- 1Engineering Research Center, Chinese Ministry of Education for Edible and Medicinal Fungi, Jilin Agricultural University, Changchun, China
- 2Institute of Special Wild Economic Animal and Plant Science, Chinese Academy of Agricultural Sciences, Changchun, China
Panax ginseng rusty root rot caused by the Ilyonectria species complex is a devastating disease, and it is one of the main factors contributing to the difficulty in continual cropping. Rusty root rot occurs in all ginseng fields, but little is known about the taxonomy of the fungal pathogen complex, especially Ilyonectria and Ilyonectria-like species. Rusty root rot samples were collected from commercial ginseng cultivation areas of China, and the pathogens were isolated and purified as single spores. Based on the combination analysis of multiple loci (rDNA-ITS, TUB, HIS3, TEF, ACT, LSU, RPB1, RPB2, and SSU) and morphological characteristics, the pathogens causing ginseng rusty root rot were determined. Fungal isolates were obtained from infected roots in 56 locations within main cultivation areas in China. A total of 766 strains were identified as Ilyonectria, Ilyonectria-like and Rhexocercosporidium species, including I. robusta (55.0%), I. communis (21.7%), I. mors-panacis (10.9%), I. pseudodestructans (2.0%), I. changbaiensis (1.3%), I. qitaiheensis (1.3%), Neonectria obtusispora (2.0%), Dactylonectria torresensis (0.5%), D. sp. (0.5%), and R. panacis (1.5%), and four novel species, Thelonectria ginsengicola (1.0%), T. jixiensis (1.0%), T. mulanensis (0.8%) and T. fusongensis (0.5%), with a total of 14 species. As the pathogen present in the highest proportion, I. robusta was the most prevalent and damaging species, unlike the pathogens reported previously. All of the examined strains were proven to cause ginseng rusty root rot. Our results indicate that the taxonomy of the fungal complex associated with ginseng rusty root rot includes Ilyonectria, Ilyonectria-like genera (Dactylonectria, Neonectria, and Thelonectria) and Rhexocercosporidium.
Introduction
Panax ginseng is one of the most cultivated medicinal plants in China, and the quality of ginseng greatly influences the quality of cosmetics, health care products and medicines that use ginseng as a raw material (Baranov, 1966; Hu, 1977). Ginseng rusty root rot is caused by Ilyonectria/Cylindrocarpon or Ilyonectria/Cylindrocarpon-like species, and it is the most devastating chronic disease and the greatest threat to ginseng (Panax ginseng) cultivation (Guan et al., 2020). Rusty root rot may cause 30% loss of ginseng, and it can be found in all ginseng planting areas. Rusty root rot has always been one of the important factors that interferes with the maintenance of continuous ginseng cropping. The incidence of rusty root rot in 2-year-old ginseng continuous cropping is 95.8% (Cho et al., 1995).
All parts of the ginseng root may be infected, and the infected root exhibits reddish brown dry rot with a gully appearing on most of the root. The infection stops with increased temperature, and some parts of the ginseng root may undergo self-healing, but the gully or dry rot scars remain. The appearance of ginseng root rot and the degree of damage greatly influence the value of ginseng (Zhou et al., 2017). Ilyonectria and Ilyonectria-like species are common soil fungi, opportunistic plant root pathogens, or asymptomatic root endophytes (Seifert et al., 2003). Ginseng seeds also carry Ilyonectria pathogens that may pose a threat to ginseng cultivation (Guan et al., 2019). Ilyonectria fungi play an important role in black foot rot in grapevines (Halleen et al., 2004; Halleen et al., 2006a,b), apple replant disease (Tewoldemedhin et al., 2011), and beech cankers (Castlebury et al., 2006); the affected plants are representative hosts of economic importance, especially the lignified roots of perennials. Thus, Ilyonectria and Ilyonectria-like species are commonly associated with rot and decay of woody and herbaceous plants (Domsch et al., 2007).
The pathogen associated with root rot disease of Panax quinguefaliam was first extracted by Zinnsmeister (1918), and it was designated Ramularia destructans. Similar diseases were found by Chou and Chung on ginseng in China and Korea (Chung, 1979; Wang, 2001). Scholten established the new species C. destructans, and R. destructans was treated as a synonym (Scholten, 1964).
Neonectria/Cylindrocarpon is a paraphyletic based on phylogenetic analysis (Mantiri et al., 2001; Halleen et al., 2004; Hirooka et al., 2005; Castlebury et al., 2006). The Neonectria complex was divided into four genera (Ilyonectria, Neonectria/Cylindrocarpon, Rugonectria and Thelonectria) based on a combination of morphological characteristics. Ilyonectria replaced the N. radicicola group based on the description by Booth (1966); Seifert et al. (2003), and the corresponding Cylindrocarpon also became Ilyonectria (Mantiri et al., 2001).
I. destructans was linked to the teleomorph I. radicicola (Booth, 1966; Samuels and Brayford, 1990; Chaverri et al., 2011). Based on a phylogenetic analysis of nuclear ribosomal internal transcribed spacer (rDNA-ITS) gene sequences, Schroers et al. concluded that the I. radicicola complex included C. destructans, C. destructans var. crassum, I. coprosmae, I. liriodendri, N. austroradicicola and N. macroconidialis (Schroers et al., 2007). The rDNA-ITS, β-tubulin (TUB), histone H3 (HIS3) and translation elongation factor 1-α (TEF) genes were used to support the I. radicicola species complex, and HIS3 was the most contributing gene (Cabral et al., 2012b). Sixty-eight strains of pathogenic fungi from ginseng and other hosts were once considered C. destructans within four groups: I. mors-panacis, I. robusta, I. panacis, and I. crassa (Cabral et al., 2012a). Lu et al. (2020) obtained 230 isolates and believed that ginseng red-sin root was caused by 12 species, including Fusarium, Dactylonectria, Ilyonectria and others.
Thelonectria was first classified in Cylindrocarpon and first described in 2011 to accommodate grouping in the Cylindrocarpon classification (Chaverri et al., 2011). However, the forms isolated from soil were the asexual states of Thelonectria. Thelonectria and related species with Cylindrocarpon-like morphology were redefined using molecular phylogenetic techniques (Chaverri et al., 2011; Salgado-Salazar et al., 2012, 2013, 2015). The observation that R. panacis causes ginseng rusted root rot was reported for the first time in 2006, after more than 70 years of the disease (Reeleder et al., 2006; Reeleder, 2007).
In this work, we report the population structure of pathogens associated with ginseng rusty root rot. In the commercial ginseng cultivation areas in China, 90% of rusty root rot is thought to be caused by I. destructans (Wang, 2001), but the experimental data support different conclusions. The present study determined that the pathogens that cause rusty root rot disease of ginseng are consistently associated with a fungal complex species, and management strategies must be developed.
Materials and Methods
Sample Collection, Fungal Isolation, and Morphological Observation
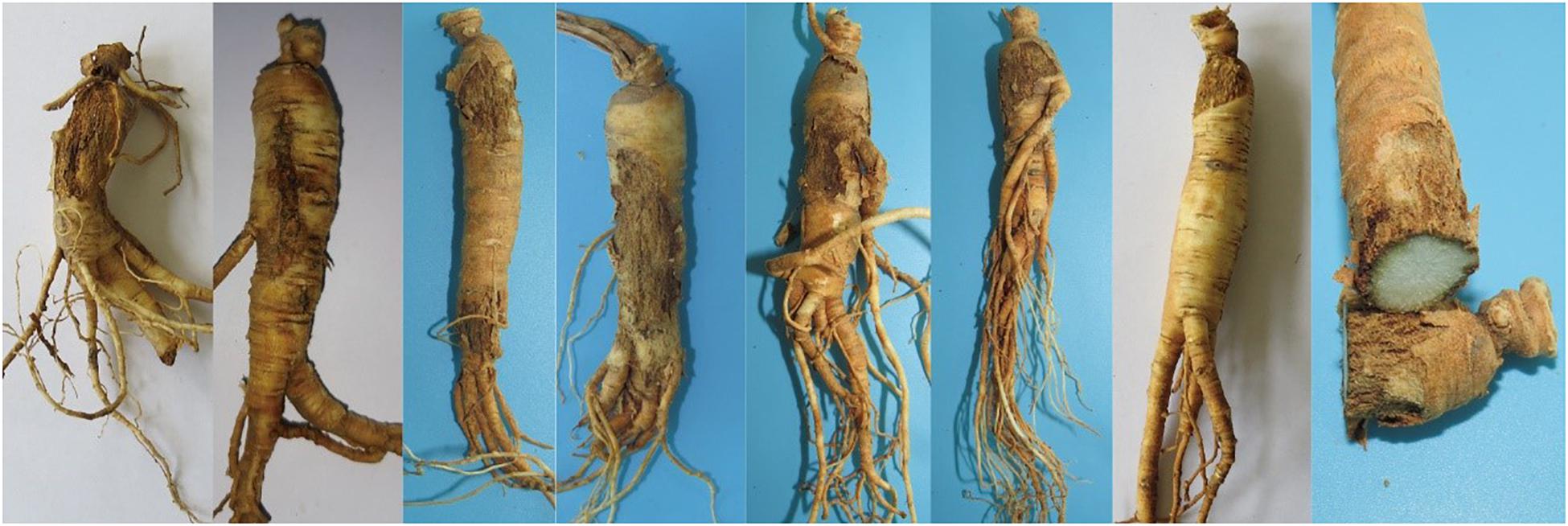
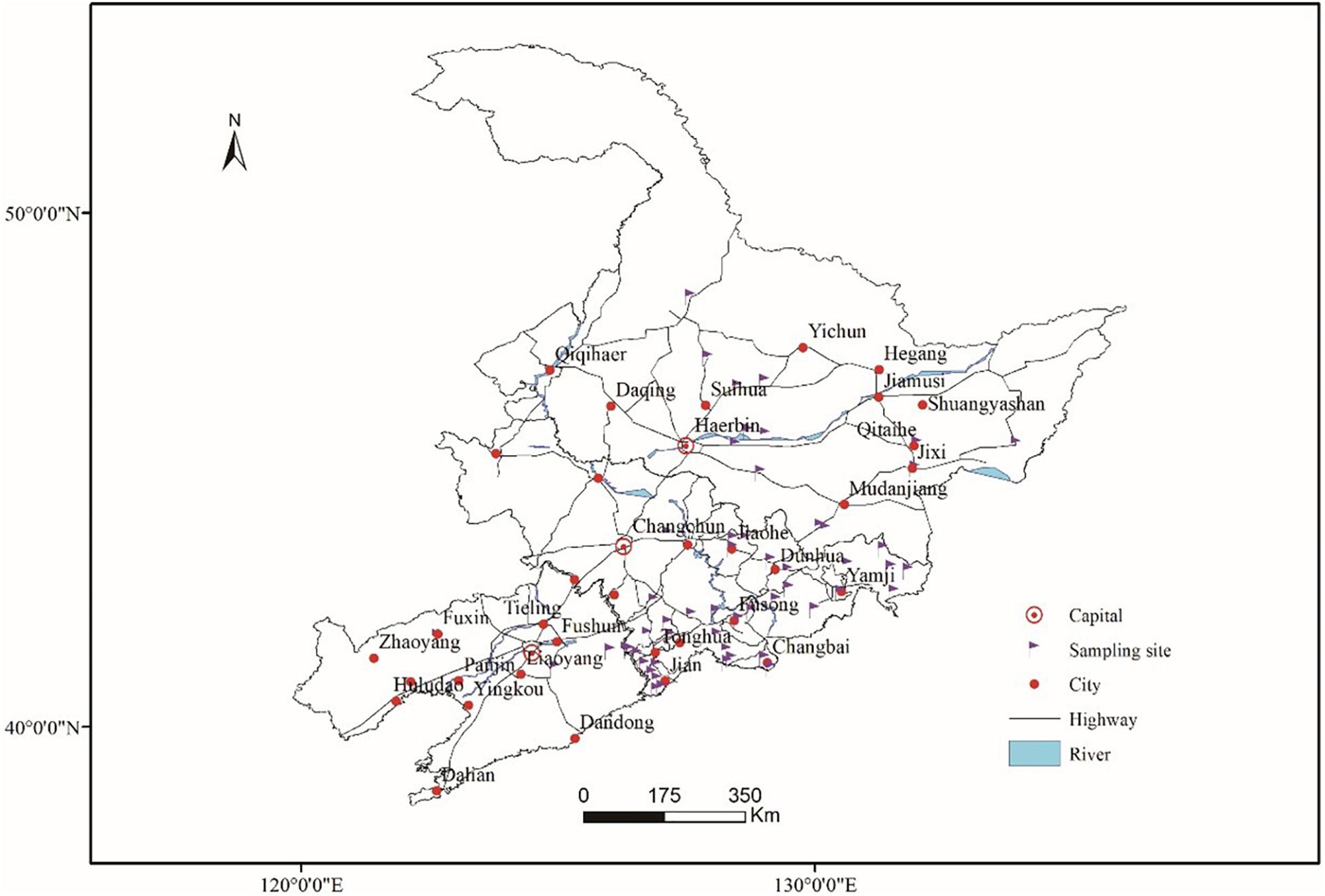
Fresh ginseng roots with rusty root rot symptoms (Figure 1) were collected between 2017 and 2019 from 56 sampling locations in northeast China that corresponded to the commercial ginseng-producing areas of China (Figure 2). The junction of the healthy and diseased parts was cut into 25-mm3 pieces, and the surfaces of the pieces were disinfected by immersion in 1% NaOCl for 2 min and washing three times with sterile water. The pieces were evenly placed on water agar (WA) medium and cultured in the dark at 22°C. Colonies were collected for 15-21 days, purified and cultured from single spores (Punja, 1997). The cultures were transferred to potato dextrose agar (PDA) medium and synthetic nutrient-poor agar (SNA) and cultured in the dark for 20-60 days, and the characteristics of the resulting cultures were recorded. The isolates were cultivated on PDA supplemented with ginseng roots under continuous near-ultraviolet light (n-UV, 315-400 nm), and the characteristics of conidiophore cells and the shape and size of chlamydospores and conidia were observed. Images and measurements were acquired using a KEYENCE VXH-5000 digital microscope system (Osaka, Japan).
