- 1Pathobiology and Population Sciences, The Royal Veterinary College, Hatfield, United Kingdom
- 2Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, United Kingdom
- 3Department of Clinical, Pharmaceutical & Biological Science, School of Life & Medical Sciences, University of Hertfordshire, Hatfield, United Kingdom
- 4Centre of Excellence for Bovine TB, Aberystwyth University, Aberystwyth, United Kingdom
- 5Department of Infection Biology, The London School of Hygiene and Tropical Medicine, London, United Kingdom
A greater understanding of the genes involved in antibiotic resistance in Mycobacterium tuberculosis (Mtb) is necessary for the design of improved therapies. Clustered regularly interspaced short palindromic repeat interference (CRISPRi) has been previously utilized in mycobacteria to identify novel drug targets by the demonstration of gene essentiality. The work presented here shows that it can also be usefully applied to the study of non-essential genes involved in antibiotic resistance. The expression of an ADP-ribosyltransferase (Arr) involved in rifampicin resistance in Mycobacterium smegmatis was silenced using CRISPRi and the impact on rifampicin susceptibility was measured. Gene silencing resulted in a decrease in the minimum inhibitory concentration (MIC) similar to that previously reported in an arr deletion mutant. There is contradictory evidence for the toxicity of Streptococcus pyogenes dCas9 (dCas9Spy) in the literature. In this study the expression of dCas9Spy in M. smegmatis showed no impact on viability. Silencing was achieved with concentrations of the aTc inducer lower than previously described and with shorter induction times. Finally, designing small guide RNAs (sgRNAs) that target transcription initiation, or the early stages of elongation had the most impact on rifampicin susceptibility. This study demonstrates that CRISPRi based gene silencing can be as impactful as gene deletion for the study of non-essential genes and further contributes to the knowledge on the design and induction of sgRNAs for CRISPRi. This approach can be applied to other non-essential antimicrobial resistance genes such as drug efflux pumps.
Introduction
Human pulmonary tuberculosis (TB) caused by Mtb remains one of the most important infectious diseases in public health worldwide. It is estimated that up to 1.5 million people die of TB annually, making it the highest cause of death from an infectious agent (Global tuberculosis report, 2020). Current TB treatments involve long regimens with four antibiotics: rifampicin, isoniazid, ethambutol and pyrazinamide and have associated side-effects. The length of the treatment and frequent patient non-compliance encourage the occurrence of multi-drug resistant (MDR) and extremely drug resistant (XDR) TB with the emergence of drug resistance in Mtb being a considerable public health crisis hindering control and eradication programs (Dheda et al., 2017). The discovery of new drugs and novel targets as well as efforts to improve the efficacy of current available drugs are crucial to control the global epidemic.
Essential genes are attractive novel drug targets, and the demonstration of essentiality can require modulating the expression of the putative essential gene in a merodiploid strain, an approach that is time consuming, costly, and inefficient in organisms such as mycobacteria that are particularly challenging to genetically modify due to low frequencies of recombination and slow growth (Choudhary et al., 2016). The CRISPRi system overcomes a number of these challenges. A catalytically inactive dCas9 protein is expressed and, guided by sgRNAs, targets a specific gene sequence. The resulting complex binds to the target sequence and sterically inhibits transcription initiation or elongation (Larson et al., 2013). The CRISPRi/dCas9 system has been utilized to study essential genes in a number of bacteria including; Escherichia coli (Qi et al., 2013), Streptococcus pneumoniae (Bikard et al., 2013), Streptomyces coelicolor (Tong et al., 2015), Bacillus subtilis (Peters et al., 2016), Clostridium acetobutylicum (Li et al., 2016), Corynebacterium glutamicum (Cleto et al., 2016), Staphylococcus aureus (Zhao et al., 2017) and Pseudomonas spp. (Tan et al., 2018).
Several papers have described the use of CRISPRi for silencing of essential genes in mycobacteria. Choudhary et al. (2015) introduced the CRISPRi approach in M. smegmatis, Mycobacterium bovis BCG Pasteur and Mtb using a codon-optimized S. pyogenes dCas9Spy. Singh et al. (2016) followed with a similar CRISPRi/dCas9 system for use in Mtb, in this case a two-plasmid system with dCas9Spy cloned into an integrative plasmid under an anhydro-tetracycline (aTc) inducible promoter, co-expressed with the sgRNA on a second plasmid. Additionally, a single plasmid system using dCas9 derived from Streptococcus thermophilus (dCas9Sth1) has also been developed and used to silence several putative essential genes allowing the verification of their suitability as drug targets (Rock et al., 2017; McNeil and Cook, 2019).
The majority of previous studies have utilized this system to silence the expression of putative essential drug-targets to demonstrate their requirement for bacterial growth by measuring the impact on the growth of the silenced strain. An alternative approach in the search for new therapeutic strategies is to make the current antibiotics more effective by targeting genes that are not essential for growth but are involved in antibiotic resistance, i.e., re-sensitizing antibiotic resistant strains. This approach has recently been used in E. coli where sequence-specific targeting an antibiotic efflux pump using peptide conjugated oligomers not only restored antibiotic sensitivity in resistant strains, but rendered strains hyper-sensitive allowing 2- to 40-fold lower antibiotic doses (Ayhan et al., 2016). Given the time and costs associated with the development of new drugs, the improvement of the efficacy of the existing antimicrobials is a useful alternative strategy. The silencing approach also holds promise as a way to understand bacterial mechanisms of resistance.
In this study, we demonstrate the utilization of the CRISPRi system to restore sensitivity to the antibiotic rifampicin in M. smegmatis, a saprophytic species that is intrinsically resistant to rifampicin due the presence of Arr, which inactivates rifampicin by ribosylation (Dabbs et al., 1995). We measure the impact of repression of the arr gene on sensitivity to rifampicin by measuring the MIC in silenced strains. In addition, we measure the effect of varying the concentration of the aTc inducer and the position of the sgRNA in relation to the translational start site on the MIC of rifampicin.
