- 1Department of Soil Ecology, Helmholtz Centre for Environmental Research, Halle (Saale), Germany
- 2Agro-Technological Institute, RUDN University, Moscow, Russia
- 3German Centre for Integrative Biodiversity Research Halle–Jena–Leipzig, Leipzig, Germany
- 4Institute of Crop Science and Resource Conservation – Molecular Biology of the Rhizosphere, University of Bonn, Bonn, Germany
- 5Institute of Bio- and Geosciences, IBG-2: Plant Sciences, Forschungszentrum Jülich GmbH, Jülich, Germany
- 6Institute of Soil Science, University of Hannover, Hanover, Germany
- 7Institute of Stochastics, Ulm University, Ulm, Germany
- 8Department Soil Science, Institute for Plant Nutrition and Soil Science, Christian-Albrechts University Kiel, Kiel, Germany
- 9Department of Soil Science, Faculty of Organic Agricultural Sciences, University of Kassel, Kassel, Germany
- 10Department of Soil and Plant Microbiome, Institute of Phytopathology, Christian-Albrechts-University of Kiel, Kiel, Germany
In this review, we introduce microbially-mediated soil processes, players, their functional traits, and their links to processes at biogeochemical interfaces [e.g., rhizosphere, detritusphere, (bio)-pores, and aggregate surfaces]. A conceptual view emphasizes the central role of the rhizosphere in interactions with other biogeochemical interfaces, considering biotic and abiotic dynamic drivers. We discuss the applicability of three groups of traits based on microbial physiology, activity state, and genomic functional traits to reflect microbial growth in soil. The sensitivity and credibility of modern molecular approaches to estimate microbial-specific growth rates require further development. A link between functional traits determined by physiological (e.g., respiration, biomarkers) and genomic (e.g., genome size, number of ribosomal gene copies per genome, expression of catabolic versus biosynthetic genes) approaches is strongly affected by environmental conditions such as carbon, nutrient availability, and ecosystem type. Therefore, we address the role of soil physico-chemical conditions and trophic interactions as drivers of microbially-mediated soil processes at relevant scales for process localization. The strengths and weaknesses of current approaches (destructive, non-destructive, and predictive) for assessing process localization and the corresponding estimates of process rates are linked to the challenges for modeling microbially-mediated processes in heterogeneous soil microhabitats. Finally, we introduce a conceptual self-regulatory mechanism based on the flexible structure of active microbial communities. Microbial taxa best suited to each successional stage of substrate decomposition become dominant and alter the community structure. The rates of decomposition of organic compounds, therefore, are dependent on the functional traits of dominant taxa and microbial strategies, which are selected and driven by the local environment.
Relevant Microbially-Mediated Soil Processes
In terrestrial ecosystems, the most critical biochemical processes are performed by soil microorganisms (Brussaard, 2012; Fierer, 2017), and a broad range of microbial functions contribute to essential ecosystem services, such as soil fertility, resilience, and resistance to abiotic and biotic stresses (Mulder et al., 2011). One major category of microbial functions in terrestrial ecosystems is the decomposition and transformation of organic compounds entering the soil, predominantly as plant materials. The majority of microorganisms are capable of breaking down labile compounds derived from fresh plant litter or rhizodeposits, thus ensuring functional redundancy. Other processes rely on more specialized microorganisms in the breakdown of recalcitrant compounds, which usually occur at later stages of organic matter decomposition (Bastian et al., 2009; Pepe-Ranney et al., 2016). Functional redundancy in the soil microbiome, or more generally biodiversity, provides ecosystem resilience (Fierer, 2017; Louca et al., 2018; Maron et al., 2018) and is crucial for ecosystem multifunctionality (Wagg et al., 2014, 2019).
The primary input organic substances in soil are microbially transformed into cell constituents or excreted by cells as labile metabolic products (Bradford, 2016). Moreover, microbial respiration during the transformation of organic material results in carbon loss from the soil and atmospheric CO2 emissions. An essential fraction of organic C that is assimilated within microbial biomass is further re-utilized after microbial death by multi-stage microbial succession (Morriën, 2016). Plant-derived resources are then transferred to the microbial food web (Kramer et al., 2016; Hünninghaus et al., 2019). Products from living microorganisms, particularly residues of dead microorganisms (necromass), also serve as a secondary source of soil organic substrates, finally resulting in sequestration of up to 40% of primary C input (Miltner et al., 2012; Kallenbach et al., 2016; Buckeridge et al., 2020).
Microorganisms decompose organic substrates to maintain their metabolic requirements and enable their growth. Microbial growth and anabolic reactions require not only C and energy, but also a general stoichiometric composition of nutrients (e.g., N and P), which microorganisms have to mobilize from a multiphase (gaseous, liquid, and solid) soil environment (Zechmeister-Boltenstern et al., 2015). If their own stoichiometric requirements are fulfilled, they can release nutrients, thereby increasing the availability to plants (Hodge et al., 2000; Griffiths et al., 2012). Thus, the metabolic activity of soil microorganisms can cause both positive and negative consequences at the ecosystem level, such as (i) C sequestration and losses during decomposition and transformation of soil organic matter (SOM) or (2) nutrient mobilization, possibly followed by losses through leaching of mineral nitrogen and phosphates. These processes can also cause greenhouse gas (N2O, CO2, and CH4) emissions. The direction and intensity of the consequences of microbial metabolic activity are dependent on the functional traits of the organisms performing the ecologically-relevant processes below-ground and the abiotic and biotic conditions these organisms encounter in their habitats.
Relevant Players, Functional Traits, and Links to Processes
Even though the active fraction of a predominantly dormant microbial community can be small in nutrient-poor or stressed environments (Jones and Lennon, 2010; Barnard et al., 2013), soil microorganisms are among the most abundant players in the process of decomposition and transformation of SOM (McGuire and Treseder, 2010). Fresh input of labile organic substrates, e.g., in the rhizosphere by rhizodeposition, may enormously increase the fraction of active microorganisms (Blagodatsky et al., 2000) and therewith the decomposition of SOM, thus causing the well-known “rhizosphere priming effect” (Arsjad and Giddens, 1966; Cheng et al., 2003). Moreover, both SOM stabilization and destabilization in the rhizosphere are driven by different processes, not only rhizodeposition, but also root turnover, as well as nutrient and water uptake by plants (Dijkstra et al., 2020).
Metrics Used to Distinguish Microbial Traits
Soil organic matter transformation processes mainly rely on the production of extracellular enzymes that facilitate the oxidation or hydrolysis of diverse and complex SOM compounds (Nannipieri et al., 2012). The decomposition rate of SOM is mediated by the molecular nature of SOM as well as by the degree of biotic interactions (Sokol et al., 2018). Moreover, it depends on microbial community traits (DeAngelis et al., 2009), which can be subdivided into three groups (Figure 1). Microbial traits in the first group are very dynamic, for example, the size of the microbial fraction maintaining activity or alert state (active biomass) and the time required for dormant microorganisms to switch to active growth (i.e., lag time). The second group represents intrinsic functional traits of the microbial population, such as maximal specific growth rate (μm), generation time (Tg), and affinity of extracellular enzyme systems (Km) to soil organic substrates used for microbial growth. The third group refers to phenotypic traits at the level of functional genes, for example, those related to internal microbial metabolism, extracellular resource acquisition, or stress tolerance.
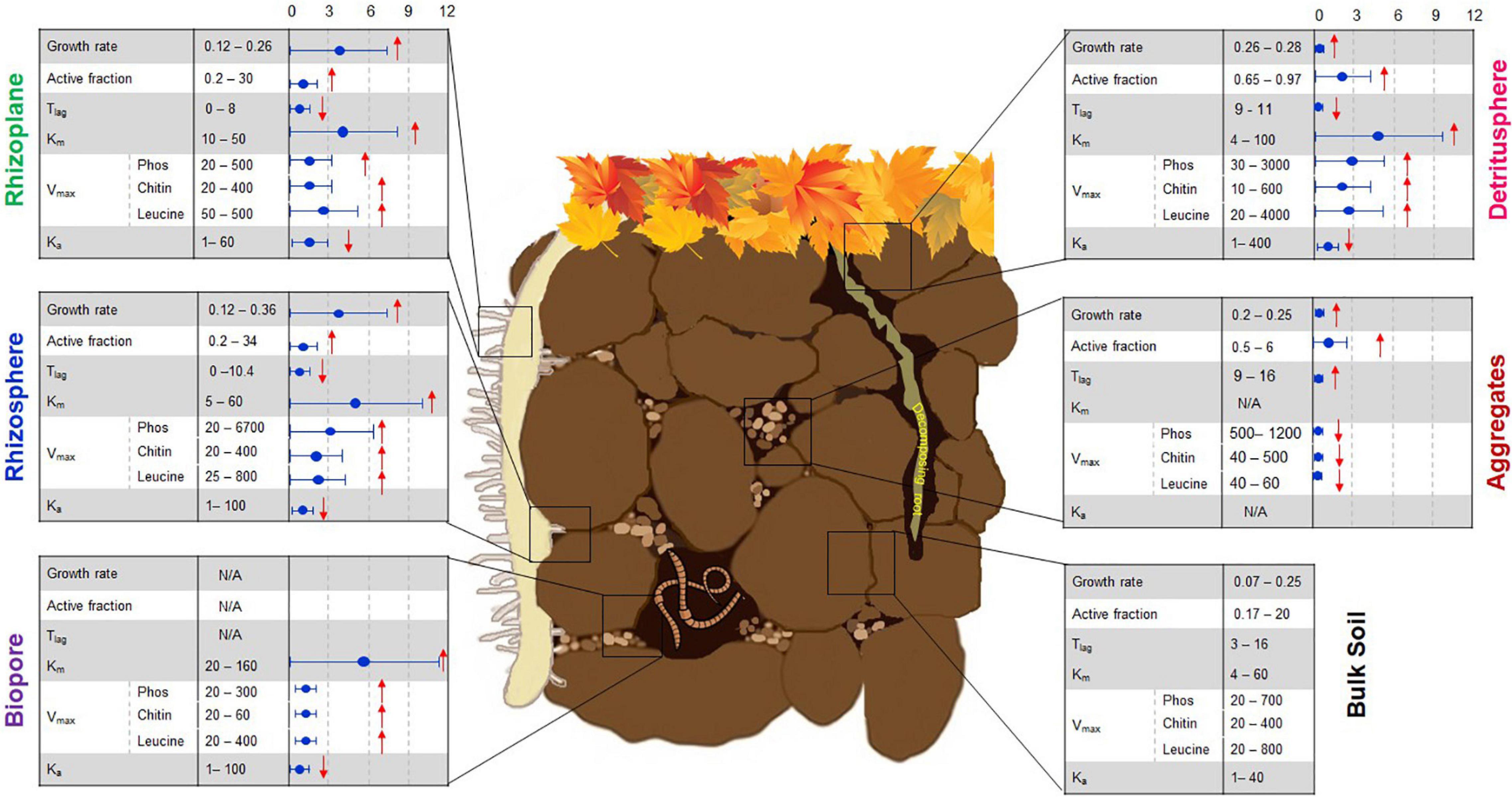
Figure 1. Linking microbial functional traits to process rates in the soil microbial hotspots. First column (left): Growth rate (h–1), active fraction (% of total biomass), lag-time, Tlag (h), Functional genes (log copies/g dry soil), Km, enzyme affinity to the substrate (μmol g–1 soil), Vmax, enzyme activity (Phos, phosphatase; Chitin, chitinase; Leucine, leucine-aminopeptidase; nmol g–1 h–1), Ka (h–1). Column (middle): ranges of original values based on literature, the column (right) shows standardized times of changes in comparison with bulk soil “0, 3, 6, 9, 12.” The arrows show the increased ↑ or decreased ↓ trend compared to bulk soil. References used for this figure have been published as Supplementary Data Sheet 1. Please note that only experiments and analyses performed in soil as matrix were included. After modification from © Nature Education 2012.
Microbial Traits and Life Strategy Concepts
Bimodal classical concepts based on functional traits (Panikov, 1995; Morris and Blackwood, 2007) exploit the general ecological principles that classify microorganisms, for example, resource requirements (copiotrophs versus oligotrophs), spatial mobility (zymogenous vs. autochtonous), or by growth and efficiency (r- versus K-strategists). These concepts are often not capable of covering a great diversity of microbial functions and life strategies. They have thus been complemented by other concepts such as the competitor-ruderal-stress-tolerator life strategy concept (Krause et al., 2014; Ho et al., 2017), which has very recently been further developed into a “high yield, resource acquisition, and stress tolerance” concept (Malik et al., 2020). However, it remains challenging to identify proxies for specific traits that can serve as quantitative measures of a category. For example, r- and K- strategists can be differentiated by the values of maximum specific growth rates (μm), and by enzyme affinities for a substrate (Km), experimentally determined under in situ soil conditions (Blagodatskaya et al., 2014; Tian et al., 2020). In contrast, quantitative estimation of traits specifically responsible for stress tolerance or resource acquisition remains challenging.
Linking Gene Phenotypic Traits and Microbial Physiology to Estimate Growth Rates
Recent developments in molecular approaches have provided potential for microbial trait differentiation based on information regarding genome size, number of ribosomal gene copies per genome, and quantification of functional marker genes or their transcripts by -omics approaches (Li et al., 2019; Malik et al., 2020). The idea that multiple versus single rrn operons in a genome corresponds to faster versus slower growing taxa raises the question of how to relate gene copy number (single vs. multi-copy genes) encoding certain functions with microbial ability to grow under in situ soil conditions. At the physiological level, growth rate is dependent on a balance between two fractions of the total proteome (Scott et al., 2014), which are growth-independent (i.e., related to maintenance function) and growth-dependent (i.e., ribosomal and metabolic proteins). In accordance with this concept, a physiological approach based on substrate-induced growth respiration (SIGR) estimates microbial-specific growth rates on glucose, considering the partitioning of microbial respiration into growth-related and growth-independent fractions (Panikov, 1995). As the SIGR approach estimates non-limited microbial growth in soil enabling access to glucose and nutrients, a complementary approach has been developed to determine bacterial and fungal growth by incorporating trace amounts of labeled 3H-leucine, 3H-thymidine, or 14C-acetate (Bååth, 2001; Bååth et al., 2001). Thus, combining molecular and respiratory approaches with isotopic labeling is promising for linking genetic and metabolic potential with microbial functions. However, synchronization of experimental design by sampling time is required for correct comparisons of microbial growth rates obtained by different approaches (see the section “Relevant Approaches for Process Localization”).
A larger genome size corresponds to slower growth induced by glucose; this relationship is weakened in both non-amended and nutrient-rich soil, thus indicating a regulatory role of the environment under natural soil conditions (Li et al., 2019). Reduced growth rates under either C or nutrient limitation may cause contrasting responses of genes involved in bacterial metabolism. Thus, the expression of C catabolic genes increases with decreasing growth rates under C limitation, but decreases under N limitation (You et al., 2013). In contrast, the expression of biosynthetic genes exhibit the opposite growth-rate dependence as catabolic genes. Under environmental control, the growth rates of individual taxa can vary by a factor of two in non-supplemented soils of contrasting ecosystems (Morrissey et al., 2018).