DNA Extraction and PCR Amplification
Colonies grown for 15 days were collected for DNA extraction. DNA was obtained by following the procedures described in the Qiagen DNeasy Plant Mini Kit (Qiagen 69104, Qiagen, Hilden, Germany). Each 50-μl PCR aliquot contained 25 ml of premix (Takara R045, Kusatsu, Japan; the error rate of the DNA polymerase is 0.02%), 2 μl of each primer (10 μM), 1 μl of DNA (500 ng/μl), and 20 μl of double-distilled H2O. The following PCR program was used to obtain the sequences of the actin gene (ACT), rDNA-ITS, the 28S ribosomal RNA gene (LSU), the gene encoding RNA polymerase II largest subunit (RPB1), the gene encoding RNA polymerase II second-largest subunit (RPB2), 18S ribosomal RNA (SSU), TEF, TUB and HIS3: 95°C for 3 min; 30 cycles of 95°C for 15 s, annealing temperature for 15 s, 72°C for 60 s, and 72°C for 10 min; the sample was then held at 4°C. The amplicons were analyzed using 0.8% agarose gel electrophoresis in 0.5 × TAE buffer (150 V for 25 min) and purified using the MinElute PCR Purification Kit (Qiagen 28004, Qiagen, Hilden, Germany). Details regarding the PCR primers and annealing temperatures are listed in Table 1.
DNA Sequencing and Phylogenetic Analysis
The purified amplicons were sequenced in both directions by Sangon Biotech (Shanghai, China), and the sequences were assembled using Seqman (v8.1). Phylogenetic trees were constructed using the combined rDNA-ITS, TUB, HIS3 and TEF genes for analyses of Dactylonectria, Ilyonectria and Neonectria, and the analysis of Thelonectria was based on a combination of the ACT, rDNA-ITS, LSU, RPB1, RPB2, SSU, TEF and TUB genes. All sequences were homologously aligned by mega 7.06. A maximum parsimony (MP) method was used to construct a phylogenetic tree in the PAUP 4.0b program. The MP analyses were performed with the heuristic search option, and 100 random sequence additions were used to find the global optimum tree. The gaps were treated as missing data, and the strength of the internal branches of the resulting trees was tested with bootstrap analysis using 1000 replications. The consistency index (CI), retention index (RI), and rescaled consistency index (RC) of the tree were also calculated. Detailed information on the sequences of the genes whose accession numbers were applied for in the NCBI database, including all strains used for phylogeny, is shown in Tables 2, 3.
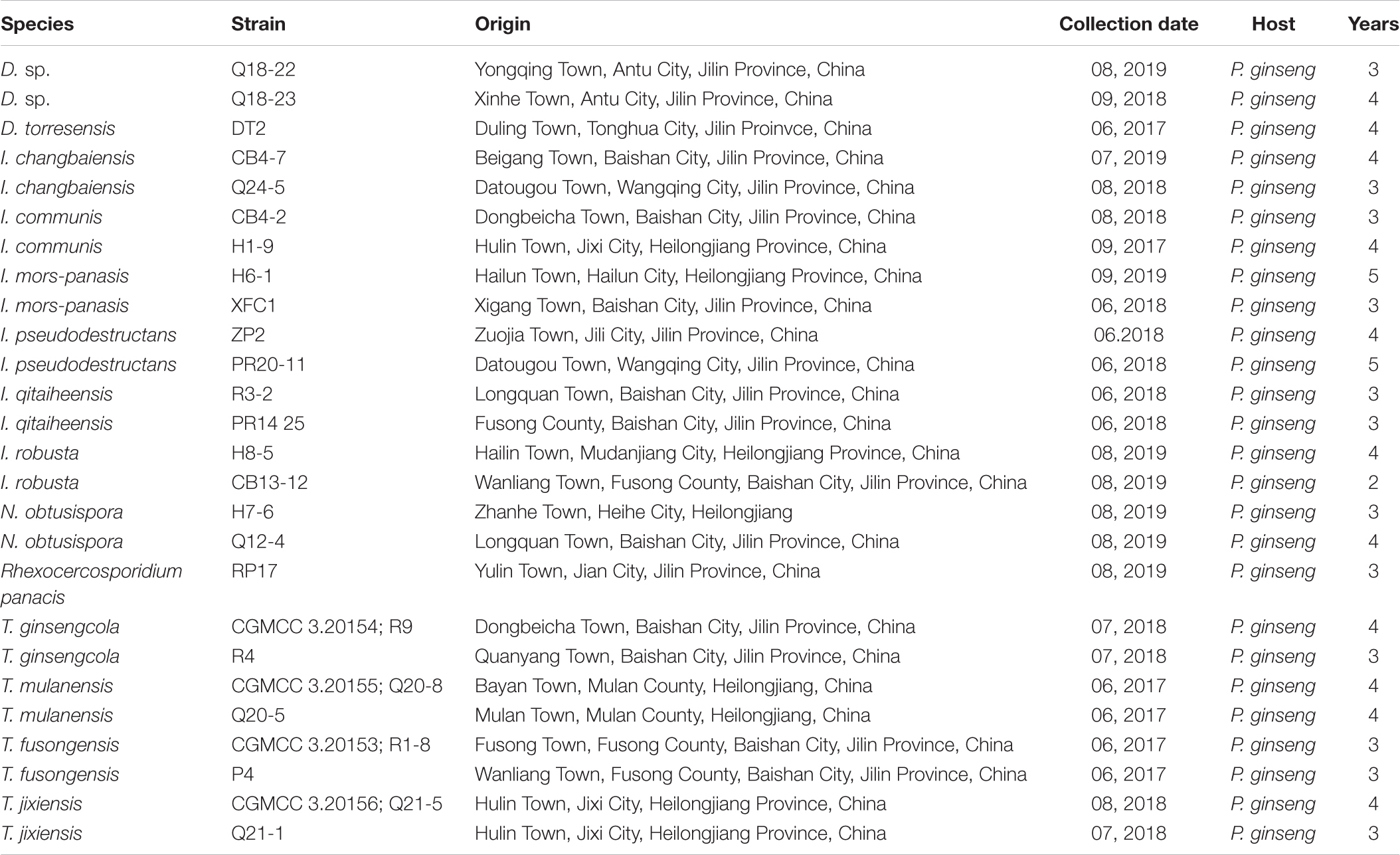
Table 3. Detailed information on the collection of isolates used in sequencing and pathogenicity testing.
Pathogenicity Tests
The 26 newly sequenced strains were also subjected to pathogenicity testing (Table 3). The pathogenicity of the isolates was evaluated on 3-year-old detached ginseng roots (cultivar: Damaya) using an improved published method in vitro (Lu et al., 2020). The fresh ginseng roots were washed, wiped with 75% alcohol, and rinsed with sterile water. Healthy ginseng roots were inoculated by placing 50 μl of 1 × 105/ml spore suspension taken from the edges of actively growing colonies on PDA plates into premade holes 4 mm in diameter and 1 mm deep. Two to three holes per root and four replicated roots were inoculated for each isolate, and sterile water was used as the control. The ginseng roots were cultured in a fresh-keeping box at 25°C in the dark and evaluated after 7 days. The same strain and disinfection method were used to inoculate whole plants in the greenhouse (25°C, 78% humidity). After disinfection, 3-year-old ginseng seedlings were dipped into a 1 × 105/ml spore suspension for 20 min and then transferred to pots containing sterile sand, with 6 plants per pot. Each group was grown in triplicate. The inoculum was treated with sterile water as a control. After 2 months, plant infection was evaluated.
Results
Population Structure of Fungal Complex Associated With Ginseng Rusty Root Rot
A total of 766 isolates were obtained from ginseng roots that showed typical signs of rusty root rot disease. All of these strains were determined by sequencing and morphological characteristics, and 26 strains from each genus were randomly selected for phylogenetic analysis. A total of 14 species (Figures 3, 4) were identified as Ilyonectria, Ilyonectria-like and Rhexocercosporidium species.
