- Department of Rheumatology and Immunology, Frontiers Science Center for Disease-Related Molecular Network, West China Hospital, Institute of Immunology and Inflammation, Sichuan University, Chengdu, China
Quorum-sensing (QS) system is a rapidly developing field in which we are gradually expanding our understanding about how bacteria communicate with each other and regulate their activities in bacterial sociality. In addition to collectively modifying bacterial behavior, QS-related autoinducers may also be embedded in the crosstalk between host and parasitic microbes. In this review, we summarize current studies on QS in the intestinal microbiome field and its potential role in maintaining homeostasis under physiological conditions. Additionally, we outline the canonical autoinducers and their related QS signal-response systems by which several pathogens interact with the host under pathological conditions, with the goal of better understanding intestinal bacterial sociality and facilitating novel antimicrobial therapeutic strategies.
Introduction
Microorganisms are omnipresent in nature and form a significant part of our micro- and macro-environment. The human body is home to several microorganisms, especially at mucosal sites. The first studies suggestive of quorum-sensing (QS) as a mode of cell to cell communication within bacterial communities were published in 1965 (Bassler and Losick, 2006). Prior to this, researchers focused on eliminating bacteria using conventional antimicrobials. In the 1950s, success in rescuing Clostridium difficile infection using fecal microbiota transplantation (FMT) introduced a new perspective, i.e., introduction of a mixed microbial community, instead of eliminating specific bacteria, may benefit the host in fighting microbial infection. Around the same time, a phenomenon of intercellular communication in specific bacterial communities was discovered (Tomasz, 1965; Bassler and Losick, 2006). Researchers began to realize that gregarious communication is common in bacterial sociality, and named it the QS system (Bassler and Losick, 2006).
QS is a cell-cell communication process that enables bacteria to feel their surrounding environment and to regulate their density and behavior, which allows them to live like multicellular organisms (Bassler and Losick, 2006). They make use of their community to manage self-competition, as well as collectively interact with their host. Gut microbiomes can be identified and characterized from feces or other samples using 16S ribosomal RNA (rRNA) gene sequencing technology. Here, we summarize the classical QS systems in intestinal bacteria and highlight the newly discovered functions mediated by QS. Moreover, the potential roles of QS in host-bacterial interaction are also discussed. It might provide a new perspective for microbiome researchers to advance toward development of new medicines to treat devastating microbial infection and associated pathologies.
Classical QS in Bacterial Communication
QS is defined as a bacterial cell-cell communication process that controls bioluminescence, competence, antibiotic production, sporulation, biofilm formation, and virulence factor secretion in bacteria. Recent research has shown that the scope of QS extends to inter-kingdom communication, mediated by several newly identified extra-cellular signaling molecules called autoinducers (AIs). Among these AIs, five main signaling molecules are involved in the classical QS system (Figure 1).
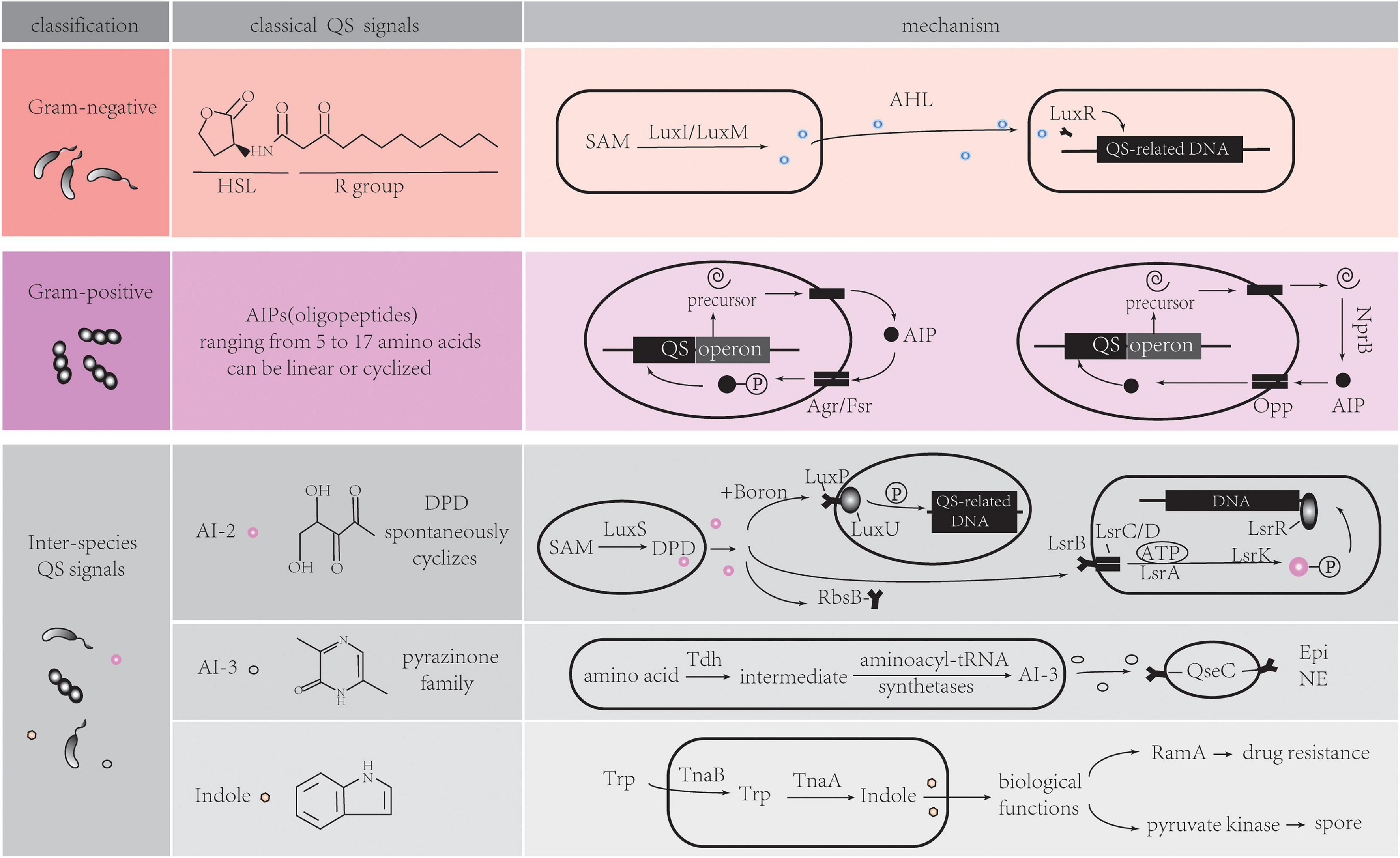
Figure 1. Five main signaling molecules in QS system. HSL, homoserine lactone; SAM, S-adenosylmethionine; AHL, acyl-homoserine lactones; AIP, autoinducing Peptides; NprB, neutral protease B; Opp, oligopeptide permease system; AI, autoinducer; DPD, 4,5-dihydroxy-2,3-pentanedione; Tdh, threonine dehydrogenase; Epi, epinephrine; NE, norepinephrine; Trp, tryptophan; Tna, tryptophanase. Detailed information can be found in section “Classical QS in Bacterial Communication.”
Acyl-Homoserine Lactones (AHL) QS System in Gram-Negative Bacteria
Both Gram-positive (Gram+) and Gram-negative (Gram–) bacteria use QS to communicate with each other. Although the types of QS pathways are different in Gram+ and Gram– bacteria, they all have fundamental biological roles. Gram– QS system has several common features (Ng and Bassler, 2009; Papenfort and Bassler, 2016). Firstly, AIs are molecules synthesized from S-adenosylmethionine (SAM) as a substrate. Acyl-homoserine lactones (AHLs) are the most common class of AIs. They have an N-acylated homoserine-lactone ring as the core and a 4–18 carbon acyl chain containing modifications. The length of the acyl chain affects their stability (von Bodman et al., 2008; Galloway et al., 2011). LuxI-type enzymes are the major, but not the sole producer of AHLs. A LuxM synthase found in Vibrio harveyi, which is not a homolog of LuxI, can produce AHLs for their intra-species communication (Bassler et al., 1993). SAM can be metabolized into special signals which can be sensed by different bacteria species. Diffusible signal factor (DSF) type molecules are synthesized by RpfF proteins in Pseudomonas aeruginosa and Burkholderia cenocepacia (Ryan et al., 2015; Zhou et al., 2015). Cholera autoinducer 1 (CAI-1) is produced by the CAI-1 AI synthase (CqsA) in Vibrio cholerae. Due to the common prevalence of homologs of CqsA in Vibrio spp., various CAI-1 are produced by this bacterial species. Additionally, Vibrio spp. may have different affinities to CAI-1 not produced by themselves, suggesting CAI-1 is a vibrio inter-genus communication molecule (Miller et al., 2002; Higgins et al., 2007; Rutherford and Bassler, 2012; Papenfort and Bassler, 2016).
