- 1School of Life Sciences, Warwick Crop Centre, University of Warwick, Wellesbourne Campus, Warwick, United Kingdom
- 2NIAB-EMR, East Malling Research, Kent, United Kingdom
- 3Natural Resources Institute, University of Greenwich, Kent, United Kingdom
- 4Computational and Analytical Sciences, Rothamsted Research, Harpenden, United Kingdom
Fusarium oxysporum is a soilborne fungal plant pathogen responsible for causing disease in many economically important crops with “special forms” (formae speciales) adapted to infect specific plant hosts. F. oxysporum f. sp. pisi (FOP) is the causal agent of Fusarium wilt disease of pea. It has been reported in every country where peas are grown commercially. Disease is generally controlled using resistant cultivars possessing single major gene resistance and therefore there is a constant risk of breakdown. The main aim of this work was to characterise F. oxysporum isolates collected from diseased peas in the United Kingdom as well as FOP isolates obtained from other researchers representing different races through sequencing of a housekeeping gene and the presence of Secreted In Xylem (SIX) genes, which have previously been associated with pathogenicity in other F. oxysporum f. spp. F. oxysporum isolates from diseased United Kingdom pea plants possessed none or just one or two known SIX genes with no consistent pattern of presence/absence, leading to the conclusion that they were foot-rot causing isolates rather than FOP. In contrast, FOP isolates had different complements of SIX genes with all those identified as race 1 containing SIX1, SIX6, SIX7, SIX9, SIX10, SIX11, SIX12, and SIX14. FOP isolates that were identified as belonging to race 2 through testing on differential pea cultivars, contained either SIX1, SIX6, SIX9, SIX13, SIX14 or SIX1, SIX6, SIX13. Significant upregulation of SIX genes was also observed in planta over the early stages of infection by different FOP races in pea roots. Race specific SIX gene profiling may therefore provide potential targets for molecular identification of FOP races but further research is needed to determine whether variation in complement of SIX genes in FOP race 2 isolates results in differences in virulence across a broader set of pea differential cultivars.
Introduction
Fusarium oxysporum is the most widely dispersed and economically important plant pathogenic species in the Fusarium genus as it infects numerous hosts and causes extensive crop losses (Leslie and Summerell, 2006). In 2012 it was identified as fifth in a list of the top ten fungal plant pathogens in terms of scientific and economic importance, just behind Fusarium graminearum (Dean et al., 2012). F. oxysporum is responsible for a wide range of plant diseases, usually causing a vascular wilt but also causes root and bulb rots (Edel-Hermann and Lecomte, 2019). F. oxysporum is a species complex with “special forms” (formae speciales; f. spp.), adapted to infect specific hosts plants. These are thought to have evolved though convergent evolution and many are therefore polyphyletic (Fourie et al., 2011; van Dam et al., 2018); hence, isolates from one f. spp. may be more closely related to isolates infecting other hosts than to each other (O’Donnell et al., 1998). In F. oxysporum, more than 150 host specific formae speciales have been described (Edel-Hermann and Lecomte, 2019), including well studied and economically important examples such as F. oxysporum f. sp. cubense (banana), f. sp. lycopersici (tomato), f. sp. cepae (onion), and f. sp. pisi (pea). F. oxysporum f. sp. lycopersici (FOL), cubense, and pisi are also examples of f. spp. which contain multiple economically damaging races, which are specialised to infect certain cultivars of a host species; for example, f. sp. cubense tropical race 4 which is devasting the popular Cavendish cultivar of banana in the tropics (Ploetz, 2015).
The emergence of new pathogenic races in F. oxysporum f. spp. is a constant threat, driven by the evolutionary adaptation of the pathogen’s complement of effector genes, and their function, to overcome host resistance (Takken and Rep, 2010). A well-documented example of this type of evolutionary adaption between pathogen and host is that of FOL and tomato. Several FOL races have evolved to overcome I (immunity) genes in tomato that confer resistance to FOL, by loss or mutation of Secreted In Xylem (SIX) genes, which code for small cysteine rich proteins secreted into the xylem sap of tomato during infection. So far, there are 14 characterised SIX genes in FOL (Houterman et al., 2007; Lievens et al., 2009; Schmidt et al., 2013), and all (other than SIX13) are located on chromosome 14, a lineage specific (LS) chromosome found only in FOL. This is also known as a pathogenicity chromosome due to its ability to confer virulence in a non-pathogenic F. oxysporum isolate when artificially transferred (Ma et al., 2010; Schmidt et al., 2013). Several SIX genes (SIX1, 3, 5, and 6) have been shown to be directly involved in pathogenicity of FOL in tomato, and have been shown to be essential for full virulence through gene knockout studies (Rep et al., 2004; Houterman et al., 2009; Gawehns et al., 2014). SIX genes have also been identified in other F. oxysporum f. spp. including f. sp. cubense, f. sp. cepae, f. sp. niveum, f. sp. pisi, f. sp. Melonis, and f. sp. vasinfectum (Chakrabarti et al., 2011; Meldrum et al., 2012; Covey et al., 2014; Fraser-Smith et al., 2014; Sasaki et al., 2015; Taylor et al., 2016). The presence/absence and sequence variation within SIX genes has been used as a new approach to distinguish between F. oxysporum f. spp. and even between races. For example, sequence differences in SIX8 have been used to distinguish race 4 from races 1 and 2 in F. oxysporum f. sp. cubense (Fraser-Smith et al., 2014).
Fusarium wilt caused by F. oxysporum f. sp. pisi (FOP) is a major disease of pea that has been reported in every country where peas are grown commercially (Kraft, 1994). F. oxysporum has also been identified as a potentially important pathogen in the pea root rot complex, in addition to Fusarium solani and Fusarium avenaceum (Chittem et al., 2015). FOP can result in severe crop losses especially when peas are included in crop rotations more often than recommended, allowing inoculum levels to build up, particularly as chlamydospores can remain viable in the soil for 10 years (Kraft et al., 1994). To date, four races of FOP have been characterised: race 1, race 2, race 5, and race 6 (Haglund and Kraft, 1970, 1979), with races 1 and 2 currently the most important globally, and the only races identified in the United Kingdom (Biddle and Cattlin, 2007). Resistance to FOP by pea cultivars is conferred by single dominant genes (one for each race) for races 1, 5, and 6; however, race 2 resistance has been shown to be quantitative with a continuous scale of disease severity (McPhee et al., 2012; Bani et al., 2018). The documented method to determine FOP race is through the host response observed following inoculation of a set of pea differential cultivars. These have been used in several studies to distinguish between the four races (Haglund and Kraft, 1979; McPhee et al., 1999; Kraft and Pfleger, 2001; Bani et al., 2012). Cultivars are assessed for wilt symptoms, which begin with the downward curling then progressive yellowing and drying of the leaves from the base of the stem to the apex of the plant and as the disease develops (Kraft, 1994).
Although SIX genes have been identified in FOP previously, this was usually as part of studies focusing on other f. spp. One study identified SIX1, SIX13, and SIX14 in FOP and suggested that they could contribute to pathogenicity (Fraser-Smith et al., 2014). Additional SIX genes were identified in three isolates of FOP representing races 1, 2, and 5 as part of a larger screening of SIX gene presence in F. oxysporum f. sp. cepae (FOC). Here, up to six SIX genes were present (SIX7, SIX10, SIX11, SIX12, SIX13, and SIX14) with different complements across the three putative FOP races tested (Taylor et al., 2016). Furthermore, a recent genome study of a FOP race 5 isolate identified four SIX genes; SIX1, SIX9, SIX13, and SIX14 (Williams et al., 2016). However, none of these studies determined the expression of these SIX genes in planta, which is an important indicator of whether they may have a role in facilitating infection.
The main aim of this study was to determine the complement of SIX genes in FOP isolates from different putative races to establish if this approach could be used to distinguish between them and provide targets for molecular diagnostics. SIX gene expression was then examined using quantitative polymerase chain reaction (qPCR) to determine whether SIX genes were expressed in the early stages of pea infection. Finally, FOP isolate race was determined using a pathogenicity test on pea differential cultivars.
Materials and Methods
Fusarium Isolate Collection
Fusarium isolates were obtained from the Processors and Growers Research Organisation (PGRO; crop clinic isolations and field sampling 2012–2014) and also from sampling of United Kingdom pea plants displaying symptoms of root rot or wilt, from the major pea growing areas of Yorkshire, Lincolnshire, and Suffolk in 2015/2016 (Supplementary Table 1). Isolations were made from plant root and stem sections which were surface sterilised in a 10% bleach/sterile water solution (v/v) for 3 min, then washed twice in sterile water to remove bleach residue. Three sections from the sterilised material were plated onto potato dextrose agar (39 g L–1 PDA; Merck, United Kingdom) containing 2 mL L–1 chlortetracycline [10 mg mL–1 in 1/1 methanol/water (v/v)] and incubated at 20°C for 5–7 days. Fusarium cultures distinct in morphology were sub-cultured onto fresh PDA plates grown for approx. 7 days at 20°C. In addition, 28 isolates of FOP were obtained from other researchers mostly from outside the United Kingdom (mainly the United States) who had pre-assigned a putative race based on their own tests using sets of differential pea cultivars or based on infection of certain cultivars in the field (Supplementary Table 2). These were sub-cultured onto PDA and grown for 5–7 days at 20°C. Spore suspensions of all isolates were made in potato dextrose broth (PDB) + 20% glycerol (v/v) for storage on ceramic beads at −80°C.
