
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 March 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.628133
This article is part of the Research Topic Plasmid Transfer: Mechanisms, Ecology, Evolution, and Applications View all 34 articles
 Rui Xiong1†
Rui Xiong1† Yuyang Liu1†
Yuyang Liu1† Jieying Pu2†
Jieying Pu2† Jianping Liu2
Jianping Liu2 Dexiang Zheng2
Dexiang Zheng2 Jianming Zeng2
Jianming Zeng2 Cha Chen2*
Cha Chen2* Yang Lu2*
Yang Lu2* Bin Huang2*
Bin Huang2*Indole works as an interspecies signal molecule to regulate multiple physiological activities, like antibiotic resistance, acid resistance, and virulence. However, the effect of indole on conjugation is unknown. Here, with Escherichia coli SM10λπ as a donor strain that carries a chromosomally integrated conjugative RP4 plasmid, we explored the effect of indole on conjugation of a mobilizable pUCP24T plasmid imparting gentamycin resistance. The results showed that exogenous indole treatment inhibited conjugative transfer of pUCP24T from SM10λπ to recipient strains, Pseudomonas aeruginosa PAO1 and E. coli EC600. Furthermore, raising endogenous indole production through overexpression of TnaA, a tryptophanase, in SM10λπ significantly inhibited both SM10λπ-PAO1 and SM10λπ-EC600 conjugation, whereas deficiency of tnaA reversed the phenotype. Subsequent mechanistic studies revealed that exogenous indole significantly inhibited the expression of mating pair formation gene (trbB) and the DNA transfer and replication gene (trfA), mainly due to the promotion of regulatory genes (korA and korB), and the result was confirmed in tnaA knockout and overexpression strains. Additionally, we found that both extracellular indole production and tnaA expression of SM10λπ were downregulated by ciprofloxacin (CIP). Intriguingly, one-eighth minimum inhibitory concentration of CIP treatment clearly facilitated both SM10λπ-PAO1 and SM10λπ-EC600 conjugation, and indole inhibited CIP-induced conjugation frequency. These data suggest that indole may play a negative role in the process of CIP-induced conjugation. This is the first study to reveal the biological function of indole-inhibiting conjugation and its role in CIP-induced conjugation, which may be developed into a new way of controlling the spread of antibiotic resistance.
Conjugation achieves the transmission of bacterial genetic materials, especially plasmids that carry antibiotic resistance genes, in a unidirectional manner from a donor cell to a recipient cell (Waksman, 2019), mediating the process of horizontal gene transfer (HGT) and inducing the spread of antibiotic resistance. It requires a type IV secretion system (T4SS), which is encoded by Mating pair formation (Mpf) genes to form the conjugative pore, and DNA transfer and replication (Dtr) genes-encoded relaxosome composed of the relaxase, which nicks at the origin of transfer (oriT) and other auxiliary proteins (Schröder and Lanka, 2005; De La Cruz et al., 2010; Koraimann and Wagner, 2014). Conjugative plasmids, such as the well-studied IncP-1α plasmid RP4, generally carry all the structural genetic elements required to perform transfer (an oriT site, Dtr, and Mpf genes) (Pansegrau et al., 1990; Frost et al., 2005; Carballeira et al., 2014). Specifically, expression of plasmid RP4 conjugation genes is regulated by KorA, KorB, and TrbA, which act by binding to conserved nucleotide sequences localized in replication and maintenance regions, as well as transfer-essential Tral and Tra2 regions (Balzer et al., 1992; Zatyka et al., 1994; Jagura-Burdzy and Thomas, 1997). In contrast, mobilizable plasmids carry partial genetic information necessary for transfer (an oriT region and Dtr), and so usually there is a need to leverage the corresponding conjugative apparatus of co-resident self-transmissible plasmids to effect transfer (Francia et al., 2004).
The process of conjugation can be affected and regulated by many factors, summed up from two perspectives: (i) the internal environment involved in the interactions between the different genetic elements and (ii) the external environment including antibiotics, metals, carbon compounds, and quorum sensing (QS) (Banuelos-Vazquez et al., 2017). Specifically, the sub-minimal inhibitory concentration (sub-MIC) of antibiotics, such as gentamicin (Gm), sulfamethoxazole, tetracycline, and ciprofloxacin (CIP), increases the frequency of conjugation (Jutkina et al., 2016, 2018; Shun-Mei et al., 2018). The SOS response was revealed to promote horizontal dissemination of antibiotic resistance genes (Beaber et al., 2004). As for the effects of QS, our previous study has showed that the QS system signal molecules N-acyl homoserine lactones (AHLs) of Pseudomonas aeruginosa inhibit conjugation by activating SdiA in Escherichia coli (Lu et al., 2017b). Because indole is a signal molecule like QS, we were intrigued to explore whether they may play a role in conjugation.
Indole is used as a biochemical identification index of bacteria, whose production is controlled by the tryptophanase operon, including a promoter, a regulatory gene tnaC, and two structural genes tnaA and tnaB encoding tryptophanase and tryptophan permease, respectively (Yanofsky et al., 1991). However, a growing number of studies elucidate the function of indole as a signal molecule: (i) indole is a plateauing signal molecule (Kobayashi et al., 2006) which acts extracellularly to activate genes of metabolic enzymes in a concentration-dependent manner (Wang et al., 2001) and (ii) indole increases drug resistance by inducing intrinsic xenobiotic exporter genes in E. coli (Hirakawa et al., 2005). But there are no reports about indole working as a signal molecule to regulate conjugation.
