
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 15 December 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.609237
This article is part of the Research TopicInterview with the Translational Apparatus: Stories of Intriguing Circuits and Mechanisms to Regulate Translation in BacteriaView all 8 articles
Prokaryotic organisms often react instantly to environmental variations to ensure their survival. They can achieve this by rapidly and specifically modulating translation, the critical step of protein synthesis. The translation machinery responds to an array of cis-acting elements, located on the RNA transcript, which dictate the fate of mRNAs. These cis-encoded elements, such as RNA structures or sequence motifs, interact with a variety of regulators, among them small regulatory RNAs. These small regulatory RNAs (sRNAs) are especially effective at modulating translation initiation through their interaction with cis-encoded mRNA elements. Here, through selected examples of canonical and non-canonical regulatory events, we demonstrate the intimate connection between mRNA cis-encoded features and sRNA-dependent translation regulation. We also address how sRNA-based mechanistic studies can drive the discovery of new roles for cis-elements. Finally, we briefly overview the challenges of using translation regulation by synthetic regulators as a tool.
Prokaryotic organisms depend on protein synthesis to grow and adjust to their surroundings. The inability to produce functional gene products could result in bacterial cell growth inhibition. At the forefront of protein synthesis is the translational machinery, which requires the following elements: an mRNA, the 30S ribosomal subunit (small), three initiation factors (IF1, IF2, and IF3), an initiator tRNA, and the 50S ribosomal subunit (large). Together, these elements form the 70S translation initiation complex (Laursen et al., 2005; Gualerzi and Pon, 2015). Whether they are proteins or RNA molecules, translational machinery components must be synthesized, assembled properly, and available (Figure 1A). In eubacteria, most ribosomal RNAs (rRNAs) are encoded in polycistronic transcripts that must be precisely processed through multistep pathways to be functional (Deutscher, 2009). The presence of mutations in rRNAs or defect in their processing, causing mis-assembly of ribosomal subunits, can lead to rapid rRNA degradation (Deutscher, 2009; Basturea et al., 2011). The same type of quality control is also applied to both synthesis and maturation of tRNAs (Shepherd and Ibba, 2015). Furthermore, translation initiation often requires formylation of the initiator tRNAMet by the methionyl-tRNAfMet transformylase (FMT) as disruption of the fmt gene leads to important growth defects (Guillon et al., 1992).
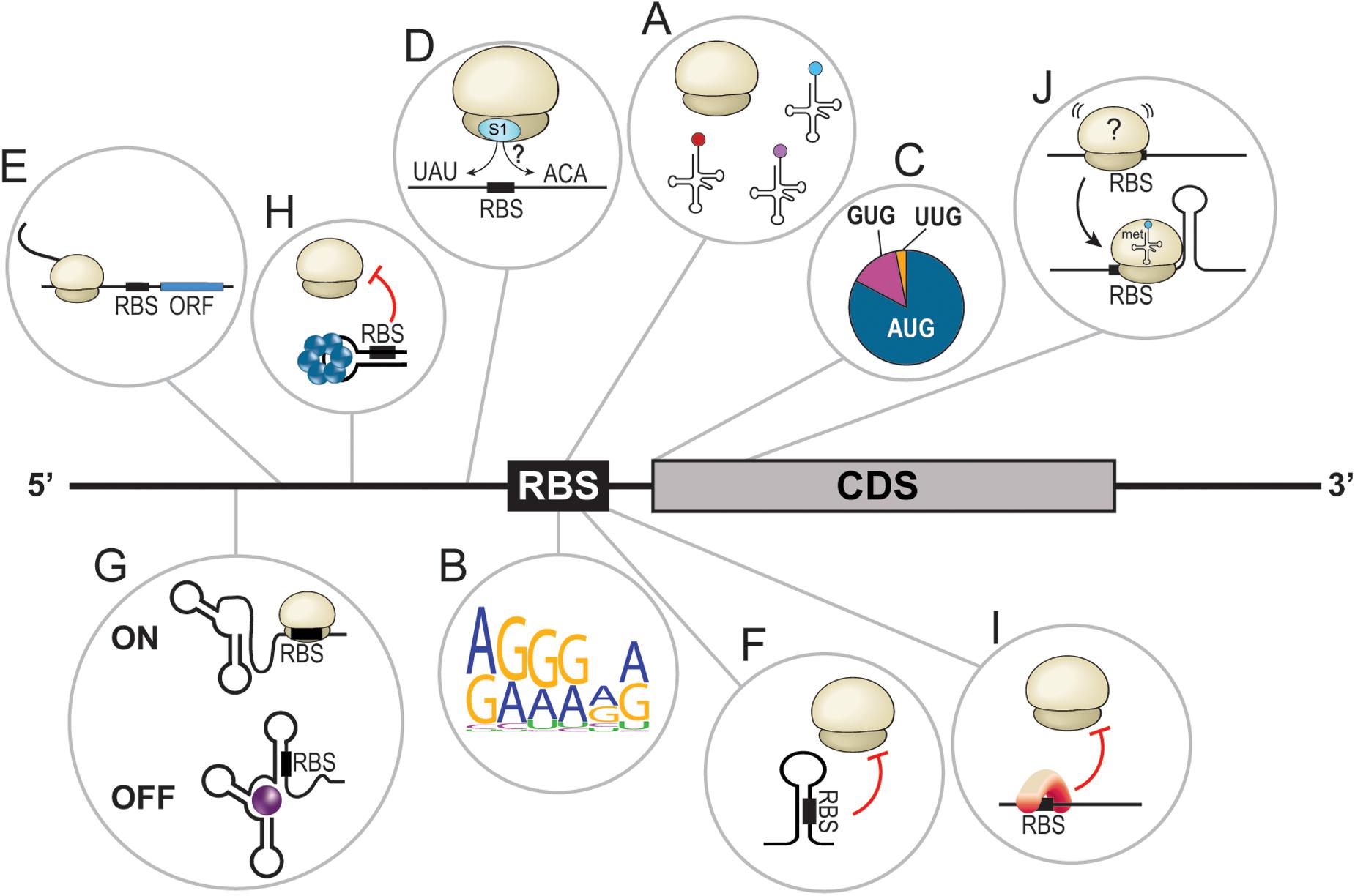
Figure 1. Overview of bacterial translational determinants. (A) Proper assembly of the translational machinery, including the ribosomes and tRNAs, is required for efficient translation. (B) Graphical representation (Crooks et al., 2004) of the Shine-Dalgarno core sequence in E. coli based on 50 random mRNAs. (C) Distribution of the different initiator codons in bacteria. AUG = 80%, GUG = 12%, and UUG = 8%. (D) Translation enhancers are often A/U- or C/A-rich sequences located upstream or downstream of the ribosome binding site (RBS). (E) Leader open reading frames dictate the translation of downstream CDS. (F) Secondary structures in the 5′UTR sequester the RBS and hinder translation by steric inhibition. (G) Riboswitches are complex RNA structures in 5′UTRs that, upon binding of a ligand, change conformation to turn the translation of the downstream open reading frame ON or OFF. (H) The binding of a protein forces the adoption of a 5′UTR structure that sequesters the RBS, preventing translation initiation. (I) Binding of a protein to the RBS prevents translation initiation. (J) A stem loop at the beginning of the coding sequence seems to stabilize the ribosome at the RBS and facilitate translation initiation.
