
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Microbiol., 28 January 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.607002
This article is part of the Research TopicThe Impact of Biofilms on Evolution and Dissemination of Antimicrobial ResistanceView all 5 articles
 Rania Nassar1,2*
Rania Nassar1,2* Mahmood Hachim1
Mahmood Hachim1 Mohannad Nassar3
Mohannad Nassar3 Eleftherios G. Kaklamanos4
Eleftherios G. Kaklamanos4 Mohamed Jamal4
Mohamed Jamal4 David Williams2
David Williams2 Abiola Senok1
Abiola Senok1Bacterial biofilms are microbial lifestyles found in all environments. Up to 80% of human infections and 60–70% of hospital-acquired infections have a biofilm origin, with Staphylococcus aureus one of the leading causes of these infections. Microorganisms in biofilms exhibit significant antimicrobial resistance which poses important treatment challenges, hence the urgent need to identify novel antibiofilm strategies. Microbes form biofilms in response to various factors, and once these 3-dimentional structures form they are highly recalcitrant to removal. The switch from planktonic lifestyle to the biofilm protected mode of growth results in a phenotypic shift in the behavior of the microorganisms in terms of growth rate and gene expression. Given these changes, investigation of microbial gene expression and their modulation at different stages of biofilm maturation is needed to provide vital insight into the behavior of biofilm cells. In this study, we analyzed publicly available transcriptomic dataset of S. aureus biofilms at different stages of maturation to identify consistently upregulated genes irrespective of the biofilm maturation stage. Our reanalysis identified a total of 6 differentially expressed genes upregulated in both 48 and 144-h old S. aureus biofilms. Functional analysis revealed that these genes encode for proteins which play a role in key microbial metabolic pathways. However, these genes, as yet, are unrelated or fully studied in the context of biofilm. Moreover, the findings of this in silico work, suggest that these genes may represent potential novel targets for the development of more effective antibiofilm strategies against S. aureus biofilm-associated infections.
Microbial biofilms are ubiquitously found on biotic and abiotic surfaces in environmental, industrial and medical settings (Kostakioti et al., 2013). The cells in these adherent microbial communities are typically embedded in self-produced extracellular polymeric substances (EPS) (Vert et al., 2012). Microbes form biofilms in response to various factors, which can include changes in nutrient availability or exposure of free living (planktonic) cells to sub-inhibitory concentrations of antibiotics (De la Fuente-Núñez et al., 2013). Biofilm development on biotic surfaces follows a universal ordered sequence of events, initiated by adherence of planktonic microorganisms to the surfaces via surface charges and specific adhesin-receptor interactions (Hinsa et al., 2003). Sustained adherence triggers a switch to the biofilm protected mode of growth, which results in a phenotypic shift in the behavior of the microorganisms in terms of growth rate and gene expression (Armbruster and Parsek, 2018). Following irreversible attachment, cells multiply and start to produce biofilm matrix components, leading to small aggregates of microorganisms called microcolonies which eventually develop into large cellular aggregates encased in the self-produced EPS. Once these 3-dimentional structures form they are highly recalcitrant to removal.
It is estimated that up to 80% of human infections (Römling and Balsalobre, 2012) and 60–70% of hospital-acquired infections have a biofilm origin, with Staphylococcus aureus one of the leading causes of such infections (Haney et al., 2018). It has been shown that microorganisms within biofilms are protected from both host defenses and administered antimicrobials, with up to 1,000-fold greater tolerance to certain antimicrobials reported (Davies, 2003), which accounts for frequent treatment failures associated with biofilm infections. Hence, there is an urgent need for the development of novel antibiofilm agents and strategies.
Given the changes in microbial gene expression that occur during the biofilm lifestyle, investigation of microbial gene expression and their modulation at different stages of biofilm maturation is needed to provide vital insight into the behavior of biofilm cells. This will indeed help in the identification of genes necessary for biofilm survival, and thus identifying attractive targets for antibiofilm strategies. Using publicly available transcriptomic datasets, this study was undertaken to identify consistent differentially expressed genes (DEGs) in S. aureus biofilms at different stages of maturation. To the best of our knowledge, this is the first reported re-analysis of publicly available S. aureus transcriptomic datasets for biofilm related gene expression and the findings demonstrate prospective genes and pathway associations that may be targeted for antibiofilm therapies.
