
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 January 2021
Sec. Food Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.634255
This article is part of the Research Topic New Advances in Identification and Quantification of Foodborne Pathogens View all 13 articles
 Fan Li1,2†
Fan Li1,2† Qinghua Ye1†
Qinghua Ye1† Moutong Chen1†
Moutong Chen1† Jumei Zhang1
Jumei Zhang1 Liang Xue1
Liang Xue1 Juan Wang3
Juan Wang3 Shi Wu1
Shi Wu1 Haiyan Zeng1
Haiyan Zeng1 Qihui Gu1
Qihui Gu1 Youxiong Zhang1
Youxiong Zhang1 Xianhu Wei1
Xianhu Wei1 Yu Ding4*
Yu Ding4* Qingping Wu1*
Qingping Wu1*Listeria spp. is an important foodborne disease agent, often found in the fresh mushroom (Flammulina velutipes) and its production environment. The aim of this study was to develop multiplex PCR for rapid identification of Listeria monocytogenes and Listeria ivanovii, and nonpathogenic Listeria in F. velutipes plants. Pan-genome analysis was first used to identify five novel Listeria-specific targets: one for the Listeria genus, one for L. monocytogenes, and three for L. ivanovii. Primers for the novel targets were highly specific in individual reactions. The detection limits were 103–104 CFU/mL, meeting the requirements of molecular detection. A mPCR assay for the identification of pathogenic Listeria, with primers targeting the novel genes specific for Listeria genus (LMOSLCC2755_0944), L. monocytogenes (LMOSLCC2755_0090), and L. ivanovii (queT_1) was then designed. The assay specificity was robustly verified by analyzing nonpathogenic Listeria and non-Listeria spp. strains. The determined detection limits were 2.0 × 103 CFU/mL for L. monocytogenes and 3.4 × 103 CFU/mL for L. ivanovii, for pure culture analysis. Further, the assay detected 7.6 × 104 to 7.6 × 100 CFU/10 g of pathogenic Listeria spiked into F. velutipes samples following 4–12 h enrichment. The assay feasibility was evaluated by comparing with a traditional culture-based method, by analyzing 129 samples collected from different F. velutipes plants. The prevalence of Listeria spp. and L. monocytogenes was 58.1% and 41.1%, respectively. The calculated κ factors for Listeria spp., L. monocytogenes, and L. ivanovii were 0.97, 0.97, and 1, respectively. The results of the novel mPCR assay were highly consistent with those of the culture-based method. The new assay thus will allow rapid, specific, and accurate detection and monitoring of pathogenic Listeria in food and its production environment.
Listeria is a genus of gram-positive facultative anaerobic bacilli that is widely distributed in food and natural environments. The genus Listeria includes many species: Listeria monocytogenes, Listeria innocua, Listeria ivanovii, Listeria seeligeri, Listeria welshimeri, Listeria grayi, Listeria rocourtiae, Listeria fleischmannii, and Listeria marthii, etc. (Vitullo et al., 2013). Among them, L. monocytogenes and L. ivanovii are human and animal pathogens (Snapir et al., 2006; Jagadeesan et al., 2019; Lee et al., 2020). These bacteria can cause severe listeriosis, leading to spontaneous abortion, neonatal sepsis, and meningoencephalitis, and the post-infection mortality rate ranges from 20 to 30% (Radoshevich and Cossart, 2018; Ranjbar and Halaji, 2018), which places this pathogen among the most frequent causes of death from foodborne illnesses (Tamburro et al., 2015b).
Listeria spp. can contaminate food at multiple stages of production and distribution because it is ubiquitous in the environment and tolerates harsh environmental conditions (Tamburro et al., 2015a). Edible mushrooms are potential vehicles for the transmission of pathogenic Listeria (Chen et al., 2014, 2018). For example, our previous study found that the prevalence of Listeria spp. and L. monocytogenes in F. velutipes production plants is 52.5% (155/295) and 18.6% (55/295), respectively (Chen et al., 2014). However, currently, the gold standard method for detection of Listeria in F. velutipes is the conventional culture method (Chen et al., 2014; Tao et al., 2017), which is labor-intensive, expensive, and time-consuming. It is therefore important to develop rapid and accurate diagnostic techniques or tools for the detection and distinguishing of pathogenic Listeria in F. velutipes, to facilitate introduction of appropriate intervention measures in the F. velutipes production chain to reduce the risk of Listeria contamination.
Polymerase chain reaction (PCR) has been widely employed as a rapid and specific method for the detection of Listeria in a variety of foods and processing environments because of its high specificity, sensitivity, time-saving and easy operation (Norton et al., 2000; Ryu et al., 2013; Rosimin et al., 2016; Sheng et al., 2018). However, most of the reported PCR-based methods for the identification and characterization of Listeria target the bacterial virulence genes (Doijad et al., 2011; Mao et al., 2016; Li et al., 2017; Tamburro et al., 2018, 2019), or 16S and 23S rRNA genes (Czajka et al., 1993; Hellberg et al., 2013), which have a limited number of targets, and some genes cannot accurately identify target bacteria. For example, avirulent L. monocytogenes strains may lack one or more virulence genes due to mutations or low gene expression under certain conditions (Nishibori et al., 1995). In addition, the highly conserved sequence of the 16S rRNA gene may fail to distinguish between Listeria species, especially L. monocytogenes, L. innocua, and L. welshimeri (Hellberg et al., 2013; Tao et al., 2017). Therefore, mining of target genes with high species-specificity is vital for improved accuracy, specificity, and efficiency of pathogen detection.
At present (as of 27 May 2019), a large number of sequenced genomes of Listeria strains are available from the Genome Database of the National Center for Biotechnology Information (NCBI)1. Using a wealth of genome data, pan-genome analysis has become a representative discipline for studying an entire repertoire of gene families in genomes of a clade of pathogenic bacteria, which provides not only the whole set of genes shared among Listeria species but also can be applied for inter-species differentiation analysis to mine species-specific genes (Kuenne et al., 2013; Kim et al., 2020). In the current study, we aimed to devise a novel mPCR assay for accurate detection and identification of pathogenic Listeria. The pan-genome analysis was performed to screen the genus- and species-specific genes. Using the identified genes, a mPCR method was developed to simultaneously detect L. monocytogenes, L. ivanovii, and nonpathogenic Listeria. Then the assay was used to evaluate the prevalence of pathogenic Listeria (including L. monocytogenes and L. ivanovii) in samples from F. velutipes plants.
A total of 205 genome sequences of pathogenic bacteria were downloaded from the NCBI Genome Database (last accessed on 27 May 2019), including 165 Listeria sequences and 40 other common foodborne pathogens sequences. The specific information for the sequences is provided in Supplementary Table S1. The Pan-genome analysis was used to identify Listeria genus- and species-specific genes. Briefly, all nucleic acid sequences were annotated using Prokka v1.11 (Seemann, 2014). The output of Prokka was then used to construct a pan-genome analysis using Roary v3.11.2 (Page et al., 2015). A core genome was determined for each strain using a 99% cutoff, with a BLASTP identity cutoff of 85% (Chen et al., 2019; Pang et al., 2019). The absence/existence profile of all genes across strains was converted into a 0/1 matrix with local script. The Listeria genus-specific (or species-specific) genes were screened according to the following criteria: 100% presence in Listeria (or L. monocytogenes, or L. ivanovii) strains and no presence in non-Listeria (or non-target Listeria species) strains. Then these candidate targets were used as further screened against the nucleotide collection (nr/nt) databases using the online BLAST program2 and PCR verification to ensure its specificity. At the same time, the specific targets reported in the previous studies [including prs specific for Listeria genus (Doumith et al., 2004), actA (Gouin et al., 1995; Zhou and Jiao, 2005), hly (Paziak-Domańska et al., 1999), and inlA (Jung et al., 2003) specific for L. monocytogenes, and iactA specific for L. ivanovii (Gouin et al., 1995)] were also analyzed in the 0/1 matrix to evaluate their absence/existence profile.
A total of 126 bacteria strains were used in the study, which were obtained respectively from the Chinese Center for Disease Control and Prevention, the College of Food Science and Technology of Nanjing Agricultural University, the Guangdong Huankai Co., Ltd., the State Key Laboratory of Food Science and Technology of Nanchang University, and our laboratory (Table 1 and Supplementary Table S2). For DNA extraction, all strains were inoculated in brain heart infusion (BHI) medium (Guangdong Huankai Co., Ltd., Guangzhou, China) and incubated for 8–12 h at 37 °C. DNA was extracted using DNeasy Blood and Tissue kit (Qiagen, Shanghai, China), according to the manufacturer’s instructions. Extracted DNA was stored at –20 °C for PCR analysis.