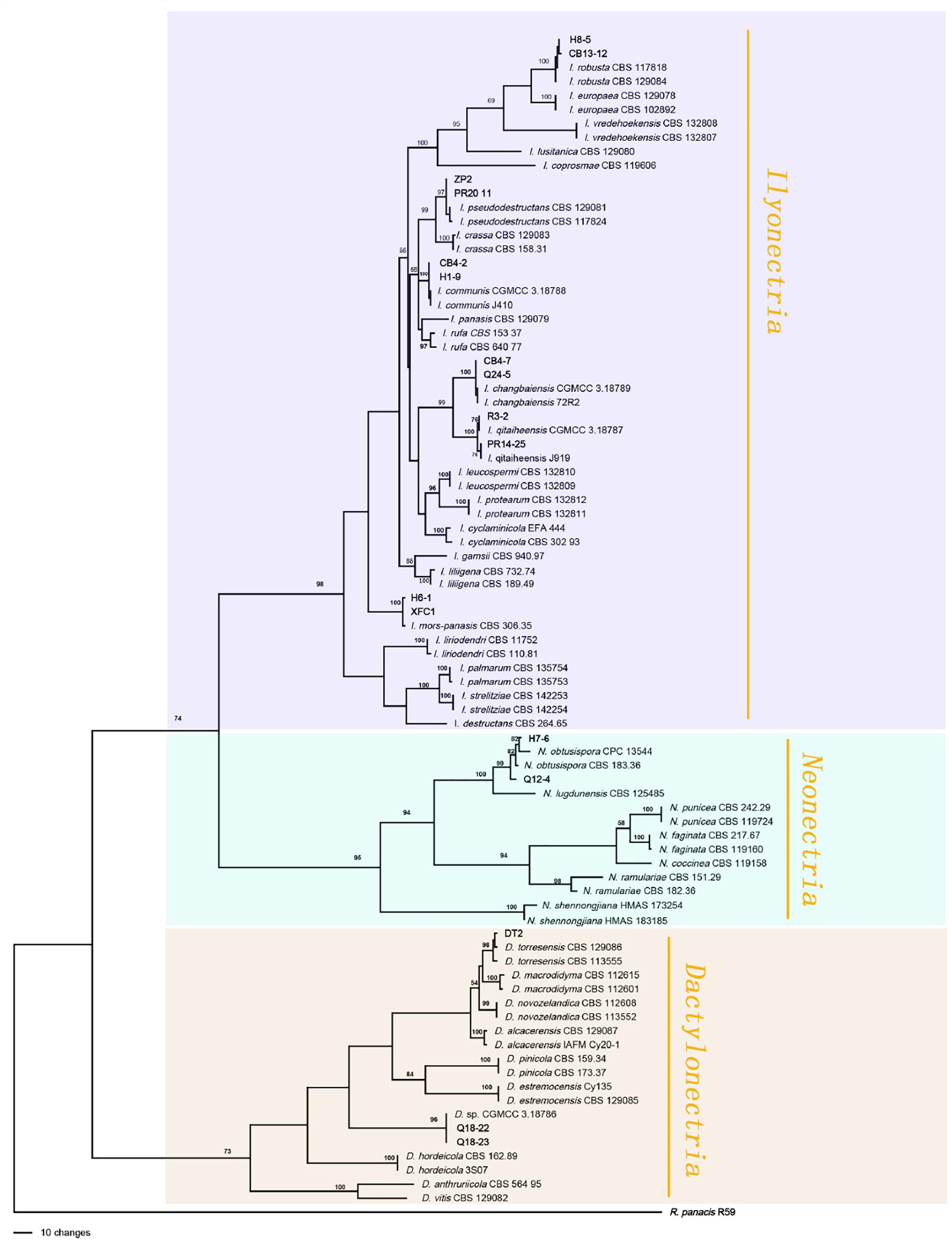
Figure 3. Phylogenetic tree based on the combined rDNA-ITS, TUB, HIS3 and TEF gene sequences constructed using the maximum parsimony method of the PAUP 4.0b program. Rhexocercosporidium panacis R59 was used as an outgroup. Bold font indicates the strains isolated in this study.
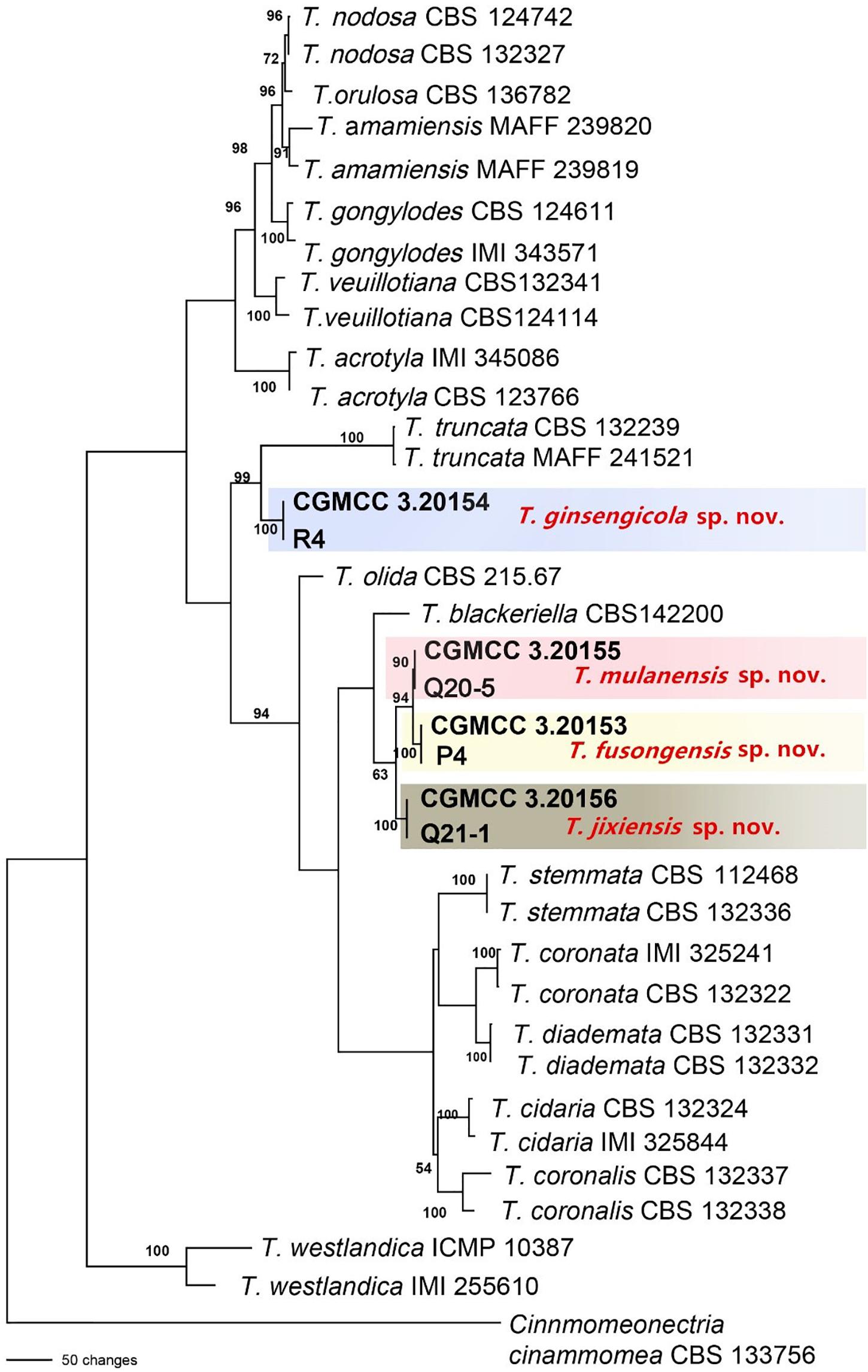
Figure 4. Phylogenetic tree based on the combined ACT, rDNA-ITS, LSU, RPB1, RPB2, SSU, TEF and TUB gene sequences constructed using the maximum parsimony method of the PAUP 4.0b program. Cinnmomeonectria cinammomea CBS 133756 was used as an outgroup. Bold font indicates the strains isolated in this study.
The Ilyonectria species complex included I. robusta, I. communis, I. mors-panacis, I. pseudodestructans, I. changbaiensis and I. qitaiheensis, and the isolate proportions were 55.0%, 21.7%, 10.9%, 2.0%, 1.3% and 1.3%, respectively. Ilyonectria-like contained three genera that were also members of the Nectriaceae family: Dactylonectria, Neonectria and Thelonectria. N. obtusispora, D. torresensis and D. sp. accounted for 2.0%, 0.5% and 0.5% of the population, respectively. Four novel species are named in this article: T. ginsengicola (1.0%), the type strain is R9; T. jixiensis (1.0%), the type strain is Q21-5; T. mulanensis (0.8%), the type strain is Q20-8 and T. fusongensis (0.5%), the type strain is R1-8. R. panacis was the only pathogen except Ilyonectria-like that was present at an abundance of 1.5%.I. communis, I. pseudodestructans, I. changbaiensis, I. qitaiheensis, N. obtusispora, D. torresensis, T. jixiensis, T. mulanensis, T. fusongensis and T. ginsengicola were first reported to cause ginseng rusty root rot. Thelonectria is also the first Ilyonectria-like genus reported to cause ginseng rusty root rot. I. robusta was the dominant pathogen and appeared in the largest proportion; this is the greatest difference between our results and previous conclusions. The number of fungal isolates recovered from each sampling location in China is shown in Table 4.
Molecular Phylogenetic Analysis
The sequences of rDNA-ITS, TEF, TUB and HIS3 were obtained using polymerase chain reaction amplicons and were analyzed in Dactylonectria, Ilyonectria, and Neonectria. Low-quality sequences were removed after base alignment to obtain the sequences of rDNA-ITS (491 bp), HIS3 (472 bp), TEF (507 bp), and TUB (467 bp). The sequences were combined, and a sequence with a total length of 2231 bp, including alignment gaps, was used to build a phylogenetic tree (Figure 3). Similarly, ACT (575 bp), rDNA-ITS (533 bp), LSU (700 bp), RPB1 (630 bp), RPB2 (856 bp), SSU (521 bp), TEF (858 bp), and TUB (479 bp) were combined, resulting in a total length of 4422 bp including alignment gaps, and analyzed for Thelonectria (Figure 4). All of the sequences were topologically congruent, and the results indicated that Dactylonectria, Ilyonectria, Neonectria and Thelonectria formed a single clade. The isolated strains were divided into 14 highly supported clades; the branches containing I. rubosta, I. mors-panacis and other known species were highly consistent with species type or verified species.
Four novel species were divided into independent clades, and they were supported with 90-100% bootstrap support for T. mulanensis, T. jixiensis, T. fusongensis and T. ginsengicola (Figure 4). For D. sp., we observed only a few macroconidia, and the specific sporulation structure and other morphological characteristics were not clear. Therefore, the species is not named here. Similarly, a separate gene phylogenetic tree was constructed for Thelonectria, with the least information obtained from the SSU and TUB genes, and the lowest contribution. ACT and RPB2 were the most informative and were key to distinguishing the phylogenetic tree of all of the known candidate strains and four novel species. The combined use of rDNA-ITS, TUB, TEF, and LSU genes will aid in the classification and analysis of all candidate species. The four novel species of Thelonectria belong to the T. coronata complex classification reported in 2016 (Salgado-Salazar et al., 2016). The three closely related species T. mulanensis, T. fusongensis, and T. jixiensis have nucleotide sequence differences in their ACT, rDNA-ITS, LSU, RPB1, RPB2, SSU, TEF and TUB genes (Table 5). There is no nucleotide difference in the SSU gene, 1 bp difference in the rDNA-ITS and LSU genes, and 2 bp differences in the RPB1 gene; thus, these three genes are ineffective for distinguishing the three species. These three species have 5 bp differences in ACT, 14 bp differences in RPB2, 19 bp differences in TEF and 4 bp differences in TUB. T. mulanensis and T. fusongensis have 3 bp differences in RPB2, 5 bp differences in TEF and 4 bp differences in TUB. Phylogenetic analysis combining the RPB2, TEF and TUB genes is most effective at distinguishing T. mulanensis and T. fusongensis. T. mulanensis and T. fusongensis were clustered on a branch with a close genetic relationship and may form a complex group. However, due to their distinct morphological characteristics of conidia size and number of septa, they were considered two novel species.