To identify datasets for assessment of DEGs in S. aureus biofilms, the data repository of the National Center for Biotechnology Information (NCBI), Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/), was searched for datasets that include in vitro planktonic cultures and biofilm cultures at different maturation stages. The single dataset (Accession number: GSE35837) which fulfilled this inclusion criteria was derived from S. aureus at three growth stages (planktonic, 48-h and 144-h biofilm), with three replicates for each growth stage (Ryder et al., 2012). Figure 1 shows simplified flowchart of the reanalysis process.
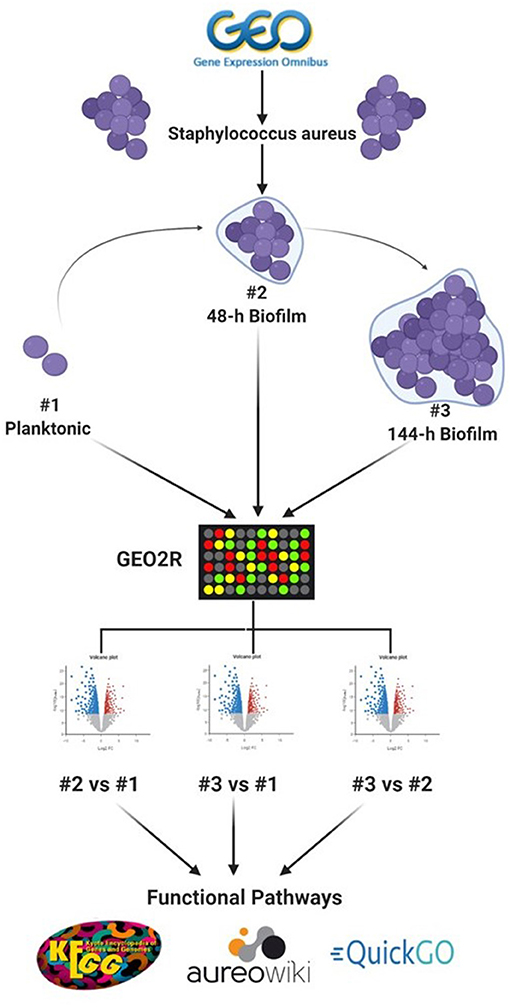
Figure 1. Schematic representation of the reanalysis process of publicly available dataset (GSE35837) retrieved from GEO Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). Created with Biorender.com.
To identify DEGs between the three growth stages, GEOquery and limma R packages were used through the GEO2R tool for each comparison, as previously described (Barrett et al., 2013; Hachim et al., 2020). Briefly, the top 5,000 probes with an adjusted p < 0.1 were selected as the differentially expressed probes in each of three comparisons (48-h old biofilm vs. planktonic, 144-h old biofilm vs. planktonic and 144-h old biofilm vs. 48-h old biofilm). To identify consistent DEGs in biofilm irrespective of the stage of biofilm maturation, the annotated genes of each comparison were intersected as shown in Figure 2.
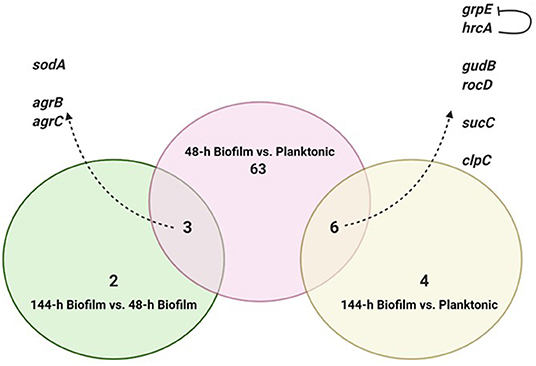
Figure 2. Intersection of differentially expressed genes from the three comparisons. Created with Biorender.com.
Publicly available domains: quickGO, aureuwiki and kyoto encyclopedia of genes and genomes (KEGG) were searched to identify the biological role and functional pathways associated with the consistently DEGs.
