- 1CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2Department of Entomology and Plant Pathology, Faculty of Agriculture, Chiang Mai University, Chiang Mai, Thailand
- 3Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand
- 4World Agroforestry Centre, East and Central Asia, Kunming, China
- 5Centre for Mountain Futures, Kunming Institute of Botany, Kunming, China
- 6Yunnan Key Laboratory for Fungal Diversity and Green Development, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 7Mushroom Research Foundation, Chiang Mai, Thailand
- 8School of Science, Mae Fah Luang University, Chiang Rai, Thailand
- 9Innovative Institute of Plant Health, Zhongkai University of Agriculture and Engineering, Guangzhou, China
- 10Department of Botany and Microbiology, College of Sciences, King Saud University, Riyadh, Saudi Arabia
- 11Department of Botany, Faculty of Science, University of Peradeniya, Peradeniya, Sri Lanka
- 12Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 13Research Center of Microbial Diversity and Sustainable Utilization, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 14Academy of Science, The Royal Society of Thailand, Bangkok, Thailand
Entomopathogenic fungi are ubiquitous in tropical rainforests and feature a high level of diversity. This group of fungi not only has important ecological value but also medicinal value. Nevertheless, they are often ignored, and many unknown species have yet to be discovered and described. The present study aims to contribute to the taxonomical and phylogenetic understanding of the genus Paraisaria by describing three new species collected from Guizhou and Yunnan Provinces in China and Krabi Province in Thailand. The three novel species named Paraisaria alba, P. arcta, and P. rosea share similar morphologies as those in the genus Paraisaria, containing solitary, simple, fleshy stroma, completely immersed perithecia and cylindrical asci with thickened caps and filiform ascospores that often disarticulate at maturity. Phylogenetic analyses of combined LSU, SSU, TEF1-α, RPB1, RPB2, and ITS sequence data confirm their placement in the genus Paraisaria. In this study, the three entomopathogenic taxa are comprehensively described with color photographs and phylogenetic analyses. A synopsis table and a key to all treated species of Paraisaria are also included.
Introduction
Entomopathogenic fungi are a group of unicellular or multicellular, heterotrophic, eukaryotic microorganisms that can enter into a parasitic relationship with parasitized insects, killing or otherwise disabling their hosts (Samson et al., 1988). They reproduce via sexual or asexual spores, or both (Mora et al., 2017). It is of global importance to survey and describe insect pathogens (Hyde et al., 2019). Entomopathogenic fungi can act as natural enemies of agricultural pests and play an important role in maintaining ecological balance (Fernández-Grandon et al., 2020; Sobczak et al., 2020). For example, fungal pathogens such as, Coelomomyces, Culicinomyces, and Lagenidium have the capacity to kill larva and adult mosquitoes, reducing their host population (Scholte et al., 2004). Some entomopathogenic fungi, e.g., Beauveria bassiana, Beauveria brongniartii, Metarhizium anisopliae, and Verticillium lecanii, have been developed as biocontrol agents usable against agricultural pests like aphids, locusts, grasshoppers and cockchafer in Africa and Europe (Roberts and Hajek, 1992; Shah and Pell, 2003). Beauveria bassiana and B. brongniartii were found to be especially safe bioinsecticides (Zimmermann, 2007). Additionally, some insect pathogens with pharmacological activities are frequently studied, such as Cordyceps militaris extract, which exhibits antitumor properties (Li et al., 2020). Cordyceps spp. have been utilized as therapeutic agents for metabolic-related disorders (Cao et al., 2020). Cordyceps cicadae has renoprotective effects on hypertensive renal injuries (Huang et al., 2020). Entomopathogenic fungi have important biotechnological applications (Hyde et al., 2019) and Paraisaria is no exception. Several studies have explored the importance of Paraisaria species, such as their antioxidative activity (Ma et al., 2012), nucleoside components (Suo et al., 2013), intracellular polysaccharide composition (Wang et al., 2019) and AGS gastric cancer cells anti-proliferation effects (Ye et al., 2015). Additionally, P. heteropoda reportedly produces anti-bacterial and anti-fungal compounds (Krasnoff et al., 2005). Experiments into optimal cultural conditions and nutritional sources were conducted by Sung et al. (2011). Applications of other species in this genus have been poorly studied.
Entomopathogenic fungi are phylogenetically diverse and taxonomically distributed in Ascomycota, Basidiomycota, Chytridiomycota, Entomophthoromycota, Microsporidia, Oomycota and Zygomycota (Vega et al., 2012; Araújo and Hughes, 2016; Mora et al., 2017). Different groups of entomopathogens usually develop respectively unique strategy to colonize their hosts (Mora et al., 2017). It is worth to mention that entomopathogenic taxa in Entomophthorales (Entomophthoromycota) enter into biotrophic relationships with their insect hosts, while those in Hypocreales (Ascomycota) can be hemibiotrophic at earlier stages and transform into saprophytism (Shah and Pell, 2003). The diversity, taxonomy and phylogeny of entomopathogenic fungi have been extensively studied recently (Aung et al., 2008; Mora et al., 2017; Hyde et al., 2018). Most insect pathogens are known from three families: Clavicipitaceae, Cordycipitaceae, and Ophiocordycipitaceae. They are found in the Hypocreales, Hypocreomycetidae, Sordariomycetes, Ascomycota (Sung et al., 2007a; Maharachchikumbura et al., 2016; Wijayawardene et al., 2018). The generic composition of Ophiocordycipitaceae underwent several changes over time (Sung et al., 2007a; Quandt et al., 2014; Maharachchikumbura et al., 2016; Shrestha et al., 2017; Wijayawardene et al., 2018), and currently ten genera are accepted (Hyde et al., 2020). New combinations of these genera were proposed for Polycephalomyces by Kepler et al. (2013), Tolypocladium by Quandt et al. (2014), Perennicordyceps by Matočec et al. (2014) and Drechmeria, Harposporium, Ophiocordyceps, and Purpureocillium by Spatafora et al. (2015). The genus Paraisaria was recently recovered in Ophiocordycipitaceae (Mongkolsamrit et al., 2019).
The genus Paraisaria was established by Samson and Brady (1983), with P. dubia as the type species, whose sexual morph was known as Ophiocordyceps gracilis (syn. Cordyceps gracilis). The sexual morph of this genus is characterized by solitary stromata with a stipe terminating in a globose or ellipsoid fertile head, completely immersed, ostiolate, gregarious perithecia, cylindrical asci and hyaline, filiform, multi-septate ascospores, which break into aseptate fragments when mature. Its asexual morphs are characterized by verticillate branched conidiophores, phialidic, flask-shaped, usually sympodially proliferating conidiogenous cells, which terminate in 1–4 necks, and aseptate, hyaline, smooth-walled conidia, which usually aggregate in slimy heads (Samson and Brady, 1983). Li et al. (2004) synonymized Isaria gracilioides under P. gracilioides and linked its sexual morph to Ophiocordyceps gracilioides. Evans et al. (2010) found the asexual morph of P. myrmicarum from a red ant host (Myrmica rubra) in a natural environment in the United Kingdom. Quandt et al. (2014) have dropped the genus Paraisaria and used its sexual genus Ophiocordyceps according to the ‘one fungus one name’ principle. Mongkolsamrit et al. (2019) resurrected Paraisaria on the basis of three new species, e.g., P. orthopterorum, P. phuwiangensis, and P. yodhathaii as well as eight new combinations, e.g. P. amazonica (Sanjuan et al., 2015), P. blattarioides (Sanjuan et al., 2015), P. coenomyiae (Ban et al., 2015), P. gracilioides (Kobayasi, 1941; Pérez-Villamares et al., 2017), P. gracilis (Samson and Brady, 1983; Pérez-Villamares et al., 2017), P. heteropoda (Sung et al., 2011; Mongkolsamrit et al., 2019), P. paramyrmicarum (= P. myrmicarum) (Evans et al., 2010) and P. tettigonia (Wen et al., 2016). So far, together with the three new species in this study, 14 species are accepted in Paraisaria.
This study is part of a larger survey of fungi in the Greater Mekong Subregion where we came across numerous new taxa (Hyde et al., 2018). In this study, three specimens of entomopathogenic fungi were collected from disturbed forests in China and Thailand, and the typical macro- and micro- morphological characteristics indicate that they are of the Paraisaria species. The multigene phylogenetic analysis of LSU, SSU, TEF1-α, RPB1, RPB2, and ITS confirmed their placement within Paraisaria as three distinct new species.
Materials and Methods
Sample Collection, Isolation, and Morphological Studies
In this study, a total of four fungal specimens were collected. One specimen (HKAS 102484) was collected from Krabi Province in Thailand on an adult cricket. Two specimens (HKAS 102553 and HKAS 102552) on dead larvae of Lepidoptera sp. were collected from Guizhou Province of China. One specimen (HKAS 102546) was collected from Yunnan Province in China on Coleoptera sp. larva. Among them, the hosts of specimens HKAS 102484, HKAS 102553 and HKAS 102552 were found completely immersed into soil with the stroma protruding from the ground in a forest. Specimen HKAS 102546 was found in a similar condition, but differed in that it was found under a karst stone formation. Macro-morphological characteristics of fresh collections were recorded with a camera (iPhone XS Max) in the field and then the specimens were transported to the laboratory in plastic boxes for subsequent studies. The culture of the specimen HKAS 102546 was created by transferring a small mass of mycelium inside the body of the host into potato dextrose agar (PDA, 1% w/v peptone) using a burned needle and incubated at room temperature (25°C). The pure culture was stored in twice-sterilized water, a 15% glycerinum solution and PDA medium, and deposited in the KUMCC culture collection of the Kunming Institute of Botany (KIB), Chinese Academy of Sciences (CAS). The fruiting bodies were dried with allochroic silica gel and deposited in KUN herbarium of KIB. Facesoffungi numbers were registered as outlined in Jayasiri et al. (2015).
