
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 15 March 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.585716
This article is part of the Research TopicCarbapenemase-Producing Organisms as Leading Cause of Hospital InfectionsView all 25 articles
 Xiaobing Guo1*†
Xiaobing Guo1*† Qian Wang1,2,3†
Qian Wang1,2,3† Hao Xu2
Hao Xu2 Xiaohong He4
Xiaohong He4 Lihua Guo2
Lihua Guo2 Shuxiu Liu1,2,3
Shuxiu Liu1,2,3 Peipei Wen1,3
Peipei Wen1,3 Jianjun Gou1*
Jianjun Gou1*The emergence of carbapenem resistance (CR) caused by hydrolytic enzymes called carbapenemases has become a major concern worldwide. So far, CR genes have been widely detected in various bacteria. However, there is no report of CR gene harboring Comamonas thiooxydans. We first isolated a strain of an IMP-8-producing C. thiooxydans from a patient with urinary tract infection in China. Species identity was determined using MALDI-TOF MS analysis and carbapenemase-encoding genes were detected using PCR. The complete genomic sequence of C. thiooxydans was identified using Illumina Novaseq and Oxford Nanopore PromethION. Antimicrobial susceptibility analysis indicated that the C. thiooxydans strain ZDHYF418 was susceptible to imipenem, intermediate to meropenem, and was resistant to aztreonam, fluoroquinolones, and aminoglycosides. The blaIMP–8 gene was chromosomally located, and was part of a Tn402-like class 1 integron characterized by the following structure: DDE-type integrase/transposase/recombinase-tniB-tniQ-recombinase family protein-aac(6′)-Ib-cr-blaIMP–8-intI1. Phylogenetic analysis demonstrated that the closest relative of ZDHYF418 is C. thiooxydans QYY (accession number: CP053920.1). We detected 330 SNP differences between ZDHYF418 and C. thiooxydans QYY. Strain QYY was isolated from activated sludge in Jilin province, China in 2015. In summary, we isolated a strain of C. thiooxydans that is able to produce IMP-8 and a novel blaOXA. This is the first time that a CR gene has been identified in C. thiooxydans. The occurrence of the strain needs to be closely monitored.
The genus Comamonas contains species that are Gram-negative, aerobic, non-pigmented, and rod-shaped bacteria that belong to β-Proteobacteria, which are motile through the use of at least one polar tuft of flagella, and has non-fermentative chemoorganotrophic metabolism. These are quite ubiquitous in the environment and have been isolated from soil, termite guts, activated sludge, humans, fresh water, sediments, and garden ponds. Comamonas strains have been found to be isolated from various clinical samples, as well as from the hospital environment. However, they are not seen as pathogenic to healthy humans (Willems and De Vos, 2006; Hatayama, 2014; Narayan et al., 2016; Subhash et al., 2016; Kämpfer et al., 2018). Despite the fact that Comamonas spp. are considered to be non-pathogenic or rare opportunistic pathogens to human, some Comamonas species have been suggested to be involved in many different infections, including Comamonas testosteroni, Comamonas kerstersii, and so on (Tsui et al., 2011; Zhou et al., 2018). Comamonas thiooxydans is a Gram-negative bacterium that belongs to the genus Comamona. Comamonas is comprised by 23 species with validly published names1. C. thiooxydans had the ability to oxidize thiosulfate under mixotrophic growth condition (Narayan et al., 2016). C. thiooxydans can grow under anoxic conditions, while other species that belong to Comamonas are strictly aerobes (Chen et al., 2016). Comamonas thiooxydans is most closely related to Comamonas testosteroni (Pandey et al., 2009). Of the all 21 C. thiooxydans in Genome of NCBI, 14 of them were originally submitted as Comamonas testosteroni but have later changed to C. thiooxydans due to ANI results2 (Supplementary Table 1). To date, infections of C. thiooxydans have not yet been reported. This current study describes the bacterium C. thiooxydans ZDHYF418, which has been isolated from a patient’s urine specimen.
