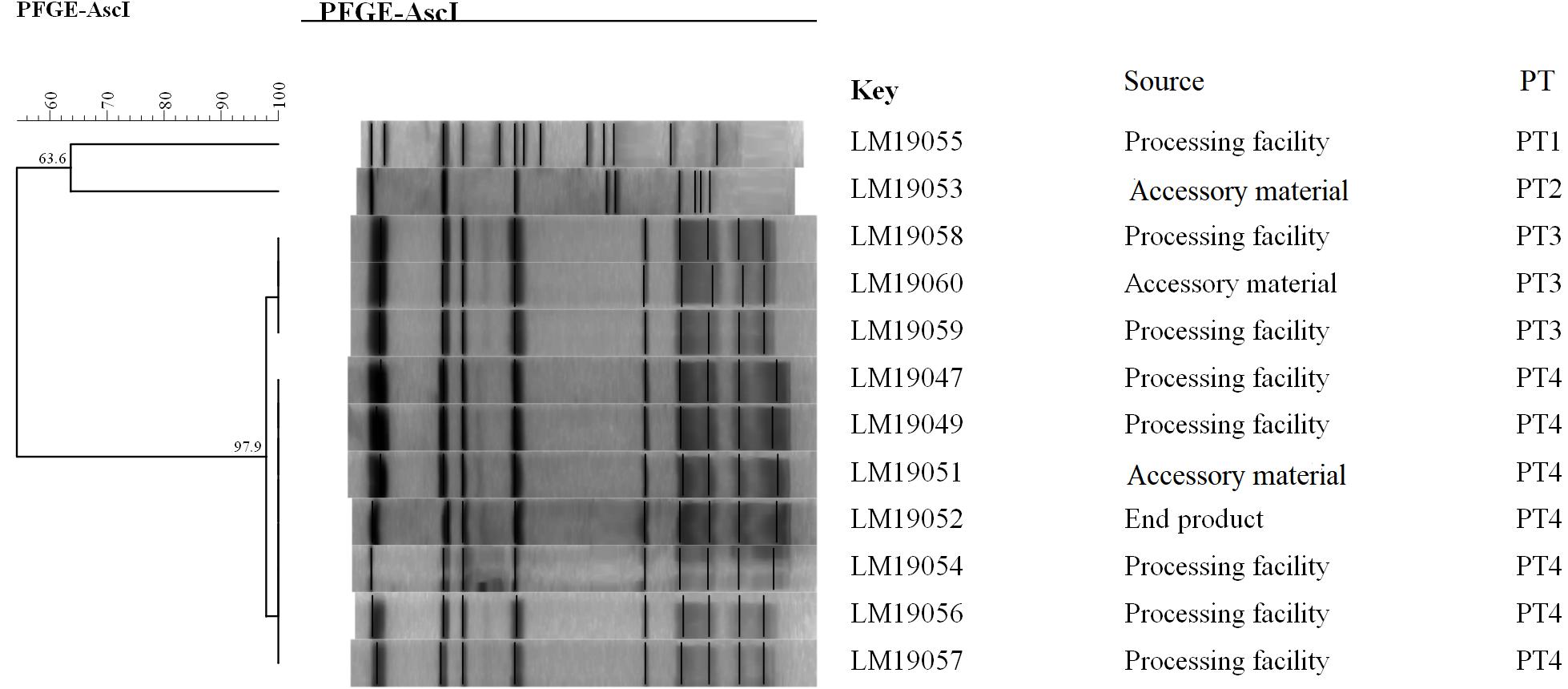
Figure 1. Relationships of the L. monocytogenes isolates based on PFGE. The 12 L. monocytogenes isolates were analyzed by PFGE using AscI.
The molecular typing using WGS data showed that 11 of 12 L. monocytogenes isolates belonged to ST5 (1/2b) with LIPI-1 (Listeria pathogenicity island-1). These ST5 isolates were compared with 10 other ST5 (1/2b) isolates from food products and patients using cgMLST. Nine clusters (CL1-CL9) were obtained (Figure 2). The L. monocytogenes isolates of three PTs (PT1, PT3 and PT4) and one clinical isolate belonged to CL8 and had 0–12 alleles based on cgMLST. However, >70 alleles were found between the isolates in CL8 and other CLs.
