Corrigendum: Rhizobium leguminosarum Glutathione Peroxidase Is Essential for Oxidative Stress Resistance and Efficient Nodulation
- Hubei Provincial Engineering and Technology Research Center for Resources and Utilization of Microbiology, College of Life Sciences, South-Central University for Nationalities, Wuhan, China
Glutathione (GSH) plays a key role in regulating the cellular Redox Homeostasis, and appears to be essential for initiation and development of root nodules. Glutathione peroxidase (Gpx) catalyzes the reduction of H2O2 and organic hydroperoxides by oxidation of GSH to oxidized GSH (GSSG), which in turn is reduced by glutathione reductase (GR). However, it has not been determined whether the Rhizobium leguminosarum Gpx or GR is required during symbiotic interactions with pea. To characterize the role of glutathione-dependent enzymes in the symbiotic process, single and double mutants were made in gpxA (encoding glutathione peroxidase) and gshR (encoding glutathione reductase) genes. All the mutations did not affect the rhizobial growth, but they increased the sensitivity of R. leguminosarum strains to H2O2. Mutant in GpxA had no effect on intracellular GSH levels, but can increase the expression of the catalase genes. The gshR mutant can induce the formation of normal nodules, while the gpxA single and double mutants exhibited a nodulation phenotype coupled to more than 50% reduction in the nitrogen fixation capacity, these defects in nodulation were characterized by the formation of ineffective nodules. In addition, the gpxA and gshR double mutant was severely impaired in rhizosphere colonization and competition. Quantitative proteomics using the TMT labeling method was applied to study the differential expression of proteins in bacteroids isolated from pea root nodules. A total of 27 differentially expressed proteins were identified in these root bacteroids including twenty down-regulated and seven up-regulated proteins. By sorting the down-regulated proteins, eight are transporter proteins, seven are dehydrogenase, deoxygenase, oxidase, and hydrolase. Moreover, three down-regulating proteins are directly involved in nodule process.
Introduction
Reactive oxygen species (ROS) are biologically important O2 derivatives that are by-products of aerobic metabolism (Köhler et al., 2014). The most important ROS include several small chemical compounds such as the superoxide anion, hydroxyl anion (OH–), hydrogen peroxide (H2O2), nitric oxide (NO), and peroxynitrite (ONOO–) (Suvorava and Kojda, 2009). In addition, ROS function as crucial second messengers, working as redox regulators in a multitude of biological processes (Ortega-Villasante et al., 2018). Excessive production of ROS or impaired ROS detoxification causes oxidative damage to lipid, protein, and DNA (Chung et al., 2010). Oxidative damage is associated with various metabolic disorders (Luo et al., 2020). Removal of ROS by antioxidants plays an important part in limiting this damage (Oláhová et al., 2008). Intracellular antioxidants include enzymes such as superoxide dismutase, catalase, and Gpx, as well as low molecular weight antioxidants like flavonoids, glutathione (GSH) and phenolics (Liu et al., 2014). Among the detoxification enzymes, glutathione peroxidase plays an important role in scavenging reactive oxygen metabolites and protection of organisms from oxidative damage (Khanyok et al., 1997).
Glutathione peroxidase (EC 1.11.1.9) is a selenium-containing antioxidant enzyme with peroxidase activity in catalyzing the reduction of hydroperoxides by GSH (Zheng et al., 2008). Gpx catalyzes the reduction of H2O2 and organic hydroperoxides by oxidation of GSH to GSSG, which in turn is reduced by GR (Heyob et al., 2008). GSH (gamma-L-glutamyl-L-cysteinyl-glycine) is a ubiquitous low-molecular-weight, thiol-containing compound inside the cells and involved in the antioxidant defense and intracellular redox signaling (Desideri et al., 2012). GSH is present in millimolar concentrations in eukaryotic and prokaryotic cells (Fujii et al., 2011). In cells, GSH biosynthesis occurs through two ATP-dependent reactions, usually involving distinct enzymes (Albino et al., 2012). Glutamic acid are combined with cysteine by γ-glutamyl cysteine synthetase. The product γ-glutamylcysteine (γ-GC) is conjugated to glycine to form GSH by the enzyme glutathione synthetase (Cheng et al., 2017).
Rhizobia are Gram-negative nitrogen-fixing soil bacteria that can form nodules on the roots of the host family Leguminosae. During infection, rhizobia are exposed to ROS released by the oxidative burst which is an early plant defense response (Temme and Tudzynski, 2009), and the nodules are also exposed to high concentrations of ROS due to the high rate of respiration necessary to supply the energy expenditure required for the reduction of dinitrogen to ammonia by nitrogenase (Harrison et al., 2005). Therefore, rhizobia require mechanisms for scavenging excess ROS that allow them to maintain sufficient levels of cellular redox homeostasis to support symbiotic processes. GSH plays a critical role by regulating redox homeostasis, and appears to be essential for initiation and development of the root nodules (Bianucci et al., 2008; Yin et al., 2018). In Rhizobium etli and Sinorhizobium meliloti, GSH produced in the bacteroids and host cells is required for symbiotic effectiveness (Harrison et al., 2005; Taté et al., 2012; Becana et al., 2018). For Rhizobium leguminosarum, GSH-deficient mutant cannot efficiently utilize various compounds and induced the formation of pea nodules with poor nitrogen-fixing capacity (Cheng et al., 2017). Notably, glutathione reductase in S. meliloti has been recently reported to be required for both redox homeostasis and symbiosis (Tang et al., 2018). In bradyrhizobial peanut symbionts, the activity of peroxidases is counteracting an oxidative burst for the successful establishment of the symbiosis (Muñoz et al., 2016), however, to date, it has not been determined whether the Gpx-catalyzed the reduction of harmful peroxides by the oxidation of GSH to GSSG is required for intracellular redox homeostasis and symbiosis. In this study, mutation in gpxA gene, gshR gene and both gpxA and gshR genes, was constructed by homologous recombination, and the role of R. leguminosarum Gpx in oxidative stress and during nitrogen-fixing symbiosis was investigated by comparing the phenotypes of wild-type and mutant strains. Proteome analysis provides a more comprehensive understanding of the differences between the wild-type and gpxA mutant nodule bacteroids.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains, oligonucleotide primers, and plasmids used in this study are listed in Supplementary Table 1. Escherichia coli strains were grown in LB medium at 37°C. R. leguminosarum strains were grown under aerobic conditions at 28°C in acid minimal salts medium (AMS) (Poole et al., 1994) or tryptone-yeast extract (TY) medium (Beringer and Hopwood, 1976) supplemented with NH4Cl (10 mM) as a nitrogen source and D-glucose (10 mM) as a carbon source (AMS Glc/NH4+). The following antibiotics were added at the following concentrations (μg/mL): streptomycin (Str), 500; ampicillin (Amp), 50; spectinomycin (Spe), 100; kanamycin (Km), 20; gentamicin (Gm), 20; neomycin (Neo), 80, or 250 (for making gshR mutant). Optical density at 600 nm (OD600) was measured throughout three independent culture growth.
Construction of the R. leguminosarum Mutants and Complementation
To mutate gpxA, primers gpxAF and gpxAR derived from the gpxA (RL1698) gene sequence were used to amplify genomic DNA specifically from R. leguminosarum 3841 (Johnston and Beringer, 1975), and the 2.0-kb product was inserted into pMD18-T vector, giving plasmid pMDgpxA. The spectinomycin resistance cassette from BamHI and PstI-digested pHP45Ωspe was first inserted into the unique BglII site of pMDgpxA, producing pMDgpxAΩSpe (Fellay et al., 1987). The BamHI/XbaI fragment from pMDgpxAΩSpe was further inserted into pJQ200SK to create pJQgpxAΩspe (Quandt and Hynes, 1993). Plasmid pJQgpxAΩspe was conjugated into the recipient strain RL3841, and gpxA mutant (RLgpxA) isolated by selecting for recombination using the spectinomycin resistant and a previously described sac mutagenesis strategy (Kumar et al., 2005). The insertion was mapped by the PCR method using primers MgpxAF or MgpxAR together with pOTF.
To mutate gshR, primers gshRF and gshRR derived from the gshR region were used to amplify genomic DNA specifically from R. leguminosarum 3841. The about 600-bp gshR PCR product was first inserted into XbaI and HindIII sites of pK19mob (Schäfer et al., 1994), giving plasmid pKgshR. Using pRK2013 as helper plasmid (Figurski and Helinski, 1979), pKgshR was further conjugated with R. leguminosarum 3841 as previously described (Karunakaran et al., 2010). The mutations RLgpxA and RLgshR were further combined to construct double mutant as described below. To introduce the spectinomycin resistance-marked gpxA:Spe mutation, the phage RL38 was used to lyse wild-type strain RLgpxA (gpxA:Spe). Using transduction of spectinomycin resistance to RLgshR (gshR), strain RLgpxAgshR (gpxA gshR) was generated.
For complementation of the gpxA mutation, the wild-type gpxA gene fragment was amplified with cgpxAF and cgpxAR and subcloned into pBBR1MCS-5 as an XbaI/PstI fragment (Kovach et al., 1994). Using the pRK2013 as helper plasmid, the resulting plasmid, pBBRgpxA, was conjugated into the RLgpxA recipient strain by triparental mating (Karunakaran et al., 2010), Complemented strain RLgpxA(pBBRgpxA) was isolated using selection for gentamicin resistance as previously described (Karunakaran et al., 2006).
