- 1College of Laboratory Medicine, Hangzhou Medical College, Hangzhou, China
- 2Institute of Microbiology and College of Life Sciences, Zhejiang University, Hangzhou, China
- 3College of Biotechnology and Bioengineering, Zhenjiang University of Technology, Hangzhou, China
To colonize in the digestive tract of animals and humans, Yersinia pseudotuberculosis has to deal with reactive oxygen species (ROS) produced by host cells and microbiota. However, an understanding of the ROS-scavenging systems and their regulation in this bacterium remains largely elusive. In this study, we identified OxyR as the master transcriptional regulator mediating cellular responses to hydrogen peroxide (H2O2) in Y. pseudotuberculosis through genomics and transcriptomics analyses. OxyR activates transcription of diverse genes, especially the core members of its regulon, including those encoding catalases, peroxidases, and thiol reductases. The data also suggest that sulfur species and manganese may play a particular role in the oxidative stress response of Y. pseudotuberculosis. Among the three H2O2-scavenging systems in Y. pseudotuberculosis, catalase/peroxidase KatE functions as the primary scavenger for high levels of H2O2; NADH peroxidase alkyl hydroperoxide reductase (AhpR) and catalase KatG together are responsible for removing low levels of H2O2. The simultaneous loss of both AhpC (the peroxidatic component of AhpR) and KatG results in activation of OxyR. Moreover, we found that AhpC, unlike its well-characterized Escherichia coli counterpart, has little effect on protecting cells against toxicity of organic peroxides. These findings provide not only novel insights into the structural and functional diversity of bacterial H2O2-scavenging systems but also a basic understanding of how Y. pseudotuberculosis copes with oxidative stress.
Introduction
Oxidative stress caused by reactive oxygen species (ROS), including superoxide (O2–), hydrogen peroxide (H2O2), and hydroxyl radical (OH⋅), is inevitable to all organisms that respire oxygen (Li et al., 2016). These strong oxidants and/or radicals could damage virtually all biomolecules, such as nucleic acids, proteins, and lipids (Imlay, 2013). Naturally, detoxification of ROS is extremely critical for survival of diverse bacteria, pathogens in particular, because the host cells release ROS as a deadly weapon to defend against bacterial infections (Fang, 2004). Among ROS, H2O2 not only can be formed very rapidly endogenously (for example, 15 μM s–1 in Escherichia coli) but also enters into cells nearly freely, where it reacts with Fe2+ to generate most deadly hydroxyl radicals (Li et al., 2016). Because of these features, most bacteria employ multiple enzymes, including catalases, various peroxidases, and rubrerythrin, to keep the intracellular concentrations of H2O2 at safe limit (nanomolar levels; Mishra and Imlay, 2012).
Catalases are principal enzymes that protect bacterial cells against H2O2 stress by catalyzing H2O2 into water and oxygen (Mishra and Imlay, 2012). Two types of catalases are present in bacteria: one is bifunctional (or called catalase-peroxidase, HPI, e.g., E. coli KatG), with both catalytic and peroxidatic activities, and the other is monofunctional (HPII, e.g., E. coli KatE), with only catalytic activity (Mishra and Imlay, 2012). Although it is well recognized that catalases serve as the primary scavenger of H2O2, they generally work best with H2O2 at high levels (millimolar levels; Mishra and Imlay, 2012). One of the best characterized peroxidases is NADH peroxidase AhpR (named from alkyl hydroperoxide reductase), which could decompose both H2O2 and organic peroxides (OPs) in diverse bacteria (Jacobson et al., 1989; Niimura et al., 1995). Compared to catalase, AhpR is more efficient in scavenging low-level H2O2 and regarded as the primary scavenger of endogenous H2O2 (Seaver and Imlay, 2001). AhpR typically consists of two cytoplasmic proteins encoded by a single operon, the peroxidase component AhpC and its cognate reductase AhpF: the former is a bacterial representative of typical 2-Cys peroxiredoxins and the latter is a flavoprotein with NADH:disulfide oxidoreductase activity (Poole, 2005). AhpC reduces H2O2 by oxidizing the two conservative cysteines to form an intermolecular disulfide bond, which is reactivated by AhpF using NADH as reducing equivalent (Perkins et al., 2015). Atypical AhpR systems, in which AhpF is absent, have been found in some bacteria (Bryk et al., 2000; Baker et al., 2001). For instance, in Helicobacter pylori, AhpC is reduced by a thioredoxin/thioredoxin reductase (TrxB) system, and in Mycobacterium tuberculosis, AhpD reduces AhpC with electrons from NADH through dihydrolipoamide dehydrogenase and dihydrolipoamide succinyltransferase (Baker et al., 2001; Jaeger et al., 2004). Because of the essential roles of catalase and AhpR in H2O2-scavenging, bacterial strains lacking both together display a drastically elevated sensitivity to H2O2 and carry an apparent aerobic growth defect (Cosgrove et al., 2007; Ezraty et al., 2017).
In many bacteria, OxyR is a primary transcriptional regulator that mediates cellular response to oxidative stress (Imlay, 2013; Fu et al., 2015). As a LysR-family DNA-binding protein, OxyR senses and responds to H2O2 stress via formation of an intramolecular disulfide bond between two conserved cysteine residues [Cys 199 and Cys 208 within E. coli OxyR (EcOxyR); Zheng et al., 1998]. The regulatory mode of OxyR varies among bacteria. For example, in E. coli, Salmonella enterica serovar Typhimurium (S. Typhi), and many other bacteria, OxyR functions as an activator only for major H2O2-scavenging proteins, such as catalases and AhpR (Imlay, 2008). However, it can also function in a dual-control manner (both a repressor and an activator for certain genes) in Neisseria and Shewanella oneidensis or as a repressor only in Corynebacterium diphtheriae (Ieva et al., 2008; Kim and Holmes, 2012; Jiang et al., 2014). OxyR proteins now have been generally regarded as a global regulator implicated in diverse biological processes, but the core members of their regulons are consistently constituted by operons responsible for H2O2 degradation such as catalases and peroxidases, iron-sequestering proteins, and thioredoxin and glutathione antioxidant systems (Imlay, 2015).
Yersinia pseudotuberculosis is a Gram-negative enteric pathogen that often causes self-limiting gastrointestinal disorders such as enteritis, diarrhea, and mesenteric lymphadenitis to animals and humans (Brady et al., 2020). During infection, environmental stress and host immunity reactions in human guts can introduce an increase in levels of ROS that Y. pseudotuberculosis cells encounter, thus requiring a sophisticated regulation of genes to reduce the intracellular H2O2 level to a safe line (Knaus et al., 2017). It has been reported that Y. pseudotuberculosis is able to combat oxidative stress by an unconventional way: it imports zinc to mitigate ROS by secreting a zincophore via the type VI secretion system (T6SS; Wang et al., 2016, 2020). However, how this bacterium copes with oxidative stress has not been systematically investigated and thus is poorly understood.
In this study, we carried out the transcriptomics analysis of Y. pseudotuberculosis YPIII in response to exogenous H2O2. We identified an OxyR analog in YPIII and found that its regulon members are highly conserved, including those encoding H2O2-scavenging enzymes, iron-sequestering proteins, and thiol-reducing systems. Our data support that OxyR of YPIII functions in an activator-only mode, and its absence causes a plating defect on LB agar, a result of insufficient production of H2O2-scavenging enzymes, including KatE, KatG, and AhpC. We further showed that KatE functions as the primary scavenger for H2O2, but both KatG and AhpC together play an essential role in decomposing low levels of H2O2. The simultaneous loss of both KatG and AhpC results in activation of OxyR, which in turn upregulates KatE production, conferring cells’ enhanced resistance to H2O2. The findings presented here reveal a novel mechanism through which bacterial cells differentially exploit individual components of the H2O2-scavenging repertoire to increase fitness in harsh living environments.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
All bacterial strains and plasmids used in this study are listed in Table 1. All chemicals were obtained from Sigma (Shanghai, China) unless otherwise noted. For genetic manipulation, E. coli and Y. pseudotuberculosis were grown in LB (containing 1% tryptone, 0.5% yeast extract, and 0.5% NaCl) under aerobic conditions at 37 and 26°C. When needed, the following chemicals were added to the growth medium: 2,6-diaminopimelic acid (DAP), 0.3 mM; ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; gentamycin, 15 μg/ml; and streptomycin, 100 μg/ml.
Transcriptomics Analysis
For transcriptomics analysis, cell samples were prepared as described previously (Jiang et al., 2014). In brief, cultures of the mid-exponential phase (∼0.4 of OD600, the same throughout the study) were subjected to the H2O2 treatment. H2O2 was added to a final concentration of 0.5 mM, and cells were collected before and 5 min after the addition. Cells were pelletted at 14,000 rpm for 30 s at room temperature and frozen immediately in liquid nitrogen. Three biological replicates under each condition were prepared. Total RNA was extracted with RNeasy Mini Kit (QIAGEN, Alameda, United States), and RNA sequencing (RNA-seq) analysis was conducted as before (Gao et al., 2004; Sun et al., 2020). Data analysis was conducted according to standard procedures used previously (Sun et al., 2020). Genes that passed statistical analysis by analysis of variance (ANOVA; p < 0.05) with Benjamini–Hochberg false discovery rate multiple-testing correction and showed two-fold difference between the H2O2-treated and untreated control samples were discussed in the study. NCBI SRA accession number is PRJNA671546 for raw transcriptomics analysis data.
Real-Time Quantitative RT-PCR
Quantitative reverse transcription-PCR (qRT-PCR) was performed to verify the expression of key OxyR regulon members with an ABI7300 96-well qRT-PCR system (Applied Biosystems) as described previously (Jiang et al., 2014). The expression of each gene was determined from four replicas in a single real-time qRT-PCR experiment. The cycle threshold (CT) values for each gene of interest were averaged and normalized against the CT value of the 16s rRNA gene, whose abundance was constant during the exponential phase. Relative abundance (RA) of each gene was standardized to the CT values of the 16s rRNA gene using the equation RA = 2–ΔCT.
Mutagenesis, Complementation of Mutant Strains
Yersinia pseudotuberculosis in-frame deletion strains were constructed by the att-based Fusion PCR method as described previously (Jin et al., 2013). In brief, two fragments flanking the target gene were generated by PCR with primers containing attB and the gene-specific sequence, which were linked by a linker sequence via second round of PCR. The fusion fragments were integrated into plasmid pHGM01 by site-specific recombination using Gateway BP clonase II enzyme mix (Invitrogen). The resulting vectors were introduced in E. coli WM3064 and transferred to Y. pseudotuberculosis by conjugation. Integration of the mutagenesis constructs into the chromosome was selected by resistance to gentamycin and confirmed by PCR. Verified trans-conjugants were grown in LB broth without NaCl and plated on LB supplemented with 10% sucrose. Gentamycin-sensitive and sucrose-resistant colonies were screened by PCR for deletion of the target gene. To facilitate growth of mutants, catalase (from bovine liver, Sigma) was added onto the plates. All mutations were verified by sequencing the mutated regions.