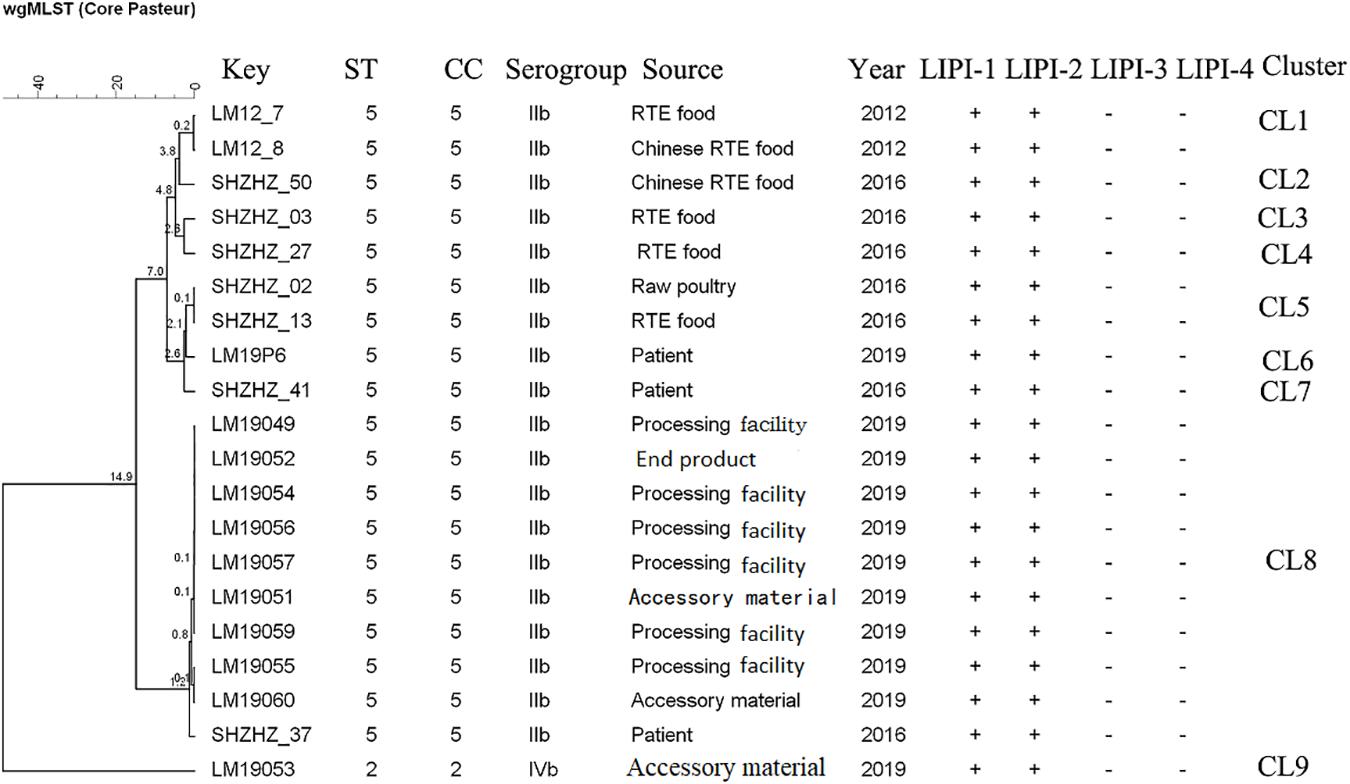
Figure 2. Minimum spanning tree of cgMLST data for 20 L. monocytogenes isolates. The multiplication by 10 in the tree represents the number of different alleles among isolates. The corresponding data, including the name of the isolate (key), MLST type (ST), MLST clonal complex (CC), source, year, Listeria pathogenicity island (LIPI-1, LIPI-2, LIPI-3, and LIPI-4), and cluster are shown alongside the dendrogram to the right.