Assay of H2O2 Sensitivity
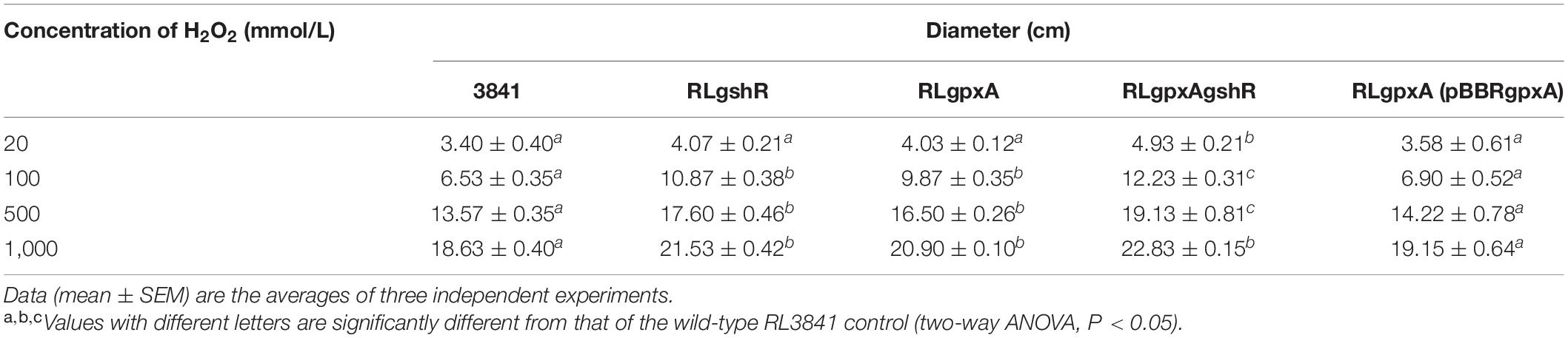
The sensitivity of R. leguminosarum to H2O2 was assessed with a filter paper disc diffusion method. The logarithmic phase (OD600 0.4–0.6) R. leguminosarum strains were collected by centrifugation, washed twice and resuspended in sterile phosphate-buffered saline (PBS, 8.0 mM Na2HPO4, 136 mM NaCl, 1.5 mM KH2PO4, 2.6 mM KCl). 100 μL of bacterial suspensions containing 108 cfu/mL was spread on agar plates made from AMS Glc/NH4+. Sterile filter paper discs (6-mm diameter) were placed on the TY media plate on which a bacterial suspension was spread, and a disc was impregnated with 30 μL of H2O2 solution. In this experiment, H2O2 was used in different concentrations (20 mmol/L, 100 mmol/L, 500 mmol/L, and 1,000 mmol/L). Water was used as a negative control. The experiment consisted of three independent experiments, and the two-way ANOVA (P < 0.05) was used to test for differences.
Assay of Glutathione Levels in R. leguminosarum
For analysis of glutathione content, logarithmic phase cultures of mutant strains RLgpxA, RLgshR, RLgpxAgshR, and wild-type RL3841 with an optical density (OD600) 0.4–0.6 were collected on ice, centrifuged (5,000 rpm) for 5 min at 4°C. The cells were sonicated in an ice-water bath for 15 min. The sonicate was centrifuged (12,000 rpm, 10 min) at 4°C. GSH contents were measured spectrophotometrically as previously described (Rahman et al., 2006). The experiment was constructed from three independent experiments, and the two-way ANOVA (P < 0.05) was used to test for differences.
Plant Experiments and Microscopy Studies of Nodules
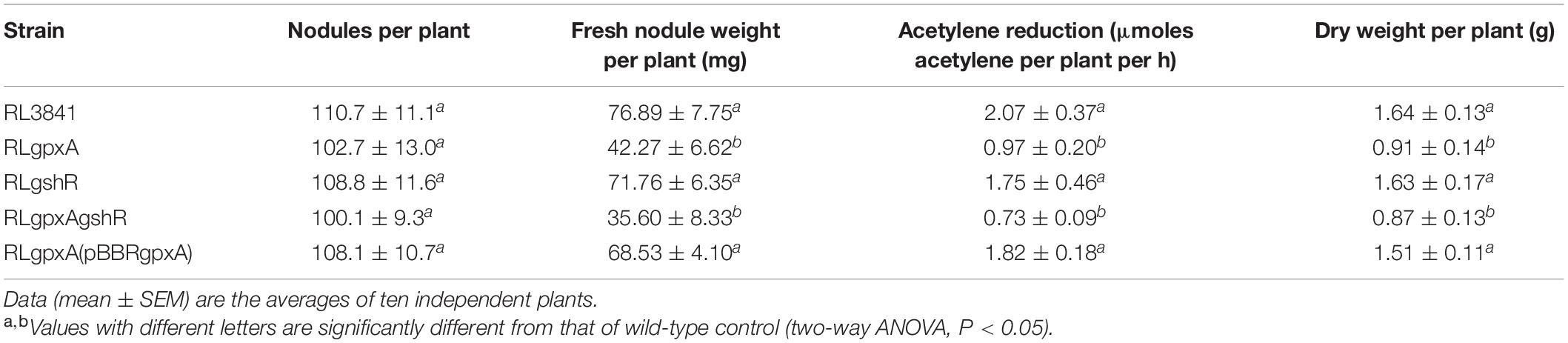
Nodulation assays on pea were carried out as previously described (Cheng et al., 2017). Pea seeds were sterilized with 5% sodium hypochlorite, placed in 1 L pots containing sterile vermiculite with nitrogen-free Fahraeus solution. At the time of sowing, R. leguminosarum strains were inoculated with 107 CFU per seed. Plants were incubated in a controlled-environment growth chamber with a 8 h at 20°C in the dark and 16 h photoperiod at 22°C in the light. For the dry weight plants were placed in 2 L beaker containing sterile vermiculite, watered with distilled nitrogen-free Fahraeus solution and harvested at 7 weeks from planting. The shoot was removed from the root and dried at 70°C in a warm incubator for 72 h (Wang et al., 2019). Nitrogenase activity of nodules at flowering (4 weeks) was assessed by acetylene reduction assay according to the method described previously (Allaway et al., 2000). The experiment was constructed from three independent experiments, and the two-way ANOVA (P < 0.05) was used to test for differences.
Four-week-old nodules were obtained and examined by the use of both light and electron microscopes. Nodules were fixed with cold 2.5% glutaraldehyde, and then postfixed in 1.5% osmium tetroxide after collection. Semi-thin sections (1–3 μm) of root nodules were obtaining, colored in toluidine blue, and evaluated with light microscope (SZX16, Olympus). For electron microscopy studies, ultra-thin sections were cut, double-stained with uranyl acetate and lead citrate, and photographed with a transmission electron microscope Hitachi H-7100.
Colonization Ability in the Pea Rhizosphere
Root colonization assays were performed according to the method described previously (Cheng et al., 2017). Pea seedlings were grown in vermiculite for 1 week, and inoculated with each mutant strain and wild-type RL3841 at the cfu ratios 1,000:1,000, 0:1,000, and 1,000:0. Shoots were cut-off after 1 week (2 weeks after sowing), and the roots were added with 20 mL of sterile PBS and vortexed for 15 min (Karunakaran et al., 2006). The samples were further serially diluted 10-fold in sterile water, and the colony number was counted on TY solid medium plates containing streptomycin (for mutant and wild-type strains together) or neomycin and streptomycin (for RLgshR) or streptomycin and spectinomycin (for RLgpxA or RLgpxAgshR), giving the number of viable root – and rhizosphere-associated bacteria (Barr et al., 2008). Each treatment included 10 replications, and the two-way ANOVA (P < 0.05) was used to test for differences.
RNA Extraction and Quantitative Reverse Transcription-Polymerase Chain Reaction
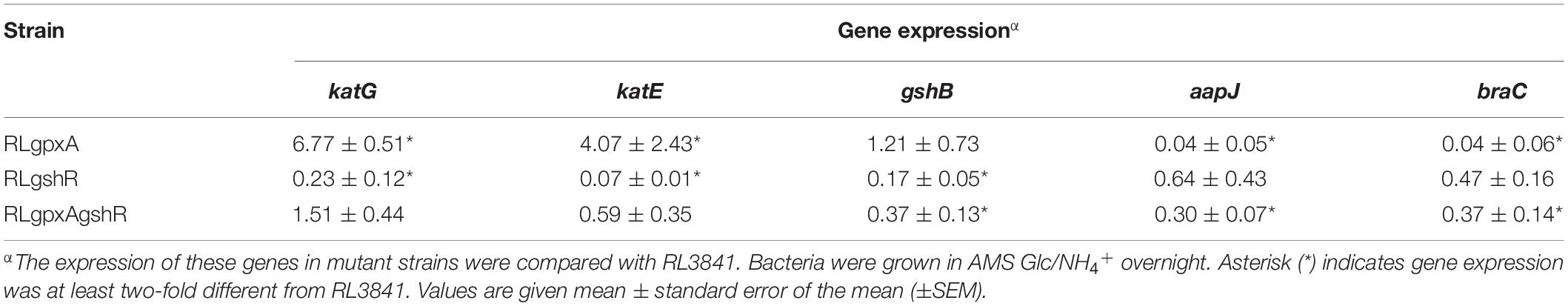
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed to investigate the differences in the expression of various genes between mutant and wild type. R. leguminosarum samples were collected in triplicates from free-living cultures in AMS liquid medium, or 2, 4, 6-week-old nodules. The pea nodules were harvested, and then finely grinded into powder in liquid nitrogen. Total RNA was isolated using TRIzol reagent (Invitrogen) and measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific) (Smith et al., 1985). cDNA was synthesized with the use of random hexamers and SuperScriptTM II reverse transcriptase. qRT-PCR assay was carried out using SYBR® premix Ex TaqTM (Takara, Dalian, China) on a CFX96 Real-Time System (Bio-Rad). Primer sequences for gpxA, gshR, gyrB1, katG, katE, gshB, aapJ, braC are listed in Supplementary Table 1. Gyrb1 was used as the reference housekeeping gene. Data were obtained and analyzed according to the previously described method (Prell et al., 2009). The one-way ANOVA (P < 0.05) was used to test for differences.
