
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 12 April 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.617349
This article is part of the Research TopicBacterial Pathogen Genomics: Recent Achievements, Current Applications and Future ChallengesView all 16 articles
Extended-spectrum beta-lactamase (ESBL)-producing bacterial isolates are emerging within the last years. To understand this emergence, a thorough genome-based analysis of ESBL isolates from different sources (One Health approach) is needed. Among these, analysis of surface water is underrepresented. Therefore, we performed a genome-based analysis of ESBL-producing Escherichia coli isolates from surface water samples. Water samples were collected from eleven different surface water sites (lakes, river). ESBL-producing E. coli were recovered from these samples using filters and chromogenic media. Whole-genome sequencing of ESBL-producing E. coli was performed followed by determination of the multilocus sequence type (ST), ESBL-type, and virulence genes. Phylogenetic analysis was done using single nucleotide analysis. From all water samples taken, nineteen ESBL-producing E. coli were recovered. All of them harbored an ESBL gene. Nine different multilocus STs were determined, among which ST-949 was the ST detected most frequently. Phylogenetic analysis of ST-949 isolates revealed that all those isolates were closely related. In addition, they harbored an identical chromosomal insertion of blaCTX–M–15, indicating a clonal relationship among these isolates. Genetic comparison with isolates from all over the world revealed that these isolates were closely related to human clinical isolates derived from New Zealand and Sweden. An ESBL-producing E. coli ST-949 clone was detected in German surface waters. Its close relationship to human clinical isolates suggests its ability to colonize or even infect humans. Our findings reveal that water sources indeed may play a hitherto underreported role in spread of ESBL-producing isolates.
Extended-spectrum beta-lactamase (ESBL)-producing bacterial isolates are emerging in the last years (Peirano and Pitout, 2019). The spread of ESBL-producers is a clear One Health issue, as they have been found to be present in different sources, animals, humans, and environment (Hooban et al., 2020). This is true for Germany as well. In Germany, 6.3% of humans are colonized with ESBL-producing Escherichia coli isolates (Valenza et al., 2014). In diseased food-producing animals, the prevalence of ESBL-producing E. coli ranges between 0.8 and 11.2% depending on the animal species (Michael et al., 2017).
The commonly accepted opinion is that all different sources play a role in the spread of ESBL-producing bacteria. To be able to track the transfer routes of ESBL-producers among different sources, a thorough understanding of the epidemiology of these bacteria is needed. The method of choice to perform an in-depth epidemiological analysis is to use whole genome sequence-based methods. They have been used a lot in human and veterinary medicine (in particular to track outbreaks), but genome-based data from water sources are still very rare. Few epidemiological studies have been performed to analyze the genomes of ESBL-producing isolates from water samples. These studies showed a high identity between ESBL producers from water samples and clinical samples indicating a spread from either clinical to water sources or vice versa (Fagerström et al., 2019).
In order to gain more insight into this topic, an investigation was performed that included water samples from official and unofficial bathing sites at lakes and a river in Hesse, Germany.
During the bathing season 2018, samples were taken from swimming lakes in Hesse (n = 10). According to the European Bathing Water Directive (BWD; EG 2006/7) the sites were checked at least monthly for the presence of coliform bacteria. Additionally, samples from unofficial bathing sites of the Hessian river Lahn around Marburg and Giessen were taken (n = 9). Procedures for sampling as well as preparation, filtration, and enumeration were performed conforming with the DIN EN ISO 9308-2 (K6-1) 07-2014, DIN EN ISO 19458 (K19), DIN EN ISO 8199 (K20) 01-2008, and DIN EN ISO 9380-1: 2014 (K12) regulations within 24 h.
For detection of ESBL-producing isolates, water samples were filtered and the filters put onto BrillianceTM ESBL chromogenic medium (OXOID, Wesel, Germany). For isolates growing on the chromogenic agar, species confirmation was performed using MALDI-TOF-MS (Biomérieux, Nürtingen, Germany). Antibiotic susceptibility testing and ESBL phenotype confirmation was performed using the VITEK 2 System (AST-N263 cards, Biomérieux, Nürtingen, Germany). Classification of the antibiotic resistance/susceptibility was performed according to EUCAST criteria1.
