
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 January 2021
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.621519
This article is part of the Research Topic Plant Pathogenic Fungi: Molecular Systematics, Genomics and Evolution View all 16 articles
A correction has been applied to this article in:
Corrigendum: The GTPase-Activating Protein FgGyp1 Is Important for Vegetative Growth, Conidiation, and Virulence and Negatively Regulates DON Biosynthesis in Fusarium graminearum
 Qiaojia Zheng1†
Qiaojia Zheng1† Zhi Yu1,2†
Zhi Yu1,2† Yanping Yuan2†
Yanping Yuan2† Danli Sun1
Danli Sun1 Yakubu Saddeeq Abubakar3
Yakubu Saddeeq Abubakar3 Jie Zhou2
Jie Zhou2 Zonghua Wang1,2
Zonghua Wang1,2 Huawei Zheng1*
Huawei Zheng1*Ypt1 is a small Rab GTPase in yeast, Gyp1 functions at the Golgi as a negative regulator of Ypt1. Gyp1 homologs are conserved in filamentous fungi. However, the roles of Gyp1 in phytopathogenic fungi are still unclear. Herein, we investigated the functions of FgGyp1 in the wheat pathogen Fusarium graminearum by live-cell imaging, genetic, and pathological analyses. Targeted gene replacement method was used to delete FgGYP1 in F. graminearum. Phenotypic analyses showed that FgGyp1 is critically important not only for the vegetative growth of F. graminearum but also its conidiation. The mutant’s vegetative growth was significantly reduced by 70% compared to the wild type PH-1. The virulence of FgGYP1 deletion mutant was significantly decreased when compared with the wild type PH-1. We further found that FgGyp1 negatively regulates DON production of the fungus. Live-cell imaging clearly demonstrated that FgGyp1 mainly localizes to the Golgi apparatus. Moreover, the TBC domain, C-terminal, and N-terminal regions of FgGyp1 are found to be indispensable for its biological functions and normal localization. The Arg357 residue of FgGyp1 is essential for its functions but dispensable for the normal localization of the protein, while the Arg284 residue is not required for both the functions and normal localization of the protein. Furthermore, we showed that FgGyp1 essentially hydrolyzes the GTP-bound FgRab1 (activated form) to its corresponding GDP-bound (inactive) form in vitro, suggesting that FgGyp1 is a GTPase-activating protein (GAP) for FgRab1. Finally, FgGyp1 was found to be important for FgSnc1-mediated fusion of secretory vesicles from the Golgi with the plasma membrane in F. graminearum. Put together, these data demonstrate that FgGyp1 functions as a GAP for FgRab1 and is important for vegetative growth, conidiation and virulence, and negatively regulates DON biosynthesis in F. graminearum.
Fusarium graminearum causes the Fusarium head blight (FHB) of wheat, barley and other cereals, globally (McMullen et al., 1997; Bushnell et al., 2003). FHB epidemics have been prevalent in China since 2010 and caused enormous yield losses so far (Chen et al., 2019). Furthermore, F. graminearum infestation has serious negative impacts on human and animal health as the pathogen renders the crops contaminated with mycotoxins, such as zearalenone (ZEA) and trichothecenes, which when consumed cause serious food poisoning (Bushnell et al., 2003; Chen et al., 2019). In recent years, a number of studies demonstrated that vesicles trafficking is indispensable for the fungal normal development, pathogenicity, and DON production (Zheng et al., 2015, 2018b,c; Zhang et al., 2016; Li et al., 2017, 2018, 2019; Xie et al., 2019; Adnan et al., 2020).
Small Rab GTPases regulate vesicle trafficking processes in eukaryotic cells, including endocytosis and exocytosis (Li and Marlin, 2015). GTPase-activating proteins (GAPs) are negative regulators of Rab GTPases, they do so by inactivating Rab GTPases via promoting their GTPase activity (Mizuno-Yamasaki et al., 2012). As regulators of Rab-dependent pathways, RabGAPs regulate many diseases including bacterial and viral infections, development of cancer, and onset of obesity in mammalian cells (Mizuno-Yamasaki et al., 2012). So far, most identified RabGAPs contain Tre2-Bub2-Cdc16 (TBC) domains (Fukuda, 2011). Gyp1 was previously shown to regulate the trafficking of vesicles from the ER (endoplasmic reticulum) to the plasma membrane (Huang et al., 2014). Gyp1 interacts in vitro with Ypt1, Ypt7, Ypt51, and Sec4 (Du et al., 1998; Albert and Gallwitz, 1999). As a GAP of the small GTPase Ypt1, Gyp1 negatively regulates Ypt1 on the Golgi membrane (Du and Novick, 2001). The protein is also a GAP for Sec4p, a protein involved in the secretion pathway (Du et al., 1998). It has an arginine residue in its motif B sequence and such residue is essential for its catalytic function (Albert and Gallwitz, 1999). Furthermore, the crystal structure of Gyp1 GAP domain reveals important insights into the mechanism of its interaction with Ypt/Rab proteins, showing that in the GAP reaction Gyp1 uses an arginine finger similar the interactions of Cdc42-GAP and Ras-GAP (Rak et al., 2000).
In budding yeast, Gyp1 is dispensable for cell viability when grown on nutrient-rich media at different temperatures (Du et al., 1998) while Δgyp1 mutant shows defect in growth on a medium at 37°C (Du and Novick, 2001). In addition, the gyp1 deletion mutant showed minimal medium-specific growth defect (Winzeler et al., 1999). A study in Candida albicans demonstrates that Gyp1 is required for hyphal growth and Golgi polarization, and it localizes to Golgi apparatus (Huang et al., 2014). However, the functions of Gyp1 in other filamentous fungi particularly in F. graminearum remain obscured.
In our previous study, we systematically characterized all the 11 putative Rab proteins in F. graminearum and found that they are essentially required for growth and pathogenicity of F. graminearum (Zheng et al., 2015). FgMon1, FgSec2A, and FgVps9 proteins are GEFs (Guanine nucleotide exchange factors) for FgRab7, FgRab8, and FgRab5, respectively (Li et al., 2015; Zheng et al., 2018a; Yang et al., 2020). They are all required for pathogenicity and development of F. graminearum. Recently, we demonstrated that FgMsb3 protein is a GAP for FgRab8 and regulates hyphal tip expansion, polarized trafficking and pathogenicity in F. graminearum (Zheng et al., 2020).
FgRab1 is critically important for F. graminearum normal development (Zheng et al., 2015), but the upstream regulators of FgRab1 are still unknown. The fact that Gyp1 serves as a GAP of Ypt1/Rab1 in yeast prompted us to dissect the relationship between FgGyp1 and FgRab1, as well as the function(s) of FgGyp1 in F. graminearum. Our in vitro analyses indicate that FgGyp1 is a GAP of FgRab1 and is important for growth, conidiation, virulence, and DON biosynthesis of F. graminearum.
The Supplementary Table 1 presents all the strains used in this study. The culture media used in the study include complete medium (CM) consisting (all in w/v) 0.6% casein acid hydrolysate, 0.6% yeast extract, 1% sucrose, 2% agar (for solid media) or starch yeast medium (SYM) consisting (all in w/v) 1% starch, 0.2% yeast extract, 0.3% sucrose, and 2% agar (Zheng et al., 2015). The incubations were done at 28 °C for 3 days. Previous protocols (Cappellini and Peterson, 1965; Zheng et al., 2015; Fan et al., 2020) were used to assay for conidiation and sexual reproduction.
Previously reported protocols (Hou et al., 2002) were used for the preparation of F. graminearum protoplasts and subsequent transformations. The split-marker approach was used to generate gene replacement constructs for the FgGYP1 (FGSG_17336) deletion mutants. The Supplementary Table 2 presents all the primers used for this purpose. Two gene deletion mutants (ΔFggyp1) were successfully generated and further confirmed by Southern blotting. To reintroduce this gene into the mutant (complementation), a vector harboring FgGyp1-GFP fusion protein was generated by amplifying the coding sequence and native promoter of FgGyp1 using FgGyp1CF and FgGyp1CR primer pair (Supplementary Table 2). A Cloning Kit (Vazyme Biotech Co., Ltd., China) was then used to insert the amplicon into a pKNTG2 vector and the product was sequenced for verification. The vector (pFgGyp1-GFP) was then transformed into the protoplast of ΔFggyp1 mutant and a complemented strain was successfully generated which has similar phenotypes to the PH-1.
