
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 February 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.621564
This article is part of the Research Topic Antimicrobial Resistance in Aquatic Environments View all 15 articles
Globally, urban water bodies have emerged as an environmental reservoir of antimicrobial resistance (AMR) genes because resistant bacteria residing here might easily disseminate these traits to other waterborne pathogens. In the present study, we have investigated the AMR phenotypes, prevalent plasmid-mediated AMR genes, and integrons in commensal strains of Escherichia coli, the predominant fecal indicator bacteria isolated from a major urban river of northern India Yamuna. The genetic environment of blaCTX–M–15 was also investigated. Our results indicated that 57.5% of the E. coli strains were resistant to at least two antibiotic classes and 20% strains were multidrug resistant, i.e., resistant to three or more antibiotic classes. The multiple antibiotic resistance index of about one-third of the E. coli strains was quite high (>0.2), reflecting high contamination of river Yamuna with antibiotics. With regard to plasmid-mediated AMR genes, blaTEM–1 was present in 95% of the strains, followed by qnrS1 and armA (17% each), blaCTX–M–15 (15%), strA-strB (12%), and tetA (7%). Contrary to the earlier reports where blaCTX–M–15 was mostly associated with pathogenic phylogroup B2, our study revealed that the CTX-M-15 type extended-spectrum β-lactamases (ESBLs) were present in the commensal phylogroups A and B1, also. The genetic organization of blaCTX–M–15 was similar to that reported for E. coli, isolated from other parts of the world; and ISEcp1 was present upstream of blaCTX–M–15. The integrons of classes 2 and 3 were absent, but class 1 integron gene intI1 was present in 75% of the isolates, denoting its high prevalence in E. coli of river Yamuna. These evidences indicate that due to high prevalence of plasmid-mediated AMR genes and intI1, commensal E. coli can become vehicles for widespread dissemination of AMR in the environment. Thus, regular surveillance and management of urban rivers is necessary to curtail the spread of AMR and associated health risks.
The gastrointestinal tract of humans and animals is regarded as the primary/natural habitat of Escherichia coli. Besides its natural habitat, E. coli is also found in secondary habitats like aquatic and terrestrial reservoirs (Méric et al., 2013). Aquatic environments, especially urban water bodies, harbor a heterogeneous collection of microorganisms originating from fecal, hospital, agricultural, and veterinary sources. Moreover, several studies have suggested that aquatic environments serve as genetic reactors that promote transfer of antimicrobial resistance (AMR) and virulence genes among bacteria (Baquero et al., 2008; Hasegawa et al., 2018).
The phylogrouping methods commonly used for population/clonal studies of E. coli include multilocus sequence typing (MLST) (Aanensen and Spratt, 2005), multilocus enzyme electrophoresis (MLEE), triplex PCR, etc. Triplex PCR is a widely used rapid and simple technique for phylotyping E. coli. In triplex PCR, three genes (chuA, yjaA, and a gene encoding a fragment of a putative lipase esterase) are PCR-amplified (Clermont et al., 2000). Based on the presence or absence of these three genetic elements, a strain can be assigned to belong to any of the four phylogroups—A, B1, B2, and D (Clermont et al., 2000). Several researchers have studied AMR in E. coli isolated from aquatic environments and have reported that these traits are easily transmissible among bacterial species with the help of several mobile genetic elements like integrons, insertion sequences (ISs), plasmids, and transposons (Su et al., 2012; Koczura et al., 2013; Liebana et al., 2013; Pereira et al., 2013; Kaushik et al., 2018). Integrons are regarded as the primary vehicles that disseminate AMR genes among bacterial species (Gillings, 2014) because they can be located on conjugative plasmids, which enhance their horizontal spread.
River Yamuna is a major river of northern India, which is associated with several anthropogenic activities of the population residing in the National Capital Region of India. It gets contaminated with effluents originating from hospital and municipal wastewaters; discharge from livestock, poultry, and agriculture production plants; industries; etc. The high levels of pollutants are expected to provide a positive selection pressure for increasing AMR in bacterial population residing there (Kümmerer, 2009; Tacão et al., 2012). Thus, it is expected to be a crucial reservoir of a diverse E. coli populations and an ideal ecological niche for studying strains with diverse phenotypes, genotypes, and AMR. In an earlier study published from our laboratory, we had described the β-lactam susceptibilities and β-lactamase genes in 61 E. coli strains of all phylogroups (A, B1, B2, and D) isolated from river Yamuna (Bajaj et al., 2015). Though AMR resistance has been investigated for commensal phylogroups of E. coli isolated from veterinary or clinical sources, only a few studies have investigated the AMR phenotypes, genes, and integrons in commensal E. coli isolated from urban rivers. The commensal strains of E. coli in urban rivers can easily disseminate AMR determinants to pathogenic E. coli or other waterborne pathogens via mobile genetic elements. Thus, it is important to study the AMR determinants and integrons in commensal strains residing in water bodies also. Our study is the first report on AMR phenotypes, plasmid-mediated AMR genes, and integrons in the strains of commensal phylogroups (A and B1) of E. coli from river Yamuna, India.
Two hundred water samples were collected from different sites along the entire stretch of the river Yamuna, which flows through the National Capital Region of India in sterile screw-capped bottles, transported to the laboratory on ice, and processed within 6 h of the sample collection. A schematic figure showing the details of the sampling sites has been published earlier (Bajaj et al., 2015). Enrichment of the samples for isolation of Escherichia coli was performed using a published method (Ram et al., 2008). Briefly, 100 ml of water sample was filtered through a 0.45 μm membrane filter (Millipore, MA, United States). The membrane filter was cut into four pieces, and each piece was incubated in 50 ml of MacConkey broth at 37°C, 220 rpm, overnight. The next day, a loopful of the broth culture was streaked on the surface of MacConkey agar plates and incubated at 35°C for 18–20 h. One hundred sixty-two typical E. coli colonies were selected and maintained as pure cultures on Luria–Bertani (LB) agar slants at 4°C. Of these, 126 isolates were presumptively identified as E. coli using API 20E strips (bioMérieux, France). API 20E is a standardized kit of biochemical tests used to identify members of the family Enterobacteriaceae and other non-fastidious Gram-negative rods.
DNA was extracted from the E. coli strains using the boilinglysis method (Rodríguez-Baño et al., 2004). The gene encoding 16S rRNA was PCR-amplified using universal eubacterial forward primer 27F (5′AGAGTTTGATCCTGGCTCAG3′) and reverse primer 1492R (5′ACGGCTACCTTGTTACGACTT3′). The contents of the PCR mixture were 1 × PCR buffer (1.5 MgCl2, 1.5 mM of KCl, 10 mM of Tris–HCl, and 0.1% Triton X-100), 200 μM of the four dNTPs, 1 U of Taq DNA polymerase (New England Biolabs, Ipswich, MA, United States), 10 pmol of forward and reverse primers, and 1 ng of genomic DNA in a final volume of 25 μl. The PCR conditions and methods for purification of PCR amplicons and sequencing have been described earlier (Singhal et al., 2019). Briefly, the PCR amplicons were purified by HiYieldTM extraction kit (RBC Bioscience, New Taipei City, Taiwan) and sequenced at a commercial facility using Sanger sequencing (Invitrogen BioServices India Pvt. Ltd., Bangalore, India). The nucleotide sequence homology was analyzed using the nucleotide BLAST (BLASTn) algorithm available at the National Center for Biotechnology Information (NCBI).
The phylogenetic profiles of all the isolates were determined by triplex PCR (Clermont et al., 2000); and 40 strains representing the commensal phylogroups (A and B1) were selected for studying the AMR phenotypes, plasmid-mediated AMR genes, and integrons.
