
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 28 January 2021
Sec. Fungi and Their Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.552301
This article is part of the Research TopicFungal Genetics in Plant Biomass ConversionView all 11 articles
 Wolfgang Hinterdobler1
Wolfgang Hinterdobler1 Guofen Li1
Guofen Li1 Katharina Spiegel1
Katharina Spiegel1 Samira Basyouni-Khamis1,2
Samira Basyouni-Khamis1,2 Markus Gorfer1
Markus Gorfer1 Monika Schmoll1*
Monika Schmoll1*Fungi of the genus Trichoderma are of high importance for biotechnological applications, in biocontrol and for production of homologous and heterologous proteins. However, sexual crossing under laboratory conditions has so far only been achieved with the species Trichoderma reesei, which was so far only isolated from tropical regions. Our isolation efforts aimed at the collection of Trichoderma strains from Austrian soils surprisingly also yielded 12 strains of the species T. reesei, which was previously not known to occur in Europe. Their identity was confirmed with tef1- and rpb2-sequencing and phylogenetic analysis. They could clearly be distinguished from tropical strains including the common laboratory wildtypes by UP-PCR and genetic variations adjacent to the mating type locus. The strains readily mated with reference strains derived from CBS999.97. Secreted cellulase and xylanase levels of these isolates were up to six-fold higher than those of QM6a indicating a high potential for strain improvement. The strains showed different responses to injury in terms of induction of sporulation, but a correlation to alterations in the nox1-gene sequence was not detected. Several synonymous SNPs were found in the sequence of the regulator gene noxR of the soil isolates compared to QM6a. Only in one strain, non-synonymous SNPs were found which impact a PEST sequence of NoxR, suggesting altered protein stability. The availability of sexually fertile strains from middle Europe naturally producing decent amounts of plant cell wall degrading enzymes opens up novel perspectives for non-GMO strain improvement and biological pretreatment of plant biomass for bioethanol production. Moreover, the varied response of these strains to injury in terms of sporulation, which is independent of Nox1 and NoxR suggests that additional regulators impact this phenomenon in T. reesei.
The genus Trichoderma comprises fungi with widespread ecological functions, which are of high interest in research. Species of this genus include a considerable number of efficient biocontrol agents (Harman et al., 2004; Harman, 2011), the biotechnological workhorse T. reesei (Bischof et al., 2016) and species know to comprise pathogenic strains impacting mushroom farming (Komon-Zelazowska et al., 2007) or even threatening human health (Hatvani et al., 2013). Roughly 260 species are documented from this genus in current literature (Atanasova et al., 2013; Bissett et al., 2015), with a total of 443 species listed in Mycobank for Trichoderma1 (access on July 31st, 2020).
Among Trichoderma spp., T. reesei is among the best studied species (Schmoll et al., 2014, 2016), predominantly due to its broad application in biotechnology (Gudynaite-Savitch and White, 2016; Arnau et al., 2020). After the isolation of the wild-type strain QM6a from the Solomon Islands during World War II, its efficient production of cellulolytic enzymes was recognized (Bischof et al., 2016). Since then, decades of research, including mutation programs were aimed at improvement of cellulase production as well as optimization of T. reesei as a host for heterologous protein production (Seiboth et al., 2011; Paloheimo et al., 2016). The enhanced strains from these efforts are all derived from the original isolate QM6a.
The natural habitat of Trichoderma reesei is known to be a tropical rain forest with abundant decaying plant material (Druzhinina et al., 2011). The teleomorph of T. reesei was initially known as Hypocrea jecorina, a name which was frequently used in literature of this species as well, until the one fungus = one name nomenclature ended the century old practice of naming sexual and asexual forms independently (Bissett et al., 2015). Since then, only the name T. reesei is accepted. Phylogenetically the species T. reesei belongs to the Longibrachiatum clade, which comprises 26 putative species (Druzhinina et al., 2012) with the closest relatives being Trichoderma parareesei, Trichoderma orientalis, and Trichoderma longibrachiatum. Most analyzed isolates originate from tropical or subtropical regions like the Solomon Islands, French Guiana, Brazil, or Peru (Druzhinina et al., 2012). Genome analysis revealed that T. reesei, representing the Longibrachiatum clade is evolutionarily one of the youngest species of Trichoderma (Kubicek et al., 2011). The same study indicated that the rather poor performance of T. reesei in most biocontrol applications compared to other Trichoderma species may be due to the loss of multiple genes associated with antagonism and mycoparasitism.
As a highly efficient producer of homologous and heterologous proteins, manipulation of the genome of T. reesei is supported by optimized tools like non-homologous end joining deficient (NHEJ) strains and genome wide deletion primer design (Schuster et al., 2012), transient knock down of NHEJ (Chum et al., 2017), or CRISPR/Cas9 mediated genome editing (Liu et al., 2015). The genome of T. reesei is sequenced in high quality (Martinez et al., 2008; Marie-Nelly et al., 2014; Li et al., 2017) and manually annotated (Druzhinina et al., 2016; Schmoll et al., 2016; Li et al., 2017). Despite the broad array of versatile tools for genome manipulation, a common drawback for strain improvement lies in the fact that either the genes responsible for an improvement has to be known or that deleterious mutations are introduced by random mutagensis along with the desired mutations leading to an overall suboptimal result. The latter drawback can be avoided by the use of sexual crossing for strain improvement (Ashton and Dyer, 2016).
Breeding of industrial fungi for high efficiency production has been an aim for decades. Although the sexually propagating H. jecorina was identified as the teleomorph of T. reesei (Kuhls et al., 1996) and fruiting bodies were seen with an isolate from French Guiana (Lieckfeldt et al., 2000), fruitless attempts to achieve mating led to the assumption that T. reesei is a clonal species. Only about 11 years ago, after the discovery of peptide pheromone precursor genes in the genome, sexual development under laboratory conditions was achieved (Schmoll et al., 2004, 2010; Seidl et al., 2009; Hinterdobler et al., 2020). T. reesei is a heterothallic fungus with a common mating type structure of three mat-genes in the MAT1-1 locus and one mat-gene in the MAT1-2 locus (Seidl et al., 2009). Its pheromone system comprises two pheromone receptors HPR1 and HPR2 as well as two associate peptide pheromone precursors, HPP1 and PPG1 (Schmoll et al., 2010; Seibel et al., 2012). Additionally, also chemical communication is involved in mate recognition (Bazafkan et al., 2015). In contrast to many other fungal species, mating in T. reesei occurs on rich media like malt extract and is facilitated by light (Schmoll, 2013; Hinterdobler et al., 2020). Unfortunately, the parental strain of all strains currently used in industry and academia, QM6a, was found to be female sterile (Seidl et al., 2009). This defect is due to mutations in the ham5 gene, causing erratic splicing and loss of function (Linke et al., 2015; Tisch et al., 2017). While this mutation can now be complemented, it still causes a considerable obstacle in industrial strain improvement by crossing.
Here, we identified and analyzed twelve T. reesei strains of both mating types unexpectedly isolated in the temperate climate zone in Austria (Central Europe), which readily mate with common T. reesei lab strains and each other. They show production of plant cell wall degrading enzymes and different inducibility of asexual development by injury. Their application for strain improvement by crossing renders them a valuable resource for diverse plant cell wall degrading enzymes using diverse substrates. Fungal strain improvement and/or GMO-free optimization of production is moreover crucial if the resulting strains are to be distributed in nature (biocontrol applications) or due to labeling requirements of products from GMO organisms by the EU2.
In the course of a collection project aimed at isolation of novel Trichoderma strains from agricultural soil, we surprisingly found T. reesei strains in four independent locations in Austria (for GPS data see Table 1). The phylogenetic species concept (PSC) determines a species as a monophyletic group based on evolutionary history (Lee et al., 2010). Hence, species identification for the isolates from Austria was performed by sequence analysis of the tef1 diagnostic region (Samuels et al., 2002; Chaverri et al., 2003) and BLAST search against the NCBI nucleotide database. Phylogenetic analysis confirmed that the tef1 sequences amplified from the 12 strains tentatively assigned to T. reesei indeed clustered with T. reesei QM6a and several other T. reesei strains. The closely related T. parareesei strains clustered separately (Figure 1). The diagnostic rpb2 gene (Jaklitsch et al., 2008; Jaklitsch and Voglmayr, 2015) was used for additional confirmation for species identity, particularly because the pattern for AIT_MS44c2 in universal primers (UP-PCR) was divergent from the other strains (see also below). This analysis confirmed the species identity as T. reesei (Supplementary Figure 1 in Supplementary File 1). The 12 isolates showed clearly distinguishable phenotypes upon growth on plates with malt extract medium (Figure 2).

Figure 1. Phylogram obtained from an alignment of the tef1 locus. The evolutionary history was inferred by using the Maximum Likelihood method and Tamura-Nei model (Tamura and Nei, 1993). The tree with the highest log likelihood (–1894.50) is shown. The bootstrap test was carried out with 1,000 replicates (Felsenstein, 1985) and values >50 are shown. Species names are given along with the GenBank accession number of the sequence (as retrieved from the NCBI nucleotide database) used for the analysis.
