
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 March 2021
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.621875
This article is part of the Research Topic Plants and Microbial Communities: Diversity, Pathogens and Biological Control View all 28 articles
Amino-phosphonate derivative R-diphenyl-1-(4-methylbenzothiazole-2-amino)-1-(thiphene-2-yl)-methylphosphonate (Q-R) has a high protective anti-tobacco mosaic virus (TMV) activity. However, the mechanism responsible for Q-R’s effect on TMV infection is largely unknown. Here, we studied the expression levels of harpin-binding protein 1 (HrBP1) and pathogenesis-related protein-1a (PR-1a) in TMV-infected tobacco plants by using reverse transcription quantitative real-time PCR. Then, we verified the interactions between Q-R and the HrBP1 protein from Escherichia coli using isothermal titration calorimetry and studied the Q-R-associated assembly of HrBP1 using size-exclusion chromatography. The results showed that the expression levels of HrBP1 and PR-1a genes were significantly increased by Q-R at the transcriptional level in TMV-infected tobacco plants, and the E. coli-expressed HrBP1 protein was assembled into oligomers by Q-R via binding to HrBP1 with a dissociation constant of 1.19 μM. We, therefore, concluded that Q-R activated the HrBP1 and PR-1a genes and enhanced the ability of HrBP1 to assemble in tobacco plants.
Harpin induces hypersensitive responses to plants and induces the defensive processes of pathogen-associated immunity in plant roots (El-Maarouf et al., 2001; Aljaafri et al., 2017; Lawaju et al., 2018).
Harpin-binding protein (HrBP1) is a protein receptor of Harpin. HrBP1 was first discovered in planting cell walls. HrBP1 induces systemic acquired resistance to plants (El-Maarouf et al., 2001). HrBP1 can be selectively up-regulate several signaling pathways, including those of salicylic acid (SA), ethylene and the jasmonic acid. In the SA pathway, the genes encoding enhanced disease susceptibility 1, non-expressor of pathogenesis- related 1, pathogenesis-related protein-1a (PR-1a), pathogenesis related- protein-2, pathogenesis-related protein-5, and alternative oxidase were activated. In the ethylene and jasmonic acid pathways, PDF1.2 and Thi2.1 were activated. In addition, HrBP1 generates disease resistance by up-regulating the harpin-induced gene related to the mitogen-activated protein kinase gene (Lee et al., 2001; Wang et al., 2002; Liu et al., 2006; Penmetsa et al., 2008; Camehl et al., 2010). HrBP1 expression results in the hypersensitive reaction and systemic acquired resistance related to the non-race-specific disease resistance and enhanced disease susceptibility-1 genes (Peng et al., 2003; Gopalan, 2008). Thus, HrBP1 can generate anti-viral responses in plants.
In a previous study, we found that dufulin-associated activation of HrBP1 leads to the SA signaling pathway and produces antiviral responses from the tobacco plants (Chen et al., 2012). Amino-phosphonate derivative (R)-diphenyl-1-(4-methylbenzothiazole-2-amino)-1-(thiphene-2-yl)-methylphosphonate (Q-R) is a derivative compound of dufulin with high bioactivities on tobacco mosaic virus (TMV) and cucumber mosaic virus (Song et al., 2009; Zhang et al., 2016, 2017). However, the mechanism of Q-R that affects TMV infection is largely unknown. The structure of Q-R is similar to that of dufulin (Figure 1). To further to investigate the protective mechanism of Q-R, we studied the expression levels of the HrBP1 and PR-1a genes in TMV-infected tobacco (Nicotiana tabacum) plant treated with Q-R. Then, we studied the mechanism of Q-R affecting by HrBP1 protein.
The chiral α-aminophosphonate derivative Q-R (99%) and dufulin (99%) was synthesized in our laboratory (Zhang et al., 2016). Benzo-1, 2, 3-thiadiazole-7-carbothioic acid S-methyl ester (BTH) (99%) was purchased from Aladdin Biochemical Technology Co. (Shanghai, China) (Sleiman et al., 2017). The structural formulae of these compounds are presented in Figure 1. In total, ∼30 Nicotiana tabacum K326 seeds were placed in a growth chamber at 25°C, after 12 weeks, the tobacco plants were used for the study. The TMV was purified as previously described by Gooding (Gooding and Hebert, 1967). The 12-weeks-old tobacco plants were sprayed with 5 mL of 500-μg/mL Q-R three times for 1-d intervals (500-μg/mL dufulin and BTH were positive control). Then, the plants were inoculated with TMV and grown for 5 days. The leaf samples of tobacco plants were harvested for further analyses. Three independent experimental replicates were conducted. The control tobacco plants, not exposed to TMV, were kept in another growth chamber.
To verify the effects of Q-R on the expression of antiviral response genes in the TMV-infected tobacco plants, the expression levels of the HrBP1 and PR-1a genes were examined using qPCR. Briefly, 100 mg total RNA from tobacco samples (treated with 500 μg/mL of an antiviral agent) was extracted using the TRIzol reagent kit, independently (TakaRa, Japan), and cDNAs were synthesized using a Reverse Transcription System kit (TakaRa). Then, qPCR was performed using SYBR Premix Ex TaqII (TaKaRa). Gene expression levels were normalized using β-actin as the internal control (Supplementary Table 1; Chen et al., 2012; Gao et al., 2019). The relative expression gene levels were calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001; Wang et al., 2019).
