
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 December 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.611912
This article is part of the Research Topic Exposure, Risks, and Drivers of the Mobile Antimicrobial Resistance Genes in the Environment – a Global Perspective View all 17 articles
Gastrointestinal bacteria that harbor antibiotic resistance genes (ARG) become enriched with antibiotic use. Livestock manure application to cropland for soil fertility presents a concern that ARG and bacteria may proliferate and be transported in the environment. In the United States, manure applications typically occur during autumn with slow mineralization until spring planting season. A laboratory soil incubation study was conducted mimicking autumn swine manure application to soils with concentrations of selected ARG monitored during simulated 120-day winter incubation with multiple freeze-thaw events. Additionally, the effects of two soil moistures [10 and 30% water holding capacity (WHC)] and two manure treatments [raw versus hydrated lime alkaline stabilization (HLAS)] were assessed. Fourteen tetracycline resistance genes were evaluated; tet(D), tet(G), and tet(L) were detected in background soil while swine manure contained tet(A), tet(B), tet(C), tet(G), tet(M), tet(O), tet(Q), and tet(X). By day 120, the manure-borne tet(M) and tet(O) were still detected while tet(C), tet(D), tet(L), and tet(X) genes were detected less frequently. Other tet resistance genes were detected rarely, if at all. The sum of unique tet resistance genes among all treatments decreased during the incubation from an average of 8.9 to 3.8 unique tet resistance genes. Four resistance elements, intI1, blactx–m–32, sul(I), erm(B), and 16s rRNA genes were measured using quantitative PCR. ARG abundances relative to 16S abundance were initially greater in the raw manure compared to background soil (−1.53 to −3.92 log abundance in manure; −4.02 to <−6.7 log abundance in soil). In the mixed manure/soil, relative abundance of the four resistance elements decreased (0.87 to 1.94 log abundance) during the incubation largely because 16S rRNA genes increased by 1.21 log abundance. Throughout the incubation, the abundance of intI1, blactx–m–32, sul(I), and erm(B) per gram in soil amended with HLAS-treated manure was lower than in soil amended with raw manure. Under low initial soil moisture conditions, HLAS treatment reduced the abundance of intI1 and resulted in loss of blactx–m–32, sul(I), and erm(B)] compared to other treatment-moisture combinations. Although one might expect antibiotic resistance to be relatively unchanged after simulated winter manure application to soil, a variety of changes in diversity and relative abundance can be expected.
Manure from animal production is a valuable source of fertilizer nutrients and organic matter for crop production offsetting the need for chemical fertilizer inputs and enhancing many soil attributes (Edmeades, 2003). However, manure management presents potential challenges, including odors, runoff, greenhouse gas emissions, and pathogens when best management practices are not used. Impairment of surface waters with zoonotic pathogens that may be associated with manure application have lent more focus toward pathogen control/inactivation prior to and during manure application to cropland. Contributions from livestock manure to antibiotic resistance in environmental matrices presents a growing concern about the fate of resistant microorganisms or the dissemination of antibiotic resistance genes (ARG) during manure treatment or following manure application to soil (Durso and Cook, 2014; Topp et al., 2018). The use of manure as a soil and crop fertility input could represent an important contribution of antibiotic resistant bacteria (ARB) and ARG to the soil environment (Franklin et al., 2016).
Croplands receiving manure application contain indigenous soil microorganisms that normally harbor a low, yet diverse, abundance of background of ARG (Durso et al., 2016). Once in the soil environment, ARGs originating from animal manures or human biosolids could potentially move into indigenous soil microbial populations though horizontal gene transfer of mobile genetic elements (von Wintersdorff et al., 2016), which are often found on particularly stable plasmids that increase the chance of inheritance between species (Heuer et al., 2009). Once acquired, the low fitness cost of antibiotic resistance enables soil bacteria harboring ARGs to persist in the environment even though antibiotic pressures are no longer present (Cook et al., 2014). Although this chain of events seems exceedingly rare, Forsberg et al. (2012) identified non-pathogenic soil microorganisms containing several long resistance genes having 100% nucleotide identity to those in human pathogens indicating a possible recent transfer event.
The abundance of specific ARGs may increase, decrease, or remain unchanged overtime with continuous/long term manure inputs (Chee-Sanford et al., 2009; Mantz et al., 2013; Peng et al., 2015; Tang et al., 2015; Durso et al., 2018; McKinney et al., 2018), indicating the fate of ARB and ARG in soils is likely dependent upon repeated manure inputs and a multitude environmental factors. Proliferation of indigenous ARB with the influx of manure nutrients and substrates can enhance ARB persistence (Heuer et al., 2011; Jechalke et al., 2013; Udikovic-Kolic et al., 2014; Fang et al., 2015) but may not be as important as the manure microbes themselves (Peng et al., 2016). Manure source (cattle, swine, or poultry), environmental factors (soil type, moisture and temperature), and management decisions (tillage practices, application timing, and method, and location of manure application) also likely influence persistence of ARG in soils.
In confined swine production systems, manure is typically collected and stored (with or without treatment) for a period of 6 months to a year to reduce the occurrence of pathogens (Hill, 2003; McLaughlin et al., 2012). During manure storage/treatment, biological, chemical, and physical processes reduce pathogen load and provide some capacity to reduce, but not eliminate, ARGs (Joy et al., 2014). Untreated human biosolids have many of the same characteristics as swine manure, and a preferred method to reduce pathogen loads in human biosolids is through alkaline stabilization using hydrated lime to raise the pH of the biosolids for a sustained period of time (US Environmental Protection Agency, 2000).
Recently, a hydrated lime alkaline stabilization (HLAS) manure treatment, modeled after the alkaline stabilization used on human biosolids, has been shown to reduce porcine epidemic diarrhea virus (PEDV), an important coronavirus disease in swine (Stevens et al., 2018). Research investigating the impact of HLAS manure treatment on ARG abundance is limited; however, a survey of three resistance genes [tet(W), tet(O), and sul(I)] in manure, soil, human biosolids, and lime-stabilized human biosolids found lower abundances of these ARG in lime-stabilized biosolids (Munir and Xagoraraki, 2011). The long-term impact of manure or biosolids application on soil microbial communities in this study was less clear, with considerable farm-to-farm variability. It should be noted that soil pH is an important driver of microbial community structure (Lauber et al., 2009; Tan et al., 2020). Indeed, agricultural lime addition to soils, a practice used to raise the pH of acidic soils, also affects soil community structure (Barth et al., 2018; Lin et al., 2018) and may impact diversity and abundance of soil ARG.
In swine production areas across the midwestern United States, stored manure slurry is land applied in the fall because manure nutrients are more easily retained in the cold soil over winter and available for new crop growth in the spring. Winter application of manure is discouraged due to contaminant runoff concerns resulting from frozen, snow-covered or saturated soil. Two manure applications methods are commonly used depending upon the solids content of the manure: surface and sub-surface application. Surface application via irrigation is common where manure is stored in treatment lagoons, as the very low solids content in lagoon effluent does not interfere with spray nozzle performance. An alternative storage method used in many swine production systems to minimize manure volume and retain valuable nitrogen for crop fertilization is collection in deep pits below the production area. This “slurry” manure has higher solids content and greater concentration of nutrients and is typically injected in narrow bands into crop soils to minimize nitrogen losses via volatilization of ammonia. Manure injection produces a unique microenvironment where manure microbes and nutrients are in close contact with soil microbes. During application of liquid or slurry manure application, soil moisture also increases based upon manure water content, soil characteristics and hydrologic conditions. Application of manure to saturated soils is both difficult and discouraged as it contributes to manure runoff. Following fall manure application, low temperatures and soil freeze-thaw during winter months may impact bacterial competition, survival, and persistence of organisms and ARG.