Short-read whole genome sequencing was performed for all E. coli isolates growing on the chromogenic medium (n = 21). DNA from overnight cultures was isolated using the Purelink genomic DNA kit (ThermoFisher, Dreieich, Germany). Short read sequencing was performed on a NextSeq 500 machine (Illumina, Eindhoven, Netherlands) using a Nextera XT sequencing library with an average read length of 115 nt and an average coverage of 33.5 x. Raw reads were processed using the ASA3P pipeline using default parameters (Schwengers et al., 2020).
Long-read sequencing of a representative E. coli ST-949 isolate (EDCC5518) was performed using the Nanopore technology. The library was prepared using the native barcoding kit (EXP-NBD103, Oxford Nanopore Technologies Ltd., Oxford, United Kingdom) and 1D chemistry (SQK-LSK108). Sequencing was performed using the SpotON Flow Cell Mk I R9 Version (FLO-MIN106) on a MinION/MinIT machine with an average read length of 4,137 nt. Basecalling was performed directly on the MinIT machine. Demultiplexing was performed using Porechop (v. 0.2.32). Hybrid assembly was performed using Unicycler (v. 0.4.7) (Wick et al., 2017) and the short and long reads with default parameters.
In silico multilocus sequence typing of E. coli isolates was performed using the scheme presented by Wirth et al. (2006). Antibiotic resistance genes, plasmid incompatibility groups and fimH types were determined using the bacterial analysis pipeline of the Center for Genomic Epidemiology3. Insertion elements were determined using ISFinder (Siguier et al., 2006). Virulence gene determination was performed using ASA3P (Schwengers et al., 2020). Comparative genome analysis was performed using the HarvestSuite package (Treangen et al., 2014). Publicly available assembled E. coli genomes of the multilocus sequence type (ST) ST-949 were downloaded using Enterobase (as of 8th June 2020, Supplementary Table 1) (Zhou et al., 2020). Geographical representation of sampling sites was visualized using MicroReact (Argimón et al., 2016).
