
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 February 2021
Sec. Biology of Archaea
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.625599
This article is part of the Research Topic Diversity, Ecology and Evolution of Archaeal Viruses View all 5 articles
 Colin Tittes1†
Colin Tittes1† Sabine Schwarzer1†
Sabine Schwarzer1† Friedhelm Pfeiffer2
Friedhelm Pfeiffer2 Mike Dyall-Smith2,3
Mike Dyall-Smith2,3 Marta Rodriguez-Franco4
Marta Rodriguez-Franco4 Hanna M. Oksanen5
Hanna M. Oksanen5 Tessa E. F. Quax1*
Tessa E. F. Quax1*Hypersaline environments are the source of many viruses infecting different species of halophilic euryarchaea. Information on infection mechanisms of archaeal viruses is scarce, due to the lack of genetically accessible virus–host models. Recently, a new archaeal siphovirus, Haloferax tailed virus 1 (HFTV1), was isolated together with its host belonging to the genus Haloferax, but it is not infectious on the widely used model euryarcheon Haloferax volcanii. To gain more insight into the biology of HFTV1 host strain LR2-5, we studied characteristics that might play a role in its virus susceptibility: growth-dependent motility, surface layer, filamentous surface structures, and cell shape. Its genome sequence showed that LR2-5 is a new strain of Haloferax gibbonsii. LR2-5 lacks obvious viral defense systems, such as CRISPR-Cas, and the composition of its cell surface is different from Hfx. volcanii, which might explain the different viral host range. This work provides first deep insights into the relationship between the host of halovirus HFTV1 and other members of the genus Haloferax. Given the close relationship to the genetically accessible Hfx. volcanii, LR2-5 has high potential as a new model for virus–host studies in euryarchaea.
Viruses outnumber their microbial hosts by about a factor of 10 (Bergh et al., 1989; Wommack and Colwell, 2000; Suttle, 2007). Consequently, viruses have an important role in many ecosystems and impact microbial communities worldwide (Fuhrman, 1999; Suttle, 2007; Danovaro et al., 2016). Archaea are ubiquitous microorganisms that thrive both in extreme habitats such as thermal hot springs and hypersaline lakes, as well as in moderate environments like the oceans, soil, and human gut (Karner et al., 2001; Lloyd et al., 2013; Lurie-Weinberger and Gophna, 2015). Many archaeal viruses differ significantly from those infecting bacteria and eukaryotes. The morphology of archaeal virions and the viral genomes are characterized by a high level of diversity. Some archaeal viruses, especially those infecting hyperthermophilic crenarchaea, have unique morphologies that are not encountered for viruses infecting bacteria and eukaryotes (Prangishvili et al., 2017; Munson-Mcgee et al., 2018). Other archaeal viruses, mainly infecting euryarchaea, display morphologies shared with some bacterial viruses (bacteriophages), such as head-tail or icosahedral shapes (Pietilä et al., 2014; Prangishvili et al., 2017). The study of archaeal viruses has been important to gain insight into the origin and evolution of viruses in general (Forterre and Prangishvili, 2009). The large majority of genes carried by archaeal virus genomes encode proteins of unknown function, and consequently many aspects of the interaction between these viruses and their hosts remain enigmatic (Prangishvili et al., 2017; Krupovic et al., 2018). Studies on infection mechanisms or host recognition complexes of archaeal viruses are rare, but have yielded surprising results showing that infection strategies can be unique or display similarities with viruses infecting other domains of life (El Omari et al., 2019; Santos-Pérez et al., 2019). For example, entry and egress mechanisms of archaeal viruses can rely on fusion with and budding of the virus through the cell membrane, respectively, as observed for eukaryotic viruses, or on the formation of archaeal specific pyramidal egress structures (Bize et al., 2009; Snyder et al., 2013; Quemin et al., 2016; El Omari et al., 2019). To advance studies on virus–host interactions and infection mechanisms in archaea, model systems consisting of a well-characterized host and virus are needed. Halophilic euryarchaea have proven a rich source of archaeal viruses. To date, more than 100 haloarchaeal viruses have been isolated of which the majority are tailed icosahedral double-stranded DNA viruses representing the order Caudovirales (Atanasova et al., 2015a). Several haloarchaea are excellent research organisms as they are straightforward to cultivate, have relatively fast doubling times, and for several of them elaborate genetic tools are available (Leigh et al., 2011; Cheng et al., 2017). Haloferax volcanii is widely used as genetically accessible model organism, and different aspects of its biology such as replication, cell division, protein turn-over, transcription, translation, and defense against viruses have been studied in detail (Eichler and Maupin-Furlow, 2013; Hawkins et al., 2013; Duggin et al., 2015; Maier et al., 2015; Pohlschroder and Schulze, 2019; Haque et al., 2020; Schulze et al., 2020). Curiously, in contrast to other haloarchaeal genera, viruses for the genus of Haloferax are extremely rare (Atanasova et al., 2012, 2015b). The first reports of Haloferax infecting viruses are those of HF1 virus infecting Haloferax lucentense and Hfx. volcanii, and a defective provirus of Haloferax mediterranei, both of which are no longer available (Nuttall and Smith, 1993; Li et al., 2013; M.L. Dyall-Smith, pers. communication). Recently, a new virus was isolated infecting a member of the genus Haloferax. Haloferax tailed virus 1 (HFTV1) and its host, Haloferax sp. LR2-5, originate from the saline Lake Retba near Dakar in Senegal (Mizuno et al., 2019).
The siphovirus HFTV1 has an icosahedral head of ∼50 nm diameter and a long non-contractile tail of ∼60 nm (Mizuno et al., 2019). Four major protein types were detected in the HFTV1 virion. The linear, circularly permuted dsDNA genome of 38 kb encodes 70 ORFs, of which half has homology to haloarchaeal viral genes, such as the archaeal siphovirus HRTV-4 isolated from Margherita di Savoia, Italy, and of uncultivated haloviruses from the solar saltern Santa Pola, Spain (Mizuno et al., 2019). The genome is likely subjected to a headful packaging mechanism initiated from a pac site (Mizuno et al., 2019). HFTV1 has a narrow host range among the Lake Retba archaeal strains tested. Besides its original host Hfx. sp. LR2-5, it infects Halorubrum sp. LR1-23 but not any of the endogenous Haloferax strains (Mizuno et al., 2019).
Currently (as of 26 January 2021), 21 species of the genus Haloferax are listed by name in the NCBI taxonomy1 of which 13 have validly published names according to LPSN2. Twenty-eight complete or draft genome sequences from the genus Haloferax are available (Lynch et al., 2012; Becker et al., 2014) including the complete sequences of Haloferax gibbonsii strain ARA6 (Pinto et al., 2015), Hfx. volcanii strain DS2T (Hartman et al., 2010), and Hfx. mediterranei strain R-4 (ATCC 33500) (Han et al., 2012).
To gain insight into the question why Haloferax strains seem so resilient to viral infection and what makes the LR2-5 strain an exception, we sequenced and annotated its genome and compared it to closely related strains. We could assign this isolate to the species Hfx. gibbonsii using a genome-based taxonomy (TYGS), and consequently LR2-5 was renamed as Hfx. gibbonsii LR2-5. In addition, we studied its biological properties, such as growth, cell shape, motility, and composition of its cell surface, which will contribute to developing the LR2-5 and HFTV1 system as an attractive model for archaeal virus–host studies.
To gain insight into the optimal cultivation conditions of the environmentally isolated strain Hfx. gibbonsii LR2-5, growth media with different composition and several incubation temperatures were tested (Supplementary Figure 1). The shortest doubling time of 3.5 h was achieved when the strain was aerobically grown in rich YPC medium with a total salt concentration of 18% (wt/vol) at 42°C (Supplementary Figure 1). The doubling time of cells grown at 37°C is about 4.5 h in MGM medium and 6.5 h in CA medium. Cultures grown at 37°C exhibit prolonged lag-phases compared to cells grown at higher temperatures, whereas cultures grown at 45°C reach lower final optical densities (Supplementary Figure 1).
Within the tested range (18 and 23%), the salt concentrations had no effect on the doubling times. The highest final optical densities under all conditions were reported in YPC and CAB medium with 18% (wt/vol) salt concentration. The optimal growth medium and conditions are similar to those of Hfx. volcanii H26 (a derivative of strain DS2T, see Supplementary Text) (Jantzer et al., 2011), facilitating use of methods established for this model strain.
Haloferax volcanii is reported to change its shape during growth in CA medium (Li et al., 2019; de Silva et al., 2021). We analyzed the cell shape of LR2-5 in CA medium by phase contrast light microscopy. This revealed that the cells were rod-shaped and usually ca. 1.5–4 μm in length in early exponential growth phase (below OD600 0.2) (Supplementary Table 1). As the cultures reached mid-exponential growth phase (OD600 0.2–0.6), a mix of rod- and plate-shaped cells was observed. In stationary phase (above OD600 0.6), all cells appeared plate-shaped (Figure 1A). This transition from rod- to plate-shaped cells is reminiscent of what was recently observed for Hfx. volcanii (Li et al., 2019; de Silva et al., 2021). However, whereas Hfx. volcanii is rod-shaped only during the very early exponential growth phase (below OD600 0.1), Hfx. gibbonsii LR2-5 maintains the rod-shape much longer during development. The differences between the strains might originate from the longer adaptation of Hfx. volcanii to laboratory culture conditions where motility is not an evolutionary advantage, as the rod-shape seems to be linked with motility in Hfx. volcanii (Duggin et al., 2015; Li et al., 2019).