Antimicrobial susceptibilities of E. coli strains for various classes of antibiotics like β-lactams, aminoglycosides, quinolones, and tetracycline were determined by Kirby–Bauer disk diffusion method. The antibiotic disks (Himedia, India) that were used in this study were (charge in μg/disk) as follows: ampicillin (10 μg), piperacillin (100 μg), amoxicillin–clavulanic acid (20/10 μg), cefazolin (30 μg), cefuroxime (30 μg), cefotaxime (30 μg), cefepime (30 μg), streptomycin (10 μg), kanamycin (30 μg), tobramycin (10 μg), netilmicin (30 μg), amikacin (30 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), ofloxacin (5 μg), and tetracycline (30 μg). The results of antimicrobial susceptibility testing were interpreted following the guidelines of Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute [CLSI], 2018). The multiple antibiotic resistance (MAR) index of each strain was calculated by dividing the number of antibiotics to which a strains was resistant by the number of antibiotics that were tested (Krumperman, 1983). Here, the number of antibiotics to which an E. coli strain exhibited resistance was divided by 16 because susceptibility of each strain was tested for 16 antibiotics. The E. coli strains were tested for production of extended-spectrum β-lactamases (ESBLs) using a phenotypic confirmatory test recommended by the CLSI (Clinical and Laboratory Standards Institute [CLSI], 2018). Briefly, cefotaxime and ceftazidime disks (30 μg) alone and in combination with clavulanic acid (30/10 μg) were placed on the surface of bacterial lawn spread over Mueller–Hinton agar petri plates. Strains whose zone diameter in the presence of antibiotic–clavulanic acid combination was ≥5 mm were considered as ESBL producers. The strains were tested for phenotypic production of AmpC using AmpC E-test strips (bioMérieux Inc., MO, United States) following the manufacturer’s instructions. Strains that showed cefotetan/cefotetan + cloxacillin (CN/CNI) ratio of ≥8 were considered as AmpC producers (Bajaj et al., 2015).
Genes encoding β-lactamases and ESBLs, viz., blaTEM, blaSHV, and blaCTX–M, were detected by PCR amplification of the AMR genes using group-specific primers that amplified the internal coding regions of the genes (Dhanji et al., 2011; Bajaj et al., 2015). The presence of plasmid-encoded AmpC enzymes of the CMY types was determined by PCR amplification of the AMR gene using published primers (Pérez-Pérez and Hanson, 2002). The promoter region and genetic environment of blaCTX–M–15 were studied in blaCTX–M–15-positive E. coli strains by PCR amplification of the corresponding regions using the primers and methods described earlier (Saladin et al., 2002; Dhanji et al., 2011). The primers and the annealing temperatures for amplification of the blaTEM, blaSHV, and blaCTX–M and genetic environment of blaCTX–M are described in Table 1. The contents of the PCR mixture and methods for purification of the PCR amplicons and sequencing were the same as used for 16s rRNA gene sequencing. The nucleotide sequence homology was analyzed using the nucleotide BLAST (BLASTn) available at NCBI.
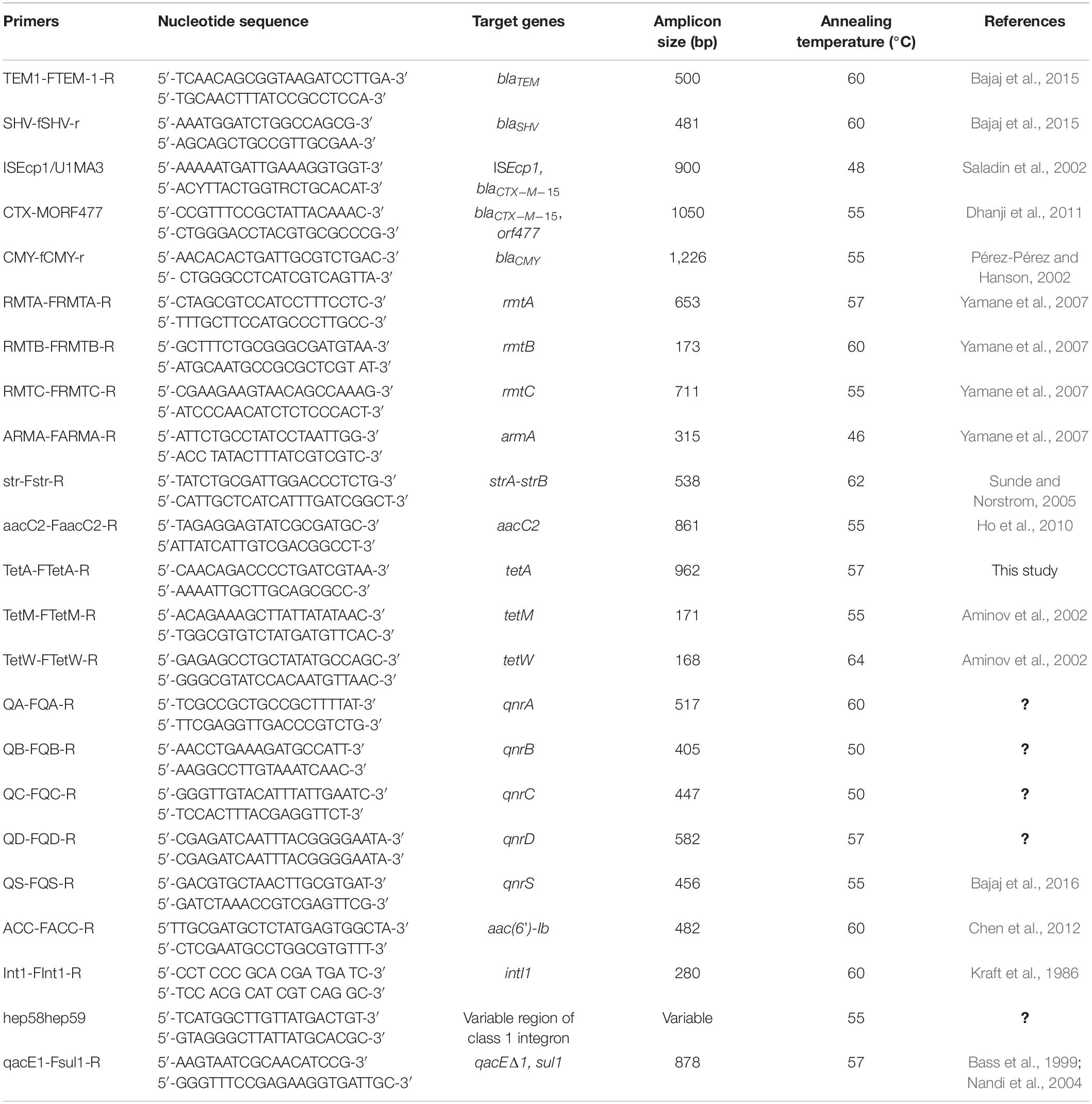
Table 1. Primers and PCR conditions for amplification of antimicrobial resistance genes, integrons and genetic environment of blaCTX–M.
The presence of genes encoding plasmid-mediated quinolone resistance (PMQR) was determined by PCR amplification of genes encoding for (i) proteins that protect DNA from quinolone binding (qnrA, qnrB, qnrC, and qnrD); (ii) aac(6’)-Ib-cr acetyltransferase (aac), which modifies fluoroquinolones like ciprofloxacin and enrofloxacin; and (iii) active efflux pump (qepA).
The presence of aminoglycoside resistance genes was determined by PCR amplification of the genes encoding for linked strA-strB genes and four types of plasmid-mediated 16S rRNA methylases—armA, rmtA, rmtB, and rmtC— using published primers and annealing temperatures described in Table 1 (Sunde and Norstrom, 2005; Yamane et al., 2007).
The presence of tetracycline resistance genes was determined by PCR amplification of the tetracycline efflux gene tetA using self-designed primers and genes encoding ribosome protective proteins tetM and tetW using published primers and annealing temperatures (Aminov et al., 2002; Table 1). The contents of the PCR mixture and protocols for purification of the PCR amplicons, sequencing of PMQR, and aminoglycoside- and tetracycline-resistance genes and homology search were the same as used for the 16S rRNA genes.
