
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 February 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.619369
This article is part of the Research Topic New Insights into the Transmission Dynamics and Control of Antimicrobial Resistance to Last-resort Antibiotics View all 22 articles
Multidrug-resistant bacteria, including carbapenem-resistant Klebsiella pneumoniae (CRKP), are becoming an increasing health crisis worldwide. For CRKP, colistin is regarded as “the last treatment option.” In this study, we isolated a clinical CRKP strain named as K. pneumoniae R10-341. Phenotyping analysis showed that this strain could transit from a colistin-sensitive to a resistant phenotype by inserting an IS4 family ISKpn72 element into the colistin-resistance associated mgrB gene. To investigate the mechanism of this transition, we performed genome sequencing analysis of the colistin-sensitive parental strain and found that 12 copies of ISKpn72 containing direct repeats (DR) are located on the chromosome and 1 copy without DR is located on a multidrug-resistant plasmid pR10-341_2. Both types of ISKpn72 could be inserted into the mgrB gene to cause colistin-resistance, though the plasmid-derived ISKpn72 without DR was in higher efficiency. Importantly, we demonstrated that colistin-sensitive K. pneumoniae strain transferred with the ISKpn72 element also obtained the ability to switch from colistin-sensitive to colistin-resistant phenotype. Furthermore, we confirmed that the ISKpn72-containing pR10-341_2 plasmid was able to conjugate, suggesting that the ability of causing colistin-resistant transition is transferable through common conjugation. Our results point to new challenges for both colistin-resistance detection and CRKP treatment.
Multidrug-resistant pathogenic bacteria, such as carbapenem-resistant Klebsiella pneumoniae (CRKP), are increasingly becoming a health crisis worldwide (Nordmann et al., 2011; Pitout et al., 2015). Polymyxins, including polymyxin B and polymyxin E (colistin), have been regarded as “the last treatment option” for CRKP (Biswas et al., 2012). Polymyxins are lipopeptide antibiotics targeting the lipopolysaccharide (LPS) of the bacterial outer membrane, the main component of the Gram-negative bacterial cell wall (Hancock, 1997; Li et al., 2006).
The increase in infections by polymyxin-resistant bacteria has become a great challenge to clinical treatment (Antoniadou et al., 2007; Bogdanovich et al., 2011; Macesic et al., 2020). In K. pneumoniae, the most common polymyxin-resistance mechanism is achieved by LPS modification, which decreases the negative charge of LPS and reduces its affinity to polymyxins (Velkov et al., 2014; Liu et al., 2017). The mobile colistin resistance gene (mcr-1) is the first reported plasmid-mediated colistin resistance gene (Liu et al., 2016), which encodes a phosphoethanolamine-lipid A transferase catalyzing the addition of phosphoethanolamine (PEtN) to lipid A (Liu et al., 2016, 2017). To date, 10 variants (mcr1-10) have been identified on a wide variety of transferable plasmids (Wang et al., 2018, 2020; Yang et al., 2018; Zhong et al., 2018; Lei et al., 2020), leading to the widespread diffusion of mcr-mediated colistin-resistance (Zhong et al., 2018; Nang et al., 2019). Chromosome-mediated colistin-resistance has also been characterized by the involvement of a small transmembrane protein MgrB and the two-component systems (TCSs) PhoPQ, PmrAB, and CrrAB (Gunn and Miller, 1996; Olaitan et al., 2014). To the best of our knowledge, in contrast to mcr genes, chromosome-mediated colistin-resistance mechanisms are considered to be stable and have not been reported to be transferred or mobile to other bacteria. However, inactivation of the mgrB gene has been widely identified from clinical colistin-resistant clinical K. pneumoniae strains (Cannatelli et al., 2013; Gaibani et al., 2014; Olaitan et al., 2014).
In this study, we isolated a multidrug resistant K. pneumoniae strain named R10-341 with high frequency (∼10–6) of colistin heteroresistance (El-Halfawy and Valvano, 2015; Halaby et al., 2016). Genetic and molecular analyses identified that insertion of an ISKpn72 element into the mgrB gene was responsible for the acquisition of colistin resistance. We further analyzed ISKpn72 copies in this strain and demonstrated that the ISKpn72 element is derived from a mobile plasmid and suggested that this mobile plasmid has the ability to render transition from colistin-sensitive to resistant phenotype in K. pneumoniae.
The K. pneumoniae strains named R10-341 and 7097 used in this study were isolated from the sputum samples collected from the Tongji Hospital, Hubei Province, China. The K. pneumoniae strains were grown in LB medium with 100 μg/mL ampicillin at 37°C.
For plasmid constructions, the p15A ori from plasmid pACYC184 (Rose, 1988) and the streptomycin-resistance gene from pTargetF (Jiang et al., 2015) were PCR amplified, respectively. These two fragments were assembled as the linearized vector p15A-Sm by overlap PCR, which was then cloned with ISpla (IS fragment from pR10-341_2) or ISchr (IS fragment from chromosome) fragment using the ClonExpress II One Step Cloning Kit (Vazyme) to generate two plasmids named p15A-Sm-ISpla and p15A-Sm-ISchr respectively. All primers used in this study are listed in Supplementary Table 1.
Multilocus sequence typing (MLST) for the K. pneumoniae R10-341 strain was performed as described (Diancourt et al., 2005).1 Colistin resistance associated genes were detected by PCR. Each of the mcr genes was amplified using two pairs of primers. Genes encoding the two-component systems were amplified by PCR and confirmed by DNA sequencing in comparison with the K. pneumoniae HS11286 strain (Accession: NC_016845). All primers are listed in Supplementary Table 1.
