
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 22 June 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.569119
 Nour Yahfoufi1
Nour Yahfoufi1 Nawal Alsadi1
Nawal Alsadi1 Jean Francois Mallet1
Jean Francois Mallet1 Garima Kulshreshtha1
Garima Kulshreshtha1 Maxwell Hincke1,2
Maxwell Hincke1,2 Nafissa Ismail3
Nafissa Ismail3 Chantal Matar1,4*
Chantal Matar1,4*A novel bacterium (Rouxiella badensis subsp. acadiensis) isolated from the microbiota of wild blueberry fruit was investigated for its immunomodulation capabilities and intestinal morpho-functional aspects. The whole-genome shotgun sequencing of this bacterium led to its new taxonomy and showed absence of pathogenicity genes. Although the bacterium was used for blueberry-fermentation and enhancing its anti-inflammatory effects on neurodegeneration, diabetes, and cancer, no study has assessed the effect of the bacterium on health. In this study, we used several in vitro and in vivo assays to evaluate the interaction of R. badensis subsp. acadiensis with the intestinal mucosa and its impact on the localized immune response. The strain antibiotic susceptibility has been investigated as well as its tolerance to gastric and intestinal environment and ability to attach to human intestinal epithelial cells (Caco-2 and HT-29). In addition, Balb/c mice were used to explore the immune-modulatory characteristics of the live bacterium at the intestinal level and its impact on the morpho-functional aspects of the intestinal mucosa. In vitro assays revealed the ability of R. badensis subsp. acadiensis to survive the gastric and intestinal simulated conditions and to satisfactorily adhere to the human intestinal epithelial cells. The bacterium was shown to be sensitive to an array of antibiotics. Immuno-modulation studies with mice orally administered with R. badensis subsp. acadiensis showed a higher number of IgA positive cells in the small intestine, a higher concentration of the anti-inflammatory cytokine IL-10 in the intestinal mucosa, as well as an increase in the number of goblet cells. The anti-inflammatory cytokine miR146a was found to be increased in the ileum and brain. Furthermore, it increases the number of goblet cells which contribute to intestinal barrier integrity. Taken together, our findings reflect the ability of the tested bacterium to modulates the intestinal homeostasis and immune response. Detailed safety unpublished studies and genome data support our finding. The strain Rouxiella badensis subsp. acadiensis has been filed in a provisional patent; a U.S. Provisional Application No. 62/916,921 entitled “Probiotics Composition and Methods.” Future studies are still needed to validate the potential utilization of this strain as functional food and its potential probiotic effect.
The bacterium Rouxiella badensis subsp. acadiensis (R. badensis subsp. acadiensis) explored in this study was previously called Serratia vaccinii (Martin and Matar, 2005). The strain was isolated from the surface of lowbush blueberries (Vaccinium angustifolium Aiton) by inoculation of a Tryptic Soy Broth (TSB) (Difco Laboratories, Detroit, MI, United States) with whole blueberry fruit as described in Martin and Matar (2005). In this study, inocula were prepared from a Research Cell Bank of the bacterium. R. badensis subsp. acadiensis is a Gram negative, catalase positive, facultatively anaerobic coccobacillus. It has a fermentative ability of D-glucose, D-fructose, D-mannose, arbutin, esculin, salicin, saccharose, and D-raffinose and under specific conditions and can ferment mannitol, lactose, and trehalose (Martin, 2003; Martin and Matar, 2005). Functionally, the protective properties of a blueberry-fermented preparation utilizing this bacterium has shown a wide range of anti-inflammatory effects on neurodegeneration, diabetes, and cancer (Vuong et al., 2016, 2007; Nachar et al., 2017; Sánchez-villavicencio et al., 2017). As per published study the bacterium previously referred to as Serratia vaccinii has been deposited under Accession Number 160 103, at the International Depositary Authority of Canada. They referred to it as Serratia based on its 16S rRNA gene sequence associated with biochemical profile and physical properties (Martin and Matar, 2005). As the 16S rRNA gene sequence analysis was shown to be insufficiently robust to distinguish closely related species and new advanced techniques have emerged, a whole-genome shotgun sequencing and assembly of bacterial strain were performed to determine the phylogenetic position of the isolate. These analyses led to the description of a novel Rouxiella badensis subspecies for which we propose the name R. badensis subsp. acadiensis, following the region where it was firstly isolated. The type strain is Canan SV-53. The novel bacterium has been deposited at the International Depositary Authority of Canada under Accession Number 160103, (Antioxydant Producing Bacterium and USES Therefore US 8,617,870). A taxonomy description paper of the strain is now under revision.
Unpublished data related to genome propose a potential probiotic activity of the bacterium since full genome analysis showed that there are no genes found coding for pathogenicity. A US Provisional Application No. 62/916,921 entitled “Probiotics Composition and Methods” has been filed related to the bacterium. Since there are more papers in the process dealing with the bacterium toxicology and safety uses, we have evaluated in this study some in vitro and in vivo characteristics of the Gram negative R badensis subsp. acadiensis and its impact on immunomodulation and intestinal homeostasis and mucosal immunity.
Our aim is to study the bacterium ability to interact with the intestinal mucosa and modulate the immune response in order to determine whether future analyses are worth it to investigate potential probiotic activity. In the future, we will be proposing probiotic activity once the safety studies will be published especially that Rouxiella belongs to the family of Yerciniacae. While recent findings reveal beneficial immunological effects of certain dead bacteria (Vinderola and Reinheimer, 2003; Mottet and Michetti, 2005), most studies emphasize the importance of colonizing the intestinal epithelium to sustain bacterial beneficial effects (Gopal et al., 2001; Kos et al., 2003; Ouwehand and Salminen, 2003; Kumura et al., 2004). Thus, the assessment of bacterial tolerance to human gastric or intestinal juice is of great importance to see if the alive bacterium can reach the intestine (Sanders and Huis In’t Veld, 1999; Shah, 2001). Furthermore, beneficial bacteria can adhere to intestinal mucosal surfaces, thus adherence is a key property that shows the ability of bacteria to avoid elimination, to better colonize the intestinal epithelium and to compete with other microorganisms comprising pathogens for nutrients and attachment sites (Kos et al., 2003). In the host, a complex relationship exists between commensal bacteria, their metabolites, the gut mucosal barrier and the immune and non-immune cells along the intestine (Yahfoufi et al., 2018). The mucosal layer is an important physical barrier; it contains goblet cells which secrete a highly glycosylated mucin and create a net-like layer playing a key role in barrier maintenance. Moreover, they can produce anti-microbial proteins, chemokines, and cytokines with major role in innate immunity. Oral administration of probiotics strengthens the intestinal barrier integrity: they can influence the gut barrier by numerous mechanisms including modulation of mucus production, enhancement of tight junctions, induction of IgA and modulation of the immune response (Hemaiswarya et al., 2013; Scully et al., 2013). Moreover bacteria can modulate proinflammatory and anti-inflammatory cytokines (Ren et al., 2013; Boonma et al., 2014; Elian et al., 2015). For example, Lacticaseibacillus rhamnosus, Bifidobacterium longum, Lactobacilus acidophilus, Lactobacillus ruteri, Lacticaseibacillus paracasei, are able to modulate the T cells response by up-regulating T reg cells and IL-10 levels in intestinal contents of mice by indirectly involving short-chain fatty acids (SCFAs; Atarashi et al., 2013) which are produced by anaerobic bacterial fermentation of prebiotics (Yahfoufi et al., 2018).
Secretory IgA is of fundamental importance in host mucosal protection against mucosal pathogens (Perdigon et al., 2001; Perdigón et al., 2002). Numerous studies have reported the role of IgA+ cells in maintaining homeostasis of the intestine (Vinderola G. et al., 2005; Galdeano et al., 2007; Mallet et al., 2014). Hence the importance to investigate the effect of R. badensis subsp. Acadiensis on the number of IgA positive cells in Peyer’s patches and in the lamina propria as well as on the production of secretory IgA.
Furthermore, small non-coding RNAs regulate gene expression post-transcriptionally (Zhou et al., 2017). miRNAs have been shown to regulate various biological processes involved in inflammation, immune homeostasis and intestinal integrity (Gaudet et al., 2017). Certain miRNAs could initiate an anti-inflammatory response such as miR145 and miR146a while other miRNAs play a role in proinflammatory response and some have mixed immunomodulatory effects (Bermudez-Humaran and Langella, 2017). Bacteria like probiotics were found able to modulate miRNAs (O’Connell et al., 2012).
Finally, it is important to address concerns that have been raised in the use of some bacterial strains that carry antibiotic resistance genes themselves, as they may pass the antibiotic resistance genes to pathogenic bacteria through horizontal gene transfer. Therefore, antibiotic resistance is often studied in bacterial characterization which can present a challenge to their safe usage. Screening bacterial strains for antibiotic resistance is important to ensure their safety and reduce their potential in contributing to the spread of antibiotic resistance genes (Imperial and Ibana, 2016).
Although the most commonly used bacteria for their potential health benefits are Gram-positive (G+) bacteria such as lactic-acid producing bacteria and other that belong to the genus Bifidobacterium, some Gram-negative (G-) bacteria like Escherichia coli Nissle 1917(EcN) can exert health efficacy (Saad et al., 2013). When comparing G- and G+ probiotics, G- probiotics show higher activity in reducing the levels of inflammatory mediators during enteric infections (Cross et al., 2004). Referring to existing studies, G- induce higher IL10-responses than G+ probiotics resulting in differential induction of antibody responses. G- EcN has more potent immuno-stimulatory effects than G+ Lactobacillus rhamnosus GG in term of inducing mucosal and systemic B cells and results in higher production of IgA at the small intestinal level (Hessle et al., 2000; Kandasamy et al., 2016). The microbe-associated molecular patterns differ between G- and G+ probiotics and can explain the difference in induction of IL-10. In addition, higher antibody response induced by G- when compared to G+ probiotics can be due to different modulation of cytokine milieu (Karlsson et al., 2002; Smits et al., 2004). It is well known that variations in immunomodulation characteristics are strain-dependent even within G+ probiotics (Christensen et al., 2002).
Therefore, we investigated the resistance of this bacterium to commonly used antibiotics, its tolerance to simulated gastric and pancreatic juices, and ability to adhere to intestinal epithelial cells. In vivo, the bacterium was administered to Balb/c mice: IL-6, IL-10, and secretory IgA concentrations in mice intestinal fluid were measured, in addition to the abundance of IL-10 positive cells in the ileum. Moreover, we investigated the morpho-functional changes in the small intestine, and the expression of selected anti-inflammatory miRNAs (miR145 and miR146a).
Stock cultures of the bacterium were maintained at −80°C in Tryptic Soy Broth (Difco Laboratories, Detroit, MI, United States) supplemented with 30% (v/v) glycerol. The bacterium was grown in Tryptic Soy Agar (TSA) (Difco Laboratories, Detroit, MI, United States) or Tryptic Soy Broth (TSB) (Difco Laboratories, Detroit, MI, United States) at 30°C for 20 h.
Overnight bacterial culture inocula were diluted 1:50 in fresh TSB and grown until the optical density at 600 nm reached 0.2 (∼108 CFUs/ml). Thereafter, the standardized inoculum (100 μl, OD600 = 0.2–108 CFUs/ml) was spread uniformly on TSA. The antibiotic sensi-disks (BD BBL, Sensi-Disc, Becton, Dickinson and Company, United States) used were the following:
(ampicillin:10 μg, Tetracyclin: 10 μg, streptomycin: 10 μg, chloramphenicol: 30 μg, erythromycin: 15 μg or neomycin: 5 μg). Blank disks used as negative control were obtained as well from (BD BBL, Sensi-Disc, Becton, Dickinson and Company, United States). While ciprofloxacin 5 μg, trimethoprim-sulfamethoxazole 25 μg were obtained from Oxoid, ON, Canada. Antibiotic disks or blank disks were placed at the center of the TSA plate (one disk per plate). The plates were incubated at 37°C until even growth of the bacterium was observed. The zones of growth inhibition were measured using a Vernier caliper to categorize the strains as susceptible or resistant to the antibiotic. Experiments were performed three times in duplicates.
Simulated gastric juices were prepared by suspending pepsin (Sigma, Saint Louis, MO, United States) to a final concentration of 3 g/l in sterile NaCl (Sigma, Saint Louis, MO, United States) 0.5% weight to volume and adjusting the pH to 2.0, 3.0 or 4.0 with concentrated HCl or sterile 0.1 mol/l NaOH (Huang and Adams, 2004).
Simulated pancreatic juice was prepared by suspending porcine pancreatin (Sigma, Saint Louis, MO, United States) in sterile saline to a final concentration of 1 g/l, with 0.45% bile salts (Sigma, Saint Louis, MO, United States), and adjusting the pH to 7 or 8 with concentrated HCl or sterile 0.1 mol/l NaOH (Huang and Adams, 2004).