The presence and distribution of integrase genes intI1, intI2, intI3, and integron class 1 gene cassette were determined by PCR amplification using published primers (Kraft et al., 1986; Goldstein et al., 2001; White et al., 2001). The variable regions (VRs) of class 1 integrons, which mainly contain an array of gene cassettes, are flanked at 3′ by a conserved segment containing qacEΔ1 and genes coding for quaternary ammonium and sulfonamide, respectively. VRs were investigated as previously reported in all isolates containing class 1 integrons (Guo et al., 2011). The PCR conditions for amplifying the VRs of integrons were the same as for amplifying the 16S rRNA gene except for the primers and annealing temperatures, which have been summarized in Table 1. The PCR amplicons were purified and sequenced as described for the 16S rRNA gene, and nucleotide sequence homology was analyzed using the nucleotide BLAST (BLASTn) available at NCBI.
Gene sequencing revealed that the blaCTX–M–15 genes of the seven blaCTX–M–15-positive strains were identical to each other; hence, the partial coding sequence (CDS) of a representative strain (KP20) was submitted to GenBank (NCBI) with the accession number KF040057. The partial CDS of blaTEM–1 was also identical to each other; hence, the sequence of one representative strain (IP1N) was submitted to GenBank (NCBI) with the accession number KF055435. The partial CDS of qnrS was also identical; hence, the CDS of a representative strain (KP20) was submitted to NCBI GenBank under accession number KF055436. The partial CDS of tetA gene of all the three strains (IS47, WB3, and KKC) was submitted under the accession numbers KJ409940–KJ409942.
The results of 16S rRNA gene sequencing and homology search using BLAST confirmed that the strains presumptively identified using API 20E strips (bioMérieux, France) were Escherichia coli. The results of the triplex PCR and Clermont classification indicated that 50% (n = 20) of the strains belonged to phylogroup A [chuA (−), yjaA (−/+), and TSPE4.C2 (−)] while 50% (n = 20) to phylogroup B1 [chuA (−), yjaA (−), and TSPE4.C2 (+)]. Earlier studies have reported that E. coli strains of all phylogroups were present in river Yamuna (Bajaj et al., 2015; Kaushik et al., 2018). Phylogroups A and B1 of E. coli represent commensal strains, while phylogroups B2 and D represent pathogenic strains (Herzer et al., 1990; Bingen et al., 1998; Lecointre et al., 1998; Picard et al., 1999). Several studies have indicated that the prevalence of virulence genes in commensal strains of E. coli was lesser than in pathogenic strains (Johnson, 1991; Boyd and Hartl, 1998; Lecointre et al., 1998; Picard et al., 1999) but that the commensal strains can easily disseminate AMR determinants to pathogenic E. coli or other waterborne pathogens via mobile genetic elements. Thus, AMR determinants and integrons were investigated in these 40 commensal E. coli strains.
Antibiotic susceptibility testing revealed that 95% (n = 38) of the commensal E. coli strains wereresistant to ampicillin, while 32% (n = 13) of the strains were resistant to piperacillin. Among the cephalosporins, 42.5% (n = 17) strains were resistant to cefazolin (first-generation cephalosporin), 17.5% (n = 7) to cefuroxime (second-generation cephalosporin), 22.5% (n = 9) to cefotaxime (third-generation cephalosporin), and 15% (n = 6) to cefepime (fourth-generation cephalosporin). The fact that commensal waterborne E. coli were less resistant to new-generation cephalosporins than ampicillin is normal because ampicillin was a widely prescribed broad-spectrum penicillin, and over time, bacteria might have developed resistance to this antibiotic. Earlier studies have also reported that ampicillin resistance was highly prevalent in commensal strains of E. coli isolated from India and from other parts of the globe like Vietnam, China, Sudan, and Thailand (Dyar et al., 2012; Abdelgader et al., 2018; Lugsomya et al., 2018; Singh A. K. et al., 2018; Purohit et al., 2019). With regard to ESBL production, 17.5% (n = 7) of the commensal E. coli strains tested positive, while none of the strain tested positive for AmpC production (Table 2). All the ESBL-producing strains were resistant to four or more β-lactam antibiotics, and 57% (n = 4) of the ESBL producers were resistant to ciprofloxacin. Since, penicillins and cephalosporins are the most frequently used antibiotics in India, it is normal that all the seven ESBL-producing E. coli strains were resistant to many antibiotics of these classes. The fact that some ESBL producers were also resistant to ciprofloxacin and streptomycin/kanamycin suggests that besides β-lactam antibiotics, resistance to other antibiotic classes also exhibited co-selection.
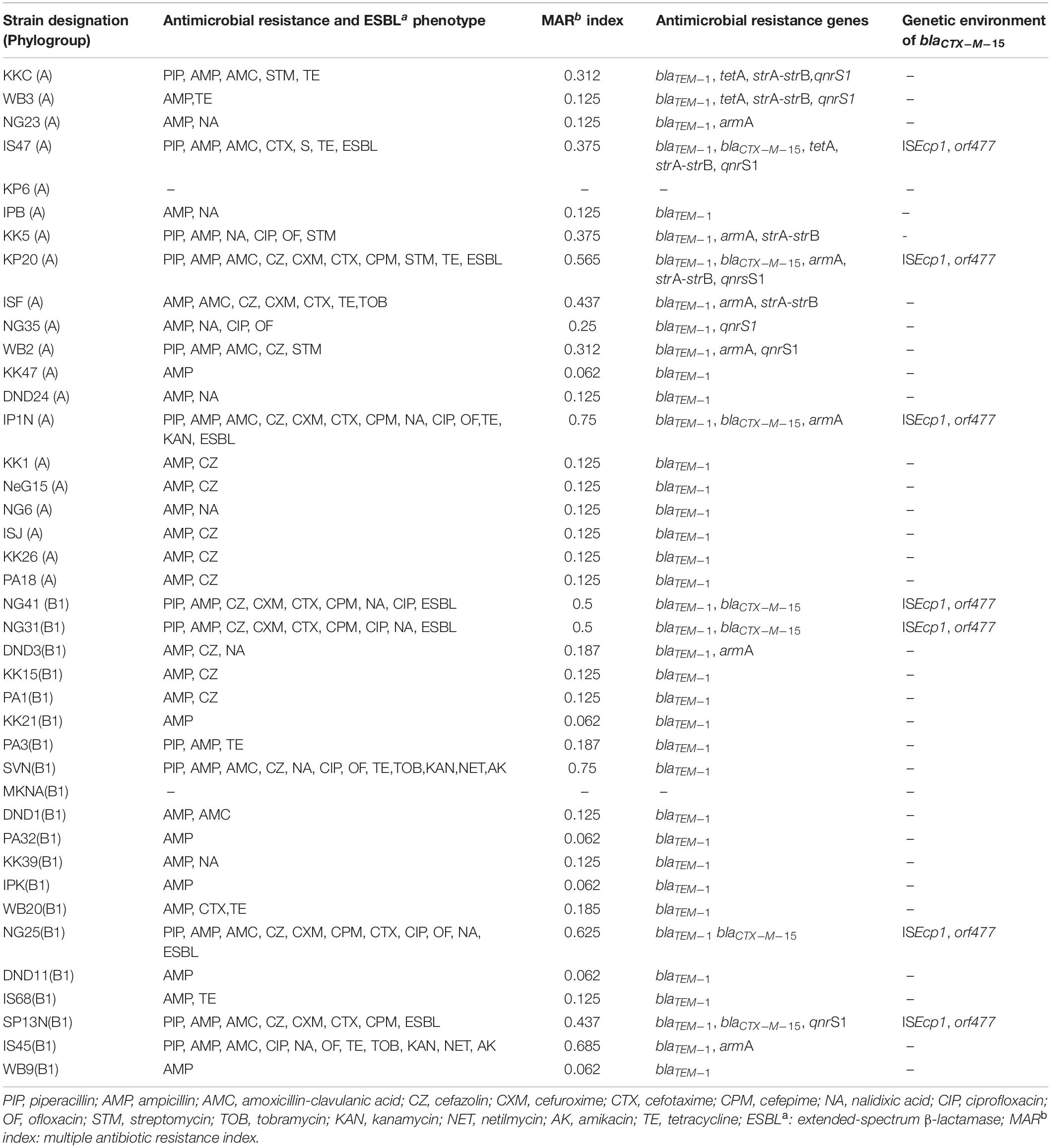
Table 2. Detailed information about the commensal strains of E. coli isolated from river Yamuna, antimicrobial resistance phenotypes, multiple antibiotic resistance index (MAR index) and antimicrobial resistance genes along with the genetic environment of blaCTX–M–15.