Secondly, AIs bind to specific membrane receptors or cytoplasmic proteins (Papenfort and Bassler, 2016). LuxR-type receptors, the cytoplasmic transcription factors, detect freely diffusible AHLs in cytoplasm and bind cognate AHLs. The stable LuxR-AHL complexes can bind to DNA while unbound LuxR proteins are rapidly degraded (Zhu and Winans, 2001; Rutherford and Bassler, 2012; Figure 1, Gram negative part). These LuxR/LuxI-type systems, including LasR/LasI and RhlR/RhlI in Pseudomonas aeruginosa, mediate inter-cellular communication (Papenfort and Bassler, 2016). Some annotated LuxR proteins belong to LuxR-solo receptor or orphan LuxR class. In the absence of LuxI synthases, they detect different AHL molecules produced by other bacterial species, thus mediating inter-species communication. Examples of these are QscR in P. aeruginosa (Zhu and Winans, 2001; Hudaiberdiev et al., 2015; Papenfort and Bassler, 2016), SdiA (LuxR homolog) in Escherichia, which can also respond to mammalian host-produced small molecules (Hughes et al., 2010; Nguyen et al., 2015; Whiteley et al., 2017).
Thirdly, combined receptors work as transcription factors to regulate dozens to hundreds of genes that affect biofilm formation, virulence, and other biological processes in bacteria. QS molecule receptors establish a feed-forward loop when regulating genes expression, which is called autoinduction. This mechanism increases the autoinducers synthesis, consequently promoting synchronous genes expression in the population (Papenfort and Bassler, 2016).
Autoinducing Peptides (AIP) QS System in Gram-Positive Bacteria
While QS circuits share some common features, there are stark distinctions between Gram+ and Gram– bacteria. The AIs in many Gram+ bacteria are oligopeptides (AIPs). There are two kinds of canonical AIP-QS circuits. One class of AIPs is encoded as a precursor from QS operon, then processed and secreted extracellularly by specialized transporters. The AIPs ranging from 5 to 17 amino acids can be linear or cyclized (Okada et al., 2005; Bouillaut et al., 2008; Thoendel et al., 2011; Rutherford and Bassler, 2012). Membrane-bound, two-component sensor histidine kinases, such as Agr system in Streptococcus aureus, and Fsr system in Enterococcus faecalis serve as AIP receptors (Waters and Bassler, 2005; Zschiedrich et al., 2016; Ali et al., 2017). The sensor kinases auto-phosphorylate after binding to AIPs, and the phosphoryl group is delivered to a cognate cytoplasmic response-regulator protein that controls the expression of QS-related genes (Rutherford and Bassler, 2012). In S. aureus, AIPs are variable and have coevolved with their receptors. Non-cognate AIPs have an inhibitory effect on QS in other strains, allowing one strain to establish its specific niche (Lyon et al., 2002; Geisinger et al., 2009).
In other canonical AIP-QS circuits, pre-AIPs are secreted by the secretory system and processed by extracellular proteases such as secreted neutral protease B (NprB). AIPs are imported and go on to bind transcription factors to regulate DNA expression through the oligopeptide permease system (Opp) (Gominet et al., 2001; Pomerantsev et al., 2009; Figure 1, Gram positive part). The PapR-PlcR system in Bacillus cereus is an example of a typical QS that functions in this way (Slamti and Lereclus, 2002). In addition, there is autoinduction that results from transcription of the QS operon which encodes pre-AIPs, transporters, receptors, regulators and proteases, leading to the synchronization of QS response (Gominet et al., 2001; Pomerantsev et al., 2009).
Inter-Species QS Signals
A Widespread Communication Signal in Bacteria Kingdom: AI-2
Bacteria direct their behavior by sensing the environment. Although many of above AIs are highly specific being produced and recognized by a single species, new studies indicate that some molecules have the potential to enable inter-species communication (Bassler et al., 1997). QS-dependent genes of luminescence in V. harveyi stains can be activated by cell-free supernatant from cultures of several unrelated bacterial species (Bassler et al., 1997). The discovery of AI-2 was the first clear indication of inter-species signaling. The release of the activated methyl group from SAM to an acceptor molecule gives rise to S-adenosylhomocysteine (SAH), which is subsequently converted into S-ribosylhomocysterine (SRH) by the enzyme 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase (Pfs). SRH is cleaved by the enzyme LuxS to produce an unstable intermediate 4,5-dihydroxy-2,3-pentanedione (DPD), which then spontaneously cyclizes into the family of active AI-2 signaling molecules (Chen et al., 2002). Hydrophilic AI-2 is secreted by exporters, but the mechanism is unclear. Extracellular AI-2 can be recognized by three specific receptors (Pereira et al., 2013). LuxP is one of receptors, which is found in Vibrionales. It is a periplasmic binding protein and can interact with LuxQ, a membrane-spanning sensor protein, to form a two-component regulatory system, LuxPQ. Boron bound-AI-2 binds to LuxP, and induces LuxQ auto-phosphorylation as well as expression of QS-related genes (Miller et al., 2004). LsrB is the second receptor, a high-affinity substrate-binding periplasmic protein in Salmonella typhimurium, Bacillus cereus, and Escherichia coli (E. coli) (Pereira et al., 2009). Non-borated AI-2 is recognized by the LsrB receptor, and internalized by a transporter that composes of LsrC, LsrD, and the ATP-binding protein LsrA. After being phosphorylated by LsrK kinase, it becomes phosphor-AI-2 (P-AI-2). P-AI-2 binds to LsrR (a repressor of the lsr operon), promoting Lsr system expression (Pereira et al., 2009). Although the overall folding of LuxP and LsrB is similar, the sequence identity is only 11%. The third receptor is RbsB, proposed as AI-2 receptors in A. actinomycetemcomitans and Haemophilus influenza strain 86-028NP. These proteins have over 70% identity in homology with the periplasmic-binding component of the ribose ABC transporter in E. coli. However, due to the lack of crystal structures of RbsB-ligand complexes, the precise amino acid residues binding to AI-2 are unclear (Armbruster et al., 2011; Figure 1, inter-species QS signals part).
A Constantly Rediscovered Molecule: AI-3
Researchers have also purified a putative autoinducer signal, AI-3, from luxS/AI-2 bacterial enterohemorrhagic E. coli (Sperandio et al., 2003). Further studies have confirmed that AI-3 synthesis is independent of LuxS (AI-2 synthase) (Walters et al., 2006). A more recent study indicates that AI-3 are several products which belong to the pyrazinone family. These AI-3 molecules are formed via a series of reactions. Among those reactions, threonine dehydrogenase (Tdh) mediated AI-3 signal production and aminoacyl-tRNA synthetases-related spontaneous cyclization are two essential reactions (Kim et al., 2020). QseC receptor, a histidine kinase sensor found in E. coli and V. cholerae, senses AI-3 signals to regulate gene expression. It also senses the host hormones epinephrine (Epi)/norepinephrine (NE) (Moreira et al., 2010). However, AI-3 analogs have no effect on adrenergic signaling in human cells in vitro (Kim et al., 2020) (Figure 1, inter-species QS signals part).