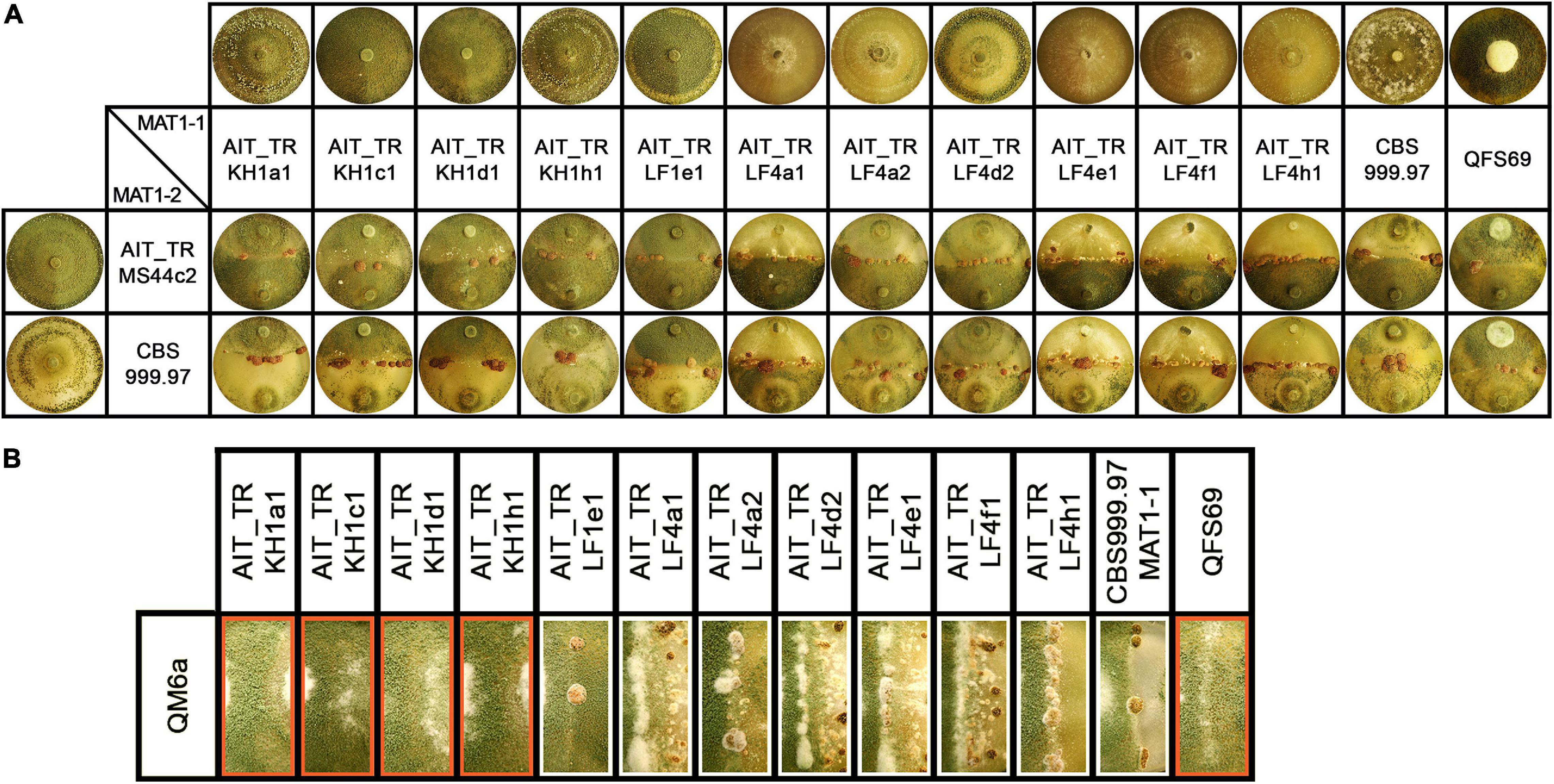
Figure 2. Growth and development of T. reesei strains isolated from Austrian soil. (A) Axenic growth and crossings of Austrian strains of MAT1-1 with AIT_TRMS44c2 (MAT1-2) and CBS999.97 MAT1-2 (wild-type control, female fertile). (B) Analysis of female fertility by crossing of Austrian soil isolates of MAT1-1 with the female sterile strain QM6a (MAT1-2). Abolished fruiting body formation indicates female sterility of the tested strains (Hinterdobler et al., 2020). Female sterile strains are indicated by red boxes. Strains were grown on malt extract agar plates (3% w/v) at 22°C with light-dark cycles. Phenotypes and fruiting body development are shown for day 14 after inoculation. One representative of three biological replicates is shown.
Previously, strains of the species T. reesei were not found in Europe and hence, T. reesei was assumed to be adapted to tropical climate. Consequently, we considered it of utmost importance to rule out any contamination by laboratory strains. In order to unequivocally distinguish these strains from our reference strains routinely used in the lab, we performed UP-PCR (Naeimi et al., 2011) using five different primers under different PCR conditions. All our isolates showed patterns distinct from the wild-type strains we are routinely using in the lab (Figure 3A, Supplementary Figure 2 in Supplementary File 1). The difference of QM6a from AIT_TRKH1a1, AIT_TRKH1c1, AIT_TRKH1d1, and AIT_TRKH1h1 is more subtle, but obvious in Figure 3A, showing one additional band migrating more slowly (green triangle) in QM6a. Moreover, these isolates also showed phenotypic differences with respect to cellulase regulation and injury response (see following sections). The differences between the individual isolates were not sufficiently clear in some cases and needed additional support (Figure 3A). Hence, we compared the banding patterns of all strains against each other and used additional criteria from our analyses for clarification (Figure 3B). Applying alternative primers, we could find characteristic patterns for most of the strains compared to the others (Supplementary Figures 3, 4 in Supplementary File 1). However, UP-PCR did not allow for clear separation of AIT_TRKH1a1, AIT_TRKH1c1, AIT_TRKH1d1, and AIT_TRKH1h1 from each other (Figure 3B). Therefore we compared the behavior of these strains in cellulase and xylanase production as well as injury response (see below). We found clear differences confirming that these strains are individuals rather than re-isolations of one and the same strain (Figure 3B). For further confirmation of the distinct origin of these strains, we screened the rapidly evolving region of the mating type locus for characteristics variations. We found a four base pair long INDEL and a SNP in the region adjacent to the mating type idiomorphs3, both of which are specific for the isolated strains and distinct from QM6a, RutC30, and CBS999.97 (Table 1 and Figure 3C).
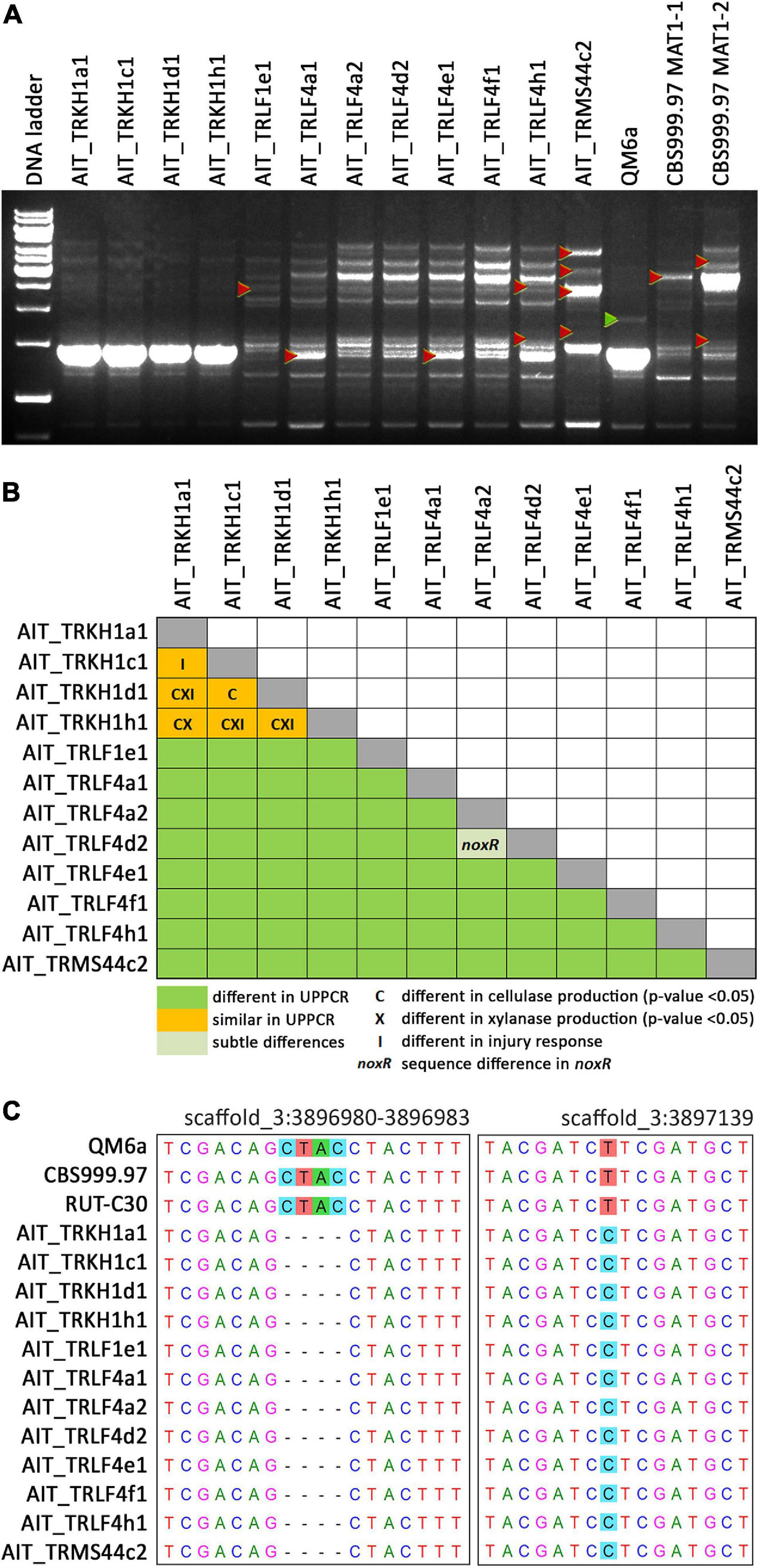
Figure 3. UP-PCR analysis, phenotypic differentiation and genetic alterations of strains. (A) UP-PCR of Austrian isolates along with QM6a and CBS999.97 MAT1-1 and MAT1-2 using as15inv as primer. For analysis with additional primers resulting in different patterns for separation of individual strains see also Supplementary Figures S2–S4 in Supplementary File 1. Triangles indicate bands indicative of differences to other strains. (B) Schematic representation of comparison of individual strains. UP-PCR band patterns and Supplementary Figures S2–S4 were used for evaluation. If no difference was found for band patterns, we considered sequence differences in noxR, altered enzyme production or injury response as well. (C) INDEL and SNP in the sequence flanking the mating type locus and specific for Austrian strains compared to laboratory strains in chromosome 3 (https://mycocosm.jgi.doe.gov/Trire_Chr/Trire_Chr.home.html; scaffold 3).