With regard to quinolone resistance, 35% (n = 14) strains were resistant to older quinolones like nalidixic acid, while 20% (n = 8) and 15% (n = 6) strains were resistant to newer quinolones like ciprofloxacin and ofloxacin, respectively. In this regard, our results are similar to those of other studies that reported lower ciprofloxacin and ofloxacin resistance in waterborne E. coli isolated from other parts of the world (Odonkor and Addo, 2018). Aminoglycoside resistance was observably less prevalent with 12.5% (n = 5) strains resistant to older aminoglycosides like streptomycin and less than 7% strains resistant to new aminoglycosides like kanamycin, tobramycin, netilmicin, and amikacin. An earlier study also reported that a low level of aminoglycoside resistance was present in E. coli strains isolated from aquatic environments of Kuala Lumpur, Malaysia (Hara et al., 2018). With regard to tetracycline, 27.5% (n = 11) of the strains exhibited resistance. The frequent use/misuse of ampicillin, streptomycin, and tetracycline due to frequent prescription, availability, and affordability might be a probable reason for higher bacterial resistance to these antibiotics (Shakya et al., 2013). Additionally, 57.5% (n = 23) E. coli strains were resistant to at least two antibiotic classes, and 20% (n = 8) of the strains were multidrug resistant (MDR), i.e., resistant to three or more antibiotic classes. MDR E. coli were defined as bacteria resistant to antibiotics belonging to three or more antimicrobial classes (Magiorakos et al., 2012). An analysis of MAR index revealed that the MAR indexes of 13 MDR E. coli strains were quite high (>0.2). Tambekar et al. (2006) reported that bacteria isolated from environments where several antibiotics are used usually show MAR index >0.2. The high MAR index of the E. coli strains observed in this study is not surprising because river Yamuna is highly contaminated with effluents originating from hospital and municipal wastewaters; discharge from livestock, poultry, and agriculture production plants; etc.
With regard to β-lactam resistance genes, blaTEM–1 was present in 95% of thestrains (n = 38) followed by blaCTX–M–15, which was present in 15% (n = 7) of the E. coli strains. The plasmid-encoded AmpC enzymes (CMY types) were not found in any strain. The presence of blaTEM–1 correlated well with ampicillin resistance in all the strains. Similarly, all the blaCTX–M–15-positive strains showed phenotypic production of ESBLs. Earlier studies had reported that blaTEM–1 was widely present in E. coli strains isolated from water bodies of India andother countries like Spain, Australia, France, China, and Poland (Lartigue et al., 2002; Tristram and Nichols, 2006; Garcia-Cobos et al., 2008; Ortega et al., 2012; Liu et al., 2014; Ojdana et al., 2014; Bajaj et al., 2015; Singh N. S. et al., 2018). CTX-M enzymes belong to the family of ESBLs and are the most widely disseminated ESBLs among Enterobacteriaceae all over the globe (Poirel et al., 2002). Among these, blaCTX–M–15 is the most widely globally disseminated CTX-M type, which was first reported from the Indian isolates in 2001 (Karim et al., 2001; Poirel et al., 2002). Later, several studies also reported the prevalence of blaCTX–M–15 in aquatic E. coli isolated from India (Bajaj et al., 2015; Singh N. S. et al., 2018; Kaushik et al., 2019). Previous studies have associated blaCTX–M–15 in aquatic E. coli with the pathogenic phylogroups B2 (especially those belonging to the genetic lineage ST131) and D (Nicolas-Chanoine et al., 2008; Coque et al., 2008). However, our study revealed that CTX-M-15 type ESBLs were present in the commensal phylogroups A and B1, also.
The linked strA-strB genes are the most widely prevalent streptomycin resistance genes in E. coli worldwide and encode for phosphotransferases (Poirel et al., 2018). However, strA-strB genes were present in only 12% (n = 5) of the waterborne E. coli. Of these, four strains exhibited phenotypic resistance to streptomycin, while one strain (ISF), despite harboring strA-strB, was phenotypically susceptible for streptomycin. The 16S rRNA methylases methylate certain amino acid residues of the 16S RNA, resulting in resistance to amikacin, tobramycin, gentamicin, and netilmicin (Griffey et al., 1999). Of the four types of plasmid-mediated 16S rRNA methylase investigated, only armA was found to be present in 17.5% (n = 7) of the strains. Earlier studies also reported that a low level of aminoglycoside resistance was present in E. coli strains isolated from aquatic environments in Kuala Lumpur, Malaysia (Hara et al., 2018). No correlation was observed in aminoglycoside resistance and presence of armA, except in the E. coli strain IS45, which also exhibited phenotypic resistance to amikacin, tobramycin, kanamycin, and netilmicin.
Of the several PMQR genes (qnrA, qnrB, qnrC, qnrD, qnrS, qep, and aac) tested, only qnrS was detected in the E. coli strains isolated from river Yamuna. The qnrS1 was detected as the predominant PMQR gene in about 17% of the aquatic strains (n = 7). Earlier studies have also reported that the qnrS type gene was the most frequently detected PMQR gene in E. coli isolated from environmental E. coli worldwide (Bonemann et al., 2006; Cattoir et al., 2008; Rodriguez-Mozaz et al., 2015; Varela et al., 2016; Hara et al., 2018). The presence of qnrS1 did not correlate with fluoroquinolone resistance, except in E. coli strain NG35. This suggests that the presence of the qnrS gene alone might not be a true indicator of fluoroquinolone resistance and the isolate despite that the presence of qnrS might exhibit phenotypic susceptibility for fluoroquinolones (Mahmud et al., 2020).
Of the three tetracycline resistance genes, tetM and tetW were absent and only tetA was present in 7.5% (n = 3) of the strains. Earlier studies have also indicated that tetracycline efflux-related genes like tetA, tetB, and tetC were more prevalent than ribosomal protection-related genes (like tetM and tetW) in waterborne E. coli (Zhang et al., 2015; Stange et al., 2016). The presence of tetA correlated well with phenotypic resistance because the three strains that harbored tetA also exhibited tetracycline resistance.
Of the 40 E. coli strains investigated in this study, only seven strains (15%) harbored the blaCTX–M–15 gene. The upstream region of the blaCTX–M–15 was analyzed in these seven strains by PCR amplification and sequencing. Gene sequencing revealed that the ISEcp1 was present in the upstream region of blaCTX–M–15 and orf477 was present in the downstream region of all the seven strains (Table 2). Several investigators have also reported the presence of orf477 in the downstream region of blaCTX–M–15 (Eckert et al., 2006; Dhanji et al., 2011; Wang et al., 2014; Ben Said et al., 2016). ISEcp1 is the most common and widely reported IS element (Dhanji et al., 2011; Liu et al., 2014; Upadhyay et al., 2015; Ben Said et al., 2016; Singh N. S. et al., 2018). In the present study, IS sequence ISEcp1 was found to be present at 48-bp upstream region of blaCTX–M–15. The 42- to 266-bp upstream region has been reported as the preferred insertion site of ISEcp1 for different blaCTX–M genes like CTX-M-1, CTX-M-2, and CTX-M-9. An analysis of the −35 and −10 promoter regions of the blaCTX–M–15 gene of the seven strains revealed that the −35 (TTGAAA) and −10 (TACAAT) regions were present within 3′ terminus end of ISEcp1 and 48 bp away from the blaCTX–M–15 start codon. The same organization was previously reported from E. coli strains isolated from different countries of the world (Saladin et al., 2002; Boyd et al., 2004; Canton and Coque, 2006; Lavollay et al., 2006; Dhanji et al., 2011; Liu et al., 2014). The presence of ISEcp1 along with blaCTX–M has been reported from E. coli strains isolated from different parts of the world, indicating that ISEcp1 might be evolutionary associated with blaCTX–M (Karim et al., 2001; Saladin et al., 2002; Dhanji et al., 2011; Liu et al., 2014; Wang et al., 2014). ISEcp1 can mobilize an adjacent gene as a part of transposition units of varying sizes (Zong et al., 2010). It has also been reported that ISEcp1 helps in improving the expression of blaCTX–M in enteric bacteria (Poirel et al., 2003). This is a matter of great concern because the subsequent transfer of ESBL genes from these commensal E. coli to pathogenic E. coli or other bacteria in aquatic water bodies might pose a serious health challenge (Figueira et al., 2011).