Materials and Methods
Bacterial Growth and Culture Conditions
M. smegmatis strains were grown in Middlebrook 7H9 broth supplemented with 0.2% glycerol, 0.05% Tween 80 (Sigma) and 10% albumin-dextrose-catalase (BD Diagnostics) at 37°C on a shaking platform or on Middlebrook 7H11 agar supplemented with 10% oleic-albumin-dextrose-catalase (BD Diagnostics) at 37°C for 3 days. For plasmid selection, antibiotics were used at the following concentrations: kanamycin 25 μg/ml (Sigma), hygromycin 50 μg/ml (Roche) and aTc 2-200 ng/ml (Sigma). Bacterial growth was measured by monitoring the OD600 over time and viability measurements were made using the Miles and Misra technique; spotting 20 μl of a series of 10-fold dilutions of culture onto agar (Miles et al., 1938).
Construction of sgRNA Expression Plasmids and Strains
The strains and plasmids used in this study are given in Table 1. pRH2502, an integrative plasmid expressing dCas9Spy and pRH2521 expressing the sgRNA scaffold, both from Tet-regulated promoters were acquired from Singh et al. (2016) currently available on Addgene. sgRNAs targeting arr were designed using a published protocol (Larson et al., 2013). Protospacer adjacent motif (PAM) sites, “NGG,” upstream and downstream of the annotated translational start site were chosen:- 20 nucleotides downstream of each PAM were selected as a potential genome specific sgRNA. The probability of complementarity to any other region of the genome was analyzed using a basic local alignment search tool (BLAST) (Altschul et al., 1990). Sequences with full length matches specific to arr were taken forward. A full length transcribed sgRNA including the terminators and dCas9 handle was designed and M-fold was used to predict the secondary structure of the full length sgRNA transcript (Zuker, 2003). Candidates were selected which were predicted to fold into the dCas9Spy and terminator hairpin loops. Complementary forward and reverse primers using the 20 nucleotide sequence (without the PAM) with appropriate ends for ligation to the pRH2521 vector were designed. All forward primers had “ggga” and all reverse primers had “aaac” added to the 5′ end. Table 2 shows the list of oligonucleotides used to construct the sgRNA plasmids and the strand targeted by each oligonucleotide. Oligos were annealed and cloned into CRISRPi plasmids using BbsI (NEB) as previously described (Choudhary et al., 2015; Singh et al., 2016). One microgram of pRH2502 (dCas9Spy integrative vector) was electroporated at 25 kV, 25 μF with 1,000 Ω resistance into electrocompetent M. smegmatis mc2155, this strain was grown and further electroporated with 1 μg of pRH2521 expressing the targeted sgRNAs (Parish and Stoker, 1998).
RNA Extraction
Total RNA was extracted as previously described (Rustad et al., 2009). Cultures were grown to OD600 0.1–0.2 and induced with 0, 50 or 200 ng/ml of aTc for an hour, followed by centrifugation at 2,000 x g at 4 °C for 5 min. Pellets were resuspended in TRIzol (Sigma) on ice and added to 1.5 ml tubes containing 0.1 mm glass beads. Cells were disrupted by three cycles of 30 s pulses at speed 6,500 rpm using a Precellys homogenizer. RNA was purified using a Qiagen RNeasy kit combined with on-column DNase digestion according to the manufacturer's instructions. Quantity and quality were determined using a DeNovix Spectrophotometer (DeNovix Inc, USA) and agarose gel electrophoresis.
RT-qPCR
To remove traces of contaminating DNA, RNA samples were treated with RNase-free DNase I (Invitrogen). cDNA was synthesized from 100 ng of RNA using Superscript III Reverse transcriptase (ThermoFisher) according to manufacturer instructions. In order to quantify the expression of arr, qPCRs were performed using PowerUp SYBR Green Master Mix (ThermoFisher) with 1 μl of cDNA and 0.3 μM of either sigA primers or arr primers (Table 2) in a final volume of 20 μl. Samples were run on a BioRad CFX96 analyser at 50 °C for 2 min, 95 °C for 2 min, followed by 39 cycles of 95 °C for 15 s, 60 °C for 1 min and 75 °C for 30 s. The mRNA levels of arr were normalized against the mRNA of the reference gene sigA. Three biological replicates were run, with technical duplicates. Melting curve analysis was carried out to verify specificity.
MIC Determinations
To quantify the susceptibility to rifampicin, a broth microdilution assay using resazurin dye was carried out (Agrawal et al., 2015). Hundred microliter of twice the required concentration of rifampicin was serial diluted across 96-well microtiter plates. Cultures were grown to exponential phase (OD600 0.4–0.6), diluted to an OD600 of 0.1 and 100 μl was added to each well. This represents ~106 cells. 2–200 ng/ml of aTc for CRISPRi/dCas9-mediated repression of arr gene expression was added where required. Plates were incubated at 37 °C for 40 h. Following incubation, 30 μl of 0.2 mg/ml resazurin was added to each well and incubated at 37 °C for an additional 24 h and fluorescence was measured using a Tecan microplate reader at excitation 560 nm and emission 590 nm. Images of the plates were also taken.