After incubation at 26°C for 24 h, the minimum of attached cells of LM19047 isolates was 5.39 Log CFU/cm2, and the maximum of attached cells of 60 LM19060 isolates was 6.50 Log CFU/cm2, which were statistically different (p < 0.05) (Figure 3). However, with increasing biofilm age, the attached cells of LM19047 isolate increased up to 6.21 Log CFU/cm2, and that of LM19060 decreased to 5.69 Log CFU/cm2. The ability of biofilm forming of other isolates at different time was different (Figure 3).
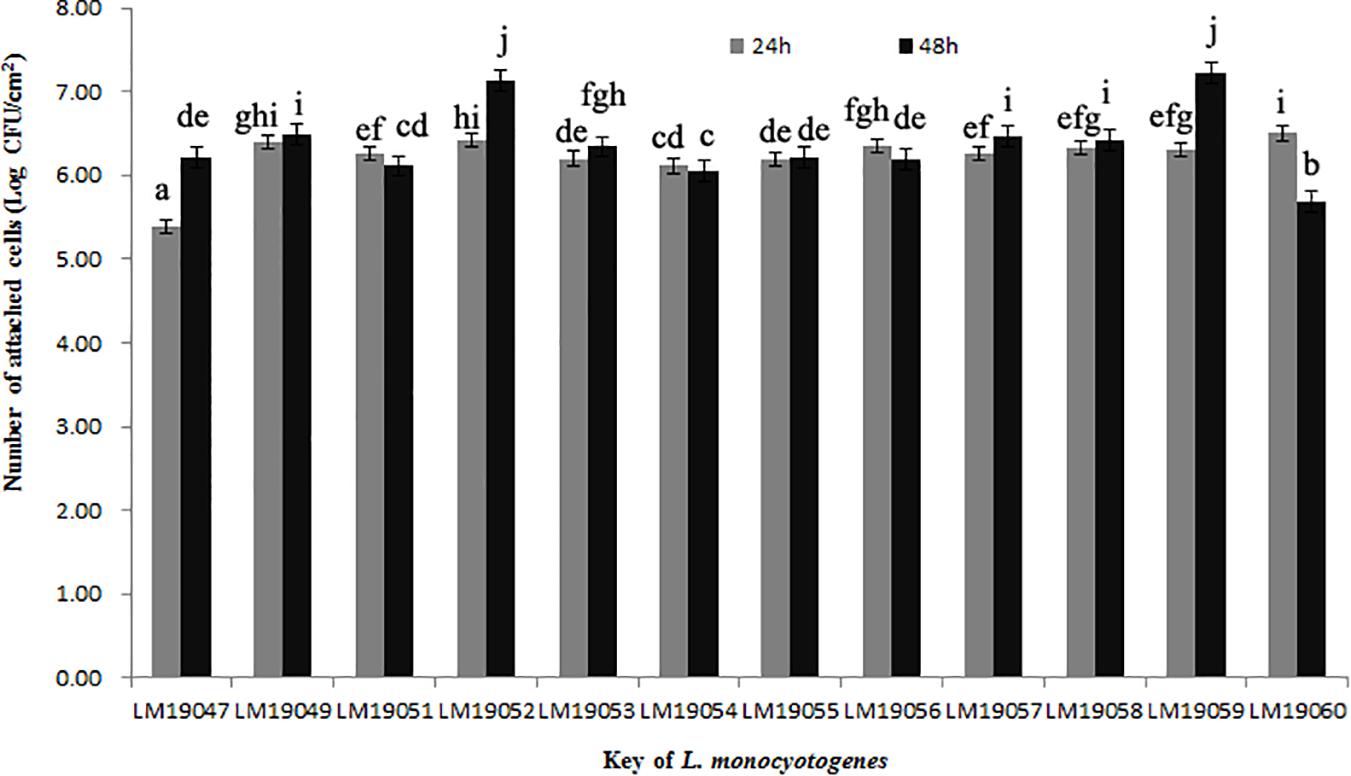
Figure 3. Population (Log CFU/cm2) of 12 L. monocytogenes isolates biofilms at 26°C on 24 and 48 h. Mean values at the different incubation time and different isolates with different lowercase letters are statistically different (p < 0.05).
