
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 November 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.610179
Halobacterium salinarum forms gas vesicles consisting of a protein wall surrounding a gas-filled space. The hydrophobic 8-kDa protein GvpA is the major constituent of the ribbed wall, stabilized by GvpC at the exterior surface. In addition, eight accessory Gvp proteins are involved, encoded by gvpFGHIJKLM that are co-transcribed in early stages of growth. Most of these proteins are essential, but their functions are not yet clear. Here we investigate whether GvpF through GvpM interact. Pull-down experiments performed in Haloferax volcanii with the cellulose-binding-domain as tag suggested many interactions, and most of these were supported by the split-GFP analyses. The latter study indicated that GvpL attracted all other accessory Gvp, and the related GvpF bound besides GvpL also GvpG, GvpH and GvpI. A strong interaction was found between GvpH and GvpI. GvpG showed affinity to GvpF and GvpL, whereas GvpJ, GvpK and GvpM bound GvpL only. Using GvpA for similar analyses yielded GvpF as the only interaction partner. The contact site of GvpF was confined to the N-terminal half of GvpA and subsequently mapped to certain amino acids. Taken together, our results support the idea that the accessory Gvp form a complex early in gas-vesicle assembly attracting GvpA via GvpF.
Halophilic archaea (haloarchaea) thrive in hypersaline environments such as salt lakes or salterns containing up to 30% NaCl. They adapt to these salty conditions by maintaining a similarly high KCl concentration in the cytoplasm. The haloarchaea Halobacterium salinarum and Haloferax mediterranei produce gas vesicles allowing the cells to float to the surface of the brine. These gas-filled vesicles consist of a wall exclusively formed by proteins. Major component is the hydrophobic, 8-kDa GvpA aggregating into a low-pitch helix seen by transmission electron microscopy as 4.6 nm wide ribs running perpendicular to the long axis of the gas vesicle (Walsby, 1994; Offner et al., 1998; Pfeifer, 2012). The protein wall is stabilized on the exterior surface by the second structural protein, GvpC (Englert and Pfeifer, 1993). Gases are able to diffuse in and out, presumably via small holes in the wall. Water vapor might enter the hollow space, but is unable to precipitate because of the hydrophobic interior surface of the wall (Walsby, 1994). Electron microscopic studies of haloarchaeal cells indicate small bicones already filled with gas in early stages of growth; these bicones are enlarged to spindle- or cylinder-shaped structures. The final diameter of the haloarchaeal gas vesicle is 200–250 nm; and the cylinder-shaped structures can grow as long as 2 μm (Walsby, 1994; Knitsch et al., 2017).
Gas vesicles of haloarchaea are encoded by a gene cluster consisting of 14 gas vesicle protein (gvp) genes arranged as gvpACNO and gvpDEFGHIJKLM, and eight of these genes are essential to produce a gas-filled particle as determined in Haloferax volcanii transformants (Englert et al., 1992; Offner et al., 2000). This moderately halophilic haloarchaeon is easy to transform, grows faster than Hbt. salinarum and lacks all of the gvp genes. The six non-essential Gvp proteins are the surface-attached GvpC, the two regulatory proteins GvpD and GvpE (Krüger et al., 1998; Zimmermann and Pfeifer, 2003; Hofacker et al., 2004), and the proteins GvpH, GvpI and GvpN (Offner et al., 2000). The absence of GvpC in Hfx. volcanii ΔC transformants (the ΔC construct contains except gvpC all gvp genes) leads to long and weak gas vesicles with altering diameters (Offner et al., 1996), whereas the absence of GvpD and GvpE causes a low expression of the gvp gene cluster. In the case of GvpH and GvpI altered gas vesicle structures are observed. ΔH transformants produce weaker gas vesicles of wild type shape, whereas extremely long and cylinder-shaped gas vesicles are found in ΔI transformants. The lack of GvpN leads to small bicones that are not enlarged (Offner et al., 2000). A deletion of any other gvp gene in the cluster leads to gas vesicle negative (Vac–) transformants (Offner et al., 2000). Isolated gas vesicles cannot be disintegrated into their protein constituents and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE); only GvpC is washed off and separated on the gel (Englert and Pfeifer, 1993). Aggregates of the hydrophobic GvpA dissolve in 80% formic acid only, and dialysis to remove the formic acids leads to amorphous precipitates of GvpA (Belenky et al., 2004). The protein constituents of the gas vesicles were determined by MS/MS-based proteome analyses or immunological methods (Shukla and DasSarma, 2004; Chu et al., 2011). Except for GvpD, GvpE, and GvpK, all Gvp proteins are present in gas vesicle preparations suggesting that they are constituents of the wall or are attached to the structure during gas-vesicle assembly.
The transcript encoding the accessory gas vesicle proteins GvpF through GvpM occurs in early exponential growth, implying that these proteins are required in early stages of gas-vesicle assembly (Offner and Pfeifer, 1995). Putative interactions of GvpM have been studied by affinity chromatography with His-tagged Gvp and Ni-NTA matrices, and the results suggest that GvpM is able to bind GvpH, GvpJ and GvpL, but not GvpG (Tavlaridou et al., 2014). For these analyses, HisGvp proteins were synthesized in Escherichia coli, purified under denaturing conditions and refolded in high salt solutions. Haloarchaea require 15–30% NaCl for growth, and adapt by maintaining a similarly high KCl concentration in the cytoplasm. The haloarchaeal proteins are usually adapted to these salt concentrations and often denature in low salt solutions. Thus, the purification of these proteins from E. coli in low-salt solutions might have influenced the protein structure and interaction. Also, other histidine-rich haloarchaeal proteins bind non-specifically to the Ni-NTA or Ni-sepharose matrices used for the binding of His-tagged Gvp to select putative binding partners and complicate the analysis.
To study putative interactions of Gvp proteins in high salt, we recently adapted the split-GFP method (Ghosh et al., 2000; Magliery et al., 2005) to high salt solutions and studied protein-protein interactions in Hfx. volcanii transformants (Winter et al., 2018). We used a derivative of the salt-adapted green fluorescent protein smRS-GFP (Reuter and Maupin-Furlow, 2004) with higher fluorescence, mGFP2 (Born and Pfeifer, 2019). The mGFP2 protein was split in the N- and C-terminal fragments (NGFP and CGFP) that were fused to the two proteins of interest. Both fragments do not assemble in trans, but will form a fluorescent GFP when the two fusion partners interact. The fluorescence of the transformants can be easily quantified (Winter et al., 2018). For each protein, four different N/CGFP fusions (N- or CGFP fused N- or C-terminally) are produced and eight combinations tested per protein pair to exclude putative steric hindrances for the assembly of GFP. Using this method, the interaction of GvpL/GvpM was confirmed, and the interaction site in GvpM confined to the N-terminal 25-amino acid (aa) (Winter et al., 2018). GvpM also interacted with GvpF and GvpH, and both proteins bound to the C-terminal 25 aa of GvpM (Winter et al., 2018).
In this report, we present a comprehensive study on the interactions of the accessory proteins GvpF through GvpM. Two different methods were applied, i.e., pull-down experiments with the cellulose-binding domain, CBD, and the in vivo analysis by split-GFP. The cellulose binding domain derives from Clostridium thermocellum and is part of the CipB protein (Poole et al., 1992; Morag et al., 1995). Proteins tagged with CBD can be selected by cellulose in high salt solutions (Ortenberg and Mevarech, 2000; Irihimovitch and Eichler, 2003; Schlesner et al., 2012). The CBDGvp proteins and their putative Gvp binding partners were selected from lysates of Hfx. volcanii producing both bait and prey proteins. Each of the accessory Gvp proteins interacted with other Gvp, and CBDGvpM attracted all of them at once. In addition, each protein pair was tested by split-GFP in Hfx. volcanii transformants, and an interaction network was deduced. GvpL and GvpF had several interaction partners, whereas GvpH and GvpI bound each other and also interacted with GvpF and GvpL. In addition, the major gas vesicle structural protein GvpA was tested by split-GFP for interactions with these accessory proteins, and GvpF appeared to be the only binding partner. More detailed analyses with fragments of GvpA and variants of GvpA carrying single substitutions confined the interaction site of GvpF in GvpA.
The Escherichia coli strains One Shot Top10 (Invitrogen by Life Technologies) and GM1674 (dam–) (Palmer and Marinus, 1994) were grown in Luria-Bertani media at 37°C overnight. For the selection of transformants, 100 μg/mL ampicillin was added. The haloarchaeon Haloferax volcanii WR340 was grown in salt media containing 3 M NaCl, 150 mM MgSO4, 50 mM KCl, 0.05% (w/v) CaCl2, 25 mM Tris–HCl pH 7.2, 10 nM MnCl2, 0.5% (w/v) tryptone, 0.3% (w/v) yeast extract and 0.02% (w/v) histidine. For solid media, 1.8% (w/v) agar was added. To select Hfx. volcanii transformants, 6 μg/mL mevinolin (for selection of pWLfdx) and 0.2 μg/mL novobiocin (for selection of pJAS35) were added. Cultures on solid medium were incubated at 42°C for 3–7 days in a humid atmosphere. Liquid cultures were grown for 3–5 days on a shaker at 180 rpm and 42°C. Cultures forming GFP were initially incubated at 37°C for 24 h before they were transferred to 30°C overnight to enhance fluorescence, always shaking at 180 rpm.