A volume of bacterial inoculum corresponding to 108 to 109 CFU/ml from an overnight culture was subjected to low-speed centrifugation 5000 × g for 5 min and the pellet was washed two times in 1X PBS (137 mM NaCl 2.7 mM KCl 10 mM Na2HPO4, 1.8mM mM KH2PO4) pH7. The starting count was determined by standard plate count prior to assay transit tolerance. When screening for gastric or intestinal transit tolerance, 1 ml of simulated gastric juice (pH 2, 3, or 4) or 1 ml of simulated pancreatic juice (pH 7 or 8) and 300 μl of NaCl (0.5% W/V) were added to washed bacterial suspension in 2 ml screw cap sterile microcentrifuge tubes. The preparations were vortexed at low speed for 10 s and incubated at 37°C. When testing for gastric transit tolerance, aliquots of 100 μl were collected after 60, 90, and 180 min for the determination of total viable counts; while aliquots of 100 μl were collected after 60, 90, 180, and 240 min when screening for intestinal transit tolerance. To better imitate the passage through the gastro-intestinal tract, bacterial inoculum exposed to simulated gastric juice (pH 2 or 4) for 180 min was centrifuged and re-exposed to simulated pancreatic juice (pH 7 or 8) for 240 min and bacterial count was done after collecting aliquots of 100 μl. Each 100 μl was serially diluted, spread on TSA plates, and incubated at 30°C for 72 h, followed by determination of CFU count. Results are expressed as Log10 of the CFU/ml.
Caco-2 cells (ATCC-HTB-37) were grown in minimum essential medium alpha (MEMα; Gibco Canada, Burlington, ON, Canada) containing 10% v/v inactivated fetal bovine serum (FBS; Thermo Fisher Scientific), 1% non-essential amino acids (Gibco) and a mixture of antibiotics at a final concentration of 50 IU/ml penicillin and 50 μg/ml streptomycin. Cells were cultured at 37°C in a humidified atmosphere of 5% v/v CO2. For adherence and invasion assays, confluent Caco-2 cells were harvested by trypsinization in 0.01% EDTA. Caco-2 cells were adjusted to 105 cells/ml by counting in a hemocytometer, seeded in a 24-well tissue culture plate and incubated at 37°C in a 5% CO2 humidified atmosphere until a confluent monolayer formed. Prior to adhesion assay, the cell monolayers were washed twice with 1X PBS (pH 7.4-Sigma, Saint Louis, MO, United States) to remove the antibiotics.
HT-29 (ATCC-HTB-38) cell line was maintained in McCoy’s medium (Gibco, United States) supplemented with 10% (v/v) heat inactivated bovine fetal serum and a mixture of antibiotics at a final concentration of 50 IU/ml penicillin, 50 μg/ml streptomycin. Cells were grown at 37°C, 5% CO2. Culture media were changed every two days and the cell line was trypsinized with 0.25% trypsin-EDTA solution (Sigma, Saint Louis, MO, United States). For experiments, 105 cells/ml were seeded in 24-well plates and incubated to confluence. Caco-2 cells were incubated till 15 days old while HT-29 were incubated as per Arboreya et al. to confluence during 13 ± 1 days.
Adhesion assay was explored as per Arboleya et al. (2011); bacterial cultures were harvested by centrifugation, washed twice with 1X PBS buffer (Sigma, Saint Louis, MO, United States) and resuspended in MEMα or McCoy’s medium supplemented with 10% heat inactivated bovine fetal serum without antibiotics. Prior to adhesion assay, the cell monolayers of Caco-2 or HT-29 were washed three times with 1X PBS (pH 7.4-Sigma, Saint Louis, MO, United States) to remove the antibiotics before adding bacterial suspensions. Bacterial cells were inoculated into wells containing confluent monolayers of Caco-2 or HT-29 cells at a targeted multiplicity of infection (MOI) of 100:1. The actual numbers of bacteria in the inoculum added to the monolayers were confirmed retrospectively by serial dilution and plate counting. Bacteria-infected cells were incubated for 4 h at 37°C and 5% CO2 to allow for bacterial adherence. Supernatants were discarded and wells were gently washed three times with 1X PBS buffer to remove the non-attached bacteria. After gentle scraping of cells, the bacterial counts were carried out by plating on TSA to determine the number of adhering bacteria. Serial dilutions were plated and incubated for 48 h at 30°C. The adherence (expressed as a percentage) was calculated by using the ratio of the number of bacterial cells that remained attached to the total number of bacterial cells added initially to each well.
Six to eight-week-old Balb/c female mice weighing 20–25 g were obtained from Charles River (Montreal, Canada). A total of 20 mice were divided into experimental and control groups consisting of 10 mice per group for all experiments except for the qPCR of brain and ileum (21 mice were used further details are provided below). Pairs of mice were housed together in plastic cages kept in a controlled atmosphere (temperature 22 ± 2°C) with a 12 h light/dark cycle. Mice received 108 CFU of R. badensis subsp. acadiensis/mouse by gavage for 7 consecutive days in 1% sucrose, 1X PBS pH 7.4 (sigma). Control mice received the same volume of 1% sucrose in 1X PBS instead. All mice received simultaneously a conventional balanced diet ad libitum and water. Test and control animals were sacrificed after 7 days of probiotic administration. All experimental procedures were approved by the Animal Care Committee of the University of Ottawa (protocol ME-2403-R3) in accordance with guidelines established by the Canadian Council of Animal Care.
After 7 days, treated and control animals were anesthetized and sacrificed by cervical dislocation; livers were immediately harvested under sterile conditions and homogenized using the Omni TH homogenizer with a 7-mm generator probe (Omni International, Kennesaw, GA, United States) in 5 ml sterile 1X PBS pH 7.4 (Sigma, St. Louis, MO, United States). One milliliter of the liver homogenate was spread onto the surface of MacConkey agar for enterobacteria (Difco Laboratories, Detroit, MI, United States). The plates were then aerobically incubated at 37°C for 48 h. Translocation was considered to have occurred when colonies were observed on the agar plates because the liver is an organ normally devoid of bacteria (Gregory et al., 1996).
The small intestines of sacrificed mice were removed, washed and the ileum parts were collected and fixed overnight in 4% PFA solution (paraformaldehyde). Afterward, the tissues were dehydrated in increasing concentrations of alcohol, cleared in xylene, and embedded in paraffin wax using conventional methods (Alturkistani et al., 2016). Then histological slices of 4μm were prepared from paraffin blocks using a rotation microtome (Leica RM2255 Automated Microtome). Processing, paraffin embedding, and histological slices were performed by The University of Ottawa histology core facility.
The number of IgA and IgG producing (IgA+ and IgG+) cells was determined on histological slices prepared from the ileum using the direct immunofluorescence method. The immunofluorescence test was performed using (α-chain) anti-mouse IgA FITC conjugate (Sigma–Aldrich, St. Louis, MO, United States) or (δ-chain) anti-mouse IgG FITC conjugate (Sigma–Aldrich, St. Louis, MO, United States). Histological sections were deparaffinized and then rehydrated in a graded ethanol series. Sections were incubated at 37°C in two different solutions of xylene (Sigma–Aldrich, St. Louis, MO, United States) successively for 15 min in each solution followed by incubation in graded ethanol series 95, 70, and 40% for 5 min each at 4°C. Deparaffinized histological samples were incubated with the appropriate antibody dilution (1:100 for IgA or 1:50 for IgG) in 1X PBS solution for 30 min at 37°C in the dark. Samples were then washed two times with PBS solution and examined using a fluorescent light microscope. Dry slides were mounted using Fluoromount (Sigma-Aldrich, St. Louis, MO, United States). The results were expressed as the number of IgA+ or IgG+ cells (positive = fluorescent cell) per 10-fields (magnification 100×). Data represent the mean of three readings for each animal.
At the end of the 7 days period, the small intestine was removed and processed for histological preparation as described above. The indirect immunofluorescence method was used to label IL-10 positive cells. After deparaffinization of the histological slices, they were rehydrated in a graded series of ethanol -as per described above- and then incubated for 30 min in a 1% blocking solution of bovine serum albumin (Jackson Immuno Research, West Grove, PA, United States) at room temperature. Histological slices mounted on slides were then incubated for 60 min at 37°C with rabbit anti-mouse IL-10 polyclonal antibodies (Peprotech, Inc., Rocky Hill, NJ, United States) at a dilution 1:50. 1X PBS solution was used for two consecutive washes, then sections were treated for 45 min at 37°C with a 1:100 dilution of a goat anti-rabbit antibody conjugated with FITC (Jackson Immuno Research, West Grove, PA, United States). Finally, sections were washed twice with 1X PBS and examined using a fluorescent light microscope. Dry slides were mounted using Fluoromount (Sigma-Aldrich, St. Louis, MO, United States). The results were expressed as the number of IL10 + cells (positive = fluorescent cell) per 10-fields (magnification 100X).
Different intestinal segments were collected from treated and control animals.
Intestinal segments were fixed in 4% PFA, dehydrated, cleared and embedded in paraffin. Serial sections were cut at 4 μm, deparaffinized in xylene, rehydrated, and stained with hematoxylin and eosin. For staining neutral glycoconjugates or acid glycoconjugate in the intestinal sections we used periodic acid Schiff (PAS) or Alcian blue (AB), pH 2.5. This staining reveals neutral PAS reactive and acid AB-reactive glycoconjugates (Smirnov et al., 2005; Martín et al., 2016). All processing, paraffin embedding and histological staining with hematoxylin, eosin, PAS or AB were performed by The University of Ottawa histology core facility.
The morphology of different intestinal segments was revealed by staining with hematoxylin and eosin for examination by light microscopy. Positive goblet cells for neutral glycoconjugates or acid glycoconjugates were counted. The results were expressed as the number of goblet cells per 10-fields (magnification 100×). Data represent the mean of three readings for each animal.
The small intestine of treated and control animals was flushed with 5 ml of 1X PBS pH7.4 (Sigma, St. Louis, MO, United States) and this fluid was centrifuged at 10,000 g at 4°C for 10 min to separate particulate material. The supernatant was collected and kept frozen at −80°C until use. IL-6 and IL-10 were determined in supernatant using the corresponding mouse ELISA Set (BD OptEIA, BD Biosciences PharMingen, San Diego, CA, United States) with catalog numbers respectively: 555240 and 555252. For IL-6, capture antibody was diluted at 1:250 in coating buffer (BD OptEIA, BD Biosciences PharMingen, San Diego, CA, United States) while the detection antibody was diluted at 1:259 in assay diluent (BD OptEIA, BD Biosciences PharMingen, San Diego, CA, United States). For Il-10, dilutions were respectively 1:250 dilution in coating buffer for capture antibody while 1:250 dilution in assay diluent for the detection antibody. For secretory IgA, the level of IgA was analyzed by DAS-ELISA using affinity-purified goat anti-mouse IgA antibodies (α-chain specific) was added at 1.25 mg/well and horseradish-peroxidase conjugated anti-IgA specific antibodies at 1.25 mg/well (Sigma Chemical Co., St Louis, MO, United States). Absorbance was read at 450 nm within 30 minutes of stopping reaction with wave length correction 570 nm.
Ileum pieces and brains from 11 control: CTR and 10 R. badensis subsp. acadiensis treated mice: R. badensis subsp. acadiensis were snap frozen after collection and kept at −80°C until RNA extraction. Samples were extracted using miRNeasy mini kit (Qiagen, Toronto, ON, Canada). Sample purity were verified with a NanoDrop 2000 (Thermo Scientific, Waltham, MA, United States). Samples underwent a reverse transcription reaction to produce cDNA using individual probes. The cDNA was synthesized by Moloney Murine Leukemia Virus (MMLV) Reverse Transcriptase (Invitrogen, Canada inc.). The expression of target anti-inflammatory miR-145 and miR-146a was measured by RT-qPCR using Taqman primers for hsa-miR-146a (Thermofisher Taqman miRNA assay ID 000468) and hsa-miR-145 (Thermofisher Taqman assay ID 002278) and a FastStart Taq Polymerase (Roche, Mississauga, ON, Canada) in a CFX96 real-time PCR detection system (Bio-RAD, Laboratories, Hercules, CA, United States). Gene expression was normalized to gene reference U6 small non-coding RNA (Thermofisher assay ID 001973).
Results are presented as means ± SEM. Results represent three independent experiments. GraphPad Prism 5.0 software (GraphPad Software Inc., San Diego, CA, United States) was employed to carry out statistical analysis. The statistical significance was determined by applying an unpaired two-tailed Student’s t-test when comparing two groups, and one-way analysis of variance (ANOVA) when comparing more than two groups. A difference was considered significant for p-values <0.05.
Rouxiella badensis subsp. acadiensis demonstrated susceptibility to different classes of antibiotics like β-lactams, fluoroquinolones, aminoglycosides, chloramphenicols, tetracyclines, and trimethoprim-sulfamethoxazole. R. badensis subsp. acadiensis is sensitive to ampicillin 10μg (a β-lactam) with a diameter of inhibition zone (di) greater than 17 mm di = 38.67 ± 0.72), the strain is sensitive to ciprofloxacin 5 μg (fluoroquinolones class) with a di = 39.16 ± 1.57 mm > 21 mm. Streptomycin 10 μg (an aminoglycoside) revealed an inhibition zone with a di = 18.08 ± 0.73 > 15 mm reflecting sensitivity of the strain. The bacterium is as well sensitive strain to: chloramphenicols 30 μg with a di = 23.5 ± 1.38 > 15 mm, tetracyclines 10 μg with di = 27.6 ± 1.1 > 15 mm and trimethoprim-sulfamethoxazole 25 μg with a di = 24.5 ± 2.1 > 16 mm while it possessed intermediate sensitivity to erythromycin15 μg (a macrolide) with a di = 20.1 ± 0.6 mm (Table 1).