The IMP-type metallo beta-lactamase (MBL) was first reported in Japan in 1991 when the blaIMP–1 was identified in a Pseudomonas aeruginosa isolate (Watanabe et al., 1991). The IMP family spread to various areas including Europe (Riccio et al., 2000), China (Hawkey et al., 2001), Australia (Peleg et al., 2004), and the United States (Hanson et al., 2006). The blaIMP–8 was first identified from Klebsiella pneumoniae in Taiwan in 2001, it is a variant of blaIMP–2 with four nucleotide differences, which resulting in two amino acid differences (Yan et al., 2001). Subsequently, it were also found in Pseudomonas mendocina in Portugal (Santos et al., 2010), Enterobacter cloacae in Argentina (Gomez et al., 2011), Klebsiella oxytoca in Spain (Vergara-Lopez et al., 2013), and Klebsiella pneumoniae in Tunisia (Chouchani et al., 2013). To date, blaIMP genes have not been identified in C. thiooxydans. In this study, we set out to describe the isolation of an IMP-8-producing C. thiooxydans from a patient with a urinary tract infection in China. This is the first time that a carbapenem resistance (CR) gene has been found in C. thiooxydans. Therefore, it is necessary to carry out further study on this strain, the genomic and phenotypic characteristics.
Species identification was performed using MALDI-TOF/MS (Bruker Daltonik GmbH, Bremen, Germany), as well as average nucleotide identity (ANI). Carbapenemase-encoding genes were detected using PCR. Supplementary Table 2 contains data regarding specific experimental conditions and primer sequence information. Antimicrobial Susceptibility Testing (AST) was carried out using agar dilution, and results were interpreted according to CLSI 2020 standards for other non-enterobacterales bacteria (Clinical and Laboratory Standards Institute, 2020). The strains Escherichia coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as controls.
Genomic DNA was extracted using a Genomic DNA Isolation Kit (QIAGEN, Hilden, Germany) and sequenced using Illumina Novaseq (Illumina, Inc., CA, United States) and Oxford Nanopore PromethION (Oxford Nanopore Technologies, Oxford, United Kingdom). Draft genomes were obtained using SPAdes version 3.9.13 and annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP)4 and RAST version 2.05 (Aziz et al., 2008). Acquired antibiotic resistance genes were identified using ResFinder version 3.26 and CARD7. Comparison of the genetic structures carrying blaIMP–8 gene were performed through Easyfig version 2.2.3 (Sullivan et al., 2011).
In order to investigate the phylogenetic relationships between C. thiooxydans ZDHYF418 and additional C. thiooxydans strains, we downloaded all 21 C. thiooxydans publicly available genomes in the NCBI Genome database8 as of October 2020 (Supplementary Table 1). Next, we used KSNP 3.0 (Gardner et al., 2015) to create a SNP-based phylogenetic tree through the use of C. thiooxydans CNB (accession number: CP001220.2) as reference strain. The phylogenetic tree was subsequently visualized and modified using iTOL version 59.
We isolated a novel strain of C. thiooxydans, designated as ZDHYF418, from the mid-section urine specimen of a 60-year-old female patient that was admitted to a public hospital in Zhengzhou, China in 2019. The patient was admitted to the hospital for treatment of left kidney stones. During hospitalization, the patient was intermittently irritable, unconscious, and went into septic shock. Additionally, the patient experienced abdominal distension, nausea, and vomiting. The blood culture results indicated the presence of an E. coli infection. The patient was administered imipenem-cilastatin 1g ivgtt Q8H for 1 month to fight the infection. The doctor plans to perform transurethral ureteroscopy with lithotripsy when her condition became stable. C. thiooxydans was detected in the patient’s urine culture the day before the surgery. The patient’s stones suddenly recurred and the pain could not be relieved, so the doctor did not postpone the operation. The patient was treated for a week until her condition stabilized and she was discharged from the hospital.
Strain ZDHYF418 was named C. thiooxydans after the observation of ANIs analysis based on BLAST. In fact, the genomic sequences of ZDHYF418 are 96.830% identical by ANI to the genome of C. thiooxydans, with 86.2% coverage of the genome. This result reveals that a phylogenetic affiliation of strain ZDHYF418 belongs to the species C. thiooxydans.