Our re-analysis selection criteria for data curation resulted in one dataset: GSE35837. Comparative analysis of S. aureus 48-h old biofilm growth state vs. planktonic cells in the dataset yielded 72 DEGs. Comparison between S. aureus 144-h old biofilm growth state vs. planktonic cells yielded 10 DEGs. While from the comparison of 144-h old biofilm and 48-h old biofilm, 5 DEGs were identified. The full list of all DEGs identified is shown in the Supplementary Tables 1–3.
To identify consistent DEGs in the two biofilm maturation stages, the annotated genes of each comparison were intersected (Figure 2). Nine genes were found to be deferentially expressed in both 48-h and 144-h old biofilms. Of these 9 DEGs, 6 were upregulated in both biofilm growth stages (Figure 2). Functional pathway analysis showed that these 6 DEGs include genes involved in cellular amino acid metabolic process (rocD and gudB), energy metabolism/tricarboxylic acid (TCA) cycle (sucC), and stress responses (grpE, hrcA, and clpC) (Table 1). Three genes were found to be up-regulated in the 48-h biofilm, but down-regulated in 144-h biofilm. These included two accessory regulatory genes (agrB, agrC) and sodA (Table 1).
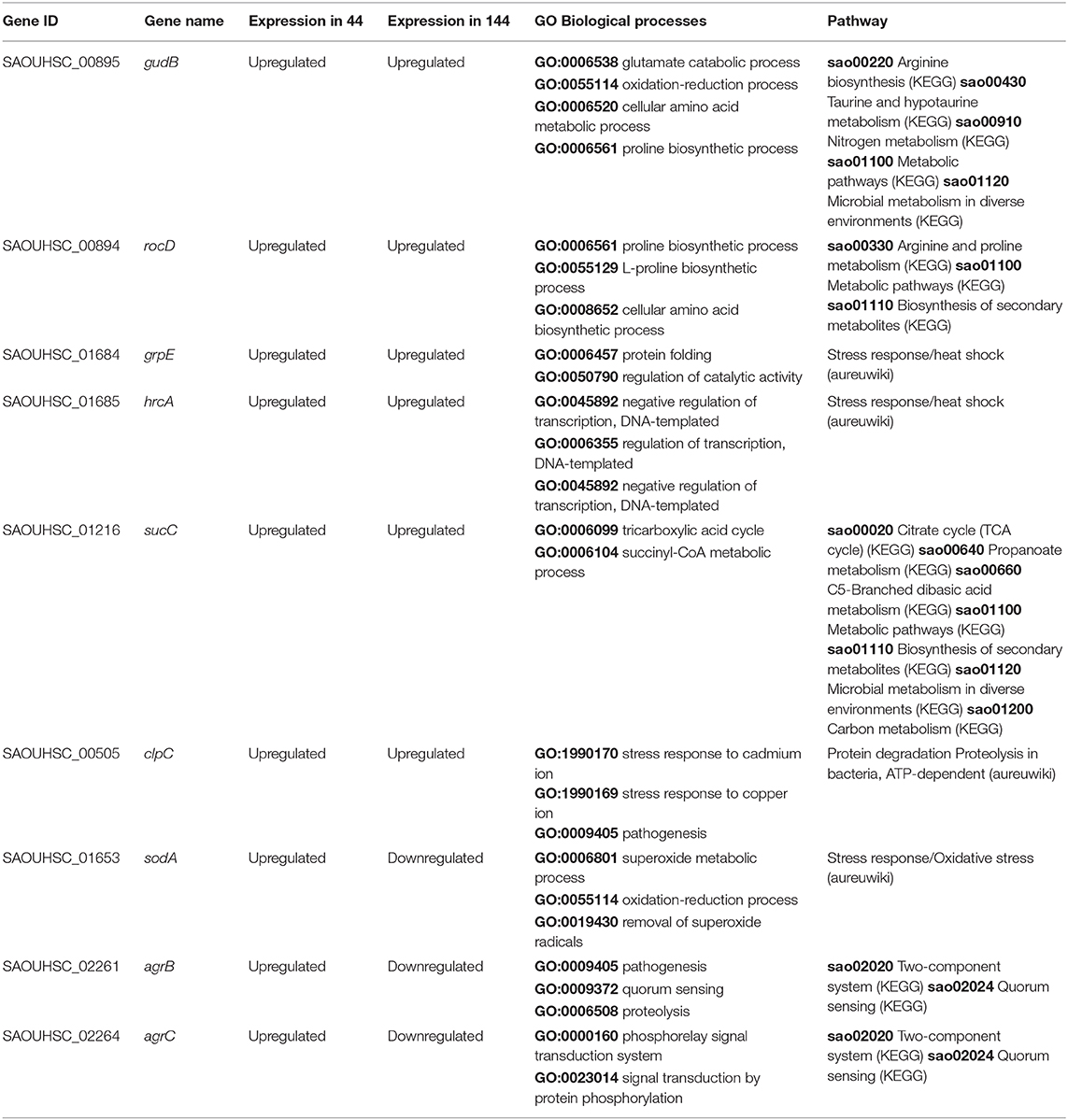
Table 1. Differentially expressed genes present in both biofilm growth stages (48 and 144-h), the biological processes they are involved in and their associated functional pathways.