The fresh fruiting bodies were examined and hand-sectioned under an Optec SZ660 stereo dissecting microscope. The key fungal structures viz. ascomata, perithecia, peridium, asci and ascospores were mounted in sterilized water or cotton blue solution slides and observed and photographed using a compound microscope (Nikon ECLIPSE Ni) with a digital camera (Canon EOS 600D) fitted on to the top of the microscope. These important fungal structures were measured with the Tarosoft (R) Image Frame Work program and the images used were processed with Adobe Photoshop CS3 Extended v. 10.0 (Adobe®, San Jose, CA, United States).
DNA Extraction, PCR Amplification, and Sequencing
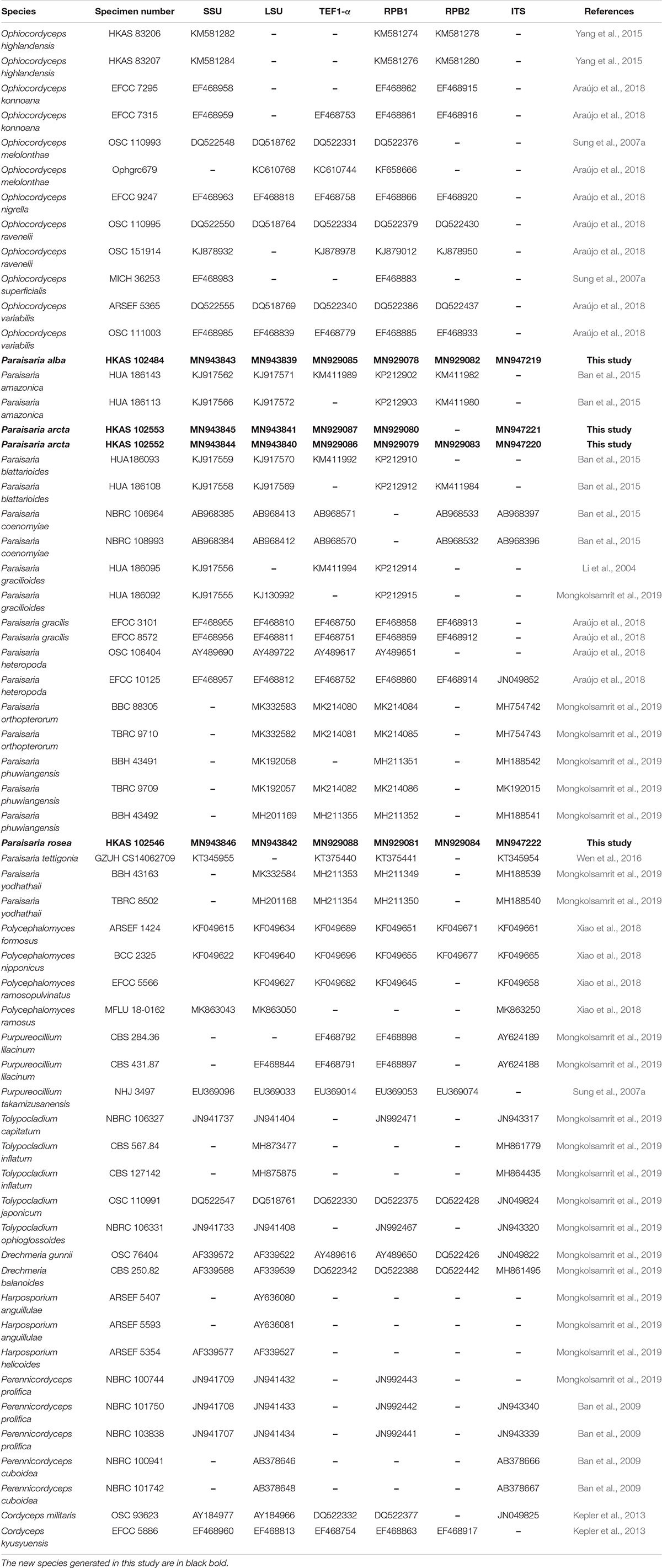
The total DNA was extracted from stromal tissue of specimens HKAS 102552, HKAS 102553, HKAS 102484 and from fresh mycelium of KUMCC 20-0001 (ex-type culture of isolate HKAS 102546) using DNA extraction kit (Omega Fungus Genomic DNA Extraction Kit, China), following the protocol of the manufacturer. The obtained DNA was stored at −20°C in a refrigerator. The PCR amplification was performed in 25 μL volumes consisting 12.5 μL PCR mixture (2 × Taq PCR Master Mix, red dye) which contains Taq DNA polymerase, dNTPs, MgCl2, a reaction buffer, a PCR reaction enhancer, an optimizer and stabilizer, 8.5 μL of twice-sterilized water, 1 μL of each primer and 2 μL of 30 μg/μl DNA template. The internal transcribed spacer (ITS1-5.8S-ITS2, ITS), large subunit ribosomal RNA (LSU rRNA), small subunit ribosomal RNA (SSU rRNA), translation elongation factor 1-alpha gene (TEF1-α) and RNA polymerase II largest subunit (RPB1) and RNA polymerase II second largest subunit (RPB2) were amplified with the primers and procedures mentioned in Table 1. The PCR products were sent to Tsingke company, Yunnan Province, China, for sequencing the above genes. The generated sequences were submitted to GenBank, and the accession numbers have been shown in Table 2.
Sequence Alignment and Phylogenetic Analyses
The generated sequences were assembled with Sequencing Project Management (SeqMan) (Clewley, 1995). The sequences for the combined alignment were selected based on the blast results of LSU, SSU, ITS, TEF, RPB1, and RPB2 as well as the recent references listed in Table 2. The individual gene alignment was aligned in MAFFT v. 7 web server1 (Kuraku et al., 2013; Katoh et al., 2019). The alignments of each locus were improved by manually removing uninformative gaps and ambiguous regions using BioEdit v. 7.0.9.1 (Hall, 1999) and were concatenated in Sequence Matrix v. 1.7.8 (Vaidya et al., 2011). The final combined alignment was converted to a NEXUS file (.nex) with ClustalX2 v. 1.83 (Thompson et al., 1997) and was used for Bayesian inference (BI) analysis and Maximum parsimony analysis (MP). The optimum nucleotide substitution model of each gene was selected by MrModeltest v.2.3 (Nylaner, 2004) using the Akaike information criterion (AIC) method and was applied to Bayesian inference (BI) analysis that was performed using MrBayes on XSEDE (2.2.7a) (Ronquist and Huelsenbeck, 2003) on CIPRES Science Gateway2. The Bayesian posterior probability (BYPP) was estimated by the Markov Chain Monte Carlo (MCMC) technique. Six simultaneous Markov Chains were run for 2,000,000 generations with sampling every 1,000 generation. The first 25% of sampled trees were discarded during the burn-in period. Maximum likelihood analysis was carried out using RAxML-HPC2 on XSEDE (8.2.10) in CIPRES Science Gateway V. 3.3 (Miller et al., 2010), with default algorithm and bootstrap iterations were set to 1,000 and substitution model was set to GTRGAMMA + I. Maximum parsimony analysis was implemented in PAUP v. 4.0b10 (Swofford, 2002) through heuristic search with 1,000 random replicates of stepwise addition and tree-bisection-reconnection (TBR) of branch-swapping algorithm. Gaps were treated as missing data and max trees was set to 1,000. Branches collapsed when minimum branch length was zero. The consistency index (CI), retention index (RI), rescaled consistency index (RC) and homoplasy index (HI) were calculated for the maximum parsimony tree. For the delimitation of new species based on nucleotide comparison, we follow the suggestion of Jeewon and Hyde (2016).
The tree topologies were visualized in FigTree v1.4.0 (Rambaut, 2006) and edited in Microsoft power point (2016) and Adobe Photoshop CS3 Extended v. 10.0 (Adobe®, San Jose, CA, United States). The final alignment and trees were submitted to TreeBASE with submission number 256643.