The sequence (GenBank accession no. 107762961) for N. tabacum HrBP1 was used to design a forward primer containing a NdeI (underlined) restriction site in (5′-GGAAT TCCATATGGCTTCTCTACTTCAGTACTCTACACT-3′) and a XhoI (underlined) restriction site in reverse primers (5′- CCGC TCGAGTGAGATAACGAAAACTCTAAGCTCTC-3′). The full- length N. tabacum HrBP1 gene (831 bp) was inserted into pET28a (Novagen, United States), resulting in the plasmid pET28a-HrBP1. The N. tabacum HrBP1 gene was confirmed by 1% agarose gel electrophoresis and the N. tabacum HrBP1 DNA sequence was confirmed at the Shanghai Sangon Company (Chen et al., 2012).
The plasmid pET28a-HrBP1 in a culture of E. coli strains BL21(DE3) (Novagen) was transferred to 1 L of Luria broth (30-μg/mL kanamycin). When the OD600 of Luria broth reached 0.65, the temperature was decreased to 16°C, and 0.8–1.0 mM isopropyl-β-D-galactopyranoside (IPTG) was supplemented in Luria broth. HrBP1 protein was expressed in Luria broth overnight. Then, the cells containing HrBP1 protein were collected, harvested, and resuspended in 50 mM Tris-HCl, 150 mM NaCl and 1 mM β-mercaptoethanol buffer (pH 7.5) and the cells were lysed at 4°C using sonication (Li et al., 2013, 2015a). The whole cells were centrifuged at 12,000 g for 30 min at 4°C, the soluble supernatants were collected and loaded onto a 5-mL HisTrap high-performance column attached to an AKTA purifier protein liquid chromatography system (GE Healthcare, United States), and the protein was eluted by 20 mM Tris-HCl and 300-mM imidazole buffer (pH 7.5). Eluate fractions were collected and analyzed by 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and the crude protein were extracted at 4°C using a desalting column (GE Healthcare), and the eluate fractions were pooled and concentrated by ultrafiltration (10-kDa cut-off). The protein amount was detected by absorbance at 280 nm using a Genequant100 (GE Healthcare).
At 4°C, SEC was performed using Superdex 200 10/300 GL column (GE Healthcare) attached to an AKTA purifier protein liquid chromatography system (GE Healthcare) (Li et al., 2015b). The column was equilibrated using a buffer containing 20 mM Tris-HCl and 100 mM NaCl (pH 7.5). Myoglobin (17 kDa), ovalbumin (43 kDa), albumin (67 kDa), IgG (158 kDa), and ferritin (440 kDa) was used as the molecular mass standards (Bio-Rad, United States). The protein content was detected at a wavelength of 280 nm. The purified protein was confirmed by 8% enriching gel and 12% SDS-PAGE, and then stained with Coomassie brilliant blue (Li et al., 2015a).
The purified HrBP1 proteins were digested using trypsin, the digested products were analyzed using a Nano LC-1DTM plus system (Eksigent Technologies, United States) and TripleTOF 5600 MS (AB SCIEX, United States) (Gopalan, 2008; MacLean et al., 2010; Chen et al., 2015; Yang et al., 2017; Shi et al., 2018; Yu et al., 2018). The achieved peptide masses were searched and blasted using UniProt database without any species or peptide limits. Multiple peptide fragments matched the reported HrBP1 segments (Uniprot accession no.: Q5QJB2). High-resolution masses spectrometry data were extracted and retrieved using PeakView (AB SCIEX) and ProteinPilot (AB SCIEX). The trypsin cleaved sites and the predicted m/z values were analyzed using Skyline (AB SCIEX) (Huntley et al., 2014).
At 25°C, ITC-based binding experiments were performed using an ITC 200 Micro Calorimeter (GE Healthcare). The HrBP1 protein was in the buffer containing 20-mM Tris-HCl and 100-mM sodium chloride (pH 7.5). The 2 mM Q-R compound was titrated into the HrBP1 protein (0.1 mM) in a 200-μL sample cell as follows: 0.4 μL for the first injection and 2 μL for the next 19 injections using a 40-μL microsyringe, the intervals of each injection was150 s. According to the manufacturer’s instructions, the integrated heat data were analyzed using the one-set-of-sites model. The first data point was not required in the analysis. The binding parameters reaction including enthalpy change, ΔH (cal⋅mol–1), binding constant K (mol–1) and number of molecules per HrBP1 protein (n) was floating in the fit. The binding free energy and reaction entropy was calculated (Kumar et al., 2014). The dissociation constant Kd was determined as 1/K (Kumar et al., 2014).
Data were submitted to an analysis of variance (ANOVA) using Duncan’s multiple range test by SPSS 19.0 software (IBM, United States). P < 0.05 was identified as the significance level.
To investigate whether Q-R inhibits the antiviral response genes’ expression levels in TMV-infected tobacco plants, we detected the relative expression of HrBP1 and PR-1a utilizing the qPCR assay. The expression levels of the HrBP1 and PR-1a genes in TMV-infected tobacco plants significantly increased compared with the CK (P < 0.05) at the transcriptional level after exposure to by 500 μg/mL of Q-R on 5 days (Figure 2). Compared with dufulin and BTH, the expression levels of the HrBP1 genes were significantly (P < 0.05) promoted by compound Q-R (Figure 2A).
