
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 26 March 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.603967
This article is part of the Research Topic Exposure, Risks, and Drivers of the Mobile Antimicrobial Resistance Genes in the Environment – a Global Perspective View all 17 articles
Antimicrobial resistance (AMR) is a global health threat. Antibiotics, heavy metals, and microplastics are environmental pollutants that together potentially have a positive synergetic effect on the development, persistence, transport, and ecology of antibiotic resistant bacteria in the environment. To evaluate this, a wide array of experimental methods would be needed to quantify the occurrence of antibiotics, heavy metals, and microplastics as well as associated microbial communities in the natural environment. In this mini-review, we outline the current technologies used to characterize microplastics based ecosystems termed “plastisphere” and their AMR promoting elements (antibiotics, heavy metals, and microbial inhabitants) and highlight emerging technologies that could be useful for systems-level investigations of AMR in the plastisphere.
The increasing resistance of pathogenic bacteria to common antibiotics (AB) found in human and veterinary settings worldwide (WHO, 2018) highlights the urgent need for improved surveillance programs (Dadgostar, 2019) and research to hinder further escalation of antimicrobial resistance (AMR) (Interagency Coordination Group on Antimicrobial Resistance, 2019). Although the number is debatable (de Kraker et al., 2016), according to O’Neill (2016), the global annual death toll due to AMR could rise to 10 million by 2050.
The emerging contaminant—plastic—has potential to further enhance AMR by providing porous micro ecosystems termed “plastisphere” (Keswani et al., 2016). In the environment, plastic does not biodegrade but fragmentizes into smaller fractions such as microplastics (MPs) (1 μm–5 mm) (Frias and Nash, 2019) or further into nanoplastics (NPs) (≤1 μm) (Gigault et al., 2018). MPs have been increasingly detected in all the ecosystems, though due to rapid microbial colonization and subsequent density changes, about 70% of the MPs in the aquatic environment sedimentates and thus the sediments, along with soils that receive MPs contamination from sludge application, have been considered as the sinks of MPs (Nizzetto et al., 2016; Corradini et al., 2019; Schmiedgruber et al., 2019). Plastic is also ingested and inhaled by humans (Cox et al., 2019; de Wit and Bigaud, 2019) as indicated by detection of plastic in stool samples (Schwabl et al., 2019) and human lung tissue (Pauly et al., 1998), respectively. Compared to MPs, NPs have been scarcely studied due to limitations of current analytical techniques (Nguyen B. et al., 2019), yet Besseling et al. (2019) have speculated future NP concentrations in mass may become 1014 times higher than currently measured MP concentrations.
The plastisphere creates a habitat that promotes attachment of and subsequent biofilm production by microbes (Zettler et al., 2013). In this habitat, the microbes are also in close vicinity of MP-associated pollutants, such as (ABs) and heavy metals (HMs) (Figure 1). This combination of being surrounded by pollutants while being protected by biofilm can lead to possible change in the microbial species distribution (Munier and Bendell, 2018; Imran et al., 2019). ABs are considered to be the primary drivers of AMR (Kraemer et al., 2019), originating largely from inefficient wastewater treatment processes and pharmaceutical discharge (Wilkinson and Boxall, 2019). HMs are accumulating in the environment via waste flows from industrial activities (mining, smelting, fertilizer use, sewage sludge application), but may also be mobilized due to natural processes (e.g., bedrock weathering) (Ali et al., 2019; Zhou et al., 2020). HM pollution drives the selection for metal resistance genes (MRGs) and correlates with increased occurrences and amount of antibiotic resistance genes (ARGs) (Figure 1; Baker-Austin et al., 2006; Li et al., 2017; Nguyen C.C. et al., 2019).
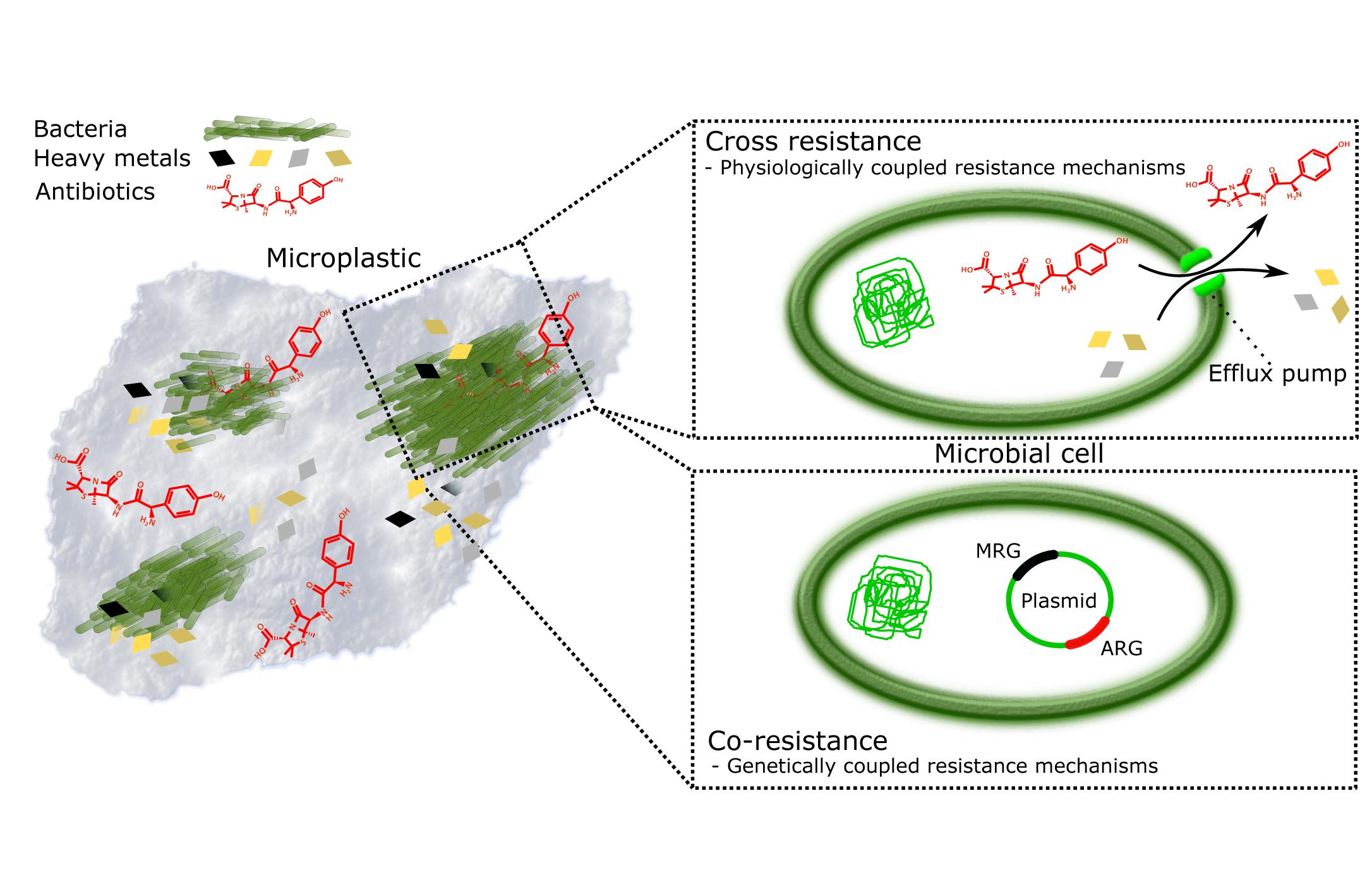
Figure 1. Plastisphere is a potential hotspot for evolution of antimicrobial resistance (AMR). Persistence and surface characteristics of microplastics make it an excellent reservoir of microbes and pollutants, such as heavy metals (HMs) and antibiotics (ABs). Together they form miniature ecosystems, plastispheres, where AMR is promoted by (i) cross-resistance where resistance mechanisms to HMs and ABs are physiologically coupled, for example, efflux pumps (Baker-Austin et al., 2006; Seiler and Berendonk, 2012); and (ii) co-resistance where antibiotic resistance genes (ARGs) and metal resistance genes (MRGs) are present on the same mobile genetic element and thus genetically coupled, whereby selection for metal resistance in, for example, animal gut, and anthropogenically contaminated soils and water bodies lead to automatic selection of ARGs (Baker-Austin et al., 2006; Seiler and Berendonk, 2012; Li et al., 2017).
