
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 January 2021
Sec. Aquatic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.581124
The paradigm of tight pelagic-benthic coupling in the Arctic suggests that current and future fluctuations in sea ice, primary production, and riverine input resulting from global climate change will have major impacts on benthic ecosystems. To understand how these changes will affect benthic ecosystem function, we must characterize diversity, spatial distribution, and community composition for all faunal components. Bacteria and archaea link the biotic and abiotic realms, playing important roles in organic matter (OM) decomposition, biogeochemical cycling, and contaminant degradation, yet sediment microbial communities have rarely been examined in the North American Arctic. Shifts in microbial community structure and composition occur with shifts in OM inputs and contaminant exposure, with implications for shifts in ecological function. Furthermore, the characterization of benthic microbial communities provides a foundation from which to build focused experimental research. We assessed diversity and community structure of benthic prokaryotes in the upper 1 cm of sediments in the southern Beaufort Sea (United States and Canada), and investigated environmental correlates of prokaryotic community structure over a broad spatial scale (spanning 1,229 km) at depths ranging from 17 to 1,200 m. Based on hierarchical clustering, we identified four prokaryotic assemblages from the 85 samples analyzed. Two were largely delineated by the markedly different environmental conditions in shallow shelf vs. upper continental slope sediments. A third assemblage was mainly comprised of operational taxonomic units (OTUs) shared between the shallow shelf and upper slope assemblages. The fourth assemblage corresponded to sediments receiving heavier OM loading, likely resulting in a shallower anoxic layer. These sites may also harbor microbial mats and/or methane seeps. Substructure within these assemblages generally reflected turnover along a longitudinal gradient, which may be related to the quantity and composition of OM deposited to the seafloor; bathymetry and the Mackenzie River were the two major factors influencing prokaryote distribution on this scale. In a broader geographical context, differences in prokaryotic community structure between the Beaufort Sea and Norwegian Arctic suggest that benthic microbes may reflect regional differences in the hydrography, biogeochemistry, and bathymetry of Arctic shelf systems.
The Arctic marine ecosystem is undergoing pronounced changes due to climbing atmospheric temperatures, occurring two to three times faster than the global average (ACIA, 2005; Walsh et al., 2011). Resulting shifts in sea-ice cover, primary production, and riverine input will likely affect the quality and quantity of organic material (OM) deposited to the seafloor and the tight pelagic-benthic coupling characteristic of Arctic ecosystems may emphasize the effects of these changes (Grebmeier, 2012; Kortsch et al., 2015). Impacts of climate change on Arctic benthic eukaryotes have varied among size classes, but benthic prokaryotic communities have not been described well enough to monitor change (Nelson et al., 2014; Kêdra et al., 2015; Renaud et al., 2019).
Benthic bacteria and archaea perform diverse metabolic functions that mediate biogeochemical processes, including degradation and early diagenesis of organic matter in marine surface sediments (Deming and Baross, 1993). Prokaryotes provide a link between the abiotic and biotic realms, such that prokaryotic community structure reflects environmental gradients in, for example, OM deposition. Studies conducted in the Norwegian Arctic suggest that prokaryotic community structure shifts with varying quality and quantity of OM inputs, with possible consequences for ecosystem function (Hoffmann et al., 2017; Braeckman et al., 2018). Certain taxonomic groups have been strongly correlated with environmental parameters, such as Chl-a and bathymetry, measured along natural environmental gradients (Bienhold et al., 2012; Jacob et al., 2013). Benthic prokaryotes are also key players in both the production and degradation of greenhouse gases such as methane and nitrous oxide and represent a first line of defense in remediating contamination by oil and other petroleum-based products (Boetius et al., 2000; Head et al., 2006; Casciotti and Buchwald, 2012). In some cases, these important functions are attributable to specific taxonomic groups (Kostka et al., 2011; Ruff et al., 2015; Otte et al., 2019). Thus, characterization of benthic prokaryotic community structure may yield valuable insights into ecosystem function in Arctic marine sediments, particularly those permeated with methane and subjected to active mineral resource exploration such as the Beaufort Sea sites examined here (Coffin et al., 2013; BOEM, 2019).
The Beaufort Sea is an Arctic marginal sea that crosses a border between the United States and Canada, stretching from Point Barrow (Alaska, United States) to Banks Island (Northwest Territories, Canada). Prior studies of benthic prokaryotes in the southern Beaufort Sea have focused on subsurface sediments (20–70 cm) to assess the influence of either methane or mud volcanoes on bacterial community structure (Hamdan et al., 2013; Kirchman et al., 2014; Treude et al., 2014; Lee et al., 2018). Communities in these deeper sediments typically differ substantially from the active surface layer (Inagaki et al., 2003; Fry et al., 2006; Roussel et al., 2009; Durbin and Teske, 2011; Jorgensen et al., 2012; Bienhold et al., 2016). Thus, there is no established baseline for benthic prokaryotes in southern Beaufort Sea surface sediments to date, and only limited data from deeper sediment horizons. Most information on Arctic benthic prokaryotes comes from the Norwegian Arctic, largely from deep-sea or fjord habitats, which differ from the Beaufort Sea in terms of primary production, terrestrial/riverine input, and bathymetry (Ravenschlag et al., 2001; Holmes et al., 2002; Carmack and Wassmann, 2006; Arnosti, 2008; Li et al., 2009; Jacob et al., 2013; Buttigieg and Ramette, 2015; Bienhold et al., 2016; Zeng et al., 2017).
Across the southern Beaufort Sea, summer depth-stratified water-mass structure includes a fresher surface Polar Mixed Layer (PML; ∼0–50 m), the Pacific-influenced Arctic Halocline Layer (AHL; ∼50 – 200 m), and warmer, more saline Atlantic Water (AW) at depths greater than 200 m (Lansard et al., 2012; Miquel et al., 2015; Smoot and Hopcroft, 2017). Eastern Beaufort Sea shelf (< 100 m water depth) and slope habitats are particularly influenced by riverine input, including the vast Mackenzie River which is the fourth largest in the Arctic in terms of freshwater discharge (330 km3/year), and the largest in terms of sediment transport (124 × 106 t/year) (Macdonald et al., 1998; Holmes et al., 2002; Rachold et al., 2004). The Mackenzie River primarily influences the water and sediment characteristics of the Canadian sector of the shelf and slope, and to a lesser extent the Amundsen Gulf and easternmost section of the Alaskan sector (Figure 1). The Mackenzie shelf functions as a vast estuary influenced by freshwater runoff, receiving inputs of both terrestrial and marine sources of organic matter (Carmack and Macdonald, 2002; Magen et al., 2010). The Amundsen Gulf, which connects the Canadian Arctic Archipelago with the southeast Beaufort Sea, is characterized by several peripheral bays, straights, and inlets and ringed by a narrow shelf surrounding a central basin (Stokes et al., 2006; Forest et al., 2010; Gamboa et al., 2017). Water mass properties in the Alaskan Beaufort Sea are influenced by warmer, more nutrient-rich waters entering from the Chukchi Sea and forming the Beaufort Shelf Jet, which flows due east from Barrow Canyon (Pickart, 2004; Weingartner et al., 2005; Dunton et al., 2006). These longitudinal environmental gradients from Point Barrow to Banks Island have been reflected in the benthic distributions of OM and certain epi-/infaunal groups (Naidu et al., 2000; Magen et al., 2010; Goñi et al., 2013; Bell et al., 2016). This benthic habitat may also be heavily influenced by methane derived from organic matter burial, mud volcanoes, subsurface permafrost, and/or gas hydrates (Paull et al., 2015; Brothers et al., 2016; Coffin et al., 2017; Lee et al., 2018; Retelletti Brogi et al., 2019).
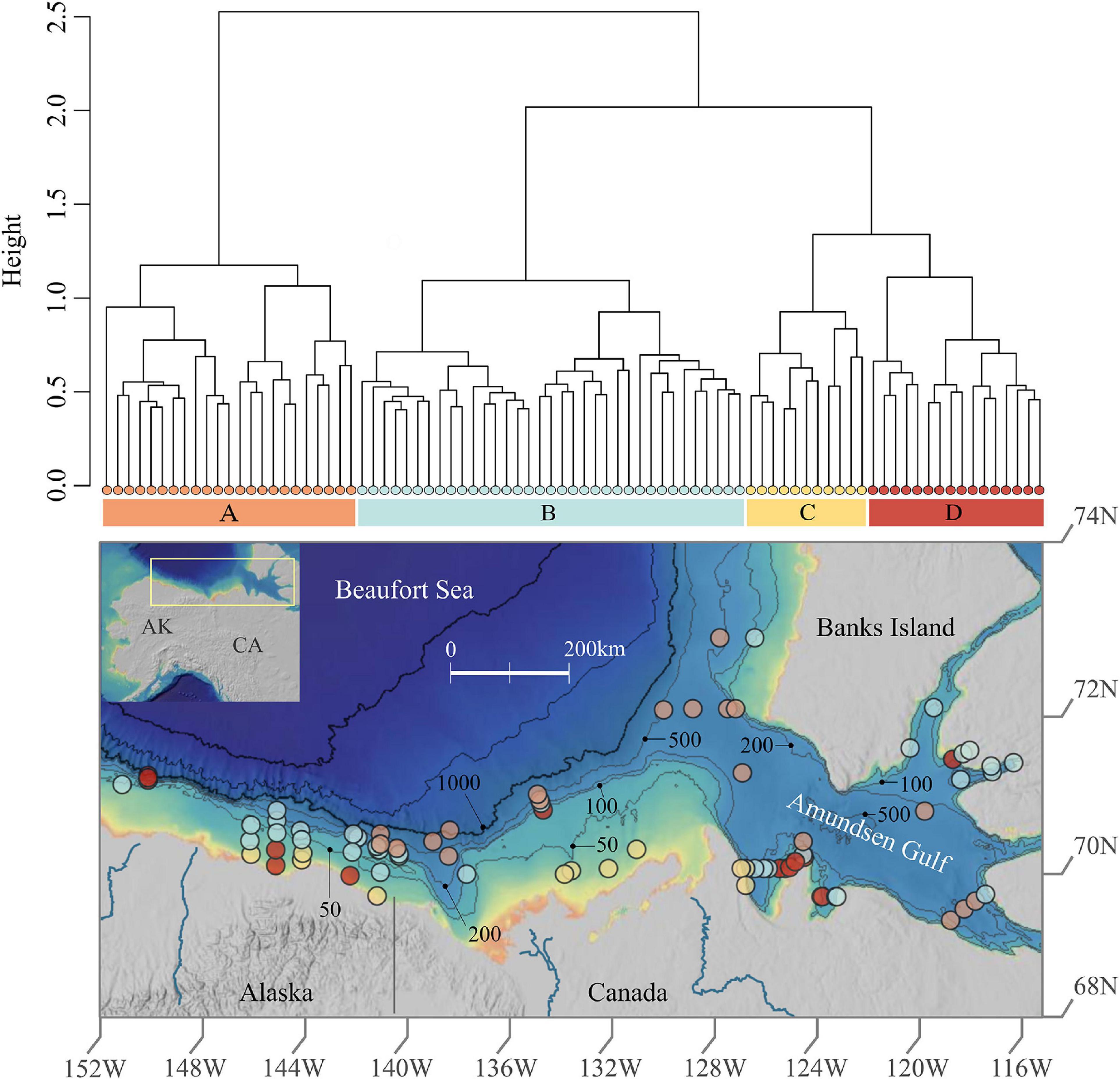
Figure 1. Prokaryotic cluster assemblages and spatial distribution in Beaufort Sea sediments. This figure shows the dendrogram of the four assemblages revealed via hierarchical clustering above the study area map which exhibits sample locations colored by assemblage. The map was created using ArcGIS online with the following layers: Northwest Territories, Esri, Garmin, FAO, NOAA, USGS, EPA, NRCan, and Parks Canada.
We sought to describe prokaryote communities across this broad, heterogeneous benthic habitat, specifically by (1) assessing the diversity, structure, and composition of archaeal and bacterial communities in the upper 1 cm of sediments, and (2) investigating environmental correlates of prokaryote community structure over a broad area of the southern Beaufort Sea shelf and slope. Expanding knowledge of functional taxonomic groups of marine prokaryotes provides insight into this unstudied microbial habitat, and a framework for developing more targeted experimental studies in this dynamic polar environment (Fuhrman, 2009; Bier et al., 2015; Fuhrman et al., 2015).
Our study area covers a broad section of the continental shelf and upper slope of the Alaskan and Canadian Beaufort Sea, from areas offshore of the Colville River in the west to Banks Island in the east, and into the Amundsen Gulf (Figure 1). A total of 85 sediment samples were collected opportunistically at water depths between 17 and 1,200 m from 2012 to 2014 as part of two international collaborative field programs: the US-Transboundary Fish and Lower Trophic Communities Project (USTB) and the Canadian Beaufort Regional Environmental Assessment Project (BREA). Fifteen sediment samples, 8 in 2012 and 7 in 2013, were collected using a 5 cm diameter sub-core from the upper 1 cm layer of a 0.25 m2 box core. In 2014, 70 sediment samples were collected using a 60-cc, 2.5 cm diameter, sterilized syringe from the upper 1 cm surface layer of a double van Veen grab (0.1 m2, USTB) or a 0.25 m2 box core (BREA). As preliminary analyses indicated that neither year or sampling gear were significant factors affecting prokaryotic community structure here, all samples were used in this study. The samples were immediately frozen at −20°C following collection aboard respective vessels, transported to the University of Alaska Fairbanks (UAF) Institute of Arctic Biology Genomics Core Laboratory, and then stored at −80°C. Environmental variables such as depth, bottom water temperature, and salinity were recorded from CTD profiles conducted at corresponding sampling locations (Eert et al., 2015; Niemi et al., 2015; Smoot and Hopcroft, 2017).
We quantified several common indicators of quality, quantity, and origin of sediment OM including chlorophyll-a concentration (Chl-a), phaeopigment concentration (Phaeo), total organic carbon (TOC), total nitrogen (TN), ratio of carbon to nitrogen (C:N), and bulk sediment δ13C and δ15N. All parameters were measured using sediment samples taken from the upper 1 cm of the same box core or grab as the genomic samples from each sampling station. Chl-a and phaeopigment concentrations were measured fluorometrically (Arar and Collins, 1997). Briefly, samples were suspended in 5 ml 100% acetone, sonicated in an ice-water bath for 10 min, and allowed to extract overnight at −20°C. Samples were then centrifuged to remove sediment, and transferred to a clean test tube. Chl-a concentration was determined using a TD-700 fluorometer (Turner Designs). After recording fluorescence values, samples were acidified with HCl, and fluorescence readings were taken of the acidified samples to produce phaeopigment values. A standard curve produced using commercially available Chl-a standard was used to convert fluorescence readings into concentrations.
Stable isotope and elemental analysis were performed on freeze-dried sediment samples at the Alaska Stable Isotope Facility (ASIF). Data for USTB samples were generated and previously reported by Bell et al. (2016); additional samples were analyzed here using the same protocol. Stable isotope data, δ13C and δ15N, were generated using a ThermoFinnigan DeltaVPlus isotope ratio mass spectrometer with Pee Dee Belemite (PDB) and atmospheric nitrogen as standards for carbon and nitrogen, respectively. Percent carbon and nitrogen content was obtained using a Costech ESC 4010 elemental analyzer, and used to calculate TOC, TN, and C:N ratios.
Sediment porosity (Φ), i.e., volume of water within sediments (Vw), was calculated as an indicator of sediment permeability/O2 penetration using Eq. 1 (Bennett and Lambert, 1971; Ullman and Aller, 1982). Sediment wet (Wsw) and dry (Wsd) weights were recorded before and after freeze-drying, and used to calculate the mass of water (Ww). Sediment density (ρs) was held constant at 2.50 g/cm3, based on Coffin et al. (2017). Seawater density (ρw) was calculated based on bottom-water temperatures and salinities recorded at sampled sites.
16S ribosomal (rRNA) gene amplicon sequencing was conducted on freeze-dried sediments in order to assess the diversity and taxonomic composition of prokaryotes. Total genomic DNA was extracted from sediment samples using the Qiagen PowerSoil kit, and revised forward (515FB) and reverse (806RB) primers from the Earth Microbiome Project (EMP) were used to amplify the V4 region of the 16S rRNA gene (Caporaso et al., 2011, 2012; Apprill et al., 2015; Parada et al., 2016). Library prep was conducted using iTru adapters for sequencing and a one-step PCR protocol with indexed primers, which is the current standard protocol used by the EMP (Caporaso et al., 2011, 2012; Apprill et al., 2015; Parada et al., 2016; Walters et al., 2016). Samples were sequenced on an Illumina MiSeq at the UAF Genomics Core Lab.
