- 1Key Laboratory of Ministry of Education for Genetics and Germplasm Innovation of Tropical Special Trees and Ornamental Plants, Key Laboratory of Germplasm Resources of Tropical Special Ornamental Plants of Hainan Province, College of Forestry, College of Tropical Crops, Hainan University, Danzhou, China
- 2Tropical Crops Genetic Resources Institute, Chinese Academy of Tropical Agricultural Sciences, Danzhou, China
This study investigated the fermentation quality of 12 varieties of Pennisetum sinese grown in different regions of Southern China. Following the production of silage from the natural fermentation of P. sinese, the interplay between the chemical composition, fermentation characteristics, environmental factors, and microbiome was examined to understand the influence of these factors on the fermentation quality of silage. The silage quality produced by most of the P. sinese was low; the pH value of the silage was high (4.26–4.86), whilst the lactic acid content was low (10.7–24.1 g/kg DM), with V-scores between 57.9 and 78.3. The bacterial alpha diversities of the 12 P. sinese silages were distinct. There was a predominance of undesirable bacteria (Pseudomonas, Massilia, and Raoultella), which likely caused the poor fermentation quality. The chemical composition and fermentation characteristics of the silage were closely correlated with the composition of the bacterial community. Furthermore, environmental factors (precipitation, temperature, humidity, location) were found to significantly influence the microbiome of the silage. The results confirmed that silage produced from the natural fermentation of 12 different P. sinese varieties had significant variation in their bacterial communities. The difference in environmental factors, due to the P. sinese being grown in various locations across south china, greatly affected the bacterial community found in the silage and thus the fermentation quality. The specific cultivar used for the silage and the environment in which the cultivar is grown must therefore be considered before the initiation of production of silage in order to ensure a higher quality product.
Introduction
Pennisetum sinese is a member of the grass family. It is adaptable and has a high biomass production rate; this has led to its widespread use in tropical/subtropical regions across the world (Zhao et al., 2019). P. sinese is primarily used for the production of feedstock for biofuels and animal feed; this is due to its high lignocellulosic biomass and excellent palatability (Li et al., 2014, 2019a, 2019b, 2020; Lu et al., 2014; Wang et al., 2016; Shah et al., 2018). The demand for P. sinese is large, but production is affected by seasonal harvest. The summer/rainy seasons see vigorous P. sinese growth, with a contrasting lack of growth during winter and times of drought; this leads to a shortage in its availability (Li et al., 2014). In order to combat this and meet the year-round demand, P. sinese needs proper pretreatment and conservation.
Pennisetum sinese has a high moisture content, making the plant susceptible to mildew and rot; this causes biomass decomposition and inefficient biotransformation. Those using P. sinese in tropical regions face considerable challenges regarding preservation and ineffective biotransformation, due to the hot and humid environment. Ensiling is a method that can conserve green plants by utilizing anaerobic fermentation. Ensiling can guarantee the quality of the feed for long term storage and has also been shown to be a promising pretreatment method in biofuel production (Wilkinson et al., 2003). Previous studies have focused primarily on the effect of additives to P. sinese silages, but little is currently understood regarding the effect of additives on the microbiome of the resultant silage (Li et al., 2014, 2018a, 2018b, 2019a, 2020; Li L. et al., 2017; Wu et al., 2020). Currently, only a limited number of studies have examined the effects of additives on the fermentation quality and microbial diversity of P. sinese silage (Dong et al., 2020; Shah et al., 2020). It is of note that these studies have focused on specific P. sinese cultivars.
As previously mentioned, uncontrollable environmental factors challenge the production of silage in tropical areas (Bernardes et al., 2018). The significance of errant weather conditions has previously been examined in silage produced from the fermentation of Napier and Italian Ryegrass (Guo et al., 2014, 2015). In addition, found that environmental factors influenced the bacterial diversity of corn silage and thus altered the fermentation quality (Guan et al., 2018). Moreover, it has been reported that temperature can influence the bacterial diversity and fermentation quality of silage produced from Moringa oleifera (Wang et al., 2019a).
Pennisetum sinese is grown across multiple regions of southern china that differ greatly in climate. Therefore, the influence of environmental factors on silage quality and microbial diversity should be examined and any effects should be investigated in the multiple P. sinese cultivars that are used across the regions.
This study aimed to examine the differing bacterial communities and the fermentation of silages produced from 12 different P. sinese varieties. The P. sinese studied were grown across multiple regions in southern China; this allowed the experimental parameters to include influence of environmental factors (precipitation, temperature, humidity, location) on the silage microbiome across a broad spectrum of varieties of P. sinese.
Materials and Methods
Silage Preparation
Twelve P. sinese varieties were grown at six different bases across southern China; the location and environmental factors related to these bases are shown in Supplementary Table 1. The P. sinese cultivars used in this study were planted on the 5th of April 2019 and then harvested on June 26th 2019. The P. sinese material was immediately chopped into small pieces (∼2 cm). Briefly, 200 g of each grass sample was vacuum-packed into plastic bags. A total of 36 bags (twelve varieties in triplicate) were prepared and incubated at 25–30°C for 30 days. The organic acid content and bacterial community of the silage was then determined.
Chemical Composition and Fermentation Analysis
Specimens were dried at 65°C for 2 days and passed through a 1.0 mm sieve before the chemical assay. The contents of dry matter (DM), crude protein (CP), organic matter (OM), and ether extracts (EE) were examined according to previously published work (AOAC, 1990). Moreover, the contents of neutral detergent fiber (NDF) and acid detergent fiber (ADF) were assessed using a previously established method (Van Soest et al., 1991). Heat-stable amylase and sodium sulfite were adopted in the determination of NDF. Water-soluble carbohydrate (WSC) was determined according to a previously described method (Murphy, 1958).
The fermentation quality of the silage was determined using distilled water extracts. 50 g of wet silage was blended with 200 mL distilled water, before 24 h of incubation (4°C) and filtration. The pH, lactic acid, acetic acid, propionic acid, butyric acid, and ammonia-N concentrations were measured as previously established (Li et al., 2019b). To evaluate the silage quality the V-score was calculated using the formula: ammonia-N/Total N and organic acid (Masaki, 2001).
Microbial Community Analysis
The above-mentioned extracts were used for molecular analysis of the silage microbiome. Briefly, 20 mL filtrate was centrifuged at 12,000 g/min for 5 min, and the sediment was collected from the bottom. Microbial DNA was isolated from silage specimens using the E.Z.N.A.®, soil DNA Kit (Omega Bio-Tek, Norcross, GA, United States), according to the manufacturer’s instructions. The concentration and purity of the extracted DNA was assessed using a NanoDrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States) and the integrity of the DNA was assessed by electrophoresis on a 1% agarose gel. A thermocycler PCR system (GeneAmp 9700, ABI, United States) and primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to amplify the V3–V4 hypervariable regions of the bacterial 16S rRNA gene. After the PCR products were purified and quantified, next-generation sequencing was carried out using an Illumina MiSeq 2500 platform (Illumina, Inc., San Diego, CA, United States). Paired-end reads of 250 bp were generated.
Tag assembly was carried out using filtered reads according to the following principles: the overlap between paired-end reads should be more than 10-bp and have less than a 2% mismatch. The unique tags were obtained by removal of redundant tags using MOTHUR software (Schloss et al., 2009). Abundancy was determined using the resultant unique tags and high-quality reads were grouped into operational taxonomic units (OTUs) defined as having 97% similarity. Diversity metrics were determined using the core-diversity plug-in within QIIME21 (Callahan et al., 2016). The microbial diversity within an individual sample was assessed using the following alpha diversity indices: the Chao 1 richness estimator, Shannon diversity index, and Faith’s phylogenetics diversity index. Beta diversity was analyzed to assess the structural variation of microbiota across P. sinese specimens. Subsequently, principal component analysis (PCA) was undertaken (Vázquez-Baeza et al., 2013). Appropriate methods were employed to identify the abundances of different bacterial strains amongst samples and groups as described previously (Segata et al., 2011). Unless specified above, parameters used in the genetic analysis were set as default. The heat map function of the R software2 and genus information for the P. sinese silage were used to generate a heat map. The data were analyzed using the free online BMKCloud Platform3. The sequencing data was submitted to the National Center for Biotechnology Information Sequence Read Archive database under the BioProject accession number PRJNA624770.
Statistics
The impact of variety was investigated using one-way analysis of variance (SAS 9.3 software, SAS Institute Inc., Cary, NC, United States). Significant differences were compared using Duncan’s multiple range tests, with P < 0.05 being regarded as statistically significant.