Protein Extraction and Trypsin Digestion
The 4-week-old nodules were dripped into liquid nitrogen and ground to a fine cell powder. The ground powder of cells was then transferred into a 5-mL ice-cold centrifuge tube, and suspended in four volumes of lysis buffer (8 M urea, 1% protease inhibitor cocktail). The slurry was sonicated for three times on ice using a high-intensity ultrasonic processor, and the lysates were cleared by centrifugation at 12,000 × g at 4°C for 10 min. The supernatant was collected and stored at −80°C. The total protein concentration in the supernatant was measured by using the bicinchoninic acid (BCA) protein assay kit (Pierce, Rockland, IL, United States). Proteins were first reduced by incubation with 5 mM dithiothreitol for 30 min at 56°C. For trypsin digestion, the protein solution were first alkylated with 11 mM iodoacetamide (IAM, Sigma) in the darkness at room temperature for 15 min. Then, each protein was dissolved by adding 100 mM tetraethylammonium bromide (TEAB) to reduce the urea concentration less than 2M. Finally, the proteins were digested with trypsin at 1:50 trypsin-to-protein mass ratio overnight at 37°C, and followed with trypsin at a trypsin-to-protein mass ratio of 1:100 for a second 4 h-digestion.
Tandem Mass Tag Labeling and LC-MS/MS Analysis
After trypsin digestion was completed, the peptides were desalted using a Strata X C18 SPE column (Phenomenex). The peptides were vacuum-dried, reconstituted in 0.5 M TEAB and processed using the 6-plex tandem mass tag (TMT) kit (Thermo Fisher Scientific, Bremen, GA, United States) according to the manufacturer’s protocol. Briefly, one unit of TMT reagent (defined as the amount of reagent required to label 100 μg of protein) was thawed and extracted with 24 μL of acetonitrile. The resulting peptide mixtures were further incubated at room temperature for 2 h, pooled, desalted, centrifuged, and dried by vacuum.
The resultant tryptic peptides were crushed into various fractions by high pH reverse-phase HPLC using an Agilent 300 Extend C18 column (150 mm × 4.6 mm, 5 μm). Briefly, the peptides were first fractionated with a gradient of 2–60% (v/v) acetonitrile in 10 mM ammonium bicarbonate at pH 10 over 80 min into 80 fractions. Then, the peptides were concatenated and combined into 18 fractions, and dried using vacuum centrifugation. LC-MS/MS analysis peptides were dissolved in solvent A (0.1% formic acid in aqueous solution), directly loaded onto a reversed phase column (Acclaim PepMap 100, Thermo Fisher Scientific). Separation of peptides was performed using a reversed-phase analytical column (Thermo Fisher Scientific, Shanghai, China). The gradient was as follows: 6–22% of solution B (0.1% FA in 98% ACN) over 26 min, 23–35% in 8 min, 80% in 3 min, and finally 80% for 3 min at a constant flow rate of 400 nL/min on an EASY-nLC 1,000 ultra-performance liquid chromatography (UPLC) system (Thermo Fisher Scientific). The resulting peptides were subjected to an NSI source, and then tested by tandem mass spectrometry (MS/MS) in a Q ExactiveTM Plus (Thermo, Shanghai, China) coupled online to UPLC. Peptides were detected by the Orbitrap analyzer at a mass resolution of 70,000. The peptides were subjected to run MS/MS analysis using a setting of 28 eV normalized collision energy (NCE), and ion fragments were detected in the Orbitrap detector at a resolution of 17,500. The MS analysis alternated between MS and data-dependent MS/MS scans using 15.0 s dynamic exclusion. The applied electrospray voltage was set to 2.0 kV. Automatic gain control (AGC) was applied to prevent overfilling of the ion trap, and 5 × 104 ions were accumulated for generation of the MS/MS spectra. The scan range of full MS was set from 350 to 1,800 m/z, and the fixed first mass was set to 100 m/z. The experiments were performed in triplicate.
Data Analysis
The MS/MS data of all experiments were processed using the MaxQuant software version 1.5.2.8 (Cox and Mann, 2008). The spectra were searched against an annotated R. leguminosarum protein database (Young et al., 2006). Trypsin/P was treated as the cleavage enzyme and the search allowed up to four missing cleavages. The mass tolerance for precursor ions was set at 20 ppm, the final tolerance was set at 5 ppm, and the mass tolerance for precursor ions was set to 0.02 Da. Alkylation of cysteine by carbamidomethylation was set as a fixed post-translational modification, and methionine oxidation was treated as variable modifications for database search. The false discovery rate (FDR) was <1%, and a minimum Andromeda score cut-off was set at 40 for modified peptides. Protein expression data indicate statistically significant differences according to Student’s t-tests (P < 0.05). Differentially expressed proteins were defined as having at least a 1.2-fold change. The mass spectrometry proteomics data and the Supplementary Data have been uploaded to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD022029.
Bioinformatics Analysis
Sequence similarity searches were performed with FASTA against current releases of the Uniprot Knowledgebase (UniProtKB). The gene ontology (GO) categories for subcellular location were derived from UniProt and Rhizobium database entries. The metabolic and biological processes were studied by using Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
Results
Growth of R. leguminosarum Mutants Under Free-Living Conditions
The whole genome sequence analysis of wild-type RL3841 revealed the presence of a glutathione peroxidase gene (gpxA) and a glutathione reductase encoding gene (gshR). gpxA (RL_RS08810, RL1698) is predicted to encode a polypeptide of 182 amino acids with a molecular mass of about 20.02 kDa and a theoretical pI value of 5.51, and gshR (RL_RS13915, RL2694) is predicted to encode a polypeptide of 461 amino acids with a molecular mass of about 50.05 kDa and a theoretical pI value of 5.87. To experimentally confirm the potential function of these two redox genes, single and double mutants were made by means of mutagenesis. The mutants were first evaluated for growth in liquid AMS minimal medium. None of the mutant strains presented a substantial reduced growth rate (Supplementary Figure 1). These results suggest neither gpxA gene nor gshR gene has effect on R. leguminosarum growth in AMS minimal medium, and contrast with the data from S. meliloti in which glutathione reductase is important for the growth (Tang et al., 2018).
Antioxidation Analysis of R. leguminosarum Mutants
In order to study the sensitivity to oxidative stress, growth of R. leguminosarum mutants and wild-type strain was monitored by carrying out disk diffusion assays (Table 1). The growth of mutants RLgpxA and RLgshR was not significantly affected by 20 mmol/L H2O2 treatment, as compared with the wt RL3841 strain. However, at concentrations of 100, 500, and 1,000 mmol/L, the RLpxA and RLgshR mutant strains presented a clear sensitivity to the H2O2 treatment, and their growth was significantly inhibited. The results suggest that the GpxA and GshR could play an important role in protecting cells from hydrogen peroxide stress. Moreover, at H2O2 concentration of 20, 100, and 500 mmol/L, the double mutant RLgpxAgshR, in which both Gpx and GR are inactivated, presented a slightly enhanced sensitivity, but significantly inhibited growth rate compared to the single mutant strains, suggesting that double mutation in Gpx and GR increased the sensitivity of R. leguminosarum strains to H2O2.
Effect of Mutant on the Concentration of GSH
The GSH contents were quantified spectrophotometrically. The data showed that the content of GSH has no significant effect in the gpxA mutant, compared to wild-type strain, while it was 2.5-fold higher in both gshR mutant and double mutant (Figure 1). These results indicate that a mutant in gshR gene increased GSH levels.