An Important Metabolite Mediating Bacteria Communication: Indole
Over time, bacteria co-evolve with the host and thus play an important role in the host metabolic processes. Bacteria are indispensable for the metabolism of some nutrients in the host; especially amino acids and carbohydrates. In addition to the involvement of bacteria-derived metabolites in regulation of host physiology and the immune system, up-to-date evidence shows that they may work as a mode of communication that affects bacterial behavior in turn. Indole is a typical example of this model. It can impact activities of indole-producing and non-indole-producing bacteria in different ways (Lee and Lee, 2010). Indole is produced when tryptophan is degraded by tryptophanase (TnaA). However, environmental factors have a critical effect on this process. The tna operon expression is elevated at extracellular tryptophan-rich conditions. This operon transcribes TnaA and TnaB, which are the permeases responsible for environmental tryptophan uptake (Gong and Yanofsky, 2002; Figure 1, inter-species QS signals part). Indole production in cells reaches a maximum and is stably maintained in the stationary phase (Mueller et al., 2009). Carbon sources such as glucose, temperature, and pH also have direct effects on the concentration of extracellular indole (Lee and Lee, 2010). Without the requirement for specific receptor binding, indole molecules can activate succinate associated genes, or act on regulatory proteins with diverse biological functions such as spore or biofilm formation, drug resistance, virulence, and plasmid stability (Martino et al., 2003; Hirakawa et al., 2005). For instance, indole works on transcriptional regulator RamA to increase drug resistance in non-indole-producing Salmonella enterica and binds pyruvate kinase to induce spore formation in Stigmatella aurantiaca. However, the effect of indole on biofilm formation in E. coli strains is different among different studies (Martino et al., 2003; Lee et al., 2007; Zhang et al., 2007).
Some bacterial metabolites can be utilized by other bacteria or pathogens as a source of nutrients or an aid for colonization. These metabolites work as cues to affect gene expression that are responsible for virulence and invasion of pathogens. For example, short-chain fatty acids (SCFAs) promote the adhesion, flagellum growth, and virulence of specific bacteria via upregulation of the expression of related genes such as T3SS in Salmonella typhimurium invasion (Lawhon et al., 2002). Secondary bile acids (BAs) generated by Clostridium scindens are able to inhibit C. difficile colonization (Buffie et al., 2015). Collectively, there is no doubt that more QS signals will be found from metabolite molecules.
In gut, components of metabolites are much complex because host cells and bacteria involving. It is easy to confuse the QS signal and metabolites which both are productions of nutrients utilization and have some similar functions. Many QS signal syntheses depend on metabolites in the environment. Both AHLs and AI-2 are intermediates of SAM metabolism which is one part of methionine metabolism and methyl cycle. When cultured in an environment devoid of methionine, E. coli demonstrates a reduction in all components of the bacterial methyl cycle (Lin and Wang, 2017). The deficit of methyl cycle elements may further affect the production of QS signals. Carbon acyl chain is an integral part of AHLs structure with some acyl chains being derived from intermediates of fatty acid biosynthesis in the host (Papenfort and Bassler, 2016). Besides indole, a metabolite derived from tryptophan, signals in the AI-3 family mainly come from threonine dehydrogenized by threonine dehydrogenase. In general, recognizing metabolites-mediated QS signals is a process in which bacteria passively adapt the environment to survive. By contrast, the classical QS is much more like an evolutionary capability of bacteria to unite as a sociality.
QS in the Gastrointestinal Tract
QS May Contribute to Spatial Distribution of Intestinal Bacteria Under Physiological Condition
Gut bacteria significantly affect the metabolic or immune status of the host (Hollister et al., 2014). In fact, the composition and relative abundance of gut bacteria is different along the length of gastrointestinal tract (Figure 2). QS is responsible for the production, extracellular accumulation, detection and response to AIs secreted by bacteria during a specific stage of growth (Winzer et al., 2002). Bacteria use QS signals as an active way to sense the environment and deliver information to its own community, as well as with other species. In the noisy environment of the gut, bacteria rely on QS to regulate survival and to compete with others for spatial dominance. However, the size of bacterial cell aggregates can influence QS signal response in turn. Smaller aggregates, but with more cells in each aggregate, showed higher bioluminescence intensity, secreted more AI-2, and had significantly higher QS-related gene expression, compared to the group with a larger aggregate but fewer cells in each aggregate (Gao et al., 2016). Another study combining micro-3D printing and scanning electrochemical microscopy revealed that QS-mediated communication can be observed in Pseudomonas aeruginosa within aggregates as small as 500 cells. However, at a defined distance of 8 μm, an aggregate containing more than 2,000 bacteria is required to stimulate QS in neighboring aggregates (Connell et al., 2014).
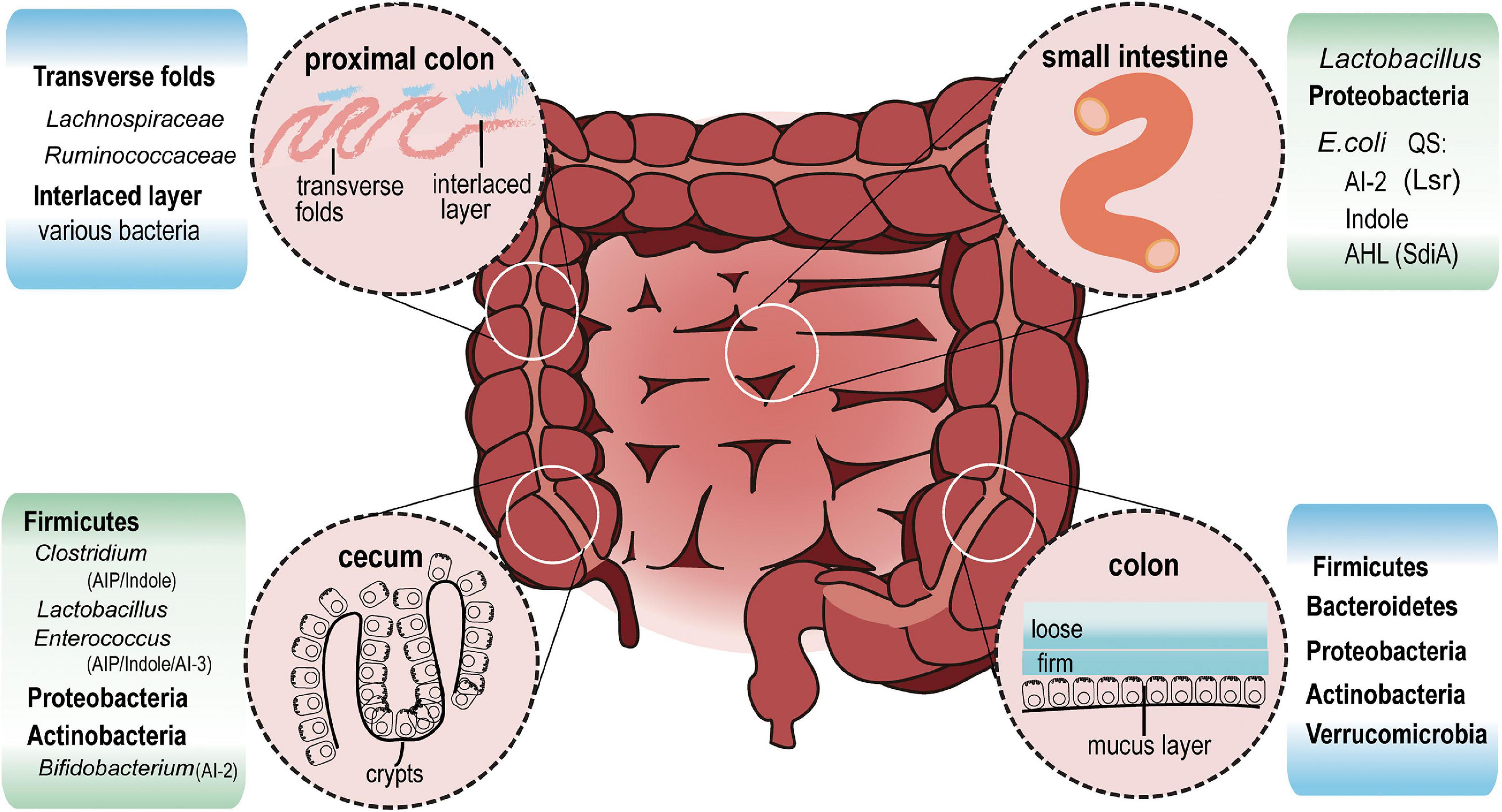
Figure 2. Intestinal bacterial distribution and QS signals. In the ileum (small intestine), Lactobacillus and Proteobacteria are the most abundant bacteria. E. coli, which belongs to Proteobacteria, communicates with other bacteria via AI-2, Indole and AHL signals. In cecum crypts, Firmicutes including Clostridium, Lactobacillus, Enterococcus, Proteobacteria, and Actinobacteria can be found. AIP, AHL, Indole, AI-2 and AI-3 may all exist in this region. At the distal end of the proximal colon, a thin but dense band called interlaced layer separates bacteria from epithelium. The interlaced layer of proximal colon is dominated by Lachnospiraceae and Ruminococcaceae. From transverse colon to rectum, mucus increases and divides into two layers. Firmicutes, Bacteroidetes, Actinobacteria, proteobacteria, and Verrucomicrobia live in the loose mucus layer.