Results
Phylogenetic Analyses
Phylogenetic analyses were constructed with combined LSU, SSU, TEF1-α, RPB1, RPB2, and ITS sequences data of 58 representative taxa in Ophiocordycipitaceae. Trees were rooted to Cordyceps militaris (OSC 93623) and C. kyusyuensis (EFCC5886) in Cordycipitaceae. The alignment contains 5239 characters, including gaps (LSU: 918, SSU: 1027, TEF1-α: 906, RPB1: 664, RPB2: 1024, ITS: 700). Parsimony analysis of this dataset produced the 20 most parsimonious trees of 4833 steps in length, of which 3436 characters were constant, 380 variable characters parsimony-uninformative and 1423 characters parsimony-informative. The first parsimonious tree was represented as the best tree, with CI = 0.549, RI = 0.777, RC = 0.426 and HI = 0.451. The RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of −30766.070218. The matrix had 2305 distinct alignment patterns, with 41.28% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.236752, C = 0.277080, G = 0.283017, T = 0.203151; substitution rates AC = 1.485223, AG = 3.851975, AT = 0.915108, CG = 1.456245, CT = 6.890167, GT = 1.000000; gamma distribution shape parameter α = 0.465094.
In the phylogenetic analyses (Figure 1), eight genera are included in Ophiocordycipitaceae labeled on the tree. With the exception of Ophiocordyceps, the other remaining genera are monophyletic and individually they received strong statistical support. The three novel entomopathogenic fungi grouped with the taxa in Paraisaria with significant statistical support (1.00 PP/100% ML/98% MP). Paraisaria alba (HKAS 102484) constitutes a sister phylogenetic affiliation to P. yodhathaii with 0.96 PP/98% MP statistical support. Paraisaria rosea (HKAS 102546) is closely related to P. amazonica and P. blattarioides, but this is statistically not supported in all three formats. Two strains of P. arcta grouped as an intermediate clade with close phylogenetic connection to P. coenomyiae, P. gracilioides, and P. heteropoda.
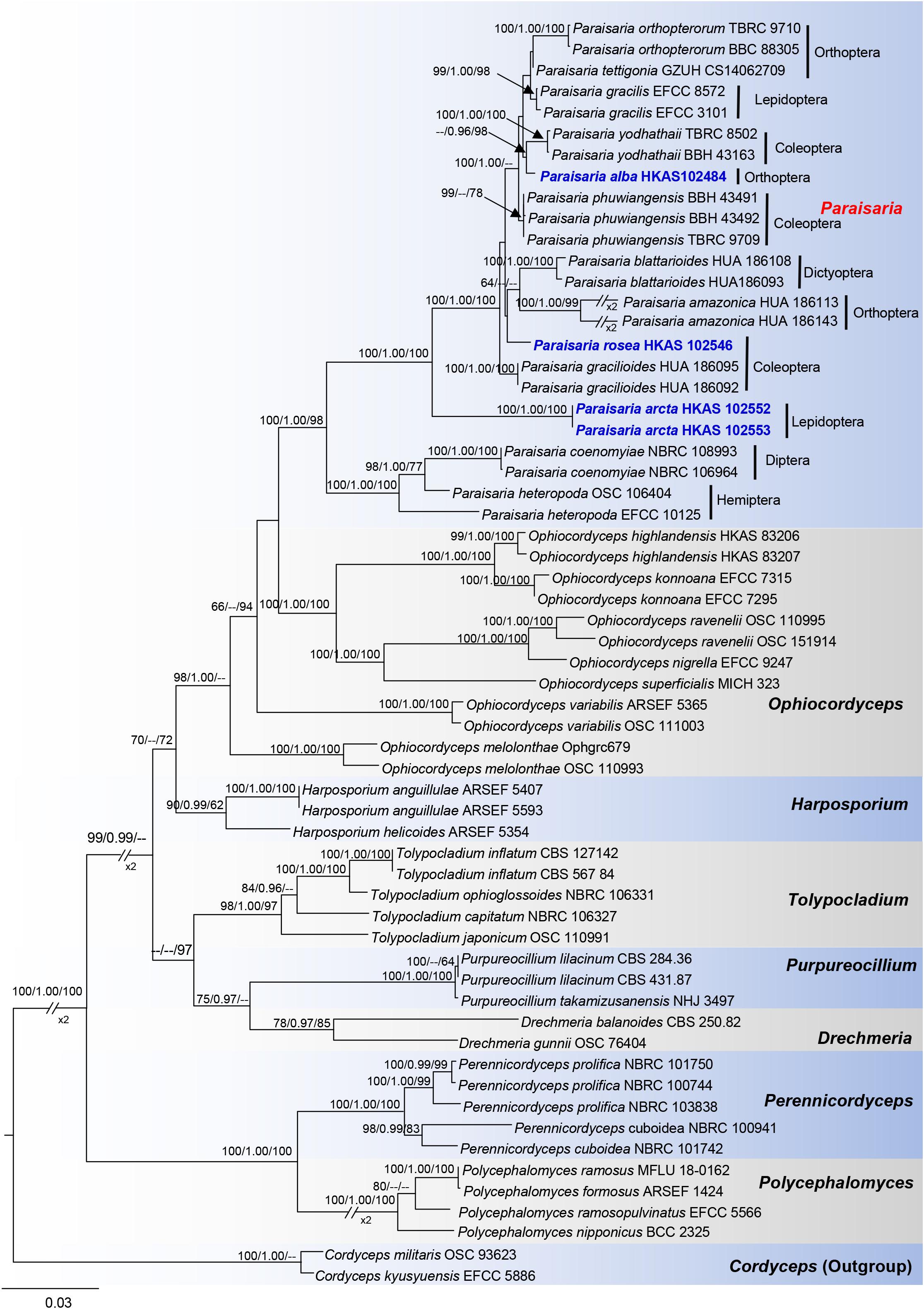
Figure 1. Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, TEF1-α, RPB1, RPB2, and ITS sequence data. Bootstrap values for BI equal to or higher than 95%, ML and MP equal to or greater than 60% are placed on the notes. The newly generated sequences are indicated in blue bold. The host order of Paraisaria species and the generic names are labeled in the right side.
Taxonomy
Paraisaria alba D. P. Wei and K. D. Hyde, sp. nov. Figure 2
Etymology: alba refers to the white fertile head.
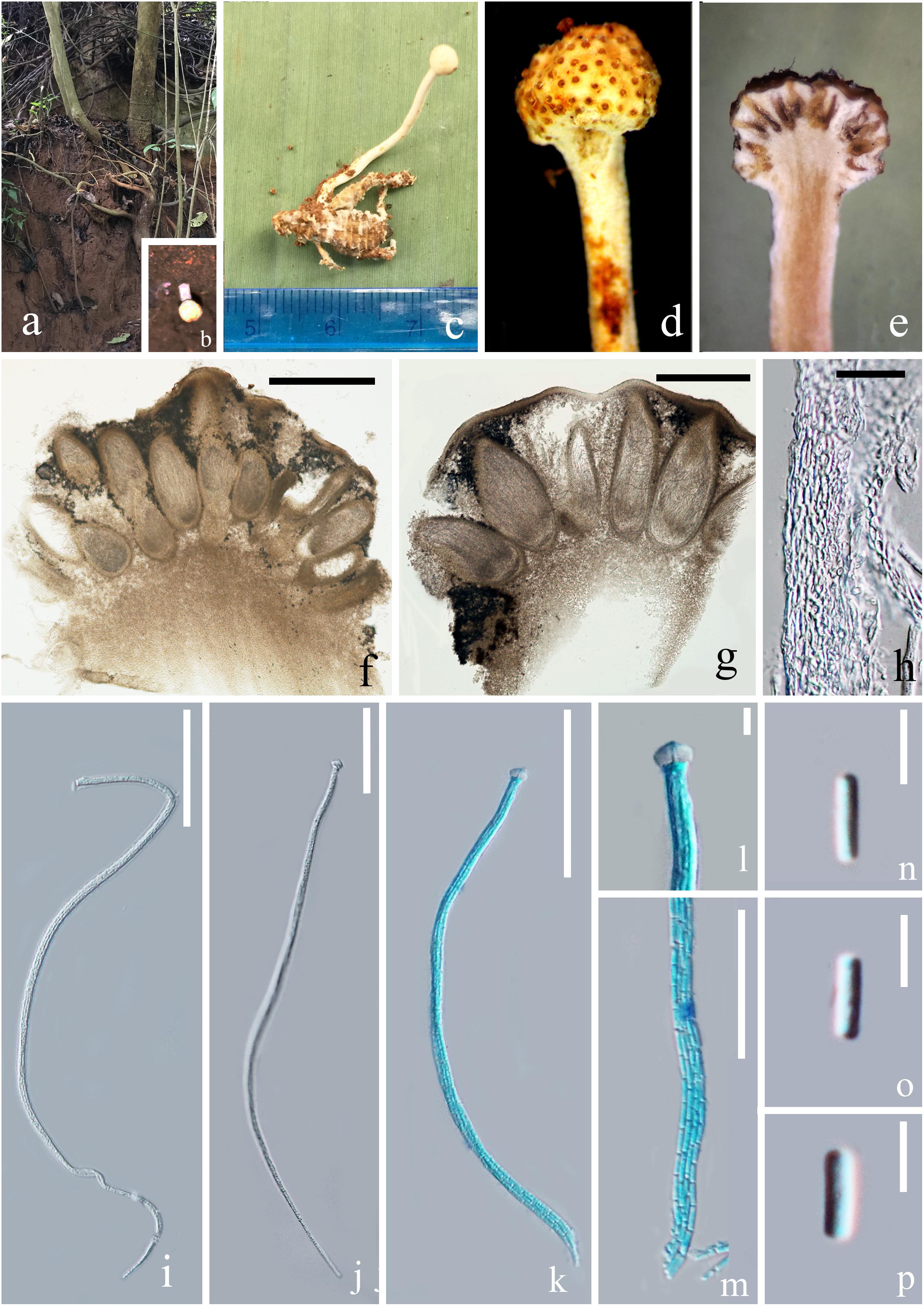
Figure 2. Paraisaria alba (HKAS 102484, holotype). (a) Habitat. (b) Host imbedded into the soil with the stroma emerging from the ground. (c) Stroma arising from host. (d) Fertile head. (e) Longitudinal section through fertile head. (f,g) Perithecia. (h) Peridium. (i–k) Asci. (l) Ascus cap. (m) Part of the asci. (n–p) Secondary ascospores. Scale bars: (f) 500 μm, (g) 200 μm, (h) 20 μm, (i–k) 50 μm, (m) 30 μm, (l,n–p) 5 μm. (k–m mounted in cotton blue reagent.).