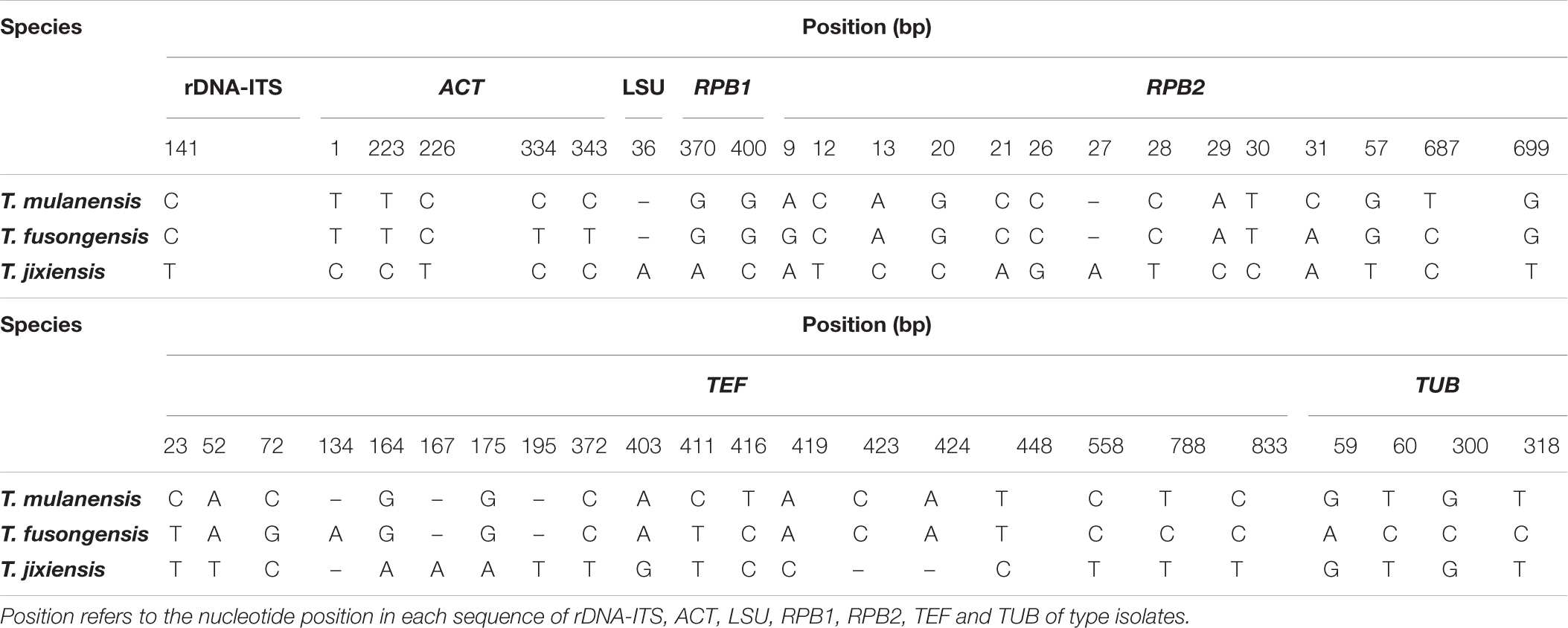
Table 5. Nucleotide differences in the partial gene sequences of rDNA-ITS, ACT, LSU, RPB1, RPB2, TEF, and TUB for T. mulanensis, T. fusongensis and T. jixiensis.
Brief Introduction to the Morphological Characteristics of Ilyonectria Species
Morphological characteristics also support the classification of phylogenetic trees. The morphological characteristics of I. robusta, I. communis, I. mors-panacis, I. pseudodestructans, I. changbaiensis, I. qitaiheensis, N. obtusispora, D. torresensis and R. panacis were consistent with previously reported strains. Four novel species were confirmed by phylogeny and are described as follows.
Thelonectria ginsengicola Y. M. Guan & Y. Li, sp. nov.
MycoBank MB836525 (Figure 5).
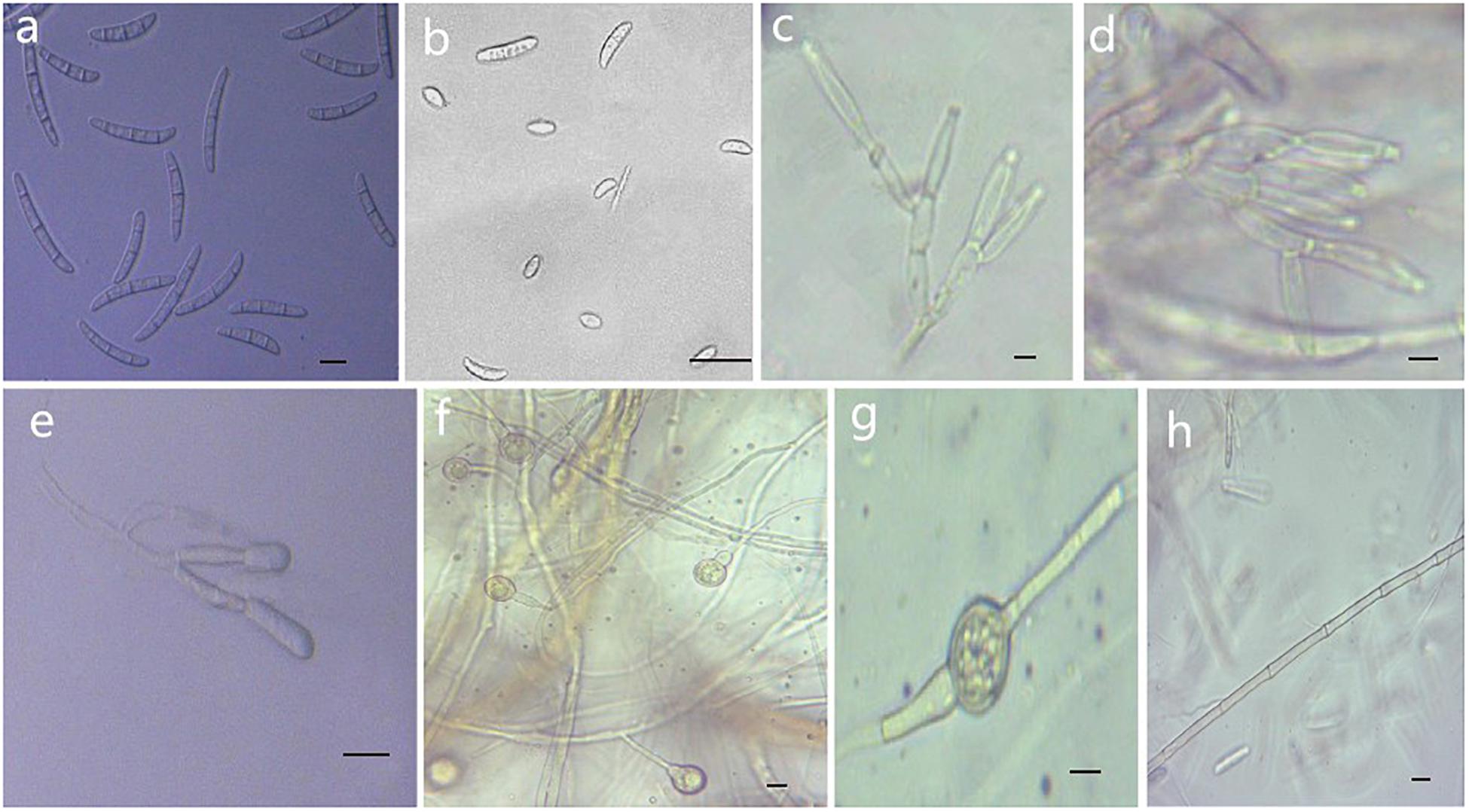
Figure 5. Morphological characteristics of Thelonectria ginsengicola (CGMCC3.20154). (a) Macroconidia. (b) Microconidia. (c-e) Conidiophores. (f,g) Chlamydospores. (h) Hypha. Bar = 10 μm.
Etymology: Named after the host Panax ginseng. “Ginsengicola” means “born on ginseng.”
Type: China, Jilin Province, Changbai City, county of Fusong, on roots of Panax ginseng, Oct 2018, Y. Guan. (CGMCC3.20154 = R9 - holotype).
Description: Sexual morph: Undetermined. Asexual morph: Conidiophores were simple or complex with 2-3 branches, monophialides. Mycelial cells were thick-walled and septate. Hyphae hyaline to yellow. Chlamydospores were borne apically or intermittently in hyphae and were more commonly single or in pairs. Macroconidia were the majority, with 1-5 septa; most had 3-4 septa. The shape was a sickle-shaped curve; the base foot was blunt and became gradually thicker toward the top, and the top was also blunt. The size was (38.4) 44.3 (57.7) μm × (4.8) 5.7 (6.9) μm, and the length/width ratio was (5.9) 8.1 (10.3). Microconidia were elliptical, stick or sickle-shaped curves with an irregular shape and 0-1 septa. The size of microconidia was (12.5) 9.69 (12.5) μm × (4.8) 4.1 (6.9) μm, and the length/width ratio was (1.4) 2.7 (3.7). The size of one septate microconidium was (19.5) 24.3 (29.3) μm × (4.1) 4.8 (6.5) μm, and its length/width ratio was (3.9) 5.5 (6.4).
Culture characteristics: On PDA, the color was initially white, later becoming dark yellow with a wavy margin; the reverse side was reddish brown. The colony grows outward in a wave shape. The mycelium grew vigorously, and it was difficult to produce conidia on PDA and SNA. After the addition of ginseng roots and continuous n-UV irradiation, pionnotes were produced on the mycelium or on the vertical wall of the petri dish. The growth of the colony formed concentric rings, and the growth was very slow. The size of the colony reached 22 mm in 10 days. On SNA, the colonies were sparse and white to transparent with regular margins and chlamydospores; macroconidia and microconidia were not observed.
Note: T. ginsengicola and T. truncata were grouped together on a branch. The greatest difference was the presence of microconidia and chlamydospores. Macroconidia were smaller than those of T. truncate (Salgado-Salazar et al., 2012).
Thelonectria jixiensis Y. M. Guan & Y. Li, sp. nov.
MycoBank MB 836520 (Figure 6).