It is hypothesized that weathering can intensify both HM (Prunier et al., 2019) and AB association (Zhou et al., 2020) with MPs and potential migration of additives (Commission Regulation, 2011) from the polymer. Indeed, the European Chemical Agency has identified 1,550 additives (European Chemical Agency, 2019), many of which are known to leach into the environment (De-la-Torre et al., 2020; Bolívar-Subirats et al., 2021) as they are generally not chemically bound to polymers and can thus potentially migrate (Hahladakis et al., 2018). Metals and metal-based additives are mostly used as colorants and fillers, and research on release rate of toxic HM additives (e.g., Cd, Pb, Sb, Sn) in plastics during recycling is ongoing (Hahladakis et al., 2018). Long-term impact of MP pollution on the development of AMR is yet unclear, but AB retention, ARG presence, and exchange of ARGs through horizontal gene transfer among bacteria on MPs has been shown (Imran et al., 2019; Yang et al., 2019; Zhu et al., 2019).
In this mini-review, we discuss the role of plastisphere in the development of AMR, and the current technologies used to address various aspects of AB-HM-MP pollution and highlight the data gaps, novel techniques, and approaches.
MP abundance and polymer type are determined by microscopy and spectroscopy methods (Figure 2). The first steps(s) in analyzing plastic from environmental samples usually comprises of different separation and/or purification procedures. Separation frequently consists of passing samples through sieves or filter membranes (Fu et al., 2020). The purification process commonly involves treatment with, for example, ethanol (Zettler et al., 2013; Dussud et al., 2018), purified sea water (Dussud et al., 2018), or strong acidic and/or alkaline solutions (Cole et al., 2014; Imhof et al., 2016). Stereomicroscopes are used for the general estimate of MPs in environmental samples but also to characterize their surface, size, and shape (Gimiliani et al., 2020; Zhang Y. et al., 2020). Roughness and hydrophobicity of MPs is evaluated by tensiometry, measuring the contact angle of water drops (Dussud et al., 2018; Hossain et al., 2019). For visualization with a greater resolving power, scanning electron microscopy (SEM) (Arias-Andres et al., 2018; Li et al., 2018), or atomic force microscopy (AFM) (Dussud et al., 2018) are used. The main difference between stereomicroscopy and SEM is their resolution limit of around 200 and 2 nm, respectively. AFM has a third dimension of magnification (the z-axis), enabling constructing landscape maps of surfaces. Spectroscopy methods based on molecular vibration such as Raman spectroscopy (Zettler et al., 2013; Amaral-Zettler et al., 2015; Imhof et al., 2016), Fourier Transform Infrared Spectroscopy (FTIR)(Bryant et al., 2016; Laganà et al., 2019; Zhang Y. et al., 2020), and attenuated total reflection-FTIR (Amaral-Zettler et al., 2015; Viršek et al., 2017) allow to decipher MPs chemical makeup for more precise identification. X-ray diffraction can provide the crystalline structure of MPs (Li et al., 2018). Nevertheless, there is a gap in research, because current technologies still have difficulties in accurately detecting and characterizing the chemical properties of extensively degraded plastics (especially MPs ≤50 μm and NPs) from complex environmental samples (Lehner et al., 2019). Examples include lacking a standard procedure for separating and/or purifying samples from different matrices and using purification steps that may damage the plastic (Lö et al., 2017), microscopy techniques not providing information on plastic composition (Müller et al., 2020), and spectroscopy techniques like Raman and FTIR not having the resolution power needed for NP characterization (Imhof et al., 2016; Mason et al., 2018; Fu et al., 2020).
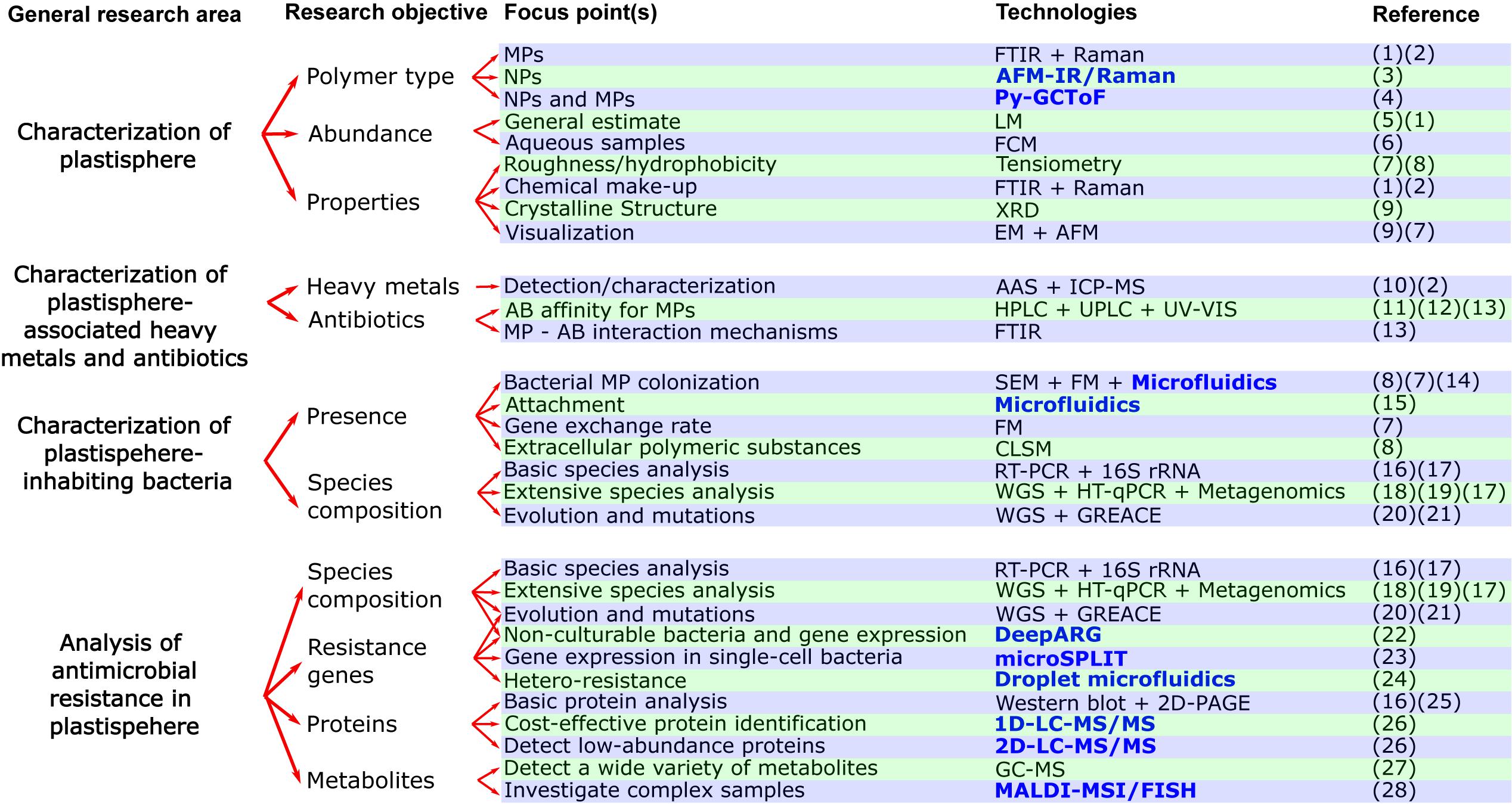
Figure 2. Flow chart with current and novel (written in bold and blue color) approaches for studying the effects of plastisphere-associated pollutants (heavy metals, antibiotics) on antimicrobial resistance. Abbreviations for technologies in alphabetical order: AAS, Atomic absorption spectroscopy; AFM, Atomic Force Microscopy; AFM-IR/Raman, Atomic force microscopy infrared/Raman; CLSM, Confocal Laser Scanning Microscopy; DeepARG, Deep learning model for antibiotic resistance genes; EM, Electron microscopy; FCM, Flow cytometry; PFGE, Pulsed-field gel electrophoresis; FM, Fluorescence microscopy; FTIR, Fourier transform infrared microscopy; GC-MS, Gas chromatography–mass spectrometry; GREACE, Genome Replication Engineering Assisted Continuous Evolution; HPLC, High-performance liquid chromatography; HT-qPCR, High-throughput qPCR; ICP-MS, Inductively coupled plasma mass spectrometry; LM, Light microscopy; MALDI-MSI/FISH, Matrix assisted laser desorption/ionization–Mass spectrometry imaging/Fluorescence in situ hybridization; microSPLIT, Microbial Split-Pool Ligation Transcriptomics; Py-GCToF, Pyrolysis–Gas Chromatography Time of Flight Mass Spectrometry; RT-PCR, Reverse transcription polymerase chain reaction; SEM, Scanning Electron Microscopy; UPLC, Ultra-performance liquid chromatography; UV-VIS, Ultraviolet–visible spectrophotometry; XRD, X-ray diffraction; WGS, Whole genome sequencing; 1D/2D-LC-MS/MS, One dimensional/Two dimensional online separation-liquid chromatography-tandem mass spectrometry; 2D-PAGE, Two-dimensional gel electrophoresis. References in numerical order: (1) = (Zhang Y. et al., 2020), (2) = (Imhof et al., 2016), (3) = (Fu et al., 2020), (4) = (Sullivan et al., 2020), (5) = (Gimiliani et al., 2020), (6) = (Kaile et al., 2020), (7) = (Dussud et al., 2018), (8) = (Hossain et al., 2019, (9) = (Li et al., 2018), (10) = (Munier and Bendell, 2018), (11) = (Bolívar-Subirats et al., 2021), (12) = (Zhang et al., 2018), (13) = (Yu et al., 2020b), (14) = (Pousti et al., 2019), (15) = (Secchi et al., 2020), (16) = (Leng et al., 2019), (17) = (Meier et al., 2020), (18) = (Pathak et al., 2020), (19) = (Zhao Y. et al., 2019), (20) = (Li X. et al., 2019), (21) = (Qin et al., 2019), (22) = (Cuadrat et al., 2020), (23) = (Kuchina et al., 2020), (24) = (Scheler et al., 2020), (25) = (Bar et al., 2007), (26) = (Hinzke et al., 2019), (27) = (Li W. et al., 2019), (28) = (Geier et al., 2020).
