
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 17 February 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.626160
This article is part of the Research TopicCarbapenemase-Producing Organisms as Leading Cause of Hospital InfectionsView all 25 articles
 Wentian Liu1,2†
Wentian Liu1,2† Huiyue Dong1,2†
Huiyue Dong1,2† Tingting Yan1,2
Tingting Yan1,2 Xuchun Liu3
Xuchun Liu3 Jing Cheng1,2
Jing Cheng1,2 Congcong Liu1,2
Congcong Liu1,2 Songxuan Zhang1,2
Songxuan Zhang1,2 Xiang Feng1,2
Xiang Feng1,2 Luxin Liu1,2
Luxin Liu1,2 Zhenya Wang1,2,4*
Zhenya Wang1,2,4* Shangshang Qin1,2*
Shangshang Qin1,2*Carbapenem-resistant Enterobacterales (CRE) pose a serious threat to clinical management and public health. We investigated the molecular characteristics of 12 IMP-4 metallo-β-lactamase-producing strains, namely, 5 Enterobacter cloacae, 3 Escherichia coli, 2 Klebsiella pneumoniae, and 2 Citrobacter freundii. These strains were collected from a tertiary teaching hospital in Zhengzhou from 2013 to 2015. The minimum inhibitory concentration (MIC) results showed that each blaIMP–4-positive isolate was multidrug-resistant (MDR) but susceptible to colistin. All of the E. coli belonged to ST167, two C. freundii isolates belonged to ST396, and diverse ST types were identified in E. cloacae and K. pneumoniae. S1-PFGE, Southern blotting, and PCR-based replicon typing assays showed that the blaIMP–4-carrying plasmids ranged from ∼52 to ∼360 kb and belonged to FII, FIB, HI2/HI2A, and N types. N plasmids were the predominant type (8/12, 66.7%). Plasmid stability testing indicated that the blaIMP–4-carrying N-type plasmid is more stable than the other types of plasmids. Conjugative assays revealed that three of the blaIMP–4-carrying N plasmids were transferrable. Complete sequence analysis of a representative N type (pIMP-ECL14–57) revealed that it was nearly identical to pIMP-FJ1503 (KU051710) (99% nucleotide identity and query coverage), an N-type blaIMP–4-carrying epidemic plasmid in a C. freundii strain. PCR mapping indicated that a transposon-like structure [IS6100-mobC-intron (K1.pn.I3)-blaIMP–4-IntI1-IS26] was highly conserved in all of the N plasmids. IS26 involved recombination events that resulted in variable structures of this transposon-like module in FII and FIB plasmids. The blaIMP–4 gene was captured by a sul1-type integron In1589 on HI2/HI2A plasmid pIMP-ECL-13–46.
The Zn(II)-containing metallo-β-lactamases (MBLs) comprise Imipenemase (IMP), New Delhi metallo-β-lactamase (NDM), and Verona Integron-encoded Metallo-β-lactamase (VIM) types that belong to class B β-lactamase according to the Ambler classification. MBLs can hydrolyze nearly all β-lactams, including carbapenems, which are important antibiotics in clinical practice and the “last line” drugs for treating infections caused by multiple drug-resistant (MDR) Gram-negative bacteria (Boyd et al., 2020). The rapid spread of MBLs among Enterobacterales has led to the increased prevalence of carbapenem-resistant Enterobacterales (CRE), and this presents a challenge for infection treatment worldwide (Nordmann and Poirel, 2019). Unlike NDMs, IMP-type β-lactamases are not often detected in CRE from China (Zhang et al., 2017; Wang et al., 2018). The most commonly encountered blaIMP–4 gene has been found captured by class 1 integrons and carried by plasmids belonging to multiple replicon types including HI2, L/M, A/C, and N for dissemination (Lai et al., 2017; Matsumura et al., 2017). An epidemic N plasmid in Enterobacterales isolates was recently recovered from Shanghai, Guangdong, and Fujian provinces of China and was responsible for the dissemination of blaIMP–4 gene (Wang et al., 2017). It is not known if this type of plasmid is prevalent in other regions of China and if it is involved in the spread of blaIMP genes. We conducted a retrospective study to investigate the prevalence and molecular characterization of IMP-positive Enterobacterales isolates in Henan Province within the north central region of China.
