- 1Shandong Province Key Laboratory of Applied Mycology, School of Life Sciences, Qingdao Agricultural University, Qingdao, China
- 2School of Chemistry and Pharmacy, Qingdao Agricultural University, Qingdao, China
Nocamycins I and II, featured with a tetramic acid scaffold, were isolated from the broth of Saccharothrix syringae NRRL B-16468. The biosynthesis of nocamycin I require an intermediate bearing a hydroxyl group at the C-10 position. A short chain dehydrogenase/reductase NcmD was proposed to catalyze the conversion of the hydroxyl group to ketone at the C-10 position. By using the λ-RED recombination technology, we generated the NcmD deletion mutant strain S. syringae MoS-1005, which produced a new intermediate nocamycin F with a hydroxyl group at C-10 position. We then overexpressed NcmD in Escherichia coli BL21 (DE3), purified the His6-tagged protein NcmD to homogeneity and conducted in vitro enzymatic assays. NcmD showed preference to the cofactor NAD+, and it effectively catalyzed the conversion from nocamyin F to nocamycin G, harboring a ketone group at C-10 position. However, NcmD showed no catalytic activity toward nocamyin II. NcmD achieved maximum catalytic activity at 45°C and pH 8.5. The kinetics of NcmD toward nocamycin F was investigated at 45°C, pH 8.5 in the presence of 2 mM NAD+. The Km and kcat values were 131 ± 13 μM and 65 ± 5 min−1, respectively. In this study, we have characterized NcmD as a dehydrogenase, which is involved in forming the ketone group at the C-10 position of nocamycin F. The results provide new insights to the nocamycin biosynthetic pathway.
Introduction
The short-chain dehydrogenases/reductases (SDRs), one of the largest protein families, distribute in all kinds of organisms. Despite low residue identities in pairwise comparisons, all SDRs share a Rossmann fold-type domain for NAD(P)+ binding (Kavanagh et al., 2008; Persson and Kallberg, 2013). SDRs have been classified into seven families: classical, extended, atypical, intermediate, divergent, complex and unassigned, and the classical type is the most prominent (Persson and Kallberg, 2013; Gräff et al., 2019). SDRs show diverse substrate spectra, including steroids, alcohols, sugars, aromatic compounds, and xenobiotics, and for this reason, more and more SDRs have been extensively explored for industrial production (Persson and Kallberg, 2013; Luo et al., 2019; Savino et al., 2019; Shanati et al., 2019; Zhou et al., 2019; Su et al., 2020). SDRs play diverse roles in core metabolism and specific metabolism pathways such as steroidal metabolism, detoxification and drug resistance (Sonawane et al., 2018; Laskar and Younus, 2019). Moreover, SDRs play important roles in biosynthetic pathways of microbial secondary metabolites (MacKenzie et al., 2007; Mattheus et al., 2010; Bown et al., 2016).
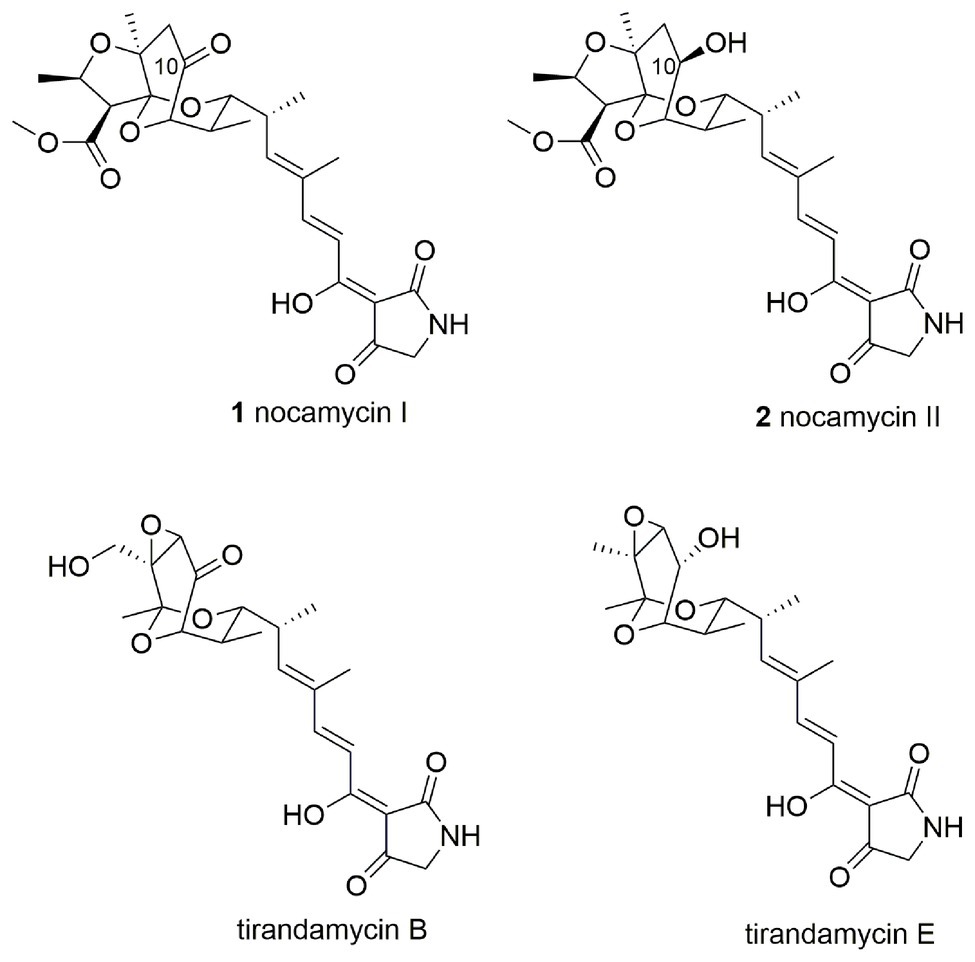
Nocamycins I and II, isolated from the broth of Saccharothrix syringae NRRL B-16468, feature two unique structural moieties, namely tetramic acid (2,4-pyrrolidinedione) and bicyclic ketal scaffolds (Figure 1; Gauze et al., 1977). Nocamycin I displays potent and broad antimicrobial activities against a panel of Gram-positive and Gram-negative bacteria, especially toward some anaerobic bacteria such as Bacteroides fragilis, Clostridium sp., Fusobacterium sp., and Sphaerophorus sp. with minimum inhibitory concentrations (MICs) in the range of 0.1–0.4 μg/ml (Tsukiura et al., 1980; Tsunakawa et al., 1980; Bansal et al., 1982). In addition, the carboxylate O-methyl group appears to be essential for nocamycins’ antibacterial property. Nocamycin E, lacking the carboxylate O-methyl group, shows less antibacterial activity (Mo et al., 2017a).
The gene cluster responsible for nocamycins biosynthesis has been identified from S. syringae NRRL B-16468 in 2017 (Mo et al., 2017b). The skeleton of nocamycins is assembled by a hybrid type I polyketide synthases (PKS) and non-ribosomal peptide synthetase (NRPS) system (Mo et al., 2017b). The gene cluster for nocamycins consists of 21 open reading frames (ORFs), which includes five genes encoding for type I PKS, one gene encoding for NRPS, one gene encoding for a Dieckmann cyclase NcmC, one gene encoding for a SDR NcmD, two genes encoding for cytochrome P450 oxidases NcmO and NcmG, one gene encoding for a glycoside dehydratase NcmE, one gene encoding for a SAM dependent methyltransferase NcmP, and five genes encoding for regulatory proteins (Mo et al., 2017b). Among the 21 ORFs, the functions of several genes have been investigated. The results of gene disruption experiment indicated that the cytochrome P450 oxidase NcmG was involved in formation of furan ring (Mo et al., 2017b). Furthermore, gene disruption and biochemical assays clearly demonstrated that the SAM-dependent methyltransferase NcmP was responsible for formation of the carboxylic methyl ester (Mo et al., 2017a).
