- 1College of Advanced Agriculture and Ecological Environment, Heilongjiang University, Harbin, China
- 2College of Life Science, Northeast Forestry University, Harbin, China
- 3Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
A novel genus Parametarhizium with two new entomopathogenic species, Parametarhizium changbaiense and Parametarhizium hingganense, was introduced based on their morphological characteristics and a multigene phylogenetic analysis, which were isolated from the forest litters collected in Northeast China. To infer their phylogenetic relationships, a six-gene dataset consisting of DNA fragments of [nuclear small subunit rDNA (SSU) + LSU + TUB + TEF + RPB1 + RPB2] was used for phylogenetic analysis, including 105 related fungi. The new genus Parametarhizium formed a monophyletic clade basal to Metarhizium and its related genera (formerly Metarhizium sensu lato). Parametarhizium can be morphologically distinguished from related genera by the combination of the following characteristics: formation of white to yellow colonies on different media, candelabrum-like arrangement of cylindrical or obpyriform phialides, and small subglobose to ellipsoidal conidia. Both P. hingganense and P. changbaiense exhibited anti-insect activities against three farmland pests Monolepta hieroglyphica, Callosobruchus chinensis, and Rhopalosiphum maidis. This is the first report of entomopathogenic fungi exhibiting the anti-insect activity against Mo. hieroglyphica.
Introduction
Fungi are diverse, with over 144,000 species (Willis, 2018), and about 2,000 new species are described every year (Hawksworth and Lücking, 2017). Fungi are a valuable natural resource and can be applied in agriculture and industry, such as biocontrol agents and antibiotic production (Aly et al., 2011; Filizola et al., 2019; Hyde et al., 2019). They can be used to control farmland pests as a promising green alternative for chemical pesticides (Tulloch, 1976; Zimmermann, 2007; Brunner-Mendoza et al., 2018). However, only a few fungal genera are commercially developed as biocontrol agents such as Metarhizium, Beauveria, and Trichoderma (Zimmermann, 1993). Besides the species from the genera Metarhizium, Beauveria, and Trichoderma, the anti-insect activities of other fungi also deserve to be investigated extensively, especially newly discovered fungal species.
The family Clavicipitaceae (Ascomycota, Hypocreales) is a large group of fungi with 48 genera (including six new genera erected in 2020) and over 500 species1 (Mongkolsamrit et al., 2020; Wijayawardene et al., 2020). Clavicipitaceae is morphologically characterized by cylindrical asci, thickened ascus apices, and filiform and multiseptate ascospores that tend to often disarticulate at maturity (Rogerson, 1970; Sung et al., 2007a). Fungi of Clavicipitaceae can grow in plants or invertebrates as symbionts or parasites, such as Epichloë spp. (symbionts of grasses), ergot fungi (Claviceps spp.) that parasitize ears of cereals, and Metarhizium spp. (parasites of insects) (Sung et al., 2007b; Schardl et al., 2013; Mongkolsamrit et al., 2020). Metarhizium is a ubiquitous genus of entomopathogenic fungi, being first discovered and erected in 1879 (Metchnikoff, 1879). Over the next 100 years, various new Metarhizium species were described based on their morphology characterized by dark spores and branched conidiophore with whorls of two to four phialides. Some Metarhizium species had been developed and become famous commercial biocontrol agents, such as Metarhizium anisopliae, Metarhizium brunneum, and Metarhizium robertsii (Meyling and Eilenberg, 2007; Zimmermann, 2007; Castrillo et al., 2011). With the increase in the fungal strain discovery of Metarhizium and its related genera, it is difficult to identify cryptic species by morphology alone (Bischoff et al., 2009; Luangsa-ard et al., 2017; Mongkolsamrit et al., 2020). Subsequently, multigene phylogenetic studies resulted in the delimitation of various lineages of Metarhizium into new species and genera (Bischoff et al., 2006, 2009; Kepler et al., 2014). Until 2014, according to the combination of phylogenetic and morphological studies, Metarhizium was expanded to over 30 species, which included members of the Metarhizium flavoviride and M. anisopliae complex as well as five additional Metarhizium spp. forming a more basal lineage within Metarhizium (Kepler et al., 2014). Since then, a dozen new species of Metarhizium were discovered including Metarhizium baoshanense (Chen et al., 2018), Metarhizium argentinense (Gutierrez et al., 2019), Metarhizium purpureogenum (Nishi et al., 2017), Metarhizium bibionidarum (Nishi et al., 2017), Metarhizium chaiyaphumense (Luangsa-ard et al., 2017), Metarhizium kalasinense (Luangsa-ard et al., 2017), Metarhizium lepidopterorum (Chen et al., 2019), Metarhizium rongjiangense (Chen et al., 2019), and Metarhizium dendrolimatilis (Chen et al., 2017). Recently, a more extensive multigene phylogenetic study of the Clavicipitaceae enabled the resolution of some of the basal lineages of Metarhizium (Mongkolsamrit et al., 2020). As a consequence, Metarhizium was redefined, leading to the erection of six new related genera, which are well-supported monophyletic groups based on the multigene phylogenetic analysis. Morphologically, the species of Metarhizium and related genera can be distinguished by the arrangement and shape of the phialides; the color, shape, and size of the conidia; and other characteristics (Mongkolsamrit et al., 2020). Although Metarhizium is one of the best-studied entomopathogenic fungi with some species being marketed as biocontrol agents, most of the newly described species related to Metarhizium in the last decade were rarely screened for their anti-insect activities.
Entomopathogenic fungi are mainly isolated from their hosts directly, which, however, are difficult to find in the field. Therefore, we started looking for alternative sources to find fungi with anti-insect activities. Forest litters naturally include fallen leaves, dead insects, feces, and diverse microorganisms such as fungi. Our previous studies showed that forest litter is a very promising substrate for the discovery of new fungal species in general as exemplified by the recently described Myxotrichum albicans (Liang et al., 2019). In this study, we identified and described a new genus related to Metarhizium represented by two new candelabrum-like species based on morphology and a multilocus phylogenetic reconstruction. In addition, their potential as biocontrol agents was evaluated by applying a spore-based anti-insect assay.
Materials and Methods
Sample Collection
Forest litters were collected in the Changbai Mountains, Jilin province, China (42°24’N, 128°06’E), in October 2017 and Greater Hinggan mountains, Heilongjiang province, China (52°20’N, 124°42’E) in September 2018. The components of forest litters were mostly fallen leaves and a few twigs. The Changbai Mountains is a typical temperate-continental climate zone with an over 300-year-old broad-leaved Korean pine mixed forest. The dominant tree species are Pinus koraiensis Siebold et Zucc., Tilia amurensis Rupr., Fraxinus mandshurica Rupr., and Quercus mongolica Firsch. ex Turcz. The Greater Hinggan mountains are located within the cold temperate continental climate zone and consist of Populus davidiana Dode, Betula platyphylla Sukaczev, Larix gmelinii Rupr., and Pinus sylvestris L. as the dominant tree species.
Fungal Medium
The fungal medium comprised potato dextrose agar (PDA; 200 g potato, 20 g glucose, 20 g agar, and 1 L distilled water); 1/4 PDA (50 g potato, 5 g glucose, 20 g agar, and 1 L distilled water); Sabouraud dextrose agar with yeast extract (SDAY; 10 g yeast, 40 g glucose, 10 g peptone, 20 g agar, and 1 L distilled water); oatmeal agar (OA; 30 g oatmeal, 20 g agar, and 1 L distilled water); malt extract agar (MEA; 30 g malt, 20 g agar, and 1 L distilled water). Malt extract powder, agar powder, and yeast extract powder were manufactured by Aoboxing Biotech, Beijing, China. Glucose and peptone were manufactured by Kermel, Tianjin, China.
Fungal Isolation
The forest litters were packed in sterile envelopes and stored at 4°C until used. Fungi were isolated according to Bills et al. (2004) and Liang et al. (2019). Briefly, a litter sample was pulverized and then subjected to a minisieve filtration. The particles in the range of 105–210 μm were transferred to 10 ml of sterile water. The 0.1 ml suspension of the particles was poured on 1/4 PDA and then cultured at 25°C for 2 weeks. During that time, newly emerging fungal colonies were directly transferred to new PDA plates. By this method, about 500 fungal isolates (data not shown) were obtained. The isolates SGSF125 and SGSF355 were suspected to be undescribed species based on their morphological traits and were used in this study. The specimens were deposited in the Herbarium of Microbiology and Phytopathology, Heilongjiang University (HMPHU 1243 and HMPHU 1244), and the living culture (preserved in a metabolically inactive state) was deposited in China General Microbiological Culture Collection Center (CGMCC 19143 and CGMCC 19144).