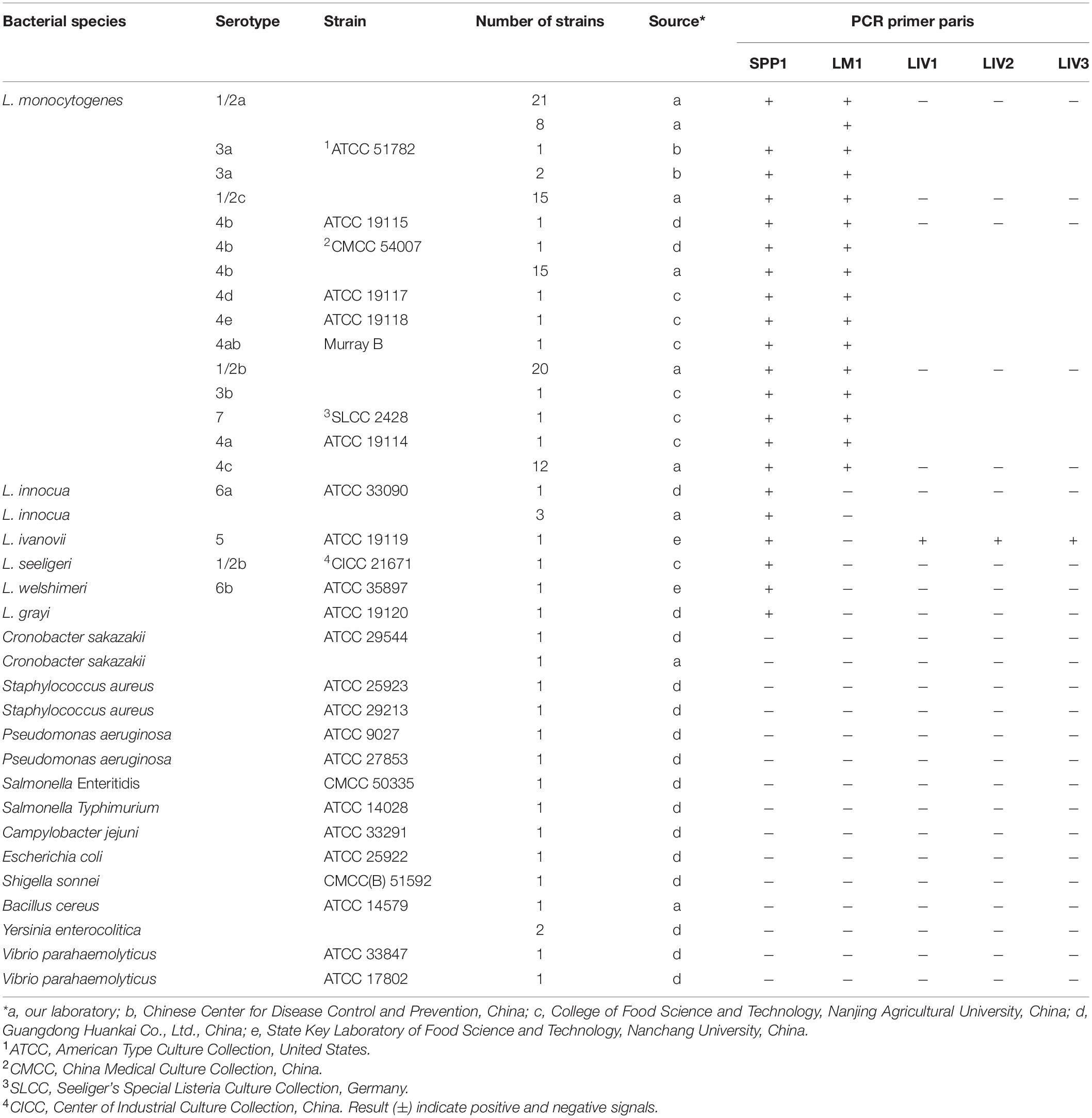
Table 1. Listeria spp. and other common foodborne pathogenic strains used in this study to verify the specificity of candidate targets.
Primers for the candidate Listeria genus- and species-specific target genes were designed using the Oligo 7.0 software. Candidate primer sets were synthesized by Generay Biotech Co., Ltd., Shanghai, China. Primer specificity was tested by PCR analysis of strains from the laboratory collection (Table 1). Detection limits were determined using serially (10-fold) diluted cell suspensions of fresh cultures of L. monocytogenes ATCC 19115 and L. ivanovii ATCC 19119. Viable cells counts were determined by plating of 100 μL of 10–6, 10–7, and 10–8 dilutions of bacterial cultures on trypticase soy-yeast extract agar plates (Guangdong Huankai Co., Ltd., Guangzhou, China) and incubating overnight at 37 °C. PCR was performed using 2 μL of genomic DNA extracted from 1 mL different dilutions of cell mixtures as a template, with genus or species-specific primers. An equal volume of sterile distilled water was used instead of the template as a negative control. PCR thermal cycling involved an initial denaturation step at 95 °C for 10 min; followed by 35 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; and a final extension step at 72 °C for 10 min. PCR products were evaluated by 1.5% agarose electrophoresis.
In the genus Listeria, only two species, L. monocytogenes and L. ivanovii, are human and animal pathogens. Therefore, a mPCR assay was devised by combining three specific primer sets targeting genes specific for the Listeria genus, L. monocytogenes, and L. ivanovii (Table 2). Multiplex PCR was performed in a volume of 25 μL; the reaction mixture contained 12.5 μL DreamTaq Green PCR Master Mix (2×), 0.1 mM dNTPs, 6 mM MgCl2, 1.5 U TaKaRa Ex Taq, 1.6 μM for Listeria genus-specific primers targeting LMOSLCC2755_0944, 0.32 μM for L. monocytogenes-specific primers targeting LMOSLCC2755_0090 and 0.32 μM for L. ivanovii-specific primers targeting queT_1, and 4 μL of template. The genomic DNA of L. monocytogenes and L. ivanovii was added as a template for the positive control, and an equal volume of sterile distilled water was used instead of the template as a negative control. Multiplex PCR thermal cycling involved an initial denaturation step at 95 °C for 10 min; followed by 35 cycles at 95 °C for 30 s, 58.1°C for 30 s, and 72 °C for 30 s; and a final extension step at 72 °C for 10 min. The mPCR products were analyzed after electrophoresis on 2.5% agarose gel under ultraviolet light illumination.
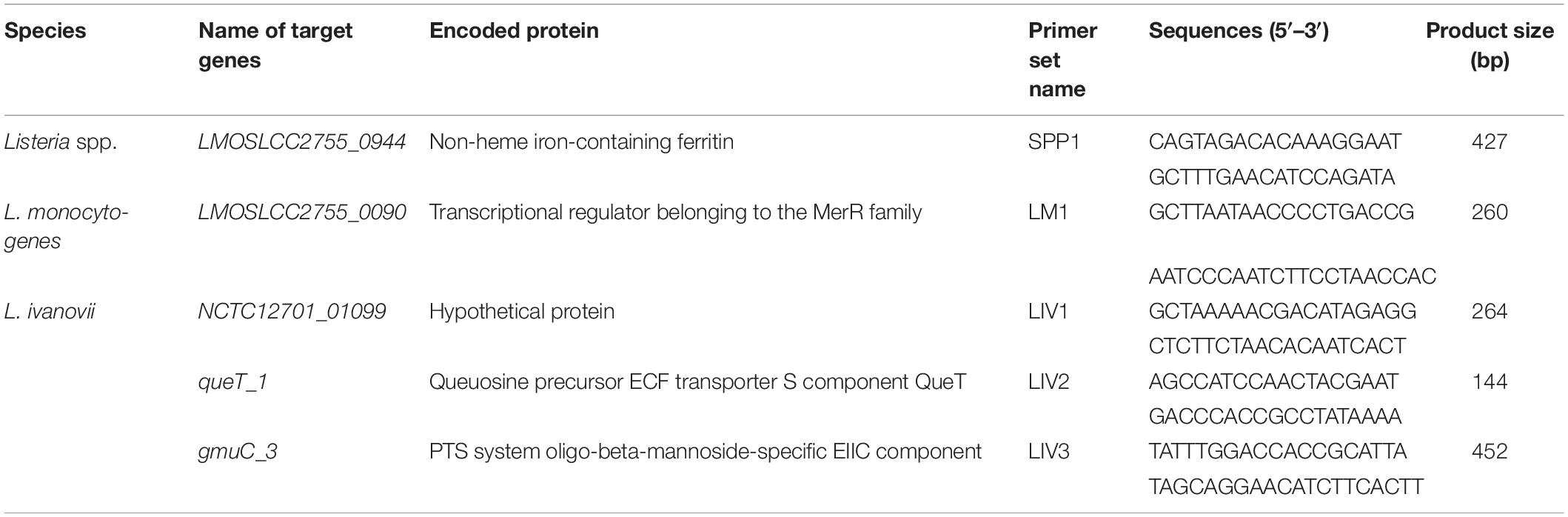
Table 2. Specific genes and primers for PCR identification of Listeria spp. and pathogenic Listeria.