Discussion
Hygiene indicators (APC and coliforms) are used to reflect specific food establishment practices and temporal influences (Weatley et al., 2014). APC was confirmed in only one processing facility, while only one end product was confirmed with APC and coliform (Table 2). Furthermore, the mean level of APC in air samples in RTE processing rooms was 33 CFU/plate, which can be permitted in the food processing environment. These results indicated that good hygiene practices were implemented in this plant.
Contrarily, 8 of the 12 L. monocytogenes isolates were confirmed in 8 processing facilities, which presented the predominant contamination scenarios. These findings were similar to a previous study that was conducted on L. monocytogenes colonized under these scenarios in a meat establishment (Nastasijevic et al., 2017). The confirmation of L. monocytogenes was irrespective of the observed hygiene levels (for example, in contrast with the confirmation of L. monocytogenes in processing facilities, only one processing facility was confirmed with APC). Therefore, it was still difficult to remove L. monocytogenes isolates in this plant although good hygienic practices were implemented.
L. monocytogenes isolates were confirmed in accessory materials, intermediate products, and an end product, but not in raw materials, which suggested that L. monocytogenes isolates might be introduced to these products from processing facilities by cross-contamination. A similar contamination model reported that when L. monocytogenes entered into food processing plants, recontamination and persistence frequently occurred (Chambel et al., 2007). L. monocytogenes isolates were able to survive in niche areas of the facilities and could adapt to stress factors such as low temperature and low pH (Melo et al., 2015). Furthermore, L. monocytogenes isolates could persist in processing equipment for a long time (Unnerstad et al., 1996). As a result, contamination could exist for an extended period.
The epidemiological data showed that 12 L. monocytogenes isolates were detected in the same RTE meat processing plant at the same time, 10 of which shared an indistinguishable PT (PT3 and PT4, respectively) (Figure 1). Furthermore, the PFGE pattern of L. monocytogenes isolates in PT3 was highly similar to those in PT4, which might suggest the same ancestor. These results indicated clone transmission of L. monocytogenes isolates occurred in processing facilities in this plant.
Though PFGE is a useful tool for the characterization of the subtypes of L. monocytogenes isolates, it lacks discriminatory power to distinguish among closely related bacterial strains, which is essential for source tracking (Lomonaco and Nucera, 2014). Thus, WGS was used for obtaining better information about the genetic similarity between isolates. The indistinguishable L. monocytogenes isolates with the same PFGE pattern could be differentiated by cgMLST (Figure 2), which has a higher discriminatory power. cgMLST analysis of L. monocytogenes isolates in three PTs (PT1, PT2, and PT3) showed 0–8 allelic differences (Figure 2), these isolates could be identified as the same clone (Ruppitsch et al., 2015). Furthermore, seven of them were from processing facility samples, two from accessory materials, and one from an end product, which confirmed the clone transmission of L. monocytogenes isolates in the processing environment. Furthermore, in-house evolution occurred in L. monocytogenes isolates with one or eight alleles, which suggested that these L. monocytogenes isolates might have existed for a long period and also might be persistent in processing environments.
WGS data showed that most L. monocytogenes isolates (11/12) were ST5, CC5, 1/2b (Table 2). Our previous study indicated that the predominant L. monocytogenes isolates from both food and clinical isolates were ST5 in Shanghai, China (data unpublished). Similarly, ST5 was the most predominant ST in RTE meat product in Nanjing, China (Wang et al., 2015). Wang et al. reported that the ST5 had been identified as an important ST in China (Wang et al., 2012). There is no obvious difference between the distribution of the frequency of CC5 in foods and patients in France (Maury et al., 2016). The ST5 strains have been globally disseminated in geographically distant areas, e.g., Austria, Canada, Australia, Switzerland, and Finland (Schmid et al., 2014; Buchanan et al., 2017; Meier et al., 2017). Several outbreaks caused by L. monocytogenes have been linked to ST5 isolates (Buchanan et al., 2017). It is worth noting that 1 clinical isolate has 12 alleles with L. monocytogenes isolates obtained in this study belonging to CL8, which were from the RTE meat processing plant. Although there is a lack of epidemiological data confirming the relationship between them, the results suggested the potential risk of pathogenicity that these isolates pose to the consumers. Further studies are needed to uncover the pathogenicity of ST5.