Results
CRISPRi Targeting of the Promoter Region of the arr Gene in M. smegmatis Increases the Susceptibility to Rifampicin
Inhibition of the expression of arr is expected to potentiate growth inhibition in the presence of rifampicin. In order to determine which region should be targeted for CRISPRi, nine sgRNAs of different lengths were designed, targeting regions in both the promoter and coding sequences of the gene, on either the coding or non-coding strand (Figure 1A). Unlike other bacteria, mycobacteria use more commonly leaderless transcripts during gene expression, however, the arr gene has been shown to contain a 5′ UTR indicating the presence of a putative Shine-Dalgarno sequence (Cortes et al., 2013; Shell et al., 2015; Martini et al., 2019). Previous RNAseq studies have shown that transcription starts 31 bp upstream of the GTG annotated translational start codon (Shell et al., 2015). Putative −35 and −10 regions were predicted using Softberry BPROM (Solovyev and Salamov, 2011) and sgRNAs were designed to include coverage of these regions. The location of the sgRNAs, alongside the transcription and translational start sites and putative promoter elements is given in Figure 1A. Growth inhibition in the presence of rifampicin after induction of the various sgRNAs is shown in Figure 1B.
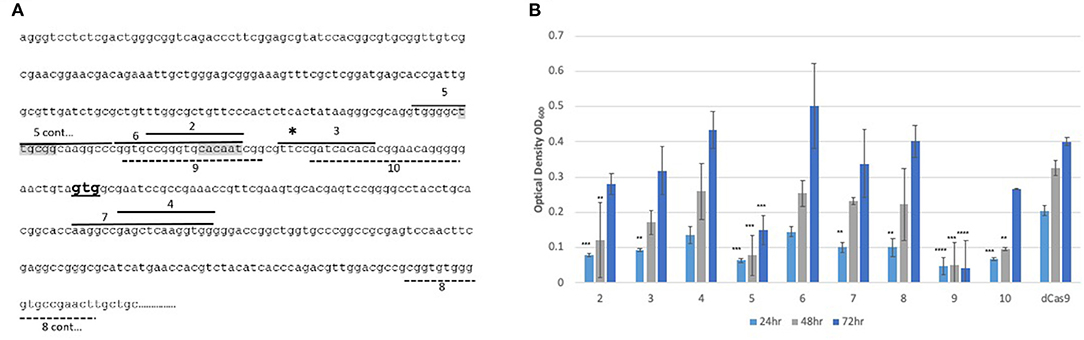
Figure 1. CRISPRi targeting of the arr gene (MSMEG_1221) over various sites in the promoter results in increased susceptibility to rifampicin (A) Position of the sgRNA targets. Nine sgRNAs (2-10) were designed to target both non-coding (sgRNAs 2-7) and coding strands (8-10). sgRNAs targeting the coding strand are indicated by a dashed line. The gene specific region has mainly 19 bp, except for sgRNAs 2 and 3 where a 13 bp region was targeted. The transcriptional start site is indicated by a * and the translation start GTG is in bold. Putative −10 and −35 promoter elements are highlighted in gray. (B) Growth of M. smegmatis sgRNA-dCas9 strains in 7H9 growth medium supplemented with aTc (200 ng/ml) and rifampicin (3 μg/ml). Optical density (OD600) was measured at 24, 48, and 72 h. Numbers refer to the sgRNA oligos as listed in Table 2. Statistical significance was determined using One-way ANOVA with Dunnett's multiple comparisons test against the dCas9-only strain for each time point (****p < 0.0001, ***p < 0.001, **p < 0.01). Error bars represent standard deviation from the mean of triplicate repeats.
At 24 h, all M. smegmatis strains containing an sgRNA targeting arr, apart from those targeting regions 4 and 6, exhibited increased susceptibility (p < 0.05) to rifampicin compared to the control strain expressing only dCas9Spy. At 48 h, M. smegmatis strains containing sgRNAs targeting regions 2, 5, 9, and 10 still displayed significantly increased susceptibility and this endured for a further 24 h for regions 5 and 9. The shorter sgRNAs which were 13 bp long, targeting regions 2 and 3 were less effective than the 19 bp long sgRNAs. sgRNAs targeting region 5 and 9, at the promoter region were the most effective, with targeting of the −10 (sgRNA 9) region being the most impactful.
CRISPRi Targeting of arr in the Absence of Rifampicin Results in Reduction of Gene Expression but Does Not Impact Growth
Three further sgRNAs were designed (arr1, arr2, and arr3) that targeted a 20 bp gene specific region designed to cover the putative −10 region (arr1), the 5′ UTR (arr2) and the 5′ end of the coding sequence (arr3) (Figure 2A). All sgRNAs were designed to target the coding strand. The impact of induction of sgRNAs arr1, arr2 and arr3 alongside dCas9 on the expression of the arr gene was measured using RT-qPCR. Expression levels were normalized to sigA and expressed relative to the uninduced cultures. The results are shown in Figure 2B. Induction of arr1 and arr3 with aTc at 50 ng/ml led to more than 50% reduction in the expression of the arr gene. Induction of arr2 with 50 ng/ml of aTc resulted in 40% reduction in its expression. Induction with higher levels of aTc (arr1, arr3 with 200 ng/ml) did not improve inhibition of arr expression.