This study examines how ARGs indigenous to either agricultural soil or swine manure change after simulated manure application and 120-day winter soil incubation with multiple freeze-thaw events, with and without manure HLAS treatment. We hypothesized that (1) HLAS treatment would have a strong immediate effect on resistance by directly limiting the survival of microorganisms; however, (2) few changes in diversity or persistence of ARG would occur over time under “winter” incubation conditions. Additionally, the effects of two initial soil moisture contents were assessed. Fourteen different tetracycline resistance genes were evaluated, and the abundance of four resistance elements both in bulk soil and relative to 16s rRNA gene abundance were measured using qPCR molecular techniques.
A silty clay loam soil was collected from a crop field in eastern Nebraska, sieved, and air dried (soil properties: 5.45 μg g–1 NO3-N, 2.43 μg g–1 NH4-N, 0.23 dS m–1 EC, 7.00 pH, 63.03 μg P g–1 Mehlich P). The WHC of the soil was determined gravimetrically (Klute, 1986) and two stock soil mixtures were prepared by thoroughly incorporating deionized water to be achieve 10 or 30% of the maximum WHC of the soil (i.e., “dry” and “moist” soil conditions, respectively). Expressed on a moisture content basis, the 10 and 30% WHC soils contained 82.6 and 247.7 g H2O kg–1 dry soil, respectively. From each soil mixture, 30 g (oven dry equivalent) soil was added to multiple 50 mL tubes. A soil cavity (simulated manure furrow) was made in the center of each soil tube by pressing a 10 mL pipet tip into the soil and tubes were immediately capped to prevent moisture loss.
Manure slurry was collected from the deep pit of a commercial swine production site in south central Nebraska and refrigerated at 4°C for less than 48 h prior to setting up the soil incubation. The day prior to setting up the incubations, manure slurry was divided into two stocks and equilibrated to room temperature (20°C). One manure slurry stock was amended with hydrated quicklime (10 g L–1 of manure slurry) yielding a final, stable pH of 11.5 and designated HLAS-treated. Although a higher pH of 12 for 2 h is recommended for biosolids treatment (US Code of Federal Regulations, 2018), an earlier study demonstrated that pH 10 for 12 h effectively controlled PEDV (Stevens et al., 2018). The other manure slurry stock (Raw) received no quicklime treatment. Both treatments were incubated at room temperature overnight since effective HLAS treatment recommends at least a 6-h exposure time at high pH. On day 0, 80 soil incubations were prepared by adding 10 g of manure slurry to the soil cavity in each vial yielding 20 replicates of each moisture (10 and 30% WHC) and manure (HLAS and raw manure) combination. Four samples of each treatment combination were immediately stored at −80°C as Day 0 samples. All other samples were capped loosely, secured in tube racks, and randomly placed in a refrigerated incubator (Fisherbrand Isotemp BOD Refrigerated Incubator, Fisher Scientific, Waltham, MA, United States). The incubator temperature was adjusted daily to simulate mean winter soil temperatures (5.1 cm depth) at the University of Minnesota Southern Research and Outreach Center in Waseca, Minnesota (Figure 1). The Waseca location was selected based upon the extensive soil temperature (daily high and low) records, Waseca’s location in an area of dense swine production, and because seasonal temperature changes spanned the range where soils froze for extended periods of time. Twice weekly during the first 10 weeks of incubation, the racks of tubes were removed from the incubator and either allowed to briefly (over 20 min) freeze in the −80°C freezer (if the current incubation temperature was above 0°C) or thaw at room temperature (if the current incubation temperature was below 0°C). The number of freeze/thaw events was selected based upon the number of freeze/thaw events observed within November, December, and March at 5 cm soil depth in soils at Waseca, MN in 2009, 2011, 2012, 2013, and 2014, and ranged from six in 2013 to 22 in 2011. At Days 30, 60, 90, and 120, four samples of each treatment combination were retrieved and stored at −80°C for subsequent DNA extraction.
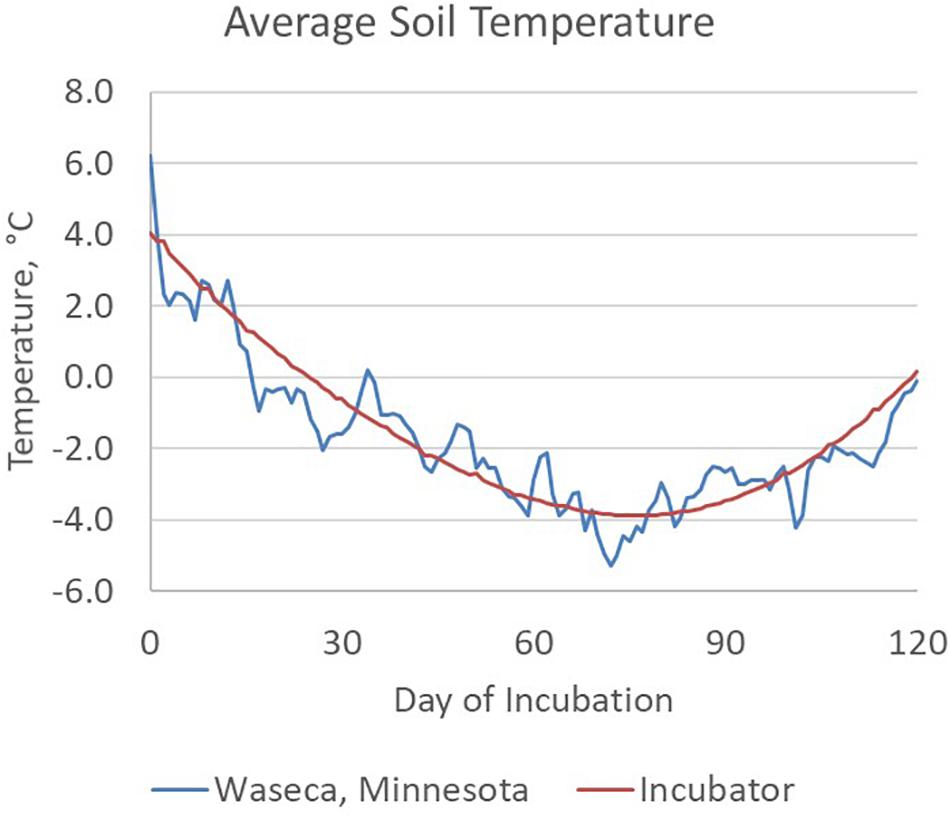
Figure 1. Average soil temperature between November 11th and March 11th calculated for a 6-year period (2009 – 2014) of field data at the Waseca, MN experimental station. Two freeze/thaw cycles were conducted each week during the first 5 weeks of the incubation, typical for surface soils at this location.
Immediately prior to DNA extraction, deionized water was thoroughly mixed into each manure/soil sample to yield a uniform sample moisture content (25% moisture content wet basis) since incubation tubes lost moisture over the 120-day incubation (determined gravimetrically). DNA was extracted from 250 mg of slurry using the PowerSoil DNA Isolation Kit (MoBio Laboratories, Solana Beach, CA, United States) according to the manufacturer’s protocol. DNA extracts were stored at −80°C until polymerase chain reaction (PCR) analysis.
A variety of target genes were assessed to gage how manure treatments and simulated winter incubation conditions affected resistance (Table 1). These target genes included “highly” and “critically important” resistance genes conferring resistance to antibiotics used in animal production. Four multiplex PCR amplification reactions for the presence/absence of fourteen tetracycline resistance genes (Table 2) were performed on each DNA extract according to the method of Ng et al. (2001), with each 25-μL reaction containing 12.5 μL Jumpstart RedTaq Master Mix and 0.5 μL of each primer. Extractions were processed in a StepOnePlus Real-Time PCR System (Applied Biosystems, Waltham, MA, United States) at settings of 5 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 55°C, 90 s at 72°C; and 5 min at 72°C. Positive controls and blanks were included in each PCR run, and PCR product was separated using agarose gel, stained (ethidium bromide), and photographed (UVP GelDoc-It TS3 Imager, Upland, CA, United States) to visualize PCR amplification products and confirm PCR product sizes.