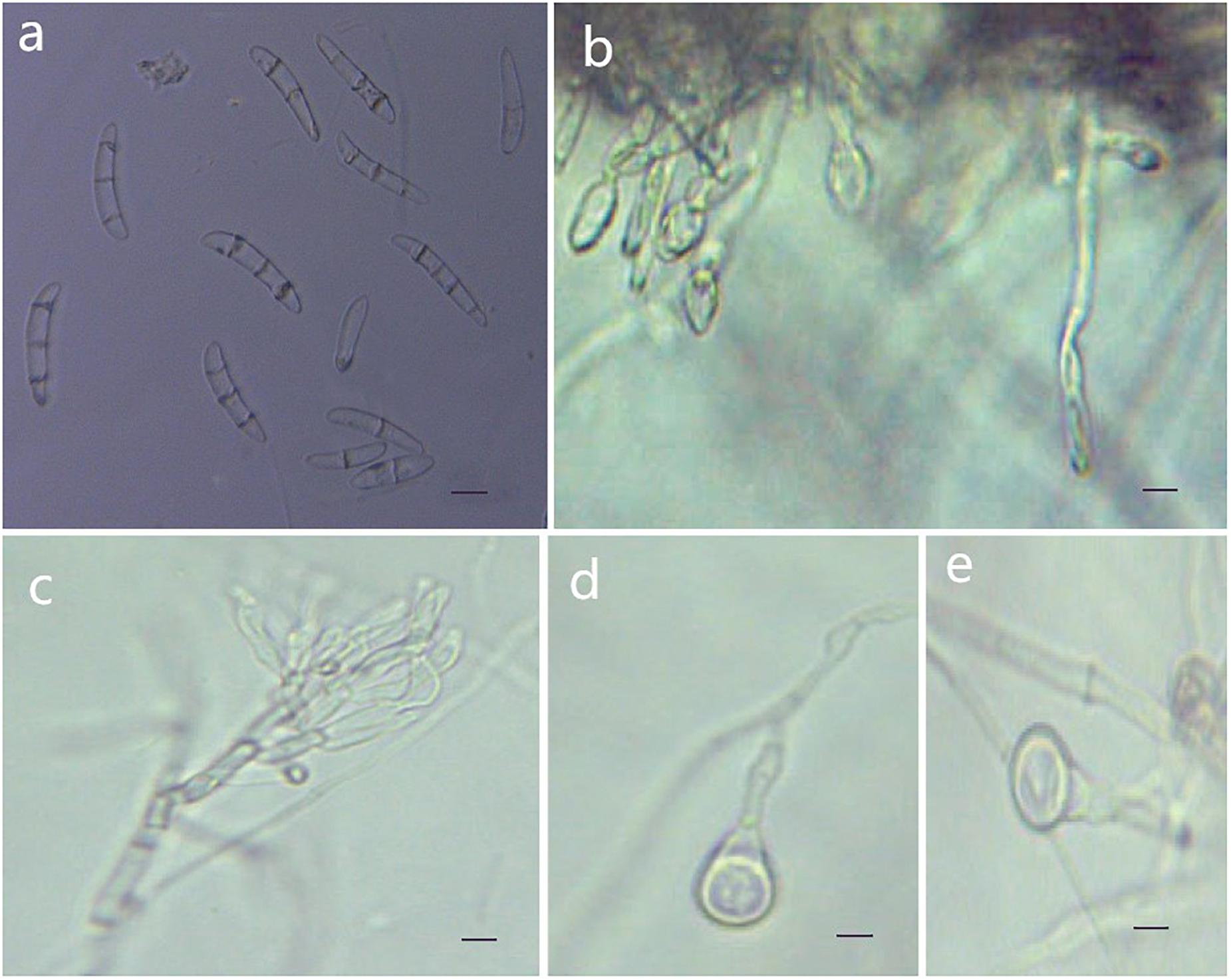
Figure 6. Morphological characteristics of Thelonectria jixiensis (CGMCC3.20156). (a) Macroconidia. (b) and (c) Conidiophores. (d) and (e) Chlamydospores. Bar = 10 μm.
Etymology: Named after the city of Jixi, Heilongjiang Province, China, where the strain type was collected.
Type: China, Heilongjiang Province, Jixi City, on roots of Panax ginseng, Oct 2018, Y. Guan. (CGMCC3.20156 = Q21-5 - holotype).
Description: Sexual morph: Undetermined. Asexual morph: Conidiophores simple or complex, spiral branches, monophialides and straight macroconidia or slightly curved head. The body is widest at one-quarter of the head; the basal cells with tips are thinner and narrower, with 0-4 septa. Most had 2-3 septa, and very few had 0 or 4 septa; sizes were in the range of (27.3) 35.8 (41.7) μm × (5.5) 6.9 (8.9) μm, with a length/width ratio of (4.3) 5.1 (7.9). Microconidia were not observed. Chlamydospores were mostly borne apically on the mycelium, and solitary was more common.
Culture characteristics: On PDA, the colonies produced were white at the beginning with a sparsely floccose to fluffy aerial mycelium and vigorous growth. The hyphae at the margins of the colony were thin, and the center hyphae were denser. Light yellow color appeared occasionally on the reverse side over time. The colony grew slowly to a size of 38 mm in 10 days. On SNA, the colonies were sparse, with regular margins, and the mycelium obviously degenerated and gradually disappeared as the number of breeding generations increased. Chlamydospores and macroconidia were not observed on SNA.
Note: The phylogenetic tree shows that T. jixiensis is closely related to T. blackeriella. T. jixiensis produced simple or complex conidiophores with spiral branches, and its macroconidia were slightly larger. The simple conidiophores of T. blackeriella and the macroconidia were relatively small. The shapes of the macroconidia of the two are very easily distinguished, and neither species produces microconidia (Carlucci et al., 2017).
Thelonectria mulanensis Y. M. Guan & Y. Li, sp. nov.
MycoBank MB 836521 (Figure 7).
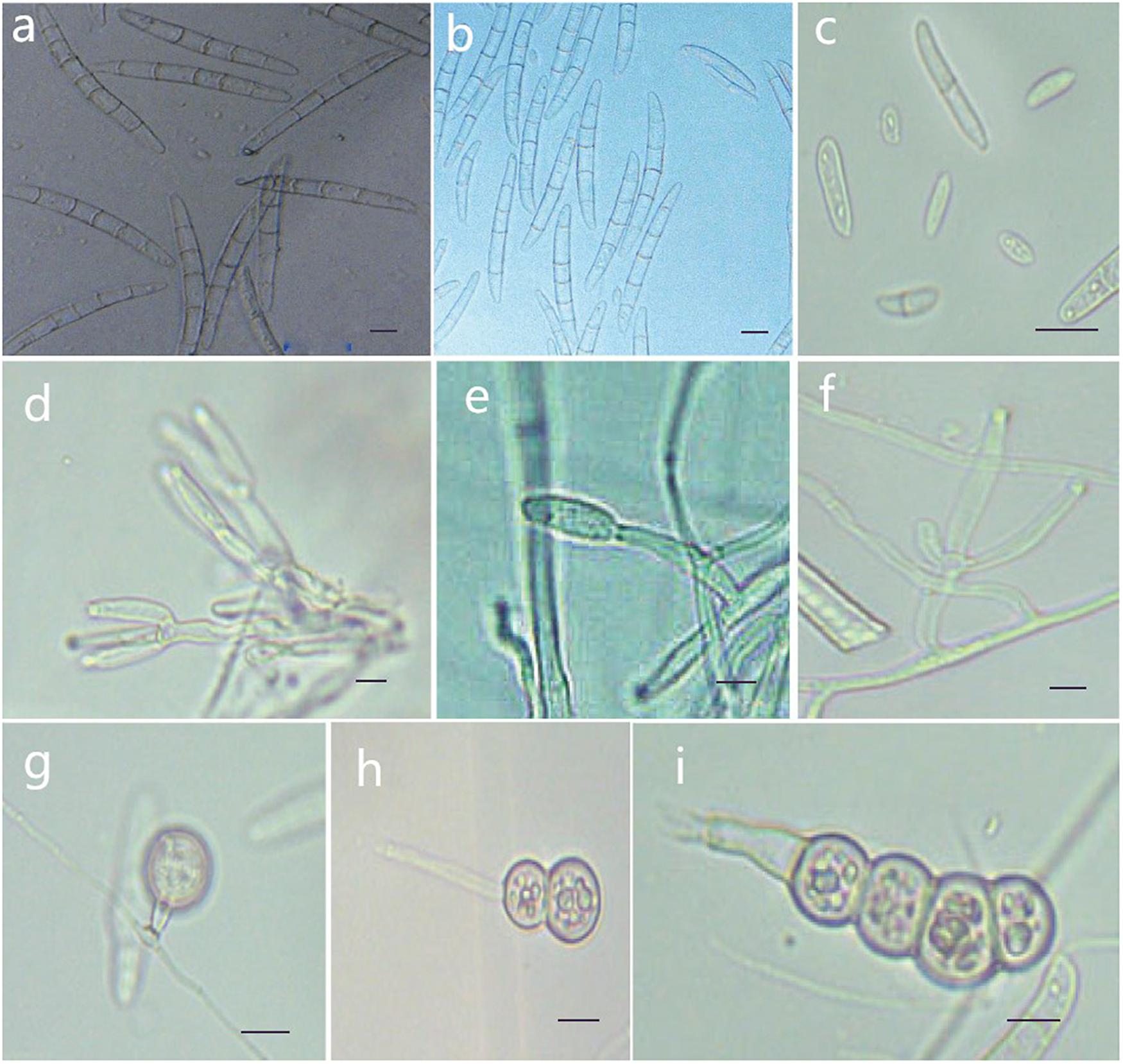
Figure 7. Morphological characteristics of Thelonectria jixiensis (CGMCC3.20155). (a,b) Macroconidia. (c) Microconidia. (d–f) Conidiophores. (g–i) Chlamydospores. Bar = 10 μm.
Etymology: Named after the County of Mulan, Heilongjiang Province, China, where the strain type was collected.
Type: China, Heilongjiang Province, Mulan County, on roots of Panax ginseng, Oct 2018, Y. Guan. (CGMCC3.20155 = Q20-8 - holotype).
Description: Sexual morph: Undetermined. Asexual morph: Conidiophores simple or complex, with 2-3 branches, monophialides. The mycelial cells were thick-walled, transparent and septate. The chlamydospores were produced from the mycelium, and spores were intercalary or terminal and single, in pairs or multiple bunches. Macroconidia were the majority, with 3-7 septa, but most were 5 septa, and lengths ranged from (59.1) 76.5 (96.6) μm × (2.7) 8.3 (5.6) μm. Fungi were a sickle-shaped curve. The base foot was blunt and became gradually thicker toward the top, and the top was also blunt. The microconidia were elliptical, stick or sickle-shaped curved of an irregular shape with 0-1 septum, and lengths ranged from (4.6) 9.5 (18.3) μm × (2.7) 3.9 (5.6) μm. Microsclerotia were produced after more than 8 weeks.
Culture characteristics: On PDA, the colonies produced a white, cottony dense floccose to fluffy aerial mycelium. The margins of the colony were irregular, and the hyphae were sparse and radial. The reverse color was white to yellow. In PDA culture, colonies grew slowly and reached 25 mm in 10 days. On SNA, the colonies were sparse, white to transparent, with regular margins and chlamydospores; macroconidia and microconidia were not observed.
Note: This species is closely related to T. jixiensis, but it produces macroconidia and microconidia, and T. jixiensis has no microconidia. The macroconidia of T. mulanensis were twice the size of those of T. jixiensis, had more septa and were thinner. The conidiophores and chlamydospores of the two species are similar.
Thelonectria fusongensis Y. M. Guan & Y. Li, sp. nov.
MycoBank MB 836524 (Figure 8).