Translational regulation through modulation of its machinery affects protein synthesis at a cellular scale. Although very effective, this is of little help when regulation of specific genes is required. To palliate this, prokaryotic organisms have developed an array of cis- and trans-acting strategies responding to environmental and cellular cues modulating translation of specific mRNAs.
The first cis-acting regulator of translation is the sequence of the mRNA itself, especially the translation initiation region (TIR; Osterman et al., 2013). Examples of TIR features include, among others: (I) the Shine-Dalgarno sequence (SD); (II) the initiation codon, with either the canonical AUG or alternative codons such as GUG or UUG (Villegas and Kropinski, 2008); (III) translational enhancers (TEs) such as A/U and C/A-rich sequences; and (IV) leader open reading frames (Figures 1B–E; de Smit and van Duin, 2003; Andreeva et al., 2018; Sterk et al., 2018; Romilly et al., 2019). Dictated by the primary sequence, the mRNA structure is also a major cis-acting regulator of translation. Simple structures, such as stem loops, prevent translation initiation through the sequestration of crucial ribosome binding site (RBS) elements, especially the SD (Figure 1F). More complex structures, called riboswitches, can also be found in 5′ untranslated regions (UTRs) of certain mRNAs. Riboswitches respond to the presence of specific molecules called ligands (e.g., metabolites, vitamins, coenzymes, ions, and uncharged tRNAs, etc.). In these cases, the interaction of a riboswitch to its ligand induces conformational changes that can turn translation ON or OFF, dictating the fate of the cognate mRNA (Figure 1G; Abduljalil, 2018; Pavlova et al., 2019; Bédard et al., 2020). Moreover, biochemical factors modulating the structure modification of riboswitches include temperature (thermosensors; Schumann, 2012; Loh et al., 2018; Mandin and Johansson, 2020) or pH (Nechooshtan et al., 2009).
Trans-acting translational regulators also play lead roles in dictating and redirecting gene expression. RNA-binding proteins (RBPs) are known to interact with various cis-elements to alter the secondary structure of an mRNA (Figure 1H) or to directly interfere with translation initiation (Figure 1I). This short review will focus on ribonucleic trans-acting regulators, particularly small regulatory RNAs (sRNAs), interacting with mRNA cis-elements to modulate their translation. Even though certain mechanisms of action are common and well described, we will overview how discovery of novel canonical regulatory events is still critical in our understanding of how bacteria adapt to their environment. Then, uncommon sRNA-dependent regulatory mechanisms targeting cis-elements will also be explored. Finally, a brief outlook on the synthetic use of cis-dependent translation regulation will be provided.
Bacterial trans-acting small regulatory RNAs are powerful regulators of gene expression. Acting through a tight network of regulation, sRNAs are responsible for the maintenance of cellular homeostasis and virulence. Their synthesis quickly responds to environmental signals, making them efficient stress-response regulators. Typically, sRNAs base-pair to their target mRNAs to repress or increase protein synthesis through various mechanisms of action, most of which have been extensively reviewed elsewhere (Gottesman and Storz, 2011; Papenfort and Vanderpool, 2015; Carrier et al., 2018; Dutta and Srivastava, 2018). These regulatory events result in modulation of mRNA stability and/or of translation efficiency. Even though sRNAs have been studied for decades now, identification of new sRNAs and sRNA targets helps understand how bacterial cells adapt to their environment. Moreover, additional regulatory mechanisms are periodically brought up to light, creating a more accurate portrait of sRNA complexity.
One of the most characterized mechanisms employed by sRNAs is the targeting of the RBS to hinder translation initiation. New examples of sRNAs using this mechanism are still discovered to this day. In Salmonella enterica serovar Typhimurium, the PinT sRNA has recently been found to regulate two mRNAs, rtsA and hilA, through a canonical mechanism (Kim et al., 2019). PinT pairs to the 5′UTR of both mRNAs, near the SD of rtsA and near the start codon of hilA. In both cases, the interaction blocks translation initiation by preventing ribosome assembly at the RBS. While PinT interaction is enough to repress hilA mRNA, the activity of the ribonuclease E (RNase E) is required to fully repress rtsA (Kim et al., 2019). The identification of these new mRNA targets regulated by the PinT sRNA added new hindsight on Salmonella transition from the invasion stage to intracellular growth during infection.