The biological species concept (BSC) supports the concept that sexually compatible individuals belong to the same species, while two populations that cannot mate represent different species (Lee et al., 2010). Sexual reproduction between strains of different species has not been shown for fungi previously and would contradict the BSC. Hence, the ability to mate with known and confirmed T. reesei strains can add further confirmation to species identity of the 12 strains investigated here.
Additionally, T. reesei strains with an origin in middle Europe and the potential of breeding for strain improvement would have a high potential for applications under natural conditions in this region. Consequently, we explored the prerequisites for that and found solid support for the strains belonging to the species T. reesei: Using mating type specific primers (Seidl et al., 2009) for T. reesei, we found that of the 12 new isolates, eleven strains comprised the MAT1-1 idiomorph and one strain was of MAT1-2. All strains showed efficient sporulation with a preference for asexual development in light, i.e., increased production of conidia in light compared to darkness. Due to the biotechnological importance of sexual development we tested all strains for mating (Schmoll, 2013) with fully fertile T. reesei strains under daylight conditions (Figure 2A). We found that all strains were able to undergo sexual development with CBS999.97 representing a fully fertile partner of opposite mating type under usual mating conditions.
Fruiting body formation of the new isolates with fully fertile strains started at 4–5 days after inoculation, which is roughly at the same time or even slightly earlier as seen with the control crossing of CBS999.97 MAT1-1 and CBS999.97 MAT1-2, our model strains for sexual development (Figure 2A). As female sterility was found to occur in natural strains of T. reesei, most importantly with QM6a (MAT1-2) (Seidl et al., 2009), we also tested whether our strains are female fertile by confrontation of the strains with the female sterile QM6a (MAT1-2). Indeed we found female sterility for four of our isolates (Figure 2B): AIT_TRKH1a1, AIT_TRKH1c1, AIT_TRKH1d1, and AIT_TRKH1h1. The only MAT1-2 strain, AIT_TRMS44c2, was crossed with a female sterile derivative of QM6a bearing the MAT1-1 locus, QFS69. Together with the fact that AIT_TRMS44c2 readily crossed with the obviously female sterile strains mentioned above, confirmed that AIT_TRMS44c2 is female fertile.
Interestingly, crossing of the MAT1-2 isolate AIT_TRMS44c2 with strains that could not form fruiting bodies with QM6a (Figure 2B, red boxes) was significantly delayed to 7–8 days for AIT_TRKH1c1, AIT_TRKH1d1 and AIT_TRKH1h1 as well as the control strain QFS69 or even 9 days with AIT_TRKH1a1. Also AIT_TRLF1e1 showed a delay to 6 to more than 7 days in all crossings. Consequently, the ability of all our new isolates to mate with different, known and confirmed T. reesei strains further confirmed species identity.
For T. reesei as an important species in biotechnological industry, production of enzymes is of particular interest. Therefore we tested our strain for production of cellulases and xylanases upon growth on Mandels Andreotti minimal medium with cellulose as carbon source. While the majority of the strains produced cellulases in the range of QM6a, some of the strains were considerably more efficient with up to a six-fold increase in cellulase efficiency (Figure 4A). Also for xylanase expression we found a considerably higher efficiency in some strains compared the QM6a (Figure 4B). As these strains showed also high cellulase production, they are the most interesting ones for further studies.
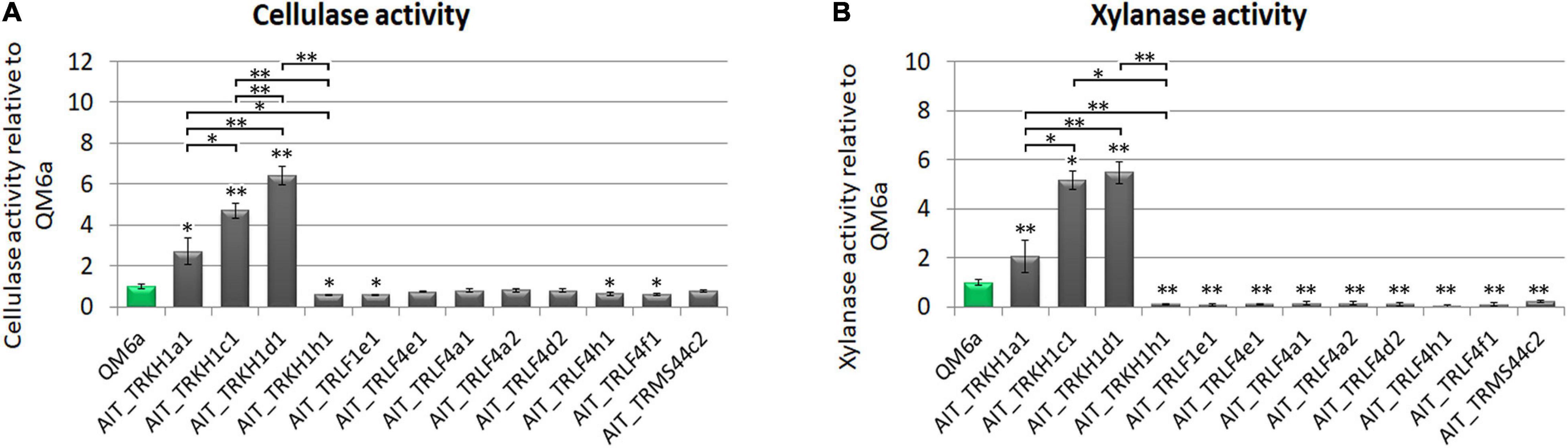
Figure 4. Enzyme production. (A) Cellulase activity and (B) xylanase activity represented by measurement of reducing sugars as released by enzymes secreted into the culture supernatant relative to wildtype strain QM6a. Strains were grown on Mandels Andreotti minimal medium with 1% (w/v) cellulose as carbon source in constant darkness for 72 h in 24-well plates with shaking. Error bars show standard deviations from three biological replicates. Statistically significant differences to wildtype are indicated by asterisks. Comparisons between individual strains are shown with brackets. *p-value < 0.05, **p-value < 0.01.
Asexual development is regulated in response to environmental conditions in fungi and is of high importance for application in biotechnology. Usually, sporulation in T. reesei is enhanced in light and diverse carbon sources modulate its efficiency (Friedl et al., 2008; Steyaert et al., 2010a,b). However, in some Trichoderma species, also injury was shown to induce sporulation (Casas-Flores et al., 2004). In the reference strain QM6a, we could not previously observe this phenomenon. Therefore we wanted to evaluate if this is a species specific characteristic or merely dependent on the strain.
The strains were grown in constant darkness for two days and then injured by cutting (Figure 5A). T. atroviride IMI206040 and P1, for which a strong response to injury in terms of conidiation is well known was used as a control (Casas-Flores et al., 2004; Hernandez-Oñate et al., 2012). In many cases, the growth front at the time of injury became clearly distinguishable, because growth after the injury commenced with mycelium, in which no premature sporulation was induced (Figures 5B,C). We observed a strong response in some, but not all strains (Figures 5–7). In QM6a and CBS999.97 we found weak, but detectable induction of sporulation by injury (Figure 5).
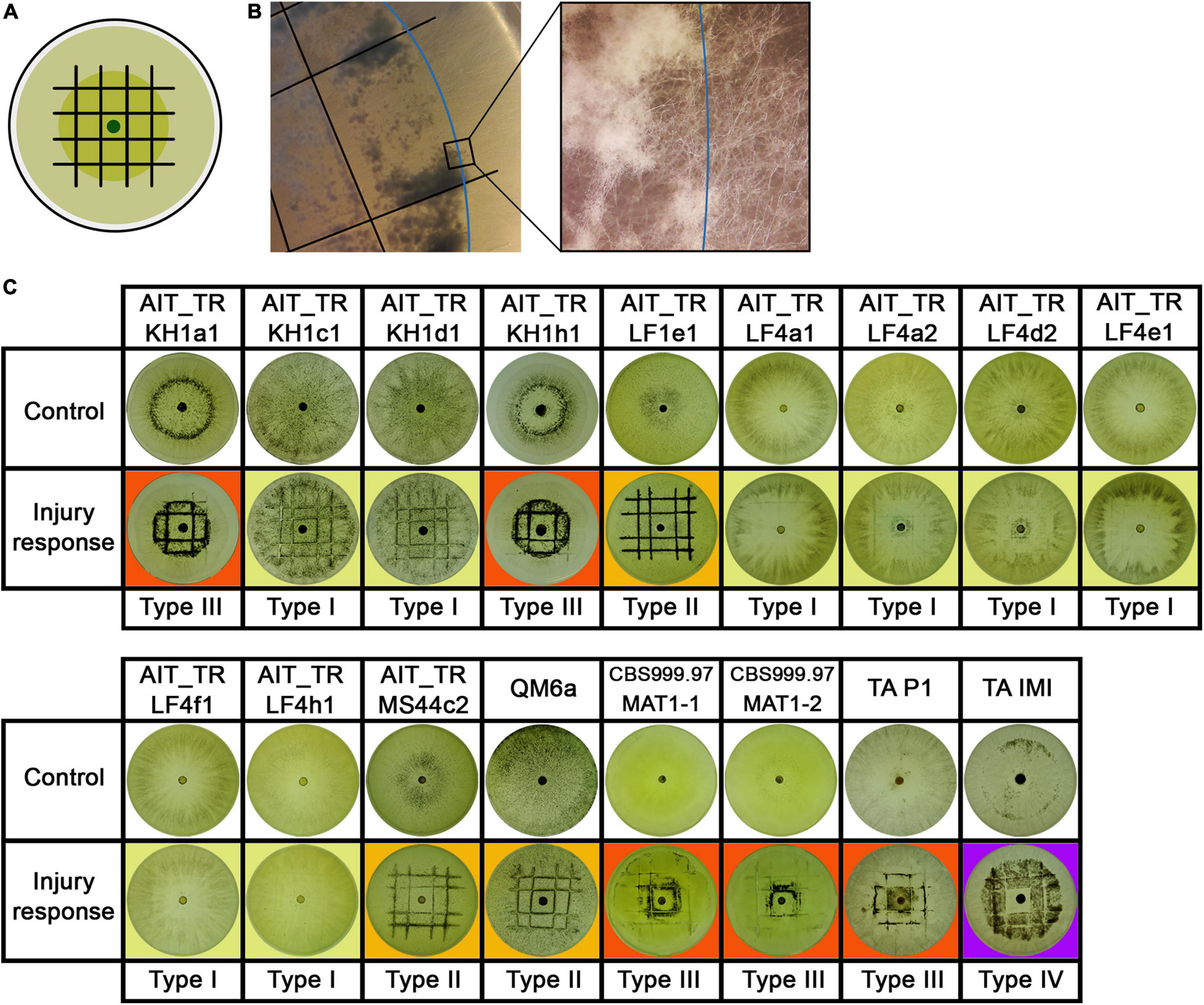
Figure 5. Injury response. (A) Illustration of the experimental setup. Strains were inoculated with an agar plug in the center of the petri dish. After 2 days of growth at 28°C in darkness, the mycelium (dark green) was injured under red safety light (black lines). Phenotypic changes of the young and developing (light green) mycelium were monitored 24 h after injury. (B) Shows the control strain T. atroviride IMI206040 (TA IMI). After injury (black lines) the young mycelium (blue line marks growth margin at timepoint of injury) grew dense, formed aerial mycelium and sporulated. The developing mycelium (right of blue line) shows undifferentiated morphology without sporulation after injury. (C) Shows response to injury of the tested strains. Four types of injury responses were observed: Type 1 (yellow background) strains show slight to no response to injury. Strains of type 2 (orange background) show clear sporulation close to the injury site. In type 3 (red background), the young mycelium shows a widespread response to the injury. Type 4 (purple background), represented only by TA IMI, shows a pattern similar to type III but with no sporulation around the inoculum.