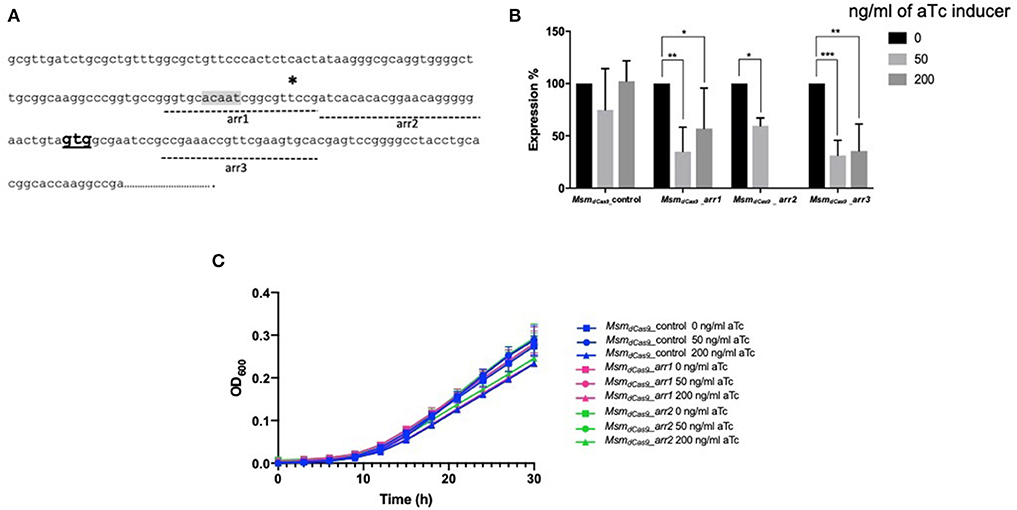
Figure 2. CRISPRi targeting of the non-essential gene arr (MSMEG_1221) in M. smegmatis results in transcriptional inhibition but does not impact growth. (A) Position of sgRNA targets on the arr gene. Three sgRNAs were designed to target arr, indicated by dashed lines. arr1 targeted −47 to −28, arr2 targeted −27 to −8 bp upstream of the annotated translational start codon. arr3 targeted +13 to +32 bp within the coding sequence. The transcriptional start site is indicated by a * and the translation start site (GTG) is in bold. The putative−10 promoter element predicted using Softberry BPROM is highlighted in gray. (B) Inhibition of arr expression in cultures expressing sgRNA targeting arr, induced with different concentration of aTc. mRNA levels are expressed relative to the control strain expressing dCas9 only. Results are the mean of three independent biological replicates and expression levels were measured in duplicate for each concentration of aTc (*p < 0.05, **p < 0.01, ***p < 0.001); (C) Growth of M. smegmatis expressing either sgRNA targeting arr or dCas9 only with differing levels of aTc. Cultures were grown in 96 well plates from a starting OD600 in a total volume of 200 μl.
As arr is a non-essential gene, inhibition of its expression is not expected to influence the growth of the bacteria. To test this, we compared the effect of the induction with aTc on the growth of MsmdCas9_arr1 and MsmdCas9_arr2. Additionally, we compared the impact of CRISPRi induction on the growth of the MsmdCas9_control strain which does not contain a targeting sgRNA. The results (Figure 2C) show that there is no growth inhibitory effect as a result of induction of the CRISPRi system
Initial studies using dCas9 from S. pyogenes to repress expression of a number of essential genes reported no toxicity in mycobacteria (Choudhary et al., 2015; Singh et al., 2016). However, a subsequent study reported toxicity (Rock et al., 2017). Given the discrepancies over the reports of toxicity in the literature, the effect of induction of dCas9Spy on the viability of M. smegmatis mc2155 was evaluated. M. smegmatis cultures expressing dCas9Spy (MsmdCas9_control) were induced with 50 and 200 ng/ml of aTc and compared to non-induced cultures and the wild type strain. The results, which are shown in Figure 3, indicate that induction of with aTc does not result in toxicity.
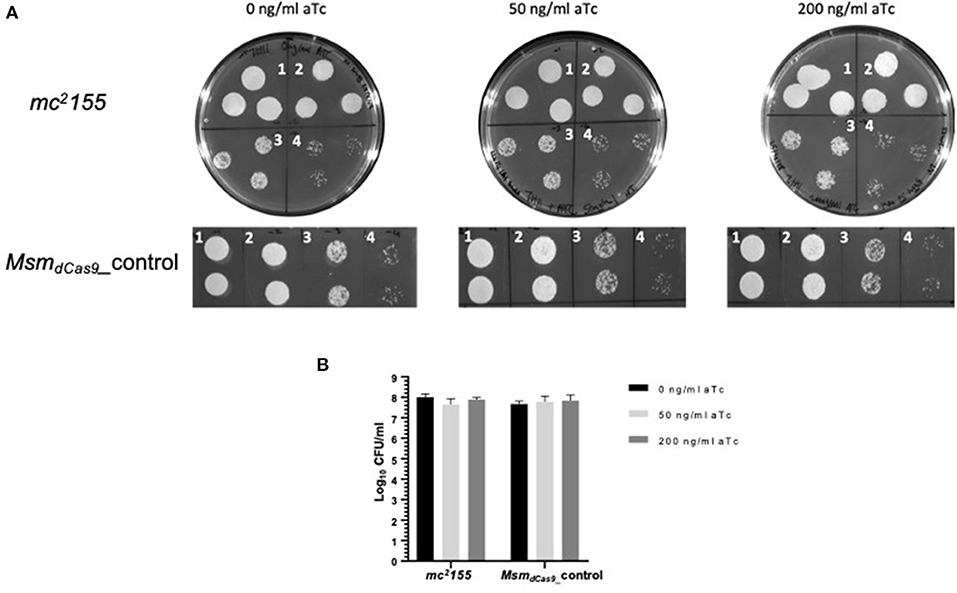
Figure 3. Induction of dCas9Spy in M. smegmatis mc2155 is not toxic to the cells. (A) Miles and Misra spotting to examine the impact of induction of dCas9 on the viability of M. smegmatis mc2155. Broth cultures of Msm_WT and MsmdCas9_control strains were grown overnight at 37 °C and diluted to OD600 0.01. Cultures were further diluted: 1- 10−1, 2- 10−2, 3- 10−3, 4- 10−4 as labeled, and spotted using Miles and Misra on 7H11 agar supplemented with 0, 50, and 200 ng/ml of aTc. (B) The CFU/ml was estimated from colony counts.