Quantitative PCR reaction amplifications of 16s rRNA, intI1, sul(I), erm(B), and blactx–m–32 were performed according to Table 2. Standards were prepared from serial dilutions of 4-gene multiplex with 16s rRNA gene (Integrated DNA Technologies, Coralville, IA, United States). Each 5-μL volume of standard contained from 101 to 108 gene copies. Standards and reagent blanks were included in each run. All samples were quantified in triplicate and averaged. Standard curve R2 ranged from 0.960 to 0.998 while efficiency ranged from 80 to 117%.
Equality of proportions tests and ANOVA were performed using SAS statistical software (SAS Institute, Cary, NC, United States). The two-sample test of equality of proportions was chosen to generate probabilities related to the significance of initial soil moisture and manure treatment on the percent of each tetracycline resistance gene. Analysis of variance was used to evaluate the significance of antecedent soil moisture and manure treatment on (i) the diversity of tetracycline resistance genes present over time (ii) the log abundance of intI1, sul(I), blaCTX–M–32, and erm(B) in soil, and (iii) the log abundance of of intI1, sul(I), blaCTX–M–32, and erm(B) normalized to 16s rRNA gene abundance. A P-value of <0.05 was considered significant.
Eight tet resistance genes were detected in the raw manure while background soil revealed three tet resistance genes (Table 3). There was no difference in the detection of tet resistance genes between HLAS-treated and raw manure. The tet(G) gene was detected in both soil and manure samples, while tet(E), tet(K), tet(S), and tet(A/P) genes were not initially detected in either soil or manure. Detections between replicate soil or manure samples did not differ; tet genes were either detected in 100% of samples or not detected at all.
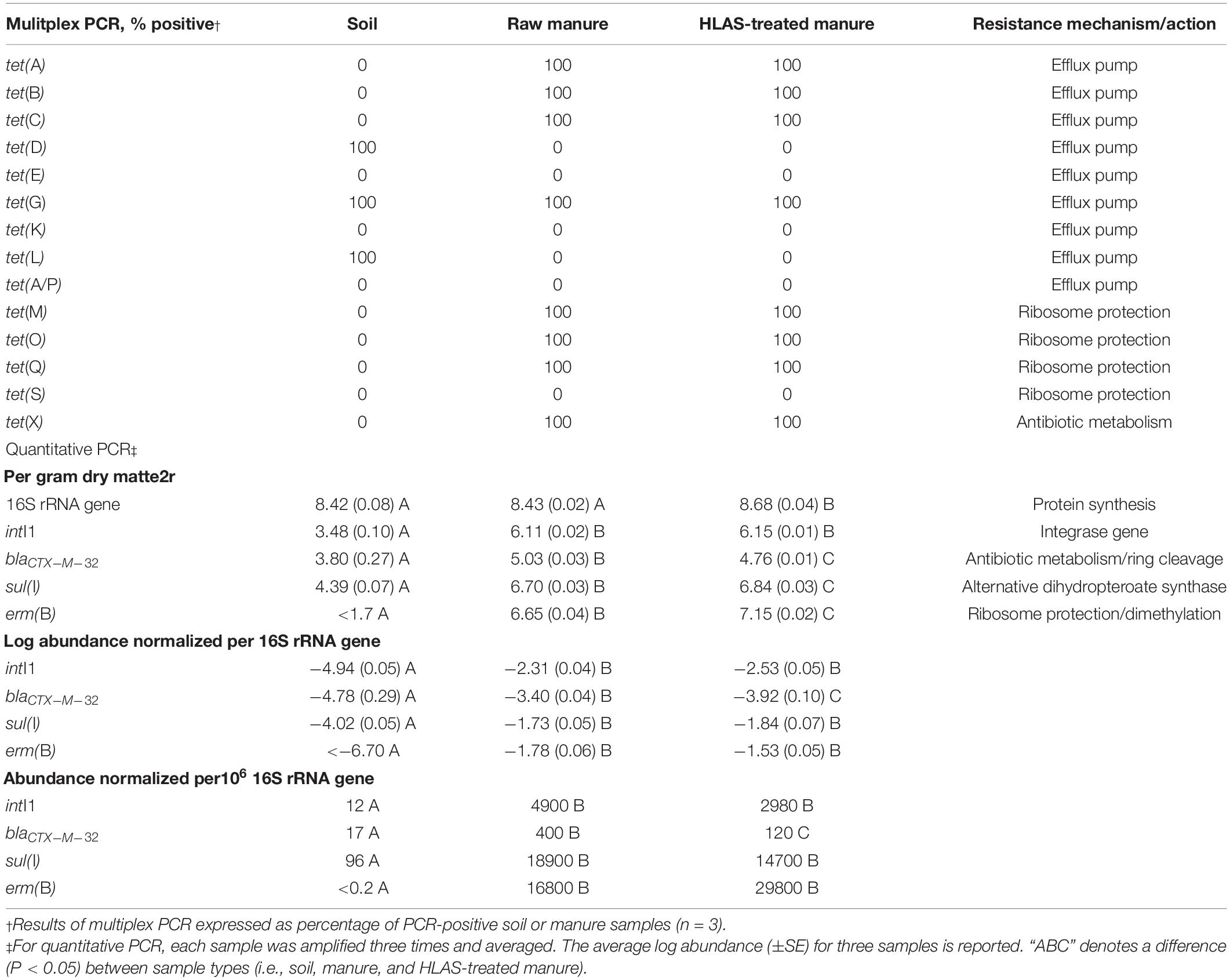
Table 3. Initial resistances in soil, manure slurry, and hydrated lime alkaline stabilized (HLAS) manure slurry assessed by multiplex PCR of tetracycline resistance genes and quantitative PCR of select resistance elements.
From a quantitative perspective (Table 3), microbial abundance (based upon 16S rRNA genes per gram) in initial sources of raw manure and soil were similar (8.43 and 8.42 log abundance, respectively) with 16S rRNA genes in HLAS-treated manure slightly higher (8.68 log abundance) than the raw manure and soil. Comparing soil to manure, soil contained much lower (10- to 100-fold) abundances of intI1, blactx–m–32, and sul(I) compared to manure (raw and HLAS-treated). Furthermore, erm(B) was not detected (50 targets per gram detection limit) in soil but was detected in the manure at 6.65 and 7.15 log abundance in raw and HLAS-treated manure, respectively. Comparing manures, the abundance of sul(I) and erm(B) were slightly higher while blactx–m–32 was slightly lower in HLAS-treated manure. Normalizing to the number of 16S rRNA genes, the differences between soil and manures remained consistent. Likewise, the significant differences observed between manures for sul(I) and erm(B), when compared on a mass basis, were insignificant when normalized to 16S rRNA gene abundance.
The multiplex PCR results revealed gene-specific persistence patterns (Table 4). Some genes were unaffected and consistently present over time. For instance, tet(M) and tet(O) genes originating in the manure were present with 100% frequency in all samples, regardless of treatment or time. Other tet genes [i.e., tet(E), tet(L), and tet(S)] were detected very infrequently or only sporadically with no obvious differences attributed to soil moisture, manure treatment, or temporal trend. Other genes [i.e., tet(G), tet(K), tet(A/P), and tet(Q)] detected initially or during the incubation were absent by Day 120 of the incubations. The frequency of tet(X) detection in soils receiving HLAS-treated manure was significantly lower than in samples receiving raw manure. Specifically, 75 to 100% of the soil reps receiving HLAS-treated manure were positive for tet(X) on Day 0, but just one of 24 reps (3.5%) were positive for tet(X) after Day 30. In comparison, the proportion of tests positive for tet(X) in soil receiving raw manure slurry after Day 30 (20 detections out of 24 tests, or 83%) was much greater (P < 0.0001). Similarly, the low-moisture soil (10% WHC) receiving raw manure retained tet(A), tet(B), and tet(C) at a greater frequency (P < 0.001) during the incubation period (69, 87, and 100%, respectively, after Day 0) when compared to higher moisture soil receiving raw manure (6, 19, and 31%, respectively, after Day 0).