DNA Extraction and PCR
Fungal genomic DNA was extracted from 100 mg of fresh mycelia. DNA extraction followed a previously published protocol (Liang et al., 2019). The DNA nuclear small subunit rDNA (SSU), internal transcribed spacer (ITS), nuclear large subunit rDNA (LSU), beta tubulin (TUB), translation elongation factor 1 alpha (TEF), RNA polymerase II largest subunit (RPB1), and RNA polymerase II second largest subunit (RPB2) were amplified by PCR. PCR was conducted in 50 μl volumes consisting of 25 μl 2 × Es Taq Master Mix, 1 μl template, 2 μl forward primer, 2 μl reverse primer, 1 μl DNA template, and 20 μl ddH2O. 2 × Es Master Mix was manufactured by CWBIO, Beijing, China. Primer sequence information is shown in Supplementary Table S1. Sequencing was performed using an ABI 3730 DNA analyzer and the BigDye Terminator mix v. 3.1 (Applied Biosystems, Foster City, California, Unites States). The sequences of the new species were submitted to GenBank (accession number is shown in Supplementary Table S2).
Physiological Assays
Each isolate was cultured in sterile 90 mm Petri dishes containing 20 ml of PDA and incubated at 20, 25, 30, and 35°C in complete darkness, for 2 weeks. The impact of different pH values (4, 5, 6, 7, 8, and 9) on fungal growth was also tested on PDA. The diameters of fungal colonies were measured every 2 days. After 14 days, 5 ml 0.05% Tween 80 was added to the PDA plates, and conidia were harvested with a sterile cotton swab and deposited into a sterile 10 ml-volume centrifuge tube. The conidial concentration was estimated using a hemocytometer.
Phylogenetic Analyses
A consensus sequence was obtained by aligning sequences from forward and reverse primers with MEGA v. 7.0 (Kumar et al., 2016) and then queried in the National Center for Biotechnology Information (NCBI) using the BLAST program2. The sequences from 105 fungi classified in Metarhizium and related genera in the Clavicipitaceae (Kepler et al., 2014; Mongkolsamrit et al., 2020) were obtained from GenBank (Supplementary Table S2) and aligned with MEGA v. 7.0 (ClustalW). Alignments were manually adjusted to allow maximum alignments and minimum gaps. The alignments were concatenated as a dataset for a multigene phylogenetic analysis (Kepler et al., 2014; Mongkolsamrit et al., 2020). The multigene phylogenetic analysis of the aligned sequences used RAxML as an optimality criterion, and statistical support was built on 1,000 bootstrap replicates (Edler et al., 2019). A general time-reversible (GTR) model with a gamma-distributed rate variation was used for the nucleotide partitions in the maximum-likelihood (ML) analysis. Markov chain Monte Carlo (MCMC) was used to estimate posterior probability by MrBayes v. 3.2.4 (Ronquist et al., 2012). Four simultaneous Markov chains were run for 3,000,000 generations (standard deviation of split frequencies less than 0.01), and trees were sampled every 1,000 generations. The first 2,000 trees representing the burn-in phase of the analysis were discarded, and the remaining trees were used to determine posterior probabilities in the majority rule consensus tree.
Collection of Insects
Adult insects of Mo. hieroglyphica, Rhopalosiphum maidis, and Callosobruchus chinensis were collected from the experimental fields of the Qiqihar Branch of Heilongjiang Academy of Agricultural Sciences from July and August in 2019. The insect adults were collected from fields, placed in a perforated plastic container, and then maintained at 25°C until use.
Anti-insect Assay
For pathogenicity assays, conidial suspensions were produced using the same method as in the physiological assays. The conidial concentration was adjusted to 1 × 108 conidia/ml using a hemocytometer. Adults of C. chinensis were immersed in conidial suspension for 5 s, and 20 adults were inoculated in each treatment (Noble et al., 2018). Adults of Mo. hieroglyphica were immersed in conidial suspension for 5 s, and 35 adults were inoculated in each treatment. The effect of fungal isolates on the mortality of R. maidis was evaluated by spraying (Hussain et al., 2015; Wu et al., 2020) 1 ml of each conidial suspension on 40 wingless adults per treatment. Tween 80 0.05% was applied as a control treatment. The treated insects were kept in 250 ml flasks with foods and maintained at 25 ± 1°C, 70 ± 5% RH in darkness for 10 days, and the survival of the insects was recorded daily. Cadavers were placed on wet filter paper disks in sterile 90 mm Petri dishes sealed with Parafilm and maintained in an incubator chamber at 25 ± 1°C and 70 ± 5% relative humidity (RH) to investigate mycosis. All the tests were performed in triplicate.
Statistical Analyses
All data were analyzed using SPSS for Windows Version 25.0 (SPSS Inc., Chicago, IL, United States). Data were shown as the mean ± standard error (SE) from three biological replicates. For multiple comparisons, Tukey’s multiple comparison test was used for significance analysis, and p-values equal to 0.05 were considered significant. The significant differences were shown as different letters (such as a, b, and c) in tables and figures.
Results
Colony Growth and Conidial Production at Different pH Values and Temperatures
There are two isolates, SGSF125 and SGSF355, isolated from the forest litters that are suspected as new species related to Metarhizium spp. according to their morphological traits. As suspected new species, the optimal pH and temperature of SGSF125 and SGSF355 were tested. The colony diameters and conidial productions of SGSF125 and SGSF355 on PDA at different temperatures, respectively, are shown in Figure 1. The optimal growth temperature of SGSF125 and SGSF355 was 25 and 30°C, respectively. At 25 and 30°C, SGSF355 had a higher growth rate and conidial production than SGSF125. SGSF355 had a wider optimal temperature range than SGSF125, and the conidial production of SGSF355 was similar at 25 and 30°C with no significant difference. The colony diameters of SGSF125 and SGSF355 were 13–18 mm on PDA on the 14th day.
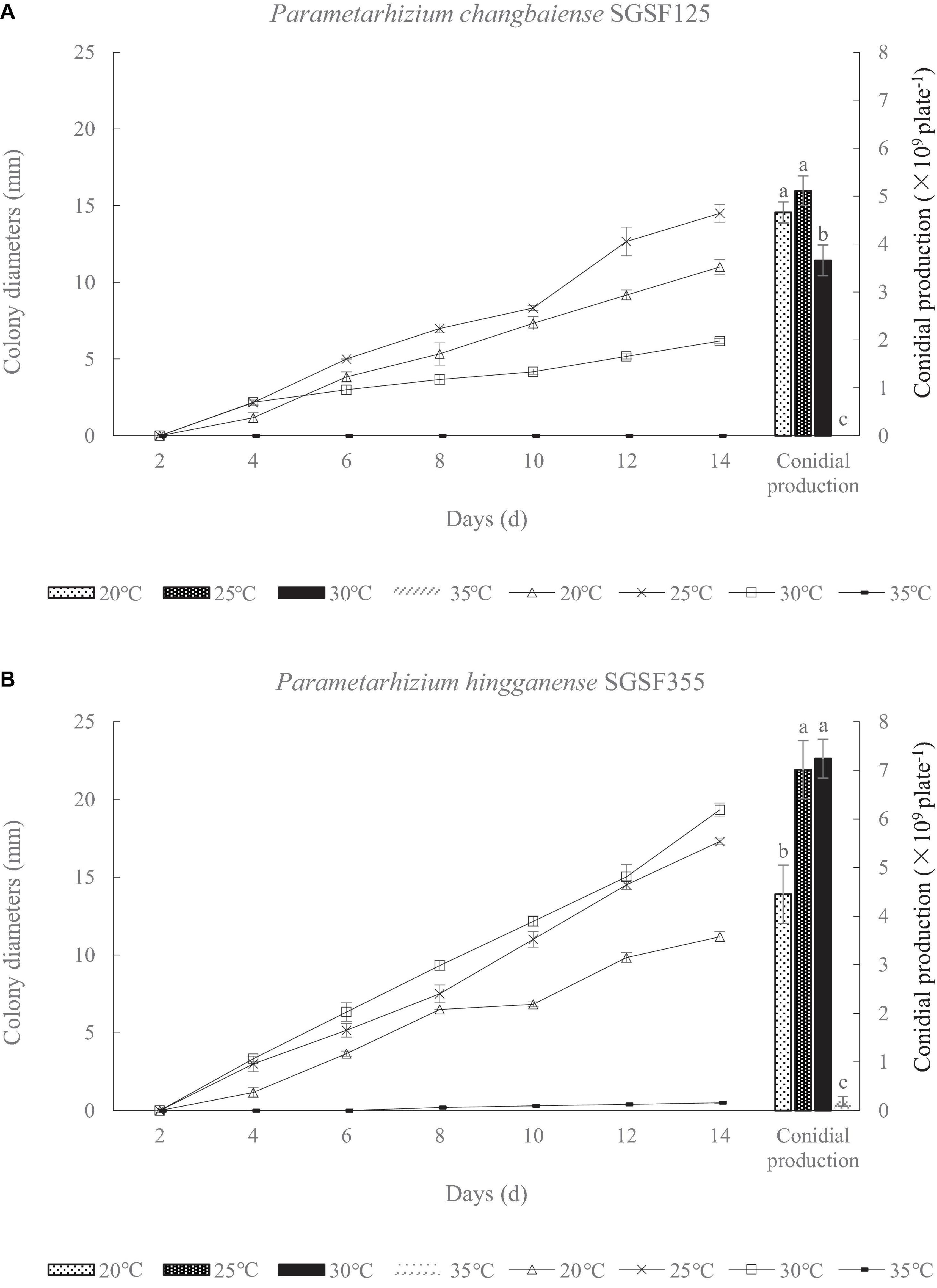
Figure 1. Colony diameter and conidial production of Parametarhizium chanbaiense SGSF125 (A) and Parametarhizium hingganense SGSF355 (B) on potato dextrose agar (PDA). Different letters (a, b, and c) above bars indicate significant differences at the 0.05 level.
