- Animal Disease Prevention and Food Safety Key Laboratory of Sichuan Province, Key Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu, China
Copper can persist stably in the environment for prolonged periods. Except for inducing antibiotic resistance in bacteria, copper ions (Cu2+) can facilitate the horizontal transfer of plasmid DNA. However, whether and how Cu2+ can accelerate the conjugative transfer of SXT/R391 integrative and conjugative element (ICE) is still largely unknown. In this study, Proteus mirabilis ChSC1905, harboring an SXT/R391 ICE that carried 21 antibiotic resistance genes (ARGs), was used as a donor, and Escherichia coli EC600 was used as a recipient. Cu2+, at subinhibitory and environmentally relevant concentrations (1–10 μmol/L), significantly accelerated the conjugative transfer of SXT/R391 ICE across bacterial genera (from P. mirabilis to E. coli) (p < 0.05). The combined analyses of phenotypic tests and genome-wide sequencing indicated that reactive oxygen species (ROS) production and cell membrane permeability were critical in the enhanced conjugative transfer of SXT/R391 ICE. Furthermore, the expression of genes related to cell adhesion and ATP synthesis was also significantly upregulated on exposure to Cu2+ at a concentration of 5 μmol/L. This study clarified the potential mechanisms of Cu2+ to promote the conjugative transfer of SXT/R391 ICE, revealing the potential risk imposed by Cu2+ on the horizontal transfer of SXT/R391 ICE-mediated ARGs.
Introduction
Currently, the extensive use of heavy metal copper in plants, livestock, and hospitals is a serious threat to public health (Zhu et al., 2013; Pal et al., 2017). Besides, the residual copper persists stably in the environment for prolonged periods (Yu et al., 2017). Copper can induce antibiotic resistance by coselection (Li et al., 2017). For instance, copper can drive the development of antibiotic resistance (Poole, 2017) owing to the mobile genetic elements carrying both antibiotic resistance genes (ARGs) and metal resistance genes (Sandegren et al., 2012; Yang et al., 2018). On the contrary, horizontal gene transfer (HGT) is another critical driver for disseminating ARGs in various environments (Wang et al., 2015; Li et al., 2020). Copper ions (Cu2+) promote the horizontal transfer of plasmid-mediated ARGs in freshwater microcosms (Wang et al., 2020). HGT includes conjugation (mediated by cell-to-cell contact), transformation (mediated by extracellular DNA), and transduction (bacteriophage mediated) (Partridge et al., 2018; McInnes et al., 2020; Nihemaiti et al., 2020). The conjugation between the donor and the recipient mostly mediates the transfer of plasmids and integrative and conjugative elements (ICEs) (Hastings et al., 2004; Redondo-Salvo et al., 2020; Rodriguez-Beltran et al., 2020). In addition, the conjugative transfer of DNA may occur within or across bacterial genera (Thomas and Nielsen, 2005; Wang et al., 2019), leading to the dissemination of ARGs among a wide range of bacterial species (Shun-Mei et al., 2018).
Nevertheless, apart from the horizontal transfer of plasmid DNA (Xie et al., 2019), increasing evidence suggested that SXT/R391 ICEs were critical drivers for the spread of ARGs, harboring the integrase gene int, a marker to define SXT/R391 ICEs in clinical strains (Wozniak et al., 2009; Bioteau et al., 2018). Recently, many clinically important ARGs were found to be located on SXT/R391 ICEs (Aberkane et al., 2016), for example, carbapenemase gene blaNDM–1 (Kong et al., 2020), fosfomycin resistance gene fosA3 (Lei et al., 2018), and tigecycline resistance gene tet(X6) (He et al., 2020). These SXT/R391 ICEs were also transferable (Badhai and Das, 2016). Antibiotics, such as ciprofloxacin, could induce SXT ICE transfer via SOS response (Beaber et al., 2004). However, studies investigating whether other non-antibiotic materials, especially Cu2+, can facilitate the conjugative transfer of SXT/R391 ICE across bacterial genera were limited. As reported, Cu2+ at subinhibitory concentrations could promote the horizontal transfer of RP4 plasmid in water environment (Zhang et al., 2018a), mainly via increased intracellular reactive oxygen species (ROS) generation, activated SOS response, and enhanced cell membrane permeability (Zhang et al., 2019). Therefore, it was hypothesized that Cu2+ could accelerate the conjugative transfer of SXT/R391 ICE in the same or a different way.
To confirm the aforementioned hypothesis, an SXT/R391 ICE that carried 21 ARGs was selected. Subsequently, the effects of Cu2+, at subinhibitory and environmentally relevant concentrations, on the conjugative transfer of this SXT/R391 ICE from Proteus mirabilis to Escherichia coli were assessed. Moreover, the mechanisms were explored by testing the intracellular ROS level, detecting cell membrane permeability, and checking changes in the expression of genes related to oxidative stress, SOS response, cell membrane, cell adhesion, and ATP synthesis. The present study was novel in exploring the effects and potential mechanisms of Cu2+ on the conjugative transfer of SXT/R391 ICE. The findings provided insights into the role of Cu2+ in the transfer of SXT/R391 ICE and emphasized the necessity for the proper use of copper in the future.
Materials and Methods
Bacterial Strains and Culture Conditions
Proteus mirabilis ChSC1905 (accession no. CP047929), isolated from the nasal swab of a diseased pig in China, was selected as the donor strain. The donor P. mirabilis harbored an SXT/R391 ICE that carried 21 ARGs, including a gene for resistance to cefotaxime (CTX). E. coli EC600, with higher resistance to rifampicin (RD), was chosen as the recipient strain. Both the donor and the recipient strains were incubated at 37°C in Luria broth (LB, Sigma, United States) medium with shaking at 180 rpm overnight. Then, the prepared bacterial strains were obtained by centrifuging (6,000 g) at 4°C for 5 min. After removing the supernatants, the pellets were washed with phosphate-buffered saline (PBS, Sangon, China) and resuspended in PBS. The CuSO4.5H2O solution was prepared with sterile water.
Minimum Inhibitory Concentrations Test
The minimum inhibitory concentrations (MICs) of Cu2+, CTX, and RD against donor P. mirabilis ChSC1905 and recipient E. coli EC600 were measured as described previously (Fang et al., 2016; Lu et al., 2020). Briefly, overnight cultures of the donor P. mirabilis and the recipient E. coli were diluted to approximately 105 cfu/ml. Then, 5 μl of the bacterial solution, 15 μl of Cu2+, CTX, or RD (at different concentrations), and 130 μl of fresh LB media were added. Sterilized LB was chosen as blank control. Subsequently, the 96-well plates were incubated at 37°C for 18 h, and then, a microplate spectrophotometer was used to obtain the optical density at 600 nm (OD600). MICs were obtained at the concentrations of Cu2+, CTX, and RD that completely inhibited the bacterial growth. All MIC tests were detected at least in triplicate.