Absorption of light and mass-to-charge ratio are used to measure the content of HMs within and on the surface of MPs via atomic absorption spectroscopy (Brennecke et al., 2016; Cabral et al., 2016; Munier and Bendell, 2018) and inductively coupled plasma mass spectrometry (ICP-MS) (Rochman et al., 2014; Cabral et al., 2016; Imhof et al., 2016), respectively (Figure 2). The latter being the gold standard for detecting and characterizing metals. AB affinity for MP has been studied in batch adsorption experiments in laboratory settings. High-performance liquid chromatography (HPLC) coupled to a diode array detector (Guo et al., 2019; Guo and Wang, 2019) and to a triple quadruple detector (Bolívar-Subirats et al., 2021) and ultra-performance liquid chromatography coupled to a photodiode array detector (Zhang et al., 2018) but also UV-visible spectroscopy have been used to determine MP-sorbed AB (Wan et al., 2019; Yu et al., 2020a,b; Figure 2). In addition, FTIR has been used to characterize the interaction mechanisms between MPs and ABs (Wan et al., 2019; Yu et al., 2020a,b; Figure 2). Although technology has allowed in-depth analysis of HMs and ABs, there is a knowledge gap on how the affinity for pollutants differs from MPs and NPs degraded from larger plastic due to weathering processes to primary MPs and NPs.
MPs in water bodies form an ideal substratum for bacterial biofilm formation as they adsorb nutrients and organic matter from the essentially nutrient-poor water habitat supporting the growth of bacteria. Generally, the colonization of MPs is a very rapid process (within 24 h) depending on a variety of factors (Oberbeckmann et al., 2015) of which environmental factors and not the plastic type have recently been shown to be the most significant influencer for microbial composition on MPs (Wright et al., 2020).
Bacterial association with MPs is analyzed by SEM and fluorescence microscopy) (Zettler et al., 2013; Bryant et al., 2016; Arias-Andres et al., 2018; Dussud et al., 2018; Hossain et al., 2019; Figure 2). Fluorescence can further examine the gene exchange rate within biofilm communities and planktonic bacteria (Arias-Andres et al., 2018), accomplished via fluorescent self-transmissible plasmids (Arias-Andres et al., 2018). To study the extracellular polymeric substances of the biofilm matrix, confocal laser scanning microscopy (CLSM) is used (Hossain et al., 2019; Figure 2). This is due to CLSM’s ability of obtaining high-resolution images in various depths of a sample, usually 50–100 μm in biological samples (Jonkman et al., 2020). Overall, research in this area has greatly expanded due to the above-mentioned technologies; however, there is a gap in analysis and modeling of microbial colonization of both MPs and NPs in different environmental settings. Change in species diversity is mostly investigated by sequencing variable regions from the conserved 16S ribosomal RNA (16S rRNA) (Knapp et al., 2017; Zhao et al., 2018; Zhao Y. et al., 2019; Chen et al., 2019; Learman et al., 2019; Meier et al., 2020; Figure 2). Sequencing methods, including whole genome sequencing (WGS), are also ideal for studying effect of HMs on resistance-related genes in bacteria (Pathak et al., 2020; Figure 2).
Metagenomics with possible combination of metatranscriptomics permits analysis of the species present in the microbial community, including non-culturable bacteria, while simultaneously studying regulation of ARGs, MRGs, and other genes at the mRNA/functional level within the whole community (Cabral et al., 2016; Meier et al., 2020; Figure 2). Functional metagenomics is useful for screening of resistance genes that are expressed in specific environments such as HM polluted sites, while further allowing discovery of possible genes with novel functions (Cheng et al., 2012; Staley et al., 2015). Further methods for routine examination are polymerase chain reaction (PCR) techniques (Medardus et al., 2014; Knapp et al., 2017; Chen et al., 2019), although here, only a limited number of genes are investigated (Zhang Y. et al., 2020). This can be overcome by high-throughput qPCR chip technologies (Zhao et al., 2018; Zhao Y. et al., 2019) and WGS (Learman et al., 2019; Figure 2). Alternations in gene expression levels caused by HM exposure can be determined by reverse transcription PCR for specific genes (Leng et al., 2019) or with metatranscriptomics for the whole transcriptome (Cabral et al., 2016). Many different ecosystems have now been investigated for ARGs and MRGs, yet a comprehensive overview of the ARG and MRG prevalence remains to be done. Another gap is single-cell bacterial research, as the effect of HMs and plastic on AMR at the level of single-bacterium is virtually non-existent. This is mainly due to lack of technologies being able to extract and analyze their genetic material (Kuchina et al., 2020).