Meanwhile, colony diameters and conidial productions of the two isolates were measured at different pH values within a period of 2 weeks (Table 1). The optimal pH values of SGSF125 and SGSF355 were 5 (Table 1). At the same pH, the colony diameter and conidial production of SGSF355 were higher than SGSF125, except at pH 4 (Table 1).
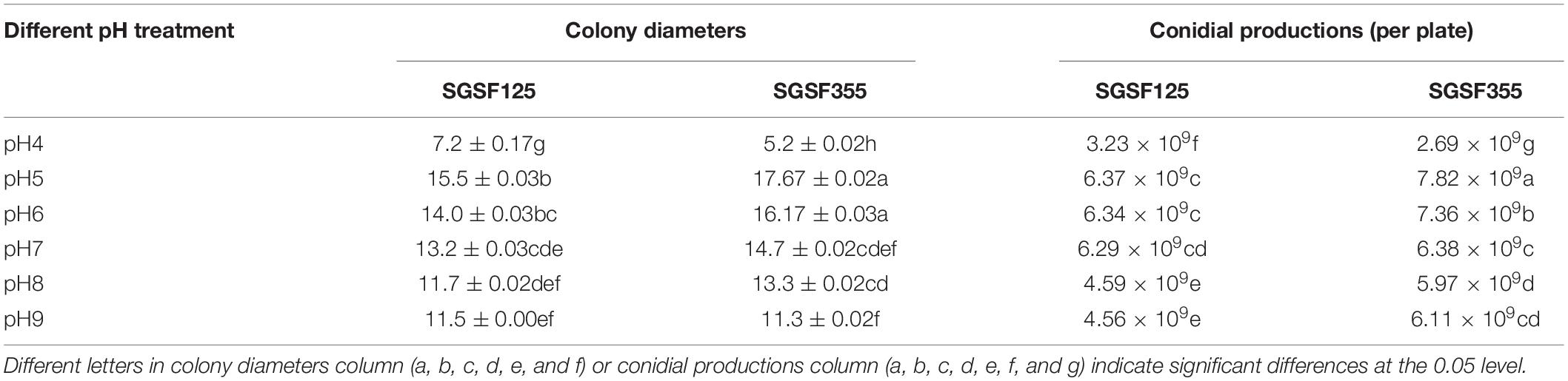
Table 1. Colony diameters and conidial productions of Parametarhizium changbaiense (SGSF125) and Parametarhizium hingganense (SGSF355).
Phylogenetic Analyses
The sequence similarity of seven molecular taxonomic markers SSU, ITS, LSU, TUB, TEF, RPB1, and RPB2 between the two new species was in the range of 93.0–99.7% (except for TUB, 86.5%), indicating their close relationship. In contrast, the similarity to Metarhizium species and taxa from other related genera was much lower in particular for the ITS (<89%), RPB sequences (<85%), and TUB (<81%) (Figure 2 and Supplementary Tables S3, S4). All available sequences of the molecular taxonomic markers from the two new and closely related species (Metarhizium and the genera related to Metarhizium) were combined for a multigene phylogenetic analysis (Supplementary Table S2). The combined dataset concatenated a six-gene dataset consisting of DNA fragments of the SSU (735 bp), LSU (627 bp), TUB (253 bp), TEF (766 bp), RPB1 (492 bp), and RPB2 (778 bp). Sequences of the genus Hypocrella were used as an outgroup. ITS sequences were generated only for barcoding purposes and not included in the multigene analyses (Kepler et al., 2014; Mongkolsamrit et al., 2020). Modeltest 3.7 resulted in the selection of the GTR model with a proportion of invariable sites (I) and the gamma distribution shape parameter (G). This model was used in MrBayes 3.2.4 and RAxML GUI 2.0.0–beta.10. Bayesian analyses resulted in 2,000 “burn-in” trees. For the ML analysis in RAxML, the GTRGAMMA model was used for the nucleotide partitions. The phylogenetic analysis (Figure 3) based on a combined dataset comprising SSU, LSU, TUB, TEF, RPB1, and RPB2 showed that the new species formed a monophyletic clade whose bootstrap values (BSs) and Bayesian posterior probability (BPP) were 100% and 1, respectively. This monophyletic was basal to the clade (BS and BPP were 67% and 1, respectively) of Metapochonia, Pochonia, Metarhizium, and other related genera. To include as many genera in Clavicipitaceae as possible, a phylogenetic analysis only based on the LSU sequences from 34 genera in Clavicipitaceae was also carried out (Supplementary Figure S1). The new species also formed a monophyletic clade in the phylogenetic tree based on the LSU sequences. The isolated position of the new fungal isolates SGSF125 and SGSF355 in the multigene phylogenetic reconstruction together with their unique combination of morphological characteristics prompted us to erect a novel genus named Parametarhizium.