Raw sequences were de-multiplexed using the Mr. Demuxy package (Cock et al., 2009). Demultiplexed sequences were run with mothur v1.40.0 on a high performance-computing cluster through UAF Research Computing Systems using a modified MiSeq standard operating procedure (Schloss et al., 2009). Operational taxonomic units (OTUs) were clustered at 100% similarity using the OptiClust option in mothur, taxonomy was assigned to OTUs using the SILVA 132 mothur formatted reference database with a bootstrap cutoff of 100%, and the samples in the resulting OTU table were normalized to 30,000 sequences and converted to relative abundances (Wang et al., 2007; Glöckner et al., 2017; Westcott and Schloss, 2017; Edgar, 2018). This OTU table (relative abundance of sequence reads per OTU for each sample) was used to conduct analyses of diversity and community structure of prokaryotes and to assess correlations with environmental variables.
All statistical analyses were conducted using R, primarily using the vegan package (Oksanen, 2015; R Core Team, 2017). Diversity was quantified using the inverse Simpson index (1/λ) which reflects evenness and richness (Morris et al., 2014). Community structure was investigated via hierarchical clustering analysis with the Ward method (ward.D2) based on a Bray-Curtis dissimilarity derived from the OTU table. Cluster tests were used to assess the validity (heterogeneity, significance) and characteristics (silhouette, stability) of the hierarchical clusters. The heterogeneity of and significance between hierarchical clusters were investigated using Bray Curtis distances with the betadisper and adonis functions, respectively (Oksanen et al., 2017). OTUs that distinguished each cluster were identified via indicator taxa analysis using the indicspecies package (De Cáceres and Legendre, 2009; De Cáceres et al., 2010). The multipatt function was used to identify taxa specific to a single cluster, or indicative of combinations of clusters, with a significance of ≤0.001 and a strength of ≥0.900 (i.e., >90% probability of occurrence within a given cluster or combination of clusters (De Cáceres and Legendre, 2009). Given the complexity of the prokaryotic species concept and the lack of taxonomic resolution for many taxa, we used this indicator analysis to identify OTUs potentially indicative of certain habitat features and/or assemblages. We assessed the distribution of these indicator taxa at varying taxonomic levels, from phylum to genus, to characterize representative assemblages for each cluster.
Differences in the median values of environmental parameters among clusters were evaluated using Kruskal-Wallis tests followed by pairwise Wilcoxon Rank Sum tests with a Benjamini and Yekutieli (BY) correction (Yekutieli and Benjamini, 2002). Relationships between prokaryotic community structure and environmental variables were modeled using Constrained Analysis of Principle Coordinates, i.e., the capscale function in the R vegan package (Oksanen, 2015; R Core Team, 2017). The best model for capscale analyses was identified based on variance inflation factors for each environmental parameter and a forward and backward stepwise model selection via permutation tests on adjusted R2 and P-values using the ordistep function. The significance of the environmental correlates yielded via Constrained Analysis of Principle Coordinates was investigated in R using the adonis function; correlation strengths were calculated using the cor function (Oksanen et al., 2017). All bioinformatic and statistical scripts can be found here on GitHub1.
In order to put Beaufort Sea surface sediment microbes into a broader geographical context, we qualitatively compared the 10 most abundant taxa at class level with those reported for global benthic surface sediments, global deep-sea sediments, and Norwegian-Arctic sediments (Zinger et al., 2011; Bienhold et al., 2016). We refer to the Arctic microbiome published by Bienhold et al. (2016) as the Norwegian Arctic microbiome because all but one dataset included in that study was generated from the Norwegian Arctic (Bienhold et al., 2016).
The total number of OTUs yielded was 295,115. No OTUs were removed (e.g., singletons or doubletons) because removals resulted in substantial changes in certain taxonomic groups (Supplementary Figure 1). Hierarchical clustering analysis identified four clusters, i.e., assemblages of prokaryotes which have significantly different (adonis: F = 12.6, P < 0.001) taxonomic composition (Figure 1). However, the silhouette test yielded an average cluster width of 0.11, indicating a high degree of similarity between clusters which suggests that the four assemblages may exist more so as a gradient rather than as discrete communities in particular locations. The number of samples within each cluster, herein referred to as assemblages A, B, C, and D, was as follows: A = 23, B = 35, C = 11, and D = 16. Assemblage A is most distinct, whereas C and D are most similar (Figure 1). The distribution of values for Inverse Simpson diversity differed for each assemblage, with the highest prokaryotic diversity exhibited in assemblage A, then B, D, and C.
Each assemblage is represented by sampling locations distributed across the study area (Figure 1), such that assemblages were not restricted to particular geographic areas that might reflect distinct environmental characteristics. However, when delving into the substructure of individual assemblages, patterns emerged at smaller spatial scales (Figure 2). Assemblage A contained two clusters, with one containing mostly Alaskan and Canadian Beaufort (AKCA) samples and some shallower (<350 m) Amundsen Gulf and Banks Island (AGBI) samples, and the other containing mostly AGBI samples with a few deeper (>350 m) AKCA samples. Assemblage B was clearly divided into AKCA and AGBI samples. Sub-structure for assemblage C was delineated by the Mackenzie River into the AK Beaufort shelf (west of the river) and the CA Beaufort with AG samples (east of the river). Assemblage D divides into two groups with no obvious geographical demarcations.
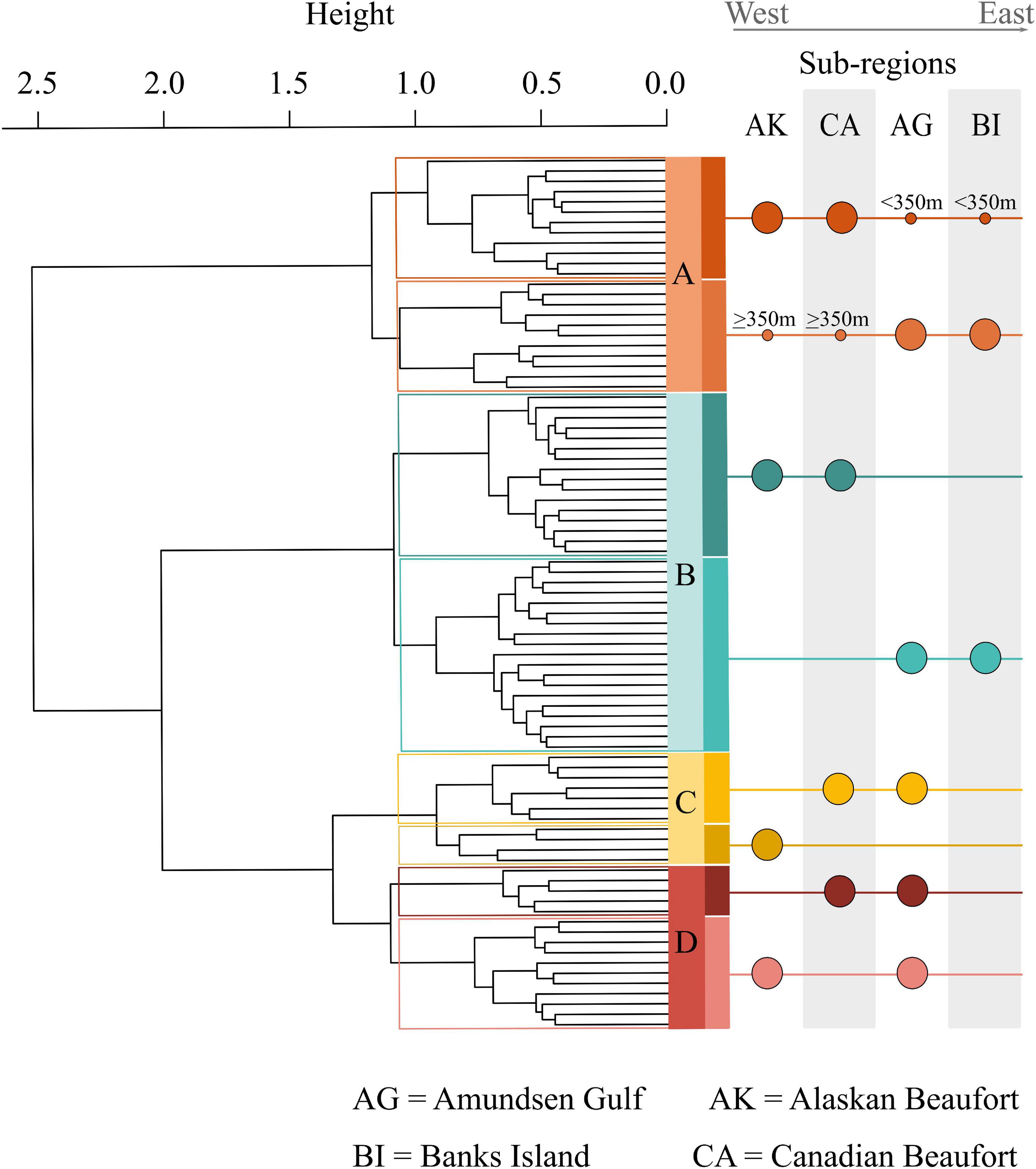
Figure 2. Substructure of prokaryotic assemblages. This dendrogram highlights the substructure within each assemblage and the differing east to west gradients reflected by the substructure. Assemblage A was divided by location and depth, with one sub-cluster dominated by Amundsen Gulf (AG) samples combined with broader Beaufort (AKCA) slope samples deeper than 350 m. The other assemblage A sub-cluster was dominated by broader Beaufort slope samples with AG samples shallower than 350 m. Assemblage B divided cleanly between broader Beaufort and Amundsen Gulf samples. Assemblage C divided between the west of the Mackenzie River, AK samples, and east of the Mackenzie River, CA Beaufort and AG samples. Assemblage D divided into combinations of AG samples with either AK or CA samples with no obvious demarcations.
There were 350 prokaryotic families represented in this dataset; we focused on the 25 most abundant families (or in some cases, orders) to assess broad-scale patterns in community structure and compare relative abundance of these dominant taxa among individual assemblages. Abundant taxa consisting of multiple unclassified OTUs within a class or phylum, specifically Alpha- and Gammaproteobacteria, were ignored. The 25 most abundant taxa accounted for 58% of the total sequence reads, and are listed in order of decreasing abundance in Figure 3. Family-level composition differed among assemblages, with inverse relationships in abundance of particular taxa in assemblages A and C. Families that were relatively abundant in assemblage A also tended to be abundant in assemblage B, whereas assemblages C and D also showed similar trends. The families Nitrosopumilaceae, Woeseiaceae, NB1-j, Actinomarinales, Kiloniellaceeae, Subgroup 22, OM190, and AT-s2-59 were particularly abundant in assemblage A. Haliaceae, BD7-8, and Methyloligellaceae were most prominent in assemblage B. In assemblage C, Flavobacteraceae, Rhodobacteraceae, Rubritalaceae, and Psychromonadaceae were more abundant. Gammaproteobacteria Incertae sedis, Desulfobulbaceae, Sva1033, Anaerolineaceae, and Desulfobacteraceae were abundant in assemblage D.
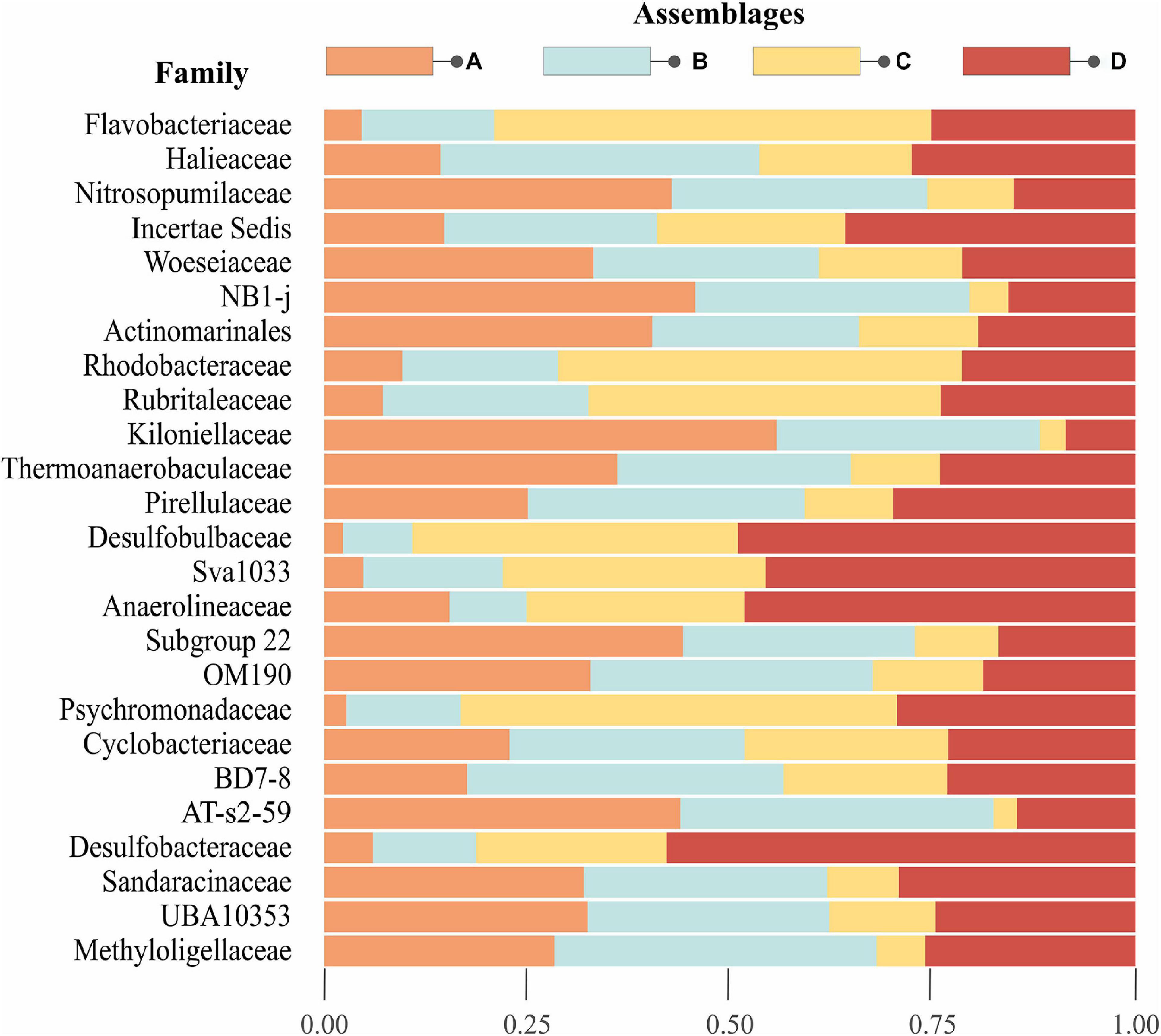
Figure 3. Top 25 abundant families in Beaufort Sea surface sediments. The stacked barplot illustrates the proportion of the top 25 families for each assemblage on the left-hand y-axis. The plot is arranged such that the most abundant taxa starts from the top and decreases toward the bottom. Note that Gammaproteobacteria Inc. sed., NB1-j, Actinomarinales, Sva1033, OM190, BD7-8, AT-s2-59, and UBA10353 are not classified to family, but order level.
Indicator taxa analyses revealed that assemblage A had the highest number of indicator OTUs (294), followed by C (61) and D (27), and no OTUs were identified as indicators for assemblage B (Supplementary Figure 2 and Supplementary Table 1). Taxa indicative of a combination of assemblages will be referred to as indicative taxa, to differentiate from assemblage-specific indicator taxa. Mirroring trends shown in Figure 3, assemblages A + B and C + D shared relatively high numbers of indicative OTUs. No indicative taxa were shared between assemblages A + C alone, but some were shared between A + B + C, suggesting B represents a transitional community with representatives from both of the distinct A and C assemblages. Assemblages B + D also shared very few indicative taxa, but more were found when combined with either assemblage A or C (A + B + D or B + C + D).
