
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 November 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.608624
This article is part of the Research Topic Bacterial Transcription Factors and the Cell Cycle View all 15 articles
The respiratory electron transport chain (ETC) of Mycobacterium smegmatis is terminated with two terminal oxidases, the aa3 cytochrome c oxidase and the cytochrome bd quinol oxidase. The bd quinol oxidase with a higher binding affinity for O2 than the aa3 oxidase is known to play an important role in aerobic respiration under oxygen-limiting conditions. Using relevant crp1 (MSMEG_6189) and crp2 (MSMEG_0539) mutant strains of M. smegmatis, we demonstrated that Crp1 plays a predominant role in induction of the cydAB operon under ETC-inhibitory conditions. Two Crp-binding sequences were identified upstream of the cydA gene, both of which are necessary for induction of cydAB expression under ETC-inhibitory conditions. The intracellular level of cAMP in M. smegmatis was found to be increased under ETC-inhibitory conditions. The crp2 gene was found to be negatively regulated by Crp1 and Crp2, which appears to lead to significantly low cellular abundance of Crp2 relative to Crp1 in M. smegmatis. Our RNA sequencing analyses suggest that in addition to the SigF partner switching system, Crp1 is involved in induction of gene expression in M. smegmatis exposed to ETC-inhibitory conditions.
The respiratory electron transport chain (ETC) of mycobacteria consists of the membrane-associated electron carriers (menaquinone/menaquinol and cytochrome c) and enzymes that catalyze electron-transfer reactions with the concomitant generation of proton motive force (Cook et al., 2014). The respiratory ETC of Mycobacterium smegmatis is terminated with two terminal oxidases like that of Mycobacterium tuberculosis, the aa3 cytochrome c oxidase and the cytochrome bd quinol oxidase, which catalyze the reduction of O2 to water molecules using the electrons from reduced cytochrome c and menaquinol, respectively (Kana et al., 2001; Matsoso et al., 2005). The aa3 cytochrome c oxidase, which belongs to the heme-copper superfamily of oxidases (HCOs) and serves as the major oxidase under aerobic conditions, forms a supercomplex with the cytochrome bc1 complex and cytochrome c (Matsoso et al., 2005; Megehee et al., 2006). Although the cytochrome bd quinol oxidase is not capable of pumping protons across the membrane during the reduction of O2, it has a higher affinity for O2 and is much less sensitive to inhibition by cyanide (CN−) than the aa3 oxidase (Puustinen et al., 1991; Cunningham et al., 1997; Kana et al., 2001; Belevich et al., 2005, 2007). It has been also demonstrated that the bd quinol oxidase is relatively insensitive to the physiologically relevant respiration-inhibiting molecules nitric oxide (NO) and hydrogen sulfide (H2S) that are produced by activated or infected host immune cells and serve as inhibitors for the bacterial and mitochondrial HCOs (Mason et al., 2009; Giuffre et al., 2012; Forte et al., 2016). From these findings, it has been suggested that the bd quinol oxidase is involved in adaptation of pathogenic bacteria such as M. tuberculosis to hostile environments created by host immunity during the infection process (Giuffre et al., 2012; Forte et al., 2016). Although the bd quinol oxidase is not essential to M. smegmatis at ambient oxygen tensions, it plays an important role in aerobic respiration under hypoxic conditions, as well as under inhibitory conditions of the bcc1-aa3 branch (Kana et al., 2001; Matsoso et al., 2005; Aung et al., 2014; Jeong et al., 2018). The bd quinol oxidase of M. smegmatis is encoded by the cydAB operon (cydA: MSMEG_3233, cydB: MSMEG_3232) (Kana et al., 2001; Aung et al., 2014). Inactivation of the bcc1-aa3 branch and hypoxic conditions were shown to result in strong upregulation of cydAB expression in M. smegmatis (Kana et al., 2001; Matsoso et al., 2005; Aung et al., 2014; Jeong et al., 2018). It was also demonstrated that albeit moderately, expression of cydA was induced in M. tuberculosis exposed to hypoxia and NO, as well as in M. tuberculosis, Mycobacterium bovis BCG, and Mycobacterium marinum treated with ETC inhibitors such as Q203, bedaquiline, and clofazimine (Shi et al., 2005; Koul et al., 2014; Boot et al., 2017; Kalia et al., 2019).
cAMP is a critical secondary messenger that controls a wide variety of cellular functions in many organisms. The cAMP receptor protein (Crp) is a transcriptional regulator that controls gene expression by recognizing altered cAMP levels in prokaryotic cells. The genome of M. smegmatis has two genes (crp1: MSMEG_6189, crp2: MSMEG_0539) encoding the Crp paralogs that show 78% sequence identity at the amino acid level (Sharma et al., 2014; Aung et al., 2015). Sequence homology and biochemical analyses revealed that Crp1 corresponds to the Crp protein (Rv3676) occurring in M. tuberculosis (Bai et al., 2005; Stapleton et al., 2010; Sharma et al., 2014; Aung et al., 2015). On the basis of high sequence similarity between the helix-turn-helix (HTH) motifs of Crp1 and Crp2, together with the results of DNA-binding analyses, it was suggested that both Crp proteins recognize and bind to the same consensus sequence (TGTGA-N6-TCACA) (Sharma et al., 2014). However, there are significant differences in amino acids forming the cAMP binding pockets of the two proteins, which accounts for their different biochemical properties such as their binding affinity for cAMP and DNA, as well as cAMP-dependent enhancement of the DNA-binding affinity (Sharma et al., 2014; Aung et al., 2015).
The expression and regulation patterns of the bd quinol oxidase genes in diverse bacteria are similar to those in mycobacteria. Expression of the genes is commonly induced under hypoxic or anaerobic conditions (Kana et al., 2001; Borisov et al., 2011; Small et al., 2013; Aung et al., 2014; Jeong et al., 2018; Mascolo and Bald, 2020). The regulation mechanism of the bd quinol oxidase genes in response to changes in oxygen availability was well-established for several bacteria. In Escherichia coli, expression of the cydAB operon is controlled by the ArcBA two-component system (TCS) and Fnr (fumarate and nitrate reduction regulatory protein) in response to changes in oxygen tensions (Cotter et al., 1990, 1997; Fu et al., 1991; Cotter and Gunsalus, 1992; Tseng et al., 1996). In Streptomyces coelicolor A3, expression of cydAB is regulated by the Rex repressor that exerts transcriptional regulation in response to changes in the cellular NADH/NAD+ ratio (Brekasis and Paget, 2003). Using both site-directed mutagenesis of a Crp-binding sequence upstream of cydA and Electrophoretic mobility shift analysis (EMSA) with purified Crp2 (MSMEG_0539), expression of the cydAB operon in M. smegmatis was suggested to be positively regulated by Crp (Aung et al., 2014). However, detailed study has not been reported regarding the regulatory mechanism for the induction of cydAB expression by inactivation of the bcc1-aa3 pathway and whether two CRP paralogs differentially contribute to the regulation of cydAB expression. Using relevant crp1 and crp2 mutant strains of M. smegmatis, we here report the roles of Crp1 and Crp2 in upregulation of cydAB expression under respiration-inhibitory conditions.
The bacterial strains and plasmids used in this study are listed in Supplementary Table 1. E. coli strains were grown in Luria-Bertani (LB) medium at 37°C. M. smegmatis strains were grown in 7H9-glucose medium [Middlebrook 7H9 medium (Difico, Sparks, MD) supplemented with 0.2% (w/v) glucose as a carbon source and 0.02% (v/v) Tween 80 as an anticlumping agent] at 37°C. M. smegmatis strains were grown aerobically in a 500-ml flask filled with 100 ml of 7H9-glucose medium on a gyratory shaker (200 rpm). For treatment of M. smegmatis cultures with potassium cyanide (KCN), M. smegmatis strains were grown until the optical density at 600 nm (OD600) reached 0.45–0.5. Following the addition of KCN to the cultures to a final concentration of 100 μM, the cultures were further grown for 15 min. For treatment of M. smegmatis cultures with sodium nitroprusside (SNP; an NO generator) or NaHS (an H2S generator), SNP and NaHS were added to the M. smegmatis cultures grown to an OD600 of 0.45–0.5 to final concentrations of 5 mM and 200 μM, respectively, and the cultures were further grown for 30 min. The SNP-treated cultures were grown under illumination of light (100 W/m2). Ampicillin (100 μg/ml for E. coli), kanamycin (50 μg/ml for E. coli and 15 or 30 μg/ml for M. smegmatis), and hygromycin (200 μg/ml for E. coli and 25 or 50 μg/ml for M. smegmatis) were added to the growth medium when required. The construction of the mutants and plasmids used in this study is described in Supplemental Material.
Standard protocols and manufacturers' instructions were followed for recombinant DNA manipulations (Sambrook and Green, 2012). Transformation of M. smegmatis with plasmids was carried out by electroporation as described elsewhere (Snapper et al., 1990). The primers used for PCR are listed in Supplementary Table 2.
To introduce point mutations into the Crp-binding sites (CBS1 and CBS2), PCR-based mutagenesis was performed using the Quick Change site-directed mutagenesis procedure (Stratagene, La Jolla, CA). Synthetic oligonucleotides 31 bases long containing the substituted nucleotides in the middle of their sequences were used to mutagenize the sequences. The primers used for mutagenesis are listed in Supplementary Table 2. Mutations were verified by DNA sequencing.
RNA isolation from M. smegmatis strains and cDNA synthesis were performed as described elsewhere (Kim et al., 2010) except for the use of a random hexamer primer (ThermoFisher, Waltham, MA) in place of the gene-specific primers in cDNA synthesis. The contamination of DNA in the isolated RNA was checked by PCR with the primers to be used in quantitative real-time PCR (qRT-PCR). To determine the transcript levels of cydA, crp2, MSMEG_3680, and sigA, qRT-PCR was performed in a 20-μl mixture containing 5 μl of the template cDNA, 15 pmol of each of two gene-specific primers, 10 μl of TB GreenTM Premix Ex TaqTM (Tli RNase Plus) (Takara, Tokyo, Japan), 0.4 μl of the ROX passive fluorescent dye, and 2.6 μl of distilled water. Thermal cycling was initiated with 1 cycle at 95°C for 2 min, followed by 40 cycles of 95°C for 5 s and 64°C for 30 s. The sigA gene encoding the principal sigma factor was used as a reference gene for qRT-PCR to normalize the expression levels of cydA, crp2, and MSMEG_3680 since our RNA sequencing analyses revealed that the sigA gene is constitutively expressed at similar levels in the wild-type (WT), Δaa3, Δcrp1, and Δcrp2 mutant strains (Supplementary Figure 1). Melting curve analysis was performed for each reaction to examine whether a single PCR product was amplified during qRT-PCR. The primers used for qRT-PCR are listed in Supplementary Table 2.