CRISPRi Targeting arr Sensitizes M. smegmatis to Rifampicin
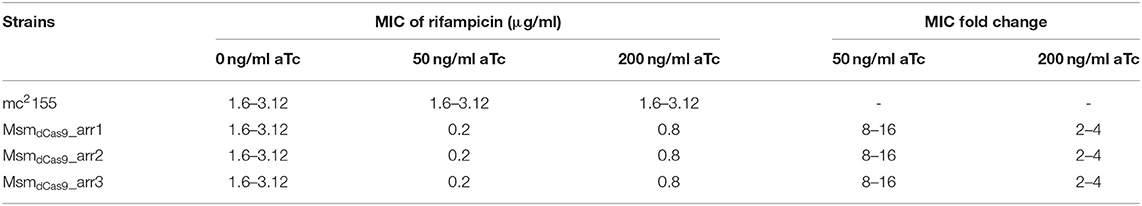
An initial qualitative screen on solid media demonstrated that silencing of arr by induction of the sgRNAs with a range of aTc concentrations (50-200 ng/ml) sensitized M. smegmatis to rifampicin (Supplementary Figure 1). In order to quantify the effect of arr silencing on rifampicin sensitivity, the MIC of rifampicin for MsmdCas9_arr1, MsmdCas9_arr2 and MsmdCas9_arr3 was determined without aTc and with aTc at 50 ng/ml and 200 ng/ml (Table 3, Supplementary Figure 2). Induction of the CRISPRi system using aTc had no impact on the MIC of the wild type strain, mc2155. The MIC obtained (1.6–3.12 μg/ml) is in-line with previously reported values [0.8–3.9 μg/ml (Agrawal et al., 2015; Gupta et al., 2015)]. The non-induced cultures and the MsmdCas9_control showed the same MIC as the wild type strain, mc2155 (Supplementary Figure 2 wells 2-5). However, at 50 ng/ml and 200 ng/ml aTc there was a clear decrease in the MIC of rifampicin for the three strains (MsmdCas9_arr1, MsmdCas9_arr2 and MsmdCas9__arr3) to 0.2 μg/ml with 50 ng/ml of aTc and 0.8 μg/ml with 200 ng/ml of aTc. The reduction in MIC due to the induction of the sgRNAs with 50 ng/ml and 200 ng/ml of aTc were 8–16 and 2–4-fold, respectively (Table 3). Deletion of the arr gene in M. smegmatis has been reported to decrease the MIC of rifampicin by 16-fold, similar to induction with 50 ng/ml of aTc (Agrawal et al., 2015).
Lower Levels of aTc Can Be Used to Sensitize the Strains to Rifampicin
To investigate whether sensitization to rifampicin could be achieved using lower concentrations of aTc, the MIC of rifampicin for the strains MsmdCas9_control, MsmdCas9_arr1 and MsmdCas9_arr2 were measured in the presence of 4 ng/ml, 6 ng/ml and 8 ng/ml of aTc and compared to non-induced cultures. The results, which are displayed as a change in fluorescence at different concentrations of rifampicin, show that without induction, all the three strains have the same MIC (1.6 μg/ml, Figure 4). However, a 2-fold reduction in the MIC is achieved at 4, 6 and 8 ng/ml of aTc. This suggests that silencing of arr by induction of the sgRNA can take place at concentrations as low as 4 ng/ml, although the impact on the level of sensitization is less than that observed at higher concentrations.
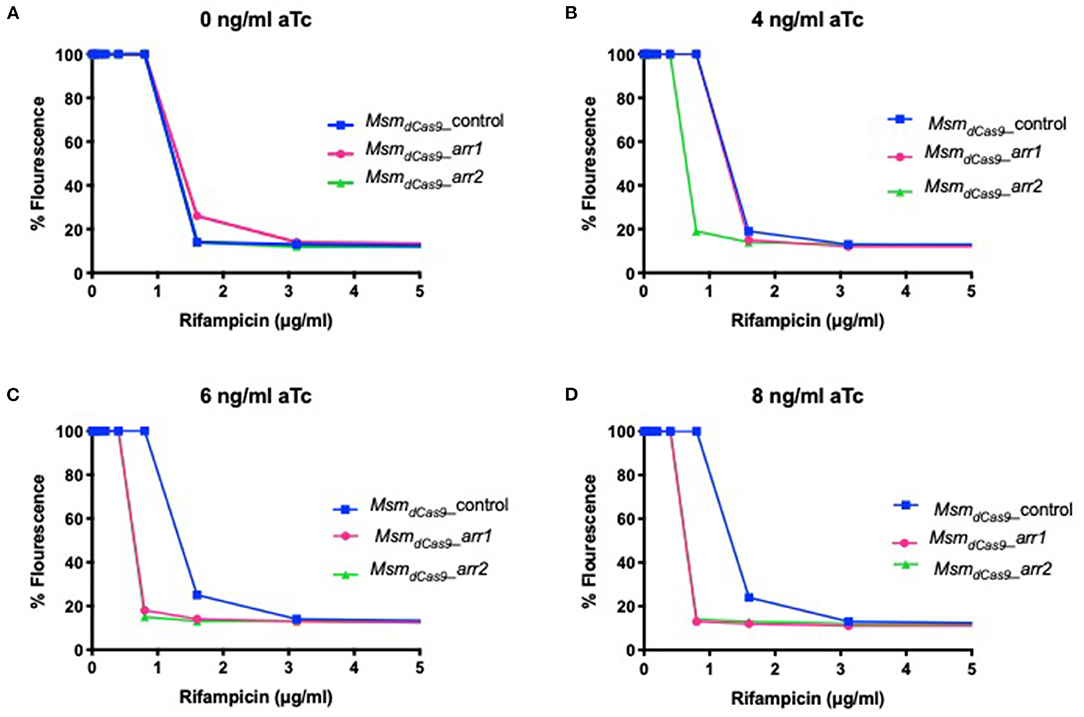
Figure 4. Induction of dCas9Spy with low concentrations of aTc sensitizes MsmdCas9_arr1 and MsmdCas9_arr2 to rifampicin. 96 well microtiter plates were filled with 100 μl of twice the required concentration of rifampicin and serial diluted in two-fold across the plates. Approximately 106 bacterial cells were added to each well with antibiotics containing (A) 0 ng/ml (B) 4 ng/ml (C) 6 ng/ml and (D) 8 ng/ml of aTc. Plates were incubated at 37°C for 40 h after which 30 μl of 0.2 mg/ml resazurin were added to each well and incubated at 37°C for an additional 24 h. Fluorescence was measured on a Tecan plate reader at 560 nm (excitation) and 590 nm (emission).