From January 2013 to December 2015, a retrospective survey for MBLs in CRE isolated from a tertiary teaching hospital of Zhengzhou University identified 12 blaIMP–4 positive isolates, which were recovered from different types of clinical specimens (Table 1). The study and consent procedure was approved by the Ethical Committee of Zhengzhou University. PCR and sequencing were used to identify MBL encoding genes, including blaIMP, blaNDM, and blaVIM, as described previously (Doyle et al., 2012). Antimicrobial susceptibility of the 12 blaIMP–4-positive isolates and their transconjugants was determined using microbroth and agar dilution methods according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2019). Escherichia coli ATCC25922 was used as the quality control.
Multilocus sequence typing (MLST) for Klebsiella pneumoniae, Enterobacter cloacae, Citrobacter freundii, and E. coli isolates were performed using previously described methods (Qin et al., 2014; Liu et al., 2015). The PCR products were purified and sequenced, and the allelic profiles and sequence types (STs) were assigned using online databases (https://pubmlst.org/ for K. pneumoniae, E. cloacae, and C. freundii, http://mlst.warwick.ac.uk/mlst/dbs/Ecoli for E. coli).
Conjugation experiments were conducted using methods described previously at 25, 30, and 37°C. Briefly, the blaIMP–4-positive isolates served as the donor, while E. coli EC600 (rifampin resistant) was used as the recipient strain. Transconjugants were selected on Mueller–Hinton (MH) agar supplemented with sodium rifampin (200 μg/ml) and meropenem (2 μg/ml). The presence of the blaIMP–4 gene and other resistance genes in transconjugants was confirmed by PCR, DNA sequencing, and antimicrobial susceptibility. S1-PFGE and Southern blotting were conducted, according to published methods, to estimate sizes of blaIMP–4 plasmids (Qin et al., 2014).
The plasmids of the blaIMP–4-positive strains were extracted using the Qiagen Midi kit (Qiagen, Hilden, Germany) and transformed into E. coli DH5α by electroporation. Transformants were selected on Luria–Bertani (LB) agar plates containing meropenem (2 μg/ml), and we confirmed the presence of the blaIMP–4 gene by using PCR and sequencing. Plasmid replicons were determined using the PCR-based replicon typing method (Carattoli et al., 2005). Plasmids were sequenced based on the Illumina HiSeq2000 platform with 2 × 100 bp paired-end reads (Majorbio Company, Shanghai, China) and the Nanopore MinION (long-read) sequencing platform. The sequencing reads were assembled de novo using SOAPdenovo v2.04. Open reading frame prediction and annotation were done with Glimmer 3.021 and BLAST at NCBI2. Plasmid comparisons were performed using BRIG3 (Alikhan et al., 2011) and Easyfig4 tools (Sullivan et al., 2011). The complete sequence of the plasmids pIMP-ECL14-57, pIMP-KP-13-9, pIMP-CF-15-127, and pIMP-CF-15-288 and the ∼46 kb fragment from pIMP-ECL-13-46 were deposited in GenBank with accession nos. MH727565 (pIMP-ECL14-57), CP068028 (pIMP-KP-13-9), CP068026 (pIMP-CF-15-127), CP068027 (pIMP-CF-15-288), and (CP068240) (pIMP-ECL-13-46, partial sequence). The final dataset of pIMP-KP-13-9, pIMP-CF-15-127, and pIMP-CF-15-288 and the ∼46 kb fragment from pIMP-ECL-13-46 is available as a fasta file from Figshare; doi: 10.6084/m9.figshare.135154825 The genetic environments surrounding the blaIMP–4 gene on the other seven N plasmids were investigated by PCR mapping and sequencing, and the plasmid pIMP-ECL14-57 and an HI2 plasmid pIMP4-SEM1 (KX810825) were used as references. PCR primers were designed from the reference sequences and are listed in Supplementary Table 1. The locations of the primers are shown in Figure 1D.