MICs of antibiotics (except colistin) for K. pneumoniae R10-341 were determined using the broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI document M100-S28)2. The susceptibility to colistin was tested according to the guidelines of European Committee on Antimicrobial Susceptibility Testing (EUCAST)3. Briefly, 100 μL Cation-adjusted Mueller-Hinton Broth containing 2-fold diluted antibiotics was added to a 96-well plate, followed by the addition of 100 μL bacterial cells (105∼106CFU/mL) to each well. The 96-well plate was incubated at 37°C for 16–24 h. The lowest concentration of antibiotic with complete inhibition (clear broth) was regarded as the MIC.
The K. pneumoniae R10-341 strain was first spread onto a LB plate. A single colony was selected and cultured in LB medium at 37°C. Genomic DNA was extracted using a bacterial genomic DNA extraction Kit (Tiangen). Genome DNA sequencing was performed by both Hiseq X Ten (Illumina) and MinION (Oxford Nanopore Technologies) platforms according to a standard protocol provided by Illumina and Oxford Nanopore Technologies. The off-machine data of Nanopore sequencing is converted to fastq format through the Albacore software in the MinKNOW software package4 (Payne et al., 2019). After filtering to obtain clean reads, these reads are randomly selected and aligned with the Nucleotide Sequence Database. De novo genome assembly was performed with Unicycler v0.4.7 (Wick et al., 2017). NCBI Prokaryotic Genomes Annotation Pipeline (PGAP) was used to annotate assembled genome sequence and to identify genes related to conjugation (Tatusova et al., 2016). Antibiotic resistance genes and plasmid replicons were respectively identified by ResFinder v3.0 (Zankari et al., 2012) and PlasmidFinder v2.0 (Carattoli et al., 2014) from the Center for Genomic Epidemiology website5. Sequence reads for the whole-genome sequencing are available from the NCBI Sequence Read Archive (PRJNA655367).
For analyzing the IS elements in K. pneumoniae R10-341, the IS element inserted into the mgrB gene in colistin resistant colonies was first identified by PCR and DNA sequencing. This IS element sequence was analyzed in ISfinder database6 and was named as ISKpn72 based on suggestion from ISfinder. The sequence of the ISKpn72 element was then aligned with the K. pneumoniae R10-341 genome sequence using BLASTn.
Amino acid sequences of transposases used in the phylogenetic tree analyses of the IS elements were downloaded from ISfinder database. Phylogenetic tree was constructed based on average distance using Jalview 2.11 (Waterhouse et al., 2009).
Since K. pneumoniae R10-341 is resistant to several antibiotics but is relatively sensitive to tetracycline, we constructed a tetracycline-resistant (TcR) E. coli K-12 strain as the recipient in conjugation experiment using a CRISPR/Cas9 system (Jiang et al., 2015). Briefly, the E. coli K-12 strain was first transformed with a Cas9 expressing plasmid pCas, and the sgRNA expressing plasmid pTargetF-EclacZ (targeting the lacZ gene) was then co-transformed with a DNA repairing fragment containing tetracycline-resistance gene from pACYC184 plasmid (Rose, 1988). The colonies resistant to tetracycline were screened. A single colony from 50 μg/ml tetracycline-containing LB agar plate was confirmed by DNA sequencing and was named as K12-TcR for subsequent conjugation tests.
Conjugation was performed by mixing an overnight donor (K. pneumoniae R10-341) and logarithmic phase recipient (K12-TcR) at a ratio of 4:1 in a total volume of 1 mL as described (Wu et al., 2019). The mixture was then concentrated and spotted onto LB agar without antibiotics at 37°C for 2–4 h to allow conjugation to occur. Since the pR10-341_2 plasmid contains streptomycin resistance gene (Table 1), the conjugated bacterial mixture was plated on LB agar containing 50 μg/ml tetracycline and 50 μg/ml streptomycin to screen for transconjugants carrying plasmid pR10-341_2. Transconjugants were confirmed by PCR using primers paired to K. pneumoniae R10-341 (Kpn-F-FR), K12-TcR (Ec-F-FR), and pR10-341_2 plasmid (P-F-FR), respectively.
The K. pneumoniae R10-341 strain was a clinical isolate from a sputum sample collected before antibiotic treatment from Tongji Hospital in Wuhan, China. This K. pneumoniae R10-341 strain was classified as ST11 and was resistant to different classes of antibiotics, including beta lactams, aminoglycosides, chloramphenicol, rifamycin, quinolones, sulfonamides and macrolides (summarized in Table 1). When testing the minimal inhibitory concentration (MIC) of colistin for this strain, we observed that some of the wells in the 96-well plate tested were resistant, while other wells showed colistin-sensitive phenotype (Figure 1). To exclude the possibility that the tested strain was a mixture of colistin sensitive and resistant, DNA sequences of known colistin resistance associated genes were tested in K. pneumoniae R10-341 parental strain. As summarized in Table 2, phoPQ, pmrAB, crrAB, and mgrB genes were the same as those in drug-sensitive strain, and the mcr1-8 genes could not be successfully amplified in K. pneumoniae R10-341 strain. To further exclude the possibility of bacterial contamination, we streaked the R10-341 strain onto LB plate and selected different single colonies. Similar results were obtained for all these single colonies (data not shown), which suggested that the appearance of colistin resistant colonies for K. pneumoniae R10-341 strain was due to a colistin heteroresistance (CHR).