MycoBank number: MB 833999
Facesoffungi number: FoF 07239
Parasitic on an adult cricket (Orthoptera). Sexual morph: Stroma up to 26 mm in tall, single, unbranched, growing from the flank of the host. Fertile head 3.5 mm in diam., globose, white when fresh, yellow brown when dry. Stipe 22.5 × 1.2 mm, slightly flexuous, fleshy, white, glossy, not hollow. Perithecia 200–500 × 100–220 ( = 325 × 145, n = 20) μm, immersed, ovoid. Asci 160–250 × 2.5–5 ( = 200 × 3.5, n = 10) μm, unitunicate, hyaline, narrow cylindrical, attenuated toward the base, with thickened cap. Peridium 10–40 ( = 20, n = 30) μm in thick, comprising hyaline, thick-walled cell of textura angularis. Apical cap 4.6–7.4 × 3.2–4.9 ( = 6 × 3.8, n = 30) μm, with a narrow tunnel throughout the center. Ascospores filiform, equal to the asci in length, when mature, breaking into numerous secondary ascospores. Secondary ascospores 3–5 × 0.5–1.5 ( = 4 × 1, n = 30) μm, cylindrical, hyaline, smooth, one-celled, straight, with truncated ends.
Material examined
Thailand, Krabi, Plai Phraya (N: 8°24′410′′, E: 98°45′34′′). On an adult cricket, 20 December 2018, Deping Wei, 211-1(HKAS 102484– holotype). We tried to culture P. alba by transferring a small piece of inner stroma tissue into a PDA medium using a sterilized needle, but growth was not observed.
Notes
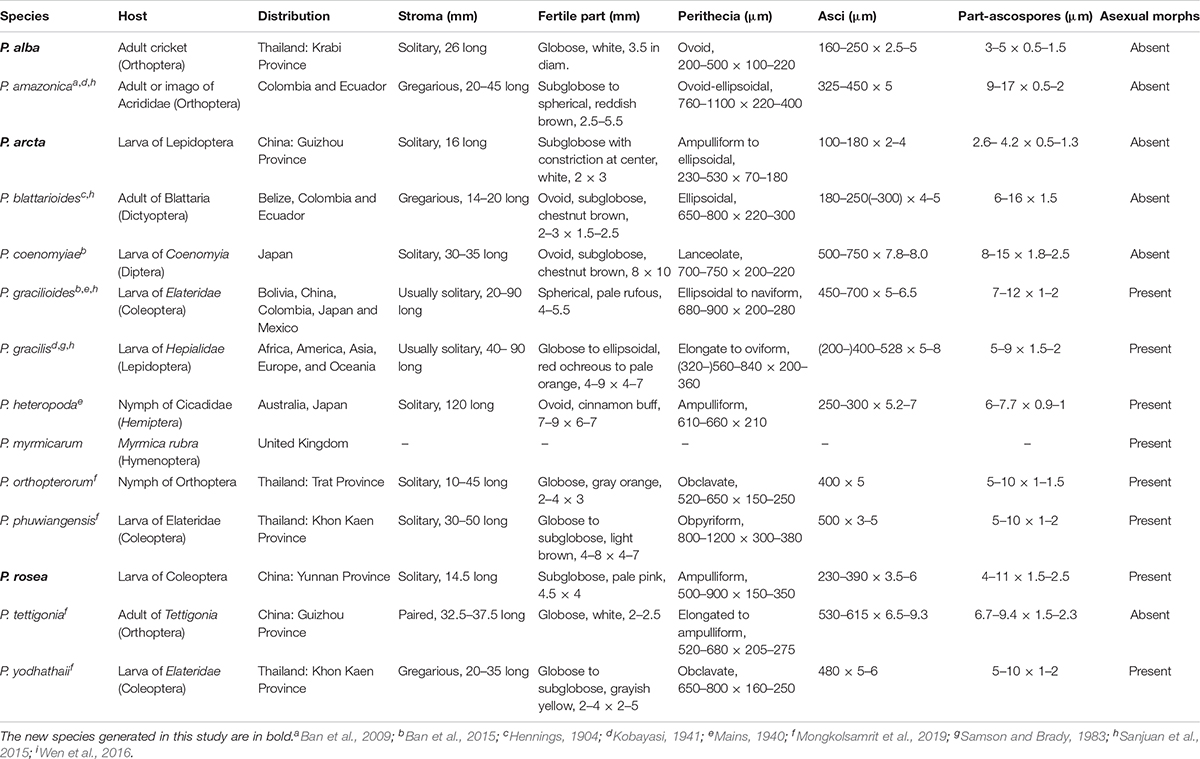
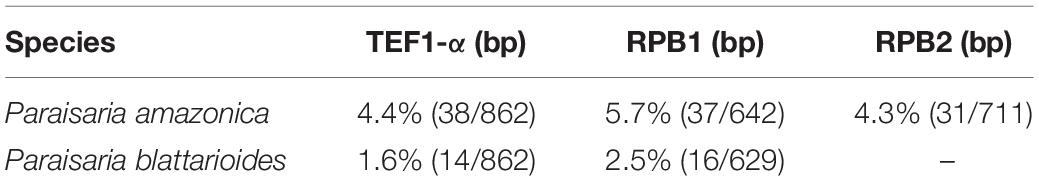
The multigene phylogenetic analysis showed that P. alba groups with P. yodhathaii with fairly good statistical support (0.96 PP/98% MP, Figure 1). This relationship is, however, not supported by the ML analysis. Paraisaria alba differs from P. yodhathaii in having solitary stroma, a white fertile head, and smaller perithecia, asci and secondary ascospores, whereas P. yodhathaii has paired stromata, grayish yellow fertile head, larger perithecia and larger asci and secondary ascospores (Table 3). The comparison of the nucleotide sequences between P. alba and P. yodhathaii show 10 (including 6 gaps) out of 410 bp (2.4%), 6 out of 746 bp (0.8%), 5 out of 881 bp (0.56%) and 8 out of 534 bp differences (1.5%) in ITS, LSU, TEF1-α and RPB1 sequences, respectively. SSU and RPB2 sequences data of P. yodhathaii are not available in GenBank. Henceforth, we describe our collection as a new species in Paraisaria according to the guidelines of Jeewon and Hyde (2016).
Paraisaria arcta D. P. Wei and K. D. Hyde, sp. nov. Figure 3
Etymology: arcta refers to the constricted fertile head.

Figure 3. Paraisaria arcta (HKAS 102553, holotype). (a) Substrate. (b–d) Stromata emerging from host mouth. (e) Fertile head. (f–h) Perithecia. (i) Ostiole. (j) Peridium. (k,l) Asci. (m) Parts of ascus. (n) Ascus cap. (o,p) Secondary ascospores. Scale bars: (d,e) 2000 μm, (f) 500 μm, (g,h) 200 μm, (i,k) 50 μm, (j,m) 15 μm, (l) 30 μm, (n–p) 3 μm.
MycoBank number: MB 834000
Facesoffungi number: FoF 07240
Parasitic on larva of Lepidopteran larva. Sexual morph: Stroma 16 mm long, single, arising from the mouth of host larva. Fertile head 2 mm long, 3 mm wide, white, nearly globose, constricted at the center, with sticky and crystal-like substance on the surface. Stipe 14 mm long, 2 mm wide, straight, fleshy, white, glossy. Perithecia 230–530 × 70–180 ( = 387 × 113, n = 20) μm, completely immersed, ampulliform to ellipsoid. Peridium 14–20 ( = 17, n = 30) μm wide, composed of hyaline, thick-walled, smooth-walled cells of textura angularis. Asci 100–180 × 2–4 μm ( = 137 × 2.9, n = 15), unitunicate, hyaline, narrow cylindrical, tapering toward the base, 8-spored, with thickened cap. Apical cap 3.5–4.5 × 2–3.6 μm thick ( = 4 × 2.8, n = 20), with a narrow tunnel throughout the center. Ascospores hyaline, narrow filiform, equal to the asci in length, when mature, breaking into numerous secondary ascospores. Secondary ascospores 2.6–4.2 × 0.5–1.3 μm ( = 3.3 × 0.9, n = 60), cylindrical, with truncated ends, hyaline, smooth, one-celled, straight.