Results
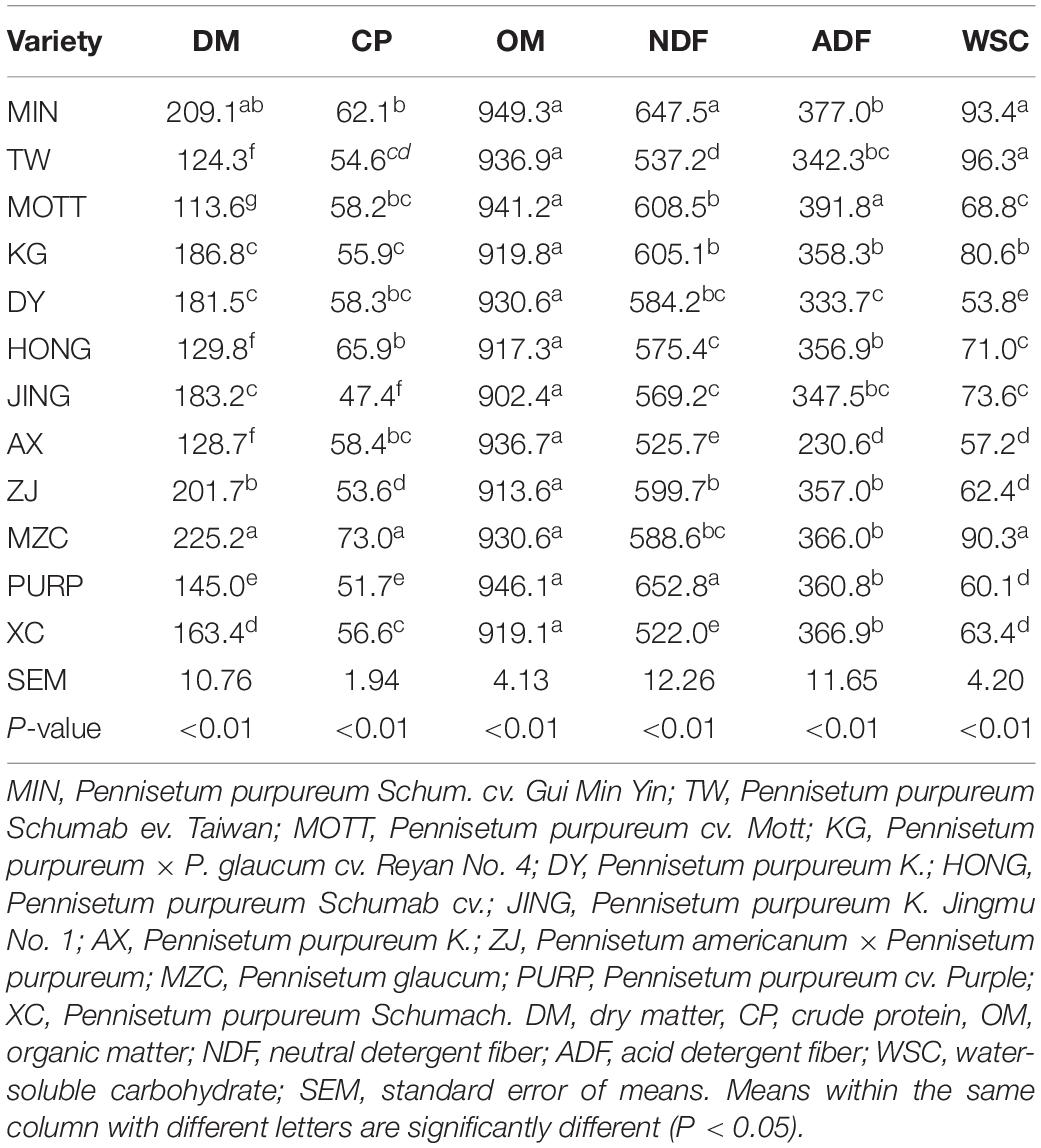
The Chemical Composition of Fresh P. sinese
The Chemical compositions of different fresh P. sinese samples are shown in Table 1. In this study, the P. sinese DM content ranged from 113.6 to 225.2 g/kg; the MZC and MOTT samples had the highest and lowest DM contents, respectively (P < 0.05). The CP contents ranged from 47.4 to 73.0 g/kg, with the MZC and JING samples (P < 0.05), having the highest and lowest CP values, respectively. The OM contents of all P. sinese samples were similar, above 900 g/kg, with no significant difference. The NDF content of all the samples ranged from 522.0 to 652.8 g/kg; the highest and lowest levels were found in the PURP and XC samples, respectively (P < 0.05). The ADF content of the P. sinese samples ranged from 230.6 to 391.8 g/kg; the highest and lowest levels were found in the MOTT and XC samples, respectively (P < 0.05). The WSC content of the P. sinese samples ranged from 53.8 to 93.4 g/kg, with the highest levels in the MIN and DY samples, respectively (P < 0.05). Overall, with the exception of the OM content, the chemical composition of the P. sinese samples varied greatly.
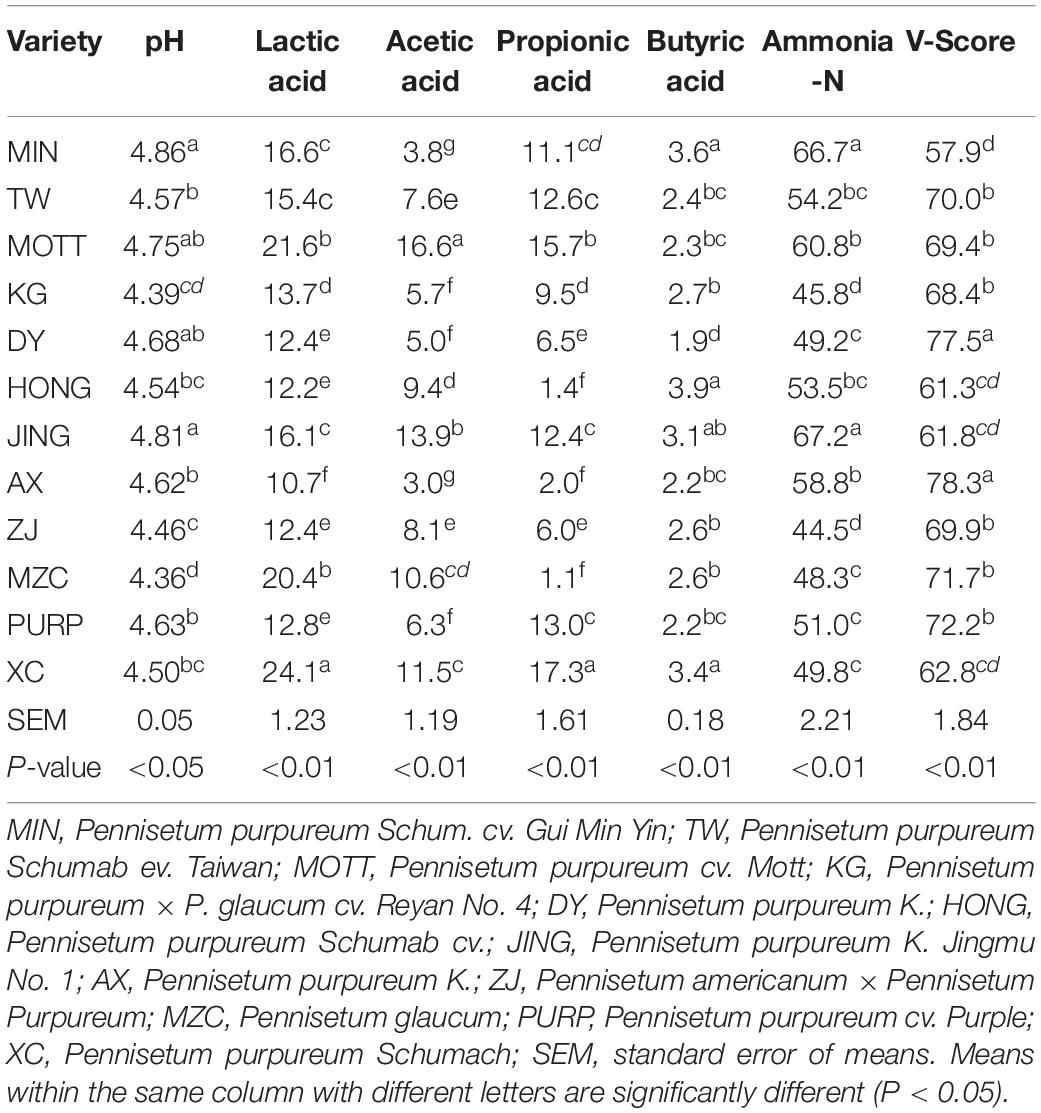
The Fermentation Property of P. sinese Silage
The fermentation properties of silage produced by different P. sinese varieties are shown in Table 2. There were significant differences in the pH levels, as well as the lactic acid, acetic acid, propionic acid, butyric acid, and ammonia-N content depending on the variety off P. sinese used to produce the silage (P < 0.05). The pH of the silages ranged from 4.36 to 4.86, with the silage produced by MIN and MZC P. sinese varieties having the highest and lowest pH levels, respectively (P < 0.05). The lactic acid content of all P. sinese silages ranged from 10.7 to 24.1 g/kg, with XC having the highest lactic acid content and AX the lowest (P < 0.05). The acetic acid content of the different silages ranged from 3.0 to 16.6 g/kg, with the silages produced by the MOTT and AX P. sinese samples having the highest and lowest levels, respectively (P < 0.05). The propionic acid content of all P. sinese silage samples ranged from 1.1 to 17.3 g/kg; the highest propionic acid content was found in the XC (P < 0.05), whilst the lowest propionic acid content was found in the MZC sample (P < 0.05). Meanwhile, the butyric acid content of all P. sinese silage samples ranged from 1.2 to 4.6 g/kg. The silage from the MIN sample had the highest butyric acid content with the PURP sample silage having the lowest (P < 0.05). Finally, the ammonia-N content of the P. sinese silages ranged from 44.5 to 67.2 g/kg, the highest ammonia-N content was found in the JING silage sample (P < 0.05), whilst the lowest in the ZJ sample (P < 0.05). The V-Scores of all the P. sinese silages ranged from 57.9 to 78.3, the highest V-Score was found in the AX sample silage with the lowest in the MIN sample (P < 0.05). All the naturally fermented P. sinese silages in this study had a low V-score, which suggests that the silage quality was unsatisfactory. In addition, the results showed large variability in the quality of fermentation and V-scores across the different P. sinese silage samples.