The biosynthesis of indole is closely related to many environmental factors, such as cell density, carbon source, temperature, pH, and antibiotics. In terms of the latter, E. coli produced a higher level of extracellular indole in the presence of ampicillin and kanamycin (Han et al., 2011). We have previously disclosed the promoting effect of CIP on conjugation (Shun-Mei et al., 2018), but the underlying mechanism, especially whether indole was involved in this process, was not explored. In this study, we aimed to determine if indole affects conjugation, if it further participates in the process of antibiotics-induced conjugation.
To elucidate the effect of indole on both intraspecies and interspecies plasmid transfer, E. coli SM10λπ-PAO1 and SM10λπ-EC600 conjugational models established in our previous study were taken into application (Lu et al., 2017b). We first determined the appropriate concentration of exogenous indole that might not threaten the growth of SM10λπ, PAO1, and EC600, since it has been reported that bacterial conjugational efficiency is closely related to the growth state of bacteria (Schuurmans et al., 2014). As shown in Supplementary Figure 1, the growth of SM10λπ, PAO1, and EC600 was only trivially affected by indole even at concentration as high as 500 μM. Then the appropriate concentrations (10, 50, 250, and 500 μM) of exogenous indole were used to treat SM10λπ-PAO1 and SM10λπ-EC600 conjugational models. The results showed that 250 μM indole was needed to start inhibiting SM10λπ-PAO1 conjugation, while 50 μM indole was sufficient to inhibit SM10λπ-EC600 conjugation. Both conjugation models were significantly depressed by indole in a dose-dependent manner (Figure 1A), although in the SM10λπ-EC600 conjugation model, the conjugation frequency of 500 μM indole treatment group was higher than that of 250 μM indole treatment group, which may be interpreted as high concentration indole, like antibiotics, increases the frequency of conjugation by promoting the growth of transconjugants (Lopatkin et al., 2016).
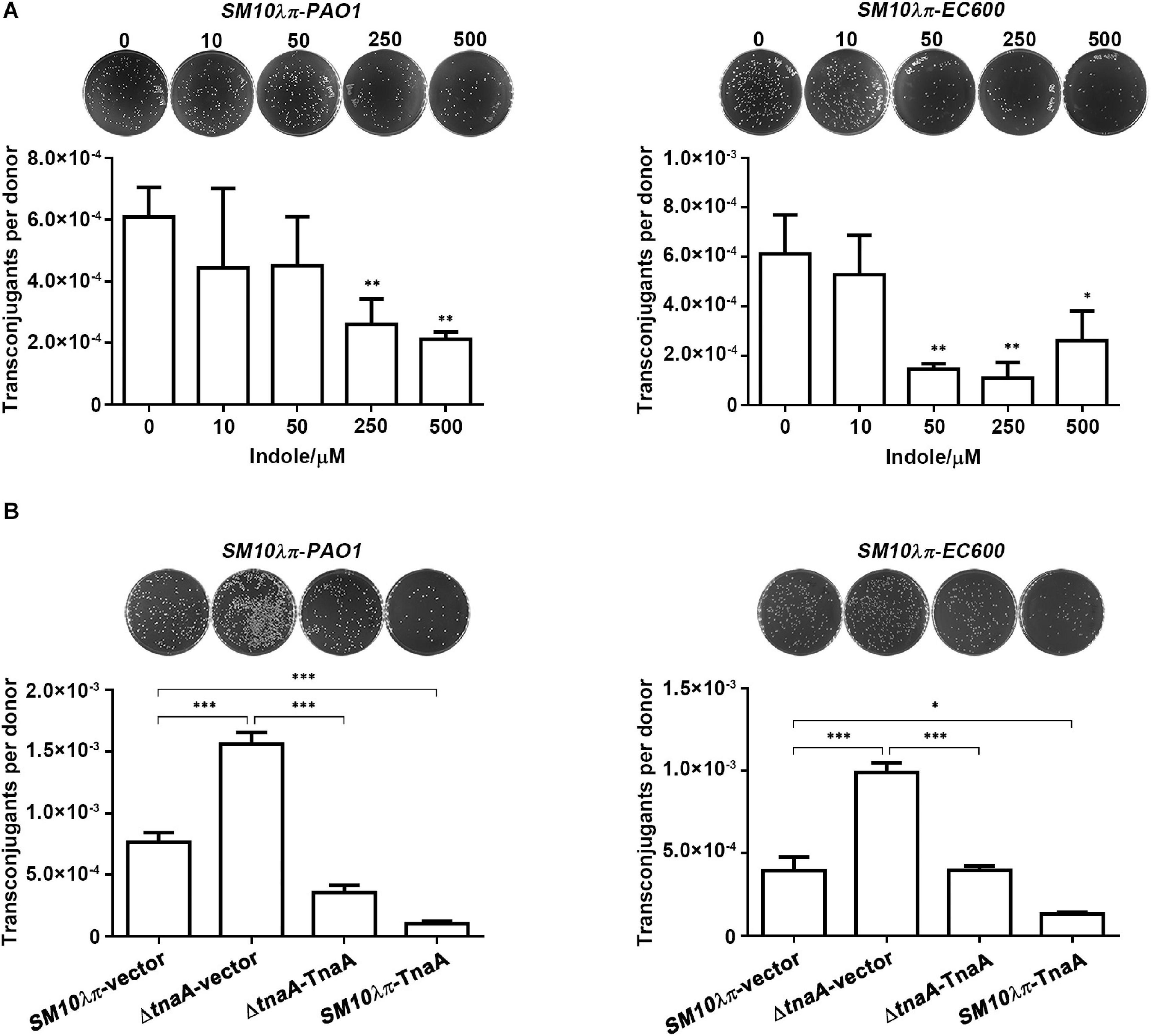
Figure 1. Indole inhibited SM10λπ-PAO1 and SM10λπ-EC600 conjugation. (A) Effect of exogenous indole on SM10λπ-PAO1 and SM10λπ-EC600 conjugation. Donor SM10λπ and recipient PAO1 or EC600 cells (1 × 107 CFU/ml each) were mated in the presence of indicated concentrations of indole at 37°C for 6 h. (B) Effect of endogenous indole on SM10λπ-PAO1 and SM10λπ-EC600 conjugation. SM10λπ wild-type strain (SM10λπ-vector), tnaA-deficient strain (ΔtnaA-vector), TnaA overexpression strain in wild-type (SM10λπ-TnaA), and tnaA mutation (ΔtnaA-TnaA) were mated with PAO1 or EC600 cells (1 × 107 CFU/ml each), respectively, at 37°C for 6 h. Values are means ± SEMs from at least three independent experiments; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