The disparity between nocamycins I and II is the ketone or hydroxyl group at C-10 position, and the similar structure has been observed in tirandamycin B and its biosynthetic intermediate tirandamycin E (Figure 1). For tirandamycins, a FAD dependent dehydrogenase TrdL/TamL has been shown to catalyze the formation of ketone group at C-10 position (Carlson et al., 2011; Mo et al., 2011). However, for nocamycins, the enzyme for formation of ketone group at C-10 position remains unclear. We initially inactivated the gene ncmL encoding for a FAD-dependent protein, and resultant research demonstrated that it exerted no impact on production of nocamycins (Mo et al., 2017b). By carefully examining the biosynthetic gene cluster, the SDR NcmD is proposed to be the candidate to catalyze the conversion from hydroxyl to ketone at C-10 position. In the current study, we have established that NcmD acts as a dehydrogenase and catalyzes the formation of C-10 ketone group in nocamycin biosynthetic pathway by using in vivo gene disruption and in vitro biochemical assays.
Materials and Methods
Bacteria, Medium, and Culture Conditions
Saccharothrix syringae NRRL B-16468 was used as producer of nocamycins (Mo et al., 2017b). Escherichia coli DH5α was used as host for general clone. Escherichia coli BL21(DE3) was used as host for protein expression. Saccharothrix syringae NRRL B-16468 and its derivative strain were maintained on ISP4 agar medium. The medium contained 1% soybean flour, 3% glycerol, 0.2% NaCl, and 0.2% CaCO3, pH 7.0 was used for fermentation of S. syringae and its derivative strain. All cultures for S. syringae were incubated at 28°C. The E. coli strains were cultured by using Luria-Bertani (LB) agar or liquid medium with appropriate antibiotics.
Generating Mutant Strain Saccharothrix syringae pMoS-1005 (ΔNcmD)
The gene NcmD was inactivated by using λ-RED recombination technology according to the literature previously reported (Mo et al., 2017b). The primers NcmD-delF (5'-CTCGCCGAGGCGTTCGCGGCCGAGGGCGCCCGAGTGGTGATTCCGGGGATCCGTCGACC-3') and NcmD-delR (5'-CCGGTTCTCCCGCACGGCGTCGAGGGTCGCCGCGGCCACTGTAGGCTGGAGCTGCTTC-3') were used to amplify the fragment oriT/acc(3)IV cassette from the plasmid pIJ773, the resultant PCR fragment was used to replace partial gene region of NcmD in cosmid p5-C-9 to generate plasmid pMoS-1005. Then, the correct mutated plasmid pMoS-1005 was introduced into E. coli ET12567/pUZ8002, which then conjugated with wild type S. syringae spores as described previously (Mo et al., 2017b). The exconjugants were firstly selected by the phenotype of kanamycin sensitive (KanS) and apramycin resistant (AprR), and then their genotypes were further verified by using PCR with the primers NcmD-tF (5'-ATGCGCGAGTTGACCGACC-3') and NcmD-tR (5'-AGCACGTCCAGGAAGTCAC-3'). The desired double cross-over mutant strain was termed as S. syringae MoS-1005.
Fermentation and Analysis of Mutant Strain Saccharothrix syringae MoS-1005
S. syringae wild type and mutant strain S. syringae MoS-1005 were cultured by using the method described previously (Mo et al., 2017b). After 7 days culture, the broth was extracted by ethyl acetate, subsequently, the organic phase was collected and evaporated into dryness, re-dissolved in methanol, and subject to high performance liquid chromatography (HPLC) analysis. Analytical HPLC was performed on Waters 2699 HPLC system (Waters Technologies Inc., United States) equipped with a PDA detector and a Welch Ultimate AQ-C18 ODS column (250 × 4.60 mm, 5 μm). The mobile phase contained solvent A and B. Solvent A consisted of 15% CH3CN in water supplemented with 0.1% formic acid. Solvent B consisted of 85% CH3CN in water supplemented with 0.1% formic acid. Samples were eluted with a linear gradient from 5 to 90% solvent B in 20 min, followed by 90 to 100% solvent B for 5 min, then 100% solvent B for 3 min, at a flow rate of 1 ml/min under UV detection at 355 nm.
Isolation of New Nocamycin Derivative Produced by Saccharothrix syringae MoS-1005
For fermentation of S. syringae MoS-1005 in a large scale, 8 L liquid media were used by using a two-step fermentation procedure as described previously (Mo et al., 2017b). After incubation, the culture broths were collected and centrifuged. The supernatant broth was extracted by ethyl acetate for three times and the mycelia were extracted by methanol for three times. Then, the entire organic solvents were evaporated into dryness to yield crude extracts. The crude extracts were dissolved in a mixture of CH2Cl2:CH3OH (1:1), and then mixed with appropriate amount of silica gel (100–200 mesh, Qingdao Marine Chemical Corporation, China). The samples were applied on normal phase silica gel chromatography column and eluted with CH2Cl2:CH3OH (100:0–50:50) to give 10 fractions, and all of them were analyzed by HPLC. Fractions 4 and 5 containing the target compound were used for further purification on reverse phase C-18 silica gel (YMC, Japan) by using medium-pressure liquid chromatography (MPLC, Agela corporation, China). The fractions containing the target compound were combined and further purified by Sephadex LH-20 (GE healthcare, Sweden) gel filtration chromatography to afford the purified nocamycin F.
Heterologous Production and Purification of Recombinant Protein NcmD
The NcmD gene was amplified from cosmid p5-C-9 by using PCR with primers NcmD-expF (5'-TAATAATCATATGCGCGAGTTGACCGACCG-3', underline is NdeI site) and NcmD-expR (5'-ATATGGATCCCTAGTCGGTCGGCGCGGCG-3', underline is BamHI site). The resultant PCR products were digested by NdeI and BamHI, and then inserted into pET-28a (+) vector digested with the same restriction enzymes to yield plasmid pMosD. After verification of the inserted gene fragment by sequencing, the plasmid pMosD was introduced into E. coli BL21(DE3) for protein expression.
Escherichia coli BL21 (DE3) strain carrying plasmid pMosD was grown in LB medium with 50 μg/ml kanamycin at 37°C to an OD600 = 0.6. Then, isopropyl-β-D-thiogalactopyranoside (IPTG) was added into the culture at a final concentration of 0.1 mM to induce the expression of NcmD for 18 h at 20°C. Subsequently, the cells were collected, centrifuged, and re-suspended in binding buffer (50 mM Tris-Cl buffer, 500 mM NaCl, and 10 mM imidazole, pH 7.9), and sonicated on ice. Cellular debris was removed by centrifugation (12,000 rpm, 30 min, 4°C). The supernatant was further purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography according to the manufacturer’s protocol (Novagen, CA, United States). The purified protein was desalted by PD-10 column (GE Healthcare, United States) according to the manufacturer’s instructions. The purified protein NcmD was finally stored in 50 mM Tris-Cl buffer (pH 8.0) with 10% glycerol at −80°C for further enzymatic assays.