Sequencing in combination with long-term experiments can detect mutations that occur in bacteria during prolonged growth in HM rich environments (Chi et al., 2017; Li X. et al., 2019; Qin et al., 2019). These experiments include serial long-term culturing of resistant mutants exposed to subtoxic levels of HMs, followed by WGS (Li X. et al., 2019). Genome Replication Engineering Assisted Continuous Evolution is another alternative, in which evolution of resistant mutants is accelerated before sequencing (Qin et al., 2019; Figure 2). In vivo experiments with mice being exposed to HMs via oral administration followed by sequencing of the gut microbiota have shed light on the effect of HMs in mammals in vivo (Chi et al., 2017). There are several ways to explore how HMs can have an effect on the protein and metabolite level in bacterial monocultures. A frequently used method for proteomics is two-dimensional gel electrophoresis (Bar et al., 2007; Figure 2). Another way to explore protein expression is through liquid chromatography-tandem mass spectrometry (LC-MS/MS) where proteins are first separated by LC and then ionized and characterized by mass-to-charge ratio and relative abundance (Li W. et al., 2019; Figure 2). Western blotting is another widely applied technique (Leng et al., 2019). For studying metabolites, gas chromatography-mass spectrometry is usually performed (Leng et al., 2019; Figure 2). Although working with monoculture bacteria does not mimic the true situation of microbial interaction in the environment, possible impact of HMs and plastics on the proteomics/metabolomics level in different types of bacteria is currently still a research gap in need of investigation. This gap needs to be addressed in the light of recent studies, such as the one by Li W. et al. (2019), showing that an alternation in bacterial metabolic pathways may affect their AMR. Exposure to HMs has already shown the ability to alter metabolic pathways of bacteria in the gut (Chi et al., 2017); nonetheless, there is still much to be learned on possible influence of HMs and perhaps plastics on the different pathways and the interplay with AMR.
The most optimal solution for future development of a standard method for quantifying and characterizing the composition of smaller fractions of MPs, including NPs (<1 μm), might be to merge completely new analytical methods with the existing technologies (Nguyen B. et al., 2019; Fu et al., 2020). For now, combining AFM with infrared spectroscopy or Raman seems promising, since AFM offers relatively simple sample preparation, and samples can be conserved during analysis (Fu et al., 2020; Figure 2). One disadvantage is, however, that obtaining quality imaging of the sample depends on how flat and smooth the sample is (Fu et al., 2020), making it difficult for simultaneous investigation of NPs and larger MPs. To obtain a more complete overview, at least in aqueous samples, we recommend analysis of plastics on PTFE membranes combined with Pyrolysis-Gas Chromatography Time of Flight Mass Spectrometry (Py-GCToF) (Sullivan et al., 2020; Figure 2). This analytical method is based on analyzing thermal degradation products, and it has shown to be fast, reliable, and have high resolution (Sullivan et al., 2020). A second option for aqueous samples that might be more easily standardized for future environmental identification and quantification of MPs and NPs (0.2–2 μm), is flow cytometry in combination with staining and cell sorting (Kaile et al., 2020).
Change in MPs and NPs composition and their affinity for pollutants and microbes can be uncovered by merging analytical and sequencing technology with in situ and ex situ experiments. Ex situ batch sorption experiments provide the opportunity to focus on specific parameters (Li et al., 2018; Zhang H. et al., 2020), while in situ studies are necessary to observe the real-life complex interaction of MPs and NPs with their surroundings (Oberbeckmann et al., 2017). Depending on the environment investigated, suitable analytical techniques for detection and characterization of MPs and NPs in such experiments could be either Py-GCToF (Sullivan et al., 2020) or micro-FTIR and Raman spectroscopy (Lö et al., 2017; Figure 2). HPLC and ICP-MS could further be used for AB and HM detection, respectively (Cabral et al., 2016; Zhang H. et al., 2020; Figure 2). Finally, microbial analysis in the experiments would need to include both analytical tools for studying biofilm (e.g., CLSM) and metagenomic sequencing for discovering possible ARGs and MRGs as well as species diversity (Cabral et al., 2016; Hossain et al., 2019).
Accumulation of MPs in the food chain and the effect on spread of AMR should be investigated by long-term in vivo studies combined with multidisciplinary tools such as NGS sequencing, ICP-MS, and vibrational spectroscopy methods. Previous in vivo studies focusing on influence of MP accumulation are inconsistent in their methods and yield conflicting results (Van Raamsdonk et al., 2020). One issue with standardizing in vivo studies is the complexity of the sample material, making it difficult to detect MPs. This could be solved by an enzymatic purification method for MPs/NPs developed by Lö et al. (2017), which can remove organic and inorganic material from different matrices while not affecting the polymers and couple it to micro-FTIR and Raman spectroscopy (Figure 2). Lö et al. (2017) provides a step-by-step guide to the enzymatic purification, which includes optional subdivision of samples, usage of specific buffers, and lipase and amylase for samples with high lipid or polysaccharide content.
Obtaining a wider overview of microbial communities, including spread of ARGs and MRGs in different habitats, is feasible with modern NGS approaches. There are two aspects that should be considered in future analyses: (1) presence of non-culturable bacteria and (2) expression level of resistant genes in the bacterial communities. Integrating metagenomics and metatranscriptomics with machine-learning tools such as DeepARG, trained to find the existing and novel ARGs and MRGs, is a suitable option for this challenge (Arango-Argoty et al., 2018; Cuadrat et al., 2020; Figure 2). Studying heterogeneous modulation of gene expression by HMs (and MPs/NPs) in a single bacterium is possible, but single-cell RNA sequencing (scRNA-seq) studies are still scarce due to differences from eukaryotic cells such as low mRNA content and lack of polyadenylation. This challenge could be overcome by the scRNA-seq platform Microbial Split-Pool Ligation Transcriptomics (Kuchina et al., 2020; Figure 2). The approach was recently adapted for Bacillus subtilis and Escherichia coli by Kuchina et al. (2020) and has advantages such as: (1) no need for single cell physical isolation, (2) compatibility with a wide range of cell shapes and sizes; and (3) enables use of un-encapsulated and fixed cells (Ma et al., 2019).
Proteomic and metabolomic pathways in bacteria play an important role in AMR (Li W. et al., 2019). 1D and 2D-LC-MS/MS spectrometry and mass spectrometry imaging (MSI) can be used for analyzing the impact that MPs and HMs potentially have on proteomics/metabolomics activity levels in the bacteria. In their extensive testing of LC-MS/MS spectrometry, Hinzke et al. (2019) suggest the most cost-effective method for maximizing the number of identified proteins by MS is online separation by 1D-LC. For a more precise guidance for specific objectives, we refer to the flow chart and overall work of Hinzke et al. (2019) (Figure 2). Though, regardless of technology used, it is essential that proteome bioinformatics progresses in parallel with the recent advances in MS methods; otherwise, proteomic analysis will remain limited (Ameen and Raza, 2017; Petriz and Franco, 2017). In congruence, technology needed for analysis of metabolites is restricted as well (Dunham et al., 2017). One promising method is MSI, because it provides chemical and spatial analysis and methods can easily be adapted to specific environmental samples. MSI works by distinguishing chemical compounds via their mass-to-charge ratio, and currently, there are three MSI methods commercially available for analyzing bacteria (Dunham et al., 2017). Though the main limiting factor is that any single MSI experiment only gives a fraction of the metabolites present in samples (Dunham et al., 2017). Nevertheless, we believe that when combined with other technologies, it could pave the way for future metabolomic research as exemplified by the work of Geier et al. (2020), where matrix-assisted laser desorption/ionization MSI was combined with FISH microscopy (Figure 2). This enabled linking metabolomes to groups of 50–100 microbial cells in complex environmental samples and be able to resolve single-cell bacteria in the near future (Geier et al., 2020).