Based on these published studies, it can be deduced that the size of aggregates in crypts and transverse folds in the cecum and proximal colon is larger than that in the lumen where feces are constantly squeezed by intestinal motility. Species of Firmicutes living here may produce enough QS signals to influence its own community as well as neighbor species via AI-2 (Jakobsson et al., 2015). About 83.33% of Firmicutes contain LuxS protein orthologs, an essential synthase for AI-2 production, which is found in only 16.83% of Bacteroidetes (Thompson et al., 2015). This significant difference in AI-2 production capability might strengthen the competitive edge of Firmicutes, allowing them to dominate the cecum and proximal colon. Among Bacteroidetes, B. thetaiotaomicron acquire carbon sources either from dietary plant polysaccharides or mucus glycans in the absence of polysaccharides. They have high metabolic capacity and replicate themselves equally in mucus and lumen (Li et al., 2015). However, due to the lack of LuxS orthologs (KEGG base), they find it hard to compete with other species such as Clostridia and Lactobacillus etc., that take advantage of AI-2 for biofilm formation or self-growth (Lebeer et al., 2008).
Mucus-resident bacteria and luminal commensal bacteria are diverse, often being distributed in distinct niches where nutrients are utilized in different ways. E. coli residing in the outer mucus replicates quickly. In the intestinal lumen, E. coli remains in the stationary phase due to the limited glycoside hydrolase (Li et al., 2015). Mucus acts as a physical protective barrier to separate bacteria and the intestinal epithelium, and is constantly renewed. The inner mucus layer has fast turnover and ultimately gets converted into the loose outer mucus layer (Johansson, 2012). The outer layer is excreted with the colon contents by intestinal movement. The next round of food, accompanied by underlying mucus, allows bacteria to recolonize. AI-2 molecules regulate biofilm formation and allows adhesion and enrichment of some bacteria such as Bifidobacterium and Lactobacillus strains (Sun et al., 2014). Instead of recolonizing from luminal contents, E. coli cells living in mucus selectively proliferate during the mucus turnover period. In addition, AI-2 signal is implicated in iron metabolic regulation in bacteria including Actinobacillus, Vibrio, and Bifidobacterium (Fong et al., 2003; Kim and Shin, 2011; Li et al., 2011; Christiaen et al., 2014). Studies have reported that E. coli colonizing the mucus can utilize more iron than their lumen counterparts (Li et al., 2015), which might occur due to higher exposure to AI-2 signals in the mucus. QS signal may help commensal intestinal bacteria cooperate to strengthen the ability to resist colonization by invaders, and keep the dynamic balance by regulating relative abundance of certain species. This is certainly an area of research which requires further exploration.
Communication via QS Between Intestinal Bacteria and Host Cells
Bacteria have co-evolved with their host for a long time. In humans, bacteria are crucial to maintain the epithelial barrier integrity and mucosal immune system via their components or bacteria-derived metabolites (Kayama et al., 2020). In addition to the classical way that microbiota influence host immune homeostasis by engaging pattern recognition receptors (PRR) at mucosal sites (Akira et al., 2006), pioneering research studies have revealed the possibility that intestinal bacteria interact with host cells by the QS system.
Bacteria-Derived QS Signaling Molecules Affect the Host Cells
Early studies on bacterial infections reported that QS signals induce host cell apoptosis and can regulate secretion of immune mediators in independent ways. This suggests that QS signals may activate more than one signal transduction pathway in host cells (Shiner et al., 2006). In one study, crosstalk between AI-2 signaling and intestinal epithelial cells (IEC) was observed. Two strains of E. coli, BL21 and W3110, were co-cultured with the IEC line HCT-8 in a transwell system. These two strains are genetically similar, but the BL21 strain is capable of producing higher levels of extracellular AI-2 without expressing appropriate receptors to uptake AI-2 molecules. Transcription sequencing analysis of HCT-8 cells showed activation of NF-κB-mediated signaling pathways. The inflammatory cytokine, IL-8, was rapidly upregulated early, from 6 to 12 h, and then downregulated at 24 h (Zargar et al., 2015). AI-2, continuously secreted by different intestinal bacteria, especially those near the epithelia, can regulate negative-feedback of inflammatory response for immune tolerance. For example, TNFSF9 gene expression was significantly upregulated in AI-2 stimulated macrophages (Li et al., 2019). Indole can be recognized by aryl hydrocarbon receptor (AHR) and bind the pregnane X receptor (PXR) in IECs (Hubbard et al., 2015). This process may enhance epithelial barrier function by upregulating the expression of cell-junction-associated molecules, and also regulate epithelial cell differentiation from crypt stem cells (Shimada et al., 2013; Venkatesh et al., 2014; Figure 3).
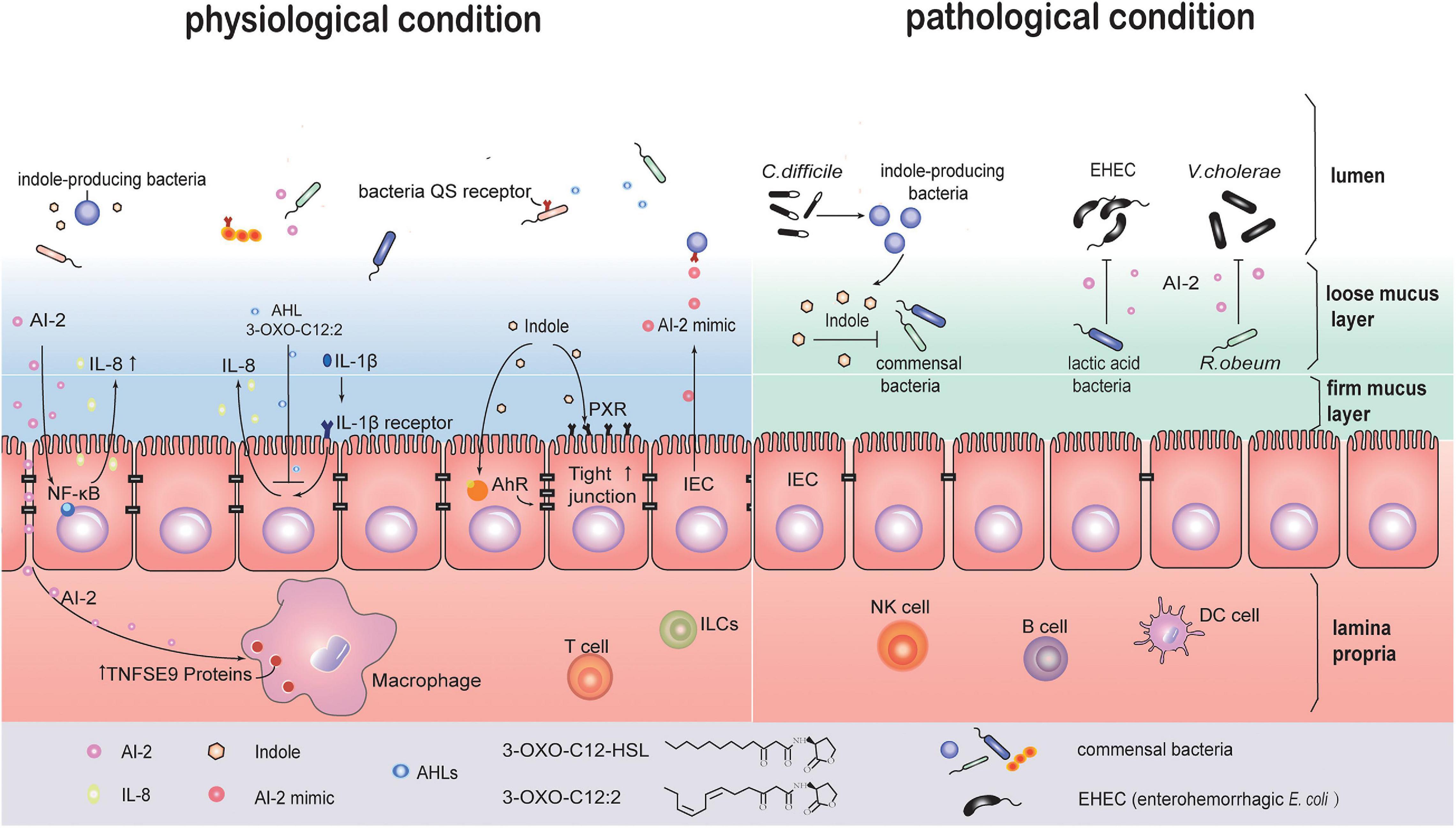
Figure 3. QS maybe involved in communication between intestinal bacteria and host cells under physiological or pathological conditions. In the physiological state, several QS signal molecules are implicated in the crosstalk between host cell and parasitic bacteria. AI-2 activates the NF-κB signaling pathway in IEC cells to upregulate the level of cytokine IL-8. AI-2 promotes TNFSF9 gene expression in macrophages. 3-oxo-C12:2 (AHLs) decreases IL-8 secretion in IL-1β-stimulated IECs. Indole enhances the epithelial barrier function via activating pregnane X receptor (PXR) and aryl hydrocarbon receptor (AHR). Meanwhile, IECs produce AI-2 mimic signals that can be recognized by AI-2 receptor, and regulate QS related genes expression in bacteria. Under pathological states, for example, Clostridium difficile infection, indole-producing bacteria sense QS signals and create a unique environment with high levels of indole, which inhibits common intestinal bacteria recovery and benefits C. difficile self-survival. “lactic acid bacteria” (LAB) such as Bifidobacterium can inhibit enterohemorrhagic Escherichia coli (EHEC) virulence by AI-2 signals. Ruminococcus. obeum restricts Vibrio cholerae colonization through AI-2 signals.