To validate the role of endogenous indole in conjugation, we subsequently constructed tnaA gene knockout and overexpression strains in SM10λπ, as tryptophanase encoded by tnaA hydrolyzes tryptophan to create indole, pyruvate, and ammonia (Newton et al., 1965; Deeley and Yanofsky, 1981). Naturally, we can barely detect indole and tnaA expression in the tnaA-deficient strain (ΔtnaA-vector), while overexpression of TnaA in wild-type (SM10λπ-TnaA) and the tnaA mutation (ΔtnaA-TnaA) strain both delivered much higher levels of indole and tnaA expression (Supplementary Figure 2). We next examined the conjugation frequency by using these constructed strains as donor cells. Compared to the wild-type strain, deficiency of tnaA in SM10λπ significantly enhanced both SM10λπ-PAO1 and SM10λπ-EC600 conjugation, whereas introducing a plasmid of TnaA overexpression reversed the phenotype. In addition, introducing a TnaA overexpression plasmid into the SM10λπ wild-type strain also remarkably inhibited conjugation frequency in both conjugation models (Figure 1B).
Collectively, these data suggest that indole plays a role in inhibiting conjugation.
To further explore the molecular mechanisms underlying the regulatory function of indole on conjugation, we measured the mRNA expression of the major global regulatory genes (korA, korB, and trbA) and conjugation genes, including Mpf gene (trbB), the Dtr gene (trfA), gene which encodes conjugative transfer relaxase (traI), and gene which activates tra gene expression (traJ). The results showed that the mRNA levels of korA and korB were unambiguously increased along with the addition of 250 μM indole (Figure 2A), and the promotion was observed as well in strains of overexpression of TnaA (ΔtnaA-TnaA and SM10λπ-TnaA) (Figure 2B), but no expression change of korA and korB was observed in tnaA-deficient strain. On the other hand, trbA was up-regulated slightly even when indole concentration was up to 500 μM (Figure 2A), and the mRNA expression level of trbA showed no significant difference in both knockout and overexpression strains (Figure 2B).

Figure 2. The mRNA expression level of conjugation-associated genes. (A) Effect of exogenous indole on the expression levels of conjugation genes (trbB, trfA, traI, and traJ) and global regulatory genes (korA, korB, and trbA). SM10λπ (1 × 107 CFU/ml) was treated with different concentrations of indole at 37°C for 3 h, followed by real-time PCR analysis. (B) Effect of endogenous indole on the expression of the aforementioned conjugation-associated genes. The indicated SM10λπ strains were mated (1 × 107 CFU/ml) at 37°C for 3 h, followed by real-time PCR analysis. The rpoD gene of SM10λπ was used as an internal control. Values are means ± SEMs from at least three independent experiments; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns, not significant.
Consequently, the mRNA expression levels of trbB and trfA decreased significantly with increasing indole concentrations (Figure 2A). Additionally, compared to the wild-type strain, deficiency of tnaA in SM10λπ significantly enhanced both the expression of trbB and trfA, whereas introducing a plasmid of TnaA overexpression reversed it, and introducing a TnaA overexpression plasmid into the SM10λπ wild-type strain also remarkably inhibited the mRNA expression levels of trbB and trfA (Figure 2B). However, the expression of traI and traJ was barely affected by exogenous indole treatment (Figure 2A) and showed no significant difference in knockout and overexpression strains (Figure 2B).
Taken together, these results suggest that indole may inhibit conjugation mainly through regulating the expression of korA and korB, further regulating the expression of trbB and trfA.