Microbial colonization and ability to form biofilm are also heterogenous characteristics of microbes, and they play a key role in AMR. Microfluidic platforms show great potential for enabling complex biofilm studies (Yawata et al., 2016; Pousti et al., 2019), including the scarcely researched effect of flow rate and motility of bacteria on attachment (Secchi et al., 2020; Figure 2). Bacteria communities, both hetero and isogenic, can contain cells with diverse range of resistance (El-Halfawy and Valvano, 2015). Droplet microfluidic technology could be the most promising tool for such investigation because it allows high-throughput culturing of bacteria at wide range of isolated conditions (Kaminski et al., 2016; Scheler et al., 2020; Figure 2).
In this mini-review, we highlighted technologies that have been used for analyzing different aspects of plastisphere-associated AMR. Although we found that many different aspects of AMR have been explored through multiple studies using advanced methods, knowledge gaps remain. To address these gaps, we summarize currently available technologies potentially suitable for future research. This should provide analytical tools for scientists of diverse backgrounds seeking answers for complex urgent problems: HM- and AB-contaminated plastisphere-associated promotion of AMR.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This research was funded by the TTÜ development program 2016-2022, project code 2014-2020.4.01.16-0032, the Estonian Research Council grants MOBJD556 (SB), MOBTP109 (OS) and PUT1512 (MH), and the European Regional Development Fund project TK134 (AK).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ali, H., Khan, E., and Ilahi, I. (2019). Environmental chemistry and ecotoxicology of hazardous heavy metals: environmental persistence, toxicity, and bioaccumulation. J. Chem. 2019:6730305. doi: 10.1155/2019/6730305
Amaral-Zettler, L. A., Zettler, E. R., Slikas, B., Boyd, G. D., Melvin, D. W., Morrall, C. E., et al. (2015). The biogeography of the plastisphere: implications for policy. Front. Ecol. Environ. 13:541–546. doi: 10.1890/150017
Ameen, A., and Raza, S. (2017). Metaproteomics approaches and techniques: a review. Int. J. Adv. Sci. Res. 3:49. doi: 10.7439/ijasr.v3i5.4167
Arango-Argoty, G., Garner, E., Pruden, A., Heath, L. S., Vikesland, P., and Zhang, L. (2018). DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 6:23. doi: 10.1186/s40168-018-0401-z
Arias-Andres, M., Klümper, U., Rojas-Jimenez, K., and Grossart, H. P. (2018). Microplastic pollution increases gene exchange in aquatic ecosystems. Environ. Pollut. 237, 253–261. doi: 10.1016/j.envpol.2018.02.058
Baker-Austin, C., Wright, M. S., Stepanauskas, R., and McArthur, J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182. doi: 10.1016/J.TIM.2006.02.006
Bar, C., Patil, R., Doshi, J., Kulkarni, M. J., and Gade, W. N. (2007). Characterization of the proteins of bacterial strain isolated from contaminated site involved in heavy metal resistance-A proteomic approach. J. Biotechnol. 128, 444–451. doi: 10.1016/j.jbiotec.2006.11.010
Besseling, E., Redondo-Hasselerharm, P., Foekema, E. M., and Koelmans, A. A. (2019). Quantifying ecological risks of aquatic micro- and nanoplastic. Crit. Rev. Environ. Sci. Technol. 49, 32–80. doi: 10.1080/10643389.2018.1531688
Bolívar-Subirats, G., Rivetti, C., Cortina-Puig, M., Barata, C., and Lacorte, S. (2021). Occurrence, toxicity and risk assessment of plastic additives in Besos river, Spain. Chemosphere 263:128022. doi: 10.1016/j.chemosphere.2020.128022
Brennecke, D., Duarte, B., Paiva, F., Caçador, I., and Canning-Clode, J. (2016). Microplastics as vector for heavy metal contamination from the marine environment. Estuar. Coast. Shelf Sci. 178, 189–195. doi: 10.1016/J.ECSS.2015.12.003
Bryant, J. A., Clemente, T. M., Viviani, D. A., Fong, A. A., Thomas, K. A., Kemp, P., et al. (2016). Diversity and activity of communities inhabiting plastic debris in the north pacific gyre. mSystems 1:e00024–16. doi: 10.1128/msystems.00024-16
Cabral, L., Júnior, G. V. L., Pereira de Sousa, S. T., Dias, A. C. F., Lira Cadete, L., Andreote, F. D., et al. (2016). Anthropogenic impact on mangrove sediments triggers differential responses in the heavy metals and antibiotic resistomes of microbial communities. Environ. Pollut. 216, 460–469. doi: 10.1016/J.ENVPOL.2016.05.078
Chen, J., Li, J., Zhang, H., Shi, W., and Liu, Y. (2019). Bacterial heavy-metal and antibiotic resistance genes in a copper tailing dam area in northern China. Front. Microbiol. 10:1916. doi: 10.3389/fmicb.2019.01916
Cheng, G., Hu, Y., Yin, Y., Yang, X., Xiang, C., Wang, B., et al. (2012). Functional screening of antibiotic resistance genes from human gut microbiota reveals a novel gene fusion. FEMS Microbiol. Lett. 336, 11–16. doi: 10.1111/j.1574-6968.2012.02647.x
Chi, L., Bian, X., Gao, B., Tu, P., Ru, H., and Lu, K. (2017). The effects of an environmentally relevant level of arsenic on the gut microbiome and its functional metagenome. Soc. Toxicol. 162, 193–204. doi: 10.1093/toxsci/kfx174
Cole, M., Webb, H., Lindeque, P. K., Fileman, E. S., Halsband, C., and Galloway, T. S. (2014). Isolation of microplastics in biota-rich seawater samples and marine organisms. Sci. Rep. 4:4528. doi: 10.1038/srep04528
Commission Regulation [EU] (2011). Commission Regulation (EU) No 10/2011 of 14 January 2011 on Plastic Materials and Articles Intended to Come into Contact with Food Text with EEA Relevance. Available online at: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32011R0010 (accessed August 14, 2020).
Corradini, F., Meza, P., Eguiluz, R., Casado, F., Huerta-Lwanga, E., and Geissen, V. (2019). Evidence of microplastic accumulation in agricultural soils from sewage sludge disposal. Sci. Total Environ. 671, 411–420. doi: 10.1016/j.scitotenv.2019.03.368
Cox, K. D., Covernton, G. A., Davies, H. L., Dower, J. F., Juanes, F., and Dudas, S. E. (2019). Human consumption of microplastics. Environ. Sci. Technol. 53, 7068–7074. doi: 10.1021/acs.est.9b01517
Cuadrat, R. R. C., Sorokina, M., Andrade, B. G., Goris, T., and Avila, A. M. R. D. (2020). Global ocean resistome revealed: exploring antibiotic resistance gene abundance and distribution in TARA oceans samples. Gigascience 9:giaa046. doi: 10.1093/gigascience/giaa046
Dadgostar, P. (2019). Antimicrobial resistance: implications and costs. Infect. Drug Resist. 12, 3903–3910. doi: 10.2147/IDR.S234610
de Kraker, M. E. A., Stewardson, A. J., and Harbarth, S. (2016). Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 13:e1002184. doi: 10.1371/journal.pmed.1002184
de Wit, W., and Bigaud, N. (2019). No Plastic in Nature: Assessing Plastic Ingestion from Nature to People. Gland: World Wide Fund For Nature.