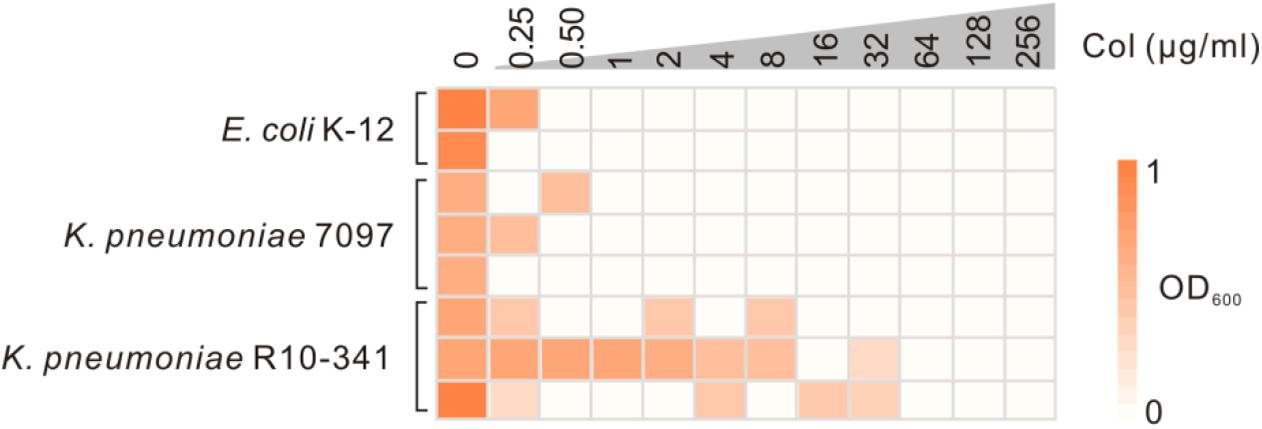
Figure 1. Sensitivity test of the K. pneumoniae R10-341 strain to colistin. E. coli K-12 and K. pneumoniae 7097 were used as control strains. The depth of the color increases with the OD600 values.
To further characterize the K. pneumoniae R10-341 strain, we tested the growth of this strain on LB plates containing 100 μg/mL colistin. Consistent with MIC testing, some colonies (∼10–6) grew on the plate containing colistin, but no colonies were obtained from E. coli K-12 nor from another clinical isolate named K. pneumoniae 7097 on the colistin-containing plate (Figure 2A). To test the mechanism of this colistin resistance, we selected two colonies of the K. pneumoniae R10-341 strain from LB plate without colistin and then streaked onto LB plates containing 100 μg/mL colistin. Again, some colonies from both strains can grow on LB plates containing colistin. We then isolated four colonies from each of these two colistin-containing plates and sequenced the phoPQ, pmrAB, and mgrB genes (Figure 2B). Surprisingly, the amplified mgrB fragments from colistin-resistant colonies were all ∼1.4 kb longer than that from parental colistin-sensitive strains (Figures 2B,C).
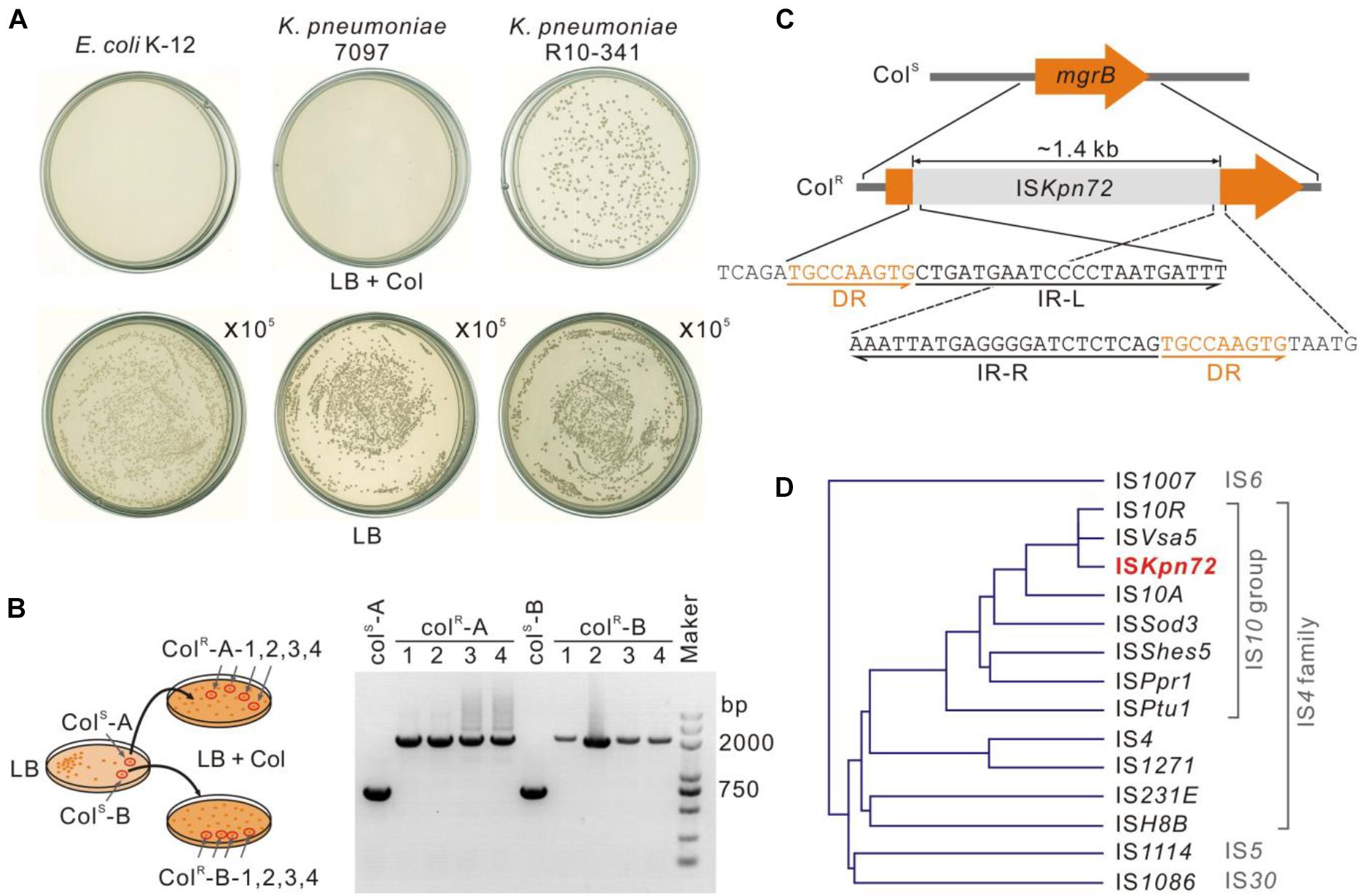
Figure 2. Insertion of an ISKpn72 transposon element into the mgrB gene in colistin-resistant strain. (A) Colonies of E. coli K-12, K. pneumoniae 7097 and K. pneumoniae R10-341 streaked on LB plates containing 100 μg/mL colistin. Each plate was streaked with 100 μL of logarithmic phase bacteria. As a control, 100 μL of 105 diluted bacteria was also spread onto LB plates without colistin. (B) Confirmation of acquisition of colistin resistance for the K. pneumoniae R10-341 strain. Two separated colistin-sensitive colonies were spread onto two LB plates containing 100 μg/mL colistin. Fragment insertions of the mgrB gene in four colonies from each plate were tested by PCR. (C) Insertion of the ISKpn72 element into the mgrB gene in K. pneumoniae R10-341 colistin-resistant colonies. (D) Phylogenetic tree of the IS elements based on amino acid sequences of transposases.