Biofilms contribute to a large proportion of hospital-acquired infections with S. aureus a key aetiological agent (Lister and Horswill, 2014). The stages of biofilm formation from attachment, growth, maturation, and the maintenance of the mature biofilm involve multiple complex pathways (O'Toole et al., 2000; Hinsa et al., 2003). Hence, the bacterial cells within a biofilm undergo gene expression and transcriptional changes in response to stressors, and metabolic and quorum sensing requirements for the different stages of biofilm growth (Whiteley et al., 2001; Schembri et al., 2003; Beloin et al., 2004; Vuong et al., 2004a). The understanding of these genetic modulations within the microbial biofilm and the key genes that play a role in both early and mature biofilms have yet to be fully elucidated. Due to the poor efficacy of currently used antimicrobial agents, the identification of novel antibiofilm targets to combat these infections is crucial. Therefore, an understanding of these genetic modulations might provide insight into novel antibiofilm targets. To address this gap in the literature, a re-analysis of publicly available transcriptomic data for S. aureus, and compared the changes in gene expression between S. aureus planktonic cells and their biofilm counterparts at different maturation stages was undertaken. Six genes namely rocD, gudB, sucC, grpE, hrcA, and clpC were upregulated at both biofilm growth stages. This suggested that these six genes played a crucial role in different biofilm maturation stages. Two of these genes, rocD and gudB, are involved in amino acid metabolism, specifically arginine metabolism and biosynthesis. rocD and gudB encode for ornithine-oxo-acid transaminase and glutamate dehydrogenase, respectively. The glutamate dehydrogenase enzyme is involved in glutamate, glutamine and ammonia metabolism (Figure 3). It has been shown that glutamine and glutamate are essential for the development and survival of Bacillus subtilis, Enterococcus faecalis, and Pseudomonas aeruginosa biofilms, and interference with their metabolism has detrimental effects on the biofilm. However, their specific contribution to biofilm development is yet to be determined (Hassanov et al., 2018). In a study using an in vivo hamster model, a mutant Clostridium difficile that was defective for the gene that encodes glutamate dehydrogenase, was unable to successfully colonize (Girinathan et al., 2016). To the best of our knowledge, no studies have been done to investigate the role of gudB product in the development and fitness of S. aureus biofilm. Of note, glutamate is also involved in ornithine and arginine biosynthesis (Figure 3) and the TCA cycle (Figure 4). rocD encodes for ornithine-oxo-acid transaminase enzyme which catalyses interconversion of ornithine to glutamate 5-semialdehyde (Figure 4). Both ornithine and glutamate 5-semialdehyde are involved in glutamate and arginine metabolism (Figures 3, 4). The role of arginine in biofilm development and pH homeostasis has previously been highlighted (Crow and Thomas, 1982; Liu et al., 1995; Zhu et al., 2007; Lindgren et al., 2014). Its role as an energy source in the absence of glucose was also investigated and it was shown that arginine, in addition to other amino acids, acts as a fuel for glutamate synthesis, in which the latter will serve as a source of carbon for the TCA cycle (Halsey et al., 2017). Therefore, given that rocD and gudB are involved in pathways of metabolites that are shown to have cardinal roles in biofilm development, we hypothesize that interference in their synthesis and use could potentially have deleterious effects on biofilm, thus making them potential targets for antibiofilm strategies. Experimental work using inhibitors of glutamine and glutamate synthesis can be used to further explore this premise (Arkin and Grossowicz, 1970; Hassanov et al., 2018; Nadar et al., 2019). To the best of our knowledge, no investigations have been done on the specific contribution of ornithine-oxo-acid transaminase enzyme on biofilm development and its metabolic contribution to biofilm development remains to be determined.