In vitro Enzymatic Assays of NcmD With Nocamycin F and Nocamycin II
The enzymatic assays were firstly conducted at 30°C in 50 mM Tris-Cl buffer (pH 8.0) containing 200 μM nocamycin F, 2 μM NcmD enzyme, 2 mM NAD+ or NADP+ for an hour. Then, the catalytic activity of NcmD toward nocamycin II was performed in 50 mM Tris-Cl buffer (pH 8.0) containing 200 μM nocamycin II, 2 μM NcmD enzyme, 2 mM NAD+ at 30°C for an hour. The reaction mixtures without NAD+ or NADP+ were set as negative controls. To investigate the optimum temperature for NcmD, the assays were carried out in 50 mM Tris-Cl buffer (pH 8.0) containing 200 μM nocamycin F, 0.8 μM NcmD, 2 mM NAD+ at various temperature (T = 25, 30, 37, 40, 45, 50, 55, and 60°C) for an hour. For probing the effect of pH on NcmD, the 50 μl reaction mixtures containing 200 μM nocamycin F, 0.8 μM NcmD, 2 mM NAD+ were performed at 45°C at different pH buffer for an hour, including Tris-Cl buffer (50 mM, pH = 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, and 9.0), NaHCO3-NaOH buffer (50 mM, pH = 10.0, 11.0). After quenching the reactions by adding 100 μl cold methanol, then, the samples were centrifuged, and the supernatants (30 μl) were subjected to HPLC analysis. For each reaction, three parallels were used.
To determine the kinetic parameters of NcmD toward nocamycin F, 100 μl reaction mixtures containing 0.2 μM NcmD, 2 mM NAD+ and varying nocamycin F (10 20, 40, 100, 150, 200, 400, and 600 μM) at pH 8.5, 45°C were conducted. After a pre-incubation at 45°C for 3 min, the reactions were initiated by adding substrate nocamycin F. At 2, 4, and 6-min, 30 μl reaction mixtures were taken, and then 60 μl ice-cold methanol was added and rigorously mixed by vortex. After centrifuge, 30 μl liquids were used for further HPLC analysis and quantified by a standard curve. The reaction rates were calculated and confirmed to be linear. The kinetics data were fitted to the Michaelis-Menten equation using Origin8.0 software. For each concentration of substrate, three replicates were conducted.
For elucidating the structure of the products obtained from the reactions with nocamycin F, 5 ml reaction mixtures containing 2 μM NcmD, 2 mM NAD+ and 6 mg nocamycin F were performed at pH 8.5, 40°C for 8 h. Then, the reaction mixtures were extracted by 10 ml ethyl acetate for three times. The target product was purified by preparative HPLC for NMR and high-resolution mass spectral (HR-MS) analysis.
Spectroscopy Analysis of New Produced Nocamycin Derivatives
1H and 13C NMR spectra were recorded at 25°C on Bruker AV 500 instruments. LC-HR-MS data were acquired on a Waters micro MS Q-Tof spectrometer or a Thermo MAT95XP high resolution mass spectrometer.
Results
Bioinformatics Analyses of NcmD
Within nocamycin biosynthetic gene cluster, the gene NcmD encoding for the SDR located next to the NRPS gene NcmB. NcmD shows identity to a series of SDRs, including 40% identity to BatM originated from kalimantacin/batumin-related polyketide antibiotic biosynthetic pathway (Mattheus et al., 2010), and 31% identity to clavulanic acid dehydrogenase (CAD) involved in clavulanic acid biosynthetic pathway (MacKenzie et al., 2007). Bioinformatics analyses revealed that NcmD shared the conserved motifs of classical SDR, namely, Rossmann fold NAD(P)-binding motif T12G13(X)3G17XG19, conserved catalytic triad S142(X)12Y155(X)3K159, N89NAG92 motif and P187G (X)3T192 motif (Figure 2; Filling et al., 2002; Persson and Kallberg, 2013; Gräff et al., 2019). In addition, an Asn115 residue (N) frequently served as an additional active site residue to make a catalytic triad is also conserved in NcmD (Figure 2). These bioinformatics analyses have demonstrated that NcmD is a classical SDR. As shown in Figure 2, an acidic residue aspartate (Asp, D) other than a basic residue occupies the position 37, indicating that the cofactor for NcmD is likely to be NAD+, which has been confirmed by other NAD+-preference SDRs such as FgaDH, PDH, 1YDE, and TQS88917 (Lukacik et al., 2007; Wallwey et al., 2010; Hofmann et al., 2016; Shanati and Ansorge-Schumacher, 2020).

Figure 2. Amino acid sequences alignment analysis of NcmD and its closely homologous proteins and characterized short-chain dehydrogenases/reductases (SDRs). BatM originated from Pseudomonas fluorescens BCCM_ID9359 shows 40% identity to NcmD. 2JAP (CAD) involved in clavulanic acid biosynthetic pathway originated from Streptomyces clavuligerus shows 31% identity to NcmD. FgaDH originated from Aspergillus fumigatus is a NAD+ binding SDR. PDH originated from Drosophila melanogaster is a NAD+ binding SDR. The ephedrine dehydrogenase TQS88917 originated from Arthrobacter sp. TS-15 is a NAD+ binding SDR. 1YDE is a human retinal SDR using NADH as cofactor. 1EDO is a β-keto acyl carrier protein reductase from Brassica napus by using NADP+ as cofactor.
Construction, Analysis of NcmD Mutant Strain Saccharothrix syringae MoS-1005 and Isolation of the New Nocamycin Analog
To investigate the exact role played by NcmD in nocamycin biosynthetic pathway, we firstly inactivated NcmD by replacing partial internal NcmD with aac(3)IV gene cassette through λ-RED recombination technology and generate ΔNcmD mutant strain (Figure 3A). After verifying the genotype of S. syringae MoS-1005 (Figure 3B), the mutant strain was cultured, and the broth was extracted for HPLC analysis. The results of HPLC revealed that three peaks with retention time of 15.1, 16.9, and 19.8 min showed UV absorption characteristics of nocamycin were detected in S. syringae MoS-1005 (Figure 3C). The peaks at 15.1 and 19.8 min showed the same retention time to nocamycin II and nocamycin I, respectively. Subsequently, the molecular mass of these three peaks were detected by LC-HR-MS. For the peak at 15.1 min, it had a molecular mass of 505 Dalton [m/z values 506.24 (M + H)+ and 528.2204 (M + Na)+; Supplementary Figure S1-A], which was identical to nocamycin II. As for the peak at 19.8 min, it had a molecular mass of 503 Dalton [m/z values 504.2295 (M + H)+ and 526.2046 (M + Na)+; Supplementary Figure S1-B], which was identical to nocamycin I. These results demonstrated the compounds in 15.01 and 19.8 min were nocamycin II and nocamycin I, respectively. However, for the peak at 16.9 min, it had a molecular mass of 461 Dalton [m/z values 462.2469 (M + H)+, 484.2298 (M + Na)+ and 945.4703 (2 M + Na)+; Supplementary Figure S1-C], which was different from the molecular mass of all the other nocamycin derivatives reported previously, indicating it was a new derivative produced by S. syringae MoS-1005.