Next, we sequenced this ∼1.4 kb inserted fragment. Sequence alignment in the NCBI database showed this fragment encodes an IS4 family transposase. Further analysis of this ∼1.4 kb inserted fragment in ISfinder suggested that this insertion sequence could be named as ISKpn72 and classified into the IS10 group in the IS4 family, as its sequence is >95% identical to IS10R (Figure 2D). In accordance with this analysis, we identified 22 bp inverted repeats (IR) at both the left and right ends of this insertion fragment and 9 bp direct repeats (DR) around the insertion site (Figure 2C).
The R10-341 strain can become colistin-resistant by inserting the ISKpn72 element into the mgrB gene, but we do not know the source of the ISKpn72 element. We therefore sequenced the genome of the original colistin-sensitive K. pneumoniae R10-341 strain. We obtained a 5.3 Mb chromosome DNA and two plasmid sequences (named pR10-341_1 and pR10-341_2, respectively), which are 5.3Mb, 10.06 kb, and 236.3 kb with G + C contents of 57.46, 55.07, and 52.72%, respectively. According to the PlasmidFinder database, pR10-341_1 and pR10-341_2 harbored ColRNAI and IncR replicon sequence, respectively. Several antibiotic resistance genes were identified both on the chromosome and plasmids, which were in consistent with our drug resistance tests (Table 1). Genome sequencing analysis confirmed that colistin-resistance related genes, including mgrB, phoPQ, and pmrAB, were all the same as the K. pneumoniae reference strain HS11286 (Genome accession: NC_016845), which further supports our hypothesis that the colistin-sensitive strain acquired resistance in the presence of colistin.
In searching for the ISKpn72 element sequence in the whole genome we obtained, we found 12 copies of this ISKpn72 element on the chromosome, and 1 copy on the plasmid pR10-341_2. Similar to that observed in the colistin-resistant R10-341 strain, all copies of the ISKpn72 element contain a pair of 22 bp-length imperfect terminal inverted repeats (IR) (Figure 3A). Surrounding the 12 copies of the ISKpn72 element located on the chromosome are 9-bp direct repeated (DR) sequences. In contrast, the plasmid encoding the ISKpn72 element only contains IR sequences but not the 9 bp-DR sequences (Figure 3A). Therefore, we assumed that the copy without DR on pR10-341_2 might be the root of all the ISKpn72 copies on the chromosome. These analyses suggested that the ISKpn72 element had already been inserted into the chromosome in the parental colistin-sensitive strain.
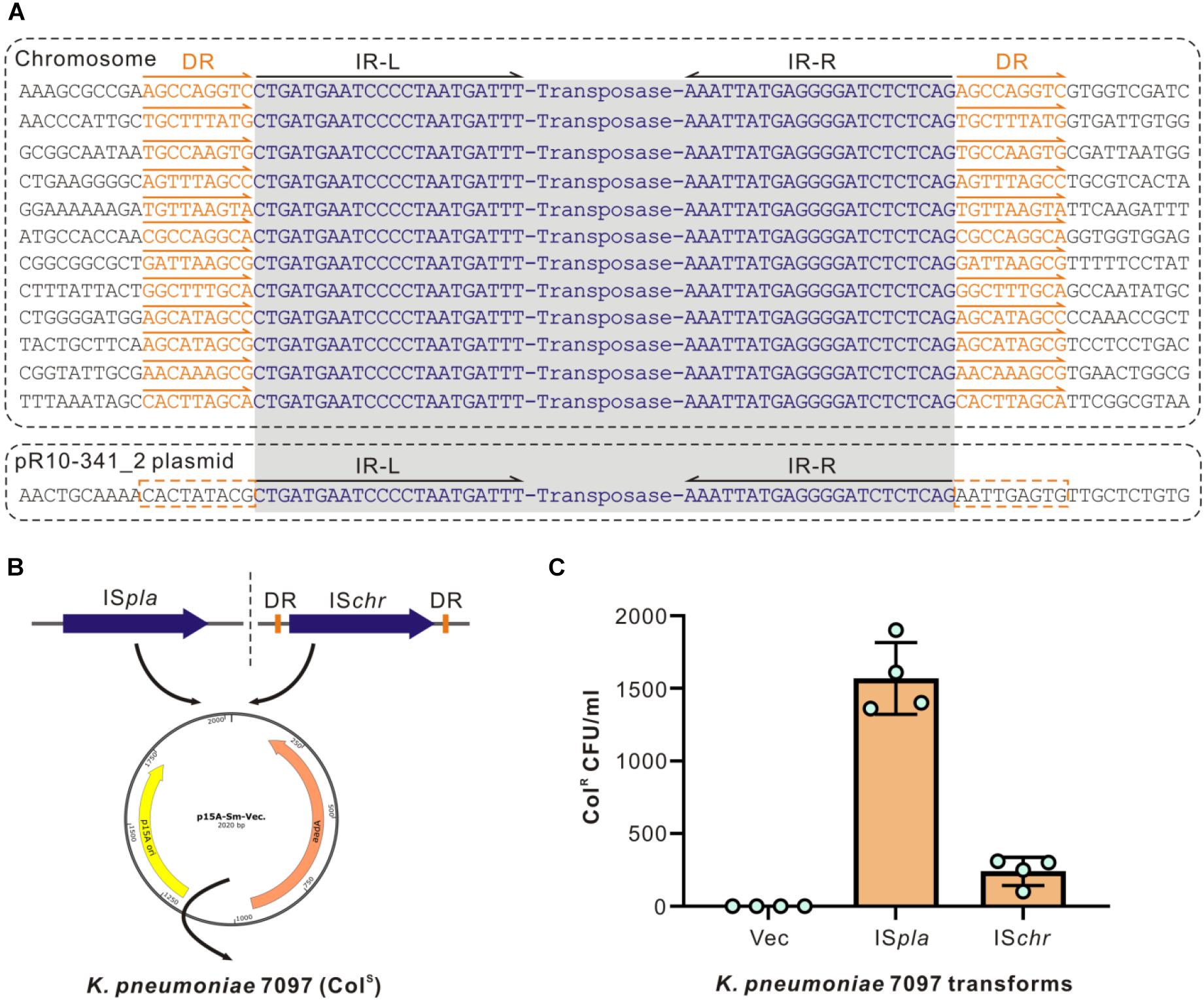