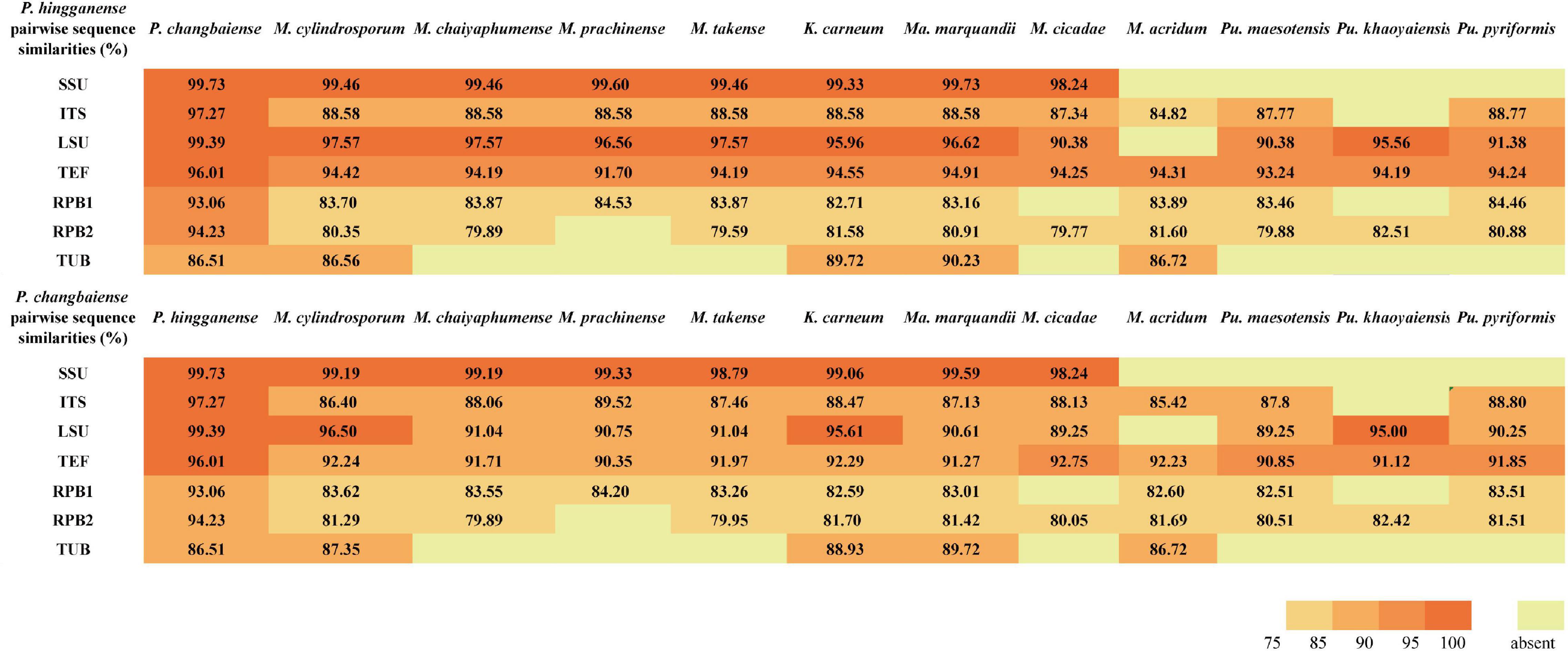
Figure 2. Pairwise nucleotide sequence similarities (in %) for 7-locus from Parametarhizium spp. and morphologically similar species.

Figure 3. Multigene phylogenetic analysis of Parametarhizium and related genera (Clavicipitaceae). Phylogenetic based on concatenated Nuclear small subunit rDNA (SSU), Nuclear large subunit rDNA (LSU), Beta tubulin (TUB), Translation elongation factor 1 alpha (TEF), RNA polymerase II largest subunit (RPB1), and RNA polymerase II second largest subunit (RPB2) using maximum likelihood as optimality criterion. ML bootstrap values/Bayesian posterior probability above 70% (MLBS)/0.7 (BPP) are shown on the nodes. *Ex-type material. Branches with 100% (MLBS) and 1.0 (BPP) support are highlighted by thickened lines.
Taxonomy
Parametarhizium S. Gao, W. Meng, Li Xiang Zhang, Q. Yue, L. J. Xu, gen. nov.
MycoBank no.: MB 837521
Etymology: based on its close morphological relationship to Metarhizium.
Type species: Parametarhizium hingganense S. Gao, W. Meng, Li Xiang Zhang, Q. Yue, L. J. Xu, sp. nov.
Diagnosis: Colonies white to yellow, velvety, exudate lemon yellow, sometimes radially sulcate, reverse pale yellow to brown. Conidiophores hyaline, arising from branches of aerial hyphae, with candelabrum-like arrangement of phialides. Phialides, 2–4 in whorls, cylindrical to obpyriform. Conidia subglobose to ellipsoidal, hyaline (1.1–) 1.2–2.2 (–2.8) × (1.0–) 1.6–1.7 (–2.6) μm.
Description: Colonies on PDA, SDAY, MEA, OA, white to yellow, never green, radially sulcate, 25°C reaching 13–20 mm in 2 weeks, velvety; exudate lemon yellow; reverse pale yellow to brown. Hyphae hyaline, septate, smooth-walled, 0.8–2.5 μm wide. Conidiophores hyaline, smooth-walled (20–) 34–62 (–100) × (0.9–) 1.5–1.9 (–2.2) μm, arising from branches of aerial hyphae, with whorls of 2–4 phialides. Phialides candelabrum-like arrangement (2.7–) 5.6–17.5 (–26) × (1.2–) 1.4–2.3 (–2.5) μm, cylindrical to obpyriform. Conidia subglobose to ellipsoidal, hyaline to yellow (1.1–) 1.2–2.2 (–2.8) × (1.0–) 1.6–1.7 (–2.6) μm (Figure 4 and Supplementary Figure S2).
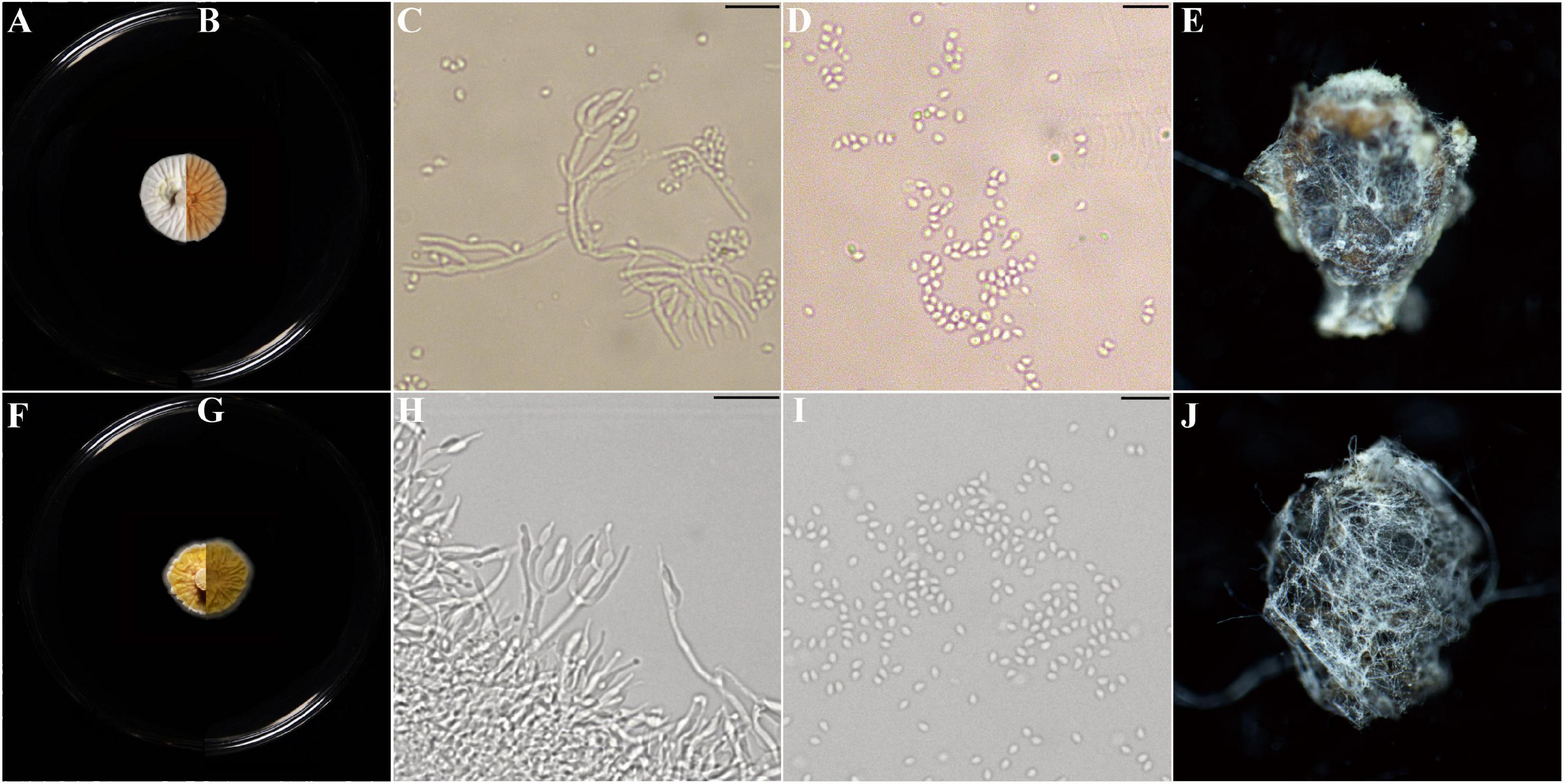
Figure 4. Morphological characters of Parametarhizium changbaiense [from ex-holotype China General Microbiological Culture Collection Center (CGMCC) 19143] and Parametarhizium hingganense (from ex-holotype CGMCC 19144). (A,B) The front and reverse of a P. changbaiense colony on PDA after 14 days at 25°C. (C,D) The phialides and conidia of P. changbaiense. (E) Rhopalosiphum maidis infected by P. changbaiense. (F,G) The front and reverse of a P. hingganense colony on PDA after 14 days at 25°C. (H,I) The phialides and conidia of P. hingganense. (J) R. maidis infected by P. hingganense.