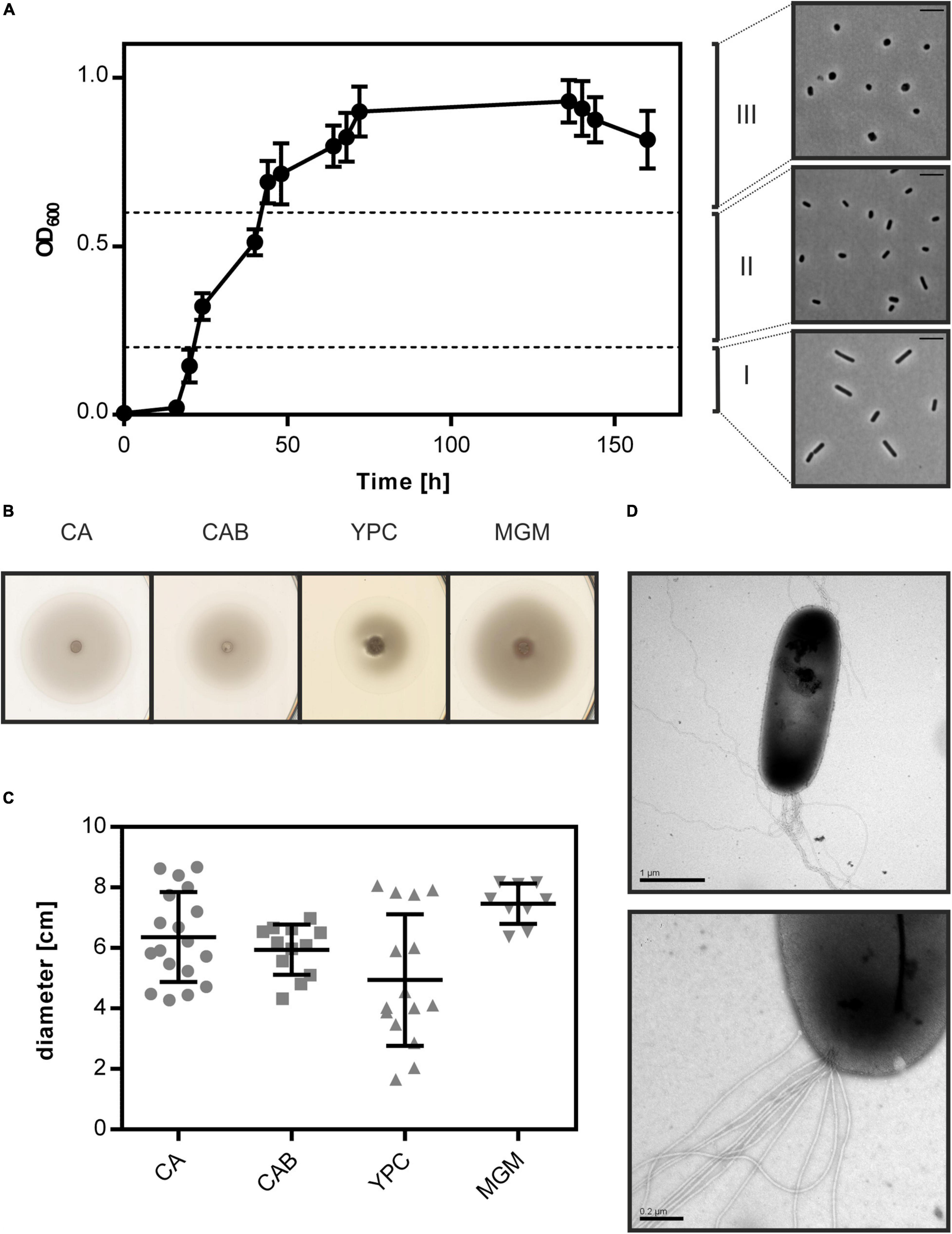
Figure 1. Cell shape change and motility of Hfx. gibbonsii LR2-5. (A) Left side: typical growth curve of Hfx. gibbonsii LR2-5 in CA medium with 18% (wt/vol) SW at 42°C. Average optical density at 600 nm (OD600) was calculated from three independent technical replicates; error bars represent the standard deviation. Right side: phase contrast images show typical cell shapes correlating with three different growth phases: early exponential (I), mid-exponential (II), and stationary (III) growth phases. (B) Representative example of motility rings from Hfx. gibbonsii LR2-5 on semi-solid agar plates with different media after 3 days of growth at 45°C. (C) Quantification of the diameter of motility rings formed on semi-solid agar plates with different media in 18% SW. Calculations were made using more than three independent experiments including three biological replicates each. Middle line indicates the mean, and lower and upper lines the standard deviation. (D) Cells from early exponential growth phase were negatively stained with 2% (wt/vol) uranyl acetate. Top: Hfx. gibbonsii LR2-5 cell with typical rod-shaped morphology and archaella at the cell pole. Scale bar, 1 μm. Bottom: Close up of a bundle of archaella filaments at the cell surface. Scale bar, 0.2 μm.
As Hfx. gibbonsii LR2-5 transitions from rod- to plate-shaped cells in a similar fashion as Hfx. volcanii, we tested the correlation of cell shape with motility. First, the strain was stab inoculated on semi-solid agar plates. This showed that LR2-5 formed motility rings of ∼6 cm diameter after 3 days in several different media (Figures 1B,C), indicative of an intact motility machinery and chemotaxis system.
Cells grown in liquid medium were observed with phase contrast microscopy at an optimal growth temperature of 42°C. Time lapse imaging showed that cells displayed swimming motility in different growth media, of which an example is shown in Supplementary Movie 1. The average swimming speed was 6.5 μm/s. Analyzing the motility using time lapse microscopy during different growth phases showed that rod-shaped cells that are present during the early exponential growth phase were highly motile, while the plate-shaped cells were immotile (Supplementary Table 1). The motile phase of Hfx. gibbonsii LR2-5 is prolonged in comparison with that of Hfx. volcanii, correlating with the rod-shape appearance of the LR2-5 cells. The transition from motile rod- to immotile plate-shaped cells seems thus not confined only to Hfx. volcanii, but might be more general for members of the genus Haloferax.
Finally, Hfx. gibbonsii LR2-5 cells from early exponential phase were examined via transmission electron microscopy, revealing large bundles of 7–10 archaellar filaments per cell (Figure 1D).
The susceptibility of Hfx. gibbonsii LR2-5, Hfx. gibbonsii Ma2.38T, and Hfx. volcanii H26 (see Supplementary Text for information on strains) to HFTV1 was examined in parallel using a spot-on-lawn assay (Juez et al., 1986; Allers et al., 2004; Mizuno et al., 2019). Serial dilutions of the HFTV1 lysate (5 × 1011 PFU/mL) were spotted onto lawns of the three strains of Haloferax and the plates incubated for 3 days (Figure 2). Clearing of the cellular lawn appeared until a 10–9 dilution of virus lysate on the original virus–host Hfx. gibbonsii LR2-5 (Figure 2A), whereas only faint spots of hazy appearance were observed when undiluted HFTV1 lysate was spotted on Hfx. gibbonsii Ma2.38 (Figure 2B). No zones of inhibition or separate plaques were observed when HFTV1 was spotted on Hfx. volcanii H26 (Figure 2C). This was confirmed by traditional plaque assay. These results show that among the analyzed Haloferax strains, HFTV1 is only capable of efficiently infecting its original host Hfx. gibbonsii LR2-5. The absence of HFTV1 plaque formation on other strains is in line with previous results showing that HFTV1 is not infectious on six environmental Haloferax strains, isolated from Lake Retba (Mizuno et al., 2019).
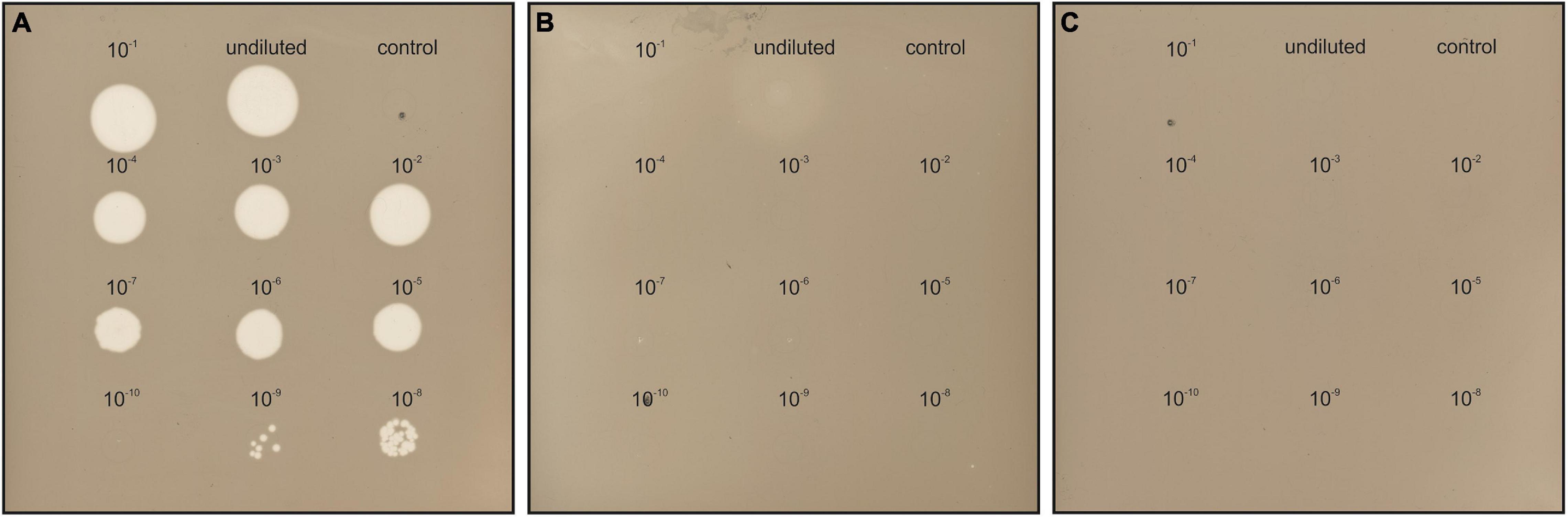
Figure 2. HFTV1 susceptibility of Haloferax strains by spot-on-lawn assay. Spot-on-lawn assay conducted with lawns of (A) Hfx. gibbonsii LR2-5, (B) Hfx. gibbonsii Ma2.38, and (C) Haloferax volcanii H26. Different dilutions of HFTV1 lysate (undiluted 5 × 1011 PFU/mL) were spotted on the host lawns and incubated for 3 days. Control spots were prepared with medium.
To sequence the genome of Hfx. gibbonsii LR2-5, DNA was extracted from exponentially growing Hfx. gibbonsii LR2-5 cells. PacBio genome sequencing and automated assembly of the sequences resulted in four circular contigs representing the four circular replicons (chromosome and three plasmids). Key characteristics of the genome are given in Table 1, and a more extensive summary is provided in Supplementary Table 2.
All replicons had at least 115-fold mean coverage. Illumina sequencing was used to validate and, if required, correct the assembled genome sequence, as described in Section “Material and Methods.” This followed an established procedure (Pfeiffer et al., 2020).