Figure 3. Different types of ISs have different insertion efficiencies. (A) Schematic diagram of ISKpn72 elements located on chromosome and P2 plasmid. IR sequences are shown in blue and the DR sequences are indicated in orange. (B) Schematic diagram for cloning two forms of the ISKpn72 element into colistin-sensitive K. pneumoniae 7097 strain. (C) Acquired colistin-resistance for K. pneumoniae 7097 strain transformed with different ISKpn72 elements. Overnight grown bacteria were spread onto plates containing 100 μg/mL colistin, and bacterial colony forming unit (CFU) numbers were calculated. Representative data from two-independent experiments with four technique replicates are shown.
Whether these existing ISKpn72 elements could be inserted into the mgrB gene to cause colistin-resistance is the next question. We transformed plasmids cloned with either the ISKpn72 element from pR10-341_2 or an ISKpn72 element from the K. pneumoniae R10-341 chromosome (named as ISpla and ISchr, respectively) into the colistin-sensitive strain K. pneumoniae 7097 (Figure 3B). In contrast to the parental K. pneumoniae 7097 strain, the transformation of the plasmid containing either ISpla or ISchr into this colistin-sensitive strain resulted in the growth of some colistin-resistant colonies. Colony forming units (CFU) on LB agar containing 100 μg/mL colistin revealed that the plasmid-derived ISKpn72 was more efficient than the chromosome-derived one in inserting into the mgrB gene to acquire colistin-resistance (Figure 3C). These data suggest that both ISpla and ISchr can be inserted into mgrB gene to acquire the colistin-resistance phenotype and ISpla without DRs had a higher efficiency.
The next question is whether the ISKpn72 element on the pR10-341_2 plasmid could be horizontally transferred to acquire the colistin-resistant phenotype. Sequence alignment of the pR10-341_2 plasmid from the NCBI database showed high similarity to the conjugative multidrug resistant plasmid pR46-270 in a K. pneumoniae R46 isolate (GenBank: CP035776.1). Accordingly, a conjugative system consisting of tra, trb, and finO genes were encoded by the plasmid (Figure 4A). To confirm the transferability of the pR10-341_2, we used K. pneumoniae R10-341 as the donor and a tetracycline-resistant E. coli K12-TcR strain as the recipient to verify plasmid conjugation (Figure 4B). As expected, the E. coli K12-TcR strain containing the pR10-341_2 plasmid was successfully obtained (Figure 4C), suggesting the pR10-341_2 plasmid was transferable. Together, these results demonstrated that the K. pneumoniae R10-341 was able to disseminate the ability to switch from colistin-sensitive to resistant phenotype by transferring an IS containing plasmid.