The class 1 integron gene intI1 was detected in 75% (n = 30) of the isolates (Table 3). None of the strains harbored class 2 and 3 integrase genes intI2 and intI3. Though some strains of E. coli reportedly harbored class 2 integrons, mostly class 1 integrons have been reported from E. coli isolated from India (Kaushik et al., 2018). Several studies have indicated that integrons of the class 3 were absent in E. coli isolated from water bodies across the globe (Laroche et al., 2009; Su et al., 2012; Pereira et al., 2013). Of the 30 intI1 harboring strains, gene cassette arrays were detected in only 10% (n = 4) of the intI1-positive E. coli strains. Three different types of gene cassette arrays of class 1 were present in the downstream region of intI1 whose size ranged from 1.7 to 2.8 kb (Table 2). Gene sequencing revealed that five gene cassettes of the dihydrofolate reductase (dfr) resistance gene family (dhfr12 and dfrA1), aminoglycoside (aad) resistance gene family (aadA1, aadA2, and aacA4), and chloramphenicol (CHL) resistance gene family (catB3) were present (Table 3). The prevalence of class 1 integron intI1 in Indian aquatic isolates was quite high (75%) and alarmingly more than that reported for E. coli isolated from global aquatic environments (Dolejska et al., 2009; Laroche et al., 2009; Pereira et al., 2013; Ghaderpour et al., 2015; Sidhu et al., 2017). E. coli isolates harboring class 1 integrons have been associated with a significantly higher probability for multidrug resistance than those devoid of class 1 integrons (Chen et al., 2011). Moreover, class 1 integron genes intI1 are accompanied by resistance genes for disinfectants and heavy metals (Partridge et al., 2001) and can also easily horizontally transfer between strains originating from different sources (Nagachinta and Chen, 2008; Zhang et al., 2009). Thus, due to high prevalence of class 1 integron gene intI1, the commensal strains of E. coli can become vehicles for widespread dissemination of antibiotic resistance to pathogenic E. coli and other waterborne bacterial pathogens.
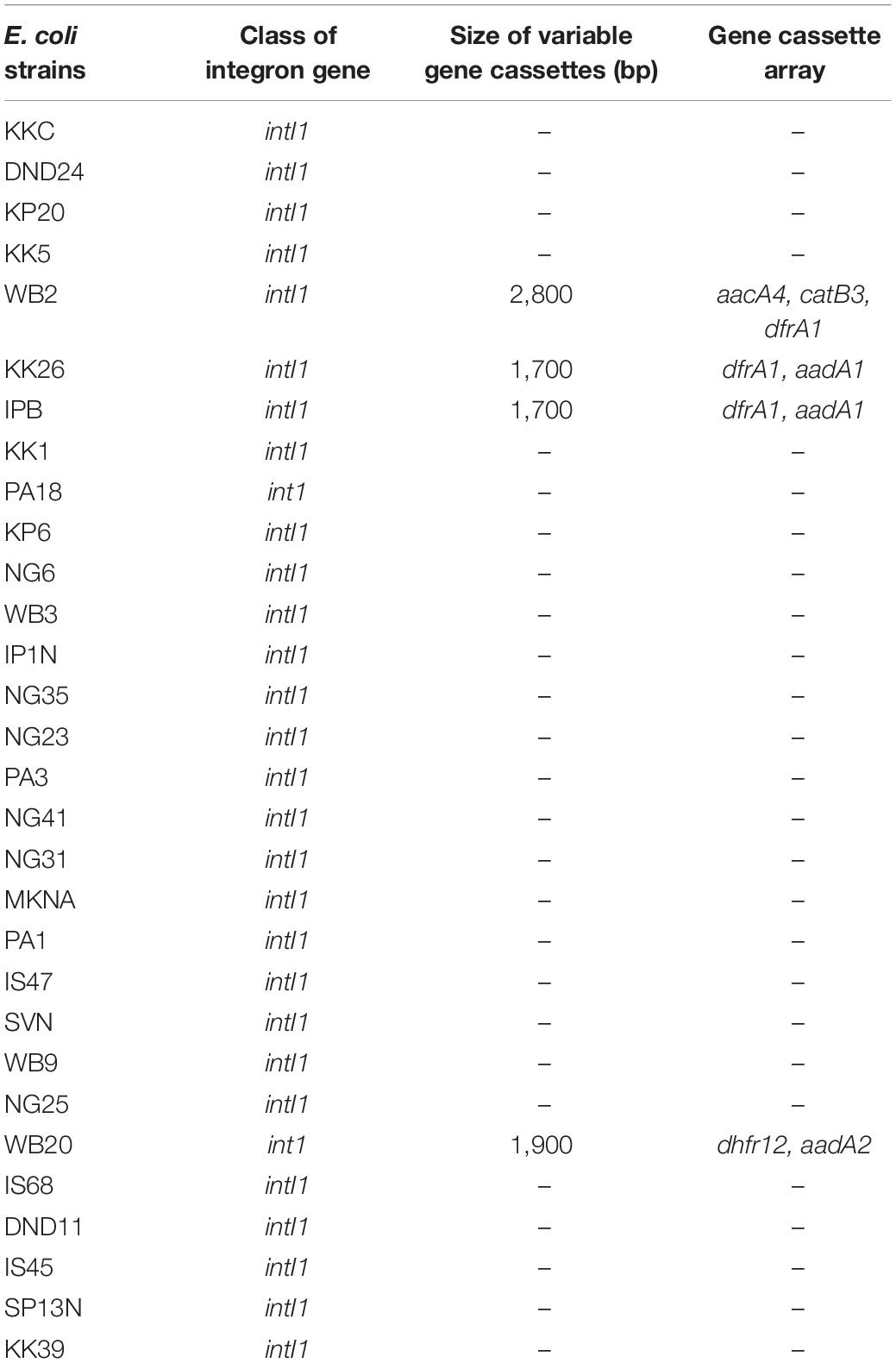
Table 3. Characteristics of integrons present in commensal E. coli strains isolated from river Yamuna, India.
Our results indicated a high prevalence of drug resistance in Escherichia coli strains of river Yamuna. With regard to plasmid-mediated AMR genes, blaTEM–1 was present in 95% strains followed by qnrS1 and armA (17% each), blaCTX–M–15 (15%), strA-strB (12%), and tetA (7%). Though most of the earlier studies have reported that blaCTX–M–15 in waterborne E. coli was mostly present in pathogenic phylogroup B2, our study revealed that CTX-M-15 type ESBLs were present in the commensal phylogroups A and B1, also. The genetic organization of blaCTX–M–15 was similar to that reported for E. coli globally, and ISEcp1 was present in the upstream region of blaCTX–M–15. Though integrons of classes 2 and 3 were absent, class 1 integron gene intI was detected in 75% of the isolates, which indicates its high prevalence in the E. coli isolates of the river Yamuna than that reported, globally. The presence of MDR phenotypes, plasmid-mediated AMR genes, and class 1 integron gene intI1 in E. coli is a serious public health risk, because these commensal strains can become potent vehicles for widespread dissemination of AMR determinants to pathogenic E. coli and other waterborne pathogens. Thus, our study suggests an urgent need for regular surveillance and management of natural water bodies to curtail the spread of antibiotic resistance in microorganisms.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
NSS, NS, MK, and JV analyzed the data. NSS and NS wrote the manuscript. JV conceptualized and supervised. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
NS was supported by the Council of Scientific and Industrial Research (CSIR) Senior Research Associateship (Scientists’ Pool Scheme) File no. 13(9089-A)/2019-POOL. NSS was thankful to the Indian Council of Medical Research (ICMR) for the Senior Research Fellowship (No. 80/919/2014-ECD-I) and Research Associateship (ISRM/12(33)/2019).