Discussion
In this study, we demonstrate the usefulness of CRISPRi silencing for the study of genes that are not essential for growth, but encode targets associated with antibiotic resistance. Induction of dCas9Spy in arr targeting strains with 50 and 200 ng/ml of aTc decreased the MIC of rifampicin and re-sensitized M. smegmatis to the antibiotic by 8-16 and 2-4-fold, respectively. M. smegmatis, which is intrinsically resistant to rifampicin due to the expression of arr, has previously been shown to be a useful surrogate for pathogenic mycobacteria to study antimicrobial susceptibility (Chaturvedi et al., 2007). Additionally, as rifampicin is used for the treatment of several mycobacterial infections, arr seemed to be a suitable target to examine the potential use of the CRISPRi/dCas9 system to explore genes which could be targeted to improve the efficacy of current antibiotics. This approach can now be used to study genes in other intrinsically rifampicin-resistant species such as Mycobacterium abscessus, a pathogenic fast growing mycobacterial species. Interestingly, M. abscessus also carries a putative arr gene and its role in intrinsic rifampicin resistance was previously demonstrated through deletion of arr (Rominski et al., 2017). The work presented here demonstrates that gene silencing can be used effectively in mycobacterial species to study intrinsic drug resistance.
In this study we show that CRISPRi/dCas9 can be used to restore sensitivity to rifampicin in M. smegmatis by silencing the expression of arr. As little as 40–60% reduction in expression of arr resulted in an 8–16-fold decrease in the MIC. Previous studies using an Δarr strain reported a 16-fold decrease in the MIC (Agrawal et al., 2015). This suggests that, for arr at least, silencing can be as effective as deletion as the levels of susceptibility reached were similar to that previously reported. Although the Δarr strain from the Agrawal study was not included here, the congruity of the MIC measurements in the wild type strains between this study and the study by Agrawal et al. validates our approach. To our knowledge there are no other isogenic Δarr mutants to which a comparison could be made. Although, another Δarr strain of M. smegmatis has been described, this is in a different genetic background, therefore not comparable to this study (Combrink et al., 2007).
Similar to previous reports (Larson et al., 2013), the findings of this study show that gene silencing using dCas9Spy is most effective if the target site is within the promoter region on the coding or non-coding strand of the gene of interest inhibiting transcription initiation. Targeting regions 5 and 9, both of which were within the promoter region, on the non-coding and coding strand respectively, was most impactful in comparison to the other target regions (Figure 1B). Additionally, targeting region 9, which include the putative −10 sequence, produced the most sustained impact on growth 24, 48, and 72 h post-induction. Strategies to inhibit the elongation of transcription (rather than transcription initiation) are reported to be most effective when targeting the coding strand of the protein coding region or the 5′ UTR (Larson et al., 2013). These are represented by sgRNAs 10 and 8 (coding) with sgRNAs 4 and 7 targeting the non-coding strand. Of those sgRNAs targeting sites beyond the transcription start site, sgRNA 10 was the most effective in improving the susceptibility to rifampicin thereby supporting the strategy of targeting the coding strand. The results further showed that the shorter sgRNAs (sgRNAs 2, 3, and 4) were less effective than longer sequences.
The three main sgRNAs used in this study were designed to target the putative −10 region (arr1), the 5′ UTR (arr2) and the 5′ end of the coding sequence (arr3), with all three being 20 bp long. The impact of induction of the system for 1 h with 0, 50 and 200 ng/ml of aTc was measured using RT-qPCR. arr expression was suppressed by 40–60% (Figure 2B). Previous studies have measured levels of silencing by CRISPRi/dCas9Spy 12–24 h post-induction with aTc. Singh et al. (2016) reported a 60–80% reduction in sigH and pknB expression following induction with 200 ng/ml of aTc for 12 h (Singh et al., 2016). This study shows that gene silencing can take place within 1 h of induction. Additionally, the data presented here show that 200 ng/ml of aTc did not improve gene silencing and sensitization compared to 50 ng/ml (Figure 2B and Table 3). In M. smegmatis, the TetR-regulated UVtetO promoter has been reported to achieve the peak of induction with 50–100 ng/ml of aTc with decreasing levels of induction at concentrations higher than 100 ng/ml (Ehrt et al., 2005) and this could explain our observations at 200 ng/ml. Furthermore, this study also shows that induction with lower concentrations (4–8 ng/ml) of aTc is sufficient to decrease the MIC of rifampicin (Figure 4). One study indicated that dCas9Spy expression is potentially toxic in mycobacteria (Rock et al., 2017). Toxicity of dCas9Spy has also been reported in E. coli upon very high levels of expression (Cho et al., 2018). However, similar to other reports (Choudhary et al., 2015, 2019; Singh et al., 2016), the work presented here shows that the induction of dCas9Spy does not impact M. smegmatis growth and viability.
These results show that the CRISPRi system can be used to target, and knock-down genes involved in antimicrobial resistance, enabling faster reverse genetics compared to gene knockout techniques. There are limitations, given the requirement for induction by aTc, however, the system has great potential for multiplexing which will be useful in the study of redundancy. In particular, it will be useful in the study of multiple drug efflux pumps in slow-growing pathogenic mycobacterial genomes where sequential mutagenesis is not efficient. The approach can improve studies on gene function and resistance to current and future antimicrobials.