Through a wide range of mechanisms, sRNAs can also positively regulate mRNA targets. The most common of those mechanisms involves the pairing of an sRNA to the 5′UTR of an mRNA, upstream from the TIR. This causes structural modifications facilitating ribosome assembly and translation initiation. Notably, sRNA-based regulation has evolved to accommodate peculiarities of different mRNAs, resulting in many variations of canonical regulatory mechanisms. For instance, in the enterohemorrhagic Escherichia coli (EHEC), the pchA mRNA, encoding a transcriptional activator, is regulated in cis by secondary structures in its own transcript. Indeed, the pchA mRNA coding sequence (CDS) presents an anti-Shine-Dalgarno (anti-SD) sequence, which forces the folding of the mRNA on itself through strong interactions with the SD. The resulting double-stranded RNA structure sequesters the RBS of pchA, thus inhibiting translation initiation (Melson and Kendall, 2019). This structure, however, is sensitive to a trans-regulator, the sRNA DicF. It has recently been found to pair within the CDS of pchA, at the anti-SD, to prevent the self-folding of pchA. This sRNA:mRNA interaction results in facilitated translation initiation by rendering the RBS of pchA accessible to ribosomes. Data indicate that the presence of DicF paired in the CDS of pchA does not impair translation elongation rates, suggesting that elongating ribosomes are able to displace the sRNA from the mRNA.
What is the cellular advantage of an mRNA being regulated in such a context? Translation inhibition often leads to mRNA degradation since it is not protected by translating ribosomes. However, the pchA mRNA does not seem to be destabilized when translation is OFF compared to a mutated version of pchA unable to form the anti-SD:SD interaction (translation ON; Melson and Kendall, 2019). Perhaps the folded structure allows translation inhibition while also protecting the transcript against degradation? If so, following DicF expression, the stable pchA mRNA could readily be translated, possibly allowing a rapid response to changes in environmental conditions.
The pairing of sRNAs in the CDS, outside of the five-codon window (Bouvier et al., 2008), is mostly known to cause destabilization of the target mRNA and lead to repression of gene expression (Pfeiffer et al., 2009; Fröhlich et al., 2012; Lalaouna et al., 2015b). The regulation of pchA by DicF is rather uncommon as the pairing of the sRNA downstream of the five-codon window directly impacts translation initiation. Moreover, another uncommon characteristic of this interaction is the fact that DicF positively regulates pchA, contrary to most of the regulatory events involving an sRNA pairing in the CDS of its target, demonstrating the versatility of sRNA mechanisms of actions.
The SD sequence and the initiator codon are critical determinants of translation. However, other cis-encoded features such as TEs can play a major role in dictating translation rates. Described as A/U- or C/A-rich sequences, TEs were found, among others, in the E. coli rnd and fepB mRNAs that present the alternative initiator codons UUG and GUG, respectively (Zhang and Deutscher, 1992; Hook-Barnard et al., 2007), or in the tuf mRNA of Mycoplasma genitalium, which lacks an SD sequence (Loechel et al., 1991). TEs were originally believed to facilitate translation initiation of mRNAs presenting suboptimal SD-AUG contexts. Surprisingly, TEs were later noticed in mRNAs with optimal RBS characteristics, for example, in the dppA mRNA of E. coli and Salmonella (Yang et al., 2014). This suggested that optimal TIR features are not necessarily sufficient to ensure required translation initiation rates. It has been proposed that A/U- and C/A-rich sequences, such as TEs, could act as binding sites for the S1 ribosomal protein, which helps position the ribosome during translation initiation (Hauryliuk and Ehrenberg, 2006; Studer and Joseph, 2006). Considering that sRNAs are known to act through non-canonical mechanisms of action, it is not surprising that they target TEs (Sharma et al., 2007). Although not the only one, GcvB is a perfect example of a TE-targeting sRNA. GcvB negatively regulates the dppA mRNA by targeting a C/A-rich stretch located immediately upstream of the SD (Yang et al., 2014). It also regulates the gltI mRNA by pairing to a TE located 40 nucleotides (nts) upstream of the RBS (Sharma et al., 2007).
While studying the regulation of the manXYZ polycistronic transcript by the SgrS sRNA, the Vanderpool lab discovered that SgrS targets manXYZ at two distinct base-pairing sites, exerting its regulatory effect through two different mechanisms of action (Figure 2A). First, SgrS pairs in the CDS of manX, recruiting the RNA chaperone Hfq to repress manX synthesis (Rice and Vanderpool, 2011; Azam and Vanderpool, 2018). Second, using a different sequence, SgrS pairs 30 nts upstream of manY SD and dictates the fate of both manY and manZ. It was elegantly demonstrated that the manY region targeted by SgrS is a U-rich TE actively participating in the translation of manY through its interaction with the S1 ribosomal protein (Azam and Vanderpool, 2020). The working model of this regulation suggests that the S1:mRNA complex remodels the SD, favoring translation. SgrS prevents this activation by masking the TE site and impairing translation initiation.