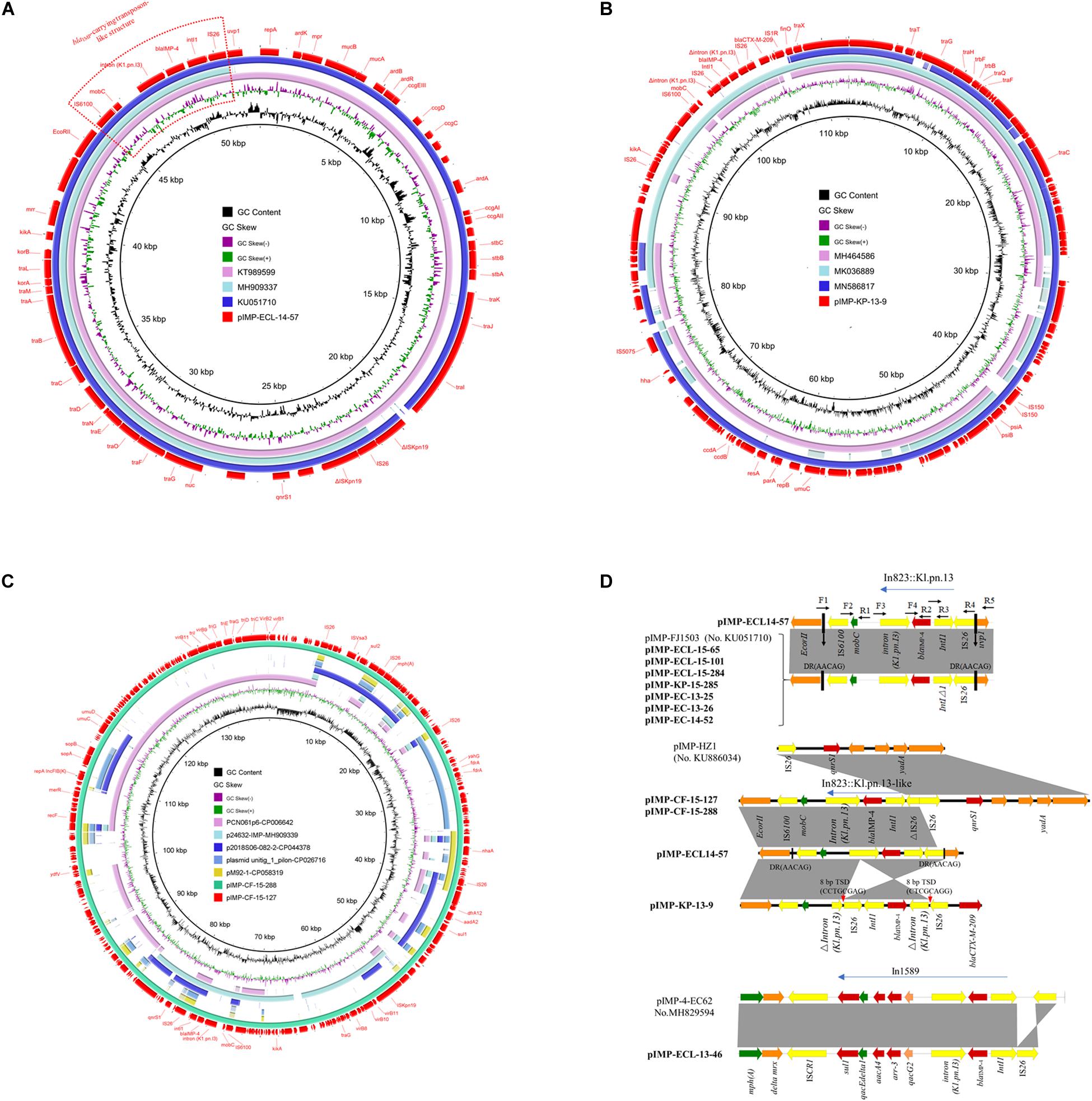
Figure 1. (A) Circular comparison between plasmid pIMP-ECL14-57 (MH727565, in this study) and other similar plasmids. Plasmid pIMP-ECL14-57 (the outer circle) was used by the BRIG software as a reference plasmid to perform the sequence alignment with BLASTN. The different colors indicated different plasmids and are listed in the color key. (B) Circular comparison between plasmid pIMP-KP-13-9 and other reported similar plasmids. Plasmid pIMP-KP-13-9 (the outer circle) was used by the BRIG software as a reference plasmid to perform the sequence alignment with BLASTN. The different colors indicate different plasmids and are listed in the color key. (C) Circular comparison between plasmids pIMP-CF-15-288 and pIMP-CF-15-127 and similar plasmids. Plasmids pIMP-CF-15-288 and pIMP-CF-15-127 (the outermost two circles) were used by the BRIG software as a reference plasmid to perform the sequence alignment with BLASTN. The different colors indicate different plasmids and are listed in the color key. (D) Comparison of the blaIMP–4 gene environments identified in this study with other publications: pIMP-FJ1503 (accession no. KU051710) and pIMP-4-EC62 (accession no. MH829594). Light gray shading indicated homologous regions (>99% DNA identity). Genetic contexts of class 1 integron carrying blaIMP–4 are shown. The different boxed arrows indicate the positions, directions of transcription, and predicted function of the genes. Positions of the primers used for PCR mapping are indicated by arrows. Genes, mobile elements, and other features are colored based on function classification.