The β-Galactosidase activity was measured spectrophotometrically as described previously (Oh and Kaplan, 1999). The protein concentration was determined using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin (BSA) as a standard protein.
Cell-free crude extracts were subjected to SDS-PAGE, and proteins on the gel were transferred to polyvinylidene fluoride membranes (Millipore, Burlington, MA). Western blotting analysis using an anti-2B8 antibody (Biojane, Pyeongtaek-si, South Korea) was performed as described previously (Mouncey and Kaplan, 1998). The anti-2B8 antibody was used at a dilution of 1:20,000. To detect GroEL, a mouse monoclonal antibody against Hsp65 (Santa Cruz Biotechnology, Dallas, TX; sc58170) was used at a 1:2,000 dilution. Alkaline phosphatase-conjugated anti-mouse IgG produced in rabbit (Sigma, St. Louis, MO; A4312) was used at a 1:10,000 dilution for the detection of the primary antibodies.
C-terminally His6-tagged Crp1 was expressed in the E. coli BL21 (DE3) strain harboring pT7-7crp1. The E. coli strain was cultivated aerobically to an OD600 of 0.4–0.6 at 37°C in LB medium containing 100 μg/ml ampicillin. Expression of the crp1 gene was induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to the cultures to a final concentration of 0.5 mM, and then cells were further grown for 4 h at 30°C. Cells were harvested from 300 ml cultures and resuspended in 10 ml of buffer A [20 mM Tris-HCl (pH 8.0) and 200 mM NaCl] containing DNase I (10 U/ml) and 10 mM MgCl2. The resuspended cells were disrupted twice using a French pressure cell, and cell-free crude extracts were obtained by centrifugation twice at 20,000 × g for 15 min. The crude extracts were loaded into a column packed with 500 μl of the 80% (v/v) slurry of Ni-Sepharose high-performance resin (GE Healthcare, Piscataway, NJ). The resin was washed with 40 bed volumes of buffer A containing 5 mM imidazole and washed further with 20 bed volumes of buffer A containing 60 mM imidazole. His6-tagged Crp1 was eluted from the resin with 6 bed volumes of buffer A containing 250 mM imidazole. The eluted protein was desalted using a PD-10 desalting column (GE Healthcare) equilibrated with appropriate buffer. The purity of Crp1 was checked by SDS-PAGE (Supplementary Figure 2).
A 99-bp DNA fragment containing the upstream region of cydA and an 80-bp control DNA fragment without the Crp-binding site were used in EMSA. The 99-bp DNA fragment was amplified by PCR using pBSIIcydA as a template and the primers F_cydAEMSA and R_cydAEMSA. The 80-bp control DNA fragment was generated by PCR using pUC19 as a template and the primers F_80_EMSA and R_80_EMSA. Purified Crp1 protein was incubated with 70 fmol of the DNA fragments containing the cydA upstream region and 100 fmol of the control DNA fragments in binding buffer [20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 2.5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol (DTT), 50 μg/ml BSA and 10% (v/v) glycerol] in a reaction volume of 10 μl for 20 min at 25°C. After addition of 2 μl of 6x loading buffer (0.25% (w/v) bromophenol blue, 0.25% (w/v) xylene cyanol and 40% (w/v) sucrose), the mixtures were subjected to non-denaturing PAGE [8% (w/v) acrylamide] using 0.5x TBE buffer (41.5 mM Tris-borate and 0.5 mM EDTA, pH8.3) at 70 V for 2 h 20 min at 4°C. The gels were stained with SYBR Green staining solution for 1 h.
DNase I Footprinting was carried out using fluorescence (TAMRA)-labeled DNA fragments and purified Crp1. TAMRA-labeled DNA fragments (271 bp) containing the cydA upstream region were generated by PCR using the primers (F_TAMRA_pUC19 and F_cydAFootR) and pUC19cydAFootR as a template. The PCR products were purified after agarose gel electrophoresis. DNA binding reaction mixtures were composed of 5 pmol of labeled DNA probes, purified Crp1 (0.15, 0.3, or 0.6 μM), 20 mM Tris-HCl (pH 8.0), 0.2 mM MgCl2, 2.1 mM KCl, 0.04 mM DTT, and 11.1% (v/v) glycerol in a final volume of 190 μl. When necessary, the Crp1 protein was mixed with 200 μM cAMP, and the mixture was incubated for 10 min at 25°C prior to binding reactions for 10 min at 25°C. DNase I (Takara) was diluted in buffer containing 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM DTT, and 10% (v/v) glycerol to reach a final concentration of 0.675 mU/μl. DNase I digestion was initiated with the addition of 10 μl of diluted DNase I to the binding reaction mixtures, conducted for 1 min at 25°C, and stopped with the addition of 400 μl of stop solution [20 mM Tris-HCl (pH 8.0) and 40 mM EDTA]. DNA was purified by phenol/chloroform/isoamyl alcohol (25:24:1) extraction and isopropyl alcohol precipitation. The pellets were dissolved in TE buffer [10 mM Tris-HCl (pH 8.0) and 1 mM EDTA]. After the addition of loading buffer [95% deionized formamide, 0.025% (w/v) bromophenol blue, 0.025% (w/v) xylene cyanol FF, and 5 mM EDTA (pH 8.0)], the samples were analyzed by electrophoresis on 6% (w/v) denaturing polyacrylamide gels with 7 M urea in 0.8x Tris-taurine-EDTA (TTE) buffer using an ABI PRISM 377 DNA sequencer (Applied Biosystems, Foster City, CA). Reference sequencing was performed by using a Thermo sequenase dye primer manual cycle sequencing kit (ThermoFisher) with the primer F_TAMRA_pUC19 and the template plasmid pUC19cydAFootR.
M. smegmatis cells corresponding to 1 ml of cultures at OD600 of 0.4 were harvested. Cell pellets were resuspended in 1 ml of 0.1 M HCl and then incubated for 10 min. Cells were disrupted once by using a Fastprep 120 beadbeater (ThermoFisher) at 5.0 m/s for 45 s. Cell-free supernatants were obtained by centrifugation at 20,000 × g for 10 min. The concentration of cAMP in the prepared supernatants was determined by using a DetectX Direct Cyclic AMP Enzyme Immunoassay kit (Arbor Assays, Ann Arbor, MI) and a microplate reader (Bio-Rad) following the manufacture's instruction.
Three biological replicate cultures of the WT and Δcrp1 strains were grown aerobically to an OD600 of 2.0–2.1 (late exponential phase). Total RNA of each culture was isolated as described previously (Kim et al., 2010). rRNA was removed from the isolated total RNA using a Ribo-Zero rRNA Removal Kit (Bacteria) (Illumina, San Diego, CA). The RNA sequencing libraries were created using a TruSeq RNA Sample Prep Kit v2 (Illumina) with the standard low-throughput protocol. Sequencing of the six libraries was conducted on an Illumina HiSeq 4000 platform at Macrogen Inc. (Seoul, South Korea) using the Hiseq 3000–4000 sequencing protocol and TruSeq 3000–4000 SBS Kit v3 reagent (Illumina). Paired-end reads (101 bp) were then mapped to the reference genome sequence of M. smegmatis mc2155 (GCF_000015005.1_ASM1500v1) with the program Bowtie 1.1.2 using default settings. Summarized statistics of RNA sequencing alignment are listed in Supplementary Table 3. The differentially expressed genes (DEGs) were subsequently identified pair-wise by the edgeR package in R language (Robinson et al., 2010). In this analysis, the genes with p < 0.05 and |FC| > 1.5 were regarded as DEGs. The RNA sequencing data have been deposited in NCBI's Gene Expression Omnibus and are accessible through the GEO Series accession number GSE158137.
Under aerobic culture conditions, the inactivation of the aa3 cytochrome c oxidase in M. smegmatis by mutation was previously shown to lead to an overall reduction in the respiration rate by ~50% and a significant increase in expression of the cydAB operon encoding the bd quinol oxidase (Jeong et al., 2018). Furthermore, we found that the genes, which belong to the SigF (an alternative sigma factor) regulon, are strongly upregulated in a mutant strain of M. smegmatis lacking the aa3 oxidase (Oh et al., 2020). To examine whether the induction of cydAB expression in the Δaa3 mutant with a deletion in ctaC encoding subunit III of the aa3 cytochrome c oxidase is a result of SigF activation, we determined expression of cydA in the WT, Δaa3 mutant, and Δaa3ΔsigF double mutant strains using the strains harboring the cydA::lacZ translational fusion plasmid pNCIIcydA. In good agreement with the previous report (Matsoso et al., 2005; Jeong et al., 2018), the expression level of cydA was increased in the Δaa3 mutant by ~107-fold relative to the WT strain (Figure 1). The expression level of cydA was not decreased in the Δaa3ΔsigF mutant compared to the Δaa3 mutant, indicating that the cydAB operon does not belong to the SigF regulon, and that the strong upregulation of the cydAB operon under respiration-inhibitory conditions is not caused by the activation of SigF.
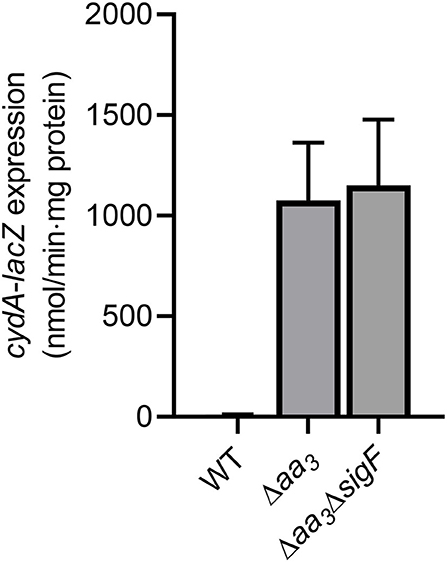
Figure 1. Expression of the cydAB operon in the WT, Δaa3, and Δaa3ΔsigF mutant strains of M. smegmatis. The WT, Δaa3, and Δaa3ΔsigF mutant strains of M. smegmatis harboring the cydA::lacZ translational fusion plasmid pNCIIcydA were grown aerobically to an OD600 of 0.45–0.5 in 7H9-glucose medium. Cell crude extracts were used to determined β-galactosidase activity. All values provided are the averages of the results from three biological replicates. The error bars indicate the standard deviations.