Bacterial Community of P. sinese Silage
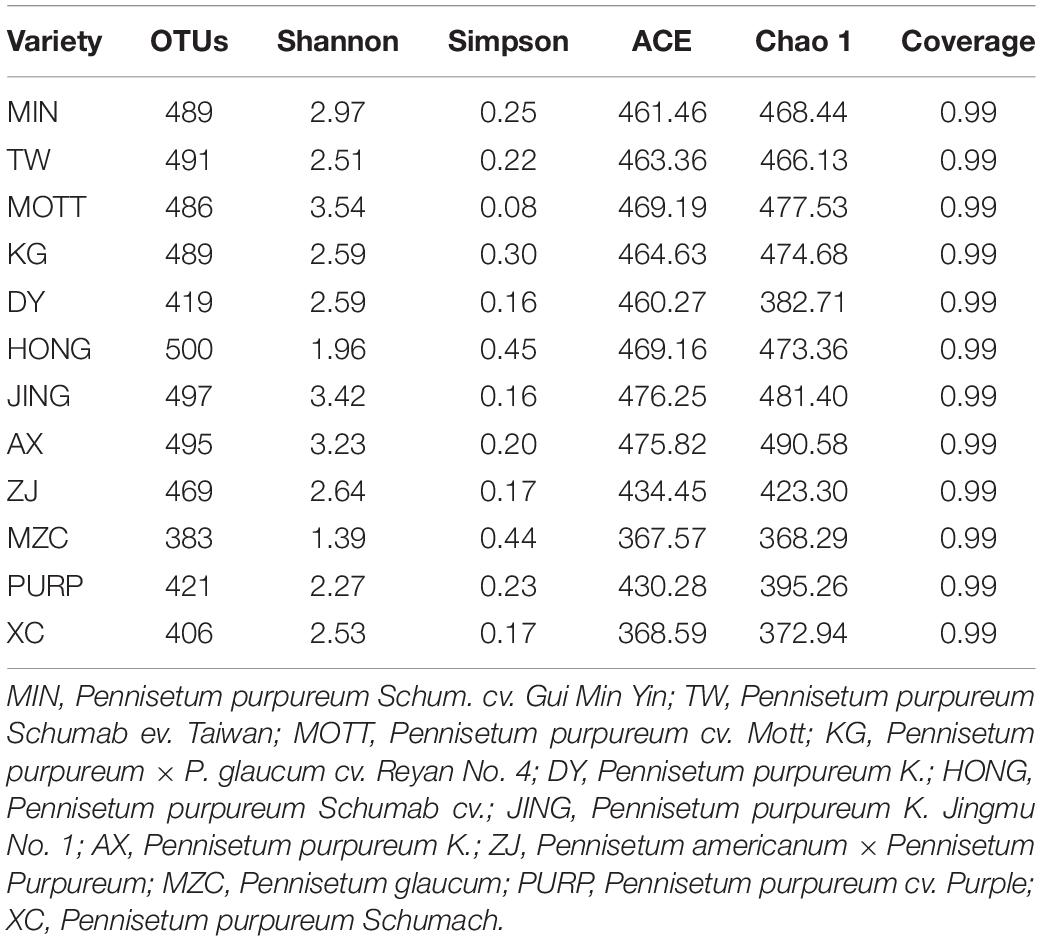
A total of 2,677,149 raw reads were generated, with an average of 71,884 raw tags per P. sinese sample. After data processing, there was an average of 71,423 clean tags and 60,717 effective tags obtained from the silage samples.
Table 3 shows the alpha diversity of the silage bacterial communities. The OTU, Shannon, Simpson, Ace, and Chao 1 indices of microbial diversity were affected by the variety of samples (Table 3). The OTUs, Shannon, Simpson, Ace, and Chao 1 values ranged from 383 to 500, 1.39 to 3.54, 0.08 to 0.45, 367.57 to 476.25, 368.29 to 490.58, respectively. In conclusion, the variety of P. sinese greatly affected the microbiome of the resultant silage. Figure 1A describes the microbial community at the phylum level. Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria were dominant in all P. sinese samples. To further investigate the effects of P. sinese variety on the resultant microbial community following ensiling, the bacterial structures of P. sinese silages at the genus level were examined (Figure 1B). The 10 genera with the highest relative abundance were: Pseudomonas, Massilia, Raoultella, Pedobacter, Sphingobacterium, Acinetobacter, Megamonas, Lactobacillus, Enterococcus, and Stenotrophomonas. The present study shows that Pseudomonas is one of the most abundant genera of bacteria present in the silages produced by differing P. sinese varieties. It is proposed that Pseudomonas contributed to the poor P. sinese silage quality.
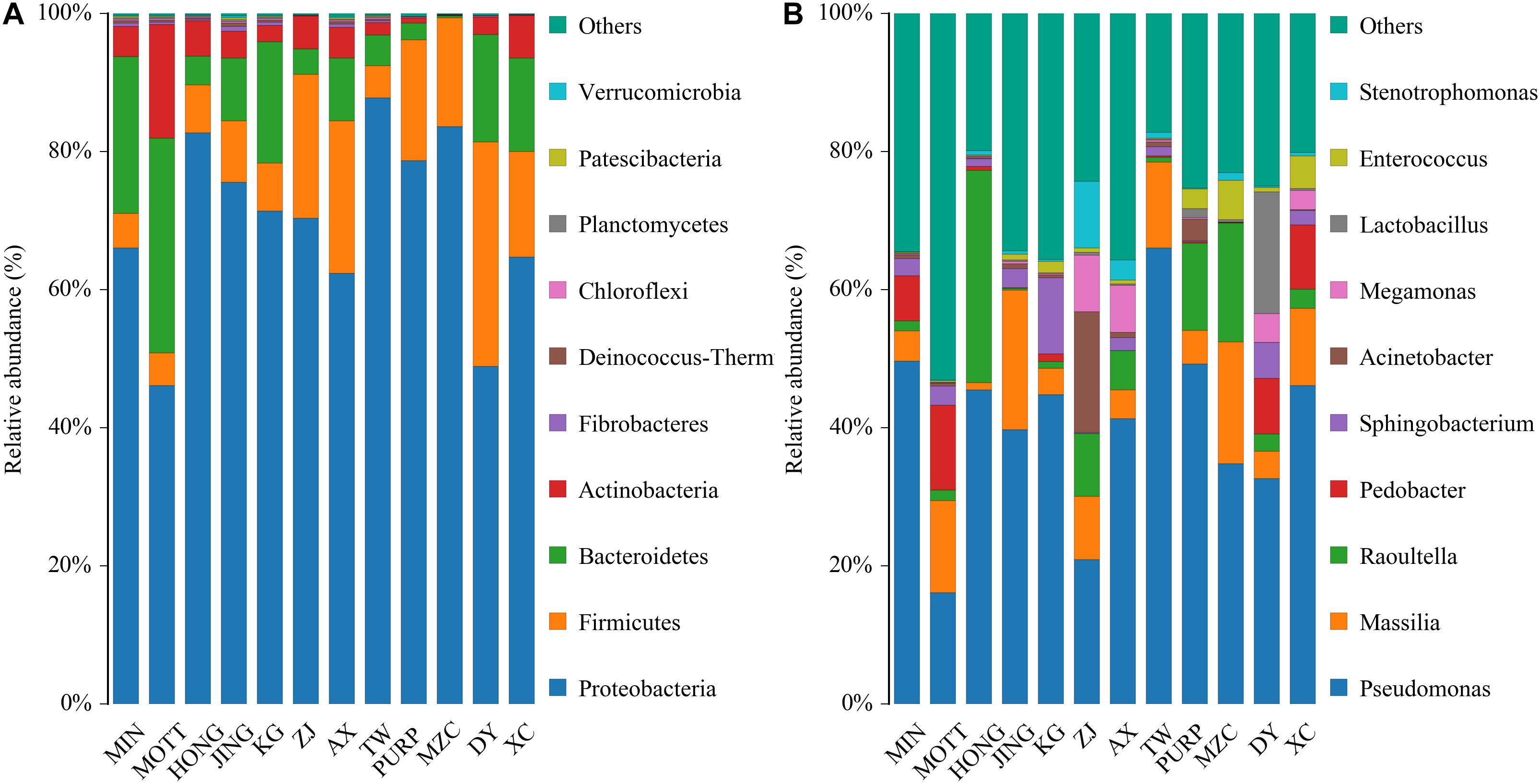
Figure 1. Relative abundance of bacterial composition in P. sinese silages at phylum (A) and genus (B) level.