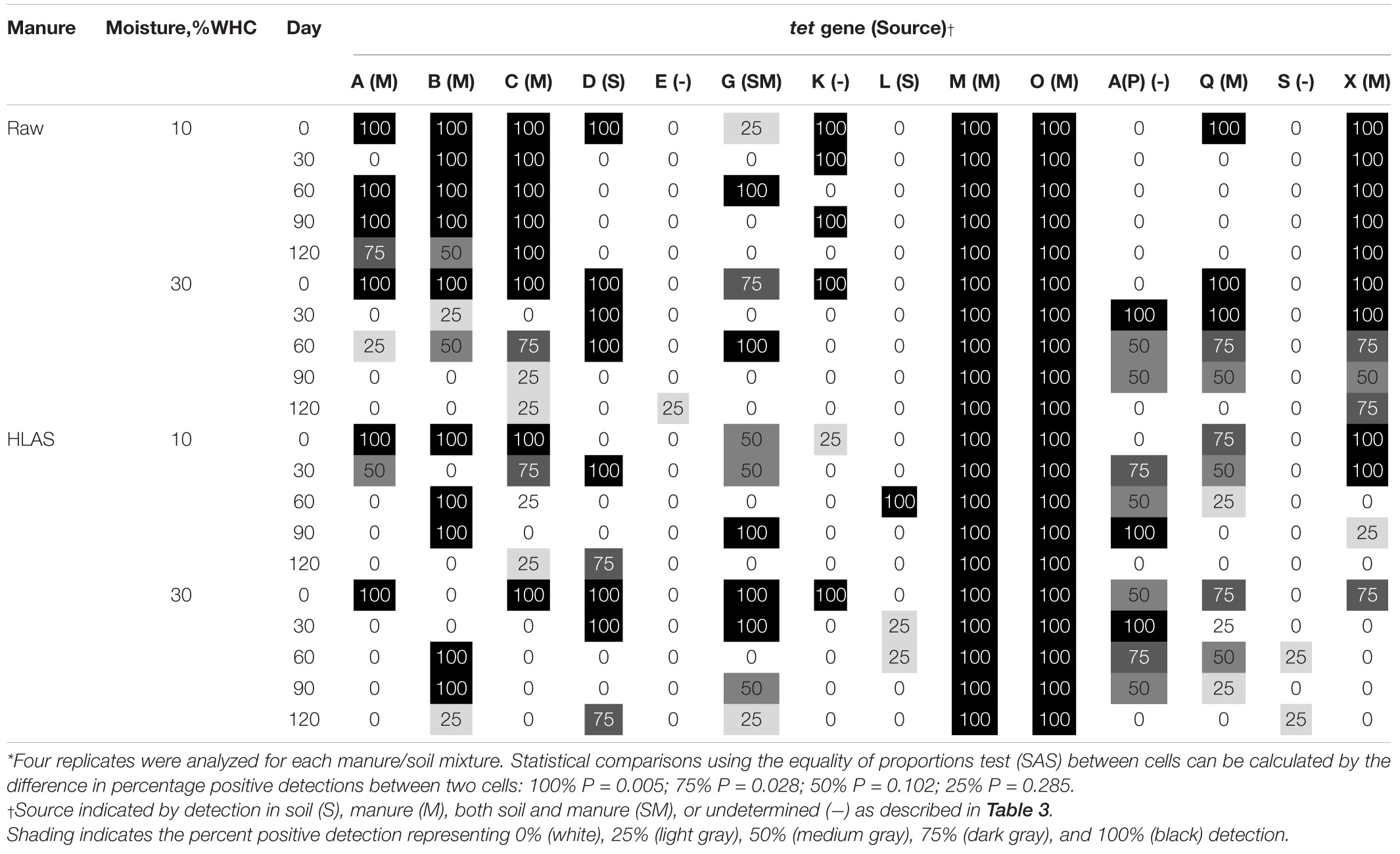
Table 4. Percent positive detection of tet genes in soil at 10 or 30% water holding capacity (WHC) with either raw manure or hydrated lime alkaline stabilized (HLAS) manure during simulated winter incubation using multiplex PCR*.
Comparing tet resistance gene prevalence on the final day of the incubation, the occurrence of tet(M) and tet(O) were unaffected by manure treatment or soil moisture (100% of samples were positive). Manure treatment affected the frequency of detection of two tet resistance genes; tet(X) gene was less prevalent (0% in soil receiving HLAS-treated manure vs. 75% and 100% in the low and high moisture content soils, respectively, receiving raw manure; P ≤ 0.028), but tet(D) was more prevalent in soil receiving HLAS-treated manure (75% detection frequency) than in soil receiving raw manure (0% detection frequency); (P = 0.028) on Day 120. Soil moisture content alone did not yield a consistent effect on the frequency of tet resistance gene detection; however, the interaction of low soil moisture (10% WHC) and application of raw manure yielded greater frequency of tet(A), tet(B), and tet(C) retention (detection frequencies of 75, 50, and 100%, respectively), in samples on Day 120. In comparison, tet(A), tet(B), and tet(C) were found at a frequency of 25% or less in other treatment combinations. Statistically significant differences between any two treatment combinations/sampling dates depended upon the difference in percentage positive. Differences between 25 or 50% detection frequencies were not significant (P = 0.285 and 0.102, respectively), while differences between 75 or 100% detection frequencies were significant (P = 0.028 and 0.005, respectively) based upon four replicates tested per treatment combination.
Finally, the overall diversity of tet resistance genes among all treatment combinations declined during the simulated incubation period (Figure 2). The average number of different tet resistance genes detected in any treatment combination ranged from 7.5 to 9.75 on Day 0 but declined to a range of 3 to 5.25 by Day 120 (P < 0.001). Comparisons made on individual sampling days revealed that soil at initial 10% WHC receiving raw manure had greater diversity of tet resistance genes (P < 0.05) on Day 90 and 120 compared to all other treatments (P = 0.021). On Day 60, the diversity of tet resistance genes in soil receiving raw manure (at initial 10 and 30% WHC) were greater (P < 0.03) than in the soil receiving HLAS-treated manure.