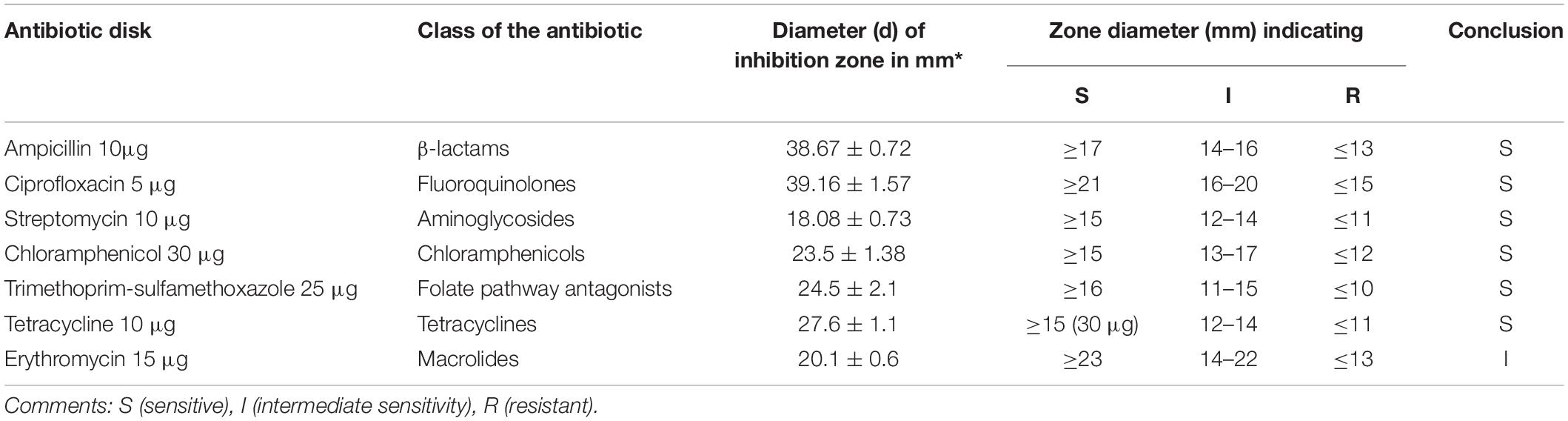
Table 1. Antibiotic susceptibility of R. badensis subsp. acadiensis to different antibiotics using the agar disk diffusion method.
Rouxiella badensis subsp. acadiensis showed high stability when incubated in simulated gastric juice. The total duration of incubation tested was 180 min at 37°C which is equivalent to time the bacteria are usually resident at the stomach level. The studies showed that the bacterium displayed high survival and high tolerance to the gastric juice at pH 3 and 4 (Figures 1B,C). After incubation for 180 min at pH 2 we observed a significant killing effect p < 0.05 (Figure 1A). However, the bacterium was still able to survive at high rate.
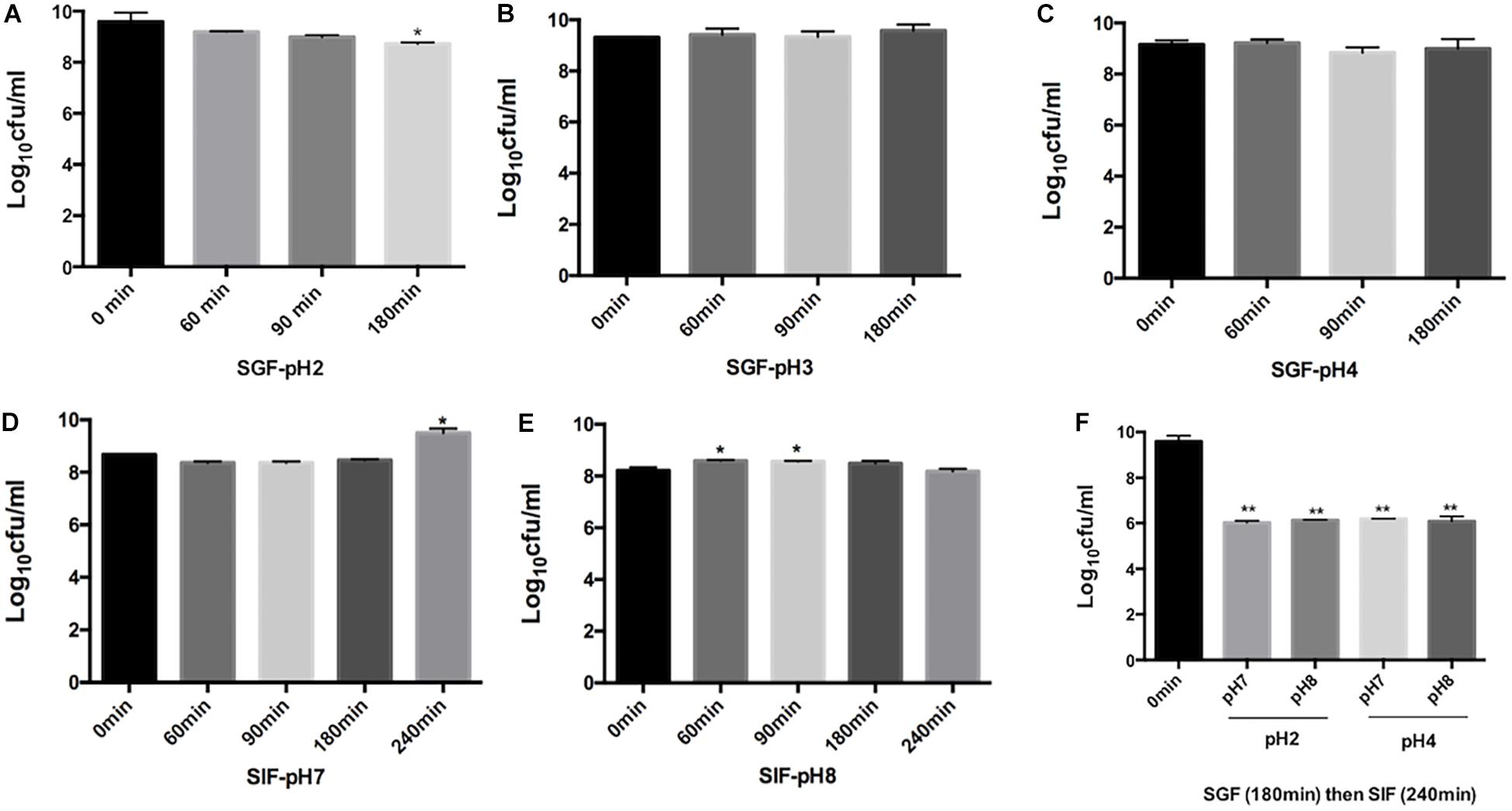
Figure 1. Effect of simulated gastric juices SGF [pH 2 (A) pH 3.0 (B), pH 4.0 (C)] and simulated intestinal juice SIF [pH7 (D), pH 8 (E)] on the viability of R. badensis subsp. acadiensis that was exposed to SGF for 60, 90,and 180 min and to SIF for 60, 90,180, and 240 min. In panel (F) R. badensis subsp. acadiensis was first exposed to SGF for 180 min then to SIF for 240 min. Data represents mean log10CFU/ml ± SEM (n = 3). Significant difference exists if *p < 0.05, **p < 0.01.
Rouxiella badensis subsp. acadiensis tolerated the simulated pancreatic juices as well; survival percentages were tested after exposure for 240 min. The bacterium showed highest survival and viability when incubated at pH 7 for 240 min. At pH 8, bacterial growth was observed after 60- and 90-min incubation at 37°C but after 3 and 4 h incubation there were no difference with t = 0 min. The bacterium maintained high survival rate at simulated pancreatic juice (Figures 1D,E). After passage of the strain through simulated gastric (pH 2 or 4) for 180 min and subsequent intestinal conditions (pH 7 and 8) for 240 min, we observed higher killing ability of the same tested pHs previously tested (Figure 1F).
Adhesion to cells of the intestinal epithelium is another property that probiotics must possess for successful colonization of the human gastrointestinal tract.
The ability of R. badensis subsp. acadiensis to adhere to intestinal epithelial cells was studied in vitro. Human epithelial colorectal adenocarcinoma Caco-2 and HT-29 cells were used. An adhesion rate of 17.8% ± 2.516 on Caco-2 cells, and 19.92% ± 2.078 on HT29 cells was found (Figure 2; Turpin et al., 2012; Argyri et al., 2013).
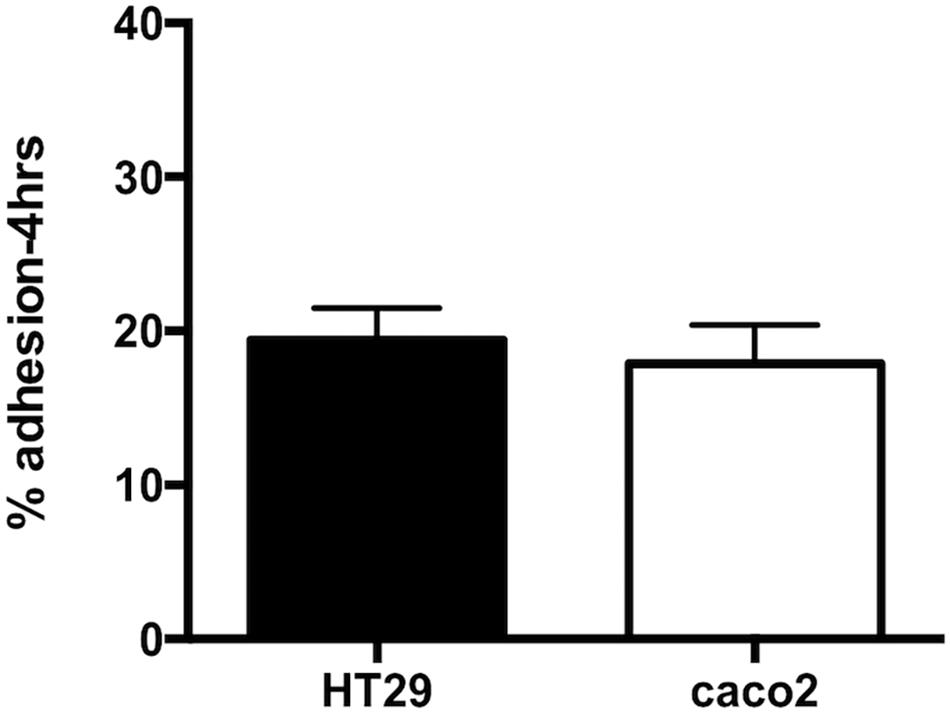
Figure 2. Data represents percentage of adhesion of R. badensis subsp. acadiensis to HT29 and Caco2 cells. Results shown are means of three independent experiments ± SEM.
Oral administration of R. badensis subsp. acadiensis did not cause translocation of Enterobacteriacae to the liver in mice treated with 108CFU/mouse. No bacterial growth on the MacConckey agar plates was observed for extracts of liver samples collected from Balb/c mice fed the bacterium.
The histological study of the small intestine of mice that received R. badensis subsp. acadiensis for 7 consecutive days showed no edema or mucosal atrophy, compared to control mice. The overall morphology of the small intestine did not show any significant changes when compared to control mice (not shown).
The mucosal immunomodulating capacity of R. badensis subsp. acadiensis was assessed in this study by examining its effects on the IgA+ and IgG+ B cell populations and selected cytokines (pro-inflammatory cytokine IL-6 and anti-inflammatory cytokine IL-10) in both the gut mucosa and the intestinal contents.
The number of IgA and IgG producing cells (IgA+ and IgG+) in the lamina propria of the small intestine of mice that received R. badensis subsp. acadiensis showed a significant increase in the number of IgA+ cells in the lamina propria (Figure 3) (96.89 ± 1.933) in comparison to the control group (74.23 ± 2.588). The number of IgG+ cells was unchanged (Figure 4).