Abdelgader, S. A., Shi, D., Chen, M., Zhang, L., Hejair, H. M. A., Muhammad, U., et al. (2018). Antibiotics resistance genes screening and comparative genomics analysis of commensal Escherichia coli isolated from poultry farms between China and Sudan. BioMed. Res. Int. 2018, 1–9. doi: 10.1155/2018/5327450
Aminov, R. I., Chee-Sanford, J. C., Garrides, N., Teferedegne, B., Krapac, I. J., White, B. A., et al. (2002). Development, validation and application of PCR primers for detection of tetracycline efflux genes of gram negative bacteria. Appl. Environ. Microbiol. 68, 1786–1793. doi: 10.1128/aem.68.4.1786-1793.2002
Aanensen, D. M., and Spratt, B. G. (2005). The multilocus sequence typing network: mlst.net. Nucleic Acids Res. 33, W728–W733. doi: 10.1093/nar/gki415
Bajaj, P., Kanaujia, P. K., Singh, N. S., Sharma, S., Kumar, S., and Virdi, J. S. (2016). Quinolone co-resistance in ESBL- or AmpC-producing Escherichia coli from an Indian urban aquatic environment and their public health implications. Environ. Sci. Pollut. Res. Int. 23, 1954–1959. doi: 10.1007/s11356-015-5609-x
Bajaj, P., Singh, N. S., Kanaujia, P. K., and Virdi, J. S. (2015). Distribution and molecular characterization of genes encoding CTX-M and AmpC beta-lactamases in Escherichia coli isolated from an Indian urban aquatic environment. Sci. Total. Environ. 505, 350–356. doi: 10.1016/j.scitotenv.2014.09.084
Baquero, F., Martinez, J. L., and Canton, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265. doi: 10.1016/j.copbio.2008.05.006
Bass, L., Liebert, C. A., Lee, M. D., Summers, A. O., White, D. G., Thayer, S. G., et al. (1999). Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob. Agents. Chemother. 43, 2925–2929. doi: 10.1128/aac.43.12.2925
Ben Said, L., Jouini, A., Alonso, C. A., Klibi, N., Dziri, R., Boudabous, A., et al. (2016). Characteristics of extended-spectrum beta-lactamase (ESBL)- and pAmpC beta-lactamase-producing Enterobacteriaceae of water samples in Tunisia. Sci. Total. Environ. 550, 1103–1109. doi: 10.1016/j.scitotenv.2016.01.042
Bingen, E., Picard, B., Brahimi, N., Mathy, S., Desjardins, P., Elion, J., et al. (1998). Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177, 642–650. doi: 10.1086/514217
Bonemann, G., Stiens, M., Puhler, A., and Schluter, A. (2006). Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob. Agents. Chemother. 50, 3075–3080. doi: 10.1128/aac.00378-06
Boyd, D. A., Tyler, S., Christianson, S., McGeer, A., Muller, M. P., Willey, B. M., et al. (2004). Complete nucleotide sequence of a 92-kilobase plasmid harbouring the CTX-M-15 extended-spectrum b-lactamase involved in an outbreak in long-term-care facilities in Toronto. Canada. Antimicrob. Agents Chemother. 48, 3758–3764. doi: 10.1128/aac.48.10.3758-3764.2004
Boyd, E. F., and Hartl, D. L. (1998). Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 180, 1159–1165. doi: 10.1128/jb.180.5.1159-1165.1998
Canton, R., and Coque, T. M. (2006). The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475.
Cattoir, V., Poirel, L., and Nordmann, P. (2008). Plasmid-mediated quinolone resistance pump QepA2 in an Escherichia coli isolate from France. Antimicrob. Agents Chemother. 52, 3801–3804.
Cavaco, L. M., Hasman, H., Xia, S., and Aarestrup, F. M. (2009). qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 53, 603–608. doi: 10.1128/AAC.00997-08
Chen, B., Zheng, W., Yu, Y., Huang, W., Zheng, S., Zhang, Y., et al. (2011). Class 1 integrons, selected virulence genes, and antibiotic resistance in Escherichia coli isolates from the Minjiang River, Fujian Province, China. Appl. Environ. Microbiol. 77, 148–155. doi: 10.1128/AEM.01676-10
Chen, X., Zhang, W., Pan, W., Yin, J., Pan, Z., Gao, S., et al. (2012). Prevalence of qnr, aac(6’)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob. Agents Chemother. 56, 3423–3427. doi: 10.1128/aac.06191-11
Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000
Clinical and Laboratory Standards Institute [CLSI] (2018). Performance Standards for Antimicrobial Susceptibility Testing; Approved Standard, 28th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Coque, T. M., Novais, A., Carattoli, A., Poirel, L., Pitout, J., Peixe, L., et al. (2008). Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 14, 195–200.
Dhanji, H., Patel, R., Wall, R., Doumith, M., Patel, B., Hope, R., et al. (2011). Variation in the genetic environments of bla(CTX-M-15) in Escherichia coli from the faeces of travellers returning to the United Kingdom. J. Antimicrob. Chemother. 66, 1005–1012. doi: 10.1093/jac/dkr041
Dolejska, M., Bierosova, B., Kohoutova, L., Literak, I., and Cizek, A. (2009). Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 106, 1941–1950. doi: 10.1111/j.1365-2672.2009.04155.x
Dyar, O. J., Hoa, N. Q., Trung, N. V., Phuc, H. D., Larsson, M., Chuc, N. T. K., et al. (2012). High prevalence of antibiotic resistance in commensal Escherichia coli among children in rural Vietnam. BMC Infect. Dis. 12:92.
Eckert, C., Gautier, V., and Arlet, G. (2006). DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57, 14–23. doi: 10.1093/jac/dki398
Figueira, V., Serra, E., and Manaia, C. M. (2011). Differential patterns of antimicrobial resistance in population subsets of Escherichia coli isolated from waste- and surface waters. Sci. Total Environ. 409, 1017–1023. doi: 10.1016/j.scitotenv.2010.12.011
Garcia-Cobos, S., Campos, J., Cercenado, E., Roman, F., Lazaro, E., Perez-Vazquez, M., et al. (2008). Antibiotic resistance in Haemophilus influenzae decreased, except for beta-lactamase-negative amoxicillin-resistant isolates, in parallel with community antibiotic consumption in Spain from 1997 to 2007. Antimicrob. Agents Chemother. 52, 2760–2766. doi: 10.1128/aac.01674-07
Ghaderpour, A., Ho, W. S., Chew, L. L., Bong, C. W., Chong, V. C., Thong, K. L., et al. (2015). Diverse and abundant multi-drug resistant E. coli in Matang mangrove estuaries, Malaysia. Front. Microbiol. 6:977.
Gillings, M. (2014). Integrons: past, present, and future. Microbiol. Mol. Biol. Rev. 78, 257–277. doi: 10.1128/mmbr.00056-13
Goldstein, C., Lee, M. D., Sanchez, S., Hudson, C., Phillips, B., Register, B., et al. (2001). Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45, 723–726. doi: 10.1128/aac.45.3.723-726.2001
Griffey, R. H., Hofstadler, S. A., Sannes-Lowery, K. A., Ecker, D. J., and Crooke, S. T. (1999). Determinants of aminoglycoside-binding specificity for rRNA by using mass spectrometry. Proc. Natl. Acad. Sci. USA 96, 10129–10133. doi: 10.1073/pnas.96.18.10129
Guo, X., Xia, R., Han, N., and Xu, H. (2011). Genetic diversity analyses of class 1 integrons and their associated antimicrobial resistance genes in Enterobacteriaceae strains recovered from aquatic habitats in China. Lett. Appl. Microbiol. 52, 667–675. doi: 10.1111/j.1472-765x.2011.03059.x
Hara, H., Yusaimi, Y. A., Zulkeflle, S. N. M., Sugiura, N., Iwamoto, K., Goto, M., et al. (2018). Molecular characterization of multi-drug resistant Escherichia coli isolates from tropical environments in Southeast Asia. J. Gen. Appl. Microbiol. 64, 284–292. doi: 10.2323/jgam.2018.02.003
Hasegawa, H., Suzuki, E., and Maeda, S. (2018). Horizontal plasmid transfer by transformation in Escherichia coli: environmental factors and possible mechanisms. Front. Microbiol. 9:2365.
