
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 April 2021
Sec. Aquatic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.628055
This article is part of the Research Topic Microbial Communities and Metabolisms Involved in the Degradation of Cellular and Extracellular Organic Biopolymers View all 8 articles
Carbohydrate-active enzymes (CAZymes) are an important feature of bacteria in productive marine systems such as continental shelves, where phytoplankton and macroalgae produce diverse polysaccharides. We herein describe Maribacter dokdonensis 62–1, a novel strain of this flavobacterial species, isolated from alginate-supplemented seawater collected at the Patagonian continental shelf. M. dokdonensis 62–1 harbors a diverse array of CAZymes in multiple polysaccharide utilization loci (PUL). Two PUL encoding polysaccharide lyases from families 6, 7, 12, and 17 allow substantial growth with alginate as sole carbon source, with simultaneous utilization of mannuronate and guluronate as demonstrated by HPLC. Furthermore, strain 62-1 harbors a mixed-feature PUL encoding both ulvan- and fucoidan-targeting CAZymes. Core-genome phylogeny and pangenome analysis revealed variable occurrence of these PUL in related Maribacter and Zobellia strains, indicating specialization to certain “polysaccharide niches.” Furthermore, lineage- and strain-specific genomic signatures for exopolysaccharide synthesis possibly mediate distinct strategies for surface attachment and host interaction. The wide detection of CAZyme homologs in algae-derived metagenomes suggests global occurrence in algal holobionts, supported by sharing multiple adaptive features with the hydrolytic model flavobacterium Zobellia galactanivorans. Comparison with Alteromonas sp. 76-1 isolated from the same seawater sample revealed that these co-occurring strains target similar polysaccharides but with different genomic repertoires, coincident with differing growth behavior on alginate that might mediate ecological specialization. Altogether, our study contributes to the perception of Maribacter as versatile flavobacterial polysaccharide degrader, with implications for biogeochemical cycles, niche specialization and bacteria-algae interactions in the oceans.
Continental shelves are productive marine systems, where photosynthesis by pelagic phytoplankton and benthic macroalgae yields considerable amounts of organic matter. Polysaccharides constitute a major fraction of the algae-derived organic matter, with important roles in nutrient cycles and microbial metabolism (Hehemann et al., 2014; Arnosti et al., 2021). Consequently, diverse bacteria are specialized for the degradation of algal polysaccharides, colonization of algal surfaces and other types of biological interactions (van der Loos et al., 2019; Ferrer-González et al., 2020; Wolter et al., 2021).
Cultured bacterial strains are a valuable resource for studying the ecological and biogeochemical implications of microbial polysaccharide degradation, complementing molecular and metagenomic approaches on community level (Arnosti et al., 2011; Wietz et al., 2015; Matos et al., 2016; Reintjes et al., 2017; Grieb et al., 2020). Culture-based studies revealed the diversity and functionality of carbohydrate-active enzymes (CAZymes), which encompass polysaccharide lyases (PL), glycoside hydrolases (GH), carbohydrate-binding modules (CBM), carbohydrate esterases (CE), glycosyl transferases (GT), and auxiliary carbohydrate-active oxidoreductases (Lombard et al., 2014). CAZyme-encoding genes are frequently transferred between microbes, providing effective mechanisms of adaptation and niche specialization (Hehemann et al., 2016).
Flavobacteria, including the families Flavobacteriaceae and Cryomorphaceae, are major contributors to marine polysaccharide degradation. Comparable to the human gut, marine flavobacteria can degrade various polysaccharides through dedicated genetic machineries (Teeling et al., 2012; Fernández-Gómez et al., 2013). Flavobacterial CAZymes are typically clustered with susCD genes in polysaccharide utilization loci (PUL) for orchestrated uptake and degradation (Grondin et al., 2017). For instance, the marine flavobacterium Zobellia galactanivorans has complex biochemical and regulatory mechanisms for degrading laminarin, alginate, agar, and carrageenan (Hehemann et al., 2012; Thomas et al., 2012, 2017; Labourel et al., 2014; Ficko-Blean et al., 2017; Zhu et al., 2017). Comparable abilities have been described in the flavobacterial genera Formosa agariphila and Gramella forsetii through diverse PUL (Mann et al., 2013; Kabisch et al., 2014; Reisky et al., 2019). Also Maribacter, the “sister genus” of Zobellia, exhibits hydrolytic activity (Bakunina et al., 2012; Zhan et al., 2017). Accordingly, both Maribacter and Zobellia are abundant on macroalgal surfaces (Martin et al., 2015), and related PUL have been detected during phytoplankton blooms in the North Sea (Kappelmann et al., 2019). Furthermore, several Maribacter and Zobellia strains stimulate algal development by producing morphogenesis factors (Matsuo et al., 2005; Weiss et al., 2017). Hence, both genera are important from ecological and biotechnological perspectives.
Here, we describe CAZyme content and hydrolytic capacities of Maribacter dokdonensis strain 62–1, isolated from an alginate-supplemented microcosm at the Patagonian continental shelf (Wietz et al., 2015). This highly productive marine region harbors frequent phytoplankton blooms and abundant coastal macroalgae, indicating regular availability of polysaccharides (Acha et al., 2004; Garcia et al., 2008). Our CAZyme characterization in the pangenomic context illustrates the role of CAZymes, PUL and exopolysaccharide-related genes in niche specialization among Maribacter and Zobellia. The finding of diverse traits for interactions with algae, together with the detection of CAZyme homologs in macroalgae-derived metagenomes, highlight the predisposition of Maribacter spp. to algae-related niches and substrates. Notably, Maribacter dokdonensis 62-1 has been isolated from the same sample as Alteromonas sp. 76–1 with shown capacities for alginate and ulvan degradation (Koch et al., 2019b), illustrating that distantly related hydrolytic strains co-occur in the same habitat. Comparison of their CAZyme machineries illuminated whether these strains might employ different ecophysiological strategies or compete for resources. These eco-evolutionary perspectives into CAZyme diversity and corresponding niche specialization contribute to the understanding of ecophysiological adaptations behind community-level polysaccharide degradation (Teeling et al., 2012, 2016). Considering the abundance and biogeochemical relevance of algal polysaccharides, our study adds further evidence to the eco-evolutionary role of CAZymes in marine flavobacteria.
Strain 62–1 was isolated in April 2012 from a microcosm with surface seawater collected at the Patagonian continental shelf (47.944722 S, 61.923056 W) amended with 0.001% sodium alginate (Wietz et al., 2015). Purity was confirmed by PCR amplification of the 16S rRNA gene after several rounds of subculturing. Alginate utilization was analyzed in seawater minimal medium (SWM) (Zech et al., 2009) supplemented with 0.2% sodium alginate (cat. no. A2158; Sigma-Aldrich, St. Louis, MO) as sole carbon source in comparison to SWM + 0.4% glucose. Precultures were grown from single colonies for 24 h, washed three times with sterile SWM, and adjusted to an optical density of 0.1 measured at 600 nm (OD600). Main cultures were inoculated with 1% (v/v) of washed preculture in triplicate, followed by cultivation at 20°C and 100 rpm with regular photometric measurements (diluted if OD600 > 0.4).
At each OD measurement, subsamples of 5 mL were filtered through 0.22 μm polycarbonate filters into combusted glass vials and stored at −20°C. Alginate concentrations were quantified by High Performance Liquid Chromatography (HPLC) of its monomers mannuronate and guluronate after chemical hydrolysis (20 h, 100°C, 0.1 M HCl) in combusted and sealed glass ampoules. Samples were neutralized with 6 N NaOH, desalted using DionexOnGuard II Ag/H cartridges (Thermo Fisher Scientific, Waltham, MA), and eluted with 100 mM sodium acetate tri-hydrate in 100 mM NaOH. Concentrations were determined in three dilutions per sample (0.01, 0.002, 0.001%) using a Carbopac PA 1 column (Thermo Fisher Scientific) and pulsed amperometric detection according to Mopper et al. (1992). A calibration curve was generated using hydrolyzed 1% alginate solution (R2 = 0.97). Glucose concentrations were measured using samples diluted to 0.001% with MilliQ followed by HPLC with NaOH (18 mM) as eluent and a Carbopac PA 1 column (Thermo Fisher Scientific). A calibration curve was generated using 24 concentrations from 0.025 to 10 μM glucose (R2 = 0.99).
Genomic DNA was extracted using the PeqGold DNA Isolation Kit (PEQLAB, Germany) according to the manufacturer’s instructions. The genome was sequenced with Illumina technology using a GAIIx sequencing platform on paired-end libraries prepared with the Nextera XT DNA Kit (Illumina, San Diego, CA). A total of 83 contigs (0.5–330 kb, average 55 kb) were assembled using SPAdes v3.0 (Bankevich et al., 2012), followed by error correction using BayesHammer (Nikolenko et al., 2013) and gene prediction using the IMG pipeline (Markowitz et al., 2012). The draft genome has been converted to EMBL format using EMBLmyGFF3 (Norling et al., 2018) and deposited at ENA under PRJEB40942. Phylogenetic analysis was carried out with 92 core genes identified using UBCG (Na et al., 2018), with Capnocytophaga ochracea DSM 7271 as outgroup. The resulting nucleotide alignment was visually confirmed for consistency and the best substitution model (GTR + G) computed using ModelTest-NG (Darriba et al., 2020). A maximum-likelihood phylogeny with 1000 bootstrap replicates was calculated using RaxML v8.2 (Stamatakis, 2014) on the CIPRES Science Gateway (Miller et al., 2010).