Plasmid pHG-101 was used in genetic complementation of mutants as described before (Wu et al., 2011). For complementation of genes next to their promoter, a fragment containing the gene of interest and its native promoter was amplified by PCR and cloned into pHG-101. After sequencing verification, the resulting vectors were transferred into the relevant strains via conjugation.
Growth and Susceptibility to H2O2 or t-BHP
The spotting assay was used to evaluate the plating defect on LB plates. Cells of the mid-exponential phase were collected by centrifugation and adjusted to 109 cells/ml, which was set as the undiluted (dilution factor 0). Ten-fold serial dilutions were prepared with fresh LB medium. Five microliter of each dilution was spotted onto LB plates. The plates were incubated for 24 h or longer in the dark before being photographed. All experiments were repeated at least three times.
Disk diffusion assays to test for sensitivity to oxidative stress conditions were performed with Y. pseudotuberculosis strains. Two hundred microliter of mid-exponential phase cultures were spread onto LB plates, 6 mm (in diameter) paper disks loaded with 10 μl H2O2 or tert-butyl hydroperoxide (t-BHP) of various concentrations were placed onto the bacterial lawn grown for 6 h, and plates were incubated 26°C for 16 h.
H2O2 Quantification
H2O2 at high concentrations (>100 μM) was quantified using the ferrous ion oxidation-xylenol orange method (Wolff, 1994). In brief, cells of the mid-exponential phase grown in liquid LB were collected by centrifugation, washed in PBS, and resuspended in the same buffer to an OD600 of 0.1. H2O2 was added to final concentrations indicated in the figure legends. Cells were filtered out at different time points, and elutions were assayed immediately for the remaining H2O2.
H2O2 at low concentrations (<50 μM) was quantified using Amplex red fluorescent method (Seaver and Imlay, 2001). In the presence of H2O2, Amplex red can be oxidized by horseradish peroxidase to the fluorescent product resorufin. To measure H2O2, 200 μl of samples from cells grown in MS medium was mixed with 100 μl of stock solutions for Amplex red and horseradish peroxidase prepared the same as described elsewhere (Seaver and Imlay, 2001). Fluorescence was then measured in a Synergy 2 Pro200 Multi-Detection Microplate Reader (Tecan) and converted to H2O2 concentration using a curve obtained from standard samples.
β-Galactosidase Activity Assay
β-Galactosidase activity assay was used to determine gene expression. The sequence in sufficient length (∼400 bp) upstream of gene of interest was amplified and inserted in front of the full-length E. coli lacZ gene in plasmid pHGEI01 (Fu et al., 2014). The resulting plasmid was verified by sequencing, introduced into E. coli WM3064, and then conjugated with relevant Y. pseudotuberculosis strains. Cultures of the mid-exponential phase were collected by centrifugation, washed with PBS, and treated with lysis buffer (0.25 M Tris/HCl, 0.5% Triton X-100, and pH 7.5). Extracts were collected by centrifugation and applied for enzyme assay by adding o-nitrophenyl-β-D-galactopyranoside (4 mg/ml). Changes in absorption over time were monitored at 420 nM with a Synergy 2 Pro200 Multi-Detection Microplate Reader (Tecan), and the results were presented as Miller units.
Bioinformatics and Statistical Analyses
Multiple sequence alignment was carried out with Clustal Omega (Madeira et al., 2019). Sequence logos were generated by using WebLogo (Crooks et al., 2004). Three-dimensional structures of YpAhpC were predicted using Phyre1 (Kelley et al., 2015). The predicted structures were then visualized by software Pymol (DeLano Scientific LLC). Genome screening for OxyR-binding sites based on established weight matrixes from various bacteria was performed using regulatory sequence analysis tools (RSATs; Medina-Rivera et al., 2015). For statistical analysis, Student’s t-test was performed for pairwise comparisons of groups, and values are presented as means ± standard deviation (SD).
Results
Genomics Analysis of Y. pseudotuberculosis YPIII With Respect to Oxidative Stress Response
To identify the oxidative stress response regulator(s) in YPIII, a BLASTp search of functional analogs of established oxidative stress-responding regulators, including EcOxyR and SoxRS as well as Bacillus subtilis PerR, against the YPIII proteome was performed. While no homolog of E. coli SoxRS or of B. subtilis PerR was found, YPIII possesses a highly confident homolog of EcOxyR, YPK_RS20585 (BLASTp E-value = 0; identities, 88%; Supplementary Figure 1). Like all 2-Cys OxyRs, YPK_RS20585 contains two conserved cysteine residues (Cys199 and Cys208 within both YPK_RS20585 and EcOxyR) implicated in the activation by disulfide bond formation, suggesting a possible role in the oxidative stress response of YPIII, and therefore we named it OxyR (YpOxyR). However, the sequence similarities between YpOxyR and dual-activity OxyRs, such as that of S. oneidensis, are substantially lower (E-value = 7e-47; identities, 33%; Supplementary Figure 2), implying that YpOxyR might function as an activator only.
The current knowledge on bacterial oxidative stress response is most well developed in E. coli, whose OxyR regulon is composed of over 20 operons (Imlay, 2015). The most important and conserved EcOxyR regulon members encode proteins involved in detoxification and prevention and/or repair of oxidative damage, such as catalases and peroxidases, iron-sequestering proteins, thioredoxin, and glutathione antioxidant systems. Similarly, the YPIII genome encodes two catalases (HPII KatE and HPI KatG), iron-sequestering protein Dps, and a complete set of thioredoxin and glutathione antioxidant proteins, including TrxA (thioredoxin), TrxB (thioredoxin-disulfide reductase), TrxC (thioredoxin), GrxA (glutaredoxin), YPK_RS20590 (glutathione peroxidase), and GorA (glutathione-disulfide reductase; Table 2).
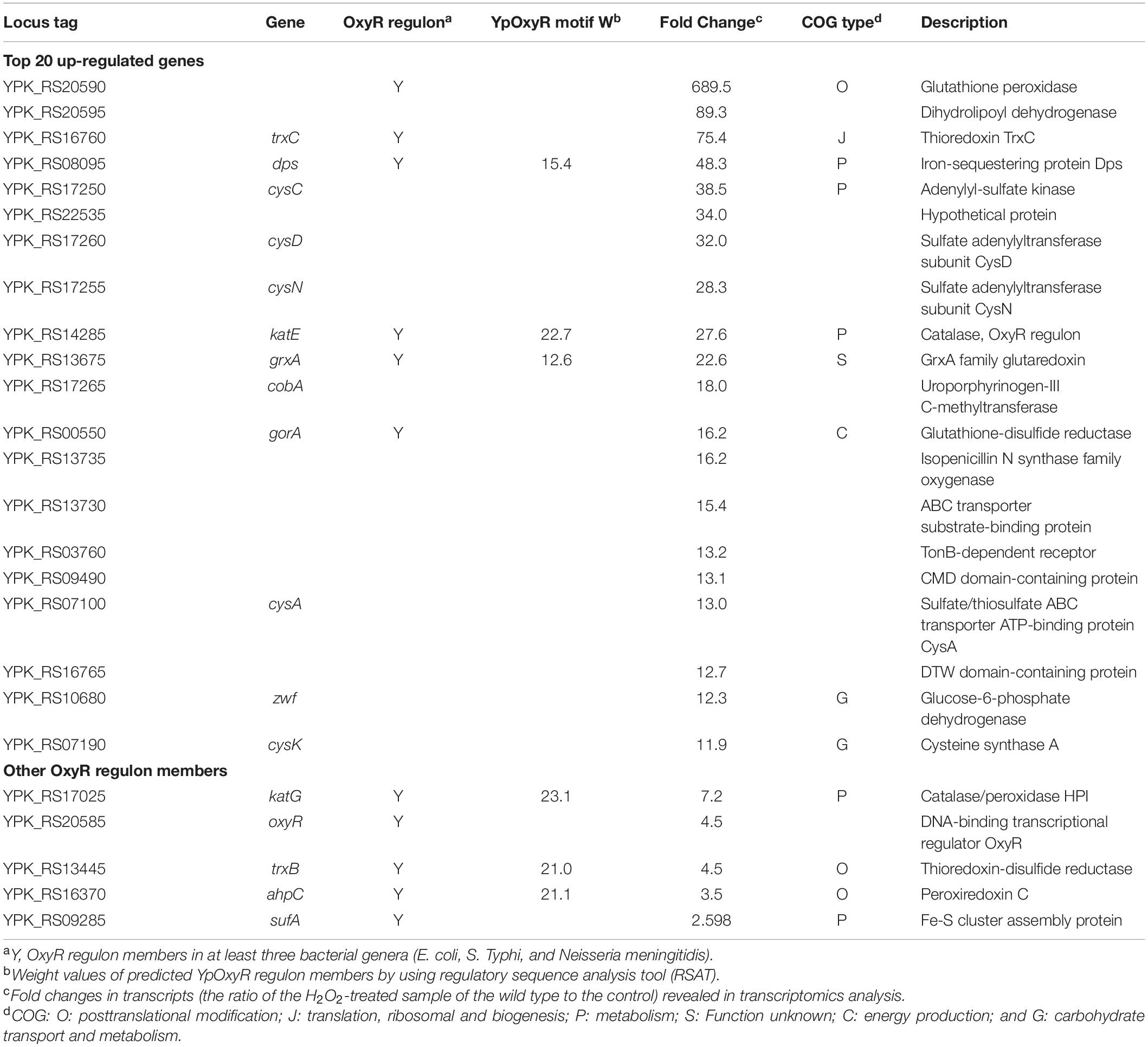
Table 2. Highly regulated genes involved in the response to H2O2 and genes encoding analogs of OxyR regulon members.
Perhaps one of the most striking observations is that YPIII seemingly lacks the counterpart of E. coli AhpF, the cognate reductase for peroxidase component AhpC of AhpR (Tartaglia et al., 1990; Poole and Ellis, 1996). A BLASTp search using E. coli AhpCF against the YPIII proteome revealed a single putative homolog for AhpC and AhpF, YPK_RS16370 (peroxiredoxin C; E-value, 3e-38) and TrxB (E-value, 3e-42), which are not in proximity on the chromosome. Combining the fact that AhpCF is encoded by a single operon in other bacteria and TrxB is clearly the counterpart of EcTrxB (E-value, 0) but not EcAhpF, these data strongly suggest that YPIII lacks a conventional AhpF. Furthermore, the YPIII genome also lacks a gene for a homolog of Ohr, the primary enzyme that decomposes OPs. Given the involvement of AhpR in scavenging OPs, thus it is particularly important and interesting to understand the physiological role of AhpC in YPIII.
