
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 February 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.596891
This article is part of the Research Topic Antimicrobials in Wildlife and the Environment View all 12 articles
 Maria J. H. O’Hagan1*
Maria J. H. O’Hagan1* Ana V. Pascual-Linaza2
Ana V. Pascual-Linaza2 Catherine Couzens3
Catherine Couzens3 Clare Holmes4
Clare Holmes4 Colin Bell3
Colin Bell3 Nessie Spence3
Nessie Spence3 Robert J. Huey5
Robert J. Huey5 Julie A. Murphy1
Julie A. Murphy1 Ryan Devaney3
Ryan Devaney3 Angela Lahuerta-Marin3
Angela Lahuerta-Marin3Antimicrobial resistant (AMR) bacteria can be shared between humans and animals, through food, water, and the environment. Wild animals are not only potential reservoirs of AMR, but are also sentinels mirroring the presence of AMR zoonotic bacteria in the environment. In Northern Ireland, little is known about levels of AMR in bacteria in wildlife, thus the current study aimed to estimate the prevalence of AMR bacteria in wildlife using wildlife species from two ongoing surveys as a proxy. Nasopharyngeal swabs and faecal samples from European badgers (Meles meles) (146 faecal samples; 118 nasal samples) and red foxes (Vulpes vulpes) (321 faecal samples; 279 nasal samples) were collected throughout Northern Ireland and were used to survey for the presence of extended spectrum beta lactamase resistant and AmpC-type beta lactamases Escherichia coli (ESBL/AmpC), Salmonella spp. (only in badgers) and methicillin resistant Staphylococcus aureus (MRSA). ESBLs were detected in 13 out of 146 badger faecal samples (8.90%) and 37 out of 321 of fox faecal samples (11.53%), all of them presenting multi-drug resistance (MDR). Fourteen out of 146 (9.59%) badger faecal samples carried Salmonella spp. [S. Agama (n = 9), S. Newport (n = 4) and S. enterica subsp. arizonae (n = 1)]. Overall, AMR was found only in the S. enterica subsp. arizonae isolate (1/14, 7.14%). No MRSA were detected in nasopharyngeal swabs from badgers (n = 118) and foxes (n = 279). This is the first attempt to explore the prevalence of AMR in the two common wildlife species in Northern Ireland. These findings are important as they can be used as a base line for further research exploring the origin of the found resistance. These results should encourage similar surveys where environmental samples are included to bring better understanding of AMR dynamics, and the impact on wildlife, domestic livestock and humans.
The phenomenon of microbes becoming resistant, due to a generalised inappropriate use of antimicrobials in humans and animals, is currently happening at a global scale for a broad range of microorganisms (Dolejska and Literak, 2019). The direct consequences of Antimicrobial Resistance (AMR) include longer illnesses, increased mortality and increased costs (World Health Organization, 2015).
AMR can occur spontaneously in nature. AMR occurring naturally in some bacterial species is denominated “intrinsic resistance” (OIE, 2012) defined as the innate ability of a bacteria species to resist the action of an antibiotic due to its structural or functional characteristics. However, high levels of AMR are a result of selective pressure exerted on the bacterial population due to the use of antimicrobial agents (Schwarz et al., 2001). Furthermore, as many antibiotics belong to the same class of medicines, resistance to one specific antibiotic agent can lead to resistance to a whole related class. Moreover, resistance that develops in one organism or location can spread rapidly through, for instance, exchange of genetic mobile material such as plasmids between different bacteria (Davies and Davies, 2010). The importance of these AMR bacteria present in humans and animals, which can be transmissible through food, water, and the environment (Arnold et al., 2016a; Dolejska and Literak, 2019), make them a focus point for a “One Health” approach. Regarding wildlife, antimicrobial resistant bacteria occurrence depends on host interaction with potentially anthropogenic impacted habitats by landfills, draining of insufficiently treated wastewaters and wastes from intensively manage livestock farms (Wellington et al., 2013; Dolejska and Literak, 2019Wellington et al., 2013). However, AMR bacteria in wildlife such as rodents has also been described in areas with low levels of anthropogenic activity (Osterbald et al., 2001). As the continuous exchange of bacteria between environmental niches contributes to their dissemination (Wellington et al., 2013), the role of the environment in relation to AMR is one of the focus areas of the European Union Action Plan (European Commission, 2017).
Little is known about whether AMR is present in the environment and if at all, the levels in zoonotic or commensal bacteria in Northern Ireland. Previous research suggests that the levels of AMR in the British Isles are relatively low compared to other areas such as Africa, parts of South America and Southern Asia and broadly similar to much of continental Europe (Hendriksen et al., 2019). As resistant bacteria are present in wild animals, they can be useful sentinels mirroring the presence of AMR bacteria in an area (Dolejska and Literak, 2019). Wildlife related data can therefore be used to gain a better understanding of the levels of AMR in environmental bacteria (Gilliver et al., 1999; Mo et al., 2018). In the current study, data collected from European badgers (Meles meles; order carnivore, omnivorous) (Roper, 1994) and red foxes (Vulpes vulpes; order carnivore, omnivorous) (Soe et al., 2017) were therefore used to estimate the prevalence of AMR in these two wildlife species in Northern Ireland.
Wildlife samples were collected as part of two ongoing surveys performed in Northern Ireland. The first of these is a road traffic accident (RTA) survey which is conducted with the aim of estimating the prevalence of bovine tuberculosis, caused by Mycobacterium bovis, in European badgers (Meles meles) (Courcier et al., 2018). This survey is conducted all year around and has been in place in Northern Ireland since 1998. It involves the collection of up to 350 badger carcasses per year. These carcasses were reported by members of the public and collected by dedicated staff and collection vehicles. Only carcasses deemed suitable for postmortem examination were taken to two veterinary diagnostic laboratories. Detailed procedures for this survey are described previously (Courcier et al., 2018). The second survey aims to establish the geographical epidemiological status of Echinococcus multilocularis using red fox (Vulpes vulpes) carcasses collected throughout Northern Ireland of which the majority are shot by hunters (for reasons of pest control), while a minority are reported after being killed by a road traffic accident (Courcier et al., 2014). This survey encompasses 325 foxes every year which are tested for E. multilocularis in faecal samples in order to demonstrate freedom from this parasite (Commission Delegated Regulation (EU) No 1152/2011 Annex I). Fox carcasses were reported and collected through the same channels as the RTA badger survey involving dedicated reporting systems, collection staff and vehicles. Faecal (badgers, n = 146; foxes, n = 321) and nasopharyngeal swabs (badgers, n = 118; foxes, n = 279) were collected from foxes and badgers from September 2018–June 2019. Badgers and foxes were collected within 24–48 h after death in order to prevent autolysis of carcasses. Post mortem procedures were not performed on carcasses in an advanced stage of autolysis.
Samples from badger and fox carcasses were processed for bacteria of interest. These were two Gram negative enterobacteriaceae [Extended spectrum beta lactamase resistant and AmpC-type beta lactamases Escherichia coli (ESBL/AmpC), Salmonella spp. (only badgers)] and one Gram positive-Methicillin resistant Staphylococcus aureus (MRSA).
E. coli (ESBL/AmpC) was tested from the faeces of foxes (n = 321) and badgers (n = 146), Salmonella spp. were tested from faeces of badgers (n = 146) and MRSA from nasopharyngeal swabs from foxes (n = 279) and badgers (n = 118). Fox samples were not tested for the presence of Salmonella spp. Both fox and badger carcasses were collected throughout Northern Ireland.
The microbiological methods applied in this study are described below and outlined in Figure 1.
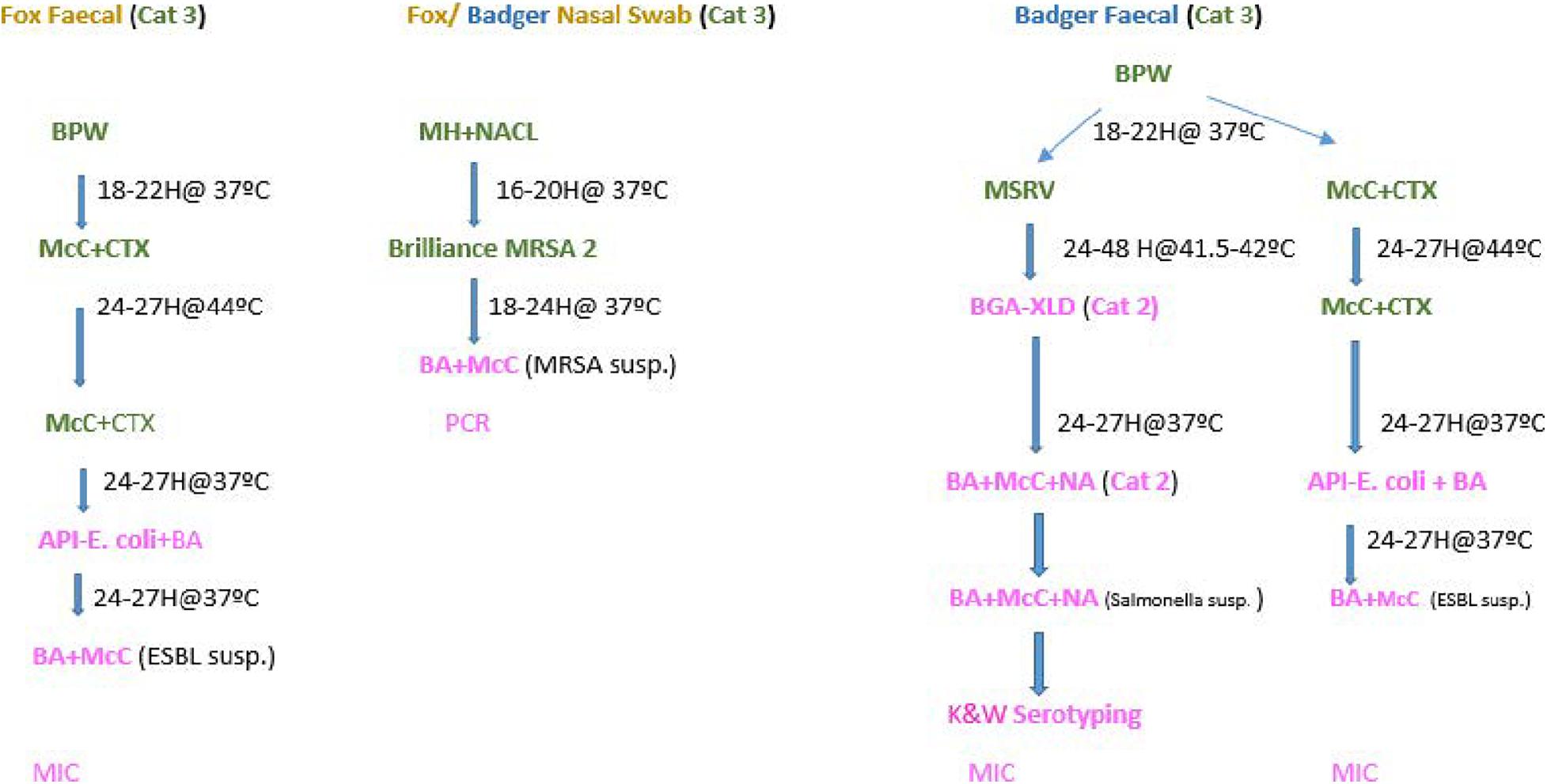
Figure 1. Flow chart- Microbiological processing of faecal and nasopharyngeal swabs collected from foxes and badgers (2018–2019).
It is well documented that badgers can carry Salmonella spp. (Wray et al., 1977; Euden, 1990; Wilson et al., 2003). Although Salmonella spp. have been occasionally isolated from foxes (Euden, 1990), at present this wildlife species can be consider an incidental host and is still not considered yet a reservoir of Salmonella spp. (Chiari et al., 2014). Hence, it was decided, for logistic and budget limitations, that only faecal samples from badgers would be tested for presence or absence of Salmonella spp.
Although the three bacteria of interest fall under hazard category two agents and can be processed in a Category two Laboratory (CL2), M. bovis and E. multilocularis (the agents that the carcasses are primarily collected for), are classified as hazard three agents. Therefore suspected samples must be processed in a Category 3 Laboratory (CL3), due to serious biosafety impact. Thus faecal and nasopharyngeal swabs were initially processed in a CL3. Once any of the microorganisms of interest were isolated, processing/confirmation continued in a CL2 (see Figure 1).
One gram of faeces was inoculated in Buffer Peptone Water (BPW) and incubated at 37°C for 18–22 h. Thereafter, a 10 μl loop was inoculated onto MacConkey agar containing 1 mg/L cefotaxime (CTX) plate. The CTX plates were incubated for 24–27 h at 44°C. Pure colonies from the CTX were then inoculated again on to MacConkey CTX plates to maintain selective pressure and incubated at 37°C for 24 h. Thereafter Analytical Profile Index (API) tests were carried for identification according to manufacturer specifications1.
Polymerase Chain Reaction (PCR) was performed according to Liu et al. (2016) on the only colistin resistant E. coli detected, to determine the presence of the plasmid-mediated gene mcr-1.
Salmonella spp. were identified and confirmed as described elsewhere (Porter et al., 2020).
Once ESBL and Salmonella spp. were identified, individual colonies were set-up in blood and McConkey plates in preparation for MIC. MIC was tested for the recommended set antimicrobials equal for E. coli and Salmonella spp. specified in Commission Decision EU/652/2013, the MIC technique is described elsewhere (Lahuerta-Marin et al., 2017). The European Committee on Antimicrobial Susceptibility Testing (EUCAST) Epidemiological cut-off values (ECOFFs) were applied as specified in EU Commission Decision EU/652/2013.
The antimicrobials for which the MICs were checked were: ampicillin, azithromycin, ceftazidime, cefepime, cefoxitin, chloramphenicol, ciprofloxacin, colistin, cefotaxime, ertapenam, gentamycin, imipenem, meropenem, nalidixic acid, sulfamethoxazole, temocillin, tetracycline, tigecycline, and trimethoprim. Cefotaxime + clavulanic acid, ceftazidime + clavulanic acid were also included to determine synergy, which allowed to classify them as phenotypic ESBLs and AmpC following growth on CTX media (Figure 1). ESBLs and AmpC were classified based on EUCAST guidelines as follows:
“Presumptive ESBL producers” refers to those isolates with MICs > 1 mg/L for cefotaxime and/or ceftazidime and a synergy test positive for any of these antimicrobials and susceptibility to meropenem (MEM ≤ 0.125 mg/L). These isolates may also harbour other resistance mechanisms (e.g., AmpC-encoding genes).
“Presumptive AmpC producers” refers to isolates with MICs > 1 mg/L for cefotaxime and/or
ceftazidime and cefoxitin MIC > 8 mg/L together with susceptibility to meropenem (MEM ≤ 0.125 mg/L). No distinction between acquired AmpC and natural AmpC was made. These isolates may also harbour other resistance mechanisms (e.g., ESBL-encoding genes).
“Presumptive ESBL + AmpC producers” refers to isolates with the ESBL + AmpC phenotype as described above.
The nasopharyngeal swabs were placed in glass universals containing 10 ml aliquots of Muller Hinton (MH) broth and 6.5% NaCl and incubated for 16–20 h at 37°C. Then a 10 μl loop of the broth was spread on Brilliance 2 MRSA agar and incubated for 24 h at 37°C. Subculture presumptive MRSA colonies were put on to blood agar and incubated for 24 h at 37°C. Presumptive colonies were then confirmed by PCR, according to the protocol for PCR Amplification of mecA, mecC, spa, and pvl validated by the European Reference Laboratory for Antimicrobial Resistance (Eurl-Ar, 2012).
Data were described and proportions along with 95% confidence intervals (95% CI) were calculated. Percentages resistance and distributions of MIC values were calculated for every antimicrobial. Significant differences of proportions were calculated applying chi-squared tests. All statistical analyses were conducted using R (version 4.0.1; The R Project for Statistical Computing2) and maps were produced using ArcMap (version 10.3.1; ESRI).
A total of 13 out of 146 badger faeces samples (8.90%; 95% CI 5.02–15.04%) carried ESBL. Fourteen of these isolates showed resistance against any of the tested antimicrobials (Table 1 and Figure 2). ECOFFs were based on European Union guidelines (European Union, 2013).
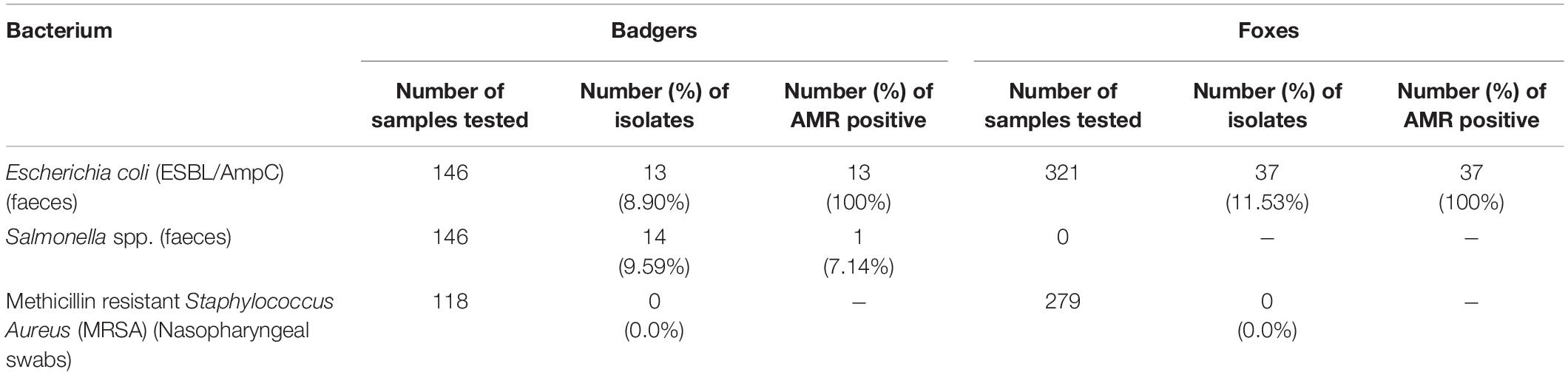
Table 1. Distribution of number of samples, isolates, and AMR positive isolates collected from foxes and badgers (2018–2019).
A total of 37 out of 321 fox faeces samples (11.53%; 95% CI 8.35–15.66%) contained ESBL. All of these 37 isolates showed resistance against any of the tested antimicrobials (Table 1). Tables 2, 3 show the distribution of resistant ESBL/AmpC type Escherichia coli found by antimicrobial. Cut off points were based on European Union guidelines (European Union, 2013).
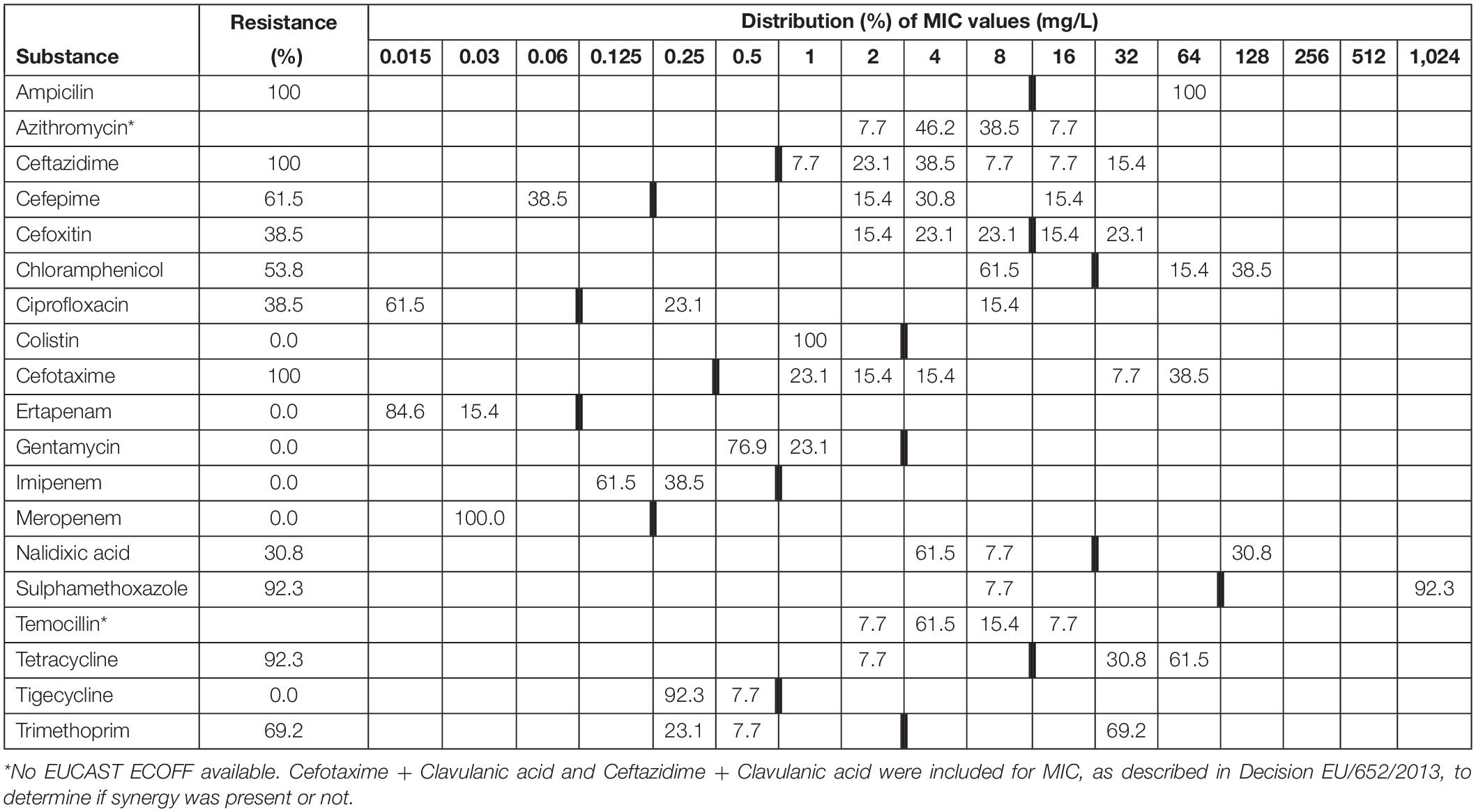
Table 2. Minimum inhibitory concentrations (MICs) and antimicrobial resistance in ESBL Escherichia coli isolated from faecal samples (n = 16) from badgers (Meles meles) in Northern Ireland in 2018–2019.
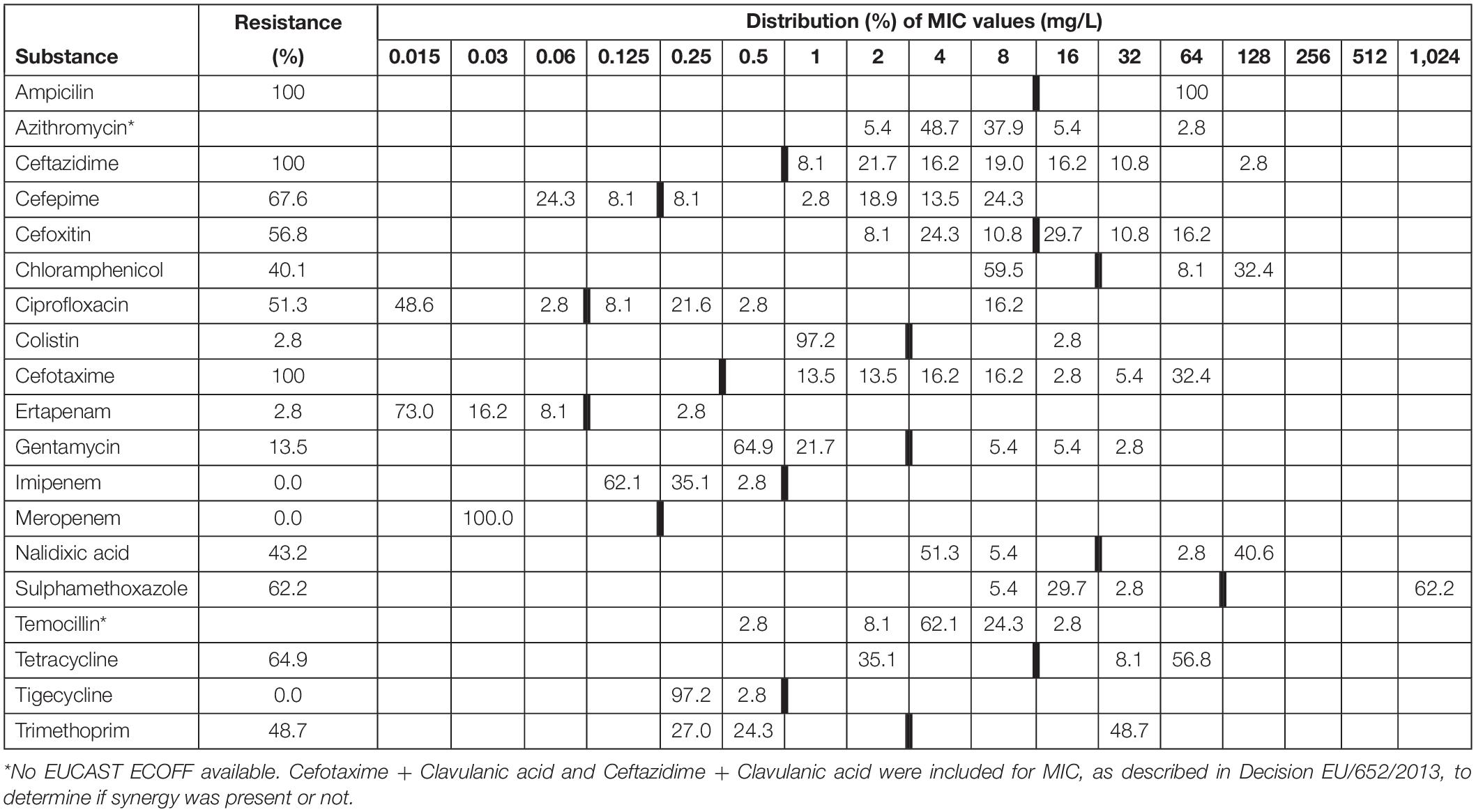
Table 3. Minimum inhibitory concentrations (MICs) and antimicrobial resistance in ESBL/AmpC isolated from faecal samples (n = 39) from red foxes (Vulpes vulpes) in Northern Ireland in 2018–2019.
There was no significant difference in the proportion of ESBL/AmpC resistant isolates found in badgers compared to foxes (Chi-squared 0.724, df = 1, p = 0.395).
One phenotypic ESBL strain was also resistant to colistin. The strain was negative to mcr-1.
Salmonella spp. was detected from 14 out of 146 badger faeces samples (9.59%; 95% CI 5.54–15.86%). S. Agama was the most prevalent serovar (n = 9) followed by S. Newport (n = 4) and S. enterica subsp. arizonae (n = 1). AMR were only observed in one out of 14 Salmonella spp. isolates (7.14%; 95% CI 0.37–35.83%), the S. enterica subsp. arizonae isolate, which showed resistance against ampicillin (MIC > 64 mg/L), ceftazidime (MIC = 64 mg/L), cefoxitin (MIC > 64 mg/L), cefotaxime (MIC = 32 mg/L), and ertapenam (MIC = 0.12 mg/L) and no synergy with clavulanate (Table 1 and Figure 2). This type of resistance is consistent with AmpC. Overall one of the 146 badger faecal samples collected contained therefore resistant Salmonella spp. isolates (0.68%; 95% CI 0.03–4.32).
No MRSA was detected in any of the nasopharyngeal swabs of badgers (n = 118) or foxes (n = 279) (Table 1).
In Northern Ireland, little is known about the prevalence of AMR in wildlife. Therefore, two common wildlife species (European badgers (Meles meles) and red foxes (Vulpes vulpes)) were used as sentinels to gain insight into this. The current study aimed to survey for the presence of resistance in these two wildlife species in ESBL/AmpC, Salmonella spp. (only in badgers) and MRSA.
In the current study 13 out of 146 badger faecal samples contained ESBL with all 13 showing resistance (all against multiple antimicrobials). Furthermore, 37 out of 321 fox faecal samples contained ESBL with all 37 showing resistance (again all against multiple antimicrobials). This is of concern as our results show high correlation between ESBL/AmpC resistance and levels of resistance to other antimicrobials including one isolate resistant to colistin. ESBL is known to be widely distributed in wildlife and is considered to be a key indicator pathogen to trace the evolution of multi-resistant bacteria in the environment and wildlife (Guenther et al., 2011). It was anticipated that ESBL in wildlife would express a multi-resistant phenotype, not due to the nearby use of antimicrobials or antimicrobials in sub-therapeutic concentrations in natural environments, but because distant use had caused a multi-resistant organism to evolve in the first place which subsequently spread to different ecological niches (O’Brien, 2002). The majority of previous research into ESBL prevalence in wildlife has focussed on birds and rodents and prevalence appears to be highest in urbanised areas (Guenther et al., 2011). However, previous research did report the presence of resistant ESBL in foxes (1 out of 7 animals sampled) (Costa et al., 2006) and in badgers (Alonso et al., 2017). It is also documented that plasmids that harbour ESBL and/or pAmpC genes may also carry other resistance genes, meaning that ESBL/pAmpC-producing pathogens can be resistant to other classes of antimicrobial agents as well (MacVane et al., 2014).
Staphylococcus aureus is a commensal bacterium with the potential to cause severe disease in humans and animals. MRSA, which is resistant to most β-lactam antibiotics, is a major cause of hospital-associated infections. Livestock-associated (LA)-MRSA has also been recognised to cause infections in humans (Köck et al., 2010) and has been detected in pigs and cattle in Northern Ireland (Hartley et al., 2014; Lahuerta-Marin et al., 2016). The first reported isolation of LA-MRSA in Northern Ireland, and indeed in the UK, was detected in a pig from a mixed swine-dairy cattle herd in 2014 (Hartley et al., 2014). Two on-farm investigations followed and environmental and animal samples were collected. LA-MRSA CC398 t034, the most common strain type in livestock in Northern Ireland (Sharma et al., 2016), was isolated from all environmental samples collected from the first infected farm and from the pig samples only. The bacterium was not detected from any other animal samples (cattle, dog, and sheep) collected form the follow-up epidemiological investigation of the index case (unpublished data). These results showed that LA-MRSA CC398 t034 was restricted to the environment and that the main animal hosts were pigs. However, in the current study no MRSA was observed in nasopharyngeal swabs from either badgers or foxes, hence more research is required and perhaps collection of wildlife around infected farms could be an option for future surveys. It is possible that sampling of badgers and foxes may not be a good proxy or indicator for environmental contamination with this LA-MRSA type. In that case, wildlife may represent an underestimation of levels of LA-MRSA in the environment (if any), thus, the use of wildlife samples as an environmental proxy could be a limitation for this particular pathogen.
Previous studies have demonstrated that a wide range of Salmonella spp. is known to be commonly present in badgers in the UK (Taylor, 1968; Wray et al., 1977; Euden, 1990; Wilson et al., 2003). The reported range of serovars is broad with S. Agama being the most commonly isolated serovar (Euden, 1990), as observed here. The only resistant serovar S. enterica subsp. arizonae detected in the current study is one of the less frequently found subspecies of Salmonella spp., most commonly detected in reptiles (especially snakes and tortoises) (Hall and Rowe, 1990; Bertrand et al., 2008; Bruce et al., 2018). Strains of Salmonella spp. with resistance to antimicrobial drugs are now widespread in both developed and developing countries (Threlfall, 2006). The only resistant isolate in the study was S. enterica subsp. arizonae showing AMR against five antimicrobials including 3rd generation of cephalosporins. ESBLs Salmonella spp. have been detected in chickens (Dierikx et al., 2010; de Souza et al., 2019) and other livestock (Riano et al., 2006). ESBL Salmonella isolations from wildlife are uncommon but ESBL S. Infantis isolated from owls have been reported (Fuentes-Castillo et al., 2019). On the other hand, human salmonellosis due to ESBL non-paratyphi Salmonella spp. have been described. The UK reported cases of clinical salmonellosis in humans due to ESBL Salmonella spp. (EFSA, 2018, 2019). However, at present, ESBL Salmonella spp. are uncommon in livestock in the UK, but have been reported by some European Member States (EFSA, 2018, 2019; Uk-Varss, 2019). Genetic characterisation of the strain will provide more clues about the potential origin. In addition, the results of the current study are very interesting as sampled badgers were carrying relatively low levels of Salmonella spp., and not many of the isolates carried any AMR compared to AMR levels observed in livestock (Porter et al., 2020). This suggests exposure and dynamics of infection with Salmonella spp. in both domestic livestock and wildlife reservoirs such as badgers are different. Nevertheless the impact of carrying ESBL Salmonella spp. may be high particularly regarding treatment, as human infections due to S. enterica subsp. arizonae can occur and have been described before (Gavrilovici et al., 2017; Lakew et al., 2013).
The reservoir of resistance genes in the environment is known to be due to a mix of naturally occurring resistance, those present in animal and human waste and the selective effects of pollutants, which can co-select for mobile genetic elements carrying multiple resistant genes. Resistance genes can be acquired from any source, but gene flow is probably structured by ecology, with species that share similar niches drawing from similar gene pools (Wellington et al., 2013). Several possible transmission routes exist including direct contact with infected individuals, their tissues or their faeces, water and soil (Vittecoq et al., 2016). Once present in the environment, the resistant bacteria can then be potentially acquired by wild animals and then reintroduced to humans. Therefore, the potential of plasmid transfer between strains of the same species or between different bacterial species or genera creates an environmental reservoir of resistance with potentially far reaching impacts for human health (Carroll et al., 2015). As there is a significant level of resistant ESBL/pAmpC- producing pathogens found in wildlife in the current study, similar to previous studies (Costa et al., 2006; Alonso et al., 2017; Darwich et al., 2019), the sources of these would need further investigation focusing on both farm animal and human related origins (Arnold et al., 2016b; Larsson et al., 2018). Potential associations that could be explored include water sources, human and farm animal population density, and proximity to hospitals and pharmaceutical industries (Arnold et al., 2016b). Further molecular characterisation into ESBL and Salmonella spp. would also be useful in order to provide insight into the possible correlation between phenotypic and genotypic patterns of resistance between isolates from wildlife, livestock and humans. The results of further research could help prioritising the development of effective One Health strategies to mitigate the spread of AMR into the environment in targeted areas in Northern Ireland, such as: pre-treatment of manure before use as fertiliser; pre-treatment of waste across the farm to slaughterhouse continuum before discharge into the general sewage system; education to increase awareness of all hospital personnel on hygiene, sanitation and safe disposal practices; insurance of the safe disposal of antimicrobial medicines and hazardous waste; consideration of pre-treatment of hospital waste before discharge into the general sewage system (FAO, 2018). Furthermore, research into the possible correlation of AMR bacteria in wildlife/livestock and their environment would be useful, because monitoring of AMR in wildlife and the environment can be used as an early detection of new AMRs (Martínez, 2009; Radhouani et al., 2014).
This survey is the first step to assess the risk of wildlife as a potential environmental reservoir of antimicrobial resistance for domestic livestock and humans. We expect that these results encourage the Environmental Agency in Northern Ireland to test bacteria isolated from environmental samples- soil, dust and water. Hence this will contribute to a better one health understanding of AMR (European Commission, 2017).
Only one ESBL strain also presented resistance to colistin. It was previously described that plasmid-mediated mcr-1 had resistance to this critically important antibiotic (Liu et al., 2016). This PCR was performed as an initial screening. Since then, 10 plasmids have been described (mcr-1–10). There is no validated PCR available to detect presence or absence of all 10 mcrs. Further analyses will be conducted in relation to testing for mcr-1. In the near future, there are plans to test for mcr-1–5 following the validated PCR protocol developed by EURL-AR3. If the colistin resistant ESBL carry any of mcr-6–10 plasmids, we will be able to detect it when we perform whole genome sequencing on this isolate. This was not performed in this initial stage of this survey.
No microbiological isolation of commensal E. coli was performed for this study. If the survey is performed again in the near future, it would be desirable to calculate the prevalence of commensal E. coli and the levels of resistance of this bacterium carried by wildlife species in Northern Ireland.
Due to the nature of the sampling within the surveys (convenience sampling) and the relatively low numbers of positive samples involved, it is not possible to identify clusters and potentially relate them to features such as rivers or drainage basins. Further research could be conducted to address this.
Wildlife sampling for AMR is important (based on the “One Health” concept), but challenging. Similar to other research (Mo et al., 2018), dead animals were therefore the sample source for this study. This provides difficulties in relation to autolysis. However, in the current study this was prevented as much as possible by collecting and processing the badgers and foxes within 24–48 h after death.
This project has provided the first insight into the prevalence of AMR in wildlife as a proxy for the environment. Results showed that ESBL/AmpC were the most prevalent type of resistance in badgers and foxes. Moreover, the ESBL/AmpC isolates recovered were also resistant to several other antimicrobial agents. AMR levels in Salmonella spp. were very low but highly resistant. The resistance pattern was unusual in that resistance against for example, AmpC and MRSA was not detected. These results suggest that the three pathogens may have different dynamics of infection and exposure from the environment into the sampled wildlife species. These findings will form a baseline for further research and are an important first step in our understanding of the levels of AMR bacteria in wildlife and potentially the environment.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethical review and approval was not required for this study because the animals were already deceased. Animal carcasses (badgers and foxes) were collected from the roadside after becoming victims of road traffic accidents or, in the case of some of the foxes, were shot by hunters (not for the purpose of the study).
AP-L and AL-M conceived and designed the study. MO’H and AL-M wrote the manuscript, with interpretation of results and discussion inputs from AP-L. JM organised collection of the samples. CC, CH, CB, NS, and RD performed laboratory work. MO’H carried out statistical analyses. RH approved the concept of the study and liaised to initiate the study. All authors read and approved the final manuscript.
This research was funded by the Department of Agriculture, Environment and Rural Affairs for Northern Ireland.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank all the DAERA and AFBI staff involved in the collection and processing of badgers and foxes during 2018/2019.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.596891/full#supplementary-material
Alonso, C. A., Alcala, L., Simon, C., and Torres, C. (2017). Novel sequence types of extended-spectrum and acquired AmpC beta-lactamase producing Escherichia coli and Escherichia clade V isolated from wild mammals. FEMS Microbiol. Ecol. 93:10.1093. doi: 10.1093/femsec/fix097
Arnold, K. E., Williams, N. J., and Bennett, M. (2016a). ‘Disperse abroad in the land’: the role of wildlife in the dissemination of antimicrobial resistance. Biol. Lett. 12:10.1098. doi: 10.1098/rsbl.2016.0137
Arnold, K. E., Williams, N. J., and Bennett, M. (2016b). Disperse abroad in the land’: the role of wildlife in the dissemination of antimicrobial resistance. Biol. Lett. 12:10.1098. doi: 10.1098/rsbl.2016.0137
Bertrand, S., Rimhanen-Finne, R., Weill, F. X., Rabsch, W., Thornton, L., Perevoscikovs, J., et al. (2008). Salmonella infections associated with reptiles: the current situation in Europe. Euro. Surveill. 13:10.2807.
Bruce, H. L., Barrow, P. A., and Rycroft, A. N. (2018). Zoonotic potential of Salmonella enterica carried by pet tortoises. Vet. Rec. 182:141, doi: 10.1136/vr.104457
Carroll, D., Wang, J., Fanning, S., and McMahon, B. J. (2015). Antimicrobial resistance in wildlife: implications for public health. Zoonoses Public Health 62, 534–542. doi: 10.1111/zph.12182
Chiari, M., Ferrari, N., Giardiello, D., Lanfranchi, P., Zanoni, M., Lavazza, A., et al. (2014). Isolation and identification of Salmonella spp. from red foxes (Vulpes vulpes) and badgers (Meles meles) in northern Italy. Act. Vet. Scan 56:10. doi: 10.1186/s13028-014-0086-7
Costa, D., Poeta, P., Saenz, Y., Vinue, L., Rojo-Bezares, B., Jouini, A., et al. (2006). Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemother. 58, 1311–1312. doi: 10.1093/jac/dkl415
Courcier, E. A., Fee, S., Malone, F., McBride, K., McKeown, I., Abernethy, D. A., et al. (2014). “Using wildlife surveillance to provide evidence of freedom from Echinococcus multilocularis in Northern Ireland,” in Proceeding of the 2nd International Conference on Animal Health Surveillance, La Havana, Cuba, 7-9 May 2014. (MDPI).
Courcier, E. A., Menzies, F. D., Strain, S. A. J., Skuce, R. A., Robinson, P. A., Patterson, I. A. P., et al. (2018). Mycobacterium bovis surveillance in Eurasian badgers (Meles meles) killed by vehicles in Northern Ireland between 1998 and 2011. Vet. Rec. 182, 259–265. doi: 10.1136/vr.103934
Darwich, L., Vidal, A., Seminati, C., Albamonte, A., Casado, A., Lopez, F., et al. (2019). High prevalence and diversity of extended spectrum β-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS One 14:10. doi: 10.1371/journal.pone.0210686
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microb. Mole. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
de Souza, M., Penha Brito, D. A., Menck-Costa, M. F., Oba, A., Kobayashi, R. K. T., Justino, L., et al. (2019). Multidrug resistant and ESBL-producing Salmonella spp. isolated from poultry. Ciências Agrárias Londrina 40, 3045–3056. doi: 10.5433/1679-0359.2019v40n6supl2p3045
Dierikx, C., Essen-Zandbergen, A., Veldman, K., Smith, H., and Mevius, D. (2010). Increased Detection of Extended Spectrum Beta-Lactamase Producing Salmonella Enterica and Escherichia Coli Isolates From Poultry. Vet. Microb. 145, 273–278. doi: 10.1016/j.vetmic.2010.03.019
Dolejska, M., and Literak, I. (2019). Wildlife is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob. Agents Chemother. 63, e1167–e1119.
EFSA (2018). (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 16:10.2903. doi: 10.2903/j.efsa.2018.5182
EFSA (2019). (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 17:10.2903. doi: 10.2903/j.efsa.2019.5598
Euden, P. R. (1990). Salmonella isolates from wild animals in Cornwall. Br. Vet. J. 146, 228–232. doi: 10.1016/s0007-1935(11)80006-0
Eurl-Ar (2012). (The European Reference Laboratory-Antimicrobial Resistance) 2012- Protocol for PCR amplification of MECA, MECC (MECALGA251), SPA and PVL, 2ST VERSION, September 2012. Available online at: https://www.eurl-ar.eu/CustomerData/Files/Folders/21-protocols/279_pcr-spa-pvl-meca-mecc- sept12.pdf [Accessed May 4, 2020]
European Commission (2017). A European One Health Action Plan against Antimicrobial Resistance (AMR).Available online at: https://ec.europa.eu/health/amr/sites/health/files/antimicrobial_resistance/docs/amr_2017_action-plan.pdf [Accessed June 10, 2020].
European Union (2013). Commission implementing decision of 12 November 2013 on the monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria, EU/2013/652. Available online at: https://www.legislation.gov.uk/eudn/2013/652/2020-12-31 [Accessed August 19, 2020]
FAO (2018). Food and Agriculture Organization of the United Nations; Antimicrobial resistance in the environment. Summary report of an FAO meeting of experts FAO Antimicrobial Resistance Working Group.Available online at: http://www.fao.org/3/BU656en/bu656en.pdf [Accessed June 29, 2020]
Fuentes-Castillo, D., Farfan-Lopez, M., Esposito, F., Moura, Q., Fernandes, M. R., Lopes, R., et al. (2019). Wild owls colonized by international clones of extended-spectrum β-lactamase (CTX-M)-producing Escherichia coli and Salmonella Infantis in the Southern Cone of America. Sci. Tot. Environ. 674, 554–562. doi: 10.1016/j.scitotenv.2019.04.149
Gavrilovici, C., Pânzaru, C. V., Cozma, S., Mârţu, C., Lupu, V. V., Ignat, A., et al. (2017). Message from a turtle: Otitis with Salmonella Arizonae in children. Case Rep. Med. 96:e8455. doi: 10.1097/MD.0000000000008455
Gilliver, M. A., Bennett, M., Begon, M., Hazel, S. M., and Hart, C. A. (1999). Antimicrobial resistance found in wild rodents. Nature 01, 233–234. doi: 10.1038/45724
Guenther, S., Ewers, C., and Wieler, L. H. (2011). Extended-Spectrum Beta-Lactamases Producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2:10.3389. doi: 10.3389/fmicb.2011.00246
Hall, M. L. M., and Rowe, B. (1990). Salmonella arizonae in United Kingdom from 1966-1990. Epidemiol. Infect. 108, 59–65. doi: 10.1017/s0950268800049505
Hartley, H., Watson, C., Nugent, P., Beggs, N., Dickson, E., and Kearns, A. (2014). Confirmation of LA-MRSA in pigs in the UK. Vet. Record 175, 74–75. doi: 10.1136/vr.g4620
Hendriksen, R. S., Munk, P., Njage, P., van Bunnik, B., McNally, L., Lukjancenko, O., et al. (2019). Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Comm. 10:10.1038. doi: 10.1038/s41467-019-08853-3
Köck, R., Becker, K., Cookson, B., van Gemert-Pijnen, J. E., Harbarth, S., Kluytmans, J., et al. (2010). Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro. Surveill. 15:10.2807. doi: 10.2807/ese.15.41.19688-en
Lahuerta-Marin, A., Guelbenzu-Gonzalo, M., Pichon, B., Allen, A., Doumith, M., Lavery, J. F., et al. (2016). First report of lukM-positive livestock-associated methicillin-resistant Staphylococcus aureus CC30 from fattening pigs in Northern Ireland”. Vet. Microb. 182, 131–134. doi: 10.1016/j.vetmic.2015.11.019
Lahuerta-Marin, A., Mu??oz-Gomez, V., Hartley, H., Guelbenzu-Gonzalo, M., Porter, R., Spence, N., et al. (2017). survey on antimicrobial resistant Escherichia coli isolated from unpasteurised cows’ milk in Northern Ireland. Vet. Record 180:426. doi: 10.1136/vr.104097
Lakew, W., Girma, A., and Triche, E. (2013). Salmonella enterica Serotype Arizonae Meningitis in a Neonate. Case Rep. Pediatrics 813495:10.1155. doi: 10.1155/2013/813495
Larsson, D. G. J., Andremont, A., Bengtsson-Palme, J., Brandt, K. K., de Roda Husman, A. M., Fagerstedt, P., et al. (2018). Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ. Internat. 117, 132–138. doi: 10.1016/j.envint.2018.04.041
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
MacVane, S. H., Tuttle, L. O., and Nicolau, D. P. (2014). Impact of extended-spectrum beta-lactamase-producing organisms on clinical and economic outcomes in patients with urinary tract infection. J. Hosp. Med. 9, 232–238. doi: 10.1002/jhm.2157
Martínez, J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Env. Pollut. 157, 2893–2902. doi: 10.1016/j.envpol.2009.05.051
Mo, S. S., Urdahl, A. M., Madslien, K., Sunde, M., Nesse, L. L., Sietemeåss, J. S., et al. (2018). What does the fox say? Monitoring antimicrobial resistance in the environment using wild red foxes as an indicator. PLoS One 13:e0198019. doi: 10.1371/journal.pone.0198019
O’Brien, T. F. (2002). Emergence, spread, and environmental effect of antimicrobial resistance: how use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin. Infect. Dis. 34, S78–S84. doi: 10.1086/340244
OIE (2012). Antimicrobial resistance in animal and public health. Rev Sci Tech. 31:24. doi: 10.20506/rst.31.1.2094
Osterbald, M., Norrdahl, K., Korpimaki, E., and Huovinen, P. (2001). Antimicrobial resistance: How wild are wild mammals? Nature 409, 37–38. doi: 10.1038/35051173
Porter, S., Strain, S. A. J., Bagdonaite, G., McDowell, S. W., Bronckaers, T., Sherrey, M., et al. (2020). Trends in Salmonella serovars and antimicrobial resistance in pigs and poultry in Northern Ireland between 1997 and 2016. Vet. Record 186:10.1136. doi: 10.1136/vr.105640
Radhouani, H., Silva, N., Poeta, P., Torres, C., Correia, S., and Igrejas, G. (2014). Potential impact of antimicrobial resistance in wildlife, environment and human health. Front. Microb. 5:10.3389. doi: 10.3389/fmicb.2014.00023
Riano, I., Moreno, M. A., Teshager, T., Saenz, Y., Dominguez, L., and Torres, C. (2006). Detection and characterization of extended-spectrum b-lactamases in Salmonella enterica strains of healthy food animals in Spain. J. Antimicrob. Chemother. 58, 844–847. doi: 10.1093/jac/dkl337
Roper, T. (1994). The European badger Meles meles: food specialist or generalist? J. Zool. 234, 437–452. doi: 10.1111/j.1469-7998.1994.tb04858.x
Schwarz, S., Kehrenberg, C., and Walsh, T. R. (2001). Use of antimicrobial agents in veterinary medicine and food animal production. Int. J. Antimicrob. Agents 17, 431–437. doi: 10.1016/s0924-8579(01)00297-7
Sharma, M., Nunez-Garcia, J., Kearns, A. M., Doumith, M., Butaye, P. R., Angeles Argudín, M., et al. (2016). Livestock Associated Methicillin Resistant Staphylococcus aureus (LAMRSA) Clonal Complex(CC)398 Isolated from UK Animals belong to European Lineages. Front. Microbiol. 7:10.3389. doi: 10.3389/fmicb.2016.01741
Soe, E., Davison, J., Suld, K., Valdmann, H., Laurimaa, L., and Saarma, U. (2017). Europe-wide biogeographical patterns in the diet of an ecologically and epidemiologically important mesopredator, the red fox Vulpes vulpes: a quantitative review. Mammal Rev. 47, 198–211. doi: 10.1111/mam.12092
Threlfall, E. J. (2006). Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microb. Rev. 26, 141–148. doi: 10.1111/j.1574-6976.2002.tb00606.x
Uk-Varss (2019). UK Veterinary Antibiotic Resistance and Sales Surveillance Report (UK-VARSS 2018). New Haw, Addlestone: Veterinary Medicines Directorate. www.gov.uk/government/collections/veterinary-antimicrobial-resistance-and-sales-surveillance [Accessed June 10, 2020]
Vittecoq, M., Godreuil, S., Prugnolle, F., Durand, P., Brazier, L., Renaud, N., et al. (2016). Antimicrobial resistance in wildlife. Journal of Applied Ecology 53, 519–529. doi: 10.1111/1365-2664.12596
Wellington, E. M. H., Boxall, A. B. A., Cross, P., Feil, E. J., Gaze, W. H., Hawkey, P. M., et al. (2013). The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 13, 155–165. doi: 10.1016/S1473-3099(12)70317-1
Wilson, J. S., Hazel, S. M., Williams, N. J., Phiri, A., French, N. P., and Hart, C. A. (2003). Nontyphoidal salmonellae in United Kingdom badgers: prevalence and spatial distribution. Appl. Environ. Microbiol. 69, 4312–4315. doi: 10.1128/aem.69.7.4312-4315.2003
World Health Organization (2015). WHO Global Action Plan on Antimicrobial Resistance. https://www.who.int/antimicrobial-resistance/publications/global-action-plan/en/. [Accessed June 4, 2020]
Keywords: Antimicrobial Resistance, wildlife, ESBL, AmpC, Salmonella, MRSA, badgers, foxes
Citation: O’Hagan MJH, Pascual-Linaza AV, Couzens C, Holmes C, Bell C, Spence N, Huey RJ, Murphy JA, Devaney R and Lahuerta-Marin A (2021) Estimation of the Prevalence of Antimicrobial Resistance in Badgers (Meles meles) and Foxes (Vulpes vulpes) in Northern Ireland. Front. Microbiol. 12:596891. doi: 10.3389/fmicb.2021.596891
Received: 20 August 2020; Accepted: 26 January 2021;
Published: 18 February 2021.
Edited by:
Ana De La Torre, Instituto Nacional de Investigación y Tecnología Agroalimentaria (INIA), SpainReviewed by:
Kinndle Blanco-Peña, National University of Costa Rica, Costa RicaCopyright © 2021 O’Hagan, Pascual-Linaza, Couzens, Holmes, Bell, Spence, Huey, Murphy, Devaney and Lahuerta-Marin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria J. H. O’Hagan, TWFyaWEuTyYjeDIwMTk7SGFnYW5AZGFlcmEtbmkuZ292LnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.