Genomes of 62–1 and related strains (Supplementary Table 1) were compared using bioinformatic software. Average nucleotide identities were calculated using the Enveomics web application (Rodriguez-R and Konstantinidis, 2016). Core, accessory and unique genes were identified from protein-translated genes using OrthoFinder (Emms and Kelly, 2019) using a 30% identity cutoff. CAZymes were identified using dbCAN2 (Zhang et al., 2018), only considering hits with e-value < 10–15 and > 65% query coverage. Gene annotations and PUL boundaries were manually curated based on the CAZy and UniprotKB-Swissprot databases (Lombard et al., 2014; Bateman et al., 2017). Sulfatases were identified using SulfAtlas v1.1 (Barbeyron et al., 2016a), only considering hits with e-value < 10–1 and > 40% query coverage. Genes were assigned to KEGG classes and pathways using KAAS and KEGG Mapper (Moriya et al., 2007; Kanehisa and Sato, 2020). PUL homologies were analyzed by custom-BLAST in Geneious Pro v71 and PULDB (Terrapon et al., 2018). Genes for downstream processing of alginate monomers (kdgA, kdgF, kgdK, and dehR) were identified by searching homologs from Gramella forsetii (NCBI assembly GCA_000060345.1). Putative kduI and kduD genes for processing unsaturated uronates were identified by searching homologs from Gramella flava (NCBI assembly GCA_001951155.1). Signal peptides were predicted using SignalP v5.0 (Almagro Armenteros et al., 2019).
Data were processed in RStudio2 using R v3.6 (R Core Team, 2018) and visualized using packages ggplot2, PNWColors and pheatmap (Wickham, 2016; Kolde, 2018; Lawlor, 2020). Code and files for reproducing the analysis are available under https://github.com/matthiaswietz/Maribacter.
Strain 62–1 was isolated from alginate-supplemented seawater collected at the Patagonian continental shelf (Wietz et al., 2015). Colonies on solid medium are round, smooth, and yellowish-colored. Genome sequencing resulted in a draft genome (83 contigs) with a cumulative length of 4.6 Mb, encoding 4,103 predicted proteins. Core genome-based phylogeny revealed clear assignment to Maribacter from the Flavobacteriaceae (Figure 1), with 99.8% 16S rRNA gene similarity and 97.8% average nucleotide identity to Maribacter dokdonensis DSW-8T (Supplementary Figure 1). Hence, strain 62–1 is a novel member of this flavobacterial species. M. dokdonensis DSW-8T originates from South Korean and hence subtropical waters, demonstrating occurrence of closely related strains on global scales.
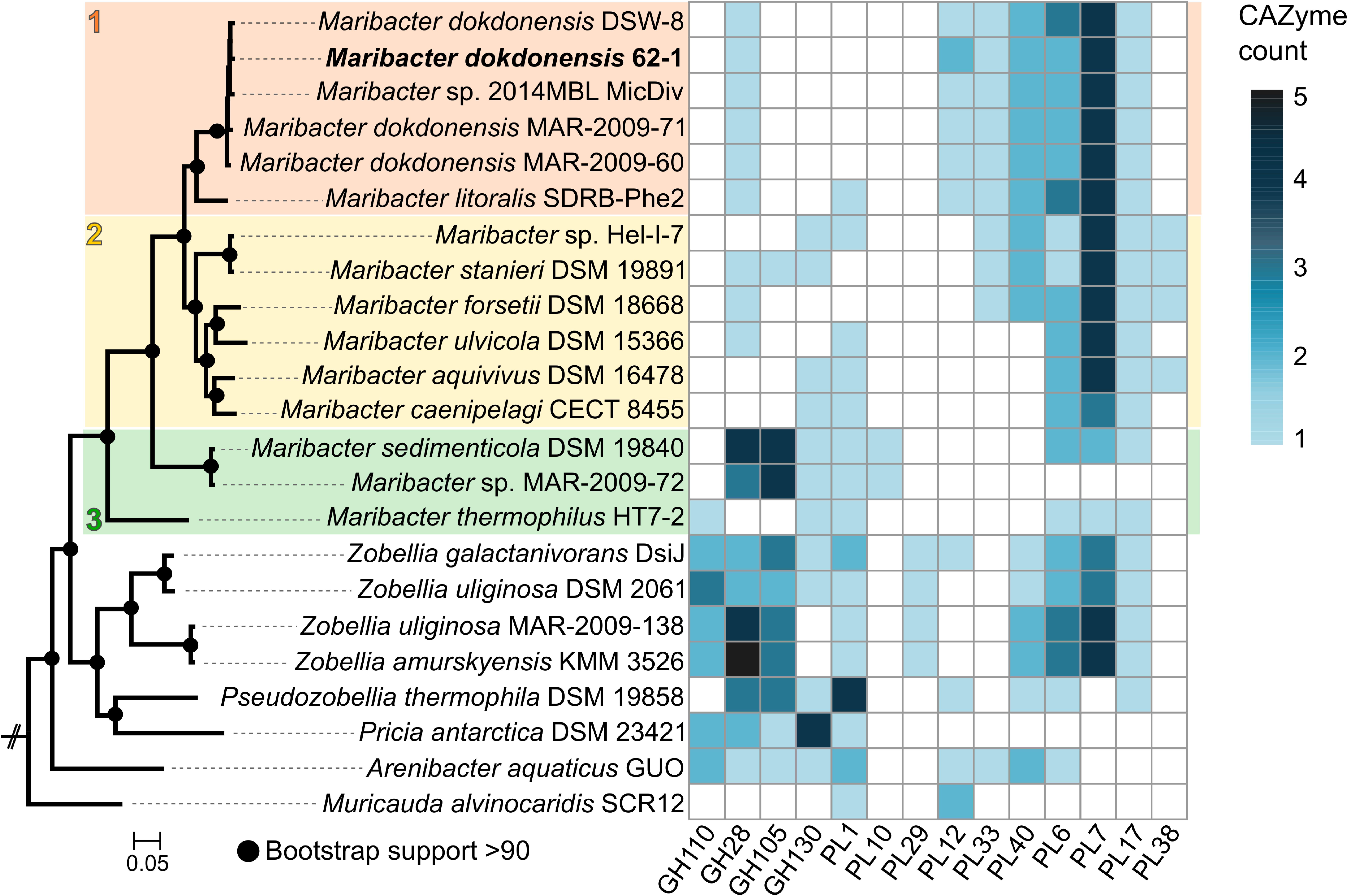
Figure 1. Maximum-likelihood phylogeny based on 82 core genes (left panel) and numbers of genes from selected CAZyme families (right panel) in Maribacter dokdonensis 62–1 and related strains. Capnocytophaga ochracea DSM 7271 served as outgroup. The three resolved Maribacter lineages are numbered and colored. PL, polysaccharide lyase; GH, glycoside hydrolase.
Maribacter dokdonensis strain 62–1 encodes 90 putative CAZymes predicted by dbCAN2, corresponding to 2% of all protein-encoding genes (Table 1 and Supplementary Table 1). As described in detail below, CAZymes commonly clustered with susCD genes, the hallmark of PUL in Bacteroidetes (Grondin et al., 2017). The presence of 12 polysaccharide lyases from PL families 6, 7, 12, and 17 illustrates specialization toward alginate, confirmed by physiological experiments (Figure 2). Most PL12 are classified as heparinases, but co-localization with known alginate lyase families indicates alginolytic activity. Strain 62–1 furthermore encodes PL33 and PL40 lyases that potentially target ulvan (Table 1). CAZyme numbers and diversity match the hydrolytic potential of related Maribacter and Zobellia strains included for comparison (Supplementary Table 1), corroborating the adaptation of these taxa to algal substrates and surfaces (Bakunina et al., 2012; Martin et al., 2015; Kwak et al., 2017; Zhan et al., 2017; Chernysheva et al., 2019).