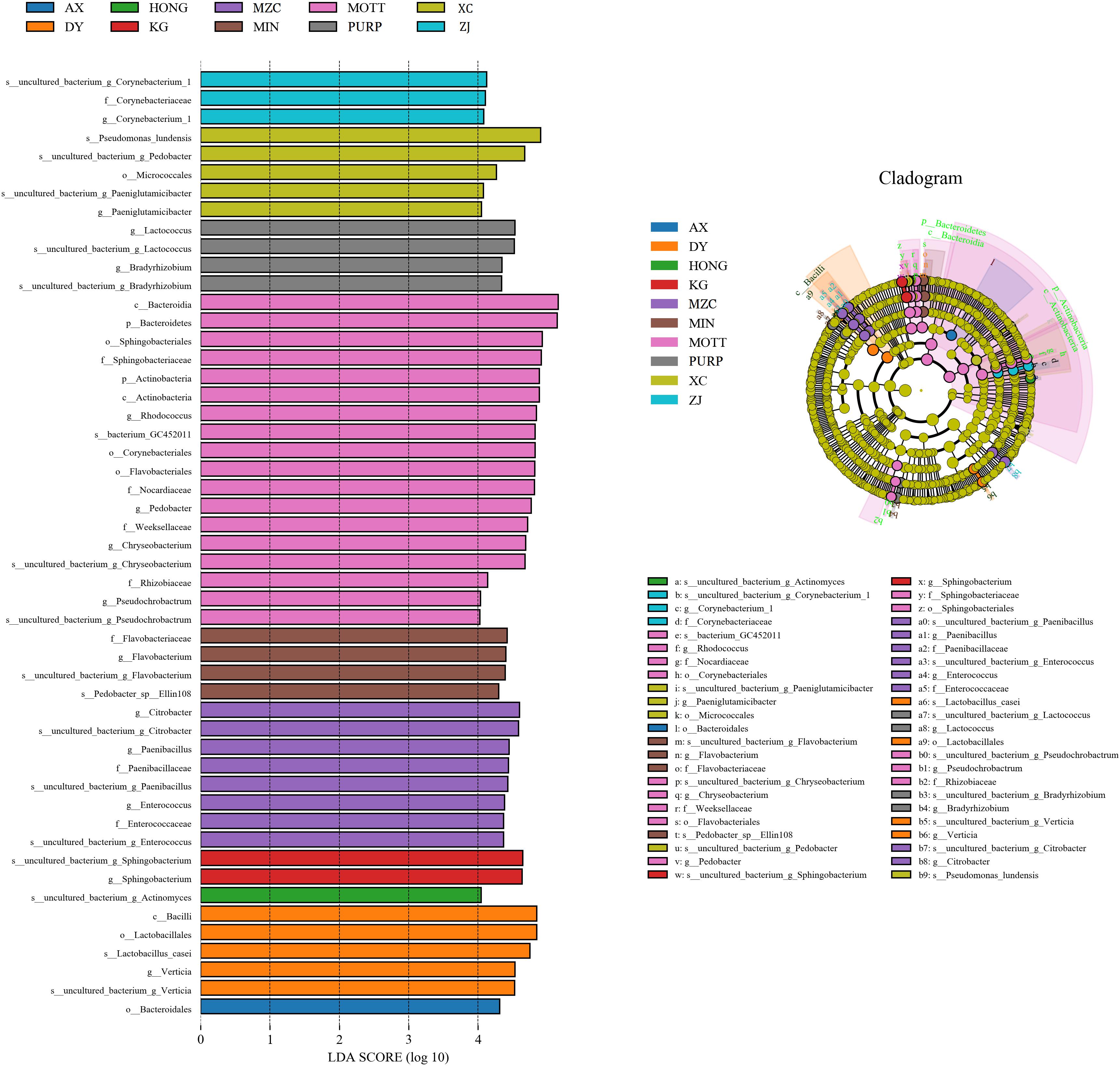
The linear discriminant analysis (LDA) effect size (LEfSe) method was used to assess the differences in the microbiome content between the silages produced by the different P. sinese varieties, as well as to explore the specific bacterial community of each silage variety (LDA score >4.0). Figure 2 shows that the variety of P. sinese used to produce the silage had a dramatic impact on the resultant microbial community. Bacteroidales was the most abundant order in the silage of the AX P. sinese variety, Verticia was the most abundant genus and Lactobacillus casei was the most abundant species in the DY silage. Actinomyces was the most abundant species in the HONG silage, Sphingobacterium was the most abundant genus in the KG silage whilst Enterococcus, Paenibacillus, and Citrobacter were the most abundant genera in the MZC silage. Flavobacterium was the most abundant genus and Pedobacter Ellin108 the most abundant species in the MIN silage. Corynebacteriales and Flavobacteriales were the most abundant orders, Sphingobacteriaceae, Nocardiopsaceae, Weeksellaceae, and Rhizobiaceae the most abundant families and Enterococcus, Pedobacter, Chryseobacterium, and Pseudochrobactrum were the most abundant genera in the MOTT silage. Conversely, Lactococcus, and Bradyrhizobium were the most abundant genera in the PURP silage, whilst in the XC silage Micrococcales was the most abundant order, Paeniglutamicibacter the most abundant genus and Pseudomonas lundensis was the most abundant species. Finally, Corynebacterium_1 was the most abundant genus in ZJ silage. It is proposed that these microorganisms could be used as biomarkers for the silages produced by the different varieties of P. sinese.
Association Analysis Between Bacterial Communities and the Chemical Composition of Raw Pennisetum sinese Before Ensiling
The correlation between chemical composition (DM, OM, CP, NDF, ADF, and WSC) and the microbiome of P. sinese silages was also assessed. At the genus level, a Spearman correlation heat map was created for the microbial communities from the silages (Figure 3). Megamonas, Pseudomonas, Bacteroides, Raoultella, Citrobacter, Stenotrophomonas, and Enterococcus abundance positively correlated with the DM, OM, CP, NDF, ADF, and WSC content (P < 0.05). In addition, Rhodococcus, Chryseobacterium, Pedobacter, and Verticia abundance was negatively correlated with these chemical compositions of raw P. sinese (P < 0.05). This present study showed significant correlation between the species of bacteria and the chemical composition of the raw P. sinese used to produce the silage. The results of this study further compound the significant relationship between the chemical composition of the raw material and the resultant silage.
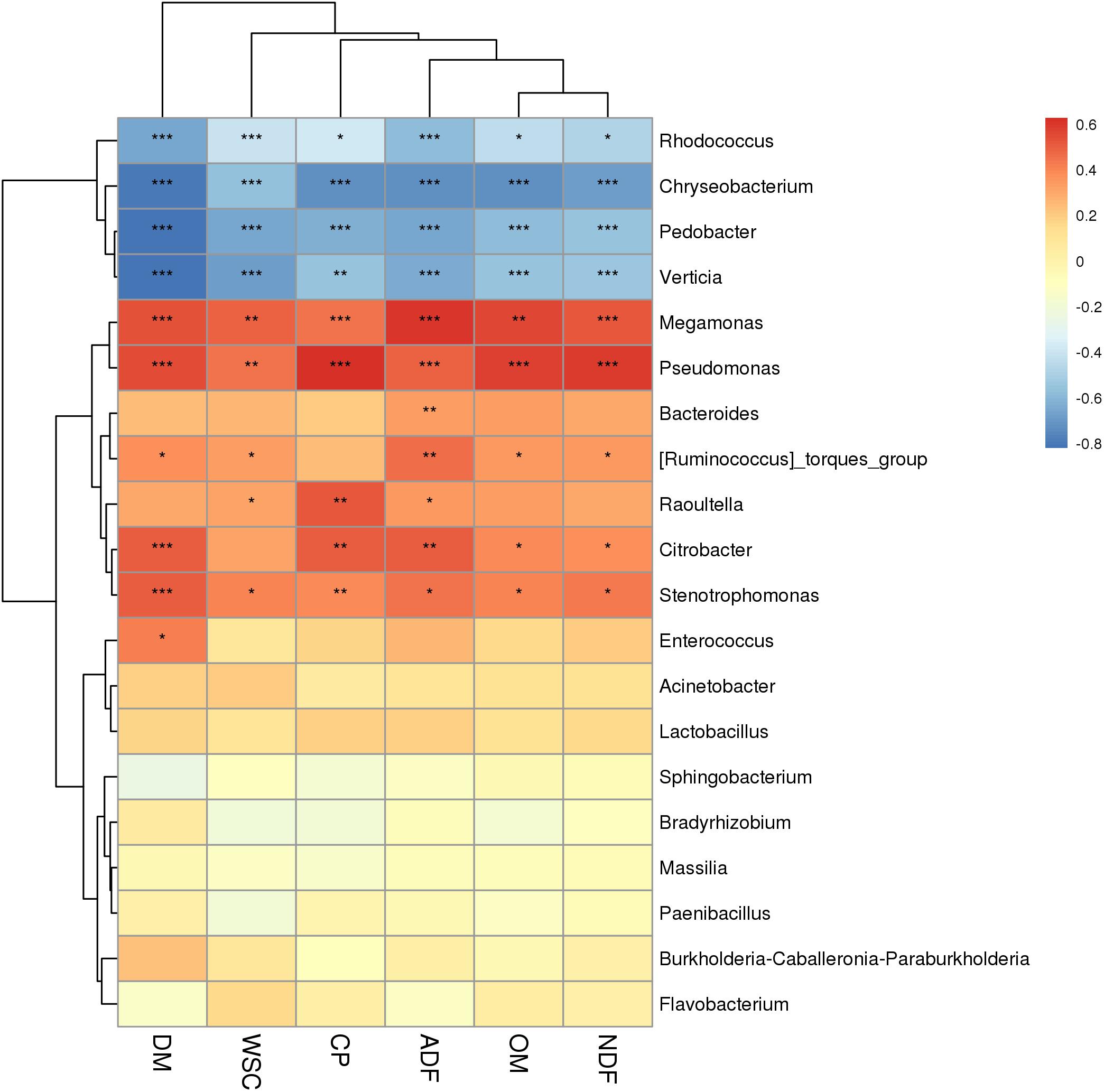
Figure 3. Correlation analysis of the bacterial community with chemical composition of P. sinese silage at genus level. The blue indicates a negative correlation, red indicates a positive correlation. “*,” “**,” and “***” represent P < 0.05, P < 0.01, and P < 0.001, respectively.