Herzer, P. J., Inouye, S., Inouye, M., and Whittam, T. S. (1990). Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172, 6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990
Ho, P. L., Wong, R. C., Lo, S. W., Chow, K. H., Wong, S. S., and Que, T. L. (2010). Genetic identity of aminoglycoside-resistance genes in Escherichia coli isolates from human and animal sources. J. Med. Microbiol. 59, 702–707. doi: 10.1099/jmm.0.015032-0
Johnson, J. R. (1991). Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4, 80–128. doi: 10.1128/cmr.4.1.80-128.1991
Karim, A., Poirel, L., Nagarajan, S., and Nordmann, P. (2001). Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201, 237–241. doi: 10.1111/j.1574-6968.2001.tb10762.x
Kaushik, M., Kumar, S., Kapoor, R. K., and Gulati, P. (2019). Integrons and antibiotic resistance genes in water-borne pathogens: threat detection and risk assessment. J. Med. Microbiol. 68, 679–692. doi: 10.1099/jmm.0.000972
Kaushik, M., Kumar, S., Kapoor, R. K., Virdi, J. S., and Gulati, P. (2018). Integrons in Enterobacteriaceae: diversity, distribution and epidemiology. Int. J. Antimicrob. Agents 51, 167–176. doi: 10.1016/j.ijantimicag.2017.10.004
Koczura, R., Mokracka, J., Barczak, A., Krysiak, N., and Kaznowski, A. (2013). Association between the presence of class 1 integrons, virulence genes, and phylogenetic groups of Escherichia coli isolates from river water. Microb. Ecol. 65, 84–90. doi: 10.1007/s00248-012-0101-3
Kraft, C. A., Timbury, M. C., and Platt, D. J. (1986). Distribution and genetic location of Tn7 in trimethoprim-resistant Escherichia coli. J. Med. Microbiol. 22, 125–131. doi: 10.1099/00222615-22-2-125
Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46, 165–170. doi: 10.1128/aem.46.1.165-170.1983
Kümmerer, K. (2009). The presence of pharmaceuticals in the environment due to human use–present knowledge and future challenges. J. Environ. Manag. 90, 2354–2366. doi: 10.1016/j.jenvman.2009.01.023
Laroche, E., Pawlak, B., Berthe, T., Skurnik, D., and Petit, F. (2009). Occurrence of antibiotic resistance and class 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France). FEMS Microbiol. Ecol. 68, 118–130. doi: 10.1111/j.1574-6941.2009.00655.x
Lartigue, M. F., Leflon-Guibout, V., Poirel, L., Nordmann, P., and Nicolas-Chanoine, M. H. (2002). Promoters P3, Pa/Pb, P4, and P5 upstream from bla(TEM) genes and their relationship to beta-lactam resistance. Antimicrob. Agents Chemother. 46, 4035–4037. doi: 10.1128/aac.46.12.4035-4037.2002
Lavollay, M., Mamlouk, K., Frank, T., Akpabie, A., Burghoffer, B., Ben Redjeb, S., et al. (2006). Clonal dissemination of a CTX-M-15 beta-lactamase-producing Escherichia coli strain in the Paris area, Tunis, and Bangui. Antimicrob. Agents Chemother. 50, 2433–2438. doi: 10.1128/aac.00150-06
Lecointre, G., Rachdi, L., Darlu, P., and Denamur, E. (1998). Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol. 15, 1685–1695. doi: 10.1093/oxfordjournals.molbev.a025895
Liebana, E., Carattoli, A., Coque, T. M., Hasman, H., Magiorakos, A. P., Mevius, D., et al. (2013). Public health risks of enterobacterial isolates producing extended-spectrum beta-lactamases or AmpC beta-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin. Infect. Dis. 56, 1030–1037. doi: 10.1093/cid/cis1043
Liu, L., Wang, X., An, S., Zhang, X., Chen, L., Li, Y., et al. (2014). Genetic environment of beta-lactamase genes of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates from patients with lower respiratory tract infection in China. Chin. Med. J. 127, 2445–2450.
Lugsomya, K., Yindee, J., Niyomtham, W., Tribuddharat, C., Tummaruk, P., Hampson, D. J., et al. (2018). Antimicrobial resistance in commensal Escherichia coli isolated from pigs and pork derived from farms either routinely using or not using in-feed antimicrobials. Mary Ann Liebert 24, 1054–1066. doi: 10.1089/mdr.2018.0154
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., and Giske, C. G. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mahmud, Z. H., Kabir, M. H., Ali, S., Moniruzzaman, M., Imran, K. M., Nafiz, T. N., et al. (2020). Extended-spectrum Beta-lactamase-producing Escherichia coli in drinking water samples Ffrom a forcibly displaced, densely populated community setting in Bangladesh. Front. Public Health 8:228.
Méric, G., Kemsley, E. K., Falush, D., Saggers, E. J., and Lucchini, S. (2013). Phylogenetic distribution of traits associated with plant colonization in Escherichia coli. Environ. Microbiol. 15, 487–501. doi: 10.1111/j.1462-2920.2012.02852.x
Nagachinta, S., and Chen, J. (2008). Transfer of class 1 integron-mediated antibiotic resistance genes from Shiga toxin-producing Escherichia coli to a susceptible E. coli K-12 strain in storm water and bovine feces. Appl. Environ. Microbiol. 74, 5063–5067. doi: 10.1128/aem.00517-08
Nandi, S., Maurer, J. J., Hofacre, C., and Summers, A. O. (2004). Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acd. Sci. 101, 7118–7122. doi: 10.1073/pnas.0306466101
Nicolas-Chanoine, M. H., Blanco, J., Leflon-Guibout, V., Demarty, R., Alonso, M. P., Canica, M. M., et al. (2008). Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61, 273–281. doi: 10.1093/jac/dkm464
Odonkor, S. T., and Addo, K. K. (2018). Prevalence of multidrug-resistant Escherichia coli isolated from drinking water sources. Int. J. Microbiol. 2018:7204013.
Ojdana, D., Sacha, P. B., and Wieczorek, P. (2014). The occurrence of blaCTX-M, blaSHV, and blaTEM genes in extended-spectrum β-lactamase-positive strains of Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis in Poland. Int. J. Antibiot. 2014:935842.
Ortega, A., Oteo, J., Aranzamendi-Zaldumbide, M., Bartolome, R. M., Bou, G., Cercenado, E., et al. (2012). Spanish multicenter study of the epidemiology and mechanisms of amoxicillin-clavulanate resistance in Escherichia coli. Antimicrob. Agents Chemother. 56, 3576–3581. doi: 10.1128/aac.06393-11
Partridge, S. R., Brown, H. J., Stokes, H., and Hall, R. M. (2001). Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45, 1263–1270. doi: 10.1128/aac.45.4.1263-1270.2001
Pereira, A., Santos, A., Tacao, M., Alves, A., Henriques, I., and Correia, A. (2013). Genetic diversity and antimicrobial resistance of Escherichia coli from Tagus estuary (Portugal). Sci. Total Environ. 1, 461–462.
Pérez-Pérez, F. J., and Hanson, N. D. (2002). Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40, 2153–2162. doi: 10.1128/jcm.40.6.2153-2162.2002
Picard, B., Garcia, J. S., Gouriou, S., Duriez, P., Brahimi, N., Bingen, E., et al. (1999). The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67, 546–553. doi: 10.1128/iai.67.2.546-553.1999
Poirel, L., Aires-de-Sousa, M., Kudyba, P., Kieffer, N., and Nordmann, P. (2018). Screening and characterization of multidrug-resistant gram-negative bacteria from a remote African area, São Tomé and Príncipe. Antimicrob. Agents Chemother. 62:e01021-18.