In light of the numerous uncultured and indicative OTUs, we limit our discussion to selected assemblage-specific indicator taxa, identified to the highest taxonomic resolution possible, for which published information could be used with confidence to infer function or distribution of a specific assemblage, and/or metabolic type or relationship to environmental variables (Figure 4). Indicator OTUs for assemblage A belonged to the class Acidobacteria (Subgroups 6, 9, 10, 21, 22, and 26), clades SAR202 and 324, and the families Thermoanaerobaculaceae, Magnetospiraceae (Magnetospira), Nitrosopumilaceae (Candidatus Nitrosopumilus), Nitrospiraceae (Nitrospira), and Scalinduaceae (Scalindua). Representative taxa under Thermoanaerobaculaceae belong to Acidobacteria subgroup 10 which was recently assigned to this family (Dedysh and Yilmaz, 2018). Indicator taxa for assemblage C included representatives from the families Flavobacteriaceae (Polaribacter and Ulvibacter), Rhodobactereacae (Loktanella and Octadecabacter), Thiovulaceae (Cocleimonas), Thiotrichaceae (Sulfurimonas), and Desulfobulbaceae (Desulfobulbus) (Figure 4). Assemblage D indicator taxa were within the families Anaerolineaceae, Desulfobacteraceae, Desulfobulbaceae, orders PHOS-HE36 and SBR1031, and phyla Bathyarchaeota, and Schekmanbacteria. The only indicator OTU identified down to genus level here was Desulfococcus (Desulfobacteraceae), therefore other abundant genus-level OTUs that belonged to the families Desulfobacteraceae and Desulfobulbaceae, were also queried and further explored with the indicator taxa (Figure 4).
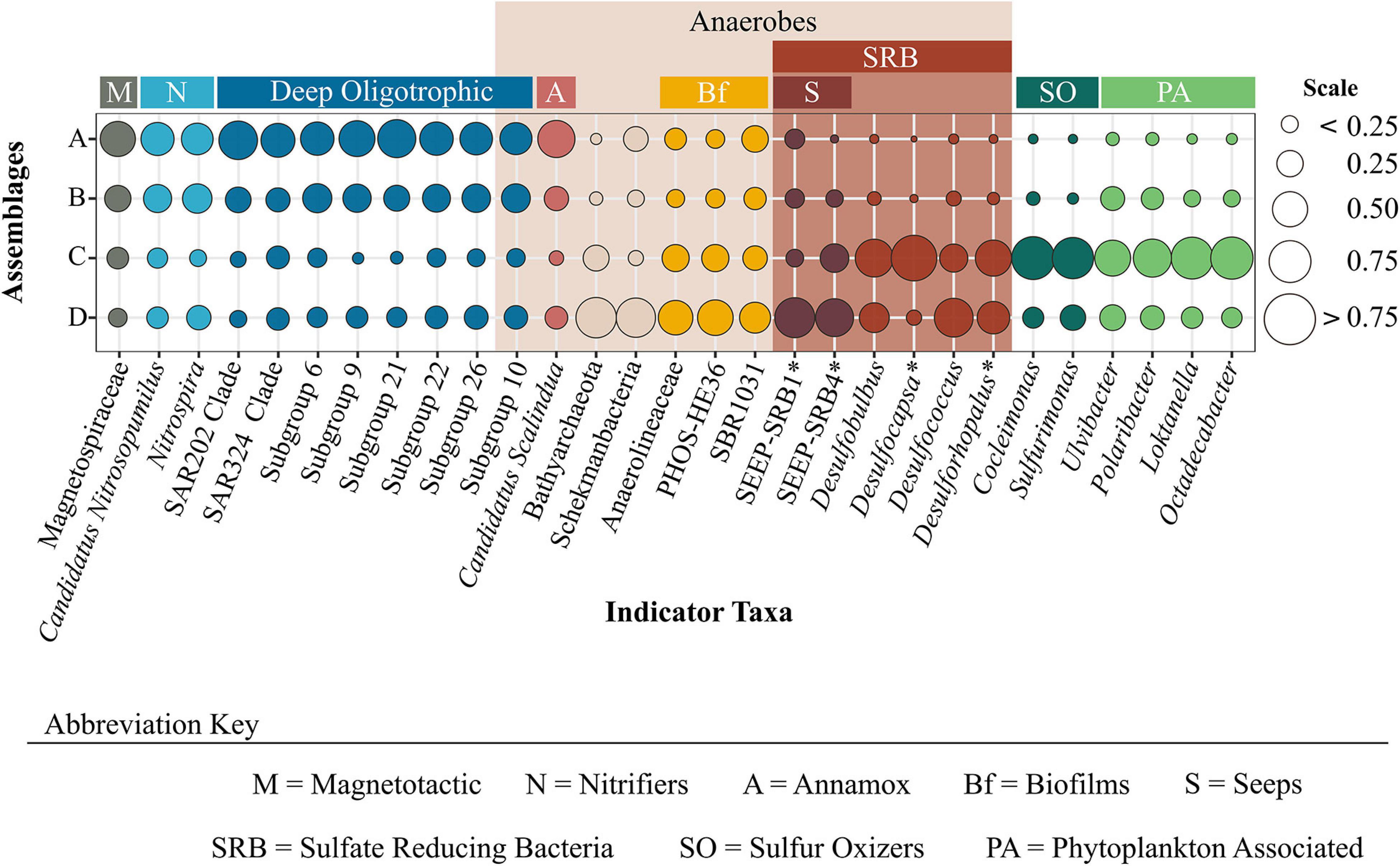
Figure 4. Proportion of assemblage-specific indicator taxa across prokaryotic assemblages. Indicator taxa are identified to the lowest taxonomic level possible. The plot is colored by functional groups which provide insight into characteristics associated with specific assemblages. *Indicates those taxa that were not yielded as indicators, but represent OTUs belonging to the same family as unidentified indicator taxa.
Overall, the suite of environmental variables measured here explained relatively little (18%; Adj. R2 = 0.18) of the variation in prokaryote community structure among sites (Figure 6). Nonetheless, results of the CAP analysis did suggest significant correlations (adonis: P < 0.001) with different habitat characteristics for each assemblage. Assemblage A was found solely on the continental slope in 23 samples ranging from 171 to 1,200 m water depth (median depth 350 m), thus generally consisting of “deep-sea” locations further offshore and below the photic zone. Sediments at these locations exhibited significantly higher δ15N and porosity, and lower values of Chl-a concentration and C:N; bottom waters were warmer and more saline (Figure 5). Assemblage A largely differentiated from the other assemblages on the CAP1 axis (Figures 6A,C). CAP1 was most strongly and positively correlated with depth, δ15N, and porosity and exhibited a moderate to weak negative correlation with Chl-a and TOC. Variation within assemblage A was most apparent on the CAP4 axis suggesting Amundsen Gulf and Banks Island samples (+CAP4) are more influenced by degraded and marine derived OM (higher δ15N and δ13C) whereas Beaufort slope samples (-CAP4) had higher porosity sediments with comparatively more phaeopigment and terrestrial OM (Figure 6C).
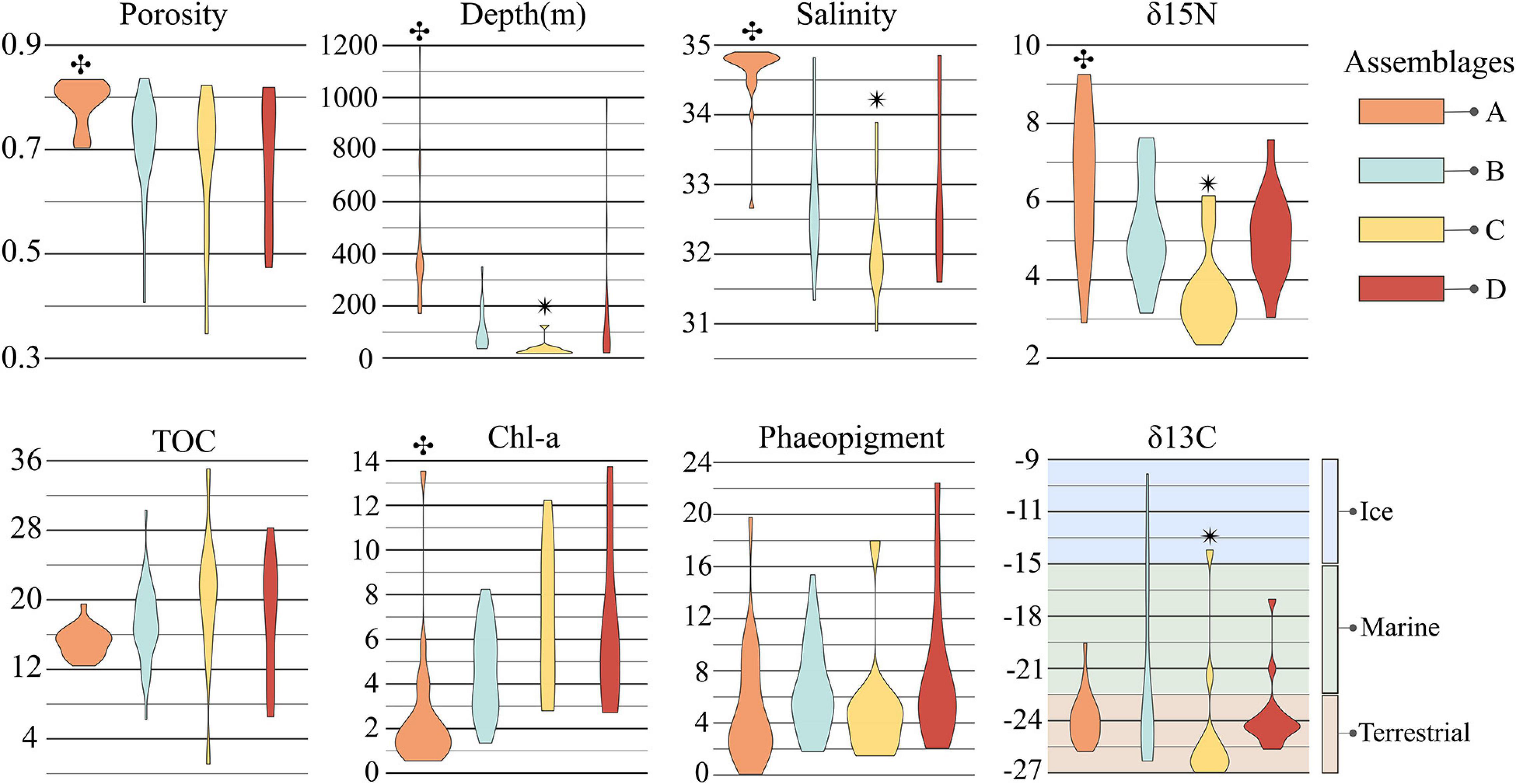
Figure 5. Distribution of environmental parameters for each assemblage. Each violin plot shows the distribution, within each assemblage, of the environmental parameters indicated as significant correlates in the ideal model for CAP analysis. Note that salinity is also included in this figure, though it was not run in CAP analysis due to the covariance inflation factor with depth. The symbols 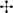 and
and  indicate significant differences between un-matching symbols with other assemblages. δ13C values for ice, marine, and terrestrial OM sources was derived from Dunton et al. (2006).
indicate significant differences between un-matching symbols with other assemblages. δ13C values for ice, marine, and terrestrial OM sources was derived from Dunton et al. (2006).
Figure 6. Environmental parameters influencing prokaryotic assemblage structure. The ordination plots above depict the results of Constrained Analysis of Principal Coordinates. All included environmental parameters are significant correlates (P < 0.001) to the first four CAP axes (A–C) and, in total, explained 18% of the variation in prokaryotic community structure (R2 = 0.18). The corresponding correlations are reported in the table for all axes.
Assemblage B consisted of 35 samples ranging between 36 and 350 m water depth (median depth 75 m), with a little more than half (19) located on the shelf and the remainder (16) on the slope. Median values of all environmental parameters for assemblage B were not significantly different than those for any of the other assemblages (Figure 5). CAP analysis showed a separation of assemblage B largely along the CAP2, axis suggesting that stations in this assemblage are characterized by more degraded OM (positive correlation with phaeopigment and δ15N) and less influence from terrestrial organic matter (higher δ13C suggesting marine or ice-algal source; Dunton et al., 2006; Figures 5, 6A). Differences among samples within assemblage B were most apparent on both the CAP3 and CAP4 axes, indicating that Amundsen Gulf and Banks Island samples (-CAP3) were generally characterized by higher TOC and lower porosity, whereas samples from the open shelf and slope (+CAP3) were characterized by higher porosity sediments with comparatively less TOC and more degraded phytodetritus (higher phaeopigment). Easternmost samples from the Amundsen Gulf (+CAP4) were separated from all other samples (-CAP4), with potentially sea-ice derived OM (higher δ13C), compared to more degraded marine and terrestrial OM (lower δ13C and higher phaeopigment) in the western Beaufort. Depth was also moderately or highly correlated with the CAP 3 and 4 axes, although this may reflect effects of other environmental variables that co-vary with depth.
The 11 samples represented by assemblage C were located solely in shallow shelf sediments ranging from 17 to 42 m water depth (median depth 21 m) with significantly lower values of δ15N, δ13C, and salinity. Assemblages C, B, and D were largely separated along the CAP2 axis, with assemblage C samples more correlated with fresh phytodetritus (higher Chl-a, lower δ15N) and terrestrial OM (lower δ13C). Samples within assemblage C varied along CAP3 axis which is most strongly correlated with TOC and porosity (Figure 6B). Nearshore samples from the Alaskan Beaufort shelf (+CAP3) were characterized by lower TOC and higher porosity, whereas those from the Canadian Beaufort and Amundsen Gulf (-CAP3) had higher TOC and lower porosity.
Assemblage D consisted of 16 samples which were collected almost exclusively at shelf locations (=100 m; median depth 72 m), except for 3 deep sites (=200 m) offshore of the Colville River plume where high concentrations of Chl-a were found. Environmental characterization for and correlations with assemblage D were not well captured with parameters measured here (Figures 5, 6). Variation within assemblage D was most apparent on the CAP3 axis, which suggests that several samples had comparatively higher organic matter input and lower porosity.
Comparisons of the top 10 class-level taxa between Beaufort Sea, global benthic, deep-sea, and Norwegian-Arctic surface sediments indicate that all four sediment microbiomes share five major taxa: Gammaproteobacteria, Alphaproteobacteria, Deltaproteobacteria, Bacteroidia, and Actinobacteria (Figure 7). The Beaufort Sea sediment microbiome included taxa that were also highly abundant in either the combination of global benthic and deep-sea microbiomes (Plantomycetia) or the Norwegian-Arctic microbiome (Phycisphaera). Interestingly, four classes prevalent in the other three microbiomes were not among the 10 most abundant in Beaufort Sea sediments: Clostridia, Betaproteobacteria, Bacilli, and Acidobacteria. Nonetheless, Acidobacteria was quite abundant in the Beaufort Sea, falling within the top 13 most abundant classes. Class Betaproteobacteria was not detected in Beaufort Sea surface sediments, whereas Clostridia and Bacilli were present but in relatively low abundances. Nitrososphearia, Verrucomicrobia, and Anaeronlinae were among the top 10 taxa of the Beaufort Sea sediment microbiome, but were not abundant in the other datasets.