The pFgBet3-mCherry vector was constructed by amplification of the 2,573-bp FgBet3 sequence (including its native promoter and coding sequence) using FgBet3CF and FgBet3CR primer pair (Supplementary Table 2). The previously stated cloning kit was used to clone the PCR product into a pKNT-mCherry plasmid and the insertion was confirmed by sequencing. pFgGyp1ΔN-GFP, pFgGyp1ΔTBC-GFP, pFgGyp1ΔC-GFP and pR357K, pR284K constructs were generated by PCR amplification of the sequences using their respective primer pairs (Supplementary Table 2) and pKNTG2 vector was used for the cloning and verified by sequencing.
Flowering wheat heads were used to test for the pathogenicity of the various strains as previously reported (Zheng et al., 2015). Wheat coleoptiles were also used for this assy. Briefly, 2 μl of 100 × 104 cells/ml conidial suspension was inoculated in each coleoptile and the observed symptoms were recorded after 7 days of inoculation. To investigate their DON production abilities, the various strains were cultured in TBI (trichothecene biosynthesis induction) liquid media and incubated for 7 days in the dark at 28°C without shaking, after which the mycelia were separated from the liquid (Gardiner et al., 2009). An ELISA (enzyme linked immunosorbent assay)-based DON assay kit (Beacon Analytical Systems, Saco, ME, United States) was used to check for DON levels in the liquids while the mycelia were quantified after drying. The experiment was repeated three times.
The sequences of FgRab1 cDNA, FgGyp1 cDNA, TBC domain and R357K point mutant of FgGyp1 were cloned into the MBP (Maltose-binding protein) vector pMAL-c2X, using their respective primers listed in Supplementary Table 2, and expressed. The protein products were isolated and purified for GAP activity assay using a GTPase assay kit (Sigma-Aldrich, Catalog Number MAK113) according to the manufacturer’s protocols. GTP hydrolysis was assayed based on previous reports (Xiong et al., 2012; Zheng et al., 2020). Briefly, the recombinant proteins MBP–FgGyp1, MBP–FgGyp1TBC (TBC), MBP–FgGyp1R357K (R357), and MBP–FgRab1 were expressed in BL21 Escherichia coli strain and isolated by Amylose resin (Sangon Biotech, NO.C500096) as per the instructions of the manufacturer. A colorimetry-based kit was used (Sigma-Aldrich, Catalog Number MAK113) to assay for the GTPase activity of Gyp1, and FgRab1 was incubated with FgGyp1, FgGyp1TBC (TBC), FgGyp1R357K (R357K), and MBP control, respectively, following the instructions of the manufacturer. The experiment was repeated three times.
Aspergillus fumigatus (AfGyp1-XP_749737.1), Candida albicans (CaGyp1-XP_717292.1), Fusarium graminearum (FgGyp1-PCD18761.1), Fusarium oxysporum (FoGyp1-EMT63394.1), Fusarium verticillioides (FvGyp1-EWG40605.1), Magnaporthe oryzae (MoGyp1-ELQ43659.1), Neurospora crassa (NcGyp1-CAC18313.2), Saccharomyces cerevisiae (ScGyp1-NP_014713.1), Schizosaccharomyces pombe (SpGyp1-NP_595314.1), and Ustilago maydis (UmGyp1-XP_011391412.1).
The live cell imaging of F. graminearum mycelia and conidia was performed using an Olympus BX51 (Olympus, Japan) microscope and a laser confocal microscope (Nikon, Japan). The following settings were used. GFP excitation: 488 nm light (Em.525/40 nm); mCherry excitation: 561 nm light (Em. 607/36 nm).
To identify the FgGYP1 gene in F. graminearum genome, we used the budding yeast Gyp1 amino acid sequences as a reference to carry out a BLAST search against the available fungal genomes. We were able to identify a homolog of Gyp1 at the FGSG_17336 locus of F. graminearum genome. FGSG_17336 encodes a protein of 601 amino acid residues and it is 40.53% similar to the yeast Gyp1, and it covers 61.00% of the total length of FgGyp1 and ScGyp1. The phylogenetic relationship of Gyp1 homologs suggest that Gyp1 is conserved in plant pathogenic fungi, especially in Fusarium oxysporum, Fusarium verticillioides, Neurospora crassa, and Magnaporthe oryzae (Supplementary Figure 1). Furthermore, targeted gene replacement strategy was used to delete FgGYP1 gene in PH-1 (wild type) where many positive mutants were identified by PCR screening (Supplementary Figure 2A). Two of these mutants (ΔFggyp1-2 and ΔFggyp1-5) were subjected to Southern blotting for confirmation (Supplementary Figure 2B).
To determine whether FgGyp1 is required for the fungal vegetative growth, we cultured the ΔFggyp1 mutant on CM (complete media) and monitored its growth rate after 3 days. Compared to the PH-1 and ΔFggyp1-C (complemented strain) strains, the ΔFggyp1 growth rate was critically reduced (Figures 1A,B and Table 1). Close microscopic examinations indicated numerous branching in the ΔFggyp1 mutant hyphae which was not observed in PH-1 and ΔFggyp1-C strains (Figure 1A). These results suggest that FgGyp1 is important for F. graminearum vegetative growth and hyphal polarity.
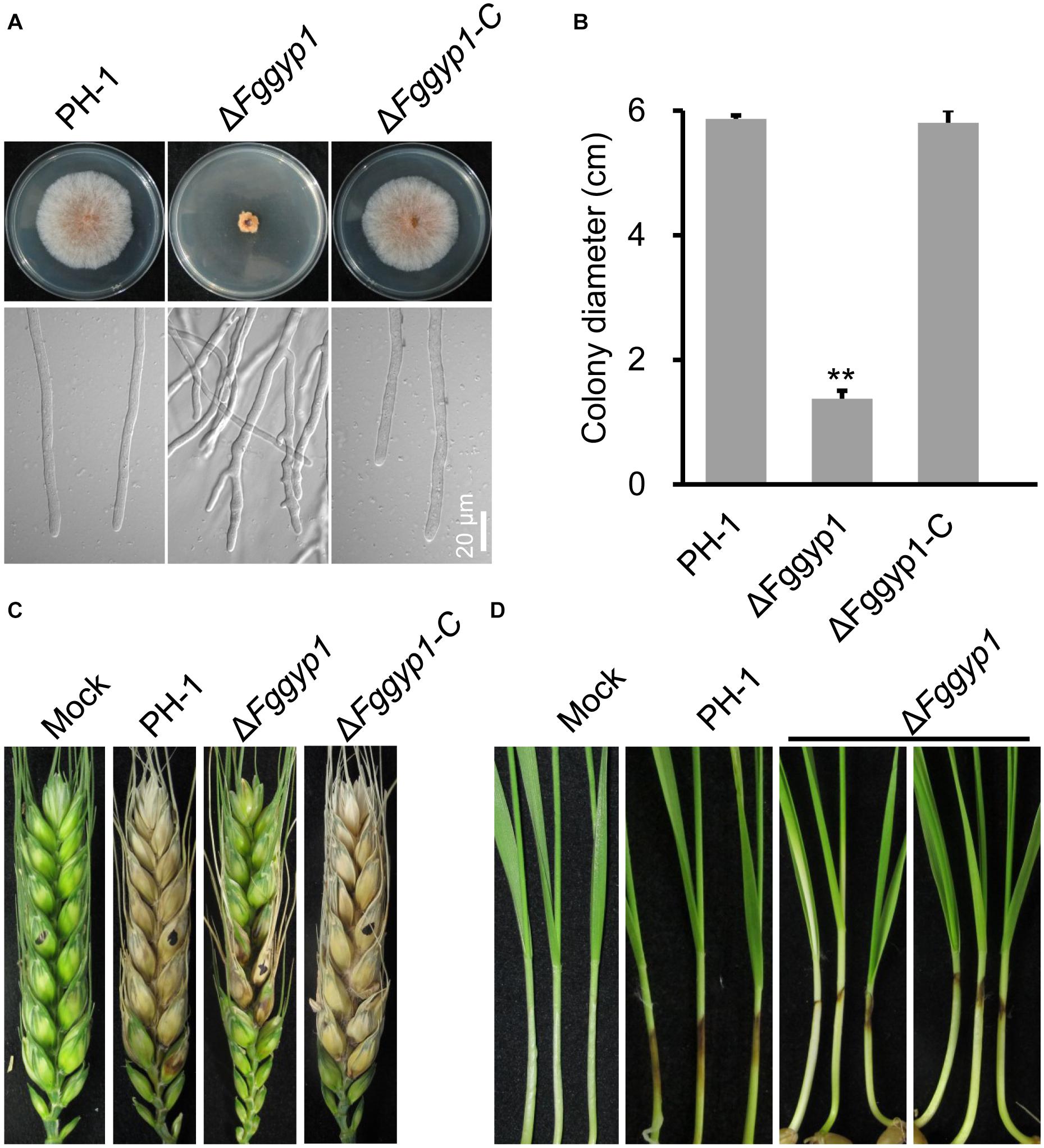
Figure 1. FgGyp1 is required for vegetative growth and virulence of Fusarium graminearum. (A) Colonies morphology of the wild type (PH-1), ΔFggyp1 mutant, and ΔFggyp1-C (the complemented strain) after growth on CM agar for 3 days. (B) Statistical analysis of the colony diameters of the PH-1, ΔFggyp1, and ΔFggyp1-C strains on CM medium after 3 days. Error bars represent the standard deviation from three replicates and Two-tailed Student’s t-test was used for paired comparison of the colony diameters between ΔFggyp1 mutant and the wild type PH-1 strain (**P < 0.01). (C) Pathogenicity test of PH-1, ΔFggyp1, and ΔFggyp1-C strains inoculated on wheat heads for 14 days. The virulence of ΔFggyp1 mutant is significantly reduced. (D) Pathogenicity of PH-1, ΔFggyp1, and ΔFggyp1-C strains on wheat coleoptiles after 7 days of inoculation. The virulence of ΔFggyp1 mutant is significantly decreased.
