Dynamic nature of SecA and its associated proteins in Escherichia coli
A Corrigendum on
Dynamic nature of SecA and Its associated proteins in Escherichia coli
by Adachi, S., Murakawa, Y., and Hiraga, S. (2015). Front. Microbiol. 6:75. doi: 10.3389/fmicb.2015.00075
In the original article, there were errors in Figures 1C,F and G as published. Images in the 1st and 2nd rows of Panel C and an image of DAPI in the 3rd row of Panel F are from different optical sections. For Panel G, the left column is from Native SecA, not Native SacA. The corrected Figure 1 appears below.
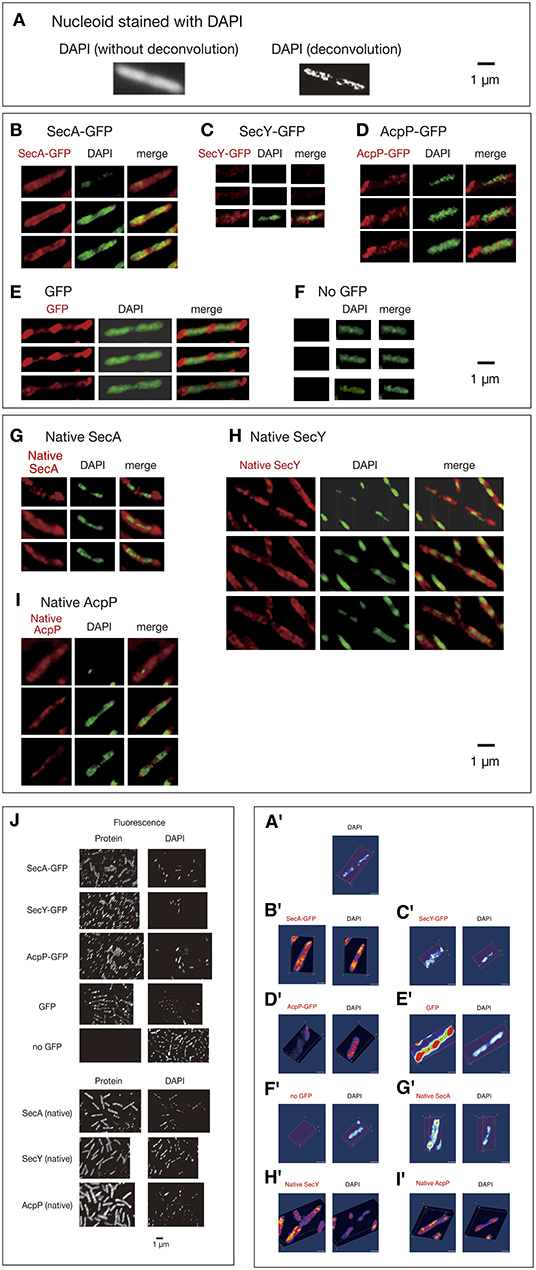
Figure 1. Three-dimensional deconvoluted images of various immunostained proteins. Cells were grown in medium L at 30°C and fixed with 70% methanol. Red, immunofluorescence of GFP or native proteins. Green, DAPI staining. Merge, merged images of red and green images. Note that synthesized yellow signal is only recognizable with eyes when the ratio of each signal is between ~4:6 and 6:4. More difference is recognizable as mere green or red. (A) Images of a DAPI-stained wild-type cell before and after 3D deconvolution in the Z-axis. (B–F) Fixed cells expressing GFPuv4-fused proteins or GFPuv4 protein (not fused with any protein) were immunostained using an anti-GFP monoclonal antibody. Fluorescence images of the proteins were analyzed with 3D deconvolution at 0.5-μm intervals in the Z-axis. Images of DAPI-stained nucleoids were also analyzed with 3D deconvolution. The yellow arrowheads indicate the clustered localization of protein molecules. (B) SecA-GFPuv4, (C) SecY-GFPuv4, (D) AcpP-GFPuv4, (E) GFPuv4 (not fused with any protein), (F) No GFPuv4. (G–I) Deconvoluted images of native proteins. Wild-type cells (PA340) were grown in medium L at 37°C, fixed, and immunostained using rabbit antisera for native proteins. Images of DAPI-stained nucleoids were also deconvoluted. (G) Native SecA, (H) Native SecY, (I) Native AcpP, (J) Three-dimensional deconvolution images of immunostained cells in wider sights corresponding to (B)–(I). (A'–I') Three-dimensional images corresponding to (A)–(I).
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Keywords: chromosome partition, SecA, SecY, AcpP, MukB, DNA topoisomerase
Citation: Adachi S, Murakawa Y and Hiraga S (2021) Corrigendum: Dynamic nature of SecA and Its associated proteins in Escherichia coli. Front. Microbiol. 11:634250. doi: 10.3389/fmicb.2020.634250
Received: 27 November 2020; Accepted: 11 December 2020;
Published: 13 January 2021.
Edited and reviewed by: Mickaël Desvaux, Institut National de la Recherche Agronomique (INRA), France
Copyright © 2021 Adachi, Murakawa and Hiraga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shun Adachi, Zi5wZXJlZ3JpbnVzbnNAbWJveC5reW90by1pbmV0Lm9yLmpw
†Present address: Shun Adachi, Department of Microbiology, Kansai Medical University, Hirakata, Japan
Yasuhiro Murakawa, RIKEN-IFOM Joint Laboratory for Cancer Genomics, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan
‡Deceased
 Shun Adachi
Shun Adachi Yasuhiro Murakawa
Yasuhiro Murakawa Sota Hiraga‡
Sota Hiraga‡