We have previously reported that sub-MIC of CIP promoted SM10λπ-PAO1 conjugation, but the underlying mechanism was not explored. Here, we measured extracellular indole production and tnaA expression of SM10λπ after cultivation in the presence of CIP at sub-inhibitory concentrations (1/8 MIC, 1/128 MIC, and 1/1024 MIC) and found that 1/8 MIC CIP significantly repressed indole production (Figure 3A) and tnaA expression (Figure 3B). It seems that indole may play a role in CIP-induced conjugation, which needs further confirmation. We next added the SM10λπ-EC600 conjugation model and further confirmed that treatment with 1/8 MIC of CIP clearly enhanced conjugation frequency in both SM10λπ-PAO1 and SM10λπ-EC600 models (Figure 3C). Exogenous indole was added after conjugation system was treated with CIP, or overexpression of TnaA, when wild-type (SM10λπ-TnaA) was used as donor cell in CIP-treated conjugation models. It turned out that indole inhibited CIP-induced conjugation frequency in both conjugation models (Figure 3D). These data suggested that indole may play a negative role in the process of CIP-induced conjugation.
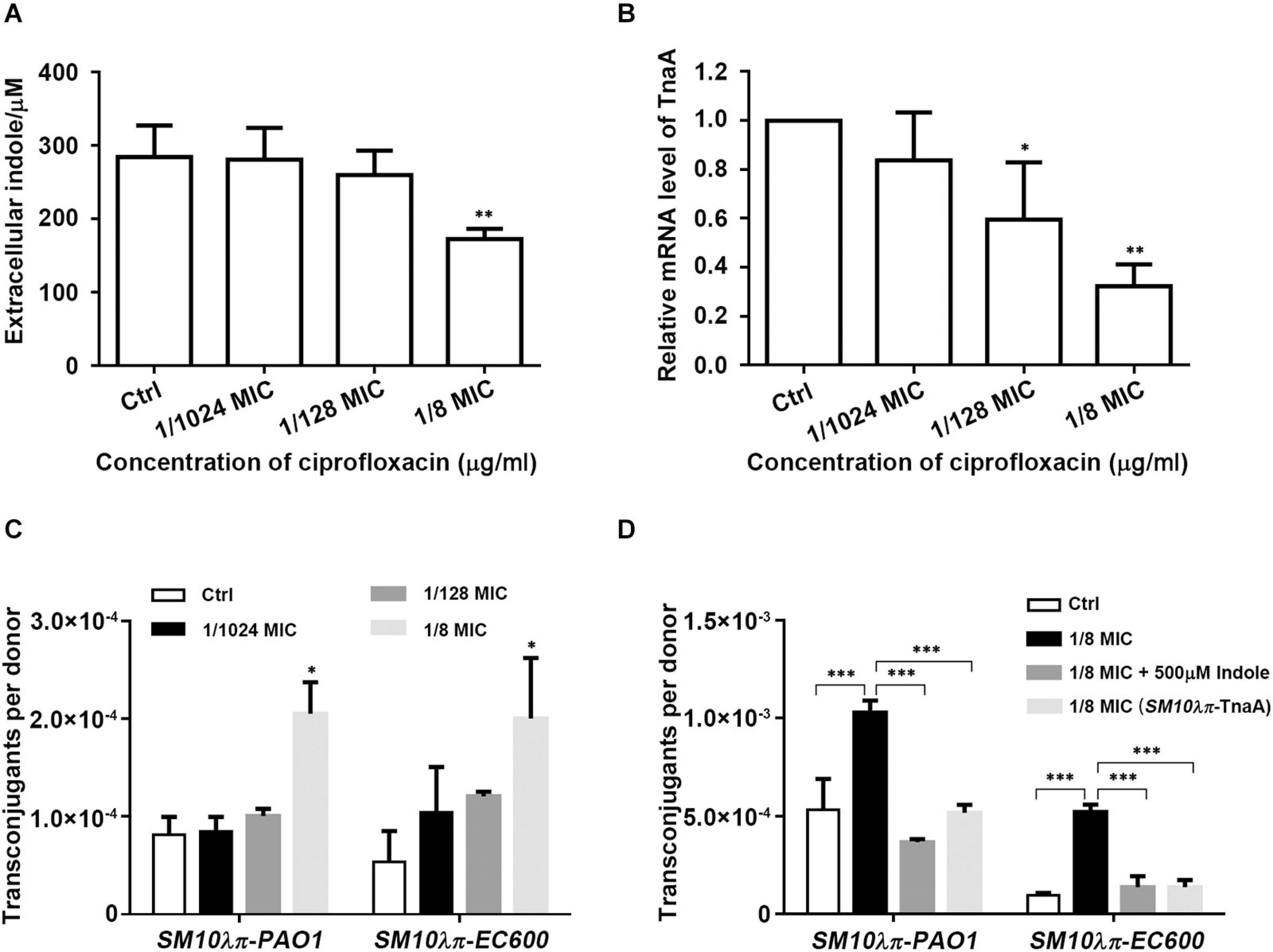
Figure 3. Indole inhibited CIP-induced conjugation. (A) Effect of CIP on indole production. SM10λπ was treated with different sub-MICs of CIP at 37°C for 5 h, followed by measurement of extracellular indole. (B) Effect of CIP on tnaA expression. SM10λπ was treated with different sub-MICs of CIP at 37°C for 4 h, followed by real-time PCR analysis of tnaA expression. (C) Effect of CIP on SM10λπ-PAO1 and SM10λπ-EC600 conjugation. SM10λπ was treated with different sub-MICs of CIP at 37°C for 8 h and then mated with PAO1 or EC600 cells at 37°C for 6 h. (D) Indole inhibited CIP-induced conjugation frequency. SM10λπ was treated with 1/8 MIC of CIP at 37°C for 8 h and then mated with PAO1 or EC600 cells in the presence of 500 μM of indole at 37°C for 6 h; meanwhile, overexpression of TnaA in wild-type (SM10λπ-TnaA) was treated with 1/8 MIC of CIP at 37°C for 8 h and then mated with PAO1 or EC600 cells at 37°C for 6 h. Values are means ± SEMs from at least three independent experiments; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
The physiological functions of indole, like maintaining plasmid stability, affecting biofilm formation, and antibiotic resistance, have been the subject of active investigation (Chant and Summers, 2007; Nishino et al., 2008; Kuczynska-Wisnik et al., 2010). Here, we explored the role of indole in the regulation of intraspecies and interspecies conjugation. Both exogenous indole treatment and gain- and loss-of-function studies showed the inhibitory effect of indole on SM10λπ-PAO1 and SM10λπ-EC600 conjugation. Compared to most research into the conjugational regulatory mechanism that has focused on self-transmissible elements (Frost et al., 2005), this study and our previous work (Lu et al., 2017b) revealed that chemical signal molecules, namely, indole and AHLs that are produced by donor and recipient cells, respectively, may inhibit conjugation.