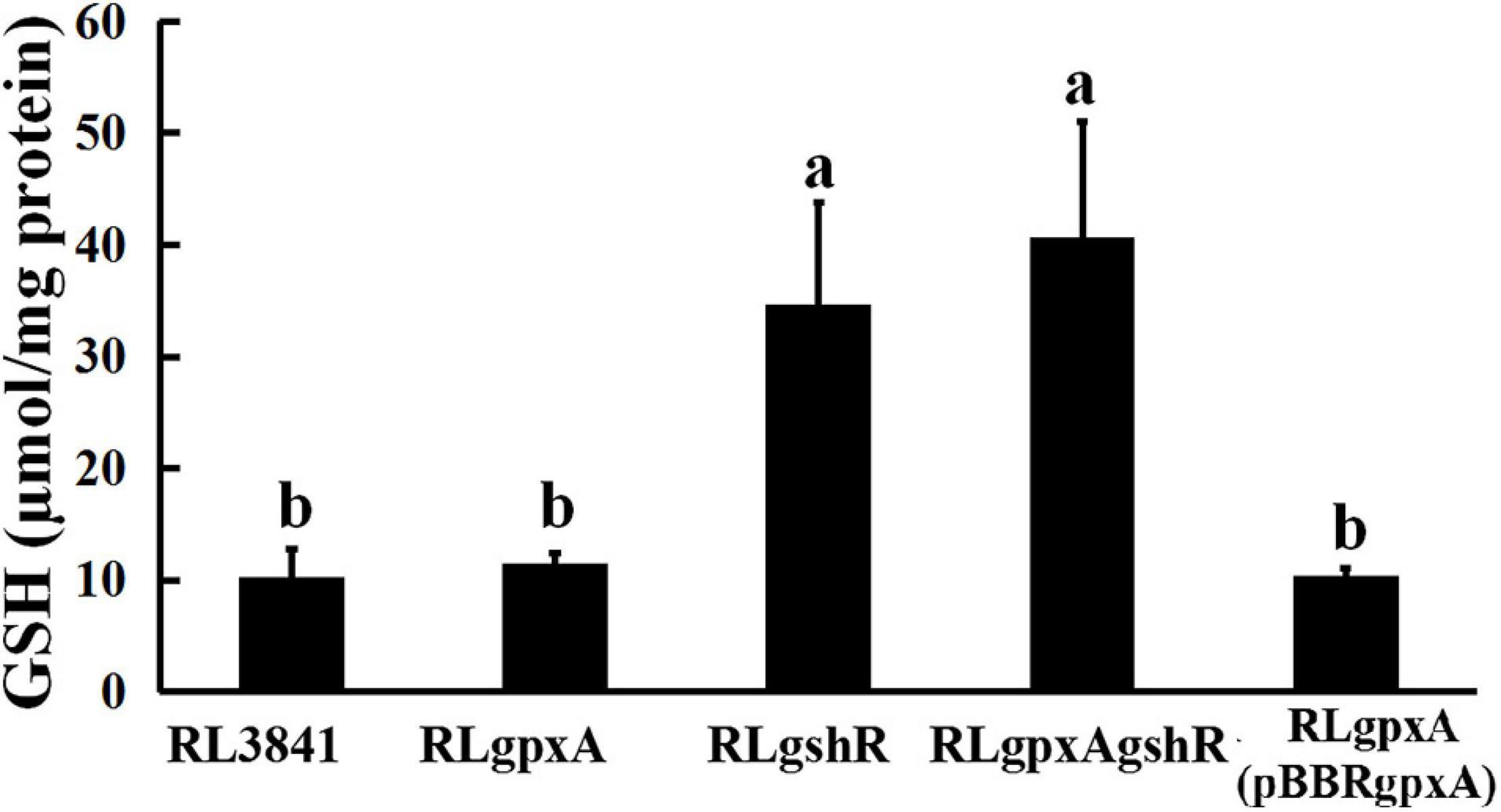
Figure 1. The accumulation of reduced glutathione in R. leguminosarum strains. Data are from three biological samples (±SEM). a,bDifferent letters indicates the value is significantly different (two-way ANOVA, P < 0.05).
Transcriptional Analysis of the Relative Gene Expression
Both Gpx and GR play an important role in anti-oxidation responses, and catalase activity can function as a marker to assess whether rhizobia are under oxidative stress (Tang et al., 2018), which prompted us to study the expression of the catalase genes by qRT-PCR. Expression of katG and katE in RLgpxA were increased 6.77- and 4.07-fold compared with levels in RL3841, while their expression in RLgshR were decreased 4.35- and 14.29-fold (Table 2). However, in the double mutant, both katG and katE expression showed no significance, suggesting a complex interrelation of the glutathione peroxidase/reductase (GSH-Px/GSSG-Rd) enzyme system. The level of reduced GSH was no significant influence in the gpxA mutant, bur strikingly increased in the gshR mutant, qRT-PCR was performed to measure the expression level of glutathione synthetase gene gshB. The data showed that the expression of gshB was increased in the gshR mutant and the double mutant compared with the parent strain but there was no statistically significant difference in the gpxA mutant (Table 2).
GSH was reported to be involved in the activity of amino acid transporters R. leguminosarum (Cheng et al., 2017). To examine its possible mechanism, expression of the two ABC transporters, the general amino acid permease (Aap) and the branched-chain amino acid permease (Bra), was detected by qRT-PCR. Expression of aapJ and braC in RLgpxA were both decreased 25-fold compared with levels in RL3841, while expression of these two genes in RLgshR did not alter significantly (Table 2). Moreover, as there was slightly but significantly decreased in expression of either aapJ or braC in double mutant RLgpxAgshR, it suggests that the transporter defect caused by the GpxA-deficiency was partly restored by mutant in the gshR gene.
Expression of the gpxA and gshR genes in root nodules collected at 14, 28, and 42 days post inoculation was also detected by qRT-PCR, but no differences were found comparison to that in free-living cells (Supplementary Table 2).
Rhizosphere Colonization and Competition by R. leguminosarum Strains
It has been reported that the R. leguminosarum gshB mutant is severely impaired in rhizosphere colonization (Cheng et al., 2017). Therefore, the colonization ability of R. leguminosarum mutants for growth in the rhizosphere of the plant was evaluated by inoculating soils of a low microbial population (103 or 104 bacteria per seedling) into the short-term colonization of the plant rhizosphere, and measuring the total count of bacteria after 1 week. When the single mutants RLgpxA, RLgshR, and the wild-type RL3841 were inoculated alone into a pea rhizosphere, there was no significant difference (P < 0.05) in the numbers of bacteria (Figure 2). However, the ratio between double mutant RLgpxAgshR and wild-type 3841 was 23.14%, and was at a significant disadvantage compared to the single mutants RLgpxA, RLgshR, and the wild-type RL3841. When inoculated together in equal proportion, mutation of gpxA did not affect R. leguminosarum rhizosphere competitive capacities. In contrast, the gshR mutant RLgshR and the double mutant RLgpxAgshR were at a significant disadvantage (16.57 and 6.10% of bacteria recovered) compared to the wild-type RL3841 (Figure 2). The results showed that GshR was essential for competition in the host plant rhizosphere by R. leguminosarum, while GpxA had an enhanced effect on the defect of the rhizosphere colonization and competition by the gpxA and gshR double mutant.
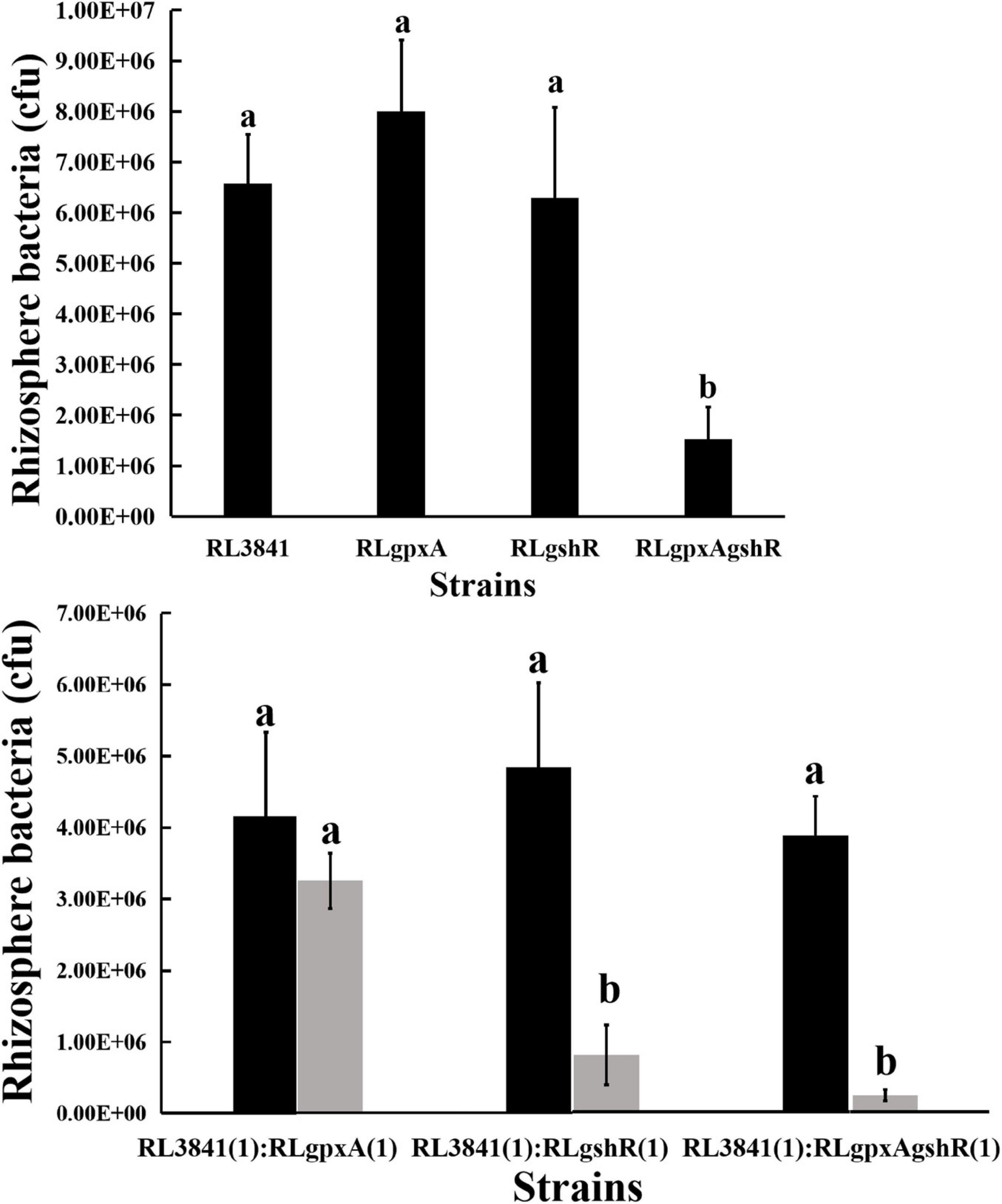
Figure 2. Competition of the wild-type RL3841 (black bars) and the mutants (gray bars) in the pea rhizospheres. The x-axis indicates the initial inoculation ratios, with 1 corresponding to 1 × 103 CFU. The number of colony-forming bacteria (per plant) from 10 plants are shown. a,bValues with different letters are significantly different between mutant and wild-type control (two-way ANOVA, P < 0.05).