Previously it has been reported that Crp is involved in the positive regulation and hypoxic induction of the cydAB operon in M. smegmatis (Aung et al., 2014). However, it remained unanswered whether two Crp paralogs (Crp1: MSMEG_6189, Crp2: MSMEG_0539) play a distinct role in induction of the cydAB operon under respiration-inhibitory conditions. To examine the involvement of Crp1 and Crp2 in the regulation of cydA expression, we determined the expression level of cydA in the Δcrp1 and Δcrp2 mutant strains. Since we failed to obtain a Δaa3Δcrp1 double mutant strain, treatment of M. smegmatis cultures with KCN, an inhibitor of aa3 cytochrome c oxidase, was applied to mimic the Δaa3 mutant conditions. Effects of KCN treatment on cydA expression were quantitatively determined by qRT-PCR in the WT, Δcrp1, and Δcrp2 mutant strains that were grown aerobically. We could not use the cydA::lacZ transcriptional fusion pNCIIcydA to determine the expression level of cydA in the M. smegmatis strains treated with KCN, since the addition of KCN interfered with expression or assay of β-galactosidase for unknown reasons. The treatment of the WT strain with 100 μM KCN led to induction of cydA expression by 388-fold compared to the untreated WT control strain (data not shown). When the aerobically grown WT, Δcrp1, and Δcrp2 mutant strains were treated with KCN, the Δcrp1 mutant showed only 30% of cydA expression observed in the WT strain, while the expression level of cydA was only slightly reduced in the Δcrp2 mutant relative to the WT strain (Figure 2A). This result indicates that Crp1 plays a predominant role in induction of cydA expression under respiration-inhibitory conditions. The ectopic expression of the intact crp1 gene in the Δcrp1 mutant using pMV306crp restored the expression level of cydA to that in the WT strain with the empty integration vector pMV306 (Figure 2B), confirming that the reduction of cydA expression in the Δcrp1 mutant is the result of crp1 inactivation. To confirm the Crp1-mediated induction of cydA expression by inactivation of the aa3 cytochrome c oxidase, effects of NO and H2S, which are the physiologically relevant inhibitors of the aa3 oxidase, on cydA expression were assessed in the WT and Δcrp1 mutant strains (Supplementary Figure 3). As in the Δaa3 mutant and the WT strain treated with KCN, expression of cydA was significantly (325-fold) increased in the WT strain treated with 5 mM SNP (NO generator) relative to that in the SNP-untreated control WT strain. Induction of cydA expression was significantly compromised in the SNP-treated Δcrp1 mutant compared to the WT strain treated with SNP. Similarly, expression of cydA was increased by 114-fold in the WT strain treated with 200 μM NaHS (H2S generator) relative to that in the NaHS-untreated control WT strain. The expression level of cydA in the NaHS-treated Δcrp1 mutant was shown to amount to ~47% of that observed for the NaHS-treated WT strain. Using KCN, we examined the aerobic growth of the WT, Δcrp1, and Δcrp2 mutant strains when the aa3 oxidase is inhibited (Supplementary Figure 4). The WT, Δcrp1, and Δcrp2 strains grew at the similar rate in 7H9-glucose medium in the absence of KCN. In contrast, the growth of the Δcrp1 mutant was severely compromised in the presence of 100 μM KCN, and that of the Δcrp2 mutant was moderately affected compared to the WT strain. The high susceptibility of Δcrp1 mutant to SNP regarding inhibition of aerobic growth was also demonstrated previously (Lee et al., 2014). Altogether, these results reinforce that inhibition of the respiratory ETC by inactivation of the aa3 oxidase leads to Crp1-mediated induction of the cydAB operon in M. smegmatis.
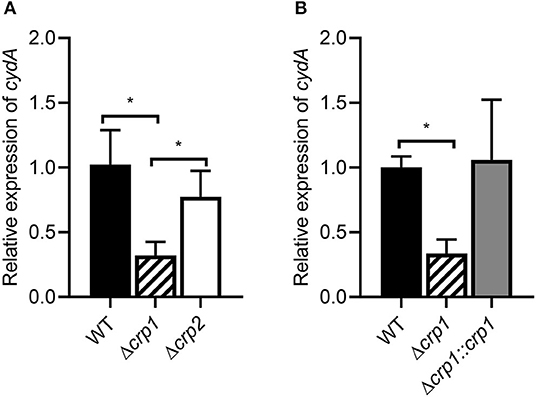
Figure 2. Expression of the cydAB operon in the WT, Δcrp1, and Δcrp2 mutant strains of M. smegmatis. (A) The relative transcript level of cydA in the WT, Δcrp1, and Δcrp2 mutant strains. (B) Complementation of the Δcrp1 mutant. For complementation of the Δcrp1 mutant, pMV306crp (a pMV306-derived plasmid carrying the intact crp1 gene and its own promoter) was introduced into the mutant. As control strains, the WT and Δcrp1 strains with the empty vector pMV306 were used in the experiment. All the strains were grown aerobically to an OD600 of 0.45–0.5 and treated with 100 μM KCN for 15 min. The expression level of cydA was quantitatively determined by qRT-PCR and normalized to sigA (the gene for the principal sigma factor) expression. The expression level of cydA in the KCN-treated WT strain is set at 1, and the relative values are expressed for the mutant strains. All values provide are the average of the results from three biological replicates. The error bars indicate the standard deviations. Statistical significance was determined by two-tailed Student's t-test. *p < 0.05.
The expression levels of crp1 and crp2 in M. smegmatis were extrapolated from the reads per kilo base pair per million mapped reads (RPKM) values obtained from RNA sequencing analysis on the WT strain of M. smegmatis that was aerobically grown to an OD600 of 0.45–0.5 (Lee et al., 2018). The RPKM values of crp1 and crp2 suggested that the transcript level of crp1 is 8-fold higher than that of crp2 in the WT strain (Figure 3A). To assess whether the estimated transcript levels of crp1 and crp2 correlate with their cellular protein levels, Western blotting analysis was performed using the Δcrp1 and Δcrp2 mutant strains expressing the C-terminally 2B8 epitope-tagged Crp1 and Crp2 proteins, respectively. For the construction of the strains, the crp1 and crp2 genes with the upstream regions encompassing their own promoters and regulatory sequences were cloned into the integration vector pMV306, and the resulting pMV306crp1_2B8 and pMV306crp2_2B8 plasmids were integrated into the chromosomes of the Δcrp1 and Δcrp2 mutant strains, respectively. Western blotting analysis revealed that Crp1 was expressed at much higher levels than Crp2 in M. smegmatis grown to various stages of exponential growth phase, while the protein level of GroEL, which was used as a loading control, was relatively constant in both strains grown to various stages of exponential growth phase (Figure 3B). This finding that Crp1 is the predominantly expressed Crp in M. smegmatis might explain the dominant role of Crp1 in the positive regulation of the cydAB operon.
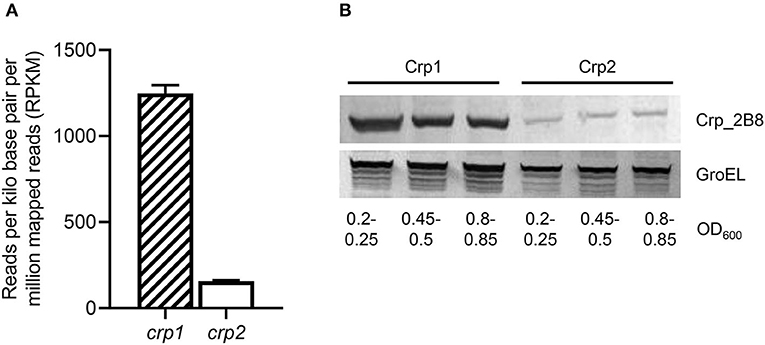
Figure 3. Expression levels of two Crp paralogs in M. smegmatis. (A) Transcript levels of crp1 and crp2 were extrapolated from the RPKM values obtained from RNA sequencing analysis on the WT strain of M. smegmatis (Lee et al., 2018). (B) Expressed protein levels of Crp1 and Crp2. To determine the protein levels of Crp1 and Crp2 expressed in M. smegmatis, the Δcrp1 and Δcrp2 mutant strains complemented with pMV306crp1_2B8 and pMV306crp2_2B8, respectively, were grown aerobically to the indicated OD600 in 7H9-glucose medium, and their crude extracts (30 μg for detection of Crp1 and Crp2; 5 μg for GroEL detection) were subjected to Western blotting analyses using a 2B8 antibody and an Hsp65 antibody to detect the C-terminally 2B8-tagged Crp and GroEL, respectively. The protein level of GroEL was determined as a loading control.
To identify the Crp-binding sequence(s) in the upstream region of cydA, DNase I footprinting analysis was performed with purified Crp1 and 271-bp TAMRA-labeled DNA fragments containing the cydA promoter region. Since Crp1 was shown to play a predominant role in the regulation of the cydAB operon and both Crp paralogs were suggested to bind to the same DNA sequence (Sharma et al., 2014), we used only Crp1 for DNA-binding analyses. As shown in Figure 4A, binding of Crp1 protected DNA from DNase I cleavage at positions between −52 and −102 with regard to the transcription start point (TSP) of cydA. The protected region contains two Crp-binding sequences (CBS1 and CBS2) that are similar to the known Crp-binding consensus sequence (TGTGA-N6-TCACA) (Figure 4B). CBS1 is located at positions between −86 and −101 relative to the TSP, and CBS2 was located between −58 and −73. The addition of cAMP to the reaction mixtures resulted in wider and more clearly protected windows for both CBS1 and CBS2, indicating that cAMP enhances the binding of Crp1 to both Crp-binding sites. At low Crp1 concentrations, CBS2 was protected better than CBS1 in the presence and absence of cAMP, indicating that Crp1 binds better to CBS2 than it does to CBS1. To confirm the result of DNase I footprinting, EMSAs were performed with purified Crp1 and 99-bp DNA fragments encompassing the cydA upstream sequence (Figure 5). An 80-bp DNA fragment without the Crp-binding sequence (non-specific DNA) was used as a negative control DNA. The binding ability of Crp1 for the DNA fragments was estimated from the band intensity of the unretarded free DNA. As shown in Figure 5A, the binding of Crp1 to the cydA regulatory region was enhanced in the presence of 200 μM cAMP, which is consistent with the DNase I footprinting result. In order to determine to what extent mutations of each Crp-binding site affect the binding of Crp1 to the cydA regulatory region, we performed EMSAs using purified Crp1 and 99-bp DNA fragments containing the WT or mutated Crp-binding sites (M1, M2, and M3) in the presence of cAMP. As shown in Figure 5B, the M1 and M2 DNA fragments containing mutations within CBS1 and CBS2, respectively, were retarded by Crp1 to a lesser extent than the WT DNA fragment as judged by the levels of free DNA. Especially the M2 mutation significantly affected the binding of Crp1 to the cydA regulatory region. Mutations of both CBS1 and CBS2 virtually abolished the binding of Crp1 to the M3 DNA fragment. Taken together, the EMSA and DNase I footprinting results indicate that Crp1 can bind to both CBS1 and CBS2 with a higher binding affinity for CBS2.