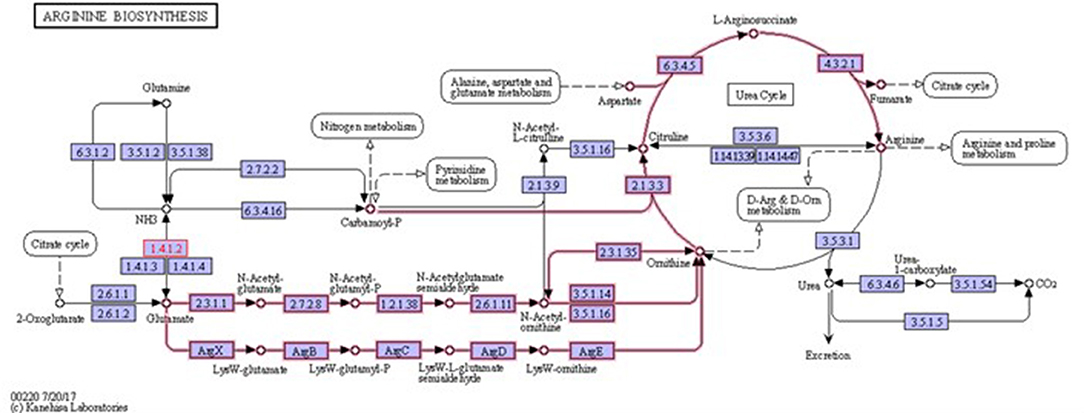
Figure 3. Arginine biosynthesis. Highlighted pathways show the involvement of glutamate in ornithine and arginine biosynthesis. It also shows the involvement of ornithine in arginine biosynthesis. 1.4.1.2 is glutamate dehydrogenase enzyme that is involved in glutamate metabolism (source: KEGG).
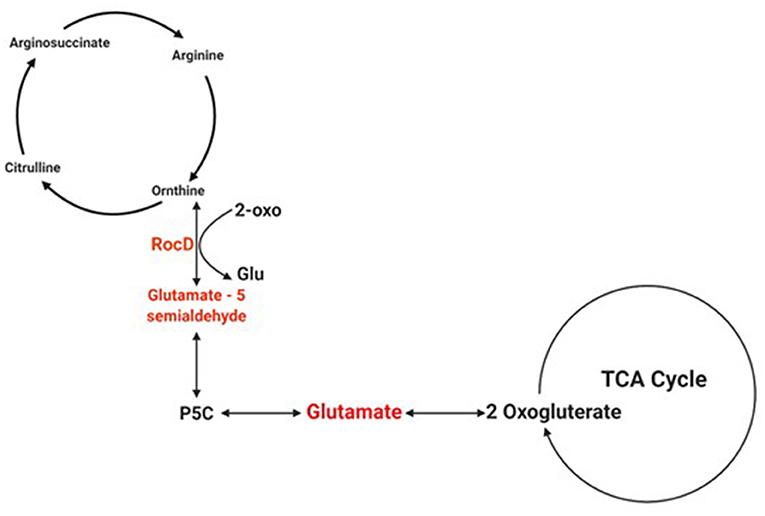
Figure 4. Schematic of the pathway of glutamate involvement in the TCA cycle. The schematic shows how the product of rocD converts ornithine to glutamate 5-semialdehyde, and how the latter is involved in glutamate synthesis. Created with Biorender.com.