According to CLSI 2020 standards for other non-enterobacterales bacteria, in vitro susceptibility tests results indicated that ZDHYF418 is a multi-drug resistant strain (Table 1). Antibiogram assays indicated that ZDHYF418 is resistant to most of the antibiotics tested in this study. ZDHYF418 was found to be resistant to ceftazidime, cefepime, levofloxacin, ciprofloxacin, amikacin, gentamicin, and aztreonam; intermediate to ceftriaxone, piperacillin/tazobactam, and meropenem; susceptible to imipenem, trimethoprim/sulfamethoxazole, and chloramphenicol. According to the analysis of ResFinder and CARD, blaIMP–8 and aac(6′)-Ib-cr are all resistance genes contained in the sequence of strain ZDHYF418. In addition, gyrA, parC, and a novel class D beta-lactamase gene blaOXA were also found in the annotation results. The blaIMP–8 and blaOXA genes confer resistance to β-lactam antibiotics, β-lactamase production is the most common resistance mechanism. The aac(6′)-Ib-cr gene mainly modifies the amino group of aminoglycosides to inactivate aminoglycoside antibiotics, thereby conferring resistance to aminoglycosides. In addition, the resistance to fluoroquinolones is mainly caused by mutations in the coding regions of the gyrase subunit (gyrA) and DNA topoisomerase IV (parC) (Hawkey and Jones, 2009). Usually, the blaIMP gene confers resistance to beta-lactam antibiotics, except monobactams. However, our data indicates that ZDHYF418 is resistant to aztreonam. According to the annotation results of ZDHYF418, a series of efflux pumps such as efflux RND transporter, multidrug effflux MFS transporter, MacB family efflux pump can be found (Cattoir, 2004; Braz et al., 2016). The efflux pump system is one of the most essential resistance mechanisms. We do not know the expression of the pumps exactly, but we suppose that the resistance of ZDHYF418 to aztreonam might be similar to that in pseudomonas.
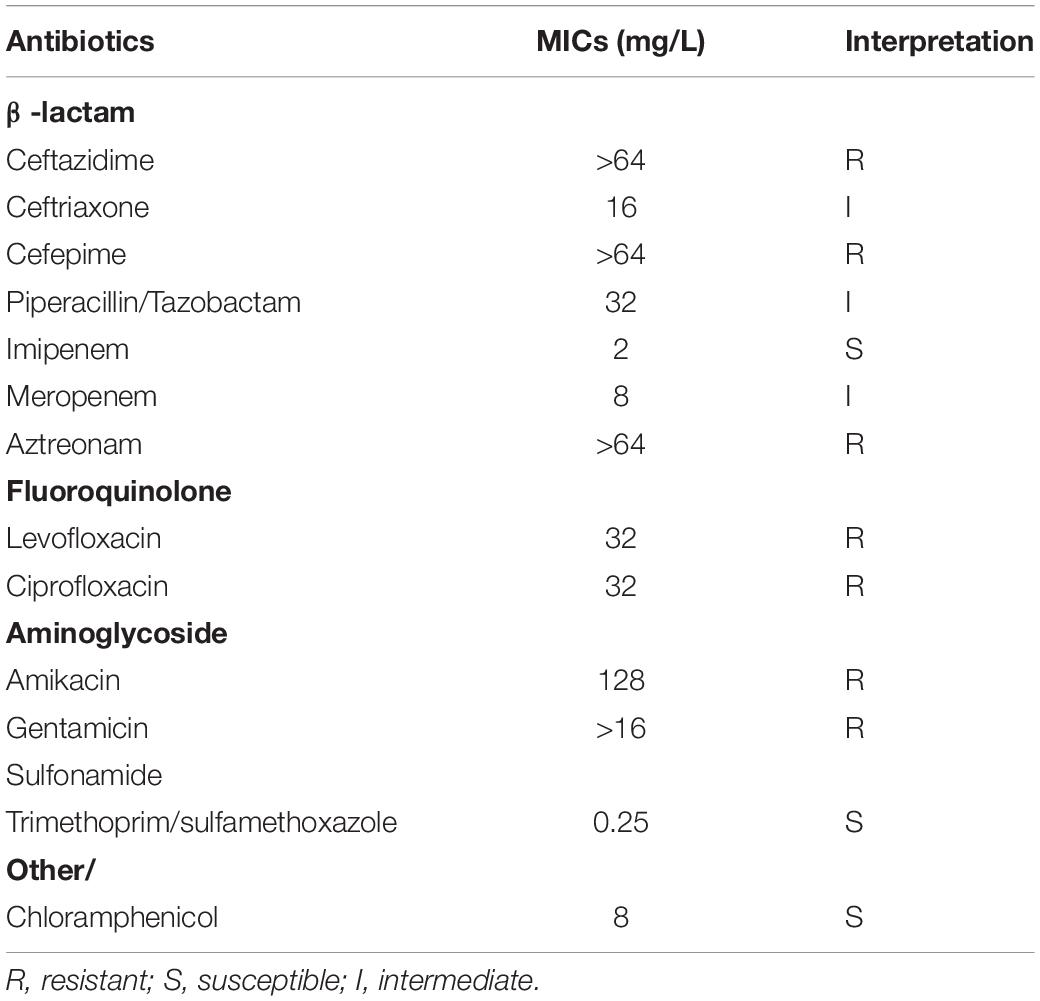
Table 1. Minimum inhibitory concentration (MIC) values of antimicrobials for Comamonas thiooxydans ZDHYF418.