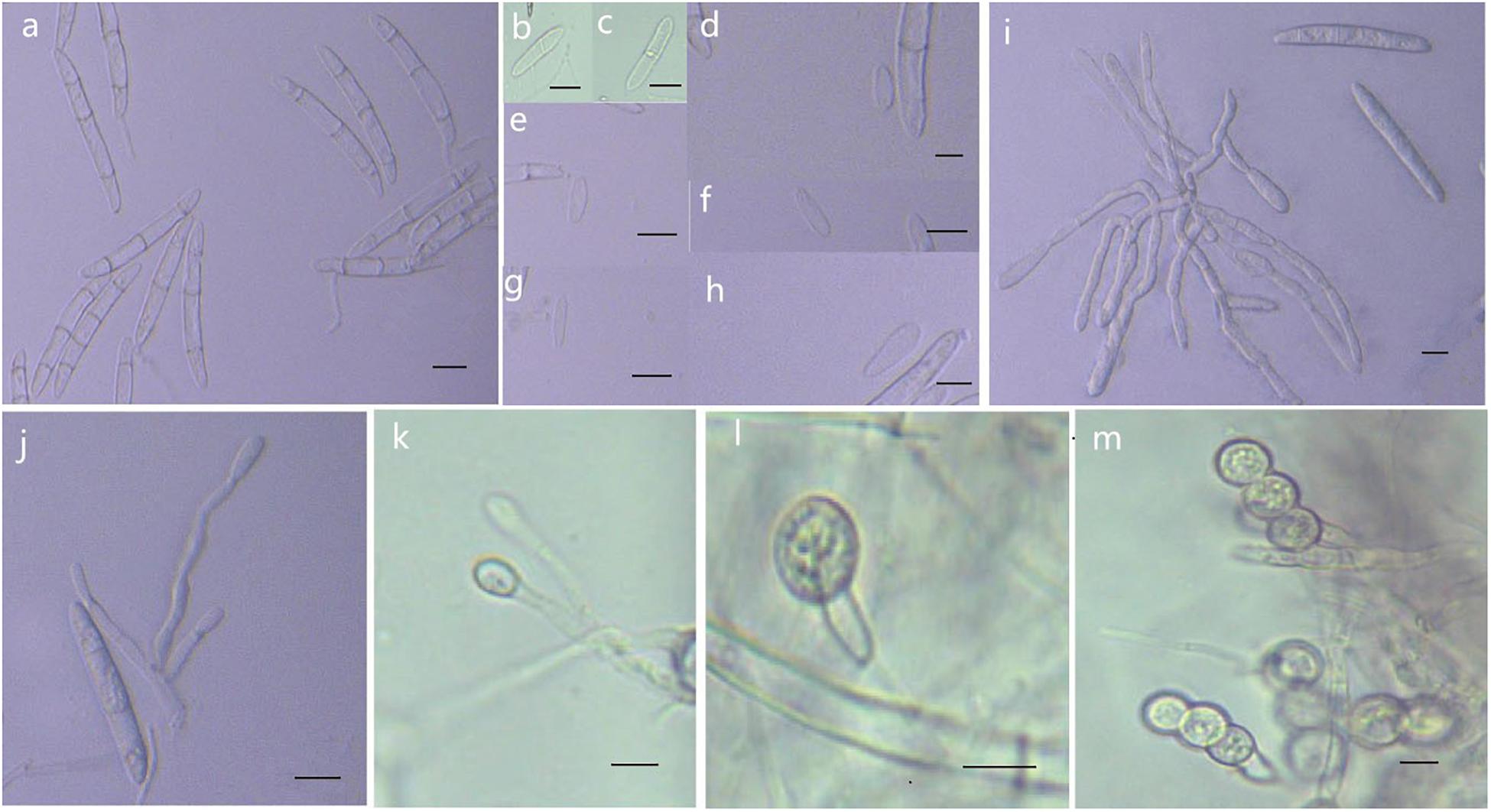
Figure 8. Morphological characteristics of Thelonectria jixiensis (CGMCC3.20154). (a) Macroconidia. (b–h) Microconidia. (i–k) Conidiophores. (l,m) Chlamydospores. Bar = 10 μm.
Etymology: Named after the county of Fusong, Changbai City, Jilin Province, China, where the strain type was collected.
Type: China, Jilin Province, Changbai City, county of Fusong, on roots of Panax ginseng, Oct 2018, Y. Guan. (CGMCC3.20153 = R1-8 - holotype).
Description: Sexual morph: Undetermined. Asexual morph: Conidiophores were simple or complex, with 2-3 branches, monophialides. The mycelial cells were thick-walled, transparent, and septate. Chlamydospores were borne apically or intercalary in hyphae and were solitary or in pairs. Macroconidia were in the majority, with 1-5 septa, but most had 2-3 septa. The body was a sickle-shaped curve, and the base was blunt and thickened gradually toward the top. The top was also blunt, not sharp, with sizes in the range of (40.2) 52.3 (62.2) μm × (4.2) 5.9 (7.9) μm. Microconidia were in the minority and exhibited irregular elliptical, stick or curved sickle shapes and 0-1 septa, with sizes in the range of (7.4) 13.5 (23.3) μm × (2.9) 4.1 (6.1) μm.
Culture characteristics: On PDA, the colonies grew in a concentric wavy pattern and presented a velvety surface; they were initially white, then brownish yellow, and brownish red on the reverse side. The mycelium became increasingly sparse from the center to the edge. It was difficult to produce conidia on PDA and SNA. The addition of ginseng roots and continuous n-UV light cultivation caused the surface of the mycelium to produce yellow pionnotes at more than 8 weeks. The colonies reached 30 mm in diameter in 10 days. On SNA, the colonies were sparse, white to transparent, with regular margins and chlamydospores; macroconidia and microconidia were not observed.
Note: Phylogenetic inference revealed that T. fusongensis is closely related to T. mulanensis, but the microconidia of the former were larger than those of the latter, there were fewer septa, the microconidia were straighter, and the bending arc was smaller.
Pathogenicity Tests
For in vitro inoculation using spore suspension, evaluation after 7 days showed that all of the strains infected ginseng roots. The mycelium grew on the ginseng root, and the longitudinal portion of the ginseng root also showed infection. Reddish-brown areas formed on the shallow ginseng roots near the inoculation point. The control showed no symptoms (Figure 9).

Figure 9. Symptoms displayed by 3-year-old ginseng roots inoculated with pathogens (longitudinal sections taken at the inoculation centerline). (a-n) I. robusta (H8-5), I. communis (CB4-2), I. mors-panacis (H6-1), I. pseudodestructans (ZP2), I. changbaiensis (CB4-7), I. qitaiheensi (R3-2), N. obtusispora (H7-6), D. torresensis (DT2), D. sp. (Q18-22), T. ginsengicola (CGMCC 3.20154), T. jixiensis (CGMCC 3.20156), T. mulanensis (CGMCC 3.20155), T. fusongensis (CGMCC 3.20153), and R. panacis (RP17). (o) control.
For whole plant inoculation in the greenhouse, all of the strains used in the test caused rusty root rot. 2 months after inoculation with the spore suspension, the inoculated 3-year-old ginseng roots showed symptoms similar to rusty root rot and to natural disease in the field. Some ginseng roots were rust-colored, but no gully like symptoms formed (Figure 10). Ginseng roots inoculated with sterile water were asymptomatic. Recovery of the diseased samples and sequencing of all of the genes described above yielded results that were the same as those obtained for the isolates used in the inoculations, satisfying Koch’s postulates, and the identity of the species was determined. Ginseng roots inoculated with R. panacis showed a darker color with grayish-black symptoms.
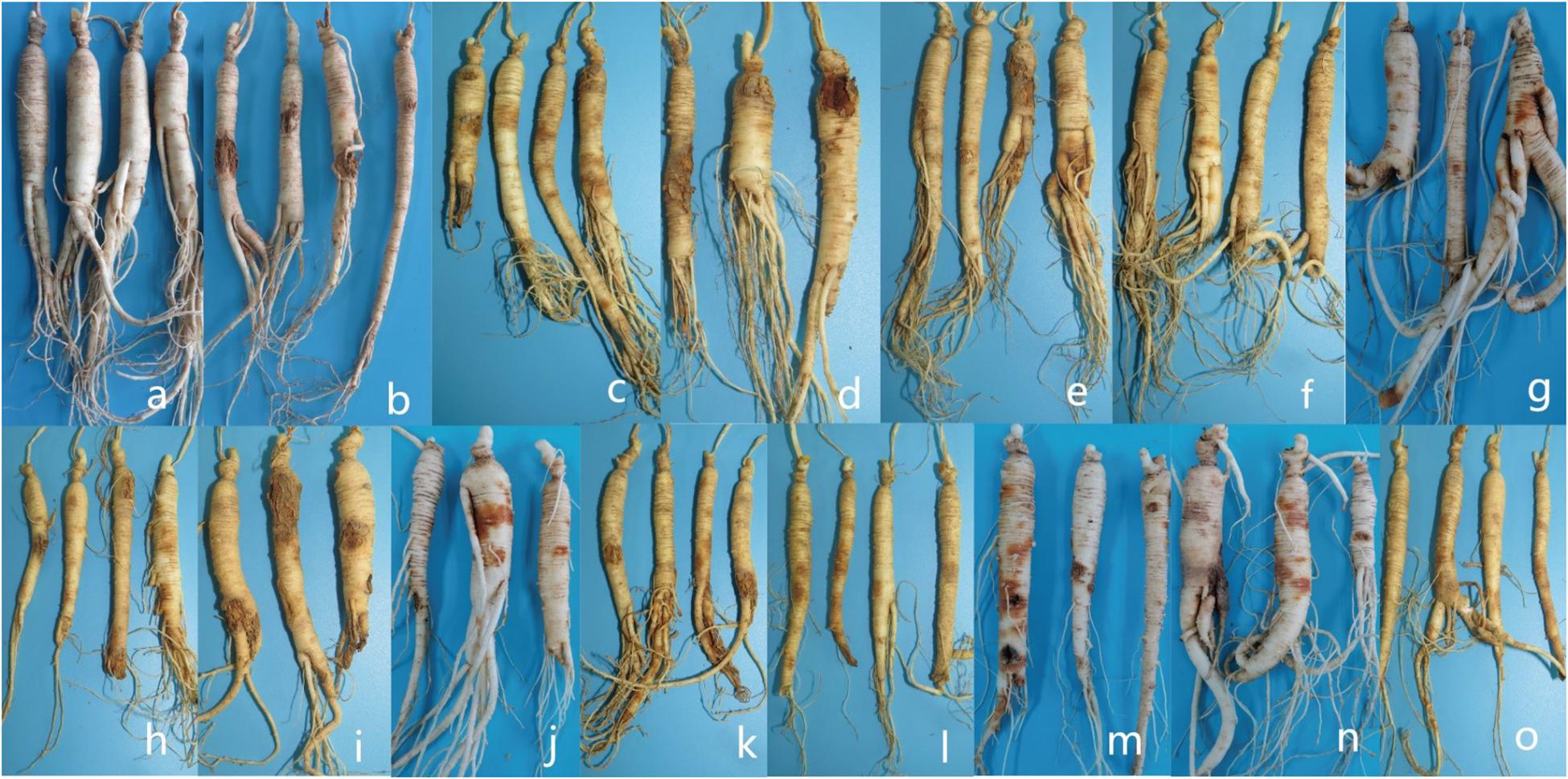
Figure 10. Symptoms displayed by 3-year-old ginseng roots inoculated with pathogens under greenhouse conditions. (a) control. (b-o) I. robusta (H8-5), I. communis (CB4-2), I. mors-panacis (H6-1), I. pseudodestructans (ZP2), I. changbaiensis (CB4-7), I. qitaiheensi (R3-2), N. obtusispora (H7-6), D. torresensis (DT2), D. sp. (Q18-22), T. ginsengicola (CGMCC 3.20154), T. jixiensis (CGMCC 3.20156), T. mulanensis (CGMCC 3.20155), T. fusongensis (CGMCC 3.20153), and R. panacis (RP17).