sucC was also upregulated in both 48-h and 144-h biofilms. The product of sucC is involved in energy metabolism via the TCA cycle. The upregulation of the TCA cycle under conditions of oxygen depletion enables better use of traces of oxygen, metabolism of excreted fermentation products like acetic or lactic acid, and catabolism of amino acids to meet the microbial need and enhance virulence (Zhu et al., 2009; Gaupp et al., 2010). Bacterial biofilms are formed in response to adverse environmental conditions including oxygen depletion and nutrient deprivation and the upregulation of this gene will be crucial to the maintenance of the biofilm lifestyle. Indeed, in a previous work, a significantly higher amount of dead cells was found in the biofilms formed by sucC mutants which is indicative of a role for sucC in biofilm maintenance (De Backer et al., 2018). Furthermore, the universal heat-shock response is also a protective mechanism that is vital for bacterial survival and adaptation in these adverse conditions. The grpE and its negative regulator hrcA, which are part of heat-shock genes (Roncarati and Scarlato, 2017), are also found to be up-regulated in both biofilm growth stages. The product of grpE plays an important role in bacterial response during environment stress by preventing the aggregation of stress-denatured proteins (Roncarati and Scarlato, 2017). Following bacterial adaptation to the stressors, the quantity of heat-shock proteins typically decreases to the steady-state needed for continued support of bacterial growth in the stressed environment (Schumann, 2016). This process is in keeping with our finding of the concomitant up-regulation of hrcA, which is a negative regulator for grpE. Both the TCA cycle and heat-shock response are crucial for overall bacterial survival and fitness which further makes these genes attractive potential antibiofilm targets as it could be postulated that the bacteria are unlikely to make genetic changes to them. As the specific role of these three genes in biofilm development, survival or fitness has hitherto not been reported, further experimental work is warranted.
Previous reports have shown that expression of clpC is upregulated in S. aureus biofilms compared to planktonic controls (Becker et al., 2001; Resch et al., 2006). This gene encodes the ClpC ATPase that has multiple functions including participation in the degradation of misfolded proteins. Frees et al. (2004) also showed that ClpC was required for S. aureus growth under thermal stress and was also required for biofilm formation (Frees et al., 2004). It also acts as ATPase partner of ClpP protease, the product of clpP that was shown to be essential for biofilm formation in Pseudomonas fluorescens clpC. Our findings further indicate the role of this clpC in different stages of biofilm formation, making it a potential antibiofilm target.
Superoxide dismutase converts deleterious superoxide to oxygen and hydrogen peroxide, and the latter is metabolized by catalase. sodA which encodes superoxide dismutase was upregulated in 48-h and down regulated in 144-h old biofilm. Similar findings were also demonstrated for the genes of the Agr quorum-sensing regulator system (agrB and agrC). Although the Agr system plays a role in the development of the biofilm architecture and structure, it has been shown that its sustained expression results in up-regulation of proteases which inhibit biofilm formation and contributes to biofilm dispersion (Boles and Horswill, 2008; Lauderdale et al., 2009; Otto, 2013). Thus, down-regulation of Agr leads to extensive biofilm maturation and enhanced bacterial colonization (Vuong et al., 2004b). Indeed, targeting these genes via the use of Agr-inhibitors on biofilm-associated staphylococcal infections has shown to have counterproductive effects (Vuong et al., 2004b). Thus, given the discrepant gene expression between biofilm growth stages these three genes (sodA, agrB, and agrC) might appear less attractive as antibiofilm targets when compared to the other genes we identified. However, it is still important to investigate them further in validation experiments.
In conclusion, we have identified six core genes crucial for mature S. aureus biofilms. As these genes encode for proteins which play a role in key microbial metabolic pathways, they represent potential novel targets for antibiofilm strategies. Experimental work to validate these findings is strongly recommended as this will be crucial for exploring any translational potential. In addition, it will be of interest to explore the contribution of host factors to biofilm formation and gene expression.
Publicly available datasets were analyzed in this study. This data can be found here: (https://www.ncbi.nlm.nih.gov/geo/ (Accession number: GSE35837).