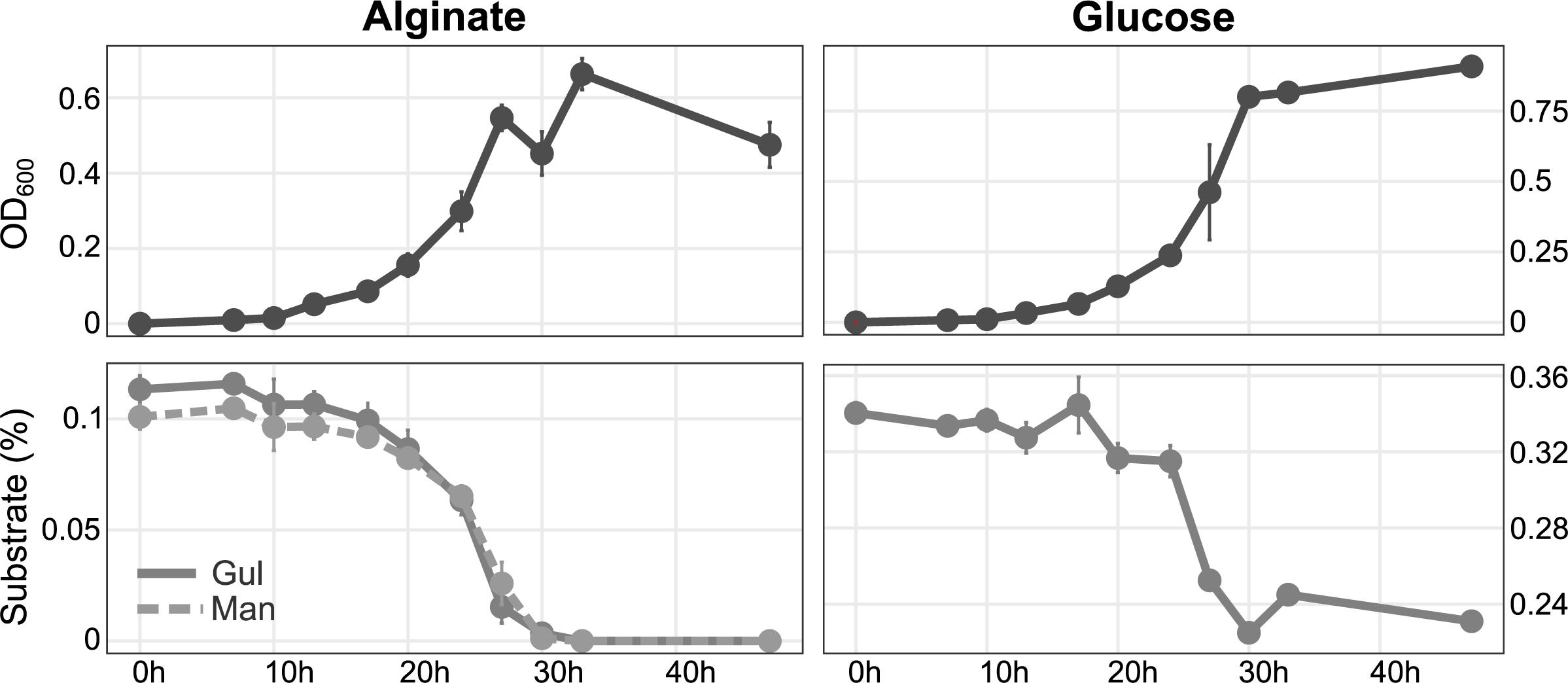
Figure 2. Growth of Maribacter dokdonensis 62–1 with alginate compared to glucose as sole carbon source over a period of 48 h, illustrated by optical density (upper panels) and substrate utilization as determined by HPLC (lower panels). Gul, guluronate; Man, mannuronate.
We contextualized CAZyme patterns with phylogenetic relationships (Figure 1 and Supplementary Table 2) and a general overview of the Maribacter pangenome (Supplementary Table 3). Core genome-based phylogeny resolved three lineages, each with distinct signatures of CAZymes and exopolysaccharide-related genes (Figure 1). Maribacter lineages 1 and 2 encode more PLs than lineage 3 (Wilcoxon rank-sum test, p = 0.01), including an entire additional PUL for alginate degradation. Moreover, one PL7 and one PL12 are unique to lineage 1 (see detailed paragraph below). In contrast, a sizeable PUL encoding a PL10 pectate lyase with CE8 methylesterase domain is restricted to lineage 3 (Figure 1). This PL10-CE8 presumably corresponds to a specific pectin-related niche, supported by elevated numbers of GH families 28 and 105 that participate in pectinolytic activity (Hobbs et al., 2019).
Notably, lineages 2 and 3 harbor distinct clusters for the biosynthesis of cell wall exopolysaccharides (EPS). Furthermore, specific O-antigen and GT variants occur on sublineage- and strain-level (Supplementary Table 3), presumably facilitating surface colonization and specific interactions with algal hosts (Deo et al., 2019). This marked variability on fine phylogenetic levels suggests EPS-related genes as adaptive feature among Maribacter strains, mediating different surface-attachment and host-interaction mechanisms (Lee et al., 2016; Decho and Gutierrez, 2017).
The array of predicted alginate lyase genes allowed considerable growth with alginate as sole nutrient source (Figure 2). Growth was only slightly lower than with the monosaccharide glucose, indicating excellent adaptation for polysaccharide degradation. HPLC demonstrated that concentrations of both monomeric building blocks of alginate, mannuronate (M) and guluronate (G), decreased at the same rate (Figure 2).
Predicted alginate lyase genes are encoded in two major PUL, plus additional, single genes dispersed throughout the genome (Figure 3 and Supplementary Table 2). AlgPUL1 harbors four adjacent PL6-12-6-17 lyase genes, co-localized with a susCD pair and all genes for downstream processing of alginate monomers (Figure 3A). Hence, AlgPUL1 presumably encodes the complete metabolic cascade from external polymer breakdown (PL6, PL12), oligosaccharide hydrolysis (PL17) to monomer processing (kdgA, kdgF, kdgK, and dehR). The co-localization of PL families with structural and catalytic diversity (Xu et al., 2017) presumably facilitates access to various alginate architectures, e.g., relating to polymer length or the ratio between M and G in poly-G, poly-M or mixed stretches. Upstream of AlgPUL1 is a cluster encoding one GH3 and two GH144 genes, together with susCD and sulfatase genes (Figure 3). The two regions are separated by a type II-C CRISPR-Cas locus, signifying a mobile genomic region that might facilitate the exchange of adjacent CAZymes. Both GH144 genes share ∼50% amino acid identity to an endo-glucanase of Chitinophaga pinensis (Abe et al., 2017), indicating glucosidase or endoglucanase activity. Their occurrence in all Maribacter and Zobellia (orthologous groups 0002512 and 0002513; Supplementary Table 3), many marine flavobacteria as determined by BLASTp (data not shown) as well as human gut microbes (McNulty et al., 2013) indicates ecological relevance across diverse habitats. The smaller AlgPUL2 encodes two PL7 lyases (Figure 3B) and might be auxiliary to AlgPUL1, potentially being activated by particular M/G architectures or environmental signals (Lim et al., 2011). The prediction of different signal peptide variants in the two PL7 (Supplementary Table 2) indicated that one PL7 is freely secreted whereas the other is anchored to the cell membrane. This complementary extracellular localization might boost alginolytic activity. Even without transcriptomic data, the combined physiological, chemical, and genomic evidence illustrates a functional and effective alginolytic pathway.
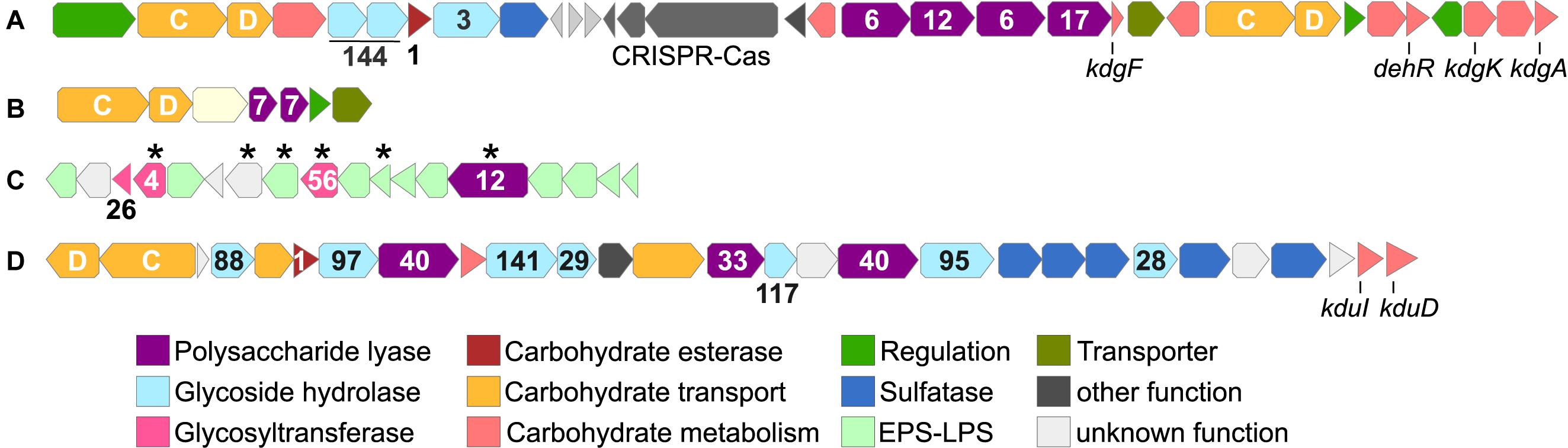
Figure 3. Gene cluster related to carbohydrate metabolism in Maribacter dokdonensis 62-1, including AlgPUL1 (A) and AlgPUL2 (B) targeting alginate; a predicted PL12 alginate lyase with adjacent EPS-related genes (C); and a putative ulvan- and fucoidan-targeting PUL (D). Numbers designate CAZyme families, letters susCD genes, and asterisks unique genes of strain 62-1. kdgA: 2-keto-3-deoxy-D-gluconate aldolase; kdgF: gene for 2-keto-3-deoxy-D-gluconate linearization; kdgK: 2-keto-3-deoxy-D-gluconate kinase; dehR: 4-deoxy-L-erythro-5-hexoseulose uronate reductase; kduI: 5-keto 4-deoxyuronate isomerase; kduD: 2-deoxy-D-gluconate 3-dehydrogenase.