We report here the genomic sequence of this strain contains 5,273,527 bp with a GC content of 61.4%. The genetic structure of the blaIMP–8 gene in ZDHYF418 and the sequences most similar to ZDHYF418 by BLAST are shown in Figure 1. The genetic structure around blaIMP–8 in C. thiooxydans ZDHYF418 has a percent identity of 99.96 and 99.94% to the p447-IMP in K. pneumoniae (accession number: KY978631) (Zhan et al., 2018) and p16005813B in Leclercia adecarboxylata, respectively (accession number: MK036884) (Yin et al., 2019). The genetic structure of blaIMP–8 gene in C. thiooxydans ZDHYF418 includes DDE-type integrase/transposase/recombinase–tniB–tniQ-recombinase family protein-aac(6′)-Ib-cr-blaIMP–8-intI1. p447-IMP and p16005813B both contain the In655 integron, which is an ancestral Tn402-associated integron. The differences among these sequences are that in ZDHYF418, the tniR module may be missing and replaced by a recombinase family protein. It has a 100% identity to MULTISPECIES: recombinase family protein (accession number: WP_003155741.1), it belongs to the serine recombinase (SR) family that can mediate site-specific recombination (Stark, 2014). The tniA module may be missing and is replaced by a DDE-type integrase/transposase/recombinase. It has a 100% identity to MULTISPECIES: DDE-type integrase/transposase/recombinase (accession number:WP_088244042.1). Both of the recombinase family protein and DDE-type integrase/transposase/recombinase are non-redundant protein sequences.
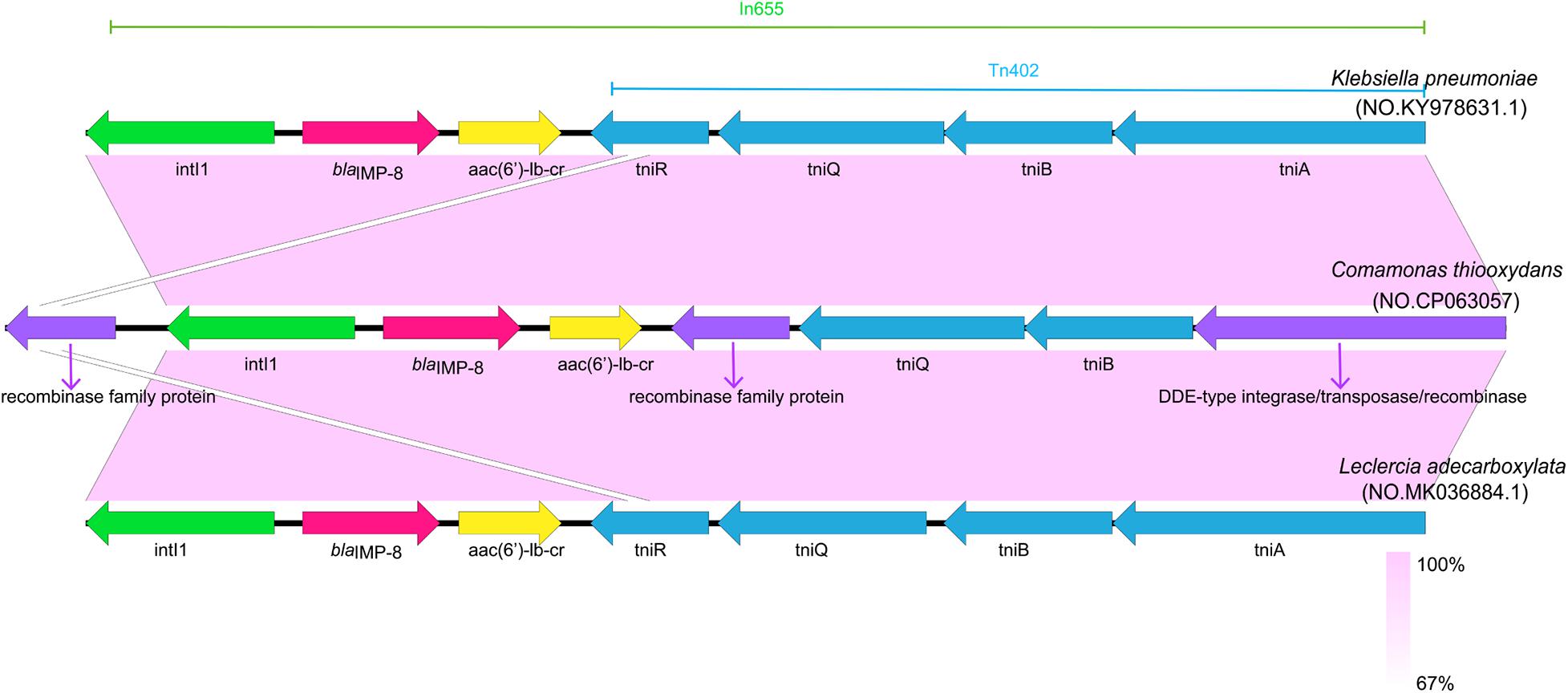