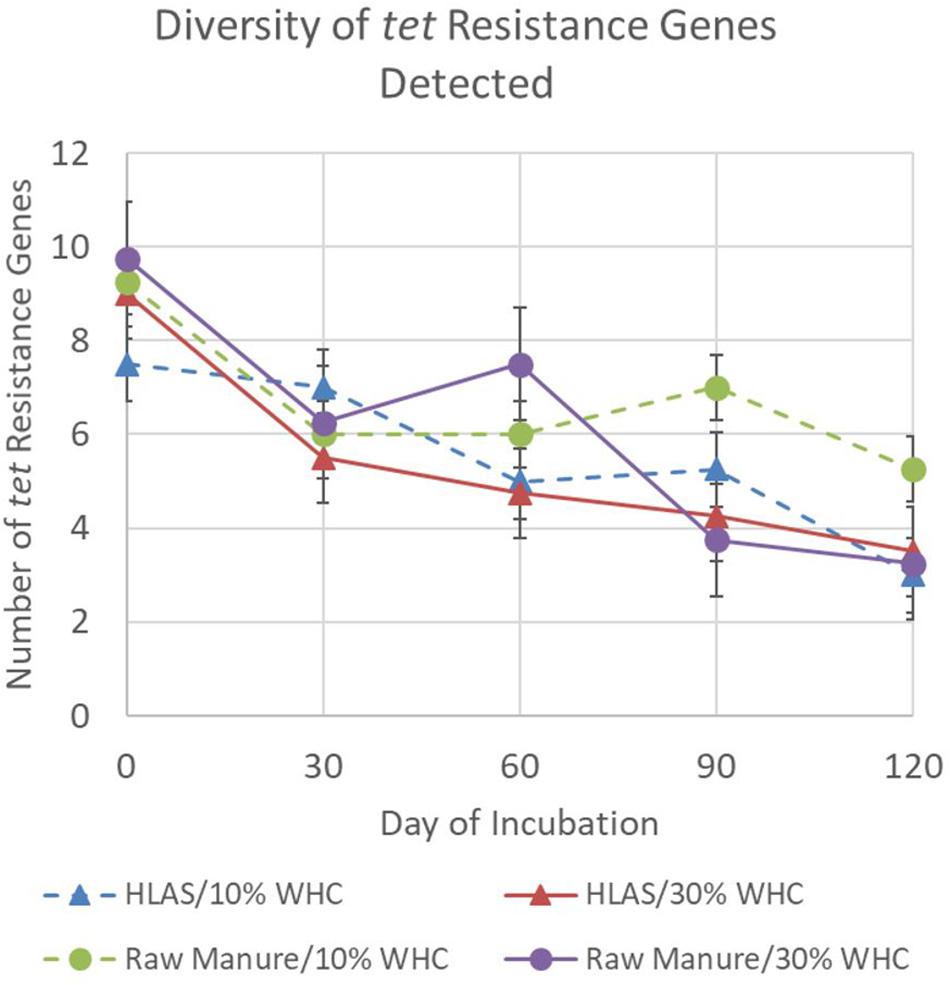
Figure 2. Number of different tetracycline resistance genes detected using the multiplex PCR method for the four treatment combinations, hydrated lime alkaline stabilized (HLAS) swine manure slurry (triangles) or raw swine manure slurry (circles) in soil originally at 10% (dashed line) or 30% (solid line) water holding capacity (WHC). Error bars = standard error.
The abundances of intI1, blaCTX–M–32, sul(I), erm(B), and 16S rRNA genes over the 120-day incubation period, when expressed on a per gram of dry slurry basis, varied among genes (Figure 3, left hand panels). Depending on the treatment combination, abundances could be (i) relatively stable [sul(I)], (ii) increasing over the incubation period (16S rRNA genes and intI1 in soil at 10% WHC receiving raw manure), (iii) decreasing slowly over the incubation period [erm(B)], or (iv) quite dynamic (blaCTX-M-32). Of the genes assessed, the abundance of the16S rRNA gene demonstrated the most consistent pattern (CV = 2.26 to 5.61%) regardless of treatments, increased through time, ranging from 7.86 to 8.11 log abundance copies per gram of dried manure/soil on Day 0 to 8.93 to 9.37 log abundance on Day 120.
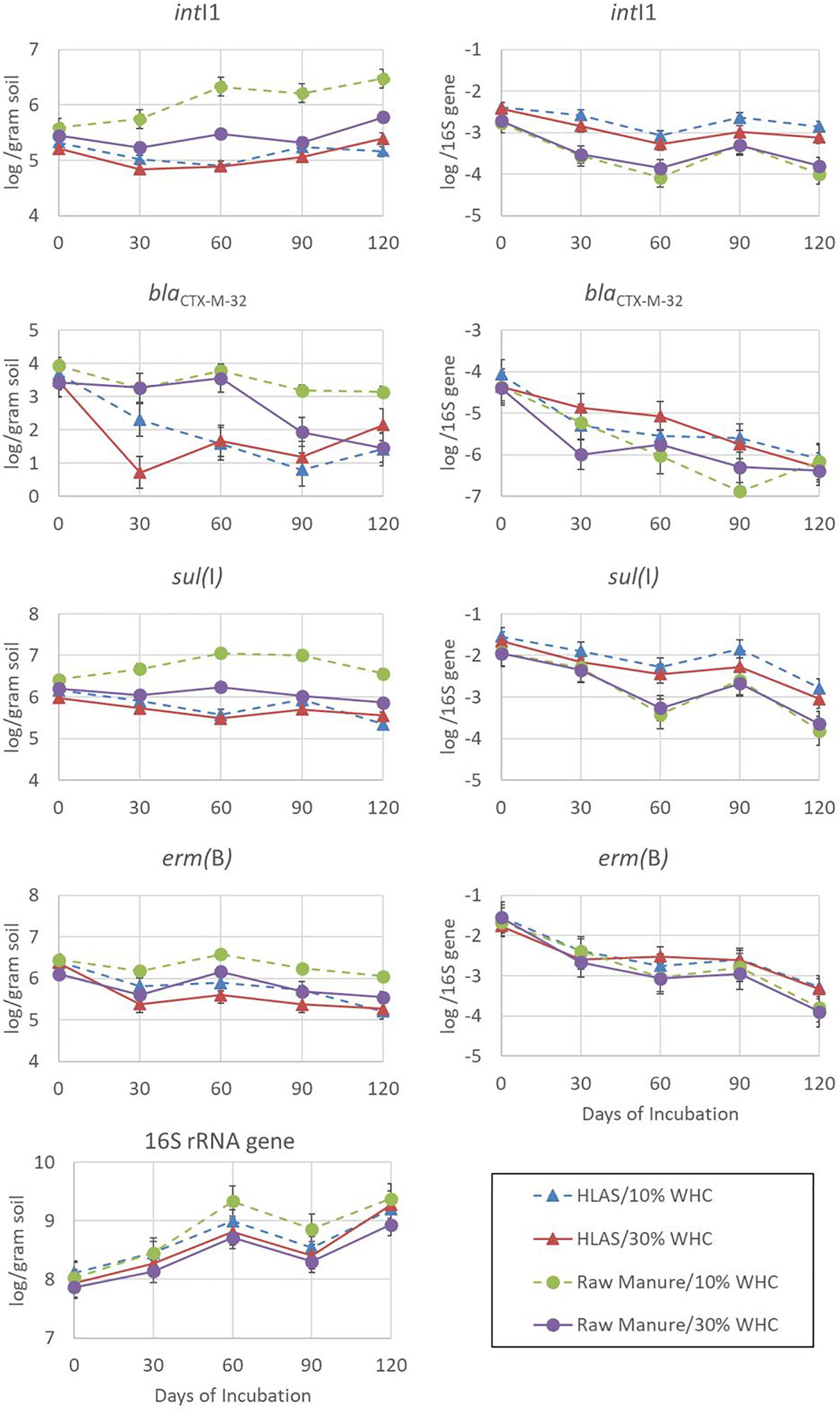
Figure 3. Abundance determined using quantitative polymerase chain reaction of resistance elements intI1, blaCTX–M–32, sul(I), and erm(B) in addition to 16S rRNA genes over the 120-day simulated winter incubation. The four treatment combinations were hydrated lime alkaline stabilized (HLAS) swine manure slurry (triangles) or raw swine manure slurry (circles) in soil originally at 10% (dashed line) or 30% (solid line) water holding capacity (WHC) and are expressed in terms of log abundance per gram of dry soil (left-hand column of figures) or log abundance per 16S rRNA gene (right-hand column of figures). Error bars = standard error.
Comparing among the four treatment combinations, soil at 10% WHC receiving raw manure yielded roughly 10-fold greater abundances of intI1, blaCTX–M–32, and sul(I) on Day 120 compared to all other treatment combinations. The result for erm(B) was consistent with other resistance genes and was 0.5 log abundance greater for soil at 10% WHC receiving raw manure than the other treatments. The abundance of resistance elements in soil receiving HLAS-treated manure was consistently lower throughout the incubation period compared to soils receiving raw manure; initial soil moisture did not have a large effect on abundance of resistance elements in soil receiving HLAS-treated manure. However, initial soil moisture did impact the abundance of resistance elements in soil receiving raw manure, with lower resistance gene abundances (P = 0.002) noted at the 30% WHC level compared to the 10% WHC level (5.54 versus 6.05 log abundance, respectively). There was no evidence of a similar synergistic effect for higher moisture soil receiving HLAS-treated manure.