During the bathing season of 2018 (June–August), fifty-five samples from nineteen sampling sites were collected. Of these samples, forty-four did not show growth of isolates on ESBL chromogenic agar. Notably, the 2018 summer was a comparatively hot summer4 resulting in low water levels. From the remaining water samples (n = 11, Figure 1), nineteen ESBL-producing bacterial isolates were detected (Table 1). Only E. coli isolates were detected. Environmental data and characterization of the sampling sites are shown in Table 1.
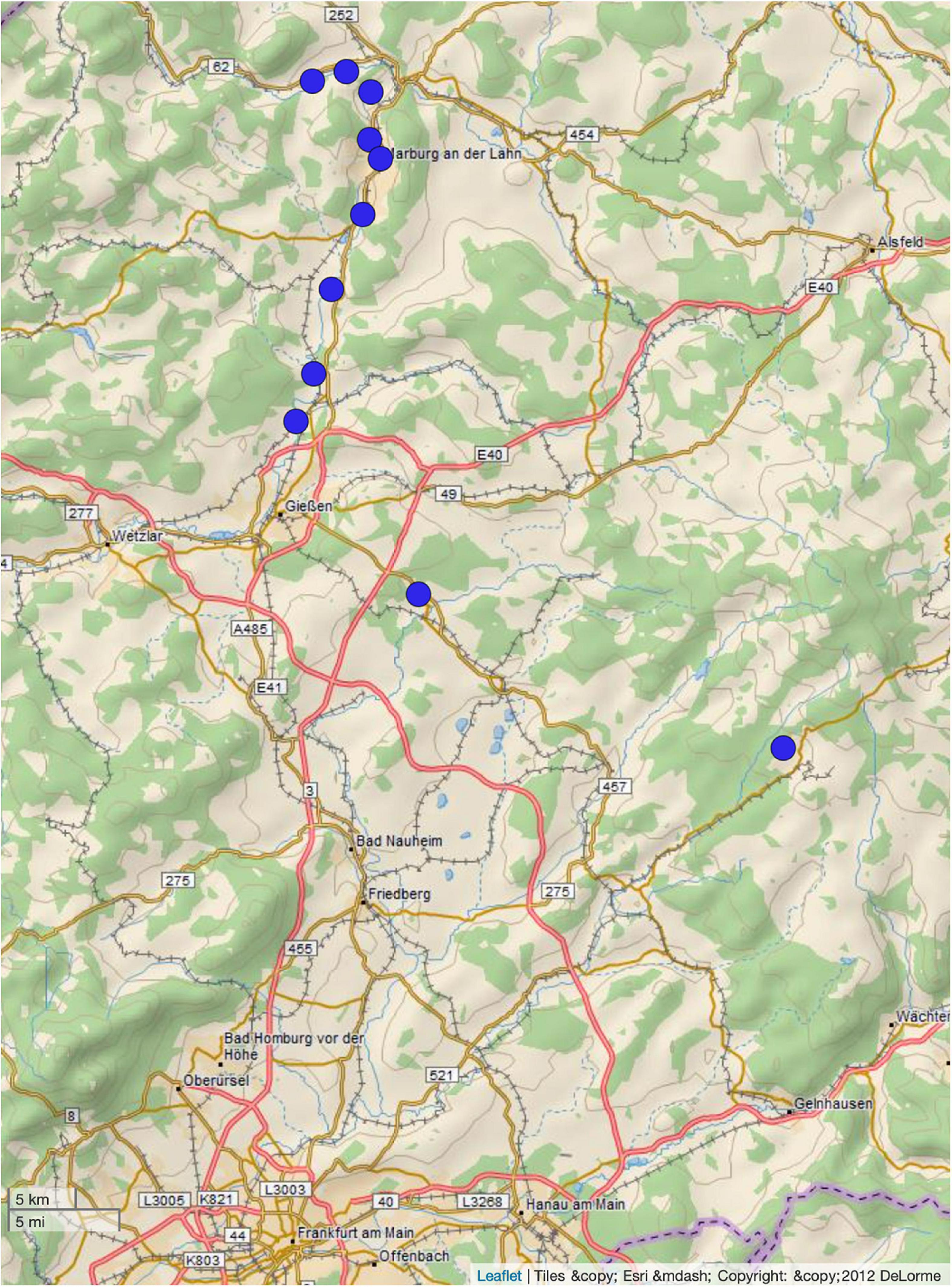
Figure 1. Distribution of sampling sites. The figure was generated using microreact (Argimón et al., 2016).
For all E. coli isolates growing on the chromogenic plates, the ESBL phenotype was confirmed. Phenotypic resistance to antibiotics other than beta-lactams was detected very seldom and included resistance to fluoroquinolones (4/21) and trimethoprim/sulfamethoxazole (5/21) (Supplementary Table 2). The isolates were not resistant to carbapenems. According to the classification proposed by Magiorakos et al. (2012), all isolates were multidrug-resistant (resistant to ≥3 different antibiotic classes; Supplementary Table 2).
All E. coli isolates harbored an ESBL gene (Table 2). The most common ESBL gene detected was blaCTX–M–15 (n = 17) followed by blaCTX–M–1 (n = 2) and blaCTX–M–27 (n = 2). The predominance of blaCTX–M–15 in our study is concordant with the results in other studies performed in Europe (Kittinger et al., 2016; Jorgensen et al., 2017). The E. coli isolates encoded other antibiotic resistance genes conferring resistance to aminoglycosides (7/21), fluoroquinolones (15/21, qnrS1), sulfonamide (5/21), trimethoprim (5/21), and tetracycline (2/21) (Table 2). In concordance with previous reports (Rodríguez-Martínez et al., 2011), the presence of qnrS1 did not lead to high-level fluoroquinolone resistance (MIC > 0.5 mg/L) in our isolates.
Multilocus sequence typing revealed that nine different STs were present (Table 2 and Figure 2). Of these, three were detected more than once: ST-949 (n = 11), ST-131 (n = 2), and ST-1431 (n = 2). E. coli ST-949 and ST-1431 isolates harbored blaCTX–M–15, while E. coli ST-131 isolates harbored blaCTX–M–15 or blaCTX–M–27.