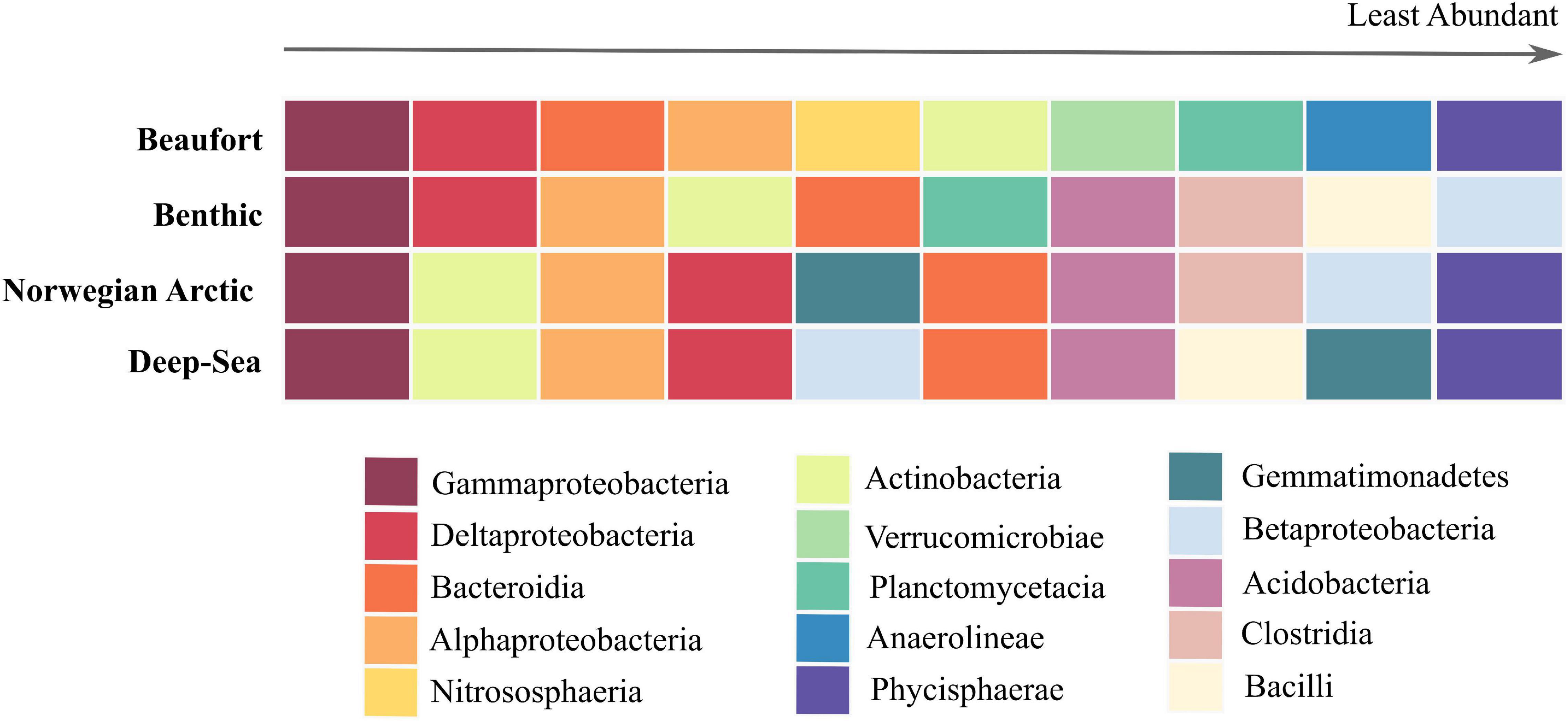
Figure 7. Top 10 abundant prokaryotes at class level exhibited in marine sediments. The colored grid above is arranged with surface sediment microbiomes on the y-axis, Beaufort, global benthic, Norwegian Arctic, and deep-sea (Zinger et al., 2011; Bienhold et al., 2016). The x-axis represents the rank from the most abundant taxa on the left to the least abundant taxa on the right exhibited by these microbiomes. The colors represent the class-level taxonomy of these prokaryotes.
We identified four distinct assemblages, A, B, C, and D, across a broad area in southern Beaufort Sea surface sediments. Assemblage A occurred only at sites on the upper continental slope with lower concentrations of more degraded OM and higher porosity sediments. A distinct community (assemblage C) was found in shallow shelf sediments, with higher OM content and terrestrial OM influence. The wide distribution of assemblage B across shelf and slope depths and overlap in taxonomic composition with assemblages A and C, may reflect a transition zone between the shelf and slope sites containing generalists found in both bathymetric zones. Assemblage D was found primarily at shelf depths, but included a few deeper slope sites in the westernmost part of the study area with high OM content. Indicator taxa with known metabolic functions and/or ecological context suggest that assemblage D was not merely comprised of generalists. In order to provide ecological context and better understand what distinguishes these assemblages, we examined taxa that were assemblage-specific and/or exhibited higher abundances associated to an individual assemblage together with environmental correlates.
The indicator taxa of assemblage A, specifically Acidobacteria subgroups and SAR clades, reflect the deeper sampling depths (171–1,200 m) typified by reduced inputs of more degraded organic matter (Figure 5). Representatives of the phylum Acidobacteria found in Arctic deep-sea sediments have been proposed as potential indicators of oligotrophic conditions, and found to be more abundant in highly permeable sub-Arctic and Arctic sediments indicating an affinity for low-molecular-weight OM; subgroups 6, 10, and 26 in particular were more active under oligotrophic conditions (Bienhold et al., 2012; Hoffmann et al., 2017; Probandt et al., 2017). Here, Acidobacteria subgroups 6, 9, 10, 21, 22, and 26 along with SAR clades 324 and 202, were prevalent in slope assemblage A. The clades SAR324 and SAR202, like the Acidobacteria subgroups, are also indicative of deep, oligotrophic environments (Morris et al., 2004; Galand et al., 2010; Orcutt et al., 2011; Li et al., 2015; Landry et al., 2017; Nunoura et al., 2018; Zinke et al., 2018).
Slope assemblage A may also reflect some of the unique biogeochemical characteristics on the Beaufort Sea slope. In the Arctic Halocline waters over the Canada Basin, SAR 202 have been implicated in terrestrial OM degradation, consistent with the relatively strong terrestrial signal exhibited throughout the Southern Beaufort, even in the upper slope sediments (Figure 5; Colatriano et al., 2018). SAR202 has also been linked with deep-sea mud volcanoes and SAR324 with methane-rich hydrothermal plumes (Sheik et al., 2014; Coelho et al., 2016). Acidobacteria subgroup 10 was recently placed in the family Themoanaerobaculaceae which encompasses thermophilic obligate anaerobes isolated from marine hydrothermal vents and freshwater hot springs (Losey et al., 2013; Dedysh and Yilmaz, 2018). Several mud volcanoes on the Beaufort Sea slope release subsurface methane and produce temperatures ≥23°C within a few centimeters of the sediment-water interface, creating suitable habitat for thermophilic anaerobes such as those within subgroup 10 (Paull et al., 2015; Gamboa et al., 2017; Lee et al., 2018; Paull, pers. comm.). Subgroup 10 is widespread across the study area, yet to date, mud volcanoes have only been reported on the Canadian Beaufort Slope. These microbes may be spore-forming, similar to other thermophilic anaerobes reported from Arctic marine sediments that produce spores that lie dormant until favorable conditions arise (Hubert et al., 2010; Müller et al., 2014). However, previous reports of thermo-anaerobic bacterial spores belonged to the phylum Firmicutes, and not Acidobacteria, and none of the current taxa within Acidobacteria subgroup 10 are known to be spore-forming (Hubert et al., 2010; Losey et al., 2013; Müller et al., 2014). Given their occurrence in Arctic sediments from other regions, it is more likely that bacteria in subgroup 10 inhabit a wider temperature range than previously recognized (Hoffmann et al., 2017).
Beaufort sediment mineralogical properties may be influenced by the abundant indicator OTUs within Magnetospiraceae, a family composed of magnetotactic bacteria. Magnetotactic bacteria are unique in that they biosynthesize a mineral nanocrystal which they can use as a “compass” to navigate toward favorable redox conditions found at the oxic-anoxic interface in marine sediments (Lefevre and Bazylinski, 2013). Though indicator OTUs were all uncultured, Magnetospira is the most abundant taxon identified to genus-level within this family. Members of the genus Magnetospira produce magnetite crystals, which are the predominant magnetic grain found in Canadian Beaufort sediments and are more prevalent on the slope (Williams et al., 2012; Lefevre and Bazylinski, 2013; Gamboa et al., 2017).
Genera tightly linked to nitrification and anaerobic ammonia oxidation were relatively more abundant in slope assemblage A. Nitrospira are exclusively chemoautotrophic nitrite-oxidizing bacteria, oxidizing nitrite to nitrate, but have recently been found to perform complete nitrification, oxidizing ammonia to nitrate (Daims, 2014; Daims et al., 2015). Candidatus Nitrosopumilus encompasses ammonia-oxidizing archaea (AOA), which are aerobic nitrifiers that exhibit an extremely high affinity for ammonia, and can oxidize ammonia to nitrate, nitrite, or nitrous oxide (Stahl and de la Torre, 2012; Könneke et al., 2014; Martens-Habbena et al., 2015; Qin et al., 2016). AOA can perform mixotrophy and heterotrophy, but are largely chemoautotrophs (Stahl and de la Torre, 2012; Offre et al., 2013; Könneke et al., 2014; Qin et al., 2014). They also exhibit photoinhibition and increased autotrophic activity, which is consistent with the distribution of this assemblage at deeper sampling stations (Alonso-Sáez et al., 2012; Qin et al., 2014). In the pelagic zone of the Beaufort Sea, AOA are relatively more abundant in bottom waters on the slope and may be capable of urea degradation to fuel nitrification, as has been observed for Candidatus Nitrosopumilus sediminis in Arctic sediments near Svalbard (Alonso-Sáez et al., 2012; Park et al., 2012; Damashek et al., 2017). In the portion of our study area characterized by assemblage A, a peak in urea concentration occurs at 250–300 m water depth due to zooplankton and fish excretion, which may fuel the activity of AOA in this region (Simpson et al., 2008).
The genus Candidatus Scalindua encompasses anaerobic ammonia oxidizing (anammox) bacteria which convert ammonia to N2 via autotrophic metabolism and can utilize nitrite, nitrate, metal oxides, and even oligopeptides and small organic molecules as electron acceptors (Penton et al., 2006; Van de Vossenberg et al., 2013). Anaerobic ammonia oxidation may be a major sink for nitrate in the marine system, and has been reported in Arctic shelf slope sediments in the Chukchi Sea, near Greenland, and in deep-sea sediments from the Arctic mid-ocean ridge (Rysgaard et al., 2004; Penton et al., 2006; Jorgensen et al., 2012). The predominance of anammox bacteria at the deeper slope sites occupied by assemblage A supports previous claims that anammox is more likely to occur in areas with less organic matter loading (Thamdrup and Dalsgaard, 2002; Rysgaard et al., 2004; Burgin and Hamilton, 2007; McTigue et al., 2016).
While no indicator taxa were identified for assemblage B, one abundant taxon included members of the bacterial family Methyloligellaceae, dominated in this study by the genus Methyloceanibacter which contains known methylotrophs (Takeuchi et al., 2014; Vekeman et al., 2016a). Some species of Methyloceanibacter oxidize methanol, while others exhibit methane oxidation and include the methane monooxygenase gene in their genome (Takeuchi et al., 2014; Vekeman et al., 2016a,b). We have detected a relatively high abundance of the methane monooxygenase gene in these sediments through metagenomics analyses, although further analyses are needed to attribute these genes to a specific taxonomic group (Walker et al. unpublished data). Nonetheless, the presence of Methyloceanibacter coupled with the abundant distribution of the methane monooxygenase gene in methane-permeated Beaufort Sea sediments suggests that aerobic methane oxidation may be an active process in this region, although further research confirming this activity would be required. As there are multiple sources of methane in these sediments, methane oxidation in the Beaufort Sea is likely an important process constraining methane concentrations reaching the sea surface (Lorenson et al., 2016). However, no evidence for methanotrophy has yet been reported in the water column or subsurface sediments, despite chemical evidence that oxidation of methane derived from ancient sources may be occurring near the sediment-water interface (Hamdan et al., 2013; Sparrow et al., 2018).
Indicator taxa for assemblage C were characteristic of a shallow-shelf environment influenced by fresh phytodetritus, riverine input, and sea ice, in keeping with elevated Chl-a and significantly lower values of δ13C and salinity observed at assemblage C locations. Many of these taxa may be deposited to the seafloor in association with sinking particles. Flavobacteriaceae occur in conjunction with phytoplankton blooms and subsequent deposition of phytodetritus (Junge et al., 2002; Bienhold et al., 2012; Teeling et al., 2012; Hoffmann et al., 2017). Genera within this family are typically aerobic, though there are some facultative anaerobic heterotrophs capable of degrading complex polysaccharides (Teeling et al., 2012; McBride, 2014; Xing et al., 2015). Two of the genus-level indicator taxa found here, Ulvibacter and Polaribacter, are often particle attached and tightly connected with phytoplankton blooms and sea ice (Junge et al., 2002; Bowman and McCuaig, 2003; Brinkmeyer et al., 2003; Teeling et al., 2012; Xing et al., 2015). Ulvibacter is one of the first and most abundant genera to respond to a phytoplankton bloom following algal lysis, whereas Polaribacter is most abundant in later stages of an algal bloom and can use sulfatases to access highly sulfated algal material (Teeling et al., 2012, 2016; Xing et al., 2015; Helbert, 2017). Polaribacter species generally include psychrophilic bacteria that are most commonly found in polar and temperate regions. Until recently, they had only been identified in sea ice and seawater, but are now described from Arctic sediments offshore of Svalbard (Gosink et al., 1998; Bowman et al., 2012; Li et al., 2014; Rapp et al., 2018).
A few of the indicator taxa for assemblage C suggest that light availability may be a major factor influencing community composition. Genomic analyses of Polaribacter species, along with Loktanella and Octadecabacter, indicator taxa from the Rhodobacteraceae family, contain various rhodopsins (light-driven proton pumps) and other light responsive genes, indicating an affinity for well-lit shallower waters (Gonzalez et al., 2008; Boeuf et al., 2013; Vollmers et al., 2013; Wu et al., 2014; Xing et al., 2015; Dogs et al., 2017). Loktanella, described as anoxygenic photoheterotrophs (AAP), have been found in Beaufort Sea surface waters including the Mackenzie River outflow where they were positively correlated with Chl-a (Boeuf et al., 2013). AAP photosynthesize but also require oxygen for breaking down organic matter with light-harvested energy, and are most abundant in coastal waters and eutrophic estuaries (Waidner and Kirchman, 2007; Boeuf et al., 2013). Photoheterotrophy could be an advantage in such environments that are heavily influenced by terrestrial material, providing an energy source to fuel more costly metabolic processes such as the degradation of humic substances (Boeuf et al., 2013). Octadecabacter species are psychrophilic, aerobic heterotrophs with gas vesicles and are abundant in sea ice, though a few taxa have been isolated from seawater and marine sediments (Gosink et al., 1997; Brinkmeyer et al., 2003; Vollmers et al., 2013; Lee et al., 2014; Billerbeck et al., 2015). Octadecabacter spp. have also been linked to phytoplankton blooms and detrital aggregates in the Arctic and elsewhere (Grossart et al., 2005; Rapp et al., 2018; Koedooder et al., 2019).
The remaining indicator taxa for assemblage C belong to the bacterial families Thiotrichaceae, Thiovulaceae, and Desulfobulbaceae, all of which are involved in the marine sulfur cycle. Members of the Desulfobulbaceae family, which are anaerobic sulfate reducers, were also found in assemblage D and discussed further below (Kuever, 2014b). Thiotrichaceae and Thiovulaceae, represented here by Cocleimonas and Sulfurimonas, respectively, are sulfur oxidizers (Tanaka et al., 2011; Garrity et al., 2015; Han and Perner, 2015). Sulfur oxidizing bacteria are generally limited by the availability of necessary electron donors (typically reduced sulfur compounds) and acceptors (oxygen, nitrate) favored in anoxic and oxic conditions, respectively (Wasmund et al., 2017; Jørgensen et al., 2019). Adaptations exhibited by Sulfurimonas and Cocleimonas provide a competitive advantage in the oxidation of elemental sulfur and other sulfur intermediates (Tanaka et al., 2011; Pjevac et al., 2014; Han and Perner, 2015; Wasmund et al., 2017).
The indicator taxa for assemblage D suggest a greater extent of anoxic sediment within the upper 1 cm layer. All indicator OTUs for Assemblage D are strict anaerobes within the families Desulfobulbaceae, Desulfobacteraceae, Anaerolineaceae, PHOS-HE36, the order SBR1033, and the phyla Bathyarcheaota and Schekmanbacteria (Kuever, 2014a,b; Evans et al., 2015; Anantharaman et al., 2018; Yamada and Sekiguchi, 2018). Members of the archaeal phylum Bathyarcheaota and candidate bacterial phylum Schekmanbacteria consisted of uncultured OTUs with less than 85% matches to published sequences in BLAST. Though inferences based on this level of taxonomy are highly speculative, representatives from both phyla have been most commonly sequenced from subsurface marine sediments (Inagaki et al., 2003; Biddle et al., 2006; Sørensen and Teske, 2006; Evans et al., 2015; Anantharaman et al., 2018; Lopez-Fernandez et al., 2018; Winkel et al., 2018).