The major F. graminearum inoculums that infect flowering wheat heads are the conidia (Francl et al., 1999). As such, we inoculated the wild type PH-1, ΔFggyp1, and ΔFggyp1-C strains on carboxymethylcellulose (CMC) media at 28°C for 3 days for conidia production. As shown in Table 1, the conidiation of ΔFggyp1 mutant decreased significantly when compared to those produced by PH-1 and ΔFggyp1-C strains, suggesting that FgGyp1 plays an important role in the fungal conidiogenesis. In addition to conidia, the ascospores of F. graminearum are also an important inoculum in its infection cycle. However, sexual reproduction and ascospore formation are not significantly affected by FgGYP1 deletion (Supplementary Figures 3A,B).
To investigate the effect of FgGYP1 deletion on the pathogenicity of the fungus, the mutant and the controls were inoculated on flowering wheat heads under moist condition for 14 days. As shown in Figure 1C, deletion of FgGYP1 gene caused significantly decreased infection capability to wheat heads. The average disease index (diseased spikelets per head) for ΔFggyp1 mutant is less than six while the disease index of PH-1 and ΔFggyp1-C strains are more than 13 (Table 1). Furthermore, infection assays on wheat coleoptiles yielded similar results (Figure 1D). Taken together, our data here demonstrate that FgGyp1 is important for F. graminearum virulence.
In F. graminearum, deoxynivalenol (DON) has been widely studied as an important mycotoxin and virulence factor (Proctor et al., 1995). To investigate whether FgGyp1 is required for DON production, we checked and compared the levels of DON in the PH-1, ΔFggyp1, and ΔFggyp1-C strains cultured in liquid TBI media for 7 days at 28°C in the dark. We found that ΔFggyp1 mutant showed strikingly higher level of DON production than the wild type PH-1 (Table 1, the standard curves for quantification of the DON were showed in Supplementary Figure 4), suggesting that FgGyp1 negatively regulates DON biosynthesis in F. graminearum. However, we found that the trichothecene biosynthesis proteins FgTri1-GFP and FgTri4-GFP retain their normal localizations at the toxisomes of both strains, meaning that FgGyp1 is dispensable for their localizations (Figure 2).
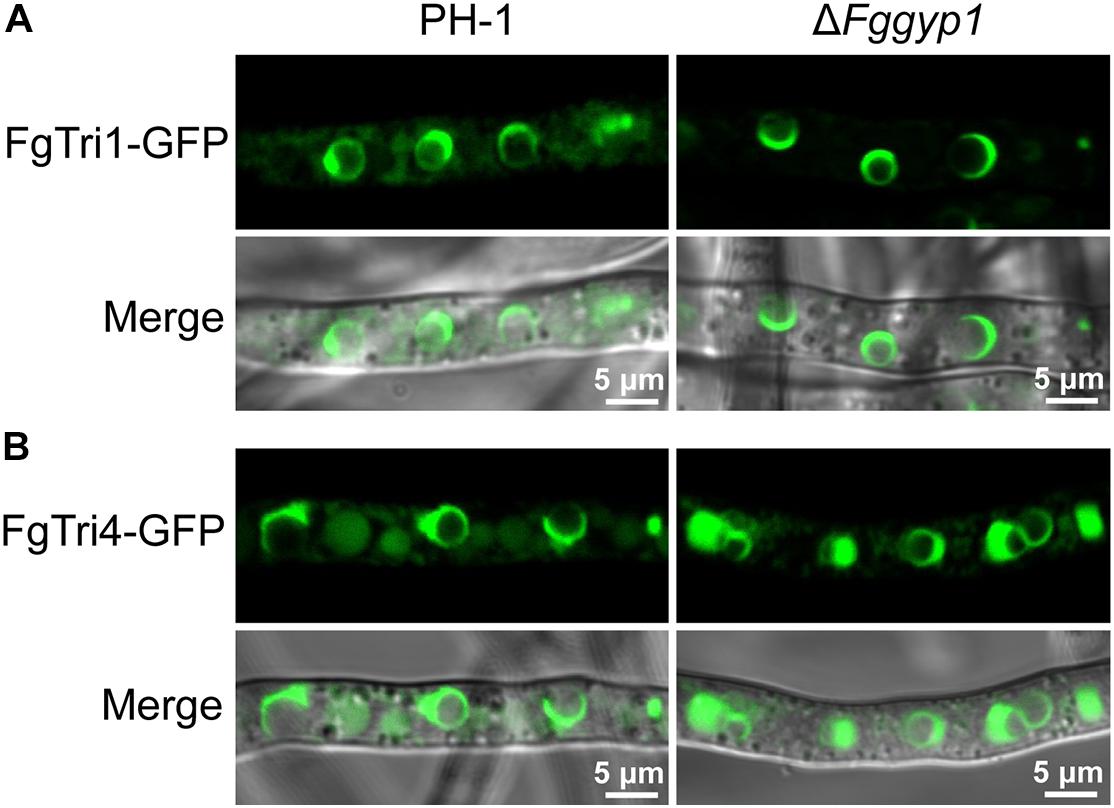
Figure 2. FgGyp1 is dispensable for FgTri1/FgTri4-mediated toxisome formation. (A) Localization of FgTri1-GFP protein in the PH-1 and ΔFggyp1 mutant. Deletion of FgGYP1 did not affect the formation of FgTri1-GFP-labeled toxisome. (B) Localization of FgTri4-GFP protein in PH-1 and ΔFggyp1 mutant. Deletion of FgGYP1 did not affect the formation of FgTri4-GFP-labeled toxisome.
To establish the sub-cellular localization of FgGyp1, we generated and transformed a FgGyp1-GFP-expressing vector into the ΔFggyp1 mutant protoplasts. The positive transformants were then subjected to live cell confocal microscopy. As shown in Figure 3, FgGyp1-GFP fluorescence were clearly visible as puncted structures distributed through the conidia and conidiophores of the fungus at all developmental stages (0.5, 2, and 18 h). In yeast, Gyp1 localizes to the Golgi apparatus (Du and Novick, 2001). To check whether the observed puncta are localized to the Golgi apparatus, several Golgi markers were generated, including the trans-Golgi network (TGN) marker, FgKex2-mCherry, cis-Golgi marker, FgBet3-mCherry, and the medial Golgi marker, mCherry-FgGos1 (Thomas et al., 2018). These constructs were co-transformed with FgGyp1-GFP, respectively, into the ΔFggyp1 mutant and their intracellular localization examined by confocal microscopy. As shown in Figure 4, we found that FgGyp1-GFP partially colocalizes with FgKex2-positive TGN (61.72 ± 4.67% colocalization), FgBet3-positive cis-Golgi (28.79 ± 12.03% colocalization) and FgGos1-positive medial Golgi (14.71 ± 6.48% colocalization) in CM medium. We therefore conclude here that FgGyp1 mainly localizes to the TGN.