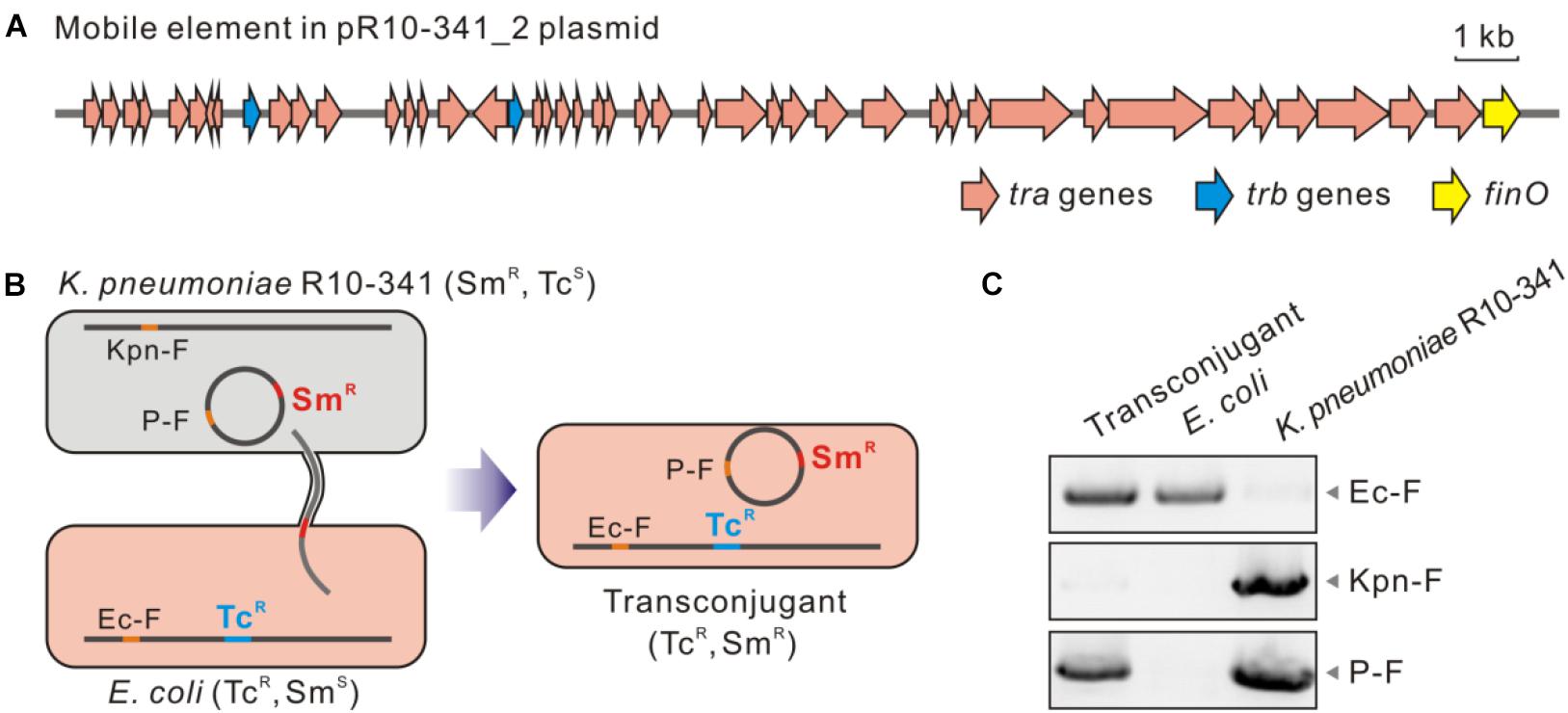
Figure 4. Transconjugation of the pR10-341_2 plasmid from K. pneumoniae R10-341. (A) Genes involved in mobility of the pR10-341_2 plasmid. (B) Strategy used in conjugation transfer analysis. (C) PCR confirmation of transconjugant. Data of amplified products for Ec-F, Kpn-F, and P2-F are shown.
Colistin-resistant bacteria are becoming an increasing threat to healthcare especially in hospitals (Antoniadou et al., 2007; Bogdanovich et al., 2011; Zhong et al., 2018; Nang et al., 2019). Previous studies have focused on mechanisms of drug-resistance and how to detect these resistant strains (Olaitan et al., 2014; Poirel et al., 2017). In this study, we identified a colistin-sensitive K. pneumoniae R10-341 strain with high frequency (∼10–6) of colistin heteroresistance. The K. pneumoniae R10-341 strain was classified as ST11, which is the most widely prevalent CRKP genotype in China and contains the blaKPC-2 gene, which encodes the KPC family carbapenem-hydrolyzing class A beta-lactamase (Qi et al., 2011; Liu et al., 2018). We showed that this K. pneumoniae R10-341 strain could transit from a colistin-sensitive to a resistant phenotype by the insertion of an ISKpn72 element into the mgrB gene. Importantly, we characterized this ISKpn72 element in a mobile plasmid, which showed high similarity to the conjugative plasmid pR46-270 (Wu et al., 2019). These analyses suggest that the ability of transiting from colistin-sensitive to resistant may be disseminated through plasmid transfer.
Heteroresistance has been reported to lead to failures in antibiotic treatment (Band et al., 2016; Band et al., 2019). In CRKP, colistin heteroresistance could be achieved by inserted inactivation and deletion or point mutations in mgrB, phoP, phoQ, lpxM, and yciM genes (Jayol et al., 2015; Halaby et al., 2016). Similar as previous studies, we found that the colistin heteroresistance in K. pneumoniae R10-341 was also caused by inactivation of chromosomal encoded mgrB gene, but we characterized that this inactivation was mediated by a mobile plasmid derived IS element. Therefore, our result is the first to report that the colistin heteroresistance is able to be transferred or spread via plasmid mobilization.
Colistin-resistance in K. pneumoniae can be achieved by chromosome- or plasmid-encoded genes (Ah et al., 2014; Olaitan et al., 2014; Rebelo et al., 2018). Chromosome-encoded mutations are considered to be stable (Gunn et al., 1998), while plasmid encoded mcr genes can be potentially transferred across bacterial species to cause direct colistin-resistance (Liu et al., 2016; Zhong et al., 2018). Being different from spreading of drug-resistance genes, we characterized the spread of the ability to become colistin-resistance via mobile plasmid. The ISKpn72-containing mobile plasmid enables the strain to transit from colistin-sensitive to resistant phenotype by inactivating the mgrB gene, which may also explain the widespread of mgrB gene inactivation in clinical samples (Cannatelli et al., 2014; Gaibani et al., 2014). Interestingly, all copies of the ISKpn72 element contain a pair of 22 bp-length imperfect terminal inverted repeats (IR), which is a characteristic feature of the IS element in bacteria (Rezsöhazy et al., 1993; Mahillon and Chandler, 1998); but only the 12 copies of the ISKpn72 element located on the chromosome contain 9-bp direct repeated (DR) sequences, which are considered as a marker of IS insertion (Mahillon and Chandler, 1998). These observations indicate that the copies of ISKpn72 on the chromosome are probably inserted from the DR-missing ISKpn72 element on the pR10-341_2 plasmid. In addition, we observed a colistin-resistance acquirement of ISchr transformers (Figure 3C), which indicated that the bacteria also can store the ability to become colistin-resistance by inserting IS in chromosome. The ability to transit to colistin-resistant can be both stored at chromosome and disseminated through mobile plasmids.