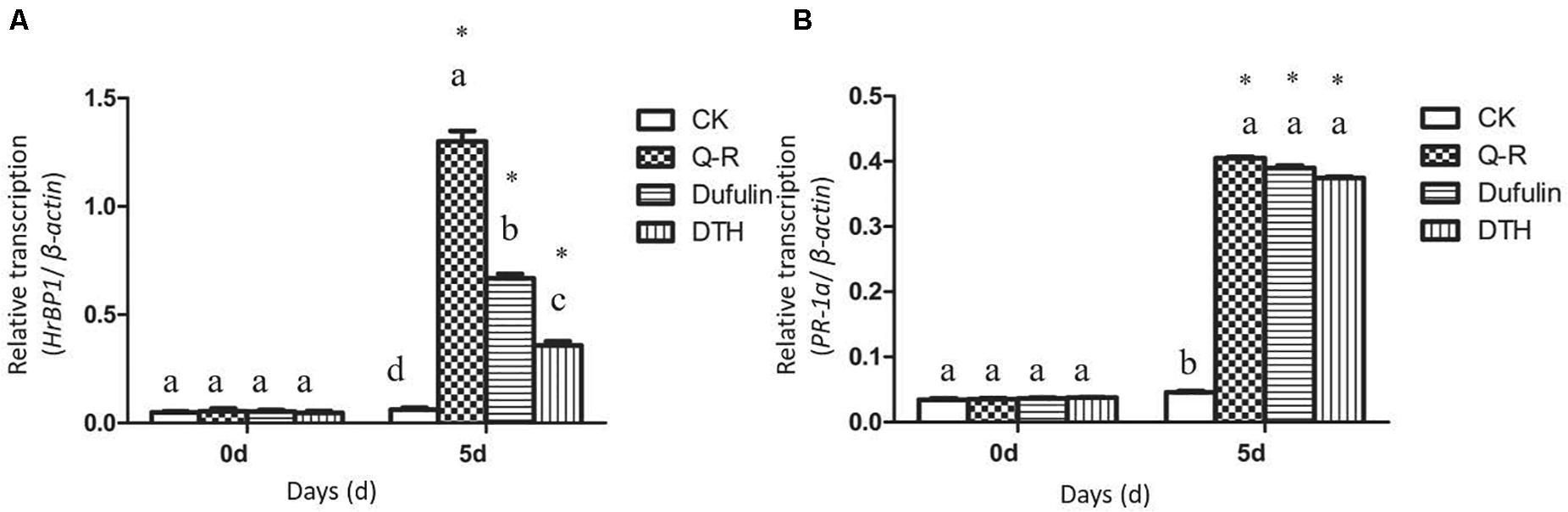
Figure 2. Antiviral response genes expression analysis by qPCR using gene-specific primers (Supplementary Table 1). Relative expression of the HrBP1 (A) and PR-1a (B) genes in TMV-infection plants treated with Q-R, dufulin and BTH at 500 μg/mL at 5 days. Asterisks represent the significantly different between agent treatment and control (P < 0.05). The different lowercase letters represent HrBP1/PR-1a genes expression values in the different treatment groups (P < 0.05).
To obtain the code sequence of the HrBP1 gene in tobacco plant, we amplified the full-length cDNA sequence of HrBP1 and cloned it into the pET28a plasmid (Figure 3A). Electrophoresis and furthering sequence of the fragment confirmed the HrBP1 gene having an expected 831-bp size and nucleotide sequence (Figure 3B and Supplementary Figure 1) as reported (Chen et al., 2012).
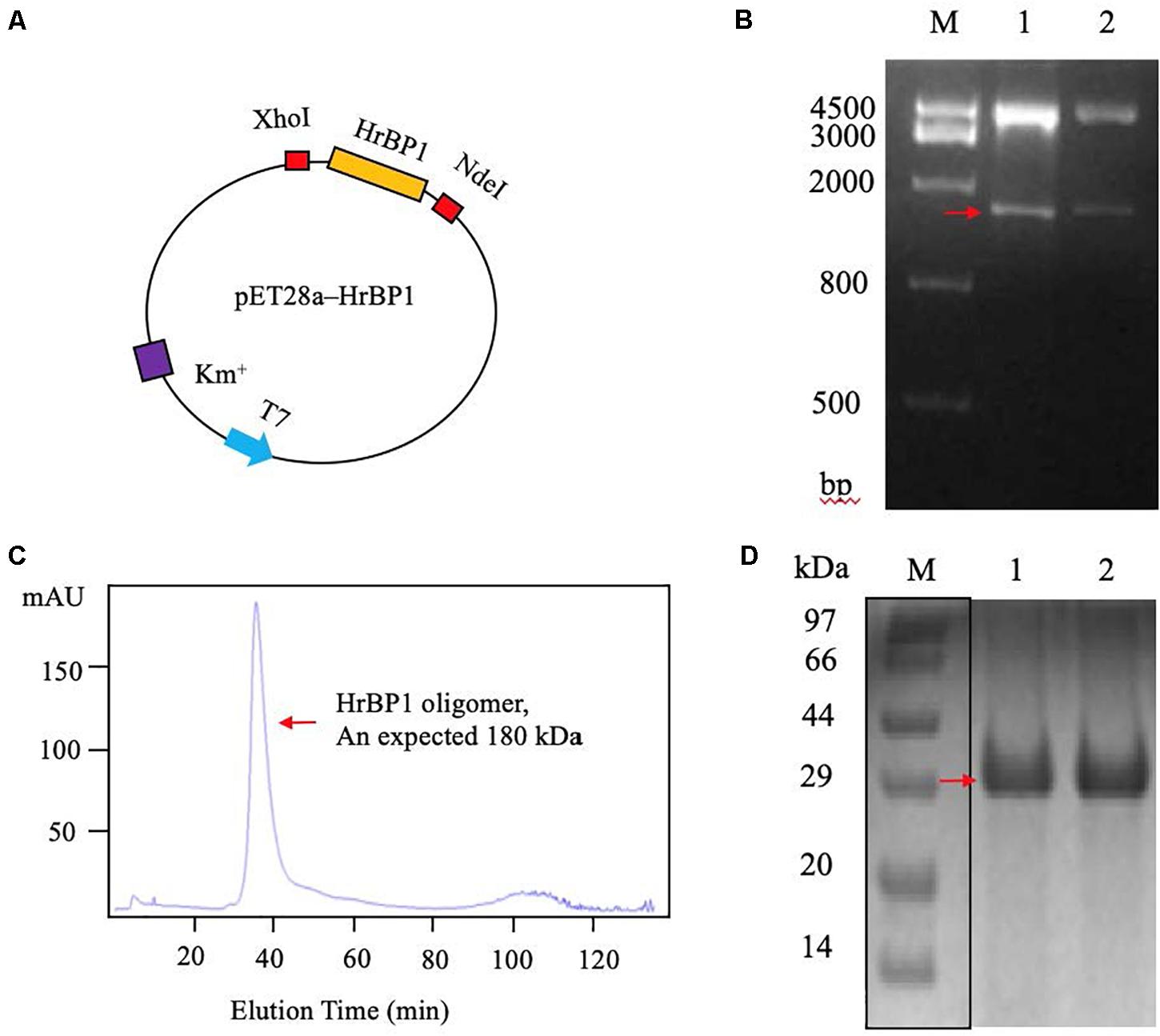
Figure 3. The HrBP1 gene determination and HrBP1 protein expression and purification. (A) Recombinant vector pET28a-HrBP1 was digested by NdeI and XhoI. (B) The HrBP1 gene size (ca. 831 bp) was confirmed by 1% agarose gel electrophoresis. Lane M is a DNA marker; Lanes 1 and 2 shows the digestion of HrBP1 gene by NdeI and XhoI in the pET28a-HrBP1 recombinant plasmid; the red arrow shows bands of HrBP1 bands. (C) HrBP1 oligomer protein with expected size 180 kDa between 440 kDa (Ferritin) and 158 kDa (IgG). (D) M is protein Marker; Lanes 1 and 2 are HrBP1 purification proteins in 12% SDS-PAGE; the red arrow shows HrBP1 bands.