ST5 isolates have been previously reported to be dominant in heavily contaminated food processing environments even after efforts on intensifying hygienic measures (Muhterem-Uyar et al., 2018). A further study indicated that ST5 plasmids harbored an efflux pump system (bcrABC cassette) and heavy metal resistance genes, which possibly provide a higher tolerance to disinfectants (Muhterem-Uyar et al., 2018). These L. monocytogenes isolates might exist in biofilm formation. L. monocytogenes biofilms could be formed on many different surfaces during food processing operations and provide a protective environment for bacterial survival and thereby increase the risk of subsequent contamination (Colagiorgi et al., 2017). Once established, L. monocytogenes biofilms act as permanent sources of contamination and dispersal in the environment and can lead to cross-contamination (Markkula et al., 2005). In this study, 12 L. monocytogenes isolates from the plant could form biofilm stainless steel coupons, which presented typical food-contact surfaces in food processing plants (Figure 3). These findings suggested that L. monocytogenes isolates in the processing equipment could not be cleaned due to biofilms formation, and L. monocytogenes biofilms might be persistent in the plant. Therefore, continuous surveillance and prevention strategies against L. monocytogenes should be implemented in this RTE meat processing plant to ensure food safety.
Conclusion
In this study, cross-contamination of L. monocytogenes in an RTE meat plant has occurred. The food processing facilities were heavily contaminated by ST5 (1/2b) isolates even though good hygienic measures had been implemented in this plant. The molecular typing and epidemiological data confirmed that clone transmission occurred in the plant. Furthermore, the clone had a strong ability to form biofilm in food-contact surfaces, which might be the reason that it could not be eradicated in the processing facilities. Furthermore, the ST5 (1/2b) isolates in this study had potential pathogenicity for having 12 alleles with clinical isolate. Therefore, continuous surveillance and effective measures to eradicate L. monocytogenes should be taken to ensure food safety.
Data Availability Statement
We have uploaded completely the WGS data of L. monocytogenes in this study in Genbank of NCBI.
Author Contributions
HZ, FQ, and BX collected the samples, analyzed the samples, performed the WGS, and analyzed the data. HZ drafted the manuscript. XZ and HL designed the study and revised this manuscript. LS tested the formation of biofilm. YZ and WC performed the PFGE. YY was involved in the collection of isolates. QD revised this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the 5th Three-year Action Plan for Public Health of Shanghai (This project No. GWV-2).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Almeida, G., Magalhaes, R., Carneiro, L., Santos, I., Silva, J., Ferreira, V., et al. (2013). Foci of contamination of Listeria monocytogenes in different cheese processing plants. Int. J. Food Microbiol. 167, 303–309. doi: 10.1016/j.ijfoodmicro.2013.09.006
Angelo, K. M., Conrad, A. R., Saupe, A., Dragoo, H., West, N., Sorenson, A., et al. (2017). Multistate outbreak of Listeria monocytogenes infections linked to whole apples used in commercially produced, prepackaged caramel apples: United States, 2014–2015. Epidemiol. Infect. 145, 848–856. doi: 10.1017/s0950268816003083
Buchanan, R. L., Gorris, L. G., Hayman, M. M., Jackson, T. C., and Whiting, R. C. (2017). A review of Listeria monocytogenes: an update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 75, 1–13. doi: 10.1016/j.foodcont.2016.12.016
Burall, L. S., Simpson, A. C., and Datta, A. R. (2011). Evaluation of a serotyping scheme using a combination of an antibody-based serogrouping method and a multilex PCR assay for identifying the major serotypes of Listeria monocyotgenes. J. Food Prot. 74, 403–409. doi: 10.4315/0362-028X.JFP-10-355
Centers for Disease Control and Prevention (2013). Standard Operating Procedure for PulseNet PFGE of Listeria Monocytogenes. PNL04 last Update July 2017. Atlanta, GA: Centers for Disease Control and Prevention. Available online at: http://www.cdc.gov/pulsenet/pathogens/index/html.