Data Availability Statement
The raw data supporting the conclusions from this article will be made available by the authors on request. The raw data used to derive the MICs in Table 3 is given in Supplementary Figure 2.
Author Contributions
VF, SG, SW, and SLK designed the study and analyzed the data. VF, AAC, SG, AB, AJG, and OL carried out the experiments. SLK and BWW did funding acquisition. VF and SLK wrote the first draft of the manuscript. All authors contributed to the manuscript revision, read, and approved the submitted version.
Funding
This work was funded by the BBSRC Grant Ref: BB/N004590/1. VF was in receipt of an RVC PhD studentship. Awarded to SLK and BWW as part of the EradbTB consortium. AJG holds a Sêr Cymru II Lectureship funded by the European Research Development Fund and Welsh Government.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Liam Good for critical review of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.619427/full#supplementary-material
Supplementary Figure 1. CRISPRi targeting of arr sensitizes M. smegmatis to rifampicin. The bacterial cultures were grown to saturation for 48 h in 7H9 and streaked on 7H11 agar plates containing different concentrations of aTc (50 ng/ml-200 ng/ml) as indicated. Plates contained rifampicin at 2 μg/ml. Plates were incubated at 37°C for 48 h before being photographed.
Supplementary Figure 2. Induction of dCas9Spy in arr targeting strains decreases the MIC of rifampicin. Ninety-Six well microtiter plates were filled with 100 μl of twice the required concentration of rifampicin and serially diluted in 2-fold across the plates. Approximately 106 bacterial cells were added to wells with antibiotics and (A) 0 ng/ml (B) 50 ng/ml (C) 200 ng/ml of aTc. Plates were incubated at 37°C for 40 h after which 30 μl of 0.2 mg/ml resazurin was added to each well and incubated at 37°C for an additional 24 h. Fluorescence was measured on a Tecan plate reader at 560 nm (excitation) and 590 nm (emission) and images taken. Pink wells indicate bacterial growth whereas blue wells indicate inhibition of growth in the presence of rifampicin. Images shown are representative of one replicate for each treatment. The MIC is determined as the concentration of rifampicin at which the first well in the row appears pink.
References
Agrawal, P., Miryala, S., and Varshney, U. (2015). Use of Mycobacterium smegmatis deficient in ADP-ribosyltransferase as surrogate for Mycobacterium tuberculosis in drug testing and mutation analysis. PLoS ONE 10:e0122076. doi: 10.1371/journal.pone.0122076
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ayhan, D. H., Tamer, Y. T., Akbar, M., Bailey, S. M., Wong, M., Daly, S. M., et al. (2016). Sequence-specific targeting of bacterial resistance genes increases antibiotic efficacy. PLoS Biol. 14:e1002552. doi: 10.1371/journal.pbio.1002552
Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., and Marraffini, L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429–7437. doi: 10.1093/nar/gkt520
Chaturvedi, V., Dwivedi, N., Tripathi, R. P., and Sinha, S. (2007). Evaluation of Mycobacterium smegmatis as a possible surrogate screen for selecting molecules active against multi-drug resistant Mycobacterium tuberculosis. J. Gen. Appl. Microbiol. 53, 333–337. doi: 10.2323/jgam.53.333
Cho, S., Choe, D., Lee, E., Kim, S. C., Palsson, B., and Cho, B.-K. (2018). High-Level dCas9 Expression induces abnormal cell morphology in Escherichia coli. ACS Synth. Biol. 7, 1085–1094. doi: 10.1021/acssynbio.7b00462
Choudhary, E., Lunge, A., and Agarwal, N. (2016). Strategies of genome editing in mycobacteria: achievements and challenges. Tuberculosis 98, 132–138. doi: 10.1016/j.tube.2016.03.005
Choudhary, E., Sharma, R., Kumar, Y., and Agarwal, N. (2019). Conditional Silencing by CRISPRi reveals the role of DNA Gyrase in formation of drug-tolerant persister population in Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 9:70. doi: 10.3389/fcimb.2019.00070
Choudhary, E., Thakur, P., Pareek, M., and Agarwal, N. (2015). Gene silencing by CRISPR interference in mycobacteria. Nat. Commun. 6:6267. doi: 10.1038/ncomms7267
Cleto, S., Jensen, J. V., Wendisch, V. F., and Lu, T. K. (2016). Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth. Biol. 5, 375–385. doi: 10.1021/acssynbio.5b00216
Combrink, K. D., Denton, D. A., Harran, S., Ma, Z., Chapo, K., Yan, D., et al. (2007). New C25 carbamate rifamycin derivatives are resistant to inactivation by ADP-ribosyl transferases. Bioorganic Med. Chem. Lett. 17, 522–526. doi: 10.1016/j.bmcl.2006.10.016
Cortes, T., Schubert, O. T., Rose, G., Arnvig, K. B., Comas, I., Aebersold, R., et al. (2013). Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 5, 1121–1131. doi: 10.1016/j.celrep.2013.10.031
Dabbs, E. R., Yazawa, K., Mikami, Y., Miyaji, M., Morisaki, N., Iwasaki, S., et al. (1995). Ribosylation by mycobacterial strains as a new mechanism of rifampin inactivation. Antimicrob. Agents Chemother. 39, 1007–1009. doi: 10.1128/AAC.39.4.1007
Dheda, K., Gumbo, T., Maartens, G., Dooley, K. E., McNerney, R., Murray, M., et al. (2017). The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir. Med. 5, 291–360. doi: 10.1016/S2213-2600(17)30079-6
Ehrt, S., Guo, X. V., Hickey, C. M., Ryou, M., Monteleone, M., Riley, L. W., et al. (2005). Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. doi: 10.1093/nar/gni013
Global tuberculosis report (2020). Available online at: https://www.who.int/publications/i/item/9789240013131 (accessed November 27, 2020).