Anaerolineaceae are comprised of anaerobic heterotrophs with a fermentative metabolism and have been sequenced in anoxic, organic-rich habitats such as sludge digesters, hydrothermal vents, and glacial fjord sediments off the coast of Svalbard (McIlroy et al., 2017; Zeng et al., 2017; Yamada and Sekiguchi, 2018). They are typically abundant in biofilms and implicated as hydrogenotrophs and alkane degraders, though few studies have examined marine representatives (Siegert et al., 2011; Sinkko et al., 2013; Al-Thani et al., 2014; Liang et al., 2015; van der Waals et al., 2017). Similarly, bacteria in the order SBR1031 have been primarily investigated in waste-water treatment plants where they produce biofilms (Xia et al., 2016; Wang X. et al., 2018). Marine representatives have been found in oil-contaminated estuarine sediments, and increase in abundance in seasonal microbial mats in the North Sea (Obi et al., 2017; Cardoso et al., 2019). Bacteria from the family PHOS-HE36 have also been predominately studied in the context of sludge digesters and biofilms, often sequenced with bacteria in the Anaerolineaceae family, but have also been found in marine sediments near methane seeps, hydrothermal vents, and within microbial mats (Dabert et al., 2001; Fujii et al., 2002; Trembath-Reichert et al., 2016; Wang L. et al., 2018; Zinke et al., 2018; Yang et al., 2019).
Indicator taxa from assemblage D are also connected to the sulfur cycle, specifically sulfate reduction, which is the dominant process responsible for anaerobic degradation of organic matter in marine sediments (Wasmund et al., 2017; Jørgensen et al., 2019). The abundance of several taxa within the families Desulfobacteraceae and Desulfobulbaceae suggest that sulfate reduction is a prevalent in both shallow shelf assemblage C and anoxic assemblage D. At family level, Desulfobacteraceae taxa were more abundant in assemblage D, whereas Desulfobulbaceae taxa were more evenly distributed between assemblages C and D, albeit with specific genera that were more abundant in shallow assemblage C. These families have shown responses to different substrates in Arctic sulfidic sediments, suggesting the taxonomic differences between assemblages C and D reflect differences in substrate availability among sites (Dyksma et al., 2018; Müller et al., 2018).
Desulfocapsa and Desulfobulbus (Desulfobulbaceae) were more abundant in shallow shelf assemblage C, and have often been sequenced in Arctic fjord sediments alongside sulfur-oxidizing bacteria (SOB) such as Sulfurimonas and Cocleimonas; which were also identified as indicator taxa for assemblage C (Zeng et al., 2017; Trivedi et al., 2018; Buongiorno et al., 2019). This repeated co-occurrence in Arctic marine sediments suggests an affinity for similar environmental/redox conditions, and further exemplifies subtle differences between sulfur cycling in assemblages C and D.
The more abundant SRB taxa in assemblage D, particularly SEEP-SRB1 and SEEP-SRB4 clades, are most commonly sequenced from cold seeps (Schreiber et al., 2010; Kleindienst et al., 2012; Ruff et al., 2015). SEEP-SRB have been implicated in coupling sulfate reduction to the anaerobic oxidation of non-methane and methane hydrocarbons, however they remain largely uncultured (Petro et al., 2019). Desulfococcus spp., though commonly found in sulfidic sediments, are also associated with marine seeps, and may be major players in anaerobic hydrocarbon degradation in association with seeps (Ravenschlag et al., 2000; Orphan et al., 2001; Kleindienst et al., 2012, 2014).
The majority of samples in this assemblage were located in shallow shelf sediments, some of which were characterized by heavy OM loading and low porosity, conducive to oxygen depletion. Samples collected on the slope in assemblage D exhibited some of the highest concentrations of Chl-a measured here, and may have received a recent pulse of fresh phytodetritus leading to shallow anoxic conditions in sediments. Moreover, multiple indicator taxa for assemblage D are also associated with biofilms, microbial mats, seep environments, or subsurface sediments, suggesting the presence of these features at some sites. Microbial mats have been reported on Beaufort Sea sediments, mostly surrounding slow gas seeps (Paull et al., 2011). The inherently patchy distribution of benthic seeps and microbial mats coupled with the varying conditions that foster anoxic sediments may explain the lack of definable substructure exhibited by assemblage D and weak correlations with environmental parameters in this study.
Benthic prokaryotic community substructure on the Beaufort Sea shelf and upper slope broadly reflected longitudinal gradients in OM quality and quantity, as well as the influence of bathymetry. While bathymetric effects were particularly apparent in distinguishing slope assemblage A, shallow shelf assemblage C, and generalist assemblage B, redox conditions likely played a role in differentiating assemblages C and D. These findings highlight the importance of assessing prokaryotic communities on a broad spatial scale, given that community structure reflected environmental variation occurring over scales of meters to kilometers. This raises the question of how prokaryotic community structure in the Beaufort Sea compares to other regions within the Arctic and beyond.
At class-level, 60% of the top 10 taxa from Beaufort Sea sediments were shared with those from the global, deep-sea, and Norwegian Arctic sediment microbiomes (Figure 7). The classes Anaerolineae, Nitrososphearia, and Verrucomicrobiae were abundant and unique to the Beaufort Sea surface sediment microbiome. The bacterial class Anaerolineae and archaeal class Nitrososphearia are typified by the families Anaerolinaceae and Nitrosopumilaceae which, as mentioned above, primarily contain anaerobic bacteria associated with heavy OM loading as was prevalent in anoxic assemblage D, and ammonia-oxidizing archaea prevalent in slope assemblage A, respectively. Not surprisingly, Nitrososphearia was not among the most abundant taxa in the global, deep-sea, or Norwegian Arctic sediment microbiomes. Though common in marine surface sediments and typically more abundant than ammonia oxidizing bacteria, archaea have been overlooked in previous studies solely focused on bacteria (Park et al., 2010; Zinger et al., 2011; Bienhold et al., 2016). Their relatively high abundance here highlights the under-appreciated role of archaea in marine surface sediments. If archaea are excluded, the bacterial taxon Thermoanaerobaculia joins the top 10 most abundant taxa in our study. This group encompasses the anaerobic, thermophilic organisms largely represented by subgroup 10 an indicator taxon for slope assemblage A. This class was not among the most abundant taxa in sediments globally, in the deep sea, or in the Norwegian Arctic. Thermoanaerobaculia together with Anaerolineae suggest comparatively shallower anoxic conditions in Beaufort Sea surface sediments, spanning both slope and shallow shelf locations.
Betaproteobacteria, Gemmatimonadetes, Bacilli, and Clostridia were among the 10 most abundant taxa from global benthic, deep-sea, and Norwegian-Arctic microbiomes, yet were not prominent in Beaufort sediments. Betaproteobacteria, which were not present even at low abundances in Beaufort Sea sediments, were discovered to be potential laboratory contaminants in other studies (Kulakov et al., 2002; Grahn et al., 2003; Laurence et al., 2014; Salter et al., 2014). On a global scale Zinger et al. (2011) observed Bacilli and Clostridia in higher abundances in coastal sediments vs. open water or deep-sea sediments. Bacilli and Clostridia can be indicators of urban fecal contamination in coastal watersheds, lagoons, and nearshore seawater (Wu et al., 2010; Dubinsky et al., 2012; Dai et al., 2018). As the Beaufort Sea is extremely remote from urban life, it is not surprising that these taxa were less prevalent. Clostridia was included in the Norwegian-Arctic microbiome and Bacilli in the deep-sea microbiome, which are also presumably more pristine than typical coastal areas, suggesting that there are bacteria in these classes which are not necessarily correlated with urban waste (Bienhold et al., 2016).
The prokaryotic community structure in the Beaufort Sea differed somewhat from the Norwegian-Arctic, even at the relatively coarse taxonomic level of class, suggesting benthic microbes may reflect regional differences in hydrography, biogeochemistry, and bathymetry of Arctic shelf systems (Carmack and Wassmann, 2006). However, samples from the Norwegian-Arctic were largely collected at depths > 1,000 m, so these regional differences need to be further explored particularly across wider depth ranges.
The occurrence of strictly anaerobic taxa in all assemblages identified here indicates that Beaufort Sea surface sediments exhibit widespread anoxia within the 0–1 cm depth horizon, consistent with oxygen penetration studies conducted in other Arctic sediments (Kostka et al., 1999; Jørgensen et al., 2005). A larger proportion of anaerobic taxa occurred in shallow shelf (C) and anoxic (D) assemblages, indicating that, in general, oxygen depletion in nearshore sediments is comparatively more widespread than offshore, particularly in areas where organic matter loading and Chl-a were higher and porosity lower. Although this is a common trend when comparing shelf to slope sediments, it may be of particular importance in the Beaufort Sea when viewed through the lens of climate change (Orcutt et al., 2011). An overall net increase of river runoff and terrestrial organic matter has already been established for the Mackenzie River, and is predicted to continue over the coming years (Holmes et al., 2012; Doxaran et al., 2015). Thus, we might expect an increase in abundance of anaerobic taxa in shallow shelf assemblage C, with potentially more stations resembling or overlapping with anoxic assemblage D. There may also be increased taxonomic overlap between microbial communities associated with the Mackenzie River outflow and shallow shelf assemblage C. In addition to riverine input, shifts in primary production and sea-ice extent will likely be most apparent in the shallow shelf sediments, as the majority of indicator taxa in assemblage C have been strongly correlated to both phytoplankton blooms and sea-ice microbial communities.
An increase in OM input, whether terrestrial or marine derived, will likely translate to an increase in OM burial and subsequent methanogenesis in the already methane-permeated Beaufort Sea sediments (Coffin et al., 2013). Characterizing the microbes involved in both methane production/degradation and associated processes in these sediments is thus of particular interest. The high relative abundance of potential methanotrophs exhibited in this study could, in addition to highlighting the presence of methane throughout sediments in the region, indicate an important constraint on methane efflux that requires further investigation.
This study also highlights the vast array of uncultured and unclassified microbes in Arctic marine sediments, particularly exemplified by slope assemblage A, which was the most divergent and diverse and contained the highest number of uncultured taxa across all assemblages. Anoxic assemblage D also exhibited a comparatively high abundance of uncultured microbes with representatives which are more rarely studied in the context of the marine system. The lack of knowledge regarding these taxa points to a need for more experimental and culture-based studies to elucidate their role in biogeochemical processes, that would provide insights into the links between microbial community structure and benthic ecosystem function. Although taxonomic classification does not directly equate to function, known microbes from slope assemblage A highlight how taxonomic composition may provide a framework for future studies. For instance, research investigating the role of nitrification and anammox in the Beaufort Sea would be most beneficial in slope sediments where microbes involved in aerobic/anaerobic ammonia oxidation and nitrite oxidation were abundant, and could reveal useful information about regional nitrogen cycling and upwelling dynamics. Additionally, microbial taxonomy can provide insights into environmental conditions that are not typically measured with surveys of larger benthic epi- and infaunal organisms, such as sediment oxygen penetration and the presence of biofilms, microbial mats, and/or seeps seemingly reflected by the taxonomic composition of anoxic assemblage D. Overall, these insights provided by prokaryotic community structure emphasize the value of incorporating microbial surveys more broadly into field sampling programs in the North American Arctic and elsewhere. This study establishes a baseline for Beaufort Sea benthic microbes, however without long-term datasets we cannot assess how these communities will respond to and reflect shifts related to climate change or other disturbances such as energy resource exploration.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA451041.
AW was involved in field work for sample collection, conducted all sample processing and data analysis, and took a lead role in the interpretation of results and manuscript preparation. ML contributed funding, advised on data analysis and interpretation, and contributed to manuscript preparation. SM conducted field work to collect samples and environmental data, provided funding, and contributed to project planning, interpretation of results, and manuscript preparation. All authors contributed to the article and approved the submitted version.
Sample sequencing, student stipend, and tuition were funded by the US Department of the Interior, Bureau of Ocean Energy Management (BOEM) through the Coastal Marine Institute graduate student fellowship program (Grant# M16AC00004). Stipend and tuition support for AW was provided in part by the North Pacific Research Board (Project #1303). Support for the use of the UAF Genomics Core lab was provided by the Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number 2P20GM103395. The content of this manuscript is solely the responsibility of the authors and does not necessarily reflect the official views of the NIH. This work was supported via in-kind match from the high-performance computing and data storage resources operated by the Research Computing Systems Group at the University of Alaska Fairbanks Geophysical Institute.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Samples for this project were collected opportunistically through field work funded by BOEM and Department of Fisheries and Oceans Canada. Sincerest gratitude goes out to Dr. Holly Bik for providing the bioinformatics and genomics foundation needed for this project to happen. Appreciation also goes to Dr. Eric Collins for ongoing bioinformatic and statistical assistance. A huge thank you to Shannon MacPhee for coordinating and collaborating with us, without you we would not have had such an expansive dataset from the Canadian Beaufort Sea. The majority of environmental variables incorporated in this study were provided through the hard work of Julia Dissen.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.581124/full#supplementary-material
Supplementary Figure 1 | Distribution of Methyloceanibacter at different levels of sequence removal. The pie chart above illustrates an example, using the genus Methyloceanibacter, of how relative abundances change with removal of singletons and doubletons compared to no removal (none). The difference in abundance is attributed to the removal of OTUs that would likely not have been removed if clustered at 97% similarity as they belong to the same genus. This is an important consideration when using amplicon sequence variants (ASVs), and software which automatically remove singletons and doubletons such as DADA2.
Supplementary Figure 2 | Indicator OTUs for individual and combinations of prokaryotic assemblages. This Venn diagram illustrates the number of indicator OTUs in parentheses identified for individual assemblages and those shared in different combinations of assemblages.
Supplementary Figure 3 | Proportion of top 10 most abundant taxa in Beaufort sediments at class level. This bubble plot illustrates the proportion of the 10 most abundant taxa at class level for Beaufort Sea surface sediments.
Supplementary Table 1 | Indicator taxa for each prokaryotic assemblage. Taxa are arranged from most abundant starting at the top to least abundant toward the bottom. Those without a semi-colon indicate OTUs which could not be IDed past the taxonomic level provided.
ACIA (2005). Arctic Climate Impact Assessment. ACIA Overview Report. Available online at: http://www.amap.no/documents/doc/arctic-arctic-climate-impact-assessment/796 (accessed March 4, 2020).
Alonso-Sáez, L., Waller, A. S., Mende, D. R., Bakker, K., Farnelid, H., Yager, P. L., et al. (2012). Role for urea in nitrification by polar marine Archaea. Proc. Natl. Acad. Sci. U.S.A. 109, 17989–17994. doi: 10.1073/pnas.1201914109
Al-Thani, R., Al-Najjar, M. A. A., Al-Raei, A. M., Ferdelman, T., Thang, N. M., Al Shaikh, I., et al. (2014). Community structure and activity of a highly dynamic and nutrient-limited hypersaline microbial mat in Um Alhool Sabkha, Qatar. PLoS One 9:e0092405. doi: 10.1371/journal.pone.0092405
Anantharaman, K., Hausmann, B., Jungbluth, S. P., Kantor, R. S., Lavy, A., Warren, L. A., et al. (2018). Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 12, 1715–1728. doi: 10.1038/s41396-018-0078-0
Apprill, A., McNally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Arar, E. J., and Collins, G. B. (1997). Method 445.0: In Vitro Determination of Chlorophyll a and Pheophytin a in Marine and Freshwater Algae by Fluorescence. Ohio: United States Environmental Protection Agency, Office of Research and Development, National Exposure Research Laboratory.