Transcriptomics Analysis of YPIII in Response to H2O2 Stress
In order to gain a comprehensive understanding of the cellular response of YPIII to H2O2, we performed an RNA-seq analysis to obtain gene expression changes resulting from the H2O2 treatment. Our early stress response studies suggest that the best concentration of H2O2 for transcriptomics analyses would be the dosage at which the agent arrests growth of cells but does show an evident killing effect on them (Gao et al., 2004; Jiang et al., 2014). To determine the concentration, the minimum inhibitory concentration (MIC) of H2O2 against the wild-type strain of YPIII was assessed. The results revealed MIC to be 4 mM, which is four times lower than that of E. coli (Figure 1A), indicating that YPIII is significantly more sensitive to H2O2 than E. coli. Then, impacts of H2O2 over a range of concentrations (under MIC) on growth and viability of actively growing cells (∼0.4 of OD600) were examined. Upon addition of H2O2 at 0.2 mM or higher, growth paused immediately and resumed after lag periods that increase with the H2O2 concentrations (Figure 1B). Additionally, viability assays revealed that H2O2 at 0.5 mM or lower did not show a significant killing effect (Supplementary Figure 3). Based on these observations, for RNA-seq transcriptomics analysis, we collected cells before and 5 min after the addition of 0.5 mM H2O2 as the untreated control and the treated samples, respectively.
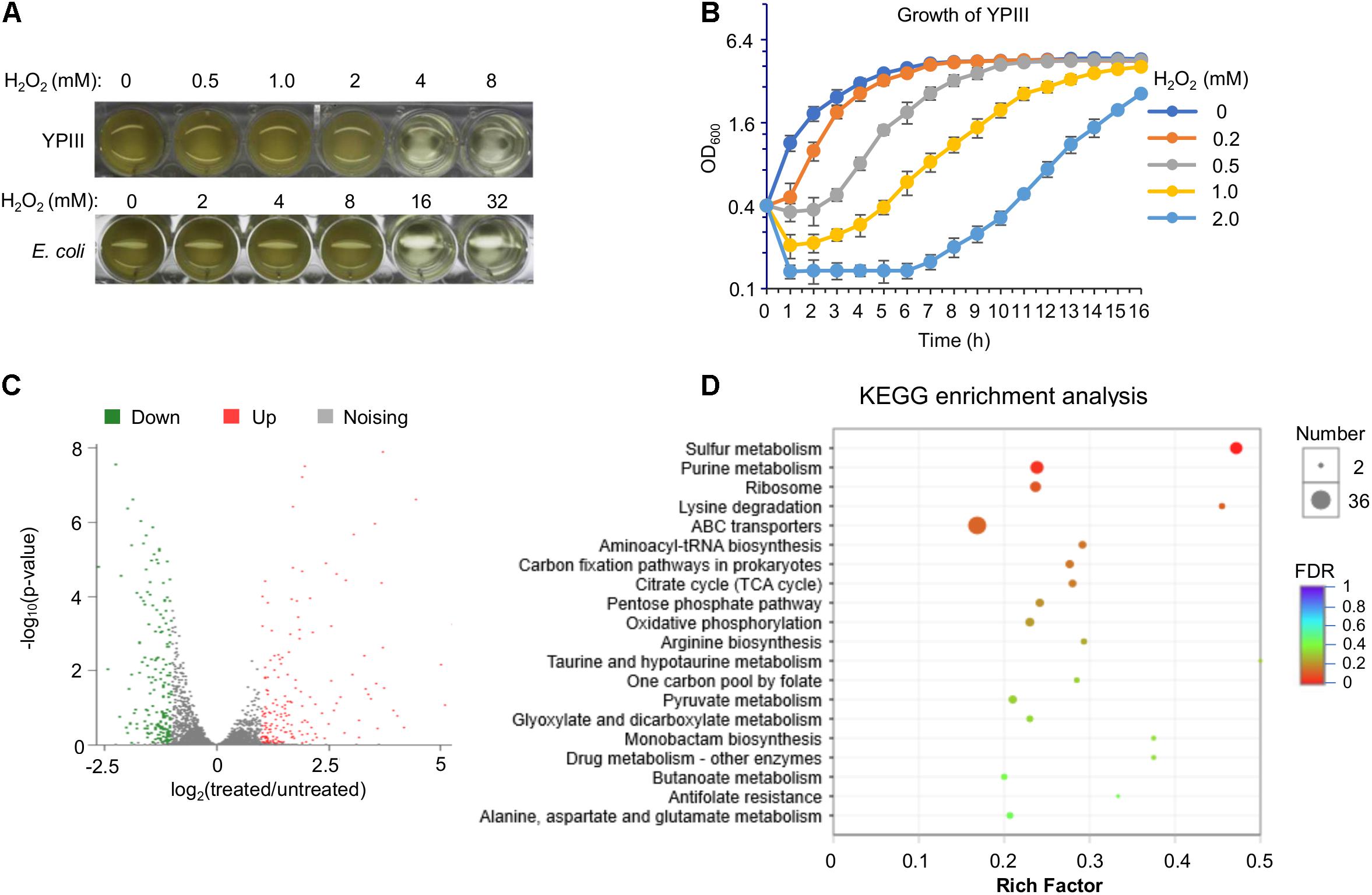
Figure 1. Characteristics of Y. pseudotuberculosis in response to H2O2. (A) Minimum inhibitory concentration (MIC) assay. Mid-exponential phase cultures (OD600 of ∼0.4) were used to inoculate each well to an OD600 of 0.01, and MIC was determined 16 h later. (B) Impact of H2O2 on growing cells. H2O2 was added to mid-exponential phase cultures of the wild type to the final concentrations as indicated. Growth was monitored by recording OD600 values. (C) Volcano plot of the different expression genes in wild-type cells between before and after the H2O2 treatment (0.5 mM). (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed genes in wild-type cells between before and after the H2O2 treatment.
In total, 364 genes displayed significant differences in transcription levels between the H2O2-treated and untreated control samples (Supplementary Table 1). Among these genes, 186 genes were up-regulated, while 178 genes were down-regulated (Figure 1C). Genes displaying significant differences in expression levels due to H2O2-induced oxidative stress were observed in almost every functional category, indicating that YPIII transcriptionally responds to H2O2 in a global scale (Supplementary Table 1). The high quality of the expression data was validated with a statistical analysis as previously described (Gao et al., 2004) and by real-time qRT-PCR. Eight genes were selected for analysis with the same RNA samples used in the RNA-seq based on the level and reproducibility of changes observed in the RNA-seq experiments. A high level of concordance (R2 = 0.96) was observed between RNA-seq and real-time qRT-PCR data despite quantitative differences in the level of change (Supplementary Figure 4), suggesting that the RNA-seq results are an accurate reflection of the gene expression profile. The highly induced included many genes encoding proteins combating oxidative stress. In contrast, genes encoding metabolic enzymes were down-regulated in general (Figure 1D), an observation that reflects a paused/reduced growth rate rather than a specific response to the stress has been commonly found in other stress response studies (Gao et al., 2004; Jiang et al., 2014). Intriguingly, many ABC transporters and sulfur metabolic enzymes are among differentially expressed genes, implicating that sulfur species and certain small molecules may have a particular significance in the oxidative stress response of YPIII.
To stay focused, here we only discussed genes whose transcription was significantly induced upon the addition of H2O2, especially those specific to H2O2-induced oxidative stress (Table 2). With respect to OxyR regulon members, our transcriptomics data revealed that YPK_RS20590 (glutathione peroxidase), trxC, dps, katE, grxA, and gorA were among the top 20 most up-regulated genes in H2O2-treated cells (Table 2). Other putative OxyR regulon members that were induced but not among the top 10 included katG, trxB, ahpC, and sufABC (Fe-S cluster assembly proteins; Table 2). Intriguingly, the entire thioredoxin and glutathione antioxidant systems, which play a supporting role in combating oxidative stress in many bacteria (Feng et al., 2019), were found to be highly induced by H2O2 (for instance, YPK_RS20590 was induced 689-fold), implying that these systems may have more significant contribution in protecting YPIII cells from oxidative damage. It is also worth mentioning that the oxyR gene was transcribed at substantially elevated levels (4.5-fold) upon H2O2 stress (Table 2), suggesting that the regulator per se is also subjected to quantity control, in addition to activity transformation upon oxidation.
Among the remaining top 20 most significantly induced genes, those for sulfur species transport and metabolism (cysC, cysD, cysN, cysA, and cysK) drew special attention. In many organisms, sulfur species are critically involved in cell protection against oxidative damage (Wu et al., 2015; Peng et al., 2017; Korshunov et al., 2020). Along with highly induced transcription of cysJ, cysW, cysT, and cysI in H2O2-challenged cells, these data suggest that sulfur species may be critical for oxidative stress response in YPIII. In addition, we also noticed that genes encoding iron/manganese ABC transport system (yfeA, yfeB, yfeC, and yfeD) were transcribed at significantly elevated levels upon the treatment. This may not be surprising given that the intracellular Mn/Fe ratios are correlated with bacterial resistance to oxidative stress (Chandrangsu et al., 2017). To maintain the activity of vulnerable mononuclear iron proteins in the presence of H2O2, many bacteria replace the iron atom with manganese as the active center (Anjem et al., 2009; Imlay et al., 2019). These data concur well with the result from the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis in which sulfur metabolic pathways are among the most enriched (Figure 1D). Together with the missing of AhpF, these results suggest that the mechanism underlying the response of YPIII to H2O2-induced oxidative stress carries novel characteristics that are different from the classical mode established in E. coli.
Characterization of YPIII oxyR Mutant
In many bacteria, OxyR-binding motifs have been determined. Given sequence and function conservation, it is not surprising that these motifs are highly similar (Wan et al., 2018). To predict the operons under the direct control of YpOxyR, we constructed a matrix for the OxyR-binding motif with verified OxyR-binding sequences from closely related bacterial species and used it to screen the entire genome of YPIII by RSAT (Medina-Rivera et al., 2015). In total, 38 putative YpOxyR-binding sites were identified with weight values greater than 7, an arbitrary cutoff implemented in RSAT (Supplementary Table 2). On the top of the list are well-established OxyR regulon members, including katG, katE, ahpC, trxB, grxA, dps, and trxC, supporting that YpOxyR recognizes a similar DNA motif (Figure 2A). For the remaining genes on the list, we supposed that many of them may not belong to the YpOxyR regulon because weight values of their binding motifs are relatively low (≤10). But this requires further verification.