Host Influence Bacteria Based on QS Signal Mimic
In addition to being regulated by bacterial QS molecules, host cells also respond to QS signaling by fighting back. When affected by bacteria-derived soluble molecules, IECs can secrete signal analogs that mimic AI-2, thus affecting gut bacteria. When LuxS mutant strains lacking AI-2 signal production were co-cultured with epithelial cells isolated from colon tissues, lsr gene transcription, usually induced by AI-2, was found to increase. This was attributed to an AI-2 mimic produced by IEC. Further, these signals were associated with epithelial tight-junction damages. The above findings suggest that the host-derived AI-2 mimic maybe associated with intestinal bacteria adhesion and epithelial barrier disruption (Ismail et al., 2016). In addition of the AI-2 mimic, bacteria can also make use of molecules secreted by the host as QS signals in the gut microenvironment. Host hormones such as catecholamines can promote bacterial growth. Epi/NE can be sensed by the QseC receptor of QS system (Clarke et al., 2006). More recent studies have shown other adrenergic receptors including QseE and CpxA that have significant differences in the phylogenetic profile with QseC also act as receptors (Reading et al., 2007; Karavolos et al., 2011; Karavolos et al., 2013). 1-octanoyl-rac-glycerol (OCL), a monoacylglycerol that is abundant in the mammalian gastrointestinal tract, forms triacylglycerols and works as a chemical chaperone placeholder to stabilize SdiA in E. coli, allowing its basal activity (Nguyen et al., 2015). Fatty acids (FAs), ubiquitously found in various organisms, have similar chemical structures with diffusible signal factors (DSF) family. DSFs are used by some Gram– bacteria as QS signals for biofilm formation and virulence. FAs, as DSFs mimic, can inhibit the bacterial biofilm or other QS-dependent gene expression, and influence AHL and AI-2 signaling. The common pathogens inhabiting the human body, including Burkholderia spp., P. aeruginosa, Vibro spp., Helicobacter pylori, and Salmonella, utilize DSFs. Some of them specifically target the gastrointestinal system (Kumar et al., 2020). In the absence of physical barrier in small intestine, chemical barriers play a critical role in separating bacterial and epithelial cells in the small intestine and thus protecting the host against pathogen infection (Kayama et al., 2020). In addition to antimicrobial peptides and Reg3 family proteins (Kayama et al., 2020), FAs present in bile juice may also mimic QS signal to regulate bacterial biofilm formation (Kumar et al., 2020).
Changes in Intestinal Bacterial Communication Under Pathological Conditions
Pseudomonas aeruginosa is a Gram– opportunistic pathogen acting on human tissues. It acts though 3 main QS systems, including two AHL dependent LuxI/LuxR-type systems and a Pseudomonas quinolone signal (PQS) system. N-3-oxo-dodecanoyl-Lhomoserine lactone (3O-C12-HSL) and N-butyryl-L-homoserine lactone (C4-HSL) from the AHL signal family are used by P. aeruginosa to control more than 300 genes, many of which are involved in virulence regulation. The PQS system is associated with biofilm formation. When P. aeruginosa infects the human body, the above mentioned QS signals interact with human cells, leading to physiological and functional changes in immune cells including neutrophils, macrophages as well as epithelial cells (Holm and Vikström, 2014). Compared to a mutant P. aeruginosa strain lacking AHL production, the wild type strain containing 3O-C12-HSL and C4-HSL promoted macrophage phagocytosis. 3O-C12-HSL caused cell-volume increase which was associated with upregulation of Aquaporin 9 (AQP9), a chronic inflammation marker in inflammatory bowel disease (IBD) (Mesko et al., 2010; Holm and Vikström, 2014).
Subsequent research analyzed feces from healthy subjects and IBD patients with flares or under remission. AHLs were detected in these samples using liquid chromatography and mass spectrometry. Among AHLs, 3-oxo-C12:2 was significantly enriched in the healthy group and was higher in the IBD group under remission compared to IBD patients with flares. Unlike 3-oxo-C12 (3O-C12-HSL), 3-oxo-C12:2 could decrease IL-8 secretion in IL-1β-stimulated IECs, but had no effect on the epithelial paracellular permeability (Figure 2). Moreover, 3-oxo-C12:2 levels positively correlated with higher counts of Faecalibacterium prausnitzii, Clostridium coccoides, and Clostridium Leptum (Landman et al., 2018). These bacterial species are rare and belong to Firmicutes, especially Gram– bacteria F. prausnitzii. In fact, an earlier study reported that oral administration of live F. prausnitzii or its supernatant could reduce the severity of 2,4,6-trinitrobenzenesulfonic acid (TNBS) colitis. The anti-inflammatory effects of F. prausnitzii are partly due to its secreted metabolites that block IL-8 production (Sokol et al., 2008).
There are high levels of indole and Clostridium difficile (C. difficile) toxin-induced QS signaling peptides in the feces from C. difficile infection (CDI) patients compared with CDI-negative diarrhea patients. It suggests that C. difficile uses QS signals to regulate its infection in the gastrointestinal tract, and this QS signal is related with indole production. However, C. difficile lacks the tryptophanase gene, which aids in indole production. Subsequent findings indicate that C. difficile may use toxin-induced QS signals to regulate the relative abundance of indole-producing bacteria and create a conducive environment for their survival (Figure 2). Meanwhile, commensal gut bacteria with a lower indole tolerance than C. difficile would be inhibited from recovering, resulting in a more beneficial environment for the colonization and expansion of C. difficile (Darkoh et al., 2019).
The production of AI-2 has a broad impact on the relative abundance of two dominant phyla of bacteria in the gut. Researchers found the abundance of Firmicutes phyla in mice can be significantly reduced by streptomycin treatment for 28 days. Consequently, Bacteroidetes become dominant and its relative abundance reaches approximately 90%. Additionally, three types of mutant E. coli are mono-colonized in those antibiotic-treated mice. One mutant strain lacks the AI-2 kinase gene lsrK, which allows extracellular AI-2 accumulation. Depletion of lsrR, a repressor of lsrK transcription, promotes another strain to deplete environmental AI-2 by internalizing them. The third mutant strain has no effect on AI-2 production and accumulation due to the removal of signal synthase LuxS. The three mutant strains could be assembled into three groups, namely ΔlsrK AI-2 secreting-, ΔlsrRΔLuxS AI-2 absorbing- and ΔlsrKΔLuxS neutralizing- group. The ΔlsrK AI-2 secreting-group showed an increasing ratio of Firmicutes to Bacteroidetes (Thompson et al., 2015).