To obtain sufficient quantities of HrBP1 protein for further analyses, we designed the pET28a expression vector including the HrBP1 gene and transformed it into E. coli strain BL21(DE3). The expressed protein was supported by SDS-PAGE analysis to be 30 kDa. The HrBP1 protein was overexpressed after exposure to 0.8 mM isopropyl-β-D-galactopyranoside (IPTG) at 16°C for 16 h (Figures 3C,D). The expressed HrBP1 proteins (approximately 90%) was purified and concentrated on subsequent analysis.
To further determine the amino acid composition of the expressed HrBP1 protein through a polypeptide analysis. The 18.1% fraction of the obtained polypeptide fragments covered the entire 276 amino acids, and the sequence was equally expected.
The SEC results showed that the HrBP1 recombinant protein was an oligomer. The HrBP1 protein was elected and calculated to be approximately 180 kDa based on a reference size (Figure 3C). The SDS-PAGE analysis results showed that the size of the collected protein was approximately 30 kDa (Figure 3D), which indicated that the HrBP1 oligomer was destroyed by SDS in vitro.
To examine the interaction between Q-R and the HrBP1 protein, we analyzed the binding affinity between Q-R to HrBP1 protein using ITC. The binding affinity with Q-R to the HrBP1 protein was stronger than that of the control of antiviral agents BTH and dufulin. Q-R on the HrBP1 protein had a Kd value of 1.19 μM (8.43 × 105 mol–1, Figures 4A,B). The titration data indicated an apparent negative enthalpy values (ΔG ≈−8.08) during the binding of Q-R to the HrBP1 protein. While no binding occurred between the BTH and HrBP1 protein (Figures 4A,C), and weak binding occurred between the dufulin and HrBP1 protein (Figures 4A,D). Thus, the HrBP1–Q-R complex was stable, implying that Q-R had the ability to bind the HrBP1 protein.