Notes: Compared with those of Metarhizium, the colonies of Parametarhizium are white to yellow (vs. green), and its subglobose-to-ellipsoidal conidia are smaller (<3.3 μm) than most of Metarhizium spp. Parametarhizium exhibits a candelabrum-like arrangement of phialides, while Metapochonia and Pochonia have Verticillium-like conidiophores, and other related genera show Nomuraea-like, Paecilomyces-like or Lecanicillium-like conidiophores.
Parametarhizium changbaiense S. Gao, W. Meng, Li Xiang Zhang, Q. Yue, L. J. Xu, sp. nov.
MycoBank no.: MB 837522
Etymology: referring to the location where the type material was collected.
Description: Colonies on PDA reaching 13–15 mm in 2 weeks, white, radially sulcate, velvety; reverse brown, radially sulcate. Colonies on SDAY reaching 15–17 mm, pale yellow, wrinkled, velvety, with undulate margin; reverse yellow. Colonies on MEA reaching 15–18 mm, white, flat; reverse brown. Colonies on OA reaching 18–19 mm, flat, initially white, turning lemon yellow due to the production of conidial masses, exudate lemon yellow; reverse pale yellow.
Hyphae hyaline, smooth-walled, 0.8–2.5 μm wide. Conidiophores arising from branches of aerial hyphae, bearing dense whorls of branches, terminating in branches with 2–4 phialides per branch, candelabrum–like arrangement of phialides. Phialides cylindrical (2.7–) 5.6–12.5 (–20) × (1.4–) 1.6–2.2 (–2.5) μm, with a short neck (0.9–) 1.2–5.5 (–7.6) × (0.6–) 0.7–1.1 (–1.3) μm. Conidia unicellular, subglobose to ellipsoidal, hyaline to yellow (1.6–) 2.2 (–2.8) × (1.3–) 1.7 (–2.2) μm (Figure 4 and Supplementary Figure S2).
Type: China, Jilin Province, Changbai Mountains, on litters of forest, October 2017, Li Zeyu and Liu Boyang (holotype HMPHU 1243, culture ex-type CGMCC 19143).
Gene sequences ex-holotype: MN589741 (ITS), MN589994 (LSU), MN590231 (SSU), MN908589 (TEF), MT921830 (TUB), MN917168 (RPB1a), MT921829 (RPB2a).
Sexual morph: not observed
Habitat: forest litters
Distribution: Changbai Mountains, Jilin province, China
Notes: P. changbaiense is compared with P. hingganense, Keithomyces aciculare, M. flavoviride, Marquandomyces marquandii, Metarhizium frigidum, and Metarhizium suchlasporia. The conidial size of P. changbaiense is smaller than those found in M. flavoviride, M. frigidum, and K. aciculare, and the color of their colonies is not white to yellow. Ma. marquandii differs from P. changbaiense by the Paecilomyces-like conidiophores and purplish colonies, while M. suchlasporia exhibits slender phialides. A detailed list of the diagnostic features and sequence similarities in comparison with related species can be found in Table 2 and Figure 2.
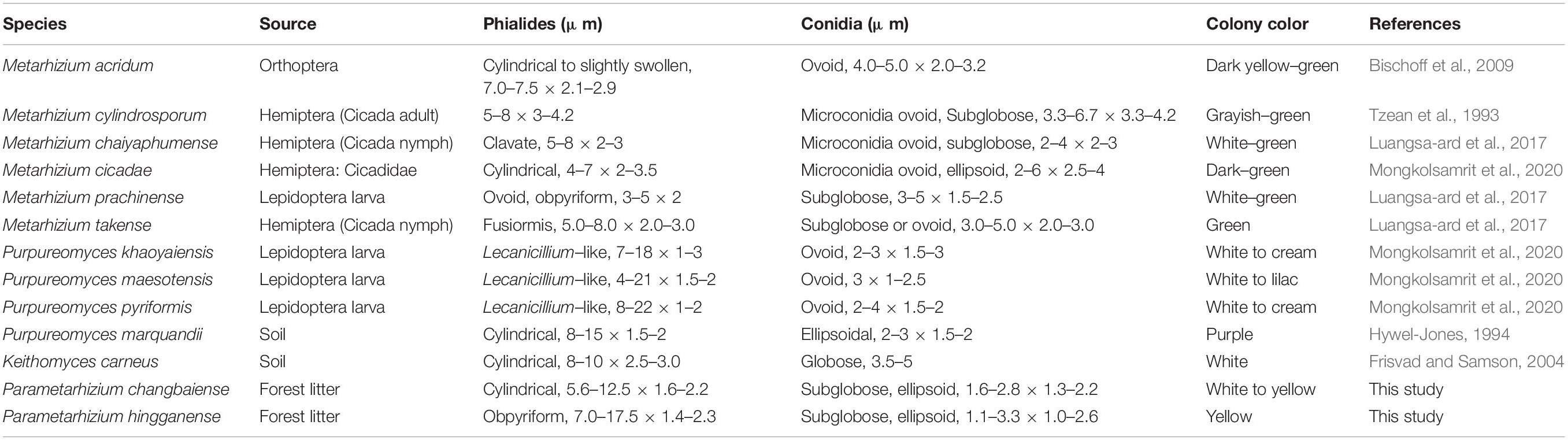
Table 2. Morphological comparison of Parametarhizium changbaiense and Parametarhizium hingganense with similar species.
Parametarhizium hingganense S. Gao, W. Meng, Li Xiang Zhang, Q. Yue, L. J. Xu, sp. nov.
MycoBank no.: MB 837523
Etymology: referring to the location where the type material was collected.
Description: Colonies on PDA reaching 17–18 mm in 2 weeks, yellow, radially sulcate, velvety, with undulate margin; exudate lemon yellow; reverse yellow, radially sulcate. Colonies on SDAY reaching 17–20 mm, pale yellow, wrinkled, velvety, with undulate margin; reverse brown, wrinkled. Colonies on MEA reaching 15–18 mm, lemon yellow, radially sulcate, velvety; reverse brown, radially sulcate. Colonies on OA reaching 18–19 mm, flat, initially cream, turning lemon yellow due to the production of conidial masses; reverse pale yellow.
Hyphae hyaline, septate, smooth-walled, 0.8–2.5 μm wide. Conidiophores arising from branches of aerial hyphae, bearing dense whorls of branches, terminating in branches with 2–4 phialides per branch, candelabrum-like arrangement of phialides. Phialides obpyriform (3.5–) 7.0–17.5 (–26) × (1.2–) 1.4–2.3 (–2.5) μm, with a long distinct neck, (1.6–) 2.5–7.1 (–8.5) × (0.5–) 0.6–1.1 (–1.2) μm. Conidia unicellular, subglobose to ellipsoidal, hyaline to yellow, (1.1–) 1.2 (–3.3) × (1.0–) 1.6 (–2.6) μm (Figure 4 and Supplementary Figure S2).
Type: China, Heilongjiang province, Greater Hinggan mountains, on litters of forest, September 2018, Li Zeyu and Liu Boyang (holotype HMPHU 1244, culture ex-type CGMCC 19144).
Gene sequences ex-holotype: MN055703 (ITS), MN061635 (LSU), MN055706 (SSU), MN065770 (TEF), MN061672 (TUB), MN917170 (RPB1a), MT939494 (RPB2a).
Sexual morph: not observed
Habitat: forest litters
Distribution: Greater Hinggan mountains, Heilongjiang province, China.