Association Analysis Between Bacterial Communities and Fermentation Characteristics
The correlation between fermentation characteristics and the microbiome of P. sinese silages was assessed (Figure 4). Silage pH was positively correlated with the abundance of Bacteroides, Megamonas, and Citrobacter (P < 0.01) in the silage microbiome, while it was negatively correlated with the abundance of Chryseobacterium and Pedobacter (P < 0.01). The silage lactic acid content was positively correlated with the abundance of Bacteroides, Megamonas, and Citrobacter (P < 0.01), while it was negatively correlated with the abundance of Chryseobacterium, Paenibacillus, Sphingobacterium, Flavobacterium, Pedobacter, and Rhodococcus (P < 0.01). The acetic acid content of the silage was positively correlated with the abundance of Citrobacter (P < 0.01), while it was negatively correlated with the abundance of Chryseobacterium, Sphingobacterium, Verticia, Pedobacter, and Rhodococcus (P < 0.01). The propionic acid content of the silage was positively correlated with the abundance of Bacteroides (P < 0.01), while it was negatively correlated with the abundance of Chryseobacterium (P < 0.01). Finally, the butyric acid content of the silage was positively correlated with the abundance of Bacteroides (P < 0.01), while it was negatively correlated with the abundance of Bradyrhizobium and Paenibacillus (P < 0.01).
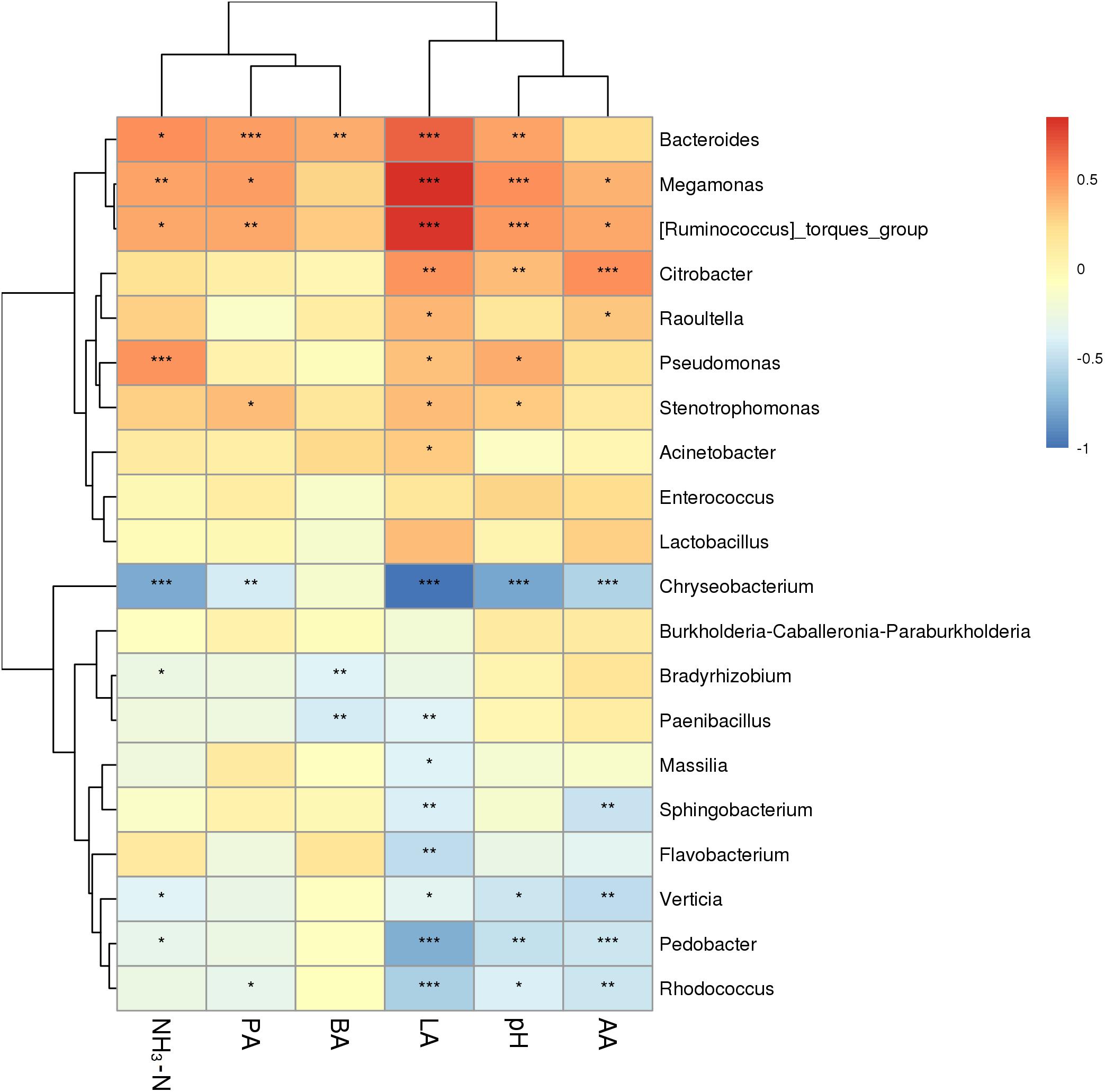
Figure 4. Correlation analysis of P. sinese silage bacterial communities and fermentation characteristics at genus level. The blue indicates a negative correlation, red indicates a positive correlation. “*,” “**,” and “***” represent P < 0.05, P < 0.01, and P < 0.001, respectively.
Association Analysis Between Bacterial Communities and Environmental Factors
The relationship between environmental factors the varying P. sinese samples were exposed to Supplementary Table 1 and the silage they produced was assessed (Figure 5). The level of precipitation the P. sinese sample was exposed to was positively correlated with the abundance of Pseudomonas, Stenotrophomonas, Citrobacter, Raoultella, Lactobacillus, Enterococcus, and Megamonas (P < 0.001), whilst it was negatively correlated with the abundance of Rhodococcus, Verticia, Chryseobacterium, Pedobacter, and Sphingobacterium (P < 0.01). The environmental temperature was positively correlated with the abundance of Pseudomonas and Raoultella (P < 0.05), whilst it was negatively correlated with the abundance of Rhodococcus, Verticia, Chryseobacterium, and Pedobacter (P < 0.01). Meanwhile, environmental humidity was positively correlated with the abundance of Pseudomonas, Stenotrophomonas, and Citrobacter (P < 0.01), whilst it was negatively correlated with the abundance of Rhodococcus, Verticia and Chryseobacterium, Pedobacter (P < 0.01). The longitude and latitude of the base in which the P. sinese sample was grown was positively correlated with the abundance of Pseudomonas, Stenotrophomonas, Citrobacter, and Raoultella (P < 0.05), whilst it was negatively correlated with the abundance of Rhodococcus, Verticia, Chryseobacterium, and Pedobacter (P < 0.05). The altitude at which the P. sinese sample was grown was positively correlated with the abundance of Pseudomonas but no other bacteria, suggesting that altitude played a only a minor role in determining the microbiome content of the silage (P < 0.05).
