
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 December 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.616364
This article is part of the Research Topic Antibiotic Resistance and its Continuity in the Environmental Niche View all 19 articles
 Jiafang Fu1,2
Jiafang Fu1,2 Chuanqing Zhong3
Chuanqing Zhong3 Peipei Zhang1,2,4
Peipei Zhang1,2,4 Qingxia Gao2
Qingxia Gao2 Gongli Zong1,2,4
Gongli Zong1,2,4 Yingping Zhou3
Yingping Zhou3 Guangxiang Cao1,2,4*
Guangxiang Cao1,2,4*Antibiotics and organoarsenical compounds are frequently used as feed additives in many countries. However, these compounds can cause serious antibiotic and arsenic (As) pollution in the environment, and the spread of antibiotic and As resistance genes from the environment. In this report, we characterized the 28.5 kb genomic island (GI), named as ICERspD18B, as a novel chromosomal integrative and conjugative element (ICE) in multidrug-resistant Rheinheimera sp. D18. Notably, ICERspD18B contains six antibiotic resistance genes (ARGs) and an arsenic tolerance operon, as well as genes encoding conjugative transfer proteins of a type IV secretion system, relaxase, site-specific integrase, and DNA replication or partitioning proteins. The transconjugant strain 25D18-B4 was generated using Escherichia coli 25DN as the recipient strain. ICERspD18B was inserted into 3'-end of the guaA gene in 25D18-B4. In addition, 25D18-B4 had markedly higher minimum inhibitory concentrations for arsenic compounds and antibiotics when compared to the parental E. coli strain. These findings demonstrated that the integrative and conjugative element ICERspD18B could mediate both antibiotic and arsenic resistance in Rheinheimera sp. D18 and the transconjugant 25D18-B4.
In aquaculture systems, the indiscriminate use of chemical additives and antimicrobials (especially antibiotics) as preventative and curative measures for diseases has resulted in antimicrobial resistance among bacteria (Buschmann et al., 2012; Sun et al., 2016; Nakayama et al., 2017; Rico et al., 2017). Additionally, the transfer of antibiotic resistance elements from aquaculture facilities into the environment could have negative impacts on environmental biodiversity and human health as a result of further antimicrobial resistance development (Garcia-Aljaro et al., 2014; Xu et al., 2017). In addition to antibiotics, the metalloid arsenic (As) has been used as a feed additive, although it was ranked first on the priority list of hazardous substances by the Agency for Toxic Substances and Disease Registry1; arsenic has a significant impact on the aquaculture environment because of its toxic, persistent, and accumulative properties in organisms, which have devastating effects on the diversity of aquatic animals and on the ecological balance of aquaculture systems (Miazek et al., 2015; Rahman and De Ley, 2017). Arsenic resistance genes, usually organized in ars operons, have been widely identified in bacteria (Fekih et al., 2018; Serrato-Gamino et al., 2018). Therefore, the aquaculture environment poses a potential risk for the dissemination of arsenic resistance genes as well as antibiotic resistance genes (ARGs) through mobile genetic elements (Abdelhamed et al., 2019).
Bacteria of the genus Rheinheimera are frequently isolated from freshwater and estuaries (Baek and Jeon, 2015; Chen et al., 2019); and saline and slightly alkaline lakes (Liu et al., 2012; Zhong et al., 2014). Currently, the genus comprises 27 species.2 Comparative genomics analysis of Rheinheimera genomes revealed that the core genome is relatively small (Presta et al., 2017), which may be related to the different ecological niches colonized by members of this genus (Wang et al., 2018; Panda et al., 2020). It has been reported that many Rheinheimera strains are multidrug-resistant (Liu et al., 2012; Mengoni et al., 2014; Suarez et al., 2014; Kumar et al., 2015), and a series of ARGs in the genomes of Rheinheimera spp. have been uncovered, such as acrD in Rheinheimera sp. EpRS3, encoding an aminoglycoside efflux pump; acrB in Rheinheimera sp. KL1, encoding a multidrug resistance-nodulation-division efflux pump; and tet(B) in Rheinheimera sp. D18, encoding a tetracycline efflux major facilitator superfamily (MFS) transporter (O’Connor et al., 2015; Presta et al., 2017; Fu et al., 2020). In addition, bioinformatics analyses have predicted the widespread presence of arsenical resistance genes in Rheinheimera. However, the transferability of ARGs and arsenic resistance genes in Rheinheimera has not been well characterized.
Rheinheimera sp. D18 strain was previously isolated from mariculture environment in the Yellow Sea, which has been reported to be polluted by notable amounts of antibiotic residues (Du et al., 2017; Han et al., 2020) and arsenic (Jiang et al., 2015; Xiao et al., 2017), and D18 was found to have high-level resistance to tetracycline, florfenicol, amikacin, and sulfamethoxazole (Fu et al., 2020). In this study, the novel integrative and conjugative element ICERspD18B was characterized in Rheinheimera sp. D18 genome. In addition to genes related to DNA replication/partitioning and conjugative transfer, ICERspD18B was found to contain three repeated copies of a chloramphenicol/florfenicol efflux MFS transporter-encoding gene (floR), and several other ARGs. An arsenic tolerance operon was also identified in ICERspD18B, indicating that ICERspD18B mediates combined resistance to antibiotics and arsenic, and further analysis indicated that ICERspD18B was transferable. This report characterized the first mobile genomic island (GI) ICERspD18B that endows both antibiotic and arsenic resistance in the genus Rheinheimera, providing new insights into antibiotic and arsenic spread in the mariculture environment.
Rheinheimera sp. D18 strain was previously isolated from maricultural environment (Fu et al., 2020). Rheinheimera sp. D18 was cultured in LB solid medium (tryptone 1%, yeast extract 0.5%, 1% sodium chloride, and agar 2%) at 28°C and was used as a donor in conjugation experiments. Escherichia coli strain 25DN was cultured at 37°C in LB medium and was used as recipient in conjugation experiments. Transconjugants from conjugation experiments were cultured on LB medium containing florfenicol (24 mg/l) and roxarsone (8 mM) at 37°C.
The Rheinheimera sp. D18 whole genome sequence has been deposited in GenBank (CP037745). The GIs were identified using Island Viewer 4 (Bertelli et al., 2017) and were further analyzed using ICEfinder (Liu et al., 2019). The genes in genomic island were annotated using the Prokaryotic Genome Annotation Pipeline on NCBI3 and RASTtk server (Overbeek et al., 2014; Brettin et al., 2015). Insertion sequence transposases were detected using IS-Finder (Siguier et al., 2012).
Pairwise alignment of ICERspD18B and other relevant genetic elements was performed using the BLAST search tool and ICEberg WU-BLAST search tool (Liu et al., 2019). Further alignment between two sequences was performed using BioXM 2.6 software.
To determine whether the antibiotic and arsenic resistance genes in ICERspD18B could be horizontal transferred among bacteria, conjugation experiments were carried out as previously described with some modification (Fu et al., 2020). Transconjugants were selected on LB agar plates with florfenicol (24 mg/l), roxarsone (8 mM), X-Gluc (5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid), and sodium azide. The donor (Rheinheimera sp. D18) and the recipient (E. coli 25DN) strains are inhibited and only the transconjugants would survive on the selective agar plates. ICERspD18B and its insertion site in the transconjugant were demonstrated by PCR and direct DNA sequencing. The ability of ICERspD18B to form a ring in Rheinheimera sp. D18 was also verified by PCR and DNA sequencing. All the primers used in this report are listed in Supplementary Table S1.
The broth microdilution method was used (CLSI, 2017) to determine the MICs for roxarsone, sodium hexafluoroarsenate and different antibiotics, including amikacin, florfenicol, and sulfamethoxazole. Escherichia coli 25DN strain was also tested for MICs.
All the experiments in this study were carried out in triplicate. The differences in MICs for the transconjugant strain and E. coli 25DN strain were analyzed using the Student’s t-test (p < 0.05).
A chromosomal GI in Rheinheimera sp. D18 was identified using Island Viewer 4 (Figure 1), while it was not predicted as a typical integrative and conjugative element (ICE) by ICEfinder software. This GI extends from position 2,629,186 to 2,657,721 in the chromosome of D18 and contains 28,536 bp. Gene annotation indicated that it contains 33 open reading frames (ORFs; Supplementary Table S2), among which six ORFs were predicted to be ARGs, including one sulfonamide resistance gene (sul2), two aminoglycoside resistance genes (aph(3'')-Ib and strB), and three repeated copies of a chloramphenicol/florfenicol resistance gene (floR); and four ORFs were predicted to be arsenic resistance genes, forming the operon arsRHCB. The GI also contains three identical copies of a relaxase-encoding gene (E0Z06_RS12465, E0Z06_RS12485, and E0Z06_RS12505) related to a type IV secretion system; three conjugative transfer protein-encoding genes (trbL, trbK, and trbJ); four genes associated with DNA replication or partitioning (repC, repA, E0Z06_RS12575, and E0Z06_RS12520); and genes encoding a site-specific integrase (int) and its transcriptional regulator (E0Z06_RS12590). Sequence examination further indicated that the GI was bordered by a 20-bp direct repeat (DR; 5'-ACAATNGAGTGGGAATNNTT-3') at both ends and that it was inserted into the guaA gene (E0Z06_RS12600) in the chromosome of D18. These findings suggest that this GI might be an ICE-like genomic island, named as ICERspD18B, and provide antibiotic and arsenic tolerance to Rheinheimera sp. D18, as we know, ICEs are now recognized as a large and diverse class of chromosomal mobile genetic elements in bacteria that can transfer between bacteria through conjugation (Baranowski et al., 2018; Partridge et al., 2018).
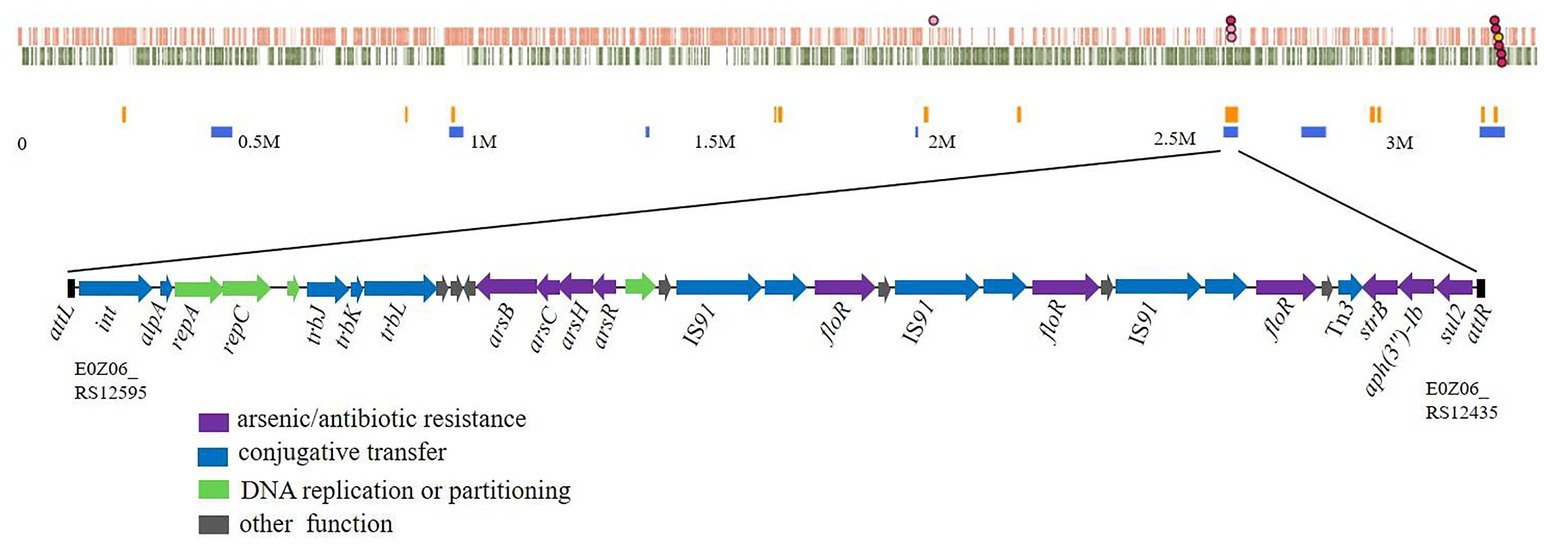
Figure 1. Schematic view of a new identified genomic island (GI) and its position in Rheinheimera sp. D18. Top image, GIs predicted by Island Viewer 4 in Rheinheimera sp. D18. Putative genomic islands were predicted by IslandPath-DIMOB method (blue squares) or SIGI-HMM method (orange squares). Bottom image, gene arrangement in the genomic island named ICERspD18B. ICERspD18B (from E0Z06_RS12595 to E0Z06_RS12435) is bordered by a 20-bp DR (5'-ACAATNGAGTGGGAATNNTT-3') in the chromosome of D18. The diagram shows the predicted classification/function of each gene (represented by arrows) as follows: violet, arsenic, or antibiotic resistance; blue, conjugative transfer; green, DNA replication or partitioning; and gray, other functions.
The whole ICERspD18B nucleotide sequence was analyzed using BLAST, and results revealed that this ICERspD18B presents only in the Rheinheimera sp. D18 genome. GC content of ICERspD18B is 58.28%, different from that of the overall GC content of Rheinheimera sp. D18 genome (44.39%), indicating that this genomic island ICERspD18B was derived from other bacteria. Pairwise alignment of ICERspD18B with other relevant DNA sequences was performed, and the sequence alignment results are shown in Figure 2. BLASTn analysis indicated that genes relating to conjugative transfer and DNA replication or partitioning (from E0Z06_RS12595 to E0Z06_RS12545) in ICERspD18B were highly similar to genes in the Klebsiella pneumoniae NCTC9180 genome (GenBank accession number LR134202.1), and these genes were also predicted to be present in the K. pneumoniae NCTC9171 genome (GenBank accession number LR588410.1). A larger region that included the above genes and the arsenic operon (arsRHCB; from E0Z06_RS12595 to E0Z06_RS12510) in ICERspD18B showed 99% identity with a genomic region of K. pneumoniae NCTC9171. In addition, the ICERspD18B arsenic operon (arsRHCB) had 100% nucleotide sequence identity to the arsenic operon located in Salmonella enterica strain 20–56 plasmid 1 (GenBank accession number LR536427.1). Of particular note, there were three tandem repeats of a set of genes that includes IS91, floR, a relaxase-encoding gene, and a LysR family transcriptional regulator-encoding gene in ICERspD18B, one or two set of these genes were also predicted in ICEVchBan5 of Vibrio cholerae O1 Ban5 (GenBank accession number GQ463140) and ICEPmiChn3 of Proteus mirabilis JN28 (GenBank accession number KY437727). The structure of the remaining part of ICERspD18B, including genes related to aminoglycoside and sulfonamide resistance, showed high similarity to genes in the Providencia rettgeri Pr-15-2-50 genome (GenBank accession number CP039844.1).
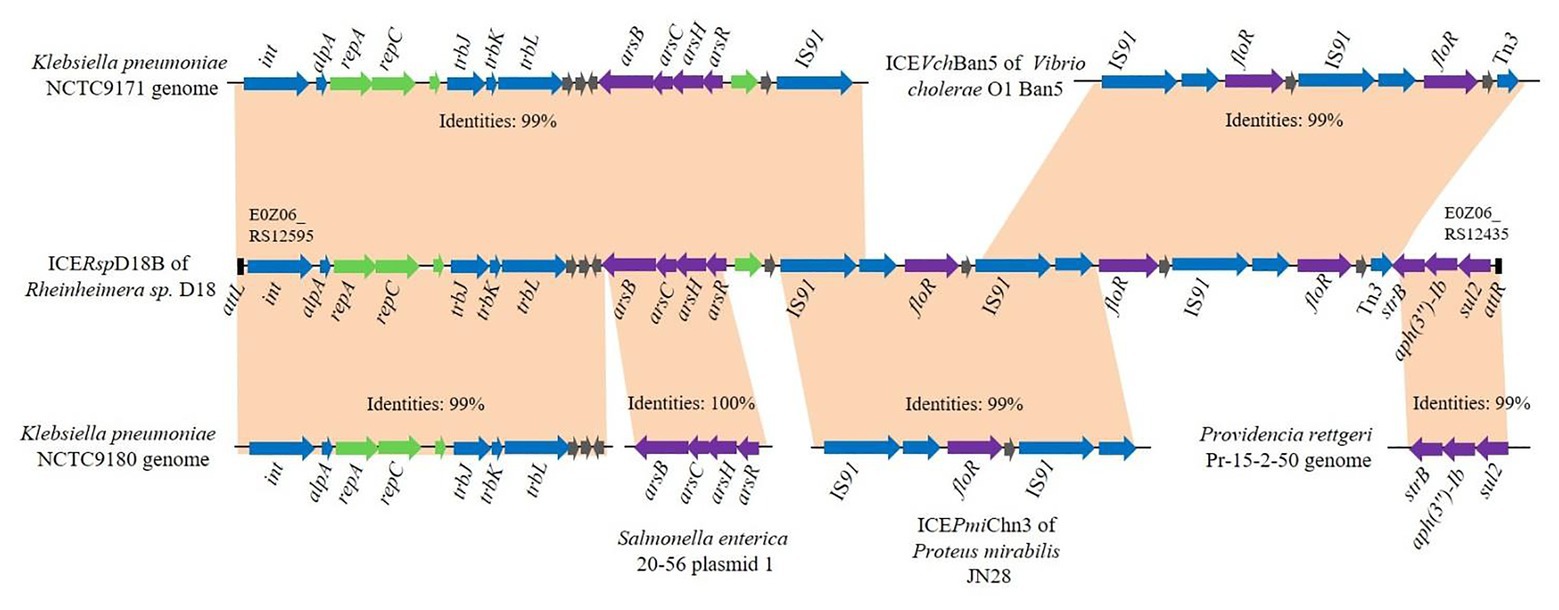
Figure 2. Schematic representation of the potential sources of genes in ICERspD18B. Pairwise alignment of ICERspD18B of Rheinheimera sp. D18 with closely related DNA sequences from ICEVchBan5 of Vibrio cholerae O1 Ban5, ICEPmiChn3 of Proteus mirabilis JN28, plasmid 1 of Salmonella enterica 20–56, and the Klebsiella pneumoniae NCTC9171, K. pneumoniae NCTC9180, and Providencia rettgeri Pr-15-2-50 genomes. Genes are indicated by arrows, and colors represent the following predicted functions: violet, arsenic, or antibiotic resistance; blue, conjugative transfer; green, DNA replication or partitioning; and gray, other functions. Orange shading matches regions with high sequence identity.
In order to determine whether the ICE-like chromosomal genomic island ICERspD18B could be horizontally transferred, conjugation experiments between the donor strain D18 and the recipient strain E. coli 25DN (sodium azide-resistant) were performed. Florfenicol and roxarsone were used as the selective pressure, and the transconjugation frequency was about 2.76 × 10−7 colony-forming units/donor. One of the transconjugants was isolated and named 25D18-B4. To determine whether ICERspD18B was inserted into the chromosome of E. coli 25D18-B4, PCR assays and DNA sequencing analysis were performed. The results demonstrated that genes strB, floR, and arsB, and the region between repC and trbJ in ICERspD18B, were present in 25D18-B4 but not in strain 25DN (Figures 3A,B). Furthermore, these sequences had 100% identity with those of Rheinheimera sp. D18, revealing that ICERspD18B had been transferred to 25D18-B4. Results also revealed that this ICERspD18B had been excised from the chromosome and was present in a circular form in Rheinheimera sp. D18 (Figure 3C), which is considered to be the first step of conjugation.
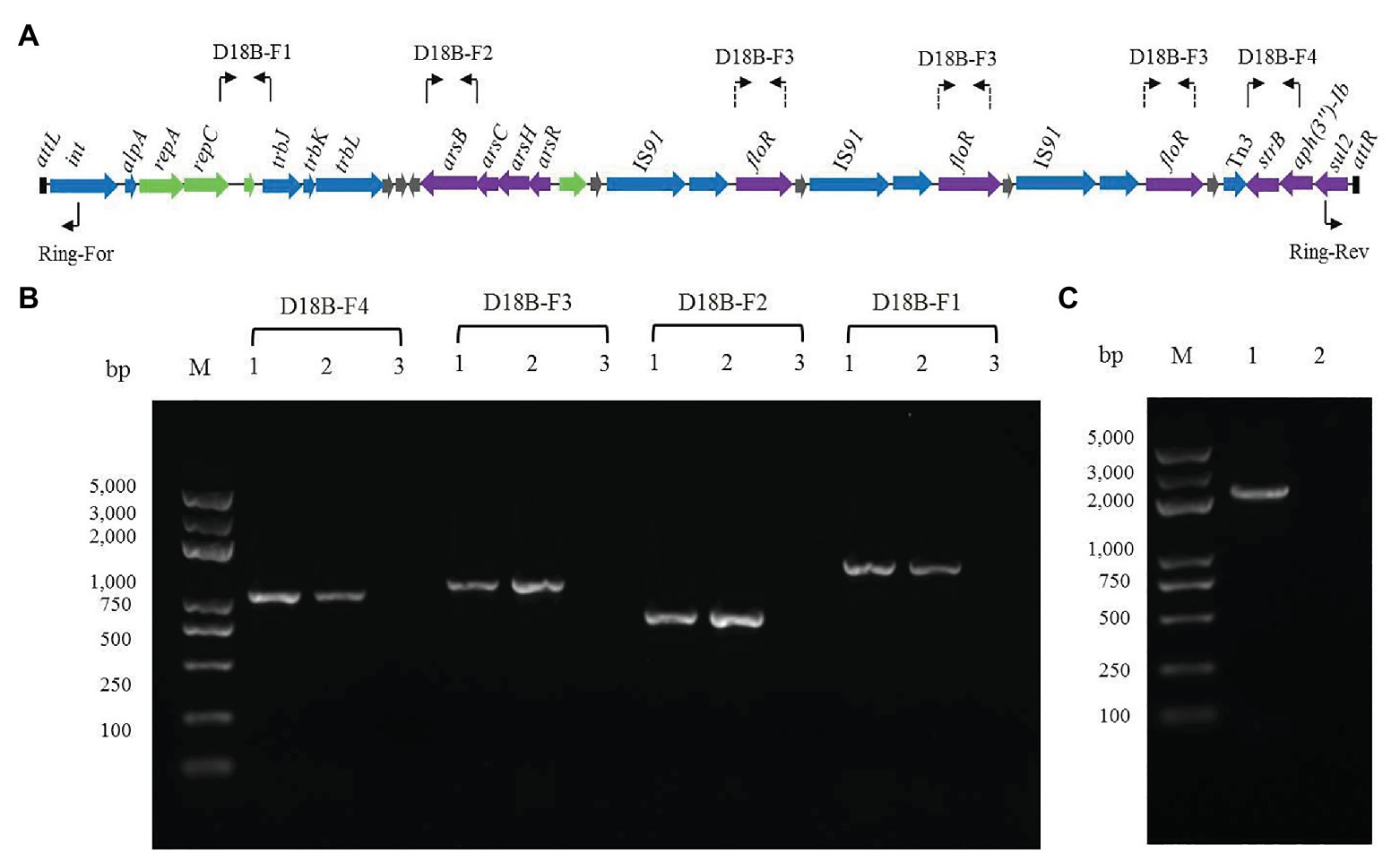
Figure 3. Verification of the presence and the circular form of ICERspD18B. (A) Primer positions in ICERspD18B are indicated using bent arrows. As ICERspD18B harbors three copies of floR, primers for determination of the presence of floR are shown in three locations by dashed arrows. (B) Four ICERspD18B fragments were amplified by PCR using total DNA of transconjugant 25D18-B4 (lanes 1), strain D18 (lanes 2), strain 25DN (lanes 3) as templates. (C) Verification of the circular form of ICERspD18B using the primer pair Ring-For/Rev. Total DNA of strain D18 (lane 1) or strain 25DN (lane 2) was used as template. M, molecular size markers.
The 3'-ends of tRNA/tmRNA genes are known attachment sites of ICEs (Williams, 2002; Liu and Zhu, 2010; Del Canto et al., 2011). However, the 3'-end of the guanosine monophosphate synthetase-encoding gene guaA has also been reported as an insertion site of genomic islands (Song et al., 2012). As bioinformatics analysis had indicated that ICERspD18B was inserted into 3'-end of guaA in the Rheinheimera sp. D18 genome, we investigated its location in the transconjugant 25D18-B4 and whether integration was orientation-specific, using PCR and DNA sequencing. 25D18-B4 was analyzed by PCR using combinations of two primer pairs: Junction L-For/Junction L-Rev and Junction R-For/Junction R-Rev, with D18 and E. coli 25DN as controls (Figure 4). It should be noted that the sequence of the Junction L-For primer is also present in the guaA gene of D18, due to the high similarity of guaA in D18 and 25DN, and that Junction L fragments were amplified in both 25D18-B4 and D18 (Figure 4B). PCR results indicated that ICERspD18B had been inserted into the 3'-end of guaA gene of the transconjugant 25D18-B4 strain, and DNA sequence analysis of PCR products confirmed that ICERspD18B was inserted at this site.
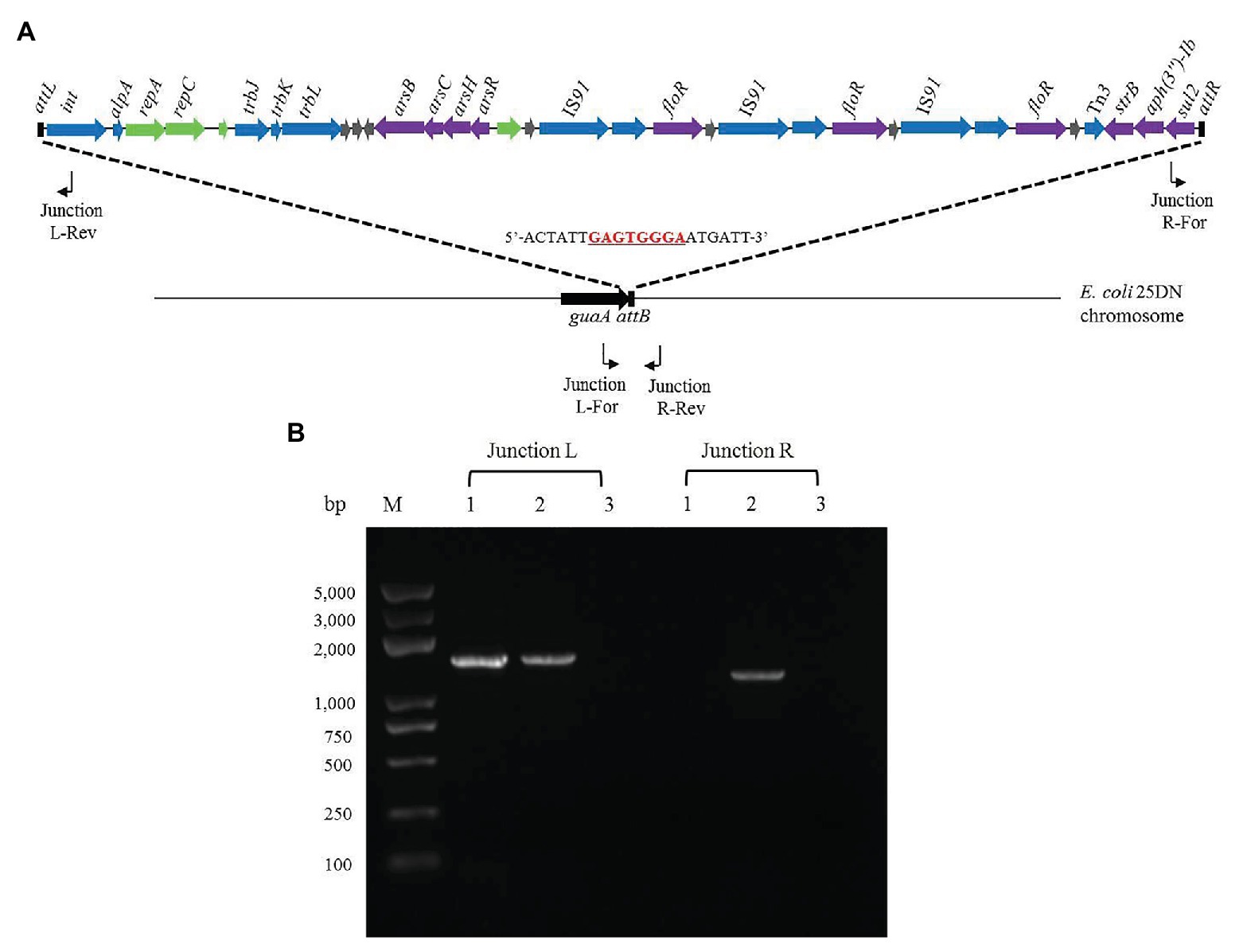
Figure 4. Analysis of the insertion site of ICERspD18B in transconjugant 25D18-B4. (A) PCR primer positions in ICERspD18B and in the strain 25DN chromosome are indicated by bent arrows. The insertion site of ICERspD18B in the strain 25DN chromosome is indicated by dashed lines, and the cutting site (5'-GAGTGGGA-3') of the integrase (Song et al., 2012) is underlined and marked in red. (B) Gel picture of the PCR products generated by the Junction L-For/Junction L-Rev and Junction R-For/Junction R-Rev primer pairs. Total DNA of strain D18 (lanes 1), transconjugant strain 25D18-B4 (lanes 2), and strain 25DN (lanes 3) was used as template. M, molecular size markers.
The susceptibility of transconjugant 25D18-B4 and Rheinheimera sp. D18 to metalloid arsenic and antibiotics was tested. As shown in Table 1, 25D18-B4 had acquired resistance to florfenicol (MIC, 92 mg/L), amikacin (MIC, 24 mg/L), sulfamethoxazole (MIC, 16 mg/L), sodium hexafluoroarsenate (MIC, 22 mM), and roxarsone (MIC, 14 mM). MIC testing revealed that the MICs for amikacin, florfenicol, sulfamethoxazole, sodium hexafluoroarsenate, and roxarsone in the transconjugant 25D18-B4 were higher than the MICs for the recipient strain 25DN (Table 1). The notable increase in antibiotic/arsenic resistance of 25D18-B4 suggested that ICERspD18B genes involved in antibiotic and arsenic resistance had been horizontally transferred to the E. coli strain.
In this study, we reported the discovery and characterization of the ICE-like chromosomal genomic island ICERspD18B in the genus Rheinheimera. BLASTn analysis indicated that only part sequence of ICERspD18B exists in other species, and mainly derived from pathogenic bacteria such as Vibrio cholerae, K. pneumoniae, and P. rettgeri (Figure 2). Further alignment with ICEberg WU-BLAST search tool revealed that the overall nucleotide sequence of ICERspD18B has low similarity to that of previously described ICEs, although a portion of ICERspD18B showed high similarity to ICEVchBan5 of Vibrio cholerae O1 Ban5 and ICEPmiChn3 of P. mirabilis JN28 (Figure 2). Additionally, our conjugation experiments indicated that ICERspD18B has the ability to transfer among bacteria. Hence, we speculate that ICERspD18B was transferred horizontally from other unsequenced strains. Moreover, ICERspD18B contains genes predicted to encode a site-specific integrase, relaxases associated with a type IV secretory pathway, conjugative transfer proteins, and DNA replication or partitioning encoding genes (Supplementary Table S2), further suggesting that ICERspD18B is an ICE.
tRNA, tmRNA, and some small RNA genes are recognized as integration hotspots of genomic islands (Williams, 2002; Del Canto et al., 2011). However, the 3'-end of the guaA gene is also an insertion site of genomic islands (Song et al., 2012). Integrases in guaA-associated genomic islands are frequently phage P4 integrases, and genes encoding AlpA (the positive regulatory protein of P4 integrases) are located near the P4 integrase genes in these genomic islands (Song et al., 2012). The 8-bp consensus sequence 5'-GAGTGGGA-3' within the DR flanking these genomic islands was reported to be the cutting site of the P4 integrases (Song et al., 2012). In our study, bioinformatics analysis revealed that the site-specific integrase in ICERspD18B belongs to the phage P4 integrases and that the AlpA-encoding gene alpA is next to the site-specific integrase-encoding gene int (Supplementary Table S1). Additionally, the 8-bp consensus sequence 5'-GAGTGGGA-3' was also found within the DR (5'-ACAATNGAGTGGGAATNNTT-3') of ICERspD18B, and ICERspD18B was confirmed to be inserted into the 3' end of guaA in the transconjugant 25D18-B4 (Figure 4). In addition, the circular, extrachromosomal form of ICERspD18B was also observed in Rheinheimera sp. D18 using PCR (Figure 3). These data suggest that ICERspD18B was first excised from the donor Rheinheimera sp. D18 chromosome, transferred via type IV secretory system-mediated conjugation and then inserted into 3'-end of guaA gene of the E. coli 25DN chromosome by site-specific recombination. These data also indicated that ICERspD18B has the ability to transfer genes horizontally from Rheinheimera sp. D18 to other bacteria. Considering that ICERspD18B is also located at the 3'-end of guaA in the Rheinheimera sp. D18 genome, our results further demonstrate that the 3'-end of guaA gene may be another integration hotspot of genomic islands.
Organoarsenic arsenical compounds (such as p-arsanilic acid and roxarsone) are widely used as feed additives in many countries, and the land application of poultry or swine litter could cause serious arsenic pollution in the environment (Liang et al., 2014; Xie and Cheng, 2019), potentially resulting in arsenic resistance among environmental bacteria and the dissemination of their arsenic resistance genes to other bacterial species. Arsenic resistance genes are usually organized in ars operons in bacteria, such as in Pseudomonas putida, which has two arsRBCH operons and which is highly resistant to organoarsenicals and inorganic arsenic (Canovas et al., 2003; Villadangos et al., 2012). The arsB gene encodes an As(III) efflux permease, arsC encodes an arsenate reductase for reduction of inorganic arsenate to As(III) and arsR encodes an As(III)-responsive transcriptional factor that controls expression of the operon (Yang et al., 2012). Arsenate [As(V)] is reduced to arsenite [As(III)] by the arsenate reductase ArsC prior to efflux, and then, arsenite is pumped out through ArsB (Shen et al., 2013). arsH encodes an organoarsenical oxidase that confers resistance to organoarsenic (Chen et al., 2015; Xie and Cheng, 2019). ICERspD18B contains one ars gene cluster, which includes arsBCHR (Figure 1). The transconjugant 25D18-B4, which acquired ICERspD18B, was found to have markedly higher MICs of roxarsone and sodium hexafluoroarsenate compared to those of the parental strain, E. coli 25DN (Table 1). These data suggest that ICERspD18B can contribute to the dissemination of arsenic resistance genes among bacteria.
Sulfonamide, chloramphenicol/florfenicol, and aminoglycoside have been used widely to treat bacterial and protozoan infections in aquaculture systems (Dang et al., 2007; Hoa et al., 2008). ICERspD18B also contains three copies of a chloramphenicol/florfenicol efflux MFS transporter-encoding gene (floR); one sulfonamide resistance gene (sul2); and two aminoglycoside resistance genes, aph(3'')-Ib, and strB. Escherichia coli is an opportunistic bacterium that can cause a wide variety of intestinal and extraintestinal infections (Riley, 2014). In this study, ICERspD18B was horizontal transferred to E. coli 25DN strain, and generated the transconjugant 25D18-B4 strain. The transconjugant 25D18-B4 was found to have notably higher MICs of amikacin, florfenicol, and sulfamethoxazole when compared to the parental strain, E. coli 25DN (Table 1), suggesting that the ARGs in ICERspD18B contribute to the antibiotic resistance profile of Rheinheimera sp. D18 as well as of E. coli 25D18-B4. These data suggest that the ICE-like genomic island ICERspD18B has the ability to disseminate these ARGs, along with arsenic resistance genes, among bacteria in the environment.
In conclusion, the findings of this study demonstrate that ICERspD18B is an ICE that increases host tolerance to arsenic and several antibiotics. Our results also reveal that this mobilizable ICERspD18B could be horizontal transferred to E. coli 25DN strain, and the transconjugant 25D18-B4 also has resistance to arsenic and antibiotic. Continuous monitoring of the antibiotic/arsenic tolerance of bacteria detected in the aquaculture industry is recommended to reduce the spread of resistance genes.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
JF: executed the experiments and manuscript preparation and submission. CZ: resources, review and editing. PZ and GZ: data curation and investigation. YZ and QG: methodology. GC: designed the work and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Shandong key research and development program (No. 2019GSF107070 and ZR2015EM018) and the Academic Promotion Programme of Shandong First Medical University (LJ001). We thank Dr. Susan T. Howard for critical reading of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.616364/full#supplementary-material
Abdelhamed, H., Ramachandran, R., Ozdemir, O., Waldbieser, G., and Lawrence, M. L. (2019). Characterization of a novel conjugative plasmid in Edwardsiella piscicida strain MS-18-199. Front. Cell. Infect. Microbiol. 9:404. doi: 10.3389/fcimb.2019.00404
Baek, K., and Jeon, C. O. (2015). Rheinheimera aestuari sp. nov., a marine bacterium isolated from coastal sediment. Int. J. Syst. Evol. Microbiol. 65, 2640–2645. doi: 10.1099/ijs.0.000312
Baranowski, E., Dordet-Frisoni, E., Sagne, E., Hygonenq, M. C., Pretre, G., Claverol, S., et al. (2018). The integrative conjugative element (ICE) of Mycoplasma agalactiae: key elements involved in horizontal dissemination and influence of coresident ICEs. mBio 9, e00873–e00918. doi: 10.1128/mBio.00873-18
Bertelli, C., Laird, M. R., Williams, K. P., Simon Fraser University Research Computing Group Lau, B. Y., Hoad, G., et al. (2017). Islandviewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Buschmann, A. H., Tomova, A., Lopez, A., Maldonado, M. A., Henriquez, L. A., Ivanova, L., et al. (2012). Salmon aquaculture and antimicrobial resistance in the marine environment. PLoS One 7:e42724. doi: 10.1371/journal.pone.0042724
Canovas, D., Cases, I., and De Lorenzo, V. (2003). Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 5, 1242–1256. doi: 10.1111/j.1462-2920.2003.00463.x
Chen, J., Bhattacharjee, H., and Rosen, B. P. (2015). ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol. Microbiol. 96, 1042–1052. doi: 10.1111/mmi.12988
Chen, W. M., Chen, W. T., Young, C. C., and Sheu, S. Y. (2019). Rheinheimera riviphila sp. nov., isolated from a freshwater stream. Arch. Microbiol. 201, 919–926. doi: 10.1007/s00203-019-01657-5
CLSI (2017). Performance standards for antimicrobial susceptibility testing. 27th Edn. Pennsylvania: CLSI.
Dang, H., Zhang, X., Song, L., Chang, Y., and Yang, G. (2007). Molecular determination of oxytetracycline-resistant bacteria and their resistance genes from mariculture environments of China. J. Appl. Microbiol. 103, 2580–2592. doi: 10.1111/j.1365-2672.2007.03494.x
Del Canto, F., Valenzuela, P., Cantero, L., Bronstein, J., Blanco, J. E., Blanco, J., et al. (2011). Distribution of classical and nonclassical virulence genes in enterotoxigenic Escherichia coli isolates from Chilean children and tRNA gene screening for putative insertion sites for genomic islands. J. Clin. Microbiol. 49, 3198–3203. doi: 10.1128/JCM.02473-10
Du, J., Zhao, H., Liu, S., Xie, H., Wang, Y., and Chen, J. (2017). Antibiotics in the coastal water of the south yellow sea in China: occurrence, distribution and ecological risks. Sci. Total Environ. 595, 521–527. doi: 10.1016/j.scitotenv.2017.03.281
Fekih, I. B., Zhang, C., Li, Y. P., Zhao, Y., Alwathnani, H. A., Saquib, Q., et al. (2018). Distribution of arsenic resistance genes in prokaryotes. Front. Microbiol. 9:2473. doi: 10.3389/fmicb.2018.02473
Fu, J., Zhong, C., Zhang, P., Zong, G., Liu, M., and Cao, G. (2020). Novel mobilizable genomic island GEI-D18A mediates conjugational transfer of antibiotic resistance genes in the multidrug-resistant strain Rheinheimera sp. D18. Front. Microbiol. 11:627. doi: 10.3389/fmicb.2020.00627
Garcia-Aljaro, C., Riera-Heredia, J., and Blanch, A. R. (2014). Antimicrobial resistance and presence of the SXT mobile element in Vibrio spp. isolated from aquaculture facilities. New Microbiol. 37, 339–346.
Han, Q. F., Zhao, S., Zhang, X. R., Wang, X. L., Song, C., and Wang, S. G. (2020). Distribution, combined pollution and risk assessment of antibiotics in typical marine aquaculture farms surrounding the yellow sea, North China. Environ. Int. 138:105551. doi: 10.1016/j.envint.2020.105551
Hoa, P. T. P., Nonaka, L., Viet, P. H., and Suzuki, S. (2008). Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of North Vietnam. Sci. Total Environ. 405, 377–384. doi: 10.1016/j.scitotenv.2008.06.023
Jiang, H., Tang, S., Qin, D., Chen, Z., Wang, J., Bai, S., et al. (2015). Heavy metals in sea cucumber juveniles from coastal areas of bohai and yellow seas, North China. Bull. Environ. Contam. Toxicol. 94, 577–582. doi: 10.1007/s00128-014-1432-1
Kumar, A., Bajaj, A., Kumar, R. M., Kaur, G., Kaur, N., Singh, N. K., et al. (2015). Taxonomic description and genome sequence of Rheinheimera mesophila sp. nov., isolated from an industrial waste site. Int. J. Syst. Evol. Microbiol. 65, 3666–3673. doi: 10.1099/ijsem.0.000471
Liang, T., Ke, Z., Chen, Q., Liu, L., and Chen, G. (2014). Degradation of roxarsone in a silt loam soil and its toxicity assessment. Chemosphere 112, 128–133. doi: 10.1016/j.chemosphere.2014.03.103
Liu, Y., Jiang, J. T., Xu, C. J., Liu, Y. H., Song, X. F., Li, H., et al. (2012). Rheinheimera longhuensis sp. nov., isolated from a slightly alkaline lake, and emended description of genus Rheinheimera Brettar et al. 2002. Int. J. Syst. Evol. Microbiol. 62, 2927–2933. doi: 10.1099/ijs.0.036020-0
Liu, M., Li, X., Xie, Y., Bi, D., Sun, J., Li, J., et al. (2019). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47, D660–D665. doi: 10.1093/nar/gky1123
Liu, H. L., and Zhu, J. (2010). Analysis of the 3' ends of tRNA as the cause of insertion sites of foreign DNA in Prochlorococcus. J. Zhejiang Univ. Sci. B 11, 708–718. doi: 10.1631/jzus.B0900417
Mengoni, A., Maida, I., Chiellini, C., Emiliani, G., Mocali, S., Fabiani, A., et al. (2014). Antibiotic resistance differentiates Echinacea purpurea endophytic bacterial communities with respect to plant organs. Res. Microbiol. 165, 686–694. doi: 10.1016/j.resmic.2014.09.008
Miazek, K., Iwanek, W., Remacle, C., Richel, A., and Goffin, D. (2015). Effect of metals, metalloids and metallic nanoparticles on microalgae growth and industrial product biosynthesis: a review. Int. J. Mol. Sci. 16, 23929–23969. doi: 10.3390/ijms161023929
Nakayama, T., Hoa, T. T. T., Harada, K., Warisaya, M., Asayama, M., Hinenoya, A., et al. (2017). Water metagenomic analysis reveals low bacterial diversity and the presence of antimicrobial residues and resistance genes in a river containing wastewater from backyard aquacultures in the Mekong Delta, Vietnam. Environ. Pollut. 222, 294–306. doi: 10.1016/j.envpol.2016.12.041
O’Connor, B. R., Perry, B. J., and Yost, C. K. (2015). Draft genome sequence of Rheinheimera sp. KL1, isolated from a freshwater lake in southern Saskatchewan, Canada. Genome Announc. 3, e01177–e01215. doi: 10.1128/genomeA.01177-15
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Panda, A. N., Ray, L., Mishra, S. R., and Raina, V. (2020). Rheinheimera pleomorphica sp. nov., a novel alkali-tolerant bacteria isolated from chilika lake, India. Curr. Microbiol. 77, 158–165. doi: 10.1007/s00284-019-01802-9
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088–e00117. doi: 10.1128/CMR.00088-17
Presta, L., Bosi, E., Fondi, M., Maida, I., Perrin, E., Miceli, E., et al. (2017). Phenotypic and genomic characterization of the antimicrobial producer Rheinheimera sp. EpRS3 isolated from the medicinal plant Echinacea purpurea: insights into its biotechnological relevance. Res. Microbiol. 168, 293–305. doi: 10.1016/j.resmic.2016.11.001
Rahman, M. T., and De Ley, M. (2017). Arsenic induction of metallothionein and metallothionein induction against arsenic cytotoxicity. Rev. Environ. Contam. Toxicol. 240, 151–168. doi: 10.1007/398_2016_2
Rico, A., Jacobs, R., Van Den Brink, P. J., and Tello, A. (2017). A probabilistic approach to assess antibiotic resistance development risks in environmental compartments and its application to an intensive aquaculture production scenario. Environ. Pollut. 231, 918–928. doi: 10.1016/j.envpol.2017.08.079
Riley, L. W. (2014). Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 20, 380–390. doi: 10.1111/1469-0691.12646
Serrato-Gamino, N., Salgado-Lora, M. G., Chavez-Moctezuma, M. P., Campos-Garcia, J., and Cervantes, C. (2018). Analysis of the ars gene cluster from highly arsenic-resistant Burkholderia xenovorans LB400. World J. Microbiol. Biotechnol. 34:142. doi: 10.1007/s11274-018-2526-4
Shen, Z., Han, J., Wang, Y., Sahin, O., and Zhang, Q. (2013). The contribution of ArsB to arsenic resistance in Campylobacter jejuni. PLoS One 8:e58894. doi: 10.1371/journal.pone.0058894
Siguier, P., Varani, A., Perochon, J., and Chandler, M. (2012). Exploring bacterial insertion sequences with ISfinder: objectives, uses, and future developments. Methods Mol. Biol. 859, 91–103. doi: 10.1007/978-1-61779-603-6_5
Song, L., Pan, Y., Chen, S., and Zhang, X. (2012). Structural characteristics of genomic islands associated with GMP synthases as integration hotspot among sequenced microbial genomes. Comput. Biol. Chem. 36, 62–70. doi: 10.1016/j.compbiolchem.2012.01.001
Suarez, C., Ratering, S., Geissler-Plaum, R., and Schnell, S. (2014). Rheinheimera hassiensis sp. nov. and Rheinheimera muenzenbergensis sp. nov., two species from the rhizosphere of Hordeum secalinum. Int. J. Syst. Evol. Microbiol. 64, 1202–1209. doi: 10.1099/ijs.0.061200-0
Sun, M., Chang, Z., Van Den Brink, P. J., Li, J., Zhao, F., and Rico, A. (2016). Environmental and human health risks of antimicrobials used in Fenneropenaeus chinensis aquaculture production in China. Environ. Sci. Pollut. Res. Int. 23, 15689–15702. doi: 10.1007/s11356-016-6733-y
Villadangos, A. F., Fu, H. L., Gil, J. A., Messens, J., Rosen, B. P., and Mateos, L. M. (2012). Efflux permease CgAcr3-1 of Corynebacterium glutamicum is an arsenite-specific antiporter. J. Biol. Chem. 287, 723–735. doi: 10.1074/jbc.M111.263335
Wang, Q., Sun, Y. W., Liu, J., and Zhang, D. C. (2018). Rheinheimera marina sp. nov., isolated from a deep-sea seamount. Int. J. Syst. Evol. Microbiol. 68, 266–270. doi: 10.1099/ijsem.0.002496
Williams, K. P. (2002). Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 30, 866–875. doi: 10.1093/nar/30.4.866
Xiao, C., Jian, H., Chen, L., Liu, C., Gao, H., Zhang, C., et al. (2017). Toxic metal pollution in the yellow sea and bohai sea, China: distribution, controlling factors and potential risk. Mar. Pollut. Bull. 119, 381–389. doi: 10.1016/j.marpolbul.2017.03.027
Xie, X., and Cheng, H. (2019). A simple treatment method for phenylarsenic compounds: oxidation by ferrate (VI) and simultaneous removal of the arsenate released with in situ formed Fe(III) oxide-hydroxide. Environ. Int. 127, 730–741. doi: 10.1016/j.envint.2019.03.059
Xu, Y., Wang, C., Zhang, G., Tian, J., Liu, Y., Shen, X., et al. (2017). ISCR2 is associated with the dissemination of multiple resistance genes among Vibrio spp. and Pseudoalteromonas spp. isolated from farmed fish. Arch. Microbiol. 199, 891–896. doi: 10.1007/s00203-017-1365-2
Yang, H. C., Fu, H. L., Lin, Y. F., and Rosen, B. P. (2012). Pathways of arsenic uptake and efflux. Curr. Top. Membr. 69, 325–358. doi: 10.1016/B978-0-12-394390-3.00012-4
Keywords: antibiotic resistance, arsenic resistance, ICERspD18B, integrative and conjugative element, Rheinheimera
Citation: Fu J, Zhong C, Zhang P, Gao Q, Zong G, Zhou Y and Cao G (2020) A Novel Mobile Element ICERspD18B in Rheinheimera sp. D18 Contributes to Antibiotic and Arsenic Resistance. Front. Microbiol. 11:616364. doi: 10.3389/fmicb.2020.616364
Received: 12 October 2020; Accepted: 30 November 2020;
Published: 18 December 2020.
Edited by:
Marina Spinu, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Jian Chen, Florida International University, United StatesCopyright © 2020 Fu, Zhong, Zhang, Gao, Zong, Zhou and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangxiang Cao, Y2FvemhvbmcwNDAyQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.