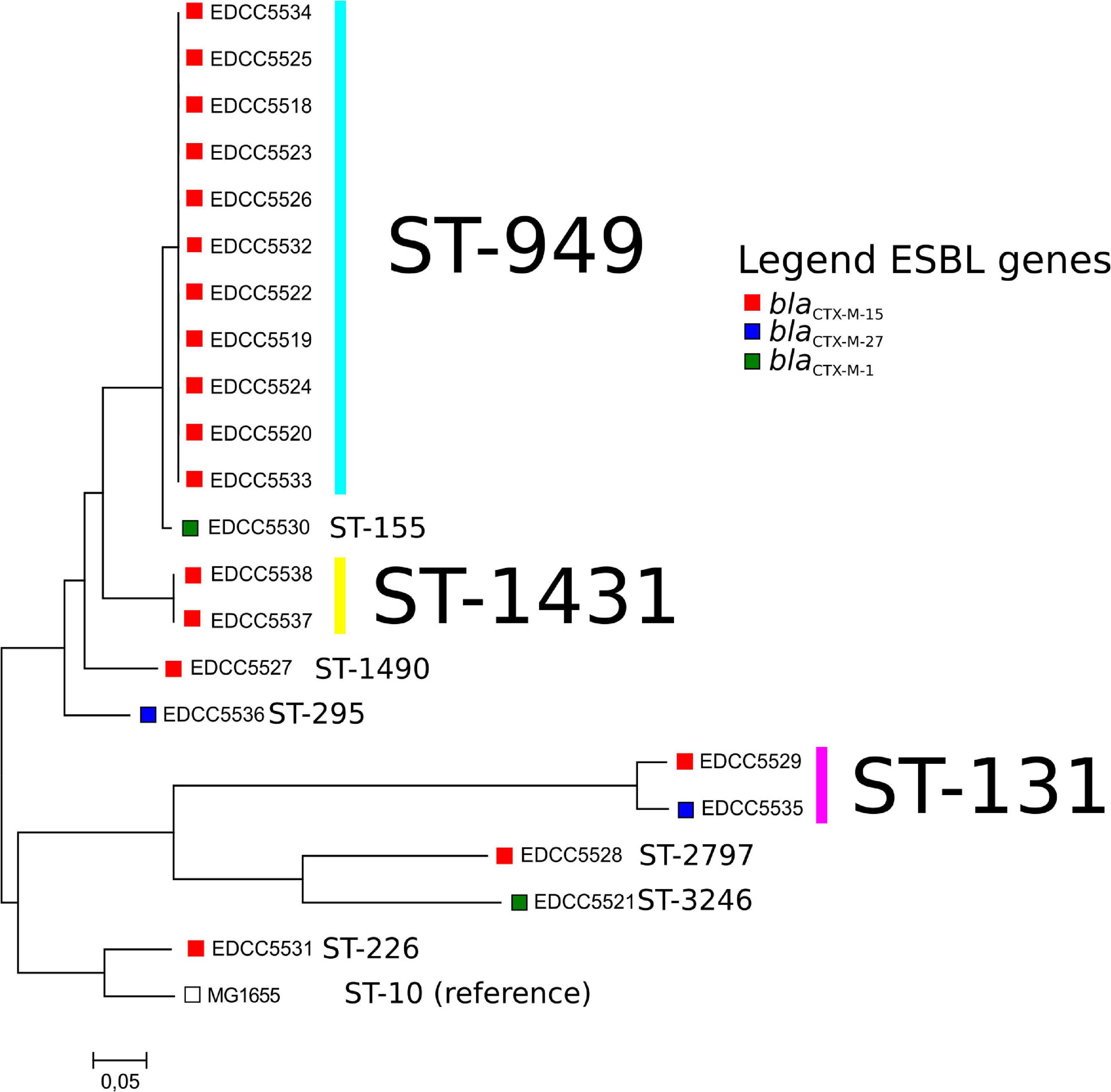
Figure 2. Core-genome-based comparison of the E. coli isolates analyzed in this study. Boxes depict the ESBL gene present in these isolates. For a representation of the isolate geographical location and the phylogeny, please see the following link: https://microreact.org/project/bELapwXL1Fi8ZdznzZHF1H.
To our knowledge, E. coli ST-949 have been reported in only five publications worldwide, indicating that this ST is less frequent and might represent an emerging clone (Oh et al., 2014; Potron et al., 2017; Potel et al., 2018; Fagerström et al., 2019; Sedrati et al., 2020). The total number of publicly available E. coli ST-949 isolates in the Enterobase database is 41 [as of 11th August 2020 (Zhou et al., 2020)], a very low number compared with frequent multilocus STs as e.g., ST-131 (n = 9202, as of 11th August 2020).