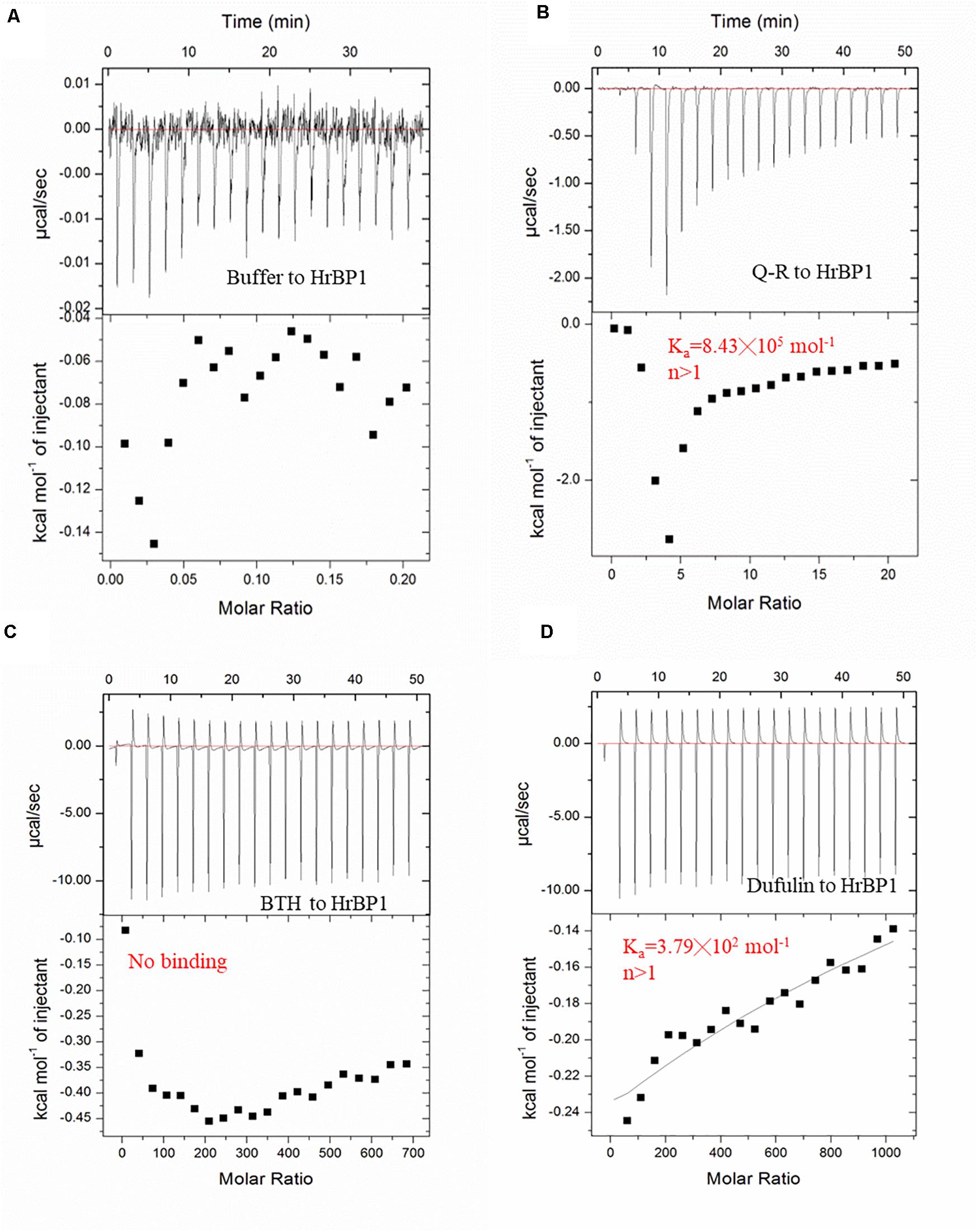
Figure 4. ITC studies of Q-R, dufulin, BTH and HrBP1 purification protein. (A) control. (B) Q-R and HrBP1. (C) BTH and HrBP1. (D) Dufulin and HrBP1.
To confirm the interaction between Q-R and the HrBP1 protein, we analyzed the HrBP1 protein’s state of aggregation after being treated by Q-R using the SEC. Q-R induced the assembly of a large HrBP1 protein oligomer. Briefly, the HrBP1 protein formed functional aggregates. After adding Q-R, the protein samples were examined by SEC. We further assessed the effects of Q-R on the biochemical properties of the HrBP1 protein. As expecting, the HrBP1 protein was present in a large aggregated state (Figure 5). Thus, we deduced that Q-R induces