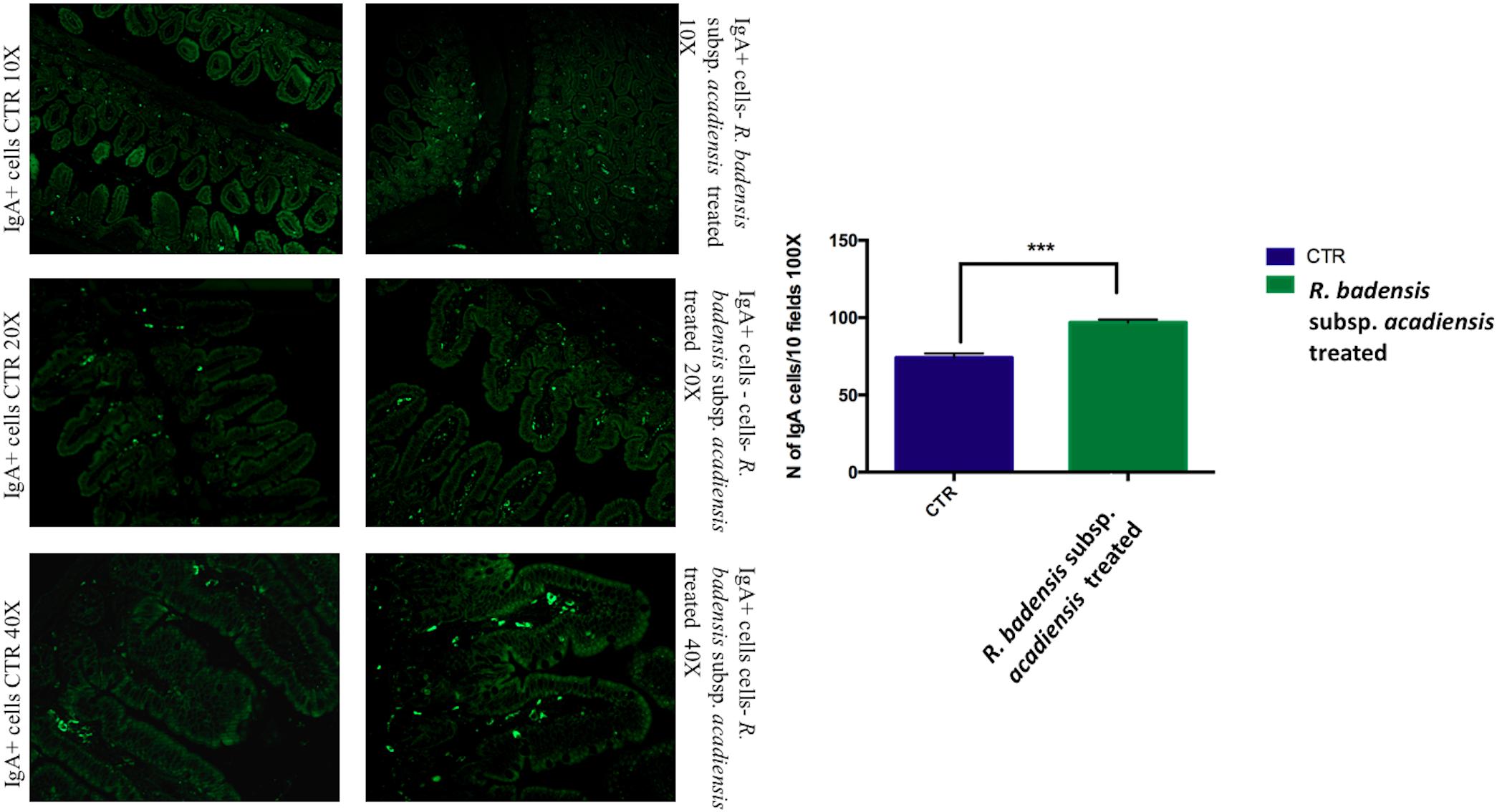
Figure 3. Mean ± SEM of the number of IgA positive cells populations in 10 fields of objective 100X in the ileum of mice fed 1% sucrose (CTR) or R. badensis subsp. acadiensis fed 108CFU/mouse/day for 7 days. Significant difference exists if *p < 0.05, representative tissue photos are taken with 10×, 20×, and 40 × magnifications.
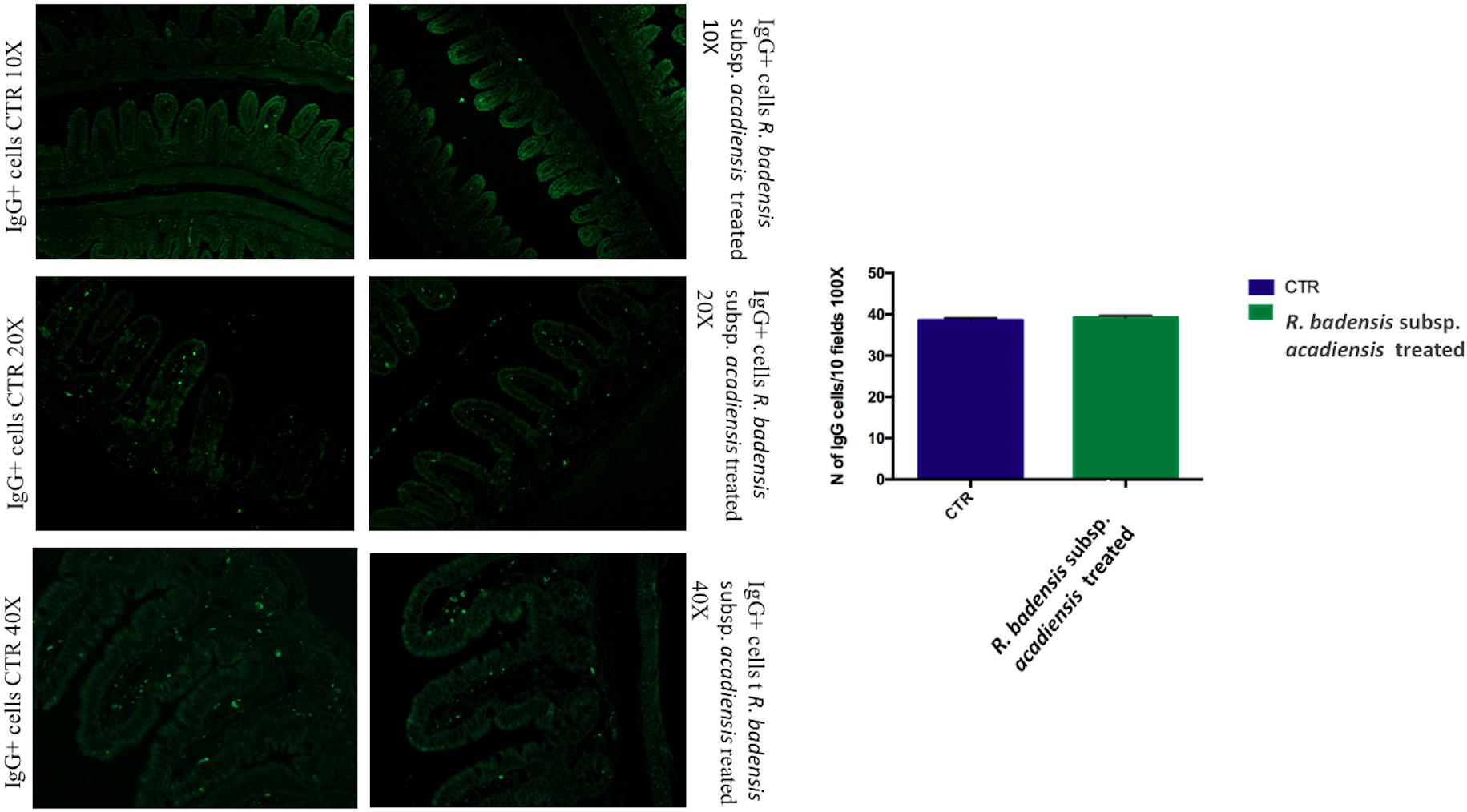
Figure 4. Mean ± SEM of the number of IgG positive cells populations in 10 fields of objective 100X in the ileum of mice fed 1% sucrose (CTR) or R. badensis subsp. acadiensis fed 108CFU/mouse/day for 7 days. Significant difference exists if *p < 0.05, representative tissue photos are taken with 10×, 20×, and 40 × magnifications.
IL10+ cells were shown to be significantly increased in the intestine of mice receiving the probiotic bacterium (74.83 ± 2.065) versus (49.43 ± 1.805) in the control group (Figure 5).
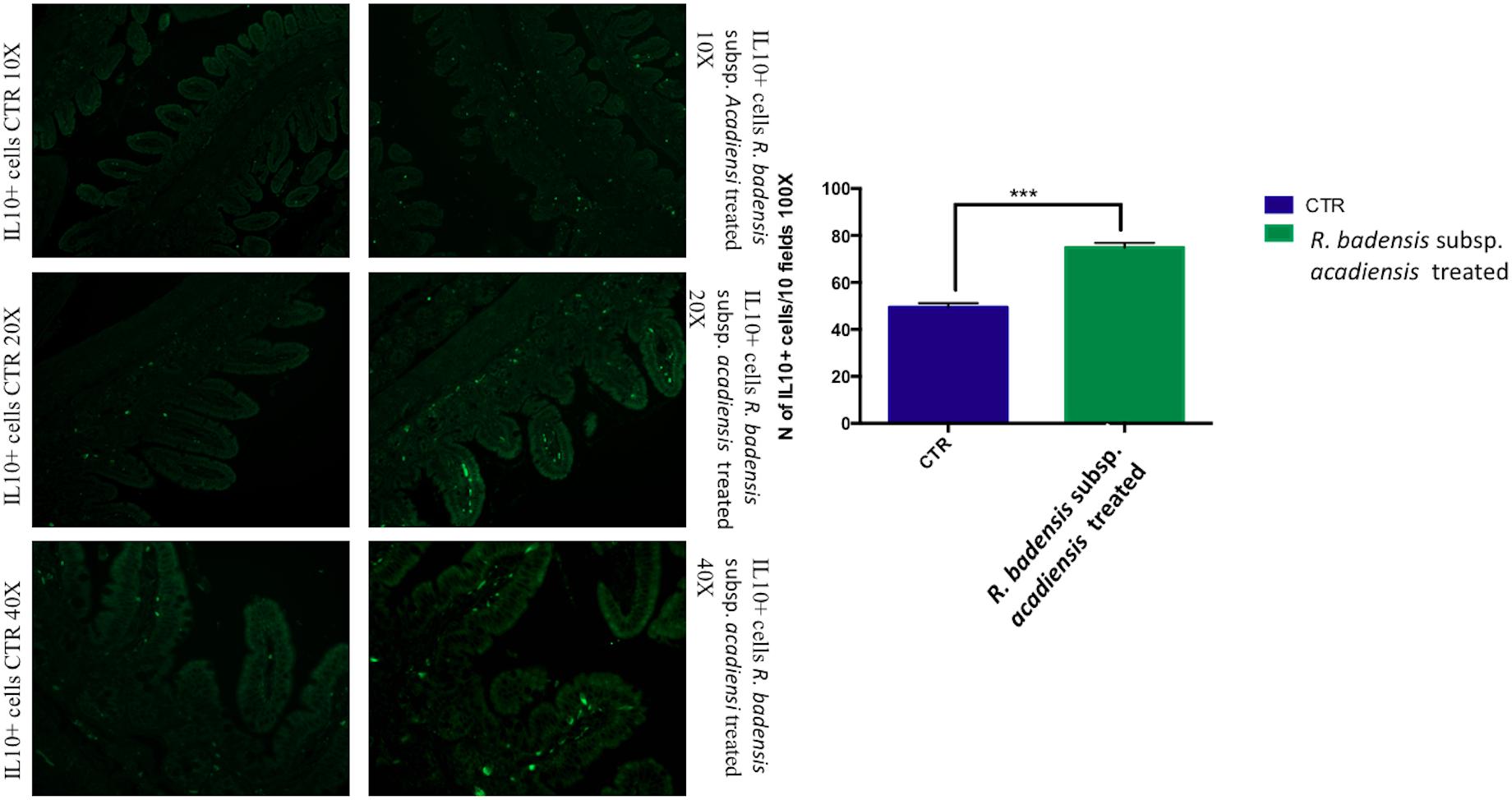
Figure 5. Mean ± SEM of the number of IL10 positive cells populations in 10 fields of objective 100X in the ileum of mice fed 1% sucrose (CTR) or R. badensis subsp. acadiensis fed 108CFU/mouse/day for 7 days. Significant difference exists if *p < 0.05- p is < 0.001 representative tissue photos of IL10 positive cells in the lamina propria are taken with 10×, 20×, and 40 × magnifications.
Considering the multiple functions of the glycoconjugates that make up intestinal mucins, positive goblet cells for neutral glycoconjugates or acid glycoconjugate were counted after staining intestinal sections with periodic acid Schiff (PAS) and AB (Smirnov et al., 2005; Martín et al., 2016). Tissue histology for small intestine of R. badensis subsp. acadiensis fed groups displayed a significant increase in the number of PAS stained goblet cells (149.1 ± 3.167) in comparison to the control group (137.9 ± 3.811) with p = 0.0355(Figure 6). However, AB staining at pH2.5 did not show any increase in the number of goblet cells positive for acid glycoconjugates (Figure 7).