Mpf gene, trbB, is responsible for the development of the conjugative pore, and the Dtr gene, trfA, encodes a trans-acting product essential for vegetative plasmid replication (Smith and Thomas, 1984; Zatyka et al., 1997). Global regulatory genes korA and korB both exert roles in repression of trfA expression, and trbA and korB expression severely represses the trbB promoter (Schreiner et al., 1985; Theophilus et al., 1985; Zatyka et al., 2001). We found that exogenous indole treatment significantly up-regulated the mRNA expression levels of korA and korB, which in turn inhibited the expression of trbB and trfA genes, thereby decreasing the conjugation frequency. Additionally, the results of the aforementioned genes’ expression in tnaA knockout and overexpression strains further confirmed the regulatory mechanism of indole on conjugation. However, it is worth noting that although korA and korB expression did not decrease in tnaA-deficient strain, the expression of trbB and trfA increased. It may be that the expression of trbB and trfA may be regulated by indole or other pathways, not only by korA and korB.
Indole production of donor cell SM10λπ was significantly repressed by CIP in sub-inhibitory concentration. This result was inconsistent with previous report that E. coli exhibited greater indole production in the presence of kanamycin which may be isolated from Streptomyces kanamyceticus (Umezawa et al., 1957; Han et al., 2011). The reason behind this phenomenon may be that E. coli may utilize indole to compete against other microorganisms that could produce antibiotics (Han et al., 2011). However, CIP is a member of synthetic quinolone antibiotics, and indole is reported to stimulate the formation of E. coli persisters against quinolone antibiotics (Zarkan et al., 2020). We referred that CIP may in turn play a role through inhibiting indole production.
The selective pressures caused by increases in the use and misuse of antibiotics in medicine and animal feedstuffs account for the spread of antibiotic resistance genes (von Wintersdorff et al., 2016). However, the underlying mechanisms for antibiotic-induced conjugative transfer remain largely unknown. Here, we found that CIP in sub-inhibitory concentration clearly enhanced conjugation frequency in both SM10λπ-PAO1 and SM10λπ-EC600 models. Combining the result that CIP inhibited indole production of donor cell and indole inhibited CIP-induced conjugation frequency, we inferred that indole may play a negative role in CIP-induced conjugation. In our previous work, AHLs secreted from recipient cell PAO1 worked to inhibit Gm-induced conjugation (Lu et al., 2017a). It seems that a different regulatory mechanism involved in the conjugation process is related to different types of antibiotics and bacteria.
In conclusion, we found that indole may function as a conjugation inhibitor and play a negative role in the process of CIP-induced conjugation. To the best of our knowledge, this is the first report to reveal the regulatory role of indole in conjugation. These results not only enrich our understanding of the biological function of indole but also inspire us to explore a new way to restrain conjugation to further control the spread of antibiotic resistance.
The bacterial strains and plasmids used in this study are listed in Supplementary Table 2. Some of these strains and plasmids have already been described in previous work (Zeng et al., 2016; Lu et al., 2017b). However, the tnaA-deficient strain (ΔtnaA-vector) and overexpression of TnaA in wild-type (SM10λπ-TnaA) and tnaA mutation (ΔtnaA-TnaA) strain were developed in this research. Bacteria were grown in a Luria–Bertani (LB) medium or on LB plates containing 1.5% agar unless otherwise indicated. If required, antibiotics were added to LB plates at the following final concentrations: ampicillin (AMP), 100 μg/ml; Gm, 30 μg/ml; rifampin (Rif), 50 μg/ml; and chloramphenicol (C), 16 μg/ml.