Therefore, environmental selection results in the activation of populations with intrinsic functional traits that are mostly suited to the individual microhabitat within heterogeneous soil pore spaces. Thus, beyond the quality and regularity of substrate input, biotic and abiotic environments (Huang et al., 2014), such as soil structure (Berg and Smalla, 2009), presence of organisms (Scheffknecht et al., 2006), and nutritional status (Hinsinger, 2001; Jones et al., 2004) affect microbial functional traits in contrasting soil habitats. In soils covered by vegetation, microbial functional traits are also affected by the physiological and morphological traits of plants.
Rhizosphere Processes and Interactions Within and Between Interfaces
Plants are the major primary producers in terrestrial ecosystems and are thus predominantly responsible for organic C input into the soil. They modulate their surrounding soil environment either actively, that is, producing exudates and exo-enzymes or passively through root and litter detritus (Kaiser et al., 2015), thus interacting with the corresponding microorganisms near the roots or of other soil interfaces [e.g., detritusphere, (bio)-pores, and aggregate surfaces]. Considering the active role of roots crossing, penetrating, and even forming aggregates, biopores, and detritus, we mainly focused on the rhizosphere and its overlap with other relevant interfaces (Figure 2).
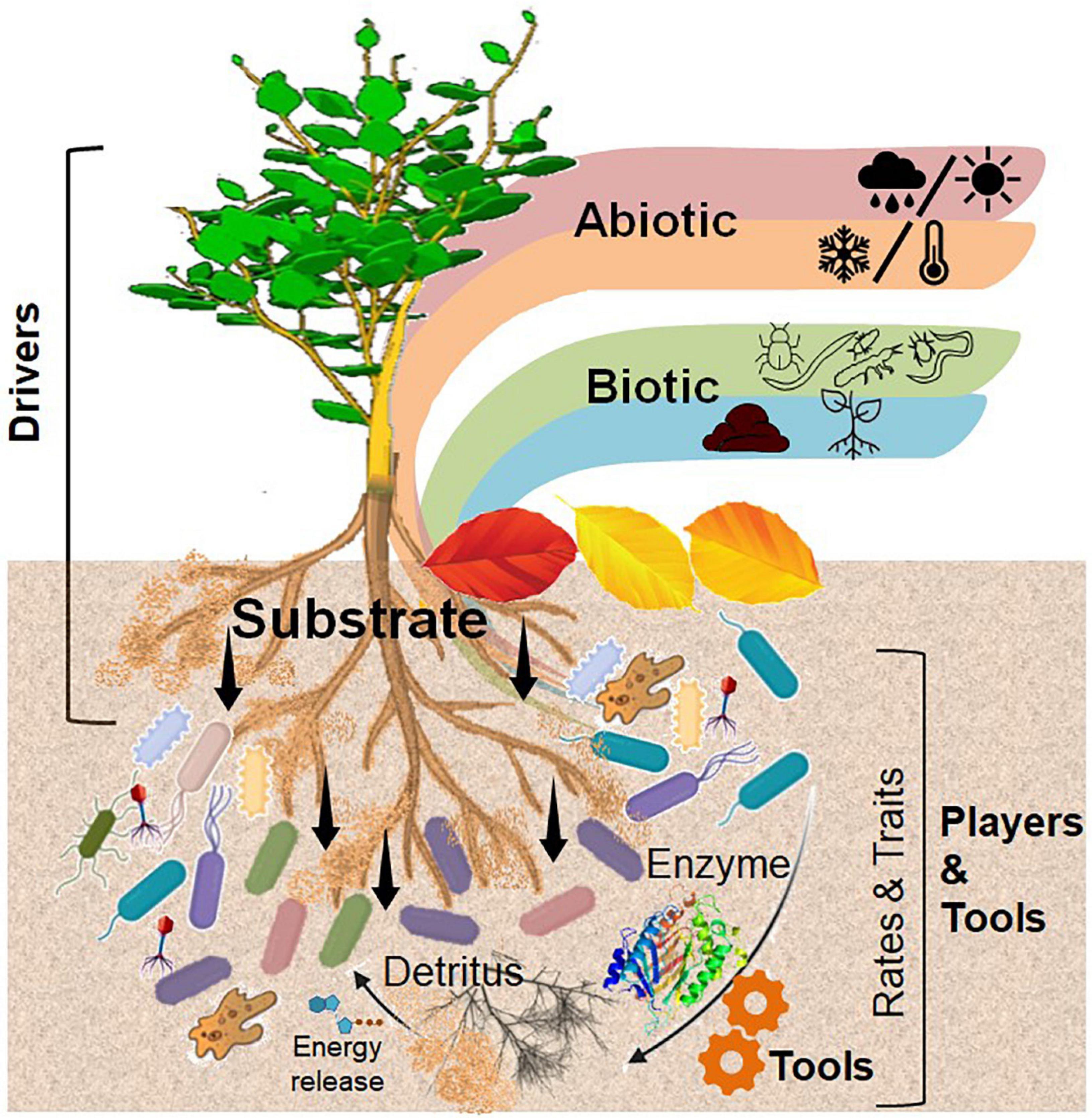
Figure 2. Conceptual illustration of central role of the rhizosphere in interactions with other biogeochemical interfaces. The main driver of plant–microbial interactions is an input of substrate through exudation, which is controlled by biotic and abiotic factors. Microorganisms are the most powerful players in the soil interfaces, using functional traits (e.g., the ability to produce specific extracellular enzymes) as a tool to develop a microbial life strategy, which in turn influences the rates of transformation of organic compounds in the soil. Self designed based on Song et al. (2020) (Figure 1).
Rhizosphere
An essential part of C assimilated via plant primary production enters the soil through roots by a process called rhizodeposition. The growing root tip and its rhizodeposits turn the bulk soil into a rhizosphere soil with its specific physical, chemical, and biological characteristics, which convert it into a hotspot of biological activity compared to the surrounding bulk soil (Goberna et al., 2007; Reinhold-Hurek et al., 2015). Thus, specific microbial communities develop in the endosphere, rhizoplane, and rhizosphere, that is, within a few millimeters from the root surface (Edwards et al., 2015; Vidal et al., 2018) and along the growing root (Cheng et al., 2017; Schmidt et al., 2018). Continuous root growth and turnover drive the spatial distribution and transformation of primary and secondary C input into the soil. This C input is determined by a combination of factors related to root type (Swinnen et al., 1994a,b, Jahnke et al., 2009), root age, root turnover rate, and specific rhizodeposition processes (Steer and Harris, 2000; Kawasaki et al., 2016). Root growth also leads to the creation of specific interfaces between the rhizosphere and other boundaries, such as the detritusphere, (bio)-pores, or aggregate surfaces (Figure 1).
Rhizodeposits range in complexity from cells and lysates to small organic molecules. Rhizodeposits are released as a consequence of tissue turnover, sloughed-off border cells, mucilage release, or the secretion of biochemically diverse root exudates (Farrar et al., 2003; Nguyen, 2003; Jones et al., 2009). The production rate and quality of rhizodeposits are governed by plant species and even genotype (Lesuffleur et al., 2007; Mönchgesang et al., 2016), the growth rate and age of an individual plant (Gransee and Wittenmayer, 2000; Zhalnina et al., 2018), and root morphology, for example, root type or root hairs (Poirier et al., 2018; Cotton et al., 2019).
Generally, plants adapt their source–sink relationships dynamically under various abiotic-biotic environments, ensuring seed production to provide offspring and supporting growth (Smith et al., 2018). Consequently, at different time scales, C allocation, and distribution patterns in the plant body and rhizosphere vary widely in terms of allocation rates, compound variety, and quality (Brüggemann et al., 2011). For example, the net C allocation from shoot to root strongly depends on the plant functional type, with a mean value of 21% for crops and 33% for grasses (Pausch and Kuzyakov, 2018), and on plant phenological stage, with a greater allocation to roots at early plant stages (40–60% of photosynthetically-fixed C) than at the reproductive and ripening stages (less than 15%) (Swinnen et al., 1994a). According to temporal profiles, C allocation within the root system is highly dynamic and differs between the internal structures of root organs (Jahnke et al., 2009). The proportion of root C moving into the rhizosphere as rhizodeposition ranges from 1.3 to 20% of photosynthetically-fixed C for crops, trees, and perennial grasses (Jones et al., 2009; Kaiser et al., 2015; Pausch and Kuzyakov, 2018) to 20–30% for cereals and mycorrhizal plants (Jakobsen and Rosendahl, 1990; Kuzyakov and Domanski, 2000), and up to 50–60% (Lynch and Whipps, 1990; van Dam and Bouwmeester, 2016), with substantial uncertainty that is still apparent with regard to ecosystem type and climatic conditions.
Soil-plant interactions in the rhizosphere extend several millimeters from the root surface to the soil (Dazzo and Gantner, 2013). The intensity of root-soil interactions is demonstrated by pronounced distance gradients from the root surface (rhizoplane) through the rhizosphere to the bulk soil (Kuzyakov and Razavi, 2019). The formation of chemical gradients in the rhizosphere is governed by the input of labile root exudation, which changes their localization in accordance with root growth. Root exudation boosts the activity and modulates the community structure of soil microorganisms, thereby explaining spatial biological gradients in the rhizosphere (Kuzyakov and Blagodatskaya, 2015). Rhizodeposition fluctuates in space and time due to root growth and stimulates the “rhizosphere priming effect” (Cheng et al., 2014; Keiluweit et al., 2015; Nie et al., 2015), which is relatively short-term because the C from rhizodeposits is rapidly converted into microbial biomass and is partly released by microbial respiration; consequently, decomposition rates are reduced in the absence of fresh C input (De Graaff et al., 2010). Furthermore, the broad spectrum of compounds in rhizodeposits (el Zahar Haichar et al., 2014; Zhalnina et al., 2018; Pett-Ridge et al., 2020) modulates biological activities in a compound mixture-dependent manner. In particular the presence of certain sugars or secondary metabolites such as phenolic compounds can exert profound negative and positive influences on community composition and the functional potential of microorganisms (Badri et al., 2009; Chaparro et al., 2013; Cotton et al., 2019), affecting microbial growth, respiration, and decomposing activity (Chigineva et al., 2009; Zwetsloot et al., 2018).
Over time, a decrease in root exudations, for example, due to a switch from vegetative to regenerative growth (Aulakh et al., 2001; De-la-Peña et al., 2010), reduces the abundance of rhizosphere microorganisms (Chaparro et al., 2014; Schmidt and Eickhorst, 2014), ultimately leading to a downregulation of enzyme production. Therefore, the temporal-spatial shifts in rhizosphere gradients of microbial activity impact soil functions such as decomposition and nutrient mobilization (Kuzyakov and Xu, 2013; Nuccio et al., 2020). As the rhizosphere is one of the most dynamic interfaces actively developing in the local environment, the interaction of the rhizosphere with other interfaces, for example, aggregates, porosphere, and detritusphere, can essentially affect the functional traits of dominant microbial populations and the rates of microbially-mediated soil processes at these interfaces.
Mycorrhizosphere
Mycorrhizal fungi form widespread symbiotic networks with the roots of most land plants, where the fungus delivers mineral nutrients to the mycorrhizal host plant and takes up plant sugars and lipids. Plant C flows to the soil through mycorrhizal roots, together with the external mycorrhizal mycelium, which are defined as “mycorrhizosphere” (van der Heijden et al., 2015), while the fungal hyphae as habitats for microorganisms are generally referred to as “hyphosphere” The external mycelium that may extend centimeters from the root surface to nutrient patches provides plant-derived C rapidly, in hours, to soil microorganisms (Drigo et al., 2010; Kaiser et al., 2015; Gorka et al., 2019), and this resource is used not only for growth by bacteria and fungi but also as a “priming” resource for decomposition (Fontaine et al., 2003). Priming via the mycorrhizosphere has been suggested for the widely distributed arbuscular mycorrhizal symbiosis (Cheng et al., 2012), and priming by arbuscular mycorrhizal fungi is associated with modifications to the soil microbial community composition (Herman et al., 2012; Nuccio et al., 2013; Gui et al., 2017). Consequently, arbuscular mycorrhizal symbiosis may influence different trophic levels by enhancing C allocation into the food web, stimulating N mobilization from SOM, and subsequent transfer to the host plant (Koller et al., 2013b; Hünninghaus et al., 2019). In contrast, ectomycorrhizal fungi produce extracellular enzymes and free radicals that release N from organic compounds (Lindahl and Tunlid, 2015; reviewed by Nicolás et al., 2019). By two distinct mechanisms related to litter decomposition stage and soil depth, they either suppress or stimulate decomposition (Brzostek et al., 2015; Sterkenburg et al., 2018). In topsoil with fresh litter, ectomycorrhizal competition for N decreases the decomposition rate (Averill et al., 2014), whereas in deeper soil layers with litter at later stages of decomposition, ectomycorrhizal fungi contribute to decomposition (Sterkenburg et al., 2018).
Rhizosphere–Detritusphere Interactions
Plant photosynthates are released into the soil in the form of soluble root exudates and plant detritus (e.g., leaf litter and rhizo-detritus). Thus, rhizodeposition processes overlap with leaf litter and dead root tissue degradation processes, forming a rhizosphere–detritusphere interface. Because of the spatial and chemical heterogeneity of rhizodeposits and rhizo-detritus in soil, the microbial communities and activities in these two spheres as well as in the overlapping sphere are specific (Marschner et al., 2012; Nuccio et al., 2020). It has been shown that root activity modulates the decomposition processes in the detritusphere by altering the structure of the microbial community feeding on detritus. Distinct microbial taxa are involved in 13C-labeled rice straw degradation in the rhizosphere compared to bulk soil (Maarastawi et al., 2018). Essentially, less 13C is assimilated by microorganisms from the straw in the rhizosphere versus bulk soil, likely due to the higher availability of labile C in the rhizosphere. The availability of detritus, in turn, reduces the consumption of root exudates by the rhizosphere microbiota (Maarastawi et al., 2019), indicating that detritusphere processes modulate rhizosphere processes and demonstrate interactions between the rhizosphere-detritusphere interface. A recent study by Nuccio et al. (2020) observed higher taxonomic and functional diversity in the combined rhizosphere–detritusphere, suggesting that the co-existence of rhizosphere guilds is facilitated by niche differentiation. This observation was based on a metatranscriptomics study where the expression of genes responsible for the decomposition of different organic substrates was studied comparatively in soils that originated either from growing roots, from decaying root material, or from overlapping spheres. Spatial and temporal niche differentiation by functional genes responsible for similar functions clearly confirmed the strong redundancy of the functions in the rhizosphere and detritusphere. For example, for various functions (genome classes), the number of active genomes shared between the rhizosphere and detritusphere is 28 (housekeeping gene gyrase A, B), 32, 67, and 50% (oligosaccharide hydrolases, cellulases, and xylanase-encoding genes, respectively) of the total number of functionally active taxa (data extracted from Figure 3 in Nuccio et al., 2020). Moreover, rhizosphere organisms express genes involved in the consumption of primary C compounds as well as breakdown products, indicating that they benefit from synergistic consumption processes. Thus, such overlapping interfaces have consequences for intra- and inter-specific interactions involving soil biota. For example, changes in litter quality in the root zone alter not only bacterial community structure and function, but also cause strong feedback from bacterial grazers, thus affecting links in the soil food webs (Koller et al., 2013a). Therefore, successional changes in bacterial and fungal populations during plant development may lead to a corresponding succession of protist communities in the rhizosphere, which translates into less complex and dense protist networks during plant senescence (Ceja-Navarro et al., 2021).