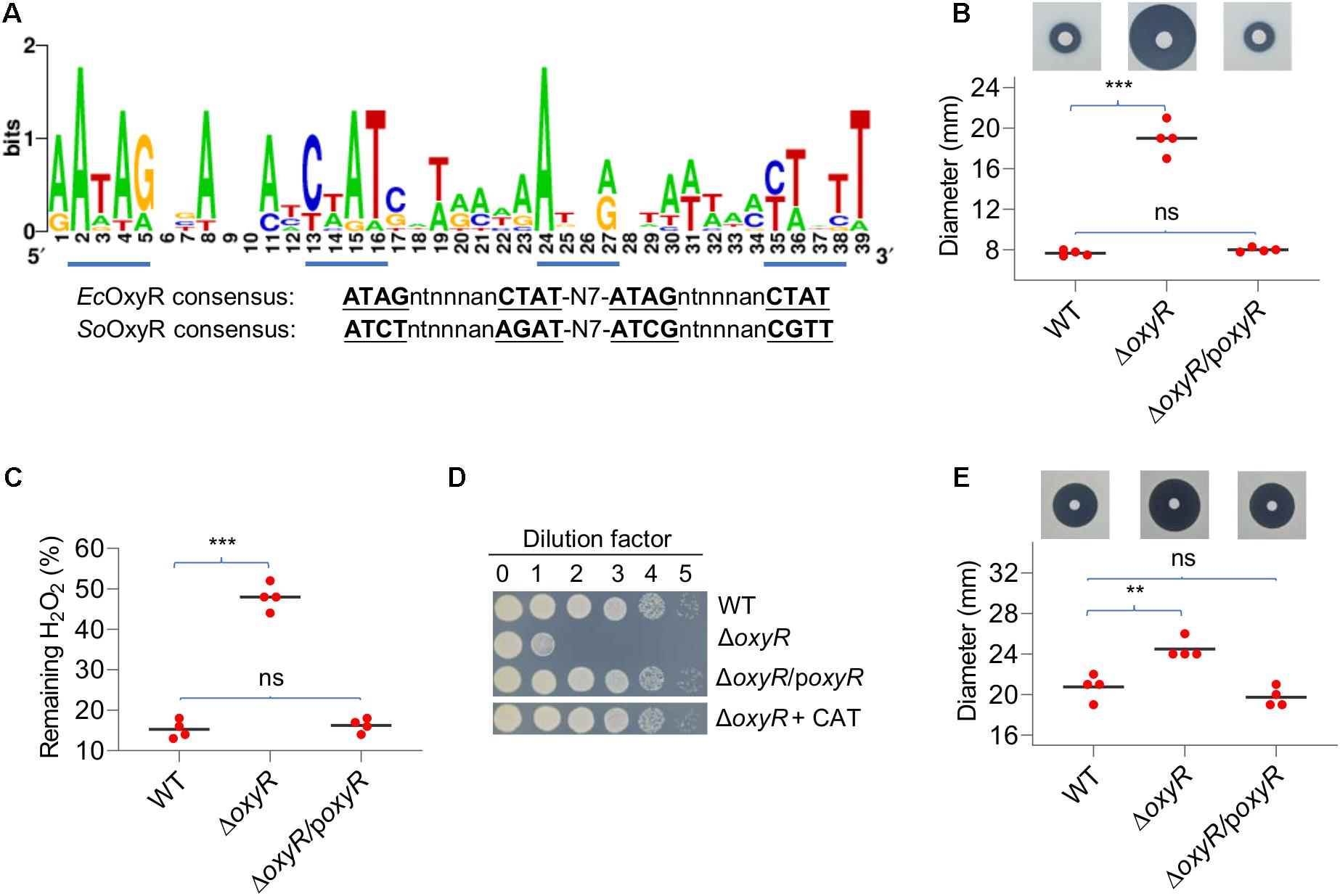
Figure 2. Characteristics of the Y. pseudotuberculosis oxyR mutant. (A) OxyR-binding motifs in YPIII derived from the top 20 members of the predicted regulon. The matrix for screening the YPIII genome was generated using most conserved members of OxyR regulons of diverse bacteria. Tetranucleotide sequences are underlined based on the E. coli and S. oneidensis OxyR consensus. (B) Disk diffusion assay for H2O2. Paper disks loaded with 10 μl of 5 M H2O2 were placed onto bacterial lawns (pregrown for 6 h). Results shown are from 24 h after the disks were placed. The sensitivity was represented by the diameter of the inhibition zone. poxyR represents a copy of oxyR to be expressed in trans for complementation. (C) Measuring H2O2-scavenging rates. Cells grown to the mid-exponential phase were collected, washed, and suspended to an OD600 of ∼0.4. 5 min after the addition of 0.5 mM H2O2, the remaining H2O2 was determined. The data were normalized to the initial concentration. (D) Plating defects of the oxyR mutant. Mid-exponential phase cultures were adjusted to ∼108 CFU/ml and diluted by dilution factor (10-fold serial dilution), and 5 μl of each dilution was spotted onto LB plates. Photos were taken 24 h after plating. CAT, catalase (2,000 Units/ml). Experiments were performed at least four times, and representative results were presented. (E) Disk diffusion assay for tert-butyl hydroperoxide (t-BHP). Paper disks loaded with 10 μl of 5 M t-BHP. In panels B, C, E, asterisks indicate statistically significant differences of the values compared (n = 4; ns, not significant; **p < 0.01; and ***p < 0.001).
To determine the impacts of OxyR on oxidative stress response, an oxyR in-frame deletion strain (ΔoxyR) was constructed and characterized in YPIII. In liquid LB, ΔoxyR grew indistinguishably from the wild type (Supplementary Figure 5), indicating that YpOxyR is not required for normal growth. We then assessed the role of YpOxyR in combating oxidative stress. Disk diffusion assays demonstrated that ΔoxyR was more sensitive to H2O2, generating inhibition zones that were substantially larger than those of the wild type (Figure 2B). The phenotype can be confidently attributed to the loss of OxyR given successful genetic complementation. To provide further support to this, we compared the mutant and the wild type with respect to H2O2 consumption. Cells at the mid-exponential phase were collected, adjusted to identical densities, and incubated with 0.2 mM H2O2. As shown in Figure 2C, the ΔoxyR strain degraded H2O2 at a rate much lower than the wild type did; 14% and 49% of H2O2 remained 2 min after the reaction started for the wild type and the mutant, respectively. These results indicate that the oxyR deletion greatly impaired the ability of YPIII to remove exogenous H2O2. Furthermore, we also confirmed that the ΔoxyR strain carried a plating defect on LB agar plates (Figure 2D), a common phenotype of the oxyR mutants in bacteria such as E. coli and S. oneidensis because of compromised H2O2-scavenging capacity (Jiang et al., 2014). As exogenous catalase completely suppressed the plate defect of the ΔoxyR strain, it is reasonable to propose that the same mechanism is responsible for the YPIII oxyR mutant (Jiang et al., 2014; Shi et al., 2015).
In many bacteria lacking Ohr, such as E. coli and S. Typhi, OxyR also mediates cellular response to OPs (Imlay, 2015). Clearly, this is also true in the case of YPIII because the oxyR mutant exhibited significantly increased sensitivity to t-BHP, a widely used representative OP (Figure 2E). All together, these data conclude that YPIII employs transcriptional regulator OxyR to mediate an oxidative stress response.
KatE Is the Primary Catalase in YPIII Positively Regulated by OxyR
In general, multiple catalases are encoded in a bacterial genome, but only one exhibits predominant H2O2-scavenging activity in vivo (Mishra and Imlay, 2012). To test whether this also holds true in YPIII, we assessed the contribution of catalases to decomposition of H2O2. Strains lacking katE and katG were generated and characterized with the disk diffusion assay and the H2O2-decomposing assay. As shown in Figure 3A, the katE mutant (ΔkatE) was hypersensitive to H2O2, generating inhibition zones that were approximately two and a half times larger than those of the wild type. On the contrary, the difference in zone sizes between the wild type and ΔkatG was insignificant. Consistent with the results in disk diffusion assay, the katE deletion resulted in severely impaired ability to decompose H2O2, with nearly 60% of H2O2 remaining 2 min after the reaction started (Figure 3B). In the case of ΔkatG, the assay revealed that this mutation slightly but still significantly compromised the ability of YPIII to decompose H2O2 when compared with that of ΔkatE, validating that KatG is also functioning in YPIII.
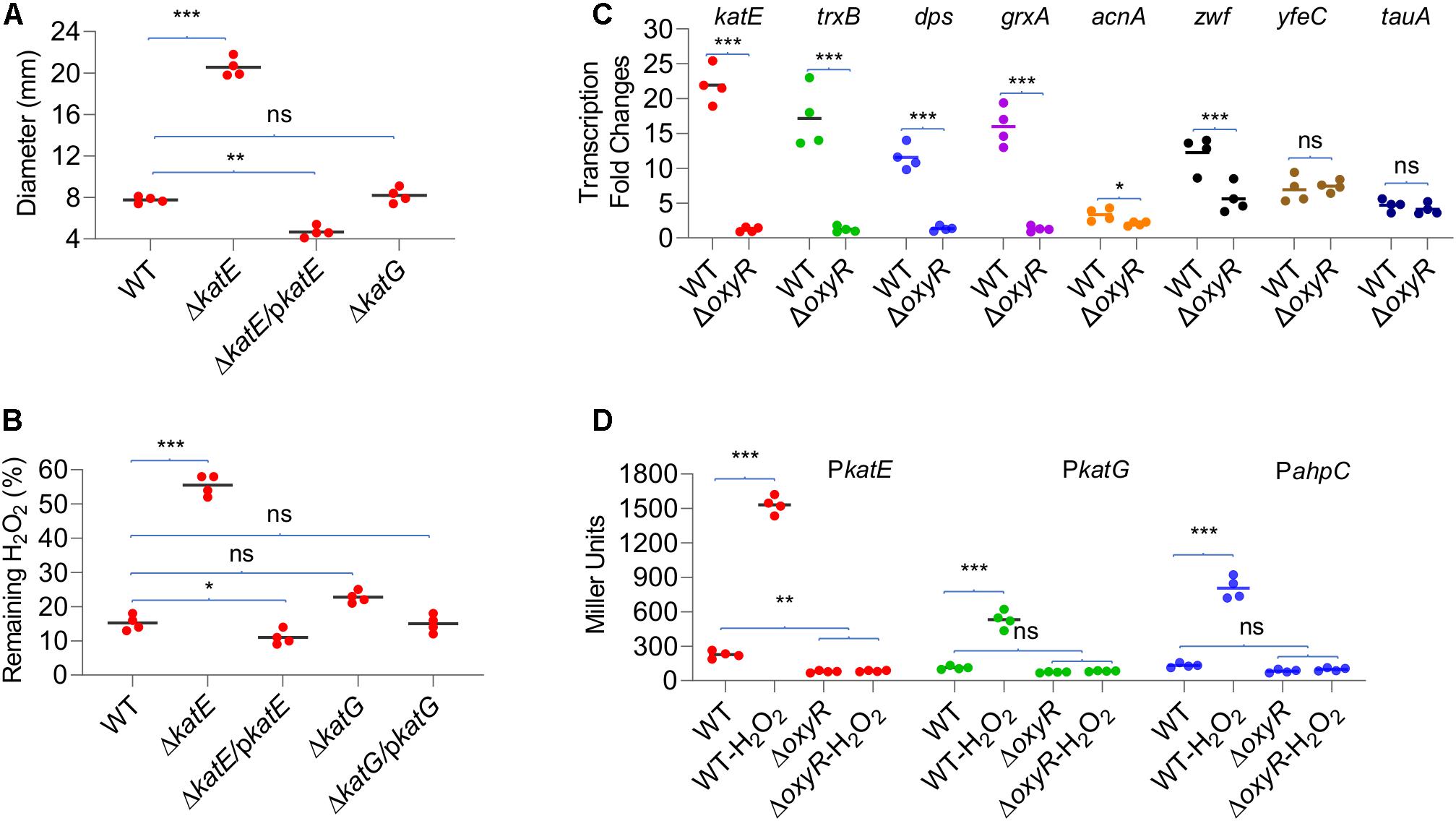
Figure 3. Role of catalases in decomposition of H2O2. (A) Disk diffusion assay of catalase mutants. (B) Measuring H2O2-scavenging rate of catalase mutants. (C) qRT-PCR analysis of transcription differences upon oxyR deletion. Genes with high-confident (katE, trxB, dps, and grxA) and low-confident (acnA and zwf) OxyR-binding motifs and without (yfeC and tauA) were examined. Signal intensities were processed, and the fold changes [values of the H2O2-treated wild type (WT) and ΔoxyR/values of the untreated WT] were calculated according to the method described in Materials and Methods. (D) Impacts of OxyR on the expression of katE, katG, and ahpC. Cells of mid-exponential phase before and 20 min after the H2O2 treatment were harvested for measuring the β-galactosidase assays using an integrative lacZ reporter. The activity of β-galactosidase represents the activity of indicated promoters. In all panels, asterisks indicate statistically significant differences of the values compared (n = 4; ns, not significant; *p < 0.05; **p < 0.01; and ***p < 0.001).