The split-GFP shuttle vectors pJAS-NGFP-Nterm and -Cterm, as well as pWL-CGFP-Nterm and -Cterm have been described previously (Winter et al., 2018). They are based on the two compatible vector plasmids pJAS35 (Pfeifer et al., 1994) and pWLfdx (Scheuch and Pfeifer, 2007). The salt adapted mGFP2 is split between the residues 157 and 158, resulting in the NGFP and CGFP fragments. Nterm describes a fusion of the respective mgfp2 fragment to the 5′-terminus of the gene of interest, whereas Cterm stands for a fusion at the 3′-terminus. The respective mgfp2 fragments plus linker region are present in these vectors. The size of the linker encoded by the pJAS-derived shuttle vector is 14 aa [(GGSGSGS)2], whereas the linker derived from the pWLfdx vector is 16 aa long [(GGSG)4] (Winter et al., 2018). The gvp reading frame under investigation was fused to ngfp or cgfp by inserting the respective fragment in these vector plasmids. The gvp reading frames were amplified by PCR using the p-vac region as template (containing the 14 gvp genes of Hbt. salinarum PHH1; Englert et al., 1992), and oligonucleotides including the desired restriction site for insertion (Supplementary Table 1). The NcoI-gvp-BlpI fragments amplified were inserted in pJAS-NGFP-Nterm or pJAS-NGFP-Cterm. For the insertion of gvp in pWL-CGFP-Nterm, BamHI and KpnI sites were used, and NcoI and BamHI for the insertion in pWL-CGFP-Cterm. In some cases, BspHI (gvpF, gvpI gvpJ, gvpL) or PciI (gvpG), was used instead of NcoI. Due to the presence of a KpnI site in gvpH, the amplified gvpH fragment was blunt-ended inserted in pWL-CGFP-Nterm.
For pull-down assays of CBD-tagged proteins, the shuttle vector pCBD was used (Supplementary Figure 1). The cbd reading frame encodes the cellulose binding domain of the CipB protein from Clostridium thermocellum (Poole et al., 1992; Morag et al., 1995). The cbd reading frame was amplified from plasmid pWL-CBD-sec11b (Fine et al., 2006) and inserted in pWLfdx via NcoI and KpnI. Two additional cloning sites are present (BamHI, XbaI) allowing the insertion of the respective gvp reading frame upstream or downstream of cbd [NcoI and BamHI upstream of cbd are used to yield XCBD, and XbaI and KpnI downstream to yield CBDX (with X = GvpF, G, H, I, J, K, L, or M)]. The expression of the respective gene fusion is under control of the strong ferredoxin promoter, Pfdx (Pfeifer et al., 1994). The gvp sequences were amplified using p-vac as template. The oligonucleotides used are listed in Supplementary Table 1. For the construction of pF-Lex, the gvpFGHIJKL reading frames of p-vac were amplified as XbaI–KpnI fragment and inserted in pJAS35 for expression under Pfdx control.
In all cases, the correct insertion was verified by DNA sequence analysis. To avoid a restriction barrier in Hfx. volcanii WR340, the plasmid DNA was demethylated by passage through E. coli GM1674 (dam–) (Palmer and Marinus, 1994). Hfx. volcanii was transformed simultaneously with the two vector plasmids as described previously (Pfeifer and Ghahraman, 1993). In each case, the presence of both plasmids was confirmed by PCR, and the presence of the respective Gvp protein determined by Western analysis.
To verify the expression of gvp-gfp or gvp-cbd fusions, total protein was isolated from 50 mL cultures of the transformants in the late exponential growth phase. Cells were harvested by centrifugation (2,370 × g, 30 min, 4°C) and re-suspended in 5 mL lysis buffer (2.5 M KCl, 50 mM MgCl2, 1 mM EDTA, 5% (v/v) glycerol, 50 mM Tris–HCl pH 8.0) + DNaseI (0.1 mg/mL). Cells were lysed by sonification on ice. The lysate was cleared from cell debris by centrifugation and dialyzed against 10 mM Tris–HCl pH 7.2 overnight at 20°C to eliminate salts. 20 μg of total protein was separated by SDS-PAGE (Schägger and von Jagow, 1987) and transferred to a PVDF membrane (Roti Fluoro PVDF, Carl Roth) using a Semidry-Blotter (PerfectBlueTM, 30 min, 2 mA). The membrane was dried for one hour, reactivated with 100% (v/v) methanol and washed two times in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) before blocking for an hour with Odyssey Blocking Buffer (Licor). The membrane was incubated with the respective antiserum raised against GvpF, GvpG, GvpH, GvpI, GvpJ, GvpK, GvpL, GvpM, or isolated gas vesicles (to detect GvpA). The membrane was washed four times in PBS + 0.1% (v/v) Tween20, incubated with the secondary antibodies labeled with IRDye 800 CW (Licor) for 1–2 h at 20°C, and washed four times for 5 min in PBS + 0.1% (v/v) Tween20. Excessive Tween20 was removed by washing the membrane with PBS. The fluorophore coupled to the secondary antibody is detectable at 800 nm with the Odyssey Fc Imager (Licor).
The fluorescence of Hfx. volcanii transformants was measured to determine the formation of fluorescent GFP as a result of an interaction of the two fused proteins of interest. Transformants were grown as described to OD600nm 1.5–2. Two mL of the culture were harvested (2 min, 9,600 x g, 20°C), the cells washed in 1 mL basal salts (3 M NaCl, 150 mM MgSO4, 50 mM KCl), and re-suspended in 500 μL basal salts. To investigate equal amounts of cells, the OD value was adjusted to OD600nm 1, and 300 μL of the culture transferred into a microtiter dish. For each Gvp protein combination, two biological and three technical replicates were investigated. The fluorescence was measured (Phosphorimager FLA-5000 and the software Fujifilm science lab image gauge ver. 4.24), and the fluorescence intensity obtained in light absorbing units (LAU) per mm2. The relative fluorescence, rf, was calculated as described (Winter et al., 2018). The p-value was determined by Student’s T-test.
Each accessory Gvp was fused to the cellulose binding domain, CBD, at the N- or C-terminus (CBDX or XCBD) and lysates of the respective CBDX/Y transformants were tested in pulldown experiments (X, Y = any accessory Gvp). In the case of CBDM, also CBDM/pF-Lex transformants were tested. For each pull-down experiment, 400 mL cultures were grown at 37°C, 180 rpm to the late exponential growth phase. The cells were harvested (12,210 × g, 20 min, 4°C), and resuspended in 5 mL lysis buffer (2.5 M KCl, 50 mM MgCl2, 1 mM EDTA, 5% (v/v) glycerol, 50 mM Tris–HCl pH 8.0) plus DNaseI (0.1 mg/mL). The cells were lysed by ultrasound (Branson Sonifier 250, duty cycle: 55%, output control: 5, 2 min) and the suspension centrifuged for 20 min at 2,370 × g, 4°C. The soluble protein fraction (7 mL) was incubated with 1 mL of a 10% (w/v) cellulose suspension (Avicel PH-101, Sigma Aldrich) for 30 min at room temperature on an overhead rotator. The suspension was centrifuged (2,370 × g, 30 s), the supernatant removed and the resulting cellulose pellet with bound proteins resuspended in 600 μL washing buffer (2.5 M KCl, 50 mM Tris–HCl pH 8.0). The solution was transferred to an empty Mobicol column (Mobitec) and centrifuged (1,000 × g, 1 min), followed by six washing steps with 600 μl washing buffer. For protein elution, the cellulose was resuspended in 500 μL 100% ethylene glycol, incubated for 1 min at room temperature and centrifuged at 4,700 × g, 5 min. All fractions were dialyzed against 10 mM Tris–HCl, pH 7.2 overnight. For the analysis of proteins, 15 μL of each fraction were separated by SDS-PAGE (Schägger and von Jagow, 1987). The Gvp proteins were detected by Western analysis using the respective antisera.
Two different methods were applied to investigate protein-protein interactions. The first approach was pull-down experiments using the cellulose-binding domain as tag, and the second approach was split-GFP to identify interaction partners in vivo.
Putative interactions of the accessory proteins GvpF through GvpM were investigated by pull-down experiments using the cellulose binding domain, CBD, allowing tagged proteins to bind cellulose at high salt concentrations (2–3 M KCl) (Ortenberg and Mevarech, 2000; Irihimovitch and Eichler, 2003; Schlesner et al., 2012). CBD was fused to the N- or C-terminus of each accessory Gvp using the vector plasmid pCBD (Supplementary Figure 1). The resulting CBDX or XCBD constructs (X = any accessory GvpF through GvpM) were used to transform Haloferax volcanii WR340. To test whether the CBD-fusion proteins can be indeed purified in 2 M KCl, lysates of the respective CBDX transformants were mixed with cellulose to bind the CBD-tagged Gvp, and the elution fractions were investigated by SDS-PAGE as well as Western analysis using antisera raised against the various Gvp proteins (Supplementary Figure 2). Except for GvpJ and GvpM, the accessory Gvp were isolated in decent amounts (Table 1), and well visible in the Coomassie-stained gels (Supplementary Figure 2). GvpJ and GvpM were obtained in much lower amounts, presumably due to their hydrophobic nature and tendency to aggregate. Aggregates of GvpJ and GvpM might be present in the solid fraction of the cell extracts, and the addition of the detergents DDM (n-dodecyl-β-D-maltoside) or OGP (octyl-β-D-glucopyranoside) to the lysate and wash buffer slightly improved the presence of these proteins in the soluble fraction (Table 1). All of the accessory Gvp proteins including GvpJ and GvpM were detectable by Western analysis (Supplementary Figure 2). A smear of larger bands is always detectable with GvpJ, indicative of its ability to aggregate (Supplementary Figure 2). Thus, each of the CBDGvp proteins was selected and could be purified. To ensure that the untagged Gvp interacted neither with the cellulose matrix nor with CBD, transformants were produced harboring the plasmids pCBD (without gvp) and pXex (gvpX-pJAS35 expressing any gvp without cbd fusion). None of the Gvp proteins tested was detectable in the respective elution fraction demonstrating that none of them bound to cellulose or to CBD itself (data not shown).
The CBD-tagged Gvp proteins were used as bait and tested for the selection of a putative Gvp interaction partner produced in the same cell. The first protein pair tested was GvpL/GvpM, where an interaction has been demonstrated using His-tagged Gvp proteins. GvpL or GvpM carried CBD fused to the N- or C-terminus (CBDL, LCBD or CBDM, MCBD), and the combinations CBDL/M, LCBD/M, CBDM/L and MCBD/L were analyzed. Lysates of the CBDL/M and LCBD/M transformants were tested for the presence of GvpM, and monomers and dimers of this protein were identified by Western analysis (Supplementary Figure 3). Also, GvpL was present in the elution fractions of the CBDM/L and MCBD/L transformants indicating that GvpL bound GvpM (Supplementary Figure 3). These results confirmed the GvpL/GvpM interaction already seen using the respective His-tagged Gvp proteins and a Ni-NTA matrix for affinity chromatography (Tavlaridou et al., 2014).