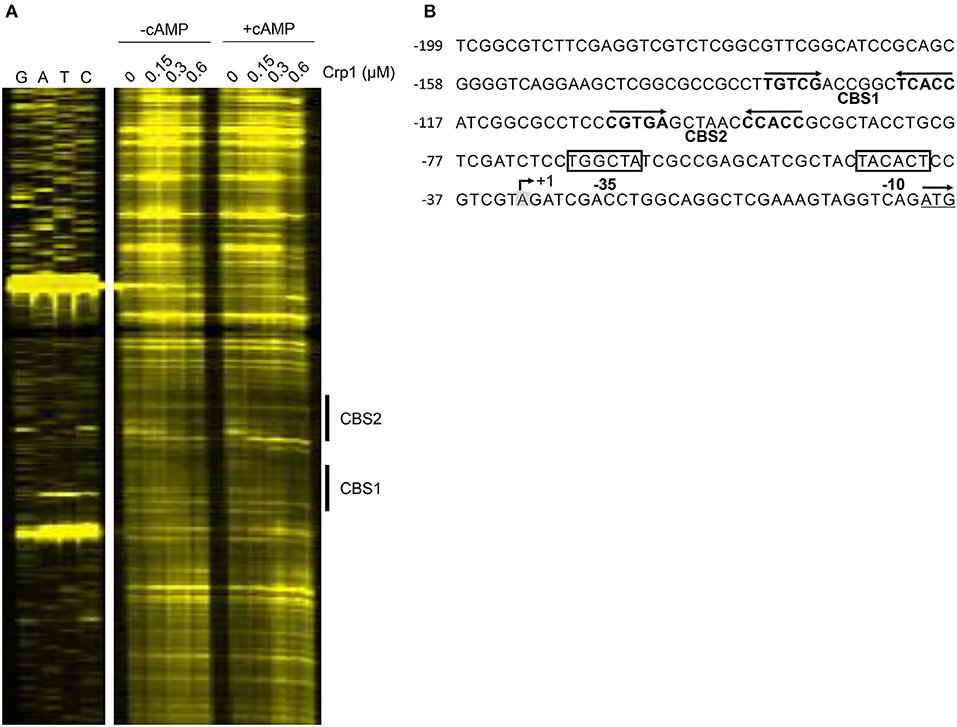
Figure 4. Identification of the Crp-binding sites in the upstream region of cydA. (A) DNase I Footprinting analysis of the cydA upstream region bound by Crp1. The DNA fragments containing the non-coding strands labeled with TAMRA at their 5′ ends were incubated with increasing concentrations of purified Crp1 in the absence or presence of 200 μM cAMP and then subjected to DNase I footprinting reactions. The concentrations of Crp1 used are given above the lanes. The Crp-binding sites (CBS1 and CBS2) are marked by the thick lines on the right. Lanes G, A, T, and C represent the sequence ladders. (B) The upstream sequence of the cydA gene. The Crp-binding sites (CBS1 and CBS2) are marked by the two head-facing arrows above the sequence. The nucleotide reported to be the TSP (+1) of cydA is shaded in gray, and the putative −10 and −35 elements extrapolated from +1 are boxed (Aung et al., 2014). The start codon of cydA is underlined and the arrow above it indicates the transcriptional direction. The numbers on the left of the sequences indicate the positions of the leftmost nucleotides relative to the cydA gene. The concentration of Crp1 refers to that of the Crp1 monomer.
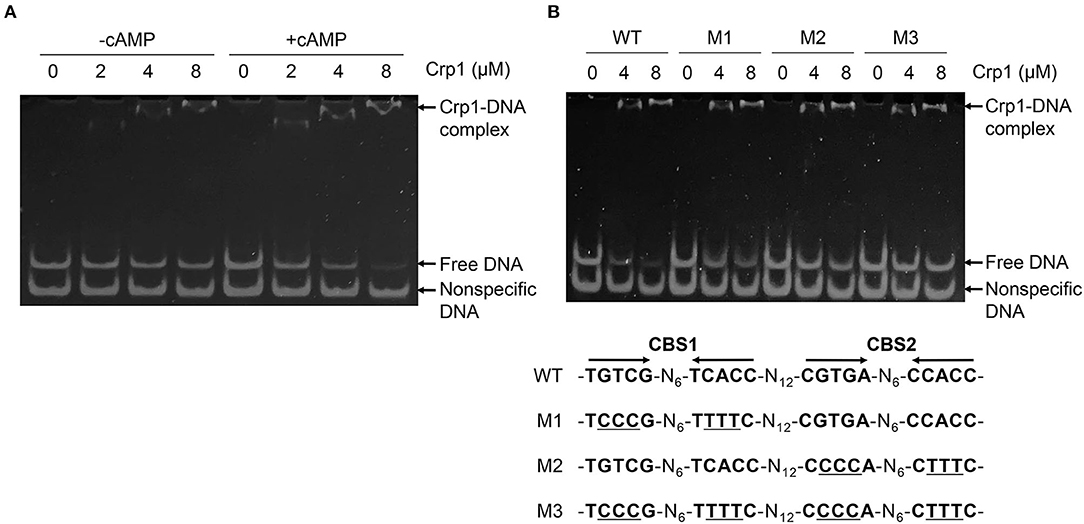
Figure 5. Binding of Crp1 to the cydA control region. (A) EMSA showing the effect of cAMP on the binding of Crp1 to the upstream region of cydA. The mixtures of 99-bp DNA fragments (70 fmol) containing two Crp-binding sites (CBS1 and CBS2) upstream of cydA and 80-bp non-specific DNA fragments without the Crp-binding site (100 fmol) were incubated with increasing concentrations of purified Crp1 in the presence and absence of 200 μM cAMP. (B) 99-bp DNA fragments (70 fmol) containing either WT or mutated Crp-binding sites (M1, M2, and M3), as well as 80-bp non-specific DNA fragments without the Crp-binding site (100 fmol), were mixed with increasing concentrations of purified Crp1 in the presence of 200 μM cAMP. The Crp1-DNA reaction mixtures were subjected to native PAGE. The concentrations of Crp1 used in EMSA are given above the lanes. The concentration of Crp1 refers to that of the Crp1 monomer.
To investigate the role of CBS1 and CBS2 in induction of cydA expression under respiration-inhibitory conditions, a series of cydA::lacZ translational fusions with 5'-serial deletions of the cydA upstream region [pNCIISD1 (SD1), pNCIISD2 (SD2), pNCIISD3 (SD3), and pNCIISD4 (SD4)] were used to determine expression of cydA in the Δaa3 mutant of M. smegmatis (Figure 6A). Consistent with the result presented in Figure 1, expression of cydA was strongly induced in the Δaa3 mutant carrying pNCIIcydA (Con) relative to the control WT strain with pNCIIcydA. The 5'-deletion up to the position−163 relative to the TSP (SD1) did not affect cydA expression in the Δaa3 mutant, and the additional 20-bp deletion of the cydA upstream region (SD2) led to a ~35% decrease in cydA expression in the Δaa3 mutant. When the cydA upstream region was further deleted to remove the 5′ half of CBS1 (SD3), the induction of cydA expression in the Δaa3 mutant was almost abolished. The deletion of both CBS1 and CBS2 (SD4) resulted in complete abolishment of cydA expression in the Δaa3 mutant. To more precisely assess the role of CBS1 and CBS2 in the regulation of cydA expression, point mutations were introduced into CBS1, CBS2, or both CBS1 and CBS2 on pNCIIcydA, and the expression level of cydA was measured using the Δaa3 mutants carrying the corresponding cydA::lacZ translational fusion plasmids [pNCIIcydA (Con), pNCIIM1 (M1), pNCIIM2 (M2), and pNCIIM3 (M3)] (Figure 6B). As observed for the Δaa3 mutants with M1 and M2, the introduction of point mutations into CBS1 (TGTCG-N6-TCACC to TCCCG-N6-TTTTC) and CBS2 (CGTGA-N6-CCACC to CCCCA -N6-CTTTC) resulted in only 15 and 23% of cydA expression observed in the Δaa3 mutant with pNCIIcydA, respectively. The effect of mutations within both CBS1 and CBS2 appeared to be cumulative as judged by the expression level of cydA in the Δaa3 strain with M3. Altogether, the results presented in Figure 6 suggest that both CBS1 and CBS2 are required for induction of cydA expression under respiration-inhibitory conditions. It is noteworthy that expression of cydA was still 7.3-fold induced in the Δaa3 mutant with M3 compared to the WT strain with pNCIIcydA (Figure 6B). From this finding together with the observed reduction in cydA expression from SD2 relative to that from SD1 (Figure 6A), we cannot rule out the possibility that there might be an additional cis-acting element within or overlapping the 20-bp sequence between SD1 and SD2, which is implicated in the induction of cydA expression in M. smegmatis under respiration-inhibitory conditions.
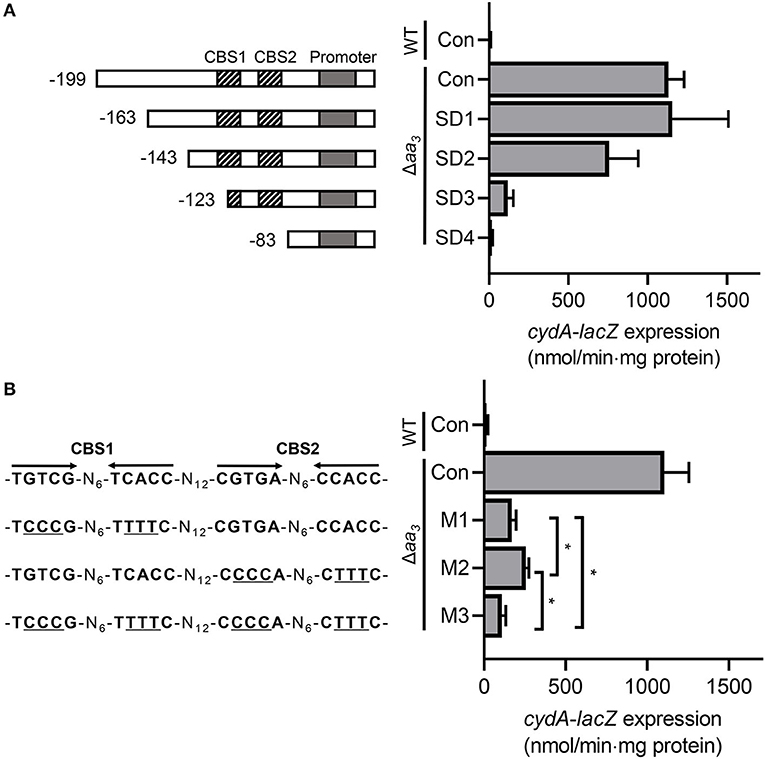
Figure 6. Effects of deletions and mutations in the Crp-binding sites (CBS1 and CBS2) on expression of the cydAB operon. (A) The cydA promoter activity was determined using the pNCIIcydA-derived cydA::lacZ translational fusions containing serial deletions of the cydA upstream region (pNCIISD1, pNCIISD2, pNCIISD3, and pNCIISD4). (B) The cydA promoter activity was determined using several pNCIIcydA derivatives containing mutations in the Crp-binding sites (pNCIIM1, pNCIIM2, and pNCIIM3). The mutations in the Crp-binding sites of the cydA::lacZ translational fusions are presented on the left. The mutations within CBS1 and CBS2 are indicated by underlines. The WT and Δaa3 mutant strains of M. smegmatis harboring the cydA::lacZ translational fusion plasmids were grown aerobically to an OD600 of 0.45–0.5 in 7H9-glucose medium. Cell crude extracts were used to determined β-galactosidase activity. All values provided are the averages of the results from three biological replicates. The error bars indicate the standard deviations. Statistical significance was determined by two-tailed Student's t-test. *p < 0.05. Con, pNCIIcydA; SD1-SD4, pNCIISD1- pNCIISD4; M1-M3, pNCIIM1-pNCIIM3.