The mechanism of a mobile plasmid-mediated transfer of the ability to transit from a colistin-sensitive to resistant phenotype brings not only challenges to colistin-resistance detection, but also raises concerns to the use of colistin in clinical treatment. Firstly, this type of colistin-resistant phenotype was generated by the insertion of an ISKpn72 element into the mgrB gene, which means the parental strain may be mis-classified as colistin-sensitive in drug-resistant genotyping or phenotyping assays. Secondly, the ISKpn72 and other IS elements widely exist in prokaryotes (Mahillon and Chandler, 1998; Frost et al., 2005), suggesting transition to colistin-resistance may be widely occurred (Cannatelli et al., 2013, 2014). Different methods have been deployed for antibiotic resistance analysis and prediction, including traditional antimicrobial susceptibility testing (AST) (Jorgensen and Ferraro, 1998) and bioinformatics tools (Zankari et al., 2012; Boolchandani et al., 2019). It is still a challenge to identify whether a strain is able to acquiring inheritable or transmissible antibiotic-resistance, or under specific conditions due to the wide existence of IS elements. Additionally, the frequency of the ISKpn72 insertion into the mgrB gene characterized in our study can reach ∼10–6, indicating that the IS insertion, especially the IS4 family, should receive more attention in the clinical use of colistin.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
MN and YH conceptualized and designed the study. BZ and BY performed the experiments. BZ, WZ, XW, and SC analyzed the data. YW and ZS provided the materials. BZ, MN, and YH drafted the manuscript. BY and SC critically revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the Open Research Fund Program of National Biosafety Laboratory, Wuhan (2018SPCAS001 to MN and YH) and the Youth Innovation Promotion Association CAS (Y201750 to YH).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Core Facility and Technical Support of Wuhan Institute of Virology for help in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.619369/full#supplementary-material
Ah, Y. M., Kim, A. J., and Lee, J. Y. (2014). Colistin resistance in Klebsiella pneumoniae. Int. J. Antimicrob. Agent. 44, 8–15.
Antoniadou, A., Kontopidou, F., Poulakou, G., Koratzanis, E., Galani, I., Papadomichelakis, E., et al. (2007). Colistin-resistant isolates of Klebsiella pneumoniae emerging in intensive care unit patients: first report of a multiclonal cluster. J. Antimicrob. Chemother. 59, 786–790. doi: 10.1093/jac/dkl562
Band, V. I., Crispell, E. K., Napier, B. A., Herrera, C. M., Tharp, G. K., Vavikolanu, K., et al. (2016). Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat. Microbiol. 1:16053.
Band, V. I., Hufnagel, D. A., Jaggavarapu, S., Sherman, E. X., Wozniak, J. E., Satola, S. W., et al. (2019). Antibiotic combinations that exploit heteroresistance to multiple drugs effectively control infection. Nat. Microbiol. 4, 1627–1635. doi: 10.1038/s41564-019-0480-z
Biswas, S., Brunel, J.-M., Dubus, J.-C., Reynaud-Gaubert, M., and Rolain, J.-M. (2012). Colistin: an update on the antibiotic of the 21st century. Expert. Rev. Anti.Infect. Ther. 10, 917–934. doi: 10.1586/eri.12.78
Bogdanovich, T., Adams-Haduch, J. M., Tian, G. B., Nguyen, M. H., Kwak, E. J., Muto, C. A., et al. (2011). Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin. Infec. Dis. 53, 373–376. doi: 10.1093/cid/cir401
Boolchandani, M., D’souza, A. W., and Dantas, G. (2019). Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 20, 356–370.
Cannatelli, A., D’andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agent. Chemother. 57, 5521–5526. doi: 10.1128/aac.01480-13
Cannatelli, A., Giani, T., D’andrea, M. M., Di Pilato, V., Arena, F., Conte, V., et al. (2014). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agent. Chemother. 58, 5696–5703. doi: 10.1128/aac.03110-14
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrobial. Agent. Chemother. 58, 3895–3903. doi: 10.1128/aac.02412-14
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A., and Brisse, S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182. doi: 10.1128/jcm.43.8.4178-4182.2005
El-Halfawy, O. M., and Valvano, M. A. (2015). Antimicrobial heteroresistance: an emerging field in need of clarity. Clin. Microbiol. Rev. 28, 191–207. doi: 10.1128/cmr.00058-14
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Gaibani, P., Lombardo, D., Lewis, R. E., Mercuri, M., Bonora, S., Landini, M. P., et al. (2014). In vitro activity and post-antibiotic effects of colistin in combination with other antimicrobials against colistin-resistant KPC-producing Klebsiella pneumoniae bloodstream isolates. J. Antimicrobial. Chemother. 69, 1856–1865. doi: 10.1093/jac/dku065
Gunn, J. S., Lim, K. B., Krueger, J., Kim, K., Guo, L., Hackett, M., et al. (1998). PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid a modification and polymyxin resistance. Mol. Microbiol. 27, 1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x
Gunn, J. S., and Miller, S. I. (1996). PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178, 6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996
Halaby, T., Kucukkose, E., Janssen, A. B., Rogers, M. R. C., Doorduijn, D. J., Van Der Zanden, A. G. M., et al. (2016). Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrobial. Agent. Chemother. 60, 6837–6843. doi: 10.1128/aac.01344-16
Jayol, A., Nordmann, P., Brink, A., and Poirel, L. (2015). Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrobial. Agent. Chemother. 59, 2780–2784. doi: 10.1128/aac.05055-14
Jiang, Y., Chen, B., Duan, C., Sun, B., Yang, J., Yang, S., et al. (2015). Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514. doi: 10.1128/aem.04023-14
Jorgensen, J. H., and Ferraro, M. J. (1998). Antimicrobial susceptibility testing: general principles and contemporary practices. Clin. Infect. Dis. 26, 973–980. doi: 10.1086/513938
Lei, C. W., Zhang, Y., Wang, Y. T., and Wang, H. N. (2020). Detection of mobile colistin resistance gene mcr-10.1 in a conjugative plasmid from Enterobacter roggenkampii of chicken origin in china. Antimicrob. Agent. Chemother. 64, e1120–e1191.