the assembly of the HrBP1 protein and the further formation of the HrBP1–Q-R complex (Figure 6), which agreed with the ITC studies.
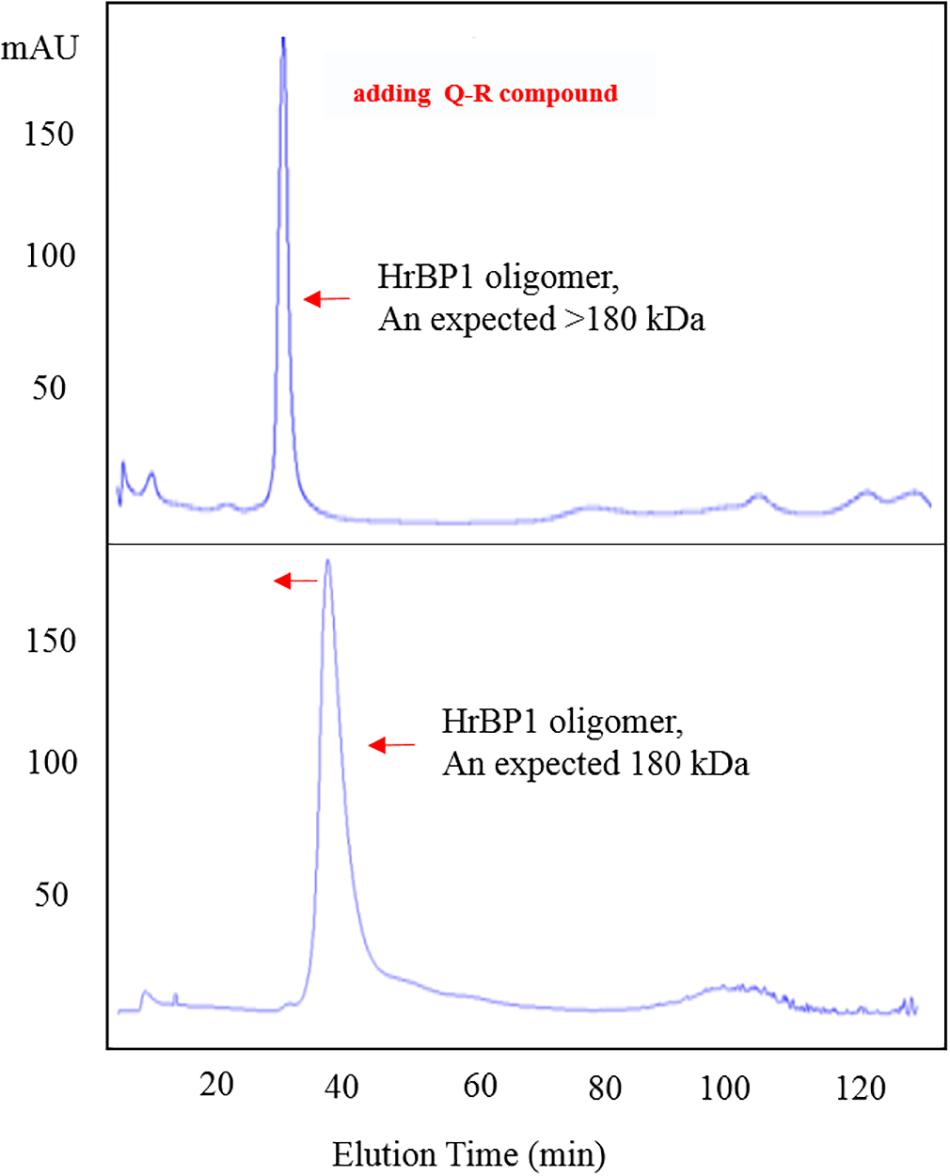
Figure 5. Q-R induced the assembly of HrBP1 large oligomer protein with expected size > 180 kDa between 440 kDa (Ferritin) and 158 kDa (IgG).
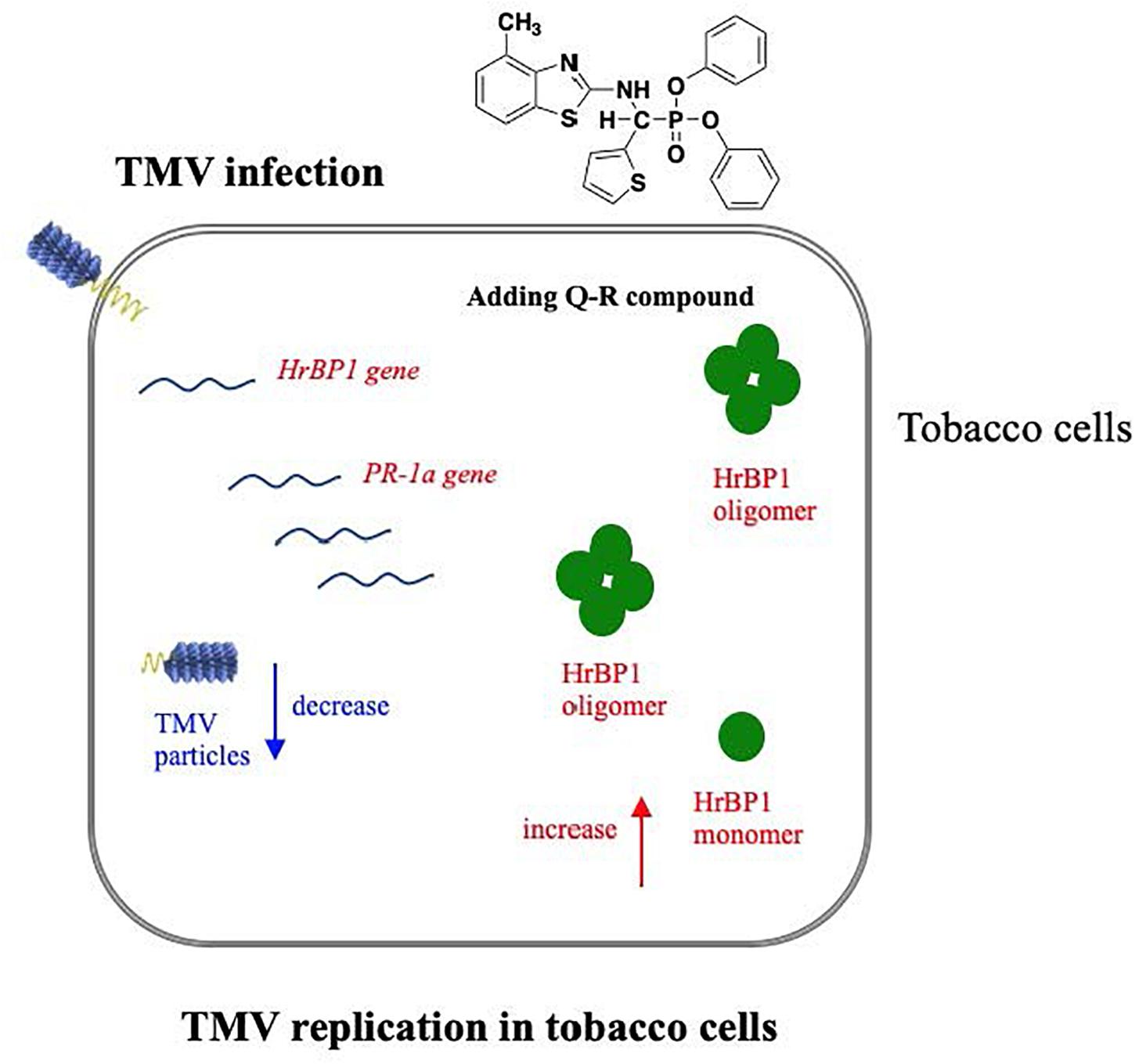
Figure 6. Hypothetical pathway wherein the HrBP1 protein of tobacco plant was induced by Q-R through promotes the expression of the HrBP1 and PR-1a genes in tobacco plants, which leads to Q-R induces antiviral responses and inhibits the duplication of TMV in TMV-infected tobacco plant. Red indicate the up-regulated genes/protein, blue indicate the down-regulated genes/protein.