Stability tests for plasmids were conducted as described previously (Wang et al., 2017). Briefly, the blaIMP–4-harboring transformants from ECL14-57, CF-15-127, and ECL-13-46, which were representative of blaIMP–4-carrying N, F, and HI2 plasmids characterized in this study, respectively, were used as the test strains. The overnight growths of the bacteria in LB broth were inoculated into 2 ml of a fresh LB broth and incubated for 12 h at 37°C (time zero). The above process was repeated every 12 h (equivalent to 10 generations each). At time zero and after passage without antibiotic for 50, 100, 150, and 200 generations, a sample of the culture was diluted and spread onto a LB plate. One hundred colonies were picked and replica plated onto a pair of plain and antibiotic-containing (0.5 μg/ml meropenem) LB plates. Plasmid stability was determined by the percentage of colonies growing on the antibiotic-containing plates.
A total of 12 (12/317, 3.79%) blaIMP–4 positive isolates, namely, 5 E. cloacae, 3 E. coli, 2 K. pneumoniae, and 2 C. freundii strains, were obtained from 317 CRE. These strains were recovered from different sample types including urine, blood, wound, abdominal drainage, sputum, and cerebrospinal fluid (Table 1). Over half of the blaIMP–4-carrying isolates (7/12, 58.33%) were collected from the ICU department, and the mortality among the patients infected with a blaIMP–4-positive isolate was 25% (3/12) (Table 1). These patients were diagnosed with different clinical diseases and none of them had a history of foreign travel.
For the antimicrobial susceptibility profiles, all the blaIMP–4-positive isolates were susceptible to colistin [minimum inhibitory concentrations (MICs) of ≤2 μg/ml]; tigecycline also had high activity against these isolates (MIC50 = 0.5 μg/ml) (Table 2). Our observation is consistent with previous data from both China and other countries which showed that colistin and tigecycline are effective for the treatment of infections caused by CRE (Wang et al., 2018).
MLST was performed for all the IMP-4-positive E. cloacae, E. coli, C. freundii, and K. pneumoniae isolates. Based on the MLST results, five E. cloacae isolates were distributed to four ST types, namely, ST133 (n = 2), ST231 (n = 1), ST754 (n = 1), and ST97 (n = 1). All of the three E. coli isolates belonged to ST167, which is regarded as the most common clone of E. coli in China (Zhang et al., 2017; Wang et al., 2018). Two C. freundii isolates belonging to ST396. ST14- and ST17-type K. pneumoniae carried blaIMP–4 in this study (Table 1). Overall, the observation of diversity in the isolates of E. cloacae for carrying blaIMP–4 indicated that the mobile genetic elements, such as conjugative plasmids and transposons, might be responsible for the horizontal transfer of blaIMP–4 among different clones.
The blaIMP–4 gene was always carried by a plasmid, so S1-PFGE and Southern blotting were performed to identify blaIMP–4 harboring plasmids. The blaIMP–4 genes in all 12 CRE isolates were located on plasmids with sizes ranging from ∼52 to ∼360 kb. The ∼52 kb plasmids were predominant among those carrying blaIMP–4 (8/12, 66.7%). Conjugative assays revealed that only three ∼52 kb blaIMP–4-carrying plasmids were successfully transferred to E. coli EC600 from the donors by conjugation at frequencies of 3.2 × 10–4–4 × 10–5 per donor cell. The other nine IMP-4-encoding plasmids which failed to transfer to the recipient strain by conjugation were electrotransformed into E. coli DH5α. PCR-based replicon typing analysis for both transconjugants and transformants showed that all the ∼52 kb blaIMP–4-carrying plasmids were distributed in four E. cloacae, three E. coli, and one K. pneumoniae isolates belonging to plasmid replicon type N (Table 1 and Figure 1A). The details concerning plasmid name, size, and replicon type are summarized in Table 1.