Arnosti, C. (2008). Functional differences between Arctic seawater and sedimentary microbial communities: contrasts in microbial hydrolysis of complex substrates. FEMS Microbiol. Ecol. 66, 343–351. doi: 10.1111/j.1574-6941.2008.00587.x
Bell, L. E., Bluhm, B. A., and Iken, K. (2016). Influence of terrestrial organic matter in marine food webs of the Beaufort Sea shelf and slope. Mar. Ecol. Prog. Ser. 550, 1–24. doi: 10.3354/meps11725
Bennett, R. H., and Lambert, D. N. (1971). Rapid and reliable technique for determining unit weight and porosity of deep-sea sediments. Mar. Geol. 11, 201–207. doi: 10.1016/0025-3227(71)90007-7
Biddle, J. F., Lipp, J. S., Lever, M. A., Lloyd, K. G., Sørensen, K. B., Anderson, R., et al. (2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U.S.A. 103, 3846–3851. doi: 10.1073/pnas.0600035103
Bienhold, C., Boetius, A., and Ramette, A. (2012). The energy–diversity relationship of complex bacterial communities in Arctic deep-sea sediments. ISME J. 6, 724–732. doi: 10.1038/ismej.2011.140
Bienhold, C., Zinger, L., Boetius, A., and Ramette, A. (2016). Diversity and biogeography of bathyal and abyssal Seafloor Bacteria. PLoS One 11:e0148016. doi: 10.1371/journal.pone.0148016
Bier, R. L., Bernhardt, E. S., Boot, C. M., Graham, E. B., Hall, E. K., Lennon, J. T., et al. (2015). Linking microbial community structure and microbial processes: an empirical and conceptual overview. FEMS Microbiol. Ecol. 113:fiv113. doi: 10.1093/femsec/fiv113
Billerbeck, S., Orchard, J., Tindall, B. J., Giebel, H.-A., Brinkhoff, T., and Simon, M. (2015). Description of Octadecabacter temperatus Sp. Nov., isolated from the Southern North sea, emended descriptions of the genus octadecabacter and its species and reclassification of octadecabacter jejudonensisPark and Yoon 2014 as pseudooctadecabacter jejudon. Int. J. Syst. Evol. Microbiol. 65, 1967–1974. doi: 10.1099/ijs.0.000205
BOEM (2019). Leasing and Plans | BOEM. Available online at: https://www.boem.gov/Alaska-Leasing-and-Plans/ (accessed January 14, 2019).
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gleseke, A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation methane. Nature 407, 623–626. doi: 10.1038/35036572
Boeuf, D., Cottrell, M. T., Kirchman, D. L., Lebaron, P., and Jeanthon, C. (2013). Summer community structure of aerobic anoxygenic phototrophic bacteria in the western Arctic Ocean. FEMS Microbiol. Ecol. 85, 417–432. doi: 10.1111/1574-6941.12130
Bowman, J. P., and McCuaig, R. D. (2003). Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69, 2463–2483. doi: 10.1128/AEM.69.5.2463-2483.2003
Bowman, J. S., Rasmussen, S., Blom, N., Deming, J. W., Rysgaard, S., and Sicheritz-Ponten, T. (2012). Microbial community structure of Arctic multiyear sea ice and surface seawater by 454 sequencing of the 16S RNA gene. ISME J. 6, 11–20. doi: 10.1038/ismej.2011.76
Braeckman, U., Janssen, F., Lavik, G., Elvert, M., Marchant, H., Buckner, C., et al. (2018). Carbon and nitrogen turnover in the Arctic deep sea: in situ benthic community response to diatom and coccolithophorid phytodetritus. Biogeosciences 15, 6537–6557. doi: 10.5194/bg-15-6537-2018
Brinkmeyer, R., Knittel, K., Jürgens, J., Weyland, H., Amann, R., and Helmke, E. (2003). Diversity and structure of bacterial communities in arctic versus Antarctic Pack Ice. Appl. Environ. Microbiol. 69, 6610–6619. doi: 10.1128/AEM.69.11.6610-6619.2003
Brothers, L. L., Herman, B. M., Hart, P. E., and Ruppel, C. D. (2016). Subsea ice-bearing permafrost on the U.S. Beaufort Margin: 1. Minimum seaward extent defined from multichannel seismic reflection data. Geochem. Geophys. Geosyst. 17, 4354–4365. doi: 10.1002/2016GC006584
Buongiorno, J., Herbert, L. C., Wehrmann, L. M., Michaud, A. B., Laufer, K., Røy, H., et al. (2019). Complex microbial communities drive iron and sulfur cycling in arctic fjord sediments. Appl. Environ. Microbiol. 85:e00949-19. doi: 10.1128/aem.00949-19
Burgin, A. J., and Hamilton, S. K. (2007). Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front. Ecol. Environ. 5, 89–96.
Buttigieg, P. L., and Ramette, A. (2015). Biogeographic patterns of bacterial microdiversity in Arctic deep-sea sediments (HAUSGARTEN, fram strait). Front. Microbiol. 6:660. doi: 10.3389/fmicb.2014.00660
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Cardoso, D. C., Cretoiu, M. S., Stal, L. J., and Bolhuis, H. (2019). Seasonal development of a coastal microbial mat. Sci. Rep. 9:9035. doi: 10.1038/s41598-019-45490-8
Carmack, E., and Wassmann, P. (2006). Food webs and physical–biological coupling on pan-Arctic shelves: unifying concepts and comprehensive perspectives. Prog. Oceanogr. 71, 446–477. doi: 10.1016/j.pocean.2006.10.004
Carmack, E. C., and Macdonald, R. W. (2002). Oceanography of the Canadian shelf of the beaufort sea: a setting for marine life. Arctic 55, 29–45. doi: 10.14430/arctic733
Casciotti, K. L., and Buchwald, C. (2012). Insights on the marine microbial nitrogen cycle from isotopic approaches to nitrification. Front. Microbiol. 3:356. doi: 10.3389/fmicb.2012.00356
Cock, P. J. A., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Coelho, F. J. R. C., Louvado, A., Domingues, P. M., Cleary, D. F. R., Ferreira, M., Almeida, A., et al. (2016). Integrated analysis of bacterial and microeukaryotic communities from differentially active mud volcanoes in the Gulf of Cadiz. Sci. Rep. 6:35272. doi: 10.1038/srep35272
Coffin, R., Smith, J., Yoza, B., Boyd, T., and Montgomery, M. (2017). Spatial variation in sediment organic carbon distribution across the alaskan beaufort Sea Shelf. Energies 10:1265. doi: 10.3390/en10091265
Coffin, R. B., Smith, J. P., Plummer, R. E., Yoza, B., Larsen, R. K., Millholland, L. C., et al. (2013). Spatial variation in shallow sediment methane sources and cycling on the Alaskan Beaufort Sea Shelf/Slope. Mar. Pet. Geol. 45, 186–197. doi: 10.1016/J.MARPETGEO.2013.05.002
Colatriano, D., Tran, P. Q., Guéguen, C., Williams, W. J., Lovejoy, C., and Walsh, D. A. (2018). Genomic evidence for the degradation of terrestrial organic matter by pelagic Arctic Ocean Chloroflexi bacteria. Commun. Biol. 1:90. doi: 10.1038/s42003-018-0086-7
Dabert, P., Sialve, B., Delgenès, J.-P., Moletta, R., and Godon, J.-J. (2001). Characterisation of the microbial 16S rDNA diversity of an aerobic phosphorus-removal ecosystem and monitoring of its transition to nitrate respiration. Appl. Microbiol. Biotechnol. 55, 500–509. doi: 10.1007/s002530000529
Dai, T., Zhang, Y., Ning, D., Su, Z., Tang, Y., Huang, B., et al. (2018). Dynamics of sediment microbial functional capacity and community interaction networks in an Urbanized Coastal Estuary. Front. Microbiol. 9:2731. doi: 10.3389/fmicb.2018.02731
Daims, H. (2014). “The family nitrospiraceae,” in The Prokaryotes, eds A. Balows, H.G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (Berlin: Springer), 733–749.
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
Damashek, J., Pettie, K. P. K., Brown, Z. W. Z., Mills, M. M. M., Arrigo, K. R. K., and Francis, C. A. C. (2017). Regional patterns in ammonia-oxidizing communities throughout Chukchi Sea waters from the bering strait to the Beaufort Sea. Aquat. Microb. Ecol. 79, 273–286. doi: 10.3354/ame01834
De Cáceres, M., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
De Cáceres, M., Legendre, P., and Moretti, M. (2010). Improving indicator species analysis by combining groups of sites. Oikos 119, 1674–1684. doi: 10.1111/j.1600-0706.2010.18334.x
Dedysh, S. N., and Yilmaz, P. (2018). Refining the taxonomic structure of the phylum Acidobacteria. Int. J. Syst. Evol. Microbiol. 68, 3796–3806. doi: 10.1099/ijsem.0.003062
Deming, J. W., and Baross, J. A. (1993). “The early diagenesis of organic matter: bacterial activity,” in Organic Geochemistry, eds M. T. J. Murphy and G. Eglinton (Berlin: Springer), 119–144.
Dogs, M., Wemheuer, B., Wolter, L., Bergen, N., Daniel, R., Simon, M., et al. (2017). Rhodobacteraceae on the marine brown alga Fucus spiralis are abundant and show physiological adaptation to an epiphytic lifestyle. Syst. Appl. Microbiol. 40, 370–382. doi: 10.1016/j.syapm.2017.05.006
Doxaran, D., Devred, E., and Babin, M. (2015). A 50% increase in the amount of terrestrial particles delivered by the Mackenzie River into the Beaufort Sea (Canadian Arctic Ocean) over the last 10 years. Biogeosci. Discuss. 12, 305–344. doi: 10.5194/bgd-12-305-2015
Dubinsky, E. A., Esmaili, L., Hulls, J. R., Cao, Y., Griffith, J. F., and Andersen, G. L. (2012). Application of phylogenetic microarray analysis to discriminate sources of fecal pollution. Environ. Sci. Technol. 46, 4340–4347. doi: 10.1021/es2040366
Dunton, K. H., Weingartner, T., and Carmack, E. C. (2006). The nearshore western Beaufort Sea ecosystem: circulation and importance of terrestrial carbon in arctic coastal food webs. Prog. Oceanogr. 71, 362–378. doi: 10.1016/j.pocean.2006.09.011
Durbin, A. M., and Teske, A. (2011). Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ. Microbiol. 13, 3219–3234. doi: 10.1111/j.1462-2920.2011.02544.x
Dyksma, S., Lenk, S., Sawicka, J. E., and Mußmann, M. (2018). Uncultured gammaproteobacteria and desulfobacteraceae account for major acetate assimilation in a Coastal Marine Sediment. Front. Microbiol. 9:3124. doi: 10.3389/fmicb.2018.03124
Edgar, R. C. (2018). Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 34, 2371–2375. doi: 10.1093/bioinformatics/bty113
Eert, J., Meisterhans, G., Michel, C., Niemi, A., Reist, J., and Williams, W. J. (2015). Physical, Chemical and Biological Oceanographic Data from the Beaufort Regional Environmental Assessment: Marine Fishes Project, August- September 2012. Ottawa: Fisheries and Oceans Canada.
Evans, P. N., Parks, D. H., Chadwick, G. L., Robbins, S. J., Orphan, V. J., Golding, S. D., et al. (2015). Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350, 434–438. doi: 10.1126/science.aac7745
Forest, A., Bélanger, S., Sampei, M., Sasaki, H., Lalande, C., and Fortier, L. (2010). Three-year assessment of particulate organic carbon fluxes in Amundsen Gulf (Beaufort Sea): satellite observations and sediment trap measurements. Deep. Res. Part I Oceanogr. Res. Pap. 57, 125–142. doi: 10.1016/j.dsr.2009.10.002
Fry, J. C., Webster, G., Cragg, B. A., Weightman, A. J., and Parkes, R. J. (2006). Analysis of DGGE profiles to explore the relationship between prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiol. Ecol. 58, 86–98. doi: 10.1111/j.1574-6941.2006.00144.x
Fuhrman, J. A. (2009). Microbial community structure and its functional implications. Nature 459, 193–199. doi: 10.1038/nature08058
Fuhrman, J. A., Cram, J. A., and Needham, D. M. (2015). Marine microbial community dynamics and their ecological interpretation. Nat. Rev. Microbiol. 13, 133–146. doi: 10.1038/nrmicro3417
Fujii, T., Sugino, H., Rouse, J. D., and Furukawa, K. (2002). Characterization of the microbial community in an anaerobic ammonium-oxidizing biofilm cultured on a nonwoven biomass carrier. J. Biosci. Bioeng. 94, 412–418. doi: 10.1016/S1389-1723(02)80218-3
Galand, P. E., Potvin, M., Casamayor, E. O., and Lovejoy, C. (2010). Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J. 4, 564–576. doi: 10.1038/ismej.2009.134
Gamboa, A., Montero-Serrano, J.-C., St-Onge, G., Rochon, A., and Desiage, P.-A. (2017). Mineralogical, geochemical, and magnetic signatures of surface sediments from the Canadian Beaufort Shelf and Amundsen Gulf (Canadian Arctic). Geochem. Geophys. Geosyst. 18, 488–512. doi: 10.1002/2016GC006477
Garrity, G. M., Bell, J. A., and Lilburn, T. (2015). “Thiotrichaceae fam. nov,” in Bergey’s Manual of Systematics of Archaea and Bacteria, eds S. B. Kim and M. Goodfellow (Chichester, UK: John Wiley & Sons, Ltd).
Glöckner, F. O., Yilmaz, P., Quast, C., Gerken, J., Beccati, A., Ciuprina, A., et al. (2017). 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 261, 169–176. doi: 10.1016/j.jbiotec.2017.06.1198
Goñi, M. A., O’Connor, A. E., Kuzyk, Z. Z., Yunker, M. B., Gobeil, C., and Macdonald, R. W. (2013). Distribution and sources of organic matter in surface marine sediments across the North American Arctic margin. J. Geophys. Res. Ocean. 118, 4017–4035. doi: 10.1002/jgrc.20286
Gonzalez, J. M., Fernandez-Gomez, B., Fernandez-Guerra, A., Gomez-Consarnau, L., Sanchez, O., Coll-Llado, M., et al. (2008). Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria). Proc. Natl. Acad. Sci. U.S.A. 105, 8724–8729. doi: 10.1073/pnas.0712027105
Gosink, J. J., Herwig, R. P., and Staley, J. T. (1997). Octadecabacter arcticus gen. nov., sp. nov., and O. antarcticus, sp. nov., nonpigmented, psychrophilic gas vacuolate bacteria from polar sea ice and water. Syst. Appl. Microbiol. 20, 356–365. doi: 10.1016/S0723-2020(97)80003-3
Gosink, J. J., Woese, C. R., and Staley, J. T. (1998). Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of Flectobacillus glomera. Int. J. Syst. Bacteriol. 48, 223–235. doi: 10.1099/00207713-48-1-223
Grahn, N., Olofsson, M., Ellnebo-Svedlund, K., Monstein, H.-J., and Jonasson, J. (2003). Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol. Lett. 219, 87–91. doi: 10.1016/S0378-1097(02)01190-4
Grebmeier, J. M. (2012). Shifting patterns of life in the pacific arctic and sub-arctic seas. Ann. Rev. Mar. Sci. 4, 63–78. doi: 10.1146/annurev-marine-120710-100926
Grossart, H.-P., Levold, F., Allgaier, M., Simon, M., and Brinkhoff, T. (2005). Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 7, 860–873. doi: 10.1111/j.1462-2920.2005.00759.x
Hamdan, L. J., Coffin, R. B., Sikaroodi, M., Greinert, J., Treude, T., and Gillevet, P. M. (2013). Ocean currents shape the microbiome of Arctic marine sediments. ISME J. 7, 685–696. doi: 10.1038/ismej.2012.143
Han, Y., and Perner, M. (2015). The globally widespread genus Sulfurimonas: versatile energy metabolisms and adaptations to redox clines. Front. Microbiol. 6:989. doi: 10.3389/fmicb.2015.00989
Head, I. M., Jones, D. M., and Röling, W. F. M. (2006). Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4, 173–182. doi: 10.1038/nrmicro1348
Helbert, W. (2017). Marine polysaccharide sulfatases. Front. Mar. Sci. 4:6. doi: 10.3389/fmars.2017.00006
Hoffmann, K., Hassenrück, C., Salman-Carvalho, V., Holtappels, M., and Bienhold, C. (2017). Response of bacterial communities to different detritus compositions in arctic deep-sea sediments. Front. Microbiol. 8:266. doi: 10.3389/fmicb.2017.00266
Holmes, R. M., Coe, M. T., Fiske, G. J., Gurtovaya, T., McClelland, J. W., Shiklomanov, A. I., et al. (2012). “Climate change impacts on the hydrology and biogeochemistry of arctic rivers,” in Climatic Change and Global Warming of Inland Waters: Impacts and Mitigation for Ecosystems and Societies, eds M. Kumagai, C. R. Goldman, and R. D. Robarts (Chichester, UK: John Wiley & Sons, Ltd), 1–26.