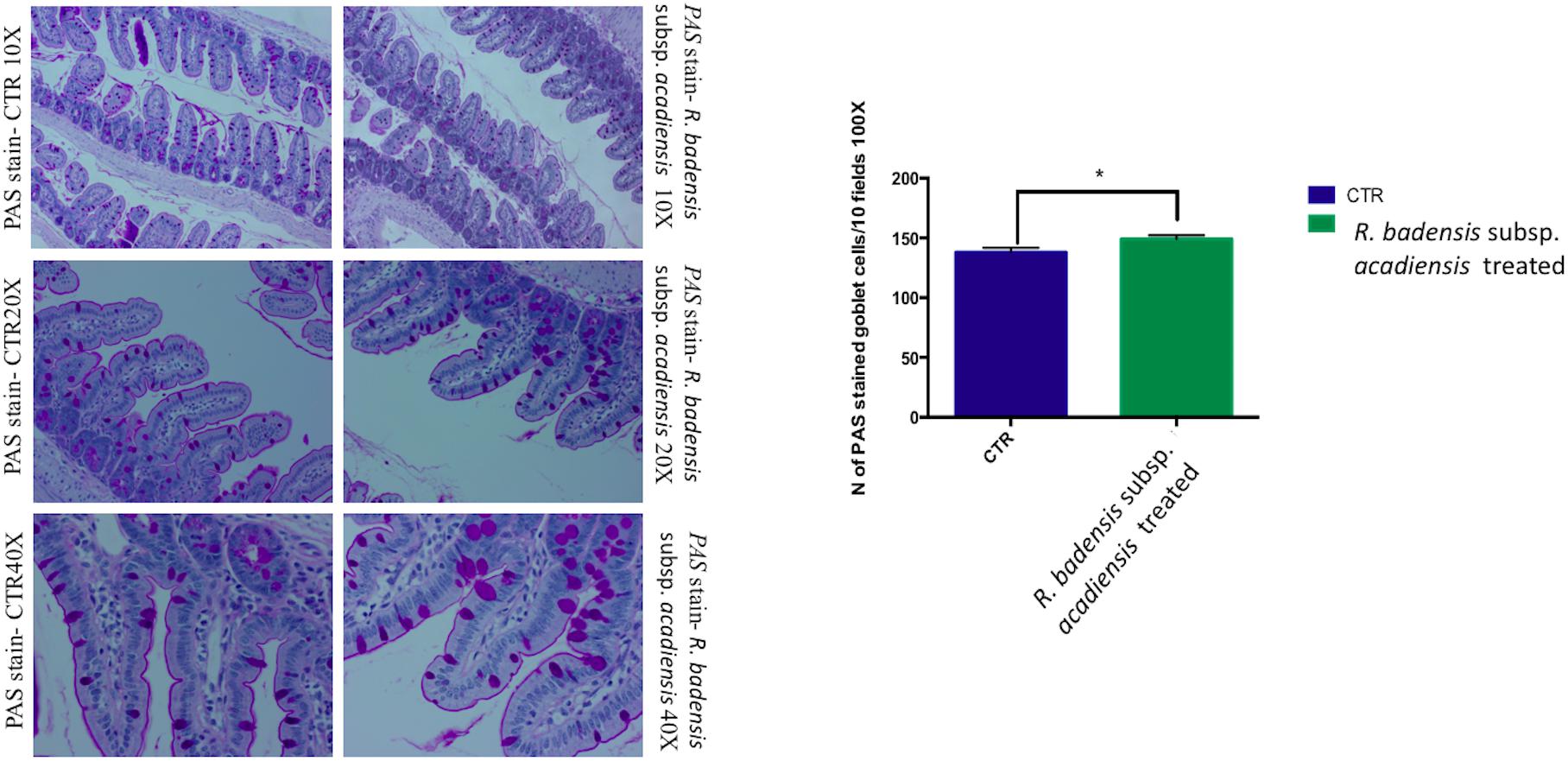
Figure 6. Mean ± SEM of the number of goblet cells stained with PAS (Periodic Acid Schiff) in 10 fields of objective 100× in the small intestine of mice fed 1% sucrose (CTR) or R. badensis subsp. acadiensis fed 108CFU/mouse/day for 7 days. Significant difference exists if *p < 0.05-p = 0.035 Representative tissue photos are taken with objective 10×, 20×, and 40× magnifications of the PAS-stained tissue.
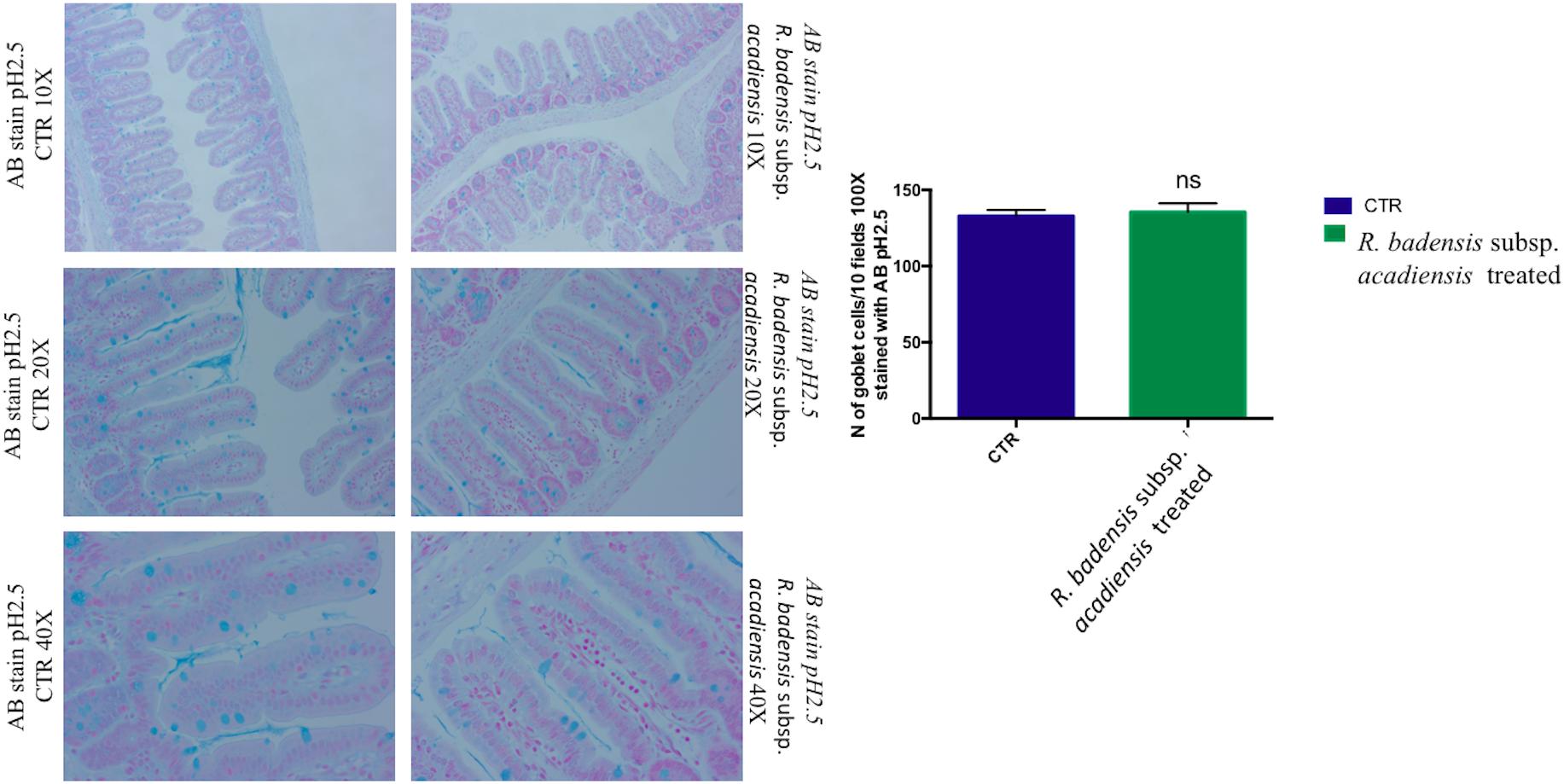
Figure 7. Mean ± SEM of the number of goblet cells stained with AB (Alcian Blue) at pH 2.5 in 10 fields of objective 100× in the small intestine of mice fed 1% sucrose (CTR) or R. badensis subsp. acadiensis fed 108CFU/mouse/day for 7 days. Significant difference exists if *p < 0.05-Representative tissue photos are taken with objective 10×, 20×, and 40× magnifications of the PAS-stained tissue. AB stains in blue acid glycoconjugates.
A significant increase in IL-10 was observed in the intestinal fluid of animals that received R. badensis subsp. acadiensis for 7-days when compared with control mice (p = 0.042) (Figure 8A). There was no change in the levels of IL-6 (Figure 8B). Secretory IgA were shown to be increased significantly in the intestinal fluid of mice fed the bacterium (19,232 ng/ml ± 2,431) compared to the control group (12,368 ± 1,429) with p = 0.0255 (Figure 8C).
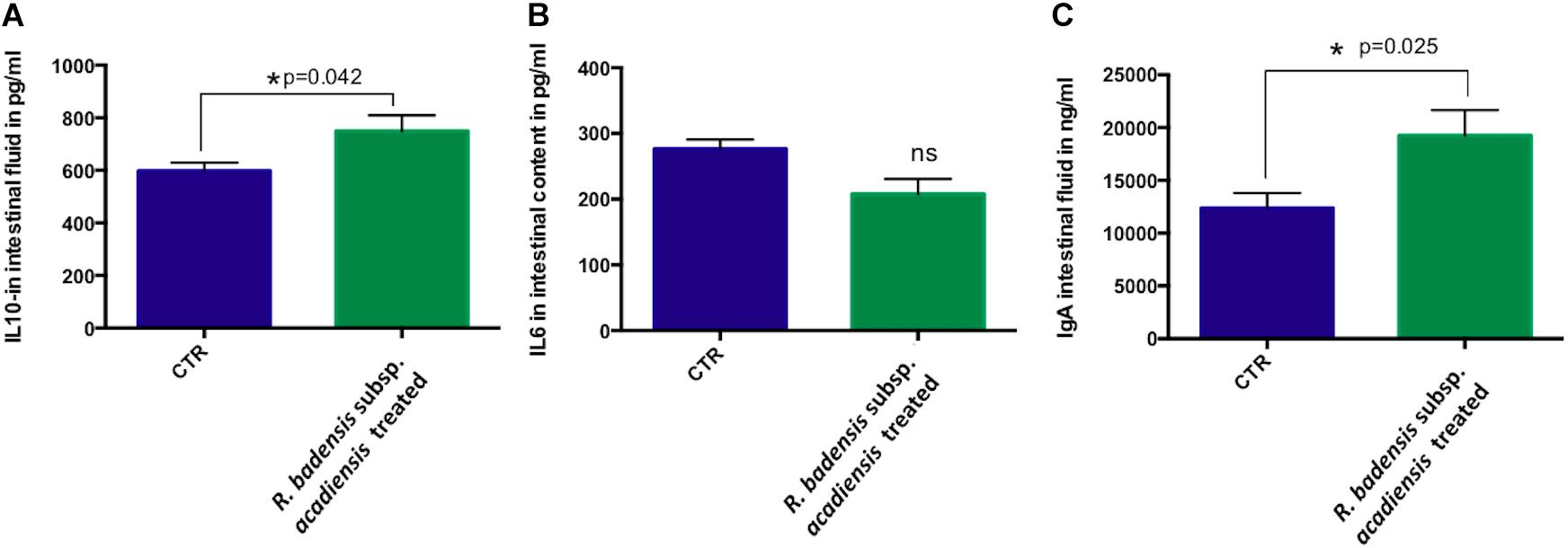
Figure 8. Data represents mean ± SEM of concentration of different (A) anti-inflammatory cytokines IL-10, (B) pro-inflammatory cytokine-IL-6 as well a (C) IgA in the intestinal fluid of (CTR) control mice fed 1% sucrose or R. badensis subsp. acadiensis treated group 108CFU/mouse/day of R. badensis subsp. acadiensis for 7 days. Number of animals per group is n = 10. Difference is considered significant between groups if *p < 0.05 ns = non-significant difference when p > 0.05.
Although no statistically significant changes were observed in the expression of miR145 in ileum or brain, we did observe a trend toward increased expression in the brain. However, a significant increase in miR146a expression was observed in ileum and brain revealing the anti-inflammatory potential of R. badensis subsp. acadiensis (Figure 9).
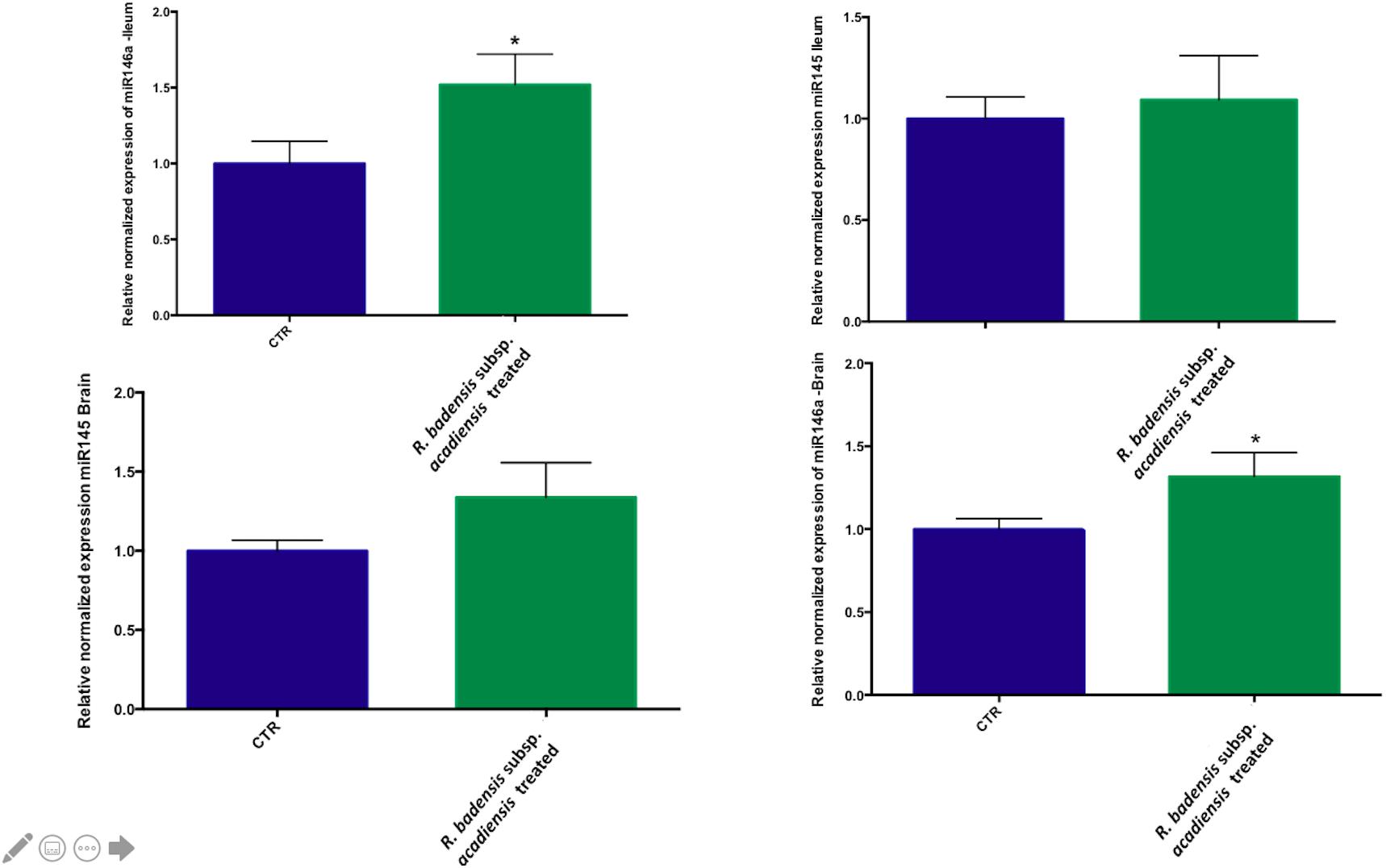
Figure 9. Mean ± SEM of relative expression of miR145 and miR146a in the brain and ileum of (CTR) control mice fed 1% sucrose or R. badensis subsp. acadiensis treated group 108CFU/mouse/day of Serratia vaccinia for 7 days. Significant difference between mice exists if *p < 0.05.