Molecular Characterisation of F. oxysporum Isolates
The identity of Fusarium isolates collected from United Kingdom fields and the additional FOP isolates with a preassigned putative race was first confirmed by sequencing part of the translation elongation factor 1α (TEF) gene. Three 5 mm agar plugs were removed from the growing edge of each actively growing culture and placed in 20 mL of 50% PDB in a 50 mL centrifuge tube. Tubes were incubated at 20°C for 5 days positioned at a 45° angle. The mycelium of each isolate was then removed from the PDB by centrifugation (3,000 rpm for 15 min) and rinsed twice with sterile water (centrifugation at 3,000 rpm for 15 min each time) before it was lyophilised.
DNA was extracted from freeze-dried mycelium using a DNeasy plant mini kit (Qiagen, Hilden, Germany), in accordance with manufacturer’s protocol, with a minor modification whereby the mycelium was first homogenised in a lysing matrix-A tube (MP Biomedicals, CA, United States) in a FastPrep-24TM machine (MP Biomedicals, United Kingdom) set at 6 ms–1 for 40 s. Identification of Fusarium isolates was carried out by polymerase chain reaction (PCR) amplification of part of the TEF gene using published primers (Supplementary Table 3). All PCR reactions were set up using REDTaq® ReadyMix® (Sigma-Aldrich, United Kingdom) in 20 μL reaction volumes containing 1 μL of DNA and a final concentration of 0.5 μM of each primer. Thermocycling conditions were: one cycle of 2 min at 94°C; 30 cycles of 45 s at 94°C, 30 s at 64°C and 1 min at 72°C, followed by one cycle of 5 min at 72°C. PCR amplicons were visualised using gel electrophoresis (1% agarose gel containing GelRedTM at 2 μL per 100 mL of gel), purified using the QIAquick PCR Purification Kit (Qiagen) and sequenced using the forward primer sequence (GATC, Germany). Sequences were subjected to basic local alignment search tool (BLAST searches; Boratyn et al., 2013) using the National Centre of Biotechnology Information (NCBI) database to identify species based on sequence identity values.
Translation elongation factor 1α sequences were aligned and trimmed in Molecular Evolutionary Genetics Analysis version 7.0 (MEGA7; Kumar et al., 2016) and used to construct a maximum-likelihood tree using a Kimura 2-parameter model (Kimura, 1980) as determined using the “Find best DNA/Proteins model (ML)” tool in the software. Sequences of other F. oxysporum f. spp. isolates from Taylor et al. (2016) and NCBI database were also included for reference (Genbank numbers in Figure 1). Bootstrap values were inferred from 1,000 replicates (Felsenstein, 1985) and percentages displayed next to the relevant branch.
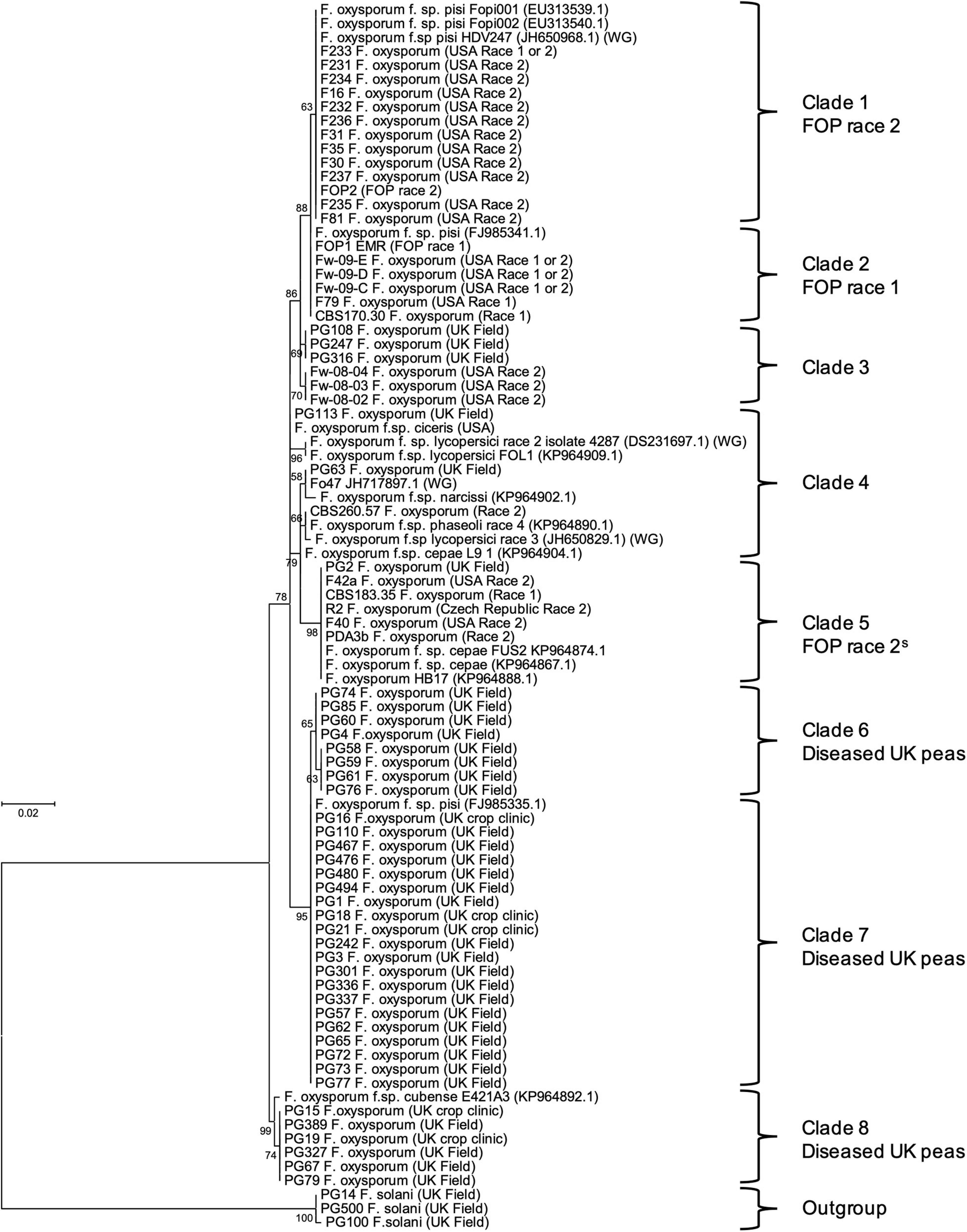
Figure 1. Maximum likelihood tree for 40 Fusarium oxysporum isolates from diseased United Kingdom peas (coded PG), 28 F. oxysporum f. sp. pisi (FOP) isolates from other researchers (with original race designation) and other F. oxysporum formae speciales, based on an alignment of translation elongation factor 1α (TEF) sequences. Numbers represent bootstrap percentage values from 1,000 replicates. Scale bar indicates 0.02 substitutions per site. The tree is rooted through three F. solani isolates (outgroup).
Identification of SIX Genes in F. oxysporum Isolates From Pea
In order to develop primers for SIX gene identification, whole genome sequencing of three pathogenic isolates of FOP (FOP1 EMR, F81, and R2) provisionally race typed by other researchers as putative race 1 (FOP1 EMR) and putative race 2 (F81 and R2), was conducted using both Illumina MiSeq and Oxford Nanopore Technology (ONT). DNA was extracted from freeze-dried mycelium using the Macherey-Nagel Nucleospin Plant II kit (Thermo Fisher Scientific, United Kingdom). For Illumina library preparation DNA was sheared in a Covaris M220 using microTUBE-50 (Covaris, United Kingdom) and size selected using the Blue Pippin (Sage Science, United Kingdom). Illumina libraries were constructed with a PCR-free method using NEBNext End Repair, NEBNext dA-tailing and Blunt T/A ligase (New England Biolabs, United Kingdom) and Illumina TruSeq barcodes. Libraries were sequenced on an Illumina MiSeq using MiSeq Reagent kit V3 (2 × 300 bp PE). For ONT library preparation, DNA was sheared using a g-TUBE (Covaris, United Kingdom) and then size selected for fragments greater than 10 kb using the Blue Pippin. Libraries were constructed using the ligation sequencing kit with NEBNext enzymes as detailed in the ONT protocol and run using a R9.5 FLO-MIN107 flow cell on the GridION.
Nanopore reads were assembled using SMARTdenovo (Ruan, 2016) and corrected using Racon for 10 rounds (Vaser et al., 2017) and Nanopolish (Loman et al., 2015), before being polished using Illumina MiSeq reads of the same isolates in Pilon (Walker et al., 2014). Quast was used to summarise assembly statistics and BUSCO v3 was used to assess completeness of gene space within the assembly (Gurevich et al., 2013; Simão et al., 2015). RepeatModeler and transposonPSI were used to identify repetitive and low complexity regions (Haas, 2007; Smit et al., 2013–2015).