The phage λ Red recombination system was employed for tnaA deletion in E. coli SM10λπ (Datsenko and Wanner, 2000). To construct TnaA overexpression, the tnaA gene was cloned into the PstI/HindIII sites of the pSTV28 vector. More details are provided in the Supplementary Materials and Methods, and corresponding validation can be found in Supplementary Figure 2.
The indicated bacterial strains were cultured in LB overnight (8–10 h) at 37°C, then diluted to 1 × 107 CFU/ml (by using the Sysmex UF-1000i automated urine particle analyzer; Tokyo, Japan) with or without treatment, further divided into 12-well plates at a volume of 1 ml per well, and finally grown at 37°C. The samples were collected at the indicated time points from each individual well, and OD600 values were determined.
Escherichia coli SM10λπ (pUCP24T) worked as donor cells, with the RP4 plasmid integrated in the chromosome, while PAO1 or EC600 worked as recipient cells. The pUCP24T plasmid was constructed by inserting the oriT fragment into pUCP24 (West et al., 1994). To achieve transfer, the mobilizable pUCP24T plasmid that contains a gene cassette (aacC1) conferring Gm resistance needed to leverage the conjugative apparatus of plasmid RP4 which was integrated in E. coli SM10λπ. For mating experiments, equal amounts of donor and recipient cells (1 × 107 CFU/ml, counted by using the Sysmex UF-1000i automated urine particle analyzer; Tokyo, Japan) were mixed in 200 μl LB with or without exogenous indole at 37°C in a 96-well plate. After 6 h of mating, the cultures were vigorously mixed, and 30 μl aliquots of each conjugation mixture were spread on screening agar plates, Gm–ampicillin and Gm–rifampicin plates, since PAO1 is ampicillin resistant and EC600 is rifampicin resistant. The numbers of transconjugant colonies were counted after an overnight incubation at 37°C.
MIC values were determined by the broth microdilution method in poly-styrene microtiter plates (no. 3599; Costar) according to CLSI protocol M07-A8 using cation-adjusted Mueller–Hinton broth (CAMHB) (no. 11865; Oxoid). MICs were interpreted visually after incubating at 37°C for 16 h.
The production of extracellular indole was measured by the modified Kawamura–Sato method (Kawamura-Sato et al., 1999; Han et al., 2011). First, Kovac’s reagent (0.4 ml) was added to cultural supernatant (1 ml), followed by sufficient mixing. Then the mixture (0.1 ml) was diluted into an HCl-amyl alcohol solution (0.9 ml, the ratio of HCl and amyl alcohol was 1:3). Finally, OD540 values were determined using synergy H1 microplate reader (BioTek; Winooski, VT, United States), and indole concentration was calculated according to the standard curve.
Total RNA was extracted using RNAiso Plus reagent (TaKaRa, Dalian, Liaoning, China). Reverse transcription (1 μg of total RNA) was performed with the PrimeScript RT reagent kit (TaKaRa, Dalian, Liaoning, China). The cDNA was subjected to quantitative PCR (qPCR) on a ViiA 7 Dx system (Applied Biosystems, Foster, CA, United States) using SYBR green qPCR master mixes (TaKaRa, Dalian, Liaoning, China). The expression levels of target genes were normalized to that of the internal control gene (rpoD) using the 2–ΔΔCt method. More details about primers are listed in Supplementary Table 3.
Data are expressed as the means ± standard errors of the means (SEMs) from at least three independent experiments. The differences between groups were analyzed using the Student’s t-test when two groups were compared or a one-way analysis of variance (ANOVA) when more than two groups were compared. All analyses were performed using GraphPad Prism, version 5 (GraphPad Software, Inc., San Diego, CA, United States). All statistical tests were two-sided; P-values of <0.05 were considered statistically significant.