The specificity of mPCR with novel specific primers was verified using 33 bacteria strains (including 17 Listeria strains and 16 non-Listeria strains) (Supplementary Table S2). Primer sets that amplified bands of the expected length from the corresponding strains of Listeria but not from non-Listeria strains were considered species-specific. For the detection limit evaluation, Listeria cultured overnight were diluted 10-fold in normal saline and mixed. Purified genomic DNA extracted from different dilutions of cell suspensions was used as the mPCR template (4 μL). An equal volume of sterile distilled water was included in the mPCR mixture instead of the template, as a negative control. The sensitivity of the PCR was determined by electrophoresis, as described in section “Multiplex PCR Conditions for Detection of Pathogenic Listeria.”
Artificial contamination experiments were performed as previously described (Chen et al., 2019). Briefly, L. monocytogenes ATCC 19115 and L. ivanovii ATCC 19119 were cultured overnight, and the cell concentration was determined by plating. F. velutipes samples (10 g) were mixed by 88 mL of the BHI medium, and then 2 mL Listeria mixtures were added with the final inoculum concentration ranging from 7.6 × 100 to 7.6 × 104 CFU/10 g. Next, the mixtures were incubated at 37 °C for 4 h, 6 h, 8 h, 10 h, or 12 h. Genomic DNA was extracted at the indicated time points from 1 mL of sample, and then analyzed by mPCR.
To validate the accuracy and scope for interference in the mPCR assay, L. monocytogenes ATCC 19115, L. ivanovii ATCC19119, and three common pathogens (Salmonella Enteritidis CMCC 50335, Staphylococcus aureus ATCC 25923, and Escherichia coli ATCC 25922) were used. The strains were cultured in the BHI broth overnight and serially diluted (10-fold) with normal saline. The density of Listeria cells was adjusted to 106 CFU/mL. Listeria cultures were individually mixed with the interference testing strain in ratios of 1:102, 1:10, 1:1, 10:1, and 102:1. Genomic DNA was extracted from the mixtures. Meanwhile, the genomic DNA from Listeria cultures without interference strains was used as a template for the positive control. The scope of mPCR assay to overcome interference was evaluated by 2.5% agarose gel electrophoresis.
To validate the detection ability of mPCR, a total of 129 samples were collected from F. velutipes plants in Guangdong Province (China). The sampled sites included composting sites (compost and sterile compost, n = 15), mycelium culture rooms (cultures and shelf surfaces, n = 18), mycelium stimulation rooms (mycelium stimulation machinery and floor, n = 18), fruiting body cultivation rooms (drains, shelf surfaces, and F. velutipes, n = 27), and harvesting rooms (packaging machinery surfaces, scales, conveyor belts, cutler surfaces, packaged F. velutipes, floor, drains, and workers, n = 51). The sites at the F. velutipes plants were randomly sampled, and the samples were transported on ice to the laboratory for immediate analysis. The conventional culture method was used to test for the presence of Listeria as previously described (Chen et al., 2019). Briefly, 25 g of each sample was randomly weighed-out, added to 225 mL of Listeria enrichment broth 1 (LB1, Guangdong Huankai Co., Ltd., Guangzhou, China), and incubated at 30 °C for 24 h. After incubation, 100 μL of LB1 enrichment culture was transferred to 10 mL of Listeria enrichment broth 2 (LB2, Guangdong Huankai Co., Ltd., Guangzhou, China), and incubated at 30 °C for 24 h. A loopful (approximately 10 μL) of the LB1 enrichment culture was streaked onto Listeria selective agar plates (Guangdong Huankai Co., Ltd., Guangzhou, China), and incubated at 37 °C for 48 h. At least three presumptive colonies were selected for the identification of Listeria using the Microgen ID Listeria identification system (Microgen, Camberley, United Kingdom), according to the manufacturer’s instructions. Meanwhile, 1 mL of LB1 enrichment culture was collected from each sample at 12 h. Genomic DNA was extracted from LB1 enrichment cultures for mPCR.
The Kappa coefficient (κ) was used to evaluate the performance of the developed mPCR assay. Since the true pathogenic Listeria status in naturally contamination samples was unknown, this calculation method assumed that conventional culture analysis was the true value. The calculation method was performed as previously described (Tao et al., 2017).
In the current study, 205 genomic sequences (Supplementary Table S1) were included in pan-genome analysis to identify novel molecular targets for the detection and differentiation of L. monocytogenes and L. ivanovii, and nonpathogenic Listeria species. After filtering using pan-genome and PCR analysis, five novel Listeria-specific targets, including LMOSLCC2755_0944 (165/165) specific for Listeria genus, LMOSLCC2755_0090 (128/128) specific for L. monocytogenes, NCTC12701_01099 (9/9), queT_1 (9/9), and gmuC_3 (9/9) specific for L. ivanovii, were uniquely present in all target strains, but not in non-target strains (Tables 1, 3 and Supplementary Figure S1). The Listeria-specific targets reported in the previous studies, including prs, actA, hly, inlA, and iactA, were used to specifically target strains and detected these targets in the current study, with 98.8% (163/165), 97.7% (125/128), 99.2% (127/128), 71.9% (92/128), and 55.6% (5/9) of target strains harboring the marker, respectively (Table 3).
As shown in Table 2, for the Listeria genus, only one gene (LMOSLCC2755_0944, encoding a non-heme iron-containing ferritin) was identified as genus-specific. The four other genes were species-specific: one gene (LMOSLCC2755_0090, encoding a transcriptional regulator from the MerR family) was specific for L. monocytogenes, and three genes (NCTC12701_01099, encoding a hypothetical protein; queT_1, encoding a queuosine precursor ECF transporter S component QueT; and gmuC_3, encoding a PTS system oligo-beta-mannoside-specific EIIC component) were specific for L. ivanovii.
Before devising a mPCR assay, the sensitivity of primers for each target gene was evaluated in an individual PCR assay. The results are shown in Supplementary Table S3 and Figure 1. The detection limit of these primer pairs was 103–104 CFU/mL. Considering the amplicon lengths for different genes, the sensitivity of species-specific primers, and the competition and inhibition of primer sets in the reaction system, a combination of suitable primer pairs (SPP1, LM1, and LIV2) targeting the LMOSLCC2755_0944, LMOSLCC2755_0090, and queT_1 genes (respectively) was selected for the mPCR assay.