Probiotics are defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (Hill et al., 2014). They are identified mainly from fermented foods. Although fermented dairy products remain a major source of probiotic bacteria such as lactic acid bacteria and bifidobacteria; some can also be found in fermented vegetable and fruits products (Battcock et al., 1998; Schrezenmeir and De Vrese, 2001). For example, the native microbiota of vegetables involve a wide variety of lactic acid bacteria species implicated in fermentation (Swain et al., 2014). In this study, we characterize the in vitro and in vivo ability of R. badensis subsp. acadiensis isolated from the surface of lowbush blueberries (Vaccinium angustifolium Aiton) and used in blueberry-fermented preparation (Antioxydant Producing Bacterium and USES Therefore US 8,617,870) to survive the gastro-intestinal conditions, interact with the lamina propria, modulate the intestinal barrier integrity, affect mucus production and modify the immune response. Since bacteria can act as potential reservoirs for antimicrobial resistance genes, potential bacterial candidates for human and animal feeds should be screened for antimicrobial resistance. The resistant micro-organisms present in food products may result in infections that are difficult to treat. The disk diffusion method showed that R. badensis subsp. acadiensis does not carry antimicrobial resistance to the various examined antibiotics. Therefore, the bacterium can be considered as safe concerning resistance to antibiotics.
Resistance to human gastro-intestinal transit represents an important in vitro selection criterion to determine the ability of the strain to survive in the intestine and interact with the intestinal mucosa (Goldin et al., 1992). The tested strain must survive passage through the upper gastrointestinal (GI) tract and arrive alive at its site of action in order to better function in the gut environment. In our study, the in vitro methodology was used to simulate in vivo gastric and small intestinal transit in the presence of bile salts. The investigated strain demonstrated high tolerance to both simulated gastric (pH 3 and 4) containing pepsin and pancreatic juices containing pancreatin and bile salts when tested separately. Nevertheless, the bacterium was more sensitive to simulated gastric juice (pH2) and the exposure to gastric conditions and subsequent intestinal fluid makes it a lot more vulnerable with a survival rate after exposure to both conditions more than 50%. These findings revealed the ability of a good portion of the delivered bacterium to survive in the intestine.
Adhesion to the intestinal epithelial cells permits to assess the ability of the investigated strain to colonize the intestinal mucosa (Fuller et al., 1992; Dunne et al., 2001; Teitelbaum and Walker, 2002). The correlation between intestinal mucosal adhesion and transient colonization is clear between in vitro and in vivo adhesion in experimental animals as found with several strains of microorganisms (Zárate et al., 2002). A good correlation was reported between the in vitro adhesion to Caco-2 cells and the in vivo persistence of Lactobacillus strains and Bifidobacterium strains (Crociani et al., 1995; Jacobsen et al., 1999). Although the ability of a microorganism to adhere to the intestinal mucosa may provide a mean by which the probiotic can modulate the immune system (Helgeland and Brandtzaeg, 2000) and are grounds for competitive exclusion between microorganisms for nutrients and binding sites (Alderberth et al., 2000; Fons et al., 2000), it is now accepted that modulation of immune system and antimicrobial effects could occur independently of the adhesion capabilities of the bacterium. Under the conditions used in our experiment, we have provided evidence that R. badensis subsp. acadiensis is an in vitro adherent strain when tested on Caco-2 and HT-29 cells.
Moreover, the introduction of an intestinal immunomodulating bacteria in the diet must be accompanied by the absence of an adverse inflammatory response (Perdigon et al., 2001). Bacterial translocation or the passage of viable indigenous bacteria from the gut to the mesenteric lymph nodes, liver and other organs can be promoted by disruption of the ecological gastrointestinal equilibrium or disrupted permeability of the intestinal barrier (Berg, 1992; Vinderola C. G. et al., 2005). In the present study, we performed the translocation assay in the liver of mice after the oral administration of R. badensis subsp. acadiensis; no translocation was observed. Non-pathogenic commensal microorganisms interact closely with the gut epithelial surface and play a key role in the physiological, anatomical and immunological development of the host. Epithelial cells are the first line of defense; they interact simultaneously with bacteria and bacterial products and neighboring immune cells on their basolateral side. They can distinguish between pathogenic and non-pathogenic microorganisms which is an essential characteristic for the tolerance of the normal microbial microbiota (Galdeano et al., 2005; Vinderola C. G. et al., 2005).
Multiple defense mechanisms are implicated in the surveillance of intestinal mucosal homeostasis. Secretory IgA antibodies and mucus secretions are examples of these mechanisms (Fagarasan and Honjo, 2003). In the gut, IgA+ cells and secretory IgA play a key role as the first line of defense in the host. IgA antibodies cooperate with the innate non-specific defense mechanisms, by exerting immune exclusion (Vinderola et al., 2006) and preventing the invasion and colonization of pathogenic microorganisms through competition for receptors and/or metabolic substrates. We observed that the oral administration of R. badensis subsp. acadiensis increased the number of IgA + cells in the small intestine lamina propria of mice as well as the secretory IgA content in the intestinal lumen. On the other hand, IgG+ cells and the pro-inflammatory cytokine IL-6 did not increase ruling out persistent immunopathology in the intestinal mucosa (Vinderola et al., 2006).
Mucosal barriers established by intestinal epithelial cells maintain gut homeostasis; to better decipher the beneficial effect of R. badensis subsp. acadiensis strain on mucus producing cells two different specific stains (AB pH2.5 and PAS) were used. PAS staining representative of tissue histology of small intestine of treated and control groups revealed the increase of goblet cell mucus producing cells in mice gavaged with R. badensis subsp. acadiensis, pointing out a positive effect of the strain on intestinal mucosa. Goblet cells are responsible for the production and preservation of a protective mucus blanket by synthesizing mucins resulting in a dynamic protective barrier. They constitute, along with epithelial cells, macrophages, and dendritic cells, the main cellular components of the innate defense system (Nakata et al., 2017). Mucin glycoproteins synthesized and secreted by goblet cells are major constituents of the mucus layer which functions as a dynamic defensive barrier (Gaskins, 1998; Mack et al., 1999).
Additionally, mucosal immune response homeostasis can be regulated by the immunoregulatory cytokine interleukin 10 (IL-10) that limits mucosal immune responses and minimizes immunopathology (Glocker et al., 2009; Christopher et al., 2013). In fact, mice with a conditional deletion of IL-10 in the CD4+ T cell subset develop spontaneous inflammation of the intestine (Roers et al., 2004; Rubtsov et al., 2008). During acute mucosal infections, the lack of IL-10 can be protective since it is accompanied with an enhanced inflammatory response and an increased production of pro-inflammatory cytokines like IL-12, TNF, and other cytokines (Arai et al., 1995; Gazzinelli et al., 1996; Dai et al., 1997; Ke and Costa, 2006; Dann et al., 2014). However, the absence of IL-10 could result in inflammation (Gazzinelli et al., 1996; Hunter et al., 1997; Reis e Sousa et al., 1999; Suzuki et al., 2000). IL-10 plays a key role in the context of acute bacterial infection of the intestine in preventing excessive inflammation by limiting innate immunity. Similarly, mice deficient for IL-10 or IL-10 receptor developed inflammation of the large intestine (Kühn et al., 1993; Spencer et al., 1998; Yen et al., 2006). Therefore, IL-10 is crucial for maintaining a balance in adaptive immune responses in the mucosae (Denning et al., 2007; Murai et al., 2009; Ueda et al., 2010; Medina-Contreras et al., 2011; Rivollier et al., 2012). Recent studies showed that IL-10 signals drive macrophages to express tolerogenic functions that prevent colitis (Shouval et al., 2014; Zigmond et al., 2014). Our data showed that R. badensis subsp. acadiensis increased the number of IL-10+ cells in the lamina propria. Secreted IL-10 was also increased in the intestinal fluid. These findings suggest that R. badensis subsp. acadiensis may actively contribute to mucosal tolerance and may have anti-inflammatory properties (Lee and Mazmanian, 2010).
The role of microRNAs in driving or controlling inflammation is now subject to intense research. More specifically, anti-inflammatory miR-146a is a key regulator of the inflammatory processes involved in Tregs and IL-10 control. The expression of anti-inflammatory molecules like IL-10 is promoted by miR146a overexpression leading to a reduction of the inflammation (Dharap et al., 2009; Liu et al., 2016). On the other hand, recent studies found that the loss of miR-145 stimulates proinflammatory signals in the innate immune system (Akao et al., 2006). In the brain, it plays a role in antioxidant defense (Witwer et al., 2010). Our data supports the ability of R. badensis subsp. acadiensis to increase the levels of miR146a at the intestine and the brain; this finding highlights its anti-inflammatory activity and its link to IL-10 expression in the ileum.
Our findings reflect the ability of the bacterium to interact with the intestinal mucosa and to modulate the immune response and the gut barrier function. Future papers assessing the bacterium safety will be published in order to assess a potential probiotic activity of R. badensis subsp. acadiensis.
We demonstrated that R. badensis subsp. acadiensis is a non-invasive bacterium. It can survive gastrointestinal tract conditions and has resistance to bile salts and digestive enzymes (pancreatin and pepsin). Under the conditions tested, the bacterium exhibits adhesion properties to Caco-2 and HT-29. The immunoregulatory effects of the novel strain was shown by an increase in IL-10, IgA and miR-146. We conclude that R. badensis subsp. acadiensis participates in specific local immune protection and that its oral administration enhances gut-associated non-specific immunity without an inflammatory outcome. This strain is a good candidate for further investigation to elucidate its potential health benefits.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by Animal Care Committee (ACC)- university of Ottawa.
NY: experimental design, experiments optimization and running, writing the manuscript, data collection and data analysis. CM: main supervisor, revision of experiments and guidance, data review, and manuscript review. MH and GK: collaboration on antibiotic experiments and correction of the manuscript. NA and JM: helps with samples collection during the Euthanasia day. All authors contributed to the article and approved the submitted version.
This publication was funded by an NSERC Collaborative Research and Development Grant (532223-18) to CM, the Nutrition and Mental Health initiative at the Faculty of Health Sciences at the University of Ottawa, Admission Scholarship by Faculty of Graduate and Postgraduate Studies at the University of Ottawa to NY, and Nutrition and Mental Health Doctoral Scholarship to NY.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Akao, Y., Yoshihito, N., and Tomoki, N. (2006). MicroRNAs 143 and 145 Are Possible Common Onco-MicroRNAs in Human Cancers. Oncol. Rep. 16, 845–850. doi: 10.3892/or.16.4.845
Alderberth, I., Cerquetti, M., Poilance, I., Wold, A., and Collignon, A. (2000). Mechanisms of Colonisation and Colonisation Resistance of the Digestive Tract. Microb. Ecol. Health Dis. 12, 223–239. doi: 10.3402/mehd.v12i2.8060
Alturkistani, H. A., Tashkandi, F. M., and Mohammedsaleh, Z. M. (2016). Histological Stains: a Literature Review and Case Study. Glob. J. Health Sci. 8, 72–79. doi: 10.5539/gjhs.v8n3p72
Arai, T., Hiromatsu, K., Nishimura, H., Kimura, Y., Kobayashi, N., Ishida, H., et al. (1995). Effects of in Vivo Administration of Anti-IL-10 Monoclonal Antibody on the Host Defence Mechanism against Murine Salmonella Infection. Immunology 85, 381–388.
Arboleya, S., Ruas-Madiedo, P., Margolles, A., Solís, G., Salminen, S., de los Reyes-Gavilán, C. G., et al. (2011). Characterization and in Vitro Properties of Potentially Probiotic Bifidobacterium Strains Isolated from Breast-Milk. Int. J. Food Microbiol. 149, 28–36. doi: 10.1016/j.ijfoodmicro.2010.10.036
Argyri, A. A., Zoumpopoulou, G., Karatzas, K., Tsakalidou, E., Nychas, G. J., Efstathios, P. Z., et al. (2013). Selection of Potential Probiotic Lactic Acid Bacteria from Fermented Olives by in Vitro Tests. Food Microbiol. 33, 282–291. doi: 10.1016/j.fm.2012.10.005
Atarashi, K., Takeshi, T., Oshima, K., Suda, W., Nagano, Y., Nishikawa, H., et al. (2013). Treg Induction by a Rationally Selected Mixture of Clostridia Strains from the Human Microbiota. Nature 500, 232–236. doi: 10.1038/nature12331
Battcock, M., Azam-Ali, S., and FAO (1998). Fermented Fruits and Vegetables-A Global Perspective. FAO Agricultural Services Bulletin. Italy: Food and Agriculture Organization.