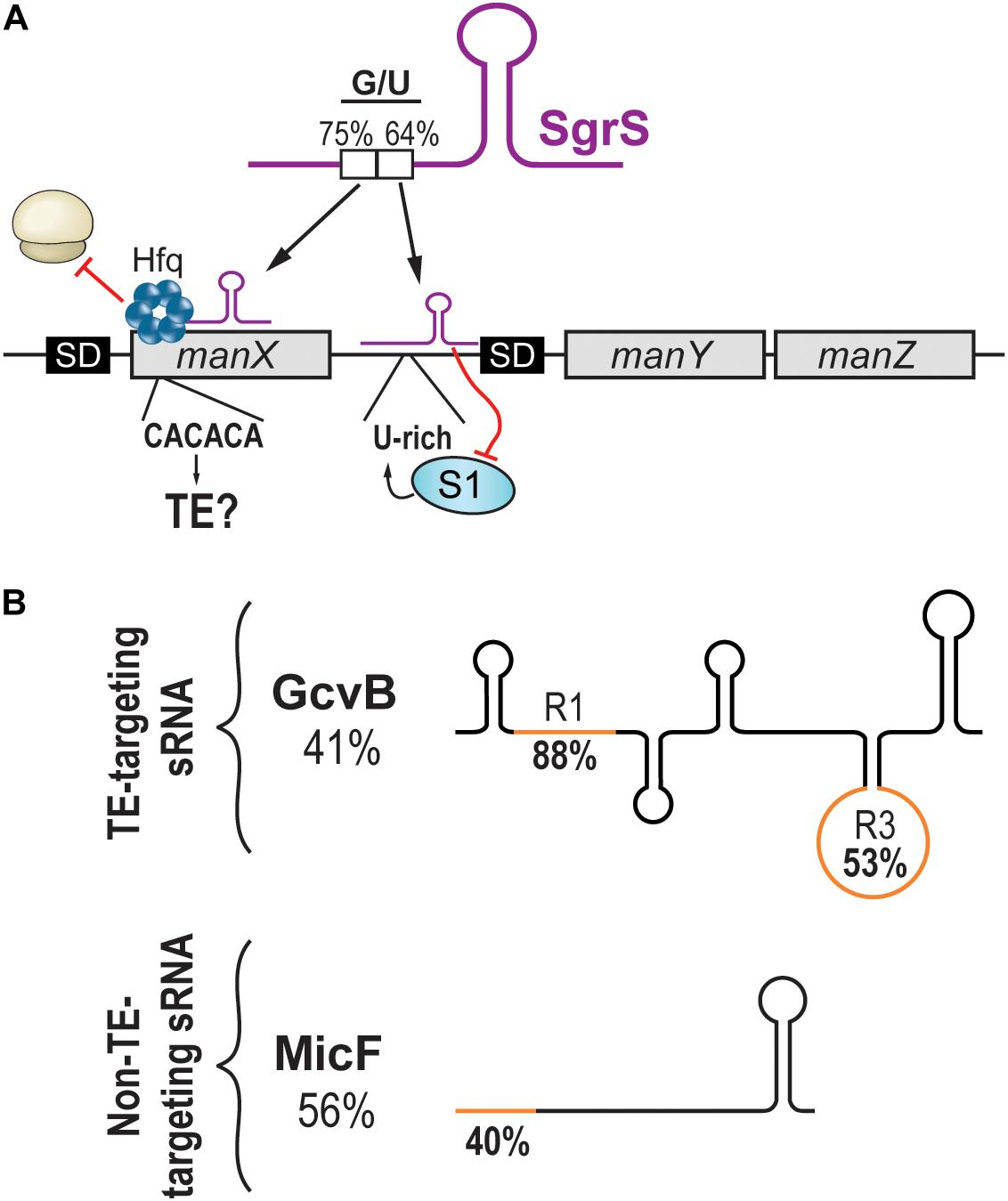
Figure 2. Targeting of translational enhancers by sRNAs. (A) The manXYZ polycistronic transcript is dually regulated by two regions of the sRNA SgrS. SgrS interacts with the coding sequence of manX and recruits Hfq to block translation. The pairing site on manX presents a C/A-rich sequence that could be a translational enhancer. On the other hand, SgrS interacts with the 5′UTR of manY and masks a U-rich translational enhancer, hindering the translation of manY and manZ. (B) Schematic representation of sRNAs targeting translational enhancers (upper panel) or not (lower panel). Percentage of G/U of the full sRNA sequence is indicated under the sRNA name. The G/U content of the seed region (orange) is indicated on the sRNA drawing.
Whereas the repression of manY occurs through the occlusion of a U-rich TE, the regulation of manX has not been shown to involve a TE. However, the high G/U content of the SgrS region interacting with the 5′UTR of manX might suggest otherwise. We wondered if the target site on manX presented features of a C/A-rich TE. A stretch of C/A-rich (CACACA) sequence was indeed found, suggesting that translation initiation of manX could depend on the S1 protein, which seems to favor A/U- and C/A-rich sequences (Figure 2A). If demonstrated, this would indicate that SgrS action on manX involves more than the recruitment of Hfq to achieve translation inhibition.
Like SgrS, which uses two different pairing sites, the sRNA GcvB possesses two distinct seed regions called R1 and R3. The regulation of both dppA and gltI mRNAs involves R1, which presents a high G/U content (88%), indicating its potential at targeting C/A-rich regions. In contrast to this, the R3 seed region of GcvB has a lower G/U content (53%), and its targets show little potential at encoding C/A-rich TEs (Figure 2B; Lalaouna et al., 2019). Therefore, would it be possible to predict an sRNA targeting C/A-rich TEs from its G/U content?
Analysis of the MicF sRNA, which is not known to target TEs, exposed a possible barrier to such predictions. Indeed, the entire sequence of MicF presents a moderate G/U content (56%), suggesting some potential at targeting TEs. However, MicF seed region (nucleotides 1–15) presents a very low G/U content (40%), indicating that actual chances of targeting C/A-rich TEs are modest (Figure 2B). This remarkable difference between whole sequence versus seed region G/U content is also observed with GcvB. The R1 seed region of GcvB has a very high G/U content (88%) compared to an overall very low G/U content (41%; Figure 2B). The question is, how would predictions be achieved for sRNAs with no obvious seed regions such as RyhB? In addition to the sequence of the binding region, would other sRNA and/or mRNA features be required for this mechanism of action to occur? Are TEs so versatile in their sequences that no prediction can be performed?