ESBL-producing ST-1431 E. coli isolates have been detected more often in animal sources (livestock, pets, wild animals) than in humans (Rocha-Gracia et al., 2015; Bachiri et al., 2017; Seiffert et al., 2017).
In this study, we detected two ST131 isolates. E. coli ST-131 are frequently associated with human clinical infections (Nicolas-Chanoine et al., 2014), in particular those depicting the fimH type H30 and harboring CTX-M-15 or CTX-M-27 (Nicolas-Chanoine et al., 2014; Stoesser et al., 2016). EDCC5529 depicted the fimH41 fimH-type and harbored blaCTX–M–15. EDCC5535 depicted a fimH30 fimH type and characteristic properties of the ST-131 C1-M27 clade (Matsumura et al., 2016): blaCTX–M–27, the GyrA S83L/D87N and ParC S80I/E84V mutations leading to fluoroquinolone resistance and the M27PP1 phage. Therefore, it is a member of the C1-M27 clade usually associated with human isolates (Matsumura et al., 2016; Ghosh et al., 2017). Thus, EDCC5535 might have originated from human sources.
The most common ST within the ESBL E. coli was ST-949 (Table 2 and Figure 2). Therefore, we analyzed these isolates in more detail. E. coli ST-949 is known to be associated with carbapenem-resistance (Potron et al., 2017; Potel et al., 2018) or ETEC pathotypes (Oh et al., 2014). They have been isolated from environmental samples (water samples) collected in Sweden, where the authors could show that the water isolates were highly related with isolates derived from a hospital that was adjoining the water source (Fagerström et al., 2019).
Because E. coli ST-949 are known to be pathogenic (Oh et al., 2014), we analyzed all available E. coli ST-949 isolates for the presence of virulence genes. The ST-949 isolates from this study harbored only ExPEC virulence genes (e.g., iron acquisition genes, Enterobactin, Supplementary Figure 2). The isolates detected in New Zealand and Sweden harbored the same sets of virulence genes. Other ST-949 harbored also toxins (Shigatoxin) and hemolysins indicating that ST-949 isolates differ widely in their virulence capabilities.
A whole-genome-based analysis of the E. coli ST-949 isolates from this study and those from Enterobase revealed two different findings (Figure 3 and Supplementary Table 1): Firstly, ST-949 isolates are divided into two different clusters. Cluster A (including our isolates) consists of isolates found in water, human, and livestock samples, while cluster B includes also isolates from companion animals. Secondly, only cluster A isolates contain CTX-M-15, while Cluster B isolates harbor either CTX-M alleles other than CTX-M-15, or AmpC beta-lactamases. Cluster B harbored two ST-949 isolates from Germany from livestock and an unknown source.