Differences in the quality of organic matter input induce contrasting competition situations within the rhizosphere and detritusphere. In the rhizosphere, the majority of easily available organic C, such as sugars, amino acids, and carboxylic acids, are released through living roots (Jones et al., 2009). In the detritusphere, such easily degraded monomers are rapidly used up, leaving behind higher molecular weight compounds such as cellulose or lignin, thereby supporting different functional guilds (Pascault et al., 2013; Pepe-Ranney et al., 2016; Nuccio et al., 2020). Generally, root morphological properties (e.g., root hairs, fine roots, mycorrhiza) intensify the release of exudates, thus increasing microbial activity, functionality, and consequently substrate utilization, thereby stimulating rhizosphere nutrient mobilization. However, at the same time, plants take up high levels of nutrients from the rhizosphere and can thus represent a strong competitor for nutrient resources, thereby reducing microbial growth (Bonkowski et al., 2000; Blagodatskaya et al., 2014). Therefore, competition for nutrients can exist in the rhizosphere, occurring mainly between plants and other soil organisms (Kuzyakov and Xu, 2013), while competition in the detritusphere occurs primarily within or between microbial species (Esperschütz et al., 2011). Due to the contrasting quality of C sources (root-derived low molecular weight C versus more macromolecular organic compounds in detritus), the abundance and successional changes of a microbial community may differ in intensity and dynamics between the rhizosphere and detritusphere. These differences and dynamics, in turn, may be a major determinant of predatory bacteria, RNA viral, and RNA phage dynamics. The functional traits of predatory versus non-predatory bacteria reveal faster growth and much faster C assimilation rates, which are comparable to the C flow through viruses and are substantially higher than those in predatory eukaryotes (Starr et al., 2020; Hungate et al., 2021). There is evidence that changes in the structure and dynamics of multi-level trophic interactions correspond to differences in energy flow between the rhizosphere and detritusphere. Thus, both the microbial community and the community of RNA eukaryotic viruses, as well as the community of phages inhabiting the detritusphere, are more distinct in structure from bulk soil than the rhizosphere community (Starr et al., 2019). As a result, successional changes in community structure are driven to a large extent by mycoviruses and phages, bacterial predators, and by protozoan grazers, demonstrating clear temporal population dynamics and patchy spatial distribution.
Rhizosphere–Porosphere Interactions
Rhizosphere processes are strongly influenced by interactions with the porosphere, forming a specific microbiome enriched in copiotrophic bacteria (Uksa et al., 2015; Blaser et al., 2016). Interactions with the porosphere alter the spatial expansion of the rhizosphere in soil, as root growth and architecture are affected by biopores (Han et al., 2015). Soil pores of different origins (e.g., root or earthworm-derived) serve as habitats for microorganisms, as well as conduits for chemical transport and water flow, and thus play a key role in controlling the rates of soil biochemical processes (Kravchenko et al., 2015; Negassa et al., 2015). Porosphere conditions influence microbial functioning due to the presence of roots, hyphae (Pagliai and De Nobili, 1993; Quigley et al., 2018), O2 levels (Keiluweit et al., 2016, 2017), or root exudate composition. Thus, it is not surprising that various groups of microorganisms are preferentially localized in pores of different sizes (Ruamps et al., 2013) or origin. C substrates localized in large pores are typically processed more rapidly than in small pores (Killham et al., 1993), and dissolved organic matter in small pores is more complex; hence, it is less decomposed than that in larger pores (Bailey et al., 2017; Toosi et al., 2017). Such a difference is not limited to size but also to the origin of the pores. For instance, biopores of decomposed roots or the drilosphere made by earthworms can host different varieties of microbes with distinct growth rates and efficiencies of growth (Ma et al., 2017; Hoang et al., 2020). The complexity of such systems increases even more when these spheres penetrate each other. For instance, earthworms reuse biopores of decomposed roots leaving behind the pore wall coatings or when roots grow within the drilosphere (Pagenkemper et al., 2013, 2015).
Rhizosphere–Aggregate Interactions
Roots and associated fungal hyphae enmesh particles and release agglutinating compounds, thus building up aggregates. Soil is defined as a group of primary soil particles (and smaller aggregates) that cohere to each other more strongly than surrounding non-cohesive particles and are considered as soil structural building units (Tisdall, 1996). A hierarchy of soil aggregates ranges from macroaggregates (>250 μm) that are unstable and susceptible to soil management to the more stable microaggregates (<250 μm) (Six et al., 2004). The group of microaggregates is not homogeneous and is organized even at the smallest scale <2 μm (Totsche et al., 2018). The primary structural units of microaggregates are composed of silicates, metal oxyhydroxides, organic matter, as well as microbial debris (Chenu and Plante, 2006). The role of roots is especially relevant in the formation of 53–250 μm micro-aggregates, while the formation of smaller aggregates (<53 μm) is mainly governed by microorganisms, clay particles, and physicochemical forces (Tisdall and Oades, 1982; Rillig and Mummey, 2006; Dultz et al., 2018). Due to relatively fast root growth and its associated rhizodeposition, aggregate formation and turnover in the rhizosphere and the root-mediated shaping of aggregate surfaces are highly dynamic processes (Wang et al., 2020).
The rhizosphere is a remarkable interface, where the aggregatosphere interacts with the detritusphere, because root detritus (sloughed root cells, dead root fragments, and residues) provide a substrate for microbial metabolism. Both, microbial metabolic products and rhizodeposits form sticky polymeric substances (Redmile-Gordon et al., 2014), which are involved in the enmeshing and gluing of aggregates (Golchin et al., 1994) by binding mineral soil particles and organic fragments in the way of homoaggregation as well as in heteroaggregation (Dultz et al., 2019). The aggregate stability at the rhizosphere–detritusphere interface is directly related to biotic factors such as root biomass, microbial, macro- and micro-faunal activity, all of those being involved in the structuring of aggregate surfaces (Golchin et al., 1994) by modifying the soil biotic and abiotic environment. Furthermore, positive correlation between soil water-stable aggregates and content of extracellular polysaccharides (EPS) indicated that soil abiotic conditions such as pH and water potential are the primary controllers of both, aggregate stability and microbial EPS production (Sher et al., 2020). This results in structural and functional self-organization of the pore space, which improves microbial habitats (Young and Crawford, 2004). In turn, aggregates, as a habitat for organisms, not only organize the soil microbiome but also serve as “concurrent incubators” that provide a refuge for microbes against predation (Hemkemeyer et al., 2014; Raynaud and Nunan, 2014).
Relevant Dynamic Drivers of Microbially-Mediated Soil Processes
Bio-Physical Conditions
The role of roots in aggregate formation has essential implications on rhizosphere physical properties such as O2 diffusion, which affects both microbiome and physiological root activities. Radial root growth and shrinkage create gaps along the root surface (Carminati et al., 2011) which may serve as conduits for preferential gas transport and enhance the replenishment of O2 consumed by aerobic respiration in deeper soil layers (Uteau et al., 2013). Growing roots also create new pores in the rhizosphere due to water extraction, which results in intensified drying and wetting cycles (Materechera et al., 1992; Rasse et al., 2000) or in local stress concentrations forming shear cracks, thus enhancing pore network connectivity in the surrounding root (Aravena et al., 2014). Air-filled porosity below a threshold of 12–15% is not sufficient to deliver enough O2 for root respiration in Zea mays, thus reducing the rates of root elongation due to low O2 levels (Grable and Siemer, 1968). This requirement of maize for air-filled porosity is higher than the 10% rule-of-thumb proposed in earlier studies (Wesseling and van Wijk, 1957; Robinson, 1964; Grable, 1966); nevertheless, the ability of the root to modify its surrounding soil structure helps to circumvent this issue (Lucas et al., 2019). The action of the root modifying its environment to ensure rapid O2 transport (Hinsinger et al., 2009) facilitates organic matter turnover in the rhizosphere (Jones and Hinsinger, 2008) because of intensified microbial activity compared to bulk soil (Nunan et al., 2003). Although most of the soil’s respiratory activity (microbial and root respiration) occurs in the rhizosphere (Kuzyakov, 2002; Raynaud, 2010), only a small number of studies have described the spatial distribution of O2 in structured aerobic rhizosphere soil, considering the water regime. A water content of approximately 30–40% of soil field capacity generally ensures high respiration rates, highlighting the importance of assessing moisture levels when estimating the required O2 supply (Balogh et al., 2011). At high moisture levels, microorganisms accelerate their metabolism, and at the same time, more pores are blocked by water bridges, limiting oxygen diffusion. Reduced redox potential at root surfaces due to high O2 consumption rates (Fischer et al., 1989) forms a gradient of oxygen concentrations, which ranges from very low at the root surface to the average soil concentration at a distance of approximately 15 mm (Keiluweit et al., 2015). Gradients of redox potential are most pronounced at the root tips, extending up to 3 mm from the root surface. Oxygen limitation can be detected at matric potentials exceeding a threshold value of approximately -3 kPa up to field capacity, showing a clear gradient while approaching the root’s surface that sharply decreases at a distance of 2–3 mm from the root surface (Uteau et al., 2015). Modeled O2 consumption in the rhizosphere demonstrates dynamic microbiome responses to O2 supply and the importance of soil structure around roots (Uteau et al., 2015).
Biochemical Conditions
Up to one-third of photosynthates allocated to roots is released to the soil, i.e., is “lost” by the plant (Pierret et al., 2007). Such losses through rhizodeposition (Lynch and Whipps, 1990) and release of protons (Ayres et al., 2009; Andrianarisoa et al., 2010; Cesarz et al., 2013) serve as plant investments to develop and modify the physical and biochemical properties of the rhizosphere environment to improve nutrient uptake (Augusto et al., 2002). Root exudates in the form of low molecular weight solutes strongly affect nutrient solubility, microbial activities, and the turnover of microbial biomass, as well as interactions between plants (Helal and Sauerbeck, 1986; Bertin et al., 2003; Vives-Peris et al., 2020), and the production of extracellular enzymes (Asmar et al., 1994), thereby indirectly influencing nutrient availability (Grayston et al., 1997; Hamilton and Frank, 2001; Herman et al., 2006; Landi et al., 2006). Roots can bypass the surrounding soil volume by self-regulation via the production of root hairs and exudates, by which more photosynthetic resources are allocated below-ground (Pages, 2002). Furthermore, the release of signaling molecules such as abscisic acid present in root exudates (Hartung, 2010) promotes the selection of particular microbial taxa within the vicinity of the root system (Marschner et al., 2004), allowing efficient complementary functioning of roots with microorganisms for nutrient mobilization. The release of H+ by roots into soils is one of the dominant mechanisms of plant nutrient mobilization and maintenance of a proper electrochemical potential in the rhizosphere (Marschner and Rengel, 2012). Among various plants, legumes strongly acidify rhizosphere soil (Israel and Jackson, 1978; Haynes, 1983), while certain other plants (e.g., most cereals) release OH– ions via the roots (Youssef et al., 1989). Overall, the ability of plant species to influence rhizosphere pH depends on the initial soil pH as well as N fertilization (Kuzyakov and Razavi, 2019).
Trophic Interactions
An additional relevant driver of microbial processes is interactions with soil organisms of higher trophic levels (Scheu et al., 2005). For example, rhizobacteria are top-down regulated by grazers, particularly by protists (Clarholm, 1985; Bonkowski, 2004). Grazing strongly affects the composition and functional evolution of microbial communities and fosters C and N mineralization from detritus for plant uptake (Alphei et al., 1996; Geisen et al., 2018). These mineralization processes depend on spatial distribution, size, and detritus quality (Bonkowski et al., 2000; Koller et al., 2013b). During decomposition of labile and recalcitrant C fractions of detritus, protist communities themselves undergo temporal succession at fine spatial and temporal scales (Hünninghaus et al., 2017). Microbial processes are also shaped by interactions with soil fauna (Bonkowski et al., 2000). For example, density-dependent and selective feeding by fungivore soil fauna affects the balance between mycorrhizal and saprotrophic fungi, nutrient mobilization, and thus plant performance (Klironomos and Ursic, 1998; Tiunov and Scheu, 2005). However, soil faunal activity affects physical soil structures, such as pores and microhabitats (Maraun et al., 1999; Eisenhauer et al., 2010). Therefore, faunal activity imposes spatial restrictions on soil organisms to sense and access food resources that shape trophic interactions (Erktan et al., 2020). The given examples above highlight the temporal and spatial complexity of multitrophic interactions as drivers of microbial processes. In turn, higher trophic level organisms, such as bacterial-feeding nematodes or protists, can provide strong feed-back in terms of bacterial respiration and nutrient mobilization, with the latter process being directly relevant for plant growth (Bonkowski, 2004; Brüggemann et al., 2011).
Relevant Scales for Process Localization
The ecological relevance of a soil process (gaseous emission, C sequestration, nutrient cycling, or leaching) is generally determined at the macro-scale, for example, on the landscape or soil profile level. Such approaches are important for global budget estimates. Understanding the mechanisms and spatial distribution of these processes requires, however, more precise mesocosm studies, while a shift to the micro- and even to the nanoscale is necessary to identify links between rates and distinct local processes in soil microhabitats.
Recent progress in process visualization at the mesoscale (root scale) was achieved using novel microsensor techniques and soil zymography. These approaches enable the monitoring of the two-dimensional distribution of soil properties, such as pH or oxygen concentration (Blossfeld and Gansert, 2007; Blossfeld et al., 2013) and the intensity of SOM decomposition (e.g., by CO2 and hydrolytic enzyme activities). Zymography coupled with X-ray CT is very promising for the 3D reconstruction of enzymatic processes within soil pore spaces, in particular, if the integration of 2D chemical imaging (in this case zymography) and 3D (μ)X-ray CT is further coupled with the modeling of pore-scale processes (Roose et al., 2016). For example, there are spatially explicit models for nutrient uptake by roots and root hairs based on addressStreetX-ray CT (Daly et al., 2016) that could be coupled with the 3D reconstruction of enzymatic processes for a comprehensive insight into enzyme-driven nutrient mobilization in the rhizosphere. A 4D visualization of dynamic developments of a process within the soil volume as well as a shift in zymography from the meso- to the microscopic scale remains a challenge. Such a challenge can be realized through the coupled visualization of soil processes and the estimation of localized process rates. This requires a methodology that considers biotic and abiotic drivers, functionally and phylogenetically diverse players, and multiple resolution scales of soil biochemical processes.