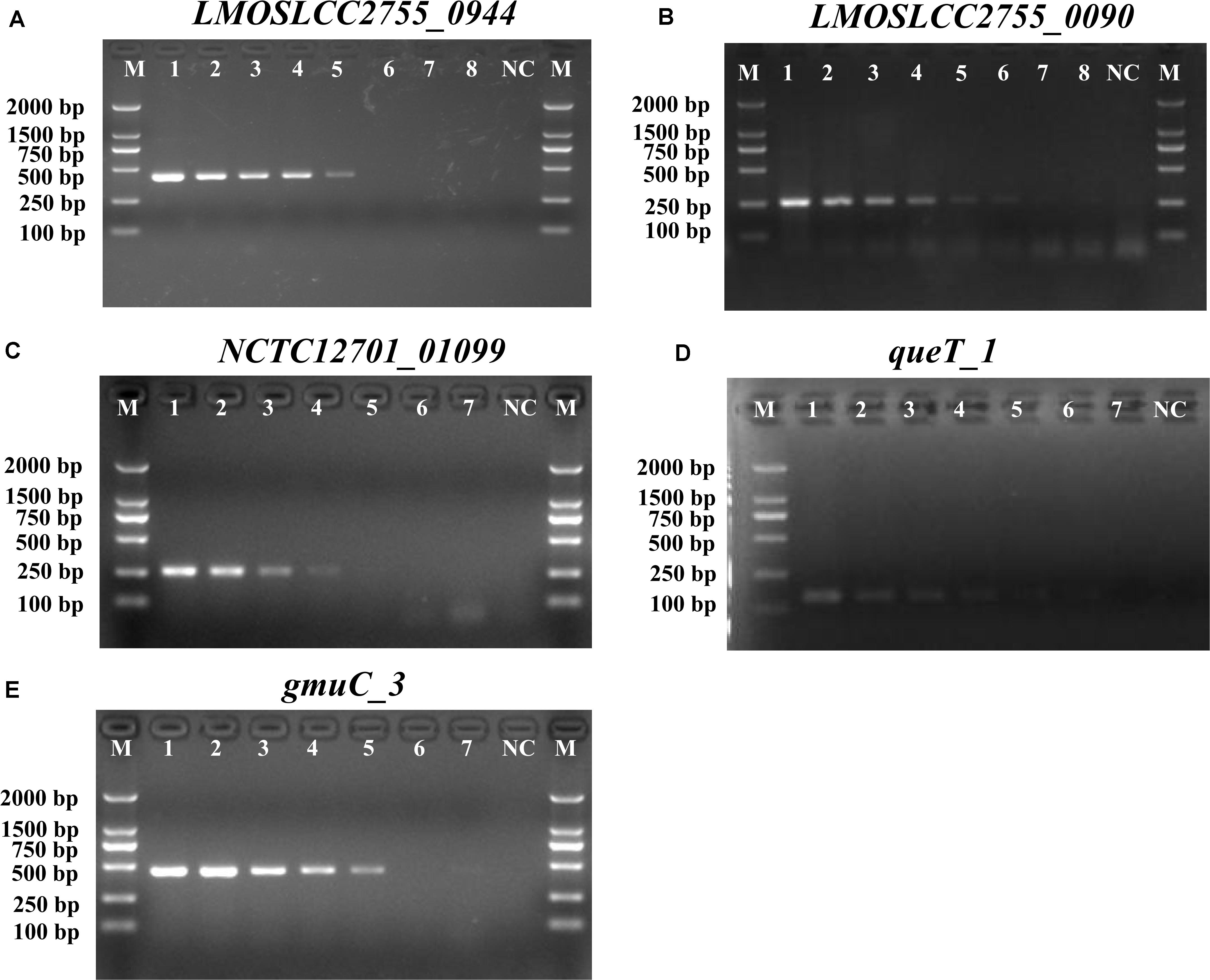
Figure 1. PCR detection sensitivity of novel targets specific for Listeria. (A) LMOSLCC2755_0944 specific for the Listeria spp.; (B) LMOSLCC2755_0090 specific for L. monocytogenes; (C) NCTC12701_01099; (D) queT_1; and (E) gmuC_3 specific for L. ivanovii. Lane M: DL2000 DNA standard marker; (A) lanes 1–8: the bacterial culture concentration per PCR assay from 107 to 100 CFU/mL; (B) lanes 1–8: the bacterial culture concentration per PCR assay from 108 to 101 CFU/mL; (C–E) lanes 1–7: the bacterial culture concentration per PCR assay from 107 to 101 CFU/mL; lane NC: negative control.
The results of the specificity of mPCR assay with novel specific primers are shown in Supplementary Table S2. The genus-specific target band of 427 bp was only obtained with Listeria spp. as the template. Further, species-specific bands were also obtained for the L. monocytogenes and L. ivanovii strains tested. No product bands were obtained with the 16 non-Listeria strains tested, and no cross-reactivity of the mPCR was observed.
To determine the detection limit of the novel assay, DNA was extracted from different dilutions of Listeria spp. cultures and used as the reaction template. Following mPCR detection, three specific fragments of 427 bp (SPP1), 260 bp (LM1), and 144 bp (LIV2) were obtained for cell concentrations of 107 to 103 CFU/mL (Figure 2). The analysis suggested that the detection limits of the mPCR assay were 2.0 × 103 CFU/mL for L. monocytogenes and 3.4 × 103 CFU/mL for L. ivanovii.
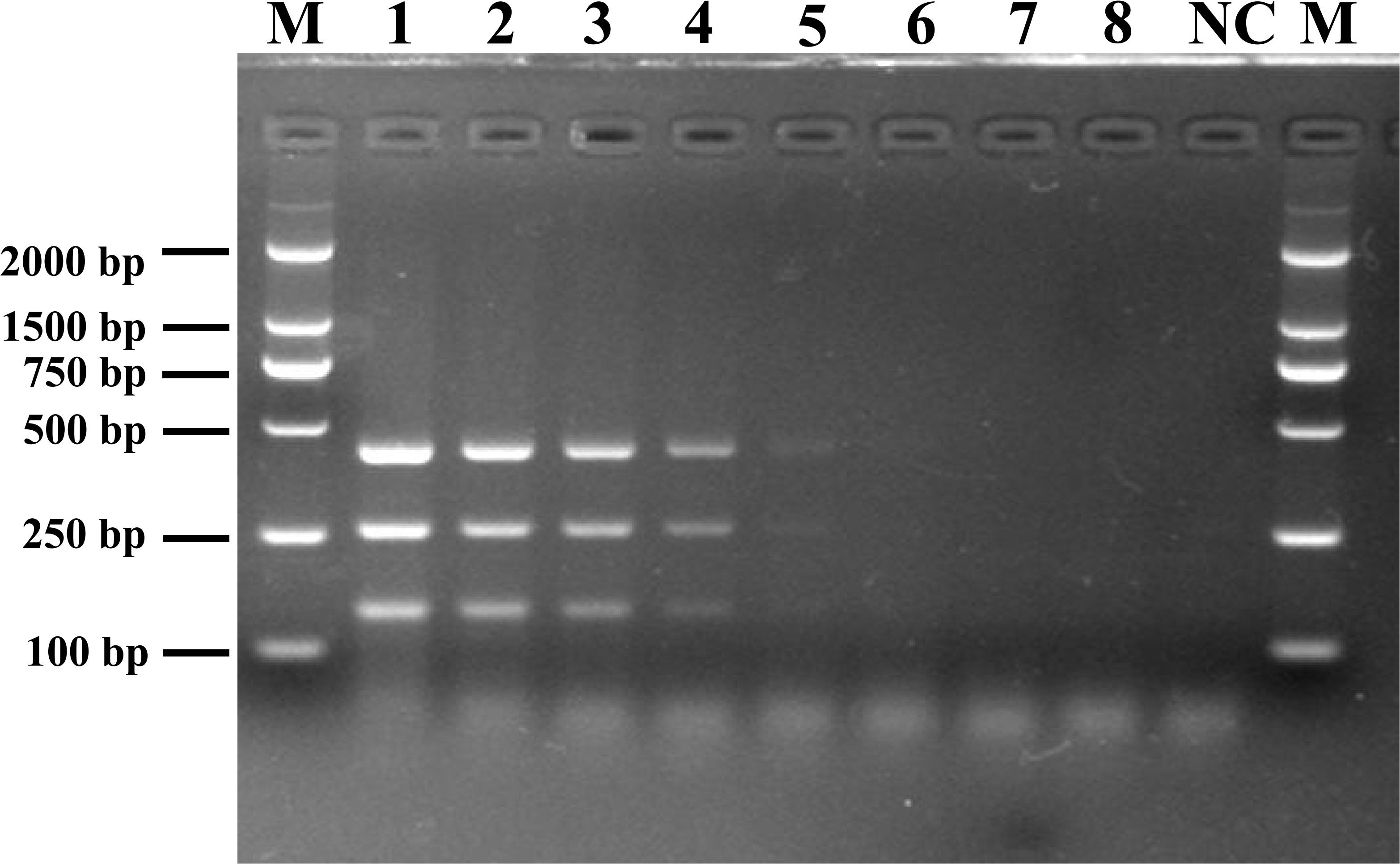
Figure 2. Detection limit of mPCR for pure culture from pathogenic Listeria. Lane M: DL2000 DNA standard marker; lanes 1–8: the bacterial culture concentration per PCR assay from 5.4 × 107 to 5.4 × 100 CFU/mL; lane NC: negative control.