First, the strand and the point of ring opening were selected. For the chromosome, we adopted the convention of choosing a start position close to a canonical replication origin. However, we used a biologically relevant variation that we have used previously for Natronomonas moolapensis (Dyall-Smith et al., 2013), Halobacterium hubeiense (Jaakkola et al., 2016), and Halobacterium salinarum strain 91-R6T (Pfeiffer et al., 2019, 2020). This point of ring opening highlights the strong syntenic association of the genes adjoining the major replication origin. On one side is a highly conserved Orc/Cdc6 family member (gene orc1 for strain LR2-5, see Supplementary Table 3). On the other side, on the opposite strand, is the oapABC cluster (oap: origin-associated protein) (Wolters et al., 2019). For the plasmids, the applied strategy is detailed in the Supplementary Material.
The genome sequence was used to assign the LR2-5 strain to a taxon using the Type Strain Genome Server (TYGS)3 (Meier-Kolthoff and Göker, 2019). It was classified within Hfx. gibbonsii with 90.6% average branch support. When using the complete genome including plasmids, the average branch support increased to 92.3%. The strain is now designated Hfx. gibbonsii LR2-5. Issues relating to the “16S rRNA based assignment” of this strain by the TYGS are described in the Supplementary Information.
The genome size and GC% are similar to the average values for other sequenced members of the genus Haloferax (3.8 Mb, 65.4% G + C)4. Two of the three plasmids (pHGLR1 and pHGLR3) show significantly lower average GC (%) compared to the main chromosome. Tetranucleotide analysis (see section “Materials and Methods”) revealed that the motif CTAG and its inverse GATC, which are usually common in haloarchaeal genomes, are significantly under-represented in the LR2-5 genome, particularly on the main chromosome (0.19 odds ratio for both) and plasmid pHGLR2 (odds ratios 0.12 and 0.18). Methylated bases (m4C and m6A) were detected within four distinct sequence motifs (Supplementary Table 4). Only one of the four motifs (CTAG) was palindromic. The most frequently modified motif (GCGm4CTG) was methylated on only one strand, while the other three were methylated on both strands.
We compared the Hfx. gibbonsii LR2-5 replicons to those of the closely related model strain Hfx. volcanii DS2T (see Supplementary Text) as well as Hfx. gibbonsii strains ARA6 and Ma2.38T. For strain ARA6, a complete genome sequence is available. For type strain Ma2.38T, only a draft genome is available, consisting of contigs with unresolved replicon structure. However, this strain is available in culture collections and could be used for virus susceptibility tests described above. BLASTn comparisons show a close similarity of the Hfx. gibbonsii LR2-5 chromosome to those of Hfx. gibbonsii strains Ma2.38T and ARA6 (the red and green BLASTn rings, Figure 3A), and to Hfx. volcanii DS2T (orange ring). Regions of sequence variation between strains (blank sections of the BLASTn rings) are frequently also regions of lower than average GC (troughs in the GC content plot) and predicted genomic islands (gray bars). These have been labeled as either integrative genetic elements (IGEs) or long variable regions (LVR), and their details are given in Supplementary Table 5.
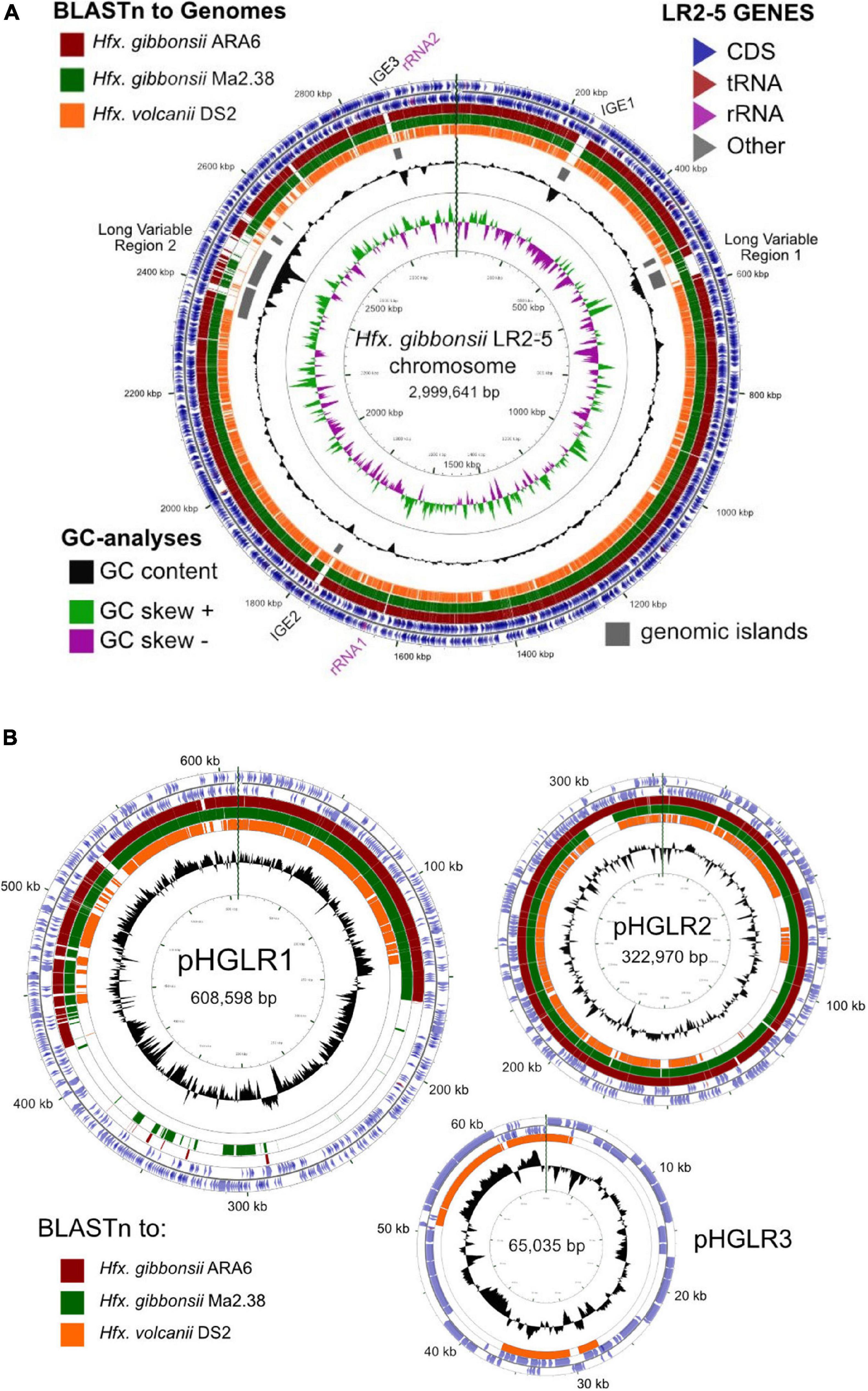
Figure 3. Comparison of the Hfx. gibbonsii LR2-5 chromosome and plasmids to those of three close relatives. (A) Hfx. gibbonsii LR2-5 chromosome map showing similarity to replicons of closely related strains. The two outermost rings depict the annotated genes (CDS, tRNA, and rRNA) of strain LR2-5, for the forward and reverse DNA strands (color key, upper right). Rings 3–5 depict BLASTn similarities between strain LR2-5 and the three strains listed in the color key (upper left). Colored bars represent regions of similarity (Expect value ≤ 10– 20, cutoff = 90% nucleotide identity), while uncolored (white) regions represent no significant similarity. Ring 6 (gray blocks) are predicted genomic islands (IslandViewer 4). The two innermost rings represent plots of GC content (black) and GC-skew (green/purple) for strain LR2-5, and the color key for these plots is given in the lower left. The GC content ring plots differences from the average GC%, with outward pointing peaks indicating higher than average, and inward pointing peaks indicating lower than average GC%. IGE1-3, integrative genetic elements (see text). rRNA1 and rRNA2 are the two ribosomal RNA operons. Tick marks around the inner-most and outer-most rings show DNA size in kb. The maps and plots were made using the CGView Server (http://stothard.afns.ualberta.ca/cgview_server). (B) Hfx. gibbonsii LR2-5 plasmid maps showing similarity to replicons of closely related strains. The color key for LR2-5 genes (outer two rings) and the inner GC content plots (black) are as described in (A). The comparison strains used to produce the BLASTn similarity rings are indicated by the colored boxes lower left. The actual replicons used are as follows. Plasmid pHGLR1 is compared to Hfx. gibbonsii ARA6 plasmid pHG1 (488,062 bp) (red), Hfx. gibbonsii Ma2.38 (green), and to Hfx. volcanii DS2T plasmid pHV3 (437,906 bp) (orange). Plasmid pHGLR2 is compared to Hfx. gibbonsii ARA6 plasmid pHG2 (335,881 bp) (red), contig 28 (AOLJ01000028) of Hfx. gibbonsii Ma2.38 (green), and to Hfx. volcanii DS2T plasmid pHV4 (635,786 bp) (orange). Plasmid pHGLR3 is compared to Hfx. volcanii DS2T plasmid pHV1 (85,092 bp) (orange).
The relation between the chromosomes of Hfx. gibbonsii LR2-5, Hfx. gibbonsii ARA6, and Hfx. volcanii DS2T has been additionally analyzed by MUMmer, presented as dotplots (Supplementary Figure 2). Both plots show a long inversion of the LR2-5 genome relative to the others, with the inversion boundaries of LR2-5 being the two inward facing rRNA operons (nt 1658979–1663984 and nt 2916773–2921778). This inversion correlates with the abrupt shifts in the GC skew which occur close to the rRNA operons (Figure 3A).
All three IGEs have integrated into tRNA genes on the chromosome, have terminal direct repeats partially duplicating the 3′ part of the tRNA, and carry a gene for XerC/D integrase near the end which is adjacent to the complete copy of the tRNA. IGEs could be provirus-related. IGE3 shows strong similarity to a betapleolipovirus-like provirus of Haloferax prahovense Arc-Hr and to betapleolipoviruses HRPV9 (Atanasova et al., 2018) and HGPV-1 (Senčilo et al., 2012) of the family Pleolipoviridae (Demina and Oksanen, 2020) (Supplementary Figure 3), although IGE3 appears to have lost many viral core genes. The other two IGEs are less clear but may also represent provirus-remnants. IGE1 carries three genes for restriction-modification proteins, as well as a gene (HfgLR_01385) that is commonly found in proviruses reminiscent of the Caudovirales type viruses of other haloarchaea, e.g., Haloferax sp. ATB1 (accession JPES01000032).