Based on the findings that the cydAB operon is strongly upregulated in the Δaa3 mutant in a Crp-dependent way and does not require SigF for its transcription, we searched for the genes that are regulated in a similar way as the cydAB operon to further exemplify the Crp-mediated induction of gene expression under respiration-inhibitory conditions. As shown in Figure 7A, our comparative RNA sequencing analysis on the WT and Δaa3 mutant strains revealed 103 DEGs whose expression is increased in the Δaa3 mutant by more than 4-fold with a p < 0.05 relative to the WT strain. Sixty-one genes among the 103 DEGs were found to belong to the known SigF regulon (Singh et al., 2015). RNA sequencing analysis on the WT and Δcrp1 mutant strains showed that among the remaining 42 genes, 25 genes was found to be less expressed in the Δcrp1 mutant by more than 1.5-fold with a p < 0.05 relative to the WT strain (Supplementary Table 4). Due to the presence of intact crp2 in the Δcrp1 mutant, the less stringent cutoff value (1.5) was applied to select the genes that are positively regulated by Crp1. Among the identified 25 DEGs showing the similar expression patterns as cydAB in terms of the strong induction of gene expression in the Δaa3 mutant in a SigF-independent way and a decrease in gene expression in the Δcrp1 mutant, MSMEG_3680 annotated as a hypothetical protein gene is such a gene that has two putative Crp-binding sites in its upstream region which are arranged similarly as those found upstream of cydA (Figure 7B). Using qRT-PCR, the expression pattern of MSMEG_3680 was verified in the WT and Δcrp1 mutant strains that were grown aerobically with or without treatment of KCN. In the same way as cydA expression, expression of MSMEG_3680 was strongly induced in the WT strain by treatment of KCN, and induction of its expression by KCN was significantly compromised in the Δcrp1 mutant. Altogether, the results from RNA sequencing analyses suggest the possibility that Crp1 is likely involved in induction of gene expression in M. smegmatis under respiration-inhibitory conditions.
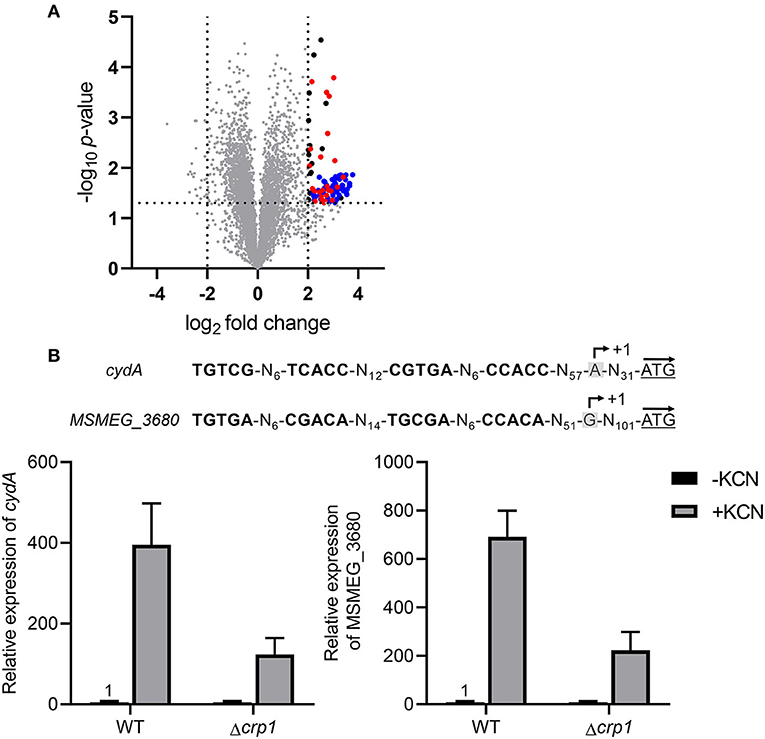
Figure 7. Overlap of the SigF and Crp1 regulons with the genes induced in the Δaa3 mutant strain of M. smegmatis. (A) Volcano plot showing the DEGs in the Δaa3 mutant strain relative to the WT strain. RNA sequencing was performed using RNA prepared from three independent replicate cultures of the WT and Δaa3 mutant strains grown aerobically in 7H9-glucose medium to an OD600 of 0.45–0.5. The x-axis displays the log2 fold change of gene expression (log2FC) in the Δaa3 mutant relative to the WT strain, and the y-axis represents -log10 p-value. The horizontal dotted line on the graph indicates the border line indicating the p-value of 0.05, and the vertical dotted lines indicate the border lines indicating the log2FC values of−2 and +2. One hundred and three genes, whose expression is increased by more than log2FC ≥ 2 with p < 0.05, are depicted by black filled circles. Among the 103 DEGs, the genes belonging to the SigF regulon are denoted by blue filled circles (Singh et al., 2015), and the genes, whose expression is reduced by FC ≥ 1.5 with p < 0.05 in the Δcrp1 mutant strain relative to the WT strain, are indicated by red filled circles. (B) The nucleotide sequences and locations of the Crp-binding sites in the upstream regions of cydA and MSMEG_3680 and the expression levels of the genes in the WT and Δcrp1 mutant strains of M. smegmatis. The Crp-binding sites are shown in bold. The previously reported TSPs of cydA and MSMEG_3680 are denoted by +1 (Aung et al., 2014; Martini et al., 2019). The start codons of cydA and MSMEG_3680 are underlined, and the arrows above them indicate the transcriptional direction. The expression levels of cydA and MSMEG_3680 in the WT and Δcrp1 mutant strains were quantitatively determined by qRT-PCR. The strains were aerobically grown to an OD600 of 0.45–0.5 and treated with 100 μM KCN for 15 min (+KCN). As controls, the WT and Δcrp1 mutant strains grown aerobically without KCN treatment were included in the experiment (-KCN). The expression levels of cydA and MSMEG_3680 determined by qRT-PCR were normalized to that of sigA. The expression level of each gene in the KCN-untreated WT strain is set at 1, and the relative values are expressed for the other strains. All values provide are the averages of the results from three independent experiments. The error bars indicate the standard deviations.
Since Crp is a regulatory protein that regulates gene expression in response to changes in cAMP levels, we determined whether the level of cAMP in M. smegmatis is changed in response to respiration inhibition. As shown in Figure 8A, the intracellular level of cAMP in the Δaa3 mutant grown aerobically was found to be 3.2-fold higher than that detected in the WT strain grown under the same conditions. The level of cAMP was even more (14-fold) increased, when the aerobically grown WT strain of M. smegmatis was treated with 100 μM KCN for 15 min relative to the KCN-untreated control WT strain (Figure 8B). The respiratory ETC in the WT strain subjected to a short period (15 min) of KCN treatment is assumed to be more inhibited, at least temporarily before the induced synthesis of the bd quinol oxidase, than that in the Δaa3 mutant in which expression of the cydAB operon is constitutively induced. The higher cAMP level in the KCN-treated WT strain than in the Δaa3 mutant likely results from the more severe inhibition of the ETC in the KCN-treated WT strain. These results suggest that inhibition of the respiratory ETC entails an increase in intracellular cAMP levels, which appears to contribute to Crp-mediated induction of cydA expression in M. smegmatis under respiration-inhibitory conditions.
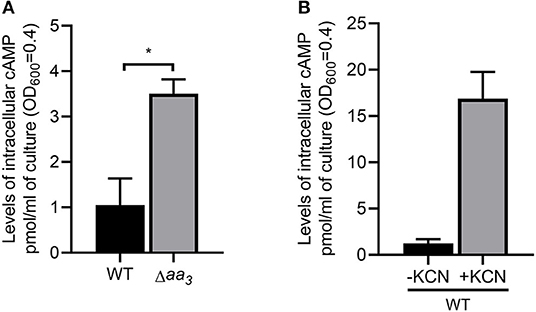
Figure 8. Intracellular levels of cAMP in M. smegmatis under respiration-inhibitory conditions. (A) Intracellular levels of cAMP in the WT and Δaa3 mutant strains of M. smegmatis. The WT and Δaa3 mutant strains of M. smegmatis were grown aerobically to an OD600 of 0.45–0.5 in 7H9-glucose medium. (B) Effect of KCN treatment on intracellular levels of cAMP in M. smegmatis. The WT strain of M. smegmatis was grown aerobically to an OD600 of 0.45–0.5 in 7H9-glucose medium, and the cultures were treated with 100 μM KCN for 15 min (+KCN). As a control, the WT strain without treatment of KCN was used in the experiment (-KCN). cAMP quantification was performed using a DetectX Direct Cyclic AMP Enzyme Immunoassay kit (Arbor Assays) as described in Materials and Methods. All values provided are the average of the results from three biological replicates. The error bars indicate the standard deviations. Statistical significance was determined by two-tailed Student's t-test. *p < 0.05.
RNA sequencing analysis showed that expression of crp2 was increased by 3.4-fold in the Δcrp1 mutant relative to the WT strain, when both strains were grown aerobically (data not shown). We found that a Crp-binding consensus sequence overlaps with the TSP of crp2 (Figure 9A). Using qRT-PCR, we determined the expression level of crp2 in the WT, Δcrp1, and Δcrp2 mutant strains after the strains had been treated with KCN (cAMP-increasing conditions). As shown in Figure 9A, expression of crp2 was increased in the Δcrp1 and Δcrp2 mutant strains by 4.1- and 1.5-fold, respectively, when compared to that in the WT strain. This result indicates that crp2 is under the negative control of Crp1 and Crp2, and that Crp1 predominates in the negative regulation of crp2 under respiration-inhibitory conditions. We also determined the expression level of crp2 in the WT and Δaa3 mutant strains that were grown aerobically (Figure 9B). The expression level of crp2 was reduced by 50% in the Δaa3 mutant compared to that in the WT strain, suggesting that expression of crp2 in M. smegmatis is even more repressed under respiration-inhibitory conditions probably via the negative regulation by Crp1.