A representative N-type blaIMP–4-carrying plasmid named pIMP-ECL14-57, which came from E. cloacae strain ECL14-57, had 51,795 bp, with an average GC content of 50.52%, encoding 54 predicted open reading frames (ORFs). It shared extensive similarity with pIMP-FJ1503 (99% nucleotide identity and query coverage) (KU051710), an N-type blaIMP–4-carrying plasmid in a carbapenem-resistant C. freundii strain CRE1503 isolated from Hong Kong (Figure 1A). Comparative genomic analysis between these two plasmids revealed only two differences: (1) the intact ISkpn19 element downstream of qnrS1 that was carried by pIMP-FJ1503 was inserted by an IS26 element in pIMP-ECL14-57 and (2) the Int1 gene immediately upstream of blaIMP–4 was complete in pIMP-ECL14-57 but was truncated in pIMP-FJ1503 (Figure 1A). Only two resistance genes, namely blaIMP–4 and qnrS1, conferring resistance to carbapenems and quinolones, respectively, were identified in each plasmid. The blaIMP–4 gene-associated class 1 integron In823 was carried by a transposon-like structure [IS6100-mobC-intron (K1.pn.I3)-blaIMP–4-IntI1-IS26] bracketed by two 5 bp direct repeats (DR: AACAG) inserted between the EcorII and uvp1 genes. In addition, this blaIMP–4-carrying transposon-like structure was also identified in the other seven N plasmids by using PCR mapping and sequencing (Figure 1D).
The FII plasmid pIMP-KP-13-9 was 112,209 bp long with an average GC content of 51.19% and encoding 138 predicted ORFs (Figure 1B). This plasmid showed 98.94% nucleotide identity and 84% query coverage with pIMP1572 (MH464586), a plasmid carrying both blaIMP–26 and tet(A) variants (Yao et al., 2020). Different from the plasmid pIMP1572, a Tn1721-like transposon structure carrying the tet(A) variant which is responsible for tigecycline was absent in pIMP-KP-13-9. Interestingly, a 3,447 bp region comprising an IS26, int1, the blaIMP–4 gene, and Δintron (K1.pn.I3) in the blaIMP–4-carrying transposon-like structure in N plasmids was reversed in pIMP-KP-13-9 due to IS26-mediated recombination indicated by the presence of target site duplications (TSD) of 8 bp (CCTGCGAG).
The two FIB plasmids pIMP-CF-15-127 and pIMP-CF-15-288 obtained from different ST396 C. freundii strains were nearly identical (96.85% nucleotide identity and 100% query coverage) (Figure 1C), both of which harbored 268 predicted ORFs. The backbone region of pIMP-CF-15-127/pIMP-CF-15-288 (∼57.6 kb) containing repA (replication), umuCD (SOS mutagenesis), sopAB (plasmid-partition), and partial type IV secretion system (T4SS) encoding gene cluster shared 51% query coverage and 99.06% nucleotide identity with PCN061p6 (CP006642) from an O9 E. coli strain. A partial, but not an intact, T4SS encoding region in the two plasmids could explain the lack of conjugation of FIB plasmids. A similar blaIMP–4-carrying transposon-like structure in N plasmids was also found in pIMP-CF-15-127/pIMP-CF-15-288, while the IS26 element located immediately downstream of the int1 gene was disrupted by a 6,689 bp pIMP-HZ1 (KU886034)-derived segment encompassing IS26, qnrS1, and multiple functional genes (Figure 1D).
Overall, the blaIMP–4-associated In823 flanked by IS6100 and IS26 in N plasmids was conserved, and IS26 involved recombination events that resulted in variable structures of this transposon-like module in the FII and FIB plasmids. Analysis of a ∼46 kb blaIMP–4-carrying segment from the HI2/HI2A plasmid pIMP-ECL-13-46 (failure to obtain complete sequence by WGS) revealed that the blaIMP–4 gene was present in the sul1-type integron In1589, which was first identified in an HI2 plasmid pIMP-4-EC62 obtained from E. cloacae EC62 of swine origin (Zhu et al., 2019).
Plasmid stability analysis revealed that the N-type plasmid pIMP-ECL14-57 in transformants from ECL14-57 could be maintained at 100% over 200 generations of multiplication in the absence of antibiotics. However, drastic loss of the F-type plasmid pIMP-CF-15-127 and HI2-type plasmid pIMP-ECL-13-46 in transformants from CF-15-127 and ECL-13-46 was observed after 50 generations of multiplication, with 35 and 3%, respectively, retaining the blaIMP–4-harboring plasmid after 150 generations. These results revealed that, among the plasmids carrying blaIMP–4, the N type is more stable than the F type and HI2 type.