F. velutipes samples were spiked with Listeria cells at the inoculum levels of 7.6 × 104, 7.6 × 103, 7.6 × 102, 7.6 × 101, and 7.6 × 100 CFU/10 g, and the enrichment cultures were followed for up to 12 h. As shown in Supplementary Table S4, the detection outcomes of mPCR varied depending on the enrichment time. The detection limit of mPCR after 4-h, 6-h, 8-h, 10-h, and 12-h enrichment was 7.6 × 104, 7.6 × 103, 7.6 × 102, 7.6 × 101, and 7.6 × 100 CFU/10 g F. velutipes, respectively (Supplementary Table S4).
The susceptibility of the mPCR assay to interference by non-target DNA was determined by mixing pathogenic Listeria and non-Listeria strains in different ratios (Figure 3). Three clear bands were generated for mixtures of all strains tested, and the brightness of these bands was comparable with that obtained from analyzing pure pathogenic Listeria cultures. This indicated that the presence of S. Enteritidis CMCC 50335, S. aureus ATCC 25923, and E. coli ATCC 25922 did not interfere with the detection of pathogenic Listeria.
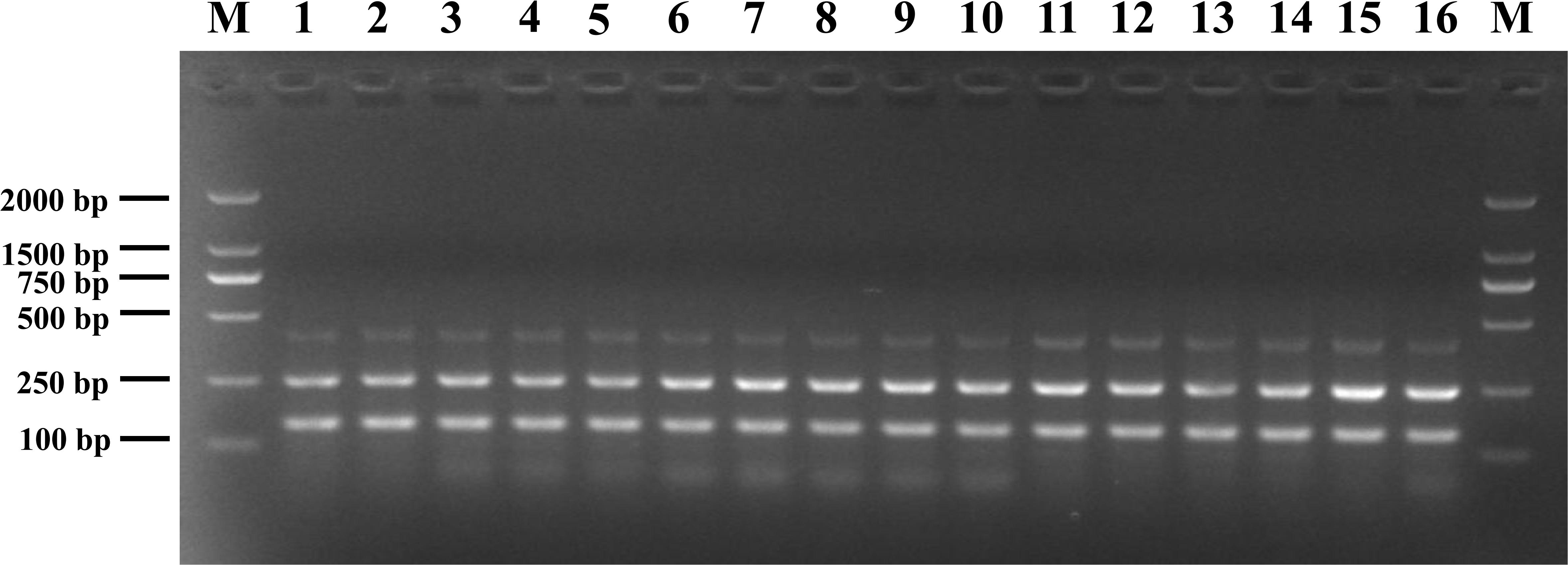
Figure 3. Anti-interference tests of mPCR detection of pathogenic Listeria with S. Enteritidis CMCC 50335 (lanes 1–5), S. aureus ATCC 25923 (lanes 6–10), and E. coli ATCC 25922 (lanes 11–15) at different mixing proportion. Lane M: DL2000 DNA standard marker; lanes 1–5, 6–10, 11–15: the ratio of target bacteria to interfering bacteria at 1:102, 1:10, 1:1, 10:1, and 102:1; lane 16: positive control.
To verify the practicality and effectiveness of the developed mPCR method, we next used the assay to detect Listeria in 129 non-spiked F. velutipes plants samples (Supplementary Table S5). In the analyzed samples, the prevalence of Listeria spp. and L. monocytogenes was 58.1% and 41.1%, respectively. The contaminated sites included the composting area, mycelium scraping equipment surfaces, floor, fruiting body cultivation room, and harvesting room. No pathogenic Listeria spp. were detected at the composting phase and mycelium culture room samples. The bacteria appeared to originate from the mycelium scraping machinery, which contaminated the product and downstream packaging equipment.
The results of positive identification of Listeria spp. by traditional methods in 75 samples also analyzed by mPCR are summarized in Figure 4. Among 174 strains identified as Listeria spp., 79 strains (45.4%) were identified as L. monocytogenes and 95 strains (54.6%) were identified as nonpathogenic Listeria (including 91 L. innocua and 4 L. grayi). In 129 F. velutipes plant samples, no L. ivanovii, L. seeligeri, and L. welshimeri were detected. A comparison of the mPCR outcomes with the results of conventional culture method is shown in Table 4. For detection of Listeria spp., the sensitivity, specificity, and efficacy of mPCR were 98.7%, 98.1%, and 98.5%, respectively. For detection of L. monocytogenes, the sensitivity, specificity, and efficacy of mPCR were 98.2%, 98.7%, and 98.5%, respectively. Only one false-positive and one false-negative result were obtained for 129 samples tested. The calculated κ factors for Listeria spp., L. monocytogenes, and L. ivanovii were 0.97, 0.97, and 1, respectively, indicating that the mPCR outcomes were highly consistent with those of the conventional culture method.