Figure 1. Comparison of the genetic structure surrounding blaIMP–8 in Comamonas thiooxydans ZDHYF418 and those observed in other blaIMP–8 positive bacteria. Open reading frames (ORFs) are indicate as arrows that show the orientation of coding sequence with the gene name. Regions with a high degree of homology are indicated by light purple shading.
The phylogenetic relationship of C. thiooxydans ZDHYF418 to the 21 C. thiooxydans are depicted in Figure 2 and Supplementary Tables 1, 3. According to Figure 2 and Supplementary Table 3, the closest relative of ZDHYF418 is C. thiooxydans QYY (accession number: CP053920.1). There are 330 SNP differences between ZDHYF418 and C. thiooxydans QYY. Strain QYY was isolated from activated sludge in Jilin province in 2015. On the other hand, C. thiooxydans has been mainly isolated from the environment, especially soil (Figure 2 and Supplementary Tables 1, 3). AWTM01, AWTP01, and AWTO01 come from the same country, as well as the same source, and their relationships are the closest. Furthermore, AWOT01, AWOU01, AWOV01, AWOS01, and VTRK01 come from the same country and the same source, and they are closely related. LIOM01 and CYHD01 come from the same country and source, and their relationship is the closest.
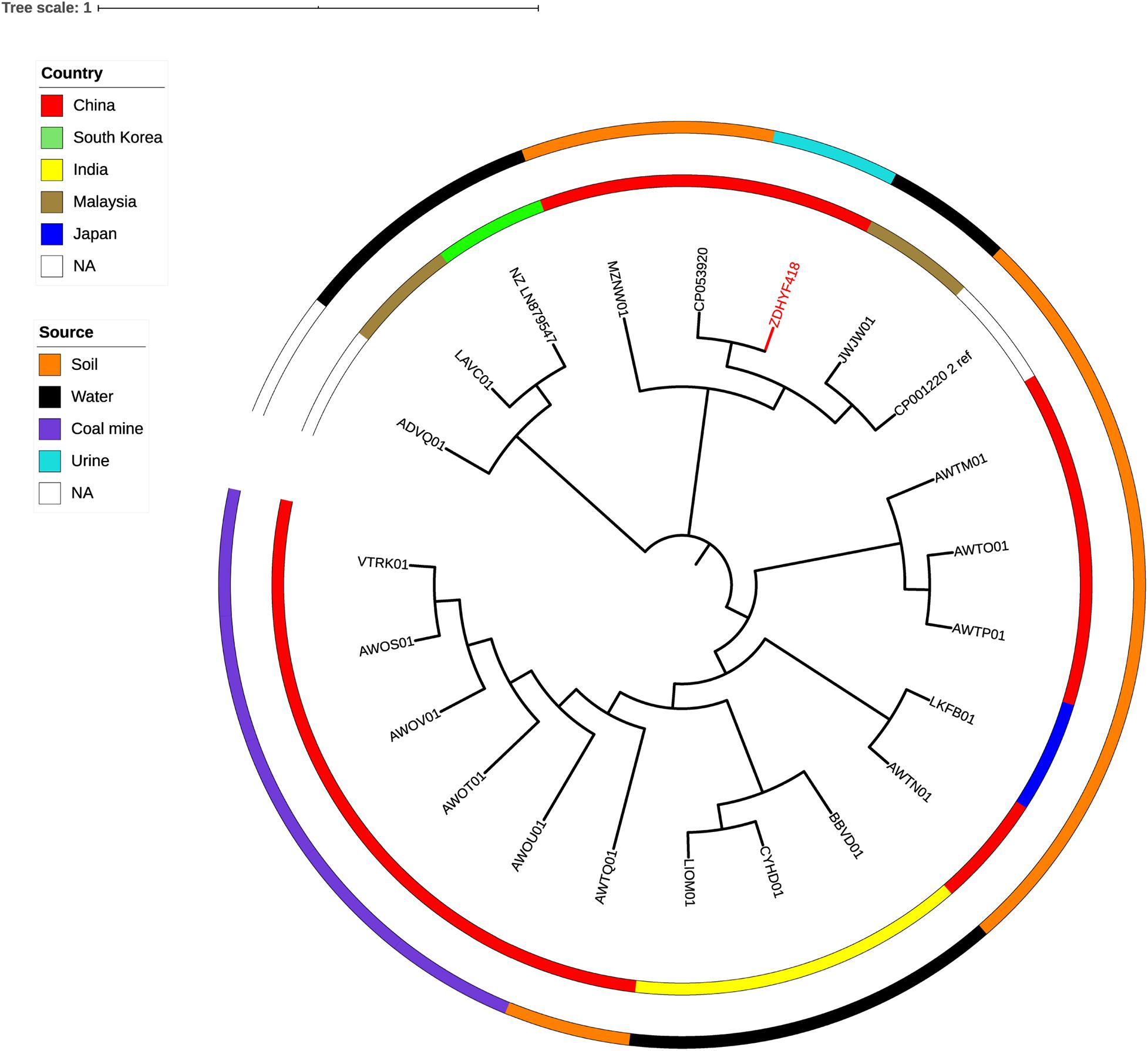
Figure 2. A phylogenetic tree showing C. thiooxydans strain ZDHYF418 along with all additional C. thiooxydans genomes are publicly available in the NCBI Genome database. The C. thiooxydans ZDHYF418 strain is indicated in red. The two circles around the phylogenetic tree indicate country of origin (inner circle) and the source (outer circle) of these strains. NA, not applicable.