Relevant Approaches for Process Localization
Existing and newly developed methods for the determination of localized process rates in soil can be differentiated into three groups based on (i) destructive sampling disturbing soil microcosms, (ii) non-destructive imaging in situ techniques, and (iii) prediction by modeling.
Destructive Approaches
Techniques to identify microorganisms are currently mostly based on DNA sequencing approaches, as DNA has high information content regarding taxonomy (Table 1). Metagenomic sequencing can be used to obtain information concerning the broad functional potential of the microbiome when DNA sequencing is not limited to specific PCR-amplified phylogenetic or functional markers. To identify or quantify more specifically active decomposers, different methods have been developed, including meta-transcriptomics (Antunes et al., 2016; Yergeau et al., 2018; Bei et al., 2019; Nuccio et al., 2020), nanoscale secondary ion mass spectrometry (nanoSIMS) (Pett-Ridge and Firestone, 2017; Vidal et al., 2018), stable isotope probing of DNA, RNA, or PLFAs (Hannula et al., 2012; Hünninghaus et al., 2019; Maarastawi et al., 2019) and other techniques that relate abundance to metabolically active microbial consortia (Baldrian et al., 2012; Emerson et al., 2017). To predict process rates, focus on the quantitative parameters of a process rather than the mere structure of the microbial community is needed (Malik et al., 2020), which can be provided by measuring expression rates of genes involved in specific processes, determined by RT-qPCR to quantify expression of specific functional genes, or by metatranscriptomics, in which transcript numbers can be taken as a proxy for gene expression rates. However, it must be kept in mind that gene expression levels do not necessarily correlate with protein abundance, enzyme activity, or respiratory-based physiological approaches (Nannipieri et al., 2003). Recently, qSIP was proposed as an approach to quantify the metabolic activity of all specific groups of microorganisms that contribute to the substrate conversion process (Hungate et al., 2015; Papp et al., 2020).
Methodological Constraints
Whereas comparisons of values obtained within specific studies can provide valuable information concerning the performance of individual taxa under specific conditions and over a given period of time, comparison of microbial growth rates between studies based on molecular and physiological approaches reveals certain methodological constraints. Microbial-specific growth rates determined by SIGR in contrasting soil microhabitats (Figure 1) are comparable, but are reasonably slower than exponential growth of E. coli in pure culture (0.2–1.1 h–1, You et al., 2013). The specific bacterial growth rates obtained by isotope trace labeling (e.g., 0.33 h–1, Meisner et al., 2013) are in good agreement with SIGR (Figure 1). However, growth rates estimated by 18O qSIP in soils without substrate input (0.0002–0.001 h–1, data extracted from Figure 4 in Morrissey et al., 2018) and after glucose addition (0.0001–0.0065 h–1, data extracted from Figure 2 in Li et al., 2019) were 2–3 orders of magnitude lower than microbial growth as assessed by SIGR (Figure 1). This underestimation of growth rates by 18O qSIP occurred despite the calculations in a study by Li et al. (2019), who considered that only 60% of the oxygen in DNA comes from water (Koch and Munks, 2018). The rates obtained for growth on glucose by qSIP were several orders of magnitude slower than those obtained by SIGR. First, the growth rate calculations by qSIP were based on an incorrect assumption of steady state after glucose addition, neglecting that microorganisms are not in a steady state during growth. Second, the application of qSIP one week after substrate input is too late compared to exponential growth usually occurring within 20–48 h after soil activation with substrate (see e.g., Meisner et al., 2013; Loeppmann et al., 2020). Therefore, microbial growth rates determined 7 days after glucose addition mirror substrate-induced successional changes and substrate re-utilization rather than bacterial growth rates. Fungal and bacterial growth rates are very dynamic and can rise or fall up to 7–10 times within one week after substrate addition; thus, a shift of several days can occur between the peak values for bacterial and fungal growth (Nannipieri et al., 1978; Silva-Sánchez et al., 2019). Thus, the task of relating gene phenotypic traits to in situ growth rates still remains a challenge and requires consideration of microbial physiology in experimental designs.
The goal of quantifying the incorporation of a stable isotope label in specific groups of microorganisms has also been achieved by combining microarray analysis with nanoSIMS (Mayali et al., 2011). Incorporation of isotopically-labeled substrates into microbial biomolecules serves as a quantitative proxy for microbial activity, contributing to the decomposition of SOM or SOM components. However, this approach requires a degree of caution when comparing quantitative data involving studies using different time intervals and various types of biomolecules, because incorporation of isotope labels is affected by the turnover times of these biomolecules; for example, nucleic acids are more rapidly labeled relative to PLFAs and membrane lipids (Malik et al., 2015).
Non-destructive Approaches
The development of new approaches and concepts is not evenly distributed among the interfaces (hotspots); in particular, modern viewpoints are mostly presented for the rhizosphere (Table 1). For instance, most in situ techniques have been adapted for imaging rhizosphere properties and processes (Oburger and Schmidt, 2016). These approaches include i) optodes for measurement of CO2, pH, O2 (Blossfeld et al., 2011; Rudolph et al., 2013), (ii) sensitive gels (pH indicators (Römheld, 1986), (iii) zymography for enzyme activity (Spohn and Kuzyakov, 2013; Razavi et al., 2019), (iv) DGT gel (Diffusive Gradient in Thin-films) for elements (Fresno et al., 2017), and (v) imaging of radioactive isotopes: 14C (Pausch and Kuzyakov, 2011), 33P, 32P; 40Ca for nutrients and neutron imaging for water (Carminati and Vetterlein, 2013), and enable visualization of spatio-temporal patterns of rhizosphere properties and rhizosphere processes (Kuzyakov and Razavi, 2019). Such novel techniques have revealed a multiscale (time and space) examination of plant-microbiome interactions and their functionality (Baveye et al., 2018).
Methodological Constraints
Although visualization techniques enable quantitative estimates based on calibration, many still remain qualitative or semi-quantitative and do not exhibit consistent correspondence with the process rates and activity obtained by destructive sampling. For example, for approaches based on the application of sensor gels or membranes to the soil surface (i.e., optodes or zymography), essential methodological uncertainties occur related to the diffusion of targeted colored or fluorescent molecules (substrates or products of reactions) between the soil and membrane as well as within the membrane (Guber et al., 2018). Possible solutions for this problem could be a combination of activity hotspot localization by zymography with precise destructive micro-sampling after visualization (Tian et al., 2020) or visualization of the processes at the microscopic scale, avoiding attachment of artificial sensors or membranes (Table 1). Certain disagreements also occur involving molecular approaches (identifying plant and microbial traits by functional genes) and estimation of process rates, for example, by enzymatic activity (Kumar et al., 2017; Nilsson et al., 2019). Such disagreement confirms that gene existence does not necessarily reflect the activity of the corresponding protein (Nannipieri et al., 2018).
Hence, a quantitative estimation of process rates and the magnitude of changes in pools and fluxes is necessary at interfaces such as the rhizosphere to clarify, for instance, how inoculants modulate the resident microbiome, how pathogenic attack affects the activity of the complex microbiota of hotspots, how grazing activities by protists, nematodes, or bacteriophages control the extinction of species, and how the rhizosphere microbiome responds to abiotic stresses (e.g., salinity, drought, heat). This remains a challenging task, considering the diversity of C compounds in the rhizosphere and the challenges regarding their analysis (van Dam and Bouwmeester, 2016; Oburger and Jones, 2018).
Prediction Based on Statistical Analysis of Process Locations
Although a range of rhizosphere process-related parameters (e.g., pH, CO2, P, Mn content, and enzyme activity) are satisfactorily visualized in 2D by application of sensor membranes to the root–soil interface (Blossfeld and Gansert, 2007; Blossfeld et al., 2013), the localization of these parameters within the soil volume requires undesirable destructive sampling (Table 1). From CT-based 3D root localization within the soil domain, the probability distribution of the distance of a randomly selected location to the nearest root (Schlüter et al., 2018) can be computed, which potentially enables the development of a probabilistic 3D model to co-localize spatially-resolved arrays of rhizosphere-relevant parameters and the 3D architecture of root systems, for example, using Gaussian random fields (Blossfeld and Gansert, 2007; Blossfeld et al., 2013; Hristopulos, 2020). Such a model is easily extensible to account for local heterogeneity in the soil as well as topological and morphological properties of root architecture, such as branching, root tips, and root age. Moreover, spatial resolution of the predicted parameters can be defined by the underlying 2D measurements, thus enabling the investigation of various soil interfaces, as outlined above. By co-registration of MRI–PET (Jahnke et al., 2009), a 3D non-invasive analysis of plant structures and recently fixed C-transport processes within a root structure that may change in response to genomic, developmental, or environmental challenges may be established.
A stereological technique based on root architecture models such as CPlantBox (Schnepf et al., 2018a,b) provides a further promising perspective to overcome the need for expensive 3D imaging of plant roots, combining extensive model-based simulation of virtual root systems in 3D with machine learning methods. Thus, the spatially-resolved distribution of processes in the rhizosphere and other soil interfaces can be simulated in the 3D soil surrounding a plant root, using 2D measurements only.
Conclusion and Outlook: Emergent Properties of Microbial Activity in Soil
Traditionally, total microbial biomass, potential enzyme activities, substrate-induced respiration, and organic matter content in a given volume of soil have been used to predict decomposition activity and to model the fate of organic matter. To assess how the microscale generates macroscopic behavior, the so-called emergent properties, microscale heterogeneity, dynamics of substrate properties, and microbial activities need to be taken into account (Baveye et al., 2018). This aim is multidisciplinary and extremely challenging. It is necessary to link the spatial distribution of SOM (Peth et al., 2014; Müller et al., 2016; Rawlins et al., 2016) with its combined biophysical and biochemical properties, as well as with decomposer microorganisms and their respective traits and activities in the contexts of space and time (Baveye et al., 2018). Promising techniques that consider soil micro-heterogeneity are reproducible systems that mimic the soil and can be used for hypothesis testing (Tecon and Or, 2017). Novel characterization techniques are increasingly being employed to systematically track the characteristics of organic C conversion at the soil micro-interface (Table 1).
The transformation process of organic matter and its influencing factors are discussed at the scale of micro-ecological systems. Progress in near-edge X-ray absorption fine structure spectroscopy (NEXAFS), scanning transmission X-ray microscopy (STXM), X-ray absorption spectroscopy, micro-fluorescence spectroscopy, and nanoSIMS, as well as combined STXM-NanoSIMS (Keiluweit et al., 2012; Remusat et al., 2012), applied to soil thin sections, have revealed distinct spatial heterogeneity in the chemical composition of soils over minute distances (Lehmann et al., 2005; Mueller et al., 2013). Pulse-labeling experiments in combination with NanoSIMS enable tracing of the uptake, storage, and translocation of stable isotopes (Vidal et al., 2018). The development of novel detection technologies, such as NEXAFS and X-ray photoelectron spectroscopy (XPS) during the last decades, has greatly enriched our understanding of the microscopic distribution characteristics of SOM (Amelung et al., 2002). XPS was successfully adapted to determine the chemical composition of SOMs occluded in different aggregate size fractions. In addition, the spatial distribution of elements at a resolution of < 3 μm can be mapped in selected regions of coatings, mineral-organic associations, and aggregates using electron probe microanalysis (EPMA). Significant advances related to molecular markers and detection sensitivity now enable better detection of specific bacteria in soils and their spatial distribution at the micrometer scale to be determined in thin sections (Eickhorst and Tippkötter, 2008; Castorena et al., 2016). All of this information can, in principle, be combined and translated into 3D distributions using recently developed statistical algorithms.
To conclude, this review suggests a conceptual view emphasizing the central role of the rhizosphere in interactions with other biogeochemical interfaces. The main drivers of plant–microbial interactions, such as substrate input through exudation and rhizodeposition, and physico-chemical conditions (e.g., proton release and oxygen diffusion and transport) are already subject to intensive research. In contrast, the driving role of trophic interactions within and between interfaces, including competition for nutrients and successional dynamics, requires more specific studies involving both higher and lower trophic levels (e.g., protists, predatory bacteria, and mycoviruses). According to our concept, microorganisms are not the drivers, but they are the most abundant and powerful players in the soil interfaces because of the great diversity and specificity of genes encoding similar functions. The combination of phylogenic specificity and functional redundancy ensures the sustainability of soil microbial communities by the use of functional traits (e.g., the ability to produce specific extracellular enzymes, rapid or slow growth, and efficiency of metabolic pathways) as a tool to develop a microbial life strategy, which in turn affects the rates of transformation of organic compounds in soil. Thus, taxa with life strategies best adapted to the environment become dominant and alter the structure of the active microbial community. This self-regulatory mechanism maintains metabolic activity by the microbial community during the successional decomposition of organic substrates entering the soil. However, the rates of substrate decomposition are dependent on the functional traits of dominant taxa and microbial life strategy, which in turn are selected according to substrate quality and local environmental constraints, for example, water and nutrient availability. The rapid development of instrumental and molecular techniques has fueled attempts to reconsider the concepts of microbial life strategies with the goal of specifying functional groups according to their ecological relevance. This requires the identification and estimation of intrinsic traits by microbial physiology or phenotypic traits at the functional gene level. The quantitative definition of functional traits based on genetic and isotopic approaches is very promising, but demands further development with caution regarding the relevant resolution time and type of biomarker. Additional technique development is needed for ground-truth measurements of microbial growth in soil, linking physiological and molecular approaches. Therefore, the current challenge in modern ecology is the further development of cutting-edge methodologies for precise localization of biochemical processes, considering interactions within and between soil interfaces, as well as identifying and linking functional traits of plants and microbial populations that contribute to the rates of soil processes relevant at the ecosystem level.
Author Contributions
EB, SS, and BR developed the concept and all the co-authors contributed to the writing and structured the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was carried out in inspiration of the framework of the priority programme 2089 “Rhizosphere spatiotemporal organization—a key to rhizosphere functions” funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) Projects: 403664478, 403670038, 403641192, and 403637614. This publication was supported by the RUDN University Strategic Academic Leadership Program. The work of VS and MW was supported by the DFG grant No. SCHM 997/33-1. RK acknowledges support from the Helmholtz Association for the Forschungszentrum Jülich. The contribution of CK was additionally supported by Germany’s Excellence Strategy—EXC 2070–390732324.