Conjugation Experiment Under Exposure of Cu2+
The conjugation mating system was established as described previously (Zhang et al., 2019). Briefly, 100 μl of the donor strain and the recipient strain were mixed at a 1:1 ratio with 108 cfu/ml. Then, the mating system was exposed to different final concentrations of Cu2+ (0, 0.5, 1, 5, 10, and 100 μmol/L, total volume of 1 ml) in the presence of DNase I (a final concentration of 285 mg/L, Sigma). After incubating for 18 h without shaking at 37°C in PBS, 50 μl of the mixtures were inoculated on eosin methylene blue (EMB) agar selection plates containing 80 mg/L RD and 16 mg/L CTX for 48 h. EMB agar plates containing 80 mg/L RD were used to determine the total recipient numbers. The conjugative transfer frequency was calculated by dividing the total numbers of transconjugants by the total numbers of recipients. Furthermore, an ROS scavenger (100 mM thiourea) was added to the aforementioned conjugation mating systems to assess whether Cu2+ accelerated conjugative transfer through inducing ROS generation. The conjugation frequency in the thiourea-treated group was compared with that of the non-thiourea-treated group. All the conjugation experiments were conducted with biological triplicates. In parallel, a 108 cfu/ml suspension of the donor and recipient strains were serially diluted of which 50 μl were plated onto EMB agar plates containing 80 mg/L RD and 16 mg/L CTX for 48 h. No colony indicating the emergence of spontaneous mutants could be observed at 108 cfu/ml, indicating that there were no spontaneous mutants of the strains in this study. Additionally, an Enhanced Cell Counting Kit-8 (Beyotime, China) was used to count living cells in the presence of Cu2+ to determine whether Cu2+ influenced cell viability of the donor and recipient strains.
Identification of Transconjugants
Transconjugants (five colonies selected randomly from each treatment group) were cultured overnight in LB media. The DNA of transconjugants was extracted using a Bacterial DNA Kit (Omega, United States) following the manufacturer’s protocols. Transconjugants were identified as E. coli using a BD PhoenixTM-100 Automated Microbiology System (Becton Dickinson, United States) (Kong et al., 2017) and 16S ribosomal DNA (rDNA) sequencing. The transconjugants were further defined by the antimicrobial resistance profile (Kirby–Bauer disk diffusion method based on the CLSI guidelines) (Clinical and Laboratory Standards Institute [CLSI], 2016). Finally, the int gene and the attachment sites attL and attR of SXT/R391 ICE in transconjugants were detected by polymerase chain reaction (Supplementary Table 1) to further confirm the presence of ICE in each transconjugant (Lei et al., 2016, 2018).
Reverse Conjugation Experiment
The reverse conjugation mating system in the presence of Cu2+ at 5 μmol/L was further used to determine whether Cu2+ could still facilitate the transfer of SXT/R391 ICE from newly generated transconjugants to another recipient. The transconjugant (E. coli EC600 carrying SXT/R391 ICE) was used as a new donor. E. coli J53, with higher resistance to sodium azide, was selected as a new recipient. EMB agar plates containing 200 mg/L sodium azide and 16 mg/L CTX were used to select the transconjugants. The conjugative transfer frequency, spontaneous mutations of the strains, and identification of transconjugants were evaluated as described earlier.
Detection of ROS and Cell Membrane Permeability
One hundred microliters of the donor P. mirabilis ChSC1905 (108 cfu/ml) and the recipient E. coli EC600 (108 cfu/ml) were separately exposed to different final concentrations of Cu2+ (0, 0.5, 1, 5, 10, and 100 μmol/L, total volume of 1 ml). For ROS detection, an ROS Assay Kit (Beyotime, China) was employed following the manufacturer’s protocols. Briefly, after incubating for 18 h, bacteria strains were incubated individually with 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA, a final concentration of 10 μmol/L) for 20 min at 37°C. Then, samples were tested by the CytoFLEX flow cytometer (Beckman, United States) at 488 nm. For cell membrane permeability test, the strains were stained with 5 μl of propidium iodide (PI, Keygen, China) and incubated for 15 min in the dark before detecting by flow cytometer at 488 nm. All data were analyzed using CytExpert. All the tests were detected at least in triplicate.
Transmission Electron Microscope
A transmission electron microscope (TEM) was employed to check whether the cell membrane or morphology was changed in the presence of Cu2+. Briefly, the mating system was exposed to Cu2+ at 5 and 0 μmol/L. After incubating for 18 h, the mixed samples were collected, fixed, dehydrated, filtered, and mounted. Ultrathin sections (50–100 nm) of each sample were applied on TEM copper grids and obtained using a JEM-1400PLUS TEM (Jeol, Japan) at 80 kV.
Whole-Genome RNA Sequencing
The mating system was exposed to Cu2+ at 5 μmol/L (treatment group) and 0 μmol/L (control group). After mating for 18 h, the cells were collected, and the total RNA of each sample was extracted. Then, all samples (three samples from the control group and three samples from the treatment group) were submitted to Novogene (Beijing, China) for strand-specific complementary DNA (cDNA) library construction and NovaSeq 6000 (Illumina, United States) Illumina paired-end sequencing (clean bases, 2G). The reads containing adapter, poly-N, and low-quality reads were removed from the raw data. The obtained clean reads (each sample) were aligned to the P. mirabilis reference genome (NC_010554), E. coli reference genome (NC_000913) and the SXT sequence (MG773277) using Bowtie2 (v2.2.3). HTSeq (v0.6.1) was used to count the read numbers mapped to each gene. The gene expression was calculated as fragments per kilobase of a gene per million mapped reads (FPKM). Differential expression analysis (with biological replicates) was performed using the DESeq R package (v1.18.0). Differences in fold changes between 0 and 5 μmol/L Cu2+-treated mating system were calculated using log2 fold change (LFC) between control and Cu2+-treated samples. Genes with | LFC| > 0 and p < 0.05 were considered as differentially expressed. The visual images were analyzed using GraphPad Prism (v8.0.1).
Statistical Analysis
Data were expressed as mean ± standard deviation. SPSS 19.0 (IBM, United States) was used for data analysis. An independent-samples t test was performed to analyze significant differences. p < 0.05 indicated a statistically significant difference (∗p < 0.05; ∗∗p < 0.01).
Results
Effects of Cu2+ on the Conjugative Transfer of SXT/R391 ICE
The results showed that the MIC of Cu2+, CTX, and RD against the donor strain was 10 mmol/L, >1,024 μg/ml, and 80 μg/ml, respectively. However, the MIC of Cu2+, CTX, and RD against the recipient strain was 10 mmol/L, <2 μg/ml, and 320 μg/ml, respectively. The mating system was exposed to different sub-MIC concentrations of Cu2+ to check whether Cu2+ could promote the conjugative transfer of SXT/R391 ICE. No spontaneous mutants were observed [frequency bellow 1/(5 × 106 cells)]. As shown in Figure 1A, the conjugative transfer frequency of SXT/R391 ICE significantly increased from 6.06 × 10–5 to 7.92 × 10–5 in the presence of Cu2+ at 1 μmol/L (p < 0.05), 5 μmol/L (p < 0.01), and 10 μmol/L (p < 0.05) compared with the control (4.70 × 10–5), respectively. Particularly, the conjugative transfer frequency displayed a maximum increase at 5 μmol/L Cu2+. On the contrary, the conjugative transfer frequency (1.17 × 10–5) on exposure to Cu2+ at 100 μmol/L significantly decreased compared with the control (p < 0.01). This was likely due to the reduced cell viability of the donor and recipient strains (Supplementary Figure 2).
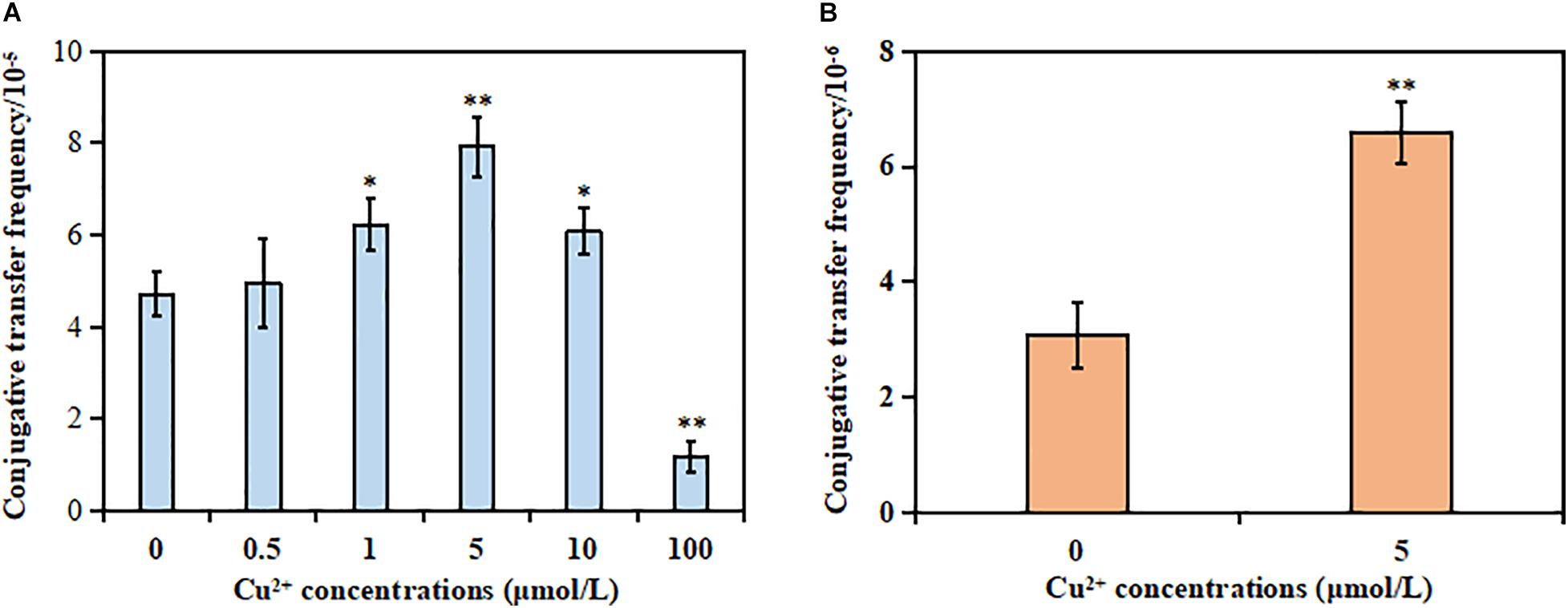
Figure 1. Effects of Cu2+ on the conjugative transfer of SXT/R391 integrative and conjugative element (ICE). (A) Conjugative transfer frequency of SXT/R391 ICE from the donor (Proteus mirabilis ChSC1905) to the recipient (Escherichia coli EC600). (B) Reverse conjugative transfer from the transconjugants (E. coli EC600) to E. coli J53. p < 0.05 indicated a statistically significant difference (*p < 0.05; **p < 0.01).
Various analyses were performed to verify that the SXT/R391 ICE was transferred successfully from the donor P. mirabilis to the recipient E. coli. First, the transconjugants were identified as E. coli species. Second, the donor P. mirabilis exhibited multidrug resistance besides being intrinsically resistant to colistin and tetracycline (Lei et al., 2018; Supplementary Table 2). However, the recipient E. coli was resistant only to rifampin. Indeed, the transconjugants displayed the same antimicrobial resistance profile compared with the donor, including resistance to cefotaxime, ceftriaxone, florfenicol, gentamicin, trimethoprim/sulfamethoxazole, fosfomycin, ampicillin, nalidixic acid, ciprofloxacin, amikacin, linezolid, and rifampin (Supplementary Table 2). Finally, both the donor strain and the transconjugants harbored the int gene, but the recipient strain did not. Besides, the attachment sites attL and attR of SXT/R391 ICE in transconjugants were detected (Supplementary Figure 1), further indicating that the transconjugants carried the SXT/R391 ICE from the donor.
The reverse conjugation experiment showed that the SXT/R391 ICE could be transferred from transconjugant (E. coli EC600) to another recipient (E. coli J53). Besides, no spontaneous mutants were observed [frequency bellow 1/(5 × 106 cells)]. The conjugative frequency (6.59 × 10–6) in the presence of Cu2+ at 5 μmol/L also significantly increased compared with the control (3.08 × 10–6; p < 0.01; Figure 1B). The azide resistance of the auxotrophic mutant strain E. coli J53 was due to a single nucleotide substitution in the secA gene, whose genome contained a large inversion and missed five prophage regions and 18 non-hypothetical genes (Yi et al., 2012). However, E. coli EC600 was a bacteriophage λ-sensitive strain, the genome of which contained six prophage-associated regions (Allué-Guardia et al., 2019). Although both the two strains were derived from progenitor strain K-12, the aspect of strain J53 was distinguishable from strain EC600 on EMB selection plates. After excluding the occurrence of spontaneous mutations of the two strains, the transconjugants in the reverse conjugation experiment could be identified as E. coli J53 carrying the SXT/R391 ICE.
Effects of Cu2+ on ROS Production
As shown in Figure 2A, the donor strain displayed a significant increase in ROS generation in the presence of Cu2+ from 1 to 100 μmol/L compared with the control (p < 0.01). Similarly, the ROS generation also significantly increased for the recipient strain in the presence of Cu2+ from 0.5 to 100 μmol/L (p < 0.01; Figure 2B). The maximum value was obtained at 100 μmol/L Cu2+.
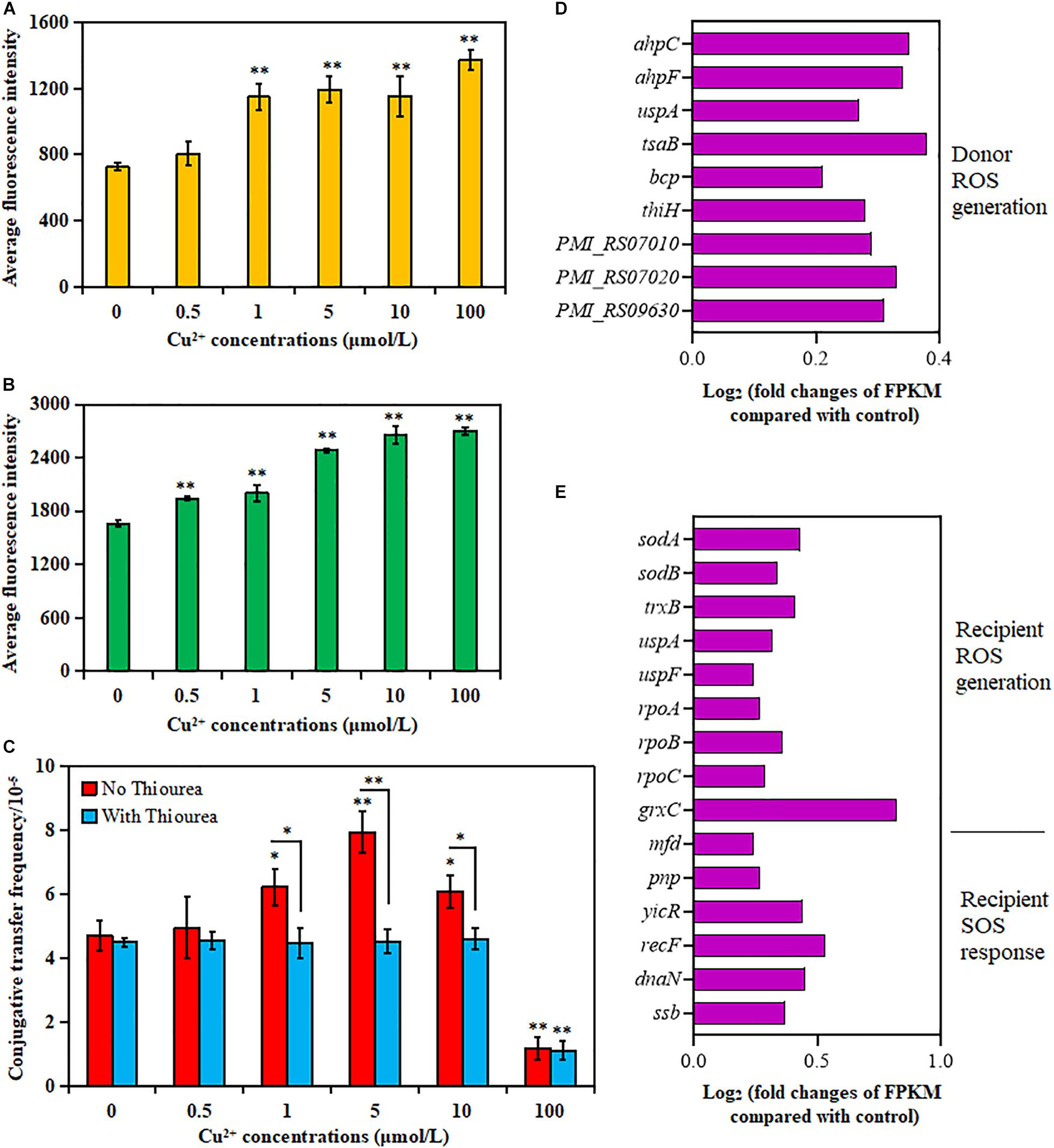
Figure 2. Effects of Cu2+ on reactive oxygen species (ROS) production in the donor (Proteus mirabilis ChSC1905) and recipient (Escherichia coli EC600). Average fluorescence intensity of ROS production in (A) the donor and (B) the recipient checked by flow cytometer. (C) Effect of ROS scavenger (thiourea) on the conjugative transfer of SXT/R391 integrative and conjugative element (ICE) from the donor to the recipient. Fold changes of the expression of genes related to ROS generation and SOS response in (D) the donor and (E) the recipient. p < 0.05 indicated a statistically significant difference (*p < 0.05; **p < 0.01).
The conjugative transfer frequency decreased significantly in the presence of Cu2+ at 1 μmol/L (p < 0.05), 5 μmol/L (p < 0.01), and 10 μmol/L (p < 0.05) on adding ROS scavenger thiourea (Figure 2C). Nevertheless, the conjugative transfer frequency showed no significant difference in the presence of Cu2+ at 0, 0.5, and 100 μmol/L (p > 0.05). The results indicated that Cu2+ might promote the conjugative transfer of SXT/R391 ICE by inducing ROS production.
The changes in gene expression also supported the aforementioned phenotypes (Supplementary Tables 3, 4). As shown in Figures 2D,E, the cellular antioxidant-related genes were overexpressed on exposure to Cu2+ at 5 μmol/L, including genes coding for alkyl hydroperoxide reductase (ahpC and ahpF) in the donor strain, as well as genes coding for superoxide dismutase (sodA and sodB) and thioredoxin reductase (trxB) in the recipient strain (Hayden et al., 2018). The expression of RNA polymerase (rpoA, rpoB, and rpoC genes) in the recipient strain was also upregulated, which was related to the induction of ROS production (Piccaro et al., 2014). The expression of the uspA gene, related to the survival of bacteria (Bandyopadhyay and Mukherjee, 2020), also increased in both donor and recipient strains. Besides, the expression of genes related to the ROS response in the donor strain (tsaB, bcp, thiH, PMI_RS07010, PMI_RS07020, and PMI_RS09630) and the recipient strain (uspF and grxC) also increased significantly. However, the genes related to the SOS response were mainly enriched in DNA repair and recombination (Figure 2E and Supplementary Table 4) in the recipient strain, including the enhanced expression of mfd, ssb, yicR, recF, dnaN, and pnp (Fan et al., 2019).
Effects of Cu2+ on Cell Membrane Permeability
As shown in Figures 3A,B, the percentage of PI-positive cells in both donor and recipient strains significantly increased in the presence of Cu2+ at 5, 10, and 100 μmol/L (p < 0.01); the maximum value was obtained at 100 μmol/L Cu2+. These results showed that Cu2+ enhanced cell membrane permeability.
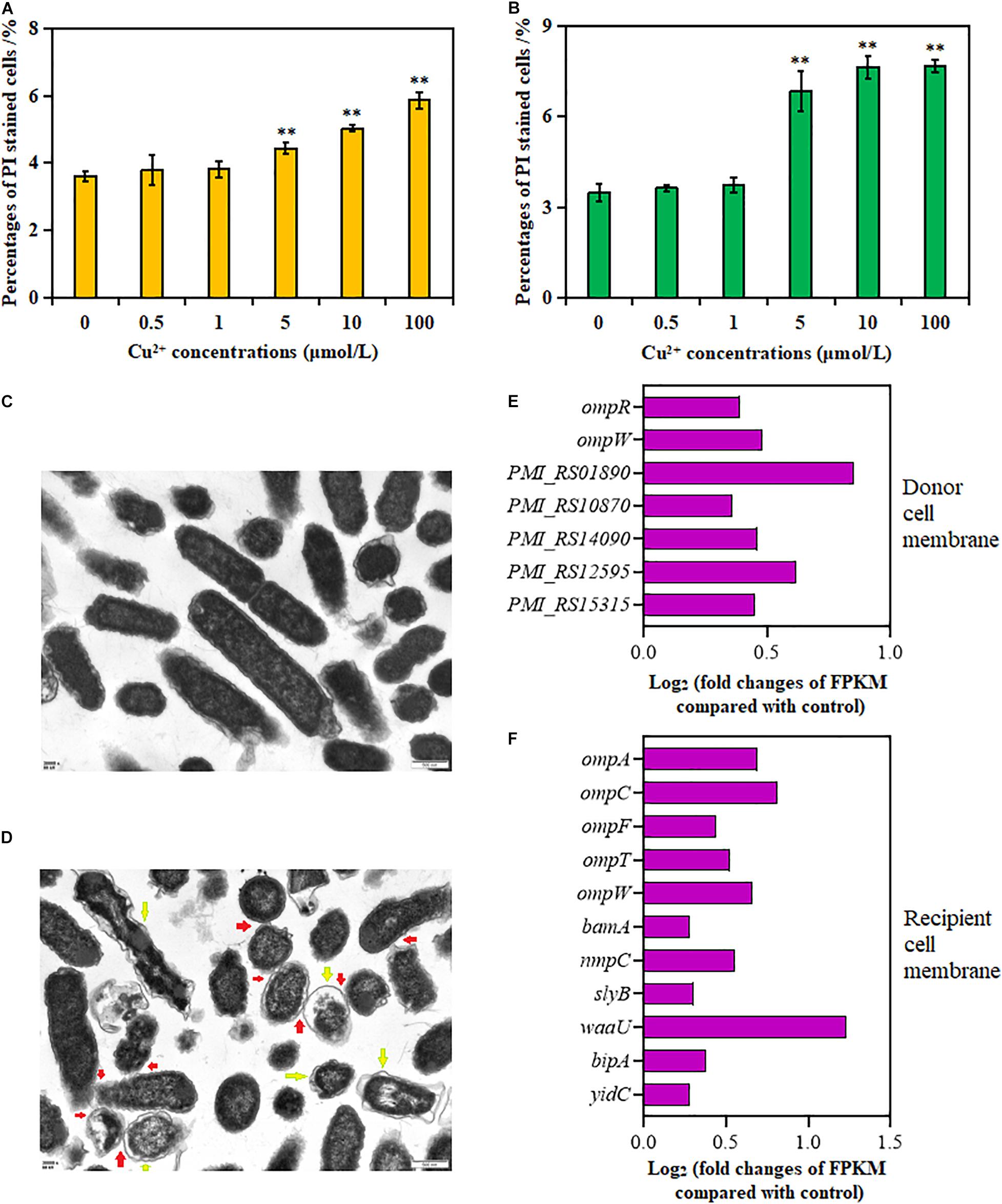
Figure 3. Effects of Cu2+ on cell membrane permeability in the donor (Proteus mirabilis ChSC1905) and recipient (Escherichia coli EC600). Percentages of PI-stained cells in (A) the donor and (B) recipient checked by flow cytometer. (C) Transmission electron microscopy (TEM) images in ultrafine slices of the control. (D) Cells exposure to Cu2+ at 5 μmol/L (scale bars, 500 nm). Yellow arrows stand for membrane damage; red arrows stand for cell-to-cell contact. Fold changes of the expression of genes related to cell membrane in the (E) donor and (F) recipient. p < 0.05 indicated a statistically significant difference (*p < 0.05; **p < 0.01).
Transmission electron microscope images of the cell morphology and membrane showed dispersed cells with less physical contact, distinct cell membranes, and compact cytoplasm in the control group (Figure 3C). On exposure to Cu2+ at 5 μmol/L (Figure 3D), more cell adhesion between these cells was observed. Besides, apparent cell membrane damage and indistinct cell borders were found.
The changes in gene expression were also consistent with the aforementioned cell membrane phenotypes (Supplementary Tables 5, 6). As shown in Figures 3E,F, the expression of genes in the donor strain encoding for membrane proteins, omp gene family (ompR and ompW), PMI_RS01890, PMI_RS10870, PMI_RS14090, PMI_RS12595, and PMI_RS15315, was significantly upregulated on exposure to Cu2+ at 5 μmol/L. Similarly, in the recipient strain, in addition to omp gene family (ompA, ompC, ompF, ompT, and ompW), the expression of lipopolysaccharide synthesis gene waa, gene bamA coding for outer membrane assembly protein, gene slyB coding for outer membrane lipoprotein (Lu et al., 2020), gene nmpC coding for outer membrane porin (Ruan et al., 2011), gene yidC involved in insertion and folding of membrane proteins (Gray et al., 2011), and gene bipA was also significantly upregulated.
Effects of Cu2+ on Cell Adhesion and ATP Synthesis
As shown in Figure 4A, the expression of adhesion-relevant genes fimA, fimC, fimG, fimH, and fimI was significantly upregulated in the recipient strain on exposure to Cu2+ at 5 μmol/L (Supplementary Table 7). Additionally, the expression of seven ATP encoding genes, atpA, atpB, atpC, atpD, atpF, atpG, and atpH, which controlled cellular energy production, was also significantly increased in the recipient strain (Figure 4B and Supplementary Table 8). Interestingly, the expression of the copA gene encoding for Cu+ translocating P-type ATPase was also significantly upregulated in the recipient strain (Supplementary Table 8).
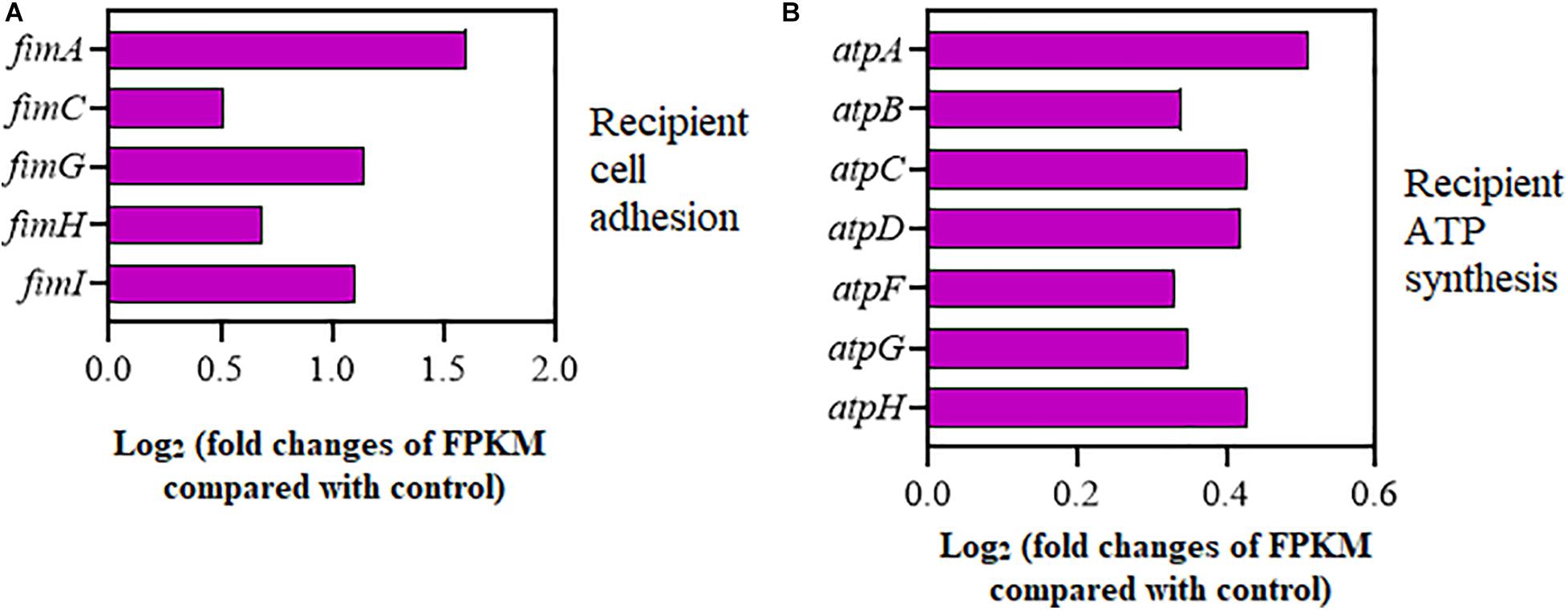
Figure 4. Effects of Cu2+ on cell adhesion and ATP synthesis in the recipient (Escherichia coli EC600). Fold changes of the expression of genes related to (A) cell adhesion and (B) ATP synthesis.
Discussion
Except for the antibiotic-driven spread of ARGs (Blazquez et al., 2012; Lopatkin et al., 2016; Liu et al., 2017; Jutkina et al., 2018), non-antibiotic materials also accelerated the dissemination of plasmid-mediated ARGs (Qiu et al., 2012; Jiao et al., 2017; Zhang et al., 2018b; Cen et al., 2020). In particular, previous studies demonstrated that Cu2+ could promote the conjugative transfer of plasmid DNA within bacterial genera (from E. coli S17-1 to E. coli K12 MG1655) (Zhang et al., 2018a) or across bacterial genera (from E. coli K-12 LE392 to P. putida KT2440) (Zhang et al., 2019). However, whether Cu2+ could facilitate the conjugative transfer of SXT/R391 ICE was rarely explored. The present study showed that the conjugative transfer of SXT/R391 ICE that carried a large number of ARGs across bacterial genera (from P. mirabilis to E. coli) could be significantly promoted by Cu2+ ranging from 1 to 10 μmol/L (Figure 1A). Noticeably, subinhibitory and environmentally relevant concentrations of Cu2+ were used in this study (Zhang et al., 2019). The antimicrobial resistance profile, the int gene, and the attachment sites attL and attR of SXT/R391 ICE in transconjugants were detected, implying that the transconjugants carried the SXT/R391 ICE from the donor. Further, 5 μmol/L Cu2+ significantly facilitated the conjugative transfer of SXT/R391 ICE from the newly generated transconjugants to another recipient (Figure 1B), indicating that these newly generated transconjugants might serve as a novel ARG source at low concentrations of Cu2+. These findings confirmed the viewpoint that Cu2+ could accelerate the conjugative transfer of SXT/R391 ICE.
The addition of ROS scavenger significantly decreased the conjugative frequency (Figure 2C), suggesting that the increased production of ROS in both donor and recipient strains (Figures 2A,B) was caused by Cu2+ and was crucial for the transfer of SXT/R391 ICE. The bacterial cells rapidly respond to oxidative stress to protect against ROS attack (Liao et al., 2019). As expected, in the present study (Figures 2D,E), the expression of antioxidant-related genes, such as ahpC and ahpF, in the donor strain and sodA, sodB, and trxB in the recipient strain, was upregulated on exposure to Cu2+ at 5 μmol/L. These antioxidant enzymes were probably expressed to protect the donor and recipient strains from the ROS attack (Zuo et al., 2014; Zhu et al., 2019) due to increased ROS generation. Therefore, it was considered that the change in ROS generation was a critical factor for Cu2+ to promote the conjugative transfer of SXT/R391 ICE. However, ROS overproduction might cause irreversible cell function damage and even cell death, making recipient cells inactive. A previous report showed that Cu2+ and CuO nanoparticles at 100 μmol/L could decrease the horizontal transfer of plasmid-mediated ARGs due to the reduced recipient number (Zhang et al., 2019). Higher sub-MIC concentrations of Cu2+ reduced conjugative transfer of plasmids (Buberg et al., 2020). As reported, 100 μmol/L Cu2+ suppressed the transfer of plasmid-mediated ARG in a sludge bacterial community, probably attributed to disrupted iron–sulfur clusters of metalloenzymes and the purified fumarase A poisoning (Lin et al., 2019). Besides, 100 μmol/L Cu2+ significantly reduced the cell viability of the donor and recipient strains compared with the control (Supplementary Figure 2). Thus, these might be the reasons for the decreased transfer frequency of SXT/R391 ICE on exposure to Cu2+ at 100 μmol/L in this study.
Reactive oxygen species generation resulted in DNA damage, thus inducing the SOS response that controlled a series of genes involved in DNA damage repair and recombination (Baharoglu et al., 2010; Baharoglu and Mazel, 2014). In this study, the differentially expressed genes related to SOS response were mainly enriched in DNA repair and recombination in the recipient E. coli when exposed to Cu2+ at 5 μmol/L (Figure 2E). The SOS response could transitorily contribute to maintain a lower pool of SetR protein (an SXT-encoded repressor), thereby increasing the expression of genes necessary for SXT transfer (Beaber et al., 2004). This included activation of the site-specific recombination system, assembly of the mating apparatus, initiation of ICE DNA transfer, and integration into chromosome of recipient cell (Poulin-Laprade et al., 2015). However, there were no significant differences in the expression of genes related to the SOS response in the donor P. mirabilis, as well as the expression of genes related to SXT/R391 ICE transfer under exposure of 5 μmol/L Cu2+ in this study. Strain genetic background, insufficient sequencing coverage to detect rare transcripts, and extended mating periods may be contributing factors to the failure of SOS response and transfer genes transcripts detection.
Increased ROS production can cause damage to the cell membrane for both the donor and recipient strains, leading to an impaired membrane barrier (Lu et al., 2018). This was consistent with the TEM images, showing that apparent cell membrane damage and indistinct cell borders were found on exposure to Cu2+ at 5 μmol/L (Figure 3D). Moreover, ROS generation was known to enhance cell membrane permeability (Liao et al., 2019), which was associated with increased conjugative transfer frequency (Zhang et al., 2018a). Indeed, the cell membrane permeability of the donor and recipient strains increased significantly in the presence of Cu2+ (Figures 3A,B). The outer membrane proteins (e.g., OmpA, OmpC, and OmpF) played important roles in forming outer membrane pores and increasing membrane permeability (Zhang et al., 2017). The overexpression of outer membrane proteins accelerated the inward or outward movement of DNA (Dreiseikelmann, 1994). The transcriptional analyses suggested that the expression of relevant genes (e.g., omp) coding for outer membrane proteins was also upregulated on exposure to Cu2+ at 5 μmol/L (Figures 3E,F). Therefore, it was presumed that the increased membrane permeability was also a pivotal factor for Cu2+ to accelerate the transfer of SXT/R391 ICE.
Physical cell-to-cell contact is essential for the plasmid DNA transfer during the conjugative process. For example, fim-like operon has been reported to be related to adhesion (Wang et al., 2019). In this study, the expression of adhesion-relevant genes (e.g., fim) was upregulated on exposure to Cu2+ at 5 μmol/L in the recipient strain (Figure 4A), which was consistent with the TEM images (Figure 3D). These findings indicated that enhanced cell adhesion might contribute to the increased transfer of SXT/R391 ICE. Besides, increased messenger RNA (mRNA) expression of ATP synthesis genes (atp) was also observed in the recipient strain (Figure 4B). As DNA movement needs energy (Lu et al., 2018), improved energy availability may be contribute to the elevated transfer of SXT/R391 ICE via providing more energy. Interestingly, the expression of copA gene (a component of the copper efflux system) was upregulated in the recipient E. coli (Supplementary Table 8). The increased expression of gene copA might contribute to the recipient E. coli to avoid any excess copper-mediated toxicity and retain adequate supply of copper for cellular processes (Pal et al., 2017). As reported, enhancement of the K+ and Na+ efflux might promote the formation of transfer channel and plasmid uptake (Liao et al., 2019). Thus, the upregulated copA gene may be involved in the uptake of SXT/R391 ICE in this study.
Conclusion
This study demonstrated that the conjugative transfer of SXT/R391 ICE across bacterial genera (from P. mirabilis to E. coli) could be significantly accelerated by Cu2+. Importantly, the SXT/R391 ICE we selected carried 21 ARGs and mediated multidrug resistance. ROS generation, cell membrane permeability, cell adhesion, and ATP synthesis were the potential mechanisms for Cu2+ to promote the conjugative transfer of SXT/R391 ICE. This study was novel in proving that Cu2+ could facilitate the conjugative transfer of SXT/R391 ICE at subinhibitory and environmentally relevant concentrations.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Sichuan University Animal Ethics Committee.
Author Contributions
ZS and HW conceived and designed the study. ZS, LZ, CL, and YT performed the experiments. ZS analyzed the data and wrote the manuscript. All authors contributed to the manuscript revision and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31830098, 31772769, and 31572547) and the National Key Research and Development Program of China (2018YFD0500305).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.616792/full#supplementary-material
References
Aberkane, S., Compain, F., Decre, D., Dupont, C., Laurens, C., Vittecoq, M., et al. (2016). High prevalence of SXT/R391-related integrative and conjugative elements carrying blaCMY-2 in Proteus mirabilis isolates from gulls in the south of France. Antimicrob. Agents Chemother. 60, 1148–1152. doi: 10.1128/aac.01654-15
Allué-Guardia, A., Nyong, E. C., Koenig, S. S. K., Vargas, S. M., Bono, J. L., and Eppinger, M. (2019). Closed genome sequence of Escherichia coli K-12 group strain C600. Microbiol. Resour. Announc. 8:e1052-18.
Badhai, J., and Das, S. K. (2016). Characterization of three novel SXT/R391 integrating conjugative elements ICEMfuInd1a and ICEMfuInd1b, and ICEMprChn1 identified in the genomes of Marinomonas fungiae JCM 18476(T) and Marinomonas profundimaris strain D104. Front. Microbiol. 7:1896.
Baharoglu, Z., and Mazel, D. (2014). SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol. Rev. 38, 1126–1145. doi: 10.1111/1574-6976.12077
Baharoglu, Z., Bikard, D., and Mazel, D. (2010). Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 6:e1001165. doi: 10.1371/journal.pgen.1001165
Bandyopadhyay, D., and Mukherjee, M. (2020). Reactive oxygen species and uspA overexpession: an alternative bacterial response toward selection and maintenance of multidrug resistance in clinical isolates of uropathogenic E. coli. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1753–1760. doi: 10.1007/s10096-020-03903-x
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427, 72–74. doi: 10.1038/nature02241
Bioteau, A., Durand, R., and Burrus, V. (2018). Redefinition and unification of the SXT/R391 family of integrative and conjugative elements. Appl. Environ. Microbiol. 84:e485-18.
Blazquez, J., Couce, A., Rodriguez-Beltran, J., and Rodriguez-Rojas, A. (2012). Antimicrobials as promoters of genetic variation. Curr. Opin. Microbiol. 15, 561–569. doi: 10.1016/j.mib.2012.07.007
Buberg, M. L., Witsø, I. L., L’Abée-Lund, T. M., and Wasteson, Y. (2020). Zinc and copper reduce conjugative transfer of resistance plasmids from extended-spectrum beta-lactamase-producing Escherichia coli. Microb. Drug Resist. 26, 842–849. doi: 10.1089/mdr.2019.0388
Cen, T., Zhang, X., Xie, S., and Li, D. (2020). Preservatives accelerate the horizontal transfer of plasmid-mediated antimicrobial resistance genes via differential mechanisms. Environ. Int. 138:105544. doi: 10.1016/j.envint.2020.105544
Clinical and Laboratory Standards Institute [CLSI] (2016). Performance Standards For Antimicrobial Susceptibility Testing. Wayne, PA: CLSI.
Dreiseikelmann, B. (1994). Translocation of DNA across bacterial membranes. Microbiol. Rev. 58, 293–316. doi: 10.1128/mmbr.58.3.293-316.1994
Fan, Z., Chen, H., Li, M., Pan, X., Fu, W., Ren, H., et al. (2019). Pseudomonas aeruginosa polynucleotide phosphorylase contributes to ciprofloxacin resistance by regulating PrtR. Front. Microbiol. 10:1762.
Fang, L., Li, X., Li, L., Li, S., Liao, X., Sun, J., et al. (2016). Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci. Rep. 6:25312.
Gray, A. N., Henderson-Frost, J. M., Boyd, D., Sharafi, S., Niki, H., and Goldberg, M. B. (2011). Unbalanced charge distribution as a determinant for dependence of a subset of Escherichia coli membrane proteins on the membrane insertase YidC. mBio 2:e238-11.
Hastings, P. J., Rosenberg, S. M., and Slack, A. (2004). Antibiotic-induced lateral transfer of antibiotic resistance. Trends Microbiol. 12, 401–404. doi: 10.1016/j.tim.2004.07.003
Hayden, H. L., Savin, K. W., Wadeson, J., Gupta, V., and Mele, P. M. (2018). Comparative metatranscriptomics of wheat rhizosphere microbiomes in disease suppressive and non-suppressive soils for rhizoctonia solani AG8. Front. Microbiol. 9:859.
He, D., Wang, L., Zhao, S., Liu, L., Liu, J., Hu, G., et al. (2020). A novel tigecycline resistance gene, tet(X6), on an SXT/R391 integrative and conjugative element in a Proteus genomospecies 6 isolate of retail meat origin. J. Antimicrob. Chemother. 75, 1159–1164. doi: 10.1093/jac/dkaa012
Jiao, Y. N., Chen, H., Gao, R. X., Zhu, Y. G., and Rensing, C. (2017). Organic compounds stimulate horizontal transfer of antibiotic resistance genes in mixed wastewater treatment systems. Chemosphere 184, 53–61. doi: 10.1016/j.chemosphere.2017.05.149
Jutkina, J., Marathe, N. P., Flach, C. F., and Larsson, D. (2018). Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci. Total Environ. 61, 172–178. doi: 10.1016/j.scitotenv.2017.10.312
Kong, L. H., Lei, C. W., Ma, S. Z., Jiang, W., Liu, B. H., Wang, Y. X., et al. (2017). Various sequence types of Escherichia coli isolates coharboring blaNDM-5 and mcr-1 genes from a commercial swine farm in China. Antimicrob. Agents Chemother. 61:e2167-16.
Kong, L. H., Xiang, R., Wang, Y. L., Wu, S. K., Lei, C. W., Kang, Z. Z., et al. (2020). Integration of the blaNDM-1 carbapenemase gene into a novel SXT/R391 integrative and conjugative element in Proteus vulgaris. J. Antimicrob. Chemother. 75, 1439–1442. doi: 10.1093/jac/dkaa068
Lei, C. W., Chen, Y. P., Kang, Z. Z., Kong, L. H., and Wang, H. N. (2018). Characterization of a novel SXT/R391 integrative and conjugative element carrying cfr, blaCTX-M-65, fosA3, and aac(6′)-Ib-cr in Proteus mirabilis. Antimicrob. Agents Chemother. 62:e849-18.
Lei, C. W., Zhang, A. Y., Wang, H. N., Liu, B. H., Yang, L. Q., and Yang, Y. Q. (2016). Characterization of SXT/R391 integrative and conjugative elements in Proteus mirabilis isolates from food-producing animals in China. Antimicrob. Agents Chemother. 60, 1935–1938. doi: 10.1128/aac.02852-15
Li, G., Chen, X., Yin, H., Wang, W., Wong, P. K., and An, T. (2020). Natural sphalerite nanoparticles can accelerate horizontal transfer of plasmid-mediated antibiotic-resistance genes. Environ. Int. 136:105497. doi: 10.1016/j.envint.2020.105497
Li, L. G., Xia, Y., and Zhang, T. (2017). Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J. 11, 651–662. doi: 10.1038/ismej.2016.155
Liao, J., Huang, H., and Chen, Y. (2019). CO2 promotes the conjugative transfer of multiresistance genes by facilitating cellular contact and plasmid transfer. Environ. Int. 129, 333–342. doi: 10.1016/j.envint.2019.05.060
Lin, H., Jiang, L. T., Li, B., Dong, Y. B., He, Y. H., and Qiu, Y. (2019). Screening and evaluation of heavy metals facilitating antibiotic resistance gene transfer in a sludge bacterial community. Sci. Total Environ. 695:133862. doi: 10.1016/j.scitotenv.2019.133862
Liu, P., Wu, Z., Xue, H., and Zhao, X. (2017). Antibiotics trigger initiation of SCCmec transfer by inducing SOS responses. Nucleic Acids Res. 45, 3944–3952. doi: 10.1093/nar/gkx153
Lopatkin, A. J., Huang, S., Smith, R. P., Srimani, J. K., Sysoeva, T. A., Bewick, S., et al. (2016). Antibiotics as a selective driver for conjugation dynamics. Nat. Microbiol. 1:16044.
Lu, J., Wang, Y., Jin, M., Yuan, Z., Bond, P., and Guo, J. (2020). Both silver ions and silver nanoparticles facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes. Water Res. 169:115229. doi: 10.1016/j.watres.2019.115229
Lu, J., Wang, Y., Li, J., Mao, L., Nguyen, S. H., Duarte, T., et al. (2018). Triclosan at environmentally relevant concentrations promotes horizontal transfer of multidrug resistance genes within and across bacterial genera. Environ. Int. 121, 1217–1226. doi: 10.1016/j.envint.2018.10.040
McInnes, R. S., McCallum, G. E., Lamberte, L. E., and van Schaik, W. (2020). Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr. Opin. Microbiol. 53, 35–43. doi: 10.1016/j.mib.2020.02.002
Nihemaiti, M., Yoon, Y., He, H., Dodd, M. C., Croue, J. P., and Lee, Y. (2020). Degradation and deactivation of a plasmid-encoded extracellular antibiotic resistance gene during separate and combined exposures to UV254 and radicals. Water Res. 182:115921. doi: 10.1016/j.watres.2020.115921
Pal, C., Asiani, K., Arya, S., Rensing, C., Stekel, D. J., Larsson, D., et al. (2017). Metal resistance and its association with antibiotic resistance. Adv. Microb. Physiol. 70, 261–313. doi: 10.1016/bs.ampbs.2017.02.001
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17.
Piccaro, G., Pietraforte, D., Giannoni, F., Mustazzolu, A., and Fattorini, L. (2014). Rifampin induces hydroxyl radical formation in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 7527–7533. doi: 10.1128/aac.03169-14
Poole, K. (2017). At the nexus of antibiotics and metals: the impact of Cu and Zn on antibiotic activity and resistance. Trends Microbiol. 25, 820–832. doi: 10.1016/j.tim.2017.04.010
Poulin-Laprade, D., Matteau, D., Jacques, P. E., Rodrigue, S., and Burrus, V. (2015). Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon. Nucleic Acids Res. 43, 2045–2056. doi: 10.1093/nar/gkv071
Qiu, Z., Yu, Y., Chen, Z., Jin, M., Yang, D., Zhao, Z., et al. (2012). Nanoalumina promotes the horizontal transfer of multiresistance genes mediated by plasmids across genera. Proc. Natl. Acad. Sci. U. S. A. 109, 4944–4949. doi: 10.1073/pnas.1107254109
Redondo-Salvo, S., Fernandez-Lopez, R., Ruiz, R., Vielva, L., de Toro, M., Rocha, E., et al. (2020). Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 11:3602.
Rodriguez-Beltran, J., Sorum, V., Toll-Riera, M., de la Vega, C., Pena-Miller, R., and San, M. A. (2020). Genetic dominance governs the evolution and spread of mobile genetic elements in bacteria. Proc. Natl. Acad. Sci. U. S. A. 117, 15755–15762. doi: 10.1073/pnas.2001240117
Ruan, L., Pleitner, A., Ganzle, M. G., and McMullen, L. M. (2011). Solute transport proteins and the outer membrane protein NmpC contribute to heat resistance of Escherichia coli AW1.7. Appl. Environ. Microbiol. 77, 2961–2967. doi: 10.1128/aem.01930-10
Sandegren, L., Linkevicius, M., Lytsy, B., Melhus, A., and Andersson, D. I. (2012). Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J. Antimicrob. Chemother. 67, 74–83. doi: 10.1093/jac/dkr405
Shun-Mei, E., Zeng, J. M., Yuan, H., Lu, Y., Cai, R. X., and Chen, C. (2018). Sub-inhibitory concentrations of fluoroquinolones increase conjugation frequency. Microb. Pathog. 114, 57–62. doi: 10.1016/j.micpath.2017.11.036
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
Wang, Q., Liu, L., Hou, Z., Wang, L., Ma, D., Yang, G., et al. (2020). Heavy metal copper accelerates the conjugative transfer of antibiotic resistance genes in freshwater microcosms. Sci. Total Environ. 717:137055. doi: 10.1016/j.scitotenv.2020.137055
Wang, Q., Mao, D., and Luo, Y. (2015). Ionic liquid facilitates the conjugative transfer of antibiotic resistance genes mediated by plasmid RP4. Environ. Sci. Technol. 49, 8731–8740. doi: 10.1021/acs.est.5b01129
Wang, Y., Lu, J., Mao, L., Li, J., Yuan, Z., Bond, P. L., et al. (2019). Antiepileptic drug carbamazepine promotes horizontal transfer of plasmid-borne multi-antibiotic resistance genes within and across bacterial genera. ISME J. 13, 509–522. doi: 10.1038/s41396-018-0275-x
Wozniak, R. A., Fouts, D. E., Spagnoletti, M., Colombo, M. M., Ceccarelli, D., Garriss, G., et al. (2009). Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5:e1000786. doi: 10.1371/journal.pgen.1000786
Xie, S., Gu, A. Z., Cen, T., Li, D., and Chen, J. (2019). The effect and mechanism of urban fine particulate matter (PM2.5) on horizontal transfer of plasmid-mediated antimicrobial resistance genes. Sci. Total Environ. 683, 116–123. doi: 10.1016/j.scitotenv.2019.05.115
Yang, Q. E., Agouri, S. R., Tyrrell, J. M., and Walsh, T. R. (2018). Heavy metal resistance genes are associated with blaNDM-1- and blaCTX-M-15-carrying Enterobacteriaceae. Antimicrob. Agents Chemother. 62:e2642-17.
Yi, H., Cho, Y. J., Yong, D., and Chun, J. (2012). Genome sequence of Escherichia coli J53, a reference strain for genetic studies. J. Bacteriol. 194, 3742–3743. doi: 10.1128/jb.00641-12
Yu, Z., Gunn, L., Wall, P., and Fanning, S. (2017). Antimicrobial resistance and its association with tolerance to heavy metals in agriculture production. Food Microbiol. 64, 23–32. doi: 10.1016/j.fm.2016.12.009
Zhang, S., Wang, Y., Song, H., Lu, J., Yuan, Z., and Guo, J. (2019). Copper nanoparticles and copper ions promote horizontal transfer of plasmid-mediated multi-antibiotic resistance genes across bacterial genera. Environ. Int. 129, 478–487. doi: 10.1016/j.envint.2019.05.054
Zhang, Y., Gu, A. Z., Cen, T., Li, X., He, M., Li, D., et al. (2018a). Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ. Pollut. 237, 74–82. doi: 10.1016/j.envpol.2018.01.032
Zhang, Y., Gu, A. Z., Cen, T., Li, X., Li, D., and Chen, J. (2018b). Petrol and diesel exhaust particles accelerate the horizontal transfer of plasmid-mediated antimicrobial resistance genes. Environ. Int. 114, 280–287. doi: 10.1016/j.envint.2018.02.038
Zhang, Y., Gu, A. Z., He, M., Li, D., and Chen, J. (2017). Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across Genera. Environ. Sci. Technol. 51, 570–580. doi: 10.1021/acs.est.6b03132
Zhu, C., Chen, J., Wang, Y., Wang, L., Guo, X., Chen, N., et al. (2019). Enhancing 5-aminolevulinic acid tolerance and production by engineering the antioxidant defense system of Escherichia coli. Biotechnol. Bioeng. 116, 2018–2028. doi: 10.1002/bit.26981
Zhu, Y. G., Johnson, T. A., Su, J. Q., Qiao, M., Guo, G. X., Stedtfeld, R. D., et al. (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U. S. A. 110, 3435–3440. doi: 10.1073/pnas.1222743110
Keywords: copper ions, SXT/R391 integrative and conjugative element, conjugative transfer, reactive oxygen species, cell membrane permeability
Citation: Song Z, Zuo L, Li C, Tian Y and Wang H (2021) Copper Ions Facilitate the Conjugative Transfer of SXT/R391 Integrative and Conjugative Element Across Bacterial Genera. Front. Microbiol. 11:616792. doi: 10.3389/fmicb.2020.616792
Received: 13 October 2020; Accepted: 22 December 2020;
Published: 02 February 2021.
Edited by:
Christophe Merlin, Université de Lorraine, FranceReviewed by:
Rob Van Houdt, Belgian Nuclear Research Centre, BelgiumDominic Poulin-Laprade, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2021 Song, Zuo, Li, Tian and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongning Wang, d2hvbmduaW5nQDE2My5jb20=
 Zhou Song
Zhou Song Lei Zuo
Lei Zuo Yiming Tian
Yiming Tian Hongning Wang
Hongning Wang