In this study, we describe a C. thiooxydans strain from the urine of a patient with urinary tract infection caused by E. coli and C. thiooxydans. The patient ultimately developed septic shock. According to a previous case report of septic shock caused by bloodstream infection (Grumaz et al., 2016), we speculate that septic shock in our patient may also be due to a bloodstream infection. Additionally, in our case, the patient was fitted with a urinary catheter during hospitalization, and therefore, the bloodstream infection may be caused by a retrograde urethral infection. However, we were unable to collect a specific catheter for testing. Therefore, it is impossible to trace the source of infection. Our experience of this case highlights the need for increased awareness with regards to hospital-acquired infections caused by C. thiooxydans.
In summary, we first identified a blaIMP–8-positive C. thiooxydans strain from a human urine sample. The isolation of C. thiooxydans from humans is very rare, and the strain we identified was clearly resistant to aztreonam, fluoroquinolones and aminoglycosides, and intermediate to meropenem. The increased resistance of bacteria to antibiotics is now starting to appear in less common bacteria, such as C. thiooxydans. This finding prompts us to standardize clinical medication and pay more attention to bacterial resistance monitoring of this species.
The datasets generated for this study can be found in NCBI under BioProject PRJNA623107, https://www.ncbi.nlm.nih.gov/bioproject/?term=623107.
Written informed consent was obtained from the participant of this study. Ethical permission was granted by the Research Ethics Committee of The First Affiliated Hospital, College of Medicine, Zhejiang University, reference number 2018#752.