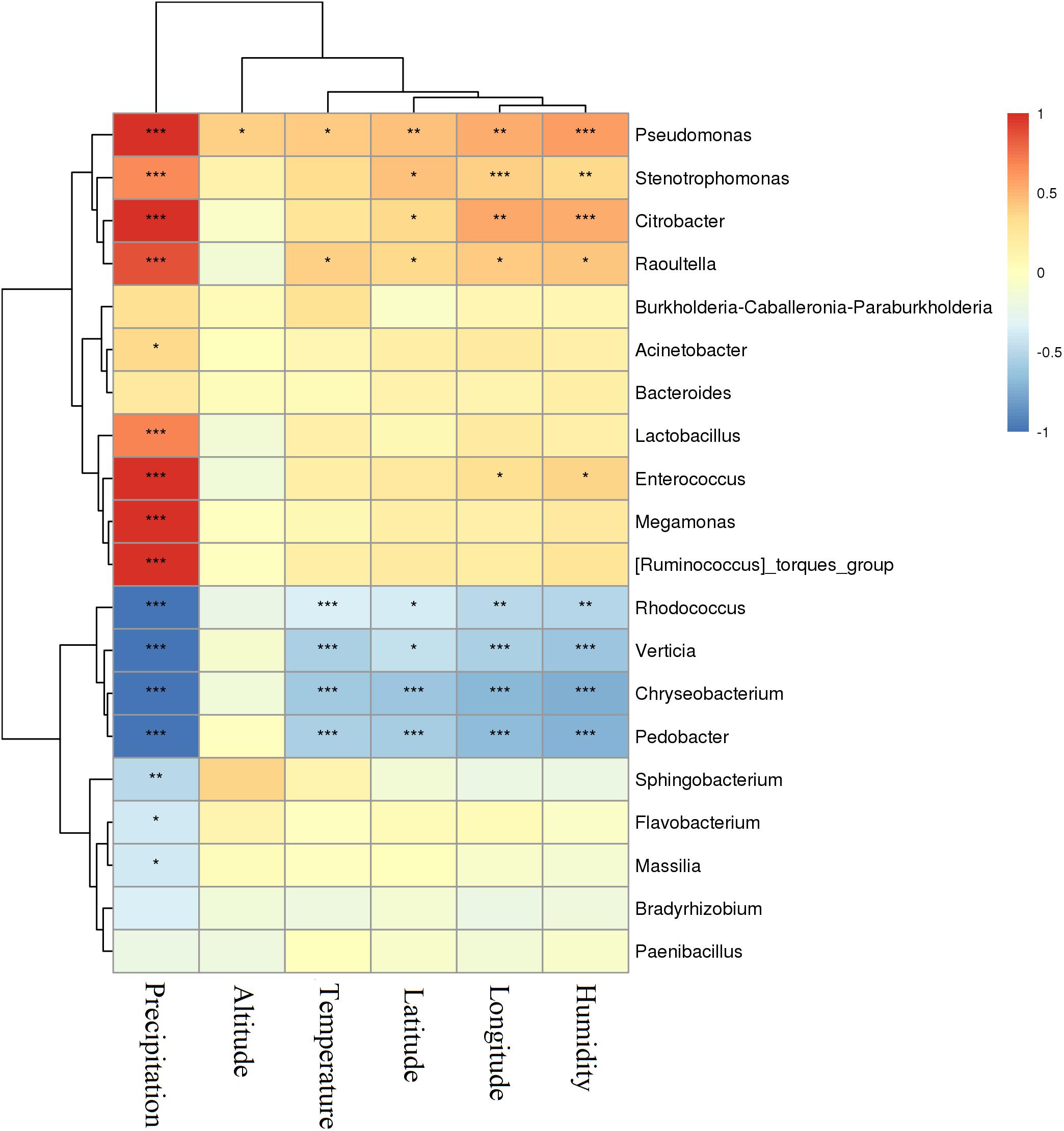
Figure 5. Correlation of P. sinese silage bacterial communities and environmental factors (longitude, latitude, temperature, humidity, and precipitation) at genus level. The blue indicates a negative correlation, red indicates a positive correlation. “*” and “**” represent P < 0.05 and P < 0.01, respectively.
Discussion
Characteristics of P. sinese Before Ensiling
The chemical composition of raw materials has an important influence on the silage quality. In this study, the chemical composition of P. sinese varieties are similar to a previous study in which king grass was examined (Li et al., 2019b). In contrast, both Mugabe et al. (2019) and Li et al. (2020) reported higher CP, NDF, and ADF content in P. sinese varieties compared to the results described. Furthermore, Desta et al. (2016) reported a higher DM, NDF, and ADF content, but lower WSC content, in Napier grass. Shah et al. (2017) also found a higher NDF content, whilst they also found a lower CP and WSC content in king grass compared to the results presented herein. These results imply that there are a large differences in the chemical compositions of different varieties of P. sinese; this could influence the silage quality. In particular, low levels of WSC are thought to contribute to the poor fermentation quality of P. sinese silage without additives (Wang et al., 2019a).
The Fermentation Properties of P. sinese Silage
Silage pH is a very important evaluation parameter to consider when assessing fermentation efficacy; well fermented silage should have a pH of 4.2 or lower (Edwards and McDonald, 1978). The results presented in this study show that all silages had pH of higher than 4.2, suggesting a lower quality of fermentation. Previous studies have reported a range of silage pH’s with some corroborating what was found in this study and other papers showing P. sinese silage with a lower pH (Li et al., 2014, 2019a, 2020; Desta et al., 2016; Mugabe et al., 2019). The pH variability may be due to different P. sinese varieties being used in this study compared to others, or differences in the silage microbiome. This study revealed relatively low lactic and propionic acid content compared to previous studies. Meanwhile, the acetic acid, butyric acid, and ammonia-N content were similar to, or lower than, the levels reported in previous papers looking at silage produced from P. sinese grass (Li et al., 2014, 2019a, 2019b, 2020; Desta et al., 2016; Shah et al., 2017; Mugabe et al., 2019). The differing chemical compositions of the raw P. sinese samples may have contributed to the substantial variation in silage fermentation characteristics; Guan et al. (2018) reported this phenomenon previously, albeit in corn silage. Desta et al. (2016) reported the V-Score of Napier grass silage without additive as 63.1, this falls within the range elucidated in this study. All the naturally fermented P. sinese silages in this study had a low V-score, which suggests that the silage quality was unsatisfactory. The poor fermentation quality may be due to the low levels of WSC. WSC is an important substrate for lactic acid bacteria (LAB); these are the dominant bacteria in the microbiome of high quality silage; they have been shown to reduce silage pH and prevent the dominance of undesirable microorganisms by producing lactic acid (Guan et al., 2018). The relatively low levels of WSC identified in the silage produced by P. sinese varieties in this study may have restricted the growth of LAB, resulting in the production of low quality silage.
However, another important reason for the low silage quality is the higher moisture content of P. sinese (77.5∼88.7%). In normal conditions, the ideal moisture content of silage materials is 65–70%, the higher moisture content is conducive to the growth of undesirable microorganisms, which decreased the silage quality. In the tropics of southern China, when wilting grass, there are often showers. In order to avoid rain damage during the silage making, in the present study, we try to prepare P. sinese silage with out wilted for investigating their silage quality, microbial community and the correlation with the environmental factor. One caveat with this study, the lower silage quality reminded us ensiled directly was not the suitable way for P. sinese preparation.
Bacterial Community of P. sinese Silage
In the present study, Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria were dominant in all P. sinese samples. These findings are consistent with previous reports examining P. sinese, corn stover and red clover silage (Xu et al., 2017; Dong et al., 2019; Wu et al., 2020) and have been corroborated by previous studies that reported Firmicutes and Proteobacteria as the most abundant phyla in silages, with their abundance increasing to more than 90% after fermentation (Li L. et al., 2017; Liu et al., 2019; Wang et al., 2019b). Normally, Lactobacillus is the major bacterial and dominant genus in well preserved silage, with Lactobacillus performing desirable functions during fermentation (Li L. et al., 2017; Liu et al., 2019; Wang et al., 2019b). However, in this study, Pseudomonas, Massilia, and Raoultella were the predominant genera, which is consistent with previous research reporting that these bacteria were associated with the unsatisfactory fermentation of silage (Li L. et al., 2017; Li et al., 2019a; Wu et al., 2020). Pseudomonas is considered especially undesirable due to the production of biogenic amines that reduce the protein content and nutritional value of silage (Silla Santos, 1996; Dunière et al., 2013). The present study shows that Pseudomonas is one of the most abundant genera of bacteria present in the silages produced by differing P. sinese varieties. It is proposed that Pseudomonas contributed to the poor P. sinese silage quality, as has been previously reported in studies examining forage silage (Xu et al., 2017; Dong et al., 2019; Wang et al., 2019a). However, Ogunade et al. (2018) found that Pseudomonas were negatively correlated with pH, ammonium nitrogen, yeast, and mold, suggesting that it may be beneficial to silage fermentation. Therefore, the underlying mechanisms of Pseudomonas in silage fermentation processing need to be further investigated.
Correlations Between Bacterial Communities and the Chemical Composition
This study found that there was a significant correlation between the chemical composition of silage raw materials and silage microorganisms. The chemical composition of different varieties of P. sinese was quite different, so the characteristics of silage microbial community were also different. This may be explained by the fact that these microorganisms are chemoorganotrophic bacteria that produce energy through the oxidation of organic matter such as starch and organic acids. Pseudomonas, Bacteroides, and Stenotrophomonas all consume protein, whilst Megamonas, Raoultella, Citrobacter, and Enterococcus ferment carbohydrates (Skerman et al., 1980; Schleifer and Kilpper-Bälz, 1984; Drancourt et al., 2001; Tian et al., 2019) and Bacteroides and Stenotrophomonas can utilize either (Palleroni and Bradbury, 1993; Silla Santos, 1996; Xu et al., 2003; Dunière et al., 2013; Li M. et al., 2017). Guan et al. (2018) reported that the WSC content of raw corn correlated with the abundance of Lactobacilli and Acetobacter in the resultant corn silage. Corroboratively, Yang et al. (2019) found the same correlation, however, there was no significant correlation between DM and microorganism abundance in alfalfa silage. The present study showed significant correlation between the species of bacteria and the chemical composition of the raw P. sinese used to produce the silage. The results of this study further compound the significant relationship between the chemical composition of the raw material and the resultant silage. Discrepancies between this study and those mentioned above are most likely due to the differences in the chemical composition of the 12 varieties of P. sinese and the forage material they used.