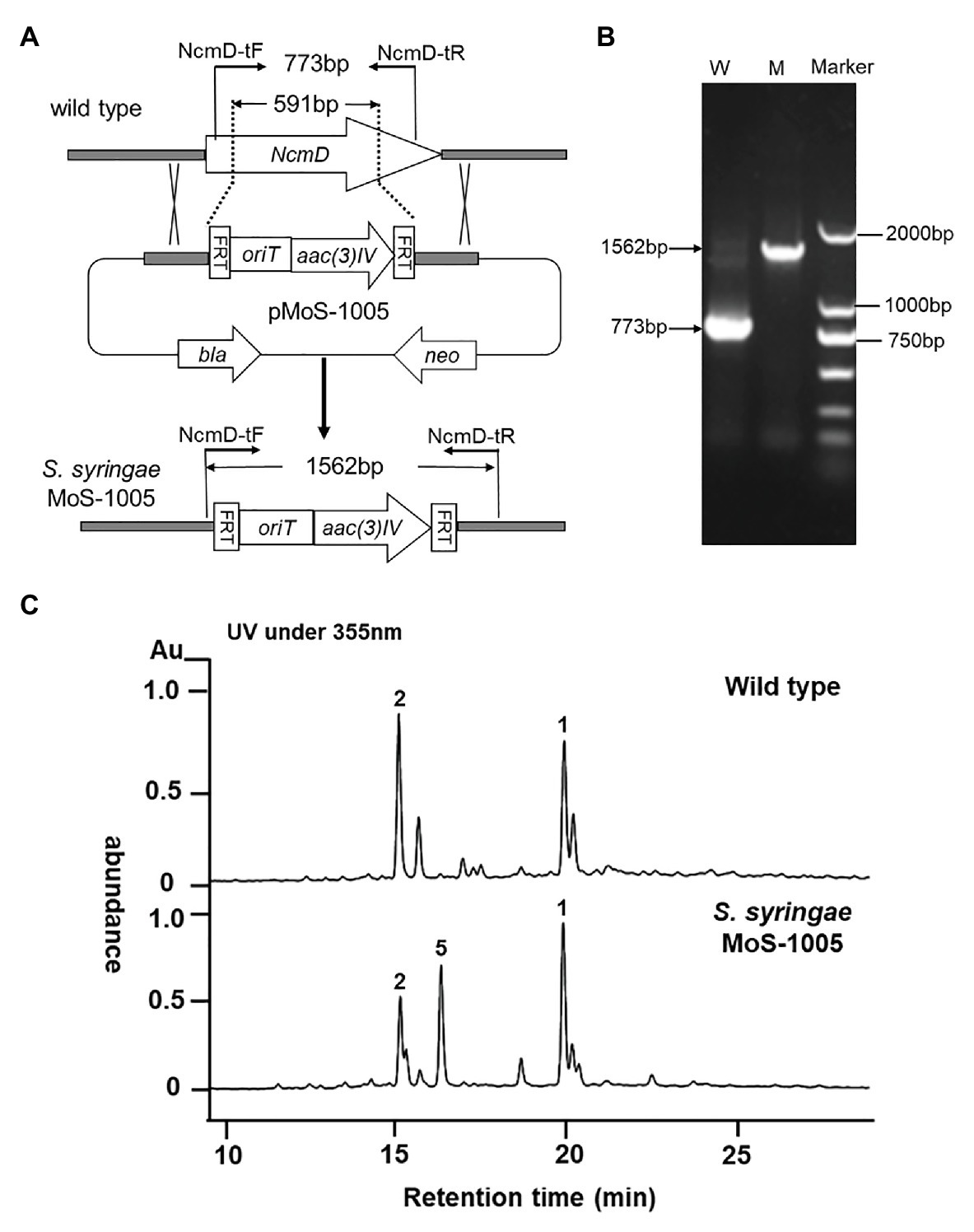
Figure 3. Generation of ΔNcmD mutant strain Saccharothrix syringae MoS-1005 and its metabolites profile analyzed by HPLC. (A) Schematic diagram of NcmD inactivation. The 591 bp internal NcmD fragment was replaced by oriT and acc3(IV) fragment in pMoS-1005. (B) PCR analysis of the double-crossover mutant S. syringae MoS-1005. W: S. syringae wild type (773 bp); M: mutant strain S. syringae MoS-1005 (1,562 bp); Marker: DNA molecular ladder. (C) Metabolite profiles of S. syringae MoS-1005 analyzed by HPLC. 1: nocamycin I, 2: nocamycin II, 5: the newly produced derivative.
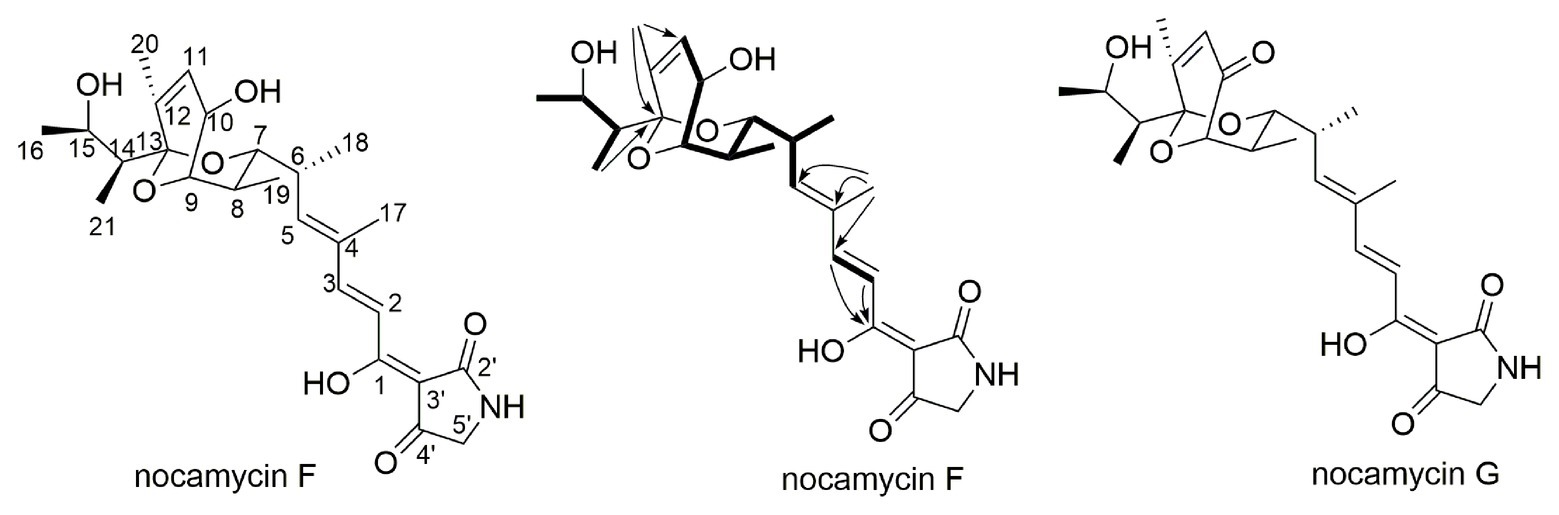
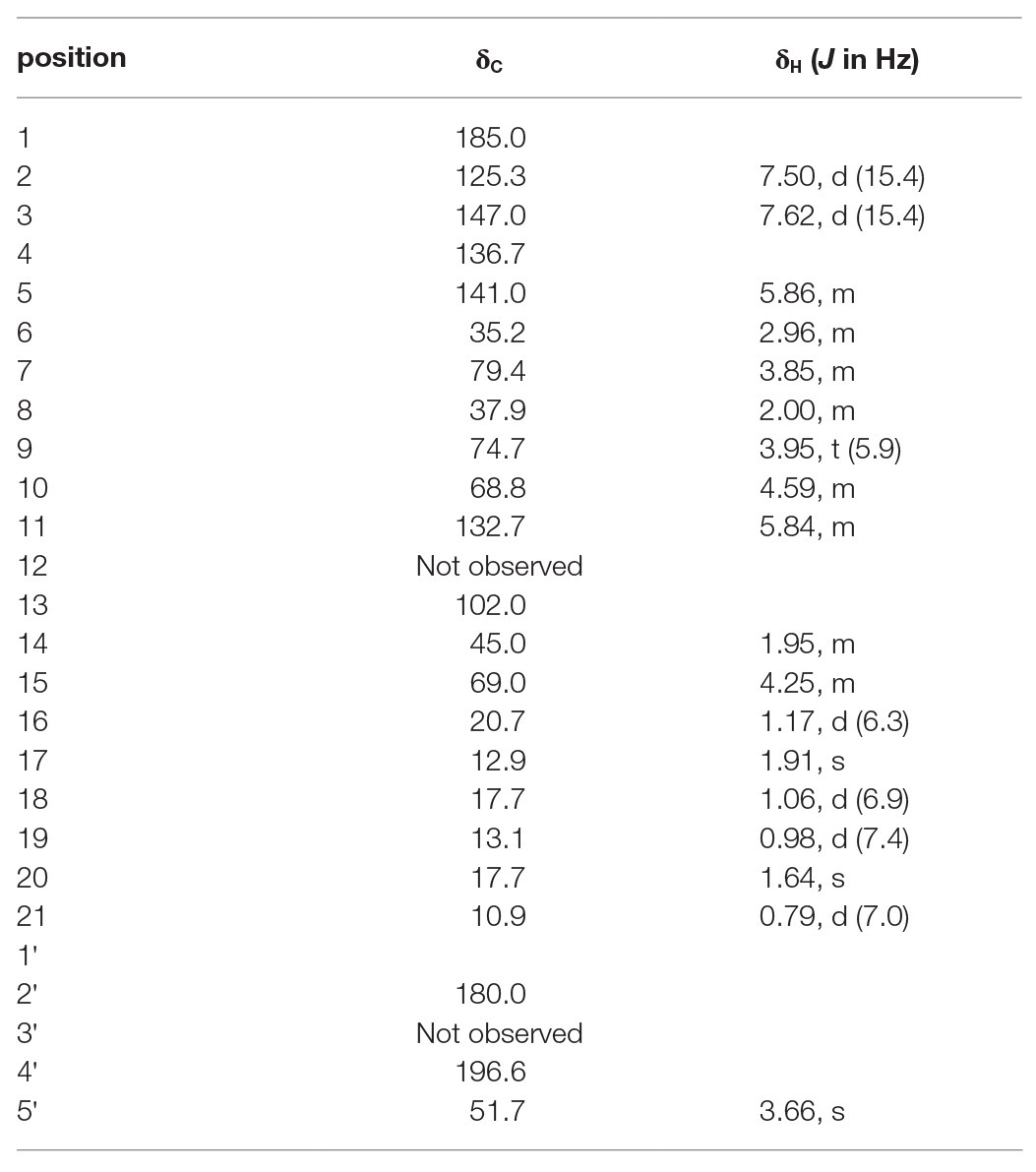
To elucidate the structure of the new nocamycin analog produced by S. syringae MoS-1005, we purified nocamycin F from 8 L culture broth by using chemical isolation methods. The structure of nocamycin F was determined by multiple spectroscopic data analyses (Figure 4). As revealed from HR-MS (Supplementary Figure S1-C), the molecular formula of nocamycin F was determined as C25H35NO7, with an oxygen atom more than that of nocamycin III (Mo et al., 2017b), suggesting that nocamycin F should be a hydroxylation congener of nocamycin III. The one-dimensional NMR data exhibited marked similarities to those of nocamycin III, except that the signal of a methylene group (CH2-10, δC 23.9) in nocamycin III was absent from the NMR spectra of nocamycin F (Table 1). Instead, another oxygenated CH group (CHOH-10, δC 68.8/δH 4.59) could be observed in the spectra of nocamycin F, also as established by the COSY and HMBC data (Supplementary Figure S2).