XG and JG conceived and designed the experiments. QW, SL, and PW collected clinical samples and performed the experiments. HX, XH, and LG analyzed the data. QW wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Key Research and Development Program of China (No. 2016YFD0501105), the Mega-projects of Science Research of China (No. 2018ZX10733402-004), the National Natural Science Foundation of China (No. 81741098), and Henan Province Medical Science and Technology Research Project Joint Construction Project (No. LHGJ20190232).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.585716/full#supplementary-material
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Braz, V. S., Furlan, J. P., Fernandes, A. F., and Stehling, E. G. (2016). Mutations in NalC induce MexAB-OprM overexpression resulting in high level of aztreonam resistance in environmental isolates of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 363:fnw166. doi: 10.1093/femsle/fnw166
Cattoir, V. (2004). [Efflux-mediated antibiotics resistance in bacteria]. Pathol. Biol. (Paris) 52, 607–616. doi: 10.1016/j.patbio.2004.09.001
Chen, Y. L., Wang, C. H., Yang, F. C., Ismail, W., Wang, P. H., Shih, C. J., et al. (2016). Identification of Comamonas testosteroni as an androgen degrader in sewage. Sci. Rep. 6:35386. doi: 10.1038/srep35386
Chouchani, C., Marrakchi, R., Henriques, I., and Correia, A. (2013). Occurrence of IMP-8, IMP-10, and IMP-13 metallo-beta-lactamases located on class 1 integrons and other extended-spectrum beta-lactamases in bacterial isolates from Tunisian rivers. Scand. J. Infect. Dis. 45, 95–103. doi: 10.3109/00365548.2012.717712
Clinical and Laboratory Standards Institute (2020). Performance Standards for Antimicrobial Susceptibility Testing, CLSI supplement M100, 30th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Gomez, S., Rapoport, M., Togneri, A., Viegas-Caetano, J., Faccone, D., Corso, A., et al. (2011). Emergence of metallo-beta-lactamases in Enterobacteriaceae from Argentina. Diagn. Microbiol. Infect. Dis. 69, 94–97. doi: 10.1016/j.diagmicrobio.2010.08.025
Grumaz, S., Stevens, P., Grumaz, C., Decker, S. O., Weigand, M. A., Hofer, S., et al. (2016). Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 8:73. doi: 10.1186/s13073-016-0326-8
Hanson, N. D., Hossain, A., Buck, L., Moland, E. S., and Thomson, K. S. (2006). First occurrence of a Pseudomonas aeruginosa isolate in the United States producing an IMP metallo-beta-lactamase, IMP-18. Antimicrob. Agents Chemother. 50, 2272–2273. doi: 10.1128/aac.01440-05
Hatayama, K. (2014). Comamonas humi sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 64(Pt 12), 3976–3982. doi: 10.1099/ijs.0.067439-0
Hawkey, P. M., and Jones, A. M. (2009). The changing epidemiology of resistance. J. Antimicrob. Chemother. 64(Suppl. 1), i3–i10. doi: 10.1093/jac/dkp256
Hawkey, P. M., Xiong, J., Ye, H., Li, H., and M’Zali, F. H. (2001). Occurrence of a new metallo-beta-lactamase IMP-4 carried on a conjugative plasmid in Citrobacter youngae from the people’s republic of China. FEMS Microbiol. Lett. 194, 53–57. doi: 10.1111/j.1574-6968.2001.tb09445.x
Kämpfer, P., Busse, H. J., Baars, S., Wilharm, G., and Glaeser, S. P. (2018). Comamonas aquatilis sp. nov., isolated from a garden pond. Int J Syst Evol Microbiol 68, 1210–1214. doi: 10.1099/ijsem.0.002652
Narayan, K. D., Badhai, J., Whitman, W. B., and Das, S. K. (2016). Draft genome sequence of Comamonas thiooxydans strain S23T (DSM 17888T), a thiosulfate-oxidizing bacterium isolated from a sulfur spring in India. Genome Announc. 4, e834–e816. doi: 10.1128/genomeA.00834-16