Comparative genomics revealed homologs of AlgPUL1 in most related Maribacter and Zobellia strains, whereas AlgPUL2 is restricted to Maribacter lineages 1 and 2. The observed structural diversity of alginolytic PUL among Maribacter and Zobellia overall corresponds to core-genome relationships, supporting the notion of lineage-specific genomic signatures (Supplementary Figure 2). Notably, a PL12 only occurs in the AlgPUL1 variant of lineage 1 (Figure 1). This PL was presumably acquired from distant Bacteroidetes taxa, considering 65% amino acid identity to FNH22_29785 from Fulvivirga sp. M361 (Cytophagales). In contrast, lineages 2 and 3 encode a PL7 in this position, or solely harbor the PL6 and PL17 (Supplementary Figure 2). An almost identical AlgPUL2 occurs in Flavivirga eckloniae (locus tags C1H87_08155-08120), with 65% amino acid identity between the respective PL7 lyases. The original isolation of F. eckloniae from the alginate-rich macroalga Ecklonia (Lee et al., 2017) highlights this PUL as adaptation to algae-related niches. In Zobellia, AlgPUL1, and AlgPUL2 occur in mixed combinations and in distant genomic regions (Thomas et al., 2012), suggesting internal recombination events (Supplementary Figure 2).
Maribacter dokdonensis 62–1 encodes a unique PL12 (locus tag 00457) within an exopolysaccharide-related cluster, including several unique GTs (Figure 3C). This cluster might represent a link between the degradation and biosynthesis of polysaccharides, considering that other bacteria regulate EPS production and release via PLs or GHs (Bakkevig et al., 2005; Köseoǧlu et al., 2015). The PL12 has 39% amino acid identity to ATE92_1054 of the North Sea isolate Ulvibacter sp. MAR-2010-11 (Kappelmann et al., 2019), indicating ecological relevance of horizontally transferred PL12 homologs in distant habitats. The PL12 is among the two lyases without predicted signal peptide (Supplementary Table 2) and hence likely retained within the cell, supporting an intracellular role in EPS metabolism. However, only transcriptomic data can confirm whether the PL12 indeed targets alginate and might be co-regulated with EPS-related genes.
Strain 62–1 harbors additional PUL for the degradation of other algal polysaccharides. The co-localization of two PL40 lyases, one PL33 from subfamily 2 as well as several sulfatases indicates activity towards a sulfated polysaccharide, presumably ulvan (Figure 3D). Although PL33 are classified as chondroitin or gellan lyases, our findings suggest an extended substrate range, potentially specific to subfamily 2. The two PL40 variants (locus tags 01347 and 01356) only have 30% identity and hence do not originate from duplication. Notably, PL40_01347 lacks a signal peptide compared to PL40_01356 and the PL33 (Supplementary Table 2), suggesting different secretory behavior and complementary functionality. The PL40_01347 homolog is conserved in most Maribacter and Zobellia genomes (Supplementary Figure 2), whereas PL40_01356 and the PL33 are missing in Zobellia. The latter PLs share 70 and 77% amino acid identity with a CAZyme pair in Formosa agariphila (locus tags BN863_10330 and BN863_10340, respectively), originally annotated as heparinase and PL8 but confirmed as PL33 and PL40 using the latest CAZy database (Mann et al., 2013; Lombard et al., 2014). Their absence in Zobellia but presence in distantly related Arenibacter spp. (Figure 1) indicates separate acquisition after speciation, potentially via transfer events between Formosa and Maribacter.
The combination of PL33 and PL40 differs from ulvanolytic PUL in other bacteria, which largely comprise ulvan lyases from PL families 24, 25, and 28 (Foran et al., 2017; Reisky et al., 2018; Koch et al., 2019b). Furthermore, the PUL in strain 62–1 also encodes α-fucosidases, including GH29, GH95, GH97, GH117, and GH141 comparable to a CAZyme plasmid in the verrucomicrobium Lentimonas (Sichert et al., 2020). Furthermore, a co-localized GH28 might remove galacturonic acid from fucoidan (Sichert et al., 2020), further metabolized by adjacent kduI and kduD genes into the Entner-Doudoroff pathway (Salinas and French, 2017). These observations indicate an alternative hydrolytic activity toward fucoidan, supported by fucose importer, fuconolactonase and fuconate dehydratase genes (locus tags 01371, 01373, 01376, respectively) encoded downstream of the PUL.
To establish a broader ecological context, we searched homologs of the PL33–PL40 pair (specific to Maribacter) and the PL12 within the EPS gene cluster (unique to strain 62–1) in 102 microbial metagenomes from marine plants. The co-detection of PL33 and PL40 homologs on diverse algae and seagrasses from global locations (Figure 4A and Supplementary Table 4) illustrates that relatives of strain 62–1 occur in such niches worldwide. The presence of PL33–PL40 homologs on brown, green and red macroalgae indicates that predicted hydrolytic capacities are functional in situ and support establishment in algal holobionts. The brown macroalgae Macrocystis and Ecklonia are rich in alginate whereas ulvan can constitute ∼40% of the green algae Ulva (Kidgell et al., 2019), highlighting the importance of related CAZymes for associated bacteria and why Maribacter spp. are common algal epibionts (Martin et al., 2015).
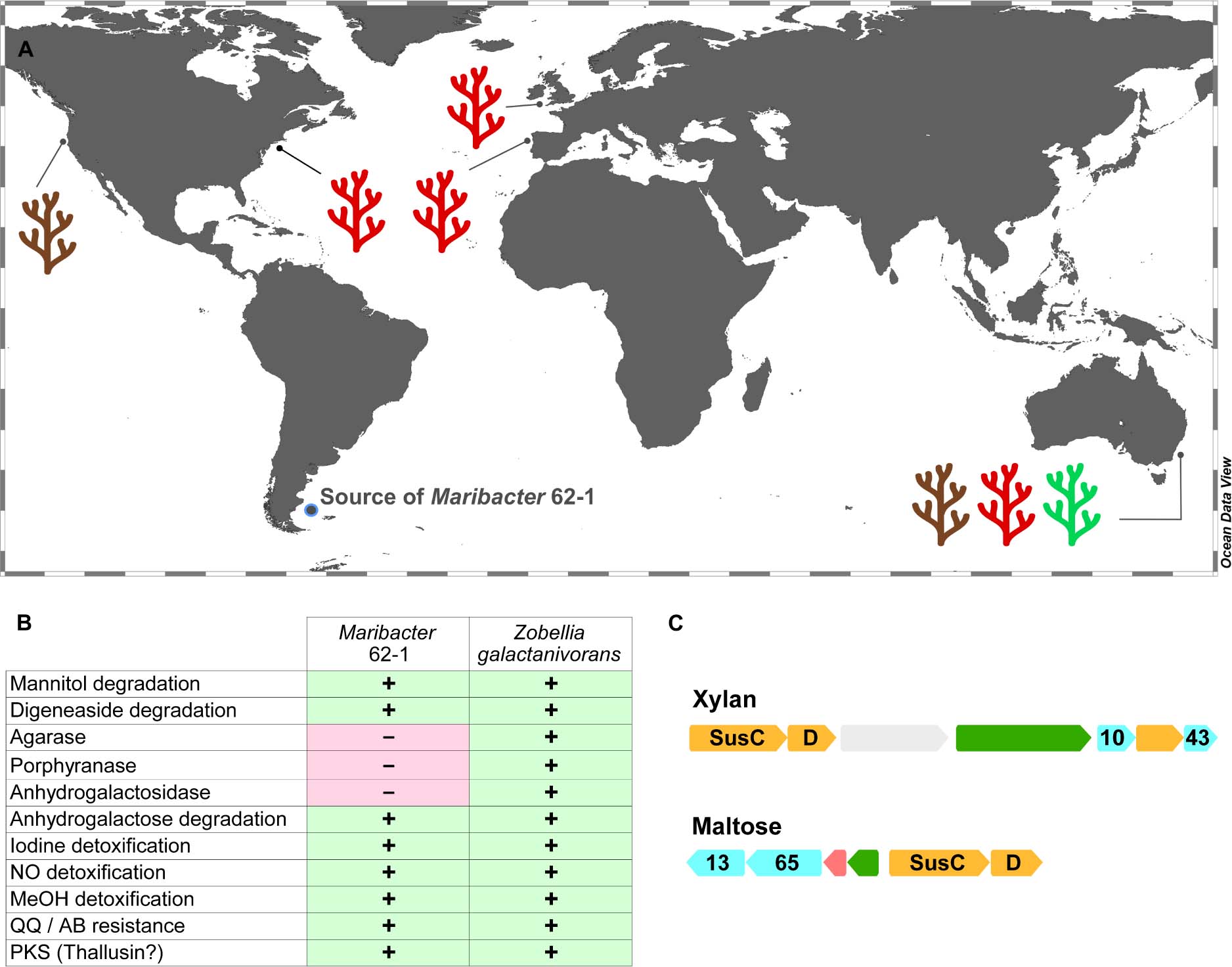
Figure 4. Ecological implications of CAZyme diversity in Maribacter dokdonensis 62–1. (A) World map illustrating the detection of polysaccharide lyase homologs in macroalgae-derived metagenomes. Colors indicate brown, red or green algal taxa (see Supplementary Table 4 for details). (B) Presence (+) or absence (-) of traits for interactions with algae compared to characterized features in Zobellia galactanivorans. MeOH, methanol, QQ, quorum quenching; AB, antibiotic; PKS, polyketide synthase. (C) Gene clusters encoding glycoside hydrolases (cyan), susCD (orange), regulators (green), and carbohydrate-processing genes (red) with homology to PUL in Z. galactanivorans targeting xylan and maltose (locus tags 0158-0164 and 1248-1253, respectively). Numbers designate GH families.