Symbiotic Phenotypes of R. leguminosarum Strains
Pea seedlings were inoculated with R. leguminosarum mutants and wild-type RL3841, and after 4 weeks, acetylene reduction activity and nodule numbers per plant were measured. RLgshR formed nodules with normal size. However, RLgpxA and double mutant RLgpxAgshR resulted in smaller nodule structures and reduction in nodule fresh weight (Table 3). There was no statistically significant difference in the number of nodules per plant between plants inoculated mutants RLgpxA, RLgshR, RLgpxAgshR and plants inoculated with wild-type RL3841 (Table 3). Mutation in gshR gene induced nodules with the same capacity for acetylene reduction as nodules infected by wild-type RL3841. In contrast, plants inoculated with RLgpxA and double mutant RLgpxAgshR showed a significantly more than 50% decrease in acetylene reduction activity compared to the wt RL3841, and no significant difference was found between these two mutant strains (Table 3). At 7 weeks post inoculation, the dry weight values of plants inoculated with RLgpxA and double mutant RLgpxAgshR was no more than 56% of that of plants inoculated with wild-type RL3841 (Table 3). When the recombinant plasmid pBBRgpxA was introduced into mutant RLgpxA, plants inoculated with the complemented strain RLgpxA(pBBRgpxA) elicited normal nodules and had the ability to fix nitrogen at the same rate as wt RL3841 inoculated plants (Figures 3M–O and Table 3).
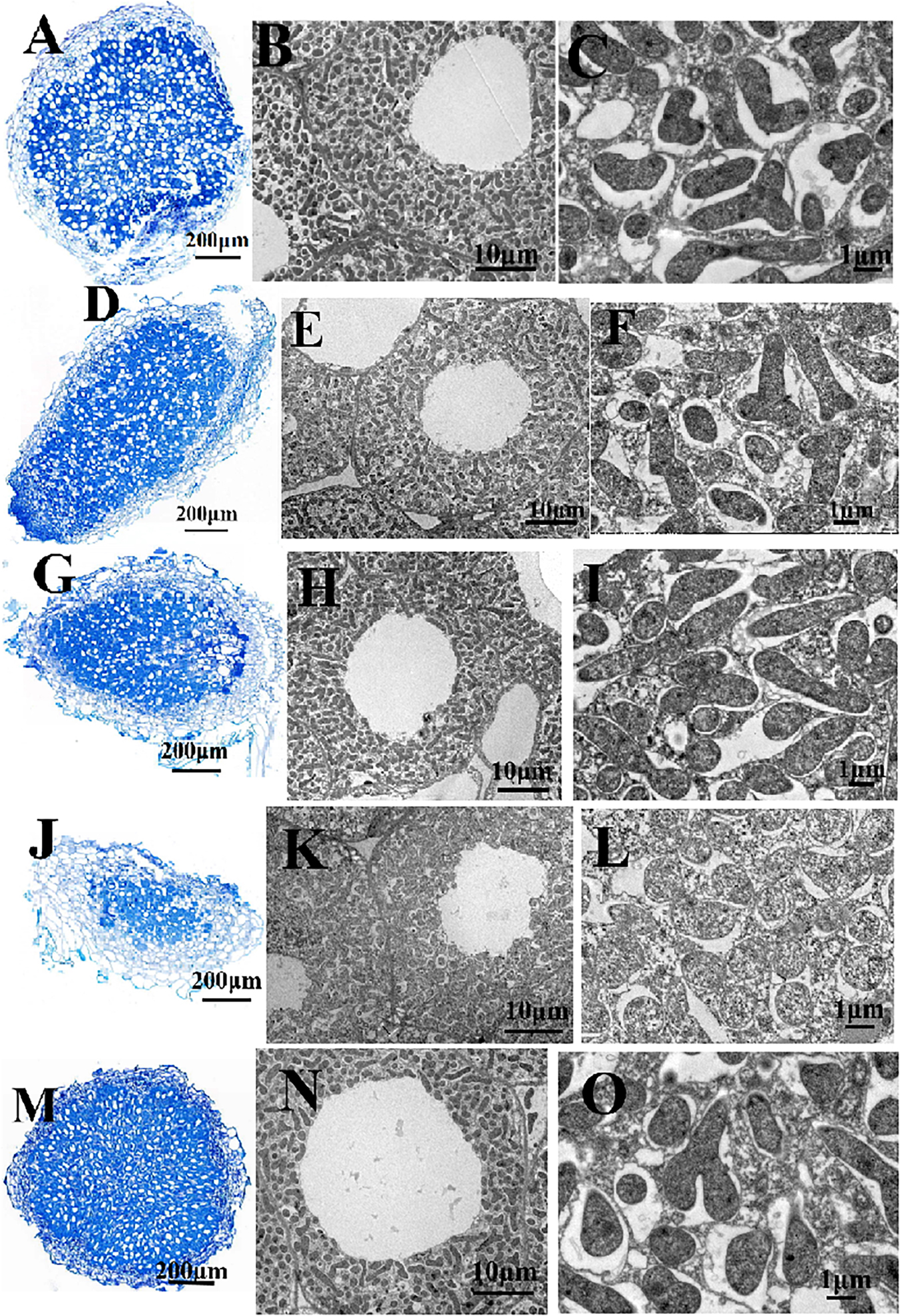
Figure 3. Structure of pea nodules and bacteroids. Root nodules were induced by RL3841 (A–C), RLgshR (D–F), RLgpxA (G–I), RLgshRgpxA (J–L), RLgpxA(pBBRgpxA) (M–O). Scale bars = 200 μm (A,D,G,J,M), 10 μm (B,E,H,K,N), 1 μm (C,F,I,L,O).
Nodules were collected and examined by combined light and transmission electron microscopy. The wt RL3841 and mutant RLgshR contained normal spherical shape of the nodules (Figures 3A,D). However, microscopic analysis of the nodules obtained with RLgpxA and double mutant RLgpxAgshR showed that they contained an abnormally thick cortex (Figures 3G,J), suggesting GpxA had an important role in the formation of normal nodules. Ultrastructural changes of the infected cells and the bacteria in pea root nodules were observed by transmission electron microscopy. Nodule cells infected by mutant RLgshR and wt RL3841 were occupied by large T-shaped, Y-shaped, and rod-Shaped differentiated bacteroids (Figures 3B,C,E,F), while nodule cells infected by mutants RLgpxA and RLgpxAgshR consisted of a small amount of rod-Shaped bacteroids, a large amount of undifferentiated spherical-shaped bacteroids (Figures 3H,I,K,L), suggesting a general lack of differentiation into mature bacteroids.
Proteomic Analysis of Differential Protein Expression
Proteomics analysis identified a total of 2,457 peptides with molecular weights ranging from 7 to 320 kDa. A total of twenty-seven proteins (P < 0.05) were identified that were differentially expressed between the mutant RlgpxA bacteroids and wild-type RL3841 bacteroids (Table 4). Among them, 20 proteins were down-regulated in gpxA mutant nodule bacteroids and seven proteins were up-regulated (Table 4). Subcellular localization of the 27 differentially expressed proteins showed that fourteen proteins were localized at the cytoplasm, nine proteins were localized in the periplasmic space, two proteins were located in the outer membrane, one protein was existed in the inner membrane, and one was extracellular protein (Table 4). Eight (29.63%) differential genes were localized on the megaplasmids pRL8 (n = 1, 3.70%), pRL9 (n = 2, 7.41%), pRL10 (n = 3, 11.11%), and pRL12 (n = 2, 7.41%). No differential genes were localized on the megaplasmids pRL11 and pRL7.