Similar pull-down experiments with CBD were performed with all other accessory Gvp proteins. The respective Hfx. volcanii transformants always carried two constructs, one for the bait protein (CBDX), and one for the prey GvpY (with X, Y = F, G, H, I, J, K, L, or M). Both reading frames encoding bait and prey were expressed under Pfdx promoter control to ensure a similarly high expression. Not all possible interactions were performed in both ways (CBDX+Y and CBDY+X), since protein interactions were already found with one protein pair. The results of these studies are presented in Figure 1 and summarized Table 2. The Western blots in Figure 1 are arranged according to the antiserum used to detect the prey protein. Using CBDX to select GvpM yielded monomeric GvpM with CBDH, and monomers as well as dimers with CBDF, CBDG, CBDJ, CBDK, and CBDL (Figure 1). Dimers and multimers of GvpM were observed with CBDI (Figure 1). Thus, GvpM was pulled-down in all these cases. GvpL was selected by CBDF, but the results obtained with CBDH or CBDI were ambiguous (Figure 1). However, when CBDL was used as bait, GvpM, GvpK, GvpJ, or GvpG were selected again suggesting that all these accessory Gvp bound GvpL. Also, GvpK interacted with any of the accessory Gvp, since CBDF through CBDJ and CBDL pulled-down GvpK, and CBDK recovered GvpM. Monomers of GvpJ were selected by CBDX, but a smear including additional GvpJ multimers were found with CBDF, CBDH, CBDI, CBDL, or CBDM (Figure 1). GvpI was only used as bait, and CBDI pulled-down any of the accessory Gvp proteins. It is interesting to note that CBDI often induced the formation of larger oligomers, especially with GvpM, GvpK or GvpJ as prey. GvpH was selected by CBDF or CBDI, and CBDH pulled down GvpG, GvpJ, GvpK, GvpL or GvpM (Figure 1 and Table 2). GvpG monomers and dimers were selected by CBDF, whereas CBDH, CBDI and CBDL selected the dimer of GvpG (and additional multimers in the case of CBDI) (Figure 1). Since CBDG bound GvpM, GvpK and GvpJ, these results demonstrated that GvpG is able to interact with all accessory proteins. Similar results were obtained with GvpF, since GvpF was pulled down by CBDI, and CBDF pulled down GvpG, GvpH, and GvpJ through GvpM (Figure 1 and Table 2). Overall, the results of these pull-down experiments implied that any accessory Gvp had multiple interaction partners and suggested that these proteins might form a larger complex.
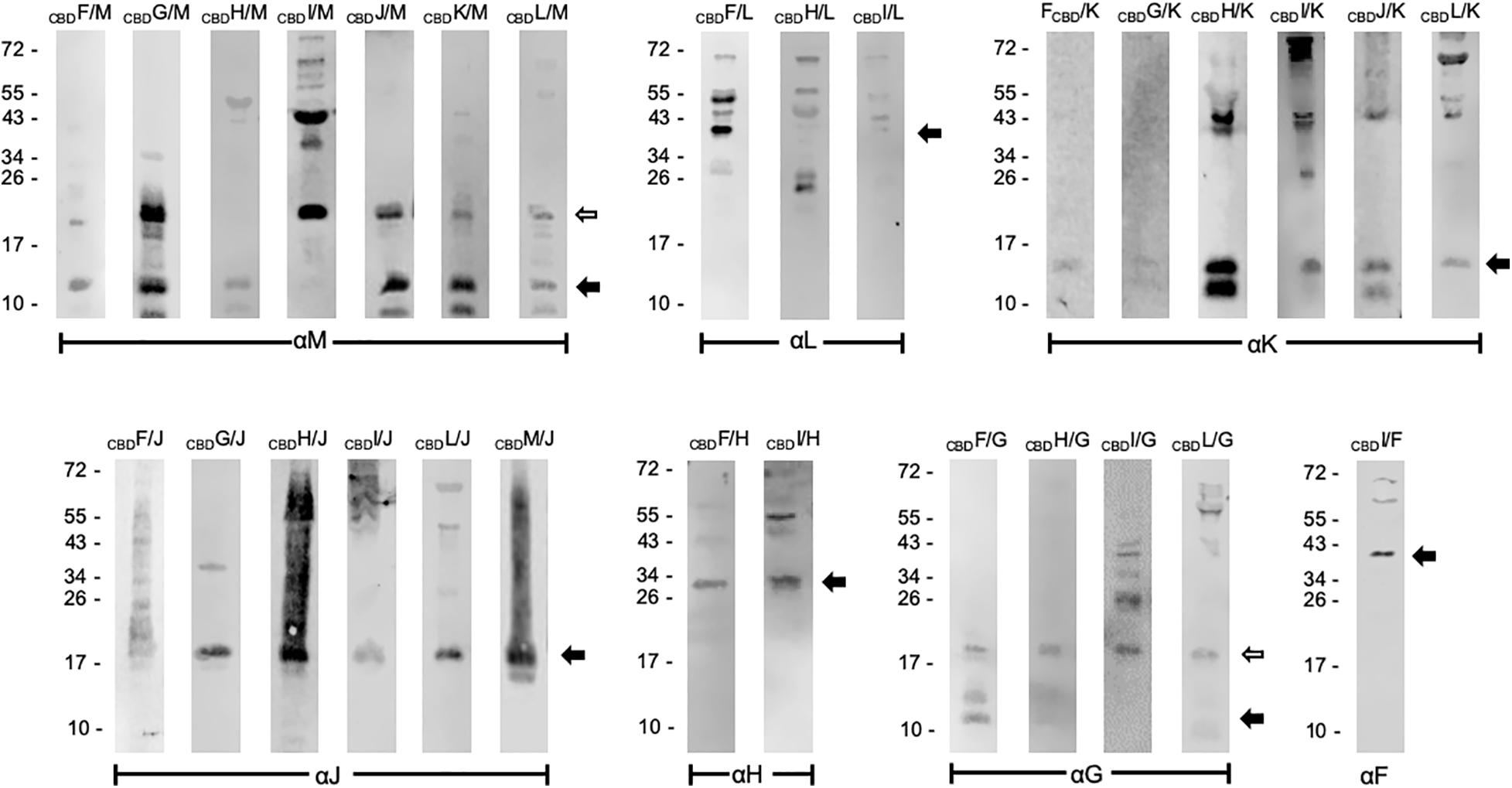
Figure 1. Western analyses of the pull-down assays using CBDX. The bait CBDX and the prey protein Y (X, Y = any GvpF through GvpM protein) were synthesized in the same Hfx. volcanii cell. Both are marked on the top of the blots. Only the elution fraction of the cellulose matrix is shown. In each case, 15 μL of the elution fraction was separated by SDS-PAGE, the proteins transferred to a PVDF membrane and incubated with the respective antiserum indicated underneath (αF through αM). The putative interaction partners were visualized with the fluorophore-labeled secondary antibody IRDye 800 CW (Licor). The blots are inverted to black-white and arranged according to the respective antiserum used. Arrows mark the expected Gvp monomer and dimer. Numbers on the left are size markers in kDa.
To investigate whether GvpM was able to select all accessory Gvp at once, Hfx. volcanii transformants were produced carrying CBDM and in addition the plasmid pF-Lex expressing gvpFGHIJKL under the strong Pfdx promoter control. The interaction partners of CBDM were selected by pull-down experiments from the lysate of this transformant, and samples were tested by Western analyses using the different antisera (Figure 2). All accessory Gvp were identified, implying that CBDM attracted all of them. In each case the respective monomeric Gvp was detected. In the case of GvpG, two 30–34 kDa bands were visible in addition to the expected 10-kDa GvpG protein, and GvpJ and GvpK also formed multimers (Figure 2). The monomer of the CBDM bait protein was also detected (Figure 2). Overall, CBDM was able to pull down GvpF through GvpL. It is possible that each of these Gvp proteins bound independently to CBDM, but it is also possible that some or all of them formed a complex that bound to GvpM.