The IMP-4-type MBL, first identified in clinical Acinetobacter spp. from Hong Kong (Chu et al., 2001), has spread to Australia but has not been frequently detected as KPC-2 and NDM among CRE in mainland China (Xiong et al., 2016). The incidence (3.79%) of IMP-4-producing Enterobacterales observed in the CRE of this study was comparable to that found in a recent nationwide survey of CRE (3.6%) (Wang et al., 2018). The blaIMP–4 gene was found in four species, namely, E. cloacae, E. coli, K. pneumoniae, and C. freundii, which are the most common species carrying blaIMP genes (Wang et al., 2018). A report from Australia indicated that IMP-4 was the predominant MBL type among CRE, particularly in carbapenem-resistant E. cloacae (CRECL) (Sidjabat et al., 2015). Our previous study together with recent findings from China revealed the dominance of NDM-type MBL among CRECL; whether IMP-4 is the second most common MBL in CRECL needs further study (Liu et al., 2015; Jin et al., 2018).
All of the blaIMP–4 genes in this study were carried by plasmids with diverse replicons. These included HI2, N, F, and especially the predominant N plasmids. The N type is a broad host range plasmid that carries a variety of resistance determinants and shows resistance to extended-spectrum-β-lactams, sulfonamides, quinolones, aminoglycosides, tetracyclines, and streptomycin (Eikmeyer et al., 2012). N plasmids are also associated with the spread of carbapenem-resistant determinants, such as blaNDM and blaKPC (Poirel et al., 2011; Partridge et al., 2012; Eilertson et al., 2017; Jiang et al., 2017; Partridge et al., 2018; Schweizer et al., 2019). This type of plasmid was recently identified as an epidemic plasmid for carrying blaIMP–4 among Enterobacterial species in China (Lai et al., 2017; Wang et al., 2017), and it was responsible for horizontal transmission of blaIMP–6 among Enterobacterales from Japan (Yamagishi et al., 2020). Our findings are consistent with these studies and indicate the prevalence of N blaIMP–4-carrying epidemic plasmids among CRE in other regions of China. Additionally, FII plasmids, which are carriers of the blaKPC gene in K. pneumoniae (Partridge et al., 2018; Yang et al., 2020), were found to carry the blaIMP gene in this study. Association with these widespread types of plasmids may accelerate dissemination of blaIMP genes among K. pneumonia.
Class 1 integrons are common vehicles for carrying the blaIMP genes. Multiple blaIMP-harboring class 1 integrons with considerable cassette array diversity, such as In992, 1312 (blaIMP–1), In809, 823, 1456, 1460, 1589 (blaIMP–4), In722, 1321 (blaIMP–6), In73 (blaIMP–8), In687 (blaIMP–14), In1310, 1386 (blaIMP–26), and 1385 (blaIMP–38), were identified in Enterobacterales and non-fermenting Gram-negative bacilli including Pseudomonas aeruginosa and Acinetobacter spp. (Lee et al., 2017; Matsumura et al., 2017; Papagiannitsis et al., 2017; Wang et al., 2017; Dolejska et al., 2018; Zhan et al., 2018; Zhu et al., 2019). Among these, the blaIMP–4-carrying In823 integron was the most frequently detected structure on N-type plasmids in isolates recovered from different regions of China including Henan Province (Feng et al., 2016; Wang et al., 2017).
In conclusion, we determined the prevalence and molecular characterization of blaIMP–4-positive Enterobacterales in clinical specimens collected at a teaching hospital in Henan Province. Previously reported epidemic N-type plasmids exhibited superior stability compared with F- and HI2-type plasmids. N-type plasmids were the predominant plasmids carrying blaIMP–4 among the collected Enterobacterales. Associated with self-transmissible N plasmids, widespread FII plasmids and a successful epidemic E. coli ST167 clone might facilitate further dissemination of blaIMP–4 among the Enterobacterales. Surveillance is needed to monitor the spread of blaIMP–4-harboring Enterobacterales.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, MH727565, CP068028, CP068026, CP068027, and CP068240.
The studies involving human participants were reviewed and approved by the Ethical Committee of Zhengzhou University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
SQ and ZW designed the study. WL, TY, CL, and SZ performed the experiments. HD, JC, LL, and XF analyzed the bioinformatics data. SQ and JC wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by grant 81501782 from the National Natural Science Foundation of China and the Outstanding Young Teacher Research Fund of Zhengzhou University (32210464) (SQ), and Fang’s family (Hong Kong) foundation (ZW).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.626160/full#supplementary-material
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Boyd, S. E., Livermore, D. M., Hooper, D. C., and Hope, W. W. (2020). Metallo-beta-lactamases: structure, function, epidemiology, treatment options, and the development pipeline. Antimicrob. Agents Chemother. 64:e00397-20. doi: 10.1128/aac.00397-20
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Chu, Y. W., Afzal-Shah, M., Houang, E. T. S., Palepou, M. F. I., Lyon, D. J., Woodford, N., et al. (2001). IMP-4, a novel Metallo-β-lactamase from nosocomial Acinetobacter spp. Collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45, 710–714. doi: 10.1128/aac.45.3.710-714.2001
CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing; 29th Informational Supplement. CLSI Document M100. Wayne, PA: CLSI.