Correlations Between Bacterial Communities and Fermentation Characteristics
Silage microorganisms and metabolites are the key factors affecting silage fermentation quality. Megamonas, Bacteroides, and Citrobacter all have the ability to ferment multiple carbohydrates and produce an array of organic acids, including lactic, acetic, and propionic acid (Skerman et al., 1980; Xu et al., 2003; Li M. et al., 2017; Tian et al., 2019). Therefore, it is unsurprising that there was a significant positive correlation between the organic acid content and the microbiome of the silage. Pseudomonas is an undesirable bacteria in silage due to its production of biogenic amines leading to a reduced protein content (Silla Santos, 1996; Dunière et al., 2013). Furthermore, higher levels of ammonium nitrate were associated with increased abundance of Pseudomonas. Silage fermentation is a complex biological process which involves a large variety of microorganisms; as such, the process produces many different metabolites which can determine the fermentation quality. Variety in the microbiome of the silage alters the metabolites produced during ensiling and may contribute to the fermentation quality. In the present study, the silage produced from the different varieties of P. sinese lacked an abundance of Lactobacillus; this may explain why the fermentation quality of the silage was poor. Other studies have corroborated these findings, by showing that fermentation characteristics are highly correlated with the microbiome of the silage, influencing the overall fermentation quality (Guan et al., 2018; Ogunade et al., 2018; Ren et al., 2019; Yang et al., 2019). Therefore, reducing the content of non-lactic acid bacteria is an effective measure to improve the fermentation quality of silage.
Correlations Between Bacterial Communities and Environmental Factors
The results of this study reveal that the microbiome of silage is affected by internal factors (chemical composition of the raw silage material) and external factors (temperature, precipitation levels). This has previously been discussed in other studies, with fermentation metabolites, precipitation, temperature, humidity, longitude, latitude and altitude all being reported to effect the microbiome of the silage and influence the fermentation quality (Muck, 2013; Bernardes et al., 2018; Guan et al., 2018). The results of this study were consistent with previous reports assessing the effect of rainfall and temperature on silage quality (Kim and Adesogan, 2006; Coblentz and Muck, 2012). Guo et al. (2014, 2015) highlighted the importance of sunlight on the fermentation of Napier grass and Italian Rye grass; the results of this study also show that sunlight affects the quality of P. sinese silage due to its influence on the silage microbiome. In addition, the storage temperatures of the silage influenced the bacterial diversity/fermentation quality, which had been previously reported in the silage of Moringa oleifera leaves (Wang et al., 2019a). Moreover, Guan et al. (2018) reported that environmental factors (temperature, humidity, and precipitation) affected the fermentation quality of silage mediated by changes in the microbiome. These results suggest that high temperature, rain, and high humidity in the south of China have a great negative influence on the microbiome of the silage from P. sinese. The reason that environmental factors affect the microbial community of silage may be that environmental differences affect the community structure of epiphytic bacteria on raw materials, which have a greater impact on the initial stage of silage fermentation. With the process of silage fermentation, the impact of environmental factors on the microbial community will be reduced, thus lightened its impact on silage quality. Interestingly, this study identified that longitude and latitude had significant effects on the bacterial diversity of the silage, leading to differences in the fermentation quality of the silage. This suggests that for silage preparation in different regions, geographical factors and their effect on silage microbiome must be considered in order to make provisions to ensure high quality production.
Conclusion
The study presented herein examined the microbiome and fermentation quality of silages from the natural fermentation of 12 varieties of P. sinese grown across southern China. The silage quality of P. sinese was generally unsatisfactory with low V-Scores. There was a predominance of undesirable genera including Pseudomonas, Massilia, and Raoultella in silage produced from P. sinese; this led to poor fermentation quality. The strong correlation between the chemical composition of the silage material and the fermentation characteristics and bacterial community of the resultant silage. In addition, precipitation, temperature, humidity, and location significantly influenced the bacterial community of the silage. The specific P. sinese cultivar used for silage production and the environmental factors it has been subject to must be considered in order to ensure a high-quality end product.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://catalog.data.gov/dataset/sequence-read-archive-sra, PRJNA624770.
Author Contributions
ML, XZ, and DY did the experimental design work. ML, XZ, DY, and JT conducted the experiments. ML, XZ, DY, JT, HZ, and YC analyzed the data. ML and XZ wrote the manuscript. All authors read and approved the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (No. 31860680), the Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (Nos. 1630032021003 and 1630032017033), the Ministry of Agriculture and Rural Affairs of the People’s Republic of China (No. 16200159), and Integrated Demonstration of Key Techniques for the Industrial Development of Featured Crops in Rocky Desertification Areas of Yunnan-Guangxi-Guizhou Provinces.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.627820/full#supplementary-material
Supplementary Table 1 | Environmental factors effecting the 12 varieties of P. sinese. MIN, Pennisetum purpureum Schum. cv. Gui Min Yin; TW, Pennisetum purpureum Schumab ev. Taiwan; MOTT, Pennisetum purpureum cv. Mott; KG, Pennisetum purpureum × P. glaucum cv. Reyan No. 4; DY, Pennisetum purpureum K.; HONG, Pennisetum purpureum Schumab cv.; JING, Pennisetum purpureum K. Jingmu No. 1; AX, Pennisetum purpureum K.; ZJ, Pennisetum americanum × Pennisetum purpureum; MZC, Pennisetum glaucum; PURP, Pennisetum purpureum cv. Purple; XC, Pennisetum purpureum Schumach.
Footnotes
References
AOAC (1990). Official Methods of Analysis, 15th Edn, ed. W. Horowitz (Arlington, VA: Association of Official Analytical Chemists).
Bernardes, T. F., Daniel, J. L. P., Adesogan, A. T., McAllister, T. A., Drouin, P., Nussio, L. G., et al. (2018). Silage review: unique challenges of silages made in hot and cold regions. J. Dairy Sci. 101, 4001–4019. doi: 10.3168/jds.2017-13703
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Coblentz, W. K., and Muck, R. E. (2012). Effects of natural and simulated rainfall on indicators of ensilability and nutritive value for wilting alfalfa forages sampled before preservation as silage. J. Dairy Sci. 95, 6635–6653. doi: 10.3168/jds.2012-5672
Desta, S. T., Yuan, X., Li, J., and Shao, T. (2016). Ensiling characteristics, structural and nonstructural carbohydrate composition and enzymatic digestibility of Napier grass ensiled with additives. Bioresour. Technol. 221, 447–454. doi: 10.1016/j.biortech.2016.09.068
Dong, Z., Li, J., Chen, L., Wang, S., and Shao, T. (2019). Effects of freeze–thaw event on microbial community dynamics during red clover ensiling. Front. Microbiol. 10:1559. doi: 10.3389/fmicb.2019.01559
Dong, Z., Shao, T., Li, J., Yang, L., and Yuan, X. (2020). Effect of alfalfa microbiota on fermentation quality and bacterial community succession in fresh or sterile Napier grass silages. J. Dairy Sci. 103, 4288–4301. doi: 10.3168/jds.2019-16961
Drancourt, M., Bollet, C., Carta, A., and Rousselier, P. (2001). Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int. J. Syst. Evol. Microbiol. 51, 925–932. doi: 10.1099/00207713-51-3-925
Dunière, L., Sindou, J., Chaucheyras-Durand, F., Chevallier, I., and Thévenot-Sergentet, D. (2013). Silage processing and strategies to prevent persistence of undesirable microorganisms. Anim. Feed Sci. Technol. 182, 1–15. doi: 10.1016/j.anifeedsci.2013.04.006
Edwards, R. A., and McDonald, P. (1978). “The chemistry of silage,” in Fermentation of Silage-a Review, ed. M. E. McCullough (West Des Moines, IA: NFIA), 27–60.
Guan, H., Yan, Y., Li, X., Li, X., Shuai, Y., Feng, G., et al. (2018). Microbial communities and natural fermentation of corn silages prepared with farm bunker-silo in Southwest China. Bioresour. Technol. 265, 282–290. doi: 10.1016/j.biortech.2018.06.018
Guo, G., Yu, C., Wang, Q., Xin, P., Shimojo, M., and Shao, T. (2014). Silage fermentation characteristics of Italian Ryegrass (Lolium multiflorum Lam.) harvested at various times on a sunny day. Crop Sci. 54, 851–858. doi: 10.2135/cropsci2012.12.0692
Guo, G., Yuan, X., Wen, A., Liu, Q., Zhang, S., and Shao, T. (2015). Silage fermentation characteristics of napiergrass harvested at various times on a sunny day. Crop Sci. 55, 458–464. doi: 10.2135/cropsci2014.03.0172
Kim, S. C., and Adesogan, A. T. (2006). Influence of ensiling temperature, simulated rainfall, and delayed sealing on fermentation characteristics and aerobic stability of corn silage. J. Dairy Sci. 89, 3122–3132. doi: 10.3168/jds.s0022-0302(06)72586-3
Li, D., Ni, K., Zhang, Y., Lin, Y., and Yang, F. (2019a). Fermentation characteristics, chemical composition and microbial community of tropical forage silage under different temperatures. Asian Australas J. Anim. Sci. 32, 665–674. doi: 10.5713/ajas.18.0085
Li, F., Ke, W., Ding, Z., Bai, J., Zhang, Y., Xu, D., et al. (2020). Pretreatment of Pennisetum sinese silages with ferulic acid esterase-producing lactic acid bacteria and cellulase at two dry matter contents: fermentation characteristics, carbohydrates composition and enzymatic saccharification. Bioresour. Technol. 295:122261. doi: 10.1016/j.biortech.2019.122261
Li, J., Yuan, X., Desta, S. T., Dong, Z., Mugabe, W., and Shao, T. (2018a). Characterization of Enterococcus faecalis JF85 and Enterococcus faecium Y83 isolated from Tibetan yak (Bos grunniens) for ensiling Pennisetum sinese. Bioresour. Technol. 257, 76–83. doi: 10.1016/j.biortech.2018.02.070
Li, J., Yuan, X., Dong, Z., Mugabe, W., and Shao, T. (2018b). The effffects of fifibrolytic enzymes, cellulolytic fungi and bacteria on the fermentation characteristics, structural carbohydrates degradation, and enzymatic conversion yields of Pennisetum sinese silage. Bioresour. Technol. 264, 123–130. doi: 10.1016/j.biortech.2018.05.059
Li, L., Yuan, Z., Sun, Y., Kong, X., Dong, P., and Zhang, J. (2017). A reused method for molasses-processed wastewater: effect on silage quality and anaerobic digestion performance of Pennisetum purpereum. Bioresour. Technol. 241, 1003–1011. doi: 10.1016/j.biortech.2017.04.117
Li, M., Zhou, H., Pan, X., Xu, T., Zhang, Z., Zi, X., et al. (2017). Cassava foliage affects the microbial diversity of Chinese indigenous geese caecum using 16S rRNA sequencing. Sci. Rep. 7:45697.