The LVRs LVR1 and LVR2 represent longer and more complex regions of variability than IGEs. They seem hotspots of recombination and their borders could not be easily defined. LVR1 carries replication genes (orc6, polB2) and may be plasmid related. A comparison between Hfx. gibbonsii LR2-5 and ARA6 across the 38 kb of LVR1 revealed the marked differences in gene composition and size (Supplementary Figure 4). LVR2 is almost 195 kb in length and also carries replication genes (e.g., orc10 and orc11) as well as many genes that could influence virus susceptibility of the host, including the genes encoding the pilin PilA2, sugar nucleotidyltransferase AglF, sugar modification enzyme AglM, and many other enzymes involved in sugar metabolism (glycosyltransferases, D-galactonate dehydratase, and sugar nucleotidyltransferases) as well as secreted glycoproteins.
All three plasmids match to plasmids of Hfx. volcanii DS2T (Figure 3B, orange rings). Plasmids pHGLR1 and pHGLR2 show strong nucleotide similarity to plasmids of Hfx. gibbonsii strains ARA6 and to contigs of strain Ma2.38T (Figure 3B; red and green BLASTn rings). Plasmid pHGLR3 does not have a counterpart in the other Hfx. gibbonsii strains. All three plasmids also have strain-specific parts. The strain-specific parts of pHGLR1 and pHGLR3 tend to have lower than average GC content, and this is particularly evident in the 150–420 kb region of pHGLR1 (56.5% G + C) compared to the rest of this plasmid (66% G + C), a difference of almost 10 percentage points (Figure 3B). This may indicate that pHGLR1 is the result of a large integration or fusion event. A similar example has been described previously in the H. salinarum plasmid pHS3, which carries a 70 kb high-GC island (Barylski et al., 2020). The borders of these two regions share a similar gene cluster of four ABC-transport genes (tsg) that show significant nucleotide similarity, e.g., tsgD5 (HfgLR_20645; HVO_A0145) and tsgD6 (HfgLR_22070; HVO_A0281), but are outwardly oriented relative to each other.
Plasmid pHGLR2 has an 11.6 kb stretch of DNA near the 300 kb mark (nt 291722–303338; HfgLR_24335 to HfgLR_24410) that is shared only with pHG2 of strain ARA6, and is absent in strain Ma2.38T and Hfx. volcanii DS2T (pHV4). This region of pHGLR2 carries genes for various transporters, phosphoribosyl-AMP cyclohydrolase, and several uncharacterized proteins.
Haloferax gibbonsii LR2-5 has two rRNA operons and 55 regular tRNA genes, of which 53 are encoded on the main chromosome (Table 1). In addition, there are 11 partial tRNAs, of which nine are on the chromosome. In several cases, the remnant is directly adjacent to a full copy of the same tRNA. Each of the three IGEs has duplicated termini, one being the tRNA and the other a tRNA remnant. For more information, see Supplementary Material.
A detailed analysis of the transposons (Supplementary Table 6) revealed that Hfx. gibbonsii strains ARA6 and LR2-5 have a low number of transposons and other mobile genetic elements compared to other haloarchaea (12 in strain ARA6 and 31 in strain LR2-5; Supplementary Table 6). For more information, see Supplementary Text.
Overall, 3204 of the 4064 proteins (∼79%) encoded in the Hfx. gibbonsii LR2-5 genome have an ortholog in Hfx. volcanii DS2T with an average of 93% protein sequence identity. One-third (37%) of these orthologs have 96–98% protein sequence identity. The distribution is slightly uneven between the chromosome (84% of proteins have an ortholog) and the plasmids (61% of proteins have an ortholog).
To gain insight into the differences between Hfx. gibbonsii LR2-5 and the other Haloferax strains, we compared protein coding genes between these species with a focus on anti-viral defense systems and genes encoding possible viral anchor points and receptors at the cell surface (Supplementary Tables 6–11). We considered Haloferax strains for which a complete genome sequence is available, Hfx. volcanii DS2 and Hfx. gibbonsii ARA6.
The arms race of bacteria and archaea with their viruses has led to the development of a plethora of defense mechanisms against viruses. These include CRISPR-Cas, toxin antitoxin (TA) systems, Restriction Modification (RM) systems, and several recently discovered new systems (Stern and Sorek, 2011; Azam and Tanji, 2019).
In bacteria, TA systems are sometimes part of an antiviral defense mechanism relying on abortive infection (Gerdes et al., 2005; Tachdjian and Kelly, 2006). Hfx. gibbonsii LR2-5 encodes a few TA systems (Supplementary Table 7), although none of the functional systems appear likely to be linked with abortive infection. Likewise, RM systems can play a role in defense against foreign genetic elements in bacteria (Tock and Dryden, 2005). Several type I RM systems were predicted in the LR2-5 genome which are only marginally related to Hfx. volcanii RM systems (Supplementary Table 8 and Supplementary Text). This, substantiated by the differences in DNA methylation (Supplementary Table 3, rebase data: http://rebase.neb.com/cgi-bin/pacbioget?5891), indicates that RM systems may be an infection barrier in Hfx. volcanii and Hfx. gibbonsii Ma2.38. Indeed, the HFTV1 genome contains 24 target sites for the Hfx. volcanii Mrr restriction endonuclease.
Surprisingly, Hfx. gibbonsii LR2-5 does not contain any CRISPR-Cas systems. These anti-viral defense systems are very common among archaea, and Hfx. volcanii contains a well-studied and functional CRISPR-Cas system (Maier et al., 2019). Hfx. gibbonsii Ma2.38 is predicted to contain two CRISPR-Cas systems, as well as an additional CRISPR array (analyzed using https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index). Neither Hfx. volcanii nor Hfx. gibbonsii Ma2.38 has any spacers matching the HFTV1 genome. On the other hand, Hfx. gibbonsii ARA6 does not contain any CRISPR-Cas systems. The lack of CRIPSR-Cas systems in LR2-5 does, however, make it an attractive model organism for the study of virus–host interactions as it is more likely to be susceptible to other viruses. Further details on defense systems can be found in the Supplementary Text.
The surface layer (S-layer) functions as a cell wall in many archaea and some bacteria and is also an attractive target for viruses at the cell surface. S-layer proteins are highly abundant and can display marked differences between strains and species. Several bacterial phages require host S-layer for infection (Mescher and Strominger, 1977; Edwards and Smit, 1991; Plaut et al., 2014). Archaeal S-layer proteins are usually heavily glycosylated (Albers and Meyer, 2011; Kaminski et al., 2013b; Kandiba and Eichler, 2014).
To determine the S-layer glycoprotein (SLG) of LR2-5, we analyzed the total cell lysates of different Haloferax strains by SDS-gel electrophoresis (Figure 4A). In several archaea, the SLG is the most abundant cellular protein. In halophilic archaea, it is easily detected by Coomassie staining as a prominent protein band running at ∼245 kDa. A prominent band in LR2-5 cell lysate migrated slightly above that of the Hfx. volcanii SLG but identically to an equally prominent band of the type strain of Hfx. gibbonsii (Ma2.38). The band from LR2-5 was excised and subjected to mass spectrometry analysis (data not shown). This showed the presence of HfgLR_04635 and HfgLR_11210 in this protein band. Both proteins are predicted to have similar molecular weights of ∼90–95 kDa, but due to their abundant predicted glycosylation sites, they run at a very different height, as is common for halophilic proteins (Shalev et al., 2018). The function of HfgLR_11210 is not clear. The detection of HfgLR_04635 is consistent with the recent identification of a homolog of HfgLR_04635 as the SLG of a Hfx. gibbonsii strain (ABY42_04395) (Shalev et al., 2018). The two proteins show 99% protein sequence identity, but are only very distantly related to the SLG of Hfx. volcanii (HVO_2072; 24% protein sequence identity) (Sumper et al., 1990).
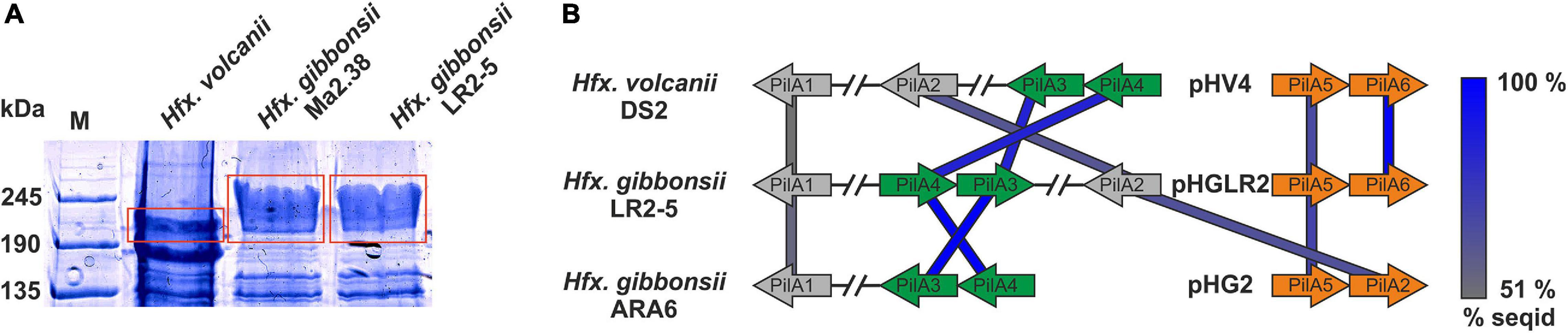
Figure 4. Surface protein and pili of Hfx. gibbonsii LR2-5. (A) Coomassie stained SDS-PAGE gel of Hfx. volcanii, Hfx. gibbonsii Ma2.38, and Hfx. gibbonsii LR2-5 whole cell lysates. The prominent bands at around 245 kDa are considered to run at the height of the S-layer glycoproteins (red boxes). The LR2-5 band was subjected to mass spectrometry. (B) Comparison of the genetic loci of Hfx. gibbonsii LR2-5 PilAs with those of Hfx. gibbonsii ARA6 and Hfx. volcanii DS2T. Homologs to all six Hfx. volcanii pilins are present in Hfx. gibbonsii LR2-5; however, particularly PilA1, PilA2, and PilA5 are more distant homologs (protein sequence identity 51–68%). PilA3 and PilA4 are organized in an operon in all three strains and the proteins have high protein sequence identity (≥85%).