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This research was funded by internal research grant from the College of Medicine, Mohammed Bin Rashid University of Medicine and Health Sciences, Dubai, UAE (Ref#: MBRU-CM-RG2020-11).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.607002/full#supplementary-material
Arkin, H., and Grossowicz, N. (1970). Inhibition by D-glutamate of growth and glutamate dehydrogenase activity of Neurospora crassa. J. Gen. Microbiol. 61, 255–261. doi: 10.1099/00221287-61-2-255
Armbruster, C. R., and Parsek, M. R. (2018). New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 115, 4317–4319. doi: 10.1073/pnas.1804084115
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2013). NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41, D991–D995. doi: 10.1093/nar/gks1193
Becker, P., Hufnagle, W., Peters, G., and Herrmann, M. (2001). Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis. Appl. Environ. Microbiol. 67, 2958–2965. doi: 10.1128/AEM.67.7.2958-2965.2001
Beloin, C., Valle, J., Latour-Lambert, P., Faure, P., Kzreminski, M., Balestrino, D., et al. (2004). Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51, 659–674. doi: 10.1046/j.1365-2958.2003.03865.x
Boles, B. R., and Horswill, A. R. (2008). Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. doi: 10.1371/journal.ppat.1000052
Crow, V. L., and Thomas, T. D. (1982). Arginine metabolism in lactic streptococci. J. Bacteriol. 150, 1024–1032. doi: 10.1128/JB.150.3.1024-1032.1982
Davies, D. (2003). Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discovery 2, 114–122. doi: 10.1038/nrd1008
De Backer, S., Sabirova, J., De Pauw, I., De Greve, H., Hernalsteens, J. P., Goossens, H., et al. (2018). Enzymes catalyzing the TCA- and urea cycle influence the matrix composition of biofilms formed by methicillin-resistant Staphylococcus aureus USA300. Microorganisms. 6:113. doi: 10.3390/microorganisms6040113
De la Fuente-Núñez, C., Reffuveille, F., Fernández, L., and Hancock, R. E. (2013). Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 16, 580–589. doi: 10.1016/j.mib.2013.06.013
Frees, D., Chastanet, A., Qazi, S., Sørensen, K., Hill, P., Msadek, T., et al. (2004). Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54, 1445–1462. doi: 10.1111/j.1365-2958.2004.04368.x
Gaupp, R., Schlag, S., Liebeke, M., Lalk, M., and Götz, F. (2010). Advantage of upregulation of succinate dehydrogenase in Staphylococcus aureus biofilms. J. Bacteriol. 192, 2385–2394. doi: 10.1128/JB.01472-09
Girinathan, B. P., Braun, S., Sirigireddy, A. R., Lopez, J. E., and Govind, R. (2016). Importance of glutamate dehydrogenase (GDH) in clostridium difficile colonization in vivo. PLoS ONE 11:e0160107. doi: 10.1371/journal.pone.0160107
Hachim, M. Y., Al Heialy, S., Hachim, I. Y., Halwani, R., Senok, A. C., Maghazachi, A. A., et al. (2020). Interferon-induced transmembrane protein (IFITM3) is upregulated explicitly in SARS-CoV-2 infected lung epithelial cells. Front. Immunol. 11:1372. doi: 10.3389/fimmu.2020.01372
Halsey, C. R., Lei, S., Wax, J. K., Lehman, M. K., Nuxoll, A. S., Steinke, L., et al. (2017). Amino acid catabolism in Staphylococcus aureus and the function of carbon catabolite repression. MBio 8:e01434-16. doi: 10.1128/mBio.01434-16
Haney, E. F., Trimble, M. J., Cheng, J. T., Vallé, Q, and Hancock, R. E. (2018). Critical assessment of methods to quantify biofilm growth and evaluate antibiofilm activity of host defence peptides. Biomolecules 8:29. doi: 10.3390/biom8020029
Hassanov, T., Karunker, I., Steinberg, N., Erez, A., and Kolodkin-Gal, I. (2018). Novel antibiofilm chemotherapies target nitrogen from glutamate and glutamine. Sci. Rep. 8, 1–12. doi: 10.1038/s41598-018-25401-z
Hinsa, S. M., Espinosa-Urgel, M., Ramos, J. L., and O'Toole, G. A. (2003). Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49, 905–918. doi: 10.1046/j.1365-2958.2003.03615.x
Kostakioti, M., Hadjifrangiskou, M., and Hultgren, S. J. (2013). Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 3:a010306–a. doi: 10.1101/cshperspect.a010306