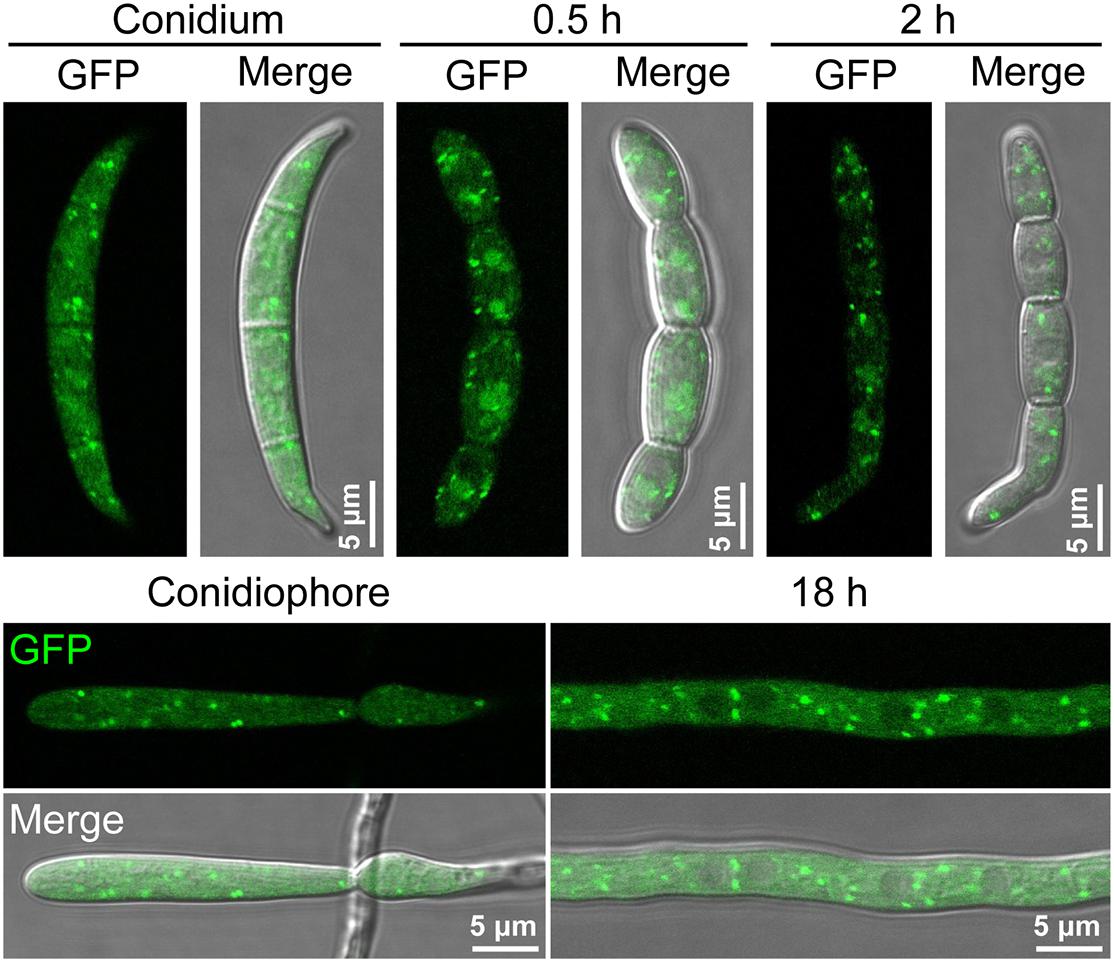
Figure 3. The localization of FgGyp1-GFP fusion protein in non-germinating and germinating conidia (0.5 and 2 h), mycelia (18 h), and conidiophore of Fusarium graminearum.

Figure 4. The co-localization of FgGyp1 with trans-Golgi network (TGN, FgKex2-mCherry), cis-Golgi marker (FgBet3-mCherry), and medial Golgi marker (mCherry-FgGos1) in Fusarium graminearum. (A) FgGyp1 partially co-localized with FgKex2-positive TGN (61.72 ± 4.67% co-localization). Arrows show co-localization. (B) FgGyp1 partially co-localized with FgBet3-positive cis-Golgi (28.79 ± 12.03% co-localization). Arrows show co-localization. (C) FgGyp1 partially co-localized with FgGos1-positive medial Golgi (14.71 ± 6.48% co-localization). Arrows show co-localization.
Ypt1/Rab1 is a substrate for Gyp1 in yeast (Albert and Gallwitz, 1999). This prompted us to check if FgGyp1 is a GTPase-activating protein (GAP) for Rabs in F. graminearum. To achieve this, we investigated its Rab GAP activity in vitro by cloning and expressing its full-length and FgRab1 in BL21 cells, respectively. We then analyzed FgGyp1 GTP-hydrolyzing potential by quantifying the amount of phosphate moiety released by activated (GTP-bound) FgRab1. As shown in Figure 5, we found that FgGyp1 has higher efficiency in hydrolyzing GTP-bound FgRab1 than the control. Furthermore, we equally found that the TBC domain of FgGyp1 more efficiently hydrolyzes GTP-bound FgRab1 than the control. The arginine 343 residue (Arg343) of the TBC domain of Gyp1 in yeast was shown to be essential for the Gyp1 GAP activity (Du and Novick, 2001). For this reason, we identified the conserved Arginine residue in FgGyp1 (Arg357) and analyzed its role in FgGyp1 GAP activity. We observed that the FgGyp1 GAP activity was significantly decreased when Arg357 residue of FgGyp1 was mutated to lysine (R357K), as shown in Figure 5. Considering these data, we conclude that FgGyp1 catalytically converts an active (GTP-bound) FgRab1 to its inactive (GDP-bound) form through its TBC domain, and Arg357 residue is an important GTP-hydrolyzing residue of FgGyp1.
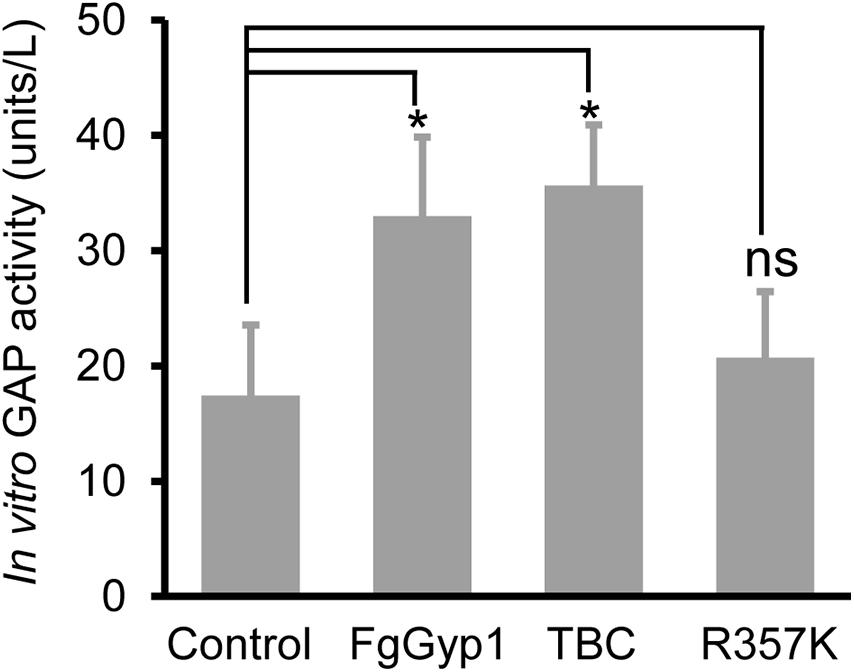
Figure 5. FgGyp1 functions in vitro as a FgRab1 GAP in Fusarium graminearum. In vitro GAP activity assay of FgGyp1. Control: MBP protein. One unit represents the amount of FgGyp1 catalyzing the release of 1 μM free phosphate/min under the experimental conditions. 1.6 μg MBP, 1.6 μg MBP-FgGyp1, 1.6 μg MBP-TBC, 1.6 μg MBP-R357K, and 0.4 μg MBP-FgRab1 proteins were used in the GAP activity assay, respectively. Two-tailed Student’s t-test was used for paired comparison of the GAP activity between MBP control and the full lengths of FgGyp1, TBC domain and R357K, respectively (*P < 0.05). ns, ‘no significant difference’.