Wide detection of the PL12 unique to strain 62–1 (Supplementary Table 4) supports the presumed role of the lyase and adjacent EPS genes in surface attachment. This gene arrangement might permit the formation of specific biofilm structures, helping to reduce competition with co-existing bacteria. EPS on algal surfaces could also provide a protective matrix, minimizing diffusion of secreted CAZymes and retaining hydrolysis products for maximal uptake (Vetter et al., 1998). In turn, EPS could also be advantageous in pelagic waters from which 62–1 has been isolated, where aggregation on self-produced EPS could constitute a protective refugium and facilitate survival when algal substrates are unavailable (Decho and Gutierrez, 2017). In general, the diversity of EPS genes on fine phylogenetic levels might mediate distinct interactions with algal hosts, considering that EPS can be strain-specific determinants of host interaction (Lee et al., 2016; Deo et al., 2019).
In addition to metagenomic analyses, we searched strain 62-1 for genes encoding characterized algae-adaptive traits in Zobellia galactanivorans, a closely related hydrolytic flavobacterium (Barbeyron et al., 2016b). This approach identified homologous genes for degradation of mannitol and digeneaside in strain 62–1 (Figure 4B and Supplementary Table 4). Furthermore, we found GH43-GH10 and GH13-GH65 gene pairs targeting the algal carbohydrates xylan and maltose (Figure 4C). We also detected most homologs for anhydrogalactose utilization and hence the second step in carrageenan metabolism, but no GH127 or GH129 anhydrogalactosidase genes for initial hydrolysis (Ficko-Blean et al., 2017). Hence, Maribacter might utilize hydrolysis products from primary degraders, employing a secondary “harvester” strategy for carrageenan on red macroalgae (Hehemann et al., 2016). However, agarase or porphyranase homologs were not detected (Figure 4B), indicating an overall narrower niche range than Zobellia.
In addition to carbohydrate utilization, we found other features typical for interactions with macroalgae. For instance, genes for the detoxification of iodine and nitrous oxide (Figure 4B and Supplementary Table 4) likely counteract algal defense mechanisms (Ogawa et al., 1995; Verhaeghe et al., 2008). Detoxification of superoxide and hydrogen peroxide is associated with two dismutases and 21 thioredoxin-like oxidoreductases (Supplementary Table 4), an almost twofold higher count despite an ∼1 Mb smaller genome than Zobellia (Barbeyron et al., 2016b). The predicted ability to detoxify methanol is likely advantageous during demethylation of pectinous algal substrates (Koch et al., 2019a). Moreover, strain 62-1 encodes traits that might be advantageous to algal hosts. For instance, a putative homoserine lactone lyase (Figure 4B and Supplementary Table 4) might interfere with communication of bacterial competitors and prevent their biofilm formation. This process, often termed quorum quenching, might antagonize resource competitors and modulate the composition of the holobiont (Wahl et al., 2012). Presence of a PKS might allow biosynthesis of thallusin, an essential algal morphogen produced by both Zobellia and Maribacter (Matsuo et al., 2003; Weiss et al., 2017). Another conserved feature is the complete pathway for biosynthesis of biotin (vitamin B7; locus tags 03353–03359 in strain 62–1), a common characteristic of macroalgal epibionts (Karimi et al., 2020). Strain 62–1 misses a single gene in biosynthetic pathways for riboflavin (vitamin B2) and pantothenate (B5), but might provide intermediates to other bacteria encoding the complete pathway (Paerl et al., 2018; Shelton et al., 2019).
The gammaproteobacterial strain Alteromonas sp. 76–1, isolated from the same seawater sample as Maribacter dokdonensis 62–1, has a comparable predisposition toward polysaccharide degradation (Koch et al., 2019b). The presence of alginolytic and ulvanolytic systems in both 62–1 and 76–1 supports the notion that alginate and ulvan commonly occur at the Patagonian continental shelf. Although the two strains might hence compete for these resources, their corresponding CAZyme repertoires substantially differ. While sharing similar numbers of PL6 and PL7 alginate lyase genes (Figure 5A), PL12 are restricted to Maribacter and PL18 to Alteromonas, indicating different alginolytic strategies. Furthermore, concerning ulvanolytic activity, Alteromonas encodes PL24 and PL25 lyases on a CAZyme plasmid opposed to PL33 and PL40 on the Maribacter chromosome (Figure 5A), potentially influencing the regulation and transfer of these genes.
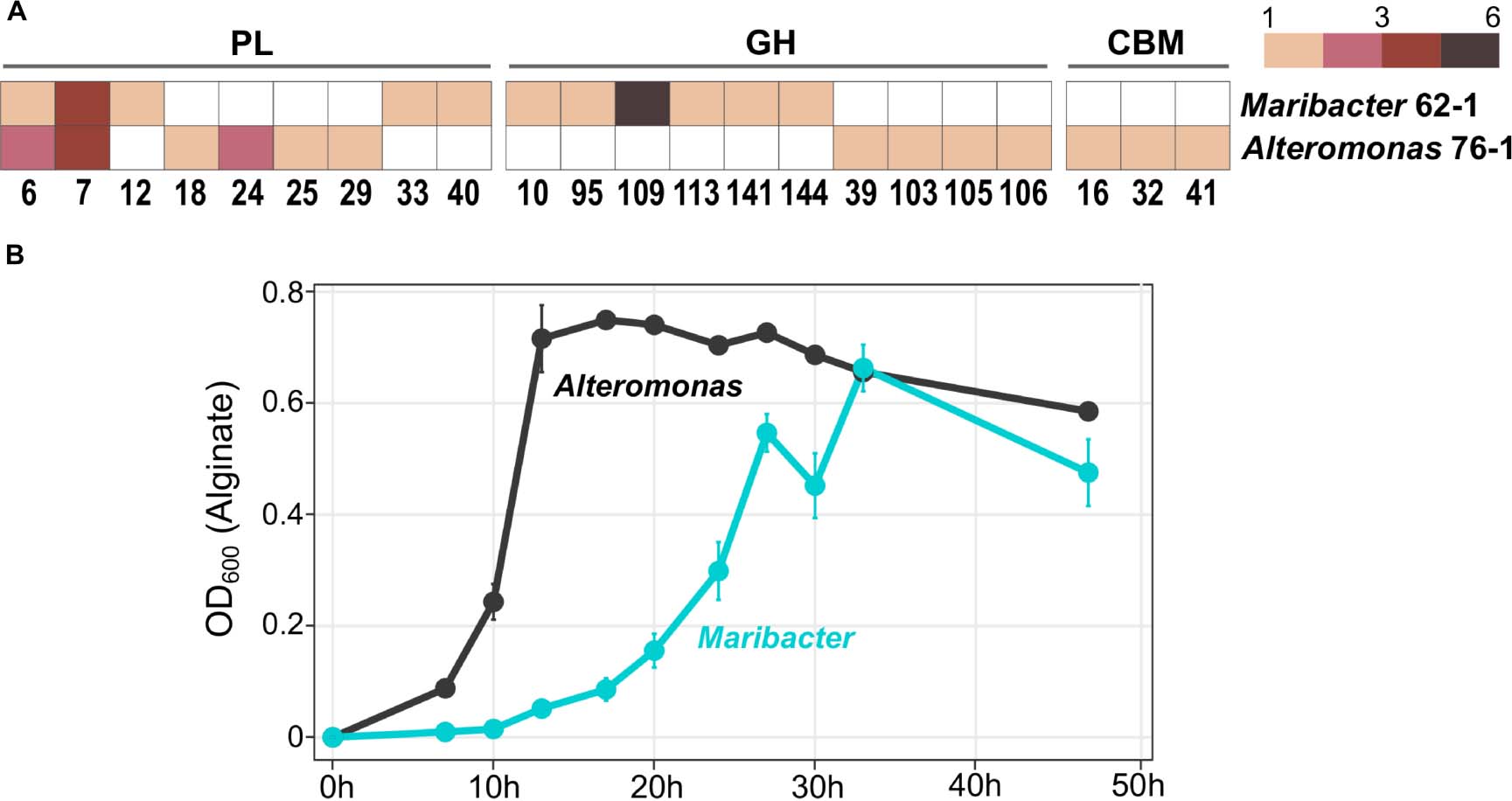
Figure 5. Comparison of Maribacter dokdonensis 62–1 with Alteromonas sp. 76–1 isolated from the same seawater sample. (A) Gene counts among shared and unique families of polysaccharide lyases (PL), glycoside hydrolases (GH) and carbohydrate-binding modules (CBM). (B) Differing growth with alginate as sole carbon source over a period of 48 h.