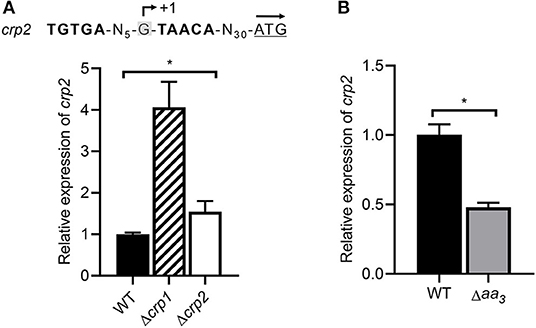
Figure 9. Expression of the crp2 gene in the WT, Δcrp1, Δcrp2, and Δaa3 mutant strains of M. smegmatis. (A) Determination of the crp2 transcript level in the WT, Δcrp1, and Δcrp2 mutant strains by means of qRT-PCR. The positions of the identified Crp-binding site and the TSP (+1) relative to the start codon of crp2 are shown above the graph (Martini et al., 2019). The start codon of crp2 is underlined and the arrow above it indicates the transcriptional direction. The strains were grown aerobically to an OD600 of 0.45–0.5 and treated with 100 μM KCN for 15 min. The expression level of crp2 in the KCN-treated WT strain is set at 1, and the relative values are expressed for the mutant strains. (B) Determination of the crp2 transcript level in the WT and Δaa3 mutant strains using qRT-PCR. The strains were grown aerobically to an OD600 of 0.45–0.5. The expression level of crp2 in the WT strain is set at 1, and the relative values are expressed for the Δaa3 mutant. All values provide are the average of the results from three biological replicates. The error bars indicate the standard deviations. Statistical significance was determined by two-tailed Student's t-test. *p < 0.05.
The prokaryotic cytochrome bd quinol oxidase is structurally and functionally distinct from the HCOs including the aa3 cytochrome c oxidase (Belevich et al., 2005; Megehee et al., 2006; Borisov et al., 2011). The bd quinol oxidase has a higher affinity for O2 than HCOs (Puustinen et al., 1991; Belevich et al., 2005, 2007) and has been suggested to possess the additional activities that decompose H2O2 and peroxynitrite which is a product of the spontaneous reaction of superoxide radical with NO (Lindqvist et al., 2000; Borisov et al., 2004, 2013; Mason et al., 2009; Giuffre et al., 2014). These properties, together with insensitivity of the bd quinol oxidase to the physiologically relevant HCO inhibitors NO and H2S, make the bd quinol oxidase beneficial for bacteria to adapt to and survive in hostile host conditions such as hypoxia and conditions exposed to reactive oxygen species, NO, and H2S (Kana et al., 2001; Matsoso et al., 2005; Giuffre et al., 2012; Small et al., 2013; Forte et al., 2016; Rahman et al., 2020; Saini et al., 2020). Accordingly, expression of the bd quinol oxidase genes is regulated to be induced under respiration-inhibitory conditions such as hypoxia and in the presence of NO and H2S that act as inhibitors of HCOs (Kana et al., 2001; Shi et al., 2005; Giuffre et al., 2012; Small et al., 2013; Aung et al., 2014; Jones-Carson et al., 2016; Jeong et al., 2018). The known regulatory systems, which are involved in the induction of gene expression under hypoxic or anaerobic conditions, generally sense either the molecular oxygen itself or cellular changes caused by the inhibition of the respiratory ETC. The DevSR TCS in M. smegmatis senses directly the molecular oxygen through the heme b in the DevS histidine kinase and upregulates its target genes under hypoxic and anaerobic conditions (Mayuri et al., 2002; O'Toole et al., 2003; Lee et al., 2008; Kim et al., 2010). The Fnr regulators found in many bacteria also sense O2 levels through their O2-labile [4Fe-4S] center and regulate gene expression in response to changes in oxygen availability (Kiley and Beinert, 1998). The inhibition of the respiratory ETC by O2 depletion or the inactivation (or inhibition) of the ETC components such as the terminal oxidases is expected to entail changes in the redox state of electron carriers to a more reduced state. The ArcB histidine kinase of the ArcBA TCS in E. coli and the RegB histidine kinase of the RegBA TCS in Rhodobacter capsulatus have been suggested to be activated by increased ubiquinol of the quinol/quinone pool under anaerobic conditions (Georgellis et al., 2001; Malpica et al., 2004; Swem et al., 2006; Bekker et al., 2010; Wu and Bauer, 2010). The Rex repressors in Gram-positive bacteria regulate gene expression in response to changes in the redox poise of the NADH/NAD+ pool (Brekasis and Paget, 2003; Schau et al., 2004; Larsson et al., 2005; Gyan et al., 2006). Under respiration-inhibitory conditions, the increased ratio of NADH to NAD+ inhibits the DNA-binding activity of Rex via the binding of NADH to the conserved Rossman fold of Rex (Brekasis and Paget, 2003). Since expression of the cydAB operon is strongly induced in the Δaa3 mutant grown under ambient air conditions, the regulatory system responsible for hypoxic induction of the cydAB operon is assumed not to be the regulator that can directly sense the molecular oxygen, but to be the regulatory system that controls gene expression through reflecting the cellular redox state or the functionality of the respiratory ETC. In this respect, the DevSR TCS appears not to be the regulatory system that is responsible for induction of cydA in M. smegmatis under respiration-inhibitory conditions. Consistent with our assumption, expression of cydA in the ΔdevR (MSMEG_5244) mutant of M. smegmatis was induced to a similar level as that in the WT strain, when growth of both strains was shifted from aerobic to hypoxic conditions (data not shown). This observation is further supported by the previous report that the cydAB operon is not included in 49 genes identified as the DevR regulon (Berney et al., 2014). Our search for the Rex, Fnr, RegB, and ArcB homologs in M. smegmatis revealed that their homologous genes are not present in the M. smegmatis genome. Our RNA sequencing analysis on Δaa3 mutant strain of M. smegmatis grown aerobically revealed that 61% of the strongly upregulated DEGs (FC ≥ 4 and p < 0.05) in the Δaa3 mutant relative to the WT belong to the SigF regulon, indicating that it is the SigF partner switching system that plays a predominant role in strong upregulation of gene expression in M. smegmatis under ETC-inhibitory conditions. Since transcription of the cydAB operon was shown to be independent of SigF in M. smegmatis (Figure 1), there must be (a) regulatory system(s) other than SigF that is (are) responsible for strong upregulation of cydAB under respiration-inhibitory conditions. Based on a previous report (Aung et al., 2014) and our results clearly showing that the cydAB operon is under the positive regulation of Crp in M. smegmatis, we assumed that Crp is involved in upregulation of cydAB expression in M. smegmatis under respiration-inhibitory conditions. This assumption is supported by our RNA sequencing result that considerable fractions of the genes, which are more than 4-fold upregulated in the Δaa3 mutant in a SigF-independent way, were shown to be statistically downregulated in the Δcrp1 mutant relative to the WT strain.
The Crp proteins are homodimeric global transcription factors that regulate expression of many genes involved in diverse metabolic and cellular processes in prokaryotes, including carbon utilization, respiration, virulence, cell cycle control, reactivation of non-replicating dormant cells, and stress responses, etc., (Utsumi et al., 1989; Rickman et al., 2005; Shimada et al., 2011; Aung et al., 2014; Green et al., 2014; Heroven and Dersch, 2014). The promoter of cydA possesses two Crp-binding sites that are centered at positions −65.5 (for CBS2) and −93.5 (for CBS1). Crp was suggested to assist the binding of RNA polymerase to the promoter through its interactions with the C-terminal domain of α-subunit of RNA polymerase, when it serves as an transcriptional activator (Busby and Ebright, 1999; Lawson et al., 2004). Mutagenesis and DNA-binding analyses on CBS1 and CBS2 revealed that mutations in the distal Crp-binding CBS1 more severely affected cydA expression than those in CBS2 with a higher binding affinity for Crp1 (Figures 4, 5, 6B), implying that Crp bound at CBS2 might either stabilize the binding of Crp to CBS1 or help the formation of a DNA loop such that Crp bound at CBS1 can participate in recruitment or activation of RNA polymerase. Recently, it was demonstrated that expression of the cydAB operon is significantly reduced in a prrA null mutant of M. smegmatis compared to the WT strain (Maarsingh et al., 2019). However, our EMSA analysis using purified PrrA, which was preincubated with acetyl phosphate for phosphorylation, did not show the binding of PrrA to the upstream region of cydA, implying that the PrrBA TCS might indirectly participate in the regulation of the cydAB operon (data not shown).
Intracellular concentrations of cAMP in mycobacteria have been found to be significantly higher than those in other bacteria grown under similar conditions (Padh and Venkitasubramanian, 1976; Lee, 1977; Shenoy and Visweswariah, 2006; Bai et al., 2011). Accordingly, it was suggested that mycobacteria have been evolved to possess the Crp proteins that have a low binding affinity for cAMP. Crp1 in M. smegmatis and Rv3676 in M. tuberculosis are the Crp proteins that have a low binding affinity for cAMP (Stapleton et al., 2010; Sharma et al., 2014). Although purified Rv3676 and Crp1 were shown to bind to their target DNA even in the absence of cAMP in contrast to Crp2 and E. coli Crp, their DNA-binding affinity was demonstrated to be enhanced by binding of cAMP (Figures 4A, 5A) (Bai et al., 2005; Rickman et al., 2005; Sharma et al., 2014). Some fast-growing mycobacteria such as Mycobacterium flavescens, Mycobacterium fortuitum, and Mycobacterium phlei and the pathogenic slow-growing Mycobacterium avium contain an additional Crp protein that corresponds to Crp2 in M. smegmatis (Sharma et al., 2014). Compared to Crp1, Crp2 of M. smegmatis has been reported to have a much higher binding affinity for cAMP (Kd = ~30 μM for Crp1 and Kd = ~3 μM for Crp2). It has been also reported that Crp2 does not bind to the target DNA in the absence of cAMP (Sharma et al., 2014). Intracellular levels of cAMP were found to be 3.2-fold increased in M. smegmatis by the inactivation of the aa3 cytochrome c oxidase and to rise even more when the aerobically grown WT strain of M. smegmatis was subjected to treatment of KCN (Figure 8), which is in line with our previous report that cAMP levels in M. smegmatis were increased by ~400- and 5.7-fold under hypoxic and SNP (5 mM)-treated conditions, respectively (Jeon et al., 2014). The genome of M. smegmatis contains eight genes encoding adenylyl cyclase (MSMEG_0228, MSMEG_3578, MSMEG_3780, MSMEG_4279, MSMEG_4477, MSMEG_4924, MSMEG_5018, and MSMEG_6154). Although expression of MSMEG_3780 and MSMEG_4279 was observed to increase under both hypoxic and SNP-treated conditions, no detailed study was performed as to which adenylyl cyclase(s) is (are) implicated in an increase in cAMP levels in M. smegmatis under respiration-inhibitory conditions (Jeon et al., 2014). Considering both considerably different cAMP-binding affinity of two Crp paralogs and high intracellular levels of cAMP in mycobacteria, it can be assumed that Crp2 with the reportedly 10-fold higher binding affinity for cAMP than Crp1 might exist in a cAMP-bound form, while Crp1 with a low cAMP-binding affinity might be as a cAMP-unbound apo-protein in M. smegmatis under normal respiration conditions. If this assumption were true, Crp2 would be inappropriate to serve as a transcription factor that reflects elevating intracellular cAMP levels under respiration-inhibitory conditions to regulate gene expression. Furthermore, both significantly low cellular abundance of Crp2 relative to Crp1 and reduced expression of crp2 under respiration-inhibitory conditions as a result of the negative regulation of crp2 by Crp1 (Figures 3, 9) renders Crp2 even more inadequate for a transcription factor that regulates gene expression in response to respiration inhibition. This assumption is consistent with our finding that Crp1, but not Crp2, plays a major role in upregulation of cydAB expression under respiration-inhibitory conditions.