Our results showed that the reaction to injury is highly strain-specific in T. reesei, and we could assign four distinguishable types of responses. We found a range from no reaction up to strong sporulation in the young mycelium present at the timepoint of injury (Figure 5). The reaction of T. atroviride IMI206040 with no sporulation close to the inoculum indicates a fast, coordinated signal transduction within the mycelium before the colony continues its radial growth (Figure 5C). While the morphological changes associated with sporulation are close to the injured mycelial areas in some strains, in other strains, injury triggers sporulation in the adjacent mycelium. Comparison between type III and type IV injury response (Figure 5C) suggests intracellular signal transduction in both directions for type III and only in direction toward the growth margin in type IV. Injury response of type IV shown here only for T. atroviride IMI206040 seems in addition to be medium dependent as a previous study (Hernandez-Oñate et al., 2012) described rather type II sporulation on PDA and minimal medium. Further research regarding the influence of the provided nutrients on intercellular injury signaling is need for a better understanding of the observed phenomenon.
Since growth and sporulation after injury were altered, we analyzed the site of injury shortly after cutting and the expected, thinner hyphae emerged from injured mycelial cells (Figure 6). Additionally, we were interested whether this growth after injury would be permanently and consistently altered. Therefore, we injured the young mycelium with one cut to be able to monitor further growth for a longer time (Figure 7A).
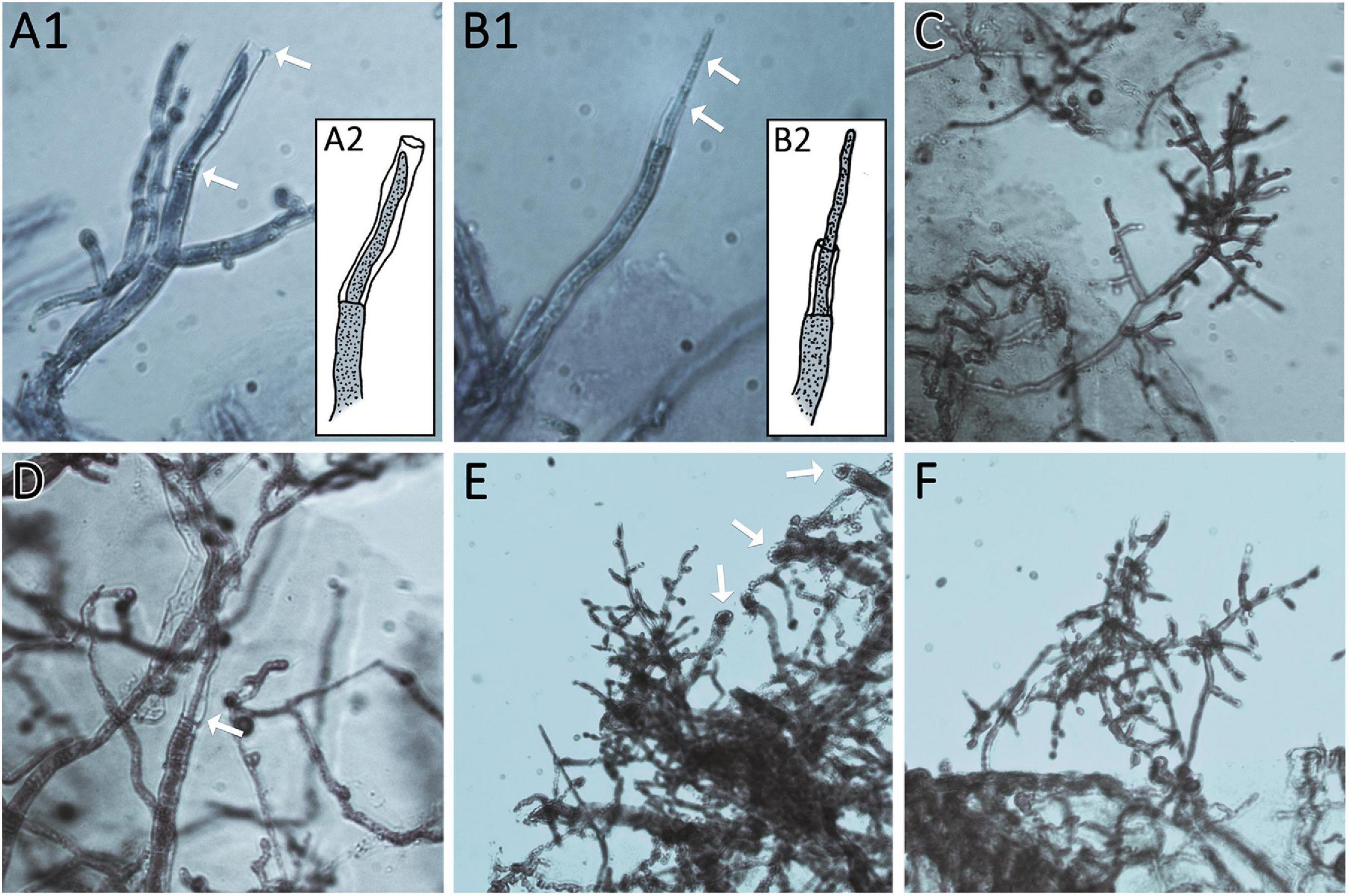
Figure 6. Thin regeneration hyphae and conidiophores emerge at the agar margin as response to previous injury. Edges of cut agar plugs are shown. (A1,2 and B1,2) Show regenerating hyphae of AIT_LF4a1 growing from the last intact septum of the cut hyphae. Arrows mark thin regenerating hyphae and empty cell walls of cut hyphae. (A2,B2) Show illustrations of (A1,B1) for better visualization. (C,D) Show AIT_LF4a2. (C) Shows an emerging conidiophore from the agar margin. (D) Shows a regenerating hypha growing in the empty cell wall of a cut hypha (arrow). (E,F) show AIT_LF4e1 with emerging conidiophores. Arrows in (E) point at the cutting sites of hyphae at the agar margin. All images were taken at 400× magnification. Samples were stained with methylene blue solution.