Unmasked assembled genome sequences were imported into Geneious (v. 6.1.5) and used to conduct BLAST (Boratyn et al., 2013) searches for homologs of the 14 SIX genes previously identified in FOL (sequences obtained from NCBI). Sequences of positive hits (plus 500 bp up and downstream) were aligned in MEGA7 and used to check the utility of previously published primers for each of the 14 SIX genes (Lievens et al., 2009; Taylor et al., 2016). Where these primers did not match target sequences in FOP, new primers were designed using Primer3Plus (Untergasser et al., 2007) and checked for self-hybridisation potential using Eurofins Oligo Analysis Tool (Eurofins, 2016). These optimised primers were then used to determine the presence of SIX genes in 40 F. oxysporum isolates from United Kingdom peas and the 28 FOP isolates obtained from other researchers with previously assigned race type. PCR reactions for each SIX gene were set up using REDTaq® ReadyMix® (Sigma-Aldrich, United Kingdom) in 20 μL reaction volumes with 1 μL of DNA and 1 μL each of 10 μM primers with thermocycling conditions as follows: one cycle for 2 min at 94°C; 30 cycles of 45 s at 94°C, 30 s at primer annealing temperature (Supplementary Table 3) and 1 min at 72°C, followed by one cycle of 7 min at 72°C. PCR products were visualised using gel electrophoresis (1% agarose gel containing GelRedTM at 2 μL per 100 mL of gel).
Secreted In Xylem gene sequences were also aligned with homologs from other F. oxysporum f. sp. in MEGA7 and used to construct maximum-likelihood trees as described above. The nucleotide substitution models used are listed in Supplementary Table 4. Additional SIX sequences were downloaded from NCBI as described above.
Expression of SIX Genes in Planta
An in vitro method where peas were grown and inoculated with FOP isolates on an agar medium was adapted from Taylor et al. (2016). Autoclaved ATS medium [1 M KNO3, 1 M KPO4, 1 M MgSO4, 1 M Ca(NO3)2, 20 mM Fe-EDTA, 70 mM H3BO3, 14 mM MnCl2, 0.5 mM CuSO4, 1 mM ZnSO4, 0.2 mM Na2MoO4, 10 mM NaCl, 0.01 mM CoCl2, and 0.45% Gelrite (Duchefa Biochemie, Haarlem, Netherlands)] was used to three-quarter fill square petri dishes (12 × 12 × 1.7 cm, Greiner Bio-One, United Kingdom) and once set, the top 4.5 cm of the gel was removed with a sterile spatula. Pea seeds (cv. Little Marvel, susceptible to all FOP races) were sterilised in a 10% bleach/sterile water (v/v) solution for 5 min, then rinsed with sterile distilled water (SDW) until no bubbles remained. Six pea seeds were spaced evenly across the plate and pushed into the cut edge of the agar before the lid was replaced and secured with tape. Stacks of 6–8 plates were wrapped in cling film and incubated at 20°C for 3 days in the dark, and then for a further 5 days in light/dark (16 h day length). Conidial suspensions of FOP1 EMR (putative race 1), F81 (putative race 2), and R2 (putative race 2 from a different TEF clade) were made by releasing spores from 2-weeks-old agar plates with 10 mL SDW and filtering through three layers of miracloth. Spore suspensions were adjusted to 1 × 106 spores mL–1 using SDW with the addition of 200 μL of Tween20 L–1 and 1 mL pipetted directly onto pea roots and spread by tilting the plate. Plates were re-sealed and returned to the 20°C (16 h photoperiod) incubator in the arrangement of a split pot design. Pea root samples were taken at 10 time points (8, 16, 24, 36, 48, 72, and 96 hours post inoculation), including a 0 h time point (pre-inoculation) and an uninoculated control (SDW/Tween only) collected at 96 hpi. Four plates per FOP isolate were sampled at each timepoint where the roots of four pea plants per plate were removed, rinsed in SDW, flash frozen in liquid nitrogen and stored at −80°C until use.
Roots were ground to a fine powder using a pestle and mortar filled with liquid nitrogen and approximately 100 mg of tissue transferred to a 2 mL tube. Frozen root material was ground further using a Dremel drill (model 398, with a rounded drill bit) and then RNA extracted using Trizol® reagent (Ambion, Thermo Fisher Scientific) following the manufacturer’s guidelines. Extracted RNA was precipitated using 900 μL of lithium chloride to 100 μL RNA (250 μL LiCl2 + 650 μL DEPC treated water) and any DNA was removed from samples using DNase 1 (Sigma-Aldrich, United Kingdom). RNA samples were visualised on a 2% agarose gel (containing GelRedTM at 2 μL per 100 mL of gel) with the addition of loading dye (Orange G, Sigma-Aldrich, United Kingdom) to check for degradation. First strand cDNA was synthesised using Superscript II reverse transcriptase (Invitrogen, Thermo Fisher Scientific, United Kingdom) following the manufacturer’s protocol.
To quantify expression of SIX genes, Real-Time RT-PCR was performed using the cDNA from each FOP isolate and a Roche Lightcycler 480 using the Lightcycler 480 SYBR Green 1 Master mix (Roche, United Kingdom), following the manufacturer’s protocol. Published primers (Taylor et al., 2016) were used for SIX7, SIX9, SIX10, SIX12, and SIX13 with the remainder (SIX1, SIX6, SIX11, and SIX14) designed in this study as described above (Supplementary Table 3). Due to the multi-copy nature of SIX1 (identified through genome sequencing) and some sequence differences between isolates, primers were designed to match one copy (copy 1, Supplementary Figure 1) of this gene in the putative race 2 isolates F81 and R2 only, and therefore no data was collected for the expression of these genes in the putative race 1 isolate FOP1 EMR. Similarly, primers were only designed for the copy of SIX6 (Supplementary Figure 1) present in all isolates. In addition to SIX genes, qPCR was also conducted for TEF using published primers (Taylor et al., 2016; Supplementary Table 3) to determine background expression levels. All primers were used at a final concentration of 0.4 μM (except qSIX11 and qSIX14 which were used at 0.2 μM and qSIX6 which was used at 0.15 μM), using the following conditions: one cycle of 95°C for 5 min, 45 cycles of 95°C for 10 s, primer annealing temp (Supplementary Table 3) for 10 s and finally 72°C for 10 s. A melt curve analysis was used to confirm the presence of a single PCR product. Standard curves were plotted for each SIX gene target by using serially diluted genomic DNA and the concentration expressed relative to TEF. All samples were run in triplicate, for each of the four replicate root samples for each time point per FOP isolate. Relative concentration values for each SIX gene were loge transformed and analysed using Analysis of variance (ANOVA) in Genstat® (release 18.1, VSN international Ltd., United Kingdom) considering the split plot blocking structure in the incubator. Significant differences between individual time points or isolates were compared with the least significant difference (LSD) at the 5% level (Supplementary Table 5).
Race Typing of FOP Isolates
Pea differential cultivars (Table 2) were inoculated using the root dipping method adapted from Bani et al. (2012) to determine the race of FOP isolates selected from different clades of the TEF phylogenetic tree and with different SIX gene profiles. Cultures of FOP1 EMR and F79 (putative race 1), FOP2 and F81 (putative race 2), and R2 and F40 (putative variant of race 2 based on variations in SIX gene complement) were initiated from glycerol bead stocks on PDA and grown for 14 days at 20°C. Meanwhile, pea seeds of differential cultivars Darkskin Perfection (DSP), Mini, Sundance II, and Little Marvel (Table 2) obtained from the Germplasm Resources Unit (GRU) at John Innes Centre (Norfolk, United Kingdom) were sown in 24 cell modular trays containing vermiculite (1–3 mm diameter) until 2–3 nodes tall (approx. 14 days), in the glasshouse (25°C day, 18°C night, 16 h photoperiod). Conidial suspensions of each FOP isolate were prepared as described above and adjusted to 1 × 106 spores mL–1 in sterile water. Pea plants from each of the differentials were removed from the vermiculite, roots washed briefly and trimmed by a third in length, before being immersed for 5 min in a spore suspension of each FOP isolate. Roots of uninoculated control plants were immersed in SDW only. Following inoculation, pea plants were transplanted into individual pots (7 × 7 × 8 cm) containing vermiculite and maintained in the glasshouse (conditions as above) where they were watered daily (or when needed) and supplemented with fertiliser (Vitax 2:1:4, diluted 100-fold). Plants were arranged in a randomised complete block design with each of the 14 blocks containing one plant of each pea cultivar. Plants were monitored for wilt symptoms each week for 41 days, commencing 7 days after inoculation, by recording the proportion of leaves showing symptoms of wilt for each plant. The number of wilted leaves at 41 dpi was calculated as a proportion of the total number and transformed using a logit transformation with an offset of one. The transformed data was analysed by ANOVA in Genstat® (release 18.1, VSN international Ltd.), considering the randomised block design in the glasshouse. Treatment means from significant ANOVA’s were compared to the control and each other using the LSD at the 5% level (Supplementary Table 6).
Results
Molecular Characterisation of F. oxysporum Isolates
Fusarium isolates collected from diseased peas from United Kingdom fields were identified using TEF gene sequencing, with the majority being confirmed as F. oxysporum, in addition to isolates of F. solani and Fusarium redolens. A selection (40 isolates) of those identified as F. oxysporum were used for phylogenetic analysis, along with 28 isolates previously identified as FOP by other researchers.