In vitro Characterization of NcmD in Nocamycin Biosynthetic Pathway
For in vitro characterization of NcmD activity, the protein NcmD was produced as a N-terminal His6-tagged protein in heterologous host E. coli BL21(DE3) carrying the plasmid pMoS-1005. The His6-tagged protein NcmD was purified to homogeneity by using Ni-NTA affinity chromatography, and SDS-PAGE analysis displayed that it had an expected molecular weight (the calculated molecular mass for NcmD is 30.078 kDa; Figure 5A). Subsequently, the catalytic properties of NcmD were investigated. At first, we detected the preference of NcmD toward cofactors NAD+ and NADP+. In the presence of NAD+, NcmD can efficiently catalyze substrate nocamycin F to a new compound with less polarity (Figure 5B). Whereas, in the reaction mixture with NADP+ instead of NAD+, only about 25% nocamycin F was transformed to a new compound. These results demonstrated that NcmD preferred NAD+ as the cofactor, which was consistent with our initial bioinformatics analyses.
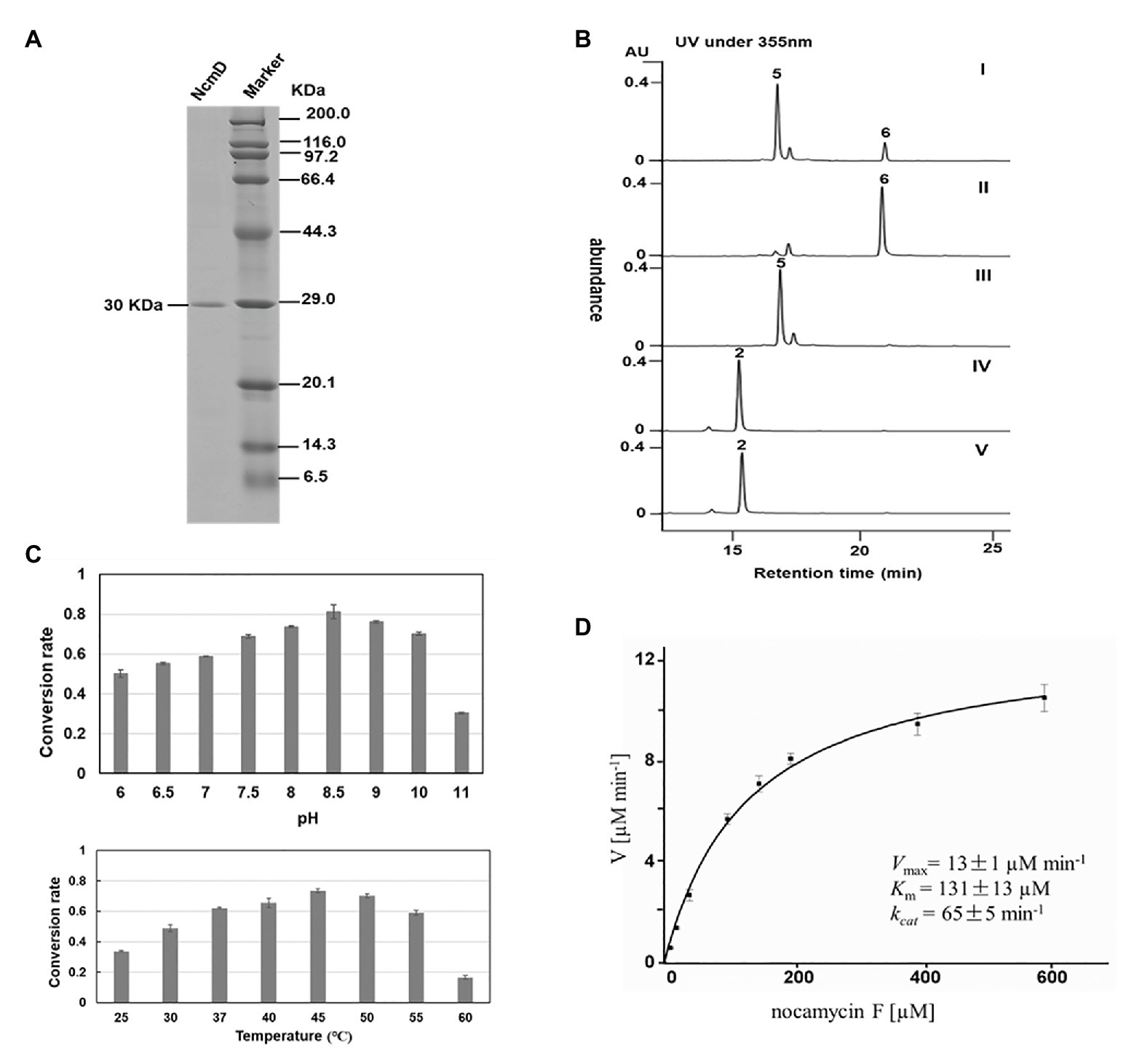
Figure 5. Biochemical characterization of NcmD. (A) Analysis of purified recombinant NcmD protein by SDS-PAGE gel. Lane 1 is the recombinant NcmD, lane 2 is protein standard. (B) HPLC analysis of NcmD with nocamycin F (5) and nocamycin II (2). I: an assay of NcmD with 5 and cofactor NADP+. II:an assay of NcmD with 5 and cofactor NAD+. III: an assay of boiled NcmD with 5 and cofactor NAD+. IV: an assay of NcmD with 2 and cofactor NAD+. V: an assay of boiled NcmD with 2 and cofactor NAD+. (C) Biochemical properties of NcmD. Upper: effects of pH on NcmD activity. Down: effects of temperature on NcmD activity. (D) Kinetic analysis of NcmD.