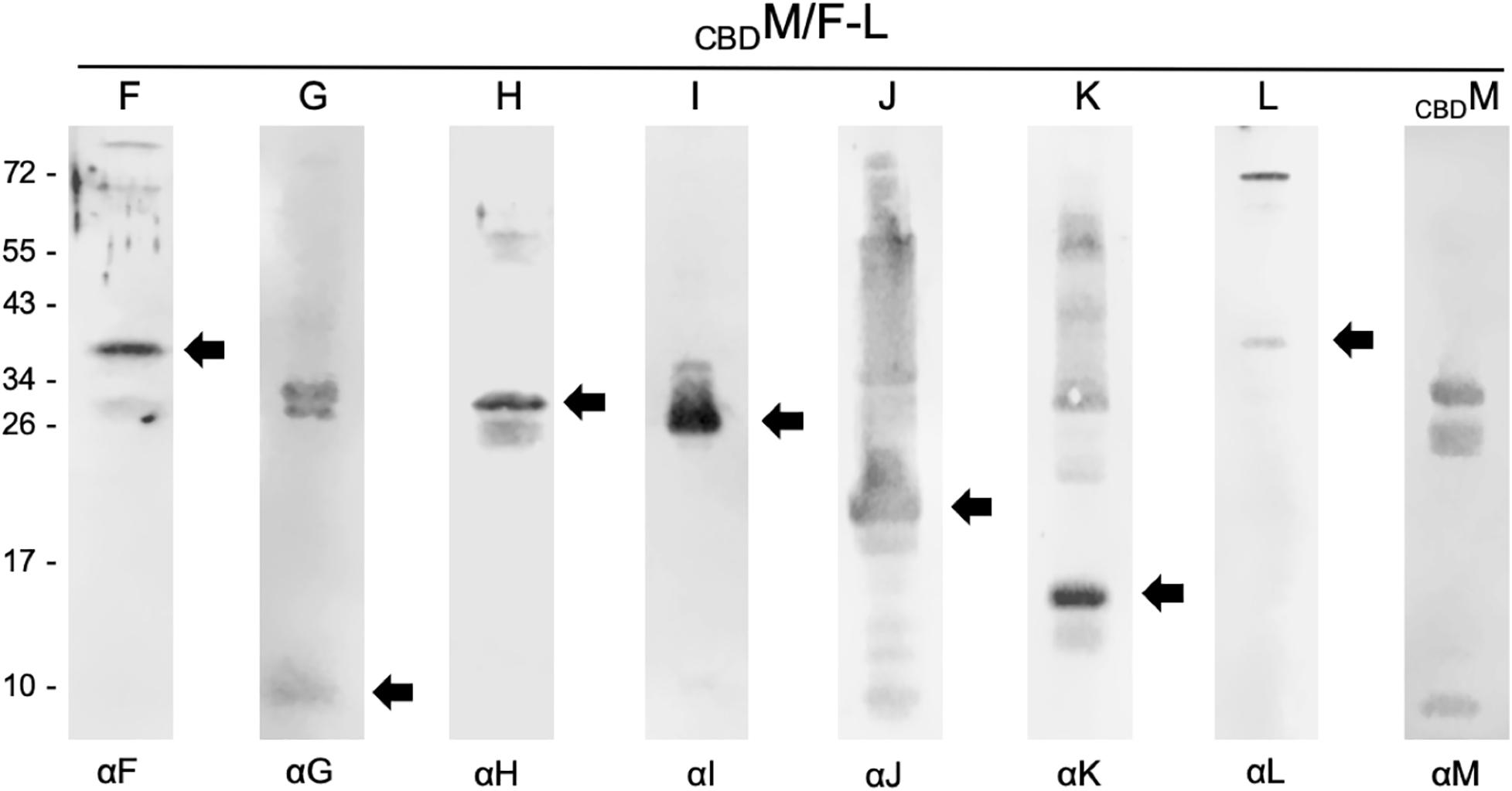
Figure 2. Western analyses of proteins selected by CBDM in CBDM/pF-Lex transformants. In each case, 15 μL of the elution fraction was separated by SDS-PAGE, transferred to a PVDF membrane and incubated with the respective antiserum marked underneath (αF through αM). Reactions are visualized with the fluorophore-labeled secondary antibody IRDye 800 CW (Licor). Arrows mark the expected Gvp monomer detected. The blots were inverted to black-white. Numbers on the left side are size markers in kDa.
Pairwise interactions of the accessory Gvp proteins were studied in Hfx. volcanii in vivo using the split-GFP method. Each gvp reading frame was amplified by PCR and inserted in the four vector plasmids to fuse the ngfp- or cgfp reading frame to the 5′ or 3′ terminus of each gvp (Winter et al., 2018). The N- and C-terminal GFP fragments NGFP and CGFP derive from the salt-adapted, green fluorescent protein mGFP2 (Born and Pfeifer, 2019). A fluorescent GFP is only formed when the two fusion partners interact and steric hindrance does not occur. The reading frame of each fusion protein is expressed under the Pfdx promoter control to yield similarly large amounts of these proteins. The four N/CGFP fusions were designated NX, or CX for the N-terminal fusion, and XC or XN for the C-terminal fusion with Gvp (with X = respective accessory Gvp; C = CGFP; N = NGFP). The eight combinations of a protein pair were tested in Hfx. volcanii transformants and the fluorescence was measured in arbitrary light absorbing units per mm2. The highest relative fluorescence (rf-values) calculated for each interaction pair are shown in Supplementary Figure 4, and the original data obtained in LAU/mm2 are presented in Supplementary Figure 5. A summary of these data is shown in Figure 3.
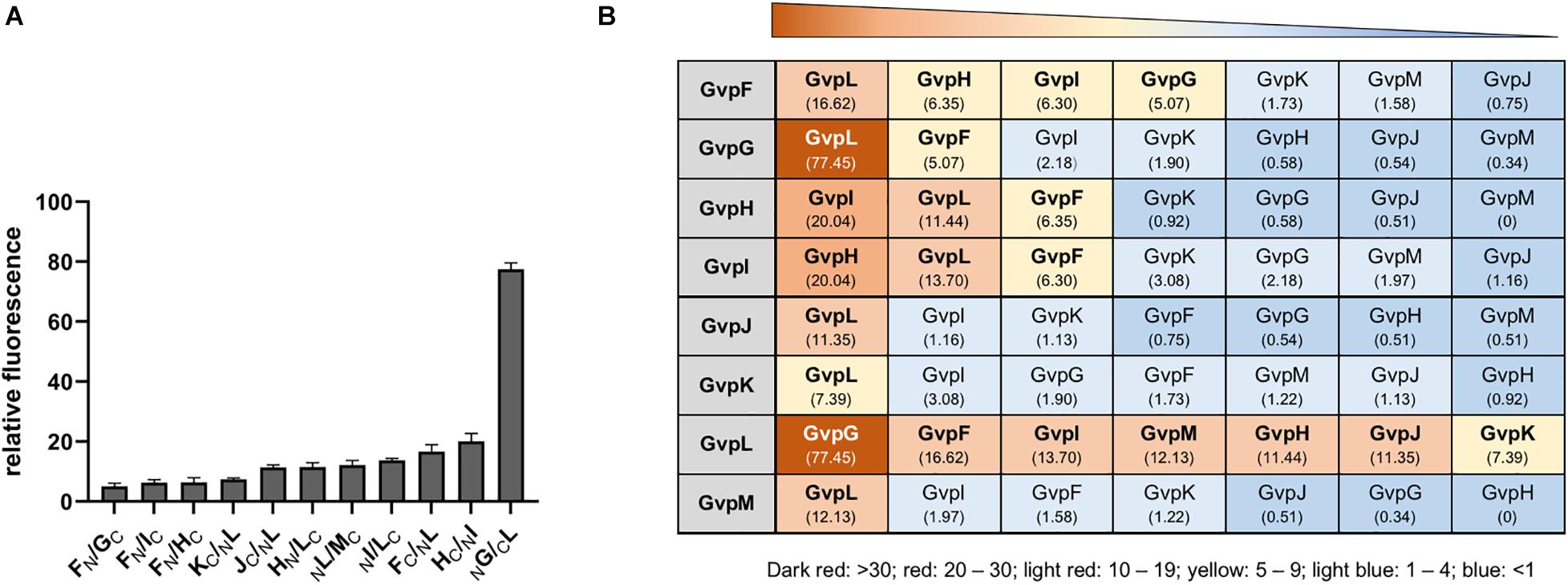
Figure 3. Interactions of the accessory Gvp determined by split-GFP. (A) Highest relative fluorescence (rf-values) determined for the protein-protein interactions calculated as described in the methods section. All experiments were performed with two biological and three technical replicates. (B) Summary of the different protein-protein interactions determined by split-GFP. The Gvp proteins tested are shown in the gray box on the left and their interaction partners are arranged according to the highest rf-value determined (given underneath). A fluorescence exceeding rf > 5 was regarded as clear interaction. Rf-values < rf 4 are regarded as weak or no interaction.
The highest relative fluorescence of all interactions was observed with the NG/CL transformant (rf 77.5) implying a strong interaction of GvpL with GvpG (Figure 3A). All other interaction partners of GvpL yielded rf-values below 20. Rf-values between 10 and 20 were observed for the interaction FC/NL, NI/LC, MC/LN, HN/LC, and JC/NL, and rf 7.4 was determined for KC/NL (Figures 3A,B and Supplementary Figure 5). These results implied that the 32-kDa GvpL interacted with all other accessory Gvp. GvpL is the largest of the accessory Gvp and might act as platform to bind all others. Also, GvpF interacted with several Gvp proteins (GvpL, GvpH, GvpI, and GvpG) (Figure 3B). The FC/LN transformant (rf 16.6) exhibited the highest relative fluorescence, whereas the other three binding partners yielded lower rf-values (rf 5.0–6.3). In the case of GvpF it should be mentioned that the highest fluorescence was always observed when N- or CGFP was fused to the C-terminus of GvpF, suggesting that a fusion to the N-terminus hinders the assembly of mGFP2. The 23-kDa GvpF is smaller than GvpL, but both proteins exhibit sequence similarities and a similar 3D-structure when modeled using the crystal structure of the cyanobacterial GvpF as template (Xu et al., 2014; Winter et al., 2018).
The second highest fluorescence (rf 20) of all combinations was observed for the HC/NI transformant (Figure 3), implying that GvpH and GvpI attract each other. Both proteins also interacted with GvpF and GvpL (rf 6.3). Two partner proteins were identified for GvpG (GvpL and GvpF), and GvpL appeared to be the only interaction partner of the three proteins GvpJ, GvpK and GvpM (Figure 3B). Overall, these results obtained by these split-GFP analyses demonstrated that all accessory Gvp proteins had at least one other Gvp protein as interaction partner. Since GvpL bound all of them it is possible that they form a larger complex. Compared to the results obtained by affinity chromatography using CBDGvp, less interactions were observed with split-GFP, especially for the two hydrophobic proteins GvpJ (12 kDa) and GvpM (9.2 kDa), and for GvpK (12.6 kDa).
To uncover interactions between the major structural gas vesicle protein GvpA and these accessory Gvp, each combination of A/X (X = GvpF through GvpM) was investigated by split-GFP. GvpA was fused at the N- or C-terminus to N- or CGFP and tested pairwise with the respective N/CGFP fusion variants of GvpF through GvpM. Eight different combinations were investigated for each pair, and the highest rf-values calculated in each case are shown in Figure 4A. The original data obtained in these experiments is presented in Supplementary Figure 6. The highest relative fluorescence was obtained for the AC/FN transformant (rf 20), whereas all other transformants yielded rf-values < 1, suggesting very weak contacts between GvpA and the other Gvp proteins (Figure 4A). Except of NA/LC, the highest fluorescence of a protein pair was always observed when N- or CGFP was fused to the C-terminus of GvpA. These results implied that GvpF is the only interaction partner of GvpA, and that the N-terminal region of GvpA might be involved in the interaction.