Since the TBC domain (with its Arg357 residue) is required for FgGyp1 GAP activity, we set to find out to the roles of this domain as well as its Arg357 residue in the biological functions of FgGyp1 in F. graminearum. To unveil this, we generated the following forms of FgGyp1-GFP fusion constructs: Fggyp1ΔTBC-GFP (lacking the TBC domain), Fggyp1R357K-GFP (mutation of the arginine 357 residue with lysine), Fggyp1R284K-GFP (having arginine 284 replaced with lysine, control), Fggyp1ΔN-GFP (lacking the N-terminal aa 1–280), and Fggyp1ΔC-GFP (lacking the C-terminal aa 543–601) (Figure 6A). We transformed these constructs into separate protoplasts of ΔFggyp1 mutant and analyzed the phenotypes of each of these mutants and further examined their respective intracellular localizations by confocal microscopy. Figures 6B–F shows that ΔTBC, ΔN, ΔC, and R357K mutants exhibit phenotypes similar to the defects observed in the ΔFggyp1 mutant, including impaired vegetative growth, conidiation, virulence, and DON production. In contrast, the phenotypes of R284K mutant remain similar to those of the controls (Figures 6B–F), suggesting that Arg284 is not required for the function of FgGyp1.
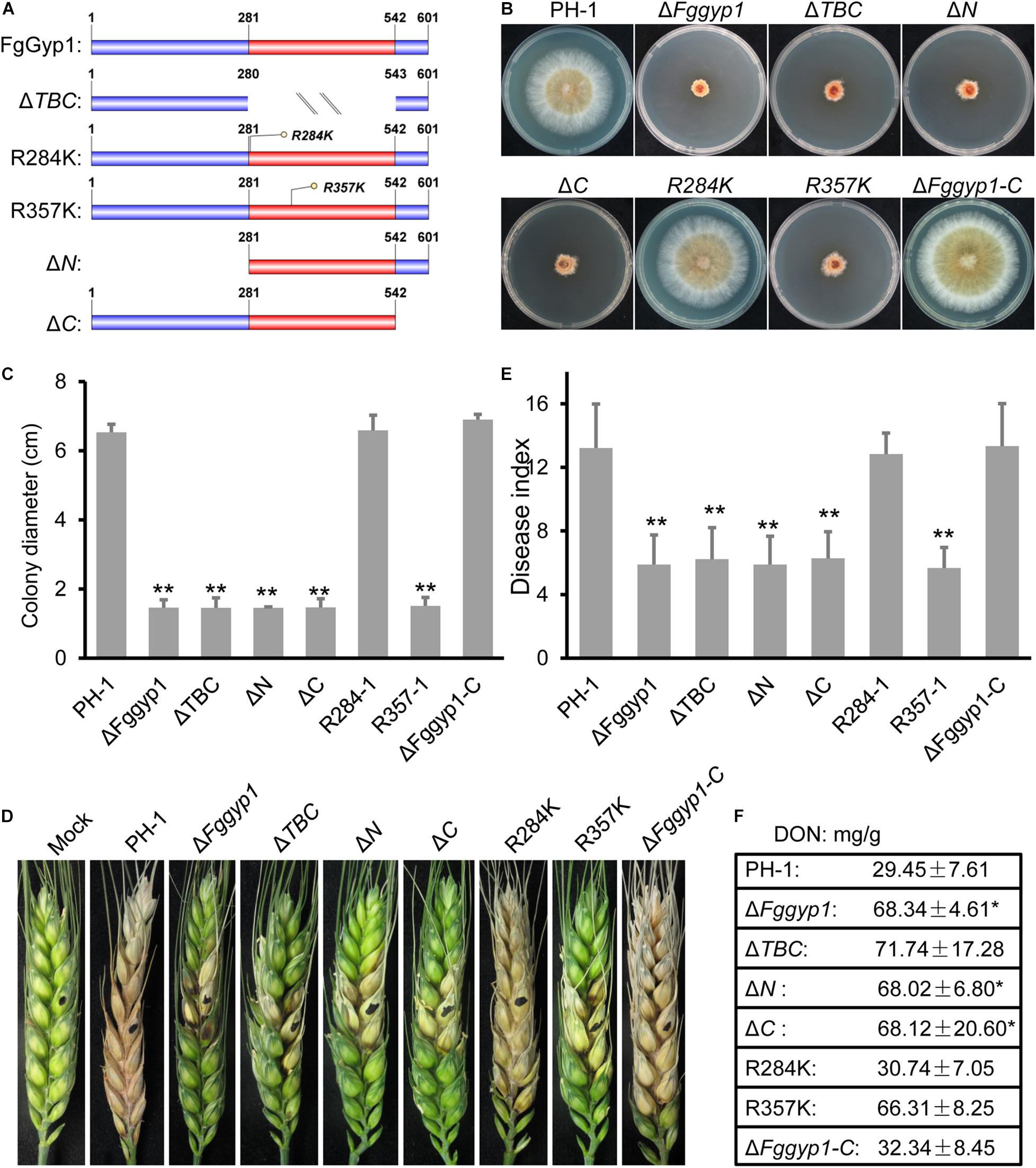
Figure 6. Functional characterization of TBC domain, Arg357/284 residues and the N/C-terminal regions of FgGyp1. (A) Schematic diagram of FgGyp1-full length, TBC domain mutant, Arg357/284 residues mutants, N-terminal, and C-terminal mutants. (B,C) Colony morphologies and diameters of PH-1, ΔFggyp1, ΔTBC,ΔN, ΔC, R284K, R357K, and ΔFggyp1-C strains on CM after 3 days. (D,E) Pathogenicity and disease indexes of the indicated strains on wheat heads. (F) DON production of the indicated strains in liquid TBI media. Two-tailed Student’s t-test was used for paired comparison of the diameter, disease index, and DON production between wild type PH-1 and the indicated strains, respectively (*P < 0.05; **P < 0.01).
Moreover, the aforementioned mutants were further analyzed for their roles in the subcellular localization of FgGyp1 protein. Figure 7 depicts that Arg284 and Arg357 residues are expendable with respect to FgGyp1 localization since their deletions do not significantly affect the localization of the protein. However, deletions of TBC domain, N-terminal, and C-terminal regions of the FgGyp1 displayed diffused GFP signals in the cytoplasm instead of the normal puncted appearance, suggesting that these domains play an important role to the subcellular localization of FgGyp1. Taken together, we conclude that the TBC domain, N-terminal, and C-terminal regions are all required for FgGyp1 functions and normal localization in F. graminearum. Arg357 residue is equally important for the functions of FgGyp1 but not for its correct localization.