Chambel, L., Sol, M., Fernandes, I., Barbosa, M., Zilhão, I., Barata, B., et al. (2007). Occurrence and persistence of Listeriosis spp. in the environment of ewe and cow’s milk cheese dairies in Portugal unveiled by an integrated analysis of identification, typing and spatial-temporal mapping along production cycle. Int. J. Food Microbiol. 116, 52–63. doi: 10.1016/j.ijfoodmicro.2006.12.035
Chen, Y., Luo, Y., Carleton, H., Timme, R., Melka, D., Muruvanda, T., et al. (2013). Whole genome and core genome multilocus sequence typing and single nucleotide polymorphism analyses of Listeria monocytogenes associated with an outbreak linked to cheese, United States. Appl. Environ. Microbiol. 83, e633–17. doi: 10.1128/AEM.00633-17
Chenal-Francisque, V., Lopez, J., Cantinelli, T., Caro, V., Tran, C., Leclercq, A., et al. (2011). Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 17, 1110–1112. doi: 10.3201/eid1706.101778
Colagiorgi, A., Bruini, I., Di Ciccio, P. A., Zanardi, E., Ghidini, S., and Ianieri, A. (2017). Listeria monocytogenes biofilms in the wonderland of food industry. Pathogens 6:41. doi: 10.3390/pathogens6030041
Currie, A., Farber, J. M., Nadon, C., Sharma, D., Whitfield, Y., Gaulin, C., et al. (2015). Multi-province listeriosis outbreak linked to contaminated Deli meat consumed primarily in institutional settings, Canada, 2008. Foodborne Patho. Dis. 12, 645–652. doi: 10.1089/fpd.2015.1939
Ferrira, V., Wiedmann, M., Teixeira, P., and Stasiewicz, M. J. (2014). Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J. Food Prot. 77, 150–170. doi: 10.4315/0362-028X.JFP-13-150
Food and Agriculture Organization of the United Nations (2016). Applications of Whole Genome Sequencing (WGS) in Food Safety Management. In: Technical Background Paper Draft Version 6:11. April 2016. Rome: Food and Agriculture Organization of the United Nations.
GB29921-2013 (2013). National Standard of Food Safety. The Permitted Levels of Pathogen in Food. National Standards of the Republic of China.
GB4789.2 (2016). National Standard of Food Safety. Detection of Aerobic Plate Count. National Standards of the Republic of China.
GB4789.3 (2016). National Standard of Food Safety. Detection of Coliforms. National Standards of the Republic of China.
GB4789.30 (2016). National Standard of Food Safety. Detection of Listeria Monocytogenes. National Standards of the Republic of China.