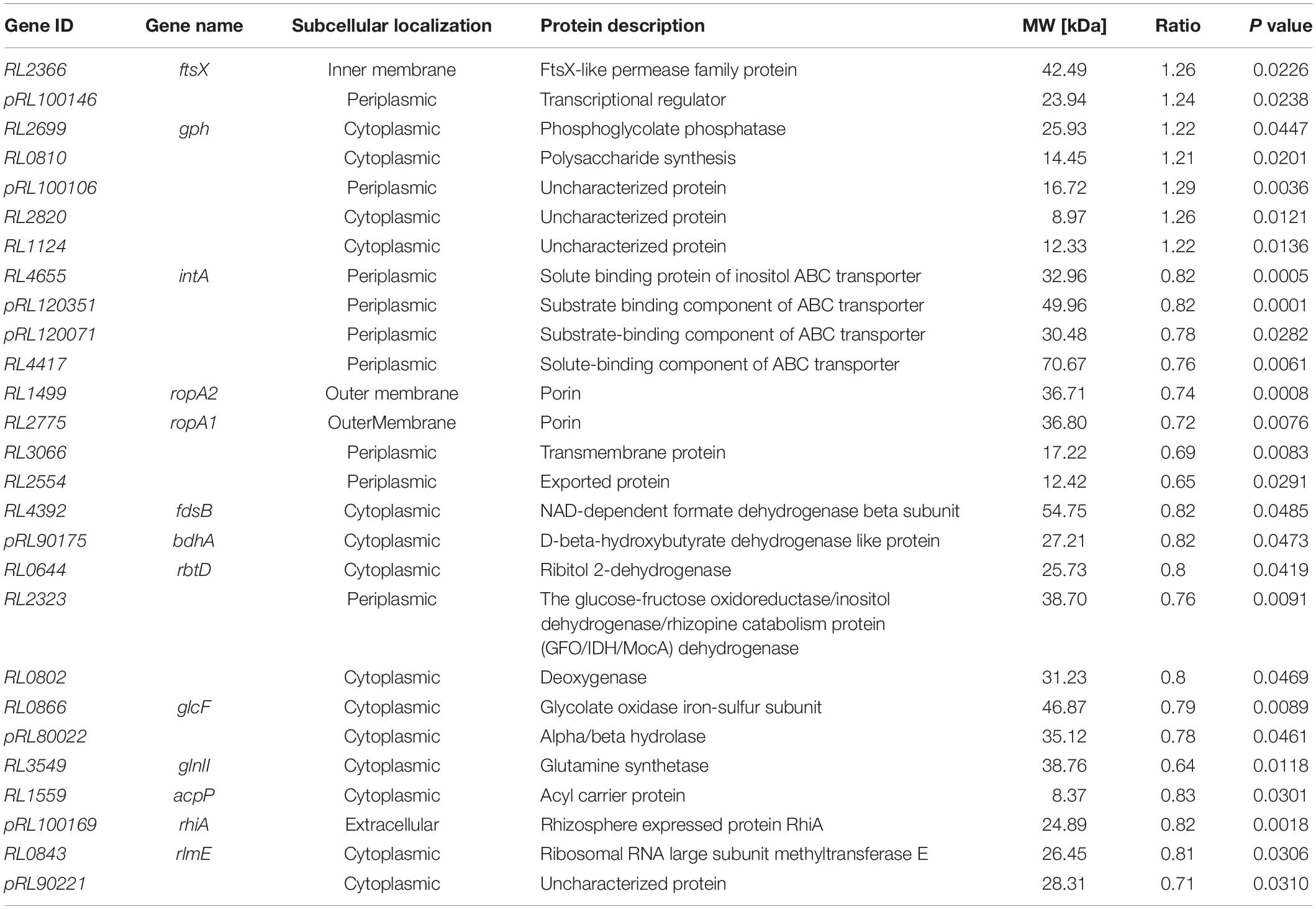
Table 4. Differential expression proteins in mutant-induced nodule bacteroids relative to wildtype-induced nodule bacteroids.
Among the seven up-regulated genes in nodules induced by the gpxA mutant, RL2366 codes for FtsX, which is an ABC transporter essential for efficient cell division in E. coli; pRL100146 codes for a transcriptional regulator; RL2699 and RL0810 code for a phosphoglycolate phosphatase and a polysaccharide synthesis, respectively (Table 4). In contrast, among the twenty down-regulated genes in nodules induced by the gpxA mutant, eight genes RL4665, pRL120351, pRL120071, RL4417, RL1499, RL2775, RL3066, and RL2554 code for membrane transport proteins, of which four were ABC-type transporters. Four genes RL4392, pRL90175, RL0644, and RL2323 code for dehydrogenases. RL0802, RL0866, and pRL80022 code for a deoxygenase, an oxidase and a hydrolase, respectively. Many of the differentially expressed genes display similarity to genes known to be involved in cellular metabolism and membrane transport processes. The number of differentially expressed metabolism-associated proteins indicated that Gpx played important roles in a wide variety of important metabolic processes by regulating the expression of genes encoding transporters, key enzymes, and other proteins involved in metabolic homeostasis. In addition, RL3549 codes for glutamine synthetase involved in assimilation of ammonium, RL1559 codes for the acyl carrier protein required for the biosynthesis of lipid A, and pRL100169 is a rhizosphere-expressed gene rhiA. These three genes have been reported by others to be functionally linked with symbiosis (Table 4) (Kannenberg and Carlson, 2001; Yadav et al., 2002; Crespo-Rivas et al., 2019). Finally, RL0843 codes Ribosomal RNA large subunit methyltransferase E, which is involved in ribosomal RNA maturation, and the loss of GpxA resulted in the differential expression of proteins (pRL100106, RL2820, RL1124, and pRL90221) of unknown function.
Discussion
The presence of GSH is likely to mean that requirement for the redox enzyme within the cell is crucially important to Rhizobium (Muglia et al., 2008; Taté et al., 2012). In R. leguminosarum, GSH plays a central role in both the growth of free-living bacteria and symbiotic nitrogen fixation (Cheng et al., 2017). Regulation of intracellular GSH/GSSG is a function of the flux through the two enzymes of the GSH redox cycle, Gpx and GR. In this study, the enzymes Gpx and GR were studied both alone and in combination. Our findings suggest as the key enzymes in the redox metabolism of GSH, both Gpx and GR are important for the cellular redox. However, the results also suggest that Gpx, but not GshR, is essential for efficient nodulation.
Glutathione has been reported to be required for the carbon uptake and has an absolute requirement for GSH for growth of R. leguminosarum 3841 (Cheng et al., 2017). Therefore, we hypothesized that Gpx or GR would have an additional function in cell growth. However, the growth rates and lag phases of R. leguminosarum 3841 were not affected by mutation in gpxA or gshR genes (Supplementary Figure 1). Although previous experimental evidences for reciprocal relationships between bacterial growth rate and Gpx ability remain unclear, mutation in glutathione reductase was able to influence markedly the growth of S. meliloti and Lactobacillus sanfranciscensis (Jänsch et al., 2007; Tang et al., 2018). According to the results of this study, we can suggest that besides through the Gpx/GR pathway, there must be another mechanism by which GSH regulates transport. In fact, it may well happen because a gor (GSH reductase) mutant of R. etli exhibited normal growth rate in medium with glucose and different amino acids (except glutamine) as sole organic source of carbon and energy (Taté et al., 2012).
Glutathione might activate a glutathione-glutathione reductase-glutathione peroxidase system to catalyze the reduction of H2O2 (Li et al., 2003). The effect of H2O2 stress under different concentration were investigated in this paper. As expected, mutants defective in both gpxA gene and gshR gene caused an increased sensitivity to H2O2 compared to wt strain RL3841. Insertion mutants of glutathione reductase gene were reported to be involved in defense against different oxides in R. etli and S. meliloti, with the former involved in H2O2 detoxification and the latter involved in diamide detoxification (Tatè et al., 2004; Tang et al., 2018). However, to date there have been no reports on the possible H2O2 detoxification for Gpx in rhizobia. Gpx is required for R. leguminosarum adaptation to oxidative stress, which could be due to the reaction of hydrogen peroxide with reduced GSH to form GSSG. To further study the underlying mechanism, intracellular levels of GSH and expression of two catalase genes (katG encoding for bifunctional catalase-peroxidase and katE encoding for monofunctional catalase) were analyzed. However, mutants in gpxA and gshR genes showed the opposite results. Mutant in GpxA had no significant effect on the content of GSH, but can increase the expression of the catalase genes, while mutant in GshR increased the content of GSH, which conversely suppresses expression of the catalase genes (Figure 1 and Table 2). Thus, there is likely to be complex influence of Gpx or GR on R. leguminosarum antioxidation, and it is not possible to conclude whether Gpx or GR is more important for antioxidation.