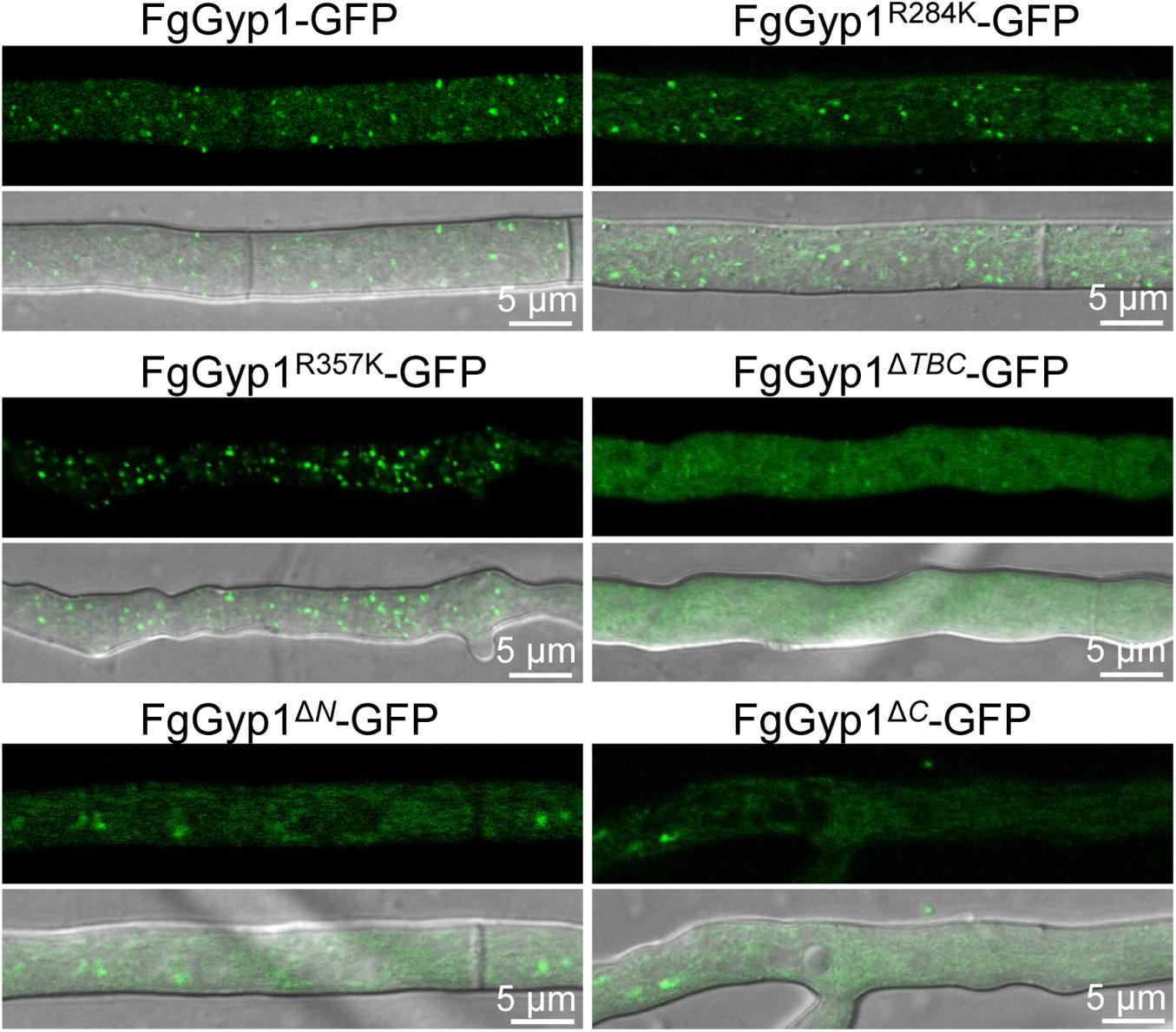
Figure 7. The normal localization of FgGyp1 requires its TBC domain, C-, and N-terminal regions. Mutations of Arg284 and Arg357 residues did not significantly affect the localization of FgGyp1. However, deletion of the TBC domain, N-terminal, and C-terminal regions of the protein caused the GFP signals to diffuse throughout the cytoplasm.
Membranes fusion and scission are known to be regulated by SNARE (soluble N-ethylmaleimide-sensitive factor attachment receptor) proteins during vesicle fission and delivery, respectively (Hong, 2005). In yeast, Snc1 (a SNARE protein) mediates the fusion of vesicles from the Golgi with the plasma membrane (Lewis et al., 2000). We previously demonstrated in F. graminearum that FgSnc1 mediates polarized secretion and fusion of vesicles (Zheng et al., 2018c). To unveil the role of FgGyp1 in vesicles fusion with the plasma membrane, we transformed GFP-FgSnc1 construct into the PH-1 and ΔFggyp1 protoplasts and subsequently observed their cellular localizations. Figure 8 clearly shows that in PH-1, GFP-FgSnc1 is localized on the cell membrane and accumulates at the spitzenkörper (SPK) of the growing hyphal tip (Figures 8A,B and Supplementary Video 1). In the ΔFggyp1 mutant, the fluorescence of GFP-FgSnc1 was similarly detected at the SPK of the growing hyphae but its plasma membrane localization was almost completely abolished (Figures 8A,B and Supplementary Video 2). These results suggest that FgGyp1 is important for FgSnc1-mediated fusion of vesicles with the plasma membrane in F. graminearum.
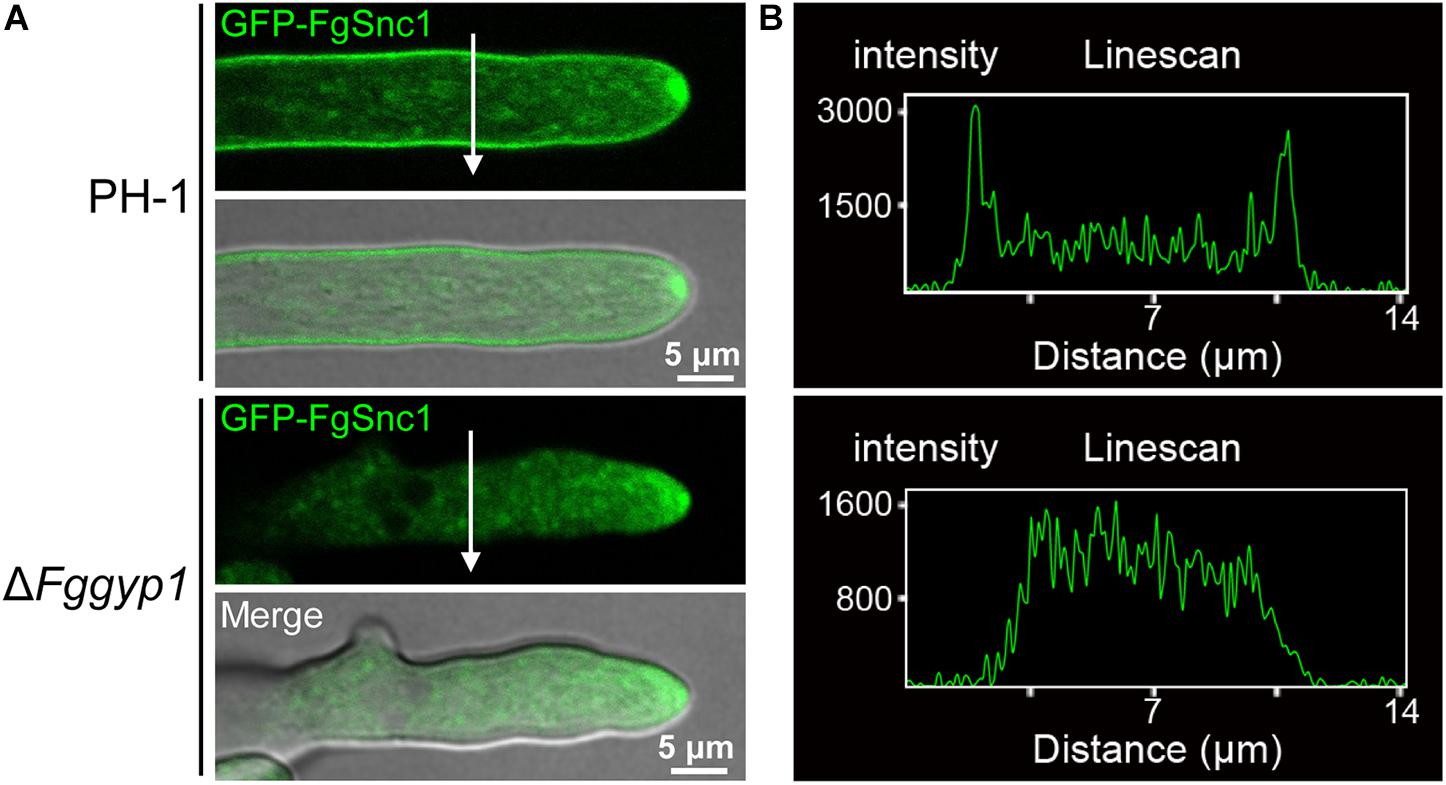
Figure 8. FgGyp1 is important for plasma membrane localization of the SNARE protein FgSnc1. (A) GFP-FgSnc1 localization in the various strains. GFP-FgSnc1 localizes to the plasma membrane of the wild type, while the membrane localization is abolished in the ΔFggyp1 mutant. (B) Line scans for GFP-FgSnc1 localization in the wild type and ΔFggyp1 mutant.