Impacts of YpOxyR on the expression of some of its regulon members predicted by the in silico analysis, including four high-confident (weight value > 12; trxB, dps, katE, and grxA) and two low-confident genes (weight value ≈9; acnA and zwf; Table 2 and Supplementary Table 2), were assessed by qRT-PCR. In cells of ΔoxyR prepared the same as for transcriptomics analysis, we found that none of the four high-confident genes were responsive to the H2O2 treatment, whereas two control genes, yfeC and tauA, which lack a predicted OxyR-binding motif, were upregulated upon exposure to H2O2 as in the wild type (Figure 3C). In the case of acnA and zwf, however, we found that the loss of OxyR negatively affected but not completely abolished their transcription in response to H2O2 (Figure 3C). Apparently, these data support that the predicted DNA motifs largely determine OxyR-dependent transcription.
Given the central role of catalases in protecting cells from H2O2 damage, we further compared the expression of katE and katG genes in the wild type and ΔoxyR strains by measuring activities of katE and katG promoters with an integrative lacZ reporter system (Figure 3D). In line with the transcriptomics data, the expression of katE and katG increased nearly six-fold in the wild-type cells when challenged by H2O2. While in the ΔoxyR strain, both genes were no longer responsive to H2O2, being expressed at levels even lower than those observed in the untreated wild-type cells. In summary, these data validate that expression of both KatE and KatG is under the positive control of OxyR, and KatE functions as the primary catalase to cope with H2O2 stress.
AhpC Possesses Scavenging Activity Against H2O2 but Not Organic Peroxides
The genomics analysis as presented above indicates that YPIII possesses an atypical AhpR, whose AhpF is missing. Like katE and katG, the ahpC gene is positively regulated by OxyR (Figure 3C), and therefore, the protein is likely produced when OxyR is activated. Given the importance of AhpR in scavenging both H2O2 and OPs, we reasoned that AhpC may be still functional in YPIII because it can also be reduced by TrxB and/or glutathione (GSH) reductase (Gor), albeit low in efficiency (Feng et al., 2020). To test this, we knocked out the ahpC gene from mutants lacking KatE, KatG, or both. The removal of AhpC alone had no detectable impact on H2O2 consumption (Figure 4A), an expected result because of the dominance of catalase in H2O2 degradation (Mishra and Imlay, 2012). In the absence of KatE, the effect of the AhpC loss became significant, indicating that AhpC does have some capacity of H2O2 degradation. Compared to ΔkatEΔahpC, the loss of both catalases (ΔkatEΔkatG) significantly impaired the ability of YPIII to decompose H2O2 (Figure 4A). Furthermore, additional removal of AhpC from ΔkatEΔkatG nearly completely abolished the H2O2-decomposing capacity of YPIII. Therefore, these data conclude that all of KatE, KatG, and AhpC are capable of decomposing H2O2 in YPIII. Intriguingly, we found that the strain lacking both KatG and AhpC had stronger H2O2-decomposing capacity than the wild type. All of these data were verified by genetic complementation, in which one of the missing genes was expressed in trans in deletion mutants (Supplementary Figure 6).
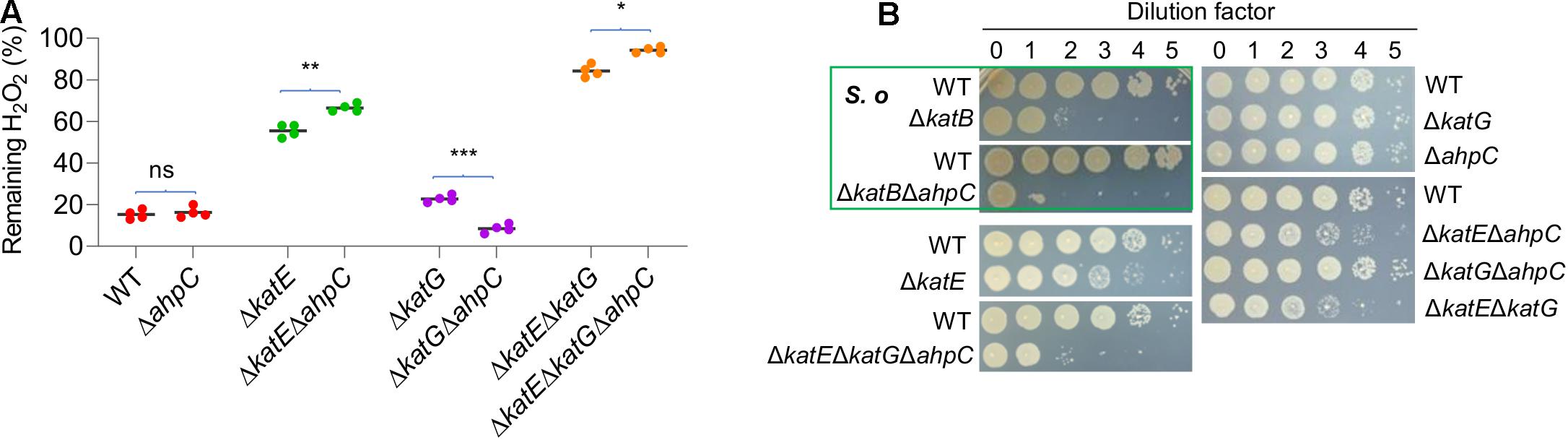
Figure 4. Role of AhpC in decomposition of H2O2. (A) Measuring the scavenging rate of H2O2 in indicated strains. Asterisks indicate statistically significant differences of the values compared (n = 4; ns, not significant; *p < 0.05; **p < 0.01; and ***p < 0.001). (B) Plating defects of the indicated strains. S. oneidensis represents S. oneidensis, which is shown for comparison. Strains outside the green box are YPIII. Experiments were performed at least four times, and representative results were presented.
To provide additional evidence, we then examined if the removal of catalases and/or AhpC would cause YPIII a plating defect, as this is also the case with S. oneidensis (Shi et al., 2015). Similar to an S. oneidensis katB (encoding dictating catalase) mutant, the ΔkatE strain of YPIII exhibited a plating defect, albeit much less severe (Figure 4B). In contrast, the removal of either katG or ahpC did not introduce a significant defect, supporting that KatE alone is sufficient to protect cells from the killing of H2O2 generated spontaneously on the plates (Shi et al., 2015). Compared to the ΔkatE strain, the additional removal of either katG or ahpC only marginally deteriorated the defect (Figure 4B). However, the triple mutant (ΔkatEΔkatGΔahpC) exhibited significantly further lowered viability, resembling the scenario observed with the S. oneidensis ΔkatBΔahpC strain. Given that S. oneidensis AhpC alone is responsible for cleaning endogenous H2O2, these data suggest that YPIII would require both KatG and AhpC to be a functional equivalence.
AhpC has been identified as a primary OP scavenger in many bacteria lacking Ohr, such as E. coli, B. subtilis, and S. Typhi (Niimura et al., 1995; Antelmann et al., 1996). We therefore predicted that AhpC is likely involved in combating against OPs in YPIII. To test this, we compared the abilities of the wild type and strains lacking catalase and/or AhpC to scavenge t-BHP. Results revealed that the removal of any of the genes under test (katE, katG, and ahpC) or all together did not introduce a significant difference in YPIII susceptibility to t-BHP (Supplementary Figure 7A). Quantification of the remaining t-BHP in the reaction also showed that the t-BHP-scavenging capacity of YPIII was not affected by any of these mutations or combined (Supplementary Figure 7B). In summary, these data suggest that AhpC in YPIII is able to decompose H2O2 but not OPs.
Loss of AhpC and KatG Together Activates OxyR
The data presented thus far have concluded that in YPIII, KatE is the primary catalase in combating H2O2-induced oxidative stress, whereas both KatG and AhpC contribute. It has been established that catalase functions to scavenge high concentrations of H2O2, whereas AhpC is a more kinetically efficient scavenger of trace H2O2 (Seaver and Imlay, 2001). To determine whether the YPIII H2O2-scavenging enzymes act in a similar manner, we measured the rates at which cells decomposed low (2 μM) and high (150 μM) concentrations of H2O2. The result showed that the katE mutant scavenged 2 μM H2O2 significantly more rapidly than ΔkatG and ΔahpC strains did (Figure 5A), indicating that in wild type, both of KatG and AhpC carry out the decomposition of low-dose H2O2 mostly. Interestingly, a ΔkatGΔahpC strain exhibited a faster rate in reducing 2 μM H2O2 than strains without either of these two proteins. Given that the additional removal of KatE totally disabled the ΔkatGΔahpC strain, this observation implies that the loss of KatG and AhpC induces the expression of KatE. On the contrary, with 150 μM H2O2, the katE mutant worked very poorly (Figure 5B), an expected result from a strain without the primary catalase. While both ΔkatG and ΔahpC strains showed normal scavenging activity, the ΔkatGΔahpC strain decomposed 150 μM H2O2 nearly two times faster than did the wild type, supporting an increased production of KatE.