Li, J., Nation, R. L., Turnidge, J. D., Milne, R. W., Coulthard, K., Rayner, C. R., et al. (2006). Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6, 589–601. doi: 10.1016/s1473-3099(06)70580-1
Liu, L., Feng, Y., Tang, G., Lin, J., Huang, W., Qiao, F., et al. (2018). Carbapenem-resistant isolates of the Klebsiella pneumoniae complex in western china: the common ST11 and the surprising hospital-specific types. Clin. Infect. Dis. 67, S263–S265.
Liu, Y. Y., Chandler, C. E., Leung, L. M., Mcelheny, C. L., Mettus, R. T., Shanks, R. M. Q., et al. (2017). Structural modification of lipopolysaccharide conferred by in gram-negative ESKAPE pathogens. Antimicrob. Agent. Chemother. 61, e517–e580.
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in china: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Macesic, N., Nelson, B., Mcconville, T. H., Giddins, M. J., Green, D. A., Stump, S., et al. (2020). Emergence of polymyxin resistance in clinical Klebsiella pneumoniae through diverse genetic adaptations: a genomic, retrospective cohort study. Clin. Infect. Dis. 70, 2084–2091. doi: 10.1093/cid/ciz623
Nang, S. C., Li, J., and Velkov, T. (2019). The rise and spread of plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841x.2018.1492902
Nordmann, P., Naas, T., and Poirel, L. (2011). Global spread of carbapenemase-producing enterobacteriaceae. Emerg. Infec. Dis. 17, 1791–1798.
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Payne, A., Holmes, N., Rakyan, V., and Loose, M. (2019). BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics 35, 2193–2198. doi: 10.1093/bioinformatics/bty841
Pitout, J. D. D., Nordmann, P., and Poirel, L. (2015). Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agent. Chemother. 59, 5873–5884. doi: 10.1128/aac.01019-15
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/cmr.00064-16
Qi, Y., Wei, Z., Ji, S., Du, X., Shen, P., Yu, Y., et al. (2011). ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66, 307–312. doi: 10.1093/jac/dkq431
Rebelo, A. R., Bortolaia, V., Kjeldgaard, J. S., Pedersen, S. K., Leekitcharoenphon, P., Hansen, I. M., et al. (2018). Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro. Surveill. 23, 17–00672.
Rezsöhazy, R., Hallet, B., Delcour, J., and Mahillon, J. (1993). The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol. Microbiol. 9, 1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x
Rose, R. E. (1988). The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355. doi: 10.1093/nar/16.1.355
Tatusova, T., Dicuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Velkov, T., Deris, Z. Z., Huang, J. X., Azad, M. A., Butler, M., Sivanesan, S., et al. (2014). Surface changes and polymyxin interactions with a resistant strain of Klebsiella pneumoniae. Innate. Immun. 20, 350–363. doi: 10.1177/1753425913493337
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020). Identification of novel mobile colistin resistance gene. Emerg. Microb. Infec. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microb. Infec. 7:122.
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, F., Ying, Y., Yin, M., Jiang, Y., Wu, C., Qian, C., et al. (2019). Molecular characterization of a multidrug-resistant strain R46 isolated from a rabbit. Int. J. Genom. 2019:5459190.
Yang, Y.-Q., Li, Y.-X., Lei, C.-W., Zhang, A.-Y., and Wang, H.-N. (2018). Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1791–1795. doi: 10.1093/jac/dky111
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhong, L. L., Phan, H. T. T., Shen, C., Vihta, K. D., Sheppard, A. E., Huang, X., et al. (2018). High rates of human fecal carriage of mcr-1-positive multidrug-resistant Enterobacteriaceae emerge in china in association with successful plasmid families. Clin. Infect. Dis. 66, 676–685. doi: 10.1093/cid/cix885
Keywords: multidrug resistance, CRKP, mgrB, colistin, phenotype transition
Citation: Zhang B, Yu B, Zhou W, Wang Y, Sun Z, Wu X, Chen S, Ni M and Hu Y (2021) Mobile Plasmid Mediated Transition From Colistin-Sensitive to Resistant Phenotype in Klebsiella pneumoniae. Front. Microbiol. 12:619369. doi: 10.3389/fmicb.2021.619369
Received: 20 October 2020; Accepted: 27 January 2021;
Published: 15 February 2021.
Edited by:
Christophe Merlin, Université de Lorraine, FranceReviewed by:
Ning Dong, City University of Hong Kong, Hong KongCopyright © 2021 Zhang, Yu, Zhou, Wang, Sun, Wu, Chen, Ni and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Ni, bmltaW5nQHRqaC50am11LmVkdS5jbg==; Yangbo Hu, eWJodUB3aC5pb3YuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.