To determine the structure of the new product converted from nocamycin F, we conducted a large-scale reaction of NcmD toward nocamycin F, leading to purification of the new compound 6. LC-HR-MS demonstrated that compound 6 had a molecular formula of C25H33NO7 with a molecular mass of 459 Dalton [m/z values 458.21(M-H)− and 460.23(M + H)+; Supplementary Figure S3], two hydrogen less than that of nocamycin F. The amount of 6 obtained here was not enough for 2D NMR analysis; however, the 13C NMR spectra still give us sufficient structural information of 6. By comparison of 13C NMR data of 6 and nocamycin F, 6 showed one more carbonyl carbon (C=O, δC 208.8) and three oxygenated carbons (δC 77.8, 79.1, and 81.4), indicating that one hydrogen group in nocamycin F should be oxidized to the carbonyl group (Supplementary Figure S4). From the biosynthesis view, the difference between nocamycin F and compound 6 should be at C-10 position. For these reasons, we proposed the structure of 6 as shown in Figure 4, and it was termed as nocamycin G. Thus, NcmD was proposed to act as a dehydrogenase to catalyze the formation of ketone moiety at C-10 position.
Considering the same hydroxyl moiety harbored by nocamycin II at C-10 position, we then investigated whether NcmD can also accept nocamycin II as substrate and catalyze the conversion from nocamycin II to nocamycin I. Unfortunately, no conversion from nocamycin II to nocamycin I was observed (Figure 5B). This result demonstrated that nocamycin II was not the substrate of NcmD.
The effect of pH and temperature on NcmD properties was also investigated. For pH in the range from 6.0 to 11.0, NcmD was found to achieve maximum catalytic activity at pH 8.5, and in the range of pH 7.5–10.0, NcmD can retain activity in a high level, indicating NcmD was tolerant to pH values (Figure 5C). As for temperature, NcmD showed robust activity in the range of 37–50°C, and the optimum temperature for NcmD was 45°C (Figure 5C). Finally, we measured the steady-state kinetic parameters of NcmD toward nocamcyin F under 45°C, pH 8.5. In the presence of 2 mM NAD+, the Km and kcat values of NcmD were 131 ± 13 μM and 65 ± 5 min−1, respectively (Figure 5D).
Discussion
Nocamycins belong to a small family of tetramic acid compounds bearing bicyclic ketone structure. Nocamycins demonstrate excellent antibacterial activity, especially against some anaerobic bacteria (Tsukiura et al., 1980; Tsunakawa et al., 1980; Bansal et al., 1982). Recently, we have identified the gene cluster responsible for nocamycin biosynthesis from a rare actinomycete S. syringae (Mo et al., 2017b). Through manipulating the gene cluster, we have generated several nocamycin analogs and characterized several gene functions involved in nocamycin biosynthetic pathway (Mo et al., 2017a,b). In this study, through in vivo gene inactivation and in vitro enzymatic assays, the SDR NcmD has been assigned to be involved in formation of ketone group at C-10 position, leading to generate nocamycin G from nocamycin F.
Short-chain dehydrogenases/reductases have been classified into seven families and the classical type is the most prominent. NcmD shows the conserved motifs belonging to the classical SDR subfamily. The majority (about 60%) of classical SDRs are expected to prefer NADP(H) (Gräff et al., 2019). For example, the SDR CAD has been shown to use NADPH as cofactor to catalyze the conversion from clavaldehyde to clavulanic acid (MacKenzie et al., 2007). The SDR Cro013448 (KP411011.1) from Catharanthus roseus recruits NADP(H) as cofactor in the biosynthetic pathway of plant monoterpene indole alkaloid vitrosamine (Stavrinides et al., 2018). However, the NAD+ dependent SDRs have also been found. FgaDH involved in ergot alkaloid fumigaclavine C biosynthetic pathway originating from Aspergillus fumigatus employs NAD+ as cofactor to catalyze the conversion from chanoclavine-I to chanoclavine-I aldehyde (Wallwey et al., 2010). Pseudoephedrine dehydrogenase (TQS88917) is a NAD+ dependent SDR and it catalyzes the oxidation of converted (S, S)-(+)-pseudoephedrine and (S, R)- (+)-ephedrine to (S)- and (R)-methcathinone (Shanati and Ansorge-Schumacher, 2020). For classical SDRs, the aspartic acid residue at standard position 37 has been described as a determinant of NAD(H) specificity (Belyaeva et al., 2015; Gräff et al., 2019). The current results confirm that Asp37-containing NcmD prefers NAD+ (Kallberg et al., 2002), which has been confirmed by resultant in vitro enzymatic assays. Generally, the catalytic activities decrease significantly when the SDRs are not compatible with cofactors, which have been observed from NcmD enzymatic assays, and similar results are also revealed from other SDRs (Moon et al., 2012; Takase et al., 2014; Cao et al., 2019; Gmelch et al., 2020).