Material examined
China, Guizhou Province, Qianxinan Buyei and Miao Autonomous Prefecture, Ceheng County, Gaofeng Villige (N: 24°57′33′′, E: 105°50′1′′), on dead larva of Lepidoptera sp., 6 August 2018, Deping Wei, GFC604 (HKAS 102553–holotype); GFC603 (HKAS 102552 – paratype). The culturing of P. arcta was tried by transferring a mass of mycelium found inside body of the larva host to a PDA medium using a sterilized needle. However, mycelium growth was not observed.
Notes
Paraisaria arcta resembles P. alba found in Krabi Province, Thailand and P. tettigonia discovered in Guizhou Province, China in having white fertile heads but differs from P. alba in its associated host and number of stromata are distinct from P. tettigonia (Wen et al., 2016). Paraisaria arcta can also be distinguished from the other species in Paraisaria by the color and shape of its fertile head. A conspicuous ravine throughout the center of the fertile head is present in P. arcta, which is lacking in the other species in this genus. The detailed comparisons are shown in Table 3. Multigene phylogenetic analysis showed P. arcta constitutes a distant clade from other species in Paraisaria, with strong statistical support (100% ML, 100% MP, 1.00 PP, Figure 1). Herein, we introduce this collection as a new species of Paraisaria.
Paraisaria rosea D. P. Wei and K. D. Hyde, sp. nov. Figures 4, 5
Etymology: rosea refers to its pink fertile head.
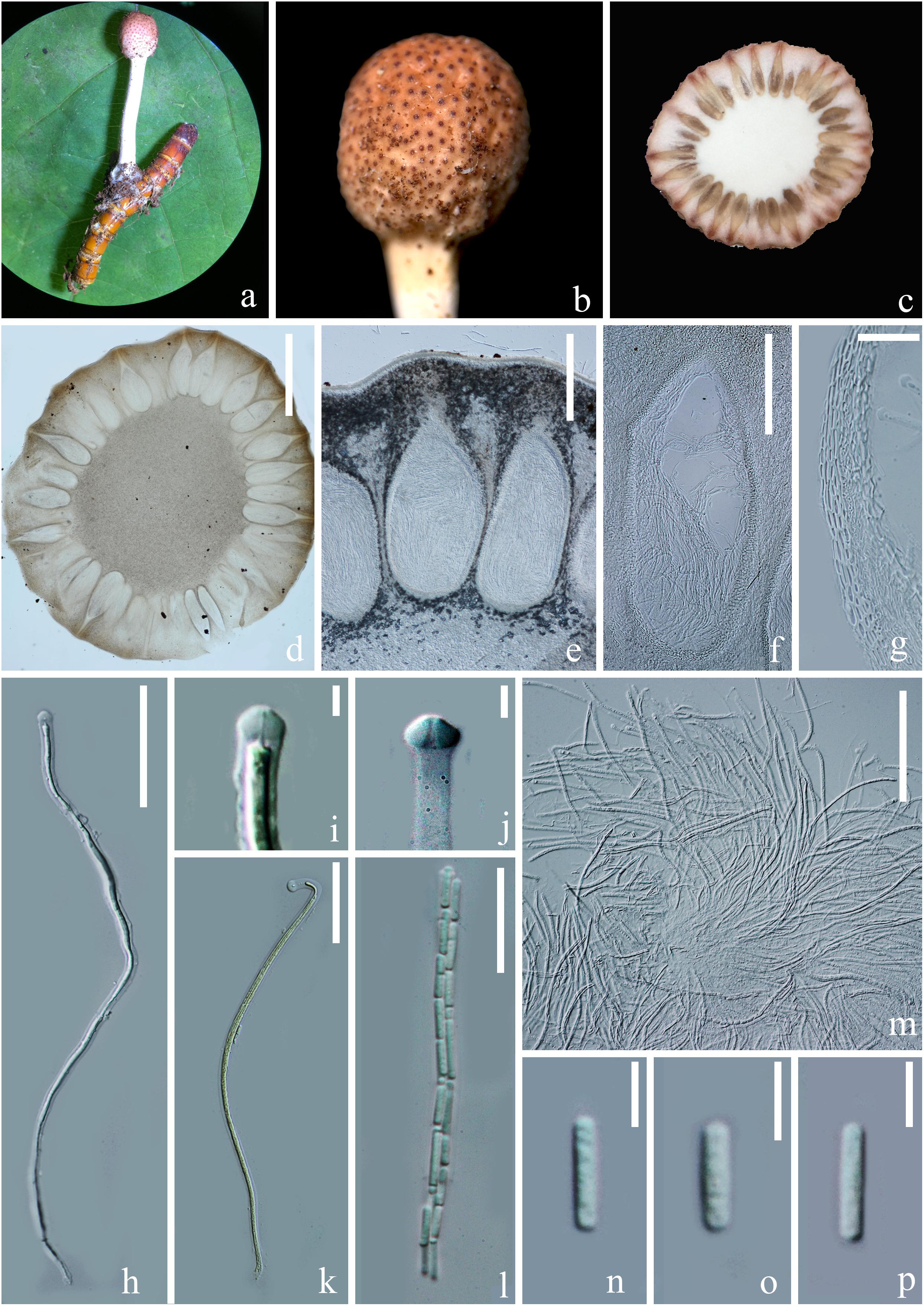
Figure 4. Sexual morph of Paraisaria rosea (HKAS 102546, Holotype). (a) Stroma emerging from host. (b) Fertile head. (c) Transverse section of the fertile head. (d–f) Perithecia. (g) Peridium. (h,k,m) Asci. (i,j) Asci cap. (l) Part of ascus. (n–p) Secondary ascospores. Scale bars: (d) 1000 μm, (e) 300 μm, (f) 200 μm, (g,m) 30 μm, (h,k) 50 μm, (l) 20 μm, (i,j, n–p) 5 μm. (k mounted in Melzer’s reagent).
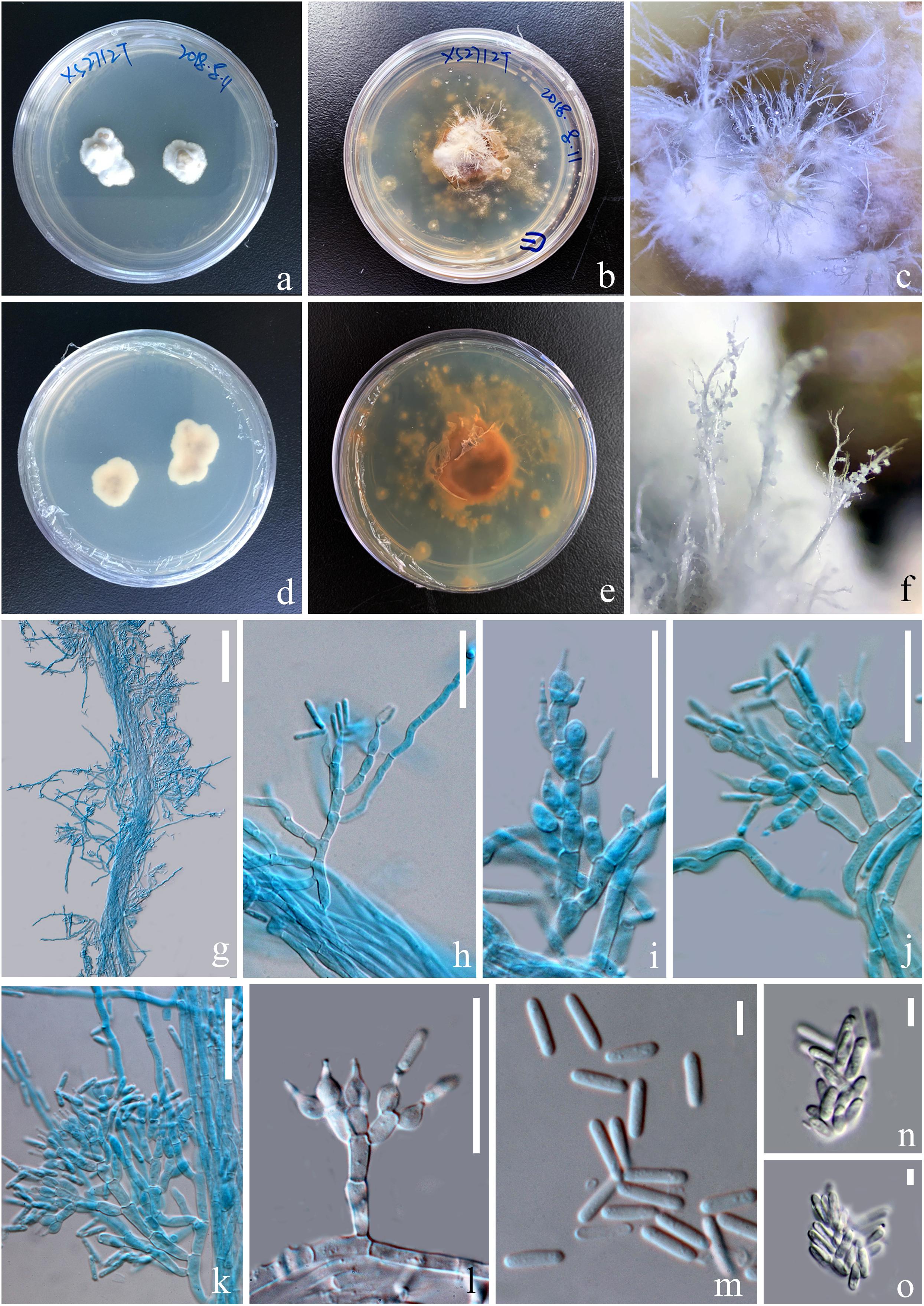
Figure 5. Asexual morph of Paraisaria rosea (KUMCC 20-0001, ex-type). (a,d) Upper and lower views of cultures on PDA after 50 days. (b,e) Upper and lower views of cultures on PDA after 16 months incubation in dark environments. (c,f) Enlargement of aerial synnemata produced on culture. (g) Synnema bearing conidiophores. (h–l) Phialides. (m) Conidia. (n,o) Irregularly aggregated conidia. Scale bars: (g) 100 μm, (h–l) 30 μm, (m–o) 5 μm. (h–k,m mounted in cotton blue reagent.).