In summary, our study clarified the mechanism by which Q-R produces antiviral responses associated with the HrBP1 protein expressed in E. coli (In vitro) and tobacco plants (In vivo). In vitro, a strong affinity existed on Q-R and the HrBP1 protein. In vivo, Q-R promoted the expression of the HrBP1 gene in tobacco plants. Thus, Q-R produces antiviral responses by activating the HrBP1 gene and protein in tobacco plants.
The data presented in the study are deposited in the NCBI repository, accession number MW256718.
XL designed this study. MH carried out the clone and protein expression studies, participated in the sequence alignment, and drafted the manuscript. YY, LW, JC, and TL performed MST and qPCR tests. XX and XL critically revised the manuscript. All authors read and approved the final version of the manuscript.
This work was supported by the National Key Research and Development Program of China (No. 2017YFD0200503-3), the National Natural Science Foundation of China (No. 31960546), and Program of Introducing Talents of Discipline to Universities of China (111 Program, D20023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.621875/full#supplementary-material
Supplementary Figure 1 | Nucleotide sequence and amino acid sequence of HrBP1 sequence from Nicotiana tabacum.
Supplementary Table 1 | Primer sequences used in qPCR.
Q-R, R-diphenyl -1-(4-methylbenzothiazole-2-amino)-1-(thiphene-2-yl)-methylphosphonate; TMV, tobacco mosaic virus; HrBP1, harpin-binding protein 1; PR-1a, pathogenesis-related protein-1a; BTH, Benzo-1, 2, 3-thiadiazole-7-carbothioic acid S-methyl ester; SA, salicylic acid; IPTG, isopropyl- β -D-galactopyranoside; SEC, Size-exclusion chromatography; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Aljaafri, W. A., McNeece, B. T., Lawaju, B. R., Sharma, K., Niruala, P. M., Pant, S. R., et al. (2017). A harpin elicitor induces the expression of a coiled-coil nucleotide binding leucine rich repeat (CC-NB-LRR) defense signaling gene and others functioning during defense to parasitic nematodes. Plant Physiol. Biochem. 121, 161–175. doi: 10.1016/j.plaphy.2017.10.004
Camehl, I., Sherameti, I., Venus, Y., Bethke, G., Varma, A., Lee, J., et al. (2010). Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytol. 185, 1062–1073. doi: 10.1111/j.1469-8137.2009.03149.x
Chen, Z., Chen, B. H., Guo, Q., Shi, L., He, M., Qin, Z., et al. (2015). A Time-course proteomic analysis of rice triggered by plant activator BTH. J. Plant Growth Regul. 34, 392–409. doi: 10.1007/s00344-015-9476-y
Chen, Z., Zeng, M. J., Song, B. A., Hou, C., Hu, D., Li, X., et al. (2012). Dufulin activates HrBP1 to produce antiviral responses in tobacco. PLoS One 7:e37944. doi: 10.1371/journal.pone.0037944
El-Maarouf, H., Barny, M. A., Rona, J. P., and Bouteau, F. (2001). Harpin, a hypersensitive response elicitor from Erwinia amylovora, regulates ion channel activities in Arabidopsis thaliana suspension cells. FEBS Lett. 497:82. doi: 10.1016/S0014-5793(01)02441-3
Gao, D., Wang, D. M., Chen, K., Huang, M., Xie, X., and Li, X. (2019). Activation of biochemical factors in CMV-infected tobacco by ningnanmycin. Pest. Biochem. Physiol. 156, 116–122. doi: 10.1016/j.pestbp.2019.02.012
Gooding, G. J., and Hebert, T. T. (1967). A simple technique for purification of tobacco mosaic virus in large quantities. Phytopathology 57:1285. doi: 10.1098/rstl.1767.0047
Gopalan, S. (2008). Reversal of an immunity associated plant cell death program by the growth regulator auxin. BMC Res. 1:126. doi: 10.1186/1756-0500-1-126
Huntley, R. P., Sawford, T., Mutowo-Meullenet, P., Shypitsyna, A., Bonilla, C., Martin, M. J., et al. (2014). The GOA database: gene ontology annotation updates for 2015. Nucleic Acid Res. 43, 1057–1063. doi: 10.1093/nar/gku1113
Kumar, R. R., Singh, G. P., Goswami, S., Pathak, H., and Rai, R. D. (2014). Proteome analysis of wheat (Triticum aestivum) for the identification of differentially expressed heat-responsive proteins. Aus. J. Crop Sci. 8, 973–986.