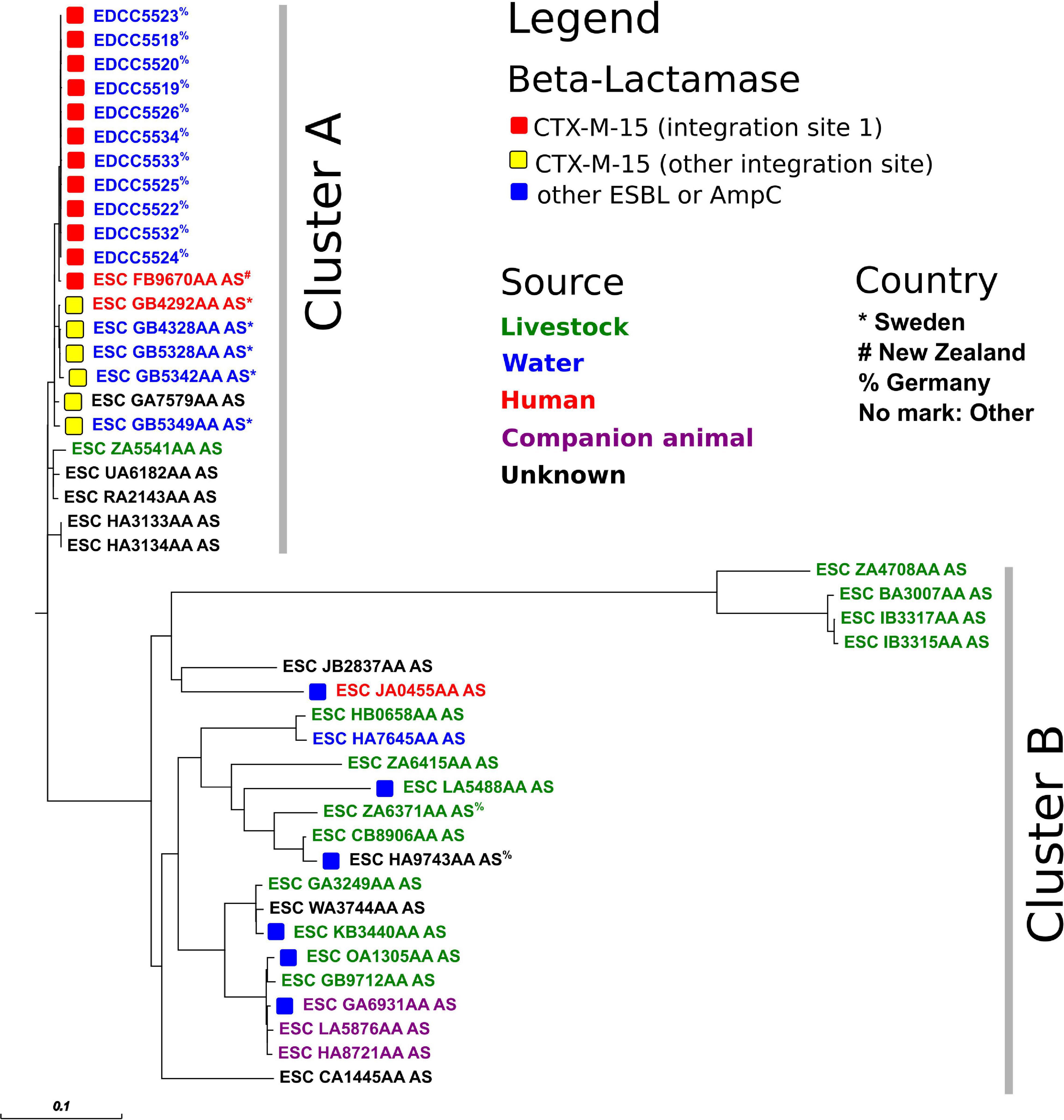
Figure 3. Core-genome-based phylogenetic tree of E. coli ST-949 isolates from this study and Enterobase (as of 8th June 2020). For a representation of the isolate sampling site, please see Supplementary Figure 1.
The E. coli ST-949 isolates from this study (Cluster A, n = 11) were highly related to ST-949 isolates found in New Zealand (ESC_FB9670AA_AS, human isolate) and Sweden (n = 4, water and human isolates, Figure 3). All these 16 isolates harbored a complex antibiotic resistance gene region including not only the blaCTX–M–15 gene, but also the fluoroquinolone resistance gene qnrS1 and several different insertion sequences (Supplementary Figure 3). The antibiotic resistance region was inserted in the chromosome at an identical location in the isolates from this study and the isolates from New Zealand, while the isolates from Sweden harbored the identical region inserted at a different location of the chromosome. This finding indicates that the acquisition of blaCTX–M–15 in the two different clones was presumably from a different source and was independent in both clones.
The epidemiological link between Germany and New Zealand is not clear. It may indicate that the ST-949 clone found in Germany is present worldwide, but this is only an assumption as the total number of ST-949 isolates throughout the world is very low. It remains to be clarified whether ST-949 is an emerging ST and whether it is present in other sources.
The epidemiological link between ST-949 isolates from our study is partly explainable. All ST-949 river isolates originate from the same river (sampling sites 10, 11, 16, and 17), indicating a common source of contamination. Possible sources of contamination along the river might be either agriculture, two large university hospitals whose cleared wastewater end up in the river itself or human influence through tourism, as the river is frequently used for recreational purposes. Sampling site 6 is located close to the sampled river, indicating a possible contamination through the river by flooding. What is not completely clear, is the epidemiological link between sampling site 1 and the other sites. They are not interconnected by any water flows. A possible connection between those might have been movement of humans or animals (in particular birds).