Rab GTPases serve as primary regulators of intracellular membrane trafficking processes (Stenmark, 2009). GTPase-activating proteins (GAPs) inactivate Rabs as when needed by hydrolyzing their GTP to GDPs (Mizuno-Yamasaki et al., 2012). Most identified RabGAPs contain Tre2-Bub2-Cdc16 (TBC) domains in mammals (Fukuda, 2011). In this study, we investigated the role of a TBC domain-containing protein FgGyp1 in the wheat pathogen F. graminearum and found that FgGyp1 acts as a GAP for FgRab1 in vitro and is importantly required for conidiation, virulence, vegetative growth and DON production in F. graminearum. Furthermore, FgGyp1 is found to regulate the FgSnc1-mediated fusion of secretory vesicles emerging from the Golgi compartment with the cell membrane.
Previously, De Antoni et al. (2002) demonstrated in vivo that GYP genes in yeast are redundant in their functions as mutants of a single GYP gene remain viable. In this study, we found that a FgRab1-GAP is critical for the development of F. graminearum and its mutants are severely impaired in intracellular vesicle trafficking processes. There are 12 TBC domain-containing proteins in F. graminearum genome, and recently, the TBC domain-containing protein FgMsb3 was shown to act as a GAP for FgRab8 and is also important for F. graminearum pathogenicity and development (Zheng et al., 2020). When compared to the observed roles in yeast, these findings imply more elaborate biological roles of GAPs during fungal evolution. Interestingly, ΔFggyp1 mutant is observed to grow even slower than the ΔFgmsb3 mutant (Zheng et al., 2020) but the latter is more impaired in virulence than the former, suggesting a less important role of FgGyp1 in virulence than FgMsb3 in F. graminearum.
Gyp1 has dual functions, as a GAP for Ypt1 and as an interacting partner of Atg8 in selective autophagy (Mitter et al., 2019). For its stability and normal subcellular localization, Gyp1 (a GAP for Ypt1) needs to interact with Ypt32 (Rivera-Molina and Novick, 2009). Gyp1 localizes to the Golgi apparatus in yeast and C. albicans (Du and Novick, 2001; Huang et al., 2014). Moreover, Gyp1 is critically required for Golgi polarization in C. albicans (Huang et al., 2014). Here, we found that FgGyp1 mainly localizes to the FgKex2-positive TGN, FgBet3-positive cis-Golgi and FgGos1-positive medial Golgi, suggesting that FgGyp1 could be involved in various Golgi-related physiological process.
Effective vesicle trafficking of a cell depends on normal localization of Rab GTPases (Thomas et al., 2019). The targeting signal for these proteins is HVD (hypervariable domain), located at their C-termini. In this study, the C- and N-terminal regions of FgGyp1 are both required for its functions and localization in F. graminearum. A previous study indicated that the polarization of Gyp1 to the growth site is regulated through its phosphorylation by PKA (protein kinase A) in C. albicans (Huang et al., 2014); four serine residues are present at the N-terminal regulatory domain of Gyp1 and are phosphorylated by PKA. These serine residues and the Arg292 residue inside the catalytic domain of CaGyp1 are required for the polarization of CaGyp1. Here, we showed that the Arg357 residue close to the N-terminus of FgGyp1 is required for its hydrolyzing activity and functions in F. graminearum but not for its correct localization. We showed previously in F. graminearum that FgMsb3 TBC domain has an Arg681 residue that is indispensable for FgMsb3 functions, but not for the normal localization of the protein (Zheng et al., 2020). The Sec2 domain and N-terminal region of FgSec2A are dispensable for its polarized localization but required for its functions (Zheng et al., 2018a).
The role of SNAREs and Rab/Ypt GTPases in targeting vesicles to their right compartments ensures trafficking pathways specificity (Segev, 2001). In yeast, recycling of Snc1 protein ensures the continuity of the secretion pathway and Gyp1 GAP activity is needed for Snc1 recycling (Lewis et al., 2000). In yeast, Ypt6 and Ric1/Rgp1 (guanine-nucleotide exchange factor for Ypt6) are also crucial for recycling of Snc1 (Siniossoglou et al., 2000). FgRab8 and its GAP FgMsb3 are required for vesicle secretion and trafficking during exocytosis mediated by FgSnc1 (Zheng et al., 2020). We found that FgGyp1 regulates the fusion of secreted vesicles from the Golgi with the cell membrane, suggesting a conserved role of Rab/Ypt GAP in vesicle transport.
In this study, DON production is significantly increased in the ΔFggyp1 mutant, suggesting that FgGyp1 negatively regulates DON metabolism. The ΔFggyp1 mutant grows very slowly while its disease index to flowering wheat heads reaches 5.88 likely due to the increasing production of DON in this mutant. In summary, the present research identified FgGyp1 as a FgRab1 GAP that is indispensable for conidiation, virulence, growth, and DON biosynthesis in F. graminearum. Furthermore, FgGyp1 is critical for the regulation of FgSnc1-mediated fusion of secreted vesicles from the Golgi with the cell membrane.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
HZ conceived and designed the experiments. HZ, QZ, ZY, YY, and DS performed the experiments. HZ wrote the manuscript. JZ, ZW, and YA critically reshaped the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31970141 and 31701742), the Natural Science Foundation of Fujian Province (2020J06047), and the Foundation of Minjiang University (MJY19019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.621519/full#supplementary-material
Supplementary Figure 1 | Phylogenetic relationship of Gyp1 proteins from different organisms.
Supplementary Figure 2 | Generation of FgGYP1 deletion mutants and confirmation by Southern blot analysis.
Supplementary Figure 3 | FgGyp1 is dispensable for sexual reproduction.
Supplementary Figure 4 | The standard curves for quantification of the DON.
Supplementary Table 1 | List of the various fungal strains used in this study.
Supplementary Table 2 | List of all primers involved in this study.
Supplementary Video 1 | GFP-FgSnc1 protein mobility in the PH-1.
Supplementary Video 2 | GFP-FgSnc1 protein mobility in the ΔFggyp1 mutant.
Adnan, M., Fang, W., Sun, P., Zheng, Y., Abubakar, Y. S., Zhang, J., et al. (2020). R-SNARE FgSec22 is essential for growth, pathogenicity and DON production of Fusarium graminearum. Curr. Genet. 66, 421–435. doi: 10.1007/s00294-019-01037-y
Albert, S., and Gallwitz, D. (1999). Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J. Biol. Chem. 274, 33186–33189. doi: 10.1074/jbc.274.47.33186
Bushnell, W., Hazen, B., Pritsch, C., and Leonard, K. (2003). “Histology and physiology of Fusarium head blight,” in Fusarium head blight of wheat and barley, eds K. J. B. Leonard and W. R. Bushnell (Minnesota: APS Press), 44–83.
Cappellini, R. A., and Peterson, J. L. (1965). Macroconidium formation in submerged cultures by a nonsporulating strain of Gibberella Zeae. Mycologia 57, 962–966. doi: 10.1080/00275514.1965.12018285
Chen, Y., Kistler, H. C., and Ma, Z. (2019). Fusarium graminearum Trichothecene Mycotoxins: Biosynthesis, Regulation, and Management. Annu. Rev. Phytopathol. 57, 15–39.
De Antoni, A., Schmitzová, J., Trepte, H. H., Gallwitz, D., and Albert, S. (2002). Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J. Biol. Chem. 277, 41023–41031. doi: 10.1074/jbc.m205783200
Du, L. L., and Novick, P. (2001). Yeast rab GTPase-activating protein Gyp1p localizes to the Golgi apparatus and is a negative regulator of Ypt1p. Mol. Biol. Cell 12, 1215–1226. doi: 10.1091/mbc.12.5.1215
Du, L. L., Collins, R. N., and Novick, P. J. (1998). Identification of a Sec4p GTPase-activating protein (GAP) as a novel member of a Rab GAP family. J. Biol. Chem. 273, 3253–3256. doi: 10.1074/jbc.273.6.3253
Fan, G., Zheng, H., Zhang, K., Devi Ganeshan, V., Opiyo, S. O., Liu, D., et al. (2020). FgHtf1 Regulates Global Gene Expression towards Aerial Mycelium and Conidiophore Formation in the Cereal Fungal Pathogen Fusarium graminearum. Appl. Environ. Microbiol. 86, 3024–3019e.