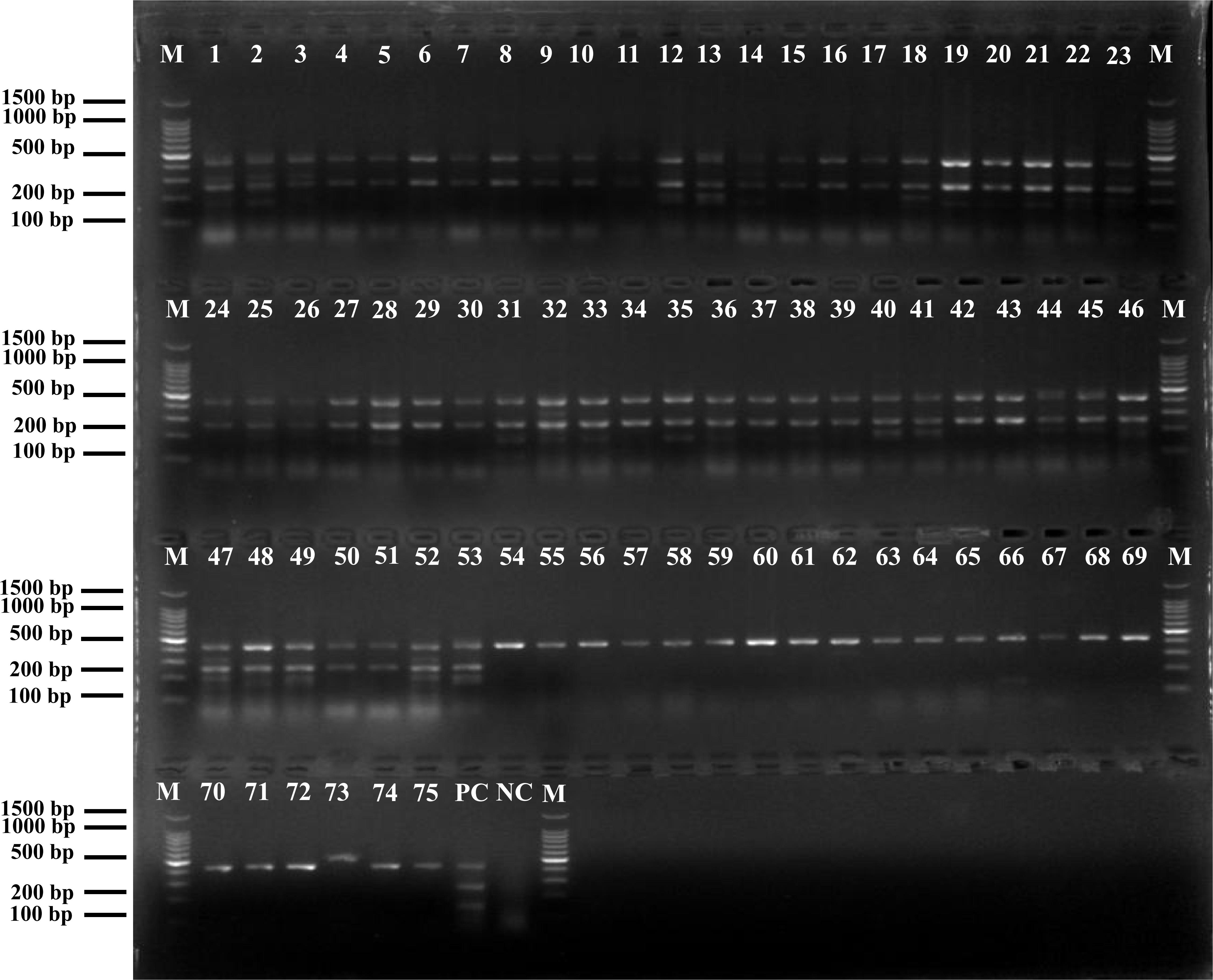
Figure 4. Positive results of mPCR in natural Flammulina velutipes plants samples. Lane M: DL2000 DNA standard marker; lanes 1–53: positive samples containing L. monocytogenes; lanes 54–75: positive samples containing non-pathogenic Listeria spp.; lane PC: positive control; lane NC: negative control.
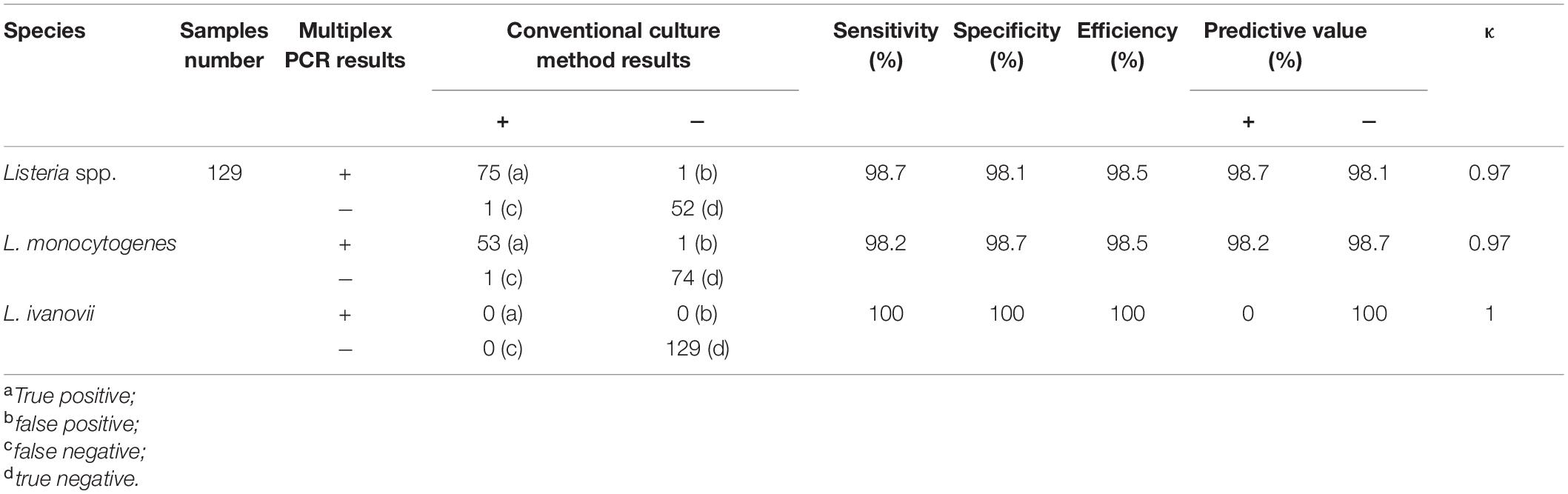
Table 4. Statistical evaluation of mPCR assay for natural Flammulina velutipes plants samples comparison with conventional culture method.
In the current study, we devised a novel mPCR method for detection of pathogenic Listeria spp. in food. The method focuses on new genus- and species-specific gene targets, and is highly specific and sensitive. It may inform the development of highly sensitive PCR approaches for the detection of Listeria in food samples and food processing plants.
Listeria spp. is widely found in food, especially in edible mushrooms, with the detection rate as high as 21.2% (Chen et al., 2018). Therefore, the development of PCR-based detection methods for species-specific classification would provide an independent means for confirming species identity (Ryu et al., 2013; Liu et al., 2015). The current PCR detection methods for Listeria species target virulence genes, or 16S rRNA and 23S rRNA genes. However, the lack of genes or mutations in Listeria strains can result in no identification or misidentification, posing a potential threat of food poisoning (Nishibori et al., 1995). As numerous complete microbial genome sequences become available with the development of sequencing technologies and bioinformatics, many researchers are committed to the search for novel specific gene targets, to replace current target genes that exhibit poor specificity (Tao et al., 2017; Sheng et al., 2018). Previously, specific target genes for Listeria were identified by DNA hybridization or using the BLAST program (Michel et al., 2004; Tao et al., 2017). Identification of a specific target by using DNA arrays involves the synthesis of probes, PCR amplification, array construction, and hybridization (Michel et al., 2004). The BLAST search method of specific target screening usually involves nucleotide sequence similarity comparisons with one or only few representative genomic sequences (Tao et al., 2017). With the rapid development of bioinformatics in recent years, using the pan-genome analysis to identify specific targets is more economical, convenient, and effective than other molecular target screening methods. In the current study, we identified gene targets specific for the Listeria genus and species via pan-genome analysis using a large number of genome sequences (n = 205). Through pan-genome and PCR analysis, five novel Listeria-specific targets were 100% specific for the targeted Listeria genomes and did not detect non-target Listeria and non-Listeria genomes. However, The Listeria-specific targets reported in the previous studies, including prs, actA, hly, inlA, and iactA, were present in 98.8%, 97.7%, 99.2%, 71.9%, and 55.6% of the target strains, respectively (Table 3). In addition, the detection limits of the corresponding primer pairs (103–104 CFU/mL) of these novel target genes were comparable with those of existing molecular detection targets (Chiang et al., 2012; Tao et al., 2017). Therefore, these Listeria-specific targets obtained by this approach have better specificity, and their sensitivity can meet the needs of existing molecular detection methods, so these novel targets can represent unique detection targets for monitoring of pathogenic Listeria in food and its production environment.
The presence of pathogenic Listeria strains in F. velutipes is an emerging public health hazard (Chen et al., 2018). Therefore, rapid detection of pathogenic Listeria is crucial for implementing optimal sterilization procedures for decontamination during the preparation of F. velutipes products. The mPCR assay developed in the current study combines three specific primer sets (SPP1, LM1, and LIV2) based on novel molecular markers (LMOSLCC2755_0944, LMOSLCC2755_0090, and queT_1, respectively) and allows simultaneous identification of pathogenic Listeria species. To increase the accuracy of Listeria identification, the Listeria genus-specific SPP1 primer set was also included. This enhances the specificity of the mPCR, especially when high concentrations of non-Listeria bacteria are present in a sample. The minimum detection limits of the assay were 2.0 × 103 CFU/mL for L. monocytogenes and 3.4 × 103 CFU/mL for L. ivanovii when pure enriched cultures were analyzed, which is comparable to those of mPCRs reported in previous studies (Chiang et al., 2012; Thapa et al., 2013; Sheng et al., 2018). In addition, the developed mPCR method detected pathogenic Listeria in artificially contaminated F. velutipes samples in the range of 7.6 × 104–7.6 × 100 CFU/10 g after 4–12 h of enrichment culture. These observations indicated that the devised mPCR assay could be used to detect pathogenic Listeria in samples more rapidly (The overall assay time, including 4–12 h pre-enrichment, DNA extraction and PCR assay, only took 5–17 h) than by using the standard culture method (4–7 days).