Pandey, S. K., Narayan, K. D., Bandyopadhyay, S., Nayak, K. C., and Das, S. K. (2009). Thiosulfate oxidation by Comamonas sp. S23 isolated from a sulfur spring. Current Microbiology 58, 516–521. doi: 10.1007/s00284-009-9357-3
Peleg, A. Y., Franklin, C., Bell, J., and Spelman, D. W. (2004). Emergence of IMP-4 metallo-beta-lactamase in a clinical isolate from Australia. J. Antimicrob. Chemother. 54, 699–700. doi: 10.1093/jac/dkh398
Riccio, M. L., Franceschini, N., Boschi, L., Caravelli, B., Cornaglia, G., Fontana, R., et al. (2000). Characterization of the metallo-beta-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of bla(IMP) allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44, 1229–1235. doi: 10.1128/aac.44.5.1229-1235.2000
Santos, C., Caetano, T., Ferreira, S., and Mendo, S. (2010). First description of bla IMP-8 in a Pseudomonas mendocina isolated at the Hospital Infante D. Pedro, Aveiro, Portugal. Res. Microbiol. 161, 305–307. doi: 10.1016/j.resmic.2010.03.004
Stark, W. M. (2014). The serine recombinases. Microbiol. Spectr. 2:6. doi: 10.1128/microbiolspec.MDNA3-0046-2014
Subhash, Y., Bang, J. J., You, T. H., and Lee, S. S. (2016). Description of Comamonas sediminis sp. nov., isolated from lagoon sediments. Int. J. Syst. Evol. Microbiol. 66, 2735–2739. doi: 10.1099/ijsem.0.001115
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tsui, T. L., Tsao, S. M., Liu, K. S., Chen, T. Y., Wang, Y. L., Teng, Y. H., et al. (2011). Comamonas testosteroni infection in Taiwan: reported two cases and literature review. J. Microbiol. Immunol. Infect. 44, 67–71. doi: 10.1016/j.jmii.2011.01.013
Vergara-Lopez, S., Dominguez, M. C., Conejo, M. C., Pascual, A., and Rodriguez-Bano, J. (2013). Wastewater drainage system as an occult reservoir in a protracted clonal outbreak due to metallo-beta-lactamase-producing Klebsiella oxytoca. Clin. Microbiol. Infect. 19, E490–E498. doi: 10.1111/1469-0691.12288
Watanabe, M., Iyobe, S., Inoue, M., and Mitsuhashi, S. (1991). Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35, 147–151. doi: 10.1128/aac.35.1.147
Willems, A., and De Vos, P. (2006). “Comamonas,” in The Prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York, NY: Springer), 723–736.
Yan, J. J., Ko, W. C., and Wu, J. J. (2001). Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-beta-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45, 2368–2371. doi: 10.1128/AAC.45.8.2368-2371.2001
Yin, Z., Hu, L., Cheng, Q., Jiang, X., Xu, Y., Yang, W., et al. (2019). First report of coexistence of three different MDR plasmids, and that of occurrence of IMP-encoding plasmid in Leclercia adecarboxylata. Front. Microbiol. 10:2468. doi: 10.3389/fmicb.2019.02468
Zhan, Z., Hu, L., Jiang, X., Zeng, L., Feng, J., Wu, W., et al. (2018). Plasmid and chromosomal integration of four novel blaIMP-carrying transposons from Pseudomonas aeruginosa, Klebsiella pneumoniae and an Enterobacter sp. J. Antimicrob. Chemother. 73, 3005–3015. doi: 10.1093/jac/dky288
Keywords: blaIMP–8, antimicrobial resistance, complete genome sequence, comparative genomic analysis, Comamonas thiooxydans
Citation: Guo X, Wang Q, Xu H, He X, Guo L, Liu S, Wen P and Gou J (2021) Emergence of IMP-8-Producing Comamonas thiooxydans Causing Urinary Tract Infection in China. Front. Microbiol. 12:585716. doi: 10.3389/fmicb.2021.585716
Received: 21 July 2020; Accepted: 19 February 2021;
Published: 15 March 2021.
Edited by:
Paul G. Higgins, University of Cologne, GermanyReviewed by:
Alberto Antonelli, University of Florence, ItalyCopyright © 2021 Guo, Wang, Xu, He, Guo, Liu, Wen and Gou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaobing Guo, Z3hiaW5nOTI4QHp6dS5lZHUuY24=; Jianjun Gou, Z291anVuNjRAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.