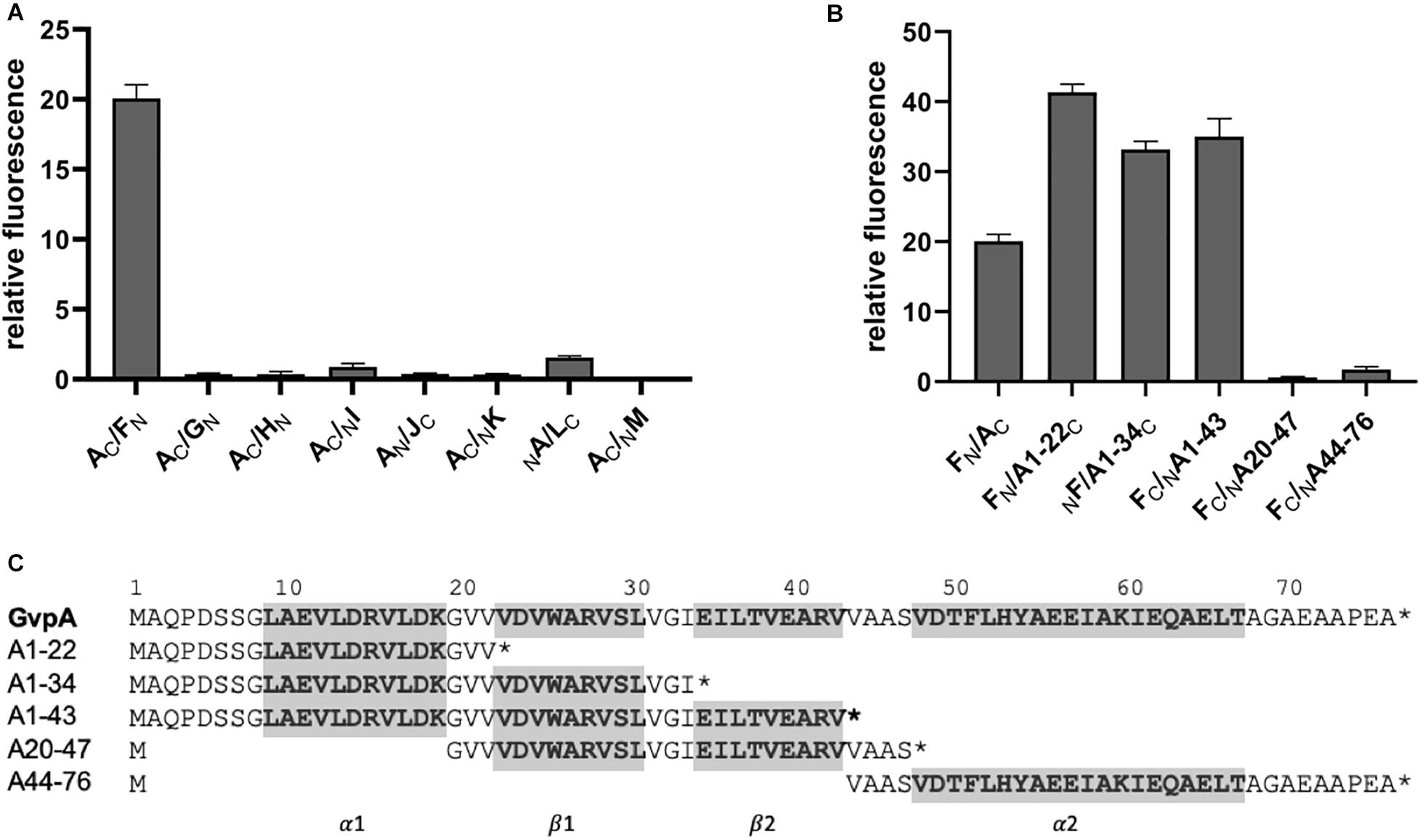
Figure 4. Split-GFP interaction studies with GvpA/GvpX. Only the highest relative fluorescence determined for each combination is given. Two biological and three technical replicates were performed in each case. (A) Interaction study of GvpA with the eight accessory proteins GvpF through GvpM. (B) Interaction of five different GvpA fragments with GvpF. (C) Sequence of the five GvpA fragments in relation to the aa sequence of GvpA shown on top. Numbers above the sequence depict the aa positions in GvpA. The helices α1 and α2 and the β-sheets β1 and β2 are in bold and marked on the bottom and also shaded in gray.
To define the interaction site of GvpF in GvpA more precisely, five GvpA fragments harboring different structural features (according to the model obtained by Strunk et al. (2011)) were studied by split-GFP. Fragment A1-22 encompasses the first 22 amino acids of GvpA including α-helix 1 (α1), fragment A1-34 contains in addition β-sheet 1 (α1-β1), and A1-43 the α1-β1-β2 elements of GvpA (Figure 4C). Fragment A20-47 contains β1-β2, and A44-76 the α-helix 2 up to the C-terminus of GvpA (α2) (Figure 4C). Each of these GvpA fragments was fused at the N- or C-terminus to N- or CGFP and tested with the respective N/CGFP fusion of GvpF. Transformant FN/A1-22C yielded the highest fluorescence (rf 41), i.e., more than twice as high as obtained for the interaction of GvpF with the entire GvpA (rf 20) (Figure 4B). Smaller protein fragments offer less steric hindrance supporting the interaction (Ghosh et al., 2000; Winter et al., 2018). An increased fluorescence was also found for the transformants harboring F/A1-34 and F/A1-43 (rf 33 – 35), whereas a very low relative fluorescence (rf < 1) was obtained for the transformants harboring F/A20-47 or F/A44-76. These results implied that the C-terminal portion of GvpA is not involved, and that the N-terminal portion contains the interaction site of GvpF.
To further confine the interaction site of GvpF in GvpA, various substitution variants of GvpA were tested by split-GFP. Many of these point mutations in GvpA are known to influence the formation of gas vesicles in ΔA+Amut transformants (Strunk et al., 2011; Knitsch et al., 2017). These transformants carry two vector plasmids, the ΔA construct (contains except gvpA all gvp genes of the p-vac region) and construct A (gvpA or mutant gvpA expressed in pMDS20 under the control of the native PA promoter). The different GvpA variants were investigated by split-GFP analysis for their potential to interact with GvpF in Hfx. volcanii. The FN fusion protein was used as interaction partner in all these cases (Figure 5A). The strongest reductions in relative fluorescence (rf < 6) compared to GvpA wild type (rf 15-18) were observed for the GvpA substitutions G20D, G20A, D24A, D24Y, R28A, R28D, or E40A, but also R15A, K19D, A27E, or G33V resulted in a low relative fluorescence of the F/Amut transformants (Figure 5A and Supplementary Figure 6). Especially the charged aa in β1 and β2 (D24, R28, E40) and G20 of GvpA had a strong influence on the interaction with GvpF (Figure 5). All these aa are located in the N-terminal half of GvpA, whereas any of the single substitutions in the C-terminal half had no effect on the interaction with GvpF, supporting the results described above. It is likely that these aa of GvpA are involved in the GvpF/GvpA interaction. In respect to the gas vesicle phenotype of the ΔA+Amut transformants, most of these mutations result in a Vac– phenotype, except for D24A (cylindrical) and R28A (mini gas vesicles) (Knitsch et al., 2017; Figure 5B). It is possible that the Vac– phenotype is caused by the lack of an interaction between GvpF and GvpA, rather than (or in addition to) an influence of the mutation on the GvpA/GvpA contact in the gas vesicle wall.