Filamentous surface structures are well-known primary anchor points for several viruses infecting bacteria (Poranen et al., 2002; Mäntynen et al., 2019). Recent data suggest that a few crenarchaeal viruses also tether to filamentous surface structures (Quemin et al., 2013; Hartman et al., 2019; Rowland et al., 2020). A hallmark of archaeal surface structures is the widespread similarity to bacterial type IV pili. Adhesive type IV pili of archaea are involved in attachment to biotic and abiotic surfaces which may lead to biofilm formation (Pohlschroder and Esquivel, 2015; van Wolferen et al., 2018). Pilins typically have a type III signal sequence, which is processed in Haloferax by the prepilin/prearchaellin peptidase PibD. After N-terminal processing, pilin subunits are inserted into surface filaments (Pohlschroder et al., 2018). The core membrane and biosynthesis complex for pilin subunit insertion is formed by the membrane platform protein PilC and the cytosolic assembly ATPase PilB (Pohlschroder et al., 2018). In Hfx. volcanii, six pilBC pairs are found.
While pilB1C1 and its associated genes are present in Hfx. gibbonsii strain ARA6 but not in LR2-5, all other pilBC pairs with their associated genes are highly conserved between Hfx. volcanii strain DS2T and Hfx. gibbonsii strains ARA6 and LR2-5 (Supplementary Table 9).
Haloferax gibbonsii LR2-5 codes for six pilins, five of which are more closely related to those of Hfx. gibbonsii ARA6 than to those of Hfx. volcanii DS2T (with PilA6 being absent from Hfx. gibbonsii ARA6, Figure 4B).
The individual roles in attachment and biofilm formation of the pilins of Hfx. volcanii, PilA1-6, seem to have slightly different functions in adhesion and microcolony formation (Esquivel et al., 2013; Legerme et al., 2016; Legerme and Pohlschroder, 2019).
The pilin genes are organized differently in the three Haloferax strains, and some Hfx. volcanii pilins have only distant homologs in Hfx. gibbonsii LR2-5 and ARA6 (Figure 4A). Strikingly, Hfx. gibbonsii LR2-5 PilA1, PilA2, and PilA5 have closer homologs in other species, particularly Hfx. sp. Atlit-19N [isolated from high salt tide-pools on the coast of Israel (Atlit, summer 2012); BioProject PRJNA431124], than in Hfx. volcanii and Hfx. gibbonsii ARA6. As mentioned above, PilA2 is located within LVR2 in Hfx. gibbonsii LR2-5.
N-glycosylation plays a crucial role in pilus-mediated surface attachment and all Hfx. volcanii PilA pilins except PilA5 are glycosylated (Esquivel et al., 2016). All Hfx. gibbonsii LR2-5 PilAs except PilA2 are predicted to contain at least one N-glycosylation site (NxS/T) (Supplementary Table 10). The predicted N-glycosylation sites in Hfx. volcanii and Hfx. gibbonsii LR2-5 are fully conserved for PilA3 but at maximum partially conserved for the other PilAs. Further details on type IV pili can be found in the Supplementary Text.
The archaeal motility structure, the archaellum (archaeal flagellum), also displays homology to type IV pili (Albers and Jarrell, 2018). The archaellum is a rotating filamentous structure which functions analogously to the bacterial flagellum, as it provides swimming motility in liquid (Alam and Oesterhelt, 1984; Kinosita et al., 2016). However, archaella and flagella have a fundamentally different structural organization and their protein components are completely unrelated (Albers and Jarrell, 2015). Proteins related to archaella biogenesis and function are typically clustered in haloarchaeal genomes (Kalmokoff and Jarrell, 1991; Patenge et al., 2001; Jarrell and Albers, 2012). This arl cluster (previously fla cluster) is highly conserved between Hfx. volcanii and Hfx. gibbonsii strain LR2-5 (84–98% protein sequence identity) with strictly conserved gene synteny.
The archaellum is involved in directional movement together with a bacterial-type chemotaxis system. The linking components have recently been identified (cheF, arlCDE) (Schlesner et al., 2009, 2012; Quax et al., 2018; Li et al., 2020). Hfx. volcanii and Hfx. gibbonsii LR2-5 have very similar che genes (89–100% protein sequence identity). Both genomes show strictly conserved gene synteny in which the che gene cluster is split and surrounds the arl cluster. Conclusively, the genomic content of LR2-5 and the observed expression of archaella and motility machinery in liquid and semi-solid agar (Figure 1B) show that LR2-5 is a highly motile strain. Due to the high level of conservation, the motility might not be responsible for the differences in virus susceptibility between strain LR2-5 and Hfx. volcanii.
N-glycosylation plays an important role in the biosynthesis and function of many surface exposed proteins such as pilins, archaellins, and S-layer glycoproteins (Esquivel et al., 2016; Tamir and Eichler, 2017). N-glycosylation pathways differ between haloarchaeal species, and glycosylation of surface proteins, particularly the S-layer, may even vary depending on differing environmental factors such as salinity (Kaminski et al., 2013c). Indeed, Hfx. volcanii uses two distinct glycosylation pathways which were initially identified as being dependent on environmental salinity: the canonical pathway (AglB-J) and the low-salt pathway (Agl5-15) (Kaminski et al., 2013a). Unexpectedly, most genes coding for the low-salt N-glycosylation pathway were recently found to be expressed under optimal growth conditions (Schulze et al., 2020).
The Hfx. gibbonsii LR2-5 glycosylation proteins are mostly found in two main chromosomal clusters (Figures 5A,B; marked I, II), with a few outliers, some being encoded on the plasmids (Figure 5C). Only five of the 20 characterized Hfx. volcanii Agl proteins have an ortholog in the LR2-5 genome (Supplementary Table 11 and Figure 5). These proteins appear to be well conserved within Haloferax, as close homologs are found in many other members of the genus. In addition, a number of LR2-5 genes in clusters I and II are distantly related to Hfx. volcanii glycosylation genes and thus are likely involved in N-glycosylation (Supplementary Table 12 and Figure 5).
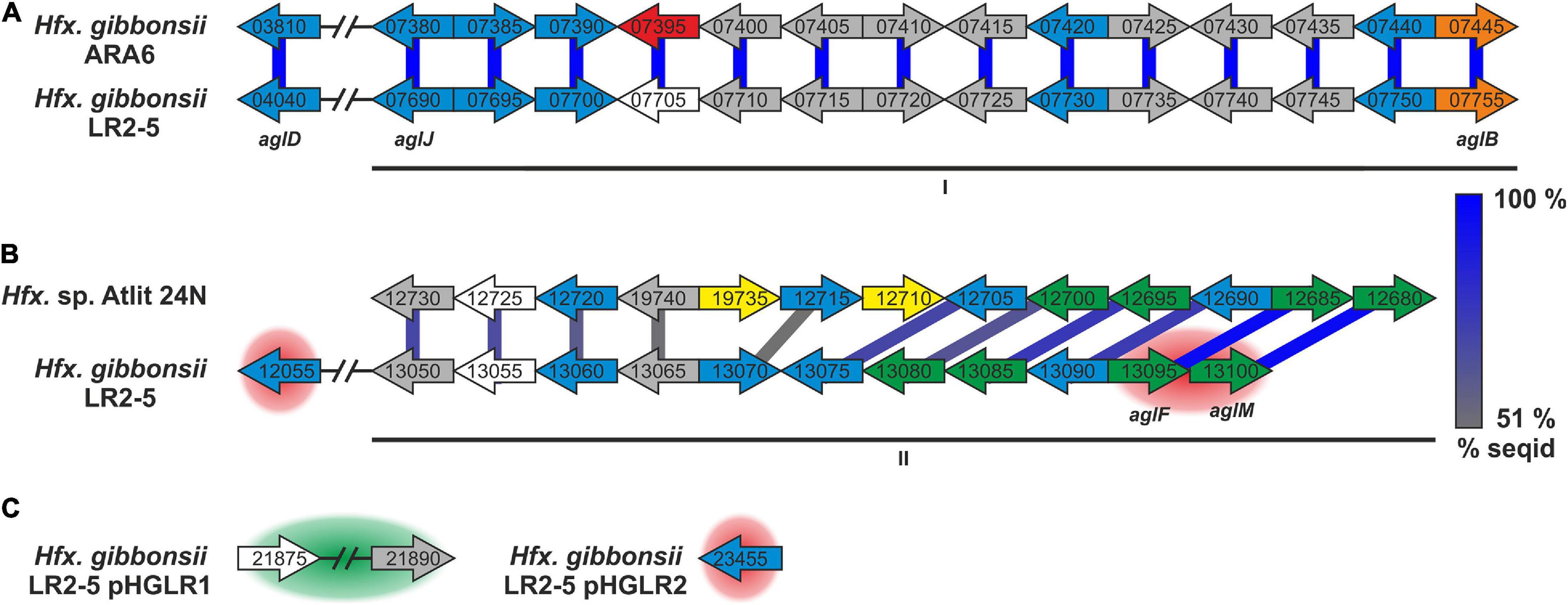
Figure 5. Hfx. gibbonsii LR2-5 N-glycosylation genes with comparison to their closest homologs. Genes are located in two clusters (marked as I and II in A,B) with some genes being located apart, e.g., on a plasmid (C). Close homologs in Hfx. volcanii are indicated with the corresponding agl name. Predicted protein function is indicated by color code; blue: glycosyltransferase, red: flippase, white: rfbX family protein (potential flippase), orange: oligosaccharyltransferase, green: sugar modification enzyme, gray: not directly related to N-glycosylation, yellow: hypothetical protein/pseudogene not apparently related to glycosylation. Protein homology and sequence identity are indicated by colored bars. Bar color indicates the level of amino acid sequence identity (see the color coding on the right). Background coloring indicates additional close homologs, red: Hfx. gibbonsii ARA6, green: Hfx. sp. Atlit-16N and Atlit-10N. Gene sizes are arbitrary.