Normalizing resistance elements to 16s rRNA genes helps to account for unequal initial microbial abundances and changes in the microbial population throughout the incubation. In this study, a very consistent pattern of decreasing resistance element abundance throughout the incubation period is evident (Figure 3, right hand panels). Although the abundance of an individual resistance element may have increased slightly during incubation (i.e., intI1 for soil at 10% WHC receiving raw manure), the proportion of the resistance element normalized to 16s rRNA genes in the microbial population decreased 10-fold (1 log). Interestingly, intI1 abundance in soil receiving HLAS-treated manure was only slightly reduced (roughly 0.5 log) relative to 16S rRNA. For resistance elements that showed decreasing abundance per gram of dried slurry [i.e., erm(B) and some blaCTX–M–32 treatments], the relative reduction was even greater (100-fold or 2 logs).
Manure injection is a common swine manure slurry application practice across the United States. When examined at a fine scale (<1–2 cm), this likely has a profound effect on the immediate soil environment and microbial processes. Considering the issue of antibiotic resistance, the narrow band of injected manure introduces a suite of very diverse tet resistance genes and resistance elements into the soil at abundances 10- to 100-fold greater than background soil concentrations. One might expect at very low winter-time temperatures that there would be limited microbial growth and very little microbial activity, having little impact on antibiotic resistance. However, studies on wheat straw decomposition at low temperatures indicate that although the rates of CO2 evolution are decreased, they are still substantial (Stott et al., 1986). In a more recent study, respiration rates measured at 0°C in agricultural and a humus forest soils from southern Sweden were positive and not predicted to reach zero respiration until −6°C (Pietikäinen et al., 2005). Supporting evidence from Arctic and Antarctic soils demonstrates that microorganisms mediate nutrient and carbon cycles at these very low temperatures (Eriksson et al., 2001; Kotsyurbenko, 2005). Stott et al. (1986) speculate that salts in soils may lower the freezing point of water enabling limited microbial decomposition. At the fine scale of manure injection bands in crop fields, this may be particularly important since swine manure slurries usually have high salinity and could depress the freezing point well below 0°C. The results of the simulated winter incubation presented herein, which included multiple freeze/thaw events, indicate that resistance in soil can be substantially altered even during the winter climate experienced in southwestern Minnesota, a region of extensive swine production.
Assessing a suite of tetracycline resistance genes through multiplex PCR reactions showed that both their occurrence and diversity were impacted by the simulated winter incubation. Ultimately, the presence of tet resistance genes on Day 120 may be the most important indicator of how soil moisture and manure treatment affect tetracycline resistance after a simulated winter incubation. From this perspective, the results are more easily interpreted. Particular tet resistance genes in the swine manure [tet(M) and tet(O)] showed no change after 120 days, yet others [tet(K), tet(L), tet(A/P), and tet(Q)] present in the initial samples or occasionally detected during the incubation were no longer detected after 120 days. Furthermore, the persistence of other tet resistance genes [tet(A), tet(B), tet(C), tet(D), and tet(X)] were affected by application of HLAS-treated manure and/or initial soil moisture and were quite dynamic, disappearing entirely from some of the treatments. Finally, assessed tet resistance gene diversity illustrated a consistent decline in all the treatments during the simulated winter incubation, but less of a decline in the drier soil receiving raw manure. It is possible that the slightly lower moisture content provided conditions where manure microbes harboring tet(A), tet(B), and tet(C) were able to persist within the soil microbial community. An alternative hypothesis is that the increased water content enhanced ice crystal formation during freezing, which increased the likelihood that manure bacteria were ruptured during multiple freeze/thaw events. Clearly additional research needs to be done to explore these hypotheses.
Converting the initial log abundances of resistance elements intI1, blaCTX–M–32, sul(I), and erm(B) to numbers of targets observed per million copies of the 16S rRNA gene revealed that resistance elements in the soil were much less abundant (<100 copies) compared to those in manures, which contained hundreds to tens of thousands more resistance elements per million 16S rRNA genes (Table 3). Although, intI1, blaCTX–M–32, sul(I), and erm(B) normalized to 16S rRNA abundance demonstrated a 10- to 100-fold reduction over the simulated winter incubation (Figure 3). A substantial portion of resistance element decrease could be attributed to the 10-fold increase in 16S rRNA abundance during the incubation. Although one would expect greatest increases in microbial abundance (i.e., 16S rRNA gene abundance) during the warmest periods (Day 0 to 30), the largest increases were observed from Day 30 to 60 and Day 90 to 120 when temperatures were slightly less than 0°C. One hypothesis that may explain these observations is that the series of freeze/thaw events (Day 0 to 30) lysed some microorganisms and/or released useful microbial substrates from manure solids. Cold tolerant species were then able to proliferate as substrates became available. A change in microbial community reflected in 16S rRNA sequence diversity would support this hypothesis but needs to be conducted.
The application of raw manure to soil at the lower WHC (10 vs. 30%) produced somewhat greater microbial abundance, which effectively diluted the relative abundance of most manure resistance elements for this treatment to a lower concentration. Somewhat to our surprise, even HLAS-treated manure also stimulated microbial growth. Although we presumed that the highly alkaline conditions in the manure would have reduced the capacity for microbial activity and survival, the soil environment was able to moderate the high pH in the treated swine slurry, and soil microbes were able to utilize the carbon and nutrient resources in the manure. For the resistance elements assessed using quantitative PCR in this study, nutrient addition did not preferentially stimulate proliferation of particular ARB over the simulated winter incubation, as was found by Udikovic-Kolic et al. (2014).
Specific tet resistance mechanisms in soil after the winter incubation were affected by swine manure application. Initially, only efflux pump types of resistance [tet(D), tet(G), and tet(L)] were detected in background soil, but application of swine manure introduced ribosome protection [tet(M), tet (O), and tet(Q)] and antibiotic metabolism [tet(X)] types of resistance to the soil environment. Although tet(Q) was no longer detectable by Day 120, the ribosomal protection resistance mechanism of tet(M) and tet(O) was still easily detected. Manure treatment also seemed to influence antibiotic metabolism resistance [tet(X)]. While tet(X) was still detected on Day 120 in soil receiving raw manure, tet(X) was no longer detectable in the soil receiving HLAS-treated manure. It is likely that the type of bacterial species harboring these tet resistances may be particularly sensitive to high pH and easily disrupted during HLAS.
The findings of these laboratory incubations present a unique look at changes in resistance at very low temperatures. However, these results are generally consistent with earlier studies investigating antibiotic resistance in swine manure slurry applied to soil and do not challenge consensus that the abundances of manure resistant genes may temporarily increase in the soil but decrease with time at seasonal scales. Even prior to swine manure application to soil, abundances of tet and erm genes in swine manure storage pits generally decrease by orders of magnitude as the fresh manure undergoes decomposition (Joy et al., 2014). A field study conducted by Garder et al. (2014) in Iowa examined swine manure injected into soil and may offer the closest comparison to the research presented here. The Iowa study collected data seasonally and found that the absolute abundance (per gram soil) of erm(B) and erm(F) decreased by 5 – 6 log abundance over a year. At the shortest time step, fall to spring, erm(F) absolute abundance decreased by 2 – 3 log per gram soil. Although over a short time period (65 days) and at higher temperature (25°C), another study conducted in a greenhouse found tet(C) and tet(Z) decreased from <0.5 to 2 log abundance after swine manure was initially added to soil (Kang et al., 2017). Finally, a recent study conducted using dairy manure investigated how resistance elements respond to freeze/thaw and various WHC (25 to 75%) at low (5°C) temperature over a 56-day incubation (McKinney and Dungan, 2020). Examining intI1, sul(I), tet(M), erm(B), and 16S rRNA genes, they were unable to detect erm(B) but found decreasing absolute abundance (per gram soil) of all other resistance elements at lowest temperature and for all resistance elements except for tet(M) which did not change at the lowest WHC. Freeze/thaw had little effect on resistance element absolute abundance except for sul(I) which declined slightly. One subtle difference with our findings was that 16S rRNA gene abundance did not change during the incubation at lowest temperatures (5°C) and WHC (25%). It is interesting to note that 16S rRNA abundance was already above 9 log abundance per gram soil while the abundance in this study started at 8 log abundance and ended near 9 log abundance. Would the 16S rRNA abundance in our study have continued to increase with a longer incubation time at 0°C or would it have plateaued?