De-la-Torre, G. E., Dioses-Salinas, D. C., Pizarro-Ortega, C. I., and Saldaña-Serrano, M. (2020). Global distribution of two polystyrene-derived contaminants in the marine environment: a review. Mar. Pollut. Bull. 161:111729. doi: 10.1016/j.marpolbul.2020.111729
Dunham, S. J. B., Ellis, J. F., Li, B., and Sweedler, J. V. (2017). Mass spectrometry imaging of complex microbial communities. Acc. Chem. Res. 50, 96–104. doi: 10.1021/acs.accounts.6b00503
Dussud, C., Hudec, C., George, M., Fabre, P., Higgs, P., Bruzaud, S., et al. (2018). Colonization of non-biodegradable and biodegradable plastics by marine microorganisms. Front. Microbiol. 9:1571. doi: 10.3389/fmicb.2018.01571
El-Halfawy, O. M., and Valvano, M. A. (2015). Antimicrobial heteroresistance: an emerging field in need of clarity. Clin. Microbiol. Rev. 28, 191–207. doi: 10.1128/CMR.00058-14
European Chemical Agency (2019). Plastic Additives Initiative Supplementary Information on Scope and Methods. Available online at: https://echa.europa.eu/echa-irs (accessed August 14, 2020).
Frias, J. P. G. L., and Nash, R. (2019). Microplastics: finding a consensus on the definition. Mar. Pollut. Bull. 138, 145–147. doi: 10.1016/j.marpolbul.2018.11.022.
Fu, W., Min, J., Jiang, W., Li, Y., and Zhang, W. (2020). Separation, characterization and identification of microplastics and nanoplastics in the environment. Sci. Total Environ. 721:137561. doi: 10.1016/j.scitotenv.2020.137561
Geier, B., Sogin, E. M., and Michellod, D. (2020). Spatial metabolomics of in situ host–microbe interactions at the micrometre scale. Nat. Microbiol. 5, 498–510. doi: 10.1038/s41564-019-0664-6
Gigault, J., ter Halle, A., Baudrimont, M., Pascal, P.-Y., Gauffre, F., Phi, T.-L., et al. (2018). Current opinion: what is a nanoplastic? Environ. Pollut. 235, 1030–1034. doi: 10.1016/j.envpol.2018.01.024
Gimiliani, G. T., Fornari, M., Redígolo, M. M., Willian Vega Bustillos, J. O., Moledo de Souza Abessa, D., and Faustino Pires, M. A. (2020). Simple and cost-effective method for microplastic quantification in estuarine sediment: a case study of the Santos and São Vicente Estuarine System. Case Stud. Chem. Environ. Eng. 2:100020. doi: 10.1016/j.cscee.2020.100020
Guo, X., Chen, C., and Wang, J. (2019). Sorption of sulfamethoxazole onto six types of microplastics. Chemosphere 228, 300–308. doi: 10.1016/j.chemosphere.2019.04.155
Guo, X., and Wang, J. (2019). Sorption of antibiotics onto aged microplastics in freshwater and seawater. Mar. Pollut. Bull. 149:110511. doi: 10.1016/j.marpolbul.2019.110511
Hahladakis, J. N., Velis, C. A., Weber, R., Iacovidou, E., and Purnell, P. (2018). An overview of chemical additives present in plastics: migration, release, fate and environmental impact during their use, disposal and recycling. J. Hazard. Mater. 344, 179–199. doi: 10.1016/j.jhazmat.2017.10.014
Hinzke, T., Kouris, A., Hughes, R.-A., Strous, M., and Kleiner, M. (2019). More is not always better: evaluation of 1D and 2D-LC-MS/MS methods for metaproteomics. Front. Microbiol. 10:238. doi: 10.3389/fmicb.2019.00238
Hossain, M. R., Jiang, M., Wei, Q., and Leff, L. G. (2019). Microplastic surface properties affect bacterial colonization in freshwater. J. Basic Microbiol. 59, 54–61. doi: 10.1002/jobm.201800174
Imhof, H. K., Laforsch, C., Wiesheu, A. C., Schmid, J., Anger, P. M., Niessner, R., et al. (2016). Pigments and plastic in limnetic ecosystems: a qualitative and quantitative study on microparticles of different size classes. Water Res. 98, 64–74. doi: 10.1016/j.watres.2016.03.015
Imran, M., Das, K. R., and Naik, M. M. (2019). Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Chemosphere 215, 846–857. doi: 10.1016/J.CHEMOSPHERE.2018.10.114
Interagency Coordination Group on Antimicrobial Resistance (2019). No Time to Wait: Securing the Future from Drug-Resistant Infections Report to the Secretary-General of the United Nations. Available online at: https://www.who.int/antimicrobial-resistance/interagency-coordination-group/final-report/en/ (accessed April 3, 2020).
Jonkman, J., Brown, C. M., Wright, G. D., Anderson, K. I., and North, A. J. (2020). Tutorial: guidance for quantitative confocal microscopy. Nat. Protoc. 15, 1585–1611. doi: 10.1038/s41596-020-0313-9
Kaile, N., Lindivat, M., Elio, J., Thuestad, G., Crowley, Q. G., and Hoell, I. A. (2020). Preliminary results from detection of microplastics in liquid samples using flow cytometry. Front. Mar. Sci. 7:856. doi: 10.3389/fmars.2020.552688
Kaminski, T. S., Scheler, O., and Garstecki, P. (2016). Droplet microfluidics for microbiology: techniques, applications and challenges. Lab Chip 16, 2168–2187. doi: 10.1039/C6LC00367B
Keswani, A., Oliver, D. M., Gutierrez, T., and Quilliam, R. S. (2016). Microbial hitchhikers on marine plastic debris: human exposure risks at bathing waters and beach environments. Mar. Environ. Res. 118, 10–19. doi: 10.1016/j.marenvres.2016.04.006
Knapp, C. W., Callan, A. C., Aitken, B., Shearn, R., Koenders, A., and Hinwood, A. (2017). Relationship between antibiotic resistance genes and metals in residential soil samples from Western Australia. Environ. Sci. Pollut. Res. 24, 2484–2494. doi: 10.1007/s11356-016-7997-y
Kraemer, S. A., Ramachandran, A., and Perron, G. G. (2019). Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms 7:180. doi: 10.3390/microorganisms7060180