Li, M., Zi, X., Tang, J., Zhou, H., and Cai, Y. (2019b). Silage fermentation, chemical composition and ruminal degradation of king grass, cassava foliage and their mixture. Grassland Sci. 64, 210–215. doi: 10.1111/grs.12235
Li, M., Zi, X., Zhou, H., Hou, G., and Cai, Y. (2014). Effects of sucrose, glucose, molasses and cellulase on fermentation quality and in vitro gas production of king grass silage. Anim. Feed Sci. Technol. 197, 206–212. doi: 10.1016/j.anifeedsci.2014.06.016
Liu, B., Huan, H., Gu, H., Xu, N., Shen, Q., and Ding, C. (2019). Dynamics of a microbial community during ensiling and upon aerobic exposure inlactic acid bacteria inoculation-treated and untreated barley silages. Bioresour. Technol. 273, 212–219. doi: 10.1016/j.biortech.2018.10.041
Lu, Q. L., Tang, L. R., Wang, S., Huang, B., Chen, Y. D., and Chen, X. R. (2014). An investigation on the characteristics of cellulose nanocrystals from Pennisetum sinese. Biomass Bioenerg. 70, 267–272. doi: 10.1016/j.biombioe.2014.09.012
Masaki, S. (2001). Silage-No-Hinshituhantei. Tokyo: Japan Grassland Agriculture and Forage Seed Association.
Muck, R. (2013). Recent advances in silage microbiology. Agric. Food Sci. 22, 3–15. doi: 10.23986/afsci.6718
Mugabe, W., Yuan, X., Li, J., Dong, Z., and Shao, T. (2019). Effects of hexanoic acid, Lactobacillus plantarum and their combination on the fermentation characteristics of Napier grass. Anim. Feed Sci. Technol. 253, 135–140. doi: 10.1016/j.anifeedsci.2019.05.005
Murphy, R. P. (1958). A method for the extraction of plant samples and the determination of total soluble carbohydrates. J. Sci. Food Agric. 9, 714–717. doi: 10.1002/jsfa.2740091104
Ogunade, I. M., Jiang, Y., Pech Cervantes, A. A., Kim, D. H., Oliveira, A. S., Vyas, D., et al. (2018). Bacterial diversity and composition of alfalfa silage as analyzed by Illumina MiSeq sequencing: effects of, Escherichia coli, O157:H7 and silage additives. J. Dairy Sci. 101, 2048–2059. doi: 10.3168/jds.2017-12876
Palleroni, N. J., and Bradbury, J. F. (1993). Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int. J. Syst. Bacteriol. 43, 606–609. doi: 10.1099/00207713-43-3-606
Ren, F., He, R., Zhou, X., Gu, Q., Xia, Z., Liang, M., et al. (2019). Dynamic changes in fermentation profiles and bacterial community composition during sugarcane top silage fermentation: a preliminary study. Bioresour. Technol. 285:121315. doi: 10.1016/j.biortech.2019.121315
Schleifer, K. H., and Kilpper-Bälz, R. (1984). Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int. J. Syst. Bacteriol. 34, 31–34. doi: 10.1099/00207713-34-1-31
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/aem.01541-09
Segata, N., Abubucker, S., Goll, J., and Schubert, A. M. (2011). Metagenomic biomarker discovery and explanation. Genom. Biol. 12:R60.
Shah, A. A., Wu, J., Qian, C., Liu, Z., Mobashar, M., Tao, Z., et al. (2020). Ensiling of whole-plant hybrid pennisetum with natamycin and Lactobacillus plantarum impacts on fermentation characteristics and meta-genomic microbial community at low temperature. J. Sci. Food Agric. 100, 3378–3385. doi: 10.1002/jsfa.10371
Shah, A. A., Xianjun, Y., Zhihao, D., Junfeng, L., and Shao, T. (2018). Isolation and molecular identification of lactic acid bacteria from King grass and their application to improve the fermentation quality of sweet Sorghum. World J. Microbiol. Biotechnol. 34:4.
Shah, A. A., Xianjun, Y., Zhihao, D., Siran, W., and Tao, S. (2017). Effects of lactic acid bacteria on ensiling characteristics, chemical composition and aerobic stability of King grass. J. Anim. Plant Sci. 27, 747–755.
Silla Santos, M. H. (1996). Biogenic amines: their importance in foods. Int. J. Food Microbiol. 29, 213–231. doi: 10.1016/0168-1605(95)00032-1
Skerman, V. B. D., McGowan, V., and Sneath, P. H. A. (1980). Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30, 225–420.
Tian, D., Xu, X., Peng, Q., Wen, Z., Zhang, Y., Wei, C., et al. (2019). In vitro fermentation of arabinoxylan from oat (Avena sativa L.) by Pekin duck intestinal microbiota. 3 Biotech 9:54.
Van Soest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and non-starch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.s0022-0302(91)78551-2
Vázquez-Baeza, Y., Pirrung, M., Gonzalez, A., and Knight, R. (2013). EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience 2:16.
Wang, J., Xin, D., Hou, X., Wu, J., Fan, X., Li, K., et al. (2016). Structural properties and hydrolysabilities of Chinese Pennisetum and Hybrid Pennisetum: effect of aqueous ammonia pretreatment. Bioresour. Technol. 199, 211–219. doi: 10.1016/j.biortech.2015.08.046
Wang, Y., Chen, X., Wang, C., He, L., Zhou, W., Yang, F., et al. (2019a). The bacterial community and fermentation quality of mulberry (Morus alba) leaf silage with or without Lactobacillus casei and sucrose. Bioresour. Technol. 293:122059. doi: 10.1016/j.biortech.2019.122059
Wang, Y., He, L., Xing, Y., Zhou, W., Pian, R., Yang, F., et al. (2019b). Bacterial diversity and fermentation quality of Moringa oleifera leaves silage prepared with lactic acid bacteria inoculants and stored at different temperatures. Bioresour. Technol. 284, 349–358. doi: 10.1016/j.biortech.2019.03.139
Wilkinson, J. M., Bolsen, K. K., and Lin, C. J. (2003). “History of silage,” in Silage Science and Technology, Vol. 42, eds D. R. Buxton, R. E. Muck, and J. H. Harrison (Madison, WI: American Society Agronomy, Crop Science Soceity of America Soil Science Society of America), 1–30. doi: 10.2134/agronmonogr42.c1
Wu, P., Li, L., Jiang, J., Sun, Y., Yuan, Z., Feng, X., et al. (2020). Effects of fermentative and non-fermentative additives on silage quality and anaerobic digestion performance of Pennisetum purpureum. Bioresour. Technol. 297:122425. doi: 10.1016/j.biortech.2019.122425
Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., et al. (2003). A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076. doi: 10.1126/science.1080029
Xu, Z., He, H., Zhang, S., and Kong, J. (2017). Effects of inoculants Lactobacillus brevis and Lactobacillus parafarraginis on the fermentation characteristics and microbial communities of corn stover silage. Sci. Rep. 7:13614.
Yang, L., Yuan, X., Li, J., Dong, Z., and Shao, T. (2019). Dynamics of microbial community and fermentation quality during ensiling of sterile and nonsterile alfalfa with or without Lactobacillus plantarum inoculant. Bioresour. Technol. 275, 280–287. doi: 10.1016/j.biortech.2018.12.067
Keywords: Pennisetum sinese, varieties, silage, fermentation quality, bacterial community, environmental factor
Citation: Zi X, Li M, Yu D, Tang J, Zhou H and Chen Y (2021) Natural Fermentation Quality and Bacterial Community of 12 Pennisetum sinese Varieties in Southern China. Front. Microbiol. 12:627820. doi: 10.3389/fmicb.2021.627820
Received: 10 November 2020; Accepted: 06 April 2021;
Published: 29 April 2021.
Edited by:
Fuyu Yang, China Agricultural University, ChinaCopyright © 2021 Zi, Li, Yu, Tang, Zhou and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mao Li, bGltYW9obkAxNjMuY29t; Yeyuan Chen, Y2hlbnl5MTk2MkAxMjYuY29t
†These authors have contributed equally to this work
 Xuejuan Zi1†
Xuejuan Zi1† Mao Li
Mao Li Hanlin Zhou
Hanlin Zhou