Conflict of Interest
RK was employed by the company Forschungszentrum Jülich GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.625697/full#supplementary-material
References
Alphei, J., Bonkowski, M., and Scheu, S. (1996). Protozoa, Nematoda and Lumbricidae in the rhizosphere of Hordelymus europeaus (Poaceae): faunal interactions, response of microorganisms and effects on plant growth. Oecologia 106, 111–126. doi: 10.1007/BF00334413
Amelung, W., Kaiser, K., Kammerer, G., and Sauer, G. (2002). Organic carbon at soil particle surfaces—evidence from x-ray photoelectron spectroscopy and surface abrasion. Soil Sci. Soc. Am. J. 66, 1526–1530. doi: 10.2136/sssaj2002.1526
Andrianarisoa, K. S., Zeller, B., Poly, F., Siegenfuhr, H., Bienaimé, S., Ranger, J., et al. (2010). Control of nitrification by tree species in a common-garden experiment. Ecosystems 13, 1171–1187. doi: 10.1007/s10021-010-9390-x
Antunes, L. P., Martins, L. F., Pereira, R. V., Thomas, A. M., Barbosa, D., Lemos, L. N., et al. (2016). Microbial community structure and dynamics in thermophilic composting viewed through metagenomics and metatranscriptomics. Sci. Rep. 6:38915. doi: 10.1038/srep38915
Aravena, J. E., Berli, M., Ruiz, S., Suárez, F., Ghezzehei, T. A., and Tyler, S. W. (2014). Quantifying coupled deformation and water flow in the rhizosphere using X-ray microtomography and numerical simulations. Plant Soil 376, 95–110. doi: 10.1007/s11104-013-1946-z
Arsjad, S., and Giddens, J. (1966). Effect of added plant tissue on decomposition of soil organic matter under different wetting and drying cycles. Soil Sci. Soc. Am. Proc. 30:457. doi: 10.2136/sssaj1966.03615995003000040018x
Asmar, F., Eiland, F., and Nielsen, N. E. (1994). Effect of extracellular-enzyme activities on solubilization rate of soil organic nitrogen. Biol. Fertility Soils 17, 32–38. doi: 10.1007/BF00418669
Augusto, L., Ranger, J., Binkley, D., and Rothe, A. (2002). Impact of several common tree species of European temperate forests on soil fertility. Ann. Forest Sci. 59, 233–253. doi: 10.1051/forest:2002020
Aulakh, M., Wassmann, R., Bueno, C., Kreuzwieser, J., and Rennenberg, H. (2001). Characterization of root exudates at different growth stages of ten rice (Oryza sativa L.) cultivars. Plant Biol. 3, 139–148. doi: 10.1055/s-2001-12905
Averill, C., Turner, B. L., and Finzi, A. C. (2014). Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature 505, 543–545. doi: 10.1038/nature12901
Ayres, E., Steltzer, H., Berg, S., and Wall, D. H. (2009). Soil biota accelerate decomposition in high-elevation forests by specializing in the breakdown of litter produced by the plant species above them. J. Ecol. 97, 901–912. doi: 10.1111/j.1365-2745.2009.01539.x
Bååth, E. (2001). Estimation of fungal growth rates in soil using C-14-acetate incorporation into ergosterol. Soil Biol. Biochem. 33, 2011–2018. doi: 10.1016/S0038-0717(01)00137-7
Bååth, E., Pettersson, M., and Söderberg, K. H. (2001). Adaptation of a rapid andeconomical microcentrifugation method to measure thymidine and leucineincorporation by soil bacteria. Soil Biol. Biochem. 33, 1571–1574.
Badri, D. V., Quintana, N., El Kassis, E. G., Kim, H. K., Choi, Y. H., Sugiyama, A., et al. (2009). An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 151, 2006–2017. doi: 10.1104/pp.109.147462
Bailey, V. L., Smith, A. P., Tfaily, M., Fansler, S. J., and Bond-Lamberty, B. (2017). Differences in soluble organic carbon chemistry in pore waters sampled from different pore size domains. Soil Biol. Biochem. 107, 133–143. doi: 10.1016/S0038-0717(01)00073-6
Baldrian, P., Kolaøík, M., Štursová, M., Kopecký, J., Valášková, V., Větrovský, T., et al. (2012). Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 6, 248–258. doi: 10.1038/ismej.2011.95
Balogh, J., Pintér, K., Fóti, Sz, Cserhalmi, D., Papp, M., and Nagy, Z. (2011). Dependence of soil respiration on soil moisture, clay content, soil organic matter, and CO2 uptake in dry grasslands. Soil Biol. Biochem. 43, 1006–1013. doi: 10.1016/j.soilbio.2011.01.017
Barnard, R. L., Osborne, C. A., and Firestone, M. K. (2013). Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 7, 2229–2241. doi: 10.1038/ismej.2013.104
Bastian, F., Bouziri, L., Nicolardot, B., and Ranjard, L. (2009). Impact of wheat straw decomposition on successional patterns of soil microbial community structure. Soil Biol. Biochem. 41, 262–275. doi: 10.1016/j.soilbio.2008.10.024
Baveye, P. C., Otten, W., Kravchenko, A., Balseiro-Romero, M., Beckers, É, Chalhoub, M., et al. (2018). Emergent properties of microbial activity in heterogeneous soil microenvironments: different research approaches are slowly converging, yet major challenges remain. Front. Microbiol. 9:1929. doi: 10.3389/fmicb.2018.01929
Bei, Q. C., Moser, G., Wu, X. H., Müller, C., and Liesack, W. (2019). Metatranscriptomics reveals climate change effects on the rhizosphere microbiomes in European grassland. Soil Biol. Biochem. 138:107604. doi: 10.1016/j.soilbio.2019.107604
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Bertin, C., Yang, X., and Weston, L. A. (2003). The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 256, 67–83. doi: 10.1023/A:1026290508166
Blagodatskaya, E., Littschwager, J., Lauerer, M., and Kuzyakov, Y. (2014). Plant traits regulating N capture define microbial competition in the rhizosphere. Eur. J. Soil Biol. 61, 41–48. doi: 10.1016/j.ejsobi.2014.01.002
Blagodatsky, S. A., Heinemeyer, O., and Richter, J. (2000). Estimating the active and total soil microbial biomass by kinetic respiration analysis. Biol. Fertility Soils 32, 73–81. doi: 10.1007/s003740000219
Blaser, M. J., Cardon, Z. G., Cho, M. K., Dangl, J. L., Donohue, T. J., Green, J. L., et al. (2016). Toward a predictive understanding of Earth’s microbiomes to address 21st century challenges. mBio 7:e00714-16. doi: 10.1128/mBio.00714-16
Blossfeld, S., and Gansert, D. (2007). A novel non-invasive optical method for quantitative visualization of pH dynamics in the rhizosphere of plants. Plant Cell Environ. 30, 176–186. doi: 10.1111/j.1365-3040.2006.01616.x
Blossfeld, S., Gansert, D., Thiele, B., Kuhn, A. J., and Losch, R. (2011). The dynamics of oxygen concentration, pH value, and organic acids in the rhizosphere of Juncus spp. Soil Biol. Biochem. 6, 1186–1197. doi: 10.1016/j.soilbio.2011.02.007
Blossfeld, S., Schreiber, C. M., Liebsch, G., Kuhn, A. J., and Hinsinger, P. (2013). Quantitative imaging of rhizosphere pH and CO2 dynamics with planar optodes. Ann. Bot. 112, 267–276. doi: 10.1093/aob/mct047
Bonkowski, M. (2004). Protozoa and plant growth: the microbial loop in soil revisited. New Phytol. 162, 617–631. doi: 10.1111/j.1469-8137.2004.01066.x
Bonkowski, M., Cheng, W., Griffiths, B. S., Alphei, J., and Scheu, S. (2000). Microbial-faunal interactions in the rhizosphere and effects on plant growth. Eur. J. Soil Biol. 36, 135–147. doi: 10.1016/S1164-5563(00)01059-1
Bradford, M. (2016). Re-visioning soil food webs. Soil Biol. Biochem. 102, 1–3. doi: 10.1016/j.soilbio.2016.08.010
Brüggemann, N., Gessler, A., Kayler, Z., Keel, S. G., Badeck, F., Barthel, M., et al. (2011). Carbon allocation and carbon isotope fluxes in the plant-soil-atmosphere continuum: a review. Biogeosciences 8, 3457–3489. doi: 10.5194/bg-8-3457-2011
Brussaard, L. (2012). “Ecosystem services provided by the soil biota,” in Soil Ecology and Ecosystem Services, ed. D. H. Wall (Oxford: Oxford University Press), 45–58. doi: 10.1093/acprof:oso/9780199575923.003.0005
Brzostek, E. R., Dragoni, D., Brown, Z. A., and Phillips, R. P. (2015). Mycorrhizal type determines the magnitude and direction of root-induced changes in decomposition in a temperate forest. New Phytol. 206, 1274–1282. doi: 10.1111/nph.13303
Buckeridge, K. M., Fabio La Rosa, A., Mason, K. E., Jeanette, W., McNamara, N. P., Grant, H. K., et al. (2020). Sticky dead microbes: rapid abiotic retention of microbial necromass in soil. Soil Biol. Biochem. 149:107929. doi: 10.1016/j.soilbio.2020.107929
Carminati, A., Schneider, C. L., Moradi, A. B., Zarebanadkouki, M., Vetterlein, D., Vogel, H.-J., et al. (2011). How the rhizosphere may favor water availability to roots. Vadose Zone J. 10, 988–998. doi: 10.2136/vzj2010.0113
Carminati, A., and Vetterlein, D. (2013). Plasticity of rhizosphere hydraulic properties as a key for efficient utilization of scarce resources. Ann. Bot. 112, 277–290. doi: 10.1093/aob/mcs262
Castorena, E. V. G., del Carmen Gutiérrez-Castorena, M., Vargas, T. G., Bontemps, L. C., Martínez, J. D., Méndez, E. S., et al. (2016). Micromapping of microbial hotspots and biofilms from different crops using digital image mosaics of soil thin sections. Geoderma 279, 11–21. doi: 10.1016/j.geoderma.2016.05.017
Ceja-Navarro, J. A., Wang, Y., Ning, D., Arellano, A., Ramanculova, L., Yuan, M. M., et al. (2021). Protist diversity and community complexity in the rhizosphere of switchgrass are dynamic as plants develop. Microbiome 9:96. doi: 10.1186/s40168-021-01042-9
Cesarz, S., Ruess, L., Jacob, M., Jacob, A., Schaefer, M., and Scheu, S. (2013). Tree species diversity versus tree species identity: driving forces in structuring forest food webs as indicated by soil nematodes. Soil Biol. Biochem. 62, 36–45. doi: 10.1016/j.soilbio.2013.02.020
Chaparro, J. M., Badri, D. V., Bakker, M. G., Sugiyama, A., Manter, D. K., and Vivanco, J. M. (2013). Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One 8:55731. doi: 10.1371/journal.pone.0055731
Chaparro, J. M., Badri, D. V., and Vivanco, J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. doi: 10.1038/ismej.2013.196
Cheng, L., Booker, F. L., Tu, C., Burkey, K. O., Zhou, L., Shew, H. D., et al. (2012). Arbuscular mycorrhizal fungi increase organic carbon decomposition under elevated CO2. Science 337, 1084–1087. doi: 10.1126/science.1224304
Cheng, L., Zhang, N., Yuan, M., Xiao, J., Qin, Y., Deng, Y., et al. (2017). Warming enhances old organic carbon decomposition through altering functional microbial communities. ISME J. 11, 1825–1835. doi: 10.1038/ismej.2017.48
Cheng, W., Johnson, D. W., and Fu, S. (2003). Rhizosphere effects on decomposition: controls of plant species, phenology, and fertilization. Soil Sci. Soc. Am. J. 67, 1418–1427. doi: 10.2136/sssaj2003.1418
Cheng, W., Parton, W. J., Gonzalez-Meler, M. A., Phillips, R., Asao, S., McNickle, G. G., et al. (2014). Synthesis and modeling perspectives of rhizosphere priming. New Phytol. 201, 31–44. doi: 10.1111/nph.12440
Chenu, C., and Plante, A. F. (2006). Clay–sized organo-mineral complexes in a cultivation chronosequence: revisiting the concept of the ‘primary organo-mineral complex’. Eur. J. Soil Sci. 57, 596–607. doi: 10.1111/j.1365-2389.2006.00834.x
Chigineva, N. I., Aleksandrova, A. V., and Tiunov, A. V. (2009). The addition of labile carbon alters litter fungal communities and decreases litter decomposition rates. Appl. Soil Ecol. 42, 264–270. doi: 10.1016/j.apsoil.2009.05.001
Clarholm, M. (1985). Interactions of bacteria, protozoa and plants leading to mineralization of soil nitrogen. Soil Biol. Biochem. 17, 181–187. doi: 10.1016/0038-0717(85)90113-0
Cotton, T. E. A., Pétriacq, P., Cameron, D. D., Meselmani, M. A., Schwarzenbacher, R., Rolfe, S. A., et al. (2019). Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J. 13, 1647–1658. doi: 10.1038/s41396-019-0375-2
Daly, K. R., Keyes, S. D., Masum, S., and Roose, T. (2016). Image-based modelling of nutrient movement in and around the rhizosphere. J. Exp. Botany 67, 1059–1070. doi: 10.1093/jxb/erv544
Dazzo, F. B., and Gantner, S. (2013). “Molecular microbial ecology of the rhizosphere,” in In Situ Calling Distances and High Population Independent N-Acylhomoserine Lactone-Mediated Communication on Plant Root Surfaces, ed. F. DeBruijn (New York: J Wiley & Sons) 1, 785–789. doi: 10.1002/9781118297674.ch74
De Graaff, M. A., Classen, A. T., Castro, H. F., and Schadt, C. W. (2010). Labile soil carbon inputs mediate the soil microbial community composition and plant residue decomposition rates. New Phytol. 188, 1055–1064. doi: 10.1111/j.1469-8137.2010.03427.x
DeAngelis, K. M., Brodie, E. L., DeSantis, T. Z., Andersen, G. L., Lindow, S. E., and Firestone, M. K. (2009). Selective progressive response of soil microbial community to wild oat roots. ISME J. 3, 168–178. doi: 10.1038/ismej.2008.103
De-la-Peña, C., Badri, D. V., Lei, Z., Watson, B. S., Brandão, M. M., Silva-Filho, M. C., et al. (2010). Root secretion of defense-related proteins is development-dependent and correlated with flowering time. J. Biol. Chem. 285, 30654–30665. doi: 10.1074/jbc.M110.119040
Dijkstra, F. A., Zhu, B., and Cheng, W. (2020). Root effects on soil organic carbon: a double-edged sword. New Phytol. 230, 60–65. doi: 10.1111/nph.17082
Drigo, B., Pijl, A. S., Duyts, H., Kielak, A. M., Gamper, H. A., Houtekamer, M. J., et al. (2010). Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proc. Natl. Acad. Sci. U S A. 107, 10938–10942. doi: 10.1073/pnas.0912421107
Dultz, S., Steinke, H., Woche, S. K., Mikutta, R., and Guggenberger, G. (2018). Impact of organic matter types on surface charge and aggregation of goethite. Coll. Surf. Phys. Engin. Aspects 554, 156–168.