Berg, R. D. (1992). “Bacterial Translocation from the Gastrointestinal Tract. Trends Microbiol. 3, 149–154. doi: 10.1016/S0966-842X(00)88906-4
Bermudez-Humaran, G., and Langella, P. (2017). Use of Traditional and Genetically Modified Probiotics in Human Health: what Does the Future Hold? Microbiol. Spectr. 5:28959936. doi: 10.1128/microbiolspec.BAD-0016-2016
Boonma, P., Spinler, J. K., Venable, S. F., Versalovic, J., and Tumwasorn, S. (2014). Lactobacillus Rhamnosus L34 and Lactobacillus Casei L39 Suppress Clostridium Difficile-Induced IL-8 Production by Colonic Epithelial Cells. BMC Microbiol. 14:177. doi: 10.1186/1471-2180-14-177
Christensen, H. R., Froakier, H., and Pestka, J. J. (2002). Lactobacilli Differentially Modulate Expression of Cytokines and Maturation Surface Markers in Murine Dendritic Cells. J. Immunol. 168, 171–178. doi: 10.4049/jimmunol.168.1.171
Christopher, M. J., Walters, T. D., Guo, C. H., Kugathasan, S., Klein, C., Turner, D., et al. (2013). IL-10R Polymorphisms Are Associated with Very-Early-Onset Ulcerative Colitis. Inflamm. Bowel Dis. 19, 115–123. doi: 10.1002/ibd.22974
Crociani, J., Grill, J. P., Huppert, M., and Ballongue, J. (1995). Adhesion of Different Bifidobacteria Strains to Human Enterocyte−like Caco−2 Cells and Comparison with in Vivo Study. Lett. Appl. Microbiol. 21, 146–148. doi: 10.1111/j.1472-765X.1995.tb01027.x
Cross, M. L., Ganner, A., Teilab, D., and Fray, L. M. (2004). Patterns of Cytokine Induction by Gram-Positive and Gram-Negative Probiotic Bacteria. FEMS Immunol. Med. Microbiol. 42, 173–180. doi: 10.1016/j.femsim.2004.04.001
Dai, E., Kohler, G., and Brombacher, F. (1997). Both Innate and Acquired Immunity to Listeria Monocytogenes Infection Are Increased in IL-10-Deficient Mice. Immunobiology 197:316.
Dann, S. M., Le, C., Choudhury, B. K., Liu, H., Saldarriaga, O., Hanson, E. M., et al. (2014). Attenuation of Intestinal Inflammation in Interleukin-10-Deficient Mice Infected with Citrobacter Rodentium. Infect. Immun. 82, 1949–1958. doi: 10.1128/IAI.00066-14
Denning, T. L., Yi, C. W., Seema, P. R., Ifor, W. R., and Bali, P. (2007). Lamina Propria Macrophages and Dendritic Cells Differentially Induce Regulatory and Interleukin 17-Producing T Cell Responses. Nat. Immunol. 8, 1086–1094. doi: 10.1038/ni1511
Dharap, A., Bowen, K., Place, R., Li, L. C., and Vemuganti, R. (2009). Transient Focal Ischemia Induces Extensive Temporal Changes in Rat Cerebral MicroRNAome. J. Cereb. Blood Flow Metab. 29, 675–687. doi: 10.1038/jcbfm.2008.157
Dunne, C., O’Mahony, L., Murphy, L., Thornton, G., Morrissey, D., O’Halloran, S., et al. (2001). In Vitro Selection Criteria for Probiotic Bacteria of Human Origin: correlation with in Vivo Findings. Am. J. Clin. Nutr. 73, 386–392. doi: 10.1093/ajcn/73.2.386s
Elian, S. D. A., Souza, E. L. S., Vieira, A. T., Teixeira, M. M., Arantes, R. M. E., Nicoli, J. R., et al. (2015). Bifidobacterium Longum Subsp. Infantis BB-02 Attenuates Acute Murine Experimental Model of Inflammatory Bowel Disease. Benef. Microbes 6, 277–286. doi: 10.3920/BM2014.0070
Fagarasan, S., and Honjo, T. (2003). Intestinal IgA Synthesis: regulation of Front-Line Body Defences. Nat. Rev. Immunol. 3, 63–72. doi: 10.1038/nri982
Fons, M., Gomez, A., and Karjalainen, T. (2000). Mechanisms of Colonisation and Colonisation Resistance of the Digestive Tract Part 2: bacteria/Bacteria Interactions. Microb. Ecol. Health Dis. 12, 240–246. doi: 10.1080/089106000750060495
Fuller, R., Havenaar, R., Ten Brink, B., and Huis In ‘t Veld, J. (1992). Selection of Strains for Probiotic Use in Probiotics:The Scientific Basis. (London: Chapman & Hall), 209–224. doi: 10.1007/978-94-011-2364-8_9
Galdeano, C. M., De Leblanc, A. D. M., Vinderola, G., Bonet, M. E. B., and Perdigón, G. (2007). Proposed Model: mechanisms of Immunomodulation Induced by Probiotic Bacteria. Clin. Vaccine Immunol. 14, 485–492. doi: 10.1128/CVI.00406-06
Galdeano, C. M., Maldonado, C., and Perdigon, G. (2005). The Probiotic Bacterium Lactobacillus Casei Induces Activation of the Gut Mucosal Immune System through Innate Immunity. Clin. Vaccine Immunol. 13, 219–226. doi: 10.1128/CVI.13.2.219
Gaskins, H. R. (1998). “Immunological Development and Mucosal Defence in the Pig Intestine,” in Progress in Pig Science, eds J. Wiseman and M. A. Varley (Nottingham, UK: Nottingham University Press), 81–100.
Gaudet, A. D., Fonken, L. K., Watkins, L. R., Nelson, R. J., and Popovich, P. G. (2017). MicroRNAs: roles in Regulating Neuroinflammation. Neuroscientist 24, 221–245. doi: 10.1177/1073858417721150
Gazzinelli, R. T., Wysocka, M., Hieny, S., Scharton-Kersten, T., Cheever, A., Kühn, R., et al. (1996). In the Absence of Endogenous IL-10, Mice Acutely Infected with Toxoplasma Gondii Succumb to a Lethal Immune Response Dependent on CD4+ T Cells and Accompanied by Overproduction of IL-12, IFN-Gamma and TNF-Alpha. J. Immunol. 157, 798–805.
Glocker, E. O., Kotlarz, D., Boztug, K., Gertz, E. M., Schäffer, A. A., Noyan, F., et al. (2009). Inflammatory Bowel Disease and Mutations Affecting the Interleukin-10 Receptor. N. Eng. J. Med. 361, 2033–2045. doi: 10.1056/NEJMoa0907206
Goldin, R., Gorbach, L., Saxelin, M., Barakat, S., Gualtieri, L., and Salminen, S. (1992). Survival of Lactobacillus Species (Strain GG) in Human Gastrointestinal Tract. Dig. Dis. Sci. 37, 121–128. doi: 10.1007/BF01308354
Gopal, P. K., Prasad, J., Smart, J., and Gill, H. S. (2001). In Vitro Adherence Properties of Lactobacillus Rhamnosus DR20 and Bifidobacterium Lactis DR10 Strains and Their Antagonistic Activity against an Enterotoxigenic Escherichia Coli. Int. J. Food Microbiol. 67, 207–216. doi: 10.1016/S0168-1605(01)00440-8
Gregory, S. H., Sagnimeni, A. J., and Wing, E. J. (1996). Bacteria in the Bloodstream Are Trapped in the Liver and Killed by Immigrating Neutrophils. J. Immunol. 157, 2514–2520.
Helgeland, L., and Brandtzaeg, P. (2000). Development and Function of Intestinal B and T Cells. Microb. Ecolo. Health Dis. 12, 110–127. doi: 10.1080/089106000750060378
Hemaiswarya, S., Raja, R., Ravikumar, R., and Carvalho, I. S. (2013). Mechanism of Action of Probiotics. Braz. Arch. Biol. Technol. 56, 113–119. doi: 10.1590/S1516-89132013000100015
Hessle, C., Andersson, B., and Wold, A. E. (2000). Gram-Positive Bacteria Are Potent Inducers of Monocytic Interleukin-12 (IL-12) While Gram-Negative Bacteria Preferentially Stimulate IL-10 Production Anaerobic. Infect. Immun. 68, 3581–3586. doi: 10.1128/iai.68.6.3581-3586.2000
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Huang, Y., and Adams, M. C. (2004). In Vitro Assessment of the Upper Gastrointestinal Tolerance of Potential Probiotic Dairy Propionibacteria. Int. J. Food Microbiol. 91, 253–260. doi: 10.1016/j.ijfoodmicro.2003.07.001
Hunter, C. A., Ellis-Neyes, L. A., Slifer, T., Kanaly, S., Grünig, G., Fort, M., et al. (1997). IL-10 Is Required to Prevent Immune Hyperactivity during Infection with Trypanosoma Cruzi. J. Immunol. 158, 3311–3316.
Imperial, I. C. V. J., and Ibana, J. A. (2016). Addressing the Antibiotic Resistance Problem with Probiotics: reducing the Risk of Its Double-Edged Sword Effect. Front. Microbiol. 7:1983. doi: 10.3389/fmicb.2016.01983
Jacobsen, C. N., Rosenfeldt Nielsen, V., Hayford, A. E., Møller, P. L., Michaelsen, K. F., Pærregaard, A., et al. (1999). Screening of Probiotic Activities of Forty-Seven Strains of Lactobacillus Spp. by in Vitro Techniques and Evaluation of the Colonization Ability of Five Selected Strains in Humans. Appl. Environ. Microbiol. 65, 4949–4956. doi: 10.1128/AEM.65.11.4949-4956
Kandasamy, S., Vlasova, A. N., Fischer, D., Kumar, A., Chattha, K. S., Lulu Shao, A. R., et al. (2016). Differential Effects of Escherichia Coli Nissle and Lactobacillus Rhamnosus Strain GG on Human Rotavirus Binding, Infection, and B Cell Immunity. J. Immunol. 196, 1780–1790. doi: 10.4049/jimmunol.1501705
Karlsson, H., Hessle, C., and Rudin, A. (2002). Innate Immune Responses of Human Neonatal Cells to Bacteria from the Normal Gastrointestinal Flora. Infect. Immun. 70, 6688–6696. doi: 10.1128/IAI.70.12.6688
Ke, Q., and Costa, M. (2006). Hypoxia-Inducible Factor-1 (HIF-1). Mol. Pharmacol. 33, 1469–1480. doi: 10.1097/01.CCM.0000191716.38566.E0
Kos, B., Šušković, J., Vuković, S., Sǐmpraga, M., Frece, J., and Matošić, S. (2003). Adhesion and Aggregation Ability of Probiotic Strain Lactobacillus Acidophilus M92. J. Appl. Microbiol. 94, 981–987. doi: 10.1046/j.1365-2672.2003.01915.x
Kühn, R., Löhler, J., Rennick, D., Rajewsky, K., and Müller, W. (1993). Interleukin-10-Deficient Mice Develop Chronic Enterocolitis. Cell 75, 263–274. doi: 10.1016/0092-8674(93)80068-P
Kumura, H., Tanoue, Y., Tsukahara, M., Tanaka, T., and Shimazaki, K. (2004). Screening of Dairy Yeast Strains for Probiotic Applications. J. Dairy Sci. 87, 4050–4056. doi: 10.3168/jds.S0022-0302(04)73546-8
Lee, Y. K., and Mazmanian, S. K. (2010). Has the Microbiota Played a Critical Role in the Evolution of the Adaptive Immune System? Science 330, 1768–1773. doi: 10.1126/science.1195568
Liu, H. J., Zhang, A. F., Zhao, N., and Li, X. Z. (2016). Role of MiR-146a in Enforcing Effect of Specific Immunotherapy on Allergic Rhinitis. Immunol. Invest. 45, 1–10. doi: 10.3109/08820139.2015.1085390
Mack, D. R., Michail, S., Wei, S., McDougall, L., and Hollingsworth, M. A. (1999). Probiotics Inhibit Enteropathogenic E. Coli Adherence in Vitro by Inducing Intestinal Mucin Gene Expression DAVID. Am. J. Physiol. 276, 941–950. doi: 10.1152/ajpgi.1999.276.4.G941
Mallet, J. F., Duarte, J., Vinderola, G., Anguenot, R., Beaulieu, M., and Matar, C. (2014). The Immunopotentiating Effects of Shark-Derived Protein Hydrolysate. Nutrition 30, 706–712. doi: 10.1016/j.nut.2013.10.025
Martin, L. J. (2003). Augmentation de La Teneur en Composés Phénoliques et de la Capacité Antioxydante du Bleuet Fermenté. Moncton, CA: Université de Moncton.