Strikingly, the five genes with strong similarity to characterized agl genes of Hfx. volcanii are not in genomic vicinity, but spread across the two clusters with one outlier. The first cluster (Figure 5A, I) encodes proteins with close homologs in Hfx. gibbonsii ARA6, Hfx. sp. Atlit 4N, 6N, 10N, 16N, and 19N as well as a few other Haloferax strains and species. The second cluster (Figure 5B, II) encodes proteins with closest homologs in Hfx. sp. Atlit 24N, 109R, and 105R. Most of these proteins (particularly HfgLR_13050 through HfgLR_13070) have very few other close homologs. Like pilA2, this entire cluster lies within LVR2.
Of the plasmid-encoded N-glycosylation proteins, HfgLR_23455 has many close homologs among proteins of other Haloferax species encoded also on plasmids, whereas HfgLR_21875 and 21890 have only four close homologs (>70% protein sequence identity, Supplementary Table 12).
We functionally characterized the euryarchaeon Hfx. gibbonsii LR2-5, the host of the siphovirus HFTV1, and sequenced its genome. Hfx. gibbonsii LR2-5 was shown to grow optimally under similar conditions as strains of the model species Hfx. volcanii, aiding in development of a genetic system for LR2-5. In addition, cells of Hfx. gibbonsii LR2-5 transition from motile rod-shaped cells to immotile plate-shape cells during growth. In comparison with the laboratory strain Hfx. volcanii, they stay rod-shaped much longer, facilitating studies of the motility machinery. Sequencing of the genome of Hfx. gibbonsii LR2-5 indicated that the cell surface of this strain likely differs from some of its close relatives. LR2-5 encodes several pilins that differ from those of Hfx. volcanii and Hfx. gibbonsii ARA6. Future work is required to analyze if the variability of these pilins is involved in the differences in virus susceptibility, as the receptor of HFTV1 has not been identified yet. The archaella of LR2-5 show high sequence conservation to other Haloferax species. Genes involved in glycosylation of surface proteins are very different between Hfx. gibbonsii and its close relatives. It is possible that the glycans of LR2-5 differ considerably from those of Hfx. volcanii and this difference might be responsible for the differences in virus susceptibility. Further analysis is needed to identify the actual glycans of LR2-5 and to establish if glycosylation is related to infection of HFTV1. Interestingly, Hfx. gibbonsii LR2-5 does not encode any CRISPR-Cas system, which is in contrast with the yet analyzed Haloferax strains that were found to be resistant to HFTV1. In addition, its R/M systems differ significantly from those in the other strains. Therefore, the absence of a CRISPR-Cas and the Mrr restriction endonuclease system might also explain the susceptibility of LR2-5 to HFTV1. This work will enable future research on HFTV1 adsorption to the host cell and interaction with anti-viral defense systems. Moreover, it significantly contributes to the development of Hfx. gibbonsii LR2-5 into a model system for the study of archaeal virus–host interactions.
Haloferax gibbonsii LR2-5 (previous Haloferax sp. LR2-5; Mizuno et al., 2019), Hfx. gibbonsii Ma2.38T (Juez et al., 1986), and Hfx. volcanii H26 (Bitan-Banin et al., 2003) cells were cultured as described previously (Nuttall and Smith, 1993; Allers et al., 2004; Duggin et al., 2015; Mizuno et al., 2019). HFTV1 virus was grown and virus stocks were prepared as described (Mizuno et al., 2019). For details, see Supplementary Information.
Haloferax gibbonsii LR2-5 cells were adsorbed to glow-discharged carbon-coated copper grids with Formvar films and imaged using a CM10 transmission electron microscope (Philips) coupled to a Gatan 792 BioScan camera. For details, see Supplementary Information.
Haloferax gibbonsii LR2-5 cells were imaged at 100× magnification using an Axio Observer.Z1 inverted microscope (Zeiss). Microscopy images were processed to analyze cell shapes using the FIJI/ImageJ plugin MicrobeJ.
Swimming analysis was performed at 63× magnification with an Axio Observer.Z1 inverted microscope (Zeiss). The movement of cells was recorded with 15 s time-lapse movies. For details, see Supplementary Information.
Motility assays were performed as described previously (Quax et al., 2018; Li et al., 2020). For details, see Supplementary Information.
A dilution series of a virus preparation was prepared and 10 μL spots of each dilution were placed on the lawn of the known or other potential host cells (Hfx. gibbonsii LR2-5, Hfx. gibbonsii Ma2.38T, and Hfx. volcanii H26). Plates were incubated for 2–5 days at 37°C and examined for the presence or absence of zones of growth inhibition. For details, see Supplementary Information.
The number of infectious viruses was determined by plaque assay by mixing 100 μL of virus dilutions with 300 μL dense host culture before plating in an overlay of MGM soft agar on MGM plates. Plates were incubated for 2–3 days at 37°C. Plaques were counted and the number of infectious viruses per unit volume, i.e., the titer (plaque-forming units/mL; PFU/mL) was determined.
A whole cell lysate of Hfx. gibbonsii LR2-5 was separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using a 7.5% polyacrylamide-SDS gel. The gel was stained using Coomassie blue stain. A major band having the appropriate molecular weight was excised and used for identification by mass spectroscopy. For details, see Supplementary Information.
Full details are provided in the Supplementary Information.
Cells of Hfx. gibbonsii LR2-5 were processed by Eurofins NGS Lab Constance (Constance) for DNA extraction. For PacBio RS sequencing, a “standard genomic library” was prepared and sequenced at Eurofins according to the manufacturer’s instructions. An automatic assembly using the HGAP3 pipeline was performed at Eurofins. To further improve the accuracy of the genome sequence, Illumina HiSeq sequencing was performed at Eurofins.
Full details are provided in the Supplementary Information.
Gene prediction was performed using the RASTtk annotation server (Overbeek et al., 2014; Brettin et al., 2015; Lomsadze et al., 2018). The resulting annotation was curated using previously established procedures (Pfeiffer and Oesterhelt, 2015; Pfeiffer et al., 2020). The Hfx. volcanii annotation referred to as “up-to-date” is that from 6 June 2019, which is the basis for the community proteome project arcPP (Schulze et al., 2020).
An effort was made to reduce missing gene calls, especially small ones, by subjecting all intergenic regions ≥ 50 bp in the LR2-5 genome to an established BLASTx analysis procedure (Babski et al., 2016).
Annotation of stable RNAs and transposon analysis is described in the Supplementary Information.
Base modifications were analyzed using the SMRT® Analysis software version 7.0.1.66975 (Base Modification and Motif Analysis tool) (Chin et al., 2013). PacBio reads and the assembled genome sequence of strain LR2-5 were used as input. Results are provided as Supplementary Table 4.
Information on bioinformatic tools (e.g., MUMmer, BLAST, and TYGS) can be found in the Supplementary Information.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP063205; https://www.ncbi.nlm.nih.gov/genbank/, CP063206; https://www.ncbi.nlm.nih.gov/genbank/, CP063207; and https://www.ncbi.nlm.nih.gov/genbank/, CP063208.
TQ, CT, and FP: conceptualization and writing—original draft. FP: data curation. FP and TQ: project administration. FP and MD-S: formal analysis. TQ: funding acquisition and supervision. SS, FP, MR-F, CT, and HO: investigation and validation. SS, CT, MR-F, and MS: visualization. FP, TQ, CT, SS, HO, and MS: writing—review and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the Deutsche Forschungsgemeins-chaft (German Research Foundation) with an Emmy Nöther grant (411069969) and by the University of Helsinki and Academy of Finland funding for FINStruct and Instruct-FI, part of Biocenter Finland and Instruct-ERIC, respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Marina Geiger and Drishya S. Gopan for the support with experiments. The TEM is operated by the University of Freiburg, Faculty of Biology, as a partner unit within the Microscopy and Image Analysis Platform, Freiburg.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.625599/full#supplementary-material
Alam, M., and Oesterhelt, D. (1984). Morphology, function and isolation of halobacterial flagella. J. Mol. Biol. 176, 459–475. doi: 10.1016/0022-2836(84)90172-4
Albers, S. V., and Jarrell, K. F. (2015). The archaellum: how archaea swim. Front. Microbiol. 6:23. doi: 10.3389/fmicb.2015.00023
Albers, S. V., and Jarrell, K. F. (2018). The Archaellum: an update on the unique archaeal motility structure. Trends Microbiol. 26, 351–362. doi: 10.1016/j.tim.2018.01.004
Albers, S. V., and Meyer, B. H. (2011). The archaeal cell envelope. Nat. Rev. Microbiol. 9, 414–426. doi: 10.1038/nrmicro2576
Allers, T., Ngo, H. P., Mevarech, M., and Lloyd, R. G. (2004). Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70, 943–953. doi: 10.1128/aem.70.2.943-953.2004
Atanasova, N. S., Bamford, D. H., and Oksanen, H. M. (2015a). Haloarchaeal virus morphotypes. Biochimie 118, 333–343. doi: 10.1016/j.biochi.2015.07.002
Atanasova, N. S., Demina, T. A., Buivydas, A., Bamford, D. H., and Oksanen, H. M. (2015b). Archaeal viruses multiply: temporal screening in a solar saltern. Viruses 7, 1902–1926. doi: 10.3390/v7041902
Atanasova, N. S., Demina, T. A., Krishnam Rajan Shanthi, S. N. V., Oksanen, H. M., and Bamford, D. H. (2018). Extremely halophilic pleomorphic archaeal virus HRPV9 extends the diversity of pleolipoviruses with integrases. Res. Microbiol. 169, 500–504. doi: 10.1016/j.resmic.2018.04.004
Atanasova, N. S., Roine, E., Oren, A., Bamford, D. H., and Oksanen, H. M. (2012). Global network of specific virus-host interactions in hypersaline environments. Environ. Microbiol. 14, 426–440. doi: 10.1111/j.1462-2920.2011.02603.x
Azam, A. H., and Tanji, Y. (2019). Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl. Microbiol. Biotechnol. 103, 2121–2131. doi: 10.1007/s00253-019-09629-x
Babski, J., Haas, K. A., Näther-Schindler, D., Pfeiffer, F., Förstner, K. U., Hammelmann, M., et al. (2016). Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq). BMC Genom. 17:629. doi: 10.1186/s12864-016-2920-y
Barylski, J., Enault, F., Dutilh, B. E., Schuller, M. B. P., Edwards, R. A., Gillis, A., et al. (2020). Analysis of spounaviruses as a case study for the overdue reclassification of tailed phages. Syst. Biol. 69, 110–123. doi: 10.1093/sysbio/syz036
Becker, E. A., Seitzer, P. M., Tritt, A., Larsen, D., Krusor, M., Yao, A. I., et al. (2014). Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. PLoS Genet. 10:e1004784. doi: 10.1371/journal.pgen.1004784
Bergh, Ø, Børsheim, K. Y., Bratbak, G., and Heldal, M. (1989). High abundance of viruses found in aquatic environments. Nature 340, 467–468. doi: 10.1038/340467a0
Bitan-Banin, G., Ortenberg, R., and Mevarech, M. (2003). Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185, 772–778. doi: 10.1128/jb.185.3.772-778.2003
Bize, A., Karlsson, E. A., Ekefjärd, K., Quax, T. E. F., Pina, M., Prevost, M. C., et al. (2009). A unique virus release mechanism in the Archaea. Proc. Natl. Acad. Sci. U.S.A. 106, 11306–11311. doi: 10.1073/pnas.0901238106
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365.