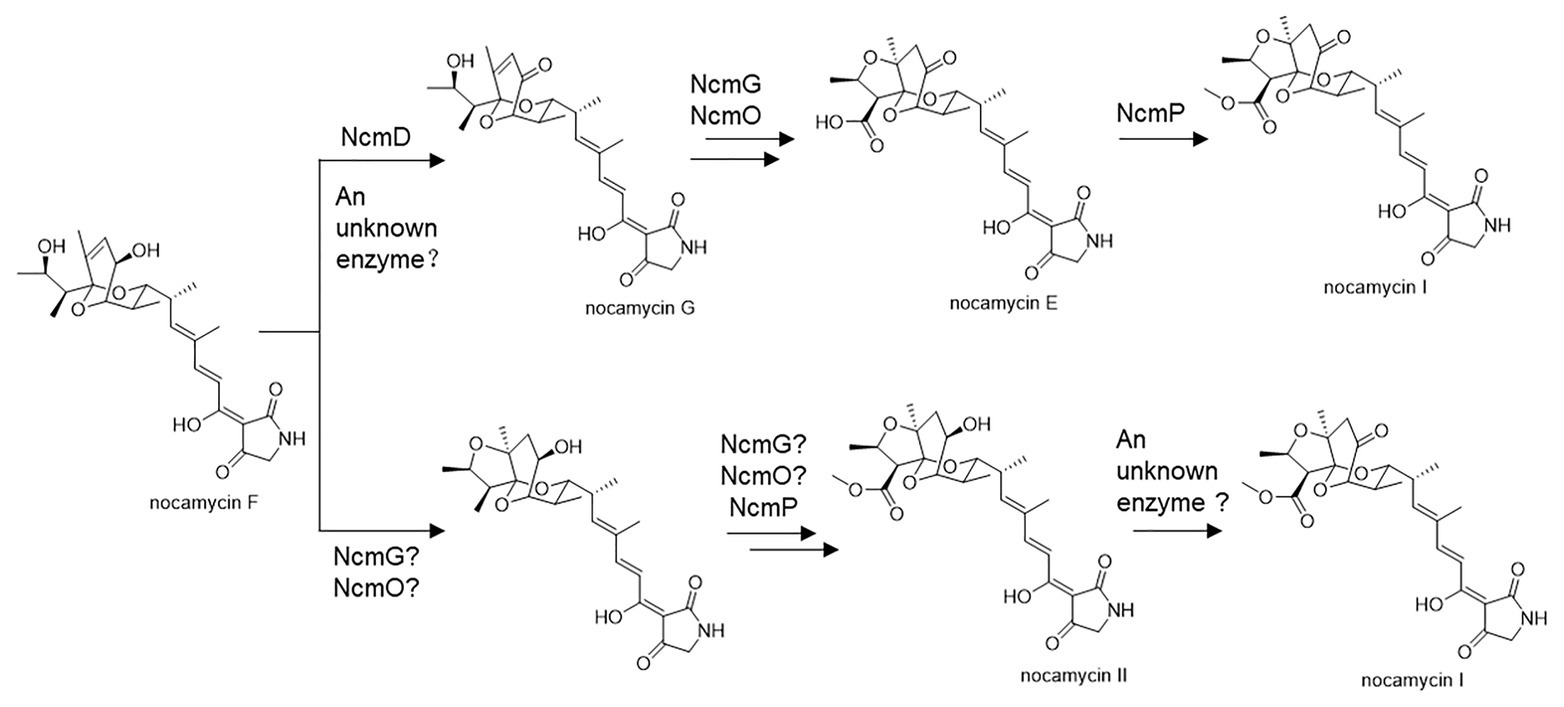
Structurally, nocamycins show high similarity to tirandamycins. Both tirandamycin B and nocamycin I harbor a ketone moiety at C-10 position. However, the proteins involved in synthesizing this moiety are significantly different. For tirandamycin B, a FAD dependent dehydrogenase TrdL catalyzes the conversion from hydroxyl moiety to ketone group at C-10 position, meanwhile, TrdL displays a flexible substrate spectrum (Mo et al., 2011). Whereas for nocamycin, the SDR NcmD shows substrate selectivity. Though NcmD was inactivated in the strain S. syringae MoS-1005, we still detected the accumulation of nocamycin I and nocamycin II, two major metabolites produced by the wild type S. syringae. Nocamyin II is a major metabolite in S. syringae wild type, which indicates the other tailoring enzymes such as NcmG, NcmO, and NcmP show flexible substrate selectivity, thus, it is rational that nocamycin II can be detected in S. syringae MoS-1005. For production of nocamycin I in S. syringae MoS1005, two possible pathways are proposed (Figure 6). Firstly, an unknown enzyme located in the genome elsewhere can compensate for the function of NcmD and catalyze the conversion from nocamycin F to nocamycin G, which then undergoes several tailoring steps to generate nocamycin I. Secondly, an unknown enzyme can catalyze the transformation from nocamycin II to nocamycin I. No matter which strategy has been employed, nocamycin I can be produced in S. syringae MoS1005. Taking these results together, we envision a plausible tailoring process from the formation of the intermediate nocamycin F, which is shown as Figure 6.
In summary, we generated NcmD deletion mutant strain S. syringae MoS-1005 and identified an important intermediate nocamycin F from this mutant strain. In vitro enzymatic assays have demonstrated that the NAD+ dependent SDR NcmD acts as a dehydrogenase and it is involved in formation of ketone moiety at C-10 position. However, NcmD shows substrate preference and it only displays catalytic activity toward nocamycin F. The results presented in this study provide new insights into nocamycin biosynthetic pathway.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
XM and SY designed the experiment. XM, SY, and FD analysis the data and wrote the paper. XM and HZ performed the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Open Research Project from Guangdong Key Laboratory of Marine Materia Medica (No. LMM2019-1), Grants from the National Natural Science Foundation of China (No. 31300063), National Undergraduate Innovation and Entrepreneurship Training Program (201910435005), Shandong Provincial Key Research and Development Project (2016GSF117026), and Shandong Provincial Natural Science Foundation (ZR2015CM017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.610827/full#supplementary-material
References
Bansal, M., Dhawan, V., and Thadepalli, H. (1982). In vitro activity of Bu-2313B against anaerobic bacteria. Chemotherapy 28, 200–203. doi: 10.1159/000238076
Belyaeva, O. V., Chang, C., Berlett, M. C., and Kedishvili, N. Y. (2015). Evolutionary origins of retinoid active short-chain dehydrogenases/reductases of SDR16C family. Chem. Biol. Interact. 234, 135–143. doi: 10.1016/j.cbi.2014.10.026
Bown, L., Altowairish, M. S., Fyans, J. K., and Bignell, D. R. (2016). Production of the Streptomyces scabies coronafacoyl phytotoxins involves a novel biosynthetic pathway with an F420-dependent oxidoreductase and a short-chain dehydrogenase/reductase. Mol. Microbiol. 101, 122–135. doi: 10.1111/mmi.13378
Cao, Z., Li, S., Lv, J., Gao, H., Chen, G., Awakawa, T., et al. (2019). Biosynthesis of clinically used antibiotic fusidic acid and identification of two short-chain dehydrogenase/reductases with converse stereoselectivity. Acta Pharm. Sin. B. 9, 433–442. doi: 10.1016/j.apsb.2018.10.007
Carlson, J. C., Li, S., Gunatilleke, S. S., Anzai, Y., Burr, D., Podust, L. M., et al. (2011). Tirandamycin biosynthesis is mediated by co-dependent oxidative enzymes. Nat. Chem. 3, 628–633. doi: 10.1038/nchem.1087
Filling, C., Berndt, K. D., Benach, J., Knapp, S., Prozorovski, T., Nordling, E., et al. (2002). Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277, 25677–25684. doi: 10.1074/jbc.M202160200
Gauze, G., Sveshnikova, M., Ukholina, R., Komarova, G., and Bazhanov, V. (1977). Formation of a new antibiotic, nocamycin, by a culture of Nocardiopsis syringae sp. nov. Antibiotiki 22, 483–486.