Notes: P. hingganense is compared with P. changbaiense, Keithomyces carneum, Ma. marquandii, Metarhizium globosum, Metarhizium minus, and Metarhizium blattodeae. The phialides arrangement of K. carneum and Ma. marquandii is Paecilomyces-like, but P. hingganense possesses candelabrum-like conidiophores. The conidia of M. blattodeae, M. globose, and M. minus are bigger than those in P. hingganense and have different shapes. P. changbaiense and P. hingganense differ mainly in the shape of their phialides and colony characteristics. The phialides of P. changbaiense are cylindrical with a short neck, while those of P. hingganense are obpyriform with a long distinct neck. Colonies of P. changbaiense are white on PDA compared to the yellow colonies of P. hingganense (Figure 4). A detailed list of the diagnostic features and sequence similarities in comparison with related species can be found in Table 2 and Figure 2.
Anti-insect Potentials of P. changbaiense and P. hingganense
Given the similarity to the core Metarhizium, the anti-insect abilities of P. changbaiense and P. hingganense were tested. P. changbaiense and P. hingganense were both pathogenic to Mo. hieroglyphica, C. chinensis, and R. maidis under laboratory conditions. The external white mycelia of P. changbaiense and P. hingganense on the cadavers of three tested insects firstly emerged from the abdomen and then gradually surrounded the insects. In contrast to green cadavers caused by most Metarhizium spp. such as M. anisopliae and M. robertsii, the color of cadavers caused by P. changbaiense and P. hingganense was white (Figure 4 and Supplementary Figure S3). The mortality of P. changbaiense and P. hingganense to above their farmland insects was higher than 85%, and there was no significant difference between P. changbaiense and P. hingganense with the exception that P. changbaiense against C. chinensis was 61.67% (Table 3).
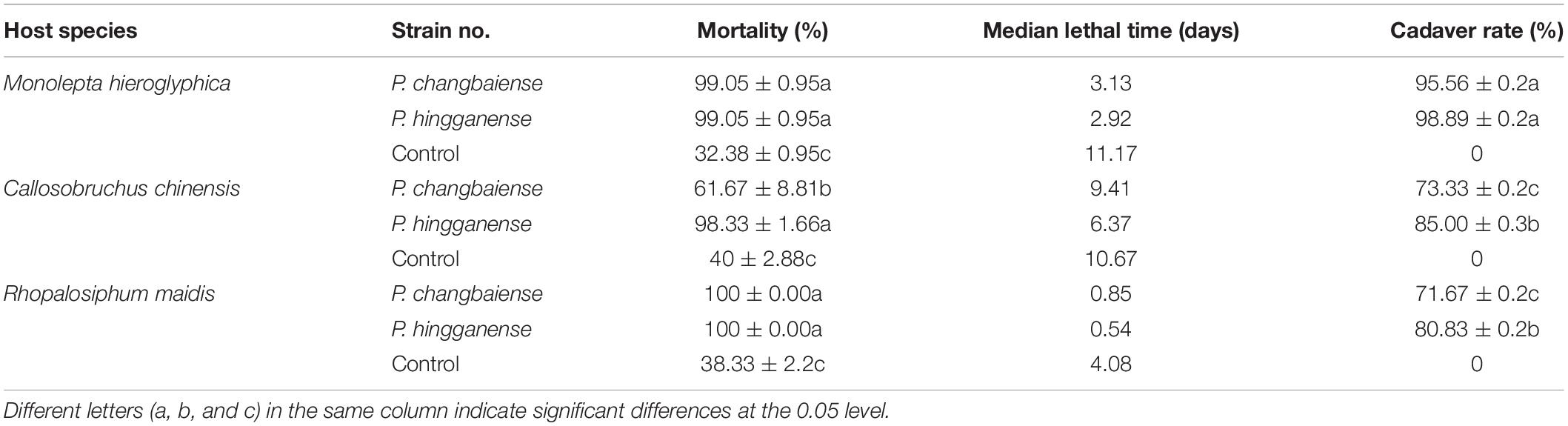
Table 3. Mortality rates, LT50 (median lethal time) and cadaver rate of Parametarhizium spp. isolates.
With respect to the cadaver rate, all of them were higher than 70%. The cadaver rate of P. hingganense to C. chinensis and R. maidis is significantly higher than that of P. changbaiense SGSF125. The median lethal time of P. changbaiense and P. hingganense strongly varied between the different insects but was significantly shorter than that of the control. Most of the R. maidis individuals died within a day after the treatment (4 days in the control), while C. chinensis individuals survived for around 6 days when treated with P. hingganense and 9 days when treated with P. changbaiense (11 days in the control). The effect on Mo. hieroglyphica was even more remarkable with a median lethal time of 3 days for both treatments compared to 11 days in the control (Figure 5). The results of the anti-insect assays implied that P. changbaiense and P. hingganense are non-host-specific entomopathogens and thus offer a biocontrol potential against farmland pests.
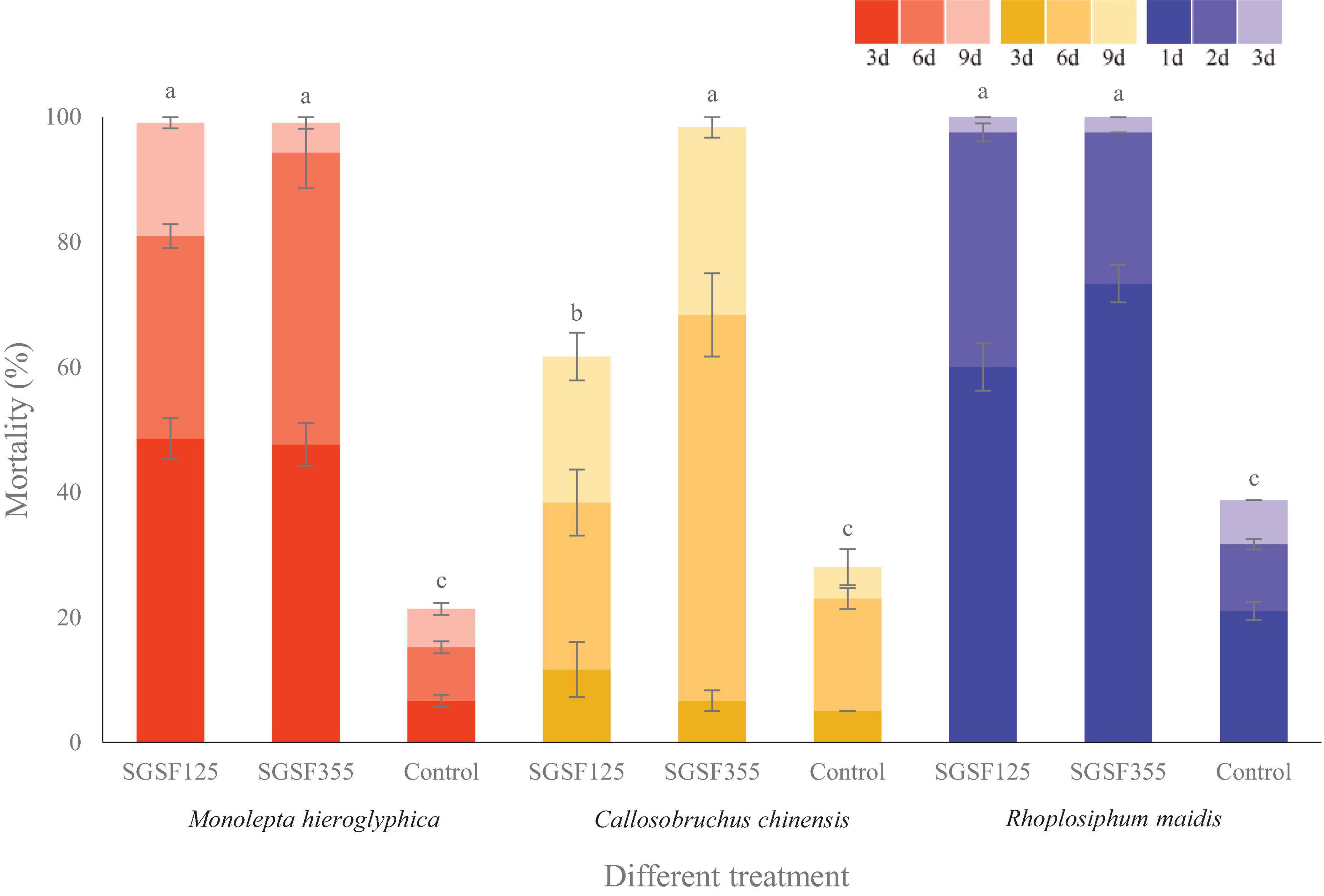
Figure 5. Anti-insect activity screening of Parametarhizium changbaiense and Parametarhizium hingganense on the different day. Red columns are Monolepta hieroglyphica, yellow columns are Callosobruchus chinensis, and blue columns are Rhopalosiphum maidis Different letters (a, b, and c) above bars indicate significant differences at the 0.05 level.