All datasets generated for this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
RX, YuL, and JP contributed to the conception and design of the study. JL organized the database. JZ performed the statistical analysis. RX wrote the first draft of the manuscript. YuL, JP, JL, and JZ wrote the sections of the manuscript. CC, YaL, and BH contributed to funding acquisition. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by the Natural Science Foundation of Guangdong Province (Grant No. 2019A1515011882), the National Natural Science Foundation of China (Grant Nos. 81772249, 81871703, and 81572058), and the Guangdong Provincial Hospital of Traditional Chinese Medicine (Grant Nos. YN2018QJ01, YN2019QJ03, and YN2019MJ01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are very grateful to B. L. Wanner (Department of Biological Sciences, Purdue University, West Lafayette, IN, United States) for generously providing the λ Red recombination system and H. P. Schweizer (Department of Microbiology and Infectious Diseases, University of Calgary Health Sciences Center, Calgary, AB, Canada) for providing the plasmid pUCP24.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.628133/full#supplementary-material
Balzer, D., Ziegelin, G., Pansegrau, W., Kruft, V., and Lanka, E. (1992). KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 20, 1851–1858. doi: 10.1093/nar/20.8.1851
Banuelos-Vazquez, L. A., Torres Tejerizo, G., and Brom, S. (2017). Regulation of conjugative transfer of plasmids and integrative conjugative elements. Plasmid 91, 82–89. doi: 10.1016/j.plasmid.2017.04.002
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427, 72–74. doi: 10.1038/nature02241
Carballeira, J. D., González-Pérez, B., Moncalián, G., and de la Cruz, F. (2014). A high security double lock and key mechanism in HUH relaxases controls oriT-processing for plasmid conjugation. Nucleic Acids Res. 42, 10632–10643. doi: 10.1093/nar/gku741
Chant, E. L., and Summers, D. K. (2007). Indole signalling contributes to the stable maintenance of Escherichia coli multicopy plasmids. Mol. Microbiol. 63, 35–43. doi: 10.1111/j.1365-2958.2006.05481.x
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U S A. 97, 6640–6645. doi: 10.1073/pnas.120163297
De La Cruz, F., Frost, L. S., Meyer, R. J., and Zechner, E. L. (2010). Conjugative DNA metabolism in gram-negative bacteria. FEMS Microbiol. Rev. 34, 18–40. doi: 10.1111/j.1574-6976.2009.00195.x
Deeley, M. C., and Yanofsky, C. (1981). Nucleotide sequence of the structural gene for tryptophanase of Escherichia coli K-12. J. Bacteriol. 147, 787–796. doi: 10.1128/jb.147.3.787-796.1981
Francia, M. V., Varsaki, A., Garcillán-Barcia, M. P., Latorre, A., Drainas, C., and de la Cruz, F. (2004). A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28, 79–100. doi: 10.1016/j.femsre.2003.09.001
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Han, T. H., Lee, J. H., Cho, M. H., Wood, T. K., and Lee, J. (2011). Environmental factors affecting indole production in Escherichia coli. Res. Microbiol. 162, 108–116. doi: 10.1016/j.resmic.2010.11.005
Hirakawa, H., Inazumi, Y., Masaki, T., Hirata, T., and Yamaguchi, A. (2005). Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 55, 1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x
Jagura-Burdzy, G., and Thomas, C. M. (1997). Dissection of the switch between genes for replication and transfer of promiscuous plasmid RK2: basis of the dominance of trfAp over trbAp and specificity for KorA in controlling the switch. J. Mol. Biol. 265, 507–518. doi: 10.1006/jmbi.1996.0747
Jutkina, J., Marathe, N. P., Flach, C. F., and Larsson, D. G. J. (2018). Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci. Total Environ. 616-617, 172–178. doi: 10.1016/j.scitotenv.2017.10.312
Jutkina, J., Rutgersson, C., Flach, C. F., and Joakim Larsson, D. G. (2016). An assay for determining minimal concentrations of antibiotics that drive horizontal transfer of resistance. Sci. Total Environ. 548-549, 131–138. doi: 10.1016/j.scitotenv.2016.01.044
Kawamura-Sato, K., Shibayama, K., Horii, T., Iimuma, Y., Arakawa, Y., and Ohta, M. (1999). Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol. Lett. 179, 345–352. doi: 10.1111/j.1574-6968.1999.tb08748.x
Kobayashi, A., Hirakawa, H., Hirata, T., Nishino, K., and Yamaguchi, A. (2006). Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J. Bacteriol. 188, 5693–5703. doi: 10.1128/jb.00217-06
Koraimann, G., and Wagner, M. A. (2014). Social behavior and decision making in bacterial conjugation. Front. Cell. Infect. Microbiol. 4:54. doi: 10.3389/fcimb.2014.00054
Kuczynska-Wisnik, D., Matuszewska, E., and Laskowska, E. (2010). Escherichia coli heat-shock proteins IbpA and IbpB affect biofilm formation by influencing the level of extracellular indole. Microbiology (Reading) 156(Pt 1), 148–157. doi: 10.1099/mic.0.032334-0
Lopatkin, A. J., Huang, S., Smith, R. P., Srimani, J. K., Sysoeva, T. A., Bewick, S., et al. (2016). Antibiotics as a selective driver for conjugation dynamics. Nat. Microbiol. 1:16044. doi: 10.1038/nmicrobiol.2016.44