Jensen, A. K., Nielsen, A. M., BjÖrkman, J. T., Jensen, T., Müller, L., Persson, S., et al. (2016). Whole-genome sequencing used to investigate a nationwide outbreak of listeriosis caused by ready-to-eat delicatessen meat, Denmark, 2014. Clin. Infect. Dis. 63, 1–23. doi: 10.1093/cid/ciw192
Leong, D. K., NicAogain, L., Luque-Sastre, O., McManamon, K., Hunt, A., Alvarez-Ordóñez, A., et al. (2017). A 3-year multi-food study of the presence and persistence of Listeria monocytogenes in 54 small food businesses in Ireland. Int. J. Food Microbiol. 249, 18–26. doi: 10.1016/j.ijfoodmicro.2017.02.015
Li, W. W., Bai, L., Ma, X. C., Zhang, X., Li, X., Yang, X., et al. (2019). Sentinel Listeriosis surveillance in selected hospitals, China, 2013-2017. Emerg. Infect. Dis. 25, 2274–2277. doi: 10.3201/eid2512.180892
Li, Z., Pérez-Osorio, A., Wang, Y., Eckmann, K., Glover, W. A., Allard, M. W., et al. (2017). Whole genome sequencing analysis of Listeria monocytogenes that persisted in a milkshake machine for a year and caused illnesses in Washington State. BMC Microbiol. 17:134. doi: 10.1186/s12866-017-1043-1
Lomonaco, S., Decastelli, L., Nucera, D., Gallina, S., Bianchi, D. M., and Civera, T. (2009). Listeria monocytogenes in Gorgonzo;a: subtypes, diversity and persistence over time. Int. J. Food Microbiol. 128, 516–520. doi: 10.1016/j.ijfoodmicro.2008.10.009
Lomonaco, S., and Nucera, D. (2014). “Molecular subtyping methods for Listeria monocytogenes: tools for tracking and control,” in DNA Methods in Food Safety: Molecular Typing of Foodborne and Waterborne Bacterial Pathogens, eds O. A. Oyarzabal and S. Kathariou (Hoboken, NJ: Wiley-Blackwell).
Malley, T. J., Butts, J., and Wiedmann, M. (2015). Seek and destroy process: Listeria monocytogenes process controls in the ready-to-eat meat and poultry industry. J. Food Prot. 78, 436–455. doi: 10.4315/0362-028X.JFP-13-507
Markkula, A., Autio, T., Lundén, F., and Korkeala, H. (2005). Raw and processed fish show identical Listeria monocytogenes genotypes with pulsed-field gel electrophoresis. J. Food Prot. 68, 1228–1231. doi: 10.1089/cmb.2005.12.554
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
McCollum, J. T., Cronquist, A. B., Silk, B. J., Jackson, K. A., O’Connor, K. A., Cosgrove, S., et al. (2013). Multistate outbreak of listeriosis associated with cantaloupe. N. Engl. J. Med. 369, 944–952. doi: 10.1056/NEJMoa1215837
Meier, A. B., Guldimann, C., Markkula, A., Pontinen, A., Korkeala, H., and Tasara, T. (2017). Comparative phenotypic and genotypic analysis of Swiss and Finnish Listeria monocytogenes isolates with respect to benzalkonium chloride resistance. Front. Microbiol. 8:397. doi: 10.3389/fmicb.2017.00397
Melo, J., Andrew, P., and Faleiro, M. (2015). Listeria monocytogenes in cheese and the dairy environment remains a food safety challenge: the role of stress responses. Food Res. Int. 67, 75–90. doi: 10.1016/j.foodres.2014.10.031
Muhterem-Uyar, M., Ciolacu, L., Wagner, K. H., Wagner, M., Schmitz-Esser, S., and Stessi, B. (2018). New aspects on Listeria monocytogenes ST5-ECVI predominant in a heavily contaminated cheese processing environment. Front. Microbiol. 9:64. doi: 10.3389/fmicb.2018.00064
Nastasijevic, I., Milanov, D., Velebit, B., Djordjevic, V., Swift, C., Painset, A., et al. (2017). Tracking of Listeria monocytogenes in meat establishment using Whole Genome Sequencing as a food safety management tool: a proof of concept. Int. J. Food Microbiol. 257, 157–164. doi: 10.1016/j.ijfoodmicro.2017.06.015
Pang, X. Y., and Yuk, H. G. (2018). Effect of Pseudomonas aeruginosa on the sanitizer sensitivity of Salmonella Enteritidis biofilm cells in chicken juice. Food Control 86, 59–65. doi: 10.1016/j.foodcont.2017.11.012
Richard Whiting, F. (2003). Quantitative Assessment of Relative Risk to Public Health from Foodborne Listeria monocytogenes Among Selected Categories of Ready-to-Eat Foods. Silver Spring, MD: Food and Drug Administration.