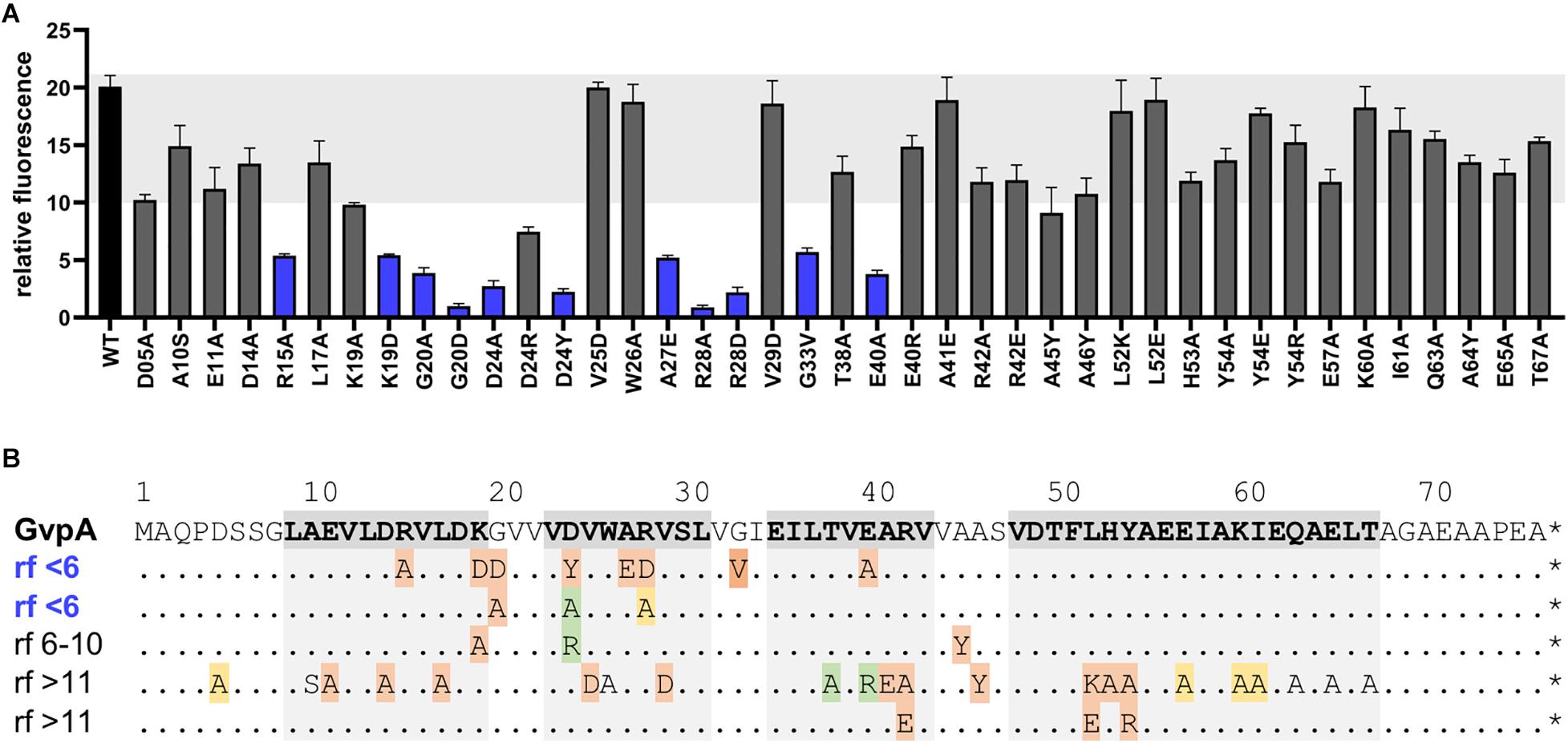
Figure 5. Split-GFP studies of GvpF and variants of GvpAmut. (A) Rf-values of the GvpF/GvpA (WT) and the various F/Amut transformants. The single substitutions in GvpA are indicated below. F/Amut transformants with relative fluorescence <6 are labeled in blue. The fluorescence was determined in LAU/mm2, and the relative fluorescence in relation to the fluorescence of Hfx. volcanii wild type calculated. Two biological and three technical replicates were performed in each case. (B). Sequence of GvpA and summary of the rf-values of the various GvpA mutants. The sequence of GvpA is presented on top and the structural features α1–β1–β2–α2 shaded in grey. The substitutions in GvpA are summarized underneath; each GvpA contains only a single substitution. Rf < 6 is regarded as weak interaction, whereas rf > 11 is similar to GvpA wild type. The Vac phenotype of the respective ΔA+Amut transformants is indicated by color: red, Vac negative; green, cylinder shaped; yellow, mini gas vesicles; without color, wild type gas vesicles.
Gas-vesicle assembly involves twelve Gvp proteins, two of which, GvpA and GvpC, form the gas vesicle wall. A third protein, GvpN, is required for the enlargement of the structure, and the function of GvpO is not yet known (Pfeifer, 2012). The accessory proteins GvpF through GvpM are encoded by an operon and produced in minor amounts in early exponential growth. All eight accessory Gvp were investigated pairwise in respect to their interaction(s) and also with the major gas vesicle structural protein, GvpA.
Earlier pull-down experiments with HisGvp proteins indicated non-specific binding of other histidine-rich proteins (such as PitA) to the Ni-NTA matrix, and also non-specific reactions of the Gvp antisera in Western analyses (Tavlaridou et al., 2014, and unpublished observations). In contrast, pull-down assays with CBDGvp on a cellulose matrix yielded a high specificity in the selection. In most cases, single protein bands were detected in the elution fraction, but also multimers were observed, especially with GvpJ, GvpK, and GvpM. None of the Gvp proteins bound to cellulose when the CBD tag was lacking, and the putative binding partner(s) of a given Gvp protein were selected in the respective CBDX/Y Hfx. volcanii transformants. Earlier studies with HisM implied an interaction of GvpM with GvpH, GvpJ, and GvpL, but not with GvpG (Tavlaridou et al., 2014). Testing the selection of these Gvp with CBDM verified these interactions, but GvpM also interacted with GvpG. A reason for this result could be that the pull-down experiments based on CBD-tagged proteins were performed in vivo with transformants synthesizing both bait and prey, whereas the earlier analyses with HisGvp were performed in vitro with proteins isolated from E. coli and refolded in high salt solutions. Only the prey protein was synthesized in Hfx. volcanii (Tavlaridou et al., 2014). The larger range of interactions observed with CBDX compared to HisX might be due to the correct folding of bait and prey under in vivo conditions. It should be noted that GvpJ and GvpM, but also GvpK oligomerized in the presence of CBDI, whereas other CBDX selected only monomers and/or dimers of these accessory Gvp. The very positively charged GvpI (pK 10.75) might be able to bind and connect the negatively charged Gvp (pK 4–5). GvpI contains 23 lysine and arginine residues in the N-terminal 53 aa, and the involvement of these aa in the oligomerization of the other proteins and also in the assembly of gas vesicles should be analyzed in further detail.
The results obtained by the pairwise CBDX/Y pull-down experiments suggested that all accessory Gvp proteins interact. In almost all cases, the prey proteins were detected by Western analyses. A quantitation of the binding efficiency is difficult, since different antisera were used. A quantitation of the interaction was easier using the split-GFP method and measuring the fluorescence of the cells in vivo. The interaction network derived from the pull-down studies suggested that all Gvp proteins attract each other and possibly form a complex. Studies with CBDM+pF-Lex transformants demonstrated that GvpM indeed selected all the Gvp proteins at once (Figure 2). It is possible that any of these proteins interacted solely with GvpM, but it is likely that they bound as a complex to the bait. GvpM is required in early steps of gas-vesicle assembly as deduced from studies with ΔM+Mmut transformants harboring construct ΔM complemented by GvpM variants with single aa substitutions (Tavlaridou et al., 2014). Only Vac+ or Vac– transformants were obtained, whereas variants in gas vesicle shape (as found with many ΔA+Amut transformants) were not observed, implying that GvpM is involved in an early step in gas-vesicle assembly.
The split-GFP method is easy to apply and the interactions can be semi-quantified. Each protein pair under investigation requires the analysis of eight combinations to exclude a possible steric hindrance in the assembly of GFP (Winter et al., 2018). Not all of the transformants tested were significantly fluorescent. The calculated relative fluorescence (rf) varied from rf-values < 1, regarded as no protein-protein interaction, over rf 1–5 regarded as weak, to rf > 5 regarded as clear interaction. The data was used to establish a network of the accessory Gvp including all interactions exceeding rf 5 (Figure 6).
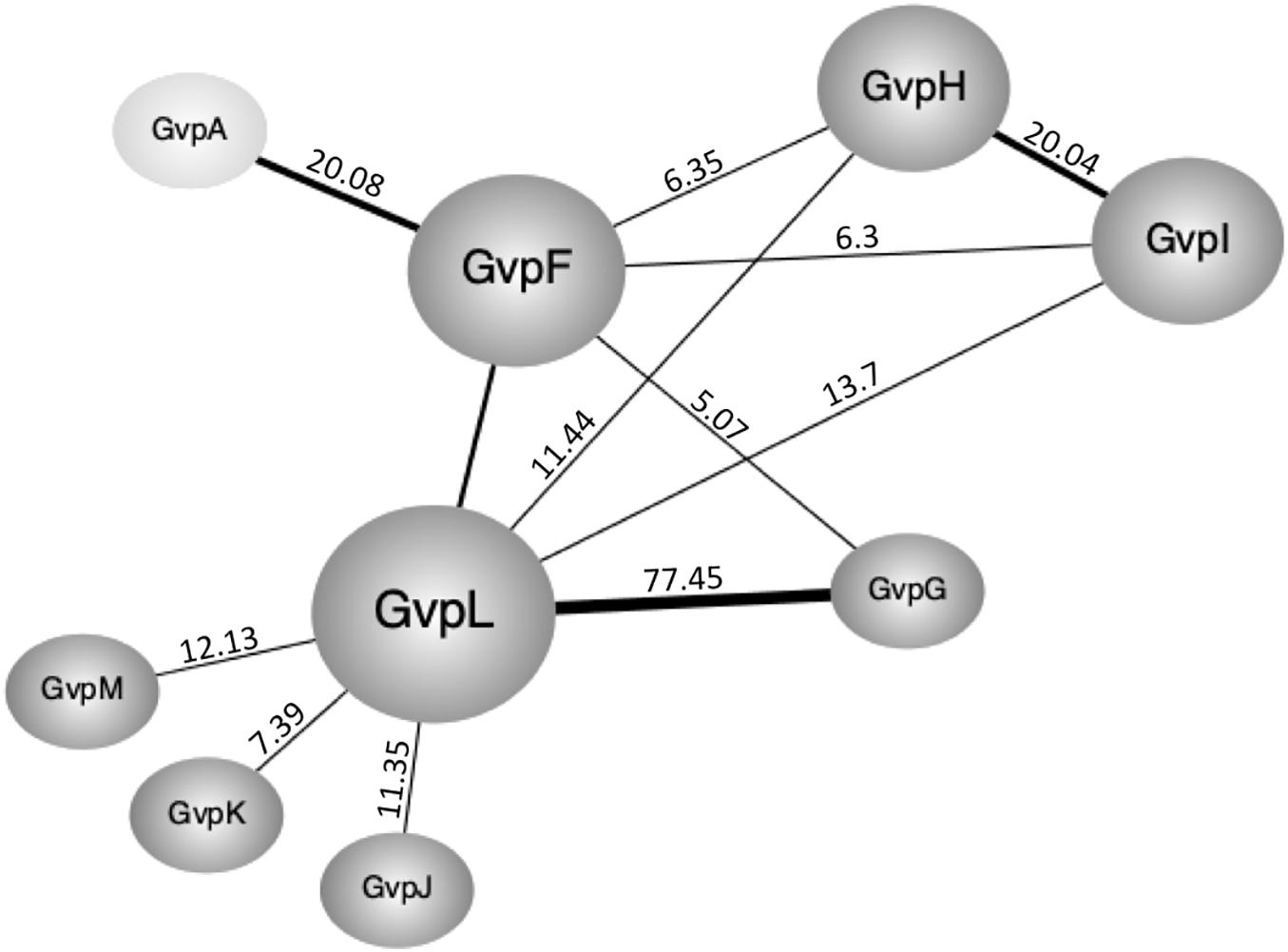
Figure 6. Interaction network of the accessory Gvp and GvpA as obtained by split-GFP. Only clear interactions (rf > 5) are incorporated. The respective rf-values are given at the connecting lines. Rf-values > 20 are indicated by thicker lines.