Dultz, S., Woche, S. K., Mikutta, R., Schrapel, M., and Guggenberger, G. (2019). Size and charge contraints in microaggregation: model experiments with particle size fractions. Appl. Clay Sci. 170, 29–40.
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U S A. 112, E911–E920.
Eickhorst, T., and Tippkötter, R. (2008). Improved detection of soil microorganisms usingfluorescence in situ hybridization (FISH) and catalyzed reporter deposition (CARD-FISH. Soil Biol. Biochem. 40, 1883–1891.
Eisenhauer, N., Beßler, H., Engels, C., Gleixner, G., Habekost, M., Milcu, A., et al. (2010). Plant diversity effects on soil microorganisms support the singular hypothesis. Ecology 91, 485–496.
el Zahar Haichar, F., Santaella, C., Heulin, T., and Achouak, W. (2014). Root exudates mediated interactions belowground. Soil Biol. Biochem. 77, 69–80.
Emerson, J. B., Adams, R. I., Román, C. M. B., Brooks, B., Coil, D. A., Dahlhausen, K., et al. (2017). Schrödinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 5:86. doi: 10.1186/s40168-017-0285-3
Erktan, A., Or, D., and Scheu, S. (2020). The physical structure of soil: determinant and consequence of trophic interactions. Soil Biol. Biochem. 148:107876. doi: 10.1002/ecy.2029
Esperschütz, J., Pérez-de-Mora, A., Schreiner, K., Welzl, G., Buegger, F., Zeyer, J., et al. (2011). Microbial food web dynamics along a soil chronosequence of a glacier forefield. Biogeosciences 8, 3283–3294.
Farrar, J., Hawes, M., Jones, D., and Lindow, S. (2003). How root control the flux of carbon to the rhizosphere. Ecology 84, 827–837.
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Fischer, W. R., Flessa, H., and Schaller, G. (1989). pH values and redox potentials in microsites of the rhizosphere. Z. Pflanzenernähr. Bodenk 152, 191–195.
Fontaine, S., Mariotti, A., and Abbadie, L. (2003). The priming effect of organic matter: a question of microbial competition? Soil Biol. Biochem. 35, 837–843.
Fresno, T., Penalosa, J. M., Santner, J., Puschenreiter, M., and Moreno-Jimenez, E. (2017). Effect of Lupinus albus L. root activities on as and Cu mobility after addition of ironbased soil amendments. Chemosphere 182, 373–381. doi: 10.1016/j.chemosphere.2017.05.034
Geisen, S., Mitchell, E. A. D., Adl, S., Bonkowski, M., Dunthorn, M., Ekelund, F., et al. (2018). Soil protists: a fertile frontier in soil biology research. FEMS Microbiol. Rev. 42, 293–323. doi: 10.1093/femsre/fuy006
Goberna, M., Pascual, J. A., Garcıa, C., and Sanchez, J. (2007). Do plant clumps constitute microbial hotspots in semiarid Mediterranean patchy landscapes? Soil Biol. Biochem. 39, 1047–1054.
Golchin, A., Oades, J. M., Skjemstad, J. O., and Clarke, P. (1994). Soil structure and carbon cycling. Aust. J. Soil Res. 32, 1043–1068.
Gorka, S., Dietrich, M., Mayerhofer, W., Gabriel, R., Wiesenbauer, J., Martin, V., et al. (2019). Rapid transfer of plant photosynthates to soil bacteria via ectomycorrhizal hyphae and its interaction with nitrogen availability. Front. Microbiol. 10:168. doi: 10.3389/fmicb.2019.00168
Grable, A. R. (1966). “Soil aeration and plant growth,” in Advances in Agronomy, ed. A. G. Norman (Cambridge, MA: Academic Press), 57–106.
Grable, A. R., and Siemer, E. G. (1968). Effects of bulk density, aggregate size, and soil water suction on oxygen diffusion, redox potentials, and elongation of corn roots. Soil Sci. Soc. Am. J. 32, 180–186.
Gransee, A., and Wittenmayer, L. (2000). Qualitative and quantitative analysis of water-soluble root exudates in relation to plant species and development. J. Plant Nutrit. Soil Sci. 163, 381–385.
Grayston, S. J., Vaughan, D., and Jones, D. (1997). Rhizosphere carbon flow in trees, in comparison with annual plants: the importance of root exudation and its impact on microbial activity and nutrient availability. Appl. Soil Ecol. 5, 29–56.
Griffiths, B., Spilles, A., and Bonkowski, M. (2012). C:N:P stoichiometry and nutrient limitation of the soil microbial biomass in a grazed grassland site under experimental P limitation or excess. Ecol. Proc. 1:6.
Guber, A., Kravchenko, A., Razavi, B. S., Uteau, D., Peth, S., Blagodatskaya, E., et al. (2018). Quantitative soil zymography: mechanisms, processes of substrate and enzyme diffusion in porous media. Soil Biol. Biochem. 127, 156–167.
Gui, H., Hyde, K., Xu, J., and Mortimer, P. (2017). Arbuscular mycorrhiza enhance the rate of litter decomposition while inhibiting soil microbial community development. Sci. Rep. 7:42184.
Hamilton, E. W. III, and Frank, D. A. (2001). Can plants stimulate soil microbes and their own nutrient supply? evidence from a grazing tolerant grass. Ecology 82, 2397–2402.
Han, E., Kautz, T., Perkons, U., Uteau, D., Peth, S., Huang, N., et al. (2015). Root growth dynamics inside and outside of soil biopores as affected by crop sequence determined with the profile wall method. Biol. Fertil. Soils 51, 847–856.
Hannula, S. E., Boschker, H. T. S., de Boer, W., and van Veen, J. A. (2012). 13C pulse-labeling assessment of the community structure of active fungi in the rhizosphere of a genetically starch-modified potato (Solanum tuberosum) cultivar and its parental isoline. New Phytol. 194, 784–799. doi: 10.1111/j.1469-8137.2012.04089.x
Hartung, W. (2010). The evolution of abscisic acid (ABA) and ABA function in lower plants, fungi and lichen. Funct. Plant Biol. 37, 806–812.
Helal, H. M., and Sauerbeck, D. (1986). Effect of plant roots on carbon metabolism of soil microbial biomass. Zeitschrift für Pflanzenernährung und Bodenkunde 149, 181–188.
Hemkemeyer, M., Pronk, G. J., Heister, K., Kögel-Knabner, I., Martens, R., and Tebbe, C. C. (2014). Artificial soil studies reveal domain-specific preferences of microorganisms for the colonisation of different soil minerals and particle size fractions. FEMS Microbiol. Ecol. 90, 770–782. doi: 10.1111/1574-6941.12436
Herman, D. J., Firestone, M. K., Nuccio, E., and Hodge, A. (2012). Interactions between an arbuscular mycorrhizal fungus and a soil microbial community mediating litter decomposition. FEMS Microbiol. Ecol. 80, 236–247.
Herman, D. J., Johnson, K. K., Jaeger, C. H., Schwartz, E., and Firestone, M. K. (2006). Root influence on nitrogen mineralization and nitrification in Avena barbata rhizosphere soil. Soil Sci. Soc. Am. J. 70, 1504–1511.
Hinsinger, P. (2001). Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237, 173–195.
Hinsinger, P., Bengough, A. G., Vetterlein, D., and Young, I. M. (2009). Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321, 117–152.
Ho, A., Lonardo, D. P. D., and Bodelier, P. L. E. (2017). Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 93, 1–14. doi: 10.1093/femsec/fix006
Hoang, D. T. T., Maranguit, D., Kuzyakov, Y., and Razavi, B. S. (2020). Accelerated microbial activity, turnover and efficiency in the drilosphere is depth dependent. Soil Biol. Biochem. 147:107852.
Hodge, A., Robinson, D., and Fitter, A. (2000). Are microorganisms more effective than plants at competing for nitrogen? Trends Plant Sci. 5, 304–308.
Huang, X. F., Chaparro, J. M., Reardon, K. F., Zhang, R., Shen, Q., and Vivanco, J. M. (2014). Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92, 267–275.
Hungate, B., Mau, R. L., Schwartz, E., Caporaso, J. G., Dijkstra, P., van Gestel, N., et al. (2015). Quantitative microbial ecology through stable isotope probing. Appl. Environ. Microbiol. 81, 7570–7581.
Hungate, B. A., Marks, J. C., Power, M. E., Schwartz, E., van Groenigen, K. J., Blazewicz, S. J., et al. (2021). The functional significance of bacterial predators. mBio 12:e00466-21. doi: 10.1128/mBio.00466-21
Hünninghaus, M., Dibbern, D., Kramer, S., Koller, R., Pausch, J., Schloter-Hai, B., et al. (2019). Disentangling carbon flow across microbial kingdoms in the rhizosphere of maize. Soil Biol. Biochem. 134, 122–130.
Hünninghaus, M., Koller, R., Kramer, S., Marhan, S., Kandeler, E., and Bonkowski, M. (2017). Changes in bacterial community composition and soil respiration indicate rapid successions of protist grazers during mineralization of maize crop residues. Pedobiologia 62, 1–8.
Israel, D. W., and Jackson, W. A. (1978). “The influence of nitrogen nutrition on ion uptake and translocation by leguminous plants,” in Mineral Nutrition of Legumes in Tropical and Subtropical Soils, eds C. S. Andrew and E. J. Kamprath (Melbourne: CSIRO), 113–129.
Jahnke, S., Menzel, M. I., van Dusschoten, D., Roeb, G. W., Bühler, J., Minwuyelet, S., et al. (2009). Combined MRI–PET dissects dynamic changes in plant structures and functions. Plant J. 59, 634–644. doi: 10.1111/j.1365-313X.2009.03888.x
Jakobsen, I., and Rosendahl, L. (1990). Carbon flow into soil and external hyphae from roots of mycorrhizal cucumber plant. New Phytol. 115, 77–83.
Jones, D., Nguyen, C., and Finlay, R. (2009). Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil 321, 5–33.
Jones, D. L., Hodge, A., and Kuzyakov, Y. (2004). Plant and mycorrhizal regulation of rhizodeposition. New Phytol. 163, 459–480.
Jones, S. E., and Lennon, J. T. (2010). Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. U S A 107, 5881–5886.
Kaiser, C., Kilburn, M. R., Clode, P. L., Fuchslueger, L., Koranda, M., Cliff, J. B., et al. (2015). Exploring the transfer of recent plant photosynthates to soil microbes: mycorrhizal pathway vs direct root exudation. New Phytol. 205, 1537–1551. doi: 10.1111/nph.13138
Kallenbach, C. M., Frey, S. D., and Grandy, A. S. (2016). Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat. Commun. 7:13630.
Kawasaki, A., Donn, S., Ryan, P. R., Mathesius, U., Devilla, R., Jones, A., et al. (2016). Microbiome and exudates of the root and rhizosphere of brachypodium distachyon, a model for wheat. PLoS One 11:e0164533. doi: 10.1371/journal.pone.0164533
Keiluweit, M., Bougoure, J. J., Nico, P. S., Pett-Ridge, J., Weber, P. K., and Kleber, M. (2015). Mineral protection of soil carbon counteracted by root exudates. Nat. Climate Change 5, 588–595.
Keiluweit, M., Bougoure, J. J., Zeglin, L. H., Myrold, D. D., Weber, P. K., Pett-Ridge, J., et al. (2012). Nano-scale investigation of the association of microbial nitrogen residues with iron (hydr)oxides in a forest soil O-horizon. Geochim. Cosmochim. Acta 95, 213–226.
Keiluweit, M., Nico, P. S., Kleber, M., and Fendorf, S. (2016). Are oxygen limitations under recognized regulators of organic carbon turnover in upland soils? Biogeochemistry 127, 157–171.
Keiluweit, M., Wanzek, T., Kleber, M., Nico, P., and Fendorf, S. (2017). Anaerobic microsites have an unaccounted role in soil carbon stabilization. Nat. Commun. 8:1771. doi: 10.1038/s41467-017-01406-6
Killham, K., Amato, M., and Ladd, J. N. (1993). Effect of substrate location in soil and soil pore-water regime on carbon turnover. Soil Biol. Biochem. 25, 57–62.
Klironomos, J. N., and Ursic, M. (1998). Density-dependent grazing on the extraradical hyphal network of the arbuscular mycorrhizal fungus, Glomus intraradices, by the collembolan, Folsomia candida. Biol. Fertility Soils 26, 250–253.
Koch, A., and Munks, S. (2018). A proposed strategy for maintaining mature forest habitat in Tasmania’s wood production forests. Ecol. Manag. Restorat. 19, 239–246.
Koller, R., Rodriguez, A., Robin, C., Scheu, S., and Bonkowski, M. (2013b). Protozoa enhance foraging efficiency of arbuscular mycorrhizal fungi for mineral nitrogen from organic matter in soil to the benefit of host plants. New Phytol. 199, 203–211. doi: 10.1111/nph.12249
Koller, R., Robin, C., Bonkowski, M., Ruess, L., and Scheu, S. (2013a). Litter quality as driving factor for plant nutrition via grazing of protozoa on soil microorganisms. FEMS Microbiol. Ecol. 85, 241–250. doi: 10.1111/1574-6941.12113
Kramer, S., Dibbern, D., Moll, J., Huenninghaus, M., Koller, R., Krueger, D., et al. (2016). Resource partitioning between bacteria, fungi, and protists in the detritusphere of an agricultural soil. Front. Microbiol. 7:1524. doi: 10.3389/fmicb.2016.01524
Krause, S., Le Roux, X., Niklaus, P. A., Van Bodegom, P. M., Lennon, J. T., Bertilsson, S., et al. (2014). Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front. Microbiol. 5:251. doi: 10.3389/fmicb.2014.00251
Kravchenko, A. N., Negassa, W. C., Guber, A. K., and Rivers, M. L. (2015). Protection of soil carbon within macro-aggregates depends on intra-aggregate pore characteristics. Sci. Rep. 5:16261. doi: 10.1038/srep16261
Kumar, A., Dorodnikov, M., Splettstößer, T., Kuzyakov, Y., and Pausch, J. (2017). Effects of maize roots on aggregate stability and enzyme activities in soil. Geoderma 306, 50–57. doi: 10.1016/j.geoderma.2017.07.007
Kuzyakov, Y. (2002). Separating microbial respiration of exudates from root respiration in non-sterile soils: a comparison of four methods. Soil Biol. Biochem. 34, 1621–1631.
Kuzyakov, Y., and Blagodatskaya, E. (2015). Microbial hotspots and hot moments in soil: concept & review. Soil Biol. Biochem. 83, 184–199.