Kuchina, A., Brettner, L. M., Paleologu, L., Roco, C. M., Rosenberg, A. B., Carignano, A., et al. (2020). Microbial single-cell RNA sequencing by split-pool barcoding. Science 371:eaba5257. doi: 10.1126/SCIENCE.ABA5257
Laganà, P., Caruso, G., Corsi, I., Bergami, E., Venuti, V., Majolino, D., et al. (2019). Do plastics serve as a possible vector for the spread of antibiotic resistance? First insights from bacteria associated to a polystyrene piece from King George Island (Antarctica). Int. J. Hyg. Environ. Health 222, 89–100. doi: 10.1016/J.IJHEH.2018.08.009
Learman, D. R., Ahmad, Z., Brookshier, A., Henson, M. W., Hewitt, V., Lis, A., et al. (2019). Comparative genomics of 16 Microbacterium spp. that tolerate multiple heavy metals and antibiotics. PeerJ 6:e6258. doi: 10.7717/peerj.6258
Lehner, R., Weder, C., Petri-Fink, A., and Rothen-Rutishauser, B. (2019). Emergence of nanoplastic in the environment and possible impact on human health. Environ. Sci. Technol. 53, 1748–1765. doi: 10.1021/acs.est.8b05512
Leng, F., Li, Y., Luo, W., Wei, Q., Jing, Y., Wang, X., et al. (2019). Cloning, expression, and bioinformatics analysis of heavy metal resistance gene afe_1862 from Acidithiobacillus ferrooxidans L1 in Escherichia coli. Biol. Trace Elem. Res. 189, 291–300. doi: 10.1007/s12011-018-1462-3
Li, J., Zhang, K., and Zhang, H. (2018). Adsorption of antibiotics on microplastics. Environ. Pollut. 237, 460–467. doi: 10.1016/J.ENVPOL.2018.02.050
Li, L.-G., Xia, Y., and Zhang, T. (2017). Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J. 11, 651–662. doi: 10.1038/ismej.2016.155
Li, W., Wang, G., Zhang, S., Fu, Y., Jiang, Y., Yang, X., et al. (2019). An integrated quantitative proteomic and metabolomics approach to reveal the negative regulation mechanism of LamB in antibiotics resistance. J. Proteomics 194, 148–159. doi: 10.1016/J.JPROT.2018.11.022
Li, X., Gu, A. Z., Zhang, Y., Xie, B., Li, D., and Chen, J. (2019). Sub-lethal concentrations of heavy metals induce antibiotic resistance via mutagenesis. J. Hazard. Mater. 369, 9–16. doi: 10.1016/J.JHAZMAT.2019.02.006
Lö, M. G. J., Imhof, H. K., Ladehoff, M., Lö, L. A., Lorenz, C., Mintenig, S., et al. (2017). Enzymatic purification of microplastics in environmental samples. Environ. Sci. Technol. 51, 14283–14292. doi: 10.1021/acs.est.7b03055
Ma, Q., Bücking, H., Hernandez, J. L. G., and Subramanian, S. (2019). Single-cell RNA sequencing of plant-associated bacterial communities. Front. Microbiol. 10:2452. doi: 10.3389/fmicb.2019.02452
Mason, S. A., Welch, V. G., and Neratko, J. (2018). Synthetic polymer contamination in bottled water. Front. Chem. 6:407. doi: 10.3389/fchem.2018.00407
Medardus, J. J., Molla, B. Z., Nicol, M., Morrow, W. M., Rajala-Schultz, P. J., Kazwala, R., et al. (2014). In-feed use of heavy metal micronutrients in U.S. swine production systems and its role in persistence of multidrug-resistant salmonellae. Appl. Environ. Microbiol. 80, 2317–2325. doi: 10.1128/AEM.04283-13
Meier, M. J., Dodge, A. E., Samarajeewa, A. D., and Beaudette, L. A. (2020). Soil exposed to silver nanoparticles reveals significant changes in community structure and altered microbial transcriptional profiles. Environ. Pollut. 258:113816. doi: 10.1016/j.envpol.2019.113816
Müller, Y. K., Wernicke, T., Pittroff, M., Witzig, C. S., Storck, F. R., Klinger, J., et al. (2020). Microplastic analysis—are we measuring the same? Results on the first global comparative study for microplastic analysis in a water sample. Anal. Bioanal. Chem. 412, 555–560. doi: 10.1007/s00216-019-02311-1
Munier, B., and Bendell, L. I. (2018). Macro and micro plastics sorb and desorb metals and act as a point source of trace metals to coastal ecosystems. PLoS One 13:e0191759. doi: 10.1371/journal.pone.0191759
Nguyen, B., Claveau-Mallet, D., Hernandez, L. M., Xu, E. G., Farner, J. M., and Tufenkji, N. (2019). Separation and analysis of microplastics and nanoplastics in complex environmental samples. Acc. Chem. Res. 52, 858–866. doi: 10.1021/acs.accounts.8b00602
Nguyen, C. C., Hugie, C. N., Kile, M. L., and Navab-Daneshmand, T. (2019). Association between heavy metals and antibiotic-resistant human pathogens in environmental reservoirs: a review. Front. Environ. Sci. Eng. 13:46. doi: 10.1007/s11783-019-1129-0
Nizzetto, L., Futter, M., and Langaas, S. (2016). Are agricultural soils dumps for microplastics of urban origin? Environ. Sci. Technol. 50, 10777–10779. doi: 10.1021/acs.est.6b04140
Oberbeckmann, S., Kreikemeyer, B., and Labrenz, M. (2017). Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 8:2709. doi: 10.3389/FMICB.2017.02709
Oberbeckmann, S., Löder, M. G. J., and Labrenz, M. (2015). Marine microplastic-associated biofilms – a review. Environ. Chem. 12, 551–562. doi: 10.1071/EN15069
O’Neill, J. (2016). Tackling Drug-Resistant Infections Globally: Final Report and Recommendations the Review on Antimicrobial Resistance. Available online at: https://amr-review.org/ (accessed April 3, 2020).
Pathak, A., Jaswal, R., and Chauhan, A. (2020). Genomic characterization of a mercury resistant Arthrobacter sp. H-02-3 reveals the presence heavy metal and antibiotic resistance determinants. Front. Microbiol. 10:3039. doi: 10.3389/fmicb.2019.03039
Pauly, J. L., Stegmeier, S. J., Allaart, H. A., Cheney, R. T., Zhang, P. J., Mayer, A. G., et al. (1998). Inhaled cellulosic and plastic fibers found in human lung tissue. Cancer Epidemiol. Biomarkers Prev. 7, 419–428.