Gmelch, T. J., Sperl, J. M., and Sieber, V. (2020). Molecular dynamics analysis of a rationally designed aldehyde dehydrogenase gives insights into improved activity for the non-native cofactor NAD. ACS Synth. Biol. 9, 920–929. doi: 10.1021/acssynbio.9b00527
Gräff, M., Buchholz, P. C. F., Stockinger, P., Bommarius, B., Bommarius, A. S., and Pleiss, J. (2019). The short-chain dehydrogenase/reductase engineering database (SDRED): a classification and analysis system for a highly diverse enzyme family. Proteins 87, 443–451. doi: 10.1002/prot.25666
Hofmann, L., Tsybovsky, Y., Alexander, N. S., Babino, D., Leung, N. Y., Montell, C., et al. (2016). Structural insights into the Drosophila melanogaster retinol dehydrogenase, a member of the short-chain dehydrogenase/reductase family. Biochemistry 55, 6545–6557. doi: 10.1021/acs.biochem.6b00907
Kallberg, Y., Oppermann, U., Jörnvall, H., and Persson, B. (2002). Short-chain dehydrogenases/reductases (SDRs). Eur. J. Biochem. 269, 4409–4417. doi: 10.1046/j.1432-1033.2002.03130.x
Kavanagh, K. L., Jörnvall, H., Persson, B., and Oppermann, U. (2008). Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65, 3895–3906. doi: 10.1007/s00018-008-8588-y
Laskar, A. A., and Younus, H. (2019). Aldehyde toxicity and metabolism: the role of aldehyde dehydrogenases in detoxification, drug resistance and carcinogenesis. Drug Metab. Rev. 51, 42–64. doi: 10.1080/03602532.2018.1555587
Lukacik, P., Keller, B., Bunkoczi, G., Kavanagh, K. L., Lee, W. H., Adamski, J., et al. (2007). Structural and biochemical characterization of human orphan DHRS10 reveals a novel cytosolic enzyme with steroid dehydrogenase activity. Biochem. J. 402, 419–427. doi: 10.1042/BJ20061319
Luo, W., Du, H. J., Bonku, E. M., Hou, Y. L., Li, L. L., Wang, X. Q., et al. (2019). An alkali-tolerant carbonyl reductase from Bacillus subtilis by gene mining: identification and application. Catal. Lett. 149, 2973–2983. doi: 10.1007/s10562-019-02873-w
MacKenzie, A. K., Kershaw, N. J., Hernandez, H., Robinson, C. V., Schofield, C. J., and Andersson, I. (2007). Clavulanic acid dehydrogenase: structural and biochemical analysis of the final step in the biosynthesis of the beta-lactamase inhibitor clavulanic acid. Biochemistry 46, 1523–1533. doi: 10.1021/bi061978x
Mattheus, W., Gao, L. J., Herdewijn, P., Landuyt, B., Verhaegen, J., Masschelein, J., et al. (2010). Isolation and purification of a new kalimantacin/batumin-related polyketide antibiotic and elucidation of its biosynthesis gene cluster. Chem. Biol. 17, 149–159. doi: 10.1016/j.chembiol.2010.01.014
Mo, X., Gui, C., and Wang, Q. (2017a). Elucidation of a carboxylate O-methyltransferase NcmP in nocamycin biosynthetic biosynthetic pathway. Bioorg. Med. Chem. Lett. 27, 4431–4435. doi: 10.1016/j.bmcl.2017.08.010
Mo, X., Huang, H., Ma, J., Wang, Z., Wang, B., Zhang, S., et al. (2011). Characterization of TrdL as a 10-hydroxy dehydrogenase and generation of new analogues from a tirandamycin biosynthetic pathway. Org. Lett. 13, 2212–2215. doi: 10.1021/ol200447h
Mo, X., Shi, C., Gui, C., Zhang, Y., Ju, J., and Wang, Q. (2017b). Identification of nocamycin biosynthetic gene cluster from Saccharothrix syringae NRRL B-16468 and generation of new nocamycin derivatives by manipulating gene cluster. Microb. Cell Fact. 16:100. doi: 10.1186/s12934-017-0718-5
Moon, H. J., Tiwari, M. K., Singh, R., Kang, Y. C., and Lee, J. K. (2012). Molecular determinants of the cofactor specificity of ribitol dehydrogenase, a short-chain dehydrogenase/reductase. Appl. Environ. Microbiol. 78, 3079–3086. doi: 10.1128/AEM.07751-11
Persson, B., and Kallberg, Y. (2013). Classification and nomenclature of the superfamily of short-chain dehydrogenases/reductases (SDRs). Chem. Biol. Interact. 202, 111–115. doi: 10.1016/j.cbi.2012.11.009
Savino, S., Borg, A. J. E., Dennig, A., Pfeiffer, M., de Giorgi, F., Weber, H., et al. (2019). Deciphering the enzymatic mechanism of sugar ring contraction in UDP-apiose biosynthesis. Nat. Catal. 2, 1115–1123. doi: 10.1038/s41929-019-0382-8
Shanati, T., and Ansorge-Schumacher, M. B. (2020). Biodegradation of ephedrine isomers by Arthrobacter sp. strain TS-15: discovery of novel ephedrine and pseudoephedrine dehydrogenases. Appl. Environ. Microbiol. 86:e02487–19. doi: 10.1128/AEM.02487-19
Shanati, T., Lockie, C., Beloti, L., Grogan, G., and Ansorge-Schumacher, M. B. (2019). Two enantiocomplementary ephedrine dehydrogenases from Arthrobacter sp. TS-15 with broad substrate specificity. ACS Catal. 9, 6202–6211. doi: 10.1021/acscatal.9b00621
Sonawane, P. D., Heinig, U., Panda, S., Gilboa, N. S., Yona, M., Kumar, S. P., et al. (2018). Short-chain dehydrogenase/reductase governs steroidal specialized metabolites structural diversity and toxicity in the genus Solanum. Proc. Natl. Acad. Sci. USA. 115, E5419–E5428. doi: 10.1073/pnas.1804835115
Stavrinides, A. K., Tatsis, E. C., Dang, T. T., Caputi, L., Stevenson, C. E. M., Lawson, D. M., et al. (2018). Discovery of a short-chain dehydrogenase from Catharanthus roseus that produces a new monoterpene indole alkaloid. Chembiochem 19, 940–948. doi: 10.1002/cbic.201700621
Su, B. M., Shao, Z. H., Li, A. P., Naeem, M., Lin, J., Ye, L. D., et al. (2020). Rational design of dehydrogenase/reductases based on comparative structural analysis of prereaction-state and free-state simulations for efficient asymmetric reduction of bulky aryl ketones. ACS Catal. 10, 864–876. doi: 10.1021/acscatal.9b04778
Takase, R., Mikami, B., Kawai, S., Murata, K., and Hashimoto, W. (2014). Structure-based conversion of the coenzyme requirement of a short-chain dehydrogenase/reductase involved in bacterial alginate metabolism. J. Biol. Chem. 289, 33198–33214. doi: 10.1074/jbc.M114.585661
Tsukiura, H., Tomita, K., Hanada, M., Kobaru, S., Tsunakawa, M., Fujisawa, M., et al. (1980). Bu-2313, a new antibiotic complex active against anaerobes I. production, isolation and properties of Bu-2313 A and B. J. Antibiot. 33, 157–165. doi: 10.7164/antibiotics.33.157
Tsunakawa, M., Toda, S., Okita, T., Hanada, M., Nakagawa, S., Tsukiura, H., et al. (1980). Bu-2313, a new antibiotic complex active against anaerobes II. Structure determination of Bu-2313 a and B. J. Antibiot. 33, 166–172. doi: 10.7164/antibiotics.33.166
Wallwey, C., Matuschek, M., and Li, S. M. (2010). Ergot alkaloid biosynthesis in Aspergillus fumigatus: conversion of chanoclavine-I to chanoclavine-I aldehyde catalyzed by a short-chain alcohol dehydrogenase FgaDH. Arch. Microbiol. 192, 127–134. doi: 10.1007/s00203-009-0536-1
Keywords: nocamycin, gene inactivation, biosynthetic pathway, Saccharothrix syringae, short chain dehydrogenase/reductase
Citation: Mo X, Zhang H, Du F and Yang S (2020) Short-Chain Dehydrogenase NcmD Is Responsible for the C-10 Oxidation of Nocamycin F in Nocamycin Biosynthesis. Front. Microbiol. 11:610827. doi: 10.3389/fmicb.2020.610827
Edited by:
Yang-Chun Yong, Jiangsu University, ChinaReviewed by:
Willem JH Van Berkel, Wageningen University and Research, NetherlandsWolfgang Buckel, University of Marburg, Germany
Copyright © 2020 Mo, Zhang, Du and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuhua Mo, eGhtb0BxYXUuZWR1LmNu; eGhtbzIwMTNAMTYzLmNvbQ==; Song Yang, eWFuZ3NvbmcxMjA5QDE2My5jb20=
 Xuhua Mo
Xuhua Mo Hui Zhang
Hui Zhang Fengyu Du
Fengyu Du Song Yang
Song Yang