Francl, L., Shaner, G., Bergstrom, G., Gilbert, J., Pedersen, W., Dill-Macky, R., et al. (1999). Daily inoculum levels of Gibberella zeae on wheat spikes. Plant Dis. 83, 662–666. doi: 10.1094/pdis.1999.83.7.662
Fukuda, M. (2011). TBC proteins: GAPs for mammalian small GTPase Rab? Biosci. Rep. 31, 159–168. doi: 10.1042/bsr20100112
Gardiner, D. M., Kazan, K., and Manners, J. M. (2009). Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum. Fungal Genet. Biol. 46, 604–613. doi: 10.1016/j.fgb.2009.04.004
Hou, Z. M., Xue, C. Y., Peng, Y. L., Katan, T., Kistler, H. C., and Xu, J. R. (2002). A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 15, 1119–1127. doi: 10.1094/mpmi.2002.15.11.1119
Huang, Z. X., Wang, H., Wang, Y. M., and Wang, Y. (2014). Novel mechanism coupling cyclic AMP-protein kinase A signaling and golgi trafficking via Gyp1 phosphorylation in polarized growth. Eukaryot Cell 13, 1548–1556. doi: 10.1128/ec.00231-14
Lewis, M. J., Nichols, B. J., Prescianotto-Baschong, C., Riezman, H., and Pelham, H. R. (2000). Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell 11, 23–38. doi: 10.1091/mbc.11.1.23
Li, B., Dong, X., Li, X., Chen, H., Zhang, H., Zheng, X., et al. (2018). A subunit of the HOPS endocytic tethering complex, FgVps41, is important for fungal development and plant infection in Fusarium graminearum. Environ. Microbiol. 20, 1436–1451. doi: 10.1111/1462-2920.14050
Li, B., Dong, X., Zhao, R., Kou, R., Zheng, X., and Zhang, H. (2019). The t-SNARE protein FgPep12, associated with FgVam7, is essential for ascospore discharge and plant infection by trafficking Ca2+ ATPase FgNeo1 between Golgi and endosome/vacuole in Fusarium graminearum. PLoS Pathog 15:e1007754. doi: 10.1371/journal.ppat.1007754
Li, B., Liu, L., Li, Y., Dong, X., Zhang, H., Chen, H., et al. (2017). The FgVps39-FgVam7-FgSso1 Complex Mediates Vesicle Trafficking and Is Important for the Development and Virulence of Fusarium graminearum. Mol. Plant Microbe Interact. 30, 410–422. doi: 10.1094/mpmi-11-16-0242-r
Li, G., and Marlin, M. C. (2015). Rab family of GTPases. Methods Mol. Biol. 1298, 1–15. doi: 10.1007/978-1-4939-2569-8_1
Li, Y., Li, B., Liu, L., Chen, H., Zhang, H., Zheng, X., et al. (2015). FgMon1, a guanine nucleotide exchange factor of FgRab7, is important for vacuole fusion, autophagy and plant infection in Fusarium graminearum. Sci. Rep. 5:18101.
McMullen, M., Jones, R., and Gallenberg, D. (1997). Scab of wheat and barley: a re-emerging disease of devastating impact. Plant Dis. 81, 1340–1348. doi: 10.1094/pdis.1997.81.12.1340
Mitter, A. L., Schlotterhose, P., and Krick, R. (2019). Gyp1 has a dual function as Ypt1 GAP and interaction partner of Atg8 in selective autophagy. Autophagy 15, 1031–1050. doi: 10.1080/15548627.2019.1569929
Mizuno-Yamasaki, E., Rivera-Molina, F., and Novick, P. (2012). GTPase Networks in Membrane Traffic. Annu. Rev. Biochem. 81, 637–659. doi: 10.1146/annurev-biochem-052810-093700
Proctor, R. H., Hohn, T. M., and Mccormick, S. P. (1995). Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant Microbe Interact. 8, 593–601. doi: 10.1094/mpmi-8-0593
Rak, A., Fedorov, R., Alexandrov, K., Albert, S., Goody, R., Gallwitz, D., et al. (2000). Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins. EMBO J. 19, 5105–5113. doi: 10.1093/emboj/19.19.5105
Rivera-Molina, F. E., and Novick, P. J. (2009). A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc. Natl. Acad. Sci. U S A. 106, 14408–14413. doi: 10.1073/pnas.0906536106
Segev, N. (2001). Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 13, 500–511. doi: 10.1016/s0955-0674(00)00242-8
Siniossoglou, S., Peak-Chew, S. Y., and Pelham, H. R. (2000). Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J. 19, 4885–4894. doi: 10.1093/emboj/19.18.4885
Stenmark, H. (2009). Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525. doi: 10.1038/nrm2728
Thomas, L. L., Joiner, A. M. N., and Fromme, J. C. (2018). The TRAPPIII complex activates the GTPase Ypt1 (Rab1) in the secretory pathway. J. Cell Biol. 217, 283–298. doi: 10.1083/jcb.201705214
Thomas, L. L., Van Der Vegt, S. A., and Fromme, J. C. (2019). A Steric Gating Mechanism Dictates the Substrate Specificity of a Rab-GEF. Dev. Cell 48:e109.
Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. doi: 10.1126/science.285.5429.901
Xie, Q., Chen, A., Zhang, Y., Yuan, M., Xie, W., Zhang, C., et al. (2019). Component Interaction of ESCRT Complexes Is Essential for Endocytosis-Dependent Growth, Reproduction, DON Production and Full Virulence in Fusarium graminearum. Front. Microbiol. 10:180. doi: 10.3389/fmicb.2019.00180
Xiong, Y., Yuan, C., Chen, R., Dawson, T. M., and Dawson, V. L. (2012). ArfGAP1 is a GTPase activating protein for LRRK2: reciprocal regulation of ArfGAP1 by LRRK2. J. Neurosci. 32, 3877–3886. doi: 10.1523/jneurosci.4566-11.2012
Yang, C., Li, J., Chen, X., Zhang, X., Liao, D., Yun, Y., et al. (2020). FgVps9, a Rab5 GEF, Is Critical for DON Biosynthesis and Pathogenicity in Fusarium graminearum. Front. Microbiol. 11:1714. doi: 10.3389/fmicb.2020.01714
Zhang, H., Li, B., Fang, Q., Li, Y., Zheng, X., and Zhang, Z. (2016). SNARE protein FgVam7 controls growth, asexual and sexual development, and plant infection in Fusarium graminearum. Mol. Plant Pathol. 17, 108–119. doi: 10.1111/mpp.12267
Zheng, H., Li, L., Miao, P., Wu, C., Chen, X., Yuan, M., et al. (2018a). FgSec2A, a guanine nucleotide exchange factor of FgRab8, is important for polarized growth, pathogenicity and deoxynivalenol production in Fusarium graminearum. Environ. Microbiol. 20, 3378–3392. doi: 10.1111/1462-2920.14373
Zheng, H., Li, L., Yu, Z., Yuan, Y., Zheng, Q., Xie, Q., et al. (2020). FgSpa2 recruits FgMsb3, a Rab8 GAP, to the polarisome to regulate polarized trafficking, growth and pathogenicity in Fusarium graminearum. N. Phytol. 2020:16935. doi: 10.1111/nph.16935
Zheng, H., Miao, P., Lin, X., Li, L., Wu, C., Chen, X., et al. (2018b). Small GTPase Rab7-mediated FgAtg9 trafficking is essential for autophagy-dependent development and pathogenicity in Fusarium graminearum. PLoS Genet. 14:e1007546. doi: 10.1371/journal.pgen.1007546
Zheng, H., Zheng, W., Wu, C., Yang, J., Xi, Y., Xie, Q., et al. (2015). Rab GTPases are essential for membrane trafficking-dependent growth and pathogenicity in Fusarium graminearum. Environ. Microbiol. 17, 4580–4599. doi: 10.1111/1462-2920.12982
Keywords: Fusarium graminearum, GTPase-activating protein, FgGyp1, DON, fungal development, pathogenicity
Citation: Zheng Q, Yu Z, Yuan Y, Sun D, Abubakar YS, Zhou J, Wang Z and Zheng H (2021) The GTPase-Activating Protein FgGyp1 Is Important for Vegetative Growth, Conidiation, and Virulence and Negatively Regulates DON Biosynthesis in Fusarium graminearum. Front. Microbiol. 12:621519. doi: 10.3389/fmicb.2021.621519
Received: 26 October 2020; Accepted: 04 January 2021;
Published: 21 January 2021.
Edited by:
Hyang Burm Lee, Chonnam National University, South KoreaReviewed by:
Wei-Hua Tang, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences (CAS), ChinaCopyright © 2021 Zheng, Yu, Yuan, Sun, Abubakar, Zhou, Wang and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huawei Zheng, emh3QG1qdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.