The role of the glutathione peroxidase/reductase enzyme system in nitrogen-fixing was investigated in nodulation of pea (Pisum sativum) after inoculation with R. leguminosarum strains. There was no statistically significant difference (P > 0.05) in numbers of pea nodules infected with the mutants RLgpxA, RLgshR, and RLgpxAgshR versus the parent strain at 21 dpi. Similarly, numbers of nodules on plants inoculated with S. meliloti gor and R. leguminosarum gshB mutants were similar to that inoculated with the wild-type strains (Cheng et al., 2017; Tang et al., 2018). The Gpx-deficient mutant elicited root nodules with a large decrease in the nitrogen-fixing activity (reduced to 53%). This defect in nodulation was characterized by the formation of the undeveloped nodules on pea, which is similar to the nodules induced by R. tropici gshB mutant on common bean (Muglia et al., 2008). Although the R. leguminosarum gshR mutant RLgshR cannot compete efficiently in the rhizosphere environment with wild-type RL3841, it formed normal nitrogen-fixing nodules on pea. The fact that mutation in gor gene of S. meliloti and gshB gene of R. leguminosarum was sufficient to affect bacterial growth and efficient nodulation, it might be hypothesized that the slow-growth defect of mutant strains can alter bacterial infection and plant nodulation (Cheng et al., 2017; Tang et al., 2018). Nevertheless, neither gpxA single mutant nor gpxA and gshR double mutant have differing effects on growth of R. leguminosarum 3841. Moreover, the R. leguminosarum gshB mutant was reported to be severely impaired in rhizosphere colonization, in contract, its gpxA mutant was normally grown in the rhizosphere environment. Furthermore, gpxA mutants of R. leguminosarum elicited the formation of nodules with lack of differentiation into mature bacteroids, while R. leguminosarum gshR formed normal-size nodules. Therefore, from the role of two glutathione enzymes investigated in the context of the differences in symbiotic phenotype, we may conclude that they might function in the different symbiotic stages, GpxA is involved in nitrogen-fixing bacteroid differentiation during pea nodule development, while GR is responsible for rhizosphere colonization at the early stage of symbiosis.
Previous reports indicated that elimination of hydrogen peroxide by glutathione peroxidase is a process of post-translational modification (Rhee et al., 2005), and many of these modifications are known to play central roles in the regulation of protein function. Henceforth, we performed a quantitative proteomic analysis to compare the protein profiles between the root bacteroids infected by gpxA mutant and wt RL3841. First, lack of GSH has been reported to lowers the transport rates of ABC transport systems (Tatè et al., 2004), similar result also happened in the gpxA mutant. Lack of GpxA reduced transcription of the Aap and Bra transporters under normal growth and down regulated eight transport proteins by proteomic analysis of nodule bacteroids (Tables 2, 4). These suggest that Gpx plays an important role in legume-rhizobial symbiosis and nodule development by maintaining the level of transport activity. Secondly, mutation in Gpx enzyme decreased the activity of redox enzymes such as dehydrogenases, deoxygenases, oxidase and hydrolase. Those enzymes were able to effectively reduced intracellular ROS production and involved in redox balance and respiration. Thirdly, three down-regulation proteins are directly involved in nodule process. Among them, glutamine synthetase has been reported to be involved in the nitric oxide signaling responses and play an important role in root nodules of Medicago truncatula by contributing to the antioxidant defenses system (Silva and Carvalho, 2013). Rhizobial acyl carrier protein (AcpP) was required for the biosynthesis of the flavonoid-inducible nodulation protein NodF (Geiger and López-Lara, 2002). In R. leguminosarum and S. meliloti, RirA participates in iron-dependent regulation, its mutant is hypersensitivity to oxidants and induced nodules accompanied by a significant reduction in the number of bacteria (Crespo-Rivas et al., 2019). Finally, RL0843 codes a ribosomal methyltransferase E, which contributes to RNA processing and modification. No studies have been reported that it was involved in rhizobial symbiosis or antioxidant function, but in thermophilic bacteria, this modification is important for environmental adaptation (Kuratani et al., 2014). Overall, these data reveal R. leguminosarum Gpx is required for redox homeostasis and symbiosis by involving the activity of transport systems, antioxidant and nodule function.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/pride/archive/projects/PXD022029.
Author Contributions
GC conceived the idea and designed the experiments. AH, XC, JX, and SL performed the experiments. GC, QZ, DH, and XL analyzed the data. GC, AH, and XC wrote the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (grant no. 31772399) and the Fundamental Research Funds for the Central Universities, South-Central University for Nationalities (grant no. CZY18022).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank PTM-Biolabs Co., Ltd. (Hangzhou, China) for mass spectrometry analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.627562/full#supplementary-material
References
Albino, A., Marco, S., Di Maro, A., Chambery, A., Masullo, M., and De Vendittis, E. (2012). Characterization of a cold-adapted glutathione synthetase from the psychrophile Pseudoalteromonas haloplanktis. Mol. Biosyst. 8, 2405–2414. doi: 10.1039/c2mb25116g
Allaway, B., Lodwig, E., Crompton, L. A., Wood, M., Parsons, R., Wheeler, T. R., et al. (2000). Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36, 508–515. doi: 10.1046/j.1365-2958.2000.01884.x
Barr, M., East, A. K., Leonard, M., Mauchline, T. H., and Poole, P. S. (2008). In vivo expression technology (IVET) selection of genes of Rhizobium leguminosarum biovar viciae A34 expressed in the rhizosphere. FEMS Microbiol. Lett. 282, 219–227. doi: 10.1111/j.1574-6968.2008.01131.x
Becana, M., Wienkoop, S., and Matamoros, M. A. (2018). Sulfur transport and metabolism in legume root nodules. Front. Plant Sci. 9:1434. doi: 10.3389/fpls.2018.01434
Beringer, J. E., and Hopwood, D. A. (1976). Chromosomal recombination and mapping in Rhizobium leguminosarum. Nature 264, 291–293. doi: 10.1038/264291a0
Bianucci, E., Tordable Mdel, C., Fabra, A., and Castro, S. (2008). Importance of glutathione in the nodulation process of peanut (Arachis hypogaea). Physiol. Plant. 134, 342–347. doi: 10.1111/j.1399-3054.2008.01126.x
Cheng, G., Karunakaran, R., East, A. K., Munoz-Azcarate, O., and Poole, P. S. (2017). Glutathione affects the transport activity of Rhizobium leguminosarum 3841 and is essential for efficient nodulation. FEMS Microbiol. Lett. 364:fnx045. doi: 10.1093/femsle/fnx045
Chung, L. Y., Chen, C. H., Wang, L. C., Chang, S. J., and Yen, C. M. (2010). Oxidative stress in mice infected with Angiostrongylus cantonensis coincides with enhanced glutathione-dependent enzymes activity. Exp. Parasitol. 126, 178–183. doi: 10.1016/j.exppara.2010.04.017
Cox, J., and Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Crespo-Rivas, J. C., Navarro-Gómez, P., Alias-Villegas, C., Shi, J., Zhen, T., Niu, Y., et al. (2019). Sinorhizobium fredii HH103 RirA is required for oxidative stress resistance and efficient symbiosis with soybean. Int. J. Mol. Sci. 20:787. doi: 10.3390/ijms20030787
Desideri, E., Filomeni, G., and Ciriolo, M. R. (2012). Glutathione participates in the modulation of starvation-induced autophagy in carcinoma cells. Autophagy 8, 1769–1781. doi: 10.4161/auto.22037
Fellay, R., Frey, J., and Krisch, H. (1987). Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52, 147–154. doi: 10.1016/0378-1119(87)90041-2
Figurski, D. H., and Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. doi: 10.1073/pnas.76.4.1648
Fujii, J., Ito, J. I., Zhang, X., and Kurahashi, T. (2011). Unveiling the roles of the glutathione redox system in vivo by analyzing genetically modified mice. J. Clin. Biochem. Nutr. 49, 70–78. doi: 10.3164/jcbn.10-138SR
Geiger, O., and López-Lara, I. M. (2002). Rhizobial acyl carrier proteins and their roles in the formation of bacterial cell-surface components that are required for the development of nitrogen-fixing root nodules on legume hosts. FEMS Microbiol. Lett. 208, 153–162. doi: 10.1111/j.1574-6968.2002.tb11075.x
Harrison, J., Jamet, A., Muglia, C. I., Van de Sype, G., Aguilar, O. M., Puppo, A., et al. (2005). Glutathione plays a fundamental role in growth and symbiotic capacity of Sinorhizobium meliloti. J. Bacteriol. 187, 168–174. doi: 10.1128/JB.187.1.168-174.2005
Heyob, K. M., Rogers, L. K., and Welty, S. E. (2008). Glutathione reductase targeted to type II cells does not protect mice from hyperoxic lung injury. Am. J. Respir. Cell Mol. Biol. 39, 683–688. doi: 10.1165/rcmb.2008-0112OC
Jänsch, A., Korakli, M., Vogel, R. F., and Gänzle, M. G. (2007). Glutathione reductase from Lactobacillus sanfranciscensis DSM20451T: contribution to oxygen tolerance and thiol exchange reactions in wheat sourdoughs. Appl. Environ. Microbiol. 73, 4469–4476. doi: 10.1128/AEM.02322-06
Johnston, A. W., and Beringer, J. E. (1975). Identification of the rhizobium strains in pea root nodules using genetic markers. J. Gen. Microbiol. 87, 343–350. doi: 10.1099/00221287-87-2-343
Kannenberg, E. L., and Carlson, R. W. (2001). Lipid A and O-chain modifications cause Rhizobium lipopolysaccharides to become hydrophobic during bacteroid development. Mol. Microbiol. 39, 379–391. doi: 10.1046/j.1365-2958.2001.02225.x
Karunakaran, R., Ebert, K., Harvey, S., Leonard, M. E., Ramachandran, V., and Poole, P. S. (2006). Thiamine is synthesized by a salvage pathway in Rhizobium leguminosarum bv. viciae strain 3841. J. Bacteriol. 188, 6661–6668. doi: 10.1128/JB.00641-06
Karunakaran, R., Haag, A. F., East, A. K., Ramachandran, V. K., Prell, J., James, E. K., et al. (2010). BacA is essential for bacteroid development in nodules of galegoid, but not phaseoloid, legumes. J. Bacteriol. 192, 2920–2928. doi: 10.1128/JB.00020-10
Khanyok, S., Liammongkolkul, S., Charoensak, Suwit, Ongajyooth, S., and Ongajyooth, L. (1997). Erythrocytic glutathione peroxidase and lipid peroxidation in chronic renal failure patients. Siriraj Med. J. 49, 758–762.