Initially, we hypothesized that HLAS treatment would have a strong effect on resistance by directly affecting the viability of microorganisms, but that incubation time would yield minimal changes in resistance after the imposition of “winter” incubation conditions due to low microbial and enzymatic activity. However, our results demonstrate that the soil community even at low temperatures is quite responsive to manure nutrient input. Furthermore, resistance elements were surprisingly dynamic during a presumed inactive period.
Swine manure application introduced a diverse set of tet resistance genes into low tet resistance diversity agricultural soil, increasing the diversity from three of fourteen assessed genes to ten different tet genes in the mixed manure/soil. During a simulated winter incubation, the diversity of tetracycline resistance genes declined. However, two swine manure associated resistance genes [tet(M) and tet(O)] persisted. The persistence of other tet resistance genes [tet(A), tet(B), tet(C), tet(D), and tet(X)] were affected by HLAS treatment and initial soil moisture. Other resistance elements [intI1, blaCTX–M–32, sul(I), and erm(B)] normalized to 16S rRNA abundance demonstrated a 10- to 100-fold reduction over the simulated winter incubation, due in part to the 10-fold increase in 16S rRNA abundance, and to a limited reduction of most resistance element abundances.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
DM was the principal scientist responsible for conducting the analysis of soil and manure samples, reviewing statistical analyses, developing figures and tables, and finishing the final drafts of the manuscript. MJ was an undergraduate student and responsible for conducting lab analyses and preparing initial statistics, figures, and drafts of the manuscript. LD contributed to laboratory analysis and manuscript preparation. AS was responsible for conducting the incubations and revising manuscripts. All authors contributed to the article and approved the submitted version.
Funding was provided by the National Pork Board Project #14-269 and #14-SR-259, NIFA AFRI-CARE #2016-68008-25043, USDA Hatch Multistate Project NC-229, and USDA Agricultural Research Service Project 3042-12630-003-00D.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to thank Bethany Brittenham and Ryan McGhee for their expert laboratory assistance.
Barraud, O., Baclet, M. C., Denis, F., and Ploy, M. C. (2010). Quantitative multiplex real-time PCR for detecting class 1, 2, and 3 integrons. J. Antimicrob. Chemother. 65, 1642–1645. doi: 10.1093/jac/dkq167
Barth, V. P., Reardon, C. R., Coffey, T., Klein, A. M., McFarland, C. L., Huggins, D. R., et al. (2018). Stratification of soil chemical and microbial properties under no-till management after lime amendment. Appl. Soil Ecol. 130, 169–177. doi: 10.1016/j.apsoil.2018.06.001
CDC, (2019). Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC.
Chee-Sanford, J. C., Mackie, R. I., Koike, S., Krapac, I. G., Lin, Y., Yannarell, A. C., et al. (2009). Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Env. Qual. 38, 1086–1108. doi: 10.2134/jeq2008.0128
Cook, K. L., Netthisinghe, A. M. P., and Gilfillen, R. A. (2014). Detection of pathogens, indicators, and antibiotic resistance genes after land application of poultry litter. J. Env. Qual. 43, 1546–1558. doi: 10.2134/jeq2013.10.0432
Denman, S., and McSweeney, C. (2006). Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 58, 572–582. doi: 10.1111/j.1574-6941.2006.00190.x
Durso, L. M., and Cook, K. L. (2014). Impacts of antibiotic use in agriculture: what are the benefits and risks? Curr. Opin. Microbiol. 19, 37–44. doi: 10.1016/j.mib.2014.05.019
Durso, L. M., Miller, D. N., and Henry, C. G. (2018). Impact of vegetative treatment systems on multiple measures of antibiotic resistance in agricultural wastewater. Int. J. Environ. Res. Public Health 15:1295.
Durso, L. M., Wedin, D. A., Gilley, J. E., Miller, D. N., and Marx, D. B. (2016). Assessment of selected antibiotic resistance in ungrazed native Nebraska prairie soils. J. Env. Qual. 45, 454–462. doi: 10.2134/jeq2015.06.0280
Edmeades, D. C. (2003). The long-term effects of manures and fertilisers on soil productivity and quality: a review. Nutr. Cycl. Agroecosys. 66, 165–180.
Eriksson, M., Ka, J. O., and Mohn, W. W. (2001). Effects of low temperature and freeze-thaw cycles on hydrocarbon biodegradation in Arctic tundra soil. Appl. Environ. Microb. 67, 5107–5112. doi: 10.1128/aem.67.11.5107-5112.2001
Fang, H., Wang, H., Cai, L., and Yu, Y. (2015). Prevalence of antibiotic resistance genes and bacterial pathogens in long-term manured greenhouse soils as revealed by metagenomics survey. Env. Sci. Tech. 49, 1095–1104. doi: 10.1021/es504157v
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O. A., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
Franklin, A. M., Aga, D. S., Cytryn, E., Durso, L. M., McLain, J. E., Pruden, A., et al. (2016). Antibiotics in agroecosystems: introduction to the special section. J. Env. Qual. 45, 377–393. doi: 10.2134/jeq2016.01.0023
Garder, J. L., Moorman, T. B., and Soupir, M. L. (2014). Transport and persistence of tylosin-resistant Enterococci, erm genes, and tylosin in soil and drainage water from fields receiving swine manure. J. Env. Qual. 43, 1484–1493. doi: 10.2134/jeq2013.09.0379
Gillings, M. R., Gaze, W. H., Pruden, A., Smalla, K., Tiedje, J. M., and Zhu, Y.-G. (2015). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279. doi: 10.1038/ismej.2014.226
Heuer, H., Kopmann, C., Binh, C. T. T., Topp, E., and Smalla, K. (2009). Spreading antibiotic resistance through spread manure: characteristics of a novel plasmid type with low %G+C content. Environ. Microb. 11, 937–949. doi: 10.1111/j.1462-2920.2008.01819.x
Heuer, H., Solehati, Q., Zimmerling, U., Kleineidam, K., Schloter, M., Muller, T., et al. (2011). Accumulation of sulfonamide resistance genes in arable soils due to repeated application of manure containing sulfadiazine. Appl. Environ. Microb. 77, 2527–2530. doi: 10.1128/aem.02577-10
Hill, V. R. (2003). Prospects for pathogen reductions in livestock wastewaters: a review. Crit. Rev. Env. Sci. Tech. 33, 187–235. doi: 10.1080/10643380390814532
Jechalke, S., Kopmann, C., Rosendahl, I., Groeneweg, J., Weichelt, V., Krogerrecklenfort, E., et al. (2013). Increased abundance and transferability of resistance genes after field application of manure from sulfadiazine-treated pigs. Appl. Environ. Microb. 79, 1704–1711. doi: 10.1128/aem.03172-12
Joy, S. R., Li, X., Snow, D. D., Gilley, J. E., Woodbury, B., and Bartelt-Hunt, S. L. (2014). Fate of antimicrobials and antimicrobial resistance genes in simulated swine manure storage. Sci. Total Env. 481, 69–74. doi: 10.1016/j.scitotenv.2014.02.027
Kang, Y., Hao, Y., Xia, D., Shen, M., Li, Q., and Hu, J. (2017). The effects of pig manure application on the spread of tetracycline resistance in bulk and cucumber rhizosphere soils: a greenhouse experiment. Can. J. Microbiol. 63, 563–572. doi: 10.1139/cjm-2016-0767
Klute, A. (1986). “Water retention: laboratory methods,” in Methods of Soil Analysis. I. Physical and Mineralogical Methods, ed. C. A. Black, (Madison, WI: ASA; SSSA), 635–662. doi: 10.2136/sssabookser5.1.2ed.c26
Koike, S., Aminov, R., Yannarell, A., Grans, H., Krapac, I., Chee-Sanford, J., et al. (2010). Molecular ecology of macrolide–lincosamide–streptogramin B methylases in water lagoons and subsurface waters associated with swine production. Microb. Ecol. 59, 487–498. doi: 10.1007/s00248-009-9610-0
Kotsyurbenko, O. R. (2005). Trophic interactions in the methanogenic microbial community of low-temperature terrestrial ecosystems. FEMS Microb. Ecol. 53, 3–13. doi: 10.1016/j.femsec.2004.12.009
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of community structure at the continental scale. Appl. Environ. Microb. 75, 5111–5120. doi: 10.1128/aem.00335-09
Lin, Y., Ye, G., Liu, D., Ledgard, S., Luo, J., Fan, J., et al. (2018). Long-term application of lime or pig manure rather than plant residues suppressed diazotroph abundance and diversity and altered community structure in an acidic Ultisol. Soil Biol. Biochem. 123, 218–228. doi: 10.1016/j.soilbio.2018.05.018
Mantz, A. R., Miller, D. N., Spiehs, M. J., Woodbury, B. L., and Durso, L. M. (2013). Persistence of erythromycin resistance genes erm(B) in cattle feedlot pens over time. Agric. Food Anal. Bacteriol. 3, 312–320.