We also used the novel mPCR assay to analyze 129 samples from F. velutipes plants. The assay was highly sensitive, specific, and efficient. High κ values of the assay (0.97 for Listeria spp., 0.97 for L. monocytogenes, and 1 for L. ivanovii) indicated good correspondence between the assay and a conventional culture method. In an earlier preliminary study, we found that the prevalence of Listeria spp. and L. monocytogenes in F. velutipes production plants is 52.5% and 18.6%, respectively (Chen et al., 2014). In the current study, we noted a relatively high incidence of Listeria contamination (58.1% for Listeria spp. and 41.1% for L. monocytogenes). A possible reason for such high incidence is that many samples were collected on the floor and drains in this study, and the cross-contamination of L. monocytogenes easily occurred in these collection sites during the production process of F. velutipes. Among 174 strains identified as Listeria spp., approximately 54.6% strains were nonpathogenic Listeria species (including 91 L. innocua and 4 L. grayi). L. innocua occurred more frequently than L. monocytogenes in the plant and related samples, which was consistent with previous findings (Chen et al., 2014). The presence of L. innocua may hamper the detectability of L. monocytogenes, by inhibiting the pathogen via excessive growth and production of inhibitory compounds (Cornu et al., 2002; Zitz et al., 2011). Therefore, the presence of nonpathogenic Listeria, especially L. innocua, may accompany L. monocytogenes contamination (Pagadala et al., 2012). Consequently, a positive band for the Listeria genus-specific target gene in the novel mPCR assay may be an indicator of the presence of L. monocytogenes. Finally, isolation of pathogenic Listeria strains from F. velutipes plants indicated a potential risk of contracting listeriosis when consuming this mushroom. In most cases, L. monocytogenes is inactivated by cooking and heating. Therefore, edible mushrooms should be fully heated before use and cross-contamination between foods should be avoided.
In conclusion, we used pan-genome analysis to identify five novel genus- and species-specific targets for Listeria detection. We then devised a mPCR assay based on these new targets for simultaneous detection of L. monocytogenes, L. ivanovii, and other Listeria spp. with high specificity and sensitivity. The results of the assay were consistent with the results of traditional culture methods. Hence, the developed assay can be applied for rapid screening and detection of pathogenic Listeria in foods and food processing environments, providing accurate results to inform effective monitoring measures to improve the microbiological safety of foods.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
QW, JZ, QY, and MC conceived and designed the experiments. FL, LX, and YZ performed the experiments. FL, JW, SW, and HZ analyzed the data. YD, QG, and XW contributed reagents, materials, and analysis tools. FL contributed to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the National Key Research and Development Program of China (2018YFD0400900), the Key Research and Development Program of Guangdong Province, China (2018B020205001), and GDAS’ Project of Science and Technology Development (2019GDASYL-0201001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank to the College of Food Science and Technology of Nanjing Agricultural University, the Chinese Center for Disease Control and Prevention, and the State Key Laboratory of Food Science and Technology of Nanchang University for donating strains, which made this study carried out successfully.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.634255/full#supplementary-material
Chen, M., Cheng, J., Pang, R., Zhang, J., Chen, Y., Zeng, H., et al. (2019). Rapid detection of Listeria monocytogenes sequence type 121 strains using a novel multiplex Pcr assay. textitLwt Food Sci. Technol. 116:108474. doi: 10.1016/j.lwt.2019.108474
Chen, M., Cheng, J., Wu, Q., Zhang, J., Chen, Y., Zeng, H.., et al. (2018). Prevalence, potential virulence, and genetic diversity of Listeria monocytogenes isolates from edible mushrooms in Chinese markets. Front. Microbiol. 9:1711. doi: 10.3389/fmicb.2018.01711
Chen, M., Wu, Q., Zhang, J., Guo, W., Wu, S., and Yang, X. (2014). Prevalence and Contamination Patterns of Listeria monocytogenes in Flammulina velutipes Plants. Foodborne Pathog. Dis. 11, 620–627. doi: 10.1089/fpd.2013.1727
Chiang, Y. C., Tsen, H. Y., Chen, H. Y., Chang, Y. H., Lin, C. K., Chen, C. Y.., et al. (2012). Multiplex PCR and a chromogenic DNA macroarray for the detection of Listeria monocytogens, Staphylococcus aureus, Streptococcus agalactiae, Enterobacter sakazakii, Escherichia coli O157:H7, Vibrio parahaemolyticus, Salmonella spp. and Pseudomonas fluorescens in milk and meat samples. J. Microbiol. Methods 88, 110–116. doi: 10.1016/j.mimet.2011.10.021
Cornu, M., Kalmokoff, M., and Flandrois, J.-P. (2002). Modelling the competitive growth of Listeria monocytogenes and Listeria innocua in enrichment broths. Int. J. Food Microbiol. 73, 261–274. doi: 10.1016/S0168-1605(01)00658-4
Czajka, J., Bsat, N., Piani, M., Russ, W., Sultana, K., Wiedmann, M.., et al. (1993). Differentiation of Listeria monocytogenes and Listeria innocua by 16S rRNA genes and intraspecies discrimination of Listeria monocytogenes strains by random amplified polymorphic DNA polymorphisms. Appl. Environ. Microb. 59, 304–308. doi: 10.1128/AEM.59.1.304-308.1993
Doijad, S., Barbuddhe, S., Garg, S., Kalekar, S., Rodrigues, J., D’Costa, D.., et al. (2011). Incidence and genetic variability of Listeria species from three milk processing plants. Food Control 22, 1900–1904. doi: 10.1016/j.foodcont.2011.05.001
Doumith, M., Buchrieser, C. P., Jacquet, C., and Martin, P. (2004). Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819. doi: 10.1128/JCM.42.8.3819-3822.2004
Gouin, E., Dehoux, P., Mengaud, J., Kocks, C., and Cossart, P. (1995). iactA of Listeria ivanovii, although distantly related to Listeria monocytogenes actA, restores actin tail formation in an L. monocytogenes actA mutant. Infect. Immun. 63, 2729–2737. doi: 10.1016/0167-5699(95)80157-X
Hellberg, R. S., Martin, K. G., Keys, A. L., Haney, C. J., Shen, Y., and Smiley, R. D. (2013). 16S rRNA partial gene sequencing for the differentiation and molecular subtyping of Listeria species. Food Microbiol. 36, 231–240. doi: 10.1016/j.fm.2013.06.001
Jagadeesan, B., Schmid, V. B., Kupski, B., McMahon, W., and Klijn, A. (2019). Detection of Listeria spp. and L. monocytogenes in pooled test portion samples of processed dairy products. Int. J. Food Microbiol. 289, 30–39. doi: 10.1016/j.ijfoodmicro.2018.08.017
Jung, Y. S., Frank, J. F., Brackett, R. E., and Chen, J. (2003). Polymerase chain reaction detection of Listeria monocytogenes on frankfurters using oligonucleotide primers targeting the genes encoding internalin AB. J. Food Protect. 66, 237–241. doi: 10.1016/S0260-8774(02)00271-6
Kim, Y., Gu, C., Kim, H. U., and Lee, S. Y. (2020). Current status of pan-genome analysis for pathogenic bacteria. Curr. Opin. Biotech. 63, 54–62. doi: 10.1016/j.copbio.2019.12.001
Kuenne, C., Billion, A., Mraheil, M. A., Strittmatter, A., Daniel, R., Goesmann, A.., et al. (2013). Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genom. 14, 1–19. doi: 10.1186/1471-2164-14-47
Lee, Y., Yoon, Y., Seo, Y., Kim, S., Ha, J., Lee, J.., et al. (2020). Combined enrichment and quantitative polymerase chain reaction to improve sensitivity and reduce time of detection of Listeria monocytogenes in mushrooms. Foodborne Pathog. Dis. 17, 276–283. doi: 10.1089/fpd.2019.2688
Li, F., Li, B., Dang, H., Kang, Q., Yang, L., Wang, Y.., et al. (2017). Viable pathogens detection in fresh vegetables by quadruplex PCR. LWT Food Sci. Technol. 81, 306–313. doi: 10.1016/j.lwt.2017.03.064
Liu, H., Lu, L., Pan, Y., Sun, X., Hwang, C., Zhao, Y.., et al. (2015). Rapid detection and differentiation of Listeria monocytogenes and Listeria species in deli meats by a new multiplex PCR method. Food Control 52, 78–84. doi: 10.1016/j.foodcont.2014.12.017
Mao, Y., Huang, X., Xiong, S., Xu, H., Aguilar, Z. P., and Xiong, Y. (2016). Large-volume immunomagnetic separation combined with multiplex PCR assay for simultaneous detection of Listeria monocytogenes and Listeria ivanovii in lettuce. Food Control 59, 601–608. doi: 10.1016/j.foodcont.2015.06.048
Michel Doumith Christel Cazalet Natalie Simoes (2004). New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72, 1072–1083. doi: 10.1128/IAI.72.2.1072-1083.2004
Nishibori, T., Cooray, K., Xiong, H., Kawamura, I., Fujita, M., and Mitsuyama, M. (1995). Correlation between the presence of virulence-associated genes as determined by PCR and actual virulence to mice in various strains of Listeria spp. Microbiol. Immun. 39, 343–349. doi: 10.1111/j.1348-0421.1995.tb02211.x
Norton, D. M., McCAMEY, M., Boor, K. J., and Wiedmann, M. (2000). Application of the BAX for screening/genus Listeria polymerase chain reaction system for monitoring Listeria species in cold-smoked fish and in the smoked fish processing environment. J. Food Protect. 63, 343–346. doi: 10.4315/0362-028x-63.3.343
Pagadala, S., Parveen, S., Rippen, T., Luchansky, J. B., Call, J. E., Tamplin, M. L.., et al. (2012). Prevalence, characterization and sources of Listeria monocytogenes in blue crab (Callinectus sapidus) meat and blue crab processing plants. Food Microbiol. 31, 263–270. doi: 10.1016/j.fm.2012.03.015
Page, A. J., Cummins, C. A., Martin, H., Wong, V. K., Sandra, R., Holden, M. T. G.., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 22:22. doi: 10.1093/bioinformatics/btv421
Pang, R., Xie, T., Wu, Q., Li, Y., Lei, T., Zhang, J.., et al. (2019). Comparative genomic analysis reveals the potential risk of Vibrio parahaemolyticus isolated from ready-to-eat foods in China. Front. Microbiol. 10:186.