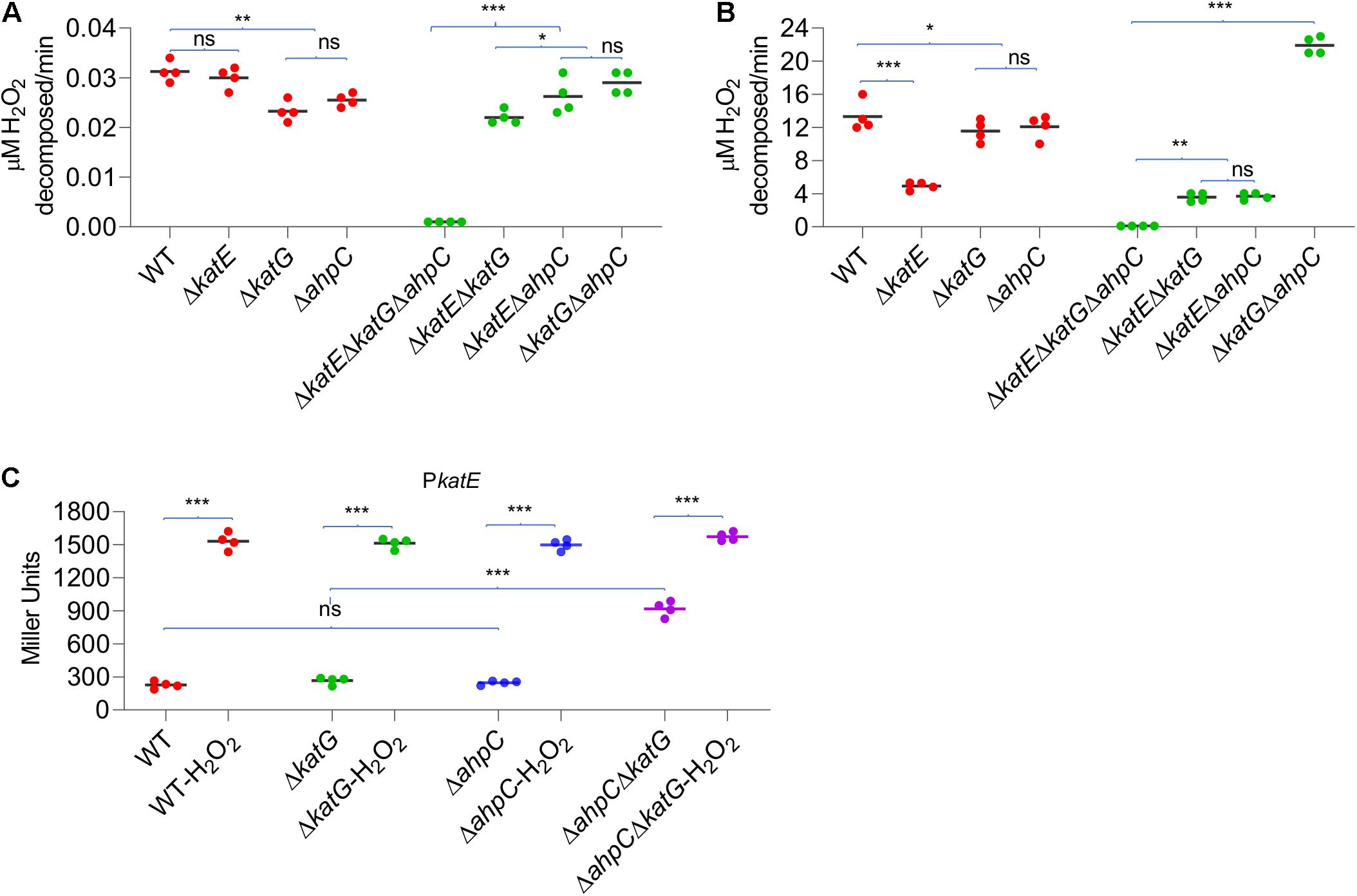
Figure 5. Simultaneous loss of AhpC and KatG increases H2O2 resistance. Efficiencies of AhpC, KatE, and KatG at different H2O2 concentrations. H2O2 was added at a final concentration of 2 μM (A) and 150 μM (B) to cultures of YPIII strains indicated. 2 min after addition of H2O2, the H2O2 concentration was measured. (C) KatE expression in strains indicated before and after the H2O2 treatment. In all panels, asterisks indicate statistically significant differences of the values compared (n = 4; ns, not significant; *p < 0.05; **p < 0.01; and ***p < 0.001).
In E. coli, the removal of AhpC results in H2O2 accumulation, leading to activation of OxyR and thereby increased catalase production (Seaver and Imlay, 2001). Although this is apparently not the case in YPIII, the data presented above imply that the loss of both KatG and AhpC is likely able to stimulate the production of KatE. To test this, we assessed the effects of depletion of KatG, AhpC, and both on KatE expression. As shown in Figure 5C, the katE gene was expressed indistinguishably in the wild-type, ΔahpC, and ΔkatG strains, and the same scenario was observed in H2O2-treated cells. These observations indicated that the loss of either AhpC or KatG alone does not significantly influence KatE production, eliminating the possibility that neither its absence is sufficient to activate OxyR in YPIII. Conversely, in the ΔkatGΔahpC cells grown under normal aerobic conditions, we found that the katE promoter activity increased 3.7 times (Figure 5C). Despite this, activity of the katE promoter can be elevated further under the treatment of H2O2 (Figure 5C), implying that the oxidative stress risen from the loss of KatG and AhpC probably activates only a portion of OxyR molecules, which is consistent with our earlier proposal that the ratio between oxidized and reduced OxyR is in a dynamic equilibrium (Wan et al., 2018, Wan et al., 2019).
Discussion
Resistance to oxidative stress resulting from ubiquitous ROS generated both endogenously and exogenously belongs to key virulent factors of bacterial pathogens. To deal with H2O2-induced oxidative stress, Gram-negative bacteria widely employ OxyR to sense the oxidant and mediate transcription of their regulon in a concerted manner (Imlay, 2013). Although regarded as a pleiotropic regulator, OxyR has evolved to consistently possess operons that are involved in H2O2 decomposition and damage control as the core member of regulons (Imlay, 2015). This is seemingly the case in Y. pseudotuberculosis YPIII. Our result in Supplementary Table 2 predicted that all H2O2-scavenging enzymes, iron-sequestering proteins, and thioredoxin and glutathione antioxidant systems are encoded by operons whose promoter regions contain most conserved OxyR-binding motifs with the highest similarities. OxyR of YPIII, highly similar to its E. coli counterpart in terms of sequence identity (88%) and the almost identical binding motifs, exhibits stimulating activity only (Zheng et al., 1998). This gains support from our transcriptomics data, which reveal elevated transcription of all predicted OxyR regulon members in the H2O2-treated cells.
In addition to OxyR regulon members, genes for sulfur species transport and metabolism and iron–manganese transport system are among the top up-regulated in the H2O2-treated cells. Sulfur species, especially hydrogen sulfide (H2S) and cysteine, are well recognized as important factors in bacterial oxidative stress response. These reductants have been shown to play a critical role in the detoxification of H2O2 in the periplasm (Ohtsu et al., 2010; Ezraty et al., 2017). Intracellular sulfur homeostasis should be carefully maintained because H2S and cysteine in vast excess promote oxidative damages by inhibiting catalases (Park and Imlay, 2003; Wu et al., 2015). Consistently, all components of thiol-based antioxidant systems of YPIII are most highly induced upon exposure to H2O2, conceivably requiring an active metabolism and fast shuttling of sulfur species (Feng et al., 2019). In parallel, genes for iron/manganese transport system are highly induced by exogenous H2O2. In bacteria, many enzymes using iron as a cofactor may become inactive due to the loss of the metal upon oxidative stress (Anjem et al., 2009; Anjem and Imlay, 2012). A common way to overcome this is to replace iron with manganese, and hence more manganese is imported when cells are challenged by H2O2 (Imlay et al., 2019). Although further investigations are needed, all of these observations suggest that sulfur species and manganese are important for YPIII to combat oxidative stress.
Like E. coli, YPIII contains two catalases: HPII KatE and HPI KatG (Mishra and Imlay, 2012). Although both KatE and KatG are highly induced upon H2O2 treatment, we first anticipate that YPIII uses KatG as the predominant H2O2-scavenging enzyme because E. coli does so (Seaver and Imlay, 2001). Unexpectedly, it is KatE that confers the H2O2-decomposing ability at high concentrations in YPIII. Instead, KatG exhibits a much minor but still significant contribution in scavenging high-dose H2O2. More importantly, KatG shows a strong scavenging activity toward low concentrations of H2O2, a role that is exclusively played by AhpC in E. coli. Despite this, given that YpOxyR becomes activated in the absence of both AhpC and KatG, but not of either one, it is clear that both KatG and AhpC, while neither is sufficient, is able to scavenge low concentrations of H2O2 in YPIII. It is well established that AhpC allows E. coli cells to degrade low concentrations of H2O2 because of its high kinetic efficiency (kcat/KM, ∼10–8 M–1 s–1; Mishra and Imlay, 2012). We believe that this is also likely true in the case of YPIII KatG. But the result of impaired H2O2-scavenging ability of ΔkatG and ΔahpC at low levels in YPIII implies that YPIII KatG possesses a kinetic efficiency similar to, or at least not significantly lower than, that of AhpC, the catalytic activity that is substantially greater than those of KatE and KatG in E. coli whose activities are similar to each other (∼10–6 M–1 s–1; Obinger et al., 1997; Hillar et al., 2000). We are working to test this notion.
Bacterial AhpC is regarded as the founding member of the peroxiredoxin family, and its homologs appear to be more widely distributed than catalase/peroxidase (Chae et al., 1994). The AhpR complex is highly conserved among diverse bacteria, with respect to not only amino acid sequence but also gene organization (Feng et al., 2020). To date, all bacteria that employ OxyR as the master H2O2-responding regulator are equipped with an ahp operon encoding both AhpC and its cognate reductase AhpF, including M. tuberculosis in which AhpF is replaced by AhpD to reduce AhpC (Baker et al., 2001; Jaeger et al., 2004). Apart from that, AhpC without a co-transcribed cognate reductase is found in H. pylori, a gastric pathogen that represents an extreme example in terms of oxidative stress response machinery because it lacks homologs of the oxidative stress regulators present in other bacteria, including OxyR, SoxR, SoxS, and PerR (Baker et al., 2001; Wang et al., 2006). A sequence alignment reveals that AhpC of Y. pseudotuberculosis is more closely related to typical AhpCs than to the H. pylori AhpC: there are significantly more identical residues in AhpCs of E. coli, S. oneidensis, and Y. pseudotuberculosis (Supplementary Figure 8). Despite this, we also observed that the linear arrangement of AhpC gene is highly conserved in Yersinia species (Supplementary Figure 9), a phenomenon that is quite rare in other bacteria. Therefore, it would be interesting to demystify how evolution has honed this particular phenomenon.