While investigating the targetome of both OmrA and OmrB sRNAs (hereby referred to as OmrA/B), Guillier and colleagues identified a negatively regulated target, the fepA mRNA (Jagodnik et al., 2017). They show that OmrA/B represses fepA by hindering translation initiation. Interestingly, both sRNAs interact with nucleotides downstream of the five-codon window usually targeted by repressing sRNAs (Bouvier et al., 2008). In this case, OmrA/B represses translation by disrupting a stem loop (SL) in the CDS of fepA, suggesting that the SL itself might favor translation. This is in direct contrast with previous observations concerning SLs. When located in 5′UTRs, SLs hinder the recognition of TIR elements through a sequestration mechanism. In many cases, this inhibition is alleviated with the help of sRNAs interacting with the SL, forcing it to open. Examples include E. coli ArcZ, DsrA, and RprA sRNAs, all increasing the translation of rpoS (Kim and Lee, 2020). Another example is the sRNA RNAIII, which disrupts hla 5′UTR inhibitory SL to increase α-hemolysin synthesis in Staphylococcus aureus (Morfeldt et al., 1995). When located in the CDS, SLs are believed to slow down the elongation rate of translating ribosomes and are known to induce frameshifts (Kim et al., 2014). Based on their observation of OmrA/B, Guillier’s group brings the novel idea that the SL structure favors translation of fepA CDS at an early stage of initiation (Jagodnik et al., 2017). They hypothesized a mechanism of activation in which the CDS-located SL acts as a starting block to help properly position the 30S ribosomal subunit and favor the formation of the translation initiation complex (Figure 1J). A similar activator SL, also targeted by OmrA/B, was found in the bamA mRNA (Jagodnik et al., 2017).
Bacterial sRNAs act through plentiful regulatory mechanisms and target different types of RNA molecules such as mRNAs or even other sRNAs (Figueroa-Bossi et al., 2009; Miyakoshi et al., 2015; Lalaouna et al., 2015a). Their impact on genomic expression and the resulting physiological effects have been extensively studied. However, to our knowledge, the core of the translation machinery, i.e., the ribosomes, is not directly targeted by sRNAs in prokaryotes. This contrasts with eukaryotic and archaeal organisms, in which direct association of non-coding RNAs to the translational apparatus has been shown. In 2012, Gebetsberger et al. (2012) demonstrated that in the archaea Haloferax volcanii, the association of a tRNA-derived fragment (tRF) to the 30S ribosomal subunit globally downregulates translation in conditions of hyperosmotic stress. In similar stressful conditions, the association of a Saccharomyces cerevisiae exon-derived RNA to the 60S ribosomal subunit can hinder translation in vitro (Pircher et al., 2014). More recently, a tRF in Trypanosoma brucei has been shown to promote protein synthesis through its direct association with the translational machinery (Fricker et al., 2019). Examples above involve RNA fragments that are relatively short (less than 50 nts) compared to the canonical bacterial sRNAs, averaging 100 nts in length. Bacterial cells, however, are not devoid of extremely short, stable RNA molecules. Many tRFs have been identified in bacteria; however, their functions remain mostly unexplored. Could their investigation reveal that, just as in eukaryotes and archaea, these short non-coding RNAs can find their way into the bacterial translational machinery?
The question of why bacterial regulatory RNAs have not yet been found to target rRNAs is still in suspense. A possible explanation could be related to experimental procedures rather than experimental limitations. Most high-throughput experiments heavily rely on the depletion of rRNAs prior to sequencing to produce analyzable data. Indeed, rRNAs are so abundant that their depletion becomes necessary to obtain enough reads from other RNAs (e.g., mRNAs, tRNAs, and sRNAs; Yang et al., 2011). However, rRNA removal also creates a bias, preventing the identification of sRNA:rRNA interactions. Future breakthroughs in RNA sequencing techniques might resolve this bias and allow the identification of a new class of small regulatory RNAs that directly target the translational machinery. Moreover, optimized techniques could lead to the identification of novel, conserved non-coding RNAs, such as those produced by pervasive transcription (Lybecker et al., 2014). For example, these transcripts could act as asRNAs, regulating components of the translational machinery.
From there, many questions might arise. Would sRNAs target all ribosomes with no selection or would they interact solely with specific specialized ribosomes? The current understanding of bacterial specialized ribosomes is still limited. These ribosomes could be generated via a modification in component stoichiometry (Chen et al., 2020) or include/exclude ribosomal proteins, such as the SRA protein (van de Waterbeemd et al., 2017). In turn, these modifications could modify the accessible parts of the rRNA, exposing novel base-pairing sites for sRNAs. However, would the base pairing of the sRNA to the rRNA be the sole determinant of the interaction? Examples of sRNAs interacting with proteins are numerous. To name only a few, their interaction with chaperone proteins such as Hfq and ProQ (Smirnov et al., 2016; Kavita et al., 2018) or with regulatory proteins such as CsrA (Liu et al., 1997; Weilbacher et al., 2003) is proof that sRNAs have protein-binding properties. Therefore, one could hypothesize that sRNAs targeting ribosomes could do so through interaction with ribosomal proteins.
Regulation of translation is a tightly controlled process essential to bacterial survival and fitness. Therefore, it offers a great opportunity to use translation regulation as a tool for diverse applications.
Currently, the massive use of antibiotics in both clinical and agricultural settings fuels the emergence of drug-resistant bacterial strains. Moreover, with the rising importance of microbiomes as beneficial health factors, the use of large spectrum antibiotics to fight bacterial infections does not appear suitable for chronic treatments as it leads to microbiome dysbiosis (Panda et al., 2014; Becattini et al., 2016), urging the scientific community to develop new antimicrobial drugs or identify new drug targets (Ventola, 2015). Unsurprisingly, a great number of antimicrobial compounds, whether they are from natural or synthetic origin, target translation (Sohmen et al., 2009; Nikolay et al., 2016; Champney, 2020). An innovative approach, termed antisense therapy, uses artificial antisense oligonucleotides (ASOs) to repress the translation of single mRNAs through base-pairing complementarity (Dias and Stein, 2002). ASO therapy strategies involve targeting antibiotic resistance genes (Daly et al., 2017; Sully et al., 2017; Kauss et al., 2020) or essential genes (Sawyer et al., 2013). It is tempting to assume that designing ASOs targeting the TIR of an mRNA would be specific enough to prevent off-target regulation. However, these elements are somewhat conserved between mRNAs and, more importantly, are conserved in closely related species. Since it is clear that inhibition of translation can occur through the targeting of cis-elements outside of the RBS by bacterial sRNAs, ASOs are now being developed to target these more complex and less conserved sequences. Even though this vision of ASOs as antibiotics is still facing major hurdles, technological breakthroughs bring us closer to this achievement every year [for a review, see Vogel (2020)].