The highest number of protein contacts was observed for GvpL that was shown to interact with all other accessory Gvp. The highest rf-value of all combinations tested was found with GvpL/GvpG (rf 77.45). The 10-kDa GvpG is relatively small, similar to GvpJ, GvpK, and GvpM (9.2–12.6 kDa). The latter three proteins yielded GvpL as sole interaction partner, whereas GvpG also contacted GvpF. It is possible that the hydrophobic nature of GvpM and GvpJ and their tendency to aggregate prevented the assembly of sufficient fluorescent GFP in some cases. GvpL (32 kDa) and GvpF (23 kDa) are structurally related as determined by homology modeling using a crystal structure of GvpF derived from the cyanobacterium Microcystis aeruginosa as template (Xu et al., 2014; Winter et al., 2018). Since GvpL attracted all other accessory Gvp, this protein might play a central role in the formation of a protein complex. However, it is not known so far whether the binding of the other Gvp occurs at the same time or sequentially.
The interaction partners of GvpF were GvpG, GvpH, GvpI, and GvpL as deduced from the split-GFP analyses (Figures 3, 6). GvpH (19.8 kDa) and GvpI (15.8 kDa) are interesting, since both are non-essential for gas vesicle formation and appear to interact with each other; a relatively high fluorescence (rf 20) was observed for GvpH/GvpI. Both proteins interacted also with GvpL and GvpF. The binding to GvpL resulted in higher rf-values (rf 11.4, GvpH and rf 13.7, GvpI) compared to the rf-values obtained with GvpF (rf 6.3 in both cases) suggesting that the preferred interaction partner of GvpH and GvpI is GvpL. It is possible that GvpH and GvpI form a heterodimer that binds only temporary to the putative complex. GvpH influences the stability of the gas vesicles, whereas a lack of the positively charged GvpI results in extremely long gas vesicles (Offner et al., 2000). Previous studies also suggest that the presence of GvpH prevents the aggregation of GvpM (Tavlaridou et al., 2014). As shown by the pull-down experiments performed in this report, CBDH selected GvpM as monomer and not as dimer, whereas all other accessory Gvp selected GvpM additionally as dimer (Figure 1). In contrast, GvpI induced the oligomerization of GvpJ, GvpK, and GvpM (Figure 1). To uncover the exact functions of GvpH and GvpI during gas vesicle formation, both proteins should be investigated in further detail.
Overall, the interaction studies by split-GFP yielded less interactions compared to the pull-down experiments, especially for the hydrophobic GvpJ and GvpM, but also for GvpK. These three proteins often form multimers as observed when investigated by SDS-PAGE. Studies on self-interactions of the Gvp proteins yielded only weak interactions for F/F (rf 1.5), or G/G (rf 3.5), whereas J/J, K/K or M/M transformants were not fluorescent (Supplementary Figure 7). The formation of additional multimers might prevent the formation of fluorescent GFP. These results implied that self-interaction of aggregating proteins is difficult to detect by split-GFP.
The analysis of the major gas vesicle structural protein, GvpA, by split-GFP uncovered GvpF as sole binding partner. This result implied that a GvpA/GvpF complex is formed, but since GvpF also attracts GvpG, GvpH, GvpI, and via GvpL all other accessory Gvp, it is also possible that a larger complex of the accessory Gvp binds GvpA via GvpF. The latter possibility is more likely, since the amount of GvpF is much smaller than the amount of the major gas vesicle protein GvpA. GvpF is only produced in early growth stages (together with GvpG through GvpM), and not in parallel to the massive production of GvpA later in growth. The PA promoter for the transcription of gvpA is 10-fold induced by GvpE, whereas the PF promoter for the transcription gvpFGHIJKLM is less active and not activated by GvpE as determined using GFP as reporter (Born and Pfeifer, 2019). Thus, the GvpF/GvpA binding cannot occur on a 1:1 basis throughout growth, and should happen in early stages of gas vesicle formation.
The contact site of GvpF in GvpA was confined to the first 40 aa of GvpA, since substitutions in the second half of this 76-aa protein had no influence on the GvpA/GvpF interaction. Several single substitutions of amino acids in the N-terminal half of GvpA influenced the GvpA/GvpF interaction, and all these positions are also important for gas vesicle formation (Knitsch et al., 2017). These aa are located in α1, β1 and β2 of GvpA, and the loop regions in between. Except for D24, the charged aa R15, K19, R28, E40 are located on the surface of GvpA (Figure 7; Strunk et al., 2011). These aa are thought to form salt bridges to adjacent GvpA molecules in the gas vesicle wall (Sivertsen et al., 2010; Knitsch et al., 2017), but as shown here they also mediate the GvpA/GvpF contact (when GvpF is available). It is possible that GvpF contacts GvpA prior to the incorporation of GvpA in the gas vesicle wall, to keep this strongly hydrophobic protein in solution (especially by shielding β1-β2) and prevent an undesirable self-aggregation. It is likely that the accessory proteins including GvpF initiate gas vesicle formation by forming a nucleation complex that binds GvpA via GvpF and forces GvpA to start aggregating into the ribs of the gas vesicle wall; this hypothesis has been raised already in Shukla and DasSarma (2004).
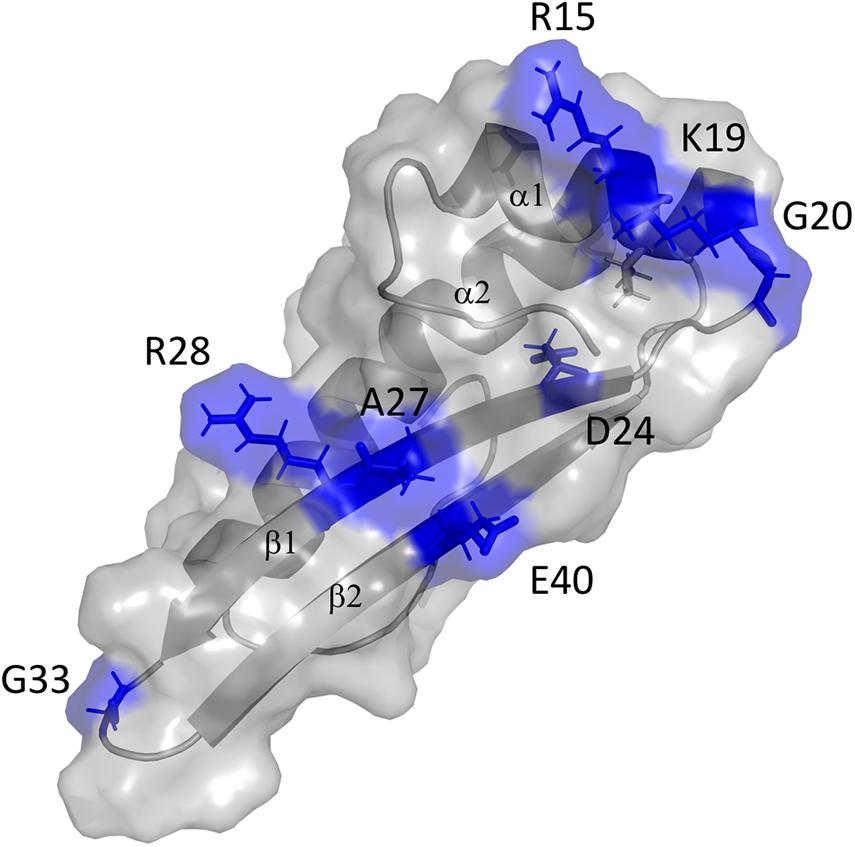
Figure 7. 3D-model of GvpA and putative interaction site of GvpF. The GvpA in silico model is according to Strunk et al. (2011). The secondary structural features α1, β1, β2, and α2 are labeled. The amino acids marked and colored in blue are positions where a substitution resulted in a low relative fluorescence of the F/Amut transformants.
A complex (or complexes) of the accessory Gvp proteins might initiate the formation of this gas-filled structure. The aggregation of GvpA to form the ribs of the gas vesicle wall most likely starts at the tips of the two conical end caps of each gas vesicle (Waaland and Branton, 1969), and the small bicones formed are enlarged by further addition of GvpA, and of GvpC at the exterior side. A larger protein complex is required at the tip that binds GvpA to initiate the formation of the gas vesicle wall. The hydrophobic interior surface of ribs formed by GvpA needs to be shielded, especially when the diameter of the conical structure is still small. A complex of the accessory Gvp in both tips could protect and ensure that the ribs become distant enough to expose the hydrophobic surface of GvpA at the interior side and form a hollow structure that rapidly fills with gas. The analysis of the cyanobacterial GvpF by immunogold labeling-based tomography suggests that GvpF is localized at the gas-facing surface of the gas vesicle wall (Xu et al., 2014), supporting this hypothesis. The accessory Gvp are presumably not only required in the tip, but also at the transition of the conical caps into the cylinder structure. In addition, helper proteins might be required at the site where the incorporation of GvpA occurs to enlarge the gas vesicle wall. Presumably this site is located in the center of the gas vesicle, and GvpN might be involved here. All these steps in gas-vesicle assembly are not yet understood, but the accessory Gvp proteins and their complex(es) certainly take part in these processes. Further studies will aim to identify the complex(es) formed by the accessory proteins and determine the location in the gas vesicles structure.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
All authors planned the study, discussed the results, wrote the manuscript, and approved the final manuscript. KV and AJ performed the analysis.