Although the reductase for YPIII AhpC remains to be determined, we may get clues from H. pylori AhpC in terms of their differences in sequence and structure from the typical ones (with AhpF as the cognate reductase). Reduction of H. pylori AhpC is carried out by thioredoxin TrxA, which is reduced by thioredoxin reductase TrxR (also called TrxB) using NADPH as the electron donor (Wang et al., 2006). A common feature that YpAhpC and HpAhpC share is that they have an additional eight-residue segment in the C-terminal end, which is missing in the typical AhpC proteins (Figure 6A and Supplementary Figure 8). In fact, the last few residues in the C terminus of AhpC have been shown to be particularly critical in maintaining an enzymatically active AhpR complex in vitro (Dip et al., 2014a). This C-terminal tail of AhpC is crucial in complexation of AhpC with AhpF, TrxR, or glutathione-disulfide reductase GorA (Dip et al., 2014a; Feng et al., 2020). Coincidently, earlier studies of certain E. coli and S. oneidensis AhpC mutants have established that certain mutations, especially in the C-terminal region, can transform AhpC to a surprisingly malleable enzyme, reducible not only by Trx system but also by Grx system (Faulkner et al., 2008; Feng et al., 2020). Therefore, we speculate that the extra eight residues at the C-terminal end of YPIII AhpC may affect the specificity of the protein for its reducing partner.
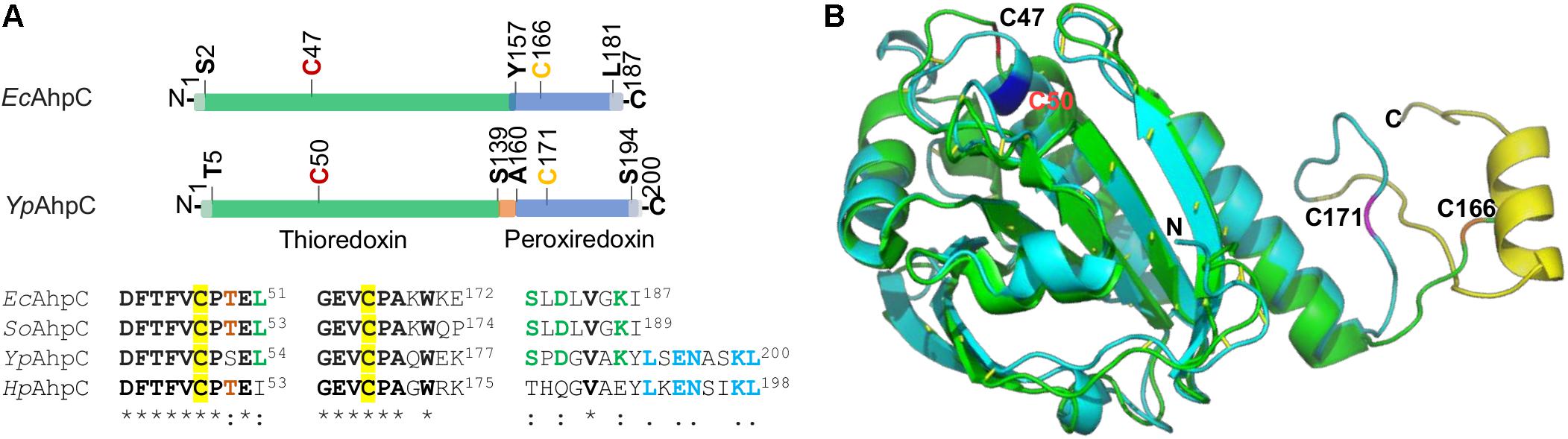
Figure 6. Comparative analysis of AhpC proteins. (A) The domain structure of EcAhpC and YpAhpC. Both proteins are composed of two domains, thioredoxin (green) and peroxiredoxin (blue), with peroxidatic Cys47 (or Cys50) and resolving Cys166 (or Cys171) in red and yellow, respectively. Sequence alignment of four representative AhpCs shown below demonstrates the conservation of regions covering two active Cys residues (identical residues are in bold with star mark) and the C-terminal tail region. Conserved residues in three out of four AhpCs are shown in red and green; the identical residues in YpAhpC and HpAhpC are shown in blue. (B) Structural comparison of YpAhpC and EcAhpC (PDB accession number 4o5r). Shown is a superimposition of YpAhpC (cyan) and EcAhpC (green). The α helix formed by the C-terminal tail of YpAhpC is shown in yellow. Active Cys residues are labeled.
The difference in the C-terminal region may also provide an explanation for the finding that YPIII AhpC does not have the ability to reduce OPs. It has been shown that catalytic efficiency of AhpC with small hydroperoxides, especially H2O2, is substantially higher (∼100-fold) than that with bulky, tertiary hydroperoxides, such as t-BHP (Parsonage et al., 2008). For both types of oxidants, the catalytic reaction adopts a binding and releasing mechanism that enables the assembly of AhpC and the reducing partner to undergo efficient catalytic cycles of transferring electrons without compromising the catalytic turnover rate (Parsonage et al., 2015). A prerequisite for such a mechanism is the correct fitting of the C-terminal tail of AhpC into the “arch-like” groove on the binding surface of the reducing partner, resulting in formation of an active-site pocket, to which small peroxides are more accessible (Dip et al., 2014b). The additional C-terminal tail of YPIII AhpC may completely prevent large OPs from entering the active-site pocket (Figure 6B). Compared to the E. coli AhpC structure (PDB accession number 4o5r; Dip et al., 2014b), Y. pseudotuberculosis AhpC has an α-helix that is formed by the extra residues. Unfortunately, how this α-helix folds remains unknown because the available H. pylori AhpC structure is truncated without this tail (PDB accession number 1zof; Papinutto et al., 2005; Supplementary Figure 10). But one may imagine that the extra C-terminus α-helix may hinder the reduction of OPs. Given that YpAhpC, unlike typical AhpCs, is not sufficient to scavenge endogenous H2O2, we speculate that the α-helix also compromises the efficiency of H2O2 reduction. If this holds, the difference in the kinetic efficiencies for H2O2 between AhpC and catalases of YPIII is likely significantly smaller than that between the E. coli counterparts.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
FW and HG designed and supported the research. FW, XF, and JY performed the research and analyzed the data. FW, XF, and HG wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31900021 to FW, 31600041 to JY, and 31930003 and 41976087 to HG).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yao Wang for providing us with the Y. pseudotuberculosis YPIII wild type.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.626874/full#supplementary-material
Footnotes
References
Anjem, A., and Imlay, J. A. (2012). Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J. Biol. Chem. 287, 15544–15556. doi: 10.1074/jbc.m111.330365
Anjem, A., Varghese, S., and Imlay, J. A. (2009). Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 72, 844–858. doi: 10.1111/j.1365-2958.2009.06699.x
Antelmann, H., Engelmann, S., Schmid, R., and Hecker, M. (1996). General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J. Bacteriol. 178, 6571–6578. doi: 10.1128/jb.178.22.6571-6578.1996
Baker, L. M., Raudonikiene, A., Hoffman, P. S., and Poole, L. B. (2001). Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J. Bacteriol. 183, 1961–1973. doi: 10.1128/jb.183.6.1961-1973.2001
Brady, M. F., Yarrarapu, S. N. S., and Anjum, F. (2020). “Yersinia pseudotuberculosis,” in StatPearls, (Treasure Island (FL): StatPearls Publishing). Available online at: https://www.ncbi.nlm.nih.gov/books/NBK430717/
Bryk, R., Griffin, P., and Nathan, C. (2000). Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407, 211–215. doi: 10.1038/35025109
Chae, H. Z., Robison, K., Poole, L. B., Church, G., Storz, G., and Rhee, S. G. (1994). Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. U S A. 91, 7017–7021. doi: 10.1073/pnas.91.15.7017
Chandrangsu, P., Rensing, C., and Helmann, J. D. (2017). Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 15, 338–350. doi: 10.1038/nrmicro.2017.15
Cosgrove, K., Coutts, G., Jonsson, I.-M., Tarkowski, A., Kokai-Kun, J. F., Mond, J. J., et al. (2007). Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189, 1025–1035. doi: 10.1128/jb.01524-06
Crooks, G. E., Hon, G., Chandonia, J.-M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Dip, P. V., Kamariah, N., Nartey, W., Beushausen, C., Kostyuchenko, V. A., Ng, T.-S., et al. (2014a). Key roles of the Escherichia coli AhpC C-terminus in assembly and catalysis of alkylhydroperoxide reductase, an enzyme essential for the alleviation of oxidative stress. Biochim. Biophys. Acta 1837, 1932–1943. doi: 10.1016/j.bbabio.2014.08.007
Dip, P. V., Kamariah, N., Subramanian Manimekalai, M. S., Nartey, W., Balakrishna, A. M., Eisenhaber, F., et al. (2014b). Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli. Acta Crystallogr D 70, 2848–2862. doi: 10.1107/s1399004714019233
Ezraty, B., Gennaris, A., Barras, F., and Collet, J. F. (2017). Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 15, 385–396. doi: 10.1038/nrmicro.2017.26
Fang, F. C. (2004). Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2, 820–832. doi: 10.1038/nrmicro1004
Faulkner, M. J., Veeravalli, K., Gon, S., Georgiou, G., and Beckwith, J. (2008). Functional plasticity of a peroxidase allows evolution of diverse disulfide-reducing pathways. Proc. Natl. Acad. Sci. U S A. 105, 6735–6740. doi: 10.1073/pnas.0801986105
Feng, X., Guo, K., and Gao, H. (2020). Plasticity of the peroxidase AhpC links multiple substrates to diverse disulfide-reducing pathways in Shewanella oneidensis. J. Biol. Chem. 295, 11118–11130. doi: 10.1074/jbc.ra120.014010
Feng, X., Sun, W., Kong, L., and Gao, H. (2019). Distinct roles of Shewanella oneidensis thioredoxin in regulation of cellular responses to hydrogen and organic peroxides. Appl. Environ. Microbiol. 85, e01700–e01719.