Kuzyakov, Y., and Domanski, G. (2000). Carbon input by plants into the soil. Rev. J. Plant Nutrition Soil Sci. 163, 421–431.
Kuzyakov, Y., and Razavi, B. S. (2019). Rhizosphere size and shape: temporal dynamics and spatial stationarity. Soil Biol. Biochem. 135, 343–360.
Kuzyakov, Y., and Xu, X. (2013). Competition between roots and microorganisms for nitrogen: mechanisms and ecological relevance. New Phytol. 198, 656–669. doi: 10.1111/nph.12235
Landi, L., Valori, F., Ascher, J., Renella, G., Falchini, L., and Nannipieri, P. (2006). Root exudate effects on the bacterial communities, CO2 evolution, nitrogen transformations and ATP content of rhizosphere and bulk soils. Soil Biol. Biochem. 38, 509–516.
Lehmann, J., Liang, B., Solomon, D., Lerotic, M., Luizão, F., Kinyangi, J., et al. (2005). Near-edge X-ray absorption fine structure (NEXAFS) spectroscopy for mapping nano-scale distribution of organic carbon forms in soil: application to black carbon particles. Global Biogeochem. Cycles 19:GB1013.
Lesuffleur, F., Paynel, F., Bataillé, M. P., Le Deunff, E., and Cliquet, J. B. (2007). Root amino acid exudation: measurement of high efflux rates of glycine and serine from six different plant species. Plant Soil 294, 235–246.
Li, J., Mau, R. L., Dijkstra, P., Koch, B. J., Schwartz, E., Liu, X. J. A., et al. (2019). Predictive genomic traits for bacterial growth in culture versus actual growth in soil. ISME J. 13, 2162–2172. doi: 10.1038/s41396-019-0422-z
Lindahl, B. D., and Tunlid, A. (2015). Ectomycorrhizal fungi–potential organic matter decomposers, yet not saprotrophs. New Phytol. 205, 1443–1447. doi: 10.1111/nph.13201
Loeppmann, S., Breidenbach, A., Spielvogel, S., Dippold, M. A., and Blagodatskaya, E. (2020). Organic nutrients induced coupled C- and P-cycling enzyme activities during microbial growth in forest soils. Front. Forests Global Change 3:100. doi: 10.3389/ffgc.2020.00100
Louca, S., Polz, M. F., Mazel, F., Albright, M. B. N., Huber, J. A., O’Connor, M. I., et al. (2018). Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2, 936–943.
Lucas, M., Schlüter, S., Vogel, H.-J., and Vetterlein, D. (2019). Roots compact the surrounding soil depending on the structures they encounter. Sci. Rep. 9:16236. doi: 10.1038/s41598-019-52665-w
Ma, X., Razavi, B. S., Holz, M., Blagodatskaya, E., and Kuzyakov, Y. (2017). Warming increases hotspot areas of enzyme activity and shortens the duration of hot moments in the root-detritusphere. Soil Biol. Biochem. 107, 226–233.
Maarastawi, S. A., Frindte, K., Bodelier, P. L. E., and Knief, C. (2019). Rice straw serves as additional carbon source for rhizosphere microorganisms and reduces root exudate consumption. Soil Biol. Biochem. 135, 235–238.
Maarastawi, S. A., Frindte, K., Geer, R., Kröber, E., and Knief, C. (2018). Temporal dynamics and compartment specific rice straw degradation in bulk soil and the rhizosphere of maize. Soil Biol. Biochem. 127, 220–212.
Malik, A. A., Dannert, H., Griffiths, R. I., Thomson, B. C., and Gleixner, G. (2015). Rhizosphere bacterial carbon turnover is higher in nucleic acids than membrane lipids: implications for understanding soil carbon cycling. Front. Microbiol. 6:268. doi: 10.3389/fmicb.2015.00268
Malik, A. A., Martiny, J. B., Brodie, E. L., Martiny, A. C., Treseder, K. K., and Allison, S. D. (2020). Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J. 14, 1–9. doi: 10.1038/s41396-019-0510-0
Maraun, M., Alphei, J., Bonkowski, M., Buryn, R., Migge, S., Peter, M., et al. (1999). Middens of the earthworm Lumbricus terrestris (Lumbricidae): microhabitats for micro- and mesofauna in forest soil. Pedobiologia 43, 276–287.
Maron, P. A., Sarr, A., Kaisermann, A., Lévêque, J., Mathieu, O., Guigue, J., et al. (2018). High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 84:e02738-17. doi: 10.1128/AEM.02738-17
Marschner, P., Crowley, D., and Yang, C. H. (2004). Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil 261, 199–208.
Marschner, P., Marhan, S., and Kandeler, E. (2012). Microscale distribution and function of soil microorganisms in the interface between rhizosphere and detritusphere. Soil Biol. Biochem. 49, 174–183.
Marschner, P., and Rengel, Z. (2012). “Nutrient availability in soils,” in Marschner’s mineral nutrition of higher plants, ed. P. Marschner (Cambridge, MA: Academic press).
Materechera, S. A., Dexter, A. R., and Alston, A. M. (1992). Formation of aggregates by plant roots in homogenised soils. Plant Soil 142, 69–79.
Mayali, X., Wber, P. K., Brodie, E. L., Mabery, S., Hoeprich, P. D., and Pett-Ridge, J. (2011). High-throughput isotopic analysis of RNA microarrays to quantify microbial resource use. ISME J. 6, 1210–1221. doi: 10.1038/ismej.2011.175
McGuire, K. L., and Treseder, K. K. (2010). Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biol. Biochem. 42, 529–535.
Meisner, A., Bååth, E., and Rousk, J. (2013). Microbial growth responses upon rewetting soil dried for four days or one year. Soil Biol. Biochem. 66, 188–192.
Miltner, A., Bombach, P., Schmidt-Brücken, B., and Kästner, M. (2012). SOM genesis: microbial biomass as a significant source. Biogeochemistry 111, 41–55.
Mönchgesang, S., Strehmel, N., Schmidt, S., Westphal, L., Taruttis, F., et al. (2016). Natural variation of root exudates in Arabidopsis thaliana-linking metabolomic and genomic data. Sci. Rep. 6:29033. doi: 10.1038/srep29033
Morriën, E. (2016). Understanding soil food web dynamics, how close do we get? Soil Biol. Biochem. 102, 10–13.
Morris, S. J., and Blackwood, C. B. (2007). “The ecology of soil organisms,” in Soil Microbiology, Ecology, and Biochemistry, ed. E. Paul (Amsterdam: Elsevier), 195–229.
Morrissey, E. M., Mau, R. L., Schwartz, E., Koch, B. J., Hayer, M., and Hungate, B. A. (2018). Taxonomic patterns in the nitrogen assimilation of soil prokaryotes. Environ. Microbiol. 20, 1112–1119.
Mueller, C. W., Weber, P. K., Kilburn, M. R., Hoeschen, C., Kleber, M., and Pett-Ridge, J. (2013). Advances in the analysis of biogeochemical interfaces: NanoSIMS to investigate soil microenvironments. Adv. Agronomy 121, 1–46. doi: 10.1016/B978-0-12-407685-3.00001-3
Mulder, V. L., De Bruin, S., Schaepman, M. E., and Mayr, T. R. (2011). The use of remote sensing in soil and terrain mapping—a review. Geoderma 162, 1–19.
Müller, M., Schwab, N., Schickhoff, U., Böhner, J., and Scholten, T. (2016). Soil temperature and soil moisture patterns in a Himalayan alpine treeline ecotone. Arctic Antarctic Alpine Res. 48, 501–521.
Nannipieri, P., Ascher, J., Ceccherini, M., Landi, L., Pietramellara, G., and Renella, G. (2003). Microbial diversity and soil functions. Eur. J. Soil Sci. 54, 655–670.
Nannipieri, P., Giagnoni, L., Renella, G., Puglisi, E., Ceccanti, B., Masciandaro, G., et al. (2012). Soil enzymology: classical and molecular approaches. Biol. Fertility Soils 48:743.
Nannipieri, P., Johnson, R. L., and Paul, E. A. (1978). Criteria for measurement of microbial growth and activity in soil. Soil Biol. Biochem. 10, 223–229.
Nannipieri, P., Trasar-Cepeda, C., and Dick, R. P. (2018). Soil enzyme activity: a brief history and biochemistry as a basis for appropriate interpretations and meta-analysis. Biol. Fertility Soils 54, 11–19.
Negassa, W., Price, R. F., Basir, A., Snapp, S. S., and Kravchenko, A. (2015). Cover crop and tillage systems effect on soil CO2 and N2O fluxes in contrasting topographic positions. Soil Tillage Res. 154, 64–74.
Nguyen, C. (2003). Rhizodeposition of organic C by plants: mechanism and controls. Agronomie 23, 375–396.
Nicolás, C., Martin-Bertelsen, T., Floudas, D., Bentzer, J., Smits, M., Johansson, T., et al. (2019). The soil organic matter decomposition mechanisms in ectomycorrhizal fungi are tuned for liberating soil organic nitrogen. ISME J. 13, 977–988. doi: 10.1038/s41396-018-0331-336
Nie, M., Bell, C., Wallenstein, M. D., and Pendall, E. (2015). Increased plant productivity and decreased microbial respiratory C loss by plant growth-promoting rhizobacteria under elevated CO2. Sci. Rep. 5:9212. doi: 10.1038/srep09212
Nilsson, R. H., Anslan, S., Bahram, M., Wurzbacher, C., Baldrian, P., and Tedersoo, L. (2019). Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 17, 95–109.
Nuccio, E. E., Hodge, A., Pett-Ridge, J., Herman, D. J., Weber, P. K., and Firestone, M. K. (2013). An arbuscular mycorrhizal fungus significantly modifies the soil bacterial community and nitrogen cycling during litter decomposition. Environ. Microbiol. 15, 1870–1881. doi: 10.1111/1462-2920.12081
Nuccio, E. E., Starr, E., Karaoz, U., Brodie, E. L., Zhou, J., Tringe, S. G., et al. (2020). Niche differentiation is spatially and temporally regulated in the rhizosphere. ISME J. 14, 999–1014. doi: 10.1038/s41396-019-0582-x
Nunan, N., Wu, K., Young, I. M., Crawford, J. W., and Ritz, K. (2003). Spatial distribution of bacterial communities and their relationships with the micro-architecture of soil. FEMS Microbiol. Ecol. 44, 203–215. doi: 10.1016/S0168-6496(03)00027-8
Oburger, E., and Jones, D. L. (2018). Sampling root exudates – mission impossible? Rhizosphere 6, 116–133.
Oburger, E., and Schmidt, H. (2016). New methods to unravel rhizosphere processes. Trends Plant Sci. 21, 243–255.
Pagenkemper, S. K., Athmann, M., Uteau, D., Kautz, T., Peth, S., and Horn, R. (2015). The effect of earthworm activity on soil bioporosity – investigated with X-ray computed tomography and endoscopy. Soil Tillage Res. 146, 79–88.
Pagenkemper, S. K., Peth, S., Uteau Puschmann, D., and Horn, R. (2013). “Effects of root-induced biopores on pore space architecture investigated with industrial X-Ray computed tomography,” in Soil-water-root Processes: Advances in Tomography and Imaging, eds S. H. Anderson and J. W. Hopmans (Madison WI: American Society of Agronomy), 69–96.
Pagliai, M., and De Nobili, M. (1993). Relationships between soil porosity, root development and soil enzyme activity in cultivated soils. Soil Structure/Soil Biota Interrelationships 56, 243–256.
Papp, K., Hungate, B. A., and Schwartz, E. (2020). Glucose triggers strong taxon-specific responses in microbial growth and activity: insights from DNA and RNA qSIP. Ecology 101:e02887. doi: 10.1002/ecy.2887
Pascault, N., Ranjard, L., Kaisermann, A., Bachar, D., Christen, R., Terrat, S., et al. (2013). Stimulation of different functional groups of bacteria by various plant residues as a driver of soil priming effect. Ecosystems 16, 810–822.
Pausch, J., and Kuzyakov, Y. (2011). Photoassimilate allocation and dynamics of hotspots in roots visualized by 14C phosphor imaging. J. Plant Nutrition Soil Sci. 174, 12–19.
Pausch, J., and Kuzyakov, Y. (2018). Carbon input by roots into the soil: quantification of rhizodeposition from root to ecosystem scale. Global Change Biol. 24, 1–12. doi: 10.1111/gcb.13850
Pepe-Ranney, C., Campbell, A. N., Koechli, C. N., Berthrong, S., and Buckley, D. H. (2016). Unearthing the ecology of soil microorganisms using a high resolution DNA-SIP approach to explore cellulose and xylose metabolism in soil. Front. Microbiol. 7:703. doi: 10.3389/fmicb.2016.00703
Peth, S., Chenu, C., Leblond, N., Mordhorst, A., Garnier, P., Nunan, N., et al. (2014). Localization of soil organic matter in soil aggregates using synchrotron-based X-ray microtomography. Soil Biol. Biochem. 78, 189–194.
Pett-Ridge, J., and Firestone, M. K. (2017). Using stable isotopes to explore root-microbe-mineral interactions in soil. Rhizosphere 3, 244–253.
Pett-Ridge, J., Shi, S., Estera-Molina, K., Nuccio, E., Yuan, M., Rijkerts, R., et al. (2020). “Rhizosphere carbon turnover from cradle to grave: the role of microbe - plant interactions,” in Rhizosphere Biology, Interactions Between, Microbes, and Plants, eds V. V. S. R. Gupta and A. K. Sharma, (Berlin: Springer Nature Singapore Pte Ltd).
Pierret, A., Doussan, C., Capowiez, Y., Bastardie, F., and Pagès, L. (2007). Root functional architecture: a framework for modeling the interplay between roots and soil. Vadose Zone J. 6, 269–281.
Poirier, V., Roumet, C., and Munson, A. D. (2018). The root of the matter: linking root traits and soil organic matter stabilization processes. Soil Biol. Biochem. 120, 246–259.
Quigley, M. Y., Negassa, W. C., Guber, A. K., Rivers, M. L., and Kravchenko, A. N. (2018). Influence of pore characteristics on the fate and distribution of newly added carbon. Front. Environ. Sci. 6:51. doi: 10.3389/fenvs.2018.00051
Rasse, D. P., Smucker, A. J. M., and Santos, D. (2000). Alfalfa root and shoot mulching effects on soil hydraulic properties and aggregation. Soil Sci. Soc. Am. J. 64, 725–731. doi: 10.2136/sssaj2000.642725x
Rawlins, B. G., Wragg, J., Reinhard, C., Atwood, R. C., Houston, A. N., Lark, R. M., et al. (2016). Three-dimensional soil organic matter distribution, accessibility and microbial respiration in macroaggregates using osmium staining and synchrotron X-ray computed tomography. Soil 2, 659–671.