This work was financially supported by the German Research Foundation, DFG (PF 165/15–1). We also acknowledged support by the German Research Foundation and the Open Access Publishing Fund of Technical University Darmstadt.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Jerry Eichler (Ben-Gurion University, Israel) providing the vector encoding the cellulose binding domain, CBD. The students Gayathiri Thavayogarajah, Regine Knitsch, and Sergio Leftheriotis (TU Darmstadt) are thanked to supply some of the vectors used in this study. Valuable discussions with Johannes Born and Arnulf Kletzin (TU Darmstadt) are gratefully acknowledged.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.610179/full#supplementary-material
Belenky, M., Meyers, R., and Herzfeld, J. (2004). Subunit structure of gas vesicles: a MALDI-TOF mass spectrometry study. Biophys. J. 86, 499–505. doi: 10.1016/S0006-3495(04)74128-4
Born, J., and Pfeifer, F. (2019). Improved GFP variants to study gene expression in haloarchaea. Front. Microbiol. 10:1200. doi: 10.3389/fmicb.2019.01200
Chu, L. J., Chen, M. C., Setter, J., Tsai, Y. S., Yang, H., Fang, X., et al. (2011). New structural proteins of Halobacterium salinarum gas vesicle revealed by comparative proteomics analysis. J. Proteome Res. 10, 1170–1178. doi: 10.1021/pr1009383
Englert, C., Krüger, K., Offner, S., and Pfeifer, F. (1992). Three different but related gene clusters encoding gas vesicles in halophilic archaea. J. Mol. Biol. 227, 586–592. doi: 10.1016/0022-2836(92)90914-6
Englert, C., and Pfeifer, F. (1993). Analysis of gas vesicle gene expression in Haloferax mediterranei reveals that GvpA and GvpC are both gas vesicle structural proteins. J. Biol. Chem. 268, 9329–9336.
Fine, A., Irihimovitch, V., Dahan, I., Konrad, Z., and Eichler, J. (2006). Cloning, expression, and purification of functional Sec11a and Sec11b, Type I signal peptidases of the archaeon Haloferax volcanii. J. Bacteriol. 188, 1911–1919.
Ghosh, I., Hamilton, A. D., and Regan, L. (2000). Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J. Am. Chem. Soc. 122, 5658–5659. doi: 10.1021/ja994421w
Hofacker, A., Schmitz, K. M., Cichonczyk, A., Sartorius-Neef, S., and Pfeifer, F. (2004). GvpE- and GvpD-mediated transcription regulation of the p-gvp genes encoding gas vesicles in Halobacterium salinarum. Microbiology 150, 1829–1838. doi: 10.1099/mic.0.27078-0
Irihimovitch, V., and Eichler, J. (2003). Post-translational secretion of fusion proteins in the halophilic archaea Haloferax volcanii. J. Biol. Chem. 278, 12881–12887. doi: 10.1074/jbc.M210762200
Knitsch, R., Schneefeld, M., Weitzel, K., and Pfeifer, F. (2017). Mutations in the major gas vesicle protein GvpA and impacts on gas vesicle formation in Haloferax volcanii. Mol. Microbiol. 106, 530–542. doi: 10.1111/mmi.13833
Krüger, K., Hermann, T., Armbruster, V., and Pfeifer, F. (1998). The transcriptional activator GvpE for the haloarchaeal gas vesicle genes resembles a basic leucine-zipper regulatory protein. J. Mol. Biol. 279, 761–771. doi: 10.1006/jmbi.1998.1795
Magliery, T. J., Wilson, C. G. M., Pan, W., Mishler, D., Ghosh, I., Hamilton, A. D., et al. (2005). Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J.Am. Chem. Soc. 127, 146–157. doi: 10.1021/ja046699g
Morag, E., Lapidot, A., Govorko, D., Lamed, R., Wilchek, M., Bayer, E. A., et al. (1995). Expression, purification, and characterization of the cellulose-binding domain of the scaffolding subunit from the cellulosome of Clostridium thermocellum. Appl. Environ. Microbiol. 61, 1980–1986. doi: 10.1128/AEM.61.5.1980-1986.1995
Offner, S., Hofacker, A., Wanner, G., and Pfeifer, F. (2000). Eight of fourteen gvp genes are sufficient for formation of gas vesicles in halophilic archaea. J. Bacteriol. 182, 4328–4336. doi: 10.1128/JB.182.15.4328-4336.2000
Offner, S., and Pfeifer, F. (1995). Complementation studies with the gas vesicle encoding p-vac region of Halobacteirum salinarium PHH1 reveal a regulatory role for the p-gvpDE genes. Mol. Microbiol. 16, 9–19. doi: 10.1111/j.1365-2958.1995.tb02387.x
Offner, S., Wanner, G., and Pfeifer, F. (1996). Functional studies of the gvpACNO operon of Halobacterium salinarium reveal that the GvpC protein shapes gas vesicles. J. Bacteriol. 178, 2071–2078. doi: 10.1128/jb.178.7.2071-2078.1996
Offner, S., Ziese, U., Wanner, G., Typke, D., and Pfeifer, F. (1998). Structural characteristics of halobacterial gas vesicles. Microbiology 144, 1331–1342. doi: 10.1099/00221287-144-5-1331
Ortenberg, R., and Mevarech, M. (2000). Evidence for post-translational membrane insertion of the intergral membrane protein bacterioopsin expressed in the heterologous halophilic archaeon Haloferax volcanii. J. Biol. Chem. 275, 22839–22846. doi: 10.1074/jbc.M908916199
Palmer, B. R., and Marinus, M. G. (1994). The dam and dcm strains of Escherichia coli - a review. Gene 143, 1–12. doi: 10.1016/0378-1119(94)90597-5
Pfeifer, F. (2012). Distribution, formation and regulation of gas vesicles. Nat. Rev. Microbiol. 10, 705–715. doi: 10.1038/nrmicro2834
Pfeifer, F., and Ghahraman, P. (1993). Plasmid pHH1 of Halobacterium salinarium: characterization of the replicon region, the gas vesicle gene cluster and insertion elements. Mol. Gen. Genet. 238, 193–200. doi: 10.1007/BF00279547
Pfeifer, F., Offner, S., Krüger, K., Ghahraman, P., and Englert, C. (1994). Transformation of halophilic archaea and investigation of gas vesicle synthesis. Syst. Appl. Microbiol. 16, 569–577. doi: 10.1016/S0723-2020(11)80327-9
Poole, D. M., Morag, E., Lamed, R., Bayer, E. A., Hazlewood, G. P., and Gilbert, H. J. (1992). Identification of the cellulose-binding domain of the cellulosome subunit S1 from Clostridium thermocellum YS. FEMS Microbiol. Lett. 99, 181–186. doi: 10.1016/0378-1097(92)90022-G
Reuter, C. J., and Maupin-Furlow, J. A. (2004). Analysis of proteasome-dependent proteolysis in Haloferax volcanii cells, using short-lived green fluorescent proteins. Appl. Environ. Microbiol. 70, 7530–7538. doi: 10.1128/AEM.70.12.7530-7538.2004
Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecylsulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. doi: 10.1016/0003-2697(87)90587-2
Scheuch, S., and Pfeifer, F. (2007). GvpD-induced breakdown of the transcriptional activator GvpE of halophilic archaea requires a functional p-loop and an arginine-rich region of GvpD. Microbiology 153, 947–958. doi: 10.1099/mic.0.2006/004499-0
Schlesner, M., Miller, A., Besir, H., Aivaliotis, M., Streif, J., Scheffer, B., et al. (2012). The protein interaction network of a taxis signal transduction system in a halophilic archaeon. BMC Microbiol. 12:272. doi: 10.1186/1471-2180-12-272
Shukla, H. D., and DasSarma, S. (2004). Complexity of gas vesicle biogenesis in Halobacterium sp. strain NRC-1: identification of five new proteins. J. Bacteriol. 186, 3182–3186. doi: 10.1128/jb.186.10.3182-3186.2004
Sivertsen, A. C., Bayro, M. J., Belenky, M., Griffin, R. G., and Herzfeld, J. (2010). Solid-state NMR characterization of gas vesicle structure. Biophys. J. 99, 1932–1939. doi: 10.1016/j.bpj.2010.06.041
Strunk, T., Hamacher, K., Hoffgaard, F., Engelhardt, H., Zillig, M. D., Faist, K., et al. (2011). Structural model of the gas vesicle protein GvpA and analysis of GvpA mutants in vivo. Mol. Microbiol. 81, 56–68. doi: 10.1111/j.1365-2958.2011.07669.x
Tavlaridou, S., Winter, K., and Pfeifer, F. (2014). The accessory gas vesicle protein GvpM of haloarchaea and its interaction partners during gas vesicle formation. Extremophiles 18, 693–706. doi: 10.1007/s00792-014-0650-0
Waaland, R. J., and Branton, D. (1969). Gas vacuole development in a bluegreen alga. Science 163, 1339–1341. doi: 10.1126/science.163.3873.1339
Winter, K., Born, J., and Pfeifer, F. (2018). Interaction of haloarchaeal gas vesicle proteins determined by split-GFP. Front. Microbiol. 9:1897. doi: 10.3389/fmicb.2018.01897
Xu, B. Y., Dai, Y. N., Zhou, K., Liu, Y. T., Sun, Q., Ren, Y. M., et al. (2014). Structure of the gas vesicle protein GvpF from the cyanobacterium Microcystis aeruginosa. Acta Crystallogr. D Biol. Crystallogr. 70, 3013–3022. doi: 10.1107/S1399004714021312
Keywords: protein-protein interaction, split-GFP, protein network, cellulose binding domain, Haloferax volcanii, Halobacterium salinarum
Citation: Völkner K, Jost A and Pfeifer F (2020) Accessory Gvp Proteins Form a Complex During Gas Vesicle Formation of Haloarchaea. Front. Microbiol. 11:610179. doi: 10.3389/fmicb.2020.610179
Received: 28 September 2020; Accepted: 23 October 2020;
Published: 12 November 2020.
Edited by:
Harold J. Schreier, University of Maryland, Baltimore County, United StatesReviewed by:
Jerry Eichler, Ben-Gurion University of the Negev, IsraelCopyright © 2020 Völkner, Jost and Pfeifer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Felicitas Pfeifer, cGZlaWZlckBiaW8udHUtZGFybXN0YWR0LmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.