Ruppitsch, W., Pietzka, A., Prior, K., Bletz, S., Fernandez, H. L., Allerberger, F., et al. (2015). Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 53, 2869–2876. doi: 10.1128/jcm.01193-15
Schmid, D., Allerberger, F., Huhulescu, S., Pietzka, A., Amar, C., Kleta, S., et al. (2014). Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011-2013. Clin. Microbiol. Infect. 20, 431–436. doi: 10.1111/1469-0691.12638
Thomas, J., Govender, N., McCarthy, K. M., Erasmus, L. K., Doyle, T. J., Allam, M., et al. (2020). Outbreak of listeriosis in South Africa associated with processed meat. N Engl. J. Med. 382, 632–643 doi: 10.1056/NEJMoa1907462
Thomas, M. K., Vriezen, R., Farber, J., Currie, A., Schlech, W., and Fazil, A. (2015). Economic cost of a Listeria monocytogenes outbreak in Canada, 2008. Foodborn Pathog. Dis. 12, 966–971. doi: 10.1089/fpd.2015.1965
Unnerstad, H., Bannerman, E., Bille, J., Danielsson-Tham, M.-L., Waak, E., and Tham, W. (1996). Prolonged contamination of a dairy with Listeria monocytogenes. Netherlands Milk Dairy J. 50, 493–499.
USDA FSIS (2003). FSIS Risk Assessment for Listeria monocytogenes in Deli Meats. Available online at: http://www.fsis.usda.gov/OPPDE/rdad/FRPUbs/97-013F/ListeriaReport.PDF (accessed May 26, 2003).
Wang, S., Weller, D., Falardeau, J., Strawn, L. K., Mardones, F. O., Adell, A. D., et al. (2016). Food safety trends: from globalization of whole genome sequencing to application of new tools to prevent foodborne disease. Trends Food Sci. Technol. 57, 188–198. doi: 10.1016/j.tifs.2016.09.016
Wang, Y., Jiao, Y., Lan, R., Xu, X., Liu, G., Wang, X., et al. (2015). Characterization of Listeria monocytogenes isolated from huan listeriosis cases in China. Emerg. Microbes. Infect. 4:e50. doi: 10.1038/emi.2015.50
Wang, Y., Zhao, A., Zhu, R., Lan, R., Jin, D., Cui, Z., et al. (2012). Genetic diversity and molecular typing of listeria monocytogenes in China. BMC Microbiol. 12:119. doi: 10.1186/1471-2180-12-119
Keywords: Listeria monocytogenes, RTE meat processing plant, PFGE, WGS, cgMLST
Citation: Zhang H, Que F, Xu B, Sun L, Zhu Y, Chen W, Ye Y, Dong Q, Liu H and Zhang X (2021) Identification of Listeria monocytogenes Contamination in a Ready-to-Eat Meat Processing Plant in China. Front. Microbiol. 12:628204. doi: 10.3389/fmicb.2021.628204
Received: 11 November 2020; Accepted: 06 January 2021;
Published: 25 February 2021.
Edited by:
Jun Lin, The University of Tennessee, Knoxville, United StatesReviewed by:
Thomas S. Hammack, United States Food and Drug Administration, United StatesGonçalo Nieto Almeida, Instituto Nacional Investigaciao Agraria e Veterinaria (INIAV), Portugal
Copyright © 2021 Zhang, Que, Xu, Sun, Zhu, Chen, Ye, Dong, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Liu, bGl1aG9uZ0BzY2RjLnNoLmNu; Xi Zhang, emhhbmd4aUBzY2RjLnNoLmNu
†These authors have contributed equally to this work
 Hongzhi Zhang
Hongzhi Zhang Fengxia Que2†
Fengxia Que2† Biyao Xu
Biyao Xu Linjun Sun
Linjun Sun Yanqi Zhu
Yanqi Zhu Wenjie Chen
Wenjie Chen Yulong Ye
Yulong Ye Qingli Dong
Qingli Dong Xi Zhang
Xi Zhang