Köhler, A. C., Sag, C. M., and Maier, L. S. (2014). Reactive oxygen species and excitation-contraction coupling in the context of cardiac pathology. J. Mol. Cell Cardiol. 73, 92–102. doi: 10.1016/j.yjmcc.2014.03.001
Kovach, M. E., Phillips, R. W., Elzer, P. H., Roop, R. M., and Peterson, K. M. (1994). pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16, 800–802.
Kumar, S., Bourdès, A., and Poole, P. (2005). De novo alanine synthesis by bacteroids of Mesorhizobium loti is not required for nitrogen transfer in the determinate nodules of Lotus corniculatus. J. Bacteriol. 187, 5493–5495. doi: 10.1128/JB.187.15.5493-5495.2005
Kuratani, M., Yanagisawa, T., Ishii, R., Matsuno, M., Si, S. Y., Katsura, K., et al. (2014). Crystal structure of tRNA m(1)A58 methyltransferase TrmI from Aquifex aeolicus in complex with S-adenosyl-L-methionine. J. Struct. Funct. Genomics 15, 173–180. doi: 10.1007/s10969-014-9183-0
Li, Y., Hugenholtz, J., Abee, T., and Molenaar, D. (2003). Glutathione protects Lactococcus lactis against oxidative stress. Appl. Environ. Microbiol. 69, 5739–5745. doi: 10.1128/aem.69.10.5739-5745.2003
Liu, L., Xu, P., Zeng, G., Huang, D., Zhao, M., and Lai, C. (2014). Inherent antioxidant activity and high yield production of antioxidants in Phanerochaete chrysosporium. Biochem. Eng. J. 90, 245–254. doi: 10.1016/j.bej.2014.06.014
Luo, S., Yin, J., Peng, Y., Xie, J., Wu, H., He, D., et al. (2020). Glutathione is involved in detoxification of peroxide and root nodule symbiosis of Mesorhizobium huakuii. Curr. Microbiol. 77, 1–10. doi: 10.1007/s00284-019-01784-8
Muglia, C., Comai, G., Spegazzini, E., Riccillo, P. M., and Aguilar, O. M. (2008). Glutathione produced by Rhizobium tropici is important to prevent early senescence in common bean nodules. FEMS Microbiol. Lett. 286, 91–198. doi: 10.1111/j.1574-6968.2008.01285.x
Muñoz, V., Ibáñez, F., Figueredo, M. S., and Fabra, A. (2016). An oxidative burst and its attenuation by bacterial peroxidase activity is required for optimal establishment of the Arachis Hypogaea-Bradyrhizobium sp. symbiosis. J. Appl. Microbiol. 121, 244–253. doi: 10.1111/jam.13149
Oláhová, M., Taylor, S. R., Khazaipoul, S., Wang, J., Morgan, B. A., Matsumoto, K., et al. (2008). A redox-sensitive peroxiredoxin that is important for longevity has tissue- and stress-specific roles in stress resistance. Proc. Natl. Acad. Sci. U.S.A. 105, 19839–19844. doi: 10.1073/pnas.0805507105
Ortega-Villasante, C., Burén, S., Blázquez-Castro, A., Barón-Sola, Á, and Hernández, L. E. (2018). Fluorescent in vivo imaging of reactive oxygen species and redox potential in plants. Free Radic. Biol. Med. 122, 202–220. doi: 10.1016/j.freeradbiomed.2018.04.005
Poole, P. S., Blyth, A., Reid, C., and Walters, K. (1994). myo-Inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv. viciae. Microbiology 140, 2787–2795. doi: 10.1099/00221287-140-10-2787
Prell, J., Bourdès, A., Karunakaran, R., Lopez-Gomez, M., and Poole, P. S. (2009). Pathway of {gamma}-aminobutyrate (GABA) metabolism in Rhizobium leguminosarum 3841 and its role in symbiosis. J. Bacteriol. 191, 2177–2186. doi: 10.1128/JB.01714-08
Quandt, J., and Hynes, M. F. (1993). Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21. doi: 10.1016/0378-1119(93)90611-6
Rahman, I., Kode, A., and Biswas, S. K. (2006). Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 1, 3159–3165. doi: 10.1038/nprot.2006.378
Rhee, S. G., Yang, K. S., Kang, S. W., Woo, H. A., and Chang, T. S. (2005). Controlled elimination of intracellular H2O2: regulation of peroxiredoxin, catalase, and glutathione peroxidase via post-translational modification. Antioxid. Redox Signal. 7:619. doi: 10.1089/ars.2005.7.619
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., and Pühler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Silva, L., and Carvalho, H. (2013). Possible role of glutamine synthetase in the NO signaling response in root nodules by contributing to the antioxidant defenses. Front. Plant Sci. 4:372. doi: 10.3389/fpls.2013.00372
Smith, P. K., Krohn, R. I., and Hermanson, G. T. (1985). Measurement of protein using bicinchoninic acid. Anal. Biochem. 163, 76–85. doi: 10.1016/0003-2697(85)90442-7
Suvorava, T., and Kojda, G. (2009). Reactive oxygen species as cardiovascular mediators: lessons from endothelial-specific protein overexpression mouse models. Biochim. Biophys. Acta 1787, 802–810. doi: 10.1016/j.bbabio.2009.04.005
Tang, G., Li, N., Liu, Y., Yu, L., Yan, J., and Luo, L. (2018). Sinorhizobium meliloti glutathione reductase is required for both redox homeostasis and symbiosis. Appl. Environ. Microbiol. 17:e01937-17. doi: 10.1128/AEM.01937-17
Taté, R., Cermola, M., Riccio, A., and Diez-Roux, G. (2012). Patriarca EJ. Glutathione is required by Rhizobium etli for glutamine utilization and symbiotic effectiveness. Mol. Plant Microbe. Interact. 25, 331–340. doi: 10.1094/MPMI-06-11-0163
Tatè, R., Ferraioli, S., Filosa, S., Cermola, M., Riccio, A., Iaccarino, M., et al. (2004). Glutamine utilization by Rhizobium etli. Mol. Plant Microbe. Interact. 17, 720–728. doi: 10.1094/MPMI.2004.17.7.720
Temme, N., and Tudzynski, P. (2009). Does botrytis cinerea Ignore H(2)O(2)-induced oxidative stress during infection? Characterization of botrytis activator protein 1. Mol. Plant Microbe. Interact. 22, 987–998. doi: 10.1094/MPMI-22-8-0987
Wang, S., Lu, T., Xue, Q., Xu, K., and Cheng, G. (2019). Antioxidation and symbiotic nitrogen fixation function of prxA gene in Mesorhizobium huakuii. Microbiologyopen 8:e889. doi: 10.1002/mbo3.889
Yadav, A. S., Vasudeva, M., Upadhyay, K. K., Sawhney, S. K., and Vashishat, R. K. (2002). Symbiotic effectiveness, rate of respiration and glutamine synthetase activity of sodium azide-resistant strains of Rhizobium leguminosarum biovar trifolii. Lett. Appl. Microbiol. 28, 466–470. doi: 10.1046/j.1365-2672.1999.00500.x
Yin, C., Tang, Y., Li, X., Yang, Z., Li, J., Li, X., et al. (2018). A single composition architecture-based nanoprobe for ratiometric photoacoustic imaging of glutathione (GSH) in living mice. Small 14:e1703400. doi: 10.1002/smll.201703400
Young, J. P., Crossman, L. C., Johnston, A. W., Thomson, N. R., Ghazoui, Z. F., Hull, K. H., et al. (2006). The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7:R34. doi: 10.1186/gb-2006-7-4-r34
Keywords: Rhizobium leguminosarum, glutathione peroxidase, antioxidant function, symbiotic nitrogen fixation, quantitative proteomics
Citation: Hu A, Chen X, Luo S, Zou Q, Xie J, He D, Li X and Cheng G (2021) Rhizobium leguminosarum Glutathione Peroxidase Is Essential for Oxidative Stress Resistance and Efficient Nodulation. Front. Microbiol. 12:627562. doi: 10.3389/fmicb.2021.627562
Received: 09 November 2020; Accepted: 04 January 2021;
Published: 09 February 2021.
Edited by:
Jacob George Malone, John Innes Centre, United KingdomReviewed by:
Barney Geddes, McMaster University, CanadaEsther Menendez, University of Evora, Portugal
Copyright © 2021 Hu, Chen, Luo, Zou, Xie, He, Li and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guojun Cheng, chengguojun@mail.scuec.edu.cn
†These authors have contributed equally to this work
 Aiqi Hu†
Aiqi Hu† Guojun Cheng
Guojun Cheng