Discussion
Ginseng rusty root rot is the root disease with the highest incidence in China’s main ginseng cultivation areas. The identities of the varieties of diseased ginseng samples collected in the experiments were not clear. Most of the currently grown Chinese cultivars are domesticated varieties, and Damaya ginseng is commercially cultivated. Therefore, the correlation between the presence of fungi on ginseng roots and ginseng variety was not analyzed. The identification and pathogenicity analysis of 766 isolates revealed that ginseng rusty root rot was caused by a complex of Ilyonectria, Ilyonectria-like, and Rhexocercosporidium fungi. The Ilyonectria-like fungi included three genera of Ilyonectria, Dactylonectria and Thelonectria, all of which belonged to Nectriaceae, except Rhexocercosporidium. I. robusta and I. communis were the dominant pathogens with the highest proportion of ginseng rusty root rot pathogens that were recovered from ginseng rusty rot roots.
Ginseng rusty root rot has been attributed to the Ilyonectria fungus in China and South Korea (Wang, 2001; Farh et al., 2018). The 766 isolates we isolated were divided into 14 species, of which R. panacis was the only pathogen other than Ilyonectria and Ilyonectria-like species previously reported to cause rusty root rot symptoms. I. robusta was the most widely isolated dominant pathogen; this is inconsistent with a previous report that 90% of Chinese ginseng rusty root rot is caused by I. radicicola/C. destructans (Wang, 2001). I. radicicola/C. destructans may be a huge complex group in terms of morphological similarity (Cabral et al., 2012a). The species found later in various plants were not closely related to the original strain type CBS 264.5 (I. destructans), and I./C. destructans has generally not been found to be as common as when it was first proposed and discovered.
Most of the Ilyonectria strains infecting ginseng were obtained from P. quinquefolius, and P. ginseng and P. quinquefolius differ significantly with respect to the proportions of pathogens present. I. crassa and I. panacis were isolated from P. quinquefolius in Canada (Cabral et al., 2012a). The sampling collections of the present study covered the main cultivated areas of Chinese ginseng, and I. panacis, I. rufa and I. crassa strains were not isolated. In contrast, the dominant pathogen I. robusta was once considered to exhibit low pathogenicity. The pathogenicity was not directly proportional to the separation ratio. I. robusta isolated from P. quinquefolium was first described by Hildebrand as Ramularia robusta (Hildebrand, 1935). I. robusta was first reported in 2014 to cause ginseng root disease in China (Lu et al., 2014). Sixty-eight strains were derived from many hosts. Twenty-one of the isolates obtained from ginseng plants were segregated phylogenetically into four different groups, each of which was considered a different species of Ilyonectria: I. mors-panacis, I. robusta, I. panacis, and I. crassa (Seifert et al., 2003; Cabral et al., 2012a). These strains were grouped according to the strength of their pathogenicity; the weakly aggressive strains also originated from other hosts, but the highly aggressive strains were derived from a specific host ginseng (Cabral et al., 2012a). I. mors-panacis is not limited to infecting ginseng (Mi et al., 2017). I. robusta is considered a weakly aggressive strain based on the phylogenetic analysis and on I. robusta’s and I. crassa’s host and other factors. It is speculated that American and Korean ginseng are not susceptible to I. robusta or I. crassa (Cabral et al., 2012a), and the present article confirmed that I. robusta was the dominant pathogen with the highest isolation ratio. The prevention and control of rusty root rot in cultivation should be dominated by attention to I. rubosta.
Ginseng red-skin root is a non-infectious disease that differs from ginseng rusty root rot in China. Lu et al. reported that ginseng red-skin root is caused by a complex group of Fusarium, Ilyonectria and Dactylonectria and that Ilyonectria fungi were the dominant pathogens. I. communis, I. changbaiensis and I. qitaiheensis were newly reported species, but the same species were obtained in the present study. The types of pathogenic populations obtained in the present research are similar to the previously identified pathogens. Red-skin root and rusty root rot may not be separated. These conditions are primarily caused by Ilyonectria and Ilyonectria-like fungi, and they may represent different infection stages or may be caused by different species of Ilyonectria and Ilyonectria-like fungi (Lu et al., 2020). However, this hypothesis must be clarified. The uniform name “rusty root rot” is more acceptable because most of the infected ginseng roots have dry rot that is similar to the symptoms of decayed wood, not merely red skin; it is possible that the pathogens that cause rusty root rot also cause red-skin root and that red-skin roots are only in the early stages of the disease. I. communis and I. robusta were also the two pathogens present in the highest proportions in red-skin root isolates. Differences in pathogenicity may be the most widely accepted cause of the different symptoms, but if the disease is inferred based only on the apparent form of the agents infecting the ginseng root, the diagnosis will not be acceptable. The lifestyle of each microorganism is different, and different effects may not result from differences in pathogenicity. Fusarium was not reported as a pathogen that causes dry rot symptoms alone. Fusarium may infect the roots of ginseng via multi-infection in tissues infected by primary intruders that are not the direct cause of rusty root rot or red-skin root (Guan et al., 2014; Gao et al., 2014; Wang et al., 2016).
I. mors-panacis was discovered on P. quinquefolium by Hildebrand (1935), who described it as “R. mors-panacis” (Hypocreales, 1935). This species was also found in Japan on P. ginseng, and it was collected as “C. destructans” f. sp. Panacis (ex-type CBS 124662) is treated as a synonym. This species is considered a strong, aggressive species in Ilyonectria, but the isolation ratio was only 10.9%. Sufficient time to complete the infection is necessary for the chronic pathogenic Ilyonectria population to cause damage to the roots of perennials (Guan et al., 2020). Of course, the relationship between the degree of epidemic disease and pathogenicity was not studied in depth, and this relationship is determined by many factors. Although I. communis is a new species that was only recently reported (Lu et al., 2020), it is one of the dominant pathogens that cause ginseng rusty root rot. I. robusta and I. communis should be the main targets for controlling ginseng rusty root rot in cultivation.
The four novel species of Thelonectria were identified based on the conclusion that closely related species were being compared. Multi-locus studies are feasible and effective for the identification of novel species. None of the individual genes studied completely matched the same gene in another species, and the combined phylogenetic analysis of multiple gene sequences may result in a suitable classification. Among the studied genes, SSU contributed very little to the strains listed in the present article, but its role cannot be ignored in Thelonectria (Salgado-Salazar et al., 2016). ACT and RPB2 provided the most information, and analysis of these genes could fully resolve all of the strains described in this article. Comparison of the RPB2, TEF and TUB genes is the most effective way to distinguish T. mulanensis from T. fusongensis. The use of all of the gene sequences studied in this work makes the phylogenetic tree more credible. For Ilyonectria, rDNA-ITS was not a good marker for distinguishing strains such as I. rufa and I. robusta, and the HIS3 gene may be used alone to initially distinguish species of Ilyonectria (Cabral et al., 2012b).
Many novel Ilyonectria and Ilyonectria-like species have been discovered recently (Carlucci et al., 2017; Lawrence et al., 2019). Due to the slow growth of some fungi, it is not easy to obtain species using separation techniques. Novel species of Ilyonectria and Ilyonectria-like fungi that infect the roots of ginseng will continue to be discovered in soil. Thelonectria fungi have saprophytic properties or weak pathogenicity, and their host specificity has not been determined. Evaluation after inoculation also showed that Thelonectria fungi had weak pathogenicity, and these fungi may also be opportunistically pathogenic, similar to Ilyonectria-like fungi.
The present research revealed pathogens that cause ginseng rusty root rot but have remained unknown for many years. These results, which were obtained using a multi-locus combined approach, update previous conclusions and identify the species that should be targeted for the prevention and control of rusty root rot in ginseng cultivation. However, due to the continuous cropping properties of ginseng and continuous land use changes, Ilyonectria and Ilyonectria-like pathogens are soil-borne diseases, and the population structure may also change.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MT678559 etc.
Author Contributions
YG conceived and designed the study and wrote the manuscript. YG, YM, and QJ performed the experiments. QW, NL, and YF analyzed the sequences. YZ and YL reviewed and edited the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the Key project at the central government level: The Establishment of Sustainable Use for Valuable Chinese Medicine Resources (2060302), the Agricultural Science and Technology Innovation Program (CAAS-XTCX20190025-6), the China Agriculture Research System (CARS-21), Central the Public-interest Scientific Institution Basal Research Fund (Nos. 1610342016010 and 1610342020004), the Science and Technology Development Plan Project of Jilin Province (20191001021XH), and the National Natural Science Foundation of China (81903755 and 31501828).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Baranov, A. (1966). Recent advances in our knowledge of the morphology, cultivation and uses of ginseng (Panax ginseng C. A. Meyer). Econ. Bot. 20, 403–406. doi: 10.1007/BF02904062
Booth, C. (1966). The genus Cylindrocarpon. Mycol. Papers 104, 1–56. doi: 10.1007/978-3-319-23534-9_1
Cabral, A., Groenewald, J. Z., Rego, C., Oliveira, H., and Crous, P. W. (2012a). Cylindrocarpon root rot: multi-gene analysis reveals novel species within the Ilyonectria radicicola species complex. Mycol. Res. 11, 655–688. doi: 10.1007/s11557-011-0777-7
Cabral, A., Rego, C., Nascimento, T., Oliveira, H., Groenewald, J. Z., Pedro, W., et al. (2012b). Multi-gene analysis and morphology reveal novel Ilyonectria species associated with black foot disease of grapevines. Fungal. Biol. 116, 62–80. doi: 10.1016/j.funbio.2011.09.010
Carbone, I., and Kohn, L. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.2307/3761358
Carlucci, A., Lops, F., Mostert, L., Halleen, F., and Raimondo, M. L. (2017). Occurrence fungi causing black foot on young grapevines and nursery rootstock plants in Italy. Phytopathol. Mediterr. 56, 10–39. doi: 10.14601/Phytopathol_Mediterr-18769
Castlebury, L. A., Rossman, A. Y., and Hyten, A. S. (2006). Phylogenetic relationships of Neonectria/Cylindrocarpon on Fagus in North America. Can. J. Bot. 84, 1417–1433. doi: 10.1139/b06-105
Castlebury, L. A., Rossman, A. Y., Sung, G. H., Joseph, A. S., and Spatafora, W. (2004). Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 108, 864–872. doi: 10.1017/S0953756204000607
Chaverri, P., Salgado, C., Hirooka, Y., Rossman, A. Y., and Samuels, G. L. (2011). Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs. Stud. Mycol. 68, 57–78. doi: 10.3114/sim.2011.68.03
Cho, D. H., Park, K. J., Yu, Y. H., Ohh, S. H., and Lee, H. S. (1995). Root-rot development of 2-year old ginseng (Panax ginseng CA Meyer) caused by Cylindrocarpon destructans (Zinssm.) Scholten in the continuous cultivation field. J. Ginseng. Res. 19, 175–180.