ST-949 isolates have never been reported in Germany. Therefore, this is the first study detecting ST-949 E. coli in Germany. Its predominance in our study indicates that either ST-949 E. coli might resemble E. coli isolates only present in water sources or a new emerging multilocus ST in Germany. To prove this hypothesis, more studies are required.
In this study, we characterized ESBL-producing E. coli isolates from water samples. Our results show that the main MLST type is ST-949, reported in only a few number of very recent publications. In addition, it has been associated with human disease. This indicates that it might be an emerging ST with human pathogenic potential that could spread through water sources.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA656216.
ED designed the study. AN and SH collected samples and data. ED performed antibiotic resistance determination. LF, AN, JF, and ED analyzed the data. LF, AN, and ED wrote the manuscript that was critically reviewed and approved by all authors.
This study has been funded through the Hessian Competence Center for Clinical Hygiene (HuKKH) funded by the Hessian Ministry for Education and Research and the German Center for Infection Research (Grant # 8032808811) funded by the Federal Ministry of Education and Research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Silke Zechel-Gran and Christina Gerstmann for excellent technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.617349/full#supplementary-material
Argimón, S., Abudahab, K., Goater, R. J. E., Fedosejev, A., Bhai, J., Glasner, C., et al. (2016). Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genomics 2:e000093. doi: 10.1099/mgen.0.000093
Bachiri, T., Bakour, S., Ladjouzi, R., Thongpan, L., Rolain, J. M., and Touati, A. (2017). High rates of CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae in wild boars and barbary macaques in Algeria. J. Glob. Antimicrob. Resist. 8, 35–40. doi: 10.1016/j.jgar.2016.10.005
Fagerström, A., Mölling, P., Khan, F. A., Sundqvist, M., Jass, J., and Söderquist, B. (2019). Comparative distribution of extended-spectrum beta-lactamase–producing Escherichia coli from urine infections and environmental waters. PLoS One 14:e0224861. doi: 10.1371/journal.pone.0224861
Ghosh, H., Doijad, S., Falgenhauer, L., Fritzenwanker, M., Imirzalioglu, C., and Chakraborty, T. (2017). blaCTX–M–27–Encoding Escherichia coli sequence type 131 lineage C1-m27 clone in clinical isolates, Germany. Emerg. Infect. Dis. 23, 1754–1756. doi: 10.3201/eid2310.170938
Hooban, B., Joyce, A., Fitzhenry, K., Chique, C., and Morris, D. (2020). The role of the natural aquatic environment in the dissemination of extended spectrum beta-lactamase and carbapenemase encoding genes: a scoping review. Water Res. 180, 1–12. doi: 10.1016/j.watres.2020.115880
Jorgensen, S. B., Soraas, A. V., Arnesen, L. S., Leegaard, T. M., Sundsfjord, A., and Jenum, P. A. (2017). A comparison of extended spectrum beta-lactamase producing Escherichia coli from clinical, recreational water and wastewater samples associated in time and location. PLoS One 12:e0186576. doi: 10.1371/journal.pone.0186576
Kittinger, C., Lipp, M., Folli, B., Kirschner, A., Baumert, R., Galler, H., et al. (2016). Enterobacteriaceae isolated from the river danube: antibiotic resistances, with a focus on the presence of ESBL and carbapenemases. PLoS One 11:e0165820. doi: 10.1371/journal.pone.0165820
Magiorakos, A.-P. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Matsumura, Y., Pitout, J. D. D., Gomi, R., Matsuda, T., Noguchi, T., Yamamoto, M., et al. (2016). Global Escherichia coli sequence type 131 clade with blaCTX–M–27 gene. Emerg. Infect. Dis. 22, 1900–1907. doi: 10.3201/eid2211.160519
Michael, G. B., Kaspar, H., Siqueira, A. K., de Freitas Costa, E., Corbellini, L. G., Kadlec, K., et al. (2017). Extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolates collected from diseased food-producing animals in the GERM-Vet monitoring program 2008–2014. Vet. Microbiol. 200, 142–150. doi: 10.1016/j.vetmic.2016.08.023