Discussion
Although the discovery of new fungal taxa is on the rise, mycotaxonomy is still an ongoing challenge when introducing new fungal species. Jeewon and Hyde (2016) provided many recommendations for the appropriate use of phylogenetic and morphological data in species delineation, and in this study, we followed the guidelines to establish our new taxa. We proposed one new genus with two new species which are related to members of the Metarhizium complex (formerly Metarhizium sensu lato). According to the pairwise nucleotide sequence similarities for seven loci (Figure 2), the close relatives of P. changbaiense and P. hingganense are genus Metarhizium and its related genera (formerly Metarhizium sensu lato). Currently, the phylogenetic relationships of Metarhizium and its related genera are inferred based on DNA sequence analyses from multigenes (Mongkolsamrit et al., 2020). Since the related genera are not in the clade of the core Metarhizium forming monophyletic clades and their morphological traits are different from core Metarhizium, they have been renamed as other new genera, respectively, such as Papiliomyces and Keithomyces (Mongkolsamrit et al., 2020). The phylogeny shown in Figure 3 was constructed from DNA sequence analyses of the SSU, LSU, TUB, TEF, RPB1, and RPB2 regions, which supported the current taxonomy of Metarhizium and its related genera erected in 2020. This phylogenetic analysis also strongly supported that P. changbaiense and P. hingganense form a monophyletic clade similar with the related genera erected in 2020 (Mongkolsamrit et al., 2020). Combined with morphological results, it is proposed that a new genus, Parametarhizium, is erected.
The morphological traits of P. changbaiense and P. hingganense are compared with their close species, including phialides arrangements, sizes and shapes of conidia, and colony colors (Table 2). The conidia of Parametarhizium are subglobose, and their size is smaller than that of Metarhizium, and the colony of Parametarhizium is yellow or white with the exudate compared with the green colony of Metarhizium. With respect to phialides arrangements, Metarhizium and its related genera have four kinds, candelabrum-like, Verticillium-like, Paecilomyces-like, and Lecanicillium-like. Parametarhizium is candelabrum-like. Metapochonia and Pochonia are Verticillium-like. Keithomyces and Marquandomyces are Paecilomyces-like. Purpureomyces and Papiliomyces are Lecanicillium-like. The conidiophores of Sungia erect with solitary, awl-shaped phialides. Furthermore, Metapochonia, Pochonia, Sungia, and Marquandomyces produce chlamydospores, but Parametarhizium on PDA, SDAY, MEA, and OA could not produce chlamydospores. In addition, the colony color of Parametarhizium is white to yellow, different from these close genera.
Since the conidia of Parametarhizium spp. are subglobose, the morphological traits of the related species producing subglobose conidia are compared and shown in Table 2. The conidia sizes of Pu. khaoyaiensis, Pu. Maesotensis, Ma. marquandii (Banihashemi, 2012) range within 2–3 × 2–3 μm and they are similar to Parametarhizium. However, the phialides arrangements of these four species are Paecilomyces-like which is different from Parametarhizium.
The anti-insect results implied the potentials of P. hingganense and P. changbaiense as new biocontrol agents against farmland pests. There are some reports about anti-insect activities against C. chinensis and R. maidis of the entomopathogenic fungi B. bassiana, M. anisopliae, and Isaria fumosorosea (Sajid et al., 2017; Khan et al., 2018). The mortality of R. maidis caused by P. hingganense and P. changbaiense is similar to M. anisopliae and higher than the others. To the best of our knowledge, no related studies about anti-insect activity of entomopathogenic fungi against Mo. hieroglyphica are published so far. Therefore, the two entomopathogens, P. hingganense and P. changbaiense, are the first fungal species reported to show anti-insect activity against Mo. hieroglyphica.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
SG isolated fungi, built up the phylogenetic tree, investigated anti-insect bioassay, and wrote the manuscript. WM designed the experiments, wrote the manuscript, and provided partial funding. LZ identified and provided insects. QY did the chemical analyses and revised the manuscript. XZ collected the insects. LX designed the experiments, identified the fungal isolates, wrote the manuscript, and provided partial funding. All authors discussed the results.
Funding
This work was funded by the National Natural Science Foundation of China (No. 31870528), the Natural Science Foundation of Heilongjiang Province (QC2015033), and Open Fund (No. 20200522-4) of Key Laboratory of Saline-alkali Vegetation Ecology Restoration, Ministry of Education (Northeast Forestry University).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate Dr. Eric Kuhnert’s help with the revision of manuscript and for his valuable suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.627744/full#supplementary-material
Abbreviations
BLAST, Basic Local Alignment Search Tool; BPP, Bayesian posterior probability; ITS, Internal transcribed spacer; LSU, Nuclear large subunit; rDNA; LT50, Median lethal time; MEA, Malt extract agar; ML, Maximum likelihood; MLBS, Maximum likelihood bootstrap values; NCBI, National Center for Biotechnology Information; OA, Oatmeal agar; PDA, Potato dextrose agar; RPB1, RNA polymerase II largest subunit; RPB2, RNA polymerase II second largest subunit; SDAY, Sabouraud dextrose agar with yeast extract; SGSF125, Parametarhizium changbaiense SGSF125 isolate; SGSF355, Parametarhizium hingganense SGSF355 isolate; SSU, Nuclear small subunit rDNA; TEF, Translation elongation factor 1 alpha; TUB, Beta tubulin.
Footnotes
References
Aly, A. H., Debbab, A., and Proksch, P. (2011). Fifty years of drug discovery from fungi. Fungal Divers. 50, 3–19. doi: 10.1007/s13225-011-0116-y
Bills, G. F., Christensen, M., Powell, M., and Thorn, G. (2004). “Saprobic soil fungi,” in Biodiversity of Fungi: Inventory and Monitoring Method, eds G. M. Mueller, G. F. Bills, and M. S. Foster (Burlington, MA: Elsevier), 271–302.
Bischoff, J., Rehner, S., and Humber, R. (2006). Metarhizium frigidum sp. nov.: a cryptic species of M. anisopliae and a member of the M. flavoviride complex. Mycologia 98, 737–745. doi: 10.1080/15572536.2006.11832645
Bischoff, J., Rehner, S., and Humber, R. (2009). A multilocus phylogeny of the Metarhizium anisopliae lineage. Mycologia 101, 512–530. doi: 10.3852/07-202
Brunner-Mendoza, C., Reyes-Montes, M., Moonjely, S., Bidochka, M., and Toriello, C. (2018). A review on the genus Metarhizium as an entomopathogenic microbial biocontrol agent with emphasis on its use and utility in Mexico. Biocontrol. Sci. Techn. 29, 83–102. doi: 10.1080/09583157.2018.1531111
Castrillo, L., Griggs, M., Ranger, C., Reding, M., and Vandenberg, J. (2011). Virulence of commercial strains of Beauveria bassiana and Metarhizium brunneum (Ascomycota: Hypocreales) against adult Xylosandrus germanus (Coleoptera: Curculionidae) and impact on brood. Biol. Control 58, 121–126. doi: 10.1016/j.biocontrol.2011.04.010
Chen, W., Han, Y., Liang, J., and Liang, Z. (2019). Morphological and phylogenetic characterization of novel Metarhizium species in Guizhou, China. Phytotaxa 419, 189–196. doi: 10.11646/phytotaxa.419.2.5
Chen, W., Han, Y., Liang, J., Liang, Z., and Jin, D. (2017). Metarhizium dendrolimatilis, a novel Metarhizium species parasitic on Dendrolimus sp. larvae. Mycosphere 8, 31–37. doi: 10.5943/mycosphere/8/1/4
Chen, Z., Xu, L., Yang, X., Zhang, Y., and Yang, Y. (2018). Metarhizium baoshanense sp. nov., a New entomopathogen fungus from Southwestern China. Pak. J. Zool. 50, 1739–1746. doi: 10.17582/journal.pjz/2018.50.5.1739.1746
Edler, D., Klein, J., Antonelli, A., and Silvestro, D. (2019). raxmlGUI 2.0 beta: a graphical interface and toolkit for phylogenetic analyses using RAxML. bioRxiv [Preprint]. doi: 10.1101/800912
Filizola, P. R. B., Luna, M. A. C., de Souza, A. F., Coelho, I. L., Laranjeira, D., and Campos-Takaki, G. M. (2019). Biodiversity and phylogeny of novel Trichoderma isolates from mangrove sediments and potential of biocontrol against Fusarium strains. Microb. Cell Fact. 18:89. doi: 10.1186/s12934-019-1108-y
Frisvad, J., and Samson, R. (2004). Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identif cation of food and airborne terverticillate Penicillia and their mycotoxins. Stud. Mycol. 2004, 1–173.