Lawaju, B. R., Lawrence, K. S., Lawrence, G. W., and Klink, V. P. (2018). Harpin-inducible defense signaling components impair infection by the ascomycete Macrophomina phaseolina. Plant Physiol. Biochem. 129, 331–348. doi: 10.1016/j.plaphy.2018.06.020
Lee, J., Klessig, D. F., and Nürnberger, T. (2001). A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. Plant Cell 13, 1079–1093. doi: 10.1105/tpc.13.5.1079
Li, X. Y., Liu, J., Yang, X., Ding, Y., Wu, J., Hu, D., et al. (2015a). Studies of binding interactions between dufulin and southern rice black-streaked dwarf virus P9-1. Bioorg. Med. Chem. 23, 3629–3637. doi: 10.1016/j.bmc.2015.04.008
Li, X. Y., Song, B. A., Chen, X., Wang, Z. C., Zeng, M. J., Yu, D. D., et al. (2013). Crystal structure of a four-layer aggregate of engineered TMV CP implies the importance of terminal residues for oligomer assembly. PLoS One 8:e77717. doi: 10.1371/journal.pone.0077717
Li, X. Y., Zhang, W. Y., Ding, Y., Wang, Z., Wu, Z., Yu, L., et al. (2015b). Characterization of the importance of terminal residues for southern rice black-streaked dwarf virus P9-1 viroplasm formations. Pro. Exp. Purif. 111, 98–104. doi: 10.1016/j.pep.2015.04.003
Liu, F. Q., Liu, H. X., Jia, Q., Wu, X., Guo, X., Zhang, S., et al. (2006). The internal glycine-rich motif and cysteine suppress several effects of the HpaGXooc protein in plants. Phytopathology 96, 1052–1059.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
MacLean, B., Tomazela, D. M., Shulman, N., Chambers, M., Finney, G. L., Frewen, B., et al. (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968.
Peng, J. L., Dong, H. S., Dong, H. P., Delaney, T. P., Bonasera, J. M., and Beer, S. V. (2003). Harpin-elicited hypersensitive cell death and pathogen resistance require the NDR1 and EDS1 genes. Physiol. Mol. Plant Pathol. 62, 317–326. doi: 10.1016/S0885-5765(03)00078-X
Penmetsa, R. V., Uribe, P., Anderson, J., Lichtenzveig, J., Gish, J. C., Woo, Y., et al. (2008). The Medicago truncatula ortholog of Arabidopsis EIN2, SICKLE, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 55, 580–595. doi: 10.1111/j.1365-313X.2008.03531.x
Shi, J., Yu, L., and Song, A. B. (2018). Proteomics analysis of xiangcaoliusuobingmi-treated Capsicum annuum L. infected with cucumber mosaic virus. Pestic. Biochem. Physiol. 149, 113–122. doi: 10.1016/j.pestbp.2018.06.008
Sleiman, M., Claire, P. D. S., and Richard, C. (2017). Heterogeneous photochemistry of agrochemicals at the leaf surface: a case study of plant activator acibenzolar-S-methyl. J. Agric. Food. Chem. 65, 7653–7660. doi: 10.1021/acs.jafc.7b02622
Song, B. A., Yang, S., Jin, L. H., and Bhadury, S. (2009). Environment Friendly Anti-plant Viral Agent. Berlin: Springer Press, 234–253.
Wang, D. M., Xie, X., Gao, D., Chen, K., Chen, Z., Jin, L., et al. (2019). Dufulin intervenes the viroplasmic proteins as the mechanism of action against southern rice black-streaked dwarf virus. J. Agric. Food Chem. 67, 11380–11387. doi: 10.1021/acs.jafc.9b05793
Wang, K. L., Li, H., and Ecker, J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14, 131–151. doi: 10.1105/tpc.001768
Yang, A. M., Yu, L., Zhuo, C., Zhang, S., Shi, J., Zhao, X., et al. (2017). Label-free quantitative proteomic analysis of chitosan oligosaccharide-treated rice infected with southern rice black-streaked dwarf virus. Viruses J. 9:115. doi: 10.3390/v9050115
Yu, L., Wang, W. L., Zeng, S., Chen, Z., Yang, A., Shi, J., et al. (2018). Label-free quantitative proteomics analysis of cytosinpeptidemycin responses in southern rice black-streaked dwarf virus-infected rice. Pestic. Biochem. Physiol. 147, 20–26. doi: 10.1016/j.pestbp.2017.06.005
Zhang, G. P., Hao, G. F., Pan, J. K., Zhang, J., Hu, D., and Song, B. (2016). Asymmetric synthesis and bioselective activities of α-amino-phosphonates based on the dufulin motif. J. Agric. Food Chem. 64, 4207–4213. doi: 10.1021/acs.jafc.6b01256
Keywords: Q-R, HrBP1, PR-1a, interaction, binding
Citation: Huang M, Yan Y, Wang L, Chen J, Liu T, Xie X and Li X (2021) Research on the Interaction Mechanism Between α Mino-Phosphonate Derivative Q-R and Harpin-Binding Protein 1 in Tobacco (Nicotiana tabacum) Plants. Front. Microbiol. 12:621875. doi: 10.3389/fmicb.2021.621875
Received: 27 October 2020; Accepted: 24 February 2021;
Published: 23 March 2021.
Edited by:
Jia Liu, Chongqing University of Arts and Sciences, ChinaReviewed by:
Orlando Borras-Hidalgo, Qilu University of Technology, ChinaCopyright © 2021 Huang, Yan, Wang, Chen, Liu, Xie and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangyang Li, eHlsaTFAZ3p1LmVkdS5jbg==; eGlhbmd5YW5nbGk4M0AxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.