The maximum likelihood phylogenetic tree based on TEF gene sequences, separated the F. oxysporum isolates from United Kingdom peas, FOP isolates and sequences from other f. spp. into a total of eight clades (based on bootstrap values > 60%; Figure 1). The majority of F. oxysporum isolates from diseased United Kingdom peas (coded PG) were typically found in three closely related clades (6, 7, and 8; Figure 1) and were distinct from the 28 FOP isolates from other researchers (clades 1, 2, and 5; Figure 1). In most cases, isolates putatively identified as FOP race 1 and race 2 grouped into separate but closely related clades in the tree (clade 1 for race 2 and clade 2 for race 1), supporting their putative race assignment. However, a small group of putative race 2 isolates were found in another clade (clade 5; Figure 1), alongside two isolates of FOC, suggesting a different origin or potentially even a different race type, and were designated a subset or variant of race 2 (2S). Generally, the 28 FOP isolates grouped separately from isolates of other F. oxysporum f. spp. obtained from NCBI, although other published FOP isolates grouped with FOP race 1 and 2 isolates from this study. The clear distinction of the isolates from diseased United Kingdom peas from previously race typed FOP isolates led us to hypothesise that they were of a different genetic origin.
Identification of SIX Genes in F. oxysporum Isolates From Pea
Genome sequence analysis revealed that FOP1 EMR (race 1), F81 (race 2), and R2 (race 2) contained homologs of eight (SIX1, SIX6, SIX7, SIX9, SIX10, SIX11, SIX12, and SIX14), five (SIX1, SIX6, SIX9, SIX13, and SIX14), and three (SIX1, SIX6, and SIX13) SIX genes, respectively. An additional copy of SIX6 (SIX62) was also identified in FOP1 EMR, while F81 and R2 contained one and two additional copies of SIX1, respectively.
Primers optimised for FOP were used to identify SIX genes within the 40 F. oxysporum isolates from diseased United Kingdom peas and the 28 previously race typed FOP isolates. Isolates previously assigned to either FOP race 1 or 2 within TEF clades 2 and 1, respectively (Figure 1) contained the same complements of SIX genes as identified through genome sequencing for isolates FOP1 EMR (race 1, Group A) and F81 (race 2, Group B), respectively (Table 1). Similarly, the subset of putative race 2 isolates from TEF clade 5 (Figure 1) contained the same complement of SIX genes as isolate R2 (race 2, Group C, Table 1). These latter race 2 isolates from TEF clade 5 differed from those in TEF clade 1 due to the absence of SIX9 and SIX14. A number of previously identified FOP isolates from other TEF clades (Figure 1) with a putative assignment of race 1 or 2 also contained few or no SIX genes (Group D, Table 1). In contrast, F. oxysporum isolates from diseased United Kingdom peas (coded PG) contained either one (SIX6 or SIX14) or no SIX genes (Group E, Table 1), suggesting that they may not be wilt causing FOP isolates.
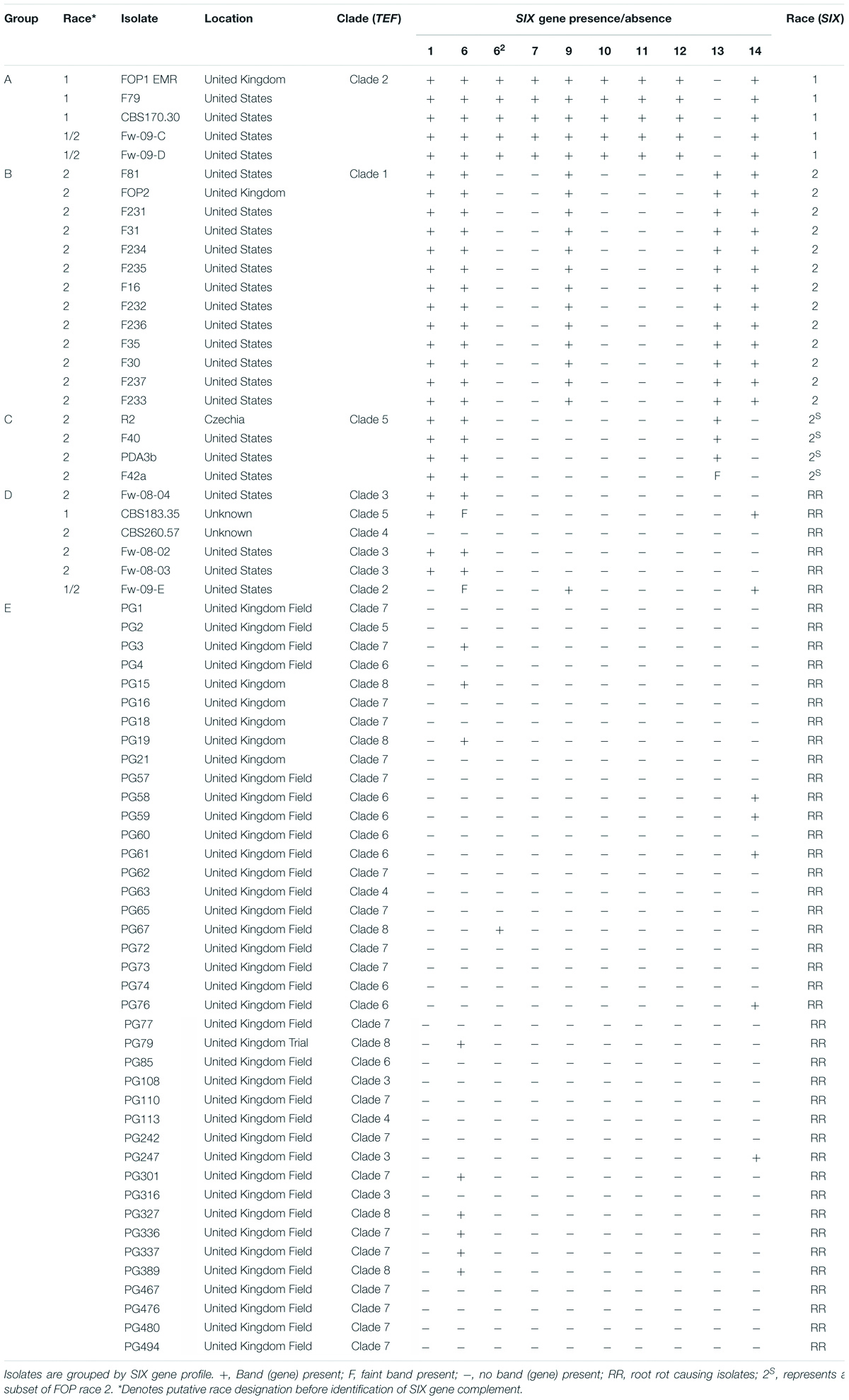
Table 1. Presence and absence of Secreted In Xylem (SIX) genes found in Fusarium oxysporum isolates from diseased United Kingdom peas and in F. oxysporum f. sp. pisi (FOP) isolates from other researchers (with previous putative race designation) as determined by PCR.
Secreted In Xylem gene sequences in FOP were also compared with SIX gene homologs in other F. oxysporum f. spp. Similar copies of SIX1 (copies 1 and 2) in F81 and R2 grouped together (Supplementary Figure 1), whereas the third copy in R2 was similar to the sequence from F. oxysporum f. sp. medicaginis (Medicago truncatula). The additional copy of SIX6 (SIX62) in FOP1 EMR grouped closer to the sequence identified in F. oxysporum f. sp. cubense, than to the copies of SIX6 found in all races (Table 1). The sequence for SIX13 in FOP was very similar to the sequence in F. oxysporum f. sp. narcissi, with isolates from both being grouped into the same clade (Supplementary Figure 1). Generally, the sequences of SIX genes in FOP differed from those identified in other F. oxysporum f. spp., with most resulting in distinct branches in the phylogenetic trees (Supplementary Figure 1).
Expression of SIX Genes in Planta
Of the SIX genes identified in FOP1 EMR (SIX1, SIX6, SIX7, SIX9, SIX10, SIX11, SIX12, and SIX14), F81 (SIX1, SIX6, SIX9, SIX13, and SIX14), and R2 (SIX1, SIX6, and SIX13), the vast majority of those tested (SIX1 in FOP1 EMR was not tested) were expressed in planta, across the early stages of infection (8–96 hpi), following inoculation of pea seedlings (Figure 2). SIX7, SIX10, SIX11, and SIX12 expression levels (present in FOP1 EMR only) increased at every time point over the experiment, with significant increases observed from 72 hpi onward (Figure 2 and Supplementary Table 5). SIX6 was expressed in all three isolates (Figure 2), although levels were significantly less in FOP1 EMR compared with the other isolates F81 and R2 (Supplementary Table 5). Although SIX14 and SIX9 were both present in FOP1 EMR and F81, these genes were only expressed in F81, with expression levels increasing over time during the early stages of infection (Figure 2). Expression of SIX13 increased over time in both F81 and R2, with similar levels in both isolates, until 96 hpi where expression in F81 was significantly higher than in R2 (Supplementary Table 5).