Petriz, B. A., and Franco, O. L. (2017). Metaproteomics as a complementary approach to gut microbiota in health and disease. Front. Chem. 5:4. doi: 10.3389/fchem.2017.00004
Pousti, M., Zarabadi, M. P., Abbaszadeh Amirdehi, M., Paquet-Mercier, F., and Greener, J. (2019). Microfluidic bioanalytical flow cells for biofilm studies: a review. Analyst 144, 68–86. doi: 10.1039/c8an01526k
Prunier, J., Maurice, L., Perez, E., Gigault, J., Pierson Wickmann, A. C., Davranche, M., et al. (2019). Trace metals in polyethylene debris from the North Atlantic subtropical gyre. Environ. Pollut. 245, 371–379. doi: 10.1016/j.envpol.2018.10.043
Qin, W., Zhao, J., Yu, X., Liu, X., Chu, X., Tian, J., et al. (2019). Improving cadmium resistance in Escherichia coli through continuous genome evolution. Front. Microbiol. 10:278. doi: 10.3389/fmicb.2019.00278
Rochman, C. M., Hentschel, B. T., and Teh, S. J. (2014). Long-term sorption of metals is similar among plastic types: implications for plastic debris in aquatic environments. PLoS One 9:85433. doi: 10.1371/journal.pone.0085433
Scheler, O., Makuch, K., Debski, P. R., Horka, M., Ruszczak, A., Pacocha, N., et al. (2020). Droplet-based digital antibiotic susceptibility screen reveals single-cell clonal heteroresistance in an isogenic bacterial population. Sci. Rep. 10:3282. doi: 10.1038/s41598-020-60381-z
Schmiedgruber, M., Hufenus, R., and Mitrano, D. M. (2019). Mechanistic understanding of microplastic fiber fate and sampling strategies: synthesis and utility of metal doped polyester fibers. Water Res. 155, 423–430. doi: 10.1016/j.watres.2019.02.044
Schwabl, P., Koppel, S., Konigshofer, P., Bucsics, T., Trauner, M., Reiberger, T., et al. (2019). Detection of various microplastics in human stool: a prospective case series. Ann. Intern. Med. 171, 453–457. doi: 10.7326/M19-0618
Secchi, E., Vitale, A., Miño, G. L., Kantsler, V., Eberl, L., Rusconi, R., et al. (2020). The effect of flow on swimming bacteria controls the initial colonization of curved surfaces. Nat. Commun. 11:2851. doi: 10.1038/s41467-020-16620-y
Seiler, C., and Berendonk, T. U. (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 3:399. doi: 10.3389/fmicb.2012.00399
Staley, C., Johnson, D., Gould, T. J., Wang, P., Phillips, J., Cotner, J. B., et al. (2015). Frequencies of heavy metal resistance are associated with land cover type in the Upper Mississippi River. Sci. Total Environ. 511, 461–468. doi: 10.1016/j.scitotenv.2014.12.069
Sullivan, G. L., Gallardo, J. D., Jones, E. W., Hollliman, P. J., Watson, T. M., and Sarp, S. (2020). Detection of trace sub-micron (nano) plastics in water samples using pyrolysis-gas chromatography time of flight mass spectrometry (PY-GCToF). Chemosphere 249:126179. doi: 10.1016/j.chemosphere.2020.126179
Van Raamsdonk, L. W. D., Van Der Zande, M., Koelmans, A. A., Hoogenboom, P. L. A., Peters, R. J. B., Groot, M. J., et al. (2020). Current insights into monitoring, bioaccumulation, and potential health effects of microplastics present in the food chain. Foods 9:72. doi: 10.3390/foods9010072
Viršek, M. K., Lovšin, M. N., Koren, Š, Kržan, A., and Peterlin, M. (2017). Microplastics as a vector for the transport of the bacterial fish pathogen species Aeromonas salmonicida. Mar. Pollut. Bull. 125, 301–309. doi: 10.1016/J.MARPOLBUL.2017.08.024
Wan, T., Lu, S., Cheng, W., Ren, J., Wang, M., Hu, B., et al. (2019). A spectroscopic and theoretical investigation of interaction mechanisms of tetracycline and polystyrene nanospheres under different conditions. Environ. Pollut. 249, 398–405. doi: 10.1016/j.envpol.2019.03.049
WHO (2018). High Levels of Antibiotic Resistance Found Worldwide, New Data Shows. Available online at: https://www.who.int/news-room/detail/29-01-2018-high-levels-of-antibiotic-resistance-found-worldwide-new-data-shows (accessed February 19, 2019).
Wilkinson, A., and Boxall, J. (2019). “The first global study of pharmaceutical contamination in riverine environments,” in Proceedings of the SETAC Europe 29th Annual Meeting, Helsinki.
Wright, R. J., Langille, M. G. I., and Walker, T. R. (2020). Food or just a free ride? A meta-analysis reveals the global diversity of the Plastisphere. ISME J. 15, 789–806. doi: 10.1038/s41396-020-00814-9
Yang, Y., Liu, G., Song, W., Ye, C., Lin, H., Li, Z., et al. (2019). Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes. Environ. Int. 123, 79–86. doi: 10.1016/J.ENVINT.2018.11.061
Yawata, Y., Nguyen, J., Stocker, R., and Rusconi, R. (2016). Microfluidic studies of biofilm formation in dynamic environments. J. Bacteriol. 198, 2589–2595. doi: 10.1128/JB.00118-16
Yu, F., Li, Y., Huang, G., Yang, C., Chen, C., Zhou, T., et al. (2020a). Adsorption behavior of the antibiotic levofloxacin on microplastics in the presence of different heavy metals in an aqueous solution. Chemosphere 260:127650. doi: 10.1016/j.chemosphere.2020.127650
Yu, F., Yang, C., Huang, G., Zhou, T., Zhao, Y., and Ma, J. (2020b). Interfacial interaction between diverse microplastics and tetracycline by adsorption in an aqueous solution. Sci. Total Environ. 721:137729. doi: 10.1016/j.scitotenv.2020.137729
Zettler, E. R., Mincer, T. J., and Amaral-Zettler, L. A. (2013). Life in the “plastisphere”: microbial communities on plastic marine debris. Environ. Sci. Technol. 47, 7137–7146. doi: 10.1021/es401288x
Zhang, H., Liu, F. F., Wang, S. C., Huang, T. Y., Li, M. R., Zhu, Z. L., et al. (2020). Sorption of fluoroquinolones to nanoplastics as affected by surface functionalization and solution chemistry. Environ. Pollut. 262:114347. doi: 10.1016/j.envpol.2020.114347
Zhang, H., Wang, J., Zhou, B., Zhou, Y., Dai, Z., Zhou, Q., et al. (2018). Enhanced adsorption of oxytetracycline to weathered microplastic polystyrene: kinetics, isotherms and influencing factors. Environ. Pollut. 243, 1550–1557. doi: 10.1016/j.envpol.2018.09.122
Zhang, Y., Lu, J., Wu, J., Wang, J., and Luo, Y. (2020). Potential risks of microplastics combined with superbugs: enrichment of antibiotic resistant bacteria on the surface of microplastics in mariculture system. Ecotoxicol. Environ. Saf. 187:109852. doi: 10.1016/j.ecoenv.2019.109852
Zhao, Y., Cocerva, T., Cox, S., Tardif, S., Su, J.-Q., Zhu, Y.-G., et al. (2019). Evidence for co-selection of antibiotic resistance genes and mobile genetic elements in metal polluted urban soils. Sci. Total Environ. 656, 512–520. doi: 10.1016/J.SCITOTENV.2018.11.372
Zhao, Y., Su, J. Q., An, X. L., Huang, F. Y., Rensing, C., Brandt, K. K., et al. (2018). Feed additives shift gut microbiota and enrich antibiotic resistance in swine gut. Sci. Total Environ. 621, 1224–1232. doi: 10.1016/j.scitotenv.2017.10.106
Zhou, Q., Yang, N., Li, Y., Ren, B., Ding, X., Bian, H., et al. (2020). Total concentrations and sources of heavy metal pollution in global river and lake water bodies from 1972 to 2017. Glob. Ecol. Conserv. 22:e00925. doi: 10.1016/j.gecco.2020.e00925
Keywords: antimicrobial resistance, microplastics, heavy metals, plastisphere, emerging technologies, antibiotics
Citation: Bartkova S, Kahru A, Heinlaan M and Scheler O (2021) Techniques Used for Analyzing Microplastics, Antimicrobial Resistance and Microbial Community Composition: A Mini-Review. Front. Microbiol. 12:603967. doi: 10.3389/fmicb.2021.603967
Received: 08 September 2020; Accepted: 10 February 2021;
Published: 26 March 2021.
Edited by:
Lisa M. Durso, Agricultural Research Service, United States Department of Agriculture, United StatesReviewed by:
Dan Miller, Agricultural Research Service, United States Department of Agriculture, United StatesCopyright © 2021 Bartkova, Kahru, Heinlaan and Scheler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simona Bartkova, c2ltb25hLmJhcnRrb3ZhQHRhbHRlY2guZWU=; c2ltb25hYmFydGtvdmE4NkBob3RtYWlsLmNvbQ==; Margit Heinlaan, bWFyZ2l0LmhlaW5sYWFuQGtiZmkuZWU=
†These authors have contributed equally and share the last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.