MycoBank number: MB834001
Facesoffungi number: FoF 07241
Parasitic on a larva of Coleoptera. Host buried in the soil, with the stroma erumpent from the ground. Sexual morph: Stroma up to 14.5 mm long, laterally emerging from the middle part of the larva body, simple, erect. Fertile head 4.5 × 4 mm, subglobose, pale pink at top and paler toward the base when fresh, pale yellow-brown when dry. Stipe 10 × 1.5 mm, white, straight, unbranched, glossy, cylindrical, inside not hollow. Perithecia 500–900 × 150–350 ( = 762 × 256, n = 30) μm, completely immersed, ampulliform, ostiolate. Peridium 9–15 ( = 12, n = 30) μm wide, composed of hyaline, thick-walled cells of textura angularis to textura globulosa to textura prismatica. Asci 230–390 × 3.5–6 ( = 280 × 5, n = 15) μm, hyaline, cylindrical, unitunicate, eight-spored, possessing a prominent apical cap. Apical cap 5–7 × 2–6 ( = 6 × 4, n = 20) μm, with a conspicuous tunnel throughout the center. Ascospores filiform, hyaline, breaking into secondary ascospores when mature. Secondary ascospores 4–11 × 1.5–2.5 ( = 7.5 × 2, n = 30) μm, hyaline, cylindrical with truncate ends, smooth-walled, aseptate. Asexual morph: Hyphomycetous. Synnemata producing from the center of culture after 16 months incubation in dark environment, composed of loose, septate hyphae, white, filamentous, aerial, straight, branched, fasciculate, bearing shining droplets and conidiophores. Mycelium 2.4–3.7 ( = 3, n = 10) μm in wide, septate, hyaline, smooth-walled. Conidiophores 33–48 ( = 41, n = 10) μm in height, irregularly differentiate from the synnemata, sparse, gregarious, branched. Phialides 5.8–11.5 × 3–5.5 ( = 8.6 × 4, n = 30) μm, ampulliform, 1-necked, hyaline, aseptate, enteroblastic, phialidic, monophialidic. Conidia 8–12 × 2–2.6 ( = 9.8 × 2.3, n = 50) μm, hyaline, cylindrical, smooth-walled, aseptate, with round ends.
Culture characteristics
Culture was made from mycelium inside body of the host larva, slowly growing on PDA, reaching 1.3 cm in diam after incubated at room temperature (25°C) for 50 days, convex, dense, with undulate edges, smooth surface become filamentous after forming aerial synnemata. The shooting conidia land on the surrounding culture and develop new colonies.
Material examined
China, Yunnan Province, Kunming, Western hill Park (N: 24°57′28′′, E: 102°38′17′′), on larva of Coleoptera sp. buried in soil, 27 July 2018, Deping Wei, XS2712 (HKAS 102546 – Holotype); (KUMCC 20-0001 – ex-type living culture).
Notes
Paraisaria rosea is closely related to P. amazonica and P. blattarioides, without any statistical support (Figure 1). However, P. rosea can be distinguished from these related species based on the number of stromata, the color of the fertile head and the size of asci and secondary ascospores (Table 3). The ITS sequence of P. amazonica and P. blattarioides are not available in GenBank database; the nucleotide differences in the TEF1-α, RPB1 and RPB2 region between P. rosea and the two above species are greater than 1.5% (Table 4). Thereby, we introduced P. rosea as a new species in this genus based on the distinctive morphology and molecular support.
Discussion
The sexual morph of Paraisaria species phenotypically share an erect or slightly flexuous, cylindrical, colorless, fleshy stipe that terminates in a subglobose to globose fertile head and completely immersed perithecia. Asci are cylindrical with a thickened apical cap. Ascospores are hyaline, multi-septate and usually break into numerous cylindrical, truncated fragments at maturity. However, they can be distinguished according to their associated host, the number of stroma and the color of the fertile head. Species in this genus usually infect several stages of insects, such as larvae of Coleoptera, Diptera, and Lepidoptera; nymphs of Hemiptera and Orthoptera; or adults of Dictyoptera, Hymenoptera (ant) and Orthoptera (Evans et al., 2010; Sanjuan et al., 2015; Mongkolsamrit et al., 2019). According to the number of stromata, species of Paraisaria can be divided into three groups: solitary stroma, paired stromata and multiple stromata (see the key below). The shape of their fertile head features little variation, though differing in color, ranging from white, pale pink, pale rufous, red ochreous to pale orange, chestnut, cinnamon buff, grayish, reddish brown to dark brown (see Table 3).
The asexual morphs of this genus are known in eight species, viz. P. myrmicarum (Evans et al., 2010), P. gracilis (Samson and Brady, 1983), P. gracilioides (Li et al., 2004), P. rosea (this study), P. heteropoda, P. orthopterorum, P. phuwiangensis, and P. yodhathaii (Mongkolsamrit et al., 2019). Their conidiophores are irregularly branched and generally develop from white, rope-like synnemata. Their phialides are flask-shaped, with a swollen base and narrow neck. Most species produce only one neck from the terminal phialides. Some species, e.g., P. gracilis, P. gracilioides, P. myrmicarum and P. orthopterorum produce 1–4 necks per phialides. Their conidia are cylindrical or ellipsoid or fusiform. Some species, e.g., P. orthopterorum and P. yodhathaii have both cylindrical and fusiform forms of conidia (Mongkolsamrit et al., 2019).
Sung et al. (2007a) have concluded that multi-gene phylogeny gave more deeper understanding of phylogenetic relationships of Cordyceps and Clavitipitaceae than that of single gene. Recently, the combined LSU-TEF1-α-RPB1 datasets (Mongkolsamrit et al., 2019), combined SSU-LSU-TEF-RPB2 datasets (Ban et al., 2015), and combined SSU-LSU-TEF1-α-RPB1-RPB2 datasets (Quandt et al., 2014; Sanjuan et al., 2015) were allowed for intraspecific and intergeneric identification within Ophiocordycipitaceae. However, individual gene phylogenies are rarely utilized for identification of species in Paraisaria.
Key to the Accepted Species in Paraisaria
(1) Host belong to Hymenoptera……………………P. myrmicarum
(1) Host not belong to Hymenoptera…………………………………….2
(2) Fertile part colorless…………………………………………………………3
(2) Fertile part pigmented……………………………………………………..4
(3) Fertile part constrict at the center………………………….P. arcta
(3) Fertile part is not constricted at the center………………………5
(4) Stromata gregarious…………………………………………………………6
(4) Stromata solitary……………………………………………………………..7
(5) Stromata branched………………………………………….P. tettigonia
(5) Stromata unbranched……………………………………………..P. alba
(6) Stromata equal or shorter than 20 mm………..P. blattarioides
(6) Stromata longer than 20 mm……………………………………………8
(7) Attack nymph stage of host……………………………………………..9
(7) Attack larva stage of host……………………………………………….10
(8) Fertile part reddish brown…………………………….P. amazonica
(8) Fertile part grayish yellow……………………………..P. yodhathaii
(9) Stromata long, 120 mm…………………………………P. heteropoda
(9) Stromata short, 10–45 mm………………………P. orthopterorum
(10) Host belong to Coleoptera……………………………………………..11
(10) Host belong to other order of insect………………………………12
(11) Stromata equal or shorter than 14.5 mm………………P. rosea
(11) Stromata longer than 14.5 mm………………………………………13
(12) Pathogenic on larva of Diptera (Coenomyia). ……………………………………………………………………P. coenomyiae
(12) Pathogenic on larva of Lepidoptera (Hepialidae) …………………………………………………………………………..P. gracilis
(13) Phialides solitary or in whorls of 2–3, with one neck………………………………………………………..P. phuwiangensis
(13) Phialides sympodially proliferating, with 1–4 necks…………………………………………………………….P. gracilioides
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MN943843, MN943839, MN929085, MN929078, MN929082, and MN947219; https://www.ncbi.nlm.nih.gov/genbank/, MN943845, MN943841, MN929087, MN929080, and MN947221; https://www.ncbi.nlm.nih.gov/genbank/, MN943844, MN943840, MN929086, MN929079, MN929083, and MN947220.