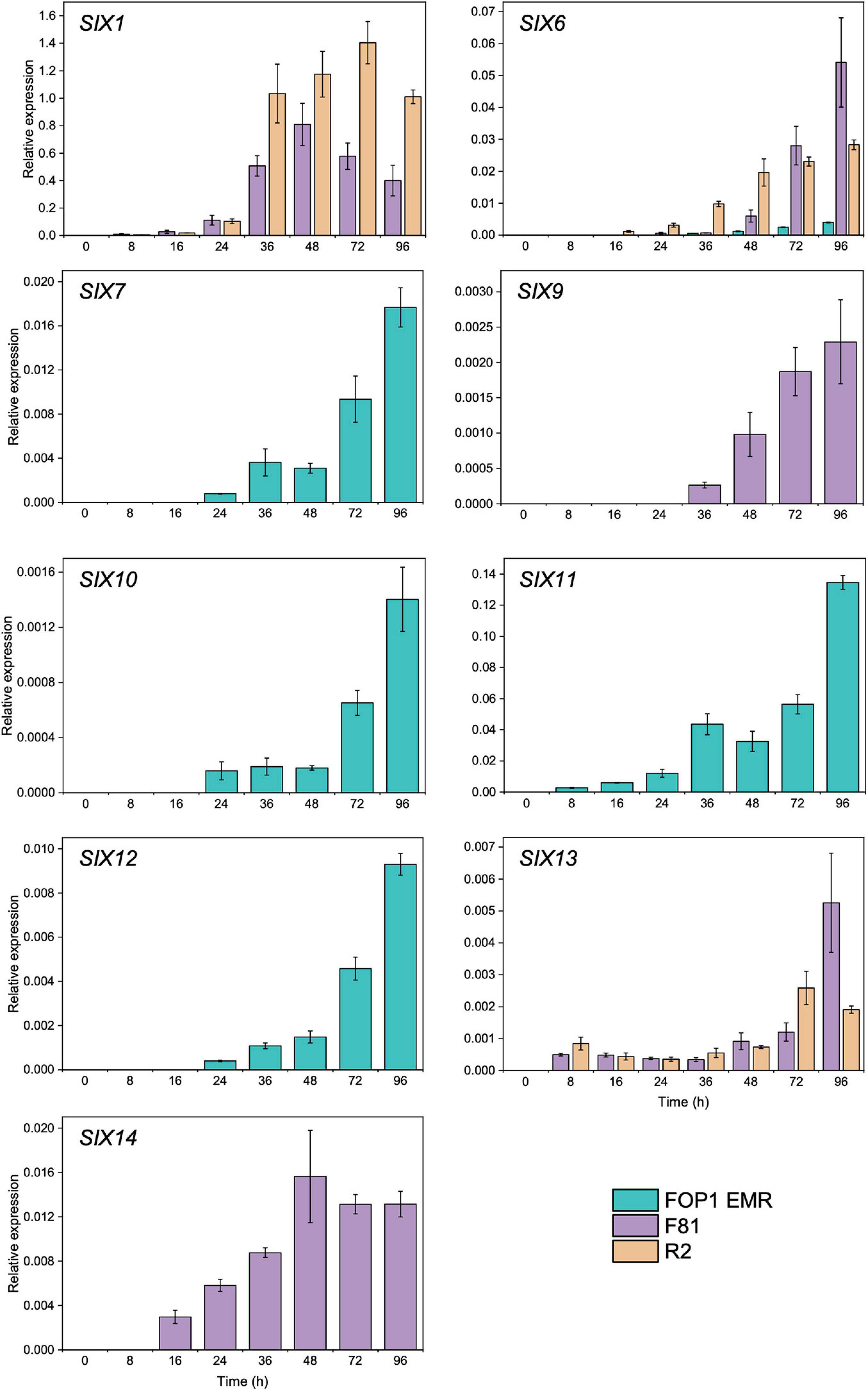
Figure 2. Expression of Secreted In Xylem (SIX) genes in pea roots infected with FOP isolates FOP1 EMR (race 1), F81 (race 2), and R2 (race 2S) as determined by reverse transcription qPCR of RNA. Expression was calculated relative to the TEF and the mean value displayed [error bars represent Standard error of the mean (SEM)].
Due to the multiple copies of SIX1, only FOP isolates F81 and R2 were included in the analysis for this gene, as the sequence of SIX1 in FOP1 EMR varied from the copies in F81 and R2. Expression of SIX1 increased over time in both isolates, with levels peaking at 48 hpi for F81 and at 72 hpi for R2 (Figure 2). However, the expression levels for SIX1 in F81 were significantly lower than in R2 at 36, 72, and 96 hpi (Supplementary Table 5). No SIX gene expression was detected for uninoculated control pea collected at 96 hpi.
Race Typing of FOP Isolates
In the root dip inoculation test there was a significant interaction observed between the six FOP isolates (FOP1 EMR, F79, FOP2, F81, R2, and F40) and the four pea cultivars (Little Marvel, DSP, Mini, and Sundance II) tested (p < 0.001; Supplementary Table 6). All F. oxysporum isolates were highly pathogenic on the universal susceptible cultivar Little Marvel, each causing over 60% leaf wilt by 41 dpi (Figure 3). Cultivar DSP (resistant to FOP race 1 and susceptible to race 2) was resistant to FOP1 EMR and F79, with less than 25% of leaves wilted, and highly susceptible to FOP2, F81, R2, and F40, with more than 81% of leaves wilted confirming these isolates as race 1 and race 2, respectively (Figure 3). This race designation was further confirmed by results on cultivar Mini which was highly susceptible to FOP1 EMR and F79 and resistant to FOP2, F81, R2, and F40. Finally, the resistance of cultivar Sundance II to FOP1 EMR and F79 confirmed these isolates as FOP race 1 (Figure 3 and Table 2). However, unexpectedly, isolates FOP2, F81, R2, and F40 caused no significant wilt compared to the control for Sundance II which is reported to be susceptible to FOP race 2.
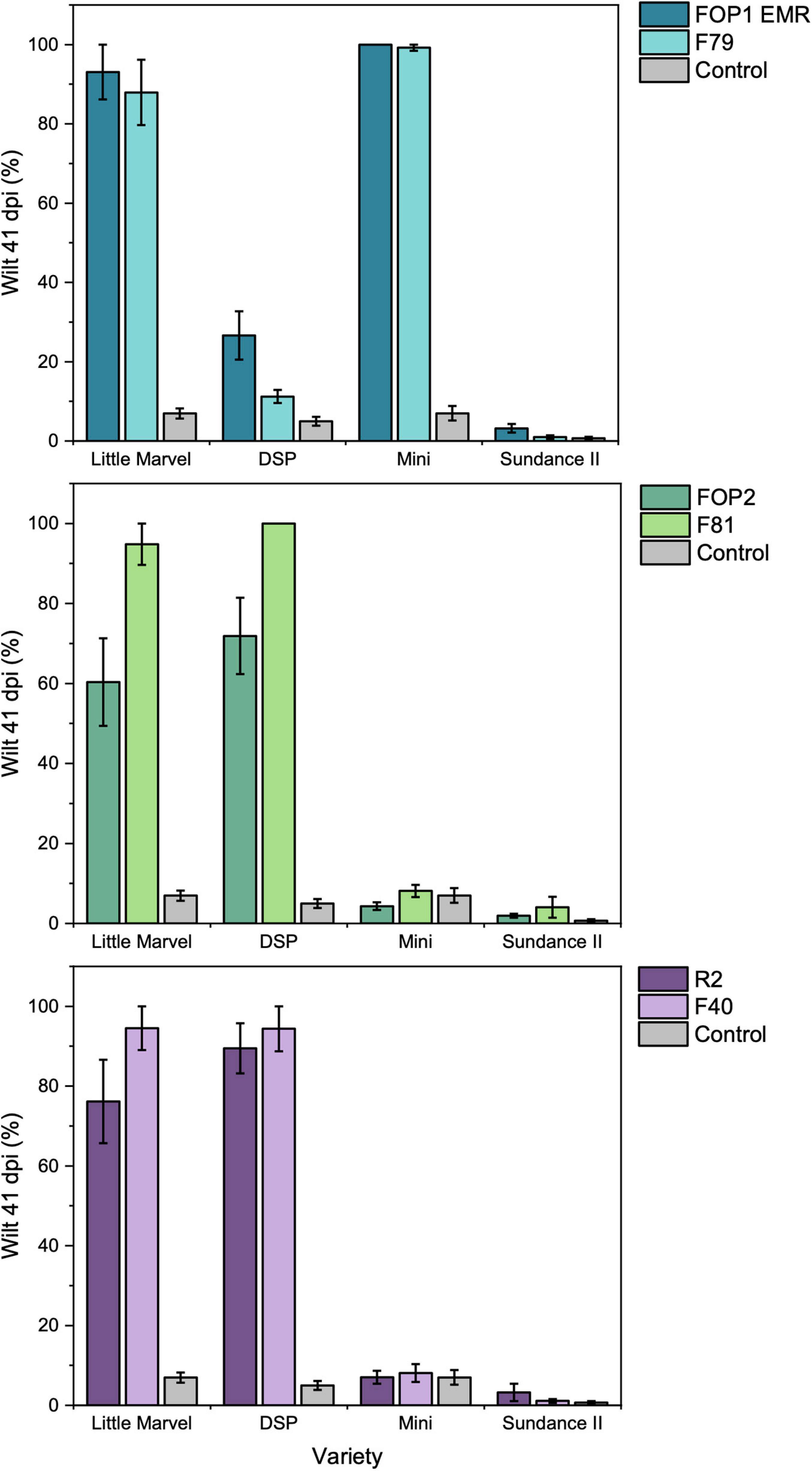
Figure 3. Pathogenicity of SIX FOP isolates on four differential cultivars of pea (Little Marvel, DSP, Mini, and Sundance II) using the root dip inoculation method. Top = putative race 1 isolates; middle = putative race 2 isolates and bottom = putative race 2S isolates. Values are average percentage wilt of 14 inoculated plants, error bars are SEM.