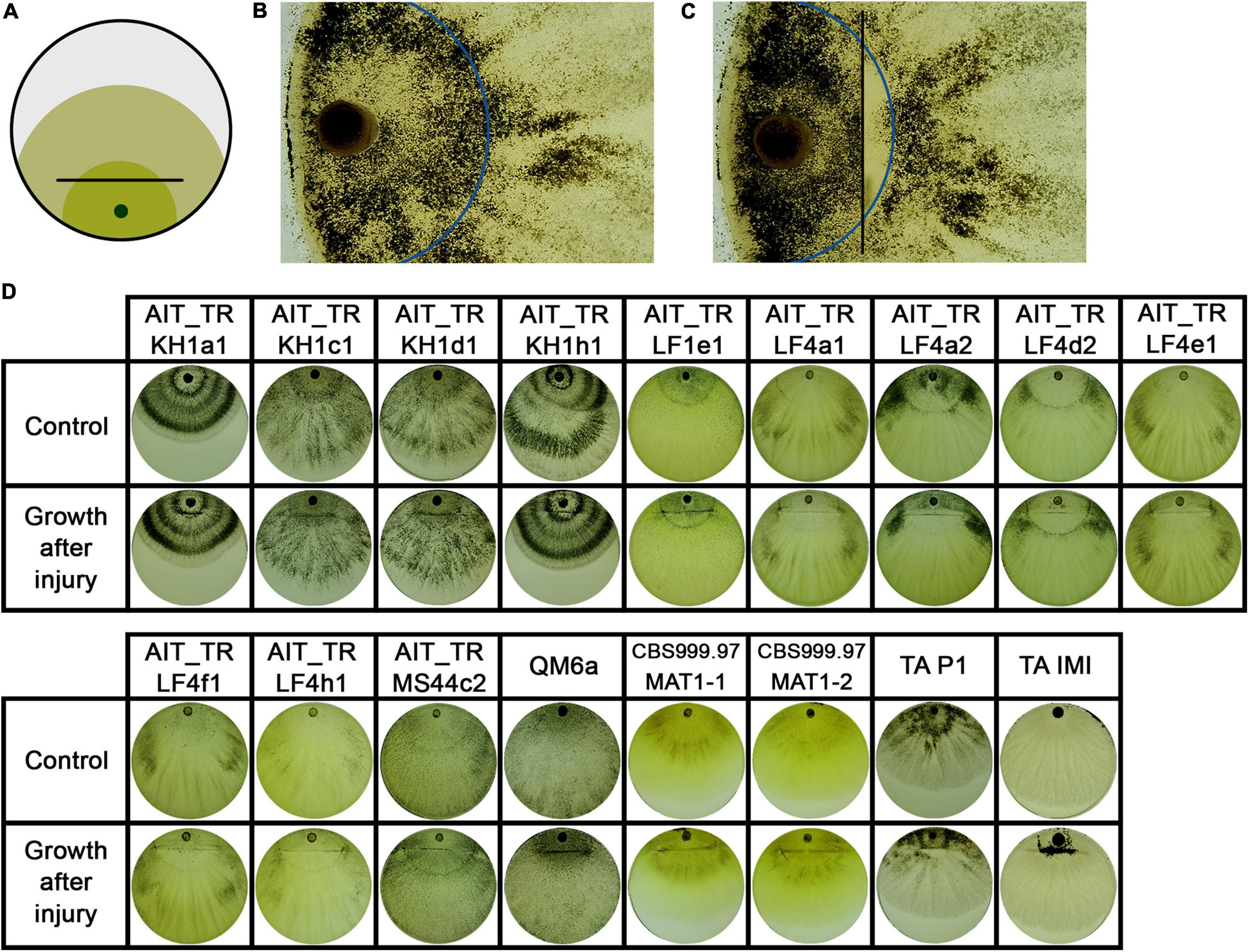
Figure 7. Growth after injury. (A) Illustration of the experimental setup. Strains were inoculated with an agar plug close to the border of the petri dish. After 2 days of growth at 28°C in darkness, the mycelium (dark green) was injured by cutting under red safety light (black line). Phenotypic changes were monitored 24 h after injury (light green). (B, C) The control strain T. atroviride P1 showed different sporulation pattern of the older compared to the younger colony area separated by the injury. Blue line indicates the growth margin at the timepoint of injury. In (C) the area between injury (black line) and the colony margin shows less sporulation compared to the old mycelium. (D) Shows phenotypic effects of early injury on the developing colonies compared to non-injured controls.
Interestingly, the separation of a small part of the young mycelium by injury as shown in Figure 7 did not affect the macroscopic phenotype of the emerging mycelium in all tested strains. In two strains, AIT_TRLF4d2 and T. atroviride P1 (Figures 7B,C), the separated, younger mycelium close to the growth margin at the timepoint of injury shows less sporulation than the emerging mycelium and the older mycelium close to the inoculum. This indicates a different influence of an injury event, either on young hyphae currently exploring new food sources compared to older hyphae, or an influence of the general size and energy reserves of the colony needed to react properly.
Since we saw differences in the injury response between many strains, also from the same locations, were interested if this might be reflected in the genome. Injury induced conidiation was found to be associated with an oxidative response and triggers NADPH oxidase (Nox)-dependent ROS production (Hernandez-Oñate et al., 2012). Fungi have between one and three NADPH oxidases (Aguirre et al., 2005), which are regulated by NoxR. Presence of the biosynthetic gene responsible for this process, noxR is required for fruiting body formation in Aspergillus nidulans (Lara-Ortiz et al., 2003). A function of nox1 in female fertility and asexual development was reported for Neurospora crassa (Cano-Dominguez et al., 2008). Therefore we evaluated whether the varied response in our strains would be due to different gene variants of the NADPH oxidase encoding nox1 or the regulator gene noxR in the strains of this study versus QM6a and CBS999.97.
For nox1, sequence analysis showed no alterations between QM6a, CBS999.97 and the 12 new isolates. However, the nucleotide sequences of noxR from the new isolates showed only a 98% identity to QM6a, but practically complete similarity with CBS999.97 (Supplementary File 1). Testing for SNPs in this sequence, which would alter the protein sequence, we found that despite numerous SNPs at the nucleotide level, the protein sequence remained the same for most strains. Or in other words, all of the SNPs were synonymous.
Interestingly, for one strain, AIT_TRLF4a2, we found alterations in the encoded protein sequence of NoxR in close proximity. In this region, no characteristic domains for NoxR are located. Also no phosphorylation sites were altered. However, testing for PEST regions, which are crucial for protein stability (Rechsteiner and Rogers, 1996) we found a potential PEST motif in QM6a and all other strains (SNAFPPTPPPEND, position 230-244, hydrophobicity index 29.94, and PEST score 8.84). In AIT_TRLF4a2, this sequence (nts marked in green in Supplementary File 1) is altered to SNVFPPTPPAEND, which causes a decrease in the PEST score to only 2.79 rendering it now a poor PEST motif. Due to the relevance of such PEST motifs for protein stability (Rechsteiner and Rogers, 1996), NoxR in AIT_TRLF4a2 may be more stable than in other strains. Since the phenotypic differences after injury are not limited to this strain, but are similar in other strains as well, the altered PEST sequence is unlikely to significantly contribute to characteristic injury response in T. reesei. Moreover, we also could not find indications that the minor sequence alterations would have an influence on sexual development in these strains.
Previously, the MAPkinase genes tmk1 and tmk3 were shown to be required for enhanced sporulation upon injury in T. atroviride (Medina-Castellanos et al., 2014). Therefore we analyzed the gene sequences of tmk1 and tmk3 for selected strains with different injury types (AIT_TRKHa1, AIT_TRKH1c1, AIT_TRLFe1, and AIT_TRMS44c2) in comparison with QM6a, RUTC30, and CBS999.97 (see Supplementary Material). Although these sequences formed groups according to their SNP patterns, they did not correlate with injury types in these strains.
We conclude that the differences in injury response that we observed for our isolates is not due to specifically altered alleles of nox1, noxR, tmk1, or tmk3.
Trichoderma spp. are among the few ascomycetes for which extensive reports on distribution in many areas all over the world are available (Druzhinina et al., 2006; Atanasova et al., 2013). These species are most commonly isolated, which has been attributed to their often mycoparasitic lifestyle and efficient enzyme production, which increase competitiveness in nature (Schuster and Schmoll, 2010; Druzhinina et al., 2011; Atanasova et al., 2013). [Early isolates originate from tropic areas, but detailed research in Europe revealed that the diversity of Trichoderma/Hypocrea in Europe consists of at least 75 holomorphic species (Jaklitsch, 2009, 2011) as well as several anamorphic species (Friedl and Druzhinina, 2012) and references therein]. Consequently, Trichoderma spp. are a major element of the mycoflora also in temperate regions. Due to their broad distribution, but also because of their efficiency in mycoparasitism and biocontrol of plant pathogens, Trichoderma spp. are included in commercial products for plant protection, which are applied in Europe as well. So far, no studies are available with investigations, whether strains applied with commercial biocontrol agents were possibly re-isolated during screening studies.
Trichoderma reesei only showed poor performance in mycoparasitism and biocontrol (Seidl et al., 2006) and is hence not commercially applied or thereby artificially distributed in nature. Consequently, isolation of T. reesei of non-natural origin would be due to an accidental contamination from labs working with this species and subsequent broad distribution, which is extremely unlikely. Actually, strains of T. reesei were not previously reported from screening studies in Austria (Wuczkowski et al., 2003; Friedl and Druzhinina, 2012) or in Europe (Jaklitsch, 2009, 2011). Unequivocal determination of species is of high and increasing importance due to the rising numbers of novel and interesting fungal isolates for diverse sources. Phylogenetic analysis using ITS sequences alone is not reliable and reproductive isolation is often difficult to confirm if sexual development cannot be achieved under laboratory conditions (Lücking et al., 2020). The strains we now describe here were confirmed to be T. reesei first by molecular methods using two diagnostic markers recommended for species identification, tef1 and rpb2 (Lücking et al., 2020) and phylogenetic analysis and second by crossing with known and accepted T. reesei strains. Although unisexual reproduction is known from some species and well-studied in Cryptococcus neoformans (Ni et al., 2011; Sun et al., 2014) there are no hints that this might occur in T. reesei or any other Trichoderma species. Complex phylogenies and reproductive strategies that render the reproductive mode unreliable for species determination are only known from lichen-associated fungi (Lücking et al., 2020), a case which is not comparable with Trichoderma spp. Hence, even if the molecular identification would leave room for interpretation, cross-species sexual development under laboratory conditions yielding fruiting bodies typical for T. reesei (Hinterdobler et al., 2020) would be an absolute novelty and can practically be excluded. Consequently, these 12 strains belong to the species T. reesei.
The fact that the 12 strains we investigated, originate from four individual samples raised the question whether they might be just re-isolations of one and the same strain from the respective location. Moreover, since T. reesei was never found in Europe before, it was important to unequivocally ensure that our screening study did not contain unwanted contaminants from T. reesei strains we routinely use in the lab. Also the potential introduction of T. reesei by the application of biocontrol products, which may be relevant for other taxonomic studies as outlined above, can rather be excluded for T. reesei, since commercial products are only known for the highly efficient Trichoderma species like T. asperellum, T. harzianum etc. UP-PCR and specific mutations clearly showed that no contaminations with lab strains had occurred and that these isolates indeed originated from Austrian agricultural soil. UP-PCR also showed that the isolates from different sites showed clearly different patterns. However, strains from the same site showed very similar patterns. In some cases, we therefore included also enzyme expression, injury response and phenotype in our analyses, which eventually allowed for discrimination between strains (Figure 2B). Therefore, we assume that the multiple strains from the separate sites are indeed individuals, albeit closely related, which suggests that sexual development and recombination likely have occurred in the past. This is particularly interesting with the female sterile strains, which are also clearly distinguishable. As they all are of the same mating type, strains of the opposite mating type can also be expected to occur in this habitat and should be female fertile.