Author Contributions
D-PW, DW, and SK: conceptualization. D-PW: data curation. D-PW and DW: formal analysis, methodology, and writing – original draft. SL, ST, and SK: funding acquisition. D-PW and DW: investigation. ST and SK: project administration. KH, J-CX, and PM: supervision. CT-a, AE, SM, ST, SK, KH, J-CX, PM, NS, and SL: writing – review and editing. All authors: contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
We appreciate Thailand Research Fund (TRF) grant no. DBG6080013 entitled “The future of specialist fungi in a changing climate: baseline data for generalist and specialist fungi associated with ants, Rhododendron species and Dracaena species” for its financial support. We are grateful for the National Science Foundation of China (NSFC) project code 31750110478 for funding the sequencing cost. DW would like to thank CAS President’s International Fellowship Initiative (PIFI) for funding his postdoctoral research (number 2019PC0008) and the 64th batch of China Postdoctoral Science Foundation (grant no. Y913083271). PM and DW thank the National Science Foundation of China for financial support under the following grants: 41761144055 and 41771063. SK thanks CAS President’s International Fellowship Initiative (PIFI) young staff under the grant number: 2020FYC0002 and the National Science Foundation of China (NSFC) for funding this work under the project code 31851110759. ST would like to thank the International Postdoctoral Exchange Fellowship Program (number Y9180822S1), CAS President’s International Fellowship Initiative (PIFI) (number 2020PC0009), China Postdoctoral Science Foundation and the Yunnan Human Resources, and Social Security Department Foundation for funding her postdoctoral research. The authors extend their appreciation to the researchers supporting project number (RSP-2021/56) King Saud University, Riyadh, Saudi Arabia. This work was partly supported by Chiang Mai University.
Acknowledgments
We acknowledge Kunming Institute of Botany, Chinese Academy of Sciences for providing the laboratories and instruments for molecular work. We appreciate the Centre of Excellence in Fungal Research (Mae Fah Luang University) for providing funding for collecting trips and Dr. Shaun Pennycook is thanked for help in naming the new fungal species. Austin Smith at World Agroforestry (ICRAF), Kunming Institute of Botany, China, is thanked for English editing.
Footnotes
- ^ http://mafft.cbrc.jp/alignment/server
- ^ https://www.phylo.org/portal2/home.action
- ^ http://purl.org/phylo/treebase/phylows/study/TB2:S25664
References
Araújo, J. P., Evans, H. C., Kepler, R., and Hughes, D. P. (2018). Zombie-ant fungi across continents: 15 new species and new combinations within Ophiocordyceps. I. Myrmecophilous hirsutelloid species. Stud. Mycol. 90, 119–160. doi: 10.1016/j.simyco.2017.12.002
Araújo, J. P., and Hughes, D. P. (2016). Diversity of entomopathogenic fungi: which groups conquered the insect body? Adv. Genet. 94, 1–39. doi: 10.1016/bs.adgen.2016.01.001
Aung, O., Soytong, K., and Hyde, K. D. (2008). Diversity of entomopathogenic fungi in rainforests of Chiang Mai Province, Thailand. Fungal Divers. 30, 15–22.
Ban, S., Sakane, T., and Nakagiri, A. (2015). Three new species of Ophiocordyceps and overview of anamorph types in the genus and the family Ophiocordyceptaceae. Mycol. Prog. 14:1017. doi: 10.1007/s11557-014-1017-8
Ban, S., Sakane, T., Toyama, K., and Nakagir, A. (2009). Teleomorph–anamorph relationships and reclassification of Cordyceps cuboidea and its allied species. Mycoscience 50, 261–272. doi: 10.1007/S10267-008-0480-Y
Cao, C., Yang, S., and Zhou, Z. (2020). The potential application of Cordyceps in metabolic-related disorders. Phytother. Res. 34, 295–305. doi: 10.1002/ptr.6536
Castlebury, L. A., Rossman, A. Y., Gi-Ho, S., Hyten, A. S., and Spatafora, J. W. (2004). Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 108, 864–872. doi: 10.1017/S0953756204000607
Clewley, J. P. (1995). Macintosh sequence analysis software. Mol. Biotechnol. 3, 221–224. doi: 10.1007/bf02789332
Evans, H. C., Groden, E., and Bischoff, J. F. (2010). New fungal pathogens of the red ant, Myrmica rubra, from the UK and implications for ant invasions in the USA. Fungal Biol. 114, 451–466. doi: 10.1016/j.funbio.2010.03.007
Fernández-Grandon, G. M., Harte, S. J., Ewany, J., Bray, D., and Stevenson, P. C. (2020). Additive effect of botanical insecticide and entomopathogenic fungi on pest mortality and the behavioral response of its natural enemy. Plants 9:173. doi: 10.3390/plants9020173
Hall, T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Huang, Y. S., Wang, X., Feng, Z., Cui, H., Zhu, Z., Xia, C., et al. (2020). Cordyceps cicadae prevents renal tubular epithelial cell apoptosis by regulating the SIRT1/p53 pathway in hypertensive renal injury. Evidence-Based Complement. Altern. Med. 1, 1–13. doi: 10.1155/2020/7202519
Hyde, K. D., Norphanphoun, C., Chen, J., Dissanayake, A. J., Doilom, M., Hongsanan, S., et al. (2018). Thailand’s amazing diversity: up to 96% of fungi in northern Thailand may be novel. Fungal Divers. 93, 215–239. doi: 10.1007/s13225-018-0415-7
Hyde, K. D., Norphanphoun, C., Maharachchikumbura, S. S. N., Bhat, D. J., Jones, E. B. G., Bundhun, D., et al. (2020). Refined families of sodariomycetes. Mycosphere 11, 305–1059. doi: 10.5943/mycosphere/11/1/7
Hyde, K. D., Xu, J. C., Rapior, S., Jeewon, R., Lumyong, S., Niego, A. G. T., et al. (2019). The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 97, 1–136. doi: 10.1007/s13225-019-00430-9
Jayasiri, S. C., Hyde, K. D., Ariyawansa, H. A., Bhat, J., Buyck, B., Cai, L., et al. (2015). The faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 74, 3–18. doi: 10.1007/s13225-015-0351-8
Jeewon, R., and Hyde, K. D. (2016). Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7, 1669–1677. doi: 10.5943/mycosphere/7/11/4
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kepler, R., Ban, S., Nakagir, A., Bischoff, J., Hywel-Jones, N., Owensby, C. A., et al. (2013). The phylogenetic placement of hypocrealean insect pathogens in the genus Polycephalomyces: an application of one fungus one name. Fungal Biol. 117, 611–622. doi: 10.1016/j.funbio.2013.06.002
Kobayasi, Y. (1941). The genus Cordyceps and its allies. Sci. Rep. Tokyo Bunrika Daigaku Sec. B 84, 53–260.
Krasnoff, S. B., Reátegui, R. F., Wagenaar, M. M., Gloer, J. B., and Gibson, D. M. (2005). Cicadapeptins I and II: new Aib-containing peptides from the entomopathogenic fungus Cordyceps heteropoda. J. Nat. Prod. 68, 50–55. doi: 10.1021/np0497189
Kuraku, S., Zmasek, C. M., Nishimura, O., and Katoh, K. (2013). aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41, 22–28.
Li, C. R., Ming, L., Fan, M. Z., and Li, Z. Z. (2004). Paraisaria gracilioides comb. nov., the anamorph of Cordyceps gracilioides. Mycosystema 23, 165–166.
Li, L. Q., Song, A. X., Yin, J. Y., Siu, K. C., Wong, W. T., and Wu, J. Y. (2020). Anti-inflammation activity of exopolysaccharides produced by a medicinal fungus Cordyceps sinensis Cs-HK1 in cell and animal models. Int. J. Biol. Macromol. 149, 1042–1050. doi: 10.1016/j.ijbiomac.2020.02.022
Liu, Y., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Ma, Y., Suo, F. Y., Wang, A. L., Lu, S., Ye, X., and Gong, J. (2012). Study on antioxidation activity of the broth of different originated Ophiocordyceps gracilis (Grev.) GH Sung, JM Sung, Hywel-Jones & Spatafora in vitro. Chin. J. Biochem. Pharm. 6:028.
Maharachchikumbura, S. S. N., Hyde, K. D., Jones, E. B. G., McKenzie, E. H. C., Bhat, J. D., Dayarathne, M. C., et al. (2016). Families of Sordariomycetes. Fungal Divers. 79, 1–317. doi: 10.1007/s13225-016-0369-6
Mains, E. B. (1940). Cordyceps species from British Honduras. Mycologia 32, 16–22. doi: 10.2307/3754313
Matočec, N., Kušan, I., and Ozimec, R. (2014). The genus Polycephalomyces (Hypocreales) in the frame of monitoring Veternica cave (Croatia) with a new segregate genus Perennicordyceps. Ascomycete 6, 125–133.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans LA, 1–8.
Mongkolsamrit, S., Noisripoom, W., Arnamnart, N., Lamlertthon, S., Himaman, W., Jangsantear, P., et al. (2019). Resurrection of Paraisaria in the Ophiocordycipitaceae with three new species from Thailand. Mycol. Prog. 18, 1213–1230. doi: 10.1007/s11557-019-01518-x
Mora, M. A. E., Castilho, A. M. C., and Fraga, M. E. (2017). Classification and infection mechanism of entomopathogenic fungi. Arq. Inst. Biol. 84, 1–10. doi: 10.1590/1808-1657000552015
Nylaner, J. A. A. (2004). MrModeltest v2. Program Distributed by the Author. Uppsala: Uppsala University.