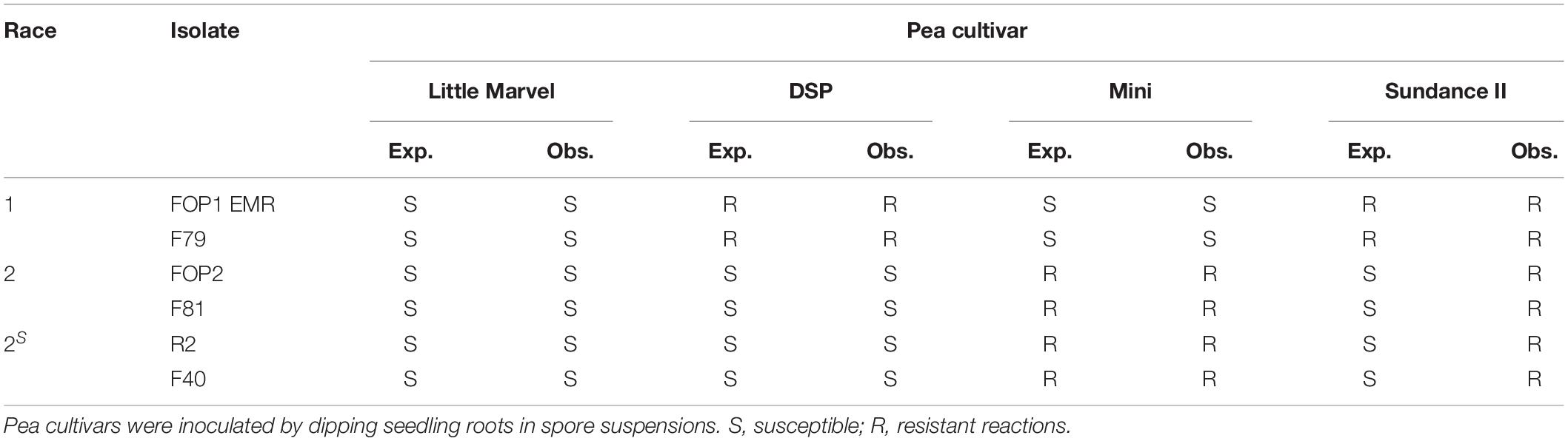
Table 2. Summary of expected (Exp.) and observed (Obs.) results for the resistance and susceptibility of pea differential cultivars to different FOP isolates representing preliminary races 1, 2, and 2S as designated based on SIX gene complement.
Discussion
In this study, we have identified for the first time, the presence of nine SIX genes in FOP, with distinct complements of genes occurring in races 1 (SIX1, SIX6, SIX7, SIX9, SIX10, SIX11, SIX12, and SIX14) and 2 (SIX1, SIX6, SIX9, SIX13, SIX14 or SIX1, SIX6, SIX13). All SIX genes tested were expressed in planta (except SIX9 and SIX14 in race 1), suggesting they could be functionally involved in the early stages of FOP infection. Race testing of FOP isolates using the pea differential cultivars confirmed the difference between the isolates putatively designated as race 1 or race 2. Therefore, the unique SIX gene complement identified in FOP race 1 isolates could be used as a way to distinguish these from race 2. Additionally, the complete absence or the presence of just one or two SIX genes in F. oxysporum isolates from diseased United Kingdom pea plants, suggests that they are not FOP isolates and preliminary tests (Jenkins, 2018) indicated that they cause foot and root rot rather than wilt in peas.
In previous research where just three FOP isolates were included in a study to identify complements of SIX genes in multiple isolates of FOC and a selection of other F. oxysporum f. spp., SIX7, SIX10, SIX11, SIX12, and SIX14 were identified in FOP race 1, SIX13 and SIX14 were identified in race 2 and SIX13 identified in race 5 leading to the conclusion that there was variation SIX gene complement between different races (Taylor et al., 2016). However, FOP race identity was not confirmed on plants and the presence of SIX genes relied on primers designed to SIX gene sequences from FOL and FOC. It is frequently documented that sequences for SIX genes vary across F. oxysporum f. spp. (Lievens et al., 2009; van Dam et al., 2016), which could account for the discrepancy in results between this and the present study where SIX gene primers were optimised for FOP. In this current study, nine SIX genes were identified in the whole genome sequences of three FOP isolates (Jenkins, 2018), which enabled previously published primers to be evaluated for their complementarity against FOP specific SIX gene sequences, and where necessary, new FOP specific primers were designed.
The variation of SIX gene complement in isolates identified previously by other researchers as race 2 was unexpected; the majority of these isolates contained five SIX genes (SIX1, SIX6, SIX9, SIX13, and SIX14) while the remainder lacked SIX9 and SIX14. These two groups of race 2 isolates also separated into different TEF clades suggesting different origins. One hypothesis is that these two groups could actually represent different races. Although FOP race 5 isolates were not represented in this study, recent genome analysis of one isolate has identified the presence of SIX1, SIX9, SIX13, and SIX14 (Williams et al., 2016), which is very close to the SIX gene profile of the majority of our race 2 isolates, lacking just SIX6. Although this further supports the idea that profiling SIX genes or other putative effector genes may be informative for distinguishing some races in FOP, the variation in SIX gene presence within race 2 isolates needs to be understood. Despite these differences, representative isolates from both groups of race 2 isolates resulted in the same reactions in the small set of differential pea cultivars tested here and were therefore confirmed as FOP race 2. This was based on expected reactions observed for all these isolates on the cultivars Little Marvel (susceptible), DSP (resistant), and Mini (resistant). Unexpectedly, however, these isolates resulted in a resistant reaction for cv. Sundance II where the expected outcome for race 2 isolates is susceptibility. There is no clear explanation for this although variation in FOP race 2 isolates has been documented previously and races originally identified as “old” race 3 and 4 have now been combined under race 2 despite some differences in pea differential response (Kraft and Haglund, 1978; Kraft, 1994). For instance, the reaction of the differential pea cultivar New Season has been reported to vary with different isolates of race 2 (Kraft, 1994; Kraft and Pfleger, 2001). To properly resolve the races in FOP and potentially separate the two groups of race 2 isolates with different SIX gene profiles, further tests are required using an improved and expanded set of pea differential cultivars. The addition of RNAseq data, as well as SIX gene profiling, might also identify presence/absence of other potential effectors which could help to explain the differences in these two groups of race 2 isolates more definitively.
Differences in SIX gene profile between F. oxysporum f. spp. races as observed in this study for FOP has been reported previously, and in cases where these have been identified as avirulence (Avr) genes, this has arisen as a means of evading detection by corresponding host R genes. For instance, SIX4 (Avr 1) which is present in FOL race 1 isolates is absent in races 2 and 3 preventing host recognition by the tomato I and I-1 resistance alleles (Takken and Rep, 2010). SIX4 is not required for general virulence, rather it suppresses the ability of other resistance alleles (I−2 and I−3) to confer resistance. In a different strategy to overcome plant resistance, single point mutation variants in SIX3 (Avr2) in FOL race 3 prevents recognition by the tomato I−2 allele (Takken and Rep, 2010). Similarly, SIX8 gene sequence can be used to distinguish race 4 from races 1 and 2 in F. oxysporum f. sp. cubense (Fraser-Smith et al., 2014). It could be hypothesised therefore, that the loss of non-essential SIX genes from FOP race 1 (SIX7, 10, 11, and 12) in race 2, and the gain of SIX13, could have contributed to the ability of race 2 to overcome resistant pea cultivars. Similarly, the loss of SIX6 in FOP race 5 (Williams et al., 2016) which is present in FOP races 1 and 2, could suggest it is non-essential for FOP virulence and may have allowed race 5 to evade host detection. In contrast, the presence of SIX1 in all FOP races could imply it is essential for virulence, although this would require testing with gene knockout studies.
The majority of F. oxysporum isolates obtained from diseased pea plants from United Kingdom fields contained no SIX genes, with only some isolates containing one SIX gene (SIX6 or SIX14). In the TEF phylogenetic tree, the majority of these isolates (with and without SIX genes) also grouped into separate clades from the FOP isolates suggesting a different origin and were shown in unpublished studies to cause foot and root rot (which results in similar symptoms to FOP in mature pea plants in the field) rather than wilt caused by FOP isolates (Jenkins, 2018). The general lack of SIX genes in foot and root-rotting F. oxysporum isolates suggests a different mode of infection to FOP resulting in slightly different disease symptoms. SIX genes (SIX1, SIX6, SIX8, SIX9, SIX11, and SIX14) have previously been identified in naturally occurring F. oxysporum isolates in Australia, which provided evidence of horizontal gene transfer (HGT) from f. spp. of an agricultural origin into the natural isolate population, especially in the case of SIX1 and SIX6 (Rocha et al., 2016). It was suspected that the SIX genes were non-functional in the natural population due to their low frequency (Rocha et al., 2016), and this could also be the case for the root rot causing F. oxysporum isolates in pea.