Already in the initial study reporting sexual development with T. reesei under laboratory conditions (Seidl et al., 2009), the issue of female sterile strains in this species was broached. Early studies with T. reesei on the topic came to the conclusion, that it would be an asexually propagating, clonal species (Kuhls et al., 1996). However, the existence and evolutionary survival of exclusively clonal species was questioned (Taylor et al., 1999) and considered a rather transient phenomenon aimed to avoid the efforts to reproduce sexually. Thereby, both sexual and asexual propagation bear costs and benefits for fungi (Nieuwenhuis and James, 2016). The strains we isolated here represent interesting examples for such a strategy. We isolated both female fertile and female sterile strains, suggesting that strategies to avoid the costs of sex are occurring naturally and are unlikely to be associated with climate or a specific habitat. However, previously also loss of female fertility after repeated asexual propagation was reported and this loss was assumed to be genetic rather than epigenetic (Saleh et al., 2012). The emerging female sterile strains then propagated more efficiently via asexual reproduction without investing energy in mating and fruiting body formation and invaded the whole population (Saleh et al., 2012). Consequently, the isolation of female sterile strains could also be due to an environment in which the conditions for sexual development were not met and female fertility was lost. Thereafter the resulting female sterile strains could have been more successful in colonization of the respective habitat.
The achievement of sexual development under laboratory conditions for T. reesei (Seidl et al., 2009) represented already an important innovation in the field of strain improvement. However, initially the application was mainly seen with proprietary industrial strains and in academia for fungal genetics and strain improvement. With the strains we introduce here, a novel opportunity opens up. We can now use natural strains from Europe, which we can improve for their degradative performance without genetic manipulation, which is still seen critically. Particularly with (decentralized) pretreatment for biogas and biofuel production from agricultural cellulosic waste these strains will be a valuable resource. Deposition of waste does then not cause environmental issues with GMO distribution and pretreatment could even be done on site on the field. Moreover, in-process production of enzymes can be supported by natural strain improvement while alleviating environmental issues of waste disposal.
Last but not least, for the broad applications of enzymes in customer products the market pull for non-GMO ingredients is rising, and EU regulations for labeling of GMO use in production processes4 increase customer awareness. Hence, production without the use of GMO organisms can be an interesting future perspective for the use of natural enzyme producer strains like those introduced in this study. We could already achieve a proof of concept for natural strain improvement by crossing of these strains for enhanced enzyme production (Basyouni-Khamis and Schmoll, unpublished), which supports the applicability of this approach. First steps in this direction were previously reported for Myceliophthora heterothallica (Aguilar-Pontes et al., 2016). Besides practical application, we can thereby also follow up which natural mechanisms become enhanced when enzyme production is increased due to natural recombination.
While the use of these strains for improvement of enzyme production is most straight forward with T. reesei, also the potential for biocontrol applications should be considered. Trichoderma spp. have a long history in safe application in agriculture worldwide and are known as beneficial plant symbionts (Guzman-Guzman et al., 2019). Also T. reesei was previously shown to be effective in plant protection (Seidl et al., 2006), albeit with lower performance than known biocontrol agents, which is why it was not used commercially for this purpose so far. Moreover, previously only tropical strains of T. reesei were known, which were not assumed to survive in the European climate. Obviously, this assumption has to be revised, since we could isolate T. reesei independently from different locations in Austria. Therefore, the possibility to adapt T. reesei to antagonism against plant pathogens by sexual development warrants exploration, particularly with respect to oomycetes for which the cellulosic cell wall should be subject to efficient degradation by the potent enzyme machinery of T. reesei. First studies with T. reesei reference strains against Pythium ultimum (Seidl et al., 2006) and Phytophthora infestans (Schmoll, unpublished), showed already promising results.
Trichoderma reesei (syn. H. jecorina) wildtype strains QM6a [female sterile (Linke et al., 2015; Tisch et al., 2017)], MAT1-2, ATCC 13631; (Martinez et al., 2008), CBS999.97 [female fertile, both mating types; (Lieckfeldt et al., 2000; Seidl et al., 2009)] and QFS69 [female sterile, MAT1-1; (Bazafkan et al., 2015)] were applied as controls and mating partners. Strains were maintained on malt extract agar.
For determination of enzyme production, strains were grown in 24-well plates with 2 mL of Mandels Andreotti minimal medium (Mandels and Andreotti, 1978) with 1% (w/v) microcrystalline cellulose as carbon source for 72 h at 28°C in darkness. Enzyme activity was determined as described previously (Mellitzer et al., 2012) from three biological and tree technical replicates with QM6a as control. Statistical significance was calculated using Student’s t-Test in RStudio (compare_means, ggpubr v0.3.0). Sexual crossing was performed as described previously (Seidl et al., 2009; Schmoll, 2013). Strains were inoculated in similar distances to enable analysis of delays in fruiting body formation. Differences in growth rates between strains, which could alter the time to fruiting body formation, were negligible. Crossings were performed in three biological replicates and fruiting body formation was monitored daily.
All chemicals mentioned were supplied by Roth Lactan (Karlsruhe, Germany) unless stated otherwise.
Soil samples in which T. reesei strains were detected originated from four different locations in Austria (Table 1). One gram of soil was suspended in 10 mL of phosphate buffered saline (PBS) containing 0.1% (w/v) Triton X-100. After homogenization of 50 rpm for 30 min, the suspension was diluted 1:100 and 1:1000 with PBS and 100 μL were applied to malt extract agar (Merck, Darmstadt, Germany) containing 50 mg/L Rose Bengal sodium salt (Sigma, Steinheim, Germany). After incubation at room temperature under daylight conditions, appearing fungal colonies were isolated and subjected to single spore separation to obtain pure cultures. The strains are available for non-commercial academic use.
DNA extraction from young mycelia grown on malt extract agar plates was performed as described previously (Liu et al., 2000). Mating type determination of isolates was performed by PCR as described previously (Seidl et al., 2009). A diagnostic fragment of the genomic region encoding translation elongation factor 1 alpha (tef1) was amplified using the primers EF1-728F (Carbone et al., 1999) and TEF1 rev (Samuels et al., 2002). Primers for amplification of rpb1 and the INDEL/SNP containing region were designed based on the genome sequence publicly available at JGI (Martinez et al., 2008; Grigoriev et al., 2014; Li et al., 2017).
Amplification of sequences of nox1 and noxR was performed using primers nox1F1, nox1R1, noxRF1, and noxRR1. For primer sequences see Table 2.
Sequences were aligned using MUSCLE and adjusted in MEGA-X [Version 10.0.5, (Kumar et al., 2018)]. Phylogenetic analysis was done using the Maximum Likelihood method and Tamura-Nei model (Tamura and Nei, 1993). Trees with highest log likelihood are shown in Figure 1 and Supplementary Figure 1 for tef1 and rpb1 marker sequences, respectively. The bootstrap test was carried out with 1,000 replicates (Felsenstein, 1985). Sequences were retrieved from the NCBI nucleotide database for T. reesei, Trichoderma thermophilum (Qin and Zhuang, 2016), T. parareesei, Trichoderma rugosum, Trichoderma beinartii, Trichoderma aethiopicum, Trichoderma gracile, Trichoderma orientale, Trichoderma pinnatum, T. longibrachiatum, Trichoderma Gamsii, and T. atroviride. The diagnostic sequences for tef1 and rpb2 of the strains investigated in this study are deposited at GenBank, for accession numbers see Table 1.
For the injury assays, round agar plugs of well grown cultures were used as inoculum. T. atroviride strains P1 (ATCC 74058) and IMI206040 (Kubicek et al., 2011) as well as T. reesei strains CBS999.97 (MAT1-2 and MAT1-1) and QM6a (Martinez et al., 2008) were used as control. For both assays, 90 mm petri dishes containing 3% (w/v) malt extract medium were used. Strains were grown in constant darkness at 28°C for two days, injured as shown in Figures 5A, 7A under red safety light. Consequences of injury were documented and analyzed 24 hours after injury. Three biological replicates were performed.
For PCR using UP-PCR, DNA was isolated using the DNAeasy plant mini kit (QIAGEN, Heidelberg, Germany). UP-PCR was performed essentially as described earlier (Naeimi et al., 2011). Primers were tested individually and in combinations using gradient PCR with annealing temperatures ranging from 50–65°C. AS15inv, L45, 3-2 and a combination of L15/AS19 showed best results and were used for PCR amplification with DreamTaq Green Polymerase (ThermoFisher Scientific, Waltham, MA, United States).
All datasets relevant for this study are included in the article/Supplementary Material. Sequence information is available either online under the provided GenBank accession numbers or in supplementary material as noted in the text.
WH contributed to injury analysis, analysis of phenotype, and sexual development, phylogenetic analyses, statistical evaluations, and MAPkinase analysis as well as to figure design, interpretation, and writing of the manuscript. GL isolated and identified strains, and contributed experimental work for UP-PCR analysis, injury analysis, and amplification of nox1 and noxR sequences. KS performed analysis of sexual development. SB-K performed analysis of enzyme production. MG performed analysis of nox1 and noxR sequences. MS conceived of the study and designed experiments, contributed to analysis of nox1, noxR, and MAPkinase sequences, to figure design, interpreted results, and wrote the manuscript. All authors read and agreed to publication of the final manuscript.