Cheng, F., Gong, L., Zhao, D., Yang, H., Zhou, J., Li, M., et al. (2017). Harnessing the native type I-B CRISPR-Cas for genome editing in a polyploid archaeon. J. Genet. Genom. 44, 541–548. doi: 10.1016/j.jgg.2017.09.010
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569.
Danovaro, R., Dell’anno, A., Corinaldesi, C., Rastelli, E., Cavicchioli, R., Krupovic, M., et al. (2016). Virus-mediated archaeal hecatomb in the deep seafloor. Sci. Adv. 2:e1600492.
de Silva, R. T., Abdul-Halim, M. F., Pittrich, D. A., Brown, H. J., Pohlschroder, M., and Duggin, I. G. (2021). Improved growth and morphological plasticity of Haloferax volcanii. Microbiology. doi: 10.1099/mic.0.001012 [Epub ahead of print].
Demina, T. A., and Oksanen, H. M. (2020). Pleomorphic archaeal viruses: the family Pleolipoviridae is expanding by seven new species. Arch. Virol. 165, 2723–2731. doi: 10.1007/s00705-020-04689-1
Duggin, I. G., Aylett, C. H. S., Walsh, J. C., Michie, K. A., Wang, Q., Turnbull, L., et al. (2015). CetZ tubulin-like proteins control archaeal cell shape. Nature 519, 362–365. doi: 10.1038/nature13983
Dyall-Smith, M. L., Pfeiffer, F., Oberwinkler, T., Klee, K., Rampp, M., Palm, P., et al. (2013). Genome of the haloarchaeon Natronomonas moolapensis, a neutrophilic member of a previously haloalkaliphilic genus. Genome Announc. 1:e0009513.
Edwards, P., and Smit, J. (1991). A transducing bacteriophage for Caulobacter crescentus uses the paracrystalline surface layer protein as a receptor. J. Bacteriol. 173, 5568–5572. doi: 10.1128/jb.173.17.5568-5572.1991
Eichler, J., and Maupin-Furlow, J. (2013). Post-translation modification in Archaea: lessons from Haloferax volcanii and other haloarchaea. FEMS Microbiol. Rev. 37, 583–606. doi: 10.1111/1574-6976.12012
El Omari, K., Li, S., Kotecha, A., Walter, T. S., Bignon, E. A., Harlos, K., et al. (2019). The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins. Nat. Commun. 10:846.
Esquivel, R. N., Schulze, S., Xu, R., Hippler, M., and Pohlschroder, M. (2016). Identification of Haloferax volcanii Pilin N-glycans with diverse roles in pilus biosynthesis, adhesion, and microcolony formation. J. Biol. Chem. 291, 10602–10614. doi: 10.1074/jbc.m115.693556
Esquivel, R. N., Xu, R., and Pohlschroder, M. (2013). Novel archaeal adhesion pilins with a conserved N terminus. J. Bacteriol. 195, 3808–3818. doi: 10.1128/jb.00572-13
Fuhrman, J. A. (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548. doi: 10.1038/21119
Gerdes, K., Christensen, S. K., and Løbner-Olesen, A. (2005). Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382. doi: 10.1038/nrmicro1147
Han, J., Zhang, F., Hou, J., Liu, X., Li, M., Liu, H., et al. (2012). Complete genome sequence of the metabolically versatile halophilic archaeon Haloferax mediterranei, a poly(3-hydroxybutyrate-co-3-hydroxyvalerate). producer. J. Bacteriol. 194, 4463–4464. doi: 10.1128/jb.00880-12
Haque, R. U., Paradisi, F., and Allers, T. (2020). Haloferax volcanii for biotechnology applications: challenges, current state and perspectives. Appl. Microbiol. Biotechnol. 104, 1371–1382. doi: 10.1007/s00253-019-10314-2
Hartman, A. L., Norais, C., Badger, J. H., Delmas, S., Haldenby, S., Madupu, R., et al. (2010). The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One 5:e9605. doi: 10.1371/journal.pone.0009605
Hartman, R., Eilers, B. J., Bollschweiler, D., Munson-McGee, J. H., Engelhardt, H., Young, M. J., et al. (2019). The molecular mechanism of cellular attachment for an archaeal virus. Structure 27, 1634–1646.e3.
Hawkins, M., Malla, S., Blythe, M. J., Nieduszynski, C. A., and Allers, T. (2013). Accelerated growth in the absence of DNA replication origins. Nature 503, 544–547. doi: 10.1038/nature12650
Jaakkola, S. T., Pfeiffer, F., Ravantti, J. J., Guo, Q., Liu, Y., Chen, X., et al. (2016). The complete genome of a viable archaeum isolated from 123-million-year-old rock salt. Environ. Microbiol. 18, 565–579.
Jantzer, K., Zerulla, K., and Soppa, J. (2011). Phenotyping in the archaea: optimization of growth parameters and analysis of mutants of Haloferax volcanii. FEMS Microbiol. Lett. 322, 123–130. doi: 10.1111/j.1574-6968.2011.02341.x
Jarrell, K. F., and Albers, S. V. (2012). The archaellum: an old motility structure with a new name. Trends Microbiol. 20, 307–312. doi: 10.1016/j.tim.2012.04.007
Juez, G., Rodriguez-Valera, F., Ventosa, A., and Kushner, D. J. (1986). Haloarcula hispanica spec. nov. and Haloferax gibbonsii spec, nov., two new species of extremely Halophilic Archaebacteria. Syst. Appl. Microbiol. 8, 75–79. doi: 10.1016/s0723-2020(86)80152-7
Kalmokoff, M. L., and Jarrell, K. F. (1991). Cloning and sequencing of a multigene family encoding the flagellins of Methanococcus voltae. J. Bacteriol. 173, 7113–7125. doi: 10.1128/jb.173.22.7113-7125.1991
Kaminski, L., Guan, Z., Yurist-Doutsch, S., and Eichler, J. (2013a). Two distinct N-glycosylation pathways process the Haloferax volcanii S-layer glycoprotein upon changes in environmental salinity. mBio 4:e0716-13.
Kaminski, L., Lurie-Weinberger, M. N., Allers, T., Gophna, U., and Eichler, J. (2013b). Phylogenetic- and genome-derived insight into the evolution of N-glycosylation in Archaea. Mol. Phylogenet. Evol. 68, 327–339. doi: 10.1016/j.ympev.2013.03.024
Kaminski, L., Naparstek, S., Kandiba, L., Cohen-Rosenzweig, C., Arbiv, A., Konrad, Z., et al. (2013c). Add salt, add sugar: N-glycosylation in Haloferax volcanii. Biochem. Soc. Trans. 41, 432–435. doi: 10.1042/bst20120142
Kandiba, L., and Eichler, J. (2014). Archaeal S-layer glycoproteins: post-translational modification in the face of extremes. Front. Microbiol. 5:661. doi: 10.3389/fmicb.2014.00661
Karner, M. B., Delong, E. F., and Karl, D. M. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510. doi: 10.1038/35054051
Kinosita, Y., Uchida, N., Nakane, D., and Nishizaka, T. (2016). Direct observation of rotation and steps of the archaellum in the swimming halophilic archaeon Halobacterium salinarum. Nat. Microbiol. 1:16148.
Krupovic, M., Cvirkaite-Krupovic, V., Iranzo, J., Prangishvili, D., and Koonin, E. V. (2018). Viruses of archaea: structural, functional, environmental and evolutionary genomics. Virus Res. 244, 181–193. doi: 10.1016/j.virusres.2017.11.025
Legerme, G., and Pohlschroder, M. (2019). Limited cross-complementation between Haloferax volcanii PilB1-C1 and PilB3-C3 paralogs. Front. Microbiol. 10:700. doi: 10.3389/fmicb.2019.00700
Legerme, G., Yang, E., Esquivel, R. N., Kiljunen, S., Savilahti, H., and Pohlschroder, M. (2016). Screening of a Haloferax volcanii transposon library reveals novel motility and adhesion mutants. Life 6:41. doi: 10.3390/life6040041
Leigh, J. A., Albers, S. V., Atomi, H., and Allers, T. (2011). Model organisms for genetics in the domain Archaea: Methanogens, Halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 35, 577–608. doi: 10.1111/j.1574-6976.2011.00265.x
Li, M., Liu, H., Han, J., Liu, J., Wang, R., Zhao, D., et al. (2013). Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei. J. Bacteriol. 195, 867–875. doi: 10.1128/jb.01688-12
Li, Z., Kinosita, Y., Rodriguez-Franco, M., Nußbaum, P., Braun, F., Delpech, F., et al. (2019). Positioning of the motility machinery in halophilic archaea. mBio 10:e0377-19.