Fu, H., Jin, M., Ju, L., Mao, Y., and Gao, H. (2014). Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ. Microbiol. 16, 3181–3195. doi: 10.1111/1462-2920.12457
Fu, H., Yuan, J., and Gao, H. (2015). Microbial oxidative stress response: novel insights from environmental facultative anaerobic bacteria. Arch. Biochem. Biophys. 584, 28–35. doi: 10.1016/j.abb.2015.08.012
Gao, H., Wang, Y., Liu, X., Yan, T., Wu, L., Alm, E., et al. (2004). Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J. Bacteriol. 186, 7796–7803. doi: 10.1128/jb.186.22.7796-7803.2004
Hillar, A., Peters, B., Pauls, R., Loboda, A., Zhang, H., Mauk, A. G., et al. (2000). Modulation of the activities of catalase-peroxidase HPI of Escherichia coli by site-directed mutagenesis. Biochemistry 39, 5868–5875. doi: 10.1021/bi0000059
Ieva, R., Roncarati, D., Metruccio, M. M. E., Seib, K. L., Scarlato, V., and Delany, I. (2008). OxyR tightly regulates catalase expression in Neisseria meningitidis through both repression and activation mechanisms. Mol. Microbiol. 70, 1152–1165. doi: 10.1111/j.1365-2958.2008.06468.x
Imlay, J. A. (2008). Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77, 755–776. doi: 10.1146/annurev.biochem.77.061606.161055
Imlay, J. A. (2013). The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454. doi: 10.1038/nrmicro3032
Imlay, J. A. (2015). Transcription factors that defend bacteria against reactive oxygen species. Annu. Rev. Microbiol. 69, 93–108. doi: 10.1146/annurev-micro-091014-104322
Imlay, J. A., Sethu, R., and Rohaun, S. K. (2019). Evolutionary adaptations that enable enzymes to tolerate oxidative stress. Free Radical. Biol. Med. 140, 4–13. doi: 10.1016/j.freeradbiomed.2019.01.048
Jacobson, F. S., Morgan, R., Christman, M., and Ames, B. (1989). An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. purification and properties. J. Biol. Chem. 264, 1488–1496. doi: 10.1016/s0021-9258(18)94214-6
Jaeger, T., Budde, H., Flohé, L., Menge, U., Singh, M., Trujillo, M., et al. (2004). Multiple thioredoxin-mediated routes to detoxify hydroperoxides in Mycobacterium tuberculosis. Arch. Biochem. Biophys. 423, 182–191. doi: 10.1016/j.abb.2003.11.021
Jiang, Y., Dong, Y., Luo, Q., Li, N., Wu, G., and Gao, H. (2014). Protection from oxidative stress relies mainly on derepression of OxyR-dependent KatB and Dps in Shewanella oneidensis. J. Bacteriol. 196, 445–458. doi: 10.1128/jb.01077-13
Jin, M., Jiang, Y., Sun, L., Yin, J., Fu, H., Wu, G., et al. (2013). Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS One 8:e75610. doi: 10.1371/journal.pone.0075610
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protocols 10, 845–858. doi: 10.1038/nprot.2015.053
Kim, J.-S., and Holmes, R. (2012). Characterization of OxyR as a negative transcriptional regulator that represses catalase production in Corynebacterium diphtheriae. PLoS One 7:e31709. doi: 10.1371/journal.pone.0031709
Knaus, U. G., Hertzberger, R., Pircalabioru, G. G., Yousefi, S. P. M., and Branco, and dos Santos, F. (2017). Pathogen control at the intestinal mucosa – H2O2 to the rescue. Gut Microbes 8, 67–74. doi: 10.1080/19490976.2017.1279378
Korshunov, S., Imlay, K. R. C., and Imlay, J. A. (2020). Cystine import is a valuable but risky process whose hazards Escherichia coli minimizes by inducing a cysteine exporter. Mol. Microbiol. 113, 22–39. doi: 10.1111/mmi.14403
Li, R., Jia, Z., and Trush, M. A. (2016). Defining ROS in biology and medicine. React. Oxygen Species 1, 9–21.
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641.
Medina-Rivera, A., Defrance, M., Sand, O., Herrmann, C., Castro-Mondragon, J. A., Delerce, J., et al. (2015). RSAT 2015: regulatory sequence analysis tools. Nucleic Acids Res. 43, W50–W56.
Mishra, S., and Imlay, J. (2012). Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem. Biophys. 525, 145–160. doi: 10.1016/j.abb.2012.04.014
Niimura, Y., Poole, L. B., and Massey, V. (1995). Amphibacillus xylanus NADH oxidase and Salmonella typhimurium alkyl-hydroperoxide reductase flavoprotein components show extremely high scavenging activity for both alkyl hydroperoxide and hydrogen peroxide in the presence of S. typhimurium alkyl-hydroperoxide reductase 22-kDa protein component. J. Biol. Chem. 270, 25645–25650. doi: 10.1074/jbc.270.43.25645
Obinger, C., Maj, M., Nicholls, P., and Loewen, P. (1997). Activity, peroxide compound formation, and heme d synthesis in Escherichia coli HPII catalase. Arch. Biochem. Biophys. 342, 58–67. doi: 10.1006/abbi.1997.9988
Ohtsu, I., Wiriyathanawudhiwong, N., Morigasaki, S., Nakatani, T., Kadokura, H., and Takagi, H. (2010). The L-cysteine/L-cystine shuttle system provides reducing equivalents to the periplasm in Escherichia coli. J. Biol. Chem. 285, 17479–17487. doi: 10.1074/jbc.m109.081356
Papinutto, E., Windle, H. J., Cendron, L., Battistutta, R., Kelleher, D., and Zanotti, G. (2005). Crystal structure of alkyl hydroperoxide-reductase (AhpC) from Helicobacter pylori. Biochim. Biophys. Acta 1753, 240–246. doi: 10.1016/j.bbapap.2005.09.001
Park, S., and Imlay, J. A. (2003). High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J. Bacteriol. 185, 1942–1950. doi: 10.1128/jb.185.6.1942-1950.2003
Parsonage, D., Karplus, P. A., and Poole, L. B. (2008). Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc. Natl. Acad. Sci. U S A. 105, 8209–8214. doi: 10.1073/pnas.0708308105
Parsonage, D., Nelson, K. J., Ferrer-Sueta, G., Alley, S., Karplus, P. A., Furdui, C. M., et al. (2015). Dissecting peroxiredoxin catalysis: separating binding, peroxidation, and resolution for a bacterial AhpC. Biochemistry 54, 1567–1575. doi: 10.1021/bi501515w
Peng, H., Zhang, Y., Palmer, L. D., Kehl-Fie, T. E., Skaar, E. P., Trinidad, J. C., et al. (2017). Hydrogen sulfide and reactive sulfur species impact proteome s-sulfhydration and global virulence regulation in Staphylococcus aureus. ACS Infect Dis. 3, 744–755. doi: 10.1021/acsinfecdis.7b00090
Perkins, A., Nelson, K. J., Parsonage, D., Poole, L. B., and Karplus, P. A. (2015). Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 40, 435–445. doi: 10.1016/j.tibs.2015.05.001
Poole, L. B. (2005). Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433, 240–254. doi: 10.1016/j.abb.2004.09.006
Poole, L. B., and Ellis, H. R. (1996). Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry 35, 56–64. doi: 10.1021/bi951887s
Rosqvist, R., Skurnik, M., and Wolf-Watz, H. (1988). Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature 334, 522–525. doi: 10.1038/334522a0
Seaver, L. C., and Imlay, J. A. (2001). Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183, 7173–7181. doi: 10.1128/jb.183.24.7173-7181.2001
Shi, M., Wan, F., Mao, Y., and Gao, H. (2015). Unraveling the mechanism for the viability deficiency of Shewanella oneidensis oxyR null mutant. J. Bacteriol. 197, 2179–2189. doi: 10.1128/jb.00154-15
Sun, Y., Meng, Q., Zhang, Y., and Gao, H. (2020). Derepression of bkd by the FadR loss dictates elevated production of BCFAs and isoleucine starvation. Biochim. Biophys. Acta 1865:158577. doi: 10.1016/j.bbalip.2019.158577
Tartaglia, L. A., Storz, G., Brodsky, M. H., Lai, A., and Ames, B. N. (1990). Alkyl hydroperoxide reductase from Salmonella typhimurium. sequence and homology to thioredoxin reductase and other flavoprotein disulfide oxidoreductases. J. Biol. Chem. 265, 10535–10540. doi: 10.1016/s0021-9258(18)86980-0
Wan, F., Kong, L., and Gao, H. (2018). Defining the binding determinants of Shewanella oneidensis OxyR: implications for the link between the contracted OxyR regulon and adaptation. J. Biol. Chem. 293, 4085–4096. doi: 10.1074/jbc.ra117.001530
Wan, F., Yin, J., Sun, W., and Gao, H. (2019). Oxidized OxyR up-regulates ahpCFexpression to suppress plating defects of oxyR- and catalase-deficient strains. Front. Microbiol. 10:439. doi: 10.3389/fmicb.2019.00439
Wang, G., Alamuri, P., and Maier, R. J. (2006). The diverse antioxidant systems of Helicobacter pylori. Mol. Microbiol. 61, 847–860. doi: 10.1111/j.1365-2958.2006.05302.x
Wang, T., Yang, X., Gao, F., Zhao, C., Kang, Y., Wang, Y., et al. (2016). Zinc acquisition via ZnuABC in Yersinia pseudotuberculosis facilitates resistance to oxidative stress. Ann. Microbiol. 66, 1189–1197. doi: 10.1007/s13213-016-1205-7
Wang, Z., Wang, T., Cui, R., Zhang, Z., Chen, K., Li, M., et al. (2020). HpaR, the repressor of aromatic compound metabolism, positively regulates the expression of T6SS4 to resist oxidative stress in Yersinia pseudotuberculosis. Front. Microbiol. 11:705. doi: 10.3389/fmicb.2020.00705
Wolff, S. P. (1994). Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 233, 182–189. doi: 10.1016/s0076-6879(94)33021-2
Wu, G., Wan, F., Fu, H., Li, N., and Gao, H. (2015). A matter of timing: contrasting effects of hydrogen sulfide on oxidative stress response in Shewanella oneidensis. J. Bacteriol. 197, 3563–3572. doi: 10.1128/jb.00603-15
Wu, L., Wang, J., Tang, P., Haijiang, C., and Gao, H. (2011). Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis. PLoS One 6:e21479. doi: 10.1371/journal.pone.0021479
Keywords: Yersinia, OxyR, AhpC, catalase, oxidative stresss response
Citation: Wan F, Feng X, Yin J and Gao H (2021) Distinct H2O2-Scavenging System in Yersinia pseudotuberculosis: KatG and AhpC Act Together to Scavenge Endogenous Hydrogen Peroxide. Front. Microbiol. 12:626874. doi: 10.3389/fmicb.2021.626874
Received: 07 November 2020; Accepted: 22 March 2021;
Published: 07 May 2021.
Edited by:
Jianping Xie, Southwest University, ChinaReviewed by:
Paula Maria Tribelli, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaXihui Shen, Northwest A and F University, China
Hui Wang, Nanjing Agricultural University, China
Copyright © 2021 Wan, Feng, Yin and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fen Wan, ZmVud0BobWMuZWR1LmNu; Haichun Gao, aGFpY2h1bmdAemp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Fen Wan
Fen Wan Xue Feng
Xue Feng Jianhua Yin
Jianhua Yin Haichun Gao
Haichun Gao