Pérez-Villamares, J., Burrola-Aguilar, C., Aguilar-Miguel, X., Sanjuan, T., and Jiménez-Sánchez, E. (2017). Nuevos registros de hongos entomopatógenos del género Cordyceps s. l. (Ascomycota: Hypocreales). del Estado de México. Rev. Mex. Biodivers. 88, 773–783. doi: 10.1016/j.rmb.2017.10.013
Quandt, C. A., Kepler, R. M., Gams, W., Araújo, J. P., Ban, S., Evans, H. C., et al. (2014). Phylogenetic-based nomenclatural proposals for Ophiocordycipitaceae (Hypocreales) with new combinations in Tolypocladium. IMA Fungus 5, 121–134. doi: 10.5598/imafungus.2014.05.01.12
Rambaut, A. (2006). FigTree. Tree Figure Drawing Tool Version 1.3.1. Edinburgh: University of Edinburgh.
Rehner, S. A., and Buckley, E. P. A. (2005). Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.3852/mycologia.97.1.84
Roberts, D. W., and Hajek, A. E. (1992). “Entomopathogenic fungi as bioinsecticides,” in Frontiers in Industrial Mycology, ed. G. F. Leatham (Boston, MA: Springer), 144–159. doi: 10.1007/978-1-4684-7112-0_10
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Samson, R. A., and Brady, B. L. (1983). Paraisaria, a new genus for Isaria dubia, the anamorph of Cordyceps gracilis. Trans. Br. Mycol. Soc. 81, 285–290. doi: 10.1016/S0007-1536(83)80081-3
Samson, R. A., Evans, H. C., and Latgé, J. P. (eds). (1988). “Taxonomy of entomopathogenic fungi,” in Atlas of Entomopathogenic Fungi, (Berlin: Springer), 5–16. doi: 10.1007/978-3-662-05890-9_2
Sanjuan, T. I., Franco-Molano, A. E., Kepler, R. M., Spatafora, J. W., Tabima, J., Vasco-Palacios, A. M., et al. (2015). Five new species of entomopathogenic fungi from the Amazon and evolution of Neotropical Ophiocordyceps. Fungal Biol. 119, 901–916. doi: 10.1016/j.funbio.2015.06.010
Scholte, E. J., Knols, B. G., Samson, R. A., and Takken, W. (2004). Entomopathogenic fungi for mosquito control: a review. J. Insect Sci. 4:19. doi: 10.1093/jis/4.1.19
Shah, P., and Pell, J. (2003). Entomopathogenic fungi as biological control agents. Appl. Microbiol. Biotecnol. 61, 413–423. doi: 10.4489/MYCO.2011.39.1.001
Shrestha, B., Sung, G. H., and Sung, J. M. (2017). Current nomenclatural changes in Cordyceps sensu lato and its multidisciplinary impacts. Mycology 8, 293–302. doi: 10.1080/21501203.2017.1386242
Sobczak, J. F., Arruda, I. D. P., Fonseca, E. O., Rabelo, P. J. Q., de Sousa Nóbrega, F. A., Pires, J. C., et al. (2020). Manipulation of wasp (Hymenoptera: Vespidae) behavior by the entomopathogenic fungus Ophiocordyceps humbertii in the Atlantic forest in Ceará, Brazil1. Entomol. News 129, 98–104. doi: 10.3157/021.129.0115
Spatafora, J. W., Quandt, C. A., Kepler, R. M., Sung, G. H., Shrestha, B., Hywel-Jones, N. L., et al. (2015). New 1F1N species combinations in Ophiocordycipitaceae (Hypocreales). IMA Fungus 6, 357–362. doi: 10.5598/imafungus.2015.06.02.07
Sung, G. H., Hyweljones, N. L., Sung, J. M., Luangsaard, J. J., Shrestha, B., and Spatafora, J. W. (2007a). Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 57, 5–59. doi: 10.3114/sim.2007.57.01
Sung, G. H., Sung, J. M., Hywel-Jones, N. L., and Spatafora, J. W. (2007b). A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 44, 1204–1223. doi: 10.1016/j.ympev.2007.03.011
Sung, G. H., Shrestha, B., Han, S. K., and Sung, J. M. (2011). Cultural characteristics of Ophiocordyceps heteropoda collected from Korea. Mycobiology 39, 1–6. doi: 10.4489/myco.2011.39.1.001
Suo, F., Guo, R., Yu, H., Zeng, W., and Yang, J. (2013). Nucleoside/nucleotide components of Ophiocordyceps gracilis and O. sinensis strains from Xinjiang and Tibet. Acta Edulis Fungi 20, 55–60.
Swofford, D. L. (2002). PAUP: Phylogenetic analysis using Parsimony, Version 4.0 b10. Sunderland, MA: Sinauer Associates.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Vaidya, G., Lohman, D. J., and Meier, R. (2011). Sequence matrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vega, F. E., Meyling, N. V., Luangsa-ard, J. J., and Blackwell, M. (2012). Fungal entomopathogens. Insect Pathol. 2, 171–220. doi: 10.1016/B978-0-12-384984-7.00006-3
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wang, Y., Li, Z. L., Suo, F. Y., and Sun, D. P. (2019). Study of mycelial polysaccharide from Paraisaria dubia of Ophiocordyceps gracilis asexual. China J. Chin. Mater. 44, 1704–1709. doi: 10.19540/j.cnki.cjcmm.20190318.201
Wen, T. C., Xiao, Y. P., Zha, L. S., Hyde, K. D., and Kang, J. C. (2016). Multigene phylogeny and morphology reveal a new species, Ophiocordyceps tettigonia, from Guizhou Province, China. Phytotaxa 280, 141–151. doi: 10.11646/phytotaxa.280.2.4
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols. A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfaud, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wijayawardene, N. N., Hyde, K. D., Lumbsch, H. T., Liu, J. K., Maharachchikumbura, S. S. N., Ekanayaka, A. H., et al. (2018). Outline of Ascomycota: 2017. Fungal Divers. 88, 167–263. doi: 10.1007/s13225-018-0394-8
Xiao, Y. P., Wen, T. C., Hongsanan, S., Jeewon, R., Luangsa-ard, J. J., Brooks, S., et al. (2018). Multigene phylogenetics of Polycephalomyces (Ophiocordycipitaceae, Hypocreales), with two new species from Thailand. Sci. Rep. 8:18087.
Yang, Z. L., Qin, J., Xia, C., Hu, Q., and Li, Q. Q. (2015). Ophiocordyceps highlandensis, a new species of a medicinal fungus from Yunnan, China. Phytotaxa 204, 287–295. doi: 10.11646/phytotaxa.204.4.5
Ye, X., Suo, F., Lu, S., and Wang, Z. (2015). Effect of Ophiocordyceps gracilis extract on the proliferation of AGS gastric cancer cells. Acta Edulis Fungi 22, 51–54.
Keywords: Insect fungi, Ophiocordycipitaceae, Paraisaria alba, Paraisaria arcta, Paraisaria rosea, taxonomy, Yunnan Province
Citation: Wei D-P, Wanasinghe DN, Xu J-C, To-anun C, Mortimer PE, Hyde KD, Elgorban AM, Madawala S, Suwannarach N, Karunarathna SC, Tibpromma S and Lumyong S (2021) Three Novel Entomopathogenic Fungi From China and Thailand. Front. Microbiol. 11:608991. doi: 10.3389/fmicb.2020.608991
Received: 13 October 2020; Accepted: 30 November 2020;
Published: 08 January 2021.
Edited by:
John R. Battista, Louisiana State University, United StatesReviewed by:
Nattawut Boonyuen, National Center for Genetic Engineering and Biotechnology (BIOTEC), ThailandYusufjon Gafforov, Institute of Botany, Academy of Sciences of the Republic of Uzbekistan, Uzbekistan
Copyright © 2021 Wei, Wanasinghe, Xu, To-anun, Mortimer, Hyde, Elgorban, Madawala, Suwannarach, Karunarathna, Tibpromma and Lumyong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saisamorn Lumyong, c2Nib2kwMDlAZ21haWwuY29t; Saowaluck Tibpromma, c2Fvd2FsdWNrZmFpQGdtYWlsLmNvbQ==; Samantha C. Karunarathna, c2FtYW50aGFAbWFpbC5raWIuYWMuY24=; c2FtYW50aGFrYXJ1bmFyYXRobmFAZ21haWwuY29t
 De-Ping Wei1,2,3
De-Ping Wei1,2,3 Dhanushka N. Wanasinghe
Dhanushka N. Wanasinghe Peter E. Mortimer
Peter E. Mortimer Kevin D. Hyde
Kevin D. Hyde Abdallah M. Elgorban
Abdallah M. Elgorban Nakarin Suwannarach
Nakarin Suwannarach Samantha C. Karunarathna
Samantha C. Karunarathna Saowaluck Tibpromma
Saowaluck Tibpromma Saisamorn Lumyong
Saisamorn Lumyong