Martin, L. J., and Matar, C. (2005). Increase of Antioxidant Capacity of the Lowbush Blueberry (Vaccinium Angustifolium) during Fermentation by a Novel Bacterium from the Fruit Microflora. J. Sci. Food Agric. 85, 1477–1484. doi: 10.1002/jsfa.2142
Martín, R., Laval, L., Chain, F., Miquel, S., Natividad, J., Cherbuy, C., et al. (2016). Bifidobacterium Animalis Ssp. Lactis CNCM-I2494 Restores Gut Barrier Permeability in Chronically Low-Grade Inflamed Mice. Front. Microbiol. 7:608. doi: 10.3389/fmicb.2016.00608
Medina-Contreras, O., Geem, D., Laur, O., Williams, I. R., Lira, S. A., Nusrat, A., et al. (2011). CX3CR1 Regulates Intestinal Macrophage Homeostasis, Bacterial Translocation, and Colitogenic Th17 Responses in Mice. J. Clin. Invest. 121, 4787–95 doi: 10.1172/JCI59150
Mottet, C., and Michetti, P. (2005). Probiotics: wanted Dead or Alive. Dig. Liver Dis. 37, 3–6. doi: 10.1016/j.dld.2004.09.010
Murai, M., Turovskaya, O., Kim, G., Madan, R., Karp, C. K., Cheroutre, H., et al. (2009). Interleukin 10 Acts on Regulatory t Cells to Maintain Expression of the Transcription Factor Foxp3 and Suppressive Function in Mice with Colitis. Nat. Immunol. 10, 1178–1184. doi: 10.1038/ni.1791
Nachar, A., Eid, H. M., Vinqvist-Tymchuk, M., Vuong, T., Kalt, W., Matar, C., et al. (2017). Phenolic Compounds Isolated from Fermented Blueberry Juice Decrease Hepatocellular Glucose Output and Enhance Muscle Glucose Uptake in Cultured Murine and Human Cells. BMC Complement. Altern. Med. 17:138. doi: 10.1186/s12906-017-1650-2
Nakata, K., Sugi, Y., Narabayashi, H., Kobayakawa, T., Nakanishi, Y., Tsuda, M., et al. (2017). Commensal Microbiota-Induced MicroRNA Modulates Intestinal Epithelial Permeability through the Small GTPase ARF4. J. Biol. Chem. 292, 15426–15433. doi: 10.1074/jbc.M117.788596
O’Connell, R. M., Rao, D. S., and Baltimore, D. (2012). MicroRNA Regulation of Inflammatory Responses. Annu. Rev. Immunol. 330, 295–312. doi: 10.1146/annurev-
Ouwehand, A. C., and Salminen, S. (2003). In Vitro Adhesion Assays for Probiotics and Their in Vivo Relevance: a Review. Microb. Ecol. Health Dis. 15, 175–184. doi: 10.1080/08910600310019886
Perdigón, G., Dde LeBlanc, A. D. M., Valdez, J., and Rachid, M. (2002). Role of Yoghurt in the Prevention of Colon Cancer. Eur. J. Clin. Nutr. 56, S65–S68. doi: 10.1038/sj.ejcn.1601490
Perdigon, G., Fuller, R., and Raya, R. (2001). Lactic Acid Bacteria and Their Effect on the Immune System. Curr. Issues. Intest. Microbiol. 2, 27–42.
Reis e Sousa, C., Yap, G., Schulz, O., Rogers, N., Schito, M., Aliberti, J., et al. (1999). Paralysis of Dendritic Cell IL-12 Production by Microbial Products Prevents Infection-Induced Immunopathology. Immunity 11, 637–647. doi: 10.1016/S1074-7613(00)80138-7
Ren, D. Y., Li, C., Qin, Y. Q., Yin, R. L., Du, S., Ye, F., et al. (2013). Lactobacilli Reduce Chemokine IL-8 Production in Response to TNF- α and Salmonella Challenge of Caco-2 Cells Lactobacilli Reduce Chemokine IL-8 Production in Response to TNF-? And Salmonella Challenge of Caco-2. Cells. Biomed. Res. Int. 2013:925219. doi: 10.1155/2013/925219
Rivollier, A., He, J., Kole, A., Valatas, V., and Kelsall, B. L. (2012). Inflammation Switches the Differentiation Program of Ly6Chi Monocytes from Antiinflammatory Macrophages to Inflammatory Dendritic Cells in the Colon. J. Exp. Med. 209, 139–155. doi: 10.1084/jem.20101387
Roers, A., Siewe, L., Strittmatter, E., Deckert, M., Schlüter, D., Stenzel, W., et al. (2004). T Cell-Specific Inactivation of the Interleukin 10 Gene in Mice Results in Enhanced T Cell Responses but Normal Innate Responses to Lipopolysaccharide or Skin Irritation. J. Exp. Med. 200, 1289–1297. doi: 10.1084/jem.20041789
Rubtsov, Y. P., Rasmussen, J. P., Chi, E. Y., Fontenot, J., Castelli, L., Ye, X., et al. (2008). Regulatory T Cell-Derived Interleukin-10 Limits Inflammation at Environmental Interfaces. Immunity 28, 546–558. doi: 10.1016/j.immuni.2008.02.017
Saad, N., Delattre, C., Urdaci, M., Schmitter, J. M., and Bressollier, P. (2013). An Overview of the Last Advances in Probiotic and Prebiotic Field. LWT Food Sci. Technol. 50, 1–16. doi: 10.1016/j.lwt.2012.05.014
Sánchez-villavicencio, M. L., Vinqvist-tymchuk, M., Kalt, W., Matar, C., Alarcón Aguilar, F. J., Escobar, C., et al. (2017). Fermented Blueberry Juice Extract and Its Specific Fractions Have an Anti-Adipogenic Effect in 3 T3-L1 Cells. BMC Complement. Altern. Med. 17:24. doi: 10.1186/s12906-016-1519-9
Sanders, M. E., and Huis In’t Veld, J. (1999). Bringing a Probiotic-Containing Functional Food to the Market: microbiological, Product, Regulatory and Labeling Issues. Antonie van Leeuwenhoek 76, 293–315 doi: 10.1007/978-94-017-2027-4_15
Schrezenmeir, J., and De Vrese, M. (2001). Probiotics, Prebiotics, and Synbiotics—Approaching a Definition 1–3. Am. J. Clin. Nutr. 73, 361S–364S. doi: 10.1093/ajcn/73.2.361s
Scully, P., Macsharry, D. O., Mahony, J., Lyons, A., O’Brien, F., Murphy, S., et al. (2013). Bifidobacterium Infantis Suppression of Peyer’s Patch MIP-1a and MIP-1b Secretion During Salmonella Infection Correlates with Increased Local CD4+CD25+ T Cell Numbers. Cell. Immunol. 281, 134–140. doi: 10.1016/j.cellimm.2013.03.008
Shouval, D. S., Biswas, A., Goettel, J. A., McCann, K., Conaway, E., Redhu, N. S., et al. (2014). Interleukin-10 Receptor Signaling in Innate Immune Cells Regulates Mucosal Immune Tolerance and Anti-Inflammatory Macrophage Function. Immunity 40, 706–719. doi: 10.1016/j.immuni.2014.03.011
Smirnov, A., Perez, P., Amit-Romach, E., Sklan, D., and Uni, Z. (2005). Mucin Dynamics and Microbial Populations in Chicken Small Intestine Are Changed by Dietary Probiotic and Antibiotic Growth Promoter Supplementation. J. Nutr. 135, 187–192. doi: 10.1093/jn/135.2.187
Smits, H. H., J Van Beelen, A., Hessle, C., Westland, R., De Jong, E., Soeteman, E., et al. (2004). Commensal Gram-Negative Bacteria Prime Human Dendritic Cells for Enhanced IL-23 and IL-27 Expression and Enhanced Th1 Development. Eur. J. Immunol. 34, 1371–1380. doi: 10.1002/eji.200324815
Spencer, D., Di Marco, F., Hooley, J., Pitts-Meek, S., Bauer, M., Ryan, A. M., et al. (1998). The Orphan Receptor CRF2-4 Is an Essential Subunit of the Interleukin 10 Receptor. J. Exp. Med. 187, 571–578. doi: 10.1084/jem.187.4.571
Suzuki, Y., Sher, A., Yap, G., Park, D., Neyer, L. E., Liesenfeld, O., et al. (2000). IL-10 Is Required for Prevention of Necrosis in the Small Intestine and Mortality in Both Genetically Resistant BALB/c and Susceptible C57BL/6 Mice Following Peroral Infection with Toxoplasma Gondii. J. Immunol. 164, 5375–5382. doi: 10.4049/jimmunol.164.10.5375
Swain, M. R., Anandharaj, M., Ray, R. C., and Rani, R. P. (2014). Fermented Fruits and Vegetables of Asia: a Potential Source of Probiotics. Biotechnol. Res. Int. 1, 1–19. doi: 10.1155/2014/250424
Teitelbaum, J. E., and Walker, W. A. (2002). Nutritional Impact of Pre- and Probiotics as Protective Gastrointestinal Organisms. Ann. Rev. Nutr. 22, 107–138. doi: 10.1146/annurev.nutr.22.110901.145412
Turpin, W., Humblot, C., Noordine, M. L., Thomas, M., and Guyot, J. P. (2012). Lactobacillaceae and Cell Adhesion: genomic and Functional Screening. PLoS One 7:e38034. doi: 10.1371/journal.pone.0038034
Ueda, Y., Kayama, H., Jeon, S. G., Kusu, T., Isaka, Y., Rakugi, H., et al. (2010). Commensal Microbiota Induce LPS Hyporesponsiveness in Colonic Macrophages via the Production of IL-10. Int. Immunol. 22, 953–962. doi: 10.1093/intimm/dxq449
Vinderola, C. G., and Reinheimer, J. A. (2003). Lactic Acid Starter and Probiotic Bacteria: a Comparative ‘in Vitro’ Study of Probiotic Characteristics and Biological Barrier Resistance. Food Res. Int. 36, 895–904. doi: 10.1016/S0963-9969(03)00098-X
Vinderola, C. G., Duarte, J., Thangavel, D., Perdigón, G., Farnworth, E., and Matar, C. (2005). Immunomodulating Capacity of Kefir. J. Dairy Res. 72, 195–202. doi: 10.1017/S0022029905000828
Vinderola, G., Matar, C., and Perdigon, G. (2005). Role of Intestinal Epithelial Cells in Immune Effects Mediated by Gram-Positive Probiotic Bacteria: involvement of Toll-Like Receptors. Clin. Vaccine Immunol. 12, 1075–1084. doi: 10.1128/CDLI.12.9.1075-1084.2005
Vinderola, G., Perdigón, G., Duarte, J., Farnworth, E., and Matar, C. (2006). Effects of the Oral Administration of the Exopolysaccharide Produced by Lactobacillus Kefiranofaciens on the Gut Mucosal Immunity. Cytokine 36, 254–260. doi: 10.1016/j.cyto.2007.01.003
Vuong, T., Mallet, J. F., Ouzounova, M., Rahbar, S., Hernandez-Vargas, H., Herceg, Z., et al. (2016). Role of a Polyphenol-Enriched Preparation on Chemoprevention of Mammary Carcinoma through Cancer Stem Cells and Inflammatory Pathways Modulation. J. Transl. Med. 14, 1–12. doi: 10.1186/s12967-016-0770-7
Vuong, T., Martineau, L. C., Ramassamy, C., Matar, C., and Haddad, P. S. (2007). Fermented Canadian Lowbush Blueberry Juice Stimulates Glucose Uptake and AMP-Activated Protein Kinase in Insulin-Sensitive Cultured Muscle Cells and Adipocytes. Can. J. Physiol. Pharmacol. 85, 956–965. doi: 10.1139/Y07-090
Witwer, K. W., Sisk, J. M., Gama, L., and Clements, J. E. (2010). MicroRNA Regulation of IFN-β Protein Expression: rapid and Sensitive Modulation of the Innate Immune Response. J. Immunol. 184, 2369–2376. doi: 10.4049/jimmunol.0902712
Yahfoufi, N., Mallet, J. F., Graham, E., and Matar, C. (2018). Role of Probiotics and Prebiotics in Immunomodulation. Curr. Opin. Food Sci. 20, 82–91. doi: 10.1016/j.cofs.2018.04.006
Yen, D., Cheung, J., Scheerens, H., Poulet, F., McClanahan, T., McKenzie, B., et al. (2006). IL-23 Is Essential for T Cell-Mediated Colitis and Promotes Inflammation via IL-17 and IL-6. J. Clin. Invest. 116, 1310–1316. doi: 10.1172/JCI21404
Zárate, G., Morata De Ambrosini, V. M., Chaia, Silvia, A. P., and González, N. (2002). Adhesion of Dairy Propionibacteria to Intestinal Epithelial Tissue in Vitro and in Vivo. J. Food Prot. 65, 534–539. doi: 10.4315/0362-028X-65.3.534
Zhou, G., Zhou, Y., and Chen, X. (2017). New Insight into Inter-Kingdom Communication: horizontal Transfer of Mobile Small RNAs. Front. Microbiol. 8:768. doi: 10.3389/fmicb.2017.00768
Keywords: probiotic, mucosal immunity, immuno-modulation, micoRNAs (miRNAs), gastro-intestinal tolerance
Citation: Yahfoufi N, Alsadi N, Mallet JF, Kulshreshtha G, Hincke M, Ismail N and Matar C (2021) Immunomodulation and Intestinal Morpho-Functional Aspects of a Novel Gram-Negative Bacterium Rouxiella badensis subsp. acadiensis. Front. Microbiol. 12:569119. doi: 10.3389/fmicb.2021.569119
Received: 03 June 2020; Accepted: 23 March 2021;
Published: 22 June 2021.
Edited by:
Haifeng Zhao, South China University of Technology, ChinaReviewed by:
Valentina Taverniti, University of Milan, ItalyCopyright © 2021 Yahfoufi, Alsadi, Mallet, Kulshreshtha, Hincke, Ismail and Matar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chantal Matar, Y2hhbnRhbC5tYXRhckB1b3R0YXdhLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.