We hypothesize that differences in CAZymes and PUL for the same substrates mediate ecological specialization. For instance, different PL combinations might specialize for different subsets of these polysaccharides, e.g., relating to polymer length, the ratio of individual carbohydrate monomers, sulfatation or the degree of side-chain decorations. For instance, the Alteromonas-specific PL18 might boost alginolytic activity by acting as bifunctional lyase on both poly-G and poly-M stretches (Li et al., 2011). Furthermore, strain-specific GH genes might allow the separation into distinct “polysaccharide niches” and minimize competition. For instance, a GH109 with presumed N-acetylgalactosaminidase activity (Lombard et al., 2014) only occurs in Maribacter (Figure 5A). Finally, ecological specialization might include “temporal niche differentiation” considering variable doubling times when degrading alginate (Figure 5B). The delayed exponential phase in Maribacter suggests K-strategy, compared to r-strategy in the faster-growing Alteromonas (Pedler et al., 2014).
The diversity of CAZymes and PUL targeting alginate, ulvan and other algal carbohydrates illustrates a marked predisposition of Maribacter dokdonensis 62–1 for polysaccharide degradation, exemplified by distinct growth with alginate as sole carbon source. The variety of EPS-related genes might facilitate switching between free-living and surface-associated lifestyles, potentially connected to the specific colonization of algal hosts. The overall conservation of adaptive features in Maribacter and the “sister genus” Zobellia highlights their importance as macroalgal epibionts, supported by detection of PL homologs in metagenomes from brown, green and red macroalgae on global scales. The presence of both adverse (utilization of algal substrates, evading algal defense) and advantageous traits (vitamin production, biofilm control) indicates versatile lifestyles, which potentially change depending on physicochemical conditions and algal health (Kumar et al., 2016). Different hydrolytic machineries and doubling times of strain 62–1 compared to the co-occurring Alteromonas sp. 76-1 might avoid competition and result in colonization of discrete niches. These insights contribute to the understanding of flavobacterial CAZymes and hydrolytic activity in context of biogeochemical cycles, niche specialization and ecological interactions in the oceans.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
LW conducted genomic analyses and contributed to writing. MM performed growth experiments and HPLC. JK contributed to genomic analyses. RD performed genome sequencing. MS co-organized the experiments yielding the isolation of strain 62–1. MW designed research and wrote the manuscript. All authors contributed to the final version of the manuscript.
This work was supported by the Japan Science and Technology Agency ERATO (JPMJER1502) and the Collaborative Research Center Roseobacter from the German Research Foundation (TRR51). MW was supported by the German Research Foundation through grant WI3888/1-2.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Rolf Weinert and Martin Mierzejewski for excellent laboratory assistance. Captain, crew and colleagues of RV Polarstern expedition ANTXXVIII-5 are thanked for their professional and friendly support. Sonja Voget is gratefully acknowledged for Illumina sequencing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.628055/full#supplementary-material
Supplementary Figure 1 | Heatmap of average nucleotide identities (ANI) between the genomes of Maribacter dokdonensis 62–1 and related strains.
Supplementary Figure 2 | Homologs of AlgPUL1 (A), AlgPUL2 (B) and the mixed-feature PUL relating to ulvan and fucoidan (C) in genomes of Maribacter and Zobellia strains. Numbers designate PL families.
Supplementary Table 1 | Strain overview. Genome characteristics of Maribacter dokdonensis 62–1 and related strains used for comparative analyses.
Supplementary Table 2 | CAZyme overview. CAZymes, PUL, sulfatases, and signal peptides in Maribacter dokdonensis 62-1 and related strains.
Supplementary Table 3 | Pangenome overview. Core, accessory and unique genes in Maribacter dokdonensis 62-1 and related strains.
Supplementary Table 4 | Ecological implications. Detection of PL homologs in microbial metagenomes from marine plants and genomic features for interactions with algae.
Abe, K., Nakajima, M., Yamashita, T., Matsunaga, H., Kamisuki, S., Nihira, T., et al. (2017). Biochemical and structural analyses of a bacterial endo-β-1,2-glucanase reveal a new glycoside hydrolase family. J. Biol. Chem. 292, 7487–7506. doi: 10.1074/jbc.M116.762724
Acha, E. M., Mianzan, H. W., Guerrero, R. A., Favero, M., and Bava, J. (2004). Marine fronts at the continental shelves of austral South America: physical and ecological processes. J. Mar. Syst. 44, 83–105. doi: 10.1016/J.JMARSYS.2003.09.005
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Arnosti, C., Steen, A. D., Ziervogel, K., Ghobrial, S., and Jeffrey, W. H. (2011). Latitudinal gradients in degradation of marine dissolved organic carbon. PLoS One 6:e28900. doi: 10.1371/journal.pone.0028900
Arnosti, C., Wietz, M., Brinkhoff, T., Hehemann, J.-H., Probandt, D., Zeugner, L., et al. (2021). The biogeochemistry of marine polysaccharides: sources, inventories, and bacterial drivers of the carbohydrate cycle. Annu. Rev. Mar. Sci. 13, 81–108.
Bakkevig, K., Sletta, H., Gimmestad, M., Aune, R., Ertesvåg, H., Degnes, K., et al. (2005). Role of the Pseudomonas fluorescens alginate lyase (AlgL) in clearing the periplasm of alginates not exported to the extracellular environment. J. Bacteriol. 187, 8375–8384. doi: 10.1128/JB.187.24.8375-8384.2005
Bakunina, I. Y., Nedashkovskaya, O. I., Kim, S. B., Zvyagintseva, T. N., and Mikhailov, V. V. (2012). Diversity of glycosidase activities in the bacteria of the phylum Bacteroidetes isolated from marine algae. Microbiology 81, 688–695. doi: 10.1134/S0026261712060033
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barbeyron, T., Brillet-Guéguen, L., Carré, W., Carrière, C., Caron, C., Czjzek, M., et al. (2016a). Matching the diversity of sulfated biomolecules: creation of a classification database for sulfatases reflecting their substrate specificity. PLoS One 11:e0164846. doi: 10.1371/journal.pone.0164846
Barbeyron, T., Thomas, F., Barbe, V., Teeling, H., Schenowitz, C., Dossat, C., et al. (2016b). Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae-associated bacterium Zobellia galactanivorans DsijT. Environ. Microbiol. 18, 4610–4627. doi: 10.1111/1462-2920.13584
Bateman, A., Martin, M. J., O’Donovan, C., Magrane, M., Alpi, E., Antunes, R., et al. (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. doi: 10.1093/nar/gkw1099
Chernysheva, N., Bystritskaya, E., Stenkova, A., Golovkin, I., Nedashkovskaya, O., and Isaeva, M. (2019). Comparative genomics and CAZyme genome repertoires of marine Zobellia amurskyensis KMM 3526T and Zobellia laminariae KMM 3676T. Mar. Drugs 17:661. doi: 10.3390/md17120661
Darriba, D., Posada, D., Kozlov, A. M., Stamatakis, A., Morel, B., and Flouri, T. (2020). ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37, 291–294. doi: 10.1093/molbev/msz189
Decho, A. W., and Gutierrez, T. (2017). Microbial extracellular polymeric substances (EPSs) in ocean systems. Front. Microbiol. 8:922. doi: 10.3389/fmicb.2017.00922
Deo, D., Davray, D., and Kulkarni, R. (2019). A diverse repertoire of exopolysaccharide biosynthesis gene clusters in Lactobacillus revealed by comparative analysis in 106 sequenced genomes. Microorganisms 7:444. doi: 10.3390/microorganisms7100444
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. doi: 10.1186/s13059-019-1832-y
Fernández-Gómez, B., Richter, M., Schüler, M., Pinhassi, J., Acinas, S. G., González, J. M., et al. (2013). Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 7, 1026–1037. doi: 10.1038/ismej.2012.169