Holmes, R. M., McClelland, J. W., Peterson, B. J., Shiklomanov, I. A., Shiklomanov, A. I., Zhulidov, A. V., et al. (2002). A circumpolar perspective on fluvial sediment flux to the Arctic ocean. Glob. Biogeochem. Cycles 16, 45–41. doi: 10.1029/2001GB001849
Hubert, C., Arnosti, C., Brüchert, V., Loy, A., Vandieken, V., and Jørgensen, B. B. (2010). Thermophilic anaerobes in Arctic marine sediments induced to mineralize complex organic matter at high temperature. Environ. Microbiol. 12, 1089–1104. doi: 10.1111/j.1462-2920.2010.02161.x
Inagaki, F., Suzuki, M., Takai, K., Oida, H., Sakamoto, T., Aoki, K., et al. (2003). Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 69, 7224–7235. doi: 10.1128/AEM.69.12.7224-7235.2003
Jacob, M., Soltwedel, T., Boetius, A., and Ramette, A. (2013). Biogeography of deep-sea benthic bacteria at regional scale (Lter Hausgarten, Fram Strait, Arctic). PLoS One 8:e72779. doi: 10.1371/journal.pone.0072779
Jørgensen, B., Glud, R., and Holby, O. (2005). Oxygen distribution and bioirrigation in Arctic fjord sediments (Svalbard, Barents Sea). Mar. Ecol. Prog. Ser. 292, 85–95. doi: 10.3354/meps292085
Jørgensen, B. B., Findlay, A. J., and Pellerin, A. (2019). The biogeochemical sulfur cycle of marine sediments. Front. Microbiol. 10:849. doi: 10.3389/fmicb.2019.00849
Jorgensen, S. L., Hannisdal, B., Lanzén, A., Baumberger, T., and Flesland, K. (2012). Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl. Acad. Sci. U.S.A. 109, 16764–16765. doi: 10.1594/PANGAEA.786687
Junge, K., Imhoff, F., Staley, T., and Deming, W. (2002). Phylogenetic diversity of numerically important arctic sea-ice bacteria cultured at subzero temperature. Microb. Ecol. 43, 315–328. doi: 10.1007/s00248-001-1026-4
Kêdra, M., Moritz, C., Choy, E. S., David, C., Degen, R., Duerksen, S., et al. (2015). Status and trends in the structure of Arctic benthic food webs. Polar Res. 34:23775. doi: 10.3402/polar.v34.23775
Kirchman, D. L., Hanson, T. E., Cottrell, M. T., and Hamdan, L. J. (2014). Metagenomic analysis of organic matter degradation in methane-rich Arctic Ocean sediments. Limnol. Oceanogr. 59, 548–559. doi: 10.4319/lo.2014.59.2.0548
Kleindienst, S., Herbst, F. A., Stagars, M., Von Netzer, F., Von Bergen, M., Seifert, J., et al. (2014). Diverse sulfate-reducing bacteria of the Desulfosarcina/Desulfococcus clade are the key alkane degraders at marine seeps. ISME J. 8, 2029–2044. doi: 10.1038/ismej.2014.51
Kleindienst, S., Ramette, A., Amann, R., and Knittel, K. (2012). Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments. Environ. Microbiol. 14, 2689–2710. doi: 10.1111/j.1462-2920.2012.02832.x
Koedooder, C., Stock, W., Willems, A., Mangelinckx, S., De Troch, M., Vyverman, W., et al. (2019). Diatom-bacteria interactions modulate the composition and productivity of benthic diatom biofilms. Front. Microbiol. 10:1255. doi: 10.3389/fmicb.2019.01255
Könneke, M., Schubert, D. M., Brown, P. C., Hügler, M., Standfest, S., Schwander, T., et al. (2014). Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc. Natl. Acad. Sci. U.S.A. 111, 8239–8244. doi: 10.1073/pnas.1402028111
Kortsch, S., Primicerio, R., Fossheim, M., Dolgov, A. V., and Aschan, M. (2015). Climate change alters the structure of arctic marine food webs due to poleward shifts of boreal generalists. Proc. R. Soc. B Biol. Sci. 282:20151546. doi: 10.1098/rspb.2015.1546
Kostka, J., Thamdrup, B., Glud, R., and Canfield, D. (1999). Rates and pathways of carbon oxidation in permanently cold Arctic sediments. Mar. Ecol. Prog. Ser. 180, 7–21. doi: 10.3354/meps180007
Kostka, J. E., Prakash, O., Overholt, W. A., Green, S. J., Freyer, G., Canion, A., et al. (2011). Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol. 77, 7962–7974. doi: 10.1128/AEM.05402-11
Kuever, J. (2014a). “The Family Desulfobacteraceae,” in The Prokaryotes, eds A. Balows, H.G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (Berlin: Springer), 45–73. doi: 10.1007/978-3-642-39044-9_266
Kuever, J. (2014b). “The Family Desulfobulbaceae,” in The Prokaryotes, eds A. Balows, H.G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (Berlin: Springer), 75–86. doi: 10.1007/978-3-642-39044-9_267
Kulakov, L. A., McAlister, M. B., Ogden, K. L., Larkin, M. J., and O’Hanlon, J. F. (2002). Analysis of bacteria contaminating ultrapure water in industrial systems. Appl. Environ. Microbiol. 68, 1548–1555. doi: 10.1128/aem.68.4.1548-1555.2002
Landry, Z., Swa, B. K., Herndl, G. J., Stepanauskas, R., and Giovannoni, S. J. (2017). SAR202 genomes from the dark ocean predict pathways for the oxidation of recalcitrant dissolved organic matter. mBio 8:e00413-17. doi: 10.1128/mBio.00413-17
Lansard, B., Mucci, A., Miller, L. A., Macdonald, R. W., and Gratton, Y. (2012). Seasonal variability of water mass distribution in the southeastern Beaufort Sea determined by total alkalinity and δ 18 O. J. Geophys. Res. Ocean 117:JC007299. doi: 10.1029/2011JC007299
Laurence, M., Hatzis, C., and Brash, D. E. (2014). Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PLoS One 9:e97876. doi: 10.1371/journal.pone.0097876
Lee, D. H., Kim, J. H., Lee, Y. M., Stadnitskaia, A., Jin, Y. K., Niemann, H., et al. (2018). Biogeochemical evidence of anaerobic methane oxidation on active submarine mud volcanoes on the continental slope of the Canadian Beaufort Sea. Biogeosciences 15, 7419–7433. doi: 10.5194/bg-15-7419-2018
Lee, Y. M., Jung, Y.-J., Hong, S. G., Kim, J. H., and Lee, H. K. (2014). Diversity and Physiological Characteristics of Culturable Bacteria from Marine Sediments of Ross Sea, Antarctica. Korean J. Microbiol. 50, 119–127. doi: 10.7845/kjm.2014.4014
Lefevre, C. T., and Bazylinski, D. A. (2013). Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol. Mol. Biol. Rev. 77, 497–526. doi: 10.1128/mmbr.00021-13
Li, H., Yu, Y., Luo, W., Zeng, Y., and Chen, B. (2009). Bacterial diversity in surface sediments from the Pacific Arctic Ocean. Extremophiles 13, 233–246. doi: 10.1007/s00792-009-0225-7
Li, H., Zhang, X.-Y., Liu, C., Lin, C.-Y., Xu, Z., Chen, X.-L., et al. (2014). Polaribacter huanghezhanensis sp. nov., isolated from Arctic fjord sediment, and emended description of the genus Polaribacter. Int. J. Syst. Evol. Microbiol. 64, 973–978. doi: 10.1099/ijs.0.056788-0
Li, Y., Liu, Q., Li, C., Dong, Y., Zhang, W., Zhang, W., et al. (2015). Bacterial and archaeal community structures in the Arctic deep-sea sediment. Acta Oceanol. Sin. 34, 93–113. doi: 10.1007/s13131-015-0624-9
Liang, B., Wang, L.-Y., Mbadinga, S. M., Liu, J.-F., Yang, S.-Z., Gu, J.-D., et al. (2015). Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation. AMB Express 5:37. doi: 10.1186/s13568-015-0117-4
Lopez-Fernandez, M., Simone, D., Wu, X., Soler, L., Nilsson, E., Holmfeldt, K., et al. (2018). Metatranscriptomes reveal that all three domains of life are active but are dominated by bacteria in the fennoscandian crystalline granitic continental deep biosphere. mBio 9:e01792-18. doi: 10.1128/mBio.01792-18
Lorenson, T. D., Grienert, J., and Coffin, R. B. (2016). Dissolved methane in the Beaufort Sea and the Arctic Ocean, 1992–2009; sources and atmospheric flux. Limnol. Oceanogr. 61, S300–S323. doi: 10.1002/lno.10457
Losey, N. A., Stevenson, B. S., Busse, H.-J., Damste, J. S. S., Rijpstra, W. I. C., Rudd, S., et al. (2013). Thermoanaerobaculum aquaticum gen. nov., sp. nov., the first cultivated member of Acidobacteria subdivision 23, isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 63, 4149–4157. doi: 10.1099/ijs.0.051425-0
Macdonald, R. W., Solomon, S. M., Cranston, R. E., Welch, H. E., Yunker, M. B., and Gobeil, C. (1998). A sediment and organic carbon budget for the Canadian Beaufort Shelf. Mar. Geol. 144, 255–273. doi: 10.1016/S0025-3227(97)00106-0
Magen, C., Chaillou, G., Crowe, S. A., Mucci, A., Sundby, B., Gao, A., et al. (2010). Origin and fate of particulate organic matter in the southern Beaufort Sea - Amundsen Gulf region, Canadian Arctic. Estuar. Coast. Shelf Sci. 86, 31–41. doi: 10.1016/j.ecss.2009.09.009
Martens-Habbena, W., Qin, W., Horak, R. E. A., Urakawa, H., Schauer, A. J., Moffett, J. W., et al. (2015). The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ. Microbiol. 17, 2261–2274. doi: 10.1111/1462-2920.12677
McBride, M. J. (2014). “The Family Flavobacteriaceae,” in The Prokaryotes, eds A. Balows, H.G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (Berlin: Springer), 643–676.
McIlroy, S. J., Kirkegaard, R. H., Dueholm, M. S., Fernando, E., Karst, S. M., Albertsen, M., et al. (2017). Culture-independent analyses reveal novel anaerolineaceae as abundant primary fermenters in anaerobic digesters treating waste activated sludge. Front. Microbiol. 8:1134. doi: 10.3389/fmicb.2017.01134
McTigue, N. D., Gardner, W. S., Dunton, K. H., and Hardison, A. K. (2016). Biotic and abiotic controls on co-occurring nitrogen cycling processes in shallow Arctic shelf sediments. Nat. Commun. 7:13145. doi: 10.1038/ncomms13145
Miquel, J. C., Gasser, B., Martín, J., Marec, C., Babin, M., Fortier, L., et al. (2015). Downward particle flux and carbon export in the Beaufort Sea, Arctic Ocean; the role of zooplankton. Biogeosciences 12, 5103–5117. doi: 10.5194/bg-12-5103-2015
Morris, E. K., Caruso, T., Buscot, F., Fischer, M., Hancock, C., Maier, T. S., et al. (2014). Choosing and using diversity indices: insights for ecological applications from the German Biodiversity Exploratories. Ecol. Evol. 4, 3514–3524. doi: 10.1002/ece3.1155
Morris, R. M., Rappé, M. S., Urbach, E., Connon, S. A., and Giovannoni, S. J. (2004). Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl. Environ. Microbiol. 70, 2836–2842. doi: 10.1128/AEM.70.5.2836-2842.2004
Müller, A. L., de Rezende, J. R., Hubert, C. R. J., Kjeldsen, K. U., Lagkouvardos, I., Berry, D., et al. (2014). Endospores of thermophilic bacteria as tracers of microbial dispersal by ocean currents. ISME J. 8, 1153–1165. doi: 10.1038/ismej.2013.225
Müller, A. L., Pelikan, C., de Rezende, J. R., Wasmund, K., Putz, M., Glombitza, C., et al. (2018). Bacterial interactions during sequential degradation of cyanobacterial necromass in a sulfidic arctic marine sediment. Environ. Microbiol. 20, 2927–2940. doi: 10.1111/1462-2920.14297
Naidu, A. S., Cooper, L. W., Finney, B. P., Macdonald, R. W., Alexander, C., and Semiletov, I. P. (2000). Organic carbon isotope ratio (δ13C) of Arctic Amerasian Continental shelf sediments. Int. J. Earth Sci. 89, 522–532. doi: 10.1007/s005310000121
Nelson, R. J., Ashjian, C. J., Bluhm, B. A., Conlan, K. E., Gradinger, R. R., Grebmeier, J. M., et al. (2014). “Biodiversity and biogeography of the lower trophic taxa of the pacific arctic region: sensitivities to climate change,” in The Pacific Arctic Region: Ecosystem Status and Trends in a Rapidly Changing Environment, eds W. Masłowski and J. M. Grebmeier (Netherlands: Springer), 269–336.
Niemi, A., Michel, C., Dempsey, M., Eert, J., Reist, J., and Williams, W. J. (2015). Physical, Chemical and Biological Oceanographic Data From the Beaufort Regional Environmental Assessment: Marine Fishes Project, August- September 2013. Ottawa: Fisheries and Oceans Canada.
Nunoura, T., Nishizawa, M., Hirai, M., Shimamura, S., Harnvoravongchai, P., Koide, O., et al. (2018). Microbial diversity in sediments from the bottom of the challenger deep, the mariana trench. Microbes Environ. 33, 186–194. doi: 10.1264/jsme2.ME17194
Obi, C. C., Adebusoye, S. A., Amund, O. O., Ugoji, E. O., Ilori, M. O., Hedman, C. J., et al. (2017). Structural dynamics of microbial communities in polycyclic aromatic hydrocarbon-contaminated tropical estuarine sediments undergoing simulated aerobic biotreatment. Appl. Microbiol. Biotechnol. 101, 4299–4314. doi: 10.1007/s00253-017-8151-6
Offre, P., Spang, A., and Schleper, C. (2013). Archaea in biogeochemical cycles. Annu. Rev. Microbiol. 67, 437–457. doi: 10.1146/annurev-micro-092412-155614
Oksanen, J. (2015). Multivariate Analysis of Ecological Communities in R: Vegan Tutorial. Available online at: http://cc.oulu.fi/~jarioksa/opetus/metodi/vegantutor.pdf (accessed December 13, 2017).
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., Mcglinn, D., et al. (2017). Title Community Ecology Package. Available online at: https://github.com/vegandevs/vegan/issues (accessed December 13, 2017).
Orcutt, B. N., Sylvan, J. B., Knab, N. J., and Edwards, K. J. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422. doi: 10.1128/MMBR.00039-10
Orphan, V. J., Hinrichs, K. U., Ussler, W., Paull, C. K., Taylor, L. T., Sylva, S. P., et al. (2001). Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67, 1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001
Otte, J. M., Blackwell, N., Ruser, R., Kappler, A., Kleindienst, S., and Schmidt, C. (2019). N2O formation by nitrite-induced (chemo)denitrification in coastal marine sediment. Sci. Rep. 9:10691. doi: 10.1038/s41598-019-47172-x
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Park, B. J., Park, S. J., Yoon, D. N., Schouten, S., Damsté, J. S. S., and Rhee, S. K. (2010). Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl. Environ. Microbiol. 76, 7575–7587. doi: 10.1128/AEM.01478-10
Park, S. J., Kim, J. G., Jung, M. Y., Kim, S. J., Cha, I. T., Ghai, R., et al. (2012). Draft genome sequence of an ammonia-oxidizing archaeon, “Candidatus Nitrosopumilus sediminis” AR2, from svalbard in the arctic circle. J. Bacteriol. 194, 6946–6947. doi: 10.1128/JB.01869-12
Paull, C., Dallimore, S., Hughes-Clarke, J., Blasco, S., Lundsten, E., Iii, W. U., et al. (2011). Tracking the Decomposition of Submarine Permafrost and Gas Hydrate Under the Shelf and Slope of the Beaufort Sea. Moss Landing, CA: Monterey Bay Aquarium Research Institute (MBARI).