Gutierrez, A., Leclerque, A., Manfrino, R., Luz, C., Ferrari, W., Barneche, J., et al. (2019). Natural occurrence in Argentina of a new fungal pathogen of cockroaches, Metarhizium argentinense sp. nov. Fungal. Biol. U.K. 123, 364–372. doi: 10.1016/j.funbio.2019.02.005
Hawksworth, D., and Lücking, R. (2017). Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 5:FUNK-0052-2016. doi: 10.1128/microbiolspec
Hussain, A., Rizwan-ul-Haq, M., Al-Ayedh, H., Ahmed, S., and Al-Jabr, A. (2015). Effect of Beauveria bassiana infection on the feeding performance and antioxidant defence of red palm weevil, Rhynchophorus ferrugineus. BioControl. 60, 849–859. doi: 10.1007/s10526-015-9682-3
Hyde, K. D., Xu, J., Rapior, S., Jeewon, R., Lumyong, S., Niego, A. G. T., et al. (2019). The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 97, 1–136. doi: 10.1007/s13225-019-00430-9
Hywel-Jones, N. (1994). Cordyceps khaoyaiensis and C. pseudomilitaris, two new pathogens of lepidopteran larvae from Thailand. Mycol. Res. 98, 939–942. doi: 10.1016/S0953-7562(09)80267-0
Jeewon, R., and Hyde, K. (2016). Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosp. 7, 1669–1677. doi: 10.5943/mycosphere/7/11/4
Kepler, R., Humber, R., Bischoff, J., and Rehner, S. (2014). Clarification of generic and species boundaries for Metarhizium and related fungi through multigene phylogenetics. Mycologia 106, 811–829. doi: 10.3852/13-319
Khan, B., Ahmad, K., Zafar, J., Shoukat, R., and Farooq, M. (2018). Efficacy of different entomopathogenic fungi on biological parameters of pulse beetle Callosobruchus chinensis L. (Coleoptera: Bruchidae). J. Entomol. Zool. Stud. 6, 1972–1976.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liang, J., Liu, B., Li, Z., Meng, W., Wang, Q., and Xu, L. (2019). Myxotrichum albicans, a new slowly-growing species isolated from forest litters in China. Mycoscience 60, 232–236. doi: 10.1016/j.myc.2019.03.002
Luangsa-ard, J. J., Mongkolsamrit, S., Thanakitpipattana, D., Khonsanit, A., Tasanathai, K., Noisripoom, W., et al. (2017). Clavicipitaceous entomopathogens: new species in Metarhizium and a new genus Nigelia. Mycol. Prog. 16, 369–391. doi: 10.1007/s11557-017-1277-1
Metchnikoff, E. (1879). Maladies des Hannetons Duble. Zapiski Imperatorskogo Obshchestua Sel’skago Khozyaistra Yuzhnoi Rossii. Odessa: Elsevier, 17–50.
Meyling, N., and Eilenberg, J. (2007). Ecology of the entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae in temperate agroecosystems: potential for conservation biological control. Biol. Control. 43, 145–155. doi: 10.1016/j.biocontrol.2007.07.007
Mongkolsamrit, S., Khonsanit, A., Thanakitpipattana, D., Tasanathai, K., Noisripoom, W., Lamlertthon, S., et al. (2020). Revisiting Metarhizium and the description of new species from Thailand. Stud. Mycol. 95, 171–251. doi: 10.1016/j.simyco.2020.04.001
Nishi, O., Shimizu, S., and Sato, H. (2017). Metarhizium bibionidarum and M. purpureogenum: new species from Japan. Mycol. Prog. 16, 987–998. doi: 10.1007/s11557-017-1333-x
Noble, R., Dobrovin-Pennington, A., Fitzgerald, J., Dew, K., Wilson, C., Ross, K., et al. (2018). Improving biocontrol of black vine weevil (Otiorhynchus sulcatus) with entomopathogenic fungi in growing media by incorporating spent mushroom compost. BioControl. 63, 697–706. doi: 10.1007/s10526-018-9877-5
Rogerson, C. T. (1970). The hypocrealean fungi (Ascomycetes, Hypocreales). Mycologia 62, 865–910. doi: 10.1080/00275514.1970.12019033
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sajid, M., Bashir, N., Batool, Q., Munir, I., Ameen, M., and Munir, S. (2017). In-vitro evaluation of biopesticides (Beauveria bassiana, Metarhizium anisopliae, Bacillus thuringiensis) against mustard aphid Lipaphis erysimi kalt. (Hemiptera: Aphididae). J. Entomol. Zool. Stud. 5, 331–335.
Schardl, C. L., Young, C. A., Hesse, U., Amyotte, S. G., Andreeva, K., Calie, P. J., et al. (2013). Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 9:e1003323. doi: 10.1371/journal.pgen.1003323
Sung, G. H., Hywel-Jones, N. L., Sung, J. M., Luangsa-ard, J. J., Shrestha, B., and Spatafora, J. W. (2007a). Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 57, 5–59. doi: 10.3114/sim.2007.57.01
Sung, G. H., Sung, J. M., Hywel-Jones, N. L., and Spatafora, J. W. (2007b). A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 44, 1204–1223. doi: 10.1016/j.ympev.2007.03.011
Tulloch, M. (1976). The genus Metarhizium. Trans. Br. Mycol. Soc. 66, 407–411. doi: 10.1016/S0007-1536(76)80209-4
Tzean, S., Hsieh, L., Chen, J., and Wu, W. (1993). Nomuraea cylindrospora comb. Nov. Mycologia 85, 514–519. doi: 10.1080/00275514.1993.12026302
Wijayawardene, N. N., Hyde, K. D., Dai, D. Q., Tang, L. Z., Aptroot, A., Castañeda-Ruiz, R. F., et al. (2020). A dynamic portal for a community-driven, continuously updated classification of Fungi and fungus-like organisms: outlineoffungi.org. Mycosphere 11, 1514–1526. doi: 10.5943/mycosphere/11/1/11
Wu, S., Sarkar, S., Lv, J., Xu, X., and Lei, Z. (2020). Poor infectivity of Beauveria bassiana to eggs and immatures causes the failure of suppression on Tetranychus urticae population. BioControl 65, 81–90. doi: 10.1007/s10526-019-09970-0
Zimmermann, G. (1993). The entornopathogenic fungus Metarhizium anisopliae and its potential as a biocontrol agent. Pestic. Sci. 37, 375–379. doi: 10.1002/ps.2780370410
Keywords: 3 new taxa, taxonomy, phylogeny, biopesticide, metarhizium, entomopathogenic fungi
Citation: Gao S, Meng W, Zhang L, Yue Q, Zheng X and Xu L (2021) Parametarhizium (Clavicipitaceae) gen. nov. With Two New Species as a Potential Biocontrol Agent Isolated From Forest Litters in Northeast China. Front. Microbiol. 12:627744. doi: 10.3389/fmicb.2021.627744
Received: 11 November 2020; Accepted: 15 January 2021;
Published: 19 February 2021.
Edited by:
Peter Edward Mortimer, Chinese Academy of Sciences, ChinaReviewed by:
Rajesh Jeewon, University of Mauritius, MauritiusNalin Nilusha Wijayawardene, Qujing Normal University, China
Copyright © 2021 Gao, Meng, Zhang, Yue, Zheng and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijian Xu, eHVsaWppYW5AaGxqdS5lZHUuY24=; eHVsaWppYW5jYXVAZ21haWwuY29t
†These authors have contributed equally to this work
 Siyu Gao1†
Siyu Gao1† Wei Meng
Wei Meng Lijian Xu
Lijian Xu