Ferrer-González, F. X., Widner, B., Holderman, N. R., Glushka, J., Edison, A. S., Kujawinski, E. B., et al. (2020). Resource partitioning of phytoplankton metabolites that support bacterial heterotrophy. ISME J. 15, 762–773. doi: 10.1038/s41396-020-00811-y
Ficko-Blean, E., Préchoux, A., Thomas, F., Rochat, T., Larocque, R., Zhu, Y., et al. (2017). Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria. Nat. Commun. 8, 1685–1685. doi: 10.1038/s41467-017-01832-6
Foran, E., Buravenkov, V., Kopel, M., Mizrahi, N., Shoshani, S., Helbert, W., et al. (2017). Functional characterization of a novel “ulvan utilization loci” found in Alteromonas sp. LOR Genome. Algal Res. 25, 39–46. doi: 10.1016/j.algal.2017.04.036
Garcia, V. M. T., Garcia, C. A. E., Mata, M. M., Pollery, R. C., Piola, A. R., Signorini, S. R., et al. (2008). Environmental factors controlling the phytoplankton blooms at the Patagonia shelf-break in spring. Deep Sea Res. Part I Oceanogr. Res. Papers 55, 1150–1166. doi: 10.1016/J.DSR.2008.04.011
Grieb, A., Francis, T. B., Krüger, K., Orellana, L. H., Amann, R., and Fuchs, B. M. (2020). Candidatus Abditibacter, a novel genus within the Cryomorphaceae, thriving in the North Sea. Syst. Appl. Microbiol. 43:126088. doi: 10.1016/j.syapm.2020.126088
Grondin, J. M., Tamura, K., Déjean, G., Abbott, D. W., and Brumer, H. (2017). Polysaccharide Utilization Loci: fuelling microbial communities. J. Bacteriol. 199:e00860-16. doi: 10.1128/JB.00860-16
Hehemann, J.-H., Arevalo, P., Datta, M. S., Yu, X., Corzett, C. H., Henschel, A., et al. (2016). Adaptive radiation by waves of gene transfer leads to fine-scale resource partitioning in marine microbes. Nat. Commun. 7:12860. doi: 10.1038/ncomms12860
Hehemann, J.-H., Boraston, A. B., and Czjzek, M. (2014). A sweet new wave: structures and mechanisms of enzymes that digest polysaccharides from marine algae. Curr. Opin. Struct. Biol. 28, 77–86. doi: 10.1016/j.sbi.2014.07.009
Hehemann, J.-H., Correc, G., Thomas, F., Bernard, T., Barbeyron, T., Jam, M., et al. (2012). Biochemical and structural characterization of the complex agarolytic enzyme system from the marine bacterium Zobellia galactanivorans. J. Biol. Chem. 287, 30571–30584. doi: 10.1074/jbc.M112.377184
Hobbs, J. K., Hettle, A. G., Vickers, C., and Boraston, A. B. (2019). Biochemical reconstruction of a metabolic pathway from a marine bacterium reveals its mechanism of pectin depolymerization. Appl. Environ. Microbiol. 85:aem.02114-18. doi: 10.1128/AEM.02114-18
Kabisch, A., Otto, A., König, S., Becher, D., Albrecht, D., Schüler, M., et al. (2014). Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes ‘Gramella forsetii’ KT0803. ISME J. 8, 1492–1502. doi: 10.1038/ismej.2014.4
Kanehisa, M., and Sato, Y. (2020). KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 29, 28–35. doi: 10.1002/pro.3711
Kappelmann, L., Krüger, K., Hehemann, J.-H., Harder, J., Markert, S., Unfried, F., et al. (2019). Polysaccharide utilization loci of North Sea Flavobacteriia as basis for using SusC/D-protein expression for predicting major phytoplankton glycans. ISME J. 13, 76–91. doi: 10.1038/s41396-018-0242-6
Karimi, E., Geslain, E., KleinJan, H., Tanguy, G., Legeay, E., Corre, E., et al. (2020). Genome sequences of 72 bacterial strains isolated from Ectocarpus subulatus: a resource for algal microbiology. Genome Biol. Evol. 12, 3647–3655. doi: 10.1093/gbe/evz278
Kidgell, J. T., Magnusson, M., de Nys, R., and Glasson, C. R. K. (2019). Ulvan: a systematic review of extraction, composition and function. Algal Res. 39:101422. doi: 10.1016/j.algal.2019.101422
Koch, H., Dürwald, A., Schweder, T., Noriega-Ortega, B., Vidal-Melgosa, S., Hehemann, J.-H., et al. (2019a). Biphasic cellular adaptations and ecological implications of Alteromonas macleodii degrading a mixture of algal polysaccharides. ISME J. 13, 92–103. doi: 10.1038/s41396-018-0252-4
Koch, H., Freese, H. M., Hahnke, R., Simon, M., and Wietz, M. (2019b). Adaptations of Alteromonas sp. 76-1 to polysaccharide degradation: a CAZyme plasmid for ulvan degradation and two alginolytic systems. Front. Microbiol. 10:504. doi: 10.3389/fmicb.2019.00504
Kolde, R. (2018). pheatmap: Pretty Heatmaps. Available online at: https://github.com/raivokolde/pheatmap (accessed July, 2020).
Köseoǧlu, V. K., Heiss, C., Azadi, P., Topchiy, E., Güvener, Z. T., Lehmann, T. E., et al. (2015). Listeria monocytogenes exopolysaccharide: origin, structure, biosynthetic machinery and c-di-GMP-dependent regulation. Mol. Microbiol. 96, 728–743. doi: 10.1111/mmi.12966
Kumar, V., Zozaya−Valdes, E., Kjelleberg, S., Thomas, T., and Egan, S. (2016). Multiple opportunistic pathogens can cause a bleaching disease in the red seaweed Delisea pulchra. Environ. Microbiol. 18, 3962–3975. doi: 10.1111/1462-2920.13403
Kwak, M.-J., Lee, J., Kwon, S.-K., and Kim, J. F. (2017). Genome information of Maribacter dokdonensis DSW-8 and comparative analysis with other Maribacter genomes. J. Microbiol. Biotechnol. 27, 591–597. doi: 10.4014/jmb.1608.08044
Labourel, A., Jam, M., Jeudy, A., Hehemann, J.-H., Czjzek, M., and Michel, G. (2014). The β-glucanase ZgLamA from Zobellia galactanivorans evolved a bent active site adapted for efficient degradation of algal laminarin. J. Biol. Chem. 289, 2027–2042. doi: 10.1074/jbc.M113.538843
Lawlor, J. (2020). PNWColors. Available online at: https://github.com/jakelawlor/PNWColors (accessed August, 2020).
Lee, I.-C., Caggianiello, G., van Swam, I. I., Taverne, N., Meijerink, M., Bron, P. A., et al. (2016). Strain-Specific features of extracellular polysaccharides and their impact on Lactobacillus plantarum-host interactions. Appl. Environ. Microbiol. 82, 3959–3970. doi: 10.1128/AEM.00306-16
Lee, J. H., Kang, J. W., Choe, H. N., and Seong, C. N. (2017). Flavivirga eckloniae sp. nov. and Flavivirga aquimarina sp. nov., isolated from seaweed Ecklonia cava. Int. J. Syst. Evol. Microbiol. 67, 3089–3094. doi: 10.1099/ijsem.0.002094
Li, J.-W., Dong, S., Song, J., Li, C.-B., Chen, X.-L., Xie, B.-B., et al. (2011). Purification and characterization of a bifunctional alginate lyase from Pseudoalteromonas sp. SM0524. Mar. Drugs 9, 109–123. doi: 10.3390/md9010109
Lim, H. N., Lee, Y., and Hussein, R. (2011). Fundamental relationship between operon organization and gene expression. Proc. Natl. Acad. Sci. U.S.A. 108, 10626–10631. doi: 10.1073/pnas.1105692108
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Mann, A. J., Hahnke, R. L., Huang, S., Werner, J., Xing, P., Barbeyron, T., et al. (2013). The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl. Environ. Microbiol. 79, 6813–6822. doi: 10.1128/AEM.01937-13
Markowitz, V. M., Chen, I. M. A., Palaniappan, K., Chu, K., Szeto, E., Grechkin, Y., et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40, D115–D122. doi: 10.1093/nar/gkr1044
Martin, M., Barbeyron, T., Martin, R., Portetelle, D., Michel, G., and Vandenbol, M. (2015). The cultivable surface microbiota of the brown alga Ascophyllum nodosum is enriched in macroalgal-polysaccharide-degrading bacteria. Front. Microbiol. 6:1487. doi: 10.3389/fmicb.2015.01487
Matos, M. N., Lozada, M., Anselmino, L. E., Musumeci, M. A., Henrissat, B., Jansson, J. K., et al. (2016). Metagenomics unveils the attributes of the alginolytic guilds of sediments from four distant cold coastal environments. Environ. Microbiol. 18, 4471–4484. doi: 10.1111/1462-2920.13433
Matsuo, Y., Imagawa, H., Nishizawa, M., and Shizuri, Y. (2005). Isolation of an algal morphogenesis inducer from a marine bacterium. Science 307, 1598–1598. doi: 10.1126/science.1105486
Matsuo, Y., Suzuki, M., Kasai, H., Shizuri, Y., and Harayama, S. (2003). Isolation and phylogenetic characterization of bacteria capable of inducing differentiation in the green alga Monostroma oxyspermum. Environ. Microbiol. 5, 25–35. doi: 10.1046/j.1462-2920.2003.00382.x
McNulty, N. P., Wu, M., Erickson, A. R., Pan, C., Erickson, B. K., Martens, E. C., et al. (2013). Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 11:e1001637. doi: 10.1371/journal.pbio.1001637
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, 1–8.