The Role of QS System in the Gut Microbiome Community Regulated by Probiotics
Probiotics, made up of “good bacteria,” help reverse gut dysbiosis. The QS system in the gut microbiome can be regulated by probiotics. In rodent models, E. coli competes with Salmonella typhimurium (S. typhimurium) for nutrient iron, resulting in reduction of S. typhimurium colonization (Deriu et al., 2013). LuxS gene plays a role in motility and virulence of S. typhimurium (Choi et al., 2007). However, AI-2 signals secreted by S. typhimurium for invasion are inhibited by E. coli. Bifidobacterium. belonging to “lactic acid bacteria” (LAB), contains the luxS gene and can therefore produce AI-2 molecules to regulate biofilm formation (Sun et al., 2014). Similarly, in weaning swine, the virulence of enterohemorrhagic E. coli (EHEC) is inhibited by the presence of AI-2 signals produced by LAB (Kim et al., 2018; Figure 3).
In a study about V. cholerae infection in Bangladesh, researchers collected feces from three cohorts including cholera patients, healthy adults and healthy children. The time-series metagenomic analysis suggested that accumulation of certain bacterial taxa is associated with recovery from a V. cholerae infection. In gnotobiotic mice, bacterial community transplantation induces increase in Ruminococcus obeum relative abundance, however V. cholerae colonization is restricted. LuxS gene expression and AI-2 production in R. obeum increase with V. cholerae invasion. These molecules target other pathways to inhibit Vibrio pathogenicity, irrespective of whether a LuxP sensor exists or not (Hsiao et al., 2014). This study indicates an unknown, novel function of the QS system that is different from the canonical pathways discussed previously (Figure 3).
QS as a Prospective Therapeutic Target for Microbial Infections
Novel therapeutic strategies can be exploited by targeting the bacterial QS system. Higher AI-2 levels were observed in colon mucosa and stools from colorectal adenoma and colorectal cancer (CRC) patients compared to healthy individuals. Importantly, AI-2 concentration was found to increase with CRC progression, suggesting its potential as a novel marker for CRC clinic screening. Another recent research on burn-site infections focused on the P. aeruginosa QS system. Burn injury is often accompanied with gut microbiome dysbiosis, impaired intestinal integrity, immune dysregulation and bacterial extra-intestinal translocation. P. aeruginosa is the main pathogen in post-burn infection. Its toxic products can prolong intestinal dysfunction and aggravate systemic infection. Researchers observed an improvement in intestinal dysfunction and a decrease in P. aeruginosa dissemination after inhibiting MvfR transcriptional factor, an important QS-associated transcriptional regulator controlling the virulence of this strains, using a MvfR antagonist (Adiliaghdam et al., 2019). In fact, antibacterial therapeutic strategies have started to expand from antibiotics usage to developing inhibitors based on QS systems such as anti-virulence or anti-biofilms. These inhibitors or analogs exist endogenously either in bacterial or eukaryotic cells. An example is Carnosic acid, a specific inhibitor of the QS system in S. aureus. It is found in rosemary leaves and can inhibit Agr expression and S. aureus virulence at a low concentration (Nakagawa et al., 2020). Taken together, these thrilling studies point to a new potential of drug development for refractory infectious diseases.
Conclusion and Further Perspectives
Researches in the field of bacterial QS have expanded rapidly. QS has a critical role in bacterial behavior during adaption and development in diverse environments like the human gut. It has the potential to affect infection status and disease development though bacterial-host crosstalk. Although the fundamental biology of bacterial sociality is complex, we have come a long way in our attempt to elucidate the roles and functions of QS in microbial communities. In addition, we have also realized the significance of understanding the interactions between microbes and their host.
Many critical questions still exist. In gut, symbiotic bacteria occupy in different areas along the gastrointestinal (GI) tract of host. Besides the influence of GI microenvironment on symbiotic bacteria occupation, it is still lack of knowledge about how QS signal systems participate in their occupation and interact with host cells. Addressing this point would be useful in exploring new methods to strengthen resistibility of commensal intestinal bacteria to colonization by invaders and maintain mucous stability in host. In addition, despite the discovered important roles of QS signals in bacterial communication, it will probably take more years to address the related molecules mechanisms in details. In the respective of problematic antibiotic resistance, the development of alternative therapies for infectious diseases is one of the major societal challenges at this moment. Probiotics has been recognized as a method of treatment. Fecal transplantation (FMT) recently drives more attention for many diseases, particularly in gastrointestinal disorders and metabolic diseases (He et al., 2017; Ding et al., 2019; Antushevich, 2020). Anti-virulence therapy by using various quorum-sensing inhibitors or agents that interfere with QS in bacterial pathogens currently is also a reasonable and promising strategy. Although we have obtained some positive results regarding to QS inhibitors in previous studies, it is still a long way to go in identifying the molecular targets of these potential agents and speeding their application in the clinic. Exploring new applications of available drugs used in the clinic for other diseases probably will be an approach to save time and money.
With the development of high-throughput-omics techniques, scientific cognition of QS signals and their regulating pathways can be clarified. Sorting out fundamental differences between diverse QS systems will give us an opportunity to understand the biology of bacterial sociality better. Moreover, it will give us the confidence to translate QS studies for potential therapeutic development for devastating infectious diseases.
Author Contributions
LW wrote the article. YL revised the manuscript. Both authors researched data for the article, made substantial contributions to the content, and edited the manuscript.
Funding
This work was carried out under the sponsorship of the National Natural Science Foundation of China (Grant Nos. 81403041 and 81770101) and Outstanding Interdisciplinary Project Award of West China Hospital, Sichuan University (Grant No. ZYJC18024).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Qiuping Zhang for editing and checking the references.
References
Adiliaghdam, F., Almpani, M., Gharedaghi, M. H., Najibi, M., Hodin, R. A., and Rahme, L. G. (2019). Targeting bacterial quorum sensing shows promise in improving intestinal barrier function following burn site infection. Mol. Med. Rep. 19, 4057–4066. doi: 10.3892/mmr.2019.10071
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Ali, L., Goraya, M. U., Arafat, Y., Ajmal, M., Chen, J. L., and Yu, D. (2017). Molecular mechanism of quorum-sensing in Enterococcus faecalis: its role in virulence and therapeutic approaches. Int. J. Mol. Sci. 18:960. doi: 10.3390/ijms18050960
Antushevich, H. (2020). Fecal microbiota transplantation in disease therapy. Clin. Chim. Acta 503, 90–98. doi: 10.1016/j.cca.2019.12.010
Armbruster, C. E., Pang, B., Murrah, K., Juneau, R. A., Perez, A. C., Weimer, K. E., et al. (2011). RbsB (NTHI_0632) mediates quorum signal uptake in nontypeable Haemophilus influenzae strain 86-028NP. Mol. Microbiol. 82, 836–850. doi: 10.1111/j.1365-2958.2011.07831.x
Bassler, B. L., Greenberg, E. P., and Stevens, A. M. (1997). Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179, 4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997
Bassler, B. L., and Losick, R. (2006). Bacterially speaking. Cell 125, 237–246. doi: 10.1016/j.cell.2006.04.001
Bassler, B. L., Wright, M., Showalter, R. E., and Silverman, M. R. (1993). Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9, 773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x