Translational regulation is a layered process depending on the translational machinery and on a variety of elements encoded in cis or in trans (Figure 1). Trans-acting regulators, especially sRNAs, have evolved to target cis-elements, some more commonly (e.g., SD, RBS) than others (e.g., translational enhancers). Thanks to rapidly evolving high-throughput RNA sequencing studies, new regulatory events are periodically brought up to light. While identification of new sRNA targets regulated through canonical mechanisms of action mainly helps to understand cellular physiology and bacterial adaptation to its environment, non-canonical events can lead to much more. Indeed, identification of new mechanisms of sRNA-dependent regulation is crucial to expand the boundaries of current regulatory networks. Moreover, in some instances, studies based on unusual regulatory events contribute to the identification of novel roles for cis-elements, strengthening the importance of studying sRNA mechanisms.
M-CC, ENKL, and GJ wrote the manuscript. M-CC prepared the figures. EM revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by Canadian Institutes of Health Research (CIHR) to EM (MOP389354).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abduljalil, J. M. (2018). Bacterial riboswitches and RNA thermometers: nature and contributions to pathogenesis. Noncoding RNA Res. 3, 54–63. doi: 10.1016/j.ncrna.2018.04.003
Andreeva, I., Belardinelli, R., and Rodnina, M. V. (2018). Translation initiation in bacterial polysomes through ribosome loading on a standby site on a highly translated mRNA. PNAS 115, 4411–4416. doi: 10.1073/pnas.1718029115
Azam, M. S., and Vanderpool, C. K. (2018). Translational regulation by bacterial small RNAs via an unusual Hfq-dependent mechanism. Nucleic Acids Res. 46, 2585–2599. doi: 10.1093/nar/gkx1286
Azam, M. S., and Vanderpool, C. K. (2020). Translation inhibition from a distance: the small RNA SgrS silences a ribosomal protein S1-dependent enhancer. Mol. Microbiol. 114, 391–408. doi: 10.1111/mmi.14514
Basturea, G. N., Zundel, M. A., and Deutscher, M. P. (2011). Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA 17, 338–345. doi: 10.1261/rna.2448911
Becattini, S., Taur, Y., and Pamer, E. G. (2016). Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol. Med. 22, 458–478. doi: 10.1016/j.molmed.2016.04.003
Bédard, A.-S. V., Hien, E. D. M., and Lafontaine, D. A. (2020). Riboswitch regulation mechanisms: RNA, metabolites and regulatory proteins. Biochim. Biophys. Acta 1863:194501. doi: 10.1016/j.bbagrm.2020.194501
Bouvier, M., Sharma, C. M., Mika, F., Nierhaus, K. H., and Vogel, J. (2008). Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol. Cell 32, 827–837. doi: 10.1016/j.molcel.2008.10.027
Carrier, M.-C., Lalaouna, D., and Massé, E. (2018). Broadening the definition of bacterial small RNAs: characteristics and mechanisms of action. Annu. Rev. Microbiol. 72, 141–161. doi: 10.1146/annurev-micro-090817-0
Champney, W. S. (2020). Antibiotics targeting bacterial ribosomal subunit biogenesis. J. Antimicrob. Chemother. 75, 787–806. doi: 10.1093/jac/dkz544
Chen, Y.-X., Xu, Z., Ge, X., Hong, J.-Y., Sanyal, S., Lu, Z. J., et al. (2020). Selective translation by alternative bacterial ribosomes. PNAS 117, 19487–19496. doi: 10.1073/pnas.2009607117
Crooks, G. E., Hon, G., Chandonia, J.-M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Daly, S. M., Sturge, C. R., Felder-Scott, C. F., Geller, B. L., and Greenberg, D. E. (2017). MCR-1 inhibition with peptide-conjugated phosphorodiamidate morpholino oligomers restores sensitivity to polymyxin in Escherichia coli. mBio 8:e1315-17. doi: 10.1128/mBio.01315-17
de Smit, M. H., and van Duin, J. (2003). Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J. Mol. Biol. 331, 737–743. doi: 10.1016/s0022-2836(03)00809-x
Deutscher, M. P. (2009). Maturation and degradation of ribosomal RNA in Bacteria. Prog. Mol. Biol. Transl. Sci. 85, 369–391. doi: 10.1016/S0079-6603(08)00809-X
Dias, N., and Stein, C. A. (2002). Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 1, 347–355.