Gupta, K., Singh, S., and van Hoek, M. L. (2015). Short, synthetic cationic peptides have antibacterial activity against Mycobacterium smegmatis by forming pores in membrane and synergizing with antibiotics. Antibiotics 4, 358–378. doi: 10.3390/antibiotics4030358
Larson, M. H., Gilbert, L. A., Wang, X., Lim, W. A., Weissman, J. S., and Qi, L. S. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196. doi: 10.1038/nprot.2013.132
Li, Q., Chen, J., Minton, N. P., Zhang, Y., Wen, Z., Liu, J., et al. (2016). CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnol. J. 11, 961–972. doi: 10.1002/biot.201600053
Martini, M. C., Zhou, Y., Sun, H., and Shell, S. S. (2019). Defining the transcriptional and post-transcriptional landscapes of Mycobacterium smegmatis in aerobic growth and hypoxia. Front. Microbiol. 10:591. doi: 10.3389/fmicb.2019.00591
McNeil, M. B., and Cook, G. M. (2019). Utilization of CRISPR interference to validate MmpL3 as a drug target in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 63:e00629-19. doi: 10.1128/AAC.00629-19
Miles, A. A., Misra, S. S., and Irwin, J. O. (1938). The estimation of the bactericidal power of the blood. J. Hyg. 38, 732–749. doi: 10.1017/S002217240001158X
Parish, T., and Stoker, N. G. (1998). “Electroporation of Mycobacteria,” in Mycobacteria Protocols, eds T. Parish and N. G. Stoker (Totowa, NJ: Humana Press), 129–144. doi: 10.1385/0-89603-471-2:129
Peters, J. M., Colavin, A., Shi, H., Czarny, T. L., Larson, M. H., Wong, S., et al. (2016). A Comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165, 1493–1506. doi: 10.1016/j.cell.2016.05.003
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence- specific control of gene expression. Curr. Opin. Cell Biol. 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Rock, J. M., Hopkins, F. F., Chavez, A., Diallo, M., Chase, M. R., Gerrick, E. R., et al. (2017). Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2:16274. doi: 10.1038/nmicrobiol.2016.274
Rominski, A., Roditscheff, A., Selchow, P., Böttger, E. C., and Sander, P. (2017). Intrinsic rifamycin resistance of Mycobacterium abscessus is mediated by ADP-ribosyltransferase MAB_0591. J. Antimicrob. Chemother. 72, 376–384. doi: 10.1093/jac/dkw466
Rustad, T. R., Roberts, D. M., Liao, R. P., and Sherman, D. R. (2009). “Isolation of mycobacterial RNA,” in Mycobacteria Protocols. Methods in Molecular Biology (Methods and Protocols), Vol. 465, eds T. Parish and A. Brown (Totowa, NJ: Humana Press). doi: 10.1007/978-1-59745-207-6_2
Shell, S. S., Wang, J., Lapierre, P., Mir, M., Chase, M. R., Pyle, M. M., et al. (2015). Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 11:e1005641. doi: 10.1371/journal.pgen.1005641
Singh, A. K., Carette, X., Potluri, L.-P., Sharp, J. D., Xu, R., Prisic, S., et al. (2016). Investigating essential gene function inMycobacterium tuberculosis using an efficient CRISPR interference system. Nucleic Acids Res. 44:e143. doi: 10.1093/nar/gkw625
Snapper, S. B., Melton, R. E., Mustafa, S., Kieser, T., and Jacobs, W. R. Jr. (1990). Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4, 1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x
Solovyev, V., and Salamov, A. (2011). Automatic annotation of microbial genomes and metagenomic sequences” in Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, ed R. W. Li (New York, NY: Nova Science Publishers), 61–78.
Tan, S. Z., Reisch, C. R., and Prather, K. L. J. (2018). A robust CRISPR interference gene repression system in Pseudomonas. J. Bacteriol. 200, e00575–e00517. doi: 10.1128/JB.00575-17
Tong, Y., Charusanti, P., Zhang, L., Weber, T., and Lee, S. Y. (2015). CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth. Biol. 4, 1020–1029. doi: 10.1021/acssynbio.5b00038
Zhao, C., Shu, X., and Sun, B. (2017). Construction of a gene knockdown system based on catalytically inactive (andquot; Deadandquot;) Cas9 (dCas9) in Staphylococcus aureus. Appl. Environ. Microbiol. 83:e00291-17. doi: 10.1128/AEM.00291-17
Keywords: antibiotic resistance, tuberculosis, CRISPRi, mycobacteria, rifampicin resistance
Citation: Faulkner V, Cox AA, Goh S, van Bohemen A, Gibson AJ, Liebster O, Wren BW, Willcocks S and Kendall SL (2021) Re-sensitization of Mycobacterium smegmatis to Rifampicin Using CRISPR Interference Demonstrates Its Utility for the Study of Non-essential Drug Resistance Traits. Front. Microbiol. 11:619427. doi: 10.3389/fmicb.2020.619427
Received: 20 October 2020; Accepted: 21 December 2020;
Published: 01 February 2021.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Peter Sander, University of Zurich, SwitzerlandDiana Machado, New University of Lisbon, Portugal
Liliana Rodrigues, New University of Lisbon, Portugal
Copyright © 2021 Faulkner, Cox, Goh, van Bohemen, Gibson, Liebster, Wren, Willcocks and Kendall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sharon L. Kendall, c2tlbmRhbGxAcnZjLmFjLnVr
 Valwynne Faulkner
Valwynne Faulkner Adrienne Adele Cox2
Adrienne Adele Cox2 Shan Goh
Shan Goh Annelies van Bohemen
Annelies van Bohemen Amanda J. Gibson
Amanda J. Gibson Oliver Liebster
Oliver Liebster Brendan W. Wren
Brendan W. Wren Sharon L. Kendall
Sharon L. Kendall