Paziak-DomaDomańska, B., Bogusławska, E., Wiêckowska-Szakiel, M., Kotłowski, R., Różalska, B., Chmiela, M.., et al. (1999). Evaluation of the API test, phosphatidylinositol-specific phospholipase C activity and PCR method in identification of Listeria monocytogenes in meat foods. Fems Microbiol. Lett. 171, 209–214. doi: 10.1111/j.1574-6968.1999.tb13434.x
Radoshevich, L., and Cossart, P. (2018). Listeria monocytogenes : towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16, 32–46. doi: 10.1038/nrmicro.2017.126
Ranjbar, R., and Halaji, M. (2018). Epidemiology of Listeria monocytogenes prevalence in foods, animals and human origin from Iran: a systematic review and meta-analysis. BMC Publ. Health 18:1057. doi: 10.1186/s12889-018-5966-8
Rosimin, A. A., Kim, M. J., Joo, I. S., Suh, S. H., and Kim, K. S. (2016). Simultaneous detection of pathogenic Listeria including atypical Listeria innocua in vegetables by a quadruplex PCR method. LWT Food Sci. Technol. 69, 601–607. doi: 10.1016/j.lwt.2016.02.007
Ryu, J., Park, S. H., Yeom, Y. S., Shrivastav, A., Lee, S.-H., Kim, Y.-R.., et al. (2013). Simultaneous detection of Listeria species isolated from meat processed foods using multiplex PCR. Food Control 32, 659–664. doi: 10.1016/j.foodcont.2013.01.048
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 14, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sheng, J., Tao, T., Zhu, X., Bie, X., Lv, F., Zhao, H.., et al. (2018). A multiplex PCR detection method for milk based on novel primers specific for Listeria monocytogenes 1/2a serotype. Food Control 86, 183–190. doi: 10.1016/j.foodcont.2017.11.028
Snapir, Y. M., Vaisbein, E., and Nassar, F. (2006). Low virulence but potentially fatal outcome - Listeria ivanovii. Eur. J. Intern. Med. 17, 286–287. doi: 10.1016/j.ejim.2005.12.006
Tamburro, M., Ripabelli, G., Vitullo, M., Dallman, T. J., Pontello, M., Amar, C. F. L.., et al. (2015a). Gene expression in Listeria monocytogenes exposed to sublethal concentration of benzalkonium chloride. Comp. Immunol. Microb. 40, 31–39. doi: 10.1016/j.cimid.2015.03.004
Tamburro, M., Sammarco, M. L., Ammendolia, M. G., Fanelli, I., Minelli, F., and Ripabelli, G. (2015b). Evaluation of transcription levels of inlA, inlB, hly, bsh and prfA genes in Listeria monocytogenes strains using quantitative reverse-transcription PCR and ability of invasion into human CaCo-2 cells. Fems Microbiol. Lett. 362:fnv018. doi: 10.1093/femsle/fnv018
Tamburro, M., Sammarco, M. L., Fanelli, I., and Ripabelli, G. (2019). Characterization of Listeria monocytogenes serovar 1/2a, 1/2b, 1/2c and 4b by high resolution melting analysis for epidemiological investigations. Int. J. Food Microbiol. 310:108289. doi: 10.1016/j.ijfoodmicro.2019.108289
Tamburro, M., Sammarco, M., and Ripabelli, G. (2018). High resolution melting analysis for the characterization of lineage II Listeria monocytogenes serovars 1/2a and 1/2c based on single nucleotide polymorphisms identification within the Listeria Pathogenicity Island−1 and inlAB operon: a novel approach for epidemiological surveillance. J. Appl. Microbiol. 125, 1920–1937. doi: 10.1111/jam.14100
Tao, T., Chen, Q., Bie, X., Lu, F., and Lu, Z. (2017). Investigation on prevalence of Listeria spp. and Listeria monocytogenes in animal-derived foods by multiplex PCR assay targeting novel genes. Food Control 73, 704–711. doi: 10.1016/j.foodcont.2016.09.026
Thapa, S. P., Han, A. R., Cho, J. M., and Hur, J. H. (2013). Multiplex PCR and DNA array for the detection of Bacillus cereus, Staphylococcus aureus, Listeria monocytogenes, Escherichia coli O157:H7, and Salmonella spp. targeting virulence-related genes. Ann. Microbiol. 63, 725–731. doi: 10.1007/s13213-012-0526-4
Vitullo, M., Grant, K. A., Sammarco, M. L., Tamburro, M., Ripabelli, G., and Amar, C. F. L. (2013). Real-time PCRs assay for serogrouping Listeria monocytogenes and differentiation from other Listeria spp. Mol. Cell. Probe. 27, 68–70. doi: 10.1016/j.mcp.2012.10.001
Zhou, X., and Jiao, X. (2005). Polymerase chain reaction detection of Listeria monocytogenes using oligonucleotide primers targeting actA gene. Food Control 16, 125–130. doi: 10.1016/j.foodcont.2004.01.001
Keywords: novel target gene, Listeria, pan-genome analysis, multiplex PCR, mushroom
Citation: Li F, Ye Q, Chen M, Zhang J, Xue L, Wang J, Wu S, Zeng H, Gu Q, Zhang Y, Wei X, Ding Y and Wu Q (2021) Multiplex PCR for the Identification of Pathogenic Listeria in Flammulina velutipes Plant Based on Novel Specific Targets Revealed by Pan-Genome Analysis. Front. Microbiol. 11:634255. doi: 10.3389/fmicb.2020.634255
Received: 27 November 2020; Accepted: 16 December 2020;
Published: 15 January 2021.
Edited by:
Dario De Medici, National Institute of Health (ISS), ItalyReviewed by:
Reza Ranjbar, Baqiyatallah University of Medical Sciences, IranCopyright © 2021 Li, Ye, Chen, Zhang, Xue, Wang, Wu, Zeng, Gu, Zhang, Wei, Ding and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, d3VxcDIwM0AxNjMuY29t; Yu Ding, ZGluZ3l1QGpudS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.