Dutta, T., and Srivastava, S. (2018). Small RNA-mediated regulation in bacteria: a growing palette of diverse mechanisms. Gene 656, 60–72. doi: 10.1016/j.gene.2018.02.068
Figueroa-Bossi, N., Valentini, M., Malleret, L., and Bossi, L. (2009). Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 23, 2004–2015. doi: 10.1101/gad.541609
Fricker, R., Brogli, R., Luidalepp, H., Wyss, L., Fasnacht, M., Joss, O., et al. (2019). A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat. Commun. 10:118. doi: 10.1038/s41467-018-07949-6
Fröhlich, K. S., Papenfort, K., Berger, A. A., and Vogel, J. (2012). A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res. 40, 3623–3640. doi: 10.1093/nar/gkr1156
Gebetsberger, J., Zywicki, M., Künzi, A., and Polacek, N. (2012). tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012:e260909. doi: 10.1155/2012/260909
Gottesman, S., and Storz, G. (2011). Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3:a003798. doi: 10.1101/cshperspect.a003798
Gualerzi, C. O., and Pon, C. L. (2015). Initiation of mRNA translation in bacteria: structural and dynamic aspects. Cell. Mol. Life Sci. 72, 4341–4367. doi: 10.1007/s00018-015-2010-3
Guillon, J. M., Mechulam, Y., Schmitter, J. M., Blanquet, S., and Fayat, G. (1992). Disruption of the gene for Met-tRNA(fMet) formyltransferase severely impairs growth of Escherichia coli. J. Bacteriol. 174, 4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992
Hauryliuk, V., and Ehrenberg, M. (2006). Two-step selection of mRNAs in initiation of protein synthesis. Mol. Cell 22, 155–156. doi: 10.1016/j.molcel.2006.04.004
Hook-Barnard, I. G., Brickman, T. J., and McIntosh, M. A. (2007). Identification of an AU-rich translational enhancer within the Escherichia coli fepB leader RNA. J. Bacteriol. 189, 4028–4037. doi: 10.1128/JB.01924-06
Jagodnik, J., Chiaruttini, C., and Guillier, M. (2017). Stem-loop structures within mRNA coding sequences activate translation initiation and mediate control by small regulatory RNAs. Mol. Cell 68, 158.e3–170.e3. doi: 10.1016/j.molcel.2017.08.015
Kauss, T., Arpin, C., Bientz, L., Vinh Nguyen, P., Vialet, B., Benizri, S., et al. (2020). Lipid oligonucleotides as a new strategy for tackling the antibiotic resistance. Sci. Rep. 10:1054. doi: 10.1038/s41598-020-58047-x
Kavita, K., de Mets, F., and Gottesman, S. (2018). New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 42, 53–61. doi: 10.1016/j.mib.2017.10.014
Kim, H.-K., Liu, F., Fei, J., Bustamante, C., Gonzalez, R. L., and Tinoco, I. (2014). A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. PNAS 111, 5538–5543. doi: 10.1073/pnas.1403457111
Kim, K., Palmer, A. D., Vanderpool, C. K., and Slauch, J. M. (2019). The small RNA PinT contributes to PhoP-mediated regulation of the Salmonella pathogenicity island 1 type III secretion system in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 201, e312–e319. doi: 10.1128/JB.00312-19
Kim, W., and Lee, Y. (2020). Mechanism for coordinate regulation of rpoS by sRNA-sRNA interaction in Escherichia coli. RNA Biol. 17, 176–187. doi: 10.1080/15476286.2019.1672514
Lalaouna, D., Carrier, M.-C., Semsey, S., Brouard, J.-S., Wang, J., Wade, J. T., et al. (2015a). A 3’ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol. Cell 58, 393–405. doi: 10.1016/j.molcel.2015.03.013
Lalaouna, D., Morissette, A., Carrier, M.-C., and Massé, E. (2015b). DsrA regulatory RNA represses both hns and rbsD mRNAs through distinct mechanisms in Escherichia coli. Mol. Microbiol. 98, 357–369. doi: 10.1111/mmi.13129
Lalaouna, D., Eyraud, A., Devinck, A., Prévost, K., and Massé, E. (2019). GcvB small RNA uses two distinct seed regions to regulate an extensive targetome. Mol. Microbiol. 111, 473–486. doi: 10.1111/mmi.14168
Laursen, B. S., Sørensen, H. P., Mortensen, K. K., and Sperling-Petersen, H. U. (2005). Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69, 101–123. doi: 10.1128/MMBR.69.1.101-123.2005
Liu, M. Y., Gui, G., Wei, B., Preston, J. F., Oakford, L., Yüksel, U., et al. (1997). The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272, 17502–17510. doi: 10.1074/jbc.272.28.17502
Loechel, S., Inamine, J. M., and Hu, P. C. (1991). A novel translation initiation region from Mycoplasma genitalium that functions in Escherichia coli. Nucleic Acids Res 19, 6905–6911. doi: 10.1093/nar/19.24.6905
Loh, E., Righetti, F., Eichner, H., Twittenhoff, C., and Narberhaus, F. (2018). RNA thermometers in bacterial pathogens. Microbiol. Spectr. 6:RWR-0012-2017. doi: 10.1128/microbiolspec.RWR-0012-2017
Lybecker, M., Bilusic, I., and Raghavan, R. (2014). Pervasive transcription: detecting functional RNAs in bacteria. Transcription 5:e944039. doi: 10.4161/21541272.2014.944039
Mandin, P., and Johansson, J. (2020). Feeling the heat at the millennium: thermosensors playing with fire. Mol. Microbiol. 113, 588–592. doi: 10.1111/mmi.14468
Melson, E. M., and Kendall, M. M. (2019). The sRNA DicF integrates oxygen sensing to enhance enterohemorrhagic Escherichia coli virulence via distinctive RNA control mechanisms. PNAS 116, 14210–14215. doi: 10.1073/pnas.1902725116
Miyakoshi, M., Chao, Y., and Vogel, J. (2015). Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 34, 1478–1492. doi: 10.15252/embj.201490546
Morfeldt, E., Taylor, D., von Gabain, A., and Arvidson, S. (1995). Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA. RNAIII. EMBO J. 14, 4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x