Poirel, L., Decousser, J. W., and Nordmann, P. (2003). Insertion sequence ISEcp1B is involved in expression and mobilization of a bla(CTX-M) beta-lactamase gene. Antimicrob. Agents Chemother. 47, 2938–2945. doi: 10.1128/aac.47.9.2938-2945.2003
Poirel, L., Kampfer, P., and Nordmann, P. (2002). Chromosome-encoded Ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 46, 4038–4040. doi: 10.1128/aac.46.12.4038-4040.2002
Purohit, M. R., Lindahl, L. F., Diwan, V., Marrone, G., and Lundborg, C. S. (2019). High levels of drug resistance in commensal E. coli in a cohort of children from rural central India. Sci. Rep. 9:6682.
Ram, S., Vajpayee, P., and Shanker, R. (2008). Contamination of potable water distribution systems by multiantimicrobial-resistant enterohemorrhagic Escherichia coli. Environ. Health Perspect. 116, 448–452. doi: 10.1289/ehp.10809
Rodríguez-Baño, J., Navarro, M. D., Romero, L., Martinez-Martinez, L., Muniain, M. A., Perea, E. J., et al. (2004). Epidemiology and clinical features of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in nonhospitalized patients. J. Clin. Microbiol. 42, 1089–1094. doi: 10.1128/jcm.42.3.1089-1094.2004
Rodriguez-Mozaz, S., Chamorro, S., Marti, E., Huerta, B., Gros, M., Sanchez-Melsio, A., et al. (2015). Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 69, 234–242. doi: 10.1016/j.watres.2014.11.021
Saladin, M., Cao, V. T., Lambert, T., Donay, J. L., Herrmann, J. L., Ould-Hocine, Z., et al. (2002). Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209, 161–168. doi: 10.1016/s0378-1097(02)00484-6
Shakya, P., Barrett, P., Diwan, V., Marothi, Y., Shah, H., Chhari, N., et al. (2013). Antibiotic resistance among Escherichia coli isolates from stool samples of children aged 3 to 14 years from Ujjain, India. BMC Infect. Dis. 13:477.
Sidhu, J. P. S., Jagals, P., Smith, A., and Toze, S. (2017). Comparative prevalence of Escherichia coli carrying virulence genes and class 1 and 2 integrons in sub-tropical and cool temperate freshwater. Environ. Sci. Pollut. Res. Int. 24, 18263–18272. doi: 10.1007/s11356-017-9497-0
Singh, A. K., Das, S., Singh, S., Gajamer, V. R., Pradhan, N., and Lepcha, Y. D. (2018). Prevalence of antibiotic resistance in commensal Escherichia coli among the children in rural hill communities of Northeast India. PLoS One 13:e0199179. doi: 10.1371/journal.pone.0199179
Singh, N. S., Singhal, N., and Virdi, J. S. (2018). Genetic environment of blaTEM-1, blaCTX-M-15, blaCMY-42 and characterization of integrons of Escherichia coli isolated from an Indian urban aquatic environment. Front. Microbiol. 9:382.
Singhal, N., Maurya, A. K., Mohanty, S., Kumar, M., and Virdi, J. S. (2019). Evaluation of bile salt hydrolases, cholesterol-lowering capabilities, and probiotic potential of enterococcus faecium isolated from rhizosphere. Front. Microbiol. 10:1567.
Stange, C., Sidhu, J. P. S., Tiehm, A., and Toze, S. (2016). Antibiotic resistance and virulence genes in coliform water isolates. Int. J. Hyg. Environ. Health. 219, 823–831. doi: 10.1016/j.ijheh.2016.07.015
Su, H. C., Ying, G. G., Tao, R., Zhang, R. Q., Zhao, J. L., and Liu, Y. S. (2012). Class 1 and 2 integrons, sul resistance genes and antibiotic resistance in Escherichia coli isolated from Dongjiang river, South China. Environ. Pollut. 169, 42–49. doi: 10.1016/j.envpol.2012.05.007
Sunde, M., and Norstrom, M. (2005). The genetic background for streptomycin resistance in Escherichia coli influences the distribution of MICs. J. Antimicrob. Chemother. 56, 87–90. doi: 10.1093/jac/dki150
Tacão, M., Correia, A., and Henriques, I. (2012). Resistance to broad-spectrum antibiotics in aquatic systems: anthropogenic activities modulate the dissemination of bla(CTX-M)-like genes. Appl. Environ. Microbiol. 78, 4134–4140. doi: 10.1128/aem.00359-12
Tambekar, D. H., Dhanorkar, D. V., Gulhane, S. R., Khandelwal, V. K., and Dudhane, M. N. (2006). Antibacterial susceptibility of some urinary tract pathogens to commonly used antibiotics. Afr. J. Biotechnol. 5:17.
Tristram, S. G., and Nichols, S. (2006). A multiplex PCR for beta-lactamase genes of Haemophilus influenzae and description of a new blaTEM promoter variant. J. Antimicrob. Chemother. 58, 183–185. doi: 10.1093/jac/dkl150
Upadhyay, S., Hussain, A., Mishra, S., Maurya, A. P., Bhattacharjee, A., and Joshi, S. R. (2015). Genetic environment of plasmid mediated CTX-M-15 extended spectrum Beta-Lactamases from clinical and food borne bacteria in North-Eastern India. PLoS One 10:e0138056. doi: 10.1371/journal.pone.0138056
Varela, A. R., Nunes, O. C., and Manaia, C. M. (2016). Quinolone resistant Aeromonas spp. as carriers and potential tracers of acquired antibiotic resistance in hospital and municipal wastewater. Sci. Total Environ. 542, 665–671. doi: 10.1016/j.scitotenv.2015.10.124
Wang, J., Stephan, R., Zurfluh, K., Hachler, H., and Fanning, S. (2014). Characterization of the genetic environment of blaESBL genes, integrons and toxin-antitoxin systems identified on large transferrable plasmids in multi-drug resistant Escherichia coli. Front. Microbiol. 5:716.
Wang, M., Guo, Q., Xu, X., Wang, X., Ye, X., Wu, S., et al. (2009). New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 53, 1892–1897. doi: 10.1128/AAC.01400-08
White, P., McIver, C. J., Deng, Y. M., and Rawlinson, W. D. (2000). Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182, 265–269. doi: 10.1111/j.1574-6968.2000.tb08906.x
White, P. A., McIver, C. J., and Rawlinson, W. D. (2001). Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45, 2658–2661. doi: 10.1128/aac.45.9.2658-2661.2001
Yamane, K. J., Wachino, S., Suzuki, H., Kato, K., Shibayama, K., Kimura, K., et al. (2007). 16S rRNA methylase-producing, gram-negative pathogens, Japan. Emerg. Infect. Dis. 13, 642–646. doi: 10.3201/eid1304.060501
Zhang, S. H., Lv, X., Han, B., Gu, X., Wang, P. F., Wang, C., et al. (2015). Prevalence of antibiotic resistance genes in antibiotic-resistant Escherichia coli isolates in surface water of Taihu Lake Basin, China. Environ. Sci. Pollut. Res. Int. 22, 11412–11421. doi: 10.1007/s11356-015-4371-4
Zhang, X. Y., Ding, L. J., and Yue, J. (2009). Occurrence and characteristics of class 1 and class 2 integrons in resistant Escherichia coli isolates from animals and farm workers in northeastern China. Microb. Drug Resist. 15, 323–328. doi: 10.1089/mdr.2009.0020
Keywords: commensal E. coli, plasmid-mediated antimicrobial resistance genes, integrons, multidrug resistance, horizontal transfer of genes
Citation: Singh NS, Singhal N, Kumar M and Virdi JS (2021) High Prevalence of Drug Resistance and Class 1 Integrons in Escherichia coli Isolated From River Yamuna, India: A Serious Public Health Risk. Front. Microbiol. 12:621564. doi: 10.3389/fmicb.2021.621564
Received: 27 October 2020; Accepted: 05 January 2021;
Published: 09 February 2021.
Edited by:
Lorena Rodriguez-Rubio, University of Barcelona, SpainReviewed by:
Mohamed Salah Abbassi, Tunis El Manar University, TunisiaCopyright © 2021 Singh, Singhal, Kumar and Virdi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Neelja Singhal, bmVlbGphMzBAZ21haWwuY29t; Jugsharan Singh Virdi, dmlyZGlfZHVzY0ByZWRpZmZtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.