Lauderdale, K. J., Boles, B. R., Cheung, A. L., and Horswill, A. R. (2009). Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77, 1623–1635. doi: 10.1128/IAI.01036-08
Lindgren, J., Thomas, V. C., Olson, M., Chaudhari, S., Nuxoll, A. S., Schaeffer, C., et al. (2014). Arginine deiminase in Staphylococcus epidermidis functions to augment biofilm maturation through pH homeostasis. J. Bacteriol. 196, 2277–2289. doi: 10.1128/JB.00051-14
Lister, J. L., and Horswill, A. R. (2014). Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 4:178. doi: 10.3389/fcimb.2014.00178
Liu, S., Pritchard, G., Hardman, M., and Pilone, G. (1995). Occurrence of arginine deiminase pathway enzymes in arginine catabolism by wine lactic Acid bacteria. Appl. Environ. Microbiol. 61, 310–316. doi: 10.1128/AEM.61.1.310-316.1995
Nadar, V. S., Chen, J., Dheeman, D. S., Galván, A. E., Yoshinaga-Sakurai, K., Kandavelu, P., et al. (2019). Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic. Commun. Biol. 2, 1–12. doi: 10.1038/s42003-019-0365-y
O'Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. doi: 10.1146/annurev.micro.54.1.49
Otto, M. (2013). Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 64, 175–188. doi: 10.1146/annurev-med-042711-140023
Resch, A., Leicht, S., Saric, M., Pásztor, L., Jakob, A., Götz, F., et al. (2006). Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics. 6, 1867–1877. doi: 10.1002/pmic.200500531
Römling, U., and Balsalobre, C. (2012). Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 272, 541–561. doi: 10.1111/joim.12004
Roncarati, D., and Scarlato, V. (2017). Regulation of heat-shock genes in bacteria: from signal sensing to gene expression output. FEMS Microbiol. Rev. 41, 549–574. doi: 10.1093/femsre/fux015
Ryder, V. J., Chopra, I., and O'Neill, A. J. (2012). Increased mutability of Staphylococci in biofilms as a consequence of oxidative stress. PLoS ONE 7:e47695. doi: 10.1371/journal.pone.0047695
Schembri, M. A., Kjærgaard, K., and Klemm, P. (2003). Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48, 253–267. doi: 10.1046/j.1365-2958.2003.03432.x
Schumann, W. (2016). Regulation of bacterial heat shock stimulons. Cell Stress Chaperones. 21, 959–968. doi: 10.1007/s12192-016-0727-z
Vert, M., Doi, Y., Hellwich, K.-H., Hess, M., Hodge, P., Kubisa, P., et al. (2012). Terminology for biorelated polymers and applications (IUPAC Recommendations 2012). Pure Appl. Chem. 84, 377–410. doi: 10.1351/PAC-REC-10-12-04
Vuong, C., Kocianova, S., Yao, Y., Carmody, A. B., and Otto, M. (2004b). Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190, 1498–1505. doi: 10.1086/424487
Vuong, C., Voyich, J. M., Fischer, E. R., Braughton, K. R., Whitney, A. R., DeLeo, F. R., et al. (2004a). Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6, 269–275. doi: 10.1046/j.1462-5822.2004.00367.x
Whiteley, M., Bangera, M. G., Bumgarner, R. E., Parsek, M. R., Teitzel, G. M., Lory, S., et al. (2001). Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864. doi: 10.1038/35101627
Zhu, Y., Weiss, E. C., Otto, M., Fey, P. D., Smeltzer, M. S., and Somerville, G. A. (2007). Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect. Immun. 75, 4219–4226. doi: 10.1128/IAI.00509-07
Keywords: mature biofilm, antibiofilm strategies, biofilm genes, staphylococcal biofilm, rocD
Citation: Nassar R, Hachim M, Nassar M, Kaklamanos EG, Jamal M, Williams D and Senok A (2021) Microbial Metabolic Genes Crucial for S. aureus Biofilms: An Insight From Re-analysis of Publicly Available Microarray Datasets. Front. Microbiol. 11:607002. doi: 10.3389/fmicb.2020.607002
Received: 16 September 2020; Accepted: 24 December 2020;
Published: 28 January 2021.
Edited by:
Hans P. Steenackers, Centre of Microbial and Plant Genetics, KU Leuven, BelgiumReviewed by:
Devendra Dusane, The Ohio State University, United StatesCopyright © 2021 Nassar, Hachim, Nassar, Kaklamanos, Jamal, Williams and Senok. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rania Nassar, rania.nassar@mbru.ac.ae; nassarrm@cardiff.ac.uk
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.