Nicolas-Chanoine, M.-H., Bertrand, X., and Madec, J.-Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-13
Oh, K. H., Kim, D. W., Jung, S. M., and Cho, S. H. (2014). Molecular characterization of enterotoxigenic Escherichia coli strains isolated from diarrheal patients in Korea during 2003-2011. PLoS One 9:e96896. doi: 10.1371/journal.pone.0096896
Peirano, G., and Pitout, J. D. D. (2019). Extended-spectrum β−Lactamase-producing Enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 79, 1529–1541. doi: 10.1007/s40265-019-01180-3
Potel, C., Ortega, A., Martinez-Lamas, L., Bautist, V., Regueiro, B., and Oteo, J. (2018). Interspecies transmission of the blaOXA–48 gene from a Klebsiella pneumoniae high-risk clone of sequence type 147 to different Escherichia coli clones in the gut microbiota. Antimicrob. Agents Chemother. 62, 2017–2019. doi: 10.1128/AAC.01699-17
Potron, A., Bernabeu, S., Cuzon, G., Pontiès, V., Blanchard, H., Seringe, E., et al. (2017). Analysis of OXA-204 carbapenemase-producing Enterobacteriaceae reveals possible endoscopyassociated transmission, France, 2012 to 2014. Eurosurveillance 22, 1–8. doi: 10.2807/1560-7917.ES.2017.22.49.17-00048
Rocha-Gracia, R. C., Cortés-Cortés, G., Lozano-Zarain, P., Bello, F., Martínez-Laguna, Y., and Torres, C. (2015). Faecal Escherichia coli isolates from healthy dogs harbour CTX-M-15 and CMY-2 β-lactamases. Vet. J. 203, 315–319. doi: 10.1016/j.tvjl.2014.12.026
Rodríguez-Martínez, J. M., Cano, M. E., Velasco, C., Martínez-Martínez, L., and Pascual, Á (2011). Plasmid-mediated quinolone resistance: an update. J. Infect. Chemother. 17, 149–182. doi: 10.1007/s10156-010-0120-2
Schwengers, O., Hoek, A., Fritzenwanker, M., Falgenhauer, L., Hain, T., Chakraborty, T., et al. (2020). ASA3P: an automatic and scalable pipeline for the assembly, annotation and higher level analysis of closely related bacterial isolates. PLoS Comput. Biol. 16:e1007134. doi: 10.1371/journal.pcbi.1007134
Sedrati, T., Menoueri, M. N., Tennah, S., Ngaiganam, E. P., Azzi, O., Chadi, H., et al. (2020). Molecular characterization of multidrug resistant Escherichia coli isolated from milk of dairy cows with clinical mastitis in Algeria. J. Food Prot. 83, 2173–2178. doi: 10.4315/JFP-20-198
Seiffert, S. N., Carattoli, A., Schwendener, S., Collaud, A., Endimiani, A., and Perreten, V. (2017). Plasmids Carrying bla(CMY –2/4) in Escherichia coli from poultry, poultry meat, and humans belong to a novel IncK subgroup designated IncK2. Front. Microbiol. 8:407. doi: 10.3389/fmicb.2017.00407
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Stoesser, N., Sheppard, A. E., Pankhurst, L., De Maio, N., Moore, C. E., Sebra, R., et al. (2016). Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio 7:e2162. doi: 10.1128/mBio.02162-15
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Valenza, G., Nickel, S., Pfeifer, Y., Eller, C., Krupa, E., Lehner-Reindl, V., et al. (2014). Extended-spectrum-β-lactamase-producing Escherichia coli as intestinal colonizers in the German community. Antimicrob. Agents Chemother. 58, 1228–1230. doi: 10.1128/AAC.01993-13
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Keywords: CTX-M-15, ST-949, ESBL-E. coli, water samples, WGS
Citation: Falgenhauer L, zur Nieden A, Harpel S, Falgenhauer J and Domann E (2021) Clonal CTX-M-15-Producing Escherichia coli ST-949 Are Present in German Surface Water. Front. Microbiol. 12:617349. doi: 10.3389/fmicb.2021.617349
Received: 14 October 2020; Accepted: 24 March 2021;
Published: 12 April 2021.
Edited by:
Edward Feil, University of Bath, United KingdomReviewed by:
Jean-Yves Madec, Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (ANSES), FranceCopyright © 2021 Falgenhauer, zur Nieden, Harpel, Falgenhauer and Domann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linda Falgenhauer, TGluZGEuZmFsZ2VuaGF1ZXJAbWlrcm9iaW8ubWVkLnVuaS1naWVzc2VuLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.