Lu, Y., Zeng, J., Wang, L., Lan, K., Shunmei, E., Wang, L., et al. (2017a). Antibiotics promote Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting quorum sensing. Antimicrob Agents Chemother 61:e01284-17. doi: 10.1128/aac.01284-1217
Lu, Y., Zeng, J., Wu, B., Shunmei, E., Wang, L., Cai, R., et al. (2017b). Quorum sensing N-acyl homoserine Lactones-SdiA suppresses Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting trai expression. Front. Cell. Infect. Microbiol. 7:7. doi: 10.3389/fcimb.2017.00007
Newton, W. A., Morino, Y., and Snell, E. E. (1965). Properties of crystalline tryptophanase. J. Biol. Chem. 240, 1211–1218. doi: 10.1016/s0021-9258(18)97562-9
Nishino, K., Senda, Y., and Yamaguchi, A. (2008). The AraC-family regulator GadX enhances multidrug resistance in Escherichia coli by activating expression of mdtEF multidrug efflux genes. J. Infect. Chemother 14, 23–29. doi: 10.1007/s10156-007-0575-y
Pansegrau, W., Balzer, D., Kruft, V., Lurz, R., and Lanka, E. (1990). In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc. Natl. Acad. Sci. U S A. 87, 6555–6559. doi: 10.1073/pnas.87.17.6555
Schreiner, H. C., Bechhofer, D. H., Pohlman, R. F., Young, C., Borden, P. A., and Figurski, D. H. (1985). Replication control in promiscuous plasmid RK2: kil and kor functions affect expression of the essential replication gene trfA. J. Bacteriol. 163, 228–237. doi: 10.1128/jb.163.1.228-237.1985
Schröder, G., and Lanka, E. (2005). The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA. Plasmid 54, 1–25. doi: 10.1016/j.plasmid.2005.02.001
Schuurmans, J. M., van Hijum, S. A. F. T., Piet, J. R., Händel, N., Smelt, J., Brul, S., et al. (2014). Effect of growth rate and selection pressure on rates of transfer of an antibiotic resistance plasmid between E. coli strains. Plasmid 72, 1–8. doi: 10.1016/j.plasmid.2014.01.002
Shun-Mei, E., Zeng, J. M., Yuan, H., Lu, Y., Cai, R. X., and Chen, C. (2018). Sub-inhibitory concentrations of fluoroquinolones increase conjugation frequency. Microb Pathog 114, 57–62. doi: 10.1016/j.micpath.2017.11.036
Smith, C. A., and Thomas, C. M. (1984). Nucleotide sequence of the trfA gene of broad host-range plasmid RK2. J. Mol. Biol. 175, 251–262. doi: 10.1016/0022-2836(84)90347-4
Theophilus, B. D., Cross, M. A., Smith, C. A., and Thomas, C. M. (1985). Regulation of the trfA and trfB promoters of broad host range plasmid RK2: identification of sequences essential for regulation by trfB/korA/korD. Nucleic Acids Res. 13, 8129–8142. doi: 10.1093/nar/13.22.8129
Umezawa, H., Ueda, M., Maeda, K., Yagishita, K., Kondo, S., Okami, Y., et al. (1957). Production and isolation of a new antibiotic: kanamycin. J. Antibiot (Tokyo) 10, 181–188.
von Wintersdorff, C. J. H., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Waksman, G. (2019). From conjugation to T4S systems in Gram-negative bacteria: a mechanistic biology perspective. EMBO Rep. 20:e47012. doi: 10.15252/embr.201847012
Wang, D., Ding, X., and Rather, P. N. (2001). Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183, 4210–4216. doi: 10.1128/jb.183.14.4210-4216.2001
West, S. E., Schweizer, H. P., Dall, C., Sample, A. K., and Runyen-Janecky, L. J. (1994). Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148, 81–86. doi: 10.1016/0378-1119(94)90237-2
Yanofsky, C., Horn, V., and Gollnick, P. (1991). Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J. Bacteriol. 173, 6009–6017. doi: 10.1128/jb.173.19.6009-6017.1991
Zarkan, A., Matuszewska, M., Trigg, S. B., Zhang, M., Belgami, D., Croft, C., et al. (2020). Inhibition of indole production increases the activity of quinolone antibiotics against E. coli persisters. Sci. Rep. 10:11742. doi: 10.1038/s41598-020-68693-w
Zatyka, M., Bingle, L., Jones, A. C., and Thomas, C. M. (2001). Cooperativity between KorB and TrbA repressors of broad-host-range plasmid RK2. J. Bacteriol. 183, 1022–1031. doi: 10.1128/jb.183.3.1022-1031.2001
Zatyka, M., Jagura-Burdzy, G., and Thomas, C. M. (1994). Regulation of transfer genes of promiscuous IncP alpha plasmid RK2: repression of Tra1 region transcription both by relaxosome proteins and by the Tra2 regulator TrbA. Microbiology (Reading) 140(Pt 11), 2981–2990. doi: 10.1099/13500872-140-11-2981
Zatyka, M., Jagura-Burdzy, G., and Thomas, C. M. (1997). Transcriptional and translational control of the genes for the mating pair formation apparatus of promiscuous IncP plasmids. J. Bacteriol. 179, 7201–7209. doi: 10.1128/jb.179.23.7201-7209.1997
Keywords: indole, conjugation, E. coli, antibiotic resistance, ciprofloxacin
Citation: Xiong R, Liu Y, Pu J, Liu J, Zheng D, Zeng J, Chen C, Lu Y and Huang B (2021) Indole Inhibits IncP-1 Conjugation System Mainly Through Promoting korA and korB Expression. Front. Microbiol. 12:628133. doi: 10.3389/fmicb.2021.628133
Received: 11 November 2020; Accepted: 12 February 2021;
Published: 19 March 2021.
Edited by:
Clay Fuqua, Indiana University Bloomington, United StatesReviewed by:
Christopher Morton Thomas, University of Birmingham, United KingdomCopyright © 2021 Xiong, Liu, Pu, Liu, Zheng, Zeng, Chen, Lu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cha Chen, Y2hlbmNoYTkwNkAxNjMuY29t; Yang Lu, aGVudGlhbmdhb3pAMTI2LmNvbQ==; Bin Huang, aHVhbmdiM0BtYWlsLnN5c3UuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.