In conclusion, we here provided the evidence that Crp1 (MSMEG_6189) of two Crp paralogs in M. smegmatis is the major transcription factor that is responsible for upregulation of the cydAB operon under ETC-inhibitory conditions. Two Crp-binding sequences were identified upstream of the cydA gene, both of which are necessary for induction of cydAB expression under ETC-inhibitory conditions. The intracellular level of cAMP in M. smegmatis was found to be increased under ETC-inhibitory conditions. The crp2 gene was found to be negatively regulated by Crp1 and Crp2, which might lead to significantly low cellular abundance of Crp2 relative to Crp1 in M. smegmatis.
The RNA sequencing data have been deposited in NCBI's Gene Expression Omnibus and are accessible through the GEO Series accession number GSE158137.
E-MK and J-IO: conception or design of the study, analysis or interpretation of the data, and writing of the manuscript. E-MK: acquisition of the data. All authors contributed to the article and approved the submitted version.
This work was supported by a 2-years Research Grant of Pusan National University to J-IO.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.608624/full#supplementary-material
Aung, H. L., Berney, M., and Cook, G. M. (2014). Hypoxia-activated cytochrome bd expression in Mycobacterium smegmatis is cyclic AMP receptor protein dependent. J. Bacteriol. 196, 3091–3097. doi: 10.1128/JB.01771-14
Aung, H. L., Dixon, L. L., Smith, L. J., Sweeney, N. P., Robson, J. R., Berney, M., et al. (2015). Novel regulatory roles of cAMP receptor proteins in fast-growing environmental mycobacteria. Microbiology 161, 648–661. doi: 10.1099/mic.0.000015
Bai, G., Knapp, G. S., and McDonough, K. A. (2011). Cyclic AMP signalling in mycobacteria: redirecting the conversation with a common currency. Cell. Microbiol. 13, 349–358. doi: 10.1111/j.1462-5822.2010.01562.x
Bai, G., McCue, L. A., and McDonough, K. A. (2005). Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J. Bacteriol. 187, 7795–7804. doi: 10.1128/JB.187.22.7795-7804.2005
Bekker, M., Alexeeva, S., Laan, W., Sawers, G., Teixeira de Mattos, J., and Hellingwerf, K. (2010). The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool. J. Bacteriol. 192, 746–754. doi: 10.1128/JB.01156-09
Belevich, I., Borisov, V. B., Bloch, D. A., Konstantinov, A. A., and Verkhovsky, M. I. (2007). Cytochrome bd from Azotobacter vinelandii: evidence for high-affinity oxygen binding. Biochemistry 46, 11177–11184. doi: 10.1021/bi700862u
Belevich, I., Borisov, V. B., Konstantinov, A. A., and Verkhovsky, M. I. (2005). Oxygenated complex of cytochrome bd from Escherichia coli: stability and photolability. FEBS Lett. 579, 4567–4570. doi: 10.1016/j.febslet.2005.07.011
Berney, M., Greening, C., Conrad, R., Jacobs, W. R. Jr., and Cook, G. M. (2014). An obligately aerobic soil bacterium activates fermentative hydrogen production to survive reductive stress during hypoxia. Proc. Natl. Acad. Sci. U. S. A. 111, 11479–11484. doi: 10.1073/pnas.1407034111
Boot, M., Jim, K. K., Liu, T., Commandeur, S., Lu, P., Verboom, T., et al. (2017). A fluorescence-based reporter for monitoring expression of mycobacterial cytochrome bd in response to antibacterials and during infection. Sci. Rep. 7:10665. doi: 10.1038/s41598-017-10944-4
Borisov, V. B., Forte, E., Davletshin, A., Mastronicola, D., Sarti, P., and Giuffre, A. (2013). Cytochrome bd oxidase from Escherichia coli displays high catalase activity: an additional defense against oxidative stress. FEBS Lett. 587, 2214–2218. doi: 10.1016/j.febslet.2013.05.047
Borisov, V. B., Forte, E., Konstantinov, A. A., Poole, R. K., Sarti, P., and Giuffre, A. (2004). Interaction of the bacterial terminal oxidase cytochrome bd with nitric oxide. FEBS Lett. 576, 201–204. doi: 10.1016/j.febslet.2004.09.013
Borisov, V. B., Gennis, R. B., Hemp, J., and Verkhovsky, M. I. (2011). The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 1807, 1398–1413. doi: 10.1016/j.bbabio.2011.06.016
Brekasis, D., and Paget, M. S. (2003). A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). EMBO J. 22, 4856–4865. doi: 10.1093/emboj/cdg453
Busby, S., and Ebright, R. H. (1999). Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293, 199–213. doi: 10.1006/jmbi.1999.3161
Cook, G. M., Hards, K., Vilcheze, C., Hartman, T., and Berney, M. (2014). Energetics of respiration and oxidative phosphorylation in mycobacteria. Microbiol. Spectr. 2:2013. doi: 10.1128/microbiolspec.MGM2-0015-2013
Cotter, P. A., Chepuri, V., Gennis, R. B., and Gunsalus, R. P. (1990). Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172, 6333–6338. doi: 10.1128/jb.172.11.6333-6338.1990
Cotter, P. A., and Gunsalus, R. P. (1992). Contribution of the fnr and arcA gene products in coordinate regulation of cytochrome o and d oxidase (cyoABCDE and cydAB) genes in Escherichia coli. FEMS Microbiol. Lett. 70, 31–36. doi: 10.1016/0378-1097(92)90558-6
Cotter, P. A., Melville, S. B., Albrecht, J. A., and Gunsalus, R. P. (1997). Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol. Microbiol. 25, 605–615. doi: 10.1046/j.1365-2958.1997.5031860.x
Cunningham, L., Pitt, M., and Williams, H. D. (1997). The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol. Microbiol. 24, 579–591. doi: 10.1046/j.1365-2958.1997.3561728.x
Forte, E., Borisov, V. B., Falabella, M., Colaco, H. G., Tinajero-Trejo, M., Poole, R. K., et al. (2016). The terminal oxidase cytochrome bd promotes sulfide-resistant bacterial respiration and growth. Sci. Rep. 6:23788. doi: 10.1038/srep23788
Fu, H. A., Iuchi, S., and Lin, E. C. (1991). The requirement of ArcA and Fnr for peak expression of the cyd operon in Escherichia coli under microaerobic conditions. Mol. Gen. Genet. 226, 209–213. doi: 10.1007/BF00273605
Georgellis, D., Kwon, O., and Lin, E. C. (2001). Quinones as the redox signal for the Arc two-component system of bacteria. Science 292, 2314–2316. doi: 10.1126/science.1059361
Giuffre, A., Borisov, V. B., Arese, M., Sarti, P., and Forte, E. (2014). Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim. Biophys. Acta 1837, 1178–1187. doi: 10.1016/j.bbabio.2014.01.016
Giuffre, A., Borisov, V. B., Mastronicola, D., Sarti, P., and Forte, E. (2012). Cytochrome bd oxidase and nitric oxide: from reaction mechanisms to bacterial physiology. FEBS Lett. 586, 622–629. doi: 10.1016/j.febslet.2011.07.035
Green, J., Stapleton, M. R., Smith, L. J., Artymiuk, P. J., Kahramanoglou, C., Hunt, D. M., et al. (2014). Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: adaptation for different ecological niches. Curr. Opin. Microbiol. 18, 1–7. doi: 10.1016/j.mib.2014.01.003
Gyan, S., Shiohira, Y., Sato, I., Takeuchi, M., and Sato, T. (2006). Regulatory loop between redox sensing of the NADH/NAD+ ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J. Bacteriol. 188, 7062–7071. doi: 10.1128/JB.00601-06
Heroven, A. K., and Dersch, P. (2014). Coregulation of host-adapted metabolism and virulence by pathogenic yersiniae. Front. Cell. Infect. Microbiol. 4:146. doi: 10.3389/fcimb.2014.00146
Jeon, H. S., Yang, K. H., Ko, I. J., and Oh, J. I. (2014). Expression of adenylyl cyclase genes in Mycobacterium smegmatis under hypoxic and nitric oxide conditions. J. Life Sci. 24, 1330–1338. doi: 10.5352/JLS.2014.24.12.1330
Jeong, J. A., Park, S. W., Yoon, D., Kim, S., Kang, H. Y., and Oh, J. I. (2018). Roles of alanine dehydrogenase and induction of its gene in Mycobacterium smegmatis under respiration-inhibitory conditions. J. Bacteriol. 200:e00152-18. doi: 10.1128/JB.00152-18
Jones-Carson, J., Husain, M., Liu, L., Orlicky, D. J., and Vazquez-Torres, A. (2016). Cytochrome bd-dependent bioenergetics and antinitrosative defenses in salmonella pathogenesis. mBio 7:e02052-16. doi: 10.1128/mBio.02052-16
Kalia, N. P., Shi Lee, B., Ab Rahman, N. B., Moraski, G. C., Miller, M. J., and Pethe, K. (2019). Carbon metabolism modulates the efficacy of drugs targeting the cytochrome bc1:aa3 in Mycobacterium tuberculosis. Sci. Rep. 9:8608. doi: 10.1038/s41598-019-44887-9
Kana, B. D., Weinstein, E. A., Avarbock, D., Dawes, S. S., Rubin, H., and Mizrahi, V. (2001). Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J. Bacteriol. 183, 7076–7086. doi: 10.1128/JB.183.24.7076-7086.2001
Kiley, P. J., and Beinert, H. (1998). Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22, 341–352. doi: 10.1111/j.1574-6976.1998.tb00375.x
Kim, M. J., Park, K. J., Ko, I. J., Kim, Y. M., and Oh, J. I. (2010). Different roles of DosS and DosT in the hypoxic adaptation of mycobacteria. J. Bacteriol. 192, 4868–4875. doi: 10.1128/JB.00550-10
Koul, A., Vranckx, L., Dhar, N., Gohlmann, H. W., Ozdemir, E., Neefs, J. M., et al. (2014). Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat. Commun. 5:3369. doi: 10.1038/ncomms4369
Larsson, J. T., Rogstam, A., and von Wachenfeldt, C. (2005). Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis. Microbiology 151, 3323–3335. doi: 10.1099/mic.0.28124-0