Mopper, K., Schultz, C. A., Chevolot, L., Germain, C., Revuelta, R., and Dawson, R. (1992). Determination of sugars in unconcentrated seawater and other natural waters by liquid chromatography and pulsed amperometric detection. Environ. Sci. Technol. 26, 133–138. doi: 10.1021/es00025a014
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Na, S.-I., Kim, Y. O., Yoon, S.-H., Ha, S., Baek, I., and Chun, J. (2018). UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 281–285. doi: 10.1007/s12275-018-8014-6
Nikolenko, S. I., Korobeynikov, A. I., and Alekseyev, M. A. (2013). BayesHammer: bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14(Suppl. 1):S7. doi: 10.1186/1471-2164-14-S1-S7
Norling, M., Jareborg, N., and Dainat, J. (2018). EMBLmyGFF3: a converter facilitating genome annotation submission to European Nucleotide Archive. BMC Res. Notes 11:584. doi: 10.1186/s13104-018-3686-x
Ogawa, K., Akagawa, E., Yamane, K., Sun, Z. W., LaCelle, M., Zuber, P., et al. (1995). The nasB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J. Bacteriol. 177, 1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995
Paerl, R. W., Sundh, J., Tan, D., Svenningsen, S. L., Hylander, S., Pinhassi, J., et al. (2018). Prevalent reliance of bacterioplankton on exogenous vitamin B1 and precursor availability. Proc. Natl. Acad. Sci. U.S.A. 115, E10447–E10456. doi: 10.1073/pnas.1806425115
Pedler, B. E., Aluwihare, L. I., and Azam, F. (2014). Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean. Proc. Natl. Acad. Sci. U.S.A. 111, 7202–7207. doi: 10.1073/pnas.1401887111
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reintjes, G., Arnosti, C., Fuchs, B. M., and Amann, R. (2017). An alternative polysaccharide uptake mechanism of marine bacteria. ISME J. 11, 1640–1650. doi: 10.1038/ismej.2017.26
Reisky, L., Préchoux, A., Zühlke, M.-K., Bäumgen, M., Robb, C. S., Gerlach, N., et al. (2019). A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan. Nat. Chem. Biol. 15, 803–812. doi: 10.1038/s41589-019-0311-9
Reisky, L., Stanetty, C., Mihovilovic, M. D., Schweder, T., Hehemann, J.-H., and Bornscheuer, U. T. (2018). Biochemical characterization of an ulvan lyase from the marine flavobacterium Formosa agariphila KMM 3901T. Appl. Microbiol. Biotechnol. 102, 6987–6996. doi: 10.1007/s00253-018-9142-y
Rodriguez-R, L. M., and Konstantinidis, K. T. (2016). The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4:e1900v1. doi: 10.7287/PEERJ.PREPRINTS.1900V1
Salinas, A., and French, C. E. (2017). The enzymatic ulvan depolymerisation system from the alga-associated marine flavobacterium Formosa agariphila. Algal Res. 27, 335–344. doi: 10.1016/j.algal.2017.09.025
Shelton, A. N., Seth, E. C., Mok, K. C., Han, A. W., Jackson, S. N., Haft, D. R., et al. (2019). Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J. 13, 789–804. doi: 10.1038/s41396-018-0304-9
Sichert, A., Corzett, C. H., Schechter, M. S., Unfried, F., Markert, S., Becher, D., et al. (2020). Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 5, 1026–1039. doi: 10.1038/s41564-020-0720-2
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Teeling, H., Fuchs, B. M., Bennke, C. M., Krüger, K., Chafee, M., Kappelmann, L., et al. (2016). Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. eLife 5:e11888. doi: 10.7554/eLife.11888
Terrapon, N., Lombard, V., Drula, É., Lapébie, P., Al-Masaudi, S., Gilbert, H. J., et al. (2018). PULDB: the expanded database of Polysaccharide Utilization Loci. Nucleic Acids Res. 46, D677–D683. doi: 10.1093/nar/gkx1022
Thomas, F., Barbeyron, T., Tonon, T., Génicot, S., Czjzek, M., and Michel, G. (2012). Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ. Microbiol. 14, 2379–2394. doi: 10.1111/j.1462-2920.2012.02751.x
Thomas, F., Bordron, P., Eveillard, D., and Michel, G. (2017). Gene expression analysis of Zobellia galactanivorans during the degradation of algal polysaccharides reveals both substrate-specific and shared transcriptome-wide responses. Front. Microbiol. 8:1808. doi: 10.3389/fmicb.2017.01808
van der Loos, L. M., Eriksson, B. K., and Falcão Salles, J. (2019). The macroalgal holobiont in a changing sea. Trends Microbiol. 27, 635–650. doi: 10.1016/j.tim.2019.03.002
Verhaeghe, E. F., Fraysse, A., Guerquin-Kern, J.-L., Wu, T.-D., Devès, G., Mioskowski, C., et al. (2008). Microchemical imaging of iodine distribution in the brown alga Laminaria digitata suggests a new mechanism for its accumulation. J. Biol. Inorg. Chem. 13, 257–269. doi: 10.1007/s00775-007-0319-6
Vetter, Y. A., Deming, J. W., Jumars, P. A., and Krieger-Brockett, B. B. (1998). A predictive model of bacterial foraging by means of freely released extracellular enzymes. Microb. Ecol. 36, 75–92. doi: 10.1007/s002489900095
Wahl, M., Goecke, F., Labes, A., Dobretsov, S., and Weinberger, F. (2012). The second skin: ecological role of epibiotic biofilms on marine organisms. Front. Microbiol. 3:292. doi: 10.3389/fmicb.2012.00292
Weiss, A., Costa, R., and Wichard, T. (2017). Morphogenesis of Ulva mutabilis (Chlorophyta) induced by Maribacter species (Bacteroidetes, Flavobacteriaceae). Bot. Mar. 60, 197–206. doi: 10.1515/bot-2016-0083
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag New York.
Wietz, M., Wemheuer, B., Simon, H., Giebel, H. A., Seibt, M. A., Daniel, R., et al. (2015). Bacterial community dynamics during polysaccharide degradation at contrasting sites in the Southern and Atlantic Oceans. Environ. Microbiol. 17, 3822–3831. doi: 10.1111/1462-2920.12842
Wolter, L. A., Wietz, M., Ziesche, L., Breider, S., Leinberger, J., Poehlein, A., et al. (2021). Pseudooceanicola algae sp. nov., isolated from the marine macroalga Fucus spiralis, shows genomic and physiological adaptations for an algae-associated lifestyle. Syst. Appl. Microbiol. 44:126166. doi: 10.1016/j.syapm.2020.126166
Xu, F., Wang, P., Zhang, Y.-Z., and Chen, X.-L. (2017). Diversity of three-dimensional structures and catalytic mechanisms of alginate lyases. Appl. Environ. Microbiol. 84:e02040-17. doi: 10.1128/AEM.02040-17
Zech, H., Thole, S., Schreiber, K., Kalhöfer, D., Voget, S., Brinkhoff, T., et al. (2009). Growth phase-dependent global protein and metabolite profiles of Phaeobacter gallaeciensis strain DSM 17395, a member of the marine Roseobacter-clade. Proteomics 9, 3677–3697. doi: 10.1002/pmic.200900120
Zhan, P., Tang, K., Chen, X., and Yu, L. (2017). Complete genome sequence of Maribacter sp. T28, a polysaccharide-degrading marine flavobacteria. J. Biotechnol. 259, 1–5. doi: 10.1016/j.jbiotec.2017.08.009
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., et al. (2018). dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101. doi: 10.1093/nar/gky418
Keywords: alginate, fucoidan, ulvan, PUL, macroalgae, EPS, pangenome, Zobellia galactanivorans
Citation: Wolter LA, Mitulla M, Kalem J, Daniel R, Simon M and Wietz M (2021) CAZymes in Maribacter dokdonensis 62–1 From the Patagonian Shelf: Genomics and Physiology Compared to Related Flavobacteria and a Co-occurring Alteromonas Strain. Front. Microbiol. 12:628055. doi: 10.3389/fmicb.2021.628055
Received: 11 November 2020; Accepted: 10 March 2021;
Published: 12 April 2021.
Edited by:
Chao Liang, Institute of Applied Ecology (CAS), ChinaReviewed by:
Abbot Okotie Oghenekaro, University of Manitoba, CanadaCopyright © 2021 Wolter, Mitulla, Kalem, Daniel, Simon and Wietz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthias Wietz, bWF0dGhpYXMud2lldHpAYXdpLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.