Li, Z., Rodriguez-Franco, M., Albers, S. V., and Quax, T. E. F. (2020). The switch complex ArlCDE connects the chemotaxis system and the Archaellum. Mol. Microbiol. 114, 468–479. doi: 10.1111/mmi.14527
Lloyd, K. G., May, M. K., Kevorkian, R. T., and Steen, A. D. (2013). Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor. Appl. Environ. Microbiol. 79, 7790–7799. doi: 10.1128/aem.02090-13
Lomsadze, A., Gemayel, K., Tang, S., and Borodovsky, M. (2018). Modeling leaderless transcription and atypical genes results in more accurate gene prediction in prokaryotes. Genome Res. 28, 1079–1089. doi: 10.1101/gr.230615.117
Lurie-Weinberger, M. N., and Gophna, U. (2015). Archaea in and on the human body: health implications and future directions. PLoS Pathog. 11:e1004833. doi: 10.1371/journal.ppat.1004833
Lynch, E. A., Langille, M. G. I., Darling, A., Wilbanks, E. G., Haltiner, C., Shao, K. S. Y., et al. (2012). Sequencing of seven haloarchaeal genomes reveals patterns of genomic flux. PLoS One 7:e41389. doi: 10.1371/journal.pone.0041389
Maier, L. K., Dyall-Smith, M., and Marchfelder, A. (2015). The adaptive immune system of Haloferax volcanii. Life 5, 521–537. doi: 10.3390/life5010521
Maier, L. K., Stachler, A. E., Brendel, J., Stoll, B., Fischer, S., Haas, K. A., et al. (2019). The nuts and bolts of the Haloferax CRISPR-Cas system I-B. RNA Biol. 16, 469–480. doi: 10.1080/15476286.2018.1460994
Mäntynen, S., Sundberg, L. R., Oksanen, H. M., and Poranen, M. M. (2019). Half a century of research on membrane-containing bacteriophages: bringing new concepts to modern virology. Viruses 11:76. doi: 10.3390/v11010076
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182.
Mescher, M. F., and Strominger, J. L. (1977). The shape-maintaining component of Halobacterium salinarium: a cell surface glycoprotein. Prog. Clin. Biol. Res. 17, 459–465.
Mizuno, C. M., Prajapati, B., Lucas-Staat, S., Sime-Ngando, T., Forterre, P., Bamford, D. H., et al. (2019). Novel haloarchaeal viruses from Lake Retba infecting Haloferax and Halorubrum species. Environ. Microbiol. 21, 2129–2147. doi: 10.1111/1462-2920.14604
Munson-Mcgee, J. H., Snyder, J. C., and Young, M. J. (2018). Archaeal viruses from high-temperature environments. Genes 9:128. doi: 10.3390/genes9030128
Nuttall, S. D., and Smith, M. L. D. (1993). HF1 and HF2: novel bacteriophages of halophilic archaea. Virology 197, 678–684. doi: 10.1006/viro.1993.1643
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214.
Patenge, N., Berendes, A., Engelhardt, H., Schuster, S. C., and Oesterhelt, D. (2001). The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum. Mol. Microbiol. 41, 653–663. doi: 10.1046/j.1365-2958.2001.02542.x
Pfeiffer, F., Losensky, G., Marchfelder, A., Habermann, B., and Dyall-Smith, M. (2020). Whole-genome comparison between the type strain of Halobacterium salinarum (DSM 3754T). and the laboratory strains R1 and NRC-1. Microbiol. Open 9:e974.
Pfeiffer, F., Marchfelder, A., Habermann, B., and Dyall-Smith, M. L. (2019). The genome sequence of the Halobacterium salinarum type strain is closely related to that of laboratory strains NRC-1 and R1. Microbiol. Resour. Announc. 8, 18–19.
Pfeiffer, F., and Oesterhelt, D. (2015). A manual curation strategy to improve genome annotation: application to a set of haloarchael genomes. Life 5, 1427–1444. doi: 10.3390/life5021427
Pietilä, M. K., Demina, T. A., Atanasova, N. S., Oksanen, H. M., and Bamford, D. H. (2014). Archaeal viruses and bacteriophages: comparisons and contrasts. Trends Microbiol. 22, 334–344. doi: 10.1016/j.tim.2014.02.007
Pinto, L. H., D’Alincourt Carvalho-Assef, A. P., Vieira, R. P., Clementino, M. M., and Albano, R. M. (2015). Complete genome sequence of Haloferax gibbonsii strain ARA6, a potential producer of polyhydroxyalkanoates and halocins isolated from Araruama. Rio Janeiro Brasil. J. Biotechnol. 212, 69–70. doi: 10.1016/j.jbiotec.2015.08.010
Plaut, R. D., Beaber, J. W., Zemansky, J., Kaur, A. P., George, M., Biswas, B., et al. (2014). Genetic evidence for the involvement of the S-Layer protein gene Sap and the sporulation genes spo0A, spo0B, and spo0F in phage AP50c infection of Bacillus anthracis. J. Bacteriol. 196, 1143–1154. doi: 10.1128/jb.00739-13
Pohlschroder, M., and Esquivel, R. N. (2015). Archaeal type IV pili and their involvement in biofilm formation. Front. Microbiol. 6:190. doi: 10.3389/fmicb.2015.00190
Pohlschroder, M., Pfeiffer, F., Schulze, S., and Halim, M. F. A. (2018). Archaeal cell surface biogenesis. FEMS Microbiol. Rev. 42, 694–717. doi: 10.1093/femsre/fuy027
Poranen, M. M., Daugelavičius, R., and Bamford, D. H. (2002). Common principles in viral entry. Annu. Rev. Microbiol. 56, 521–538. doi: 10.1146/annurev.micro.56.012302.160643
Prangishvili, D., Bamford, D. H., Forterre, P., Iranzo, J., Koonin, E. V., and Krupovic, M. (2017). The enigmatic archaeal virosphere. Nat. Rev. Microbiol. 15, 724–739. doi: 10.1038/nrmicro.2017.125
Quax, T. E. F., Altegoer, F., Rossi, F., Li, Z., Rodriguez-Franco, M., Kraus, F., et al. (2018). Structure and function of the archaeal response regulator CheY. Proc. Natl. Acad. Sci. U.S.A. 115, E1259–E1268.
Quemin, E. R. J., Chlanda, P., Sachse, M., Forterre, P., Prangishvili, D., and Krupovic, M. (2016). Eukaryotic-like virus budding in archaea. mBio 7:e01439-16.
Quemin, E. R. J., Lucas, S., Daum, B., Quax, T. E. F., Kuhlbrandt, W., Forterre, P., et al. (2013). First insights into the entry process of hyperthermophilic archaeal viruses. J. Virol. 87, 13379–13385. doi: 10.1128/jvi.02742-13
Rowland, E. F., Bautista, M. A., Zhang, C., and Whitaker, R. J. (2020). Surface resistance to SSVs and SIRVs in pilin deletions of Sulfolobus islandicus. Mol. Microbiol. 113, 718–727. doi: 10.1111/mmi.14435
Santos-Pérez, I., Charro, D., Gil-Carton, D., Azkargorta, M., Elortza, F., Bamford, D. H., et al. (2019). Structural basis for assembly of vertical single β-barrel viruses. Nat. Commun. 10:1184.
Schlesner, M., Miller, A., Besir, H., Aivaliotis, M., Streif, J., Scheffer, B., et al. (2012). The protein interaction network of a taxis signal transduction system in a Halophilic archaeon. BMC Microbiol. 12:272. doi: 10.1186/1471-2180-12-272
Schlesner, M., Miller, A., Streif, S., Staudinger, W. F., Müller, J., Scheffer, B., et al. (2009). Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiol. 9:56. doi: 10.1186/1471-2180-9-56
Schulze, S., Adams, Z., Cerletti, M., De Castro, R., Ferreira-Cerca, S., Fufezan, C., et al. (2020). The Archaeal proteome project advances knowledge about archaeal cell biology through comprehensive proteomics. Nat. Commun. 11:3145.
Senčilo, A., Paulin, L., Kellner, S., Helm, M., and Roine, E. (2012). Related haloarchaeal pleomorphic viruses contain different genome types. Nucleic Acids Res. 40, 5523–5534. doi: 10.1093/nar/gks215
Shalev, Y., Soucy, S. M., Papke, R. T., Gogarten, J. P., Eichler, J., and Gophna, U. (2018). Comparative analysis of surface layer glycoproteins and genes involved in protein glycosylation in the genus haloferax. Genes 9:172. doi: 10.3390/genes9030172
Snyder, J. C., Samson, R. Y., Brumfield, S. K., Bell, S. D., and Young, M. J. (2013). Functional interplay between a virus and the ESCRT machinery in Archaea. Proc. Natl. Acad. Sci. U.S.A. 110, 10783–10787. doi: 10.1073/pnas.1301605110
Stern, A., and Sorek, R. (2011). The phage-host arms race: shaping the evolution of microbes. Bioessays 33, 43–51. doi: 10.1002/bies.201000071
Sumper, M., Berg, E., Mengele, R., and Strobel, I. (1990). Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J. Bacteriol. 172, 7111–7118. doi: 10.1128/jb.172.12.7111-7118.1990
Suttle, C. A. (2007). Marine viruses - Major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. doi: 10.1038/nrmicro1750
Tachdjian, S., and Kelly, R. M. (2006). Dynamic metabolic adjustments and genome plasticity are implicated in the heat shock response of the extremely thermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 188, 4553–4559. doi: 10.1128/jb.00080-06
Tamir, A., and Eichler, J. (2017). N-Glycosylation is important for proper Haloferax volcanii S-layer stability and function. Appl. Environ. Microbiol. 83:e03152-16.
Tock, M. R., and Dryden, D. T. F. (2005). The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8, 466–472. doi: 10.1016/j.mib.2005.06.003
van Wolferen, M., Orell, A., and Albers, S. V. (2018). Archaeal biofilm formation. Nat. Rev. Microbiol. 16, 699–713. doi: 10.1038/s41579-018-0058-4
Wolters, M., Borst, A., Pfeiffer, F., and Soppa, J. (2019). Bioinformatic and genetic characterization of three genes localized adjacent to the major replication origin of Haloferax volcanii. FEMS Microbiol. Lett. 366:fnz238.
Keywords: haloarchaea, archaeal virus, type IV pili, S-layer, archaellum, N-glycosylation
Citation: Tittes C, Schwarzer S, Pfeiffer F, Dyall-Smith M, Rodriguez-Franco M, Oksanen HM and Quax TEF (2021) Cellular and Genomic Properties of Haloferax gibbonsii LR2-5, the Host of Euryarchaeal Virus HFTV1. Front. Microbiol. 12:625599. doi: 10.3389/fmicb.2021.625599
Received: 03 November 2020; Accepted: 28 January 2021;
Published: 16 February 2021.
Edited by:
Francisco Rodriguez-valera, Miguel Hernández University of Elche, SpainReviewed by:
Aharon Oren, The Hebrew University of Jerusalem, IsraelCopyright © 2021 Tittes, Schwarzer, Pfeiffer, Dyall-Smith, Rodriguez-Franco, Oksanen and Quax. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tessa E. F. Quax, VGVzc2EucXVheEBiaW9sb2dpZS51bmktZnJlaWJ1cmcuZGU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.