This current study showed that of the SIX genes identified in different races of FOP (SIX1, SIX6, SIX7, SIX9, SIX10, SIX11, SIX12, SIX13, and SIX14), all of those tested were expressed (except SIX9 and SIX14 in race 1) during the early stages of pea infection, hence suggesting a role in virulence and providing targets for future functional studies. Expression of SIX genes in other F. oxysporum f. spp. has been examined previously; for instance, a similar study in FOC confirmed expression in planta of all seven SIX genes present (SIX3, SIX5, SIX7, SIX9, SIX10, SIX12, and SIX14) following inoculation of onion seedling roots (Taylor et al., 2016).
Secreted In Xylem 1 gene expression was observed in FOP race 2 isolates F81 and R2 but was not confirmed for race 1 isolate FOP1 EMR due to variation in the sequence. However, additional RNAseq analysis (Jenkins, 2018) showed that SIX1 was expressed in FOP1 EMR at 96 hpi. SIX1 sequences have been shown to vary between races of F. oxysporum f. sp. cubense, with the sequence of SIX1 in sub-tropical race 4 differing from the sequence found in race 1, 2, and tropical race 4 (Meldrum et al., 2012). SIX1 has previously been shown to be essential for virulence in FOL (Rep et al., 2005), F. oxysporum f. sp. conglutinans (Li et al., 2016) and F. oxysporum f. sp. cubense tropical race 4 (Widinugraheni et al., 2018).
Secreted In Xylem 6 has also been shown to be expressed in the early stages of FOL infection on tomato seedlings (Gawehns et al., 2014) and was also found to be expressed from 3 dpi in watermelon roots infected with F. oxysporum f. sp. niveum (Niu et al., 2016). SIX6 has also been shown to be necessary for full virulence in FOL (Gawehns et al., 2014). Only one copy of SIX6 was used for qPCR analysis due to the similarities in sequences between the two copies in FOP1 EMR. Additional RNAseq analysis (Jenkins, 2018) showed that there was no expression for SIX62 in FOP1 EMR, suggesting this gene is non-functional, although tests would be needed to confirm this.
Although SIX9 and SIX14 were present in the genome of FOP1 EMR, and were detected via PCR, they were not highly expressed in planta, yielding expression values lower than the limit of detection, suggesting they could be pseudogenes and therefore not important in infection. However, SIX14 was highly expressed in FOP race 2 isolates where it was present. Both SIX9 and SIX14 were expressed during infection of onion seedlings with FOC in a similar study (Taylor et al., 2016). SIX9 was identified in the 20 highest expressed genes of a pathogenic FOC isolate during infection, whereas SIX14 was expressed at much lower levels and was ranked at 3,277th (Armitage et al., 2018). Interestingly, SIX9 was also identified in a non-pathogenic F. oxysporum isolate from onion (Armitage et al., 2018), suggesting it may not be directly related to virulence. This was also shown in F. oxysporum f. sp. radicis-cucumerinum where disruptions in SIX9 did not significantly affect virulence (van Dam et al., 2017).
Further research is now required to understand the role of SIX genes in FOP pathogenicity through knockout studies as implemented in F. oxysporum f. spp. (Rep et al., 2005; Houterman et al., 2009; Gawehns et al., 2014). Additionally, proteomic studies could also help determine the presence of functional proteins in the sap of pea plants (Houterman et al., 2007; Lievens et al., 2009).
Overall, our findings provide a greater understanding of SIX genes in FOP and their presence and absence in different races. This provides a foundation for a better understanding of the evolution of virulence in FOP races and also potentially improve the diagnosis and control of FOP in pea in the future, as the current method of differential testing to race type FOP isolates is not always reliable. Using effectors to understand plant–pathogen gene interactions could also lead to more robust plant resistance to FOP, which is less likely to breakdown with the evolution of new races.
Data Availability Statement
Oxford Nanopore and Illumina reads for the three sequenced isolates (FOP1 EMR, F81, and R2) were submitted to the NCBI Sequence Read Archive with accessions SRX8613878–SRX8613883 and are available under bioproject PRJNA641642.
Author Contributions
SJ planned and performed the experiments, analysed the data, created the figures, and drafted and edited the manuscript. AT and AJ helped to plan experiments and analyse data. HB carried out the sequencing of FOP genomes while AA carried out the genomic analyses. AM created the experimental designs and provided statistical analyses. RH obtained funding, planned experiments, and edited the manuscript. JC obtained funding, planned experiments, and wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was carried out as part of a Ph.D. studentship funded by the Biotechnology and Biological Sciences Research Council Grant BB/M50290X/1.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The content of this manuscript has been published as part of the thesis of SJ (Jenkins, 2018). We would like to acknowledge Lea Herold and Becky Howard from the Processors and Growers Research Organisation (PGRO) and Jane Thomas (NIAB) for providing expertise on peas and isolates. We are also very grateful to Lyndon Porter (USDA) and Rebecca McGee (USDA) for providing the FOP isolates used in this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.593140/full#supplementary-material
References
Armitage, A. D., Taylor, A., Sobczyk, M. K., Baxter, L., Greenfield, B. P. J., Bates, H. J., et al. (2018). Characterisation of pathogen-specific regions and novel effector candidates in Fusarium oxysporum f. sp. cepae. Sci. Rep. 8:13530. doi: 10.1038/s41598-018-30335-7
Bani, M., Perez-De-Luque, A., Rubiales, D., and Rispail, N. (2018). Physical and chemical barriers in root tissues contribute to quantitative resistance to Fusarium oxysporum f. sp pisi in Pea. Front. Plant Sci. 9:199. doi: 10.3389/fpls.2018.00199
Bani, M., Rubiales, D., and Rispail, N. (2012). A detailed evaluation method to identify sources of quantitative resistance to Fusarium oxysporum f. sp pisi race 2 within a Pisum spp. germplasm collection. Plant Pathol. 61, 532–542. doi: 10.1111/j.1365-3059.2011.02537.x
Biddle, A. J., and Cattlin, N. D. (2007). Pests, Diseases, and Disorders of Peas and Beans: A Colour Handbook. Cambridge, MA: Academic Press.
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Chakrabarti, A., Rep, M., Wang, B., Ashton, A., Dodds, P., and Ellis, J. (2011). Variation in potential effector genes distinguishing Australian and non-Australian isolates of the cotton wilt pathogen Fusarium oxysporum f.sp vasinfectum. Plant Pathol. 60, 232–243. doi: 10.1111/j.1365-3059.2010.02363.x
Chittem, K., Mathew, F. M., Gregoire, M., Lamppa, R. S., Chang, Y., Markell, S. G., et al. (2015). Identification and characterization of Fusarium spp. associated with root rots of field pea in North Dakota. Eur. J. Plant Pathol. 143, 641–649. doi: 10.1007/s10658-015-0714-8
Covey, P. A., Kuwitzky, B., Hanson, M., and Webb, K. M. (2014). Multilocus analysis using putative fungal effectors to describe a population of Fusarium oxysporum from sugar beet. Phytopathology 104, 886–896. doi: 10.1094/PHYTO-09-13-0248-R
Dean, R., Van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Edel-Hermann, V., and Lecomte, C. (2019). Current Status of Fusarium oxysporum formae speciales and races. Phytopathology 109, 512–530. doi: 10.1094/phyto-08-18-0320-rvw
Eurofins (2016). Eurofins Oligo Analysis Tool [Online]. Available: http://www.operon.com/tools/oligo-analysis-tool.aspx (accessed September, 2018).
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evol. Int. J. Org. Evol. 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fourie, G., Steenkamp, E. T., Ploetz, R. C., Gordon, T. R., and Viljoen, A. (2011). Current status of the taxonomic position of Fusarium oxysporum formae specialis cubense within the Fusarium oxysporum complex. Infect. Genet. Evol. 11, 533–542. doi: 10.1016/j.meegid.2011.01.012
Fraser-Smith, S., Czislowski, E., Meldrum, R. A., Zander, M., O’Neill, W., Balali, G. R., et al. (2014). Sequence variation in the putative effector gene SIX8 facilitates molecular differentiation of Fusarium oxysporum f. sp cubense. Plant Pathol. 63, 1044–1052. doi: 10.1111/ppa.12184
Gawehns, F., Houterman, P. M., Ichou, F. A., Michielse, C. B., Hijdra, M., Cornelissen, B. J., et al. (2014). The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2-mediated cell death. Mol. Plant Microbe Interact. 27, 336–348. doi: 10.1094/MPMI-11-13-0330-R
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Haas, B. (2007). TransposonPSI [Online]. Available: http://transposonpsi.sourceforge.net (accessed September, 2018).
Haglund, W. A., and Kraft, J. M. (1970). Fusarium oxysporum f. pisi race 5. Phytopathology 60, 1861–1862.