Paull, C. K., Dallimore, S. R., Caress, D. W., Gwiazda, R., Melling, H., Riedel, M., et al. (2015). Active mud volcanoes on the continental slope of the Canadian Beaufort Sea. Geochem. Geophys. Geosyst. 16, 3160–3181. doi: 10.1002/2015GC005928
Penton, C. R., Devol, A. H., and Tiedje, J. M. (2006). Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl. Environ. Microbiol. 72, 6829–6832. doi: 10.1128/AEM.01254-06
Petro, C., Jochum, L. M., Schreiber, L., Marshall, I. P. G., Schramm, A., and Kjeldsen, K. U. (2019). Single-cell amplified genomes of two uncultivated members of the deltaproteobacterial SEEP-SRB1 clade, isolated from marine sediment. Mar. Genomics 46, 66–69. doi: 10.1016/j.margen.2019.01.004
Pickart, R. S. (2004). Shelfbreak circulation in the Alaskan Beaufort Sea: mean structure and variability. J. Geophys. Res. C Ocean. 109, 1–14. doi: 10.1029/2003JC001912
Pjevac, P., Kamyshny, A., Dyksma, S., and Mußmann, M. (2014). Microbial consumption of zero-valence sulfur in marine benthic habitats. Environ. Microbiol. 16, 3416–3430. doi: 10.1111/1462-2920.12410
Probandt, D., Knittel, K., Tegetmeyer, H. E., Ahmerkamp, S., Holtappels, M., and Amann, R. (2017). Permeability shapes bacterial communities in sublittoral surface sediments. Environ. Microbiol. 19, 1584–1599. doi: 10.1111/1462-2920.13676
Qin, W., Amin, S. A., Martens-Habbena, W., Walker, C. B., Urakawa, H., Devol, A. H., et al. (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc. Natl. Acad. Sci. U.S.A. 111, 12504–12509. doi: 10.1073/pnas.1324115111
Qin, W., Martens-Habbena, W., Kobelt, J. N., and Stahl, D. A. (2016). “Candidatus nitrosopumilales,” in Bergey’s Manual of Systematics of Archaea and Bacteria, eds S. B. Kim and M. Goodfellow (Chichester, UK: John Wiley & Sons, Ltd), 1–2.
R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rachold, V., Eicken, H., Gordeev, V. V., Grigoriev, M. N., and Hubberten, H. (2004). The organic carbon cycle in the Arctic Ocean. Org. Carbon Cycle Arct. Ocean 363, 33–55. doi: 10.1007/978-3-642-18912-8
Rapp, J. Z., Fernández-Méndez, M., Bienhold, C., and Boetius, A. (2018). Effects of Ice-Algal Aggregate Export on the Connectivity of Bacterial Communities in the Central Arctic Ocean. Front. Microbiol. 9:1035. doi: 10.3389/fmicb.2018.01035
Ravenschlag, K., Sahm, K., and Amann, R. (2001). Quantitative molecular analysis of the microbial community in marine arctic sediments (svalbard) quantitative molecular analysis of the microbial community in marine arctic sediments (Svalbard). Appl. Environ. Microbiol. 67, 387–395. doi: 10.1128/AEM.67.1.387
Ravenschlag, K., Sahm, K., Knoblauch, C., Jørgensen, B. B., and Amann, R. (2000). Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine arctic sediments. Appl. Environ. Microbiol. 66, 3592–3602. doi: 10.1128/aem.66.8.3592-3602.2000
Renaud, P. E., Wallhead, P., Kotta, J., Włodarska-Kowalczuk, M., Bellerby, R. G. J., Rätsep, M., et al. (2019). Arctic Sensitivity? Suitable habitat for benthic taxa is surprisingly robust to climate change. Front. Mar. Sci. 6:538. doi: 10.3389/fmars.2019.00538
Retelletti Brogi, S., Kim, J.-H., Ryu, J.-S., Jin, Y. K., Lee, Y. K., and Hur, J. (2019). Exploring sediment porewater dissolved organic matter (DOM) in a mud volcano: clues of a thermogenic DOM source from fluorescence spectroscopy. Mar. Chem. 211:9. doi: 10.1016/J.MARCHEM.2019.03.009
Roussel, E. G., Sauvadet, A.-L., Chaduteau, C., Fouquet, Y., Charlou, J.-L., Prieur, D., et al. (2009). Archaeal communities associated with shallow to deep subseafloor sediments of the New Caledonia Basin. Environ. Microbiol. 11, 2446–2462. doi: 10.1111/j.1462-2920.2009.01976.x
Ruff, S. E., Biddle, J. F., Teske, A. P., Knittel, K., Boetius, A., and Ramette, A. (2015). Global dispersion and local diversification of the methane seep microbiome. Proc. Natl. Acad. Sci. U.S.A. 112, 4015–4020. doi: 10.1073/pnas.1421865112
Rysgaard, S., Glud, R. N., Risgaard-Petersen, N., and Dalsgaard, T. (2004). Denitrification and anammox activity in Arctic marine sediments. Limnol. Oceanogr. 49, 1493–1502. doi: 10.4319/lo.2004.49.5.1493
Salter, S. J., Cox, M. J., Turek, E. M., Calus, S. T., Cookson, W. O., Moffatt, M. F., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12:87. doi: 10.1186/s12915-014-0087-z
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schreiber, L., Holler, T., Knittel, K., Meyerdierks, A., and Amann, R. (2010). Identification of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade. Environ. Microbiol. 12, 2327–2340. doi: 10.1111/j.1462-2920.2010.02275.x
Sheik, C. S., Jain, S., and Dick, G. J. (2014). Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ. Microbiol. 16, 304–317. doi: 10.1111/1462-2920.12165
Siegert, M., Krüger, M., Teichert, B., Wiedicke, M., and Schippers, A. (2011). Anaerobic oxidation of methane at a marine methane seep in a forearc sediment basin off sumatra, Indian Ocean. Front. Microbiol. 2:249. doi: 10.3389/fmicb.2011.00249
Simpson, K. G., Tremblay, J. -É, Gratton, Y., and Price, N. M. (2008). An annual study of inorganic and organic nitrogen and phosphorus and silicic acid in the southeastern Beaufort Sea. J. Geophys. Res. 113:C07016. doi: 10.1029/2007JC004462
Sinkko, H., Lukkari, K., Sihvonen, L. M., Sivonen, K., Leivuori, M., Rantanen, M., et al. (2013). Bacteria contribute to sediment nutrient release and reflect progressed eutrophication-driven hypoxia in an organic-rich continental Sea. PLoS One 8:e67061. doi: 10.1371/journal.pone.0067061
Smoot, C. A., and Hopcroft, R. R. (2017). Depth-stratified community structure of Beaufort Sea slope zooplankton and its relations to water masses. J. Plankton Res. 39, 79–91. doi: 10.1093/plankt/fbw087
Sørensen, K. B., and Teske, A. (2006). Stratified communities of active archaea in deep marine subsurface sediments. Appl. Environ. Microbiol. 72, 4596–4603. doi: 10.1128/AEM.00562-06
Sparrow, K. J., Kessler, J. D., Southon, J. R., Garcia-Tigreros, F., Schreiner, K. M., Ruppel, C. D., et al. (2018). Limited contribution of ancient methane to surface waters of the U.S. Beaufort Sea shelf. Sci. Adv. 4:eaao4842. doi: 10.1126/sciadv.aao4842
Stahl, D. A., and de la Torre, J. R. (2012). Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol. 66, 83–101. doi: 10.1146/annurev-micro-092611-150128
Stokes, C. R., Clark, C. D., and Winsborrow, M. C. M. (2006). Subglacial bedform evidence for a major palaeo-ice stream and its retreat phases in Amundsen Gulf, Canadian arctic archipelago. J. Quat. Sci. 21, 399–412. doi: 10.1002/jqs.991
Takeuchi, M., Katayama, T., Yamagishi, T., Hanada, S., Tamaki, H., Kamagata, Y., et al. (2014). Methyloceanibacter caenitepidi gen. nov., sp. nov., a facultatively methylotrophic bacterium isolated from marine sediments near a hydrothermal vent. Int. J. Syst. Evol. Microbiol. 64, 462–468. doi: 10.1099/ijs.0.053397-0
Tanaka, N., Romanenko, L. A., Iino, T., Frolova, G. M., and Mikhailov, V. V. (2011). Cocleimonas flava gen. nov., sp. nov., a gammaproteobacterium isolated from sand snail (Umbonium costatum). Int. J. Syst. Evol. Microbiol. 61, 412–416. doi: 10.1099/ijs.0.020263-0
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Teeling, H., Fuchs, B. M., Bennke, C. M., Krüger, K., Chafee, M., Kappelmann, L., et al. (2016). Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. eLife 5:e11888. doi: 10.7554/eLife.11888
Thamdrup, B., and Dalsgaard, T. (2002). Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl. Environ. Microbiol. 68, 1312–1318. doi: 10.1128/AEM.68.3.1312-1318.2002
Trembath-Reichert, E., Case, D. H., and Orphan, V. J. (2016). Characterization of microbial associations with methanotrophic archaea and sulfate-reducing bacteria through statistical comparison of nested Magneto-FISH enrichments. PeerJ 2016:e1913. doi: 10.7717/peerj.1913
Treude, T., Krause, S., Maltby, J., Dale, A. W., Coffin, R., and Hamdan, L. J. (2014). Sulfate reduction and methane oxidation activity below the sulfate-methane transition zone in Alaskan Beaufort Sea continental margin sediments: implications for deep sulfur cycling. Geochim. Cosmochim. Acta 144, 217–237. doi: 10.1016/J.GCA.2014.08.018
Trivedi, C. B., Lau, G. E., Grasby, S. E., Templeton, A. S., and Spear, J. R. (2018). Low-temperature sulfidic-ice microbial communities, borup fiord pass, canadian high arctic. Front. Microbiol. 9:1622. doi: 10.3389/fmicb.2018.01622
Ullman, W. J., and Aller, R. C. (1982). Diffusion coefficients in nearshore marine sediments. Limnol. Oceanogr. 27, 552–556. doi: 10.4319/lo.1982.27.3.0552
Van de Vossenberg, J., Woebken, D., Maalcke, W. J., Wessels, H. J. C. T., Dutilh, B. E., Kartal, B., et al. (2013). The metagenome of the marine anammox bacterium “Candidatus Scalindua profunda” illustrates the versatility of this globally important nitrogen cycle bacterium. Environ. Microbiol. 15, 1275–1289. doi: 10.1111/j.1462-2920.2012.02774.x
van der Waals, M. J., Atashgahi, S., da Rocha, U. N., van der Zaan, B. M., Smidt, H., and Gerritse, J. (2017). Benzene degradation in a denitrifying biofilm reactor: activity and microbial community composition. Appl. Microbiol. Biotechnol. 101, 5175–5188. doi: 10.1007/s00253-017-8214-8
Vekeman, B., Kerckhof, F.-M., Cremers, G., de Vos, P., Vandamme, P., Boon, N., et al. (2016a). New Methyloceanibacter diversity from North Sea sediments includes methanotroph containing solely the soluble methane monooxygenase. Environ. Microbiol. 18, 4523–4536. doi: 10.1111/1462-2920.13485
Vekeman, B., Speth, D., Wille, J., Cremers, G., De Vos, P., Op den Camp, H. J. M., et al. (2016b). Genome characteristics of two novel type I methanotrophs enriched from north sea sediments containing exclusively a lanthanide-dependent XoxF5-type methanol dehydrogenase. Microb. Ecol. 72, 503–509. doi: 10.1007/s00248-016-0808-7
Vollmers, J., Voget, S., Dietrich, S., Gollnow, K., Smits, M., Meyer, K., et al. (2013). Poles apart: arctic and antarctic octadecabacter strains share high genome plasticity and a new type of xanthorhodopsin. PLoS One 8:e63422. doi: 10.1371/journal.pone.0063422
Waidner, L. A., and Kirchman, D. L. (2007). Aerobic anoxygenic phototrophic bacteria attached to particles in turbid waters of the Delaware and Chesapeake estuaries. Appl. Environ. Microbiol. 73, 3936–3944. doi: 10.1128/AEM.00592-07
Walsh, J. E., Overland, J. E., Groisman, P. Y., and Rudolf, B. (2011). Ongoing climate change in the arctic. Ambio 40, 6–16. doi: 10.1007/s13280-011-0211-z
Walters, W., Hyde, E. R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A., et al. (2016). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15
Wang, L., Yu, M., Liu, Y., Liu, J., Wu, Y., Li, L., et al. (2018). Comparative analyses of the bacterial community of hydrothermal deposits and seafloor sediments across Okinawa Trough. J. Mar. Syst. 180, 162–172. doi: 10.1016/j.jmarsys.2016.11.012
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, X., Yan, Y., and Gao, D. (2018). The threshold of influent ammonium concentration for nitrate over-accumulation in a one-stage deammonification system with granular sludge without aeration. Sci. Total Environ. 634, 843–852. doi: 10.1016/j.scitotenv.2018.04.053
Wasmund, K., Mußmann, M., and Loy, A. (2017). The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ. Microbiol. Rep. 9, 323–344. doi: 10.1111/1758-2229.12538
Weingartner, T., Okkonen, S., and Danielson, S. (2005). Circulation and water property variations in the nearshore Alaskan Beaufort Sea. US Miner. Manag. Serv. Outer Cont. Shelf Study 28, 1–103.
Westcott, S. L., and Schloss, P. D. (2017). OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. mSphere 2:e00073-17. doi: 10.1128/mSphereDirect.00073-17
Williams, T. J., Lefèvre, C. T., Zhao, W., Beveridge, T. J., and Bazylinski, D. A. (2012). Magnetospira thiophila gen. nov., sp. nov., a marine magnetotactic bacterium that represents a novel lineage within the Rhodospirillaceae (Alphaproteobacteria). Int. J. Syst. Evol. Microbiol. 62, 2443–2450. doi: 10.1099/ijs.0.037697-0
Winkel, M., Mitzscherling, J., Overduin, P. P., Horn, F., Winterfeld, M., Rijkers, R., et al. (2018). Anaerobic methanotrophic communities thrive in deep submarine permafrost. Sci. Rep. 8:1291. doi: 10.1038/s41598-018-19505-9
Wu, C. H., Sercu, B., van de Werfhorst, L. C., Wong, J., deSantis, T. Z., Brodie, E. L., et al. (2010). Characterization of coastal urban watershed bacterial communities leads to alternative community-based indicators. PLoS One 5:e0011285. doi: 10.1371/journal.pone.0011285
Wu, H., Liu, M., Zhang, W., and Xiao, T. (2014). Phylogenetic analysis of epibacterial communities on the surfaces of four red macroalgae. J. Ocean Univ. China 13, 1025–1032. doi: 10.1007/s11802-014-2325-y
Xia, Y., Wang, Y., Wang, Y., Chin, F. Y. L., and Zhang, T. (2016). Cellular adhesiveness and cellulolytic capacity in Anaerolineae revealed by omics-based genome interpretation. Biotechnol. Biofuels 9:111. doi: 10.1186/s13068-016-0524-z
Xing, P., Hahnke, R. L., Unfried, F., Markert, S., Huang, S., Barbeyron, T., et al. (2015). Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom. ISME J. 9, 1410–1422. doi: 10.1038/ismej.2014.225
Yamada, T., and Sekiguchi, Y. (2018). “Anaerolineaceae,” in Bergey’s Manual of Systematics of Archaea and Bacteria, eds S. B. Kim and M. Goodfellow (Chichester, UK: John Wiley & Sons, Ltd), 1–5.
Yang, S., Guo, B., Shao, Y., Mohammed, A., Vincent, S., Ashbolt, N. J., et al. (2019). The value of floc and biofilm bacteria for anammox stability when treating ammonia-rich digester sludge thickening lagoon supernatant. Chemosphere 233, 472–481. doi: 10.1016/j.chemosphere.2019.05.287
Yekutieli, D., and Benjamini, Y. (2002). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188. doi: 10.1214/aos/1013699998
Zeng, Y.-X., Yu, Y., Li, H.-R., and Luo, W. (2017). Prokaryotic community composition in arctic kongsfjorden and sub-arctic northern bering sea sediments as revealed by 454 pyrosequencing. Front. Microbiol. 8:2498. doi: 10.3389/fmicb.2017.02498
Zinger, L., Amaral-Zettler, L. A., Fuhrman, J. A., Horner-Devine, M. C., Huse, S. M., Welch, D. B. M., et al. (2011). Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS One 6:e0024570. doi: 10.1371/journal.pone.0024570
Keywords: bacteria, archaea, marine sediment, arctic, microbial ecology, methane, anoxic
Citation: Walker AM, Leigh MB and Mincks SL (2021) Patterns in Benthic Microbial Community Structure Across Environmental Gradients in the Beaufort Sea Shelf and Slope. Front. Microbiol. 12:581124. doi: 10.3389/fmicb.2021.581124
Received: 07 July 2020; Accepted: 05 January 2021;
Published: 28 January 2021.
Edited by:
David A. Walsh, Concordia University, CanadaReviewed by:
Huiluo Cao, The University of Hong Kong, Hong KongCopyright © 2021 Walker, Leigh and Mincks. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexis M. Walker, YW13YWxrZXI4QGFsYXNrYS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.