Nechooshtan, G., Elgrably-Weiss, M., Sheaffer, A., Westhof, E., and Altuvia, S. (2009). A pH-responsive riboregulator. Genes Dev. 23, 2650–2662. doi: 10.1101/gad.552209
Nikolay, R., Schmidt, S., Schlömer, R., Deuerling, E., and Nierhaus, K. H. (2016). Ribosome assembly as antimicrobial target. Antibiotics 5:18. doi: 10.3390/antibiotics5020018
Osterman, I. A., Evfratov, S. A., Sergiev, P. V., and Dontsova, O. A. (2013). Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res. 41, 474–486. doi: 10.1093/nar/gks989
Panda, S., Khader, I. E., Casellas, F., Vivancos, J. L., Cors, M. G., Santiago, A., et al. (2014). Short-term effect of antibiotics on human gut microbiota. PLoS One 9:e95476. doi: 10.1371/journal.pone.0095476
Papenfort, K., and Vanderpool, C. K. (2015). Target activation by regulatory RNAs in bacteria. FEMS Microbiol. Rev. 39, 362–378. doi: 10.1093/femsre/fuv016
Pavlova, N., Kaloudas, D., and Penchovsky, R. (2019). Riboswitch distribution, structure, and function in bacteria. Gene 708, 38–48. doi: 10.1016/j.gene.2019.05.036
Pfeiffer, V., Papenfort, K., Lucchini, S., Hinton, J. C. D., and Vogel, J. (2009). Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 16, 840–846. doi: 10.1038/nsmb.1631
Pircher, A., Bakowska-Zywicka, K., Schneider, L., Zywicki, M., and Polacek, N. (2014). An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol. Cell 54, 147–155. doi: 10.1016/j.molcel.2014.02.024
Rice, J. B., and Vanderpool, C. K. (2011). The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res. 39, 3806–3819. doi: 10.1093/nar/gkq1219
Romilly, C., Deindl, S., and Wagner, E. G. H. (2019). The ribosomal protein S1-dependent standby site in tisB mRNA consists of a single-stranded region and a 5′ structure element. PNAS 116, 15901–15906. doi: 10.1073/pnas.1904309116
Sawyer, A. J., Wesolowski, D., Gandotra, N., Stojadinovic, A., Izadjoo, M., Altman, S., et al. (2013). A peptide-morpholino oligomer conjugate targeting Staphylococcus aureus gyrA mRNA improves healing in an infected mouse cutaneous wound model. Int J. Pharm. 453, 651–655. doi: 10.1016/j.ijpharm.2013.05.041
Schumann, W. (2012). Thermosensor systems in eubacteria. Adv. Exp. Med. Biol. 739, 1–16. doi: 10.1007/978-1-4614-1704-0_1
Sharma, C. M., Darfeuille, F., Plantinga, T. H., and Vogel, J. (2007). A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 21, 2804–2817. doi: 10.1101/gad.447207
Shepherd, J., and Ibba, M. (2015). Bacterial transfer RNAs. FEMS Microbiol. Rev. 39, 280–300. doi: 10.1093/femsre/fuv004
Smirnov, A., Förstner, K. U., Holmqvist, E., Otto, A., Günster, R., Becher, D., et al. (2016). Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. PNAS 113, 11591–11596. doi: 10.1073/pnas.1609981113
Sohmen, D., Harms, J. M., Schlünzen, F., and Wilson, D. N. (2009). SnapShot: antibiotic inhibition of protein synthesis I. Cell 138, 1248.e1–1248.e1. doi: 10.1016/j.cell.2009.08.001
Sterk, M., Romilly, C., and Wagner, E. G. H. (2018). Unstructured 5’-tails act through ribosome standby to override inhibitory structure at ribosome binding sites. Nucleic Acids Res. 46, 4188–4199. doi: 10.1093/nar/gky073
Studer, S. M., and Joseph, S. (2006). Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell 22, 105–115. doi: 10.1016/j.molcel.2006.02.014
Sully, E. K., Geller, B. L., Li, L., Moody, C. M., Bailey, S. M., Moore, A. L., et al. (2017). Peptide-conjugated phosphorodiamidate morpholino oligomer (PPMO) restores carbapenem susceptibility to NDM-1-positive pathogens in vitro and in vivo. J. Antimicrob. Chemother. 72, 782–790. doi: 10.1093/jac/dkw476
van de Waterbeemd, M., Fort, K. L., Boll, D., Reinhardt-Szyba, M., Routh, A., Makarov, A., et al. (2017). High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Methods 14, 283–286. doi: 10.1038/nmeth.4147
Villegas, A., and Kropinski, A. M. (2008). An analysis of initiation codon utilization in the Domain Bacteria – concerns about the quality of bacterial genome annotation. Microbiology 154, 2559–2661. doi: 10.1099/mic.0.2008/021360-0
Vogel, J. (2020). An RNA biology perspective on species-specific programmable RNA antibiotics. Mol. Microbiol. 113, 550–559. doi: 10.1111/mmi.14476
Weilbacher, T., Suzuki, K., Dubey, A. K., Wang, X., Gudapaty, S., Morozov, I., et al. (2003). A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48, 657–670. doi: 10.1046/j.1365-2958.2003.03459.x
Yang, L., Duff, M. O., Graveley, B. R., Carmichael, G. G., and Chen, L.-L. (2011). Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 12:R16. doi: 10.1186/gb-2011-12-2-r16
Yang, Q., Figueroa-Bossi, N., and Bossi, L. (2014). Translation enhancing ACA motifs and their silencing by a bacterial small regulatory RNA. PLoS Genet. 10:e1004026. doi: 10.1371/journal.pgen.1004026
Keywords: translational regulation, translational determinants, cis regulatory elements, small regulatory RNAs, regulatory mechanisms, translational enhancers
Citation: Carrier MC, Ng Kwan Lim E, Jeannotte G and Massé E (2020) Trans-Acting Effectors Versus RNA Cis-Elements: A Tightly Knit Regulatory Mesh. Front. Microbiol. 11:609237. doi: 10.3389/fmicb.2020.609237
Received: 22 September 2020; Accepted: 10 November 2020;
Published: 15 December 2020.
Edited by:
Anna Maria Giuliodori, University of Camerino, ItalyReviewed by:
Isabelle Caldelari, Université de Strasbourg, FranceCopyright © 2020 Carrier, Ng Kwan Lim, Jeannotte and Massé. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eric Massé, RXJpYy5NYXNzZUBVU2hlcmJyb29rZS5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.