WH was supported by the NFB (Science Fund of Lower Austria; grant LC16-04 to MS). Work of GL was supported by the Austrian Science Fund (FWF; projects P26935). KS was supported by a fellowship of the Austrian research promotion agency (FFG; FEM-tech fellowship for female talents). SB-K was supported by the FFG, the Austrian Research Promotion Agency (e!MISSION project 848780 to MS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Christina Paiato, Ida Aglaia Gisella Scalmani, and Sabrina Beier for technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.552301/full#supplementary-material
Aguilar-Pontes, M. V., Zhou, M., van der Horst, S., Theelen, B., de Vries, R. P., and van den Brink, J. (2016). Sexual crossing of thermophilic fungus Myceliophthora heterothallica improved enzymatic degradation of sugar beet pulp. Biotechnol. Biofuels 9:41. doi: 10.1186/s13068-016-0460-y
Aguirre, J., Rios-Momberg, M., Hewitt, D., and Hansberg, W. (2005). Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13, 111–118. doi: 10.1016/j.tim.2005.01.007
Arnau, J., Yaver, D., and Hjort, M. C. (2020). “Strategies and challenges for the development of industrial enzymes using fungal cell factories,” in Grand challenges in Fungal Biotechnology, ed. H. Nevalainen (Cham: Springer Nature Switzerland AG), 179–210. doi: 10.1007/978-3-030-29541-7_7
Ashton, G. D., and Dyer, P. (2016). “Sexual development in fungi and its uses in gene expression systems,” in Gene Expression Systems in Fungi: Advancements and Applications, eds M. Schmoll and C. Dattenböck (Cham: Springer International Publishing), 335–350. doi: 10.1007/978-3-319-27951-0_15
Atanasova, L., Druzhinina, I., and Jaklitsch, W. M. (2013). “Two hundred Trichoderma species recognized on the basis of molecular phylogeny,” in Trichoderma – Biology and Applications, eds P. K. Mukherjee, B. A. Horwitz, U. S. Singh, M. Mukherjee, and M. Schmoll (Wallingford: CAB International), 10–42. doi: 10.1079/9781780642475.0010
Bazafkan, H., Dattenböck, C., Böhmdorfer, S., Tisch, D., Stappler, E., and Schmoll, M. (2015). Mating type dependent partner sensing as mediated by VEL1 in Trichoderma reesei. Mol. Microbiol. 96, 1103–1118. doi: 10.1111/mmi.12993
Bischof, R. H., Ramoni, J., and Seiboth, B. (2016). Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell. Fact 15:106. doi: 10.1186/s12934-016-0507-6
Bissett, J., Gams, W., Jaklitsch, W., and Samuels, G. J. (2015). Accepted Trichoderma names in the year 2015. IMA Fungus 6, 263–295. doi: 10.5598/imafungus.2015.06.02.02
Bulat, S. A., Lübeck, M., Mironenko, N. V., Jensen, D. F., and Lübeck, P. S. (1998). UP-PCR analysis and ITS1 ribotyping of Trichoderma and Gliocladium fungi. Mycol. Res. 102, 933–943. doi: 10.1017/s0953756297005686
Cano-Dominguez, N., Alvarez-Delfin, K., Hansberg, W., and Aguirre, J. (2008). NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot Cell 7, 1352–1361. doi: 10.1128/EC.00137-08
Carbone, I., Anderson, J. B., and Kohn, L. M. (1999). Patterns of descent in clonal lineages and their multilocus fingerprints are resolved with combined gene genealogies. Evolution 53, 11–21. doi: 10.1111/j.1558-5646.1999.tb05329.x
Casas-Flores, S., Rios-Momberg, M., Bibbins, M., Ponce-Noyola, P., and Herrera-Estrella, A. (2004). BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology 150(Pt 11), 3561–3569. doi: 10.1099/mic.0.27346-0
Chaverri, P., Castlebury, L. A., Overton, B. E., and Samuels, G. J. (2003). Hypocrea/Trichoderma: species with conidiophore elongations and green conidia. Mycologia 95, 1100–1140. doi: 10.1080/15572536.2004.11833023
Chum, P. Y., Schmidt, G., Saloheimo, M., and Landowski, C. P. (2017). Transient silencing of DNA repair genes improves targeted gene integration in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 83:e00535-17. doi: 10.1128/AEM.00535-17
Druzhinina, I. S., Komon-Zelazowska, M., Ismaiel, A., Jaklitsch, W., Mullaw, T., Samuels, G. J., et al. (2012). Molecular phylogeny and species delimitation in the section Longibrachiatum of Trichoderma. Fungal. Genet. Biol. 49, 358–368. doi: 10.1016/j.fgb.2012.02.004
Druzhinina, I. S., Kopchinskiy, A. G., and Kubicek, C. P. (2006). The first 100 Trichoderma species characterized by molecular data. Mycoscience 47, 55–64. doi: 10.1007/s10267-006-0279-7
Druzhinina, I. S., Kopchinskiy, A. G., Kubicek, E. M., and Kubicek, C. P. (2016). A complete annotation of the chromosomes of the cellulase producer Trichoderma reesei provides insights in gene clusters, their expression and reveals genes required for fitness. Biotechnol. Biofuels 9:75. doi: 10.1186/s13068-016-0488-z
Druzhinina, I. S., Seidl-Seiboth, V., Herrera-Estrella, A., Horwitz, B. A., Kenerley, C. M., Monte, E., et al. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 9, 749–759. doi: 10.1038/nrmicro2637
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Friedl, M. A., and Druzhinina, I. S. (2012). Taxon-specific metagenomics of Trichoderma reveals a narrow community of opportunistic species that regulate each other’s development. Microbiology 158(Pt 1), 69–83. doi: 10.1099/mic.0.052555-0
Friedl, M. A., Kubicek, C. P., and Druzhinina, I. S. (2008). Carbon source dependence and photostimulation of conidiation in Hypocrea atroviridis. Appl. Environ. Microbiol. 74, 245–250. doi: 10.1128/aem.02068-07
Grigoriev, I. V., Nikitin, R., Haridas, S., Kuo, A., Ohm, R., Otillar, R., et al. (2014). MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42, D699–D704. doi: 10.1093/nar/gkt1183
Gudynaite-Savitch, L., and White, T. C. (2016). “Fungal biotechnology for industrial enzyme production: focus on (hemi)cellulase production strategies, advances and challenges,” in Gene Expression Systems in Fungi: Advancements and Applications, eds M. Schmoll and C. Dattenböck (Cham: Springer International Publishing Switzerland), 395–435. doi: 10.1007/978-3-319-27951-0_19
Guzman-Guzman, P., Porras-Troncoso, M. D., Olmedo-Monfil, V., and Herrera-Estrella, A. (2019). Trichoderma species: versatile plant symbionts. Phytopathology 109, 6–16. doi: 10.1094/PHYTO-07-18-0218-RVW
Harman, G. E. (2011). Multifunctional fungal plant symbionts: new tools to enhance plant growth and productivity. New Phytol. 189, 647–649. doi: 10.1111/j.1469-8137.2010.03614.x
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species–opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Hatvani, L., Manczinger, L., Vagvölgyi, C., and Kredics, L. (2013). “Trichoderma as a human pathogen,” in Trichoderma – Biology and Applications, eds P. K. Mukherjee, B. A. Horwitz, U. S. Singh, M. Mukherjee, and M. Schmoll (Wallingford: CAB International), 202–313.
Hernandez-Oñate, M. A., Esquivel-Naranjo, E. U., Mendoza-Mendoza, A., Stewart, A., and Herrera-Estrella, A. H. (2012). An injury-response mechanism conserved across kingdoms determines entry of the fungus Trichoderma atroviride into development. Proc. Natl. Acad. Sci. U.S.A. 109, 14918–14923. doi: 10.1073/pnas.1209396109
Hinterdobler, W., Beier, S. S. K., and Schmoll, M. (2020). “Sexual development, its determinants and regulation in Trichoderma reesei,” in Recent Developments in Trichoderma Research, eds S. Zeilinger, I. Druzhinina, H. B. Singh, and V. K. Gupta (Amsterdam: Elsevier), 185–206. doi: 10.1016/b978-0-12-819453-9.00009-x
Jaklitsch, W. M. (2009). European species of Hypocrea Part I. The green-spored species. Stud. Mycol. 63, 1–91. doi: 10.3114/sim.2009.63.01
Jaklitsch, W. M. (2011). European species of Hypocrea part II: species with hyaline ascospores. Fungal Divers. 48, 1–250. doi: 10.1007/s13225-011-0088-y
Jaklitsch, W. M., Gruber, S., and Voglmayr, H. (2008). Hypocrea seppoi, a new stipitate species from Finland. Karstenia 48, 1–11. doi: 10.29203/ka.2008.423
Jaklitsch, W. M., and Voglmayr, H. (2015). Biodiversity of Trichoderma (Hypocreaceae) in Southern Europe and Macaronesia. Stud. Mycol. 80, 1–87. doi: 10.1016/j.simyco.2014.11.001
Komon-Zelazowska, M., Neuhof, T., Dieckmann, R., von Dohren, H., Herrera-Estrella, A., Kubicek, C. P., et al. (2007). Formation of atroviridin by Hypocrea atroviridis is conidiation associated and positively regulated by blue light and the G protein GNA3. Eukaryot Cell 6, 2332–2342. doi: 10.1128/ec.00143-07
Kubicek, C. P., Herrera-Estrella, A., Seidl-Seiboth, V., Martinez, D. A., Druzhinina, I. S., Thon, M., et al. (2011). Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12:R40. doi: 10.1186/gb-2011-12-4-r40
Kuhls, K., Lieckfeldt, E., Samuels, G. J., Kovacs, W., Meyer, W., Petrini, O., et al. (1996). Molecular evidence that the asexual industrial fungus Trichoderma reesei is a clonal derivative of the ascomycete Hypocrea jecorina. Proc. Natl. Acad. Sci. U.S.A. 93, 7755–7760. doi: 10.1073/pnas.93.15.7755
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lara-Ortiz, T., Riveros-Rosas, H., and Aguirre, J. (2003). Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50, 1241–1255. doi: 10.1046/j.1365-2958.2003.03800.x
Lee, S. C., Ni, M., Li, W., Shertz, C., and Heitman, J. (2010). The evolution of sex: a perspective from the fungal kingdom. Microbiol. Mol. Biol. Rev. 74, 298–340. doi: 10.1128/MMBR.00005-10
Li, W. C., Huang, C. H., Chen, C. L., Chuang, Y. C., Tung, S. Y., and Wang, T. F. (2017). Trichoderma reesei complete genome sequence, repeat-induced point mutation, and partitioning of CAZyme gene clusters. Biotechnol. Biofuels 10:170. doi: 10.1186/s13068-017-0825-x
Lieckfeldt, E., Kullnig, C. M., Samuels, G. J., and Kubicek, C. P. (2000). Sexually competent, sucrose- and nitrate-assimilating strains of Hypocrea jecorina (Trichoderma reesei) from South American soils. Mycologia 92, 374–380. doi: 10.2307/3761493
Linke, R., Thallinger, G. G., Haarmann, T., Eidner, J., Schreiter, M., Lorenz, P., et al. (2015). Restoration of female fertility in Trichoderma reesei QM6a provides the basis for inbreeding in this industrial cellulase producing fungus. Biotechnol. Biofuels 8:155. doi: 10.1186/s13068-015-0311-2
Liu, D., Coloe, S., Baird, R., and Pederson, J. (2000). Rapid mini-preparation of fungal DNA for PCR. J. Clin. Microbiol. 38:471.