Dolejska, M., Papagiannitsis, C., Medvecky, M., Davidova-Gerzova, L., and Valcek, A. J. A. (2018). Characterization of the complete nucleotide sequences of IMP-4-encoding plasmids, belonging to diverse inc families, recovered from Enterobacteriaceae isolates of wildlife origin. Chemotherapy 62:e02434-17. doi: 10.1128/aac.02434-17
Doyle, D., Peirano, G., Lascols, C., Lloyd, T., Church, D. L., and Pitout, J. D. D. (2012). Laboratory detection of Enterobacteriaceae that produce carbapenemases. J. Clin. Microbiol. 50, 3877–3880. doi: 10.1128/jcm.02117-12
Eikmeyer, F., Hadiati, A., Szczepanowski, R., Wibberg, D., Schneiker-Bekel, S., Rogers, L. M., et al. (2012). The complete genome sequences of four new IncN plasmids from wastewater treatment plant effluent provide new insights into IncN plasmid diversity and evolution. Plasmid 68, 13–24. doi: 10.1016/j.plasmid.2012.01.011
Eilertson, B., Chen, L., Chavda, K. D., and Kreiswirth, B. N. (2017). Genomic characterization of two KPC-producing klebsiella isolates collected in 1997 in New York City. Antimicrob. Agents Chemother. 61:e02458-16. doi: 10.1128/AAC.02458-16
Feng, W., Zhou, D., Wang, Q., Luo, W., Zhang, D., Sun, Q., et al. (2016). Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci. Rep. 6:33419. doi: 10.1038/srep33419
Jiang, X., Yin, Z., Yin, X., Fang, H., Sun, Q., Tong, Y., et al. (2017). Sequencing of blaIMP-carrying IncN2 plasmids, and comparative genomics of IncN2 plasmids harboring class 1 integrons. Front. Cell Infect. Microbiol. 7:102. doi: 10.3389/fcimb.2017.00102
Jin, C., Zhang, J., Wang, Q., Chen, H., Wang, X., Zhang, Y., et al. (2018). Molecular characterization of carbapenem-resistant Enterobacter cloacae in 11 chinese cities. Front. Microbiol. 9:1597. doi: 10.3389/fmicb.2018.01597
Lai, K., Ma, Y., Guo, L., An, J., Ye, L., and Yang, J. (2017). Molecular characterization of clinical IMP-producing Klebsiella pneumoniae isolates from a Chinese Tertiary Hospital. Ann. Clin. Microbiol. Antimicrob. 16:42. doi: 10.1186/s12941-017-0218-9
Lee, J. H., Bae, I. K., Lee, C. H., and Jeong, S. (2017). Molecular characteristics of first IMP-4-producing Enterobacter cloacae sequence type 74 and 194 in Korea. Front. Microbiol. 8:2343. doi: 10.3389/fmicb.2017.02343
Liu, C., Qin, S., Xu, H., Xu, L., Zhao, D., Liu, X., et al. (2015). New Delhi metallo-beta-lactamase 1(NDM-1), the dominant carbapenemase detected in carbapenem-resistant Enterobacter cloacae from henan province, China. PLoS One 10:e0135044. doi: 10.1371/journal.pone.0135044
Matsumura, Y., Peirano, G., Motyl, M. R., Adams, M. D., Chen, L., Kreiswirth, B., et al. (2017). Global molecular epidemiology of IMP-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 61:e02729-16. doi: 10.1128/aac.02729-16
Nordmann, P., and Poirel, L. (2019). Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin. Infect. Dis. 69, 521–528. doi: 10.1093/cid/ciz824
Papagiannitsis, C., Kutilova, I., Medvecky, M., Hrabak, J., and Dolejska, M. J. A. (2017). Characterization of the complete nucleotide sequences of IncA/C plasmids carrying In809-like integrons from Enterobacteriaceae isolates of wildlife origin. Chemotherapy 61:e01093-17. doi: 10.1128/aac.01093-17
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17. doi: 10.1128/CMR.00088-17
Partridge, S. R., Paulsen, I. T., and Iredell, J. R. (2012). pJIE137 carrying blaCTX-M-62 is closely related to p271A carrying blaNDM-1. Antimicrob. Agents Chemother. 56, 2166–2168. doi: 10.1128/AAC.05796-11
Poirel, L., Bonnin, R. A., and Nordmann, P. (2011). Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob. Agents Chemother. 55, 4224–4229. doi: 10.1128/AAC.00165-11