Chung, H. S. (1979). Ginseng Disease. Research Reports of the Korean Society of Plant Protection. Seoul: Korean Society of Plant Protection.
Crous, P. W., Groenewald, J. Z., Risede, J., and Hyweljones, N. (2004). Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Stud. Mycol. 50, 415–429. doi: 10.1023/B:MYCO.0000012225.79969.29
de Hoog, G. S., and van den Ende, A. H. G. G. (1998). Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses 41, 183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x
Domsch, K. H., Gams, W., and Anderson, T. H. (2007). Compendium of Soil Fungi, 2nd Edn. Eching: IHW-Verlag.
Farh, E. A., Kim, Y. J., Kim, Y. J., and Yang, D. C. (2018). Cylindrocarpon destructans/Ilyonectria radicicola-species complex: causative agent of ginseng root-rot disease and rusty symptoms. J. Ginseng. Res. 42, 9–15. doi: 10.1016/j.jgr.2017.01.004
Gao, J., Wang, Y., Guan, Y. M., and Chen, C. Q. (2014). Fusarium cerealis, a new pathogen causing ginseng (Panax ginseng) root rot in China. Plant Dis. 98:1433. doi: 10.1094/PDIS-03-14-0328-PDN
Gargas, A., and Taylor, J. (1992). Polymerase chain reaction (PCR) primers for amplifying and sequencing 18S rDNA from lichenized fungi. Mycologia 84, 589–592. doi: 10.1080/00275514.1992.12026182
Groenewald, J. Z., Nakashima, C., Nishikawa, J., Shin, H. D., Park, J. H., Jama, A. N., et al. (2013). Species concepts in Cercospora: spotting the weed among the roses. Stud. Mycol. 75, 115–170. doi: 10.3114/sim0012
Guan, Y. M., Chen, M. L., Ma, Y., Du, Z., Yuan, N., Li, Y., et al. (2020). Whole-genome and time-course dual RNA-Seq analyses reveal chronic pathogenicity-related gene dynamics in the ginseng rusty root rot pathogen Ilyonectria robusta. Sci. Rep. 10:1586. doi: 10.1038/s41598-020-58342-7
Guan, Y. M., Deng, J. C., Ma, Y. Y., Li, Y., and Zhang, Y. Y. (2019). Seed-associated fungal diversity and the molecular identification of Fusarium with potential threat to ginseng (Panax ginseng) in China. Plant Dis. 104, 330–339. doi: 10.1094/PDIS-09-19-1817-RE
Guan, Y. M., Lu, B. H., Wang, Y., Gao, J., and Wu, L. J. (2014). First report of root rot caused by Fusarium redolens on ginseng (Panax ginseng) in Jilin province of China. Plant Dis. 98:844. doi: 10.1094/PDIS-09-13-0922-PDN
Halleen, F., Fourie, P. H., and Crous, P. W. (2006a). A review of black-foot disease of grapevines. Phytopathol. Mediterr. 45, S55–S67. doi: 10.1520/STP37480S
Halleen, F., Schroers, H. J., Groenewald, J. Z., Rego, C., Oliveira, H., and Crous, P. W. (2006b). Neonectria liriodendri sp. nov., the main causal agent of black foot disease of grapevines. Stud. Mycol. 55, 227–234. doi: 10.3114/sim.55.1.227
Halleen, F., Schroers, H. J., Groenewald, J. Z., and Crous, P. W. (2004). Novel species of Cylindrocarpon (Neonectria) and Campylocarpon gen. nov. associated with black foot disease. Stud. Mycol. 50, 431–445. doi: 10.1023/B:MYCO.0000012225.7996.29
Hildebrand, A. (1935). Root rot of ginseng in Ontario caused by members of the genus Ramularia. Can. J. Res. 12, 82–114. doi: 10.1139/cjr35-007
Hirooka, Y., Kobayashi, T., and Natsuaki, K. T. (2005). Neonectria castaneicola and Neo. rugulosa in Japan. Mycologia 97, 1058–1066. doi: 10.1080/15572536.2006.11832755
Hu, S. Y. (1977). A contribution to our knowledge of ginseng. Am. J. Chinese Med. 5, 1–23. doi: 10.1142/S0192415X77000026
Lawrence, D. P., Nouri, M. T., and Trouillas, F. P. (2019). Taxonomy and multi-locus phylogeny of cylindrocarpon-like species associated with diseased roots of grapevine and other fruit and nut crops in California. Fungal Syst. Evol. 4, 59–75. doi: 10.3114/fuse.2019.04.06
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Lu, X. H., Jiao, X. L., Chen, A. J., Luo, Y., and Gao, W. W. (2014). First report of Ilyonectria robusta causing rusty root of Asian Ginseng in China. Plant Dis. 99:156. doi: 10.1094/PDIS-06-14-0663-PDN
Lu, X. H., Zhang, X. M., Jiao, X. L., Hao, J. J., Zhang, X. S., Luo, Y., et al. (2020). Taxonomy of fungal complex causing red-skin root of Panax ginseng in China. J. Ginseng Res. 44, 506–518. doi: 10.1016/j.jgr.2019.01.006
Mantiri, F. R., Samuels, G. J., Rahe, J. E., and Honda, B. M. (2001). Phylogenetic relationships in Neonectria species having Cylindrocarpon anamorphs inferred from mitochondrial ribosomal DNA sequences. Can. J. Bot. 79, 334–340. doi: 10.1139/b01-002
Mi, C., Yang, R., Rao, J., Yang, S., Wei, F., Li, O., et al. (2017). Unveiling of dominant fungal pathogens associated with rusty root rot of Panax notoginseng based on multiple methods. Plant Dis. 101, 2046–2052. doi: 10.1094/PDIS-01-17-0135-RE
Moncalvo, J. M., Lutzoni, F. M., Rehner, S. A., Johnson, J., and Vilgalys, R. (2000). Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Syst. Biol. 49, 278–305. doi: 10.1093/sysbio/49.2.278
O’Donnell, K., and Cigelnik, E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7, 103–116. doi: 10.1006/mpev.1996.0376
Punja, Z. K. (1997). Fungal pathogens of American ginseng (Panax quinquefolium) in British Columbia. Can. J. Plant Pathol. 19, 301–306. doi: 10.1080/07060669709500528
Reeleder, R. D. (2007). Rhexocercosporidium panacis sp. nov. a new anamorphic species causing rusted root of ginseng (Panax quinquefolius). Mycologia 99, 91–98. doi: 10.1080/15572536.2007.11832604
Reeleder, R. D., Hoke, S. M. T., and Zhang, Y. (2006). Rusted root of ginseng (Panax quinquefolius) is caused by a species of Rhexocercosporidium. Phytopathology 96:1243. doi: 10.1094/PHYTO-96-1243
Salgado-Salazar, C., Rossman, A. Y., and Chaverri, P. (2013). Not as ubiquitous as we thought: taxonomic crypsis, hidden diversity and cryptic speciation in the cosmopolitan fungus Thelonectria discophora (Nectriaceae, Hypocreales, Ascomycota). PLoS One 8:e76737. doi: 10.1371/journal.pone.0076737
Salgado-Salazar, C., Rossman, A. Y., and Chaverri, P. (2016). The genus Thelonectria (Nectriaceae, Hypocreales, Ascomycota) and closely related species with Cylindrocarpon-like asexual states. Fungal Divers 80, 411–455. doi: 10.1007/s13225-016-0365-x
Salgado-Salazar, C., Rossman, A. Y., Samuels, G. J., Capdet, M., and Chaverri, P. (2012). Multigene phylogenetic analyses of the Thelonectria coronata and T. veuillotiana species complexes. Mycologia 104, 1325–1350. doi: 10.3852/12-055
Salgado-Salazar, C., Rossman, A. Y., Samuels, G. J., Hirooka, Y., Sanchez, R. M., and Chaverri, P. (2015). Phylogeny and taxonomic revision of Thelonectria discophora (Ascomycota, Hypocreales, Nectriaceae) species complex. Fungal Divers. 70, 1–29. doi: 10.1007/s13225-014-0280-y
Samuels, G. J., and Brayford, D. (1990). Variation in Nectria radicicola and its anamorph, Cylindrocarpon destructans. Mycol. Res. 94, 433–442. doi: 10.1016/S0953-7562(10)80001-2
Scholten, G. (1964). Nectria radicicola en Thielaviopsis basicola als parasieten van Cyclamen persicum. Neth. J. Plant Pathol. 70, 1–68. doi: 10.1007/BF02001771
Schroers, H. J., Žerjav, M., Munda, A., Halleen, F., and Crous, P. W. (2007). Cylindrocarpon pauciseptatum sp. nov. with notes on Cylindrocarpon species with wide, predominantly 3-septate macroconidia. Mycol. Res. 112, 82–92. doi: 10.1016/j.mycres.2007.10.004
Seifert, K. A., McMullen, C. R., Yee, D., Reeleder, R. D., and Dobinson, K. F. (2003). Molecular differentiation and detection of ginseng-adapted isolates of the root rot fungus Cylindrocarpon destructans. Phytopathology 93, 1533–1542. doi: 10.1094/PHYTO.2003.93.12.1533
Tewoldemedhin, Y. T., Mazzola, M., Mostert, L., and McLeod, A. (2011). Cylindrocarpon species associated with apple tree roots in South Africa and their quantification using real-time PCR. Eur. J. Plant Pathol. 129, 637–651. doi: 10.1007/s10658-010-9728-4
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification, and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246
Wang, Y., Guan, Y. M., Lu, B. H., and Gao, J. (2016). First report of ginseng (Panax ginseng) root rot caused by Fusarium acuminatum in China. Plant Dis. 100:525. doi: 10.1094/PDIS-03-15-0273-PDN
Zhou, Y., Yang, Z., Gao, L., Liu, W., Liu, R., Zhao, J., et al. (2017). Changes in element accumulation, phenolic metabolism, and antioxidative enzyme activities in the red-skin roots of Panax ginseng. J. Ginseng Res. 41, 307–315. doi: 10.1016/j.jgr.2016.06.001
Keywords: Dactylonectria, Ilyonectria, Ilyonectria-like, Neonectria, Panax ginseng, Rhexocercosporidium, rusty root rot, Thelonectria
Citation: Guan YM, Ma YY, Jin Q, Wang QX, Liu N, Fu YP, Zhang YY and Li Y (2020) Multi-Locus Phylogeny and Taxonomy of the Fungal Complex Associated With Rusty Root Rot of Panax ginseng in China. Front. Microbiol. 11:618942. doi: 10.3389/fmicb.2020.618942
Received: 20 October 2020; Accepted: 26 November 2020;
Published: 16 December 2020.
Edited by:
Eeva Johanna Vainio, Natural Resources Institute Finland (Luke), FinlandReviewed by:
Julia Christine Meitz-Hopkins, Stellenbosch University, South AfricaZheng Wang, Yale University, United States
Copyright © 2020 Guan, Ma, Jin, Wang, Liu, Fu, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Li, ZnVuZ2k5NjZAMTI2LmNvbQ==
 Yi Ming Guan1,2
Yi Ming Guan1,2 Qiu Xia Wang
Qiu Xia Wang Ning Liu
Ning Liu Ya Yu Zhang
Ya Yu Zhang Yu Li
Yu Li