Liu, R., Chen, L., Jiang, Y., Zhou, Z., and Zou, G. (2015). Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 1:15007. doi: 10.1038/celldisc.2015.7
Lübeck, P. S., Alekhina, I. A., Lübeck, M., and Bulat, S. A. (1998). UP-PCR genotyping and rDNA analysis of Asochyta pisi Lib. J. Phytopathol. 148, 109–115. doi: 10.1046/j.1439-0434.2000.00467.x
Lücking, R., Aime, M. C., Robbertse, B., Miller, A. N., Ariyawansa, H. A., Aoki, T., et al. (2020). Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 11. doi: 10.1186/s43008-020-00033-z
Mandels, M., and Andreotti, R. (1978). Problems and challenges in the cellulose to cellulase fermentation. Proc. Biochem. 13, 6–13.
Marie-Nelly, H., Marbouty, M., Cournac, A., Flot, J. F., Liti, G., Parodi, D. P., et al. (2014). High-quality genome (re)assembly using chromosomal contact data. Nat. Commun. 5:5695. doi: 10.1038/ncomms6695
Martinez, D., Berka, R. M., Henrissat, B., Saloheimo, M., Arvas, M., Baker, S. E., et al. (2008). Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26, 553–560.
Medina-Castellanos, E., Esquivel-Naranjo, E. U., Heil, M., and Herrera-Estrella, A. (2014). Extracellular ATP activates MAPK and ROS signaling during injury response in the fungus Trichoderma atroviride. Front. Plant Sci. 5:659. doi: 10.3389/fpls.2014.00659
Mellitzer, A., Glieder, A., Weis, R., Reisinger, C., and Flicker, K. (2012). Sensitive high-throughput screening for the detection of reducing sugars. Biotechnol. J. 7, 155–162. doi: 10.1002/biot.201100001
Naeimi, S., Kocsube, S., Antal, Z., Okhovvat, S. M., Javan-Nikkhah, M., Vagvolgyi, C., et al. (2011). Strain-specific SCAR markers for the detection of Trichoderma harzianum AS12-2, a biological control agent against Rhizoctonia solani, the causal agent of rice sheath blight. Acta Biol. Hung 62, 73–84. doi: 10.1556/ABiol.61.2011.1.8
Ni, M., Feretzaki, M., Sun, S., Wang, X., and Heitman, J. (2011). Sex in fungi. Annu. Rev. Genet. 45, 405–430. doi: 10.1146/annurev-genet-110410-132536
Nieuwenhuis, B. P., and James, T. Y. (2016). The frequency of sex in fungi. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150540. doi: 10.1098/rstb.2015.0540
Paloheimo, M., Haarmann, T., Mäkinen, S., and Vehmaanperä, J. (2016). “Production of industrial enzymes in Trichoderma reesei,” in Gene Expression Systems in Fungi: Advancements and Applications, eds M. Schmoll and C. Dattenböck (Heidelberg: Springer International), 23–58. doi: 10.1007/978-3-319-27951-0_2
Qin, W., and Zhuang, W. (2016). Four species of Trichoderma with hyaline ascospores from central China. Mycol. Prog. 15, 811–815. doi: 10.1007/s11557-016-1211-y
Rechsteiner, M., and Rogers, S. W. (1996). PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21, 267–271. doi: 10.1016/s0968-0004(96)10031-1
Saleh, D., Milazzo, J., Adreit, H., Tharreau, D., and Fournier, E. (2012). Asexual reproduction induces a rapid and permanent loss of sexual reproduction capacity in the rice fungal pathogen Magnaporthe oryzae: results of in vitro experimental evolution assays. BMC Evol. Biol. 12:42. doi: 10.1186/1471-2148-12-42
Samuels, G. J., Dodd, S. L., Gams, W., Castlebury, L. A., and Petrini, O. (2002). Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia 94, 146–170. doi: 10.2307/3761854
Schmoll, M. (2013). “Sexual development in Trichoderma – scrutinizing the aspired phenomenon,” in Trichoderma: Biology and Applications, eds P. K. Mukherjee, B. A. Horwitz, U. S. Singh, M. Mukherjee, and M. Schmoll (Wallingford: CAB International), 67–86. doi: 10.1079/9781780642475.0067
Schmoll, M., Dattenböck, C., Carreras-Villasenor, N., Mendoza-Mendoza, A., Tisch, D., Aleman, M. I., et al. (2016). The genomes of three uneven siblings: footprints of the lifestyles of three Trichoderma species. Microbiol. Mol. Biol. Rev. 80, 205–327. doi: 10.1128/MMBR.00040-15
Schmoll, M., Seibel, C., Tisch, D., Dorrer, M., and Kubicek, C. P. (2010). A novel class of peptide pheromone precursors in ascomycetous fungi. Mol. Microbiol. 77, 1483–1501. doi: 10.1111/j.1365-2958.2010.07295.x
Schmoll, M., Seiboth, B., Druzhinina, I., and Kubicek, C. P. (2014). “Genomics analysis of biocontrol species and industrial enzyme producers from the genus Trichoderma,” in The Mycota XIII, eds K. Esser and M. Nowrousian (Berlin: Springer), 233–266. doi: 10.1007/978-3-642-45218-5_10
Schmoll, M., Zeilinger, S., Mach, R. L., and Kubicek, C. P. (2004). Cloning of genes expressed early during cellulase induction in Hypocrea jecorina by a rapid subtraction hybridization approach. Fungal Genet. Biol. 41, 877–887. doi: 10.1016/j.fgb.2004.06.002
Schuster, A., Bruno, K. S., Collett, J. R., Baker, S. E., Seiboth, B., Kubicek, C. P., et al. (2012). A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol. Biofuels 5:1. doi: 10.1186/1754-6834-5-1
Schuster, A., and Schmoll, M. (2010). Biology and biotechnology of Trichoderma. Appl. Microbiol. Biotechnol. 87, 787–799. doi: 10.1007/s00253-010-2632-1
Seibel, C., Tisch, D., Kubicek, C. P., and Schmoll, M. (2012). The role of pheromone receptors for communication and mating in Hypocrea jecorina (Trichoderma reesei). Fungal Genet. Biol. 49, 814–824. doi: 10.1016/j.fgb.2012.07.004
Seiboth, B., Ivanova, C., and Seidl-Seiboth, V. (2011). “Trichoderma reesei: a fungal enzyme producer for cellulosic biofuels,” in Biofuel Production – Recent Developments and Prospects, ed. M. A. dos Santos Bernardes (Rijeka: Intech), 309–340.
Seidl, V., Schmoll, M., Scherm, B., Balmas, V., Seiboth, B., Migheli, Q., et al. (2006). Antagonism of Pythium blight of zucchini by Hypocrea jecorina does not require cellulase gene expression but is improved by carbon catabolite derepression. FEMS Microbiol. Lett. 257, 145–151. doi: 10.1111/j.1574-6968.2006.00157.x
Seidl, V., Seibel, C., Kubicek, C. P., and Schmoll, M. (2009). Sexual development in the industrial workhorse Trichoderma reesei. Proc. Natl. Acad. Sci. U.S.A. 106, 13909–13914. doi: 10.1073/pnas.0904936106
Steyaert, J. M., Weld, R. J., and Stewart, A. (2010a). Ambient pH intrinsically influences Trichoderma conidiation and colony morphology. Fungal Biol. 114, 198–208. doi: 10.1016/j.funbio.2009.12.004
Steyaert, J. M., Weld, R. J., and Stewart, A. (2010b). Isolate-specific conidiation in Trichoderma in response to different nitrogen sources. Fungal Biol. 114, 179–188. doi: 10.1016/j.funbio.2009.12.002
Sun, S., Billmyre, R. B., Mieczkowski, P. A., and Heitman, J. (2014). Unisexual reproduction drives meiotic recombination and phenotypic and karyotypic plasticity in Cryptococcus neoformans. PLoS Genet. 10:e1004849. doi: 10.1371/journal.pgen.1004849
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. doi: 10.1093/oxfordjournals.molbev.a040023
Taylor, J., Jacobson, D., and Fisher, M. (1999). The evolution of asexual fungi: reproduction, speciation and classification. Annu. Rev. Phytopathol. 37, 197–246.
Tisch, D., Pomraning, K. R., Collett, J. R., Freitag, M., Baker, S. E., Chen, C. L., et al. (2017). Omics Analyses of Trichoderma reesei CBS999.97 and QM6a indicate the relevance of female fertility to carbohydrate-active enzyme and transporter levels. Appl. Environ. Microbiol. 83:e01578–17. doi: 10.1128/AEM.01578-17
Keywords: Trichoderma reesei, Hypocrea jecorina, sexual development, pretreatment for biofuels and biogas, cellulases and xylanases, biocontrol, plant cell wall degradation, strain improvement
Citation: Hinterdobler W, Li G, Spiegel K, Basyouni-Khamis S, Gorfer M and Schmoll M (2021) Trichoderma reesei Isolated From Austrian Soil With High Potential for Biotechnological Application. Front. Microbiol. 12:552301. doi: 10.3389/fmicb.2021.552301
Received: 15 April 2020; Accepted: 12 January 2021;
Published: 28 January 2021.
Edited by:
Jiwei Zhang, University of Minnesota Twin Cities, United StatesReviewed by:
Xinqing Zhao, Shanghai Jiao Tong University, ChinaCopyright © 2021 Hinterdobler, Li, Spiegel, Basyouni-Khamis, Gorfer and Schmoll. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monika Schmoll, bW9uaWthLnNjaG1vbGxAYWl0LmFjLmF0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.