Qin, S., Fu, Y., Zhang, Q., Qi, H., Wen, J. G., Xu, H., et al. (2014). High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob. Agents Chemother. 58, 4275–4282. doi: 10.1128/AAC.02813-13
Schweizer, C., Bischoff, P., Bender, J., Kola, A., Gastmeier, P., Hummel, M., et al. (2019). Plasmid-mediated transmission of KPC-2 carbapenemase in Enterobacteriaceae in critically Ill patients. Front. Microbiol. 10:276. doi: 10.3389/fmicb.2019.00276
Sidjabat, H. E., Townell, N., Nimmo, G. R., George, N. M., Robson, J., Vohra, R., et al. (2015). Dominance of IMP-4-Producing Enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob. Agents Chemother. 59, 4059–4066. doi: 10.1128/aac.04378-14
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Wang, Q., Wang, X., Wang, J., Ouyang, P., Jin, C., Wang, R., et al. (2018). Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data From a longitudinal large-scale CRE Study in China (2012-2016). Clin. Infect. Dis. 67, S196–S205. doi: 10.1093/cid/ciy660
Wang, Y., Lo, W.-U., Lai, R. W.-M., Tse, C. W.-S., Lee, R. A., Luk, W.-K., et al. (2017). IncN ST7 epidemic plasmid carrying blaIMP-4 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. J. Antimicrob. Chemother. 72, 99–103. doi: 10.1093/jac/dkw353
Xiong, J., Deraspe, M., Iqbal, N., Ma, J., Jamieson, F. B., Wasserscheid, J., et al. (2016). Genome and plasmid analysis of blaIMP-4-carrying Citrobacter freundii B38. Antimicrob. Agents Chemother. 60, 6719–6725. doi: 10.1128/AAC.00588-16
Yamagishi, T., Matsui, M., Sekizuka, T., Ito, H., Fukusumi, M., Uehira, T., et al. (2020). A prolonged multispecies outbreak of IMP-6 carbapenemase-producing Enterobacterales due to horizontal transmission of the IncN plasmid. Sci. Rep. 10:4139. doi: 10.1038/s41598-020-60659-2
Yang, X., Dong, N., Chan, E. W.-C., Zhang, R., and Chen, S. (2020). Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 29, 65–83. doi: 10.1016/j.tim.2020.04.012
Yao, H., Cheng, J., Li, A., Yu, R., Zhao, W., Qin, S., et al. (2020). Molecular characterization of an IncFIIk plasmid co-harboring blaIMP-26 and tet(A) variant in a clinical Klebsiella pneumoniae isolate. Front. Microbiol. 11:1610. doi: 10.3389/fmicb.2020.01610
Zhan, Z., Hu, L., Jiang, X., Zeng, L., Feng, J., Wu, W., et al. (2018). Plasmid and chromosomal integration of four novel blaIMP-carrying transposons from Pseudomonas aeruginosa, Klebsiella pneumoniae and an Enterobacter sp. J. Antimicrob. Chemother. 73, 3005–3015. doi: 10.1093/jac/dky288
Zhang, R., Liu, L., Zhou, H., Chan, E. W., Li, J., Fang, Y., et al. (2017). Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. Ebiomedicine 19, 98–106. doi: 10.1016/j.ebiom.2017.04.032
Keywords: blaIMP–4, transposon-like structure, class 1 integron, carbapenem-resistant Enterobacterales, N plasmid
Citation: Liu W, Dong H, Yan T, Liu X, Cheng J, Liu C, Zhang S, Feng X, Liu L, Wang Z and Qin S (2021) Molecular Characterization of blaIMP–4-Carrying Enterobacterales in Henan Province of China. Front. Microbiol. 12:626160. doi: 10.3389/fmicb.2021.626160
Received: 05 November 2020; Accepted: 18 January 2021;
Published: 17 February 2021.
Edited by:
Raffaele Zarrilli University of Naples Federico II, ItalyReviewed by:
Yunsong Yu, Zhejiang University, ChinaCopyright © 2021 Liu, Dong, Yan, Liu, Cheng, Liu, Zhang, Feng, Liu, Wang and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenya Wang, emhlbnlhd2FuZ0B6enUuZWR1LmNu; Shangshang Qin, cWluc2hhbmdzaGFuZ0AxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.