Bouillaut, L., Perchat, S., Arold, S., Zorrilla, S., Slamti, L., Henry, C., et al. (2008). Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Res. 36, 3791–3801. doi: 10.1093/nar/gkn149
Buffie, C. G., Bucci, V., Stein, R. R., McKenney, P. T., Ling, L., Gobourne, A., et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. doi: 10.1038/nature13828
Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L., et al. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549. doi: 10.1038/415545a
Choi, J., Shin, D., and Ryu, S. (2007). Implication of quorum sensing in Salmonella enterica serovar typhimurium virulence: the luxS gene is necessary for expression of genes in pathogenicity island 1. Infect. Immun. 75, 4885–4890. doi: 10.1128/iai.01942-06
Christiaen, S. E., O’Connell Motherway, M., Bottacini, F., Lanigan, N., Casey, P. G., Huys, G., et al. (2014). Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PLoS One 9:e98111. doi: 10.1371/journal.pone.0098111
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C., and Sperandio, V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10420–10425. doi: 10.1073/pnas.0604343103
Connell, J. L., Kim, J., Shear, J. B., Bard, A. J., and Whiteley, M. (2014). Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc. Natl. Acad. Sci. U.S.A. 111, 18255–18260. doi: 10.1073/pnas.1421211111
Darkoh, C., Plants-Paris, K., Bishoff, D., and DuPont, H. L. (2019). Clostridium difficile modulates the gut microbiota by inducing the production of Indole, an Interkingdom signaling and antimicrobial molecule. mSystems 4:e00346-18. doi: 10.1128/mSystems.00346-18
Deriu, E., Liu, J. Z., Pezeshki, M., Edwards, R. A., Ochoa, R. J., Contreras, H., et al. (2013). Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37. doi: 10.1016/j.chom.2013.06.007
Ding, X., Li, Q., Li, P., Zhang, T., Cui, B., Ji, G., et al. (2019). Long-term safety and efficacy of fecal microbiota transplant in active ulcerative colitis. Drug Saf. 42, 869–880. doi: 10.1007/s40264-019-00809-2
Fong, K. P., Gao, L., and Demuth, D. R. (2003). luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect. Immun. 71, 298–308. doi: 10.1128/iai.71.1.298-308.2003
Galloway, W. R., Hodgkinson, J. T., Bowden, S. D., Welch, M., and Spring, D. R. (2011). Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 111, 28–67. doi: 10.1021/cr100109t
Gao, M., Zheng, H., Ren, Y., Lou, R., Wu, F., Yu, W., et al. (2016). A crucial role for spatial distribution in bacterial quorum sensing. Sci. Rep. 6:34695. doi: 10.1038/srep34695
Geisinger, E., Muir, T. W., and Novick, R. P. (2009). agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc. Natl. Acad. Sci. U.S.A. 106, 1216–1221. doi: 10.1073/pnas.0807760106
Gominet, M., Slamti, L., Gilois, N., Rose, M., and Lereclus, D. (2001). Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 40, 963–975. doi: 10.1046/j.1365-2958.2001.02440.x
Gong, F., and Yanofsky, C. (2002). Analysis of tryptophanase operon expression in vitro: accumulation of TnaC-peptidyl-tRNA in a release factor 2-depleted S-30 extract prevents Rho factor action, simulating induction. J. Biol. Chem. 277, 17095–17100. doi: 10.1074/jbc.M201213200
He, Z., Cui, B. T., Zhang, T., Li, P., Long, C. Y., Ji, G. Z., et al. (2017). Fecal microbiota transplantation cured epilepsy in a case with Crohn’s disease: the first report. World J. Gastroenterol. 23, 3565–3568. doi: 10.3748/wjg.v23.i19.3565
Higgins, D. A., Pomianek, M. E., Kraml, C. M., Taylor, R. K., Semmelhack, M. F., and Bassler, B. L. (2007). The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450, 883–886. doi: 10.1038/nature06284
Hirakawa, H., Inazumi, Y., Masaki, T., Hirata, T., and Yamaguchi, A. (2005). Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 55, 1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x
Hollister, E. B., Gao, C., and Versalovic, J. (2014). Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 146, 1449–1458. doi: 10.1053/j.gastro.2014.01.052
Holm, A., and Vikström, E. (2014). Quorum sensing communication between bacteria and human cells: signals, targets, and functions. Front. Plant Sci. 5:309. doi: 10.3389/fpls.2014.00309
Hsiao, A., Ahmed, A. M., Subramanian, S., Griffin, N. W., Drewry, L. L., Petri, W. A., et al. (2014). Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426. doi: 10.1038/nature13738
Hubbard, T. D., Murray, I. A., and Perdew, G. H. (2015). Indole and tryptophan metabolism: endogenous and dietary routes to Ah receptor activation. Drug Metab. Dispos. 43, 1522–1535. doi: 10.1124/dmd.115.064246
Hudaiberdiev, S., Choudhary, K. S., Vera Alvarez, R., Gelencsér, Z., Ligeti, B., Lamba, D., et al. (2015). Census of solo LuxR genes in prokaryotic genomes. Front. Cell. Infect. Microbiol. 5:20. doi: 10.3389/fcimb.2015.00020
Hughes, D. T., Terekhova, D. A., Liou, L., Hovde, C. J., Sahl, J. W., Patankar, A. V., et al. (2010). Chemical sensing in mammalian host-bacterial commensal associations. Proc. Natl. Acad. Sci. U.S.A. 107, 9831–9836. doi: 10.1073/pnas.1002551107
Ismail, A. S., Valastyan, J. S., and Bassler, B. L. (2016). A host-produced autoinducer-2 mimic activates bacterial quorum sensing. Cell Host Microbe 19, 470–480. doi: 10.1016/j.chom.2016.02.020
Jakobsson, H. E., Rodríguez-Piñeiro, A. M., Schütte, A., Ermund, A., Boysen, P., Bemark, M., et al. (2015). The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 16, 164–177. doi: 10.15252/embr.201439263
Johansson, M. E. (2012). Fast renewal of the distal colonic mucus layers by the surface goblet cells as measured by in vivo labeling of mucin glycoproteins. PLoS One 7:e41009. doi: 10.1371/journal.pone.0041009
Karavolos, M. H., Bulmer, D. M., Spencer, H., Rampioni, G., Schmalen, I., Baker, S., et al. (2011). Salmonella Typhi sense host neuroendocrine stress hormones and release the toxin haemolysin E. EMBO Rep. 12, 252–258. doi: 10.1038/embor.2011.4
Karavolos, M. H., Winzer, K., Williams, P., and Khan, C. M. (2013). Pathogen espionage: multiple bacterial adrenergic sensors eavesdrop on host communication systems. Mol. Microbiol. 87, 455–465. doi: 10.1111/mmi.12110
Kayama, H., Okumura, R., and Takeda, K. (2020). Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu. Rev. Immunol. 38, 23–48. doi: 10.1146/annurev-immunol-070119-115104
Kim, C. M., and Shin, S. H. (2011). Modulation of iron-uptake systems by a mutation of luxS encoding an autoinducer-2 synthase in Vibrio vulnificus. Biol. Pharm. Bull. 34, 632–637. doi: 10.1248/bpb.34.632
Kim, C. S., Gatsios, A., Cuesta, S., Lam, Y. C., Wei, Z., Chen, H., et al. (2020). Characterization of Autoinducer-3 structure and biosynthesis in E. coli. ACS Cent. Sci. 6, 197–206. doi: 10.1021/acscentsci.9b01076
Kim, J., Kim, J., Kim, Y., Oh, S., Song, M., Choe, J. H., et al. (2018). Influences of quorum-quenching probiotic bacteria on the gut microbial community and immune function in weaning pigs. Anim. Sci. J. 89, 412–422. doi: 10.1111/asj.12954
Kumar, P., Lee, J. H., Beyenal, H., and Lee, J. (2020). Fatty acids as antibiofilm and antivirulence agents. Trends Microbiol. 28, 753–768. doi: 10.1016/j.tim.2020.03.014
Landman, C., Grill, J. P., Mallet, J. M., Marteau, P., Humbert, L., Le Balc’h, E., et al. (2018). Inter-kingdom effect on epithelial cells of the N-Acyl homoserine lactone 3-oxo-C12:2, a major quorum-sensing molecule from gut microbiota. PLoS One 13:e0202587. doi: 10.1371/journal.pone.0202587
Lawhon, S. D., Maurer, R., Suyemoto, M., and Altier, C. (2002). Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46, 1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x
Lebeer, S., Claes, I. J., Verhoeven, T. L., Shen, C., Lambrichts, I., Ceuppens, J. L., et al. (2008). Impact of luxS and suppressor mutations on the gastrointestinal transit of Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 74, 4711–4718. doi: 10.1128/aem.00133-08
Lee, J., Jayaraman, A., and Wood, T. K. (2007). Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7:42. doi: 10.1186/1471-2180-7-42
Lee, J. H., and Lee, J. (2010). Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 34, 426–444. doi: 10.1111/j.1574-6976.2009.00204.x
Li, H., Limenitakis, J. P., Fuhrer, T., Geuking, M. B., Lawson, M. A., Wyss, M., et al. (2015). The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 6:8292. doi: 10.1038/ncomms9292
Li, L., Xu, Z., Zhou, Y., Li, T., Sun, L., Chen, H., et al. (2011). Analysis on Actinobacillus pleuropneumoniae LuxS regulated genes reveals pleiotropic roles of LuxS/AI-2 on biofilm formation, adhesion ability and iron metabolism. Microb. Pathog. 50, 293–302. doi: 10.1016/j.micpath.2011.02.002
Li, Q., Peng, W., Wu, J., Wang, X., Ren, Y., Li, H., et al. (2019). Autoinducer-2 of gut microbiota, a potential novel marker for human colorectal cancer, is associated with the activation of TNFSF9 signaling in macrophages. Oncoimmunology 8:e1626192. doi: 10.1080/2162402x.2019.1626192
Lin, C. J., and Wang, M. C. (2017). Microbial metabolites regulate host lipid metabolism through NR5A-Hedgehog signalling. Nat. Cell Biol. 19, 550–557. doi: 10.1038/ncb3515
Lyon, G. J., Wright, J. S., Muir, T. W., and Novick, R. P. (2002). Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry 41, 10095–10104. doi: 10.1021/bi026049u
Martino, P. D., Fursy, R., Bret, L., Sundararaju, B., and Phillips, R. S. (2003). Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49, 443–449. doi: 10.1139/w03-056
Mesko, B., Poliska, S., Szegedi, A., Szekanecz, Z., Palatka, K., Papp, M., et al. (2010). Peripheral blood gene expression patterns discriminate among chronic inflammatory diseases and healthy controls and identify novel targets. BMC Med. Genomics 3:15. doi: 10.1186/1755-8794-3-15I
Miller, M. B., Skorupski, K., Lenz, D. H., Taylor, R. K., and Bassler, B. L. (2002). Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110, 303–314. doi: 10.1016/s0092-8674(02)00829-2
Miller, S. T., Xavier, K. B., Campagna, S. R., Taga, M. E., Semmelhack, M. F., Bassler, B. L., et al. (2004). Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15, 677–687. doi: 10.1016/j.molcel.2004.07.020
Moreira, C. G., Weinshenker, D., and Sperandio, V. (2010). QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. Infect. Immun. 78, 914–926. doi: 10.1128/iai.01038-09
Mueller, R. S., Beyhan, S., Saini, S. G., Yildiz, F. H., and Bartlett, D. H. (2009). Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J. Bacteriol. 191, 3504–3516. doi: 10.1128/jb.01240-08
Nakagawa, S., Hillebrand, G. G., and Nunez, G. (2020). Rosmarinus officinalis L. (Rosemary) Extracts containing carnosic acid and carnosol are potent quorum sensing inhibitors of Staphylococcus aureus virulence. Antibiotics 9:149. doi: 10.3390/antibiotics9040149
Ng, W. L., and Bassler, B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43, 197–222. doi: 10.1146/annurev-genet-102108-134304
Nguyen, Y., Nguyen, N. X., Rogers, J. L., Liao, J., MacMillan, J. B., Jiang, Y., et al. (2015). Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. mBio 6:e02429-14.
Okada, M., Sato, I., Cho, S. J., Iwata, H., Nishio, T., Dubnau, D., et al. (2005). Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat. Chem. Biol. 1, 23–24. doi: 10.1038/nchembio709
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Pereira, C. S., de Regt, A. K., Brito, P. H., Miller, S. T., and Xavier, K. B. (2009). Identification of functional LsrB-like autoinducer-2 receptors. J. Bacteriol. 191, 6975–6987. doi: 10.1128/jb.00976-09
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Pomerantsev, A. P., Pomerantseva, O. M., Camp, A. S., Mukkamala, R., Goldman, S., and Leppla, S. H. (2009). PapR peptide maturation: role of the NprB protease in Bacillus cereus 569 PlcR/PapR global gene regulation. FEMS Immunol. Med. Microbiol. 55, 361–377. doi: 10.1111/j.1574-695X.2008.00521.x
Reading, N. C., Torres, A. G., Kendall, M. M., Hughes, D. T., Yamamoto, K., and Sperandio, V. (2007). A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J. Bacteriol 189, 2468–2476. doi: 10.1128/jb.01848-06
Rutherford, S. T., and Bassler, B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. doi: 10.1101/cshperspect.a012427
Ryan, R. P., An, S. Q., Allan, J. H., McCarthy, Y., and Dow, J. M. (2015). The DSF family of cell-cell signals: an expanding class of bacterial virulence regulators. PLoS Pathog. 11:e1004986. doi: 10.1371/journal.ppat.1004986
Shimada, Y., Kinoshita, M., Harada, K., Mizutani, M., Masahata, K., Kayama, H., et al. (2013). Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One 8:e80604. doi: 10.1371/journal.pone.0080604
Shiner, E. K., Terentyev, D., Bryan, A., Sennoune, S., Martinez-Zaguilan, R., Li, G., et al. (2006). Pseudomonas aeruginosa autoinducer modulates host cell responses through calcium signalling. Cell. Microbiol. 8, 1601–1610. doi: 10.1111/j.1462-5822.2006.00734.x
Slamti, L., and Lereclus, D. (2002). A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 21, 4550–4559. doi: 10.1093/emboj/cdf450
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermúdez-Humarán, L. G., Gratadoux, J. J., et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105, 16731–16736. doi: 10.1073/pnas.0804812105
Sperandio, V., Torres, A. G., Jarvis, B., Nataro, J. P., and Kaper, J. B. (2003). Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U.S.A. 100, 8951–8956. doi: 10.1073/pnas.1537100100
Sun, Z., He, X., Brancaccio, V. F., Yuan, J., and Riedel, C. U. (2014). Bifidobacteria exhibit LuxS-dependent autoinducer 2 activity and biofilm formation. PLoS One 9:e88260. doi: 10.1371/journal.pone.0088260
Thoendel, M., Kavanaugh, J. S., Flack, C. E., and Horswill, A. R. (2011). Peptide signaling in the staphylococci. Chem. Rev. 111, 117–151. doi: 10.1021/cr100370n
Thompson, J. A., Oliveira, R. A., Djukovic, A., Ubeda, C., and Xavier, K. B. (2015). Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 10, 1861–1871. doi: 10.1016/j.celrep.2015.02.049
Tomasz, A. (1965). Control of the competent state in Pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature 208, 155–159. doi: 10.1038/208155a0
Venkatesh, M., Mukherjee, S., Wang, H., Li, H., Sun, K., Benechet, A. P., et al. (2014). Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41, 296–310. doi: 10.1016/j.immuni.2014.06.014
von Bodman, S. B., Willey, J. M., and Diggle, S. P. (2008). Cell-cell communication in bacteria: united we stand. J. Bacteriol. 190, 4377–4391. doi: 10.1128/jb.00486-08
Walters, M., Sircili, M. P., and Sperandio, V. (2006). AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 188, 5668–5681. doi: 10.1128/jb.00648-06
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Whiteley, M., Diggle, S. P., and Greenberg, E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320. doi: 10.1038/nature24624
Winzer, K., Hardie, K. R., and Williams, P. (2002). Bacterial cell-to-cell communication: sorry, can’t talk now - gone to lunch! Curr. Opin. Microbiol. 5, 216–222. doi: 10.1016/s1369-5274(02)00304-1
Zargar, A., Quan, D. N., Carter, K. K., Guo, M., Sintim, H. O., Payne, G. F., et al. (2015). Bacterial secretions of nonpathogenic Escherichia coli elicit inflammatory pathways: a closer investigation of interkingdom signaling. mBio 6:e00025-15.
Zhang, X. S., García-Contreras, R., and Wood, T. K. (2007). YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J. Bacteriol. 189, 3051–3062. doi: 10.1128/jb.01832-06
Zhou, L., Yu, Y., Chen, X., Diab, A. A., Ruan, L., He, J., et al. (2015). The multiple DSF-family QS signals are synthesized from carbohydrate and branched-chain amino acids via the FAS elongation cycle. Sci. Rep. 5:13294. doi: 10.1038/srep13294
Zhu, J., and Winans, S. C. (2001). The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. U.S.A. 98, 1507–1512. doi: 10.1073/pnas.98.4.1507
Keywords: bacterial communication, quorum sensing, autoinducers, metabolites, host cells
Citation: Wu L and Luo Y (2021) Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front. Microbiol. 12:611413. doi: 10.3389/fmicb.2021.611413
Received: 29 September 2020; Accepted: 12 January 2021;
Published: 28 January 2021.
Edited by:
Robert Czajkowski, University of Gdańsk, PolandReviewed by:
Randy Ortiz-Castro, National Council of Science and Technology (CONACYT), MexicoGregory F. Payne, University of Maryland, College Park, United States
Copyright © 2021 Wu and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yubin Luo, bHVveXViaW4yMDE2QDE2My5jb20=
 Liang Wu
Liang Wu Yubin Luo
Yubin Luo