Raynaud, X. (2010). Soil properties are key determinants for the development of exudate gradients in a rhizosphere simulation model. Soil Biol. Biochem. 42, 210–219.
Raynaud, X., and Nunan, N. (2014). Spatial ecology of bacteria at the microscale in soil. PLoS One 9:e87217. doi: 10.1371/journalpone.0087217
Razavi, B. S., Zhang, X., Bilyera, N., Guber, A., and Zarebanadkouki, M. (2019). Soil zymography: simple and reliable? review of current knowledge and optimization of the method. Rhizosphere 11:100161.
Redmile-Gordon, M. A., Brookes, P. C., Evershed, R. P., Goulding, K. W. T., and Hirsch, P. R. (2014). Measuring the soil-microbial interface: extraction of extracellular polymeric substances (EPS) from soil biofilms. Soil Biol. Biochem. 72, 163–171.
Reinhold-Hurek, B., Bünger, W., Burbano, C. S., Sabale, M., and Hurek, T. (2015). Roots shaping their microbiome: global hotspots for microbial activity. Ann. Rev. Phytopathol. 53, 403–424. doi: 10.1146/annurev-phyto-082712-102342
Remusat, L., Hatton, P.-J., Nico, P. S., Zeller, B., Kleber, M., and Derrien, D. (2012). NanoSIMS study of organic matter associated with soil aggregates: advantages, limitations, and combination with STXM. Environ. Sci. Technol. 46, 3943–3949. doi: 10.1021/es203745k
Robinson, F. E. (1964). Required per cent air space for normal growth of sugar cane. Soil Sci. 98, 206–207.
Römheld, V. (1986). pH-Veranderungen in der rhizosphare verschiedener kulturpflanzenarten in abhangigkeit vom nahrstoffangebot. Potash Rev. 55, 1–8.
Roose, T., Keyes, S. D., Daly, K. R., Carminati, A., Otten, W., Vetterlein, D., et al. (2016). Challenges in imaging and predictive modeling of rhizosphere processes. Plant Soil 407, 9–38. doi: 10.1021/acs.accounts.0c00128
Ruamps, L. S., Nunan, N., Pouteau, V., Leloup, J., Raynaud, X., Roy, V., et al. (2013). Regulation of soil organic C mineralisation at the pore scale. FEMS Microbiol. Ecol. 86, 26–35. doi: 10.1111/1574-6941.12078
Rudolph, N., Voss, S., Moradi, A. B., Nagl, S., and Oswald, S. E. (2013). Spatio-temporal mapping of local soil pH changes induced by roots of lupin and soft-rush. Plant Soil 369, 669–680.
Scheffknecht, S., Mammerler, R., Steinkellner, S., and Vierheilig, H. (2006). Root exudates of mycorrhizal tomato plants exhibit a different effect on microconidia germination of Fusarium oxysporum f. sp. lycopersici than root exudates from non-mycorrhizal tomato plants. Mycorrhiza 16, 365–370. doi: 10.1007/s00572-006-0048-7
Scheu, S., Ruess, L., and Bonkowski, M. (2005). “Interactions between microorganisms and soil micro-and mesofauna,” in Microorganisms in Soils: Roles in Genesis and Functions, eds F. Buscot and A. Varma (Berlin, NY: Springer), 253–275.
Schlüter, S., Blaser, S., Weber, M., Schmidt, V., and Vetterlein, D. (2018). Quantification of root growth patterns from the soil perspective via root distance models. Front. Plant Sci. 9:1084. doi: 10.3389/fpls.2018.01084
Schmidt, H., and Eickhorst, T. (2014). Detection and quantification of native microbial populations on soil-grown rice roots by catalyzed reporter deposition-fluorescence in situ hybridization. FEMS Microbiol. Ecol. 87, 390–402. doi: 10.1111/1574-6941.12232
Schmidt, H., Nunan, N., Höck, A., Eickhorst, T., Kaiser, C., Woebken, D., et al. (2018). Recognizing patterns: spatial analysis of observed microbial colonization on root surfaces. Front. Environ. Sci. 6:61. doi: 10.3389/fenvs.2018.00061
Schnepf, A., Huber, K., Landl, M., Meunier, F., Petrich, L., and Schmidt, V. (2018a). Statistical characterization of the root system architecture model CRootBox. Vadose Zone J. 17:170212.
Schnepf, A., Leitner, D., Landl, M., Lobet, G., Hieu Mai, T. H., Morandage, S., et al. (2018b). CRootBox: a structural–functional modelling framework for root systems. Ann. Botany 121, 1033–1053. doi: 10.1093/aob/mcx221
Scott, M., Klumpp, S., Mateescu, E. M., and Hwa, T. (2014). Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Systems Biol. 10:747. doi: 10.15252/msb.20145379
Sher, Y., Baker, N. R., Herman, D., Fossum, C., Hale, L., Zhang, X., et al. (2020). Microbial extracellular polysaccharide production and aggregate stability controlled by switchgrass (Panicum virgatum) root biomass and soil water potential. Soil Biol. Biochem. 143:107742.
Silva-Sánchez, A., Soares, M., and Rousk, J. (2019). Testing the dependence of microbial growth and carbon use efficiency on nitrogen availability, pH, and organic matter quality. Soil Biol. Biochem. 134, 25–35.
Six, J., Bossuyt, H., Degryze, S., and Denef, K. (2004). A history of research on the link between (micro)aggregates, soil biota, and soil organic matter dynamics. Soil Till. Res. 79, 7–31.
Smith, M. R., Rao, I. M., and Merchant, A. (2018). Source-Sink relationships in crop plants and their influence on yield development and nutritional quality. Front. Plant Sci. 9:1889. doi: 10.3389/fpls.2018.01889
Sokol, N. W., Sanderman, J., and Bradford, M. A. (2018). Pathways of mineral-associated soil organic matter formation: integrating the role of plant carbon source, chemistry, and point of entry. Glob. Change Biol. 25, 12–24. doi: 10.1111/gcb.14482
Song, C., Zhu, F., Carrión, V. J., and Cordovez, V. (2020). Beyond plant microbiome composition: exploiting microbial functions and plant traits via integrated approaches. Front. Bioeng. Biotechnol. 8:896. doi: 10.3389/fbioe.2020.00896
Spohn, M., and Kuzyakov, Y. (2013). Distribution of microbial- and root-derived phosphatase activities in the rhizosphere depending on P availability and C allocation – coupling soil zymography with 14C imaging. Soil Biol. Biochem. 67, 106–113.
Starr, E. P., Nuccio, E. E., Pett-Ridge, J. J., Banfield, J. F., and Firestoned, M. K. (2019). Metatranscriptomic reconstruction reveals RNA viruses with the potential to shape carbon cycling in soil. PNAS 116:51. doi: 10.1073/pnas.1908291116
Starr, E. P., Shi, S., Blazewicz, S. J., Koch, B. J., Probst, A. J., Hungate, B. A., et al. (2020). Stable isotope informed genomeresolved metagenomics uncovers potential trophic interactions in rhizosphere soil. bioRxiv [preprint] doi: 10.1101/2020.08.21.262063
Steer, J., and Harris, J. A. (2000). Shifts in the microbial community in rhizosphere and non-rhizosphere soils during the growth of Agrostis stolonifera. Soil Biol. Biochem. 32, 869–878.
Sterkenburg, E., Clemmensen, K. E., Ekblad, A., Finlay, R. D., and Lindahl, B. D. (2018). Contrasting effects of ectomycorrhizal fungi on early and late stage decomposition in a boreal forest. ISME J. 12, 2187–2197. doi: 10.1038/s41396-018-0181-2
Swinnen, J., van Veen, J., and Merckx, R. (1994a). 14C pulse-labeling of field-grown spring wheat: an evaluation of its use in rhizosphere carbon budget estimations. Soil Biol. Biochem. 26, 161–170.
Swinnen, J., van Veen, J., and Merckx, R. (1994b). Rhizosphere carbon fluxes in field-grown spring wheat: model calculations based on 14C partitioning after pulse-labeling. Soil Biol. Biochem. 26, 171–182.
Tecon, R., and Or, D. (2017). Biophysical processes supporting the diversity of microbial life in soil. FEME Microb. Rev. 41, 599–623.
Tian, P., Razavi, B. S., Zhang, X., Wang, Q., and Blagodatskaya, E. (2020). Microbial growth and enzyme kinetics in rhizosphere hotspots are modulated by soil organics and nutrient availability. Soil Biol. Biochem. 141:107662. doi: 10.1016/j.soilbio.2019.107662
Tisdall, J. M. (1996). “Formation of soil aggregates and accumulation of soil organic matter,” in Structure and Organic Matter Storage in Agricultural Soils, eds M. R. Carter and B. A. Stewart (Boca Raton, FL: CRC Press), 57–96.
Tisdall, J. M., and Oades, J. M. (1982). Organic matter and water-stable aggregates in soils. J. Soil Sci. 33, 141–163.
Tiunov, A. V., and Scheu, S. (2005). Arbuscular mycorrhiza and Collembola interact in affecting community composition of saprotrophic microfungi. Oecologia 142, 636–642. doi: 10.1007/s00442-004-1758-1
Toosi, E. R., Kravchenko, A. N., Mao, J., Quigley, M. Y., and Rivers, M. L. (2017). Effects of management and pore characteristics on organic matter composition of macroaggregates: evidence from characterization of organic matter and imaging. Eur. J. Soil Sci. 68, 200–211.
Totsche, K. U., Amelung, W., Gerzabek, M. H., Guggenberger, G., Klumpp, E., Knief, C., et al. (2018). Microaggregates in soils. J. Plant Nutrition Soil Sci. 181, 104–136.
Uksa, M., Schloter, M., Endesfelder, D., Kublik, S., Engel, M., Kautz, T., et al. (2015). Prokaryotes in subsoil—evidence for a strong spatial separation of different phyla by analysing co-occurrence networks. Front. Microbiol. 6:1269. doi: 10.3389/fmicb.2015.01269
Uteau, D., Hafner, S., Pagenkemper, S. K., Peth, S., Wiesenberg, G. L. B., Kuzyakov, Y., et al. (2015). Oxygen and redox potential gradients in the rhizosphere of alfalfa grown on a loamy soil. J. Plant Nutr. Soil Sci. 178, 278–287.
Uteau, D., Pagenkemper, S. K., Peth, S., and Horn, R. (2013). Root and time dependent soil structure formation and its influence on gas transport in the subsoil. Soil Tillage Res. 132, 69–76.
van Dam, N. M., and Bouwmeester, H. J. (2016). Metabolomics in the rhizosphere: tapping into belowground chemical communication. Trends Plant Sci. 21, 256–265. doi: 10.1016/j.tplants.2016.01.008
van der Heijden, M. G., Martin, F. M., Selosse, M. A., and Sanders, I. R. (2015). Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol. 205, 1406–1423. doi: 10.1111/nph.13288
Vidal, A., Hirte, J., Bender, S. F., Mayer, J., Gattinger, A., Höschen, C., et al. (2018). Linking 3D soil structure and plant-microbe-soil carbon transfer in the rhizosphere. Front. Environ. Sci. 6:9. doi: 10.3389/fenvs.2018.00009
Vives-Peris, V., de Ollas, C., Gómez-Cadenas, A., and Pérez-Clemente, R. M. (2020). Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17.
Wagg, C., Bendera, S. F., Widmer, F., and van der Heijden, M. G. A. (2014). Soil biodiversity and soil community composition determine ecosystem multifunctionality. PNAS 111, 5266–5270.
Wagg, C., Schlaeppi, K., Banerjee, S., Kuramae, E. E., and van der Heijden, M. G. A. (2019). Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 10:4841. doi: 10.1038/s41467-019-12798-y
Wang, X., Yin, L., Dijkstra, F. A., Lu, J., Wang, P., and Cheng, W. (2020). Rhizosphere priming is tightly associated with root-driven aggregate turnover. Soil Biol. Biochem. 20:107964.
Wesseling, J., and van Wijk, W. R. (1957). “Land drainage in relation to soils and crops. I. soil physical conditions in relation to drain depth,” in Drainage of Agricultural Land, ed. J. D. Luthin (Madison, WI: American Society of Agronomy), 461–504.
Yergeau, E., Tremblay, J., Joly, S., Labrecque, M., Maynard, C., Pitre, F. E., et al. (2018). Soil contamination alters the willow root and rhizosphere metatranscriptome and the root-rhizosphere interactome. ISME J. 12, 869–884. doi: 10.1038/s41396-017-0018-4
You, C., Okano, H., Hui, S., Zhang, Z., Kim, M., Gunderson, C. W., et al. (2013). Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 500, 301–306.
Young, I. M., and Crawford, J. W. (2004). Interactions and self-organization in the soil-microbe complex. Science 304, 1634–1637.
Youssef, R. A., Kanazawa, S., and Chino, M. (1989). Distribution of microbial biomass in the rhizosphere of barley in soil. Biol. Fertil. Soils 7, 341–345. doi: 10.1111/j.1574-6941.2007.00383.x
Zechmeister-Boltenstern, S., Keiblinger, K. M., Mooshammer, M., Penuelas, J., Richter, A., Sardans, J., et al. (2015). The application of ecological stoichiometry to plant-microbial-soil organic matter transformations. Ecol. Monographs 85, 133–155.
Zhalnina, K., Louie, K. B., ‘Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Keywords: rhizosphere, mycorrhizosphere, detritusphere, (bio)-pores, soil aggregates, soil priming, trophic interactions, statistical analysis of process locations
Citation: Blagodatskaya E, Tarkka M, Knief C, Koller R, Peth S, Schmidt V, Spielvogel S, Uteau D, Weber M and Razavi BS (2021) Bridging Microbial Functional Traits With Localized Process Rates at Soil Interfaces. Front. Microbiol. 12:625697. doi: 10.3389/fmicb.2021.625697
Received: 03 November 2020; Accepted: 01 September 2021;
Published: 28 October 2021.
Edited by:
Nuria Ferrol, Experimental Station of Zaidín (EEZ), SpainReviewed by:
Jennifer Pett-Ridge, Lawrence Livermore National Laboratory, United States Department of Energy (DOE), United StatesPhilippe Constant, Université du Québec, Canada
Copyright © 2021 Blagodatskaya, Tarkka, Knief, Koller, Peth, Schmidt, Spielvogel, Uteau, Weber and Razavi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Evgenia Blagodatskaya, amFuZWJsYWdAbWFpbC5ydQ==
 Evgenia Blagodatskaya
Evgenia Blagodatskaya Mika Tarkka
Mika Tarkka Claudia Knief
Claudia Knief Robert Koller
Robert Koller Stephan Peth
Stephan Peth Volker Schmidt
Volker Schmidt Sandra Spielvogel
Sandra Spielvogel Daniel Uteau
Daniel Uteau Matthias Weber
Matthias Weber Bahar S. Razavi
Bahar S. Razavi