Lawson, C. L., Swigon, D., Murakami, K. S., Darst, S. A., Berman, H. M., and Ebright, R. H. (2004). Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 14, 10–20. doi: 10.1016/j.sbi.2004.01.012
Lee, C. H. (1977). Identification of adenosine 3′,5′-monophosphate in Mycobacterium smegmatis. J. Bacteriol. 132, 1031–1033. doi: 10.1128/JB.132.3.1031-1033.1977
Lee, H. N., Ji, C. J., Lee, H. H., Park, J., Seo, Y. S., Lee, J. W., et al. (2018). Roles of three FurA paralogs in the regulation of genes pertaining to peroxide defense in Mycobacterium smegmatis mc2 155. Mol. Microbiol. 108, 661–682. doi: 10.1111/mmi.13956
Lee, H. N., Lee, N. O., Han, S. J., Ko, I. J., and Oh, J. I. (2014). Regulation of the ahpC gene encoding alkyl hydroperoxide reductase in Mycobacterium smegmatis. PLoS ONE 9:e111680. doi: 10.1371/journal.pone.0111680
Lee, J. M., Cho, H. Y., Cho, H. J., Ko, I. J., Park, S. W., Baik, H. S., et al. (2008). O2- and NO-sensing mechanism through the DevSR two-component system in Mycobacterium smegmatis. J. Bacteriol. 190, 6795–6804. doi: 10.1128/JB.00401-08
Lindqvist, A., Membrillo-Hernandez, J., Poole, R. K., and Cook, G. M. (2000). Roles of respiratory oxidases in protecting Escherichia coli K12 from oxidative stress. Antonie Van Leeuwenhoek 78, 23–31. doi: 10.1023/a:1002779201379
Maarsingh, J. D., Yang, S., Park, J. G., and Haydel, S. E. (2019). Comparative transcriptomics reveals PrrAB-mediated control of metabolic, respiration, energy-generating, and dormancy pathways in Mycobacterium smegmatis. BMC Genom. 20:942. doi: 10.1186/s12864-019-6105-3
Malpica, R., Franco, B., Rodriguez, C., Kwon, O., and Georgellis, D. (2004). Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. U.S.A. 101, 13318–13323. doi: 10.1073/pnas.0403064101
Martini, M. C., Zhou, Y., Sun, H., and Shell, S. S. (2019). Defining the transcriptional and post-transcriptional landscapes of Mycobacterium smegmatis in aerobic growth and hypoxia. Front. Microbiol. 10:591. doi: 10.3389/fmicb.2019.00591
Mascolo, L., and Bald, D. (2020). Cytochrome bd in Mycobacterium tuberculosis: a respiratory chain protein involved in the defense against antibacterials. Prog. Biophys. Mol. Biol. 152, 55–63. doi: 10.1016/j.pbiomolbio.2019.11.002
Mason, M. G., Shepherd, M., Nicholls, P., Dobbin, P. S., Dodsworth, K. S., Poole, R. K., et al. (2009). Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat. Chem. Biol. 5, 94–96. doi: 10.1038/nchembio.135
Matsoso, L. G., Kana, B. D., Crellin, P. K., Lea-Smith, D. J., Pelosi, A., Powell, D., et al. (2005). Function of the cytochrome bc1-aa3 branch of the respiratory network in mycobacteria and network adaptation occurring in response to its disruption. J. Bacteriol. 187, 6300–6308. doi: 10.1128/JB.187.18.6300-6308.2005
Mayuri, B., agchi, G., Das, T. K., and Tyagi, J. S. (2002). Molecular analysis of the dormancy response in Mycobacterium smegmatis: expression analysis of genes encoding the DevR-DevS two-component system, Rv3134c and chaperone alpha-crystallin homologues. FEMS Microbiol. Lett. 211, 231–237. doi: 10.1111/j.1574-6968.2002.tb11230.x
Megehee, J. A., Hosler, J. P., and Lundrigan, M. D. (2006). Evidence for a cytochrome bcc-aa3 interaction in the respiratory chain of Mycobacterium smegmatis. Microbiology 152, 823–829. doi: 10.1099/mic.0.28723-0
Mouncey, N. J., and Kaplan, S. (1998). Redox-dependent gene regulation in rhodobacter sphaeroides 2.4.1(T): effects on dimethyl sulfoxide reductase (dor) gene expression. J. Bacteriol. 180, 5612–5618. doi: 10.1128/JB.180.21.5612-5618.1998
Oh, J. I., and Kaplan, S. (1999). The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry 38, 2688–2696. doi: 10.1021/bi9825100
Oh, Y., Song, S. Y., Kim, H. Y., Han, G., Hwang, J., Kang, H. Y., et al. (2020). The partner switching system of the SigF sigma factor in Mycobacterium smegmatis and induction of the SigF regulon under respiration-inhibitory conditions. Front. Microbiol. 11:588487. doi: 10.3389/fmicb.2020.588487
O'Toole, R., Smeulders, M. J., Blokpoel, M. C., Kay, E. J., Lougheed, K., and Williams, H. D. (2003). A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J. Bacteriol. 185, 1543–1554. doi: 10.1128/jb.185.5.1543-1554.2003
Padh, H., and Venkitasubramanian, T. A. (1976). Cyclic adenosine 3′, 5′-monophosphate in mycobacteria. Indian J. Biochem. Biophys. 13, 413–414
Puustinen, A., Finel, M., Haltia, T., Gennis, R. B., and Wikstrom, M. (1991). Properties of the two terminal oxidases of Escherichia coli. Biochemistry 30, 3936–3942. doi: 10.1021/bi00230a019
Rahman, M. A., Cumming, B. M., Addicott, K. W., Pacl, H. T., Russell, S. L., Nargan, K., et al. (2020). Hydrogen sulfide dysregulates the immune response by suppressing central carbon metabolism to promote tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 117, 6663–6674. doi: 10.1073/pnas.1919211117
Rickman, L., Scott, C., Hunt, D. M., Hutchinson, T., Menendez, M. C., Whalan, R., et al. (2005). A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 56, 1274–1286. doi: 10.1111/j.1365-2958.2005.04609.x
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Saini, V., Chinta, K. C., Reddy, V. P., Glasgow, J. N., Stein, A., Lamprecht, D. A., et al. (2020). Hydrogen sulfide stimulates Mycobacterium tuberculosis respiration, growth and pathogenesis. Nat. Commun. 11:557. doi: 10.1038/s41467-019-14132-y
Sambrook, J., and Green, M. R. (2012). Molrcular Cloning: A Laboratory Manual. 4th Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Schau, M., Chen, Y., and Hulett, F. M. (2004). Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon. J. Bacteriol. 186, 4585–4595. doi: 10.1128/JB.186.14.4585-4595.2004
Sharma, R., Zaveri, A., Gopalakrishnapai, J., Srinath, T., Varshney, U., and Visweswariah, S. S. (2014). Paralogous cAMP receptor proteins in Mycobacterium smegmatis show biochemical and functional divergence. Biochemistry 53, 7765–7776. doi: 10.1021/bi500924v
Shenoy, A. R., and Visweswariah, S. S. (2006). New messages from old messengers: cAMP and mycobacteria. Trends. Microbiol. 14, 543–550. doi: 10.1016/j.tim.2006.10.005
Shi, L., Sohaskey, C. D., Kana, B. D., Dawes, S., North, R. J., Mizrahi, V., et al. (2005). Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl. Acad. Sci. U. S. A. 102, 15629–15634. doi: 10.1073/pnas.0507850102
Shimada, T., Fujita, N., Yamamoto, K., and Ishihama, A. (2011). Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS ONE 6:e20081. doi: 10.1371/journal.pone.0020081
Singh, A. K., Dutta, D., Singh, V., Srivastava, V., Biswas, R. K., and Singh, B. N. (2015). Characterization of Mycobacterium smegmatis sigF mutant and its regulon: overexpression of SigF antagonist (MSMEG_1803) in M. smegmatis mimics sigF mutant phenotype, loss of pigmentation, and sensitivity to oxidative stress. Microbiologyopen 4, 896–916. doi: 10.1002/mbo3.288
Small, J. L., Park, S. W., Kana, B. D., Ioerger, T. R., Sacchettini, J. C., and Ehrt, S. (2013). Perturbation of cytochrome c maturation reveals adaptability of the respiratory chain in Mycobacterium tuberculosis. mBio 4:e00475-13. doi: 10.1128/mBio.00475-13
Snapper, S. B., Melton, R. E., Mustafa, S., Kieser, T., and Jacobs, W. R. Jr. (1990). Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4, 1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x
Stapleton, M., Haq, I., Hunt, D. M., Arnvig, K. B., Artymiuk, P. J., Buxton, R. S., et al. (2010). Mycobacterium tuberculosis cAMP receptor protein (Rv3676) differs from the Escherichia coli paradigm in its cAMP binding and DNA binding properties and transcription activation properties. J. Biol. Chem. 285, 7016–7027. doi: 10.1074/jbc.M109.047720
Swem, L. R., Gong, X., Yu, C. A., and Bauer, C. E. (2006). Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J. Biol. Chem. 281, 6768–6775. doi: 10.1074/jbc.M509687200
Tseng, C. P., Albrecht, J., and Gunsalus, R. P. (1996). Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J. Bacteriol. 178, 1094–1098. doi: 10.1128/jb.178.4.1094-1098.1996
Utsumi, R., Noda, M., Kawamukai, M., and Komano, T. (1989). Control mechanism of the Escherichia coli K-12 cell cycle is triggered by the cyclic AMP-cyclic AMP receptor protein complex. J. Bacteriol. 171, 2909–2912. doi: 10.1128/jb.171.5.2909-2912.1989
Keywords: aa3 cytochrome c oxidase, cAMP, Crp, electron transport chain, Mycobacterium, regulation of gene expression, respiration
Citation: Ko E-M and Oh J-I (2020) Induction of the cydAB Operon Encoding the bd Quinol Oxidase Under Respiration-Inhibitory Conditions by the Major cAMP Receptor Protein MSMEG_6189 in Mycobacterium smegmatis. Front. Microbiol. 11:608624. doi: 10.3389/fmicb.2020.608624
Received: 21 September 2020; Accepted: 06 November 2020;
Published: 30 November 2020.
Edited by:
Morigen Morigen, Inner Mongolia University, ChinaReviewed by:
Greg Cook, University of Otago, New ZealandCopyright © 2020 Ko and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeong-Il Oh, am9oQHB1c2FuLmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.