McKinney, C. W., and Dungan, R. S. (2020). Influence of environmental conditions on extracellular and intracellular antibiotic resistance genes in manure-amended soil: a microcosm study. Soil Sci. Soc. Am. J. 84, 747–759. doi: 10.1002/saj2.20049
McKinney, C. W., Dungan, R. S., Moore, A., and Leytem, A. B. (2018). Occurrence and abundance of antibiotic resistance genes in agricultural soil receiving dairy manure. FEMS Microb. Ecol. 94:fiy010.
McLaughlin, M. R., Brooks, J. P., and Adeli, A. (2012). Temporal flux and spatial dynamics of nutrients, fecal indicators, and zoonotic pathogens in anaerobic swine manure lagoon water. Water Res. 46, 4949–4960. doi: 10.1016/j.watres.2012.06.023
Munir, M., and Xagoraraki, I. (2011). Levels of antibiotic resistance genes in manure, biosolids, and fertilized soil. J. Env. Qual. 40, 248–255. doi: 10.2134/jeq2010.0209
Ng, L. K., Martin, I., Alfa, M., and Mulvey, M. (2001). Multiplex PCR for the detection og tetracycline resistant genes. Mol. Cell. Probe. 15, 209–215. doi: 10.1006/mcpr.2001.0363
Pei, R., Kim, S., Carlson, K. H., and Pruden, A. (2006). Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 40, 2427–2435. doi: 10.1016/j.watres.2006.04.017
Peng, S., Wang, Y., Zhou, B., and Lin, X. (2015). Long-term application of fresh and composted manure increase tetracycline resistance in the arable soil of eastern China. Sci. Total Environ. 506, 279–286. doi: 10.1016/j.scitotenv.2014.11.010
Peng, S., Zhou, B., Wang, Y., Lin, X., Wang, H., and Qiu, C. (2016). Bacteria play a more important role than nutrients in the accumulation of tetracycline resistance in manure-treated soil. Biol. Fert. Soils 52, 655–663. doi: 10.1007/s00374-016-1105-9
Pietikäinen, J., Pettersson, M., and Bååth, E. (2005). Comparison of temperature effects on soil respiration and bacterial and fungal growth rates. FEMS Microbiol. Ecol. 52, 49–58. doi: 10.1016/j.femsec.2004.10.002
Stevens, E. E., Miller, D. N., Brittenham, B. A., Vitosh-Sillman, S. J., Brodersen, B. W., Jin, V. L., et al. (2018). Alkaline stabilization of manure slurry inactivates porcine epidemic diarrhea virus. J. Swine Health Prod. 26, 95–100.
Stott, D. E., Elliott, L. F., Papendick, R. I., and Campbell, G. S. (1986). Low temperature or low water potential effects on the microbial decomposition of wheat residue. Soil Biol. Biochem. 18, 577–582.
Szczepanowski, R., Linke, B., Krahn, I., Gartemann, K., Gutzkow, T., Eicher, W., et al. (2009). Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 155, 2306–2319. doi: 10.1099/mic.0.028233-0
Tan, W., Wang, W., Bai, W., Qi, J., and Chen, W. (2020). Soil bacterial diversity correlates with precipitation and soil pH in long-term maize cropping systems. Sci. Rep. 10:6012.
Tang, X., Lou, C., Wang, S., Lu, Y., Liu, M., Hashmi, M. Z., et al. (2015). Effects of long-term manure applications on the occurrence of antibiotics and antibiotic resistance genes (ARGs) in paddy soils: evidence from four field experiments in south of China. Soil Biol. Biochem. 90, 179–187. doi: 10.1016/j.soilbio.2015.07.027
Topp, E., Larsson, D. G. J., Miller, D. N., Van den Eede, C., and Virta, M. P. J. (2018). Antimicrobial resistance and the environment: assessment of advances, gaps and recommendations for agriculture, aquaculture and pharmaceutical manufacturing. FEMS Microb. Ecol. 94:fix185.
Udikovic-Kolic, N., Wichmann, F., Broderick, N. A., and Handelsman, J. (2014). Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. PNAS 111, 15202–15207. doi: 10.1073/pnas.1409836111
US Code of Federal Regulations (2018). Standards for the Use or Disposal of Sewage Sludge, 40 C.F.R. §503 Appendix B. Available online at: https://www.govinfo.gov/content/pkg/CFR-2018-title40-vol32/xml/CFR-2018-title40-vol32-part503.xml#seqnum503.25 (accessed 1 July, 2018).
US Environmental Protection Agency (2000). Biosolids Technology Fact Sheet. Alkaline stabilization of biosolids. EPA 832-F-00-052. Available online at: https://www.epa.gov/biosolids/fact-sheet-alkaline-stabilization-biosolids (accessed September, 2000)
US Food and Drug Administration, (2019). Summary Report on Antimicrobials Sold or Distributed fo Use in Food-Producing Animals. Silver Spring, MD: US Food & Drug Administration.
von Wintersdorff, C. J. H., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Keywords: antibiotic resistance, freeze-thaw, manure application, manure treatment, soil moisture, swine
Citation: Miller DN, Jurgens ME, Durso LM and Schmidt AM (2020) Simulated Winter Incubation of Soil With Swine Manure Differentially Affects Multiple Antimicrobial Resistance Elements. Front. Microbiol. 11:611912. doi: 10.3389/fmicb.2020.611912
Received: 29 September 2020; Accepted: 25 November 2020;
Published: 16 December 2020.
Edited by:
Christophe Merlin, Université de Lorraine, FranceReviewed by:
Yuyi Yang, Chinese Academy of Sciences, ChinaCopyright © 2020 Miller, Jurgens, Durso and Schmidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel N. Miller, ZGFuLm1pbGxlckB1c2RhLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.