Haglund, W. A., and Kraft, J. M. (1979). Fusarium-oxysporum f-sp pisi, race-6 - occurrence and distribution. Phytopathology 69, 818–820. doi: 10.1094/Phyto-69-818
Houterman, P. M., Ma, L., van Ooijen, G., de Vroomen, M. J., Cornelissen, B. J. C., Takken, F. L. W., et al. (2009). The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 58, 970–978. doi: 10.1111/j.1365-313X.2009.03838.x
Houterman, P. M., Speijer, D., Dekker, H. L., de Koster, C. G., Cornelissen, B. J. C., and Rep, M. (2007). The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol. Plant Pathol. 8, 215–221. doi: 10.1111/j.1364-3703.2007.00384.x
Jenkins, S. (2018). Examining Identity, Phylogeny and Pathogenicity Factors in Fusarium Species Affecting Pea. PhD thesis, University of Warwick, Coventry.
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kraft, J., and Haglund, W. (1978). A reappraisal of the race classification of Fusarium oxysporum f. sp. pisi. Phytopathology 68, 273–276.
Kraft, J. M. (1994). Fusarium-wilt of peas (a review). Agronomie 14, 561–567. doi: 10.1051/agro:19940901
Kraft, J. M., Haware, M. P., Jiménez-Díaz, R. M., Bayaa, B., and Harrabi, M. (1994). Screening techniques and sources of resistance to root rots and wilts in cool season food legumes. Euphytica 73, 27–39. doi: 10.1007/BF00027179
Kraft, J. M., and Pfleger, F. L. (2001). Compendium of Pea Diseases and Pests, 2nd Edn. St. Paul, MN: The American Phytopathological Society.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, E. F., Wang, G., Xiao, J. L., Ling, J., Yang, Y. H., and Xie, B. Y. (2016). A SIX1 homolog in Fusarium oxysporum f. sp conglutinans is required for full virulence on cabbage. PLoS One 11:e0152273. doi: 10.1371/journal.pone.0152273
Lievens, B., Houterman, P. M., and Rep, M. (2009). Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 300, 201–215. doi: 10.1111/j.1574-6968.2009.01783.x
Loman, N. J., Quick, J., and Simpson, J. T. (2015). A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 12, 733–735. doi: 10.1038/nmeth.3444
Ma, L. J., van der Does, H. C., Borkovich, K. A., Coleman, J. J., Daboussi, M. J., Di Pietro, A., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373. doi: 10.1038/nature08850
McPhee, K. E., Inglis, D. A., Gundersen, B., and Coyne, C. J. (2012). Mapping QTL for Fusarium wilt Race 2 partial resistance in pea (Pisum sativum). Plant Breed. 131, 300–306. doi: 10.1111/j.1439-0523.2011.01938.x
McPhee, K. E., Tullu, A., Kraft, J. M., and Muehlbauer, F. J. (1999). Resistance to Fusarium wilt race 2 in the Pisum core collection. J. Am. Soc. Hortic. Sci. 124, 28–31.
Meldrum, R. A., Fraser-Smith, S., Tran-Nguyen, L. T. T., Daly, A. M., and Aitken, E. A. B. (2012). Presence of putative pathogenicity genes in isolates of Fusarium oxysporum f. sp. cubense from Australia. Austral. Plant Pathol. 41, 551–557. doi: 10.1007/s13313-012-0122-x
Niu, X. W., Zhao, X. Q., Ling, K. S., Levi, A., Sun, Y. Y., and Fan, M. (2016). The FonSIX6 gene acts as an avirulence effector in the Fusarium oxysporum f. sp niveum - watermelon pathosystem. Sci. Rep. 6:28146. doi: 10.1038/srep28146
O’Donnell, K., Kistler, H. C., Cigelnik, E., and Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U.S.A. 95, 2044–2049. doi: 10.1073/pnas.95.5.2044
Ploetz, R. C. (2015). Management of Fusarium wilt of banana: a review with special reference to tropical race 4. Crop Protect. 73, 7–15. doi: 10.1016/j.cropro.2015.01.007
Rep, M., Meijer, M., Houterman, P. M., van der Does, H. C., and Cornelissen, B. J. C. (2005). Fusarium oxysporum evades I-3-mediated resistance without altering the matching avirulence gene. Mol. Plant Microbe Interact. 18, 15–23. doi: 10.1094/mpmi-18-0015
Rep, M., van der Does, H. C., Meijer, M., van Wijk, R., Houterman, P. M., Dekker, H. L., et al. (2004). A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383. doi: 10.1111/j.1365-2958.2004.04177.x
Rocha, L. O., Laurence, M. H., Ludowici, V. A., Puno, V. I., Lim, C. C., Tesoriero, L. A., et al. (2016). Putative effector genes detected in Fusarium oxysporum from natural ecosystems of Australia. Plant Pathol. 65, 914–929. doi: 10.1111/ppa.12472
Ruan, J. (2016). Ultra-Fast De Novo Assembler Using Long Noisy Reads [Online]. Available: https://github.com/ruanjue/smartdenovo (accessed September, 2018).
Sasaki, K., Nakahara, K., Tanaka, S., Shigyo, M., and Ito, S.-I. (2015). Genetic and pathogenic variability of Fusarium oxysporum f. sp cepae isolated from onion and Welsh onion in Japan. Phytopathology 105, 525–532. doi: 10.1094/phyto-06-14-0164-r
Schmidt, S. M., Houterman, P. M., Schreiver, I., Ma, L., Amyotte, S., Chellappan, B., et al. (2013). MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genomics 14:119. doi: 10.1186/1471-2164-14-119
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Smit, A. F. A., Hubley, R., and Green, P. (2013–2015). RepeatMasker Open-4.0 [Online]. Available: http://www.repeatmasker.org (accessed September, 2018).
Takken, F., and Rep, M. (2010). The arms race between tomato and Fusarium oxysporum. Mol. Plant Pathol. 11, 309–314. doi: 10.1111/j.1364-3703.2009.00605.x
Taylor, A., Vagany, V., Jackson, A. C., Harrison, R. J., Rainoni, A., and Clarkson, J. P. (2016). Identification of pathogenicity-related genes in Fusarium oxysporum f. sp cepae. Mol. Plant Pathol. 17, 1032–1047. doi: 10.1111/mpp.12346
Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R., and Leunissen, J. A. M. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74. doi: 10.1093/nar/gkm306
van Dam, P., de Sain, M., ter Horst, A., van der Gragt, M., and Rep, M. (2018). Use of comparative genomics-based markers for discrimination of host specificity in Fusarium oxysporum. Appl. Environ. Microbiol. 84:e01868-17. doi: 10.1128/aem.01868-17
van Dam, P., Fokkens, L., Ayukawa, Y., van der Gragt, M., ter Horst, A., Brankovics, B., et al. (2017). A mobile pathogenicity chromosome in Fusarium oxysporum for infection of multiple cucurbit species. Sci. Rep. 7:9042. doi: 10.1038/s41598-017-07995-y
van Dam, P., Fokkens, L., Schmidt, S. M., Linmans, J. H. J., Kistler, H. C., Ma, L. J., et al. (2016). Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 18, 4087–4102. doi: 10.1111/1462-2920.13445
Vaser, R., Sović, I., Nagarajan, N., and Šikić, M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746. doi: 10.1101/gr.214270.116
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Widinugraheni, S., Nino-Sanchez, J., van der Does, H. C., van Dam, P., Garcia-Bastidas, F. A., Subandiyah, S., et al. (2018). A SIX1 homolog in Fusarium oxysporum f.sp cubense tropical race 4 contributes to virulence towards Cavendish banana. PLoS One 13:e0205896. doi: 10.1371/journal.pone.0205896
Williams, A. H., Sharma, M., Thatcher, L. F., Azam, S., Hane, J. K., Sperschneider, J., et al. (2016). Comparative genomics and prediction of conditionally dispensable sequences in legume–infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genomics 17:191. doi: 10.1186/s12864-016-2486-8
Keywords: Fusarium oxysporum f. sp. pisi, PEA, Fusarium wilt, formae speciales, race, Secreted In Xylem, gene expression, qPCR
Citation: Jenkins S, Taylor A, Jackson AC, Armitage AD, Bates HJ, Mead A, Harrison RJ and Clarkson JP (2021) Identification and Expression of Secreted In Xylem Pathogenicity Genes in Fusarium oxysporum f. sp. pisi. Front. Microbiol. 12:593140. doi: 10.3389/fmicb.2021.593140
Received: 09 August 2020; Accepted: 17 March 2021;
Published: 09 April 2021.
Edited by:
Stefania Tegli, University of Florence, ItalyReviewed by:
Gopal Subramaniam, Agriculture and Agri-Food Canada (AAFC), CanadaNicolas Rispail, Institute for Sustainable Agriculture, Spanish National Research Council, Spain
Copyright © 2021 Jenkins, Taylor, Jackson, Armitage, Bates, Mead, Harrison and Clarkson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sascha Jenkins, c2FzY2hhLmplbmtpbnMuMUB3YXJ3aWNrLmFjLnVr
 Sascha Jenkins
Sascha Jenkins Andrew Taylor
Andrew Taylor Alison C. Jackson1
Alison C. Jackson1 Andrew D. Armitage
Andrew D. Armitage Andrew Mead
Andrew Mead